ABSTRACT
Pseudorabies virus (PRV) is the causative pathogen of Aujeszky’s disease in pigs. Although vaccination is currently applied to prevent the morbidity of PRV infection, new applications are urgently needed to control this infectious disease. Poly(ADP-ribose) polymerase 1 (PARP1) functions in DNA damage repair. We report here that pharmacological and genetic inhibition of PARP1 significantly influenced PRV replication. Moreover, we demonstrate that inhibition of PARP1 induced DNA damage response and antiviral innate immunity. Mechanistically, PARP1 inhibition-induced DNA damage response resulted in the release of double-stranded DNA (dsDNA) into the cytosol, where dsDNA interacted with cyclic GMP-AMP (cGAMP) synthase (cGAS). cGAS subsequently catalyzed cGAMP production to activate the STING/TBK1/IRF3 innate immune signaling pathway. Furthermore, challenge of mice with PARP1 inhibitor stimulated antiviral innate immunity and protected mice from PRV infection in vivo. Our results demonstrate that PARP1 inhibitors may be used as a new strategy to prevent Aujeszky’s disease in pigs.
IMPORTANCE Aujeszky’s disease is a notifiable infectious disease of pigs and causes economic losses worldwide in the pig industry. The causative pathogen is PRV, which is a member of the subfamily Alphaherpesvirinae of the family Herpesviridae. PRV has a wide range of hosts, such as ruminants, carnivores, and rodents. More seriously, recent reports suggest that PRV can cause human endophthalmitis and encephalitis, which indicates that PRV may be a potential zoonotic pathogen. Although vaccination is currently the major strategy used to control the disease, new applications are also urgently needed for the pig industry and public health. We report here that inhibition of PARP1 induces DNA damage-induced antiviral innate immunity through the cGAS-STING signaling pathway. Therefore, PARP1 is a therapeutic target for PRV infection as well as alphaherpesvirus infection.
KEYWORDS: innate immunity, cyclic GMP-AMP synthase, DNA damage, poly(ADP-ribose) polymerase 1, pseudorabies virus, stimulator of interferon genes
INTRODUCTION
Pseudorabies virus (PRV) is an enveloped, double-stranded DNA (dsDNA) virus that belongs to the subfamily Alphaherpesvirinae (1). PRV infection causes Aujeszky’s disease in pigs, leading to high mortality in suckling pigs and respiratory disease and latent infection in adult pigs. Although pigs are the natural carriers of PRV, the virus can also infect numerous other species, including ruminants, carnivores, and rodents (2). In recent years, several reports have suggested that PRV can cause human endophthalmitis and encephalitis (3–5). These findings indicate that PRV infection is a potential public health risk and not limited to the swine industry. Therefore, new applications are urgently needed to prevent PRV infection.
Poly(ADP-ribosyl)ation is a posttranslational modification of proteins that is induced by DNA damage responses (6). Poly(ADP-ribose) polymerase 1 (PARP1) is the founding member of the PARP superfamily, which comprises 18 proteins encoded by different genes (7). PARP1 utilizes NAD+ as a substrate to catalyze linear or branched poly(ADP-ribose) (PAR) polymer on itself or nuclear proteins to facilitate the process of DNA repair (8). PARP1 is rapidly recruited to sites of damaged DNA through its DNA-binding ability (9). The residues Glu, Asp, and Lys in acceptor proteins are covalently attached to PAR by transesterification of PARP1. Proteins responsible for DNA damage repair noncovalently interact with PAR on PARP proteins through PAR-binding modules, such as PAR-binding consensus motifs and PAR-binding zinc finger motifs, for recruitment to sites of DNA damage (10). PAR-interacting proteins can participate in DNA binding, protein-protein interactions, nuclear localization, and other functions (11, 12).
Cyclic GMP-AMP (cGAMP) synthase (cGAS) is a germ line-encoded DNA sensor that detects cytosolic DNA to initiate innate immune responses (13). Binding of cGAS to DNA is necessary for the activation of enzymatic activity of cGAS to generate cGAMP (14). cGAMP is an intrinsic agonist of stimulator of interferon (IFN) genes (STING) and induces conformational change of STING (15). STING then serves as an adaptor that recruits TANK binding kinase 1 (TBK1), IFN-regulated factor 3 (IRF3), and NF-κB to activate innate immune responses (16). Our previous study demonstrated that inhibition of bromodomain protein 4 (BRD4) induces DNA damage response and activates cGAS-STING-dependent antiviral activity (17).
Here, we demonstrate that inhibition of PARP1 induces DNA-damage-response-triggered activation of antiviral innate immunity through cGAS-STING. Thus, PARP1 inhibitors may be developed as promising antivirals against PRV infection.
RESULTS
Effects of PARP1 inhibitors on cell viability, apoptosis, and cell cycle.
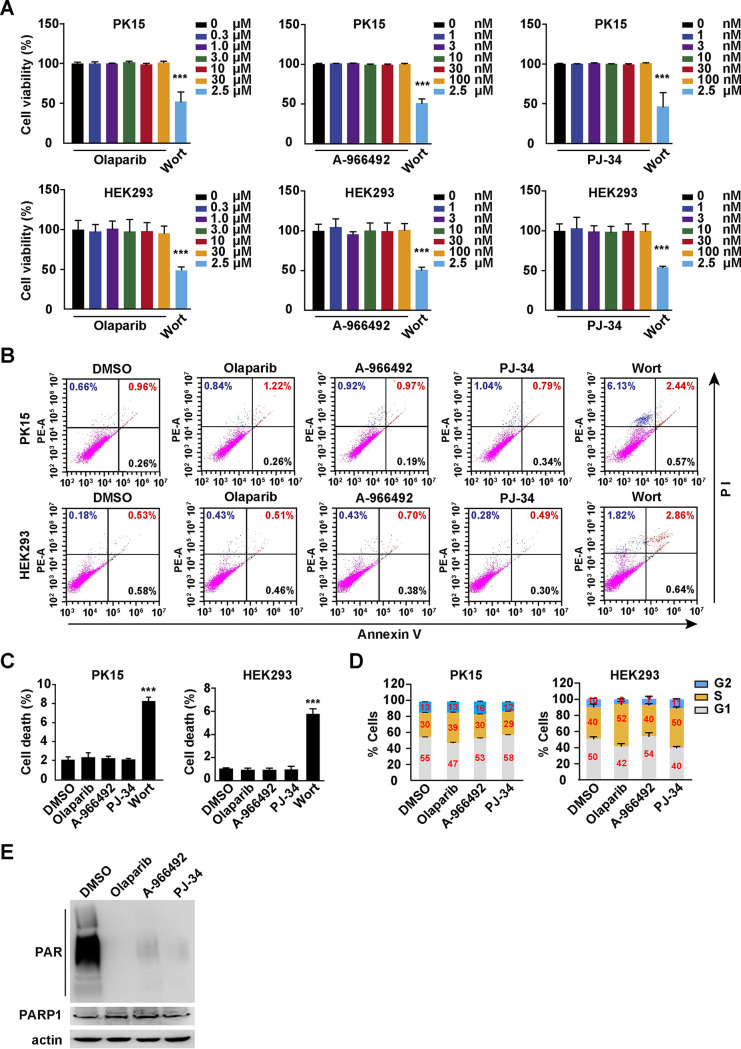
We first analyzed the effects of PARP1 inhibitors on cell viability, apoptosis, and cell cycle in vitro. PK15 and HEK293 cells were treated with the PARP1 inhibitors olaparib (0 to 30 μM), A-966492 (0 to 100 nM), and PJ-34 (0 to 100 nM) or the positive control wortmannin (Wort; 2.5 μM) for 48 h, and cell viability was determined with Cell Counting Kit-8 (CCK-8) assays. As a positive control, Wort showed toxicity to PK15 and HEK293 cells (Fig. 1A). However, PARP1 inhibitors were harmless to both types of cells (Fig. 1A). Wort, a proapoptosis compound (18), significantly induced apoptosis, but olaparib (10 μM), A-966492 (30 nM), and PJ-34 (30 nM) did not induce apoptosis in PK15 and HEK293 cells, as indicated by annexin V and propidium iodide (PI) staining (Fig. 1B and C). We also analyzed the effect of PARP1 inhibitors on the cell cycle. Olaparib treatment resulted in cell cycle arrest in S phase in PK15 and HEK293 cells (Fig. 1D). PJ-34 induced cell cycle arrest in S phase only in HEK293 cells, not in PK15 cells (Fig. 1D). However, the cell cycle was unaltered when both cells were treated with A-966492 (Fig. 1D). We treated PK15 cells with dimethyl sulfoxide (DMSO), olaparib (10 μM), A-966492 (30 nM), and PJ-34 (30 nM) for 24 h and examined PAR formation by immunoblotting analysis. Inhibition of PARP1 by olaparib, A-966492, and PJ-34 decreased the formation of PAR (Fig. 1E).
FIG 1.
Cytotoxicity and cell cycle analysis of PARP1 inhibitors in vitro. (A) PK15 and HEK293 cells were treated with olaparib (0 to 30 μM), A-966492 (0 to 100 nM), PJ-34 (0 to 100 nM), or Wort (2.5 μM) for 48 h. Cell viability was assessed with CCK-8 assays. (B and C) PK15 and HEK293 cells were treated with DMSO, olaparib (10 μM), A-966492 (30 nM), PJ-34 (30 nM), or Wort (2.5 μM) for 24 h. Apoptosis was assessed with annexin V-FITC and PI staining in flow cytometry (B). Percent cell death was quantified (C). (D) PK15 and HEK293 cells were treated with DMSO, olaparib (10 μM), A-966492 (30 nM), or PJ-34 (30 nM) for 24 h. The cell cycle was assessed with Hoechst 33342 staining in flow cytometry. (E) PK15 cells were treated with DMSO, olaparib (10 μM), A-966492 (30 nM), or PJ-34 (30 nM) for 24 h. PAR, PARP1, and actin were assessed by immunoblotting analysis. Data are means and standard deviations (SD) based on three independent experiments. ***, P < 0.001, determined by two-tailed Student's t test.
PARP1 inhibitors influence PRV infection.
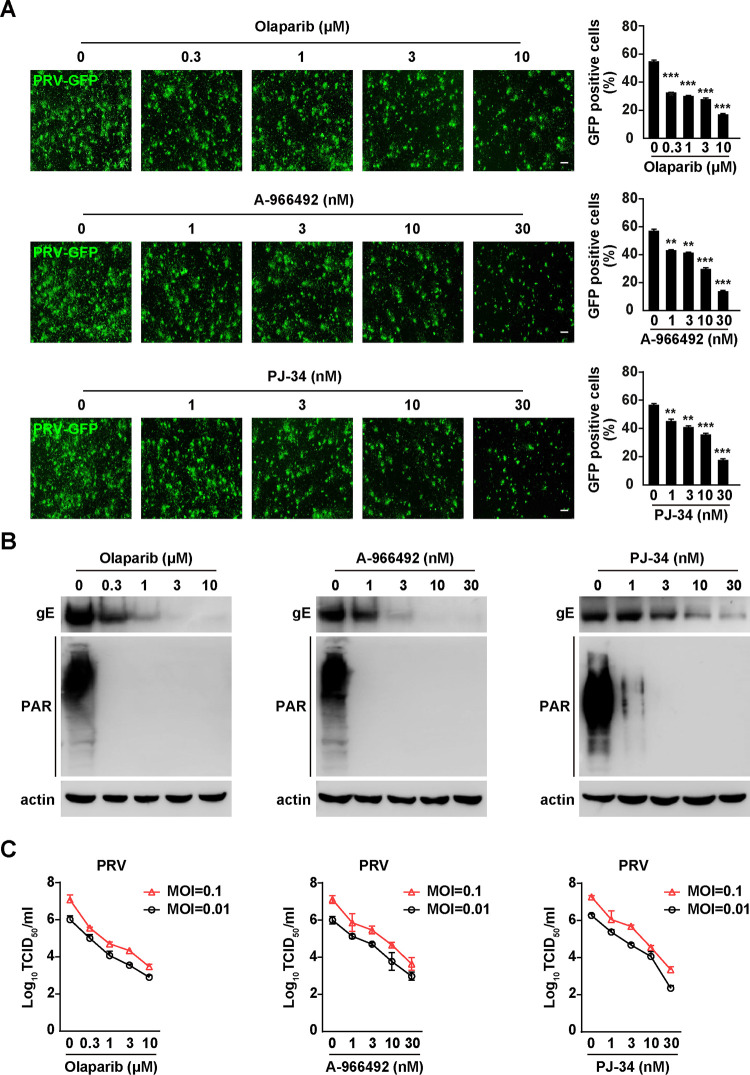
We used a previously established green fluorescent protein (GFP) reporter (PRV-GFP) assay to analyze the anti-PRV effect of PARP1 inhibitors in vitro (17). PK15 cells were infected with PRV-GFP (multiplicity of infection [MOI] = 0.01) and treated with olaparib (0 to 10 μM), A-966492 (0 to 30 nM), or PJ-34 (0 to 30 nM) for 36 h. PRV replication was monitored by fluorescence microscopy and flow cytometry. PARP1 inhibitors significantly inhibited PRV-GFP replication in a dose-dependent manner (Fig. 2A). We also detected PRV gE expression to evaluate the inhibitory effect of olaparib, A-966492, or PJ-34 on replication of the virulent strain PRV-QXX. Immunoblotting analysis indicated that expression of PRV gE and PAR formation were decreased by the increased concentration of PARP1 inhibitors (Fig. 2B). We infected PK15 cells with PRV-QXX to examine the effects of PARP1 inhibitors on virus replication using viral titer assays. In agreement with GFP reporter assay and immunoblotting analysis of PRV gE expression, PARP1 inhibitors did affect virus replication, as indicated by the decreased production of progeny virus (Fig. 2C). Taken together, these results demonstrate that PARP1 inhibitors influence PRV infection in vitro.
FIG 2.
PARP1 inhibitors exhibit inhibitory effect on PRV infection. (A) PK15 cells were infected with PRV-GFP (MOI = 0.01) and treated with olaparib (0 to 10 μM), A-966492 (0 to 30 nM), or PJ-34 (0 to 30 nM) for 36 h. Viral replication was analyzed by fluorescence microscopy, and GFP-positive cells were analyzed by flow cytometry. Bars, 100 μm. (B) PK15 cells were infected with PRV-QXX (MOI = 0.1) and treated with olaparib (0 to 10 μM), A-966492 (0 to 30 nM), or PJ-34 (0 to 30 nM) for 24 h. PRV gE, PAR, and actin were assessed by immunoblotting analysis. (C) PK15 cells were infected with PRV-QXX (MOI = 0.01 and 0.1) and treated with olaparib (0 to 10 μM), A-966492 (0 to 30 nM), or PJ-34 (0 to 30 nM) for 24 h. PRV titers were assessed by TCID50 assay. Data are means and SD based on three independent experiments. **, P < 0.01, and ***, P < 0.001, determined by two-tailed Student's t test.
Interference of PARP1 interrupts PRV infection.
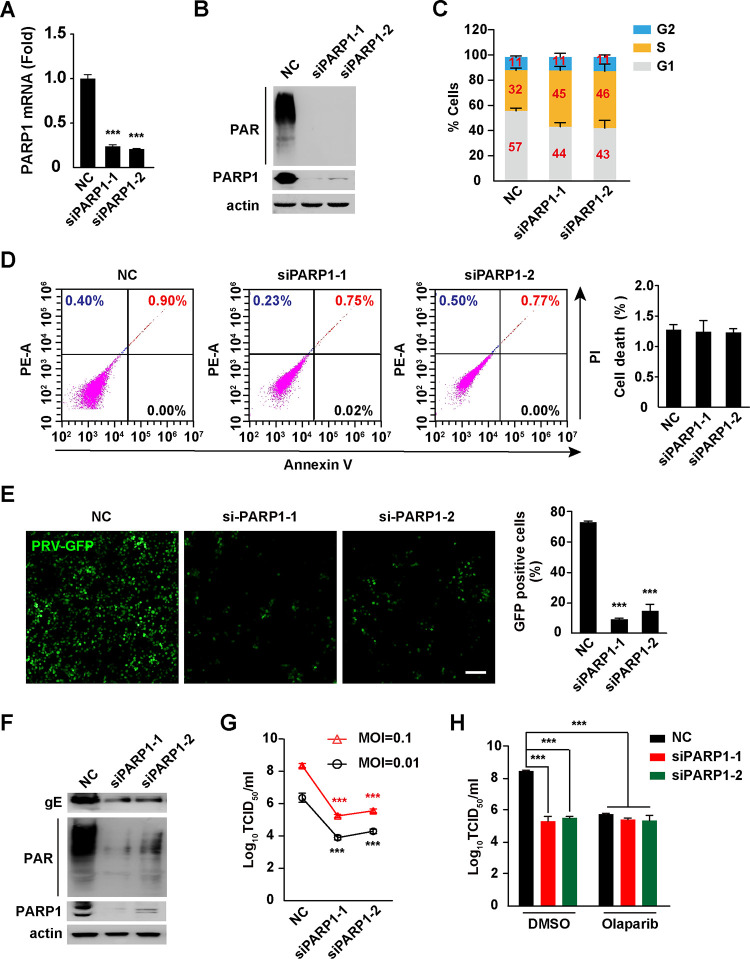
To confirm that inhibition of PARP1 disturbed PRV infection, we knocked down PARP1 expression in PK15 cells by RNA interference (RNAi). Cells were transfected with negative-control (NC) small interfering RNA (siRNA) or siRNAs specifically targeting PARP1 (siPARP1-1 and siPARP1-2). At 48 h posttransfection, the mRNA level of PARP1 in siPARP1-1- and siPARP1-2-transfected cells dropped to ∼25% compared with that in NC-transfected cells (Fig. 3A). Immunoblotting analysis indicated that both siRNAs against PARP1 significantly decreased PARP1 expression and formation of PAR polymer, suggesting that PARP1 was efficiently knocked down (Fig. 3B). Similar to the effect of olaparib on cell cycle arrest, PK15 cells with PARP1 knockdown accumulated in S phase (Fig. 3C). However, apoptosis did not occur, due to PARP1 interference (Fig. 3D).
FIG 3.
Knockdown of PARP1 inhibits PRV infection. (A) PK15 cells were transfected with NC, siPARP1-1, and siPARP1-2 for 48 h. The PARP1 mRNA level was assessed by qRT-PCR analysis. (B) PK15 cells were transfected with NC, siPARP1-1, and siPARP1-2 for 48 h. PAR, PARP1, and actin were assessed by immunoblotting analysis. (C) PK15 cells were transfected with NC, siPARP1-1, and siPARP1-2 for 48 h. The cell cycle was assessed with Hoechst 33342 staining in flow cytometry. (D) PK15 cells were transfected with NC, siPARP1-1, and siPARP1-2 for 48 h. Apoptosis was assessed with annexin V-FITC and PI staining in flow cytometry (left). Quantification of the percentage of cell death is shown on the right. (E) PK15 cells were transfected with NC, siPARP1-1, and siPARP1-2 for 24 h and infected with PRV-GFP (MOI = 0.01) for 36 h. Viral replication was analyzed by fluorescence microscopy (left), and GFP-positive cells were analyzed by flow cytometry (right). Bar, 100 μm. (F) PK15 cells were transfected with NC, siPARP1-1, and siPARP1-2 for 24 h and infected with PRV-QXX (MOI = 0.1) for 24 h. PRV gE, PAR, PARP1, and actin were assessed by immunoblotting analysis. (G) PK15 cells were transfected with NC, siPARP1-1, and siPARP1-2 for 24 h and infected with PRV-QXX (MOI = 0.01 and 0.1) for 24. PRV titers were assessed by TCID50 assay. (H) PK15 cells were transfected with NC, siPARP1-1, and siPARP1-2 for 24 h. Cells were infected with PRV-QXX (MOI = 0.01 and 0.1) and simultaneously treated with DMSO or olaparib (10 μM) for 24 h. PRV titers were assessed by TCID50 assay. Data are means and SD based on three independent experiments. ***, P < 0.001, determined by two-tailed Student's t test (A, E, and G) or one-way ANOVA (H).
We then assessed whether PRV replication was influenced by interference of PARP1 expression. The analysis of fluorescence microscopy and flow cytometry indicated that the replication of PRV-GFP was lower in siPARP1-1- and siPARP1-2-transfected than in NC-transfected PK15 cells (Fig. 3E). The percentage of cells expressing GFP dropped from ∼70% to <20% (Fig. 3E). Decreased PRV gE expression and PAR formation were detected by immunoblotting analysis in cells expressing low levels of PARP1 (Fig. 3F). Viral titer assays indicated that knockdown of PARP1 inhibited PRV replication (Fig. 3G). Moreover, olaparib had no additional effects on inhibition of PRV replication in cells with PARP1 knockdown, suggesting that olaparib specifically acted on PARP1 (Fig. 3H). These results indicate that knockdown of PARP1 inhibits PRV replication.
Inhibition of PARP1 induces DNA damage response.
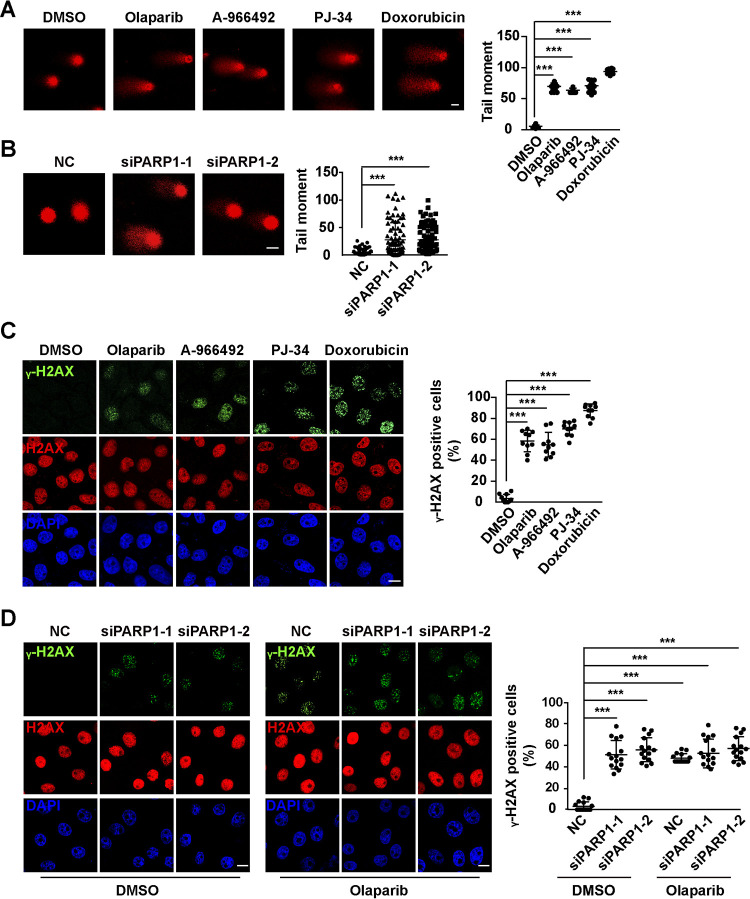
Because PARP1 is involved in DNA repair (19), we investigated whether PARP1 inhibition induced a DNA damage response. Doxorubicin, an inhibitor of DNA topoisomerase II that can induce DNA damage response (20), was used as a positive control. We performed a comet assay to determine DNA damage by PARP1 inhibition. PARP1 inhibitors or PARP1 knockdown significantly induced DNA damage response (Fig. 4A and B). Immunofluorescence analysis was carried out to detect the phosphorylation of H2AX at serine 139 (γ-H2AX), the most sensitive marker of DNA damage response (21). PK15 cells were treated with DMSO, olaparib (10 μM), A-966492 (30 nM), PJ-34 (30 nM), or doxorubicin (100 nM). At 24 h posttreatment, the percentage of γ-H2AX-positive cells was significantly increased (Fig. 4C). Transfection of siPARP1-1 and siPARP1-2 also significantly enhanced phosphorylation of H2AX (Fig. 4D). These results suggest that inhibition of PARP1 induces DNA damage response.
FIG 4.
Inhibition of PARP1 induces DNA damage response. (A) PK15 cells were treated with DMSO, olaparib (10 μM), A-966492 (30 nM), PJ-34 (30 nM), or doxorubicin (100 nM) for 24 h. DNA damage response was detected with comet assays (left), and tail moment was quantified (right). (B) PK15 cells were transfected with NC, siPARP1-1, and siPARP1-2 for 48 h. DNA damage response was detected with comet assays (left), and tail moment was quantified (right). Bar, 20 μm. (C) PK15 cells were treated with DMSO, olaparib (10 μM), A-966492 (30 nM), PJ-34 (30 nM), or doxorubicin (100 nM) for 24 h. γ-H2AX and H2AX were assessed with immunofluorescence analysis (left), and γ-H2AX-positive cells were quantified (right). Bar, 10 μm. (D) PK15 cells were transfected with NC, siPARP1-1, and siPARP1-2 and simultaneously treated with DMSO or olaparib (10 μM) for 48 h. γ-H2AX and H2AX were assessed with immunofluorescence analysis (left), and γ-H2AX-positive cells were quantified (right). Bar, 10 μm. Data are means and SD from three independent experiments. ***, P < 0.001, determined by two-tailed Student's t test.
Inhibition of PARP1 activates innate immune signaling.
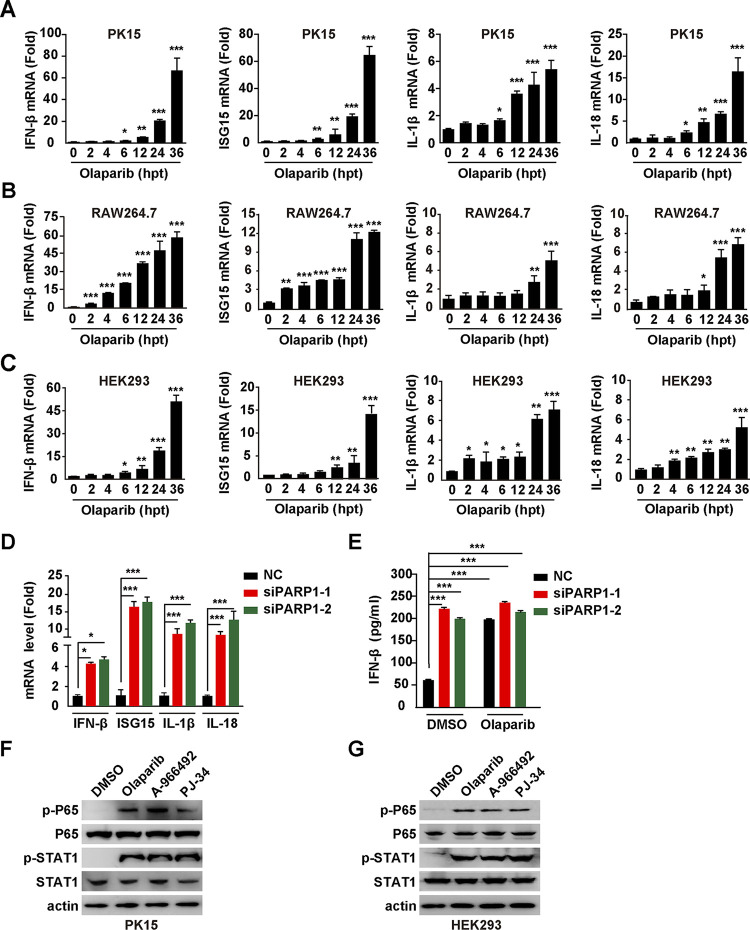
Previous studies have demonstrated that DNA damage responses can evoke innate immune signaling (22), so we examined whether PARP1 inhibition-induced DNA damage response stimulated innate immune activation. We detected transcription of IFN-β, IFN-stimulated gene 15 (ISG15), and NF-κB target genes encoding interleukin 1β (IL-1β) and IL-18 in PK15, RAW264.7, and HEK293 cells. Quantitative reverse transcription-PCR (qRT-PCR) showed that all four genes tested were upregulated in response to olaparib in a time-dependent manner in PK15, RAW264.7, and HEK293 cells, suggesting that PARP1 inhibition-induced innate immune activation was not cell type specific (Fig. 5A to C). Similarly, knockdown of PARP1 in PK15 cells significantly stimulated transcription of IFN-β, ISG15, IL-1β, and IL-18 genes (Fig. 5D). Enzyme-linked immunosorbent assay (ELISA) of IFN-β from the culture medium indicated that IFN-β secretion was enhanced in siPARP1-1- and siPARP1-2-transfected PK15 cells, compared with NC-transfected PK15 cells (Fig. 5E). We treated PK15 and HEK293 cells with DMSO, olaparib (10 μM), A-966492 (30 nM), and PJ-34 (30 nM) for 24 h and assessed innate immune activation by immunoblotting analysis. NF-κB subunit P65 and signal transducer and activator of transcription 1 (STAT1) were phosphorylated when cells were challenged with PARP1 inhibitors, suggesting that innate immunity and IFN signaling were activated (Fig. 5F and G). Collectively, these results indicate that inhibition of PARP1 evokes the innate immune signaling pathway.
FIG 5.
Inhibition of PARP1 induces innate immune response. (A to C) PK15 (A), RAW264.7 (B), and HEK293 (C) cells were treated with olaparib (10 μM) for 0 to 36 h. IFN-β, ISG15, IL-1β, and IL-18 mRNA levels were assessed by qRT-PCR analysis. (D) PK15 cells were transfected with NC, siPARP1-1, and siPARP1-2 for 48 h. IFN-β, ISG15, IL-1β, and IL-18 mRNA levels were assessed by qRT-PCR analysis. (E) PK15 cells were transfected with NC, siPARP1-1, and siPARP1-2 and simultaneously treated with DMSO or olaparib (10 μM) for 48 h. IFN-β secretion was assessed by ELISA. (F and G) PK15 (F) and HEK293 (G) cells were treated with DMSO, olaparib (10 μM), A-966492 (30 nM), or PJ-34 (30 nM) for 24 h. p-P65, P65, p-STAT1, STAT1, and actin were assessed by immunoblotting analysis. Data are means and SD from three independent experiments. *, P < 0.05, **, P < 0.01, and ***, P < 0.001, determined by two-tailed Student's t test. hpt, hours posttreatment.
Inhibition of PARP1 activates the cGAS-STING pathway.
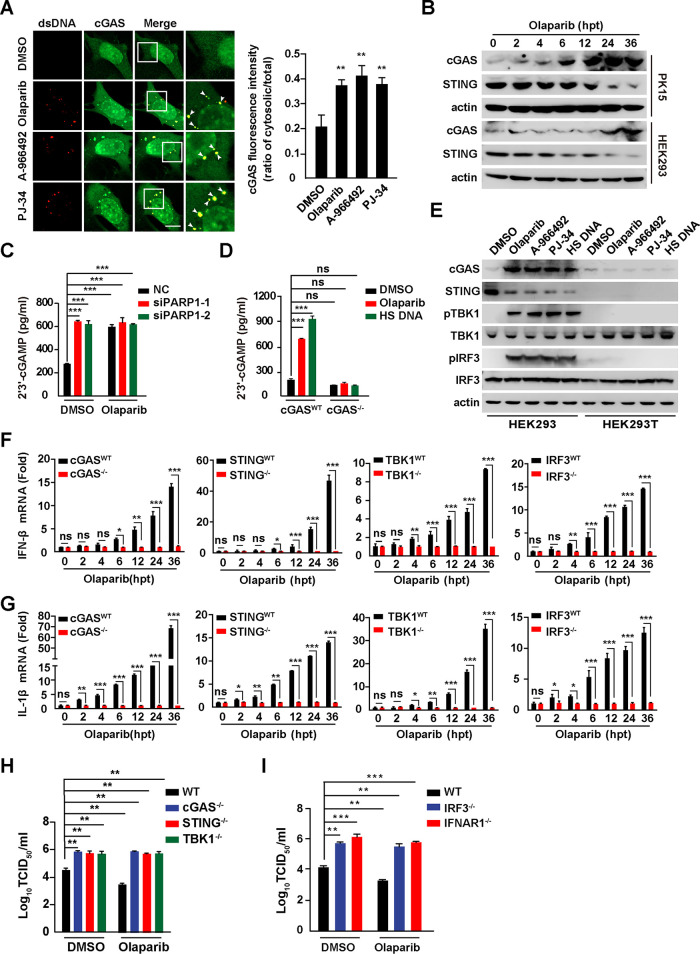
Our previous study suggested that inhibition of BRD4 exerts antiviral innate immunity through DNA-damage-induced activation of the cGAS-STING signaling pathway (17), so we examined whether PARP1 inhibition could achieve a similar phenomenon. By immunofluorescence analysis, we found that PARP1 inhibitors condensed the structure of cGAS, some of which colocalized with cytosolic dsDNA (Fig. 6A). The ratio of cytosolic to total cGAS was significantly increased, which was possibly caused by activation of IFN signaling (23). We further detected cGAS and STING expression in olaparib-treated PK15 and HEK293 cells by immunoblotting analysis. During prolonged treatment with olaparib, cGAS was gradually upregulated and STING was gradually downregulated (Fig. 6B). Because cGAS catalyzes cGAMP production to activate STING, we quantified cGAMP in the cells to determine whether cGAS was activated by PARP1 inhibition. Knockdown of PARP1 promoted cGAMP generation in PK15 cells (Fig. 6C). Olaparib treatment enhanced cGAMP level in wild-type (WT) PK15 cells but failed in cGAS−/− cells (Fig. 6D). As a positive control, herring sperm (HS) DNA stimulated cGAMP production in PK15 cells (24) (Fig. 6D).
FIG 6.
Inhibition of PARP1 activates cGAS-STING pathway. (A) PK15 cells were treated with DMSO, olaparib (10 μM), A-966492 (30 nM) or PJ-34 (30 nM) for 24 h. cGAS and dsDNA were detected by immunofluorescence. Colocalization of cGAS with dsDNA is indicated by arrowheads (left). (Right) Quantification of the ratio of cytosolic versus total cGAS. Bar, 10 μm. (B) PK15 and HEK293 cells were treated with olaparib (10 μM) for 0 to 36 h. cGAS, STING, and actin were assessed by immunoblotting analysis. (C) PK15 cells were transfected with NC, siPARP1-1, and siPARP1-2 and simultaneously treated with DMSO or olaparib (10 μM) for 48 h. 2′,3′-cGAMP in the cells was assessed by ELISA. (D) WT and cGAS−/− PK15 cells were treated with DMSO or olaparib (10 μM) or transfected with HT DNA (0.3 μg) for 24 h. 2′,3′-cGAMP in the cells was assessed by ELISA. (E) HEK293 and HEK293T cells were treated with DMSO, olaparib (10 μM), A-966492 (30 nM), or PJ-34 (30 nM) or transfected with HT DNA (0.3 μg) for 24 h. cGAS, STING, p-TBK1, TBK1, p-IRF3, IRF3, and actin were assessed by immunoblotting. (F and G) WT, cGAS−/−, STING−/−, TBK1−/−, and IRF3−/− PK15 cells were treated with olaparib (10 μM) for 0 to 36 h. IFN-β (F) and IL-1β (G) mRNA levels were assessed by qRT-PCR analysis. (H) WT, cGAS−/−, STING−/−, and TBK1−/− PK15 cells were infected with PRV-QXX (MOI = 0.1) and simultaneously treated with DMSO or olaparib (10 μM) for 24 h. PRV titer was assessed by TCID50 assay. (I) WT, IFR3−/−, and IFNAR1−/− PK15 cells were infected with PRV-QXX (MOI = 0.1) and simultaneously treated with DMSO or olaparib (10 μM) for 24 h. PRV titer was assessed by TCID50 assay. Data are means and SD from three independent experiments. ns, no significance. *, P < 0.05, **, P < 0.01, and ***, P < 0.001, determined by two-tailed Student's t test. hpt, hours posttreatment.
We next determined the activation of cGAS/STING/TBK1/IRF3 axis after PARP1 inhibition. We treated HEK293 and HEK293T (STING-deficient) cells with PARP1 inhibitors for 24 h. Challenge of HEK293 cells with olaparib, A-966492, PJ-34, or HS DNA exhibited STING-dependent phosphorylation of TBK1 and IRF3 (Fig. 6E). However, pharmacological inhibition of PARP1 failed to phosphorylate TBK1 and IRF3 in HEK293T cells, which indicated that PARP1 inhibitors activated the STING/TBK1/IRF3 axis (Fig. 6E). In addition, WT, cGAS−/−, STING−/−, TBK1−/−, and IRF3−/− PK15 cells were treated with olaparib for 0 to 36 h. IFN-β and IL-1β mRNAs were analyzed by qRT-PCR analysis. Knockout of cGAS, STING, TBK1, and IRF3 in PK15 cells abolished olaparib-stimulated IFN-β and IL-1β transcription, suggesting that olaparib activated the cGAS/STING/TBK1/IRF3 axis (Fig. 6F and G). We analyzed PRV replication in cGAS−/−, STING−/−, TBK1−/−, IRF3−/−, and IFNAR1−/− PK15 cells. Knockout of cGAS, STING, TBK1, IRF3, and IFNAR1 enhanced PRV replication (Fig. 6H and I). Olaparib treatment inhibited PRV replication only in WT PK15 cells, not in cGAS−/−, STING−/−, TBK1−/−, IRF3−/−, or IFNAR1−/− PK15 cells (Fig. 6H and I). These results demonstrate that inhibition of PARP1 activates antiviral innate immunity through the cGAS/STING/TBK1/IRF3 axis.
Olaparib inhibits PRV infection in vivo.
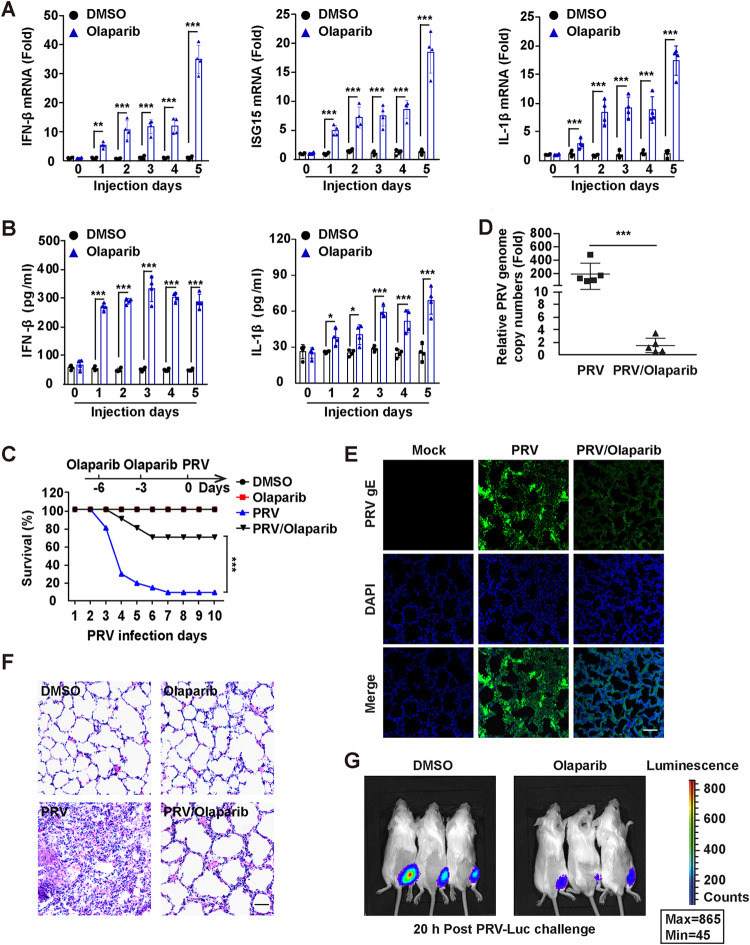
We determined whether olaparib exerted anti-PRV activity in vivo. Mice were intraperitoneally injected with olaparib on days 0 and 3. Transcription of IFN-β, ISG15, and IL-1β mRNA in the lungs was assessed by qRT-PCR analysis. IFN-β, ISG15, and IL-1β mRNA were all upregulated from day 1 after olaparib injection (Fig. 7A). ELISA showed that IFN-β and IL-1β levels in the serum were significantly higher than in DMSO-treated mice (Fig. 7B). These results suggest that olaparib activates innate immunity in vivo.
FIG 7.
Olaparib restricts PRV infection in vivo. (A) Mice were intraperitoneally injected with DMSO or olaparib (50 mg/kg) on day 0 and 3. IFN-β, ISG15, and IL-1β mRNA levels in the lungs were assessed by qRT-PCR analysis (n = 4 per group). (B) Mice were intraperitoneally injected with DMSO or olaparib (50 mg/kg) for 0 to 5 days. IFN-β and IL-1β in the serum were assessed by ELISA (n = 4 per group). (C) Mice were intraperitoneally injected with DMSO or olaparib (50 mg/kg) on days −6 and −3. On day 0, mice were mock infected or intranasally infected with PRV-QXX (5 × 103 TCID50 per mouse). The survival rate was monitored daily for 10 days (n = 12 per group). (D) PRV genome copy numbers in the lungs from C at 3 days after mock infection or PRV-QXX infection were assessed by qRT-PCR analysis (n = 5 per group). (E) PRV gE in the lung sections used for panel C at 3 days after mock infection or PRV-QXX infection were assessed by immunofluorescence. Bar, 100 μm. (F) H&E-stained images of lung sections used for panel C at 3 days after mock infection or PRV-QXX infection. Bar, 100 μm. (G) Mice were intraperitoneally injected with DMSO or olaparib (50 mg/kg) on days −6 and −3 as indicated in panel C. On day 0, mice were intramuscularly infected with PRV-Luc (1 × 104.5 TCID50). Viral growth was measured in real time by bioluminescence at 20 h after PRV-Luc challenge (n = 3). Data are means and SD from three independent experiments. *, P < 0.05, **, P < 0.01, and ***, P < 0.001, determined by two-tailed Student's t test.
We next examined the protective effect of olaparib on the mortality of mice. Mice were intraperitoneally injected with olaparib on days −6 and −3. On day 0, mice were intranasally infected with PRV-QXX for 10 days. Mice challenged with PRV were more susceptible than those infected with PRV and treated with olaparib (PRV/olaparib) (Fig. 7C). qRT-PCR analysis of PRV genome in lungs indicated that olaparib induced an ∼100-fold decrease in PRV genomes compared with PRV infection alone (Fig. 7D). Immunohistochemistry of PRV gE showed that olaparib treatment decreased PRV gE expression in lungs, suggesting that olaparib inhibited PRV replication in vivo (Fig. 7E). Histological analysis by hematoxylin-and-eosin (H&E) staining indicated that olaparib ameliorated the lung injury caused by PRV infection (Fig. 7F). Mice were also intramuscularly infected with PRV expressing luciferase (PRV-Luc), and viral replication was monitored in real time by bioluminescence. Olaparib significantly inhibited PRV-Luc replication, as indicated by a weaker bioluminescence signal in olaparib-treated than DMSO-treated mice (Fig. 7G). These results suggest that olaparib prevents PRV infection in vivo.
DISCUSSION
Harnessing innate immunity has potential for antiviral therapeutics. In this study, we demonstrated that inhibition of PARP1 by pharmacological inhibitors (olaparib, A-966492, and PJ-34) and by RNAi significantly inhibited PRV infection in vitro. This antiviral effect resulted from DNA damage-dependent antiviral innate immunity that relied on cGAS-STING. We also demonstrated that treatment of mice with olaparib activated innate immunity and inhibited PRV infection in vivo. These results suggest that using PARP1 inhibitors could be an effective strategy to prevent PRV infection.
cGAS can be activated by cytosolic DNA derived from a large variety of DNA-containing pathogens and from damage-associated release of DNA from the mitochondria or nuclei (25–27). Our previous study indicated that downregulation of porcine cGAS by RNAi markedly reduced IFN-β expression after PRV infection, suggesting that PRV infection can activate cGAS (28). We also found that inhibition of BRD4 induces DNA damage response and antiviral innate immunity against PRV through cGAS-STING (17). Here, we showed that inhibition of PARP1 exerted antiviral activity through a mechanism similar to BRD4 inhibition. However, Ghosh and colleagues reported that PARP1 depletion induces RIG-I-dependent signaling in human cancer cells (29). It was also reported that etoposide-induced DNA damage induces noncanonical activation of STING that is mediated by the DNA-binding protein IFI16, together with the DNA damage response factors ATM and PARP-1 (30). Our results showed that ablation of cGAS completely abolished PRV-infection- and olaparib-induced IFN-β and IL-1β expression, suggesting that RIG-I and noncanonical activation of STING were not involved in PARP1 inhibition-activated innate immunity.
Suppression of PARP1 results in cell cycle arrest. PARP1 siRNA suppresses human prostate cancer cell growth and progression (31). PARP1 inhibitor PJ-34 suppresses cell cycle progression in neural stem/progenitor cells (32). We noted that olaparib treatment resulted in cell cycle arrest in S phase in PK15 and HEK293 cells, and A-966492 had no effect on cell cycle progression. This difference might be because olaparib completely inhibited formation of PAR, compared to A-966492. Furthermore, knockdown of PARP1 by RNAi with high efficiency completely inhibited formation of PAR and induced cell cycle arrest in S phase.
Several lines of evidence have suggested that inhibition of PARP1 activates STING in human cancer cells. Ding and colleagues report that PARP inhibition elicits STING-dependent antitumor immunity in BRCA1-deficient ovarian cancer (33). Other researchers have demonstrated that BRCA2 abrogation triggers innate immune responses potentiated by treatment with PARP inhibitors (34). However, Shen and colleagues suggested that PARP inhibition triggers the STING-dependent immune response that is independent of BRCA1/2 mutations (35). We found that PARP1 inhibition activated STING in several normal cell lines, such as HEK293 and PK15. Given that BRCA1/2 mutations can result in homologous recombination pathway alterations and consequently lead to a clinical benefit from PARP inhibitors (36), novel combinations of PARP inhibitors with BRCA inhibitors are promising effective antiviral therapeutics.
MATERIALS AND METHODS
Reagents.
Olaparib (HY-10162), A-966492 (HY-10614), PJ-34 (HY-13688A), wortmannin (HY-10197), and doxorubicin (HY-15142) were from MedChemExpress. The Dead Cell apoptosis kit with annexin V-fluorescein isothiocyanate (FITC) and PI (V13242) was from Thermo Fisher. Hoechst 33342 (561908) was from BD. Anti-PARP1 (13371-1-AP) was from Proteintech; anti-poly(ADP-ribose) (4355-MC-100) was from Trevigen. Anti-γ-H2AX, anti-H2AX, anti-p-TBK1, anti-TBK1, anti-p-IRF3, anti-IRF3, anti-p-P65, anti-P65, anti-p-STAT1, anti-STAT1, anti-STING, anti-cGAS, anti-dsDNA, anti-actin, and anti-PRV gE were used as previously described (17). Herring sperm (HS) DNA (D3459; Sigma–Aldrich) was used as previously described (24).
Cells and viruses.
HEK293, HEK293T, Vero, and RAW264.7 cells and WT, cGAS−/−, STING−/−, TBK1−/−, IRF3−/−, and IFNAR1−/− PK15 cells were used and cultivated as previously described (17). PRV-GFP and PRV-QXX were used as previously described (17, 37).
Generation of recombinant PRV-Luc.
The recombinant PRV strain of PRV-GFP was used for generation of recombinant PRV-Luc (17). The GFP expression cassette was replaced with the firefly luciferase expression cassette from plasmid pEGFP-N1. The recombinant virus was further purified by endpoint dilution.
Mice.
Female 6- to 8-week-old BALB/c mice were purchased from the Experimental Animal Center of Zhengzhou University (Zhengzhou, China) and maintained in a specific-pathogen-free animal facility according to the Guide for the Care and Use of Laboratory Animals (38) and the related ethical regulations of Henan Agricultural University.
Cell viability analysis.
Cell viability was evaluated with CCK-8 assays (GK3607; Dingguo). Cells were seeded at 1 × 104 per well in 96-well plates. On the next day, the medium was changed to Dulbecco’s modified Eagle’s medium (DMEM)–10% fetal bovine serum (FBS) supplemented with various concentrations of inhibitors for 48 h. CCK-8 (10 μl) was then added to each well, and the cells were incubated for 3 h at 37°C. The absorbance was detected at 450 nm with a microplate reader (Varioskan Flash; Thermo Fisher).
Cell cycle analysis.
Cells were digested with trypsin-EDTA and resuspended in phosphate-buffered saline (PBS) containing 5 μg/ml Hoechst 33342 at a concentration of 1 × 106 cells/ml. After incubation for 1 h at 37°C, cell cycle profiles were collected by flow cytometry on a CytoFLEX instrument (Beckman Coulter). Data were analyzed with FlowJo software.
Apoptosis analysis.
Annexin V and PI staining was performed with a Dead Cell apoptosis kit with annexin V-FITC and PI. The percent dead cells (positive for both annexin V and PI) was measured by flow cytometry on a CytoFLEX instrument. Data were analyzed with FlowJo software.
RNAi.
Cells were seeded at 4 × 105 per dish in 60-mm dishes and transfected with siRNA (GenePharma) at a final concentration of 0.12 nM. Transfections were performed with Lipofectamine RNAiMAX reagent (13778500; Invitrogen) in Opti-MEM reduced-serum medium (31985062; Gibco). The medium was replaced with DMEM containing 10% FBS at 8 h posttransfection. The knockdown efficacy was assessed by qRT-PCR and immunoblotting analysis at 48 h posttransfection. The siRNA sequences were as follows: NC, 5′-UUCUCCGAACGUGUCACGUTT-3′; siPARP1-1, 5′-CCGAGUAUGCCAAGUCCAATT-3′; siPARP1-2, 5′-GCACCGUAAUUGGUAGUAATT-3′.
qRT-PCR analysis.
Total RNA was isolated with TRIzol reagent (9108; TaKaRa). One microgram of total RNA was prepared for cDNA synthesis with a PrimeScript RT reagent kit with gDNA Eraser (RR047; TaKaRa). qRT-PCR was carried out with the QuantStudio 6 Flex real-time PCR system (Applied Biosystems) using TB Green Premix Ex Taq (Tli RNase H Plus) (RR420; TaKaRa). Data were normalized to the expression level of β-actin in each individual sample. Transcripts were quantified with the 2−ΔΔCT method. Primers were designed in primer3 software and are shown in Table 1.
TABLE 1.
Primers used for qRT-PCR analysis
| Name | Sequence (5′–3′) | Product size (bp) |
|---|---|---|
| Q-Sus actin | CTGAACCCCAAAGCCAACCGT | 317 |
| TTCTCCTTGATGTCCCGCACG | ||
| Q-human actin | GCACAGAGCCTCGCCTT | 112 |
| CCTTGCACATGCCGGAG | ||
| Q-Mus actin | CCCCATTGAACATGGCATTG | 272 |
| ACGACCAGAGGCATACAGG | ||
| Q-Sus PARP1 | CAGGCAAGCAAGGCGGCGGC | 152 |
| CGCCCACTCTTAGCATACTC | ||
| Q-Sus IFN-β | AGTTGCCTGGGACTCCTCAA | 60 |
| CCTCAGGGACCTCAAAGTTCAT | ||
| Q-human IFN-β | CAGGAGAGCAATTTGGAGGA | 132 |
| CTTTCGAAGCCTTTGCTCTG | ||
| Q-Mus IFN-β | ATGAGTGGTGGTTGCAGGC | 82 |
| TGACCTTTCAAATGCAGTAGATTCA | ||
| Q-Sus IL-1β | CCATCCACTGAGCCAGCCTT | 103 |
| TGCCAAGGACAGAGGACTGC | ||
| Q-human IL-1β | CCTGAAGCCCTTGCTGTAGT | 284 |
| AGCTGATGGCCCTAAACAGA | ||
| Q-Mus IL-1β | GCAGAGCACAAGCCTGTCTTCC | 198 |
| ACCTGTCTTGGCCGAGGACTAAG | ||
| Q-Sus IL-18 | AGGGACATCAAGCCGTGTTT | 66 |
| CGGTCTGAGGTGCATTATCTGA | ||
| Q-human IL-18 | GACCAAGTTCTCTTCATTGAC | 144 |
| GATAGTTACAGCCATACCTCTA | ||
| Q-Sus ISG15 | ATGCCCCCTTGCCCTCTCCAGTG | 235 |
| TCCGATGCCATCATGCAGTCCCT | ||
| Q-Mus ISG15 | GACGCAGACTGTAGACACGCTTAA | 281 |
| CGTTTACATTTCCAATGCTATCCC | ||
| Q-PRV gH | CTCGCCATCGTCAGCAA | 187 |
| GCTGCTCCTCCATGTCCTT |
Immunoblotting analysis.
Cells were lysed in lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, and 2 mM MgCl2) supplemented with protease and phosphatase inhibitor cocktail (HY-K0010 and HY-K0022; MedChemExpress). The protein concentrations of the lysates were quantified with a bicinchoninic acid (BCA) protein assay kit (BCA01; Dingguo Biotechnology). Protein samples (30 μg) were denatured for 10 min at 99°C, then separated by SDS-PAGE, and transferred to polypropylene fluoride membranes (C3117; Millipore). The membranes were blocked in 5% nonfat milk (A600669; Sangon Biotech) at room temperature for 1 h, washed three times with Tris-buffered saline–Tween (TBST) and incubated with the primary antibodies overnight at 4°C. The membranes were incubated with goat anti-mouse and anti-rabbit IgG conjugated to horseradish peroxidase (HRP) (IH-0031and IH-0011; Dingguo) at room temperature for 1 h. The target proteins were detected with Luminata Crescendo Western HRP substrate (180545; Millipore) on a GE AI600 imaging system.
Immunofluorescence analysis.
Cells seeded on coverslips were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature and washed three times with PBS. The cells were permeabilized in PBS containing 0.1% Triton X-100 and blocked with 10% FBS in PBS. The primary antibodies were diluted with 10% FBS in PBS and incubated with the cells for 1 h at room temperature. After being washed with PBS, cells were incubated with the appropriate Alexa Fluor-conjugated secondary antibodies (Alexa Fluor 488-labeled goat anti-mouse immunoglobulin [A-11029; Invitrogen] or Alexa Fluor 568-labeled goat anti-rabbit immunoglobulin [A-11036; Invitrogen]) for 1 h at room temperature. Images were captured on a Zeiss LSM 800 confocal microscope.
ELISA.
Cell culture supernatants were tested for IFN-β (ABCE-EL-P1819; Advanced BioChemicals) and 2′,3′-cGAMP (501700; Cayman Chemical) with ELISA kits, according to the manufacturers’ instructions. Mouse serum was tested for IFN-β (42400; PBL Assay Science) and IL-1β (893829; R&D Systems) with ELISA kits.
Comet assays.
Normal-melting-point agarose (NMA; 0.5%) was used to coat frosted microscope slides. Approximately 10,000 cells in 10 μl DMEM were mixed with 75 μl low-melting-point agarose (LMA; 0.7%), and the mixture was dripped onto the precoated NMA layers. The third layers were prepared with 75 μl 0.7% LMA. The cells were lysed in lysis buffer (2.5 M NaCl, 100 mM Na2EDTA, 10 mM Tris [pH 10.0], 1% Triton X-100, and 10% DMSO) for 2 h at 4°C. After lysis, the slides were placed in electrophoresis solution (300 mM NaOH, 1 mM Na2EDTA [pH >13]) for 40 min, subjected to electrophoresis at 20 V (∼300 mA) for 25 min, and neutralized with 0.4 mM Tris-HCl (pH 7.5). Finally, the cells were stained with PI (5 μg/ml) and evaluated on a Zeiss LSM 800 confocal microscope. DNA damage was measured in terms of tail moment in cometscore software.
TCID50 assay.
A 50% tissue culture infective dose (TCID50) assay was performed to quantify the viral titers. Briefly, 10-fold dilutions of PRV-QXX were inoculated into Vero cells grown in 96-well plates at 1 × 104 cells/well. The plate was incubated at 37°C for a further 3 to 5 days, followed by observation of the cytopathic effect of each well under a light microscope. The TCID50 was calculated using the Reed-Muench method.
Histological analysis.
Tissues dissected from mice were fixed in 4% paraformaldehyde (158127; Sigma-Aldrich) overnight, embedded in paraffin, sectioned, and stained with H&E solution.
Statistical analysis.
All data were analyzed in Prism 7 software (GraphPad Software) with two-tailed Student's t test or one-way analysis of variance (ANOVA). P values of <0.05 were considered statistically significant. For mouse survival studies, Kaplan-Meier survival curves were generated and analyzed for statistical significance.
ACKNOWLEDGMENTS
This work was supported by grants from the Natural Science Foundation of Henan (202300410213), the 10 Thousand Talents Program for Young Talents (W03070106), and Outstanding Talents of Henan Agricultural University (30600773).
We declare that we have no competing interests.
Contributor Information
Jiang Wang, Email: wangjiang@henau.edu.cn.
Bei-Bei Chu, Email: chubeibei@henau.edu.cn.
Jae U. Jung, Lerner Research Institute, Cleveland Clinic
REFERENCES
- 1.Pomeranz LE, Reynolds AE, Hengartner CJ. 2005. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev 69:462–500. 10.1128/MMBR.69.3.462-500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woźniakowski G, Samorek-Salamonowicz E. 2015. Animal herpesviruses and their zoonotic potential for cross-species infection. Ann Agric Environ Med 22:191–194. 10.5604/12321966.1152063. [DOI] [PubMed] [Google Scholar]
- 3.Ai JW, Weng SS, Cheng Q, Cui P, Li YJ, Wu HL, Zhu YM, Xu B, Zhang WH. 2018. Human endophthalmitis caused by pseudorabies virus infection, China, 2017. Emerg Infect Dis 24:1087–1090. 10.3201/eid2406.171612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang X, Guan H, Li C, Li Y, Wang S, Zhao X, Zhao Y, Liu Y. 2019. Characteristics of human encephalitis caused by pseudorabies virus: a case series study. Int J Infect Dis 87:92–99. 10.1016/j.ijid.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Wong G, Lu J, Zhang W, Gao GF. 2019. Pseudorabies virus: a neglected zoonotic pathogen in humans? Emerg Microbes Infect 8:150–154. 10.1080/22221751.2018.1563459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pirrotta V. 2004. The ways of PARP. Cell 119:735–736. 10.1016/j.cell.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Ame JC, Spenlehauer C, de Murcia G. 2004. The PARP superfamily. Bioessays 26:882–893. 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 8.Kim MY, Zhang T, Kraus WL. 2005. Poly(ADP-ribosyl)ation by PARP-1: 'PAR-laying' NAD+ into a nuclear signal. Genes Dev 19:1951–1967. 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- 9.Huambachano O, Herrera F, Rancourt A, Satoh MS. 2011. Double-stranded DNA binding domain of poly(ADP-ribose) polymerase-1 and molecular insight into the regulation of its activity. J Biol Chem 286:7149–7160. 10.1074/jbc.M110.175190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krietsch J, Rouleau M, Pic E, Ethier C, Dawson TM, Dawson VL, Masson JY, Poirier GG, Gagne JP. 2013. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol Aspects Med 34:1066–1087. 10.1016/j.mam.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. 2008. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res 36:6959–6976. 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. 2000. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem 275:40974–40980. 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 13.Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791. 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, Zuo X, Kao CC, Herr AB, Li P. 2013. Cyclic GMP-AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39:1019–1031. 10.1016/j.immuni.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. 2013. cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING. Nature 498:380–384. 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka Y, Chen ZJ. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5:ra20. 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Li GL, Ming SL, Wang CF, Shi LJ, Su BQ, Wu HT, Zeng L, Han YQ, Liu ZH, Jiang DW, Du YK, Li XD, Zhang GP, Yang GY, Chu BB. 2020. BRD4 inhibition exerts anti-viral activity through DNA damage-dependent innate immune responses. PLoS Pathog 16:e1008429. 10.1371/journal.ppat.1008429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun J, Lv YG, Yao Q, Wang L, Li YP, Yi J. 2012. Wortmannin inhibits proliferation and induces apoptosis of MCF-7 breast cancer cells. Eur J Gynaecol Oncol 33:367–369. [PubMed] [Google Scholar]
- 19.Ray Chaudhuri A, Nussenzweig A. 2017. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol 18:610–621. 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodley A, Liu LF, Israel M, Seshadri R, Koseki Y, Giuliani FC, Kirschenbaum S, Silber R, Potmesil M. 1989. DNA topoisomerase II-mediated interaction of doxorubicin and daunorubicin congeners with DNA. Cancer Res 49:5969–5978. [PubMed] [Google Scholar]
- 21.Sharma A, Singh K, Almasan A. 2012. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol Biol 920:613–626. 10.1007/978-1-61779-998-3_40. [DOI] [PubMed] [Google Scholar]
- 22.Bednarski JJ, Sleckman BP. 2019. At the intersection of DNA damage and immune responses. Nat Rev Immunol 19:231–242. 10.1038/s41577-019-0135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma F, Li B, Liu SY, Iyer SS, Yu Y, Wu A, Cheng G. 2015. Positive feedback regulation of type I IFN production by the IFN-inducible DNA sensor cGAS. J Immunol 194:1545–1554. 10.4049/jimmunol.1402066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Ba G, Han YQ, Ming SL, Wang MD, Fu PF, Zhao QQ, Zhang S, Wu YN, Yang GY, Chu BB. 2020. Cyclic GMP-AMP synthase is essential for cytosolic double-stranded DNA and fowl adenovirus serotype 4 triggered innate immune responses in chickens. Int J Biol Macromol 146:497–507. 10.1016/j.ijbiomac.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ. 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341:1390–1394. 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS. 2015. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520:553–557. 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luthra P, Aguirre S, Yen BC, Pietzsch CA, Sanchez-Aparicio MT, Tigabu B, Morlock LK, Garcia-Sastre A, Leung DW, Williams NS, Fernandez-Sesma A, Bukreyev A, Basler CF. 2017. Topoisomerase II inhibitors induce DNA damage-dependent interferon responses circumventing Ebola virus immune evasion. mBio 8:e00368-17. 10.1128/mBio.00368-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Chu B, Du L, Han Y, Zhang X, Fan S, Wang Y, Yang G. 2015. Molecular cloning and functional characterization of porcine cyclic GMP-AMP synthase. Mol Immunol 65:436–445. 10.1016/j.molimm.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh R, Roy S, Franco S. 2018. PARP1 depletion induces RIG-I-dependent signaling in human cancer cells. PLoS One 13:e0194611. 10.1371/journal.pone.0194611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunphy G, Flannery SM, Almine JF, Connolly DJ, Paulus C, Jonsson KL, Jakobsen MR, Nevels MM, Bowie AG, Unterholzner L. 2018. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-kappaB signaling after nuclear DNA damage. Mol Cell 71:745–760.E5. 10.1016/j.molcel.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai Y, Kong Z, Zeng T, Xu S, Duan X, Li S, Cai C, Zhao Z, Wu W. 2018. PARP1-siRNA suppresses human prostate cancer cell growth and progression. Oncol Rep 39:1901–1909. 10.3892/or.2018.6238. [DOI] [PubMed] [Google Scholar]
- 32.Kurokawa S, Okuda A, Nishizawa Y, Furukawa K, Sumihiro A, Nakaji Y, Tanaka S, Takehashi M. 2019. Suppression of cell cycle progression by poly(ADP-ribose) polymerase inhibitor PJ34 in neural stem/progenitor cells. Biochem Biophys Res Commun 510:59–64. 10.1016/j.bbrc.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 33.Ding L, Kim HJ, Wang Q, Kearns M, Jiang T, Ohlson CE, Li BB, Xie S, Liu JF, Stover EH, Howitt BE, Bronson RT, Lazo S, Roberts TM, Freeman GJ, Konstantinopoulos PA, Matulonis UA, Zhao JJ. 2018. PARP inhibition elicits STING-dependent antitumor immunity in Brca1-deficient ovarian cancer. Cell Rep 25:2972–2980.E5. 10.1016/j.celrep.2018.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reislander T, Lombardi EP, Groelly FJ, Miar A, Porru M, Di Vito S, Wright B, Lockstone H, Biroccio A, Harris A, Londono-Vallejo A, Tarsounas M. 2019. BRCA2 abrogation triggers innate immune responses potentiated by treatment with PARP inhibitors. Nat Commun 10:3143. 10.1038/s41467-019-11048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen J, Zhao W, Ju Z, Wang L, Peng Y, Labrie M, Yap TA, Mills GB, Peng G. 2019. PARPi triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCAness. Cancer Res 79:311–319. 10.1158/0008-5472.CAN-18-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boussios S, Karihtala P, Moschetta M, Karathanasi A, Sadauskaite A, Rassy E, Pavlidis N. 2019. Combined strategies with poly(ADP-ribose) polymerase (PARP) inhibitors for the treatment of ovarian cancer: a literature review. Diagnostics (Basel) 9:87. 10.3390/diagnostics9030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Lu SF, Wan B, Ming SL, Li GL, Su BQ, Liu JY, Wei YS, Yang GY, Chu BB. 2018. Maintenance of cyclic GMP-AMP homeostasis by ENPP1 is involved in pseudorabies virus infection. Mol Immunol 95:56–63. 10.1016/j.molimm.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 38.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed. National Academies Press, Washington, DC. [Google Scholar]