ABSTRACT
HIV frequently escapes CD8 T cell responses, leading to the accumulation of viral adaptations. We recently demonstrated that during chronic HIV infection, adapted epitopes can promote maturation of dendritic cells (DCs) through direct CD8 T cell interactions and lead to enhanced HIV trans-infection of CD4 T cells. Here, we sought to determine the role of such adaptations following HIV MRKAd5 vaccination. We observed that vaccine-induced adapted epitope-specific CD8 T cells promoted higher levels of DC maturation than nonadapted ones and that these matured DCs significantly enhanced HIV trans-infection. These matured DCs were associated with higher levels of interleukin 5 (IL-5) and IL-13 and a lower level of CXCL5, which have been shown to impact DC maturation, as well as a lower level of CXCL16. Finally, we observed that vaccine recipients with high HLA-I-associated adaptation became HIV infected more quickly. Our results offer another possible mechanism for enhanced infection among MRKAd5 vaccinees.
IMPORTANCE Despite the well-established contribution of CD8 T cells in HIV control, prior CD8 T cell-based HIV vaccines have failed to demonstrate any efficacy in preventing viral infection. One such vaccine, known as the MRKAd5 vaccine, showed a potential increased risk of viral infection among vaccine recipients. However, the underlying mechanism(s) remains unclear. In this study, we observed that vaccine recipients with high adaptation to their HLA-I alleles were associated with an increased HIV infection risk in comparison to the others. Similar to what we observed in HIV infection in the prior study, adapted epitope-specific CD8 T cells obtained from vaccine recipients exhibit a greater capacity in facilitating viral infection by promoting dendritic cell maturation. Our findings provide a possible explanation for the enhanced viral acquisition risk among MRKAd5 vaccine recipients and highlight the importance of optimizing vaccine design with consideration of HLA-I-associated HIV adaptation.
KEYWORDS: MRKAd5 vaccine, STEP study, HIV vaccine, HLA-I-associated HIV adaptation, viral trans-infection, CD8 T cells, monocyte-derived dendritic cells
INTRODUCTION
Human immunodeficiency virus (HIV) remains a significant global health issue, and it is widely recognized that an effective vaccine would be the most potent tool to combat this infection. To date, only one vaccine, tested in the RV144 trial, has demonstrated any efficacy, which was presumed to be due to the induction of nonneutralizing antibodies (1, 2); however, this protection was not seen in a recent similarly designed study (https://www.niaid.nih.gov/news-events/experimental-hiv-vaccine-regimen-ineffective-preventing-hiv). CD8 T cells have been well established as a critical component of natural viral control, in both simian immunodeficiency virus (SIV)/nonhuman primate models (3–5) and studies of HIV-infected humans (6–8). While this abundance of literature suggests that robust, vaccine-induced CD8 T cells might play a pivotal role in HIV vaccine efficacy, there has been no clinical evidence that such responses have prevented infection or impacted natural disease progression in those infected. In fact, the MRKAd5 trial, also known as the “Step Study” or HVTN 502, testing an adenovirus serotype 5 (Ad5) vectored vaccine encoding HIV Gag, Nef, and Pol, was prematurely ended due to futility in impacting vaccine efficacy. More concerning, post hoc subgroup analyses demonstrated an elevated risk of HIV acquisition among vaccine recipients, specifically Ad5-seropositive men and uncircumcised men, in comparison to placebo recipients (9).
Several groups have attempted to define the biological mechanism(s) underlying the elevated infection risk in MRKAd5 vaccine recipients. Because an increased number of infections were observed in individuals with preexisting Ad5 titers, one commonly accepted hypothesis is that the vaccine activated preexisting Ad5-specific CD4 T cells, which increased the number of targets for HIV infection (10). However, subsequent studies by multiple groups failed to show an association between Ad5-specific CD4 T cells and HIV acquisition in MRKAd5 vaccine recipients (11, 12). Additional studies have also attempted to address this question from different angles; for instance, Barnabas et al. reported an association between preexisting herpes simplex virus 2 (HSV-2) immunity and HIV acquisition (13). However, this effect was observed in both vaccine recipients and placebo recipients and therefore could not explain the enhanced risk associated with vaccination. Another study suggested that Ad5 immune complexes could mature dendritic cells (DCs), which then enhanced HIV infection of CD4 T cells, but this study was also unable to link the in vitro findings with vaccine-induced responses in vivo (14).
Recently, the dual role played by DCs during viral infection has emerged (15). While DCs classically initiate and activate efficient immune responses against viral infections (16, 17), several studies have highlighted a less desirable and possibly pathogenic role for DCs whereby they facilitate HIV trans-infection among target cells (18–20). We recently described a role for DCs in the negative effect that HLA-I-associated adaptation has in HIV infection. HLA-I-associated polymorphisms, or adaptations, are defined based on their statistical association with certain HLA-I alleles in chronic infection, and the accumulation of adaptations in a viral strain specific for any given HLA-I repertoire can be statistically quantified (21). HLA-I-associated HIV polymorphisms largely occur from CD8 T cell immune pressure forcing viral adaptations. Poor clinical outcomes are associated with infection by HIV strains that are, by chance, preadapted to the host’s HLA-I alleles, and HLA-I-associated adapted epitopes are poorly immunogenic in acute HIV infection (21). Our group recently reported that CD8 T cell responses frequently target adapted epitopes in chronic HIV infection, and these adapted epitope-specific CD8 T cell responses appear to be exploited by HIV through the promotion of DC maturation to fuel viral infection (22). Given these data, we reasoned that CD8 T cells induced by vaccine-encoded adapted epitopes may also promote DC-mediated viral trans-infection to CD4 T cells.
Many candidate HIV vaccines are derived from natural strains, which likely harbor adaptations to their hosts’ HLA-I alleles (23). Thus, vaccine immunogens used in these candidate HIV vaccines by default encode epitopes encompassing adaptations unless they are purposefully removed. In our recent work, we found that the HIV MRKAd5 vaccine not only encoded adapted epitopes but also elicited CD8 T cell responses specific to them (24). However, it remains unclear whether such responses can lead to DC maturation, enhanced viral trans-infection of CD4 T cells, and increased infection risk.
In the present study, we explore the biological relevance of HLA-I-associated adapted epitopes encoded in MRKAd5 vaccine inserts. Based on each individual’s HLA-I alleles, we stratified vaccinees according to the degree to which the vaccine insert was adapted to their potential CD8 T cell response (24). We found that as with chronic HIV infection, CD8 T cells induced by adapted vaccine epitopes promoted viral DC maturation and viral trans-infection of CD4 T cells. Moreover, recipients of the MRKAd5 vaccine with high levels of adaptation became HIV infected at a higher rate than both placebo recipients and other vaccine recipients. These data suggest that HLA-I-associated adaptations encoded by HIV vaccine might be detrimental to vaccine efficacy.
RESULTS
No impact of vaccine-encoded adaptation on viral suppression capacity of CD8 T cells.
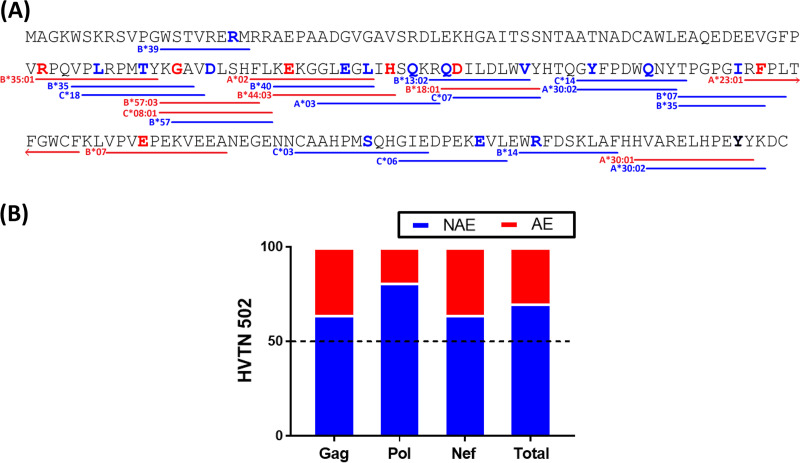
Because HIV has circulated and adapted in humans for many years, individuals can be infected with HIV that is, by chance, preadapted to the HLA-I restricted immune system of the new host (21). A similar concept can be applied to HIV vaccines. Although each vaccine recipient receives the same immunogen, the extent to which the immunogen is adapted to each individual is dependent on their unique HLA-I allelic repertoire. To illustrate this point, the adapted epitopes (AE) and nonadapted epitopes (NAE) within the Nef protein included in the MRKAd5 vaccine and their corresponding HLA-I alleles are shown in Fig. 1A. Overall, 30% of these HLA-I-restricted epitopes contained adapted polymorphisms, demonstrating the importance of establishing the relevance of HLA-I-associated adaptation in HIV vaccines (Fig. 1B).
FIG 1.
Adapted epitopes are commonly encoded in the MRKAd5 vaccine inserts. (A) Nef protein sequence encoded by the MRKAd5 vaccine is shown. Bold amino acid residues in red and blue indicate adapted and nonadapted polymorphisms, respectively. Lines underlining amino acid residues represent an adapted (red) or nonadapted (blue) epitope with their respective HLA-I restricting allele given. (B) Proportion of adapted versus nonadapted epitopes encoded within each vaccine immunogen of the MRKAd5 vaccine, and overall, is shown.
While previous work from our group showed that vaccine-encoded adapted epitopes are poorly immunogenic (24), it remains unclear how the overall adaptation impacts vaccine-induced CD8 T cell function. To address this question, we used data from the modified intent-to-treat arm of the MRKAd5 trial and categorized individuals as having a high adaptation score based on the MRKAd5 vaccine insert (vaccine recipient 1 [VR1] to VR7; nhigh [adaptation score range of 0.18 to 0.28] = 7), or a low one (VR8 to VR16; nlow [adaptation score range of −0.52 to −0.68] = 9), as previously established by Carlson et al. (Table 1) (21).
TABLE 1.
Demographics of vaccine recipients whose PBMC samples were used in this study
| Vaccine recipienta | Wkb | Age (yrs) | HLA-I allelesc |
|||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | ||||||
| VR1 | 30 | 21 | A2402 | A0210 | B3505 | B3543 | C0102 | C0401 |
| VR2 | 30 | 26 | A0201 | A3201 | B3508 | B4402 | C0202 | C0401 |
| VR3 | 30 | 36 | A0213 | A6802 | B1503 | B3543 | C0102 | C0210 |
| VR4 | 30 | 27 | A0201 | A0201 | B1504 | B3506 | C0102 | C0401 |
| VR5 | 30 | 26 | A0301 | A7401 | B1503 | B4403 | C0210 | C0303 |
| VR6 | 30 | 23 | A0201 | A0201 | B1504 | B3508 | C0401 | C0401 |
| VR7 | 30 | 25 | A0301 | A2402 | B0702 | B3543 | C0102 | C0702 |
| VR8 | 30 | 40 | A0206 | A7401 | B4001 | B4001 | C0702 | C0702 |
| VR9 | 30 | 41 | A1101 | A1101 | B3501 | B3501 | C0401 | C0602 |
| VR10 | 30 | 25 | A1101 | A3101 | B2704 | B3905 | C0702 | C1202 |
| VR11 | 30 | 33 | A0101 | A2601 | B1401 | B5701 | C0701 | C0802 |
| VR12 | 30 | 23 | A3001 | A3301 | B1302 | B1402 | C0602 | C0802 |
| VR13 | 30 | 30 | A3002 | A6602 | B4501 | B5801 | C0701 | C1601 |
| VR14 | 30 | 23 | A0201 | A1101 | B4002 | B5101 | C0304 | C1402 |
| VR15 | 30 | 33 | A0101 | A7401 | B5101 | B5703 | C0701 | C1601 |
| VR16 | 30 | 36 | A3201 | A7401 | B5101 | B5301 | C0401 | C1402 |
| VR17 | 8 | 27 | A0205 | A0201 | B4402 | B4901 | C0501 | C0701 |
| VR18 | 8 | 22 | A0101 | A0201 | B0801 | B4402 | C0501 | C0701 |
| VR19 | 8 | 26 | A0301 | A2402 | B1501 | B4402 | C0303 | C0501 |
| VR20 | 30 | 39 | A0201 | A2902 | B4403 | B4402 | C0501 | C1601 |
| VR21 | 30 | 41 | A0201 | A6801 | B4402 | B5301 | C0401 | C0501 |
| VR22 | 30 | 44 | A0201 | A2402 | B4402 | B5101 | C0501 | C1502 |
| VR23 | 30 | 29 | A0201 | A2501 | B1801 | B4001 | C0304 | C1203 |
| VR24 | 52 | 20 | A3002 | A6602 | B5702 | B5801 | C0701 | C1801 |
All vaccine recipients were male.
Wk, visit week. Week 8 is 4 weeks following the second vaccination; week 30 and week 52 are 4 and 26 weeks, respectively, following the third vaccination.
A, B, and C refer to HLA-A, HLA-B, and HLA-C, respectively.
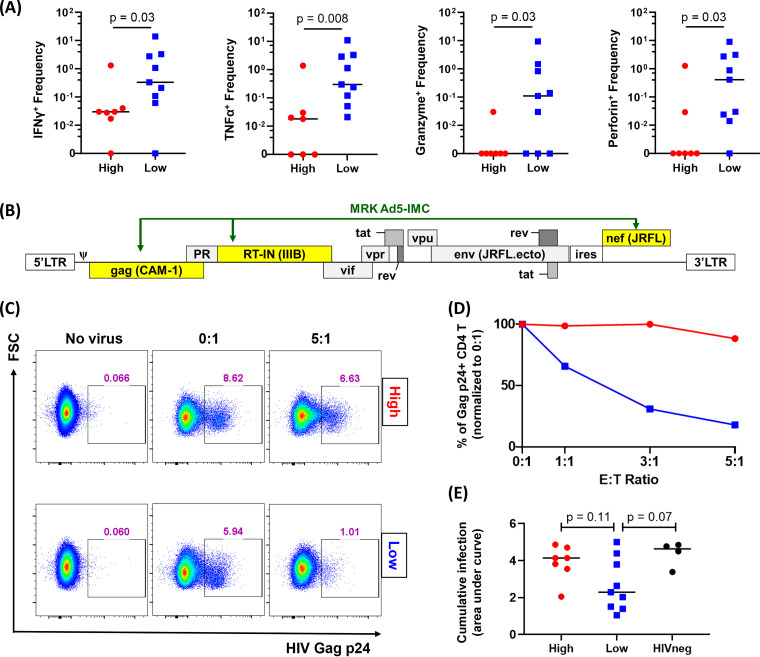
Using a CD3/CD4 (anti-CD3/anti-CD4)-bispecific monoclonal antibody, we expanded CD8 T cells from each individual and measured effector molecule production by intracellular cytokine staining with flow cytometry. We found that individuals with high adaptation scores had a lower frequency of HIV-specific CD8 T cells producing gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), granzyme A/B, and perforin induced by a Gag peptide pool (Fig. 2A). No such differences were observed for the Pol- and Nef-specific CD8 T cell responses (data not shown).
FIG 2.
CD8 T cells from recipients with high adaptation demonstrate lower effector functions. (A) Cytokine/effector molecule production levels by CD8 T cells responding to Gag peptide pool are shown. ICS staining showed poorer functionality of CD8 T cells derived from vaccinees with high adaptation scores (n = 7) than low adaptation scores (n = 9). (B) Genomic composition of MRKAd5-IMC is shown; yellow highlights indicate those sequences derived from the vaccine insert. (C) Representative flow cytometry plots showing IMC suppression data from vaccine recipients with high and low adaptation. CD8 T cells from PBMCs after nonspecific expansion in the presence of CD3/CD4 antibody were cocultured with autologous CD4 T cells infected with MRKAd5-IMC at different E:T ratios in duplicate. (D) For each E:T ratio, the average percentage of p24+ CD4 target cells was plotted for each coculture and normalized to the 0:1 E:T ratio (no CD8 T cells) and was normalized to 100% p24+ expression. (E) Cumulative data of the IMC suppression assay is shown. The area under the curve (AUC) is used to quantify viral suppression of infected CD4 T cells. The Mann-Whitney U test was used in panels A and E.
We next measured the ability of these CD8 T cells to suppress virus replication in vitro. In order to closely mimic the vaccination setting, we engineered an HIV-1 infectious molecular clone (IMC) that encodes Gag, RT-IN, and Nef sequences that exactly match those encoded by the MRKAd5 vaccine (Fig. 2B). Using nonspecifically expanded CD8 T cells as effectors and autologous CD4 T cells as targets, we cocultured infected target cells with CD8 T cells for 96 h at various effector-to-target cell (E:T) ratios. Representative Gag-p24 staining of CD4 target cells in individuals with either high or low adaptation is shown in Fig. 2C, and normalized data are shown in Fig. 2D. CD8 T cells from individuals with high adaptation tended to suppress the vaccine-matched IMC more poorly than CD8 T cells from individuals with low adaptation, but these differences did not reach statistical significance (Fig. 2E).
Differential gene expression profile of CD8 T cells responding to vaccine-encoded adapted and nonadapted epitope.
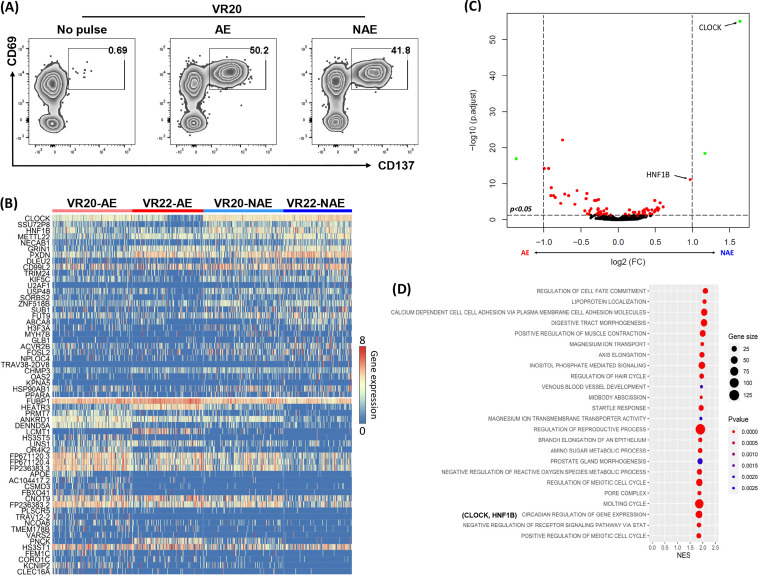
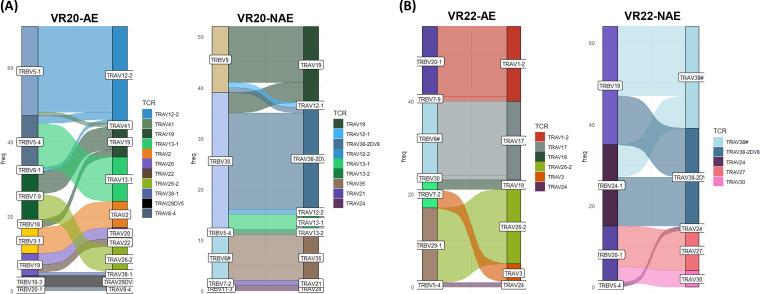
Although we did not see significant differences in viral suppression, it is still possible that CD8 T cells from vaccinees with low versus high adaptation scores have distinct cellular functions associated with viral control that can be observed at a transcriptional level. Since bulk transcriptional analyses often mask functional heterogeneity, we assessed the impact of epitope adaptation to CD8 T cells by generating single epitope (adapted or nonadapted)-specific cell lines using CD8 T cells isolated from MRKAd5 vaccine recipients (Table 2). These epitope-specific CD8 T cells were expanded for 2 weeks by using autologous peptide pulsed monocytes as antigen-presenting cells (APCs). Next, epitope-specific CD8 T cells were single cell sorted based on the expression of T cell-specific activation-induced markers (AIM), CD69 and CD137, following peptide stimulation (Fig. 3A). We sorted CD8 T cells from two vaccine recipients; for each individual, we assessed one adapted and one nonadapted epitope-specific CD8 T cell response. We performed single-cell RNA sequencing (scRNAseq) to obtain paired T cell receptor alpha/beta (TCR-α/β) and transcriptomic profiles of these CD8 T cells. Our TCR sequencing results showed no overlap in the TCR clones making up the responses to the adapted and nonadapted epitopes (Fig. 4 and Table 3). Based on the transcriptomic data, we identified 68 differentially expressed genes by comparing adapted versus nonadapted epitope-specific CD8 T cells (adjusted P value, <0.05) (Fig. 3B and C). Interestingly, CLOCK and HNF1B, which are critical for circadian rhythm, were found to be upregulated in both recipients with the nonadapted epitope-specific CD8 T cell responses. It is well known that CLOCK can activate the NF-κB pathway, which is an initial step in both pro- and antiinflammatory pathways (25, 26). To further identify the biological relevance of the differentially expressed genes, we interrogated our transcriptomic data by gene set enrichment analysis (GSEA) (Fig. 3D). Unsurprisingly, this analysis showed an enriched gene set relevant to circadian regulation in the nonadapted epitope group (Fig. 3D). We did not observe any known immunological pathways relevant to viral suppression enriched between adapted and nonadapted epitope-specific CD8 T cells. These findings suggest that the decreased function seen in CD8 T cells from individuals with high vaccine adaptation scores could be due to less in vitro proliferation of CD8 T cells specific to adapted epitopes (Fig. 2A); however, individual vaccine-induced CD8 T cells were not different with respect to these functional characteristics (Fig. 3), consistent with the similar in vitro viral inhibition capacities of CD8 T cells in those with low and high adaptation scores (Fig. 2E). Although individuals with high vaccine adaptation scores mounted less-cytotoxic CD8 T-cell responses, we did not observe differential gene expression in canonical immunological pathways in adapted versus nonadapted epitope-specific CD8 T cells.
TABLE 2.
Epitope specificity of immunogenic NAE- and AE-specific CD8 T cell responses examined in this study
| Vaccine recipient | Epitope |
||||
|---|---|---|---|---|---|
| HLA | Adapted | Protein | Sequence | Net SFUa | |
| VR17 | B49 | NAE | Gag | GELDKWEKI | 1,210.0 |
| B44 | AE | Pol | KEKGGLEGLIH | 127.5 | |
| VR18 | B44 | NAE | Pol | AEIQKQGQGQW | 95.0 |
| B44 | AE | Gag | AEQASQEVKNW | 100.0 | |
| VR19 | A03 | NAE | Pol | RLRPGGKKKYK | 757.5 |
| A03 | AE | Pol | QIYPGIKVR | 172.5 | |
| VR20 | B44 | NAE | Pol | AEIQKQGQGQW | 177.5 |
| B44 | AE | Gag | AEQASQEVKNW | 337.5 | |
| VR21 | B44 | NAE | Pol | AEIQKQGQGQW | 395.0 |
| B44 | AE | Gag | AEQASQEVKNW | 840.0 | |
| VR22 | A24 | NAE | Pol | KYKLKHIVW | 75.0 |
| B44 | AE | Gag | AEQASQEVKNW | 80.0 | |
| VR23 | B40 | NAE | Nef | KEKGGLEGL | 255.0 |
| A02 | AE | Nef | FLKEKGGLEGL | 230.0 | |
| VR24 | B57 | NAE | Gag | TSTLQEQIGW | 360.8 |
| B57 | AE | Nef | KGAVDLSHF | 220.8 | |
Net SFU, background-subtracted spot-forming units per million cells (measured by ex vivo IFN-γ ELISpot assay).
FIG 3.
Single-cell RNA sequencing shows different transcriptomic profiles between adapted and nonadapted epitope-specific CD8 T cells. (A) Representative flow chart of vaccine recipient 20 (VR20) showing the identification of antigen-specific CD8 T cells using activation-induced marker expression (upregulation of CD69 and CD137) for single cell sorting. (B) Heat map showing differential transcriptomic profile of NAE (n = 2)- and AE (n = 2)-specific CD8 T cells from two recipients (VR20 and VR22). In each group, 158 (VR20-NAE), 133 (VR22-NAE), 157 (VR20-AE), and 139 (VR22-AE) total antigen-specific CD8 T cells, respectively, were analyzed. (C) Volcano plot of combined data of AE (left) for VR20 and VR22 versus corresponding NAE-specific T cells (right). (D) GSEA of the scRNAseq data with reference to the Gene Ontology Database, with the top 25 enriched pathways in NAE-specific CD8 T cells (FDR < 25%) shown.
FIG 4.
Distinct TCR-α/β usage between NAE- and AE-specific CD8 T cells. Single-cell TCR sequencing results reveal the TCR clonotype of antigen-specific CD8 T cells sorted by AIM. The frequency of TCR expression is shown for both VR20 (A) and VR22 (B). Each TCR-α/β is a distinct color. TCR-α is shown as TRA, while TCR-β is shown as TRB.
TABLE 3.
TCR-α/β combinations and overlap within vaccine recipients
| Vaccine recipient | Adapted | Epitope sequence | No. (%) of wells with a productive TCR | No. of unique CDR3-α/β combinations (total) | Dominant CDR3-α/β clone (%)a | CDR3-α/β overlap (%)b |
|---|---|---|---|---|---|---|
| VR20 | AE | AEQASQEVKNW | 153 (80) | 75 (213) | 10.80 | 0 |
| NAE | AEIQKQGQGQW | 135 (70) | 44 (118) | 16.10 | 0 | |
| VR22 | AE | AEQASQEVKNW | 128 (67) | 13 (62) | 25.81 | 0 |
| NAE | KYKLKHIVW | 118 (61) | 19 (121) | 26.45 | 0 |
Percentage of the most frequent CDR3-α/β clone observed in each epitope-specific CD8 T cell population.
Percentage of CDR3-α/β shared between AE- and NAE-specific CD8 T cells within each vaccine recipient.
Higher levels of DC maturation and enhanced viral trans-infection induced by adapted epitope-specific CD8 T cells.
Although cytotoxicity and viral suppression are two of the most commonly measured metrics of HIV vaccine-induced CD8 T cell responses, CD8 T cells play a more nuanced role in the overall immune response than that measured by these traditional readouts. There is growing evidence that memory CD8 T cells can exhibit a viral helper phenotype fueling viral spread in the context of HIV infection and other pathogens (14, 20, 27). Along those lines, we recently reported on how memory CD8 T cells responding to adapted epitopes can promote HIV infection by inducing higher DC maturation (22). We were therefore interested in assessing whether HIV vaccine-induced adapted epitope-specific CD8 T cell responses might also have a similar helper effect.
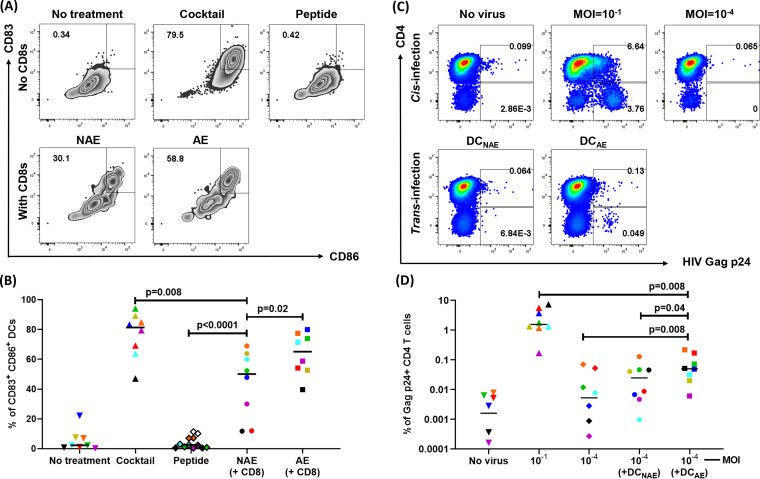
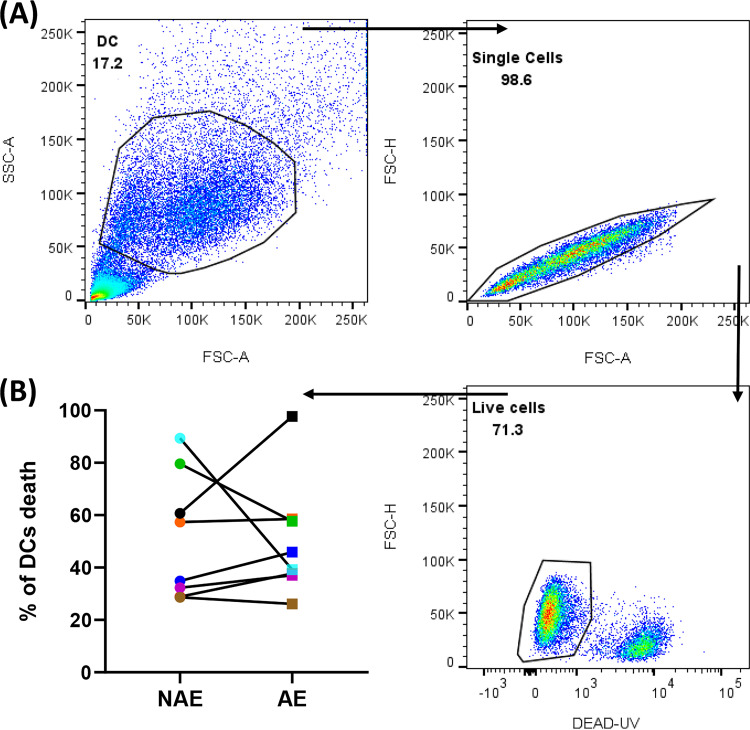
To assess the impact of adapted epitope-specific CD8 T cells on DC maturation and CD4 trans-infection, we identified eight MRKAd5 vaccine recipients who mounted both an adapted epitope-specific T cell response and a nonadapted epitope-specific CD8 T cell response (Table 2). We generated epitope-specific CD8 T cell lines and confirmed their functionality by an IFN-γ enzyme-linked immunosorbent spot (ELISpot) assay (Fig. 5). We then measured the extent of DC maturation in cocultures of adapted or nonadapted epitope-specific CD8 T cells and found that in all but one case (VR24), DCs cocultured with adapted epitope-specific CD8 T cells demonstrated a higher level of maturation as measured by CD86 and CD83 expression (n = 8, P = 0.02) (Fig. 6A, representative flow plots; Fig. 6B, compiled data). The cytotoxicity of epitope-specific CD8 T cells and the level of DC cell death were not significantly different among CD8 T cell/DC cocultures with adapted versus nonadapted epitopes (Fig. 7), further supporting our findings that vaccine-encoded adapted epitope-specific CD8 T cells drive DC maturation.
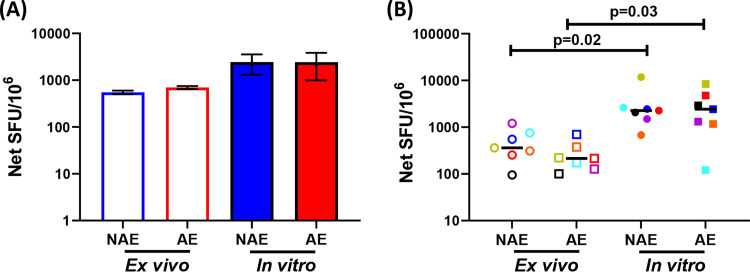
FIG 5.
Similar functionality of antigen-specific CD8 T cells when assessed in ex vivo- and in vitro-based assays. (A) Representative IFN-γ ELISpot assay using ex vivo PBMCs versus in vitro-generated CD8 T cell lines. (B) Cumulative data (n = 7) of IFN-γ production by either PBMCs (ex vivo) or antigen-specific CD8 T cells postexpansion (in vitro) are shown. Each symbol and color represent data from the same vaccine. The Wilcoxon matched-pairs signed rank test was used in panel B to determine statistical significance.
FIG 6.
Adapted epitope-specific CD8 T cells induce DC maturation and increase HIV trans-infection of CD4 T cells. (A) Representative flow plots of DC maturation assay showing frequency of mature DCs (CD83+ CD86+) for negative controls (no treatment, DC-positive peptide only) and positive control (maturation cocktail) as well as NAE- and AE-pulsed DCs cocultured with autologous CD8 T cells. (B) Cumulative data (n = 8), with each dot representing the frequency of mature DCs for each condition. (C) Representative example of viral trans-infection. Cis infection (MOI = 10−1) is shown as a positive control. The productive HIV infection was quantified by intracellular HIV Gag p24 expression within CD4 T cells (p24+ CD4−) using flow cytometry. (D) Cumulative data (n = 8) of viral trans-infection are shown. Each colored symbol represents a single vaccine recipient. The Wilcoxon matched-pairs signed rank test was used in panels B and D to determine statistical significance.
FIG 7.
Similar frequencies of cell death in DCs were observed in NAE and AE conditions following coculture. Flow cytometry viability assessment using a LIVE/DEAD UV kit was performed to measure the death of iDCs after 48 h of coculturing with CD8 T cells at a 3:1 (CD8:DC) ratio in the presence of either NAE or AE peptide (n = 8). (A) Representative flow plots of DC maturation assay showing frequency of live DCs. (B) Cumulative data demonstrate similar levels of DC viability after coculturing with NAE- or AE-specific CD8 T cells. The Wilcoxon matched-pairs signed rank test was used in panel B to determine statistical significance.
Next, we cocultured CD4 T cells with HIV presented by those DCs that had been exposed to CD8 T cells specific for adapted and nonadapted epitopes. CD4 T cells cultured with virus (multiplicity of infection [MOI] = 10−1) and in the absence of any virus were used as positive and negative controls, respectively (Fig. 6C, representative flow plots). We found that, except for VR24, DCs cocultured with adapted epitope-specific CD8 T cells demonstrated an enhanced ability to trans-infect virus to CD4 T cells in comparison with DCs cocultured with nonadapted epitope-specific CD8 T cells (n = 8, P = 0.04) (Fig. 6D). This one exception was seen in the same individual in which the adapted epitope-specific CD8 T cells did not enhance DC maturation (Fig. 6B). Our findings demonstrate that CD8 T cells elicited by vaccine-encoded adapted epitopes greatly promoted DC maturation, which was linked to viral trans-infection to CD4 T cells.
Adapted epitope-specific CD8 T cells associated with a unique microenvironment conducive to DC maturation.
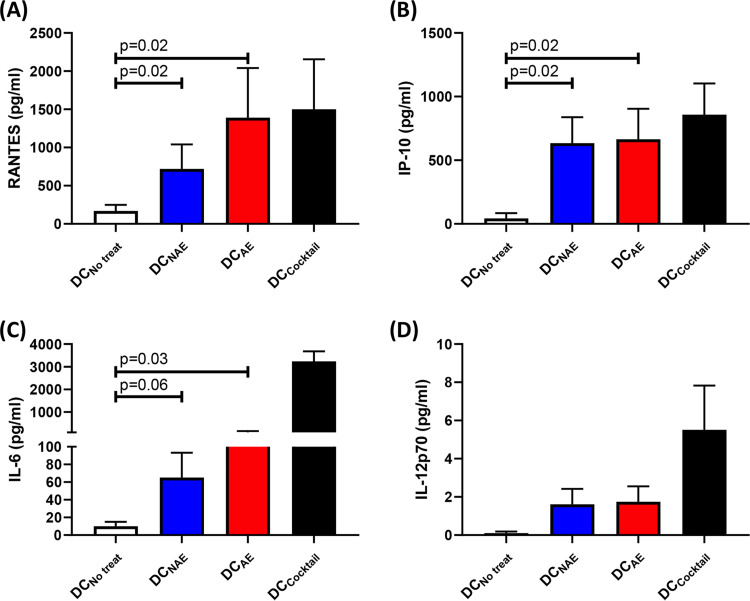
It has been previously demonstrated that DCs matured by antigen-specific CD8 T cells develop a proinflammatory phenotype with greater capacity to produce several factors, including interleukin 12p70 (IL-12p70), IL-6, IP10, and RANTES, and suggested that this type of DC might efficiently attract activated CD4 T cells as potential targets for viral infection (20). We thus examined supernatants collected from our DC maturation cocultures and observed increased levels of these same four proinflammatory factors in peptide-pulsed CD8 T cell and DC cocultures, regardless of antigen specificity, relative to those of the negative control lacking any peptide stimulation (Fig. 8). However, we observed no significant difference between adapted and nonadapted epitope-specific cultures for the four factors mentioned above.
FIG 8.
Mature DCs programmed by CD8 T cells secrete proinflammatory factors. Shown are the levels of proinflammatory factors RANTES (A), IP-10 (B), IL-6 (C), and IL-12p70 (D) in DC culture with no treatment (DCNo treat), DC-CD8 coculture with NAE (DCNAE) or AE (DCAE), or maturation cocktail (DCCocktail) as determined by multiplex array. The Wilcoxon matched-pairs signed rank test was used to determine statistical significance.
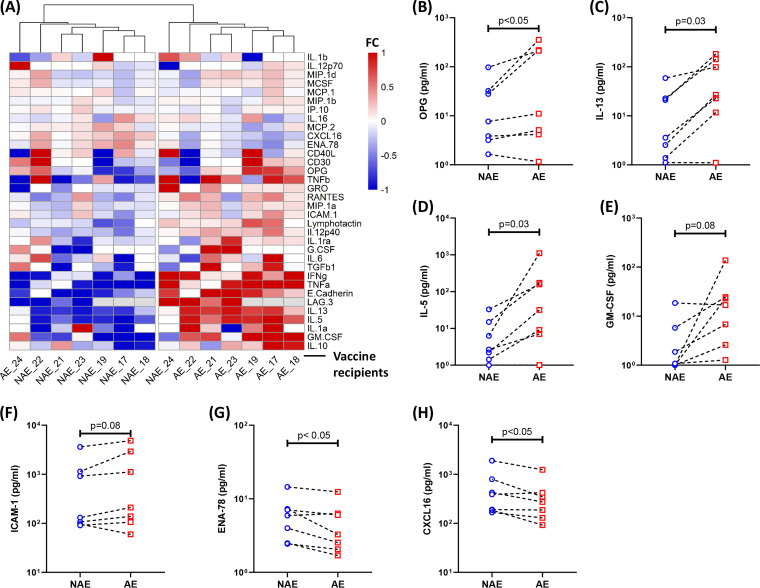
We next expanded our proteomics panel to include several other factors that could contribute to an environment favoring DC maturation (panel given in Table 4). We observed significant differences across this panel between cocultures containing adapted and nonadapted epitope-specific CD8 T cells (Fig. 9A; Table 4). Three factors, osteoprotegerin (OPG), IL-5, and IL-13, were found at higher concentrations in adapted epitope-specific cultures (Fig. 9B to D). In these cultures, we also observed a trend toward higher levels of granulocyte-macrophage colony-stimulating factor (GM-CSF) and ICAM-1 (Fig. 9E and F).
TABLE 4.
Differential levels of soluble factor in NAE- and AE-specific CD8/DC culturea
| Soluble factor | P valueb |
|---|---|
| CD30 | 0.1250 |
| CD40L | 0.9375 |
| CXCL16 | 0.0469 |
| E-cadherin | 0.2188 |
| ENA-78 | 0.0469 |
| G-CSF | 0.5000 |
| GM-CSF | 0.0781 |
| GRO | 0.1250 |
| ICAM-1 | 0.0781 |
| IFN-γ | 0.1094 |
| IL-10 | 0.1563 |
| IL-12p40 | 0.0781 |
| IL-12p70 | 0.7500 |
| IL-13 | 0.0313 |
| IL-16 | 0.9375 |
| IL-1α | 0.3125 |
| IL-1β | 0.8125 |
| IL-1rα | 0.0938 |
| IL-5 | 0.0313 |
| IL-6 | 0.4375 |
| IP-10 | 0.4688 |
| LAG-3 | 0.6250 |
| Lymphotactin | 0.1563 |
| MCP-1 | 0.8125 |
| MCP-2 | 0.1563 |
| MCSF | 0.1094 |
| MIP-1α | 0.0781 |
| MIP-1β | 0.8125 |
| MIP-1d | 0.0781 |
| OPG | 0.0469 |
| RANTES | 0.0781 |
| TGF-β1 | 0.1250 |
| TNF-α | 0.2188 |
| TNF-β | 0.8125 |
The Wilcoxon signed-rank test was used to determine the statistical significance.
Significant P values are in bold.
FIG 9.
Proteomics analyses show a unique cytokine/chemokine profile associated with promoted DC maturation and enhanced viral trans-infection. (A) Hierarchical cluster analysis of the differential production of multiple soluble factors in the tissue culture supernatant collected following DC/CD8 coculture (n = 7) is shown. Each number represents a different vaccine recipient. Comparisons between NAE and AE cultures demonstrating higher levels of OPG (B), IL-5 (C), and IL-13 (D) as well as a trend toward a higher level of GM-CSF (E) and ICAM-1 (F) are shown. The concentrations of ENA-78 (G) and CXCL16 (H) are lower in AE-specific supernatants. The Wilcoxon matched-pairs signed rank test was used in panels B to H to determine statistical significance.
On the other hand, ENA-78, also known as CXCL5, was found at lower levels in adapted epitope-specific cultures (Fig. 9G). We also saw a decreased amount of CXCL16, which is specifically produced by DCs, in adapted epitope-specific CD8 T cell cocultures (Fig. 9H). Taken together, these data suggest that adapted epitope-specific CD8 T cell responses in MRKAd5 recipients induce a unique microenvironment through interactions with DCs, which appears to favor DC maturation.
Vaccine recipients with high adaptation associated with an elevated HIV infection risk.
Our in vitro functional data suggest that CD8 T cells induced by adapted epitopes in the MRKAd5 vaccine insert might have contributed to the increased risk of infection among vaccine recipients by increasing the number of activated dendritic cells that could facilitate trans-infection of CD4 T cells upon virus exposure. Notably, such a mechanism may increase the risk of HIV infection among vaccine recipients for whom the vaccine insert contains a high burden of adapted epitopes.
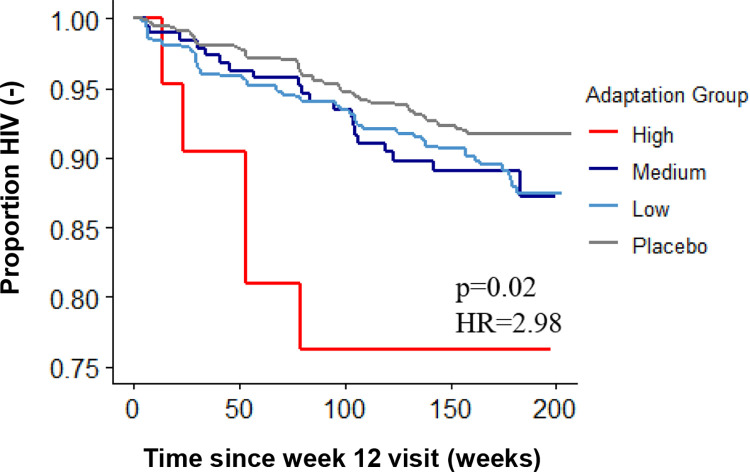
To investigate this, we stratified vaccine recipients by adaptation into high (n = 21), medium (n = 639), and low (n = 192) groups, using thresholds observed in chronic natural infection (21). We then compared the rates of infection within these groups using the Cox proportional-hazards regression analysis. Overall, compared to the other two vaccine recipient groups and the placebo recipient group, the vaccinees with a high vaccine adaptation score demonstrated an increased risk of HIV infection (Fig. 10) (P = 0.02, hazard ratio [HR] = 2.98). These findings indicate that HLA-I-associated adaptation encoded by the vaccine insert may have contributed to an increased risk of HIV acquisition, as observed among the MRKAd5 vaccine recipients.
FIG 10.
Higher HLA-I adaptation to the MRKAd5 insert increases risk of infection among vaccine recipients. Kaplan-Meier curves are shown for vaccine recipients with high, medium, or low adaptation scores to the MRKAd5 vaccine insert as well as placebo recipients (nhigh = 21, nmedium = 639, nlow = 192, nplacebo = 865; the Wald test was used to determine the statistical significance).
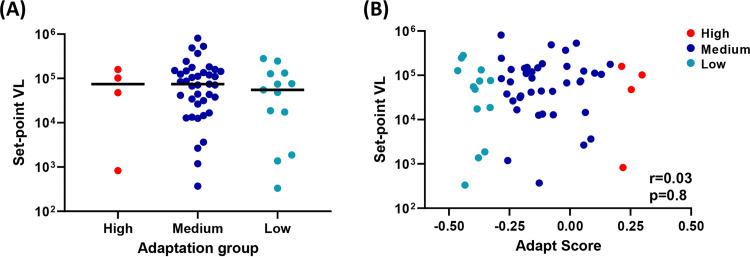
Since our prior study showed a strong correlation between the level of HLA-I-associated adaptation of the founder virus and an early set point viral load of the infected MRKAd5 vaccine recipients, we next questioned whether vaccine-encoded adaptation would have a similar correlation with set point viral load (21). We first compared the levels of set point viral load among the above-mentioned three groups of vaccine recipients who became infected. We observed no difference in set point viral load across these groups (Fig. 11A). In addition, no correlation was found between vaccine-encoded adaptation and set point viral load (Fig. 11B). These data suggest that in contrast to the negative impact of HLA-I preadaptation of the transmitted founder virus on early viral control (21), overall vaccine adaptation level does not significantly impact CD8 T cell-mediated viral control in vaccine recipients who get infected. This may be due to the fact that the infecting virus induces a broader immune response than does the HIV vaccination. In summary, the level of vaccine-encoded HLA-I-associated adaptation appears to negatively impact the risk of infection, while viral control is dominantly impacted by adaptation encoded by transmitted founder virus (21).
FIG 11.
No association between vaccine-encoded adaptation and set point viral load of infected vaccine recipients. (A) Similar levels of set point viral load among vaccine recipients who became infected in the high, medium, and low adaptation groups (nhigh = 4, nmedium = 39, nlow = 13). The Mann-Whitney U test was used to determine statistical significance. (B) Spearman's rank correlation analysis between adaptation score and set point viral load.
DISCUSSION
The MRKAd5 HIV vaccine study was halted for futility and showed an unexpectedly high number of HIV infections in vaccine recipients (9). While a prior study suggested that this elevated infection risk was associated with the activation of preexisting Ad5-specific CD4 T cells by the vaccine vector, no study has provided solid biological support for this hypothesis to date (11, 12). Here, we demonstrate that HLA-I-associated adaptation commonly encoded in the MRKAd5 vaccine insert is associated with increased HIV infections and that this infection risk might be caused by CD8 T cell-driven maturation of DCs and enhanced viral trans-infection. Given that MRKAd5 heavily focused on cellular immunity, it is worth noting that inclusion of env sequences in other vaccine regimens might alleviate the observed untoward effects of adaptation, since antibody responses induced by env immunogens have been shown to promote CD8 T cell functionality (28). Nevertheless, these data are critical to consider in ongoing and future HIV vaccine trials, as many of these vaccines are not designed with HLA-I-associated adaptation in mind and often encode high frequencies of adapted epitopes, the inclusion of which, based on our data, is unlikely to confer significant benefits and might hinder vaccine efficacy.
The idea that DCs played a role in MRKAd5-associated infection risk is not new. One previous study focused on the DC-T cell axis and the maturation of DCs by Ad5 immune complexes but was unable to establish a link between in vitro data in unvaccinated individuals and vaccine-induced immune responses (14). Moreover, extensive data suggest that although both CD8 T cells and DCs are generally thought to play a protective role against pathogens, they can also cause undesirable outcomes in the host immune response. For example, Dejnirattisai et al. described an association between preexisting responses, mediated by DC-T cell cross talk, and disease severity in dengue virus infection (27). In HIV infection, recent studies from our group and others suggest that a “helper” CD8 T cell phenotype can fuel HIV infection via interaction with DCs (14, 20, 22). Our current findings build upon these prior studies, suggesting that HIV vaccines can also induce a proinflammatory DC phenotype through CD8 T cells responding to HLA-I-associated adapted epitopes. Notably, in all but one vaccine recipient assayed, adapted epitope-specific CD8 T cells induced more DC maturation. The exception was vaccine recipient 24 (VR24), in whom we tested B*57-restricted adapted and nonadapted epitope-specific responses. In contrast to what we saw in the other recipients, nonadapted epitope-specific CD8 T cells in this individual induced a higher frequency of mature DCs and an increased level of trans-infected CD4 T cell targets. Consistent with these results, cluster analysis of our proteomics data also indicated that VR24 has a rather “swapped” adapted and nonadapted epitope-specific proteomics profile compared to the other recipients (Fig. 9A). HLA-I allele B*57 is well known to be associated with improved viral control in individuals with HIV-1 infection, so it is possible that the opposite results seen in this individual are a result of the B*57 restriction. Due to sample limitation, we were unable to test more vaccine recipients with CD8 T cell responses restricted by protective HLA-I alleles; however, HLA-I adaptation presented by these protective alleles in vaccination is worth investigating in the future.
In this study, we also identified a unique proteomics profile associated with adapted epitope-specific CD8 T cell-DC interactions. Factors that promote DC differentiation (IL-13) and DC maturation (GM-CSF) (29–32) and facilitate viral replication (ICAM-1) (33) were enriched in adapted epitope cultures. Past studies from different groups suggested various mechanisms by which CD8 T cells shape DC phenotype and function. In addition, past studies demonstrated that T cells expressing LAG-3 have been shown to promote DC maturation and activation (34, 35). Consistent with these studies, in all cases for which we evaluated LAG-3 production, we observed higher levels of LAG-3 concentration in adapted epitope cultures (n = 4) (Fig. 9A). On the other hand, some factors, including CXCL5 and CXCL16, were reduced in adapted epitope cultures. A prior study showed that CXCL5 was upregulated in tumor conditioned medium, and pretreatment with such a medium effectively inhibited the maturation of monocyte-derived dendritic cells (36). Thus, lower levels of CXCL5 in adapted epitope cultures likely contribute to the enhanced maturation of DCs. CXCL16 is a small chemokine specifically produced by DCs. It interacts with the chemokine receptor CXCR6, which has been suggested to serve as a coreceptor for cellular entry of HIV and SIV (37–40). Therefore, lower levels of CXCL16 produced by DCs cultured with adapted epitope-specific CD8 T cells might leave more CXCR6 molecules available on the surface of CD4 T cells, allowing HIV entry. Although we did not observe overlap between the transcriptomics and proteomics readouts, the latter analysis is more likely to provide pertinent insights as it takes into account CD8-DC interactions while the transcriptomic data were derived exclusively from expanded CD8 T cells.
One limitation of our in vitro DC assays was a small sample size, due to the infrequent vaccine-encoded adapted epitope-specific CD8 T cell responses. Despite limited peripheral blood mononuclear cells (PBMCs), we were able to generate an adequate number of epitope-specific CD8 T cells for use in CD8-DC coculture assays. However, after CD8-DC coculturing, inherent technical difficulties in enriching for nonadapted/adapted epitope-specific CD8 T cells in sufficient numbers precluded us from performing single-cell RNA sequencing studies on these cells. These “conditioned” CD8 T cells could potentially harbor unique and important transcriptomic signatures associated with DC maturation, information that will likely be different when examining nonadapted/adapted epitope-specific CD8 T cell lines stimulated with cognate antigen without interaction with DCs. While we measured proteins in the supernatant of the CD8 T cell-DC cocultures, future studies to identify which cell types are producing these factors and an assay that blocks these molecules via antibodies would be useful to confirm their role in DC maturation and viral trans-infection.
Although our study provides one possible mechanism by which adapted epitope-specific CD8 T cell may contribute to an enhanced risk of HIV acquisition among MRKAd5 vaccine recipients, other possible mechanisms for this exist. For example, an increased risk of infection among individuals with high adaptation might also be due to an ineffectual viral control at the mucosal portal of viral entry due to the elicitation of suboptimal function of HIV-specific CD8 T cells.
The enhanced risk of infection seen in MRKAd5 vaccinees wanes over time (10), which is consistent with the vaccine-induced immune phenomenon seen in this study. Because adapted epitope-specific CD8 T cell responses have decreased durability in comparison to nonadapted responses (24), it is likely such negative effects would not persist long term; however, even in the short term, vaccines should be designed to avoid any increase in infection risk. Furthermore, inclusion of env sequences might enhance the function of adapted epitope-specific CD8 T cells, so future analysis of these responses in more contemporary vaccine regimens is important, especially those that increase the number of vaccine-encoded adapted epitopes (41, 42).
In summary, we show that high levels of HLA-I-associated adaptation to the MRKAd5 vaccine insert are associated with increased HIV infection risk and that this is likely due to interactions of CD8 T cells with DCs, which leads to a microenvironment and enhanced viral trans-infection of CD4 T cells. These data illustrate the importance of not only measuring their functionality but also more holistically characterizing CD8 T cell responses via their interactions with other immune cells. Moreover, this finding suggests that HIV adaptation in vaccine inserts might be detrimental to vaccine efficacy and highlights the need to design HIV vaccines through the lens of HLA-I-associated adaptation.
MATERIALS AND METHODS
Ethics statement.
All subjects included in this study were adults. Institutional review board approvals from the University of Alabama at Birmingham (no. X981027004, X160125005, and X140612002) were obtained to analyze previously collected samples from the HIV Vaccine Trials Network (HVTN) 502 study participants and from HIV-seronegative donors recruited from the Alabama Vaccine Research Clinic.
Patient cohorts.
HVTN 502 participants (MRKAd5; ClinicalTrials.gov identifier NCT00095576) were randomized to receive an adenovirus serotype 5-vectored vaccine with HIV gene inserts (gag, pol, nef) or placebo (9). Peripheral blood mononuclear cells (PBMCs) from vaccine recipients (n = 24) who were previously found to have positive responses to vaccine-encoded epitopes were used for this study (24). None of the vaccine recipients studied were HIV infected at the time points tested. PBMCs from HIV-seronegative donors (n = 11) from the Alabama Vaccine Research Clinic were used as controls.
Survival analysis with HIV infection in vaccine recipients.
We used the existing data from the modified intent-to-treat arm of the MRKAd5 study, which were obtained from the HVTN through a formal data request. Analysis included male participants that were HIV uninfected at the week 12 visit of the study protocol. Adaptation scores were generated using the online PhyloD tool by inputting HLA-I alleles for each vaccine recipient and the MRKAd5 vaccine sequence (gag from strain CAM-1, GenBank accession no. D10112.1; pol from strain IIIB, accession no. KJ925006.1; nef from strain JRFL, accession no. U63632.1). Groups were defined using adaptation score thresholds that were established by tertiles of the distribution of scores in a large chronic-infection cohort in British Columbia (21). Vaccine recipients were categorized as having a high (greater than or equal to 0.17), medium (less than 0.17 and greater than or equal to −0.32), or low (less than −0.32) adaptation score to the vaccine. Survival curves (Kaplan-Meier and Cox proportional hazard) were built in R using the survminer and survival packages. HIV infection was defined at the endpoint, time was defined as weeks from week 12 visit to HIV infection or end of follow-up, and events were censored if the participants were uninfected at the last time point of follow-up.
Epitope selection and peptide synthesis.
Nonadapted and adapted epitopes were determined in a previous study (24). Briefly, optimal sequences (8- to 11-mer) of HLA-I-restricted nonadapted and adapted epitopes were predicted based on HLA-I-associated polymorphisms and using Microsoft Research’s EPIPRED software. All peptides were synthesized in a 96-well array format (New England Peptide). Each peptide was reconstituted at 40 mM in 100% dimethyl sulfoxide (DMSO) and stored at −70°C. HIV protein-specific pools of overlapping 15-mer peptides were obtained from the HVTN. These peptide pools matched the MRKAd5 insert sequence and included 122 Gag, 103 Pol-1, 107 Pol-2, and 51 Nef peptides.
Vaccine-matched IMC suppression.
An HIV-1 infectious molecular clone (IMC) that matched the HIV sequences of the MRKAd5 vaccine insert was constructed. The gag (from strain CAM-1), RT-IN (from strain IIIB), and nef or env (from strain JRFL) gene sequences were inserted into a lab-adapted NL4-3 (GenBank accession no. M19921) backbone. Specifically, the NL4-3 gag sequence, nucleotides (nt) 798 to 2292, was replaced with nt 794 to 2294 from strain CAM-1; the NL4-3 pol sequence encoding RT-IN, nt 2550 to 5093, was replaced with the pol (RT-IN) sequence, nt 2586 to 5129, from strain IIIB; and the NL4-3 nef sequence, nt 8787 to 9407, was replaced with the nef sequence, nt 8086 to 8736, from strain JRFL. In addition, to render the chimeric virus R5 tropic and provide nef strain-matched env sequences, we replaced NL4-3 nt 6308 to 8767, encoding the envelope ectodomain and membrane-spanning domain, with env nt 5625 to 7628 from strain JRFL, essentially as described previously (43). The respective gag, pol, and nef sequences matching the vaccine insert were obtained as synthesized DNA fragments ligated into a plasmid vector (GenScript Corp., Piscataway NJ), from which they were amplified by PCR for InFusion ligation into the proviral backbone. env sequences were PCR amplified from a JRFL env-containing plasmid template prior to InFusion ligation. The entire chimeric provirus designated pNL-B.JRFL.ecto-CAM1.gag-IIIB.RT.IN-JRFL.nef (unique identifier K5482), and referred to herein as MRKAd5-IMC, was confirmed by Sanger sequencing. Infectious virus stocks were produced by proviral transfection of 293T cells. Virus was titered on TZM-bl cells before the replication kinetics of the novel MRKAd5-IMC was assessed in CD4 T cells, which confirmed its ability to cause productive HIV infection in primary cell culture before its use in the HIV trans-infection assay.
To study the impact of adaptation of vaccine-induced CD8 T cell viral suppression, PBMCs from vaccine recipients with either high (n = 7) or low (n = 9) adaptation scores were used to generate CD8 T cell effectors and CD4 T cell targets, as previously described (44) Briefly, CD8 T cells were generated by culturing thawed PBMCs in RPMI medium plus 10% fetal bovine serum (FBS) (R-10 medium) with IL-2 (50 U/ml) and CD3/CD4-bispecific antibody (0.5 μg/ml; NIH AIDS Reagent Program) for 10 days (45). The culture was maintained with fresh medium containing IL-2 (50 U/ml) every 3 days. In parallel, CD4 T cell targets were generated by first isolating CD4 T cells using magnetic beads (Miltenyi Biotec) and then expanding these cells in R-10 medium with IL-2 (50 U/ml) and CD3.28 beads (25 μg/ml; Miltenyi Biotec). Expanded CD8 T cells were washed and rested for 24 h without IL-2 before functional analysis and IMC suppression. CD4 T cells were then activated by CD3/CD28-bispecific antibody (25 ng/ml) for 4 days and then infected with the MRKAd5-IMC at an MOI of 10 for 4 h. The effector CD8 T cells were cultured with infected CD4 T cell targets at various effector-to-target cell (E:T) ratios (0:1, 1:1, 3:1, 5:1) for 4 days. The number of effectors was controlled across all individuals. After 4 days, cells were labeled with dead cell dye (Invitrogen), anti-CD3–Pacific Blue (BD Pharmingen), and anti-CD4–Alexa 780 (BD Pharmingen) at 4°C for 30 min. The cells were fixed and permeabilized and then labeled intracellularly with anti-Gag p24–phycoerythrin (PE) (BD Pharmingen). Events were acquired on an LSR II flow cytometer, and Gag-p24 expression was quantified in CD3+ T cells.
ICS.
CD8 T cell effector function was assessed by measuring effector molecule production using intracellular cytokine staining (ICS) flow cytometry as described previously (24). Briefly, 1 × 106 CD8 T cells were stimulated with peptide pools in the presence of anti-CD28 and anti-CD49d as well as anti-CD107a–fluorescein isothiocyanate (FITC) antibodies (all from BD Biosciences). Monensin and brefeldin A (BD Biosciences) were added 1 h after peptide stimulation, and the cells were incubated for an additional 11 h at 37°C, in 5% CO2. Following incubation, cells were stained with dead cell dye (Invitrogen), anti-CD3–Alexa 780 (eBioscience), anti-CD4–Qdot655 (Invitrogen), anti-CD8–V500 (BD Pharmingen), anti-CD14–peridinin chlorophyll protein (PerCP)/CY5.5 (BD Pharmingen), and anti-CD19–PerCP/CY5.5 (BD Pharmingen) at 4°C for 30 min. The cells were then fixed and permeabilized using Cytofix/Cytoperm (BD Biosciences) and intracellularly stained with anti-IFN-γ–Alexa 700 (BD Biosciences), anti-IL-2–allophycocyanin (APC) (BD Biosciences), anti-TNF-α–PECy7 (BD Biosciences), anti-perforin–PE (eBioscience), and anti-granzyme A/B–V450 (BD Biosciences) at 4°C for 30 min. At least 300,000 total events were acquired on an LSR II flow cytometer (BD Immunocytometry Systems), and data were analyzed using FlowJo (version 10.6.1; TreeStar, Inc.).
In vitro expansion of epitope-specific CD8 T cells.
Epitope-specific CD8 T cell lines were expanded in vitro as previously described (21, 46). Briefly, cryopreserved PBMCs (obtained from HVTN 502 recipients) were thawed and plated in a 48-well plate at 1.2 × 106 cells/ml in serum-free RPMI medium. Plates were incubated at 37°C in 5% CO2 for 2 h, after which the medium containing nonadherent cells was removed. Adherent cells were irradiated at 3,000 rads and pulsed with the appropriate peptide at 10 μM for 2 h. Autologous CD8 T cells were isolated from the same PBMC sample using the CD8 untouched isolation kit (MACS Miltenyi Biotec). CD8 T cells were then plated at 0.5 × 106 cells/well onto the peptide-pulsed monocytes in the presence of R-10 medium containing IL-7 (25 ng/ml). IL-2 (50 U/ml) was added to the culture on the second day. CD8 T cells were restimulated on day 7 (and weekly thereafter) with peptide-pulsed monocytes.
Activation-induced marker (AIM)-based single-cell sorting and cDNA preparation.
Epitope-specific CD8 T cells with a positive response to vaccine-encoded epitopes were pulsed with the peptide of interest at 10 μM in the presence of anti-CD28 and anti-CD49d. The cells were incubated in R-10 AB medium (RPMI medium plus 10% human AB serum) for 18 h. Following incubation, cells were surface stained for 30 min at 4°C with LIVE/DEAD aqua stain (Invitrogen), anti-CD3–Alexa 780 (eBioscience), anti-CD4–BV785 (BioLegend), anti-CD8–FITC (BioLegend), anti-CD14–PerCP/CY5.5 (BD Pharmingen), anti-CD19–PerCP/CY5.5 (BD Pharmingen), anti-CD137–PE (BD Pharmingen), and anti-CD69–APC (BD Pharmingen). CD69+ CD137+ CD8 T cells were identified as activated CD8 T cells (47), and single cells were sorted directly into 96-well plates containing 3 μl lysis buffer (RNAseOUT, deoxynucleoside triphosphates [dNTPs], and X-100) using a fluorescence-activated cell sorter (FACS) (BD Immunocytometry Systems). After sorting, cells were flash-frozen on dry ice and stored at −70°C until sequencing.
The single-cell TCR/RNA sequencing was performed at the Institute for Immunology and Infectious Diseases in Perth, Australia. Single-cell lysates underwent oligo(dT)-primed reverse transcription. The assay utilizes uniquely tagged primers for reverse transcription and template switching with a preamplification step to increase the yield and transcript length of the single-cell cDNA library. During the initial reverse transcription step, cDNA is tagged with well-specific barcodes coupled with a unique molecular identifier (UMI). Samples are then pooled and amplified using the KAPA HiFi HotStart ReadyMix (Roche, Basel, Switzerland) for first-round PCR.
Single-cell TCR sequencing.
Following the first round PCR, all sorted wells were pooled prior to a second-round nested PCR using template switch oligonucleotide (ISTSO) forward primers and barcoded reverse primers specific to the alpha and beta chains of the TCR. PCR products are purified using Agencourt AMPure XP (Beckman Coulter, CA, USA) and pooled at equimolar concentrations. Indexed libraries were created for sequencing using TruSeq adapters and quantified using the KAPA universal qPCR library quantification kit (KAPA Biosystems, Inc., MA, USA), per the manufacturer’s instructions. Samples were sequenced on an Illumina MiSeq using a 2 × 300 bp paired-end chemistry kit (Illumina, Inc., CA, USA), per the manufacturer’s instructions. Reads were quality filtered and passed through a demultiplexing tool to assign reads to individual wells and mapped to the TCRB and TCRA loci. TCR clonotypes were assigned using the MIXCR software prior to analysis using the VGAS software (Visual Genomics Analysis Suite), a program developed at the Institute for Immunology and Infectious Diseases (IIID) for visualizing single-cell RNA sequencing (scRNAseq) data (IIID, Murdoch, Western Australia).
Single-cell RNA sequencing.
Briefly, gene-specific read counts were calculated using HTSeq-count with the latest Gencode annotations, and the 3′ and 5′ counts were summed. Cells with fewer than 200 genes and more than 5% mitochondrial content were removed. Furthermore, genes with >0 counts in fewer than three cells were also removed. Normalization and scaling were performed using Seurat v2.3.4 R package. Differential expression analysis and visualization were performed using the VGAS software.
IFN-γ ELISpot.
Nitrocellulose plates (Millipore) were coated overnight with anti-IFN-γ antibody and subsequently blocked with R-10 medium for 2 h (21). Epitope-specific CD8 T cells were washed and rested overnight with R-10 medium without IL-2 at 37°C in 5% CO2. Then, these CD8 T cells (105 cells/well) were cultured in triplicate with the peptide of interest at 10 μM in R-10 medium for 22 to 24 h. Cells cultured in medium without peptide and in medium with phytohemagglutinin (PHA) were used as negative and positive controls, respectively. Following incubation, the plates were washed and treated with biotinylated anti-IFN-γ antibody for 2 h, followed by streptavidin-alkaline phosphatase for 1 h, and finally developed with the nitroblue tetrazolium (NBT)/5-bromo-4-chloro-3-indolylphosphate (BCIP) substrate for 5 to 10 min. Plates were read, and counts were determined by use of a CTL ImmunoSpot analyzer (version 5). The number of spots was averaged and normalized to spot-forming units per 106 cells (SFU/106). A positive response was defined as ≥55 SFU/106 cells and ≥4 times background (negative control).
DC maturation.
The DC maturation and viral trans-infection assay was conducted similarly to that previously described, with a few key modifications (22). We used HLA-single-allele-matched allogeneic DCs as APCs. In addition, for the first two experiments testing antigen-specific CD8 T cell lines derived from vaccine recipients, we tested multiple sets of DCs derived from different HIV-naive donors. Our data showed consistent results among different donors, which further supported the reliability of this assay.
We derived DCs by first isolating monocytes from PBMCs by using human CD14 microbeads (MACS Miltenyi Biotec) and culturing for 7 days in Iscove modified Dulbecco medium (IMDM; Invitrogen) containing 10% FBS in the presence of GM-CSF and IL-4 (both at 1,000 IU/ml; R&D Systems) to generate immature DCs (iDCs).
Epitope-specific CD8 T cells, from the antigen-specific CD8 T cell lines described above, were added directly to iDCs at a 3:1 effector-to-target cell (E:T) ratio in the presence or absence of the peptide of interest (1 μM) and cocultured for 48 h. DCs matured in the presence of TNF-α (50 ng/ml), IFN-α (3,000 U/ml), IFN-γ (1,000 U/ml), IL-1B (25 ng/ml), and polyinosinic:polycytidylic acid (pI:C) (20 μg/ml) were used as a positive control (20). After 2 days, cells were stained with dead cell dye (Invitrogen), anti-CD3–Pacific Blue, anti-CD8–V500, anti-CD14–Alexa700, anti-CD83–PE, and anti-CD86–FITC (all from BD Pharmingen) at 4°C for 30 min. The cells were then washed, events were acquired on an LSR II flow cytometer, and CD83/CD86 coexpression was assessed to quantify DC maturation.
HIV trans-infection.
To assess for HIV trans-infection, we used an HIV infectious molecular clone (IMC) that was generated using the sequence of a R5-tropic transmitted founder virus (HIV-TRJO) as previously described (22). DCs were cocultured with epitope-specific CD8 T cells in the presence of the peptide of interest or maturation cocktail (see above) for 48 h. CD8 T cells were then removed from the culture using CD8 Dynabeads (Invitrogen), and DCs were loaded with a low MOI of virus (10−4) for 2 h at 37°C in 5% CO2. DCs were then washed three times with fresh medium to remove excess virus. Autologous CD4 T cell targets isolated from PBMCs (obtained from HIV-seronegative donors) were activated with IL-2 and PHA as described above and were added to the DC culture at a DC-to-CD4 ratio of 1:10. After 4 days, cells were labeled with dead cell dye (Invitrogen), anti-CD3–Pacific Blue (BD Pharmingen), and anti-CD4–Alexa 780 (BD Pharmingen) at 4°C for 30 min. The cells were fixed and permeabilized and were intracellularly labeled with anti-Gag p24–PE (BD Pharmingen). Events were acquired on an LSR II flow cytometer, and Gag-p24 expression was quantified in CD3+ CD4− T cells to reflect productive HIV infection.
Quantitative multiplex ELISA array.
Cell-free supernatants from the DC maturation assay were collected and stored at −70°C prior to analysis. Quantitative analysis for human cytokines produced in the culture using the human cytokine array was performed per the manufacturer’s instructions (Ray Biotech, Peachtree Corners, GA). The concentration of molecules for each condition was first normalized to the limit of detection (LOD). To compare the difference between NAE and AE, fold changes of NAE and AE tested for each vaccine recipient were calculated by dividing the mean value of each NAE and AE pair using the following equation: FC = (C − M)/M, where FC represents the fold change of either NAE or AE, C represents the concentration of either NAE or AE, and M represents the mean value of the concentration of NAE and AE for each vaccine recipient. Visualization was then performed using Pretty Heatmaps v1.0.12 R package.
Statistical analysis.
Data were analyzed using the Mann-Whitney U test for unpaired comparisons, the Wilcoxon ranked test (two-tailed) for paired comparisons, and Spearman’s correlation analysis. R (version 3.6.1), GraphPad Prism (version 8.0), GSEA (version 4.0.3), and VGAS software were used to perform these analyses. Significance was determined as a P value of <0.05, unless otherwise stated. In scRNAseq analysis, P values were adjusted using the false discovery rate (FDR) to account for multiple comparisons.
Data availability.
The single-cell RNA sequence data (GEO accession number GSE163669) are available for download from the GEO database.
ACKNOWLEDGMENTS
We thank NIAID and the NIAID-funded HIV Vaccine Trials Network (HVTN) for providing specimens and data from HVTN 502. We thank staff across the HVTN Leadership Operations Center, the Statistical Data Management Center, and the Laboratory Center for their assistance with this project. The following reagent(s) was obtained through the NIH Tetramer Core Facility: CD3/CD4 (anti-CD3:anti-CD4)-bispecific monoclonal antibodies.
Author contributions were as follows: conceptualization, K.Q., A.B., and P.G.; methodology, K.Q., S.B., and A.B.; investigation, K.Q. and S.B.; formal analysis, K.Q., S.B., and J.M.C.; funding acquisition, S.B. and P.G.; resources, J.M.C., J.Z., J.E., C.O., and P.G.; supervision, A.B. and P.G.; visualization, K.Q.; writing (original draft preparation), K.Q.; writing (review and editing), K.Q., S.B., J.M.C., A.F.-G., J.F., R.B.M., C.O., A.B., and P.G.
This work was supported by the National Institutes of Health (NIH) under grant no. RO1 AI112566 (P.G.), R56 AI143482 (P.G.), F30 AI140829 (S.B.), UM1 AI068614 to the HVTN Leadership Operations Center, UM1 AI068618 to the HVTN Laboratory Center, and UM1 AI068635 to the HVTN Statistical Data Management Center. Parts of the work were performed in the UAB HIV containment facility and by the UAB CFAR Flow Cytometry Core/Joint UAB Flow Cytometry Core, which are funded by NIH/NIAID P30 AI027767, NIH 5P30 AR048311, and P30 AI110527.
Contributor Information
Anju Bansal, Email: pgoepfert@uabmc.edu.
Paul Goepfert, Email: pgoepfert@uabmc.edu.
Guido Silvestri, Emory University.
REFERENCES
- 1.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH, MOPH-TAVEG Investigators. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, Kostrikis LG, Zhang L, Perelson AS, Ho DD. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med 189:991–998. 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendoza D, Migueles SA, Rood JE, Peterson B, Johnson S, Doria-Rose N, Schneider D, Rakasz E, Trivett MT, Trubey CM, Coalter V, Hallahan CW, Watkins D, Franchini G, Lifson JD, Connors M. 2013. Cytotoxic capacity of SIV-specific CD8(+) T cells against primary autologous targets correlates with immune control in SIV-infected rhesus macaques. PLoS Pathog 9:e1003195. 10.1371/journal.ppat.1003195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cartwright EK, Spicer L, Smith SA, Lee D, Fast R, Paganini S, Lawson BO, Nega M, Easley K, Schmitz JE, Bosinger SE, Paiardini M, Chahroudi A, Vanderford TH, Estes JD, Lifson JD, Derdeyn CA, Silvestri G. 2016. CD8(+) Lymphocytes are required for maintaining viral suppression in SIV-infected macaques treated with short-term antiretroviral therapy. Immunity 45:656–668. 10.1016/j.immuni.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ndhlovu ZM, Kamya P, Mewalal N, Kloverpris HN, Nkosi T, Pretorius K, Laher F, Ogunshola F, Chopera D, Shekhar K, Ghebremichael M, Ismail N, Moodley A, Malik A, Leslie A, Goulder PJ, Buus S, Chakraborty A, Dong K, Ndung'u T, Walker BD. 2015. Magnitude and kinetics of CD8+ T cell activation during hyperacute HIV infection impact viral set point. Immunity 43:591–604. 10.1016/j.immuni.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards BH, Bansal A, Sabbaj S, Bakari J, Mulligan MJ, Goepfert PA. 2002. Magnitude of functional CD8+ T-cell responses to the Gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J Virol 76:2298–2305. 10.1128/jvi.76.5.2298-2305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goulder PJ, Phillips RE, Colbert RA, McAdam S, Ogg G, Nowak MA, Giangrande P, Luzzi G, Morgan B, Edwards A, McMichael AJ, Rowland-Jones S. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med 3:212–217. 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 9.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN, Step Study Protocol Team. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372:1881–1893. 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duerr A, Huang Y, Buchbinder S, Coombs RW, Sanchez J, del Rio C, Casapia M, Santiago S, Gilbert P, Corey L, Robertson MN, Step/HVNT Study Team. 2012. Extended follow-up confirms early vaccine-enhanced risk of HIV acquisition and demonstrates waning effect over time among participants in a randomized trial of recombinant adenovirus HIV vaccine (Step Study). J Infect Dis 206:258–266. 10.1093/infdis/jis342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hutnick NA, Carnathan DG, Dubey SA, Makedonas G, Cox KS, Kierstead L, Ratcliffe SJ, Robertson MN, Casimiro DR, Ertl HC, Betts MR. 2009. Baseline Ad5 serostatus does not predict Ad5 HIV vaccine-induced expansion of adenovirus-specific CD4+ T cells. Nat Med 15:876–878. 10.1038/nm.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Brien KL, Liu J, King SL, Sun YH, Schmitz JE, Lifton MA, Hutnick NA, Betts MR, Dubey SA, Goudsmit J, Shiver JW, Robertson MN, Casimiro DR, Barouch DH. 2009. Adenovirus-specific immunity after immunization with an Ad5 HIV-1 vaccine candidate in humans. Nat Med 15:873–875. 10.1038/nm.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnabas RV, Wasserheit JN, Huang Y, Janes H, Morrow R, Fuchs J, Mark KE, Casapia M, Mehrotra DV, Buchbinder SP, Corey L, NIAID HIV Vaccine Trials Network. 2011. Impact of herpes simplex virus type 2 on HIV-1 acquisition and progression in an HIV vaccine trial (the Step study). J Acquir Immune Defic Syndr 57:238–244. 10.1097/QAI.0b013e31821acb5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perreau M, Pantaleo G, Kremer EJ. 2008. Activation of a dendritic cell-T cell axis by Ad5 immune complexes creates an improved environment for replication of HIV in T cells. J Exp Med 205:2717–2725. 10.1084/jem.20081786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin-Moreno A, Munoz-Fernandez MA. 2019. Dendritic cells, the double agent in the war against HIV-1. Front Immunol 10:2485. 10.3389/fimmu.2019.02485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banchereau J, Steinman RM. 1998. Dendritic cells and the control of immunity. Nature 392:245–252. 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 17.Surenaud M, Montes M, Lindestam Arlehamn CS, Sette A, Banchereau J, Palucka K, Lelievre JD, Lacabaratz C, Levy Y. 2019. Anti-HIV potency of T-cell responses elicited by dendritic cell therapeutic vaccination. PLoS Pathog 15:e1008011. 10.1371/journal.ppat.1008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cameron PU, Freudenthal PS, Barker JM, Gezelter S, Inaba K, Steinman RM. 1992. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science 257:383–387. 10.1126/science.1352913. [DOI] [PubMed] [Google Scholar]
- 19.Pope M, Betjes MG, Romani N, Hirmand H, Cameron PU, Hoffman L, Gezelter S, Schuler G, Steinman RM. 1994. Conjugates of dendritic cells and memory T lymphocytes from skin facilitate productive infection with HIV-1. Cell 78:389–398. 10.1016/0092-8674(94)90418-9. [DOI] [PubMed] [Google Scholar]
- 20.Mailliard RB, Smith KN, Fecek RJ, Rappocciolo G, Nascimento EJ, Marques ET, Watkins SC, Mullins JI, Rinaldo CR. 2013. Selective induction of CTL helper rather than killer activity by natural epitope variants promotes dendritic cell-mediated HIV-1 dissemination. J Immunol 191:2570–2580. 10.4049/jimmunol.1300373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlson JM, Du VY, Pfeifer N, Bansal A, Tan VY, Power K, Brumme CJ, Kreimer A, DeZiel CE, Fusi N, Schaefer M, Brockman MA, Gilmour J, Price MA, Kilembe W, Haubrich R, John M, Mallal S, Shapiro R, Frater J, Harrigan PR, Ndung'u T, Allen S, Heckerman D, Sidney J, Allen TM, Goulder PJ, Brumme ZL, Hunter E, Goepfert PA. 2016. Impact of pre-adapted HIV transmission. Nat Med 22:606–613. 10.1038/nm.4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin K, Boppana S, Du VY, Carlson JM, Yue L, Dilernia DA, Hunter E, Mailliard RB, Mallal SA, Bansal A, Goepfert PA. 2019. CD8 T cells targeting adapted epitopes in chronic HIV infection promote dendritic cell maturation and CD4 T cell trans-infection. PLoS Pathog 15:e1007970. 10.1371/journal.ppat.1007970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monaco DC, Dilernia DA, Fiore-Gartland A, Yu T, Prince JL, Dennis KK, Qin K, Schaefer M, Claiborne DT, Kilembe W, Tang J, Price MA, Farmer P, Gilmour J, Bansal A, Allen S, Goepfert P, Hunter E. 2016. Balance between transmitted HLA preadapted and nonassociated polymorphisms is a major determinant of HIV-1 disease progression. J Exp Med 213:2049–2063. 10.1084/jem.20151984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boppana S, Sterrett S, Files J, Qin K, Fiore-Gartland A, Cohen KW, De Rosa SC, Bansal A, Goepfert PA. 2019. HLA-I associated adaptation dampens CD8 T-cell responses in HIV Ad5-vectored vaccine recipients. J Infect Dis 220:1620–1628. 10.1093/infdis/jiz368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence T. 2009. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol 1:a001651. 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spengler ML, Kuropatwinski KK, Comas M, Gasparian AV, Fedtsova N, Gleiberman AS, Gitlin II, Artemicheva NM, Deluca KA, Gudkov AV, Antoch MP. 2012. Core circadian protein CLOCK is a positive regulator of NF-kappaB-mediated transcription. Proc Natl Acad Sci U S A 109:E2457–65. 10.1073/pnas.1206274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dejnirattisai W, Duangchinda T, Lin CL, Vasanawathana S, Jones M, Jacobs M, Malasit P, Xu XN, Screaton G, Mongkolsapaya J. 2008. A complex interplay among virus, dendritic cells, T cells, and cytokines in dengue virus infections. J Immunol 181:5865–5874. 10.4049/jimmunol.181.9.5865. [DOI] [PubMed] [Google Scholar]
- 28.Nishimura Y, Gautam R, Chun TW, Sadjadpour R, Foulds KE, Shingai M, Klein F, Gazumyan A, Golijanin J, Donaldson M, Donau OK, Plishka RJ, Buckler-White A, Seaman MS, Lifson JD, Koup RA, Fauci AS, Nussenzweig MC, Martin MA. 2017. Early antibody therapy can induce long-lasting immunity to SHIV. Nature 543:559–563. 10.1038/nature21435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webb DC, Cai Y, Matthaei KI, Foster PS. 2007. Comparative roles of IL-4, IL-13, and IL-4Ralpha in dendritic cell maturation and CD4+ Th2 cell function. J Immunol 178:219–227. 10.4049/jimmunol.178.1.219. [DOI] [PubMed] [Google Scholar]
- 30.Alters SE, Gadea JR, Holm B, Lebkowski J, Philip R. 1999. IL-13 can substitute for IL-4 in the generation of dendritic cells for the induction of cytotoxic T lymphocytes and gene therapy. J Immunother 22:229–236. 10.1097/00002371-199905000-00005. [DOI] [PubMed] [Google Scholar]
- 31.Lutz MB, Schnare M, Menges M, Rossner S, Rollinghoff M, Schuler G, Gessner A. 2002. Differential functions of IL-4 receptor types I and II for dendritic cell maturation and IL-12 production and their dependency on GM-CSF. J Immunol 169:3574–3580. 10.4049/jimmunol.169.7.3574. [DOI] [PubMed] [Google Scholar]
- 32.Min L, Mohammad Isa SA, Shuai W, Piang CB, Nih FW, Kotaka M, Ruedl C. 2010. Cutting edge: granulocyte-macrophage colony-stimulating factor is the major CD8+ T cell-derived licensing factor for dendritic cell activation. J Immunol 184:4625–4629. 10.4049/jimmunol.0903873. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Plata MT, Puigdomenech I, Izquierdo-Useros N, Puertas MC, Carrillo J, Erkizia I, Clotet B, Blanco J, Martinez-Picado J. 2013. The infectious synapse formed between mature dendritic cells and CD4(+) T cells is independent of the presence of the HIV-1 envelope glycoprotein. Retrovirology 10:42. 10.1186/1742-4690-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andreae S, Piras F, Burdin N, Triebel F. 2002. Maturation and activation of dendritic cells induced by lymphocyte activation gene-3 (CD223). J Immunol 168:3874–3880. 10.4049/jimmunol.168.8.3874. [DOI] [PubMed] [Google Scholar]
- 35.Avice MN, Sarfati M, Triebel F, Delespesse G, Demeure CE. 1999. Lymphocyte activation gene-3, a MHC class II ligand expressed on activated T cells, stimulates TNF-alpha and IL-12 production by monocytes and dendritic cells. J Immunol 162:2748–2753. [PubMed] [Google Scholar]
- 36.Michielsen AJ, Hogan AE, Marry J, Tosetto M, Cox F, Hyland JM, Sheahan KD, O'Donoghue DP, Mulcahy HE, Ryan EJ, O'Sullivan JN. 2011. Tumour tissue microenvironment can inhibit dendritic cell maturation in colorectal cancer. PLoS One 6:e27944. 10.1371/journal.pone.0027944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng HK, Unutmaz D, KewalRamani VN, Littman DR. 1997. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296–300. 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 38.Alkhatib G, Liao F, Berger EA, Farber JM, Peden KW. 1997. A new SIV co-receptor, STRL33. Nature 388:238. 10.1038/40789. [DOI] [PubMed] [Google Scholar]
- 39.Matloubian M, David A, Engel S, Ryan JE, Cyster JG. 2000. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat Immunol 1:298–304. 10.1038/79738. [DOI] [PubMed] [Google Scholar]
- 40.Liao F, Alkhatib G, Peden KW, Sharma G, Berger EA, Farber JM. 1997. STRL33, A novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med 185:2015–2023. 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baden LR, Walsh SR, Seaman MS, Cohen YZ, Johnson JA, Licona JH, Filter RD, Kleinjan JA, Gothing JA, Jennings J, Peter L, Nkolola J, Abbink P, Borducchi EN, Kirilova M, Stephenson KE, Pegu P, Eller MA, Trinh HV, Rao M, Ake JA, Sarnecki M, Nijs S, Callewaert K, Schuitemaker H, Hendriks J, Pau MG, Tomaka F, Korber BT, Alter G, Dolin R, Earl PL, Moss B, Michael NL, Robb ML, Barouch DH, IPCAVD006/RV380/HIV-V-A002 Study Group. 2018. First-in-human randomized, controlled trial of mosaic HIV-1 immunogens delivered via a modified vaccinia Ankara vector. J Infect Dis 218:633–644. 10.1093/infdis/jiy212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baden LR, Stieh DJ, Sarnecki M, Walsh SR, Tomaras GD, Kublin JG, McElrath MJ, Alter G, Ferrari G, Montefiori D, Mann P, Nijs S, Callewaert K, Goepfert P, Edupuganti S, Karita E, Langedijk JP, Wegmann F, Corey L, Pau MG, Barouch DH, Schuitemaker H, Tomaka F, Traverse/HVTN 117/HPX2004 Study Team. 2020. Safety and immunogenicity of two heterologous HIV vaccine regimens in healthy, HIV-uninfected adults (TRAVERSE): a randomised, parallel-group, placebo-controlled, double-blind, phase 1/2a study. Lancet HIV 7:e688–e698. 10.1016/S2352-3018(20)30229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edmonds TG, Ding H, Yuan X, Wei Q, Smith KS, Conway JA, Wieczorek L, Brown B, Polonis V, West JT, Montefiori DC, Kappes JC, Ochsenbauer C. 2010. Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC. Virology 408:1–13. 10.1016/j.virol.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spentzou A, Bergin P, Gill D, Cheeseman H, Ashraf A, Kaltsidis H, Cashin-Cox M, Anjarwalla I, Steel A, Higgs C, Pozniak A, Piechocka-Trocha A, Wong J, Anzala O, Karita E, Dally L, Gotch F, Walker B, Gilmour J, Hayes P. 2010. Viral inhibition assay: a CD8 T cell neutralization assay for use in clinical trials of HIV-1 vaccine candidates. J Infect Dis 201:720–729. 10.1086/650492. [DOI] [PubMed] [Google Scholar]
- 45.Wong JT, Colvin RB. 1991. Selective reduction and proliferation of the CD4+ and CD8+ T cell subsets with bispecific monoclonal antibodies: evidence for inter-T cell-mediated cytolysis. Clin Immunol Immunopathol 58:236–250. 10.1016/0090-1229(91)90139-2. [DOI] [PubMed] [Google Scholar]
- 46.Du VY, Bansal A, Carlson J, Salazar-Gonzalez JF, Salazar MG, Ladell K, Gras S, Josephs TM, Heath SL, Price DA, Rossjohn J, Hunter E, Goepfert PA. 2016. HIV-1-specific CD8 T cells exhibit limited cross-reactivity during acute infection. J Immunol 196:3276–3286. 10.4049/jimmunol.1502411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfl M, Kuball J, Ho WY, Nguyen H, Manley TJ, Bleakley M, Greenberg PD. 2007. Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities. Blood 110:201–210. 10.1182/blood-2006-11-056168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The single-cell RNA sequence data (GEO accession number GSE163669) are available for download from the GEO database.