Abstract
Background
Various mathematical models were published to predict the epidemiological consequences of the COVID-19 pandemic. This systematic review has studied the initial epidemiological models.
Methods
Articles published from January to June 2020 were extracted from databases using search strings and those peer-reviewed with full text in English were included in the study. They were analysed as to whether they made definite predictions in terms of time and numbers, or contained only mathematical assumptions and open-ended predictions. Factors such as early vs. late prediction models, long-term vs. curve-fitting models and comparisons based on modelling techniques were analysed in detail.
Results
Among 56,922 hits in 05 databases, screening yielded 434 abstracts, of which 72 articles were included. Predictive models comprised over 70% (51/72) of the articles, with susceptible, exposed, infectious and recovered (SEIR) being the commonest type (mean duration of prediction being 3 months). Common predictions were regarding cumulative cases (44/72, 61.1%), time to reach total numbers (41/72, 56.9%), peak numbers (22/72, 30.5%), time to peak (24/72, 33.3%), hospital utilisation (7/72, 9.7%) and effect of lockdown and NPIs (50/72, 69.4%). The commonest countries for which models were predicted were China followed by USA, South Korea, Japan and India. Models were published by various professionals including Engineers (12.5%), Mathematicians (9.7%), Epidemiologists (11.1%) and Physicians (9.7%) with a third (32.9%) being the result of collaborative efforts between two or more professions.
Conclusion
There was a wide diversity in the type of models, duration of prediction and the variable that they predicted, with SEIR model being the commonest type.
Keywords: Mathematical models, COVID-19, Systematic review, Nonpharmaceutical interventions
Introduction
The first case of COVID-19 was detected in Wuhan on 17 November 2019.1 On 11 Mar 2020, the WHO declared COVID-19 as a pandemic.2 This pandemic is the defining global health crisis of our time and the greatest challenge we have faced since World War II. Since its emergence in Asia late last year, the virus has spread to every continent except Antarctica.3 The total number of cases worldwide on 14 April 2021 was 138,297,267 with 2,976,297 deaths and 111,208,976 recoveries.3
The seminal paper of Kermack et al. in 1927 introduced the Susceptible, Infectious and Recovered (SIR) model for infectious diseases.4 Since then, with advances in information technology and fast computing methods, many variants of the SIR model have been developed. Mathematical models predicting the impact of COVID-19 have burgeoned since the onset of pandemic at a global level, with publications on models doubling almost every 20 days.5 Published predictive models have looked at various aspects of the pandemic. The models have used specific modelling techniques, assumptions and data gathered from real cases. The predicted outcomes depend on these variables. Models developed early in the epidemic might have had greater impact in planning and allocation of healthcare resources than the later ones.
Numerous mathematical models with varied techniques and predictions often leave the readers confused. This systematic review was carried out to analyse the initial epidemiological predictive models of the COVID-19 pandemic.
Materials and methods
The protocol for this systematic review was registered at www.osf.io on 30/07/2020.6 The population for the review comprised all studies on mathematical modelling of the COVID-19 epidemic. The outcomes of interest were epidemiological predictions of the model including the techniques used for mathematical predictions. Only those mathematical models that were published in peer-reviewed journals were included in the study. Studies without explicit mathematical modelling, reviewing COVID-19 with guesstimates, with no full text freely available, with only model description but no predictions and pre-print articles, were excluded. Narrative reviews and commentaries, perspective articles too were excluded, unless they provided novel modelling analyses or outcomes. Publication date range included in our review was from January to June 2020, in the predefined databases to ensure that we considered the initial models.
A detailed literature search was carried out and the following databases were searched; Medline through PubMed, Web of Science, medRxiv, bioRxiv and arXiv. Key words used were mathematical modelling, predictive modelling, COVID-19, SARS-CoV-2, peak infected cases and total deaths. The search strategy followed a two-stage approach. In the first stage, databases were searched using the keywords, and in the second stage, a manual search of articles from the references of the selected article was carried out. The literature search was conducted by two researchers independently. The searches were then reviewed by two other authors.
A data extraction form was designed for the study. Two authors independently extracted the data from all the studies and then compared them. In case of any disagreement, a designated third author again extracted the data separately and the discrepancy was resolved through mutual consultation.
Models were assessed for various prediction characteristics such as peak numbers, total infections, point of time where the pandemic was predicted to peak at a particular location, time to end of pandemic, effectiveness of nonpharmaceutical interventions (NPIs) and hospital admissions. The models were classified as ‘early’ if they had made their predictions before cases reached 2000 at their respective geographical location. Others were classified as ‘late’ models. Short-term models were those that predicted up to two weeks while long-term models predicted beyond this period.
Risk of bias in mathematical model is difficult to assess including with the use of PROBAST checklist for predictive models in medical science, as there is no selection and measurement bias in mathematical models, only the approach and type of models may differ.7 Quality assessment of the study articles was carried out based on five questions adapted from Holmdahl et al. published in June 2020.8 Narrative synthesis was performed as per the synthesis without meta-analysis guideline.9
Results
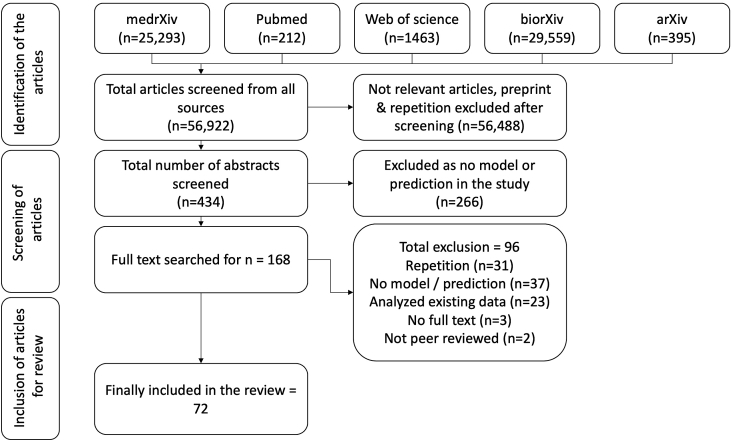
The chosen search strings yielded 56,922 hits in the five databases. The screening of titles yielded 434 abstracts, of which 266 did not contain predictive models. Another 96 were excluded due to other reasons listed in the PRISMA chart (Fig. 1). Finally, 72 articles were included in the systematic review.10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81
Fig. 1.
PRISMA: Selection of articles.
Over 70% (51/72) of the articles contained predictive mathematical models, with susceptible, exposed, infectious and recovered (SEIR) being the commonest one (41/72, 56.9%). The common predictions were regarding cumulative cases (44/72, 61.1%), time to reach total numbers (41/72, 56.9%), peak numbers (22/72, 30.5%), time to peak (24/72, 33.3%), hospital utilisation (7/72, 9.7%) and impact of lockdown and NPIs (50/72, 69.4%). Prediction characteristics studied are listed in Supplementary Table 1. It was found that of the 72 articles, 20 articles were based on hypothetical scenarios which predicted outcomes in numbers, but these outcomes (number of asymptomatic cases and effect of screening, etc) could not be compared with real data. Among those which did (n = 52), the median duration of prediction was 3 months (interquartile range, 2–4 months; range: 10 days–48 months). Curve-fitting models invariably made short-term predictions.
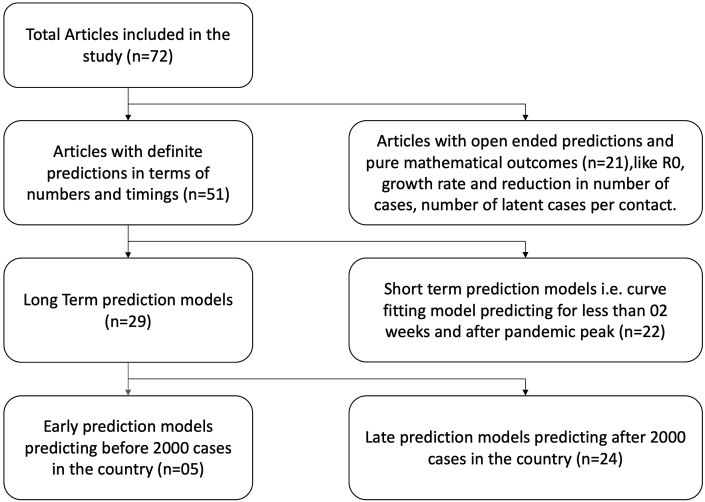
While certain models predicted definite outcomes in terms of time and numbers (51/72, 71%), others were pure mathematical models (21/72, 29%) which studied the impact of various parameters on the pandemic such as basic reproduction number (R0), variable contact rates of infectious individuals, time spent in crowded zones and population density.22,28,31,34,35,43
Of the 51 models which predicted definite outcomes, models were divided into long-term (n = 29) and short-term (n = 22) models. Furthermore, the long-term models were classified into early predictor models (n = 5) and late predictor models (n = 24) (Fig. 3). Among articles with definite predictions in terms of numbers or time, 22 were curve-fitting models with short-term outcomes, which modelled the epidemiological characteristics around the peak in their region.11,13,14,20,22,23,25,26,34,37,40,42,48, 49, 50,53,57,60, 61, 62,64,74
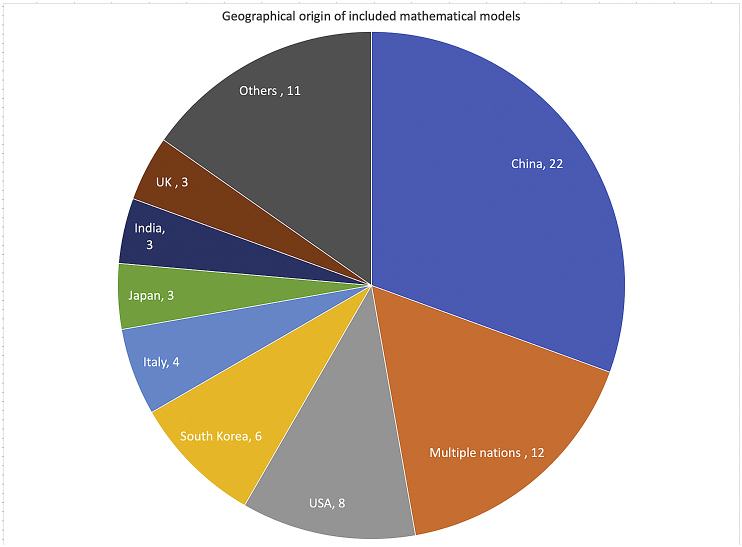
Fig. 3.
Geographical origin of included mathematical models.
While one model looked into the effects of lockdown and other NPIs like screening at airports another looked at specific impact of lockdown in India at 21, 42 and 60 days.39,44 Many models predicted the estimated burden on health care system and utilisation.10,23,30,38,51,52,74
The countrywise distribution of the studies is shown in Fig. 2. Most of the models predicted outcomes for China (22/71, 31%) followed by USA, South Korea, Japan, India, Iran, UK and Canada (Fig. 2). While most models predicted for a single country or city, there were 12 models that predicted outcomes for more than one country and 3 models that made worldwide predictions. Predictions were also made for the following regions—Hubei Province in China, California, Michigan and Utah in USA, Ontario in Canada, cities such as New York, London and Wuhan and cities in India such as New Delhi, Kolkata, Mumbai and Bangalore. Most studies were published in March (26/72, 36.1%), followed by April (21/72, 29.2%).
Fig. 2.
Characteristics of included mathematical models.
Mathematical models were published by various professionals including Engineers (12.5%), Mathematicians (9.7%), Epidemiologists (11.1%), Physicians (9.7%), Biostatisticians (5.5%) and others (18%). A third of the articles (32.9%) were collaborative efforts between two professions (e.g., Mathematicians and Physicians).
Quality assessment of the models is depicted in Table 1. All the models have clearly defined the purpose in their study. Only 11 (15.27%) considered the population density in the region studied.
Table 1.
Qualitative analysis of mathematical models.8
| S No | Question | Yes (%) |
|---|---|---|
| 1 | Has the purpose of model been clearly depicted in the study? | 72 (100%) |
| 2 | Has immunity been taken into account In the model? | 54 (75%) |
| 3 | Has asymptomatic transmission been taken into account into the model? | 37 (52%) |
| 4 | Has contact transmission been taken into account in the model? | 72 (100%) |
| 5 | Has the statistical model displayed confidence intervals? (n = 22) | 22 (100%) |
| 6 | Has the mechanistic model depicted various parameters, ranges? (n = 50) | 50 (100%) |
| 7 | Has the prediction been made for a defined geographical region? | 58 (81%) |
| 8 | Is population density taken into account in the model? | 11 (16%) |
A table of narrative synthesis of all the articles is provided depicting authors, title, geographical area of predictions, mathematical modelling technique and outcomes evaluated (Suppl Table 1). The characteristics of early and late predictors are also shown (Suppl Table 1).
Discussion
“All models are wrong, but some are useful” said George Fox.82 In a situation like the COVID-19 epidemic where uncertainty is rife, predictions from epidemiological models are one of the key tools available for early decision-making.
This review of all the published initial peer-reviewed mathematical models of COVID-19 attempts to summarise and synthesise their findings. The question of prediction arises in the initial stages of any situation (especially a pandemic like COVID-19) that is fraught with ambiguity, chaos and uncertainty. With the passage of time, most situations become clearer and hard data become available for computation. Hence, we did not consider subsequent models from July 2020 onwards. Our review showed that of the large numbers of models published, only a few were both predictive and peer reviewed. Because we studied only the peer-reviewed models, it is likely that models which were published largely for planning purposes by governments or models from news or social media are not included. It is also possible that some authors chose to forgo peer review and submitted their articles directly to various databases.
Systematic review of mathematical models has earlier been carried out in the context of other infectious diseases. Prieto et al. performed a systematic review to identify areas of enhancement of pandemic simulation models of Influenza epidemics for operational use at provincial and local levels.83 A study by Harris et al. has reviewed mathematical models exploring the epidemiological impact of future TB vaccines.84
There was a wide heterogeneity seen in the mathematical models evaluating epidemiology of the COVID epidemic (Table 2). The commonest one was SIR model and its variant. The model divided the population into different compartments and the movement of population from one compartment to another is predicted by differential equations. The approach is flexible as more number of compartments may be added. Both stochastic and deterministic model are possible. There were many non–SEIR-based models based on regression models and techniques such as ARIMA (based on time series). However, the model used depends on the aim of the researcher and data available. None of the methods is established as superior to another.85
Table 2.
Types of mathematical models.
| Model types | Characteristics | Strengths | Weaknesses |
|---|---|---|---|
| Epidemiological models (n = 41) | Compartmental models divide the population into different compartments. Movement from one compartment to another is predicted by differential equations. Can be stochastic or deterministic. Approach is flexible as number of compartments can be varied. e.g, SEIR or variations (SIR, SIRD etc). |
Take into account dynamics of spread of infectious disease in a population. Ability to model numerous variables affecting spread like quarantine, isolation, vaccination, re-infection etc. Good for predicting worst-case scenarios and aggregate effect of interventions. |
Highly dependent on estimation of parameters. Do not, usually, take into account variability of parameters during the course of the epidemic. |
| Data-driven models (n = 31) | Usually curve-fitting in nature. Can be predictive or pure mathematical (Open-ended). Used to evaluate concepts for validation or preparedness e.g. Regression model, ARIMA, Log logistic model etc. |
Generally have a good fit to retrospective data. Good for short-term projections based on current estimated parameters. |
Do not take into account dynamics of disease spread. Lack reliability for long-term predictions. |
While short-term models that predicted over just two weeks were more accurate, they were not particularly useful due to the short prediction horizon. These curve-fitting models predicted close to actual outcomes, but with minimal preparedness benefits. However, in special situations where short-term predictions are desired, these might still have a role. These models can be used for validating a new set of assumptions or a new modelling technique.
A few long-term models were constructed early in the epidemic. What they lacked in precision, they made up in usefulness.86 Later models were more precise due to larger data sets of more cases, but their usefulness progressively declined. Pure mathematical models (open-ended models) were used to evaluate concepts for validation or preparedness, without predicting any particular quantifiable outcome.
Mathematical modelling was done by a variety of professionals from engineers and mathematicians to doctors and biologists. Epidemiologists comprised about a tenth of all modellers. This demonstrated the interest that the epidemic had generated among various scientists traversing traditional professional silos. Even more interesting was the fact that almost a third of all models were collaborations between multiple professions.
Our study is one of the first systematic reviews of mathematical models of COVID-19 and seeks to synthesise the key characteristics. A limitation of our study was that we were unable to combine the various modelling results into a common numerical estimate. We attempted to estimate the Mean Absolute Percentage Error (MAPE) of the models with real-time data but were constrained by the predicted numbers not being available for different time-points. Another possible limitation was that we have not included articles whose free full text was not available.
An ideal model would be one which factors in the maximum number of relevant variables which could possibly affect study outcomes and predicts with closest proximity to the real outcomes. In this systematic review, the chosen mathematical models had different modelling and prediction characteristics, precluding such an analysis.
There was a wide range of variation in the outcomes predicted in various studies. Which is the model that a country should follow remains an unresolved question till date. It would be ideal for a model to have inputs from mathematicians, epidemiologists, and health care workers (involved in dealing with COVID). Inputs from varied professions help ensure the widest range of reasonable assumptions and make the model more robust. This epidemic is an opportunity for those interested in mathematical modelling (mathematicians, epidemiologists, HCWs and others) to collaborate and improve the models for their regions, considering various parameters based on their specific experience and training. It is also an opportunity for planners and administrators to utilise comprehensive models for informing the planning and preparedness for their regions.
The advent of newer technologies has made mathematical calculation relatively easy. Many free software such as ‘R’, ‘PYTHON’ etc. are now available for mathematical modelling. In the future, the use of more advanced and refined technologies such as artificial intelligence will allow more accurate real-time predictions.
Mathematical models have used varied assumptions and have predicted various outcomes. Actual data for comparisons remains dynamic as the pandemic is still evolving. Data are changing rapidly depending on time, place and effects of NPIs. Hence, it remains difficult to compare predictions with actual outcomes and also to compare all available models at a single platform. A similar review may require to be carried out after some time, when more epidemic data are available and more models are published.
Conclusion
This systematic review analyses all the initial epidemiological models of the COVID-19 pandemic from January to June 2020. The analysis of mathematical models was constrained by varied prediction parameters of different models and differing time horizons of predictions among different models. There was a wide range of assumptions and implications. Thus, no particular model was substantially superior to others.
This review revealed that majority of the mathematical models studied the effect of NPIs, which helped administrators to plan preventive measures. Early predictor models were possibly of greatest utility to administrators in taking planning decisions. SEIR variants were the commonest modelling technique. This systematic review has utility in helping future modellers choose among assumptions to incorporate in their models and also decide on preferred prediction parameters for a particular location or region.
Disclosure of competing interest
Four of the authors have previously authored a mathematical model for the COVID epidemic.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mjafi.2021.05.005.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Characteristics of mathematical models included in the study
References
- 1.Bryne J. Live Science; 2020 Mar 14. 1st known case of coronavirus traced back to November in China. Editor-in-Chief. [Google Scholar]
- 2.Coronavirus Disease (COVID-19) - events as they happen [Internet]. [cited 2020 Jun 19]. Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen.
- 3.Coronavirus Update (Live): 8,649,131 Cases and 458,122 Deaths from COVID-19 Virus Pandemic - Worldometer [Internet]. [cited 2020 Jun 19]. Available from: https://www.worldometers.info/coronavirus/.
- 4.Kermack W.O., McKendrick A.G., Walker G.T. A contribution to the mathematical theory of epidemics. Proc R Soc Lond - Ser A Contain Pap a Math Phys Character. 1927 Aug 1;115(772):700–721. [Google Scholar]
- 5.Brainard J. Scientists are drowning in COVID-19 papers. Can new tools keep them afloat? Science. May. 13, 2020 https://www.sciencemag.org/news/2020/05/scientists-are-drowning-covid-19-papers-can-new-tools-keep-them-afloat [Internet] AAAS. 2020 [cited 2020 Jun 20]. Available from: [Google Scholar]
- 6.OSF | My Projects. [cited 2020 Jun 19]; Available from: https://osf.io/myprojects/#registrations.
- 7.Wolff R.F., Moons K.G.M., Riley R.D. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann Intern Med. 2019 Jan 1;170(1):51–58. doi: 10.7326/M18-1376. [DOI] [PubMed] [Google Scholar]
- 8.Holmdahl I., Buckee C. Wrong but useful — what Covid-19 epidemiologic models can and cannot tell us. N Engl J Med. 2020 May 15 doi: 10.1056/NEJMp2016822. 0(0):null. [DOI] [PubMed] [Google Scholar]
- 9.Campbell M., McKenzie J.E., Sowden A. Synthesis without meta-analysis (SWiM) in systematic reviews: reporting guideline. BMJ. 2020 Jan 16;368 doi: 10.1136/bmj.l6890. https://www.bmj.com/content/368/bmj.l6890 [Internet]. [cited 2020 Jun 20]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moghadas S.M., Shoukat A., Fitzpatrick M.C. Projecting hospital utilization during the COVID-19 outbreaks in the United States. Proc Natl Acad Sci U S A. 2020 Apr 21;117(16):9122–9126. doi: 10.1073/pnas.2004064117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Y., Wang S. Mathematic modeling of COVID-19 in the United States. Emerg Microb Infect. 2020 Dec;9(1):827–829. doi: 10.1080/22221751.2020.1760146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qeadan F., Honda T., Gren L.H. Naive forecast for COVID-19 in Utah based on the South Korea and Italy models-the fluctuation between two extremes. Int J Environ Res Public Health. 2020 Apr 16;17(8) doi: 10.3390/ijerph17082750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petropoulos F., Makridakis S. Forecasting the novel coronavirus COVID-19. PloS One. 2020 Mar 31;15(3) doi: 10.1371/journal.pone.0231236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.A conceptual model for the coronavirus disease 2019 (COVID-19) outbreak in Wuhan, China with individual reaction and governmental action - ScienceDirect [Internet]. [cited 2021 Apr 11]. Available from: https://www.sciencedirect.com/science/article/pii/S120197122030117X. [DOI] [PMC free article] [PubMed]
- 15.Anastassopoulou C., Russo L., Tsakris A., Siettos C. Data-based analysis, modelling and forecasting of the COVID-19 outbreak. PloS One. 2020 Mar 31;15(3) doi: 10.1371/journal.pone.0230405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manchein C., Brugnago E.L., da Silva R.M., Mendes C.F.O., Beims M.W. Strong correlations between power-law growth of COVID-19 in four continents and the inefficiency of soft quarantine strategies. Chaos. 2020 Apr;30(4) doi: 10.1063/5.0009454. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7192349/ [Internet]. [cited 2021 Apr 12]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zareie B., Roshani A., Mansournia M.A., Rasouli M.A., Moradi G. A model for COVID-19 prediction in Iran based on China parameters. Arch Iran Med. 2020 Apr 1;23(4):244–248. doi: 10.34172/aim.2020.05. [DOI] [PubMed] [Google Scholar]
- 18.Moftakhar L., Seif M. The exponentially increasing rate of patients infected with COVID-19 in Iran. Arch Iran Med. 2020 Apr 1;23(4):235–238. doi: 10.34172/aim.2020.03. [DOI] [PubMed] [Google Scholar]
- 19.Wan K., Chen J., Lu C., Dong L., Wu Z., Zhang L. When will the battle against novel coronavirus end in Wuhan: a SEIR modelinganalysis. J Glob Health. 2020 Jun;10(1) doi: 10.7189/jogh.10.011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S., Seo Y.B., Jung E. Prediction of COVID-19 transmission dynamics using a mathematical model considering behavior changes in Korea. Epidemiol Health. 2020;42 doi: 10.4178/epih.e2020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang S., Diao M., Yu W., Pei L., Lin Z., Chen D. Estimation of the reproductive number of novel coronavirus (COVID-19) and the probable outbreak size on the Diamond Princess cruise ship: a data-driven analysis. Int J Infect Dis. 2020 Apr;93:201–204. doi: 10.1016/j.ijid.2020.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ngonghala C.N., Iboi E., Eikenberry S. Mathematical assessment of the impact of non-pharmaceutical interventions on curtailing the 2019 novel Coronavirus. Math Biosci. 2020 Jul;325:108364. doi: 10.1016/j.mbs.2020.108364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peirlinck M., Linka K., SahliCostabal F., Kuhl E. Outbreak dynamics of COVID-19 in China and the United States. Biomech Model Mechanobiol. 2020 Dec;19(6):2179–2193. doi: 10.1007/s10237-020-01332-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L., Yang Z., Dang Z. Propagation analysis and prediction of the COVID-19. Infect Dis (Except HIV/AIDS) 2020 doi: 10.1016/j.idm.2020.03.002. http://medrxiv.org/lookup/doi/10.1101/2020.03.14.20036202 [Internet] Mar [cited 2021 Apr 11]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z., Magal P., Seydi O., Webb G. Predicting the cumulative number of cases for the COVID-19 epidemic in China from early data. medRxiv. 2020 Mar 13 doi: 10.3934/mbe.2020172. 2020.03.11.20034314. [DOI] [PubMed] [Google Scholar]
- 26.Zhao S., Chen H. Modeling the epidemic dynamics and control of COVID-19 outbreak in China. medRxiv. 2020 Mar 9 doi: 10.1007/s40484-020-0199-0. 2020.02.27.20028639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shim E., Tariq A., Choi W., Lee Y., Chowell G. Transmission potential of COVID-19 in South Korea. medRxiv. 2020 Mar 2 doi: 10.1016/j.ijid.2020.03.031. 2020.02.27.20028829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryu S., Ali S.T., Lim J.-S., Chun B.C. Estimation of the excess COVID-19 cases in Seoul, South Korea by the students arriving from China. Int J Environ Res Publ Health. 2020 Jan;17(9):3113. doi: 10.3390/ijerph17093113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuite A.R., Fisman D.N., Greer A.L. Mathematical modelling of COVID-19 transmission and mitigation strategies in the population of Ontario, Canada. CMAJ (Can Med Assoc J) 2020 May 11;192(19):E497–E505. doi: 10.1503/cmaj.200476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou C., Chen J., Zhou Y. The effectiveness of quarantine of Wuhan city against the Corona Virus Disease 2019 (COVID-19): a well-mixed SEIR model analysis. J Med Virol. 2020;92(7):841–848. doi: 10.1002/jmv.25827. [DOI] [PubMed] [Google Scholar]
- 31.Chintalapudi N., Battineni G., Amenta F. COVID-19 virus outbreak forecasting of registered and recovered cases after sixty day lockdown in Italy: a data driven model approach. J Microbiol Immunol Infect. 2020 Jun 1;53(3):396–403. doi: 10.1016/j.jmii.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuniya T. Prediction of the epidemic peak of coronavirus disease in Japan, 2020. J Clin Med. 2020 Mar;9(3):789. doi: 10.3390/jcm9030789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivorra B., Ferrández M.R., Vela-Pérez M., Ramos A.M. Mathematical modeling of the spread of the coronavirus disease 2019 (COVID-19) taking into account the undetected infections. The case of China. Commun Nonlinear Sci Numer Simulat. 2020 Sep 1;88:105303. doi: 10.1016/j.cnsns.2020.105303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou T., Liu Q., Yang Z. Preliminary prediction of the basic reproduction number of the Wuhan novel coronavirus 2019-nCoV. J Evid Base Med. 2020;13(1):3–7. doi: 10.1111/jebm.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang R., Liu M., Ding Y. Spatial-temporal distribution of COVID-19 in China and its prediction: a data-driven modeling analysis. J Infect Dev Ctries. 2020 Mar 31;14(3):246–253. doi: 10.3855/jidc.12585. [DOI] [PubMed] [Google Scholar]
- 36.Chatterjee K., Chatterjee K., Kumar A., Shankar S. Healthcare impact of COVID-19 epidemic in India: a stochastic mathematical model. Med J Armed Forces India. 2020 Apr 1;76(2):147–155. doi: 10.1016/j.mjafi.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandal S., Bhatnagar T., Arinaminpathy N. Prudent public health intervention strategies to control the coronavirus disease 2019 transmission in India: a mathematical model-based approach. Indian J Med Res. 2020;151(2–3):190–199. doi: 10.4103/ijmr.IJMR_504_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z., Zeng Z., Wang K. Modified SEIR and AI prediction of the epidemics trend of COVID-19 in China under public health interventions. J Thorac Dis. 2020 Mar;12(3):165–174. doi: 10.21037/jtd.2020.02.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi S., Ki M. Estimating the reproductive number and the outbreak size of COVID-19 in Korea. Epidemiol Health. 2020;42 doi: 10.4178/epih.e2020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciufolini I., Paolozzi A. Mathematical prediction of the time evolution of the COVID-19 pandemic in Italy by a Gauss error function and Monte Carlo simulations. Eur Phys J Plus. 2020;135(4):355. doi: 10.1140/epjp/s13360-020-00383-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiang Y., Jia Y., Chen L., Guo L., Shu B., Long E. COVID-19 epidemic prediction and the impact of public health interventions: a review of COVID-19 epidemic models. Infect Dis Model. 2021;6:324–342. doi: 10.1016/j.idm.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karako K., Song P., Chen Y., Tang W. Analysis of COVID-19 infection spread in Japan based on stochastic transition model. Biosci Trends. 2020 May 21;14(2):134–138. doi: 10.5582/bst.2020.01482. [DOI] [PubMed] [Google Scholar]
- 43.Wei Y.Y., Lu Z.Z., Du Z.C. Fitting and forecasting the trend of COVID-19 by SEIR(+CAQ) dynamic model. Zhonghua Liuxingbingxue Zazhi. 2020 Apr 10;41(4):470–475. doi: 10.3760/cma.j.cn112338-20200216-00106. [DOI] [PubMed] [Google Scholar]
- 44.Ambikapathy B., Krishnamurthy K. Mathematical modelling to assess the impact of lockdown on COVID-19 transmission in India: model development and validation. JMIR Public Health Surveill. 2020 May 7;6(2) doi: 10.2196/19368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rocchi E., Peluso S., Sisti D., Carletti M. A possible scenario for the Covid-19 epidemic, based on the SI(R) model. SN Compr Clin Med. 2020 May 12:1–3. doi: 10.1007/s42399-020-00306-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Early prediction of the 2019 novel coronavirus outbreak in the mainland China based on simple mathematical model. IEEE Access. 2020 Mar 9;8:51761–51769. doi: 10.1109/ACCESS.2020.2979599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanchez-Caballero S., Selles M.A., Peydro M.A., Perez-Bernabeu E. An efficient COVID-19 prediction model validated with the cases of China, Italy and Spain: total or partial lockdowns? J Clin Med. 2020 May 20;9(5) doi: 10.3390/jcm9051547. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7290738/ [Internet]. [cited 2021 Apr 11]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou L., Wu K., Liu H., Gao Y., Gao X., CIRD-F Spread and influence of COVID-19 in China. J Shanghai Jiao Tong Univ (Sci) 2020 Apr 1;25(2):147–156. doi: 10.1007/s12204-020-2168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cássaro F.A.M., Pires L.F. Can we predict the occurrence of COVID-19 cases? Considerations using a simple model of growth. Sci Total Environ. 2020 Aug 1;728:138834. doi: 10.1016/j.scitotenv.2020.138834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chowdhury R., Heng K., Shawon M.S.R. Dynamic interventions to control COVID-19 pandemic: a multivariate prediction modelling study comparing 16 worldwide countries. Eur J Epidemiol. 2020 May 19:1–11. doi: 10.1007/s10654-020-00649-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Putra M., Kesavan M., Brackney K., Hackney D.N., Roosa K.M. Forecasting the impact of coronavirus disease during delivery hospitalization: an aid for resource utilization. Am J ObstetGynecol MFM. 2020 Aug;2(3):100127. doi: 10.1016/j.ajogmf.2020.100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goscé L., Phillips P.A., Spinola P., Gupta D.R.K., Abubakar P.I. Modelling SARS-COV2 spread in London: approaches to lift the lockdown. J Infect. 2020 Aug;81(2):260–265. doi: 10.1016/j.jinf.2020.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bayham J., Fenichel E.P. Impact of school closures for COVID-19 on the US health-care workforce and net mortality: a modelling study. Lancet Public Health. 2020 May 1;5(5):e271–e278. doi: 10.1016/S2468-2667(20)30082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prem K., Liu Y., Russell T.W. The effect of control strategies to reduce social mixing on outcomes of the COVID-19 epidemic in Wuhan, China: a modelling study. Lancet Public Health. 2020 May 1;5(5):e261–e270. doi: 10.1016/S2468-2667(20)30073-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Al-Rousan N., Al-Najjar H. The correlation between the spread of COVID-19 infections and weather variables in 30 Chinese provinces and the impact of Chinese government mitigation plans. Eur Rev Med Pharmacol Sci. 2020 Apr;24(8):4565–4571. doi: 10.26355/eurrev_202004_21042. [DOI] [PubMed] [Google Scholar]
- 56.Pearson C.A., Van Schalkwyk C., Foss A.M., O'Reilly K.M. SACEMA Modelling and Analysis Response Team, CMMID COVID-19 working group, et al. Projected early spread of COVID-19 in Africa through 1 June 2020. Euro Surveill. 2020 May;25(18) doi: 10.2807/1560-7917.ES.2020.25.18.2000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turk P.J., Chou S.-H., Kowalkowski M.A. Modeling COVID-19 latent prevalence to assess a public health intervention at a state and regional scale: retrospective cohort study. JMIR Public Health Surveill. 2020 Jun 19;6(2) doi: 10.2196/19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khajji B., Kada D., Balatif O., Rachik M. A multi-region discrete time mathematical modeling of the dynamics of Covid-19 virus propagation using optimal control. J Appl Math Comput. 2020 May 8:1–27. doi: 10.1007/s12190-020-01354-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao Y, Zhang Z, Yao W, Ying Q, Long C, Fu X. Forecasting the cumulative number of COVID-19 deaths in China: a Boltzmann function-based modeling study. Infect Control Hosp Epidemiol. 1–3. Published online Apr 2, 10.1017/ice.2020.101. [DOI] [PMC free article] [PubMed]
- 60.Singh S., Parmar K.S., Kumar J., Makkhan S.J.S. Development of new hybrid model of discrete wavelet decomposition and autoregressive integrated moving average (ARIMA) models in application to one month forecast the casualties cases of COVID-19. Chaos, Solit Fractals. 2020 Jun;135:109866. doi: 10.1016/j.chaos.2020.109866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh R.K., Rani M., Bhagavathula A.S. Prediction of the COVID-19 pandemic for the top 15 affected countries: advanced autoregressive integrated moving average (ARIMA) model. JMIR Public Health Surveill. 2020 May 13;6(2) doi: 10.2196/19115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao Z., Li X., Liu F., Zhu G., Ma C., Wang L. Prediction of the COVID-19 spread in African countries and implications for prevention and control: a case study in South Africa, Egypt, Algeria, Nigeria, Senegal and Kenya. Sci Total Environ. 2020 Aug 10;729:138959. doi: 10.1016/j.scitotenv.2020.138959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu Z., Cui Q., Han J., Wang X., Sha W.E.I., Teng Z. Evaluation and prediction of the COVID-19 variations at different input population and quarantine strategies, a case study in Guangdong province, China. Int J Infect Dis. 2020 Jun;95:231–240. doi: 10.1016/j.ijid.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim S., Kim Y.-J., Peck K.R., Jung E. School opening delay effect on transmission dynamics of coronavirus disease 2019 in Korea: based on mathematical modeling and simulation study. J Korean Med Sci. 2020 Apr 1;35(13) doi: 10.3346/jkms.2020.35.e143. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7131906/ [Internet]. [cited 2021 Apr 11]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang B., Wang X., Li Q. Estimation of the transmission risk of the 2019-nCoV and its implication for public health interventions. J Clin Med. 2020 Feb 7;9(2) doi: 10.3390/jcm9020462. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7074281/ [Internet]. [cited 2021 Apr 11]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilbert M., Pullano G., Pinotti F. Preparedness and vulnerability of African countries against importations of COVID-19: a modelling study. Lancet. 2020 Mar 14;395(10227):871–877. doi: 10.1016/S0140-6736(20)30411-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rocklöv J., Sjödin H., Wilder-Smith A. COVID-19 outbreak on the Diamond Princess cruise ship: estimating the epidemic potential and effectiveness of public health countermeasures. J Travel Med. 2020 May 18;27(3) doi: 10.1093/jtm/taaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Victor Okhuese A. Estimation of the probability of reinfection with COVID-19 by the susceptible-exposed-infectious-removed-undetectable-susceptible model. JMIR Public Health Surveill. 2020 May 13;6(2) doi: 10.2196/19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang G., Pan Q., Zhao S., Gao Y., Gao X. Prediction of COVID-19 outbreak in China and optimal return date for university students based on propagation dynamics. J Shanghai JiaotongUniv Sci. 2020;25(2):140–146. doi: 10.1007/s12204-020-2167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu Y., Chen Y.Q. On a statistical transmission model in analysis of the early phase of COVID-19 outbreak. Stat Biosci. 2020 Apr 2:1–17. doi: 10.1007/s12561-020-09277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iwata K., Miyakoshi C. A simulation on potential secondary spread of novel coronavirus in an exported country using a stochastic epidemic SEIR model. J Clin Med. 2020 Mar 30;9(4) doi: 10.3390/jcm9040944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hellewell J., Abbott S., Gimma A. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Global Health. 2020 Apr 1;8(4):e488–e496. doi: 10.1016/S2214-109X(20)30074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carcione J.M., Santos J.E., Bagaini C., Ba J. A Simulation of a COVID-19 Epidemic Based on a Deterministic SEIR Model. Front Public Health. 2020 May 28;8 doi: 10.3389/fpubh.2020.00230. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7270399/ [Internet]. [cited 2021 Apr 11]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Crokidakis N. COVID-19 spreading in Rio de Janeiro, Brazil: do the policies of social isolation really work? Chaos, Solit Fractals. 2020 Jul;136:109930. doi: 10.1016/j.chaos.2020.109930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dropkin G. COVID-19 UK lockdown forecasts and R 0. Front Public Health. 2020;8:256. doi: 10.3389/fpubh.2020.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kochanczyk M., Grabowski F., Lipniacki T. Dynamics of COVID-19 pandemic at constant and time-dependent contact rates. Math Model Nat Phenom. 2020 Apr 8;15 [Google Scholar]
- 77.Munayco C.V., Tariq A., Rothenberg R. Early transmission dynamics of COVID-19 in a southern hemisphere setting: Lima-Peru: February 29th–March 30th, 2020. Infect Dis Model. 2020 May 12;5:338–345. doi: 10.1016/j.idm.2020.05.001. eCollection 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Salje H., Tran Kiem C., Lefrancq N. Estimating the burden of SARS-CoV-2 in France. Science. 2020 Jul 10;369(6500):208–211. doi: 10.1126/science.abc3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou X., Wu Z., Yu R. Modelling-based evaluation of the effect of quarantine control by the Chinese government in the coronavirus disease 2019 outbreak. Sci China Life Sci. 2020 May 8:1–4. doi: 10.1007/s11427-020-1717-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsallis C., Tirnakli U. Predicting COVID-19 peaks around the World. Front Phys. 2020;8 https://www.frontiersin.org/articles/10.3389/fphy.2020.00217/full [Internet]. [cited 2021 Apr 11]. Available from: [Google Scholar]
- 81.Diop B.Z., Ngom M., PouguéBiyong C., PouguéBiyong J.N. The relatively young and rural population may limit the spread and severity of COVID-19 in Africa: a modelling study. BMJ Glob Health. 2020 May 25;5(5) doi: 10.1136/bmjgh-2020-002699. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7252974/ [Internet]. [cited 2021 Apr 11]. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Box G.E.P. Science and statistics. J Am Stat Assoc. 1976;71(356):791–799. [Google Scholar]
- 83.Prieto D.M., Das T.K., Savachkin A.A., Uribe A., Izurieta R., Malavade S. A systematic review to identify areas of enhancements of pandemic simulation models for operational use at provincial and local levels. BMC Publ Health. 2012 Mar 30;12(1):251. doi: 10.1186/1471-2458-12-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Harris R.C., Sumner T., Knight G.M., White R.G. Systematic review of mathematical models exploring the epidemiological impact of future TB vaccines. Hum Vaccin Immunother. 2016 Nov;12(11):2813–2832. doi: 10.1080/21645515.2016.1205769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kotwal A., Yadav A.K., Yadav J., Kotwal J., Khune S. Predictive models of COVID-19 in India: a rapid review. Med J Armed Forces India. 2020 Oct 1;76(4):377–386. doi: 10.1016/j.mjafi.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Levins-1966-Model_Building.pdf [Internet]. [cited 2021 Apr 11]. Available from: https://uberty.org/wp-content/uploads/2015/07/Levins-1966-Model_Building.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of mathematical models included in the study