Abstract
Background
The systematic review was conducted to summarize and synthesize evidence from all available case series and case reports published on re-positive COVID-19 cases.
Methods
The systematic review was registered with Prospero (CRD42020210446). PRISMA guidelines were followed for conducting the systematic review. Inclusion criteria for studies included case reports and case series which have documented cases of positive reverse transcriptase polymerase chain reaction (RT-PCR) after a period of clinical improvement or a negative RT-PCR report. Reviews, opinions, and animal studies were excluded. Methodological quality was assessed using the modified Murad scale.
Results
A total of 30 case reports/case series were included in the study, wherein a total of 219 cases were included. In re-positive cases, the age range varied from 10 months to 91 years. The pooled proportion of positive cases after follow-up using random-effects was 12% (95% confidence interval [CI]: 09%–15%). Among the re-positives, a total of 57 cases (26%) had comorbidities. A total of 51 (23.3%) and 17 (7.8%) re-positive cases had been treated with antivirals and corticosteroids, respectively. Only a few studies have confirmed the presence of antibodies after the first episode. Studies that included contact tracing of re-positives did not find any positive cases among close contacts of re-positive cases.
Conclusion
The systemic review found that reinfection is a possibility within 123 days of a negative RT-PCR test in a small number of cases of COVID-19. This has wider ramifications in framing clinical, preventive, and public health policy guidelines.
Keywords: COVID-19, Re-positives, Reinfection, Systematic review
Introduction
Clusters of atypical pneumonia cases were reported from Wuhan city, China, in December 2019 in the Hubei province.1 The agent was identified as severe acute respiratory syndrome corona virus 2 (SARS-CoV-2) and the disease was named as COVID-19.2 World Health Organization declared it as Public Health Emergency of International Concern on 30 January 20 and subsequently as a pandemic on 11 March 20.3
Although scientific knowledge of the novel SARS-CoV-2 in the context of characteristics, transmission dynamics, pathophysiology, and clinical spectrum of disease manifestations has considerably increased over the past one year, knowledge gaps continue to persist in the natural history of the disease. The immune response to the infection (humoral versus cellular Immunity, the persistence of acquired immunity, and natural immunity to the disease) are still plagued with uncertainty.
Case reports and case series have documented COVID-19 cases with reverse transcriptase polymerase chain reaction (RT-PCR)–positive test reports at two different time frames following a symptom free period and/or RT-PCR–negative test. These cases may include re-positives, reactivated, and reinfection cases. It is unknown whether these cases share common characteristics or features that may help identify re-positive cases before discharge. The systematic review of the case reports and case series of the re-positives may help in better understanding of the natural history of the disease. Hence, a systematic review to summarize and synthesize evidence from all the published case series and case reports was conducted.
Materials and methods
The present systematic review was registered with Prospero with registration number CRD42020210446. We followed PRISMA guidelines for conducting the systematic review. A detailed literature search was carried out until 12 November 2020 for studies with reported cases of COVID-19 after a symptom-free interval. The databases that were searched included Medline through Pubmed and Cochrane databases. The key terms used were COVID-19, severe acute respiratory syndrome corona virus, relapse, re-activation, re-positive, and re-infection. The detailed search for Pubmed is given in Supplementary Table 1. Hand searches of the references of articles were also carried out. Observational studies, including case reports and case series, which had reported COVID-19 cases positive for RT-PCR on different occasions following a symptom-free interval and/or negative RT-PCR test were considered for the systematic review. Studies published in English language only were considered for the systematic review. Inclusion criteria for studies included case reports and case series that have documented positive RT-PCR cases after a period of clinical improvement or after a negative RT-PCR report. Review, opinions, and animal studies were excluded. Case reports which described clinical presentation or manifestations of COVID-19 cases were also excluded from the studies if they did not specify the positive molecular test after a symptom-free period or negative RT-PCR test.
Case definition
For this systematic review, the words relapse, re-activation, and re-positives were used interchangeably to include anyone who had become RT-PCR positive again after a symptom-free interval or negative RT-PCR test. Reinfection was restricted to only those studies where genomic characterization of the virus at two different time frames following a negative RT-PCR test proved fresh infection. The term “Recurrence” was used for encompassing both reinfection and re-positive/relapse/reactivation.
A data extraction form was developed, and data were extracted by two authors independently. The data items consisted of age and sex of the patients, clinical comorbidities, date of initial positive RT-PCR test, date of negative RT-PCR test based on which the patient was declared as cured, and date of positive RT-PCR test in recovered individuals who reported with new onset of symptoms suggestive of COVID-19 reinfection after a disease-free interval. Data on serology (if performed) and the clinical outcome of patients were also collated. If there was a mismatch in data extraction by the two authors, the same was resolved through discussion with a senior epidemiologist.
Methodological quality was assessed using the existing Murad scale.4 The scale consists of eight items that converge into four domains: selection, ascertainment, causality, and reporting. Two items pertaining to adverse drug events (dose–response effect and challenge and rechallenge phenomenon) were not considered relevant. The data were extracted for remaining six items by two independent authors, and in case of mismatch, consensus was made in consultation with a senior epidemiologist. Narrative synthesis of the results was carried out. Random-effects model was used for the pooling of results. The description of variable was carried out as mean and standard deviation for continuous variables and proportion for categorical variables. 95% confidence interval (95% CI) was calculated. The statistical analysis was carried out using StataCorp. 2019. Stata Statistical Software: Release 16. College Station, TX: StataCorp LLC.
Results
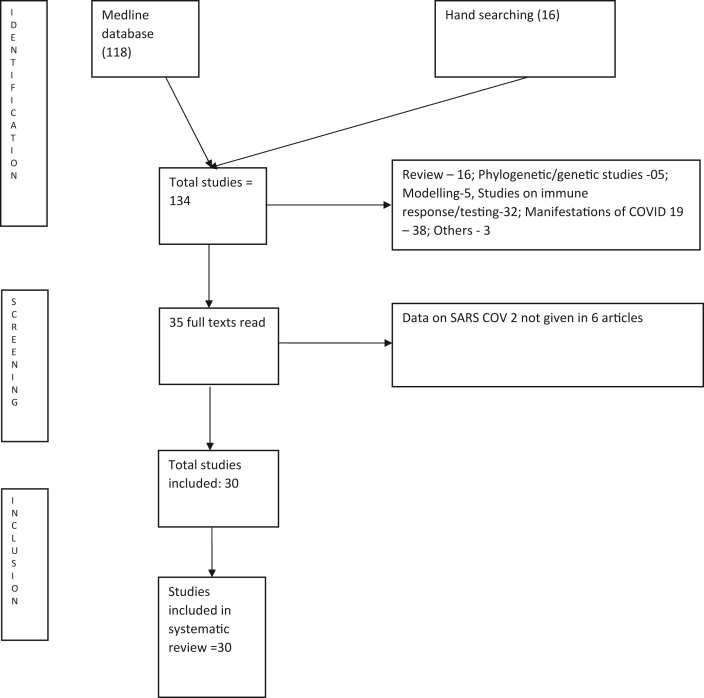
The selection for the study is shown as PRISMA Chart in Fig. 1. A total of 30 case reports/case series with 219 cases were included in the study. The patients' details and characteristics in the case series and case reports are shown in Table 1.5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33 A study carried out in China among children with a median age of age of 5.7 years which studied recurrence in 14 children22 and another Chinese study among 10 elderly subjects which did not mention the age and gender of the participants20 were also included in the study. The pooled mean age of 195 cases was 44.3 ± 19.2 years. A total of 111 (50.68%) of 195 were women. The age range of the recurrence cases varied from 10 months to 91 years of age.
Fig. 1.
Prisma chart for the inclusion of studies in the systematic review.
Table 1.
Characteristics of studies.
| S no | Study | Age and sex | Country | Sympt-omatic | Comor-bidity | Clinical severity | First COVID 19 (PCR) | Test Done | Serological test done after first episode | RT PCR negative after first episode | Symptomatic again after period of weeks | Date of Second COVID 19 | Test done | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Batisse et al.6 | |||||||||||||
| 1 | 19, F | France | Yes | 7 Cormo-bidity: 4 No co-morbitiy |
Mild | D2 | RT-PCRa | Available for 9 patients 5 were positive, one slightly positive and three negatives | NM | Yes | D29, | RT- PCRa | 3 Dead and 8 Alive | |
| 2 | 32, F | Yes | Mild | D18 | NM | Yes | D36,55 | |||||||
| 3 | 33, F | Yes | Mild | D3 | NM | Yes | D28 | |||||||
| 4 | 43, M | Yes | Mild | D1 | NM | Yes | D38 | |||||||
| 5 | 85, M | Yes | Mild | D16 | NM | Yes | D46 | |||||||
| 6 | 54, M | Yes | Mild | D38,44 | NM | Yes | D45 | |||||||
| 7 | 91, F | Yes | Mild | D3 | NM | Yes | D26 | |||||||
| 8 | 55, M | Yes | Mild | D6 | NM | Yes | D31 | |||||||
| 9 | 72, M | Yes | Mild | D7 | NM | Yes | D23, 32, 36 | |||||||
| 10 | 73, M | Yes | Mild | D6 | NM | Yes | D35 | |||||||
| 11 | 84, F | Yes | Mild | D11 | NM | Yes | D50 | |||||||
| 2 | Lafai et al.7 | |||||||||||||
| 1 | 84, F | France | Yes | Yes | Severe | 26 March | PCR | Yes∗∗ | No | Yes | 26 days | RT PCRa | Death | |
| 2 | 90, F | Yes | Yes | Severe | 05 April | PCR | No | No | Yes | 15 days | Death | |||
| 3 | 84, F | Yes | Yes | Severe | 15 April | PCRa (neg) | Yes∗∗ | Yes | Yes | 11 days | Death | |||
| 3 | Enrico et al.8 | 69, F | Italy | Yes | Yes | Mild | 24 March | RT-PCR | Yes IgG Positive | Yes (two) | Yes | 32 days | RT PCR | Alive |
| 4 | Ye et al.9 | |||||||||||||
| 1 | 30, M | China | Yes | No | Mild | NM | NM | NM | Yes | NM | 4–17 days after negative test | RT PCRa | Alive | |
| 2 | 42, M | Yes | No | Mild | NM | NM | NM | Yes | NM | Alive | ||||
| 3 | 32, F | Yes | No | Mild | NM | NM | NM | Yes | NM | Alive | ||||
| 4 | 27, F | No | No | Mild | NM | NM | NM | Yes | NM | Alive | ||||
| 5 | 31, F | Yes | No | Mild | NM | NM | NM | Yes | NM | Alive | ||||
| 5 | Ravioli et al.10 | |||||||||||||
| 1 | 81, F | Switzerland | Yes | Yes | Moderate | 09 March | RT-PCRa | NM | Yes | Yes | 21 | RT-PCRa | Died | |
| 2 | 77, F | Yes | Yes | 23 March | NM | Yes | Yes | 14 | Alive | |||||
| 6 | Loconsole et al.11 | 48, M | Italy | Yes | No | Severe | 17 March | RT-PCR | Yes | Yes | Yes | 30 | RT PCR | Alive |
| 7 | Jiang et al.12 | |||||||||||||
| 1 | 35 F | China | Yes | No | Mild | 30 January | RT-PCRa | No | Yes | Yes | 9 days | RT-PCRa | Re-hosp | |
| 2 | 56 F | Yes | Yes | Mild | 30 January | No | Yes | No | 14 days | Alive | ||||
| 3 | F | Yes | No | Mild | 03 February | No | Yes | Yes | 8 days | Alive | ||||
| 4 | F | Yes | No | Mild | 03 February | No | Yes | No | 7 days | Alive | ||||
| 5 | F | Yes | Yes | Mild | 05 February | No | Yes | No | 9 days | Alive | ||||
| 6 | F | Yes | No | Mild | 06 February | No | Yes | No | 5 days | Alive | ||||
| 8 | Chang et al.13 | |||||||||||||
| 1 | 14M | China | No | No | Mild-6 Moderate - 1 |
01 February | RT-PCRa | No | Yes | No | 7 | 4RS 2 RT-PCRa 1RT-PCRa and 1 Rs |
Alive | |
| 2 | 13M | No | No | 01 February | No | Yes | No | 11 | Alive | |||||
| 3 | 0.8F | Yes | No | 05 February | No | Yes | No | 9 | Alive | |||||
| 4 | 35M | Yes | No | 02 February | No | Yes | No | 9 | Alive | |||||
| 5 | 35M | No | No | 31 January | No | Yes | No | 8 | Alive | |||||
| 6 | 33M | No | No | 27 January | No | Yes | No | 5 | Alive | |||||
| 7 | 26M | Yes | No | 26 January | No | yes | No | 11 | Alive | |||||
| 9 | Yoo et al.14 | 8M | Korea | Yes | No | Mild | 03 March | RT-PCR | No | Yes | Yes | 14 | RT-PCR | Alive |
| 10 | Liu et al.15 | 35 M | China | yes | No | Mild | 30 January | RT-PCR | Yes | Yes | Yes | 15 | RT-PCR | Alive |
| 11 | Yuan et al.16 | |||||||||||||
| 1 | 38M | China | 19- Yes 1 - No |
6 people had comorbidities | Mild to moderate | NM for all | RT-PCRa | 14 were tested and all of them have antibodies | Yes | No for all | 13- retested at 07 days 7 retested 14 days |
14 nasopharyngeal and 7 anal swabs | Alive (all) | |
| 2 | 53M | |||||||||||||
| 3 | 40F | |||||||||||||
| 4 | 61F | |||||||||||||
| 5 | 64F | |||||||||||||
| 6 | 53F | |||||||||||||
| 7 | 33F | |||||||||||||
| 8 | 1F | |||||||||||||
| 9 | 34F | |||||||||||||
| 10 | 43M | |||||||||||||
| 11 | 34F | |||||||||||||
| 12 | 38M | |||||||||||||
| 13 | 50F | |||||||||||||
| 14 | 50F | |||||||||||||
| 15 | 5F | |||||||||||||
| 16 | 55F | |||||||||||||
| 17 | 72F | |||||||||||||
| 18 | 54M | |||||||||||||
| 19 | 8M | |||||||||||||
| 20 | 12M | |||||||||||||
| 12 | Lan et al.17 | |||||||||||||
| 1 | 30-36, 2 M | China | 3-Yes 1- No |
NM | Mild to moderate | NM | RT-PCRa | NM | Yes | No | 5–13 days after discharge | RT-PCRa | Alive | |
| 2 | NM | NM | NM | Yes | No | Alive | ||||||||
| 3 | NM | NM | Nm | Yes | No | Alive | ||||||||
| 4 | NM | NM | NM | Yes | No | Alive | ||||||||
| 13 | Cao et al.18 | |||||||||||||
| 1 | 54F | China | Yes | No | Severe | NM | RT-PCRa | NM | Yes | No | 12 | RT-PCRa | Alive | |
| 2 | 72F | Yes | No | Moderate | NM | NM | Yes | No | 14 | Alive | ||||
| 3 | 60F | Yes | No | Moderate | NM | NM | Yes | No | 09 | Alive | ||||
| 4 | 65F | Yes | Yes | Moderate | NM | NM | Yes | No | 12 | Alive | ||||
| 5 | 58M | Yes | No | Moderate | NM | NM | Yes | No | 16 | Alive | ||||
| 6 | 64M | Yes | No | Severe | NM | NM | Yes | No | 29 | Alive | ||||
| 7 | 36F | Yes | No | Moderate | NM | NM | Yes | No | 06 | Alive | ||||
| 8 | 26M | No | No | Moderate | NM | NM | Yes | No | 06 | Alive | ||||
| 14 | Deng et al.19 | Age - 54.8 years, F- 36 | China | NM | 24 (39.3%) | Severe-3 (4.9%) | NM | RT-PCRa | Not done | Yes | 38-No | 0 (7–13) | 36-RT-PCR 17- AS; 8- sputum |
Alive (All) |
| 15 | Peng et al.20 | |||||||||||||
| 1 | 67M | China | Yes | NM | Mild | 24 January | PCR | NM | Yes | No | 4 | RT-PCR | Alive | |
| 2 | - M | Yes | NM | Mild | 24 January | PCR | NM | Yes | No | 6 | RT-PCR | Alive | ||
| 3 | - F | Yes | NM | Mild | 27 January | PCR | NM | Yes | No | 3 | RT-PCR | Alive | ||
| 4 | - M | Yes | NM | Mild | 28 January | PCR | NM | Yes | No | 7 | RT-PCR | Alive | ||
| 5 | 38F | Yes | NM | Mild | 24 January | PCR | NM | Yes | No | 6 | AS | Alive | ||
| 6 | 29M | Yes | NM | Mild | 29 January | PCR | NM | Yes | No | 6 | AS | Alive | ||
| 7 | 21F | Yes | NM | Mild | 31 January | PCR | NM | Yes | No | 5 | RT-PCR | Alive | ||
| 16 | Wu et al.21 | |||||||||||||
| 1 | >70 | China | NM | Yes | NM | 01 February | NM | NM | NM | Yes | 3 | RT-PCR | Alive | |
| 2 | >70 | NM | Yes | NM | 02 February’ | NM | NM | NM | Yes | 5 | RT-PCR/AS | Alive | ||
| 3 | NM | NM | NM | NM | 02February | NM | NM | NM | No | 6 | AS | Alive | ||
| 4 | NM | NM | NM | NM | 23 January | NM | NM | NM | No | 25 | RT-PCR | Alive | ||
| 5 | NM | NM | NM | NM | 27 January | NM | NM | NM | No | 16 | RT-PCR | Alive | ||
| 6 | NM | NM | NM | NM | 30 January | NM | NM | NM | No | 9 | RT-PCR | Alive | ||
| 7 | NM | NM | NM | NM | 29 January | NM | NM | NM | No | 22 | AS | Alive | ||
| 8 | NM | NM | NM | NM | 28 January | NM | NM | NM | No | 23 | AS | Alive | ||
| 9 | NM | NM | NM | NM | 07 February | NM | NM | NM | No | 11 | AS | Alive | ||
| 10 | NM | NM | NM | NM | 07 February | NM | NM | NM | No | 07 | AS | Alive | ||
| 17 | Zhou et al.22 | 40M | China | Yes | Yes | Severe | 23 January | RT-PCR | Yes | Yes | Yes | 5 days after discharge | RT-PCR | Alive |
| 18 | Zhao et al23 | |||||||||||||
| (7/14) | 5.7 (Median) (2.9–7.3)Range F-4 |
China | 5 Yes 2 No |
No Co-morbidity | Mild (All) | NM (All) | RT-PCRa | NM (All) | Yes(all) | 6-No 1- Yes |
14 days from discharge (7–17) | RT-PCRa | Alive (All) | |
| 19 | Li et al.24 | 50M | China | Yes | Yes | Mild | D13 | RT-PCR | Yes on D 40. IgM and IgG positive | Yes | No | 14 | RT-PCR | Alive |
| 20 | Chen et al.25 | |||||||||||||
| 1 | 29M | China | Yes | NM | Mild | 01 February | RT-PCRa | NM | Yes | No | 3 | RT-PCRa | Alive | |
| 2 | 49F | Yes | NM | Mild | 02 February | NM | Yes | No | 3 | Alive | ||||
| 3 | 12F | No | NM | Mild | 05 February | NM | Yes | No | 3 | Alive | ||||
| 4 | 38M | Yes | NM | mild | 30 January | NM | Yes | No | 3 | Alive | ||||
| 21 | Hu et al26 (11) | median age 27, range 4–58 years F-4 |
China | Yes (All) | 3-Co-morbidities | Mild-1 Moderate- 9 Severe-1 |
NM(All) | RT-PCRa | NM(All) | Yes (All) | No (All) | 14 (9–17) | RT-PCRa | Alive (All) |
| 22 | Jianghong An et al.27 | Median age 20 (5–64) 7-F (Mild) 38 (2–60) 15-F |
China | Yes | 1/11 1/27 |
Mild −11 Moderate 27 |
Patient were discharged, January 23 to February 25 (14 days) | RT PCR, Anal swab | Yes no difference between the two groups | Yes (All) | No (All) | Weekly after discharge | RT PCRa | Alive (All) |
| 23 | Chen et al.28 | 46 F | China | Yes | No | Mild | 24 January | RT-PCR | No | Yes | No | 03 days after last negative test | RT-PCR | Alive |
| 24 | Duggan et al.29 | 82 M | USA | Yes | Yes | Severe | Early April | RT-PCR | No | Yes | No | 10 days post discharge | RT-PCR | Alive |
| 25 | Ye-min et al.30 | 49 M | China | Yes | NM | Mild | 22 January | RT-PCR | NM | Yes | No | 3 days after discharge | Sputum positive PCR -ve | Alive |
| 26 | To et al.31 | 33M | Hong kong | Yes | No Co-morbidity | Mild | 29 March | RT- PCR | NM | Yes | No | 123 days after discharge | RT-PCR | Alive |
| 27 | Tillet et al.32 | 25M | USA | Yes | No | Mild | 18 April | RT- PCR | Yes | Yes | Yes | 10 days after last negative test | RT-PCR | Alive |
| 28 | Elslande et al.33 | 51F | Belgium | Yes | Asthma | Moderate | March 20 | RT-PCR | Yes (second time) | No | Yes | 10 weeks after home quarantine | RT-PCR | Alive |
| 29 | Prado-Vivar B et al.34 | 46M | Eucadorian | Yes | NM | Mild | May 12 | RT-PCR | Yes | Yes | Yes | 6 weeks after being negative | RT-PCR | Alive |
| 30 | Gupta et al.35 | 25M | India | Yes | No | No | 05 May | RT-PCRa | NM NM |
Yes | No | 100 days after tested negative | RT-PCR | Alive |
| 28F | No | 17 May | Yes | No | 101 days | RT-PCR | Alive | |||||||
AS, anal swab; F, female; M, male; NM, not mentioned; RT-PCR, reverse transcriptase polymerase chain reaction (naso-pharyngeal swab).
Same for all.
Molecular test for COVID-19 among discharged patients had been performed on sputum (lower respiratory tract), nasopharyngeal and anal swab. The details are shown in Table 1.
The majority of the cases (197, 89.9%) had mild to moderate clinical presentation. The clinical severity at initial presentation was not specified for 10 cases. Only 12 cases (5.5%; 95% CI: 2.8%–9.4%) had severe disease manifestation at initial presentation. A total of 64 (29.2%) reported cases were symptomatic during the second episode with the majority of them having less severe disease manifestation compared with the first episode. One hundred fifty (68.5%) cases were asymptomatic, and the status of five was unknown.
A total of 57 cases (26%) among the re-positives cases had comorbidities. A total of 51 and 17 re-positive cases had received antivirals and corticosteroids, respectively. Time interval between discharge/preceding a RT-PCR–negative report and a positive molecular test report ranged from 03 days to 123 days.
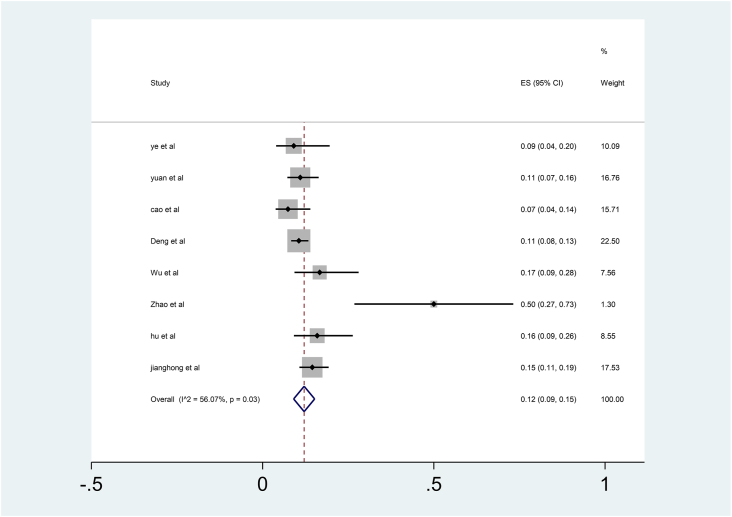
Eight studies have mentioned the proportion of cases that became re-positives after a negative RT-PCR test during follow-up period. The summary of proportions and their pooled ratio is given in Fig. 2. The pooled proportion using random-effects was 12% (95% CI: 09%–15%). All studies had a follow-up period in the range of 4–17 days except one which had a follow-up period of 14–46 days.26
Fig. 2.
Pooled proportions of re-positives from studies.
Only a few studies confirmed the presence of antibodies after the first episode of clinical illness (Table 1). Even after the development of antibodies, studies had reported re-positivity (Table 1). A few studies had conducted contact tracing of re-positives. The studies did not find any positive cases among high risk contact with re-positives (Table 1). Mortality was reported in seven re-positive cases. The age range of these cases ranges from 73 to 91 years. All of them had multiple comorbidities.
Only a few studies had looked into the genetic analysis of the SARS-COV-2 to confirm reinfection.30, 31, 32, 33, 34, 35 These studies had found reinfection to occur even after a period of 123 days after the last RT-PCR negative test.
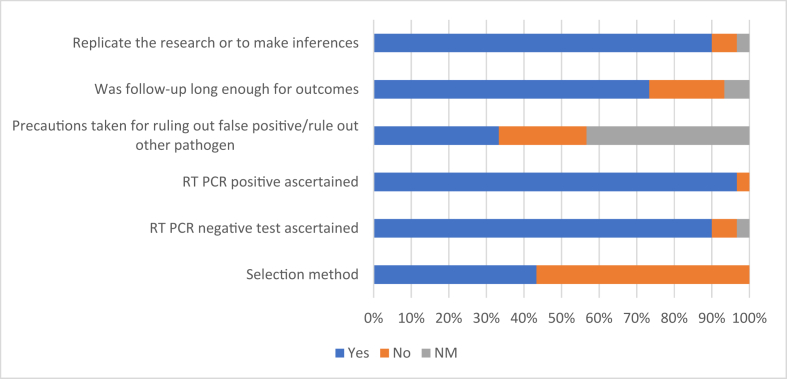
The quality of studies was assessed by using the modified Murad et al scale as shown in Fig. 3. In most of the studies, selection methods of COVID-19 cases were not clear; in addition, there were no precautions taken for ruling out false positives or rule out an alternate pathogen, which could produce similar signs and symptoms.
Fig. 3.
Quality of study as assessed using the modified Murad scale. ∗NM- Not Mentioned.
Korea Centers for Disease Control and Prevention reported 141 cases positive by RT-PCR after they recovered from COVID-19.34 However, the probable reason given was relapse or inconsistent tests. The details were not available on the site.
Discussion
The systematic review was carried out for all case reports and case series to identify common characteristics and evidence available for re-positive cases. Although during review of available literature, we found evidence of re-positives after symptom free and negative RT-PCR test, yet it is difficult to ascertain whether it was due to continuous shedding of the virus, relapse, or reinfection by the virus. Only six studies that have carried out the genetic analysis of the COVID-19 virus in re-positives found genomic diversity, thus establishing reinfection.
Recurrence has been observed across all ages, from 10 months to 91 years of age. Mortality after reinfection is seen in the older age group with multiple comorbidities which is consistent with primary infection. Innate and acquired immunity of the individual may also influence recurrences.35 Hence, immune-senescence of the old age and immunosuppressant drugs may affect recurrence. However, the majority (92.2%) of the COVID-19 re-positive cases had not been given corticosteroids for management during the primary episode of illness. Many re-positive cases were also given antivirals. However, in absence of control group, it is difficult to draw any inference for association of corticosteroids or antivirals. Second, the denominator in case reports or case series is difficult to ascertain, hence rate can also be not calculated. The effect of other immunomodulators and antiviral drugs on recurrence may be studied in a well-designed study with control group.
Pooled proportion of studies that have specified the proportion of COVID-19 re-positives was carried out. Approximately 12% of discharged COVID-19 cases after the first episode of infection were detected positive during subsequent molecular testing. The reasons may be related to Intermittent shedding of virus, the persistence of the virus, testing technique including sampling, or host characteristics. There was no evidence of secondary cases arising from these re-positives. Study carried out on nine patients of COVID-19 cases noted prolonged viral shedding in sputum.36 However, there is a little residual risk of infectivity with viral load less than 100,000 viral RNA copies per ml of sputum.36 This viral shedding in sputum needs to be further explored for infectivity of virus during recurrences as infectiousness of recurrence cases would have major implication on public health policy.
A notable area of scientific interest is the role of seroconversion among re-positives. Although animal studies suggest that antibody formation is protective against reinfection, yet in present systematic review we found that re-positives can occur even after seroconversion.37 The relation between seroconversion and re-positives further need to be explored.
Different anatomical sampling sites may also have some effect on viral detection. In many cases, even if the sample from the nasopharyngeal is negative, the samples from sputum (lower respiratory tract) and anal swab have been positive. There is evidence that the virus may be shed longer from the extrapharyngeal sites. There are reports that virus shedding from asymptomatic patients may continue from extrapulmonary sites in various bodily fluids (saliva, tears, faeces, throat, or nasal discharge) for a longer duration of time.38,39 Its role in reinfection is still not known.
Antibody-dependent enhancement is a known phenomenon in viral disease and responsible for increased severity of subsequent infections.40 However, in this systematic review, we found that clinical manifestations in majority of re-positive cases were milder than the initial infection. This may be because most of the cases were not true reinfections but persistence of the same infection or due to intermittent virus shedding. Even in the six studies with documented genomic analysis, clinical manifestations in the reinfection cases were mild to moderate. A model for reinfection has concluded that the rate of reinfection in the recovered population would decline to zero over time as the virus is cleared clinically from the system of the recovered cases.41
Risk of bias
Although there are no set guidelines for estimating the risk of bias in case reports and case series, the authors feel that initial RT-PCR positive, subsequent RT-PCR negative, serological testing, and RT-PCR positive after symptom-free period are essential for drawing conclusion about relapse or reinfection. Few case reports did not mention a negative RT-PCR test after the first COVID-19 infection.5,9
One of the limitations of our study is that the literature search has been restricted to only English language and to Medline and Cochrane database. Hence, we may have missed articles published in Chinese and other non-English languages.
Since these patients of recurrence may represent a special subset of COVID-19 cases, the findings may not be generalizable to all COVID-19 cases. More research is needed to delineate the factors responsible for recurrence in recovered cases. As the pandemic progresses, more conclusive evidence in this context would be gathered. Nevertheless, there is a strong case for proper documentation of all the cases to further refute or confirm the findings.
Disclosure of competing interest
The authors have none to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mjafi.2021.05.025.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.She J., Jiang J., Ye L., Hu L., Bai C., Song Y. Novel coronavirus of pneumonia in Wuhan, China: emerging attack and management strategies. Clin Transl Med. 2019;2020:9. doi: 10.1186/s40169-020-00271-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naming the coronavirus disease (COVID-19) and the virus that causes it. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it [accessed August 17, 2020].
- 3.Coronavirus (COVID-19) events as they happen. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen [accessed August 17, 2020].
- 4.Murad M.H., Sultan S., Haffar S., Bazerbachi F. Methodological quality and synthesis of case series and case reports. BMJ Evid-Based Med. 2018;23:60–63. doi: 10.1136/bmjebm-2017-110853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batisse D., Benech N., Botelho-Nevers E. Clinical recurrences of COVID-19 symptoms after recovery: viral relapse, reinfection or inflammatory rebound? J Infect. 2020 doi: 10.1016/j.jinf.2020.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lafaie L., Célarier T., Goethals L., et al. Recurrence or relapse of COVID-19 in older patients: a description of three cases. J Am Geriatr Soc. n.d.;n/a. doi: 10.1111/jgs.16728.. [DOI] [PMC free article] [PubMed]
- 7.Bentivegna E., Sentimentale A., Luciani M., Speranza ML., Guerritore L., Martelletti P. New IgM seroconversion and positive RT-PCR test after exposure to the virus in recovered COVID-19 patient. J Med Virol. n.d.;n/a. doi: 10.1002/jmv.26160.. [DOI] [PMC free article] [PubMed]
- 8.Ye G., Pan Z., Pan Y. Clinical characteristics of severe acute respiratory syndrome coronavirus 2 reactivation. J Infect. 2020;80:e14–e17. doi: 10.1016/j.jinf.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravioli S., Ochsner H., Lindner G. Reactivation of COVID-19 pneumonia: a report of two cases. J Infect. 2020;81:e72–e73. doi: 10.1016/j.jinf.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loconsole D., Passerini F., Palmieri V.O. Recurrence of COVID-19 after recovery: a case report from Italy. Infection. 2020 doi: 10.1007/s15010-020-01444-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang M., Li Y., Han M., Wang Z., Zhang Y., Du X. Recurrent PCR positivity after hospital discharge of people with coronavirus disease 2019 (COVID-19) J Infect. 2020;81:147–178. doi: 10.1016/j.jinf.2020.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang S.-C., Wang J.-T., Huang L.-M. Longitudinal analysis of severe acute respiratory syndrome (SARS) coronavirus-specific antibody in SARS patients. Clin Diagn Lab Immunol. 2005;12:1455–1457. doi: 10.1128/CDLI.12.12.1455-1457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo S.Y., Lee Y., Lee G.H., Kim D.H. Reactivation of SARS-CoV-2 after recovery. Pediatr Int. 2020;62:879–881. doi: 10.1111/ped.14312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu F., Cai Z., Huang J. Positive SARS-CoV-2 RNA recurs repeatedly in a case recovered from COVID-19: dynamic results from 108 days of follow-up. Pathog Dis. 2020 doi: 10.1093/femspd/ftaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan B., Liu H.-Q., Yang Z.-R. Recurrence of positive SARS-CoV-2 viral RNA in recovered COVID-19 patients during medical isolation observation. Sci Rep. 2020;10 doi: 10.1038/s41598-020-68782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan L., Xu D., Ye G. Positive RT-PCR test results in patients recovered from COVID-19. J Am Med Assoc. 2020;323:1502–1503. doi: 10.1001/jama.2020.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao H., Ruan L., Liu J., Liao W. The clinical characteristic of eight patients of COVID-19 with positive RT-PCR test after discharge. J Med Virol. 2020 doi: 10.1002/jmv.26017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng W., Guang T., Yang M. Positive results for patients with COVID-19 discharged form hospital in Chongqing, China. BMC Infect Dis. 2020;20 doi: 10.1186/s12879-020-05151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng J., Wang M., Zhang G., Lu E. Seven discharged patients turning positive again for SARS-CoV-2 on quantitative RT-PCR. Am J Infect Contr. 2020;48:725–726. doi: 10.1016/j.ajic.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu J., Liu X., Liu J. Coronavirus disease 2019 test results after clinical recovery and hospital discharge among patients in China. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.9759. e209759–e209759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou W., Xu X., Chang Z., et al. The dynamic changes of serum IgM and IgG against SARS-CoV-2 in patients with COVID-19. J Med Virol. n.d.;n/a. doi: 10.1002/jmv.26353.. [DOI] [PMC free article] [PubMed]
- 22.Zhao W., Wang Y., Tang Y. Characteristics of children with reactivation of SARS-CoV-2 infection after hospital discharge. Clin Pediatr (Phila). 2020;59:929–932. doi: 10.1177/0009922820928057. [DOI] [PubMed] [Google Scholar]
- 23.Li J., Long X., Fang X. SARS-CoV-2 positivity in a discharged COVID-19 patient: a case report. Clin Microbiol Infect. 2020;26:1115–1117. doi: 10.1016/j.cmi.2020.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen D., Xu W., Lei Z. Recurrence of positive SARS-CoV-2 RNA in COVID-19: a case report. Int J Infect Dis. 2020;93:297–299. doi: 10.1016/j.ijid.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu R., Jiang Z., Gao H. Recurrent positive Reverse transcriptase–polymerase Chain reaction results for coronavirus disease 2019 in patients discharged from a hospital in China. JAMA Netw Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.10475. e2010475–e2010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.An J., Liao X., Xiao T. Clinical characteristics of recovered COVID-19 patients with re-detectable positive RNA test. Ann Transl Med. 2020;8:1084. doi: 10.21037/atm-20-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duggan N.M., Ludy S.M., Shannon B.C., Reisner A.T., Wilcox S.R. A case report of possible novel coronavirus 2019 reinfection. Am J Emerg Med. 2020 doi: 10.1016/j.ajem.2020.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Positive Result of Sars-Cov-2 in Sputum from a Cured Patient with COVID-19 | Elsevier Enhanced Reader. doi: 10.1016/j.tmaid.2020.101619.. [DOI] [PMC free article] [PubMed]
- 29.To K.K.-W., Hung I.F.-N., Ip J.D. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin Infect Dis Off Publ Infect Dis Soc Am. 2020 doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tillett R.L., Sevinsky J.R., Hartley P.D. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Elslande J., Vermeersch P., Vandervoort K. Symptomatic SARS-CoV-2 reinfection by a phylogenetically distinct strain. Clin Infect Dis Off Publ Infect Dis Soc Am. 2020 doi: 10.1093/cid/ciaa1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prado-Vivar B., Becerra-Wong M., Guadalupe J.J. Social Science Research Network; Rochester, NY: 2020. COVID-19 Re-infection by a Phylogenetically Distinct SARS-CoV-2 Variant, First Confirmed Event in South America. [Google Scholar]
- 33.Gupta V., Bhoyar R.C., Jain A. Asymptomatic reinfection in two healthcare workers from India with genetically distinct SARS-CoV-2. Clin Infect Dis Off Publ Infect Dis Soc Am. 2020 doi: 10.1093/cid/ciaa1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.www.ETHealthworld.com. Why are some S.Koreans who recovered from the coronavirus testing positive again ? - ET HealthWorld. ETHealthworld.Com. Available at: https://health.economictimes.indiatimes.com/news/industry/why-are-some-s-koreans-who-recovered-from-the-coronavirus-testing-positive-again-/75178047 [accessed August 15, 2020].
- 35.Rouse B.T., Sehrawat S. Immunity and immunopathology to viruses: what decides the outcome? Nat Rev Immunol. 2010;10:514–526. doi: 10.1038/nri2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wölfel R., Corman V.M., Guggemos W. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 37.Bao L., Deng W., Gao H. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. BioRxiv. 2020 doi: 10.1101/2020.03.13.990226. 2020.03.13.990226. [DOI] [Google Scholar]
- 38.Kalkeri R., Goebel S., Sharma G.D. SARS-CoV-2 shedding from asymptomatic patients: contribution of potential extrapulmonary tissue reservoirs. Am J Trop Med Hyg. 2020;103:18–21. doi: 10.4269/ajtmh.20-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du W., Yu J., Liu X., Chen H., Lin L., Li Q. Persistence of SARS-CoV-2 virus RNA in feces: a case series of children. J Infect Public Health. 2020;13:926–931. doi: 10.1016/j.jiph.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulkarni R. Antibody-dependent enhancement of viral infections. Dyn Immune Act Viral Dis. 2019:9–41. doi: 10.1007/978-981-15-1045-8_2. [DOI] [Google Scholar]
- 41.Okhuese A.V. Estimation of the probability of reinfection with COVID-19 by the susceptible-exposed-infectious-removed-undetectable-susceptible model. JMIR Public Health Surveill. 2020;6 doi: 10.2196/19097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.