Banana (Musa spp.) is an important staple food crop and a source of income for resource‐poor farmers in more than 136 tropical and sub‐tropical countries with an annual production of 155 million tons (FAOSTAT, 2018). Many diseases severely constrain banana production, particularly where many pathogens co‐exist (Tripathi et al., 2020). Banana Xanthomonas wilt (BXW) caused by Xanthomonas campestris pv. musacearum (Xcm) is considered among the most destructive banana diseases in East and Central Africa (Tripathi et al., 2009). All the cultivated banana varieties are susceptible, and only the wild‐type progenitor, Musa balbisiana, is resistant to BXW disease (Tripathi et al., 2019). Overall economic losses from BXW were estimated at US$ 2–8 billion over a decade. The use of disease‐resistant varieties is one of the most effective strategies to manage diseases. Recent advances in CRISPR/Cas‐based genome editing can accelerate banana improvement. The availability of reference genome sequences and the CRISPR/Cas9‐editing system has made it possible to develop disease‐resistant banana by precisely editing the endogenous genes (Ntui et al., 2020).
The downy mildew resistance 6 (DMR6) is a susceptibility gene encoding 2‐oxoglutarate Fe(II)‐dependent oxygenase (2OGO), which is up‐regulated during pathogen infection (Low et al., 2020). DMR6 and its paralog DMR6‐Like Oxygenase1 (DLO1) are suppressors of plant immunity and co‐expressed during pathogen infection (Zeilmaker et al., 2015). In barley, the dmr6 mutants demonstrated a higher level of resistance than the dlo1 mutant, whereas the dmr6dlo1 double‐mutants showed complete resistance to downy mildew but exhibited dwarf phenotype. The dmr6 mutants could provide broad‐spectrum disease resistance to bacterial pathogens. Considering that dmr6 mutants can provide broad‐spectrum resistance to different pathogens, developing CRISPR/Cas9‐edited banana with the targeted mutations in the MusaDMR6 orthologues represents a strategy for controlling BXW.
Phylogenetic analysis of the 2OGO gene family with Musa acuminata, Musa balbisiana, Arabidopsis thaliana, Solanum lycopersicum and Nicotiana tabacum identified seven AtDMR6 orthologues in Musa acuminata and two in Musa balbisiana belonging to the same clade as Q9FLV0.1 (Figure 1a). No DLO paralog was observed in banana clustering with Q9ZSA8 (DLO1) and Q9ZSA7 (DLO2). One of the MusaDMR6 orthologues, Ma04_p20880.1 (annotated as flavanone 3‐dioxygenase‐like), clustered with Q9FLV0.1 (AtDMR6), was selected for further analysis as a putative candidate for enhancing disease resistance in banana. The differential expression of Ma04_p20880.1 was investigated upon Xcm infection in banana. The qRT‐PCR analysis of the in vitro plantlets showed a 60‐fold higher expression of the Ma04_p20880.1 in response to Xcm in the BXW‐susceptible cultivar Sukali Ndiizi, compared to the non‐infected plants (Figure 1b), suggesting that Ma04_p20880.1 might exhibit similar functions as AtDMR6. However, up‐regulation of the Ma04_p20880.1 gene was not observed in the BXW‐resistant genotype Musa balbisiana, confirming the activation of MusaDMR6 only in the compatible plant–pathogen interaction. Based on this result, the Ma04_p20880.1 gene from Musa accuminata (AA genome) and its corresponding gene ITC1587_Bchr1_T00081 (annotated as oxoglutarate 3‐dioxygenase‐like) from Musa balbisiana (BB genome) were selected for the editing of BXW‐susceptible banana cultivar Sukali Ndiizi (AAB genome). The gene sequences downloaded from http://banana‐genome‐hub.southgreen.fr were aligned, and two gRNAs, 114 bp apart, were designed from the conserved region in exon4 (Figure 1c). A plasmid construct, pMDC32‐Cas9‐MusaDMR6, containing a plant‐codon‐optimized Cas9 gene driven by a 2XCaMV35S promoter, two gRNAs targeting MusaDMR6 under OsU6 promoter individually, and a plant selectable‐marker hpt gene regulated by CaMV35S promoter was prepared (Figure 1d).
Figure 1.
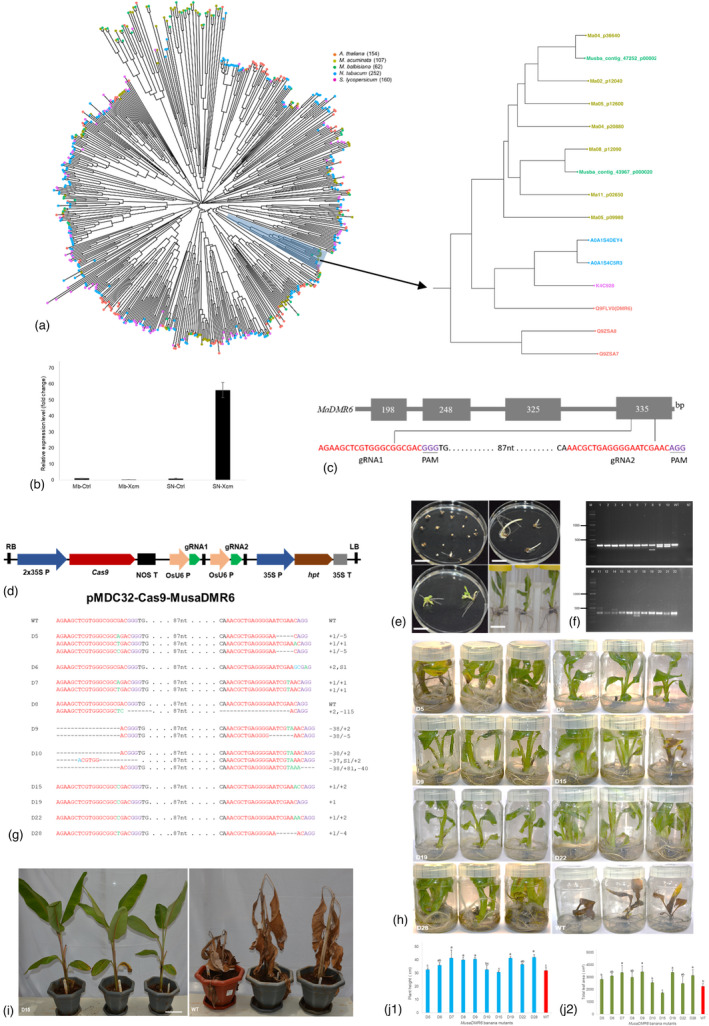
Generation and evaluation of Musadmr6 events for enhanced disease resistance. (a) Phylogenetic tree of 2‐oxoglutarate Fe(II)‐dependent oxygenase (2OGO) gene family. Protein sequences belonging to the 2OGO gene family (PFAM ID‐PF03171) for Arabidopsis thaliana, Nicotiana tabacum and Solanum lycopersicum were downloaded from the PFAM database (http://pfam.xfam.org/). The predicted protein sequence for Musa acuminata and Musa balbisiana were downloaded from the PanMusa database (https://panmusa.greenphyl.org/) for the Interpro family Oxoglutarate/iron‐dependent dioxygenase (IPR005123). The protein sequences were aligned using the Clustal Omega multiple sequence alignment program, and a phylogenetic tree displayed using the ggtree package in R. The DMR6 orthologues found in the same clade with Q9FLV0.1 are highlighted in blue, and a zoomed view of the clade is shown. (b) Expression of MusaDMR6 orthologue (Ma04_p20880.1) in Musa balbisiana and Sukali Ndiizi in response to Xanthomonas campestris pv. musacearum. (c) Gene structure of MusaDMR6 orthologue (Ma04_p20880.1) showing the gRNA target sites. Black blocks indicate exons of the gene, PAM sequences in purple, and protospacer sequences in red. (d) Schematic presentation of construct pMDC32‐Cas9‐MusaDMR6. (e) Regeneration of transgenic banana mutants. Size bar 5 cm. (f) Band‐shift‐PCR analysis of banana mutants. 1‐22‐banana mutants, WT‐wild‐type, M‐molecular marker. (g) Sequencing‐based detection of targeted mutations induced by two gRNAs in exon4 of MusaDMR6 orthologue. PAM is in purple, and protospacers in red. Deletions are denoted by black dashes, insertion by green and substitution by blue. (h) Rapid bioassay of mutants (D5‐28) and wild‐type (WT) plants for resistance against Xanthomonas campestris pv. musacearum. Photographs were taken at 6 weeks post‐inoculation. Size bar 5 cm. (i) Greenhouse evaluation of mutant (D15) and wild‐type (WT) plants for resistance against Xanthomonas campestris pv. musacearum. Photographs were taken at 8 weeks post‐inoculation. Size bar 10 cm. (j) Evaluation of mutants for plant growth; (j1) plant height; (j2) total leaf area. The data were analysed using Minitab14 and presented as mean and standard deviation of three replicates. Multiple comparisons of mutants with control by Dunnett’s test were performed, and bars with the same alphabet are not significantly different.
The plasmid pMDC32‐Cas9‐MusaDMR6 was introduced into the embryogenic cell suspension (ECS) of Sukali Ndiizi through Agrobacterium‐mediated transformation as described by Tripathi et al. (2015). Thirty transgenic events were generated (Figure 1e) and validated for the presence of the Cas9 gene by PCR. The events were checked by band shift PCR using the primers flanking both target sites. The amplicons of D8 and D17 showed a predicted PCR band shift of 114 bp, suggesting deletions in the region between both gRNAs as expected when Cas9 cuts both target sites simultaneously (Figure 1f). D8 and D17 also showed a second amplicon similar to that of WT, suggesting heterozygous mutations. D11 and D14 showed an amplicon identical to WT and a second bigger amplicon, suggesting a possible addition of a large fragment.
The targeted mutations in the events were confirmed by amplifying the target region (304 bp) of MusaDMR6 and directly Sanger sequencing of 10 replicates of purified amplicons. Sequencing of all the 30 events showed different indels proving to be independent and giving a mutation frequency of 100% (Figure 1g). The indels were observed at both target sites for gRNA1 and gRNA2 in most events (26/30) within 3–4 bp upstream of the PAM sequences. All types of mutations, deletions (∆4 bp to ∆115 bp), insertions (+1 to +172), substitutions (S1) or deletions accompanied by insertions were observed. All three different WTs, generated from the same ECS line used in the experiments along with the mutants, showed wild‐type sequences with no indels confirming that all the mutations among the various events were due to CRISPR/Cas9 action.
Three replicates of each event were evaluated for resistance against the Ugandan isolate of Xcm using the in vitro and potted plants. Xcm is a genetically homogenous pathogen, though genome sequencing has revealed six closely related clades (Nakato et al., 2021). No virulence difference has been reported among different isolates of Xcm; therefore, only one isolate of Xcm was used in this study. Rapid in vitro screening (Tripathi et al., 2008) demonstrated enhanced resistance to BXW in most of the events (25/30). The in vitro plantlets of four events (D6, D9, D19 and D22) challenged with Xcm did not show any BXW symptom up to 45 days post‐inoculation (dpi). In contrast, WT displayed BXW symptoms as browning and necrosis in 14.7 ± 1.2 dpi and finally complete wilting of the plants within 26.7 ± 1.2 dpi (Figure 1h). D5, D15 and D28 showed significantly delayed symptoms with lower disease incidence (33%–66%) compared to 100% disease incidence in WT. About 52% of events (12/23) further confirmed the enhanced resistance (25%–100%) to BXW in the greenhouse evaluation using potted plants. The inoculated plants of D15 did not show any BXW symptoms up to 60 dpi; however, the WT completely wilted within 29.3 ± 4.2 dpi (Figure 1i). The suckers of the ratoon crop produced from the potted plants of D15 were initiated into the tissue culture to test the stability of mutations in the micropropagated plantlets. Sequencing of the three micropropagated plantlets showed the same indels (+1–2 bp) as the mother plant, confirming the mutations’ stability.
Growth trial of three replicates of the potted plants of all the edited events under the greenhouse conditions showed normal growth with no morphological differences when compared to WT. No adverse effect was observed on the plant height and total leaf area of the 90‐day‐old potted plants of mutants compared to WT (Figure 1j), confirming no detrimental impact of knocking‐out one DMR6 orthologue.
In summary, Musadmr6 mutants of banana showed enhanced resistance to important disease, BXW, and did not show any detrimental effect on plant growth. The performance of these banana mutants needs to be evaluated in field conditions.
Conflict of interest
Authors declare no conflict of interest.
Authors’ contributions
LT conceived the idea; JNT generated mutants and performed disease resistance evaluations; VON designed gRNAs, made construct and performed molecular analysis; TS performed phylogenetic analysis; all authors wrote the manuscript.
Acknowledgement
This research was supported by the CGIAR program for roots, tubers and banana. The assistance provided by Sarah Macharia and Mark Adero is appreciated.
Tripathi, J. N. , Ntui, V. O. , Shah, T. and Tripathi, L. (2021) CRISPR/Cas9‐mediated editing of DMR6 orthologue in banana (Musa spp.) confers enhanced resistance to bacterial disease. Plant Biotechnol J, 10.1111/pbi.13614
References
- FAOSTAT . (2018). http://www.fao.org/FAOSTAT./en/#data/QC (Accessed: 26.10.20).
- Low, Y.C. , Lawton, M.A. and Di, R. (2020) Validation of barley 2OGO gene as a functional orthologue of Arabidopsis DMR6 gene in Fusarium head blight susceptibility. Sci. Rep. 10, 9935. 10.1038/s41598-020-67006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakato, G.V. , Studholme, D.J. , Blomme, G. , Grant, M. , Coutinho, T.A. , Were, E.M. , Wicker, E. et al. (2021) SNP‐based genotyping and whole‐genome sequencing reveal previously unknown genetic diversity in Xanthomonas vasicola pv. musacearum, causal agent of banana xanthomonas wilt, in its presumed Ethiopian origin. Plant. Pathol. 70, 534–543. 10.1111/ppa.13308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntui, V.O. , Tripathi, J.N. and Tripathi, L. (2020) Robust CRISPR/Cas9 mediated genome editing tool for banana and plantain (Musa spp.). Curr. Plant Biol. 21, 100128, 10.1016/j.cpb.2019.100128. [DOI] [Google Scholar]
- Tripathi, J.N. , Oduor, R.O. and Tripathi, L. (2015) A high‐throughput regeneration and transformation platform for production of genetically modified banana. Front. Plant Sci. 6, 1025. 10.3389/fpls.2015.01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi, L. , Mwangi, M. , Abele, S. , Aritua, V. , Tushemereirwe, W.K. and Bandyopadhyay, R. (2009) Xanthomonas Wilt: A threat to banana production in East and central Africa. Plant Dis. 93, 440–451. 10.1094/PDIS-93-5-0440. [DOI] [PubMed] [Google Scholar]
- Tripathi, L. , Ntui, V.O. and Tripathi, J.N. (2020) CRISPR/Cas9‐based genome editing of banana for disease resistance. Curr. Opin. Plant Biol. 56, 118–126. 10.1016/j.pbi.2020.05.003. [DOI] [PubMed] [Google Scholar]
- Tripathi, L. , Odipio, J. , Tripathi, J.N. and Tusiime, G. (2008) A rapid technique for screening banana cultivars for resistance to Xanthomonas wilt. Eur. J. Plant Pathol. 121, 9–19. 10.1007/s10658-007-9235-4. [DOI] [Google Scholar]
- Tripathi, L. , Tripathi, J.N. , Shah, T. , Muiruri, S.K. and Katari, M. (2019) Molecular basis of disease resistance in banana progenitor Musa balbisiana against Xanthomonas campestris pv. musacearum . Sci. Rep. 9, 7007, 10.1038/s41598-019-43421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilmaker, T. , Ludwig, N.R. , Elberse, J. , Seidl, M.F. , Berke, L. , Van Doorn, A. , Schuurink, R.C. et al. (2015) DOWNY MILDEW RESISTANT 6 and DMR6‐LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 81, 210–222. 10.1111/tpj.12719. [DOI] [PubMed] [Google Scholar]


