Summary
Abiotic stress resistance traits may be especially crucial for sustainable production of bioenergy tree crops. Here, we show the performance of a set of rationally designed osmotic‐related and salt stress‐inducible synthetic promoters for use in hybrid poplar. De novo motif‐detecting algorithms yielded 30 water‐deficit (SD) and 34 salt stress (SS) candidate DNA motifs from relevant poplar transcriptomes. We selected three conserved water‐deficit stress motifs (SD18, SD13 and SD9) found in 16 co‐expressed gene promoters, and we discovered a well‐conserved motif for salt response (SS16). We characterized several native poplar stress‐inducible promoters to enable comparisons with our synthetic promoters. Fifteen synthetic promoters were designed using various SD and SS subdomains, in which heptameric repeats of five‐to‐eight subdomain bases were fused to a common core promoter downstream, which, in turn, drove a green fluorescent protein (GFP) gene for reporter assays. These 15 synthetic promoters were screened by transient expression assays in poplar leaf mesophyll protoplasts and agroinfiltrated Nicotiana benthamiana leaves under osmotic stress conditions. Twelve synthetic promoters were induced in transient expression assays with a GFP readout. Of these, five promoters (SD18‐1, SD9‐2, SS16‐1, SS16‐2 and SS16‐3) endowed higher inducibility under osmotic stress conditions than native promoters. These five synthetic promoters were stably transformed into Arabidopsis thaliana to study inducibility in whole plants. Herein, SD18‐1 and SD9‐2 were induced by water‐deficit stress, whereas SS16‐1, SS16‐2 and SS16‐3 were induced by salt stress. The synthetic biology design pipeline resulted in five synthetic promoters that outperformed endogenous promoters in transgenic plants.
Keywords: synthetic biology, synthetic promoters, de novo motif analysis, computational design, abiotic stress, cis‐regulatory elements, poplar, bioenergy
Introduction
Abiotic stresses such as water‐deficit stress and salinity are major challenges in agriculture. Rising temperatures associated with climate change can exacerbate these challenges by increasing evaporation rates and plant transpiration. Thus, there is great interest and demand for improving abiotic stress tolerance through biotechnology. One approach is to introduce transgenes into crops to endow stress resistance using constitutive promoters, for example the Cauliflower mosaic virus (CaMV) 35S promoter (Wang et al., 2016). However, constitutive expression of stress resistance genes can result in unintended pleiotropic effects on plant growth when stressors are not present (Lee and Pijut, 2018; Li et al., 2012; Okumura et al., 2016; Wei et al., 2017; Yamasaki and Randall, 2016).
Expressing transgenes under the control of stress‐specific inducible promoters may be useful to minimize negative pleiotropic effects (Das et al., 2011; Datta et al., 2012; Saint Pierre et al., 2012; Wei et al., 2017), but native plant promoters often result in relatively low and complex patterns of induced gene expression (Halpin, 2005; Hou et al., 2012; Peramuna et al., 2018). Synthetic promoters may be designed to overcome shortcomings of native promoters in genetic engineering (Mehrotra et al., 2011; Liu and Stewart, 2015; Rushton, 2016).
Rational design of synthetic promoters typically starts with appropriate ‐omics data to identify cis‐regulatory elements (CREs). CREs are composed of non‐coding DNA that is recognized by transcription factor(s) that specifically regulate downstream genes. Key motifs from these elements can be multimerized upstream of a core promoter sequence (Liu et al., 2014). The simplest architecture consists of a single short DNA motif that is multimerized and fused upstream of core promoter, which results in low transcriptional complexity (Rushton, 2016). Spatial and temporal transgene expression can be made more complex by integrating additional CREs, synthetic motifs and enhancers into the design (Hernandez‐Garcia and Finer, 2014; Kassaw et al., 2018). In addition, synthetic promoter CREs may be composed of purely native sequences, those from in silico design, or a mix (Roberts, 2011; Wu et al., 2018). While synthetic promoters have been quite useful in plant biology (Dey et al., 2015; Hernandez‐Garcia and Finer, 2014; Liu and Stewart, 2016), their construction is challenged by our limited knowledge of appropriate CREs and their relationship in controlling gene expression.
Populus is an economically important genus and one of the top tree models in research. Poplar is a feedstock for paper, cellulose, wood and fibre and a candidate for next‐generation biofuels (Sannigrahi et al., 2009). Populus genomes have been sequenced (Tuskan et al., 2006) and are routinely genetically engineered (Han et al., 2000). Because of their experimental tractability, poplar species have been widely used in physiological studies to better understand how deciduous trees respond to changing environments (Jia et al., 2017; Li et al., 2014).
Here, we describe the performance of rationally designed synthetic osmotic stress‐inducible promoters in response to treatments including osmotic stress, salinity and water‐deficit stress. Published poplar transcriptomic data were mined to discover CREs and motifs that were used to design and construct synthetic promoters. The resulting synthetic promoters were tested for stress induction using poplar leaf protoplasts, transiently transformed Nicotiana benthamiana leaves and in whole, stably‐engineered, Arabidopsis thaliana plants using reporter genes.
Results
Co‐expressed gene analysis from water‐deficit or salt‐stressed poplar transcriptomes
Synthetic promoter design was performed using de novo cis‐regulatory DNA motif discovery (Figure 1). Synthetic promoters were built using a unified architecture by multimerizing candidate motifs upstream of the CaMV 35S −46 core promoter sequence, which was, in turn, fused to GFP.
Figure 1.
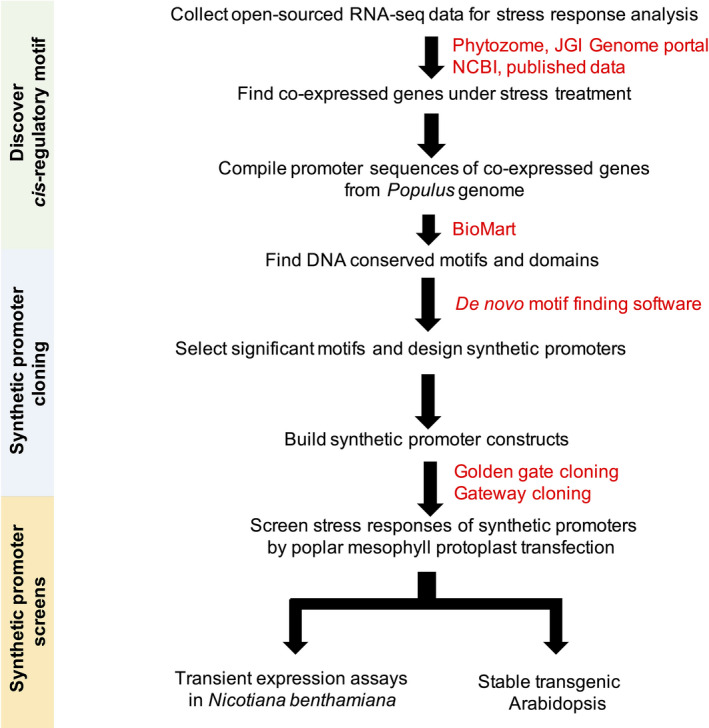
Research scheme to develop synthetic promoters for sensing osmotic stress in poplar.
As a first step to identify core motifs for designing synthetic promoters, we analysed gene expression patterns from published RNA‐seq data of different Populus species. A subset of water‐deficit or salt‐responsive genes were identified from six different transcriptomes (four from water‐deficit and two from salt stress experiments) (Cossu et al., 2014; Filichkin et al., 2018; Jia et al., 2017; Tang et al., 2013; Figure S1). In water‐deficit‐treated poplar transcriptomes, a total of 210 genes were selected to be mined for motifs (Figure S1). The respective 210 genes were detected in at least three out of four transcriptomic studies, in which there was at least a fourfold increase in gene expression in leaves under stress conditions (Table S1). To identify salt stress‐inducible genes, we compiled up‐regulated genes from two different root RNA‐seq data sets from salt‐treated plants (Filichkin et al., 2018; Zheng et al., 2015; Figure S1). Fourteen genes were commonly detected with over fourfold increase in both, which likewise was too few to result in significant motif hits. Thus, we expanded the candidate pool to 340 salt‐induced genes from the two transcriptomes that had at least a fourfold increase in gene expression from salt treatments (Table S2). In all cases, we assumed each gene promoter was contained in the immediate 2 kb DNA sequence upstream of the ATG translation initiation codon.
De novo DNA motif selection from promoters of expressed genes
Promoters from 210 water stress‐related co‐expressed and 340 salt stress‐related expressed genes were collected from P. trichocarpa genome annotation (v 3.0) and analysed for common motifs using MEME Suite (Bailey et al., 2015), MotifSampler (Thijs et al., 2002) and Weeder (Pavesi et al., 2004). Thirty and 34 DNA motifs (E < 0.001) were predicted by MEME to be water‐deficit‐ and salt stress‐inducible motifs, respectively (Tables S3 and S4). The same input was used to run MotifSampler and Weeder whose overlapped motifs with MEME output are summarized together in Tables S3 and S4.
Among the predicted DNA motifs from water‐deficit stress‐induced genes, 9 motifs (motifs 27, 19, 18, 13, 25, 9, 24, 1 and 4) were conserved sequentially on a 110‐bp domain in the final localization image of predicted motifs by motif location diagram integrated in MEME Suite. We denote this 110‐bp domain as the A domain (Figure 2a,b). The A domain was identified in gene promoters of 16 of the 210 water stress‐related genes. Their locations were variably distributed over 2‐kb upstream sequence from the start codon among the 16 promoters (Figure 2c; Table S5). All motifs, except motif 25, were predicted simultaneously by both MEME and MotifSampler (Table S3).
Figure 2.
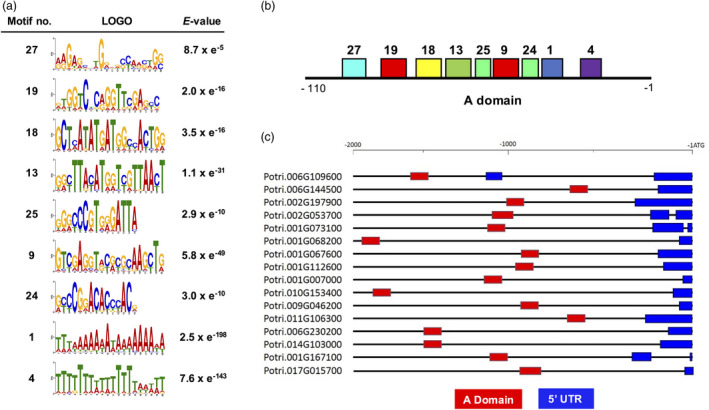
Bioinformatics characterization of SD motifs. (a) The LOGO sequence results generated by MEME Suite of 9 predicted SD motifs that were used to construct the domains shown in panel (b). Motifs 1 and 4 contain many T and A repeats; therefore, they were excluded from further analysis. Note that motifs 18, 13 and 9 were selected based on the top scored E‐values for subsequent experiments. (b) The ‘A domain’ included conserved nine SD motifs. The motifs and their locations were predicted by the MEME Suite. (c) Location of the conserved A domain on −2 kb promoter sequences of 16 co‐expressed genes.
To find CREs over the 9 long motifs of interest that were putatively related to water stress, we submitted these 9 long motifs to the database of plant cis‐acting regulatory DNA elements (PLACE; Higo et al., 1999). Interestingly, an auxin‐responsive motif was identified in motifs 19 and 18 (Table 1). Motif 18 also contained sequences for cold and freezing stress‐responsive factors such as MYC and an ICE binding factor (Ohta et al., 2018). A metal‐responsive factor‐binding sequence was detected in motif 9 (Table 1). Furthermore, these 9 motifs had higher statistical significance among 30 predicted DNA motifs (Table S3). However, motifs 1 and 4 had abundant A and T repeats (Table 1), suggesting that these two motifs may not be stress‐specific even though their E ‐values were significant (Figure 2a). Since these motifs came from water‐deficit stress‐inducible promoters and were subsequently used to make synthetic promoters, hereafter these elements are referred to as SD (synthetic promoter from drought stress‐inducible promoters) motifs.
Table 1.
Plant CREs in the SD A domain and SS motif predicted by PLACE
| ID | Motif sequence | Predicted element in PLACE | Sequence | Position (strand) | |
|---|---|---|---|---|---|
| SD | 27 | AAGAGCTTGGGACCAAGAAG | root nodule‐specific element | CTCTT | 1(−) |
| 19 | GTGGTCTCAGGTTCGAGCC | auxin‐responsive factor | GAGAC | 4(−) | |
| 18 | GCTCATATGATGGCCACTGG | MYC, ICE1 binding | CANNTG | 4(−) | |
| auxin‐responsive factor | CATATG | 4(+) | |||
| light induced | GCCAC | 13(+) | |||
| 13 | GGCTTACATGGTCGTTAACT | N.D. | N.D. | ||
| 25 | GGGCCCGTGGGATTA | phyA‐induced motif | GGGCC | 1(+) | |
| 9 | GTCGAGGTACGCGCAAGCTG | copper and oxygen responsive | GTAC | 7(+) | |
| calmodulin binding | VCGCGB | 9(−) | |||
| 24 | GCCCGGACACCCACG | N.D. | |||
| 1 | TTTAAAAAAAWAAAAAAAWA | T‐box | TTWTWTTWTT | 6(−) | |
| 4 | TTTTTTTTTTTTTTTAATTT | T‐box | TTWTWTTWTT | 6(+) | |
| SS16 | CGGGTCCTGAAGTTAACGAC | N.D. | N.D. | N.D. | |
N.D., not detected.
Thirty‐four DNA motifs were predicted by MEME from 340 gene promoter ‘hits’ from salt treatment studies (Tables S2 and S4). These predicted DNA motifs appeared well dispersed throughout candidate endogenous promoters. Most predicted motifs had many A/T or several sequence repeats (Table S4). Nonetheless, we found two GC‐rich motif candidates (motifs 10 and 16) that were salt stress‐responsive in protoplast transfection assays. Since 4 copies of motif 16 resulted in high GFP induction in transfected protoplast, we selected 20‐base motif 16 (SS16, synthetic promoter from salt stress‐inducible promoter 16) to make the respective synthetic promoter. SS16 has not been reported in the literature as a CRE (Table 1). The SS16 motif was predicted from information in 34 expressed gene promoters (Figure 3). For our application, we selected two genes, Potri.002G039300 and Potri.006G148800, annotated as basic helix–loop–helix family protein and ethylene‐responsive transcription factor genes in P. trichocarpa genome for further analysis (Table S5).
Figure 3.
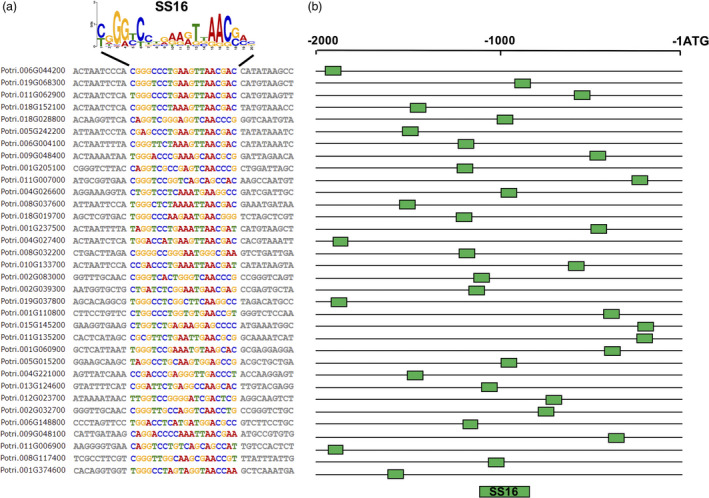
Bioinformatic determination of the cis‐regulatory SS16 motif that is putatively responsible for salt‐inducibility in native poplar promoters. (a) Alignment of promoter regions among poplar genes that contain the conserved SS motif 16 (SS16). Published root RNA‐seq data from salt‐treated poplar were analysed. A conserved 20‐base motif was found in 34 expressed genes by MEME. (b) Variable relative location of the SS16 motif on the −2 kb promoter sequences of 34 co‐expressed genes.
Osmotic stress‐related responses of native poplar promoters
To verify that the 16 water‐deficit‐inducible promoters containing SD motifs and 34 salt stress‐inducible promoters containing SS16 motif were stress‐inducible, we performed quantitative reverse transcriptase–PCR (qRT‐PCR) assays to measure gene expression of randomly selected downstream genes in stress‐treated poplar. The gene expression was assessed in wilted 717‐1B4 leaves by water cessation for 10 days or salt stress by watering 200 mM NaCl for 3 days. Neither stress treatment resulted in mortality nor discernable plant damage (Figure 4a). The expression of four water‐deficit‐induced genes was up‐regulated significantly over 1.2‐fold (log2) higher in water‐stressed plant leaves as compared to those grown under normal conditions (Figure 4b). Interestingly, all of these genes, except Potri.017G015700, were also highly expressed in leaves from salt‐treated plants. Six randomly selected salt stress‐responsive genes from 34 genes containing SS16 motif in their promoter were induced in the range of 1.5‐ to 4.3‐fold change (log2) by the salt treatment; the expression of Potri.018G019700 increased 4.3‐fold (log2) in salt treatment and was similar in water‐deficit stress (Figure 4b). Potri.002G029300 had a 2.1‐fold (log2) increase under salt treatment, whereas its expression was reduced under water cessation (Figure 4b). Taken together, the expression of each gene was specifically induced during water‐deficit stress and salt stress, respectively. Four genes responded to both water‐deficit and salt stress.
Figure 4.
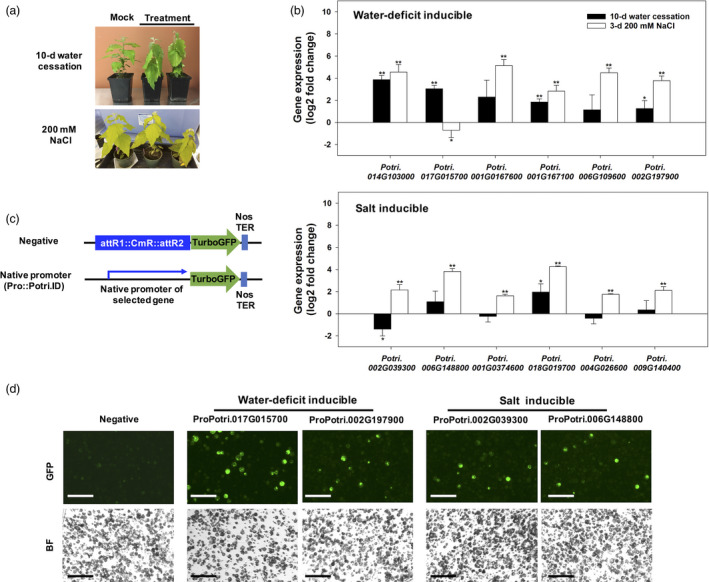
Stress induction of native poplar promoters. (a) Three‐month‐old poplar 717‐1B4 before and after 10‐d water cessation and 200 mM NaCl treatments. (b) Stressed poplar plant leaves were sampled to yield qRT‐PCR relative expression data. The candidate genes were randomly selected among 16 genes whose promoters possessed the conserved A domain from water‐deficit stress data and 34 genes whose promoters included the SS16 motif in salt stress RNA‐seq data (Figures 2c and 3a). PtUBCc was used as the internal reference gene. Values are mean ± standard deviation (SD, n = 3). Asterisks denote significant differences of gene expression between stress‐treated and untreated samples via t‐tests (*P < 0.05, **P < 0.01). (c) Gene constructs of native promoters fused with TurboGFP for poplar mesophyll protoplast transfection screens. Approximately 1 kb upstream of the putative ATG start codon was used to represent each native stress‐induced gene. The negative control was a promoterless GFP. (d) Promoter screens from (c) in poplar mesophyll protoplasts. Each plasmid was PEG‐transfected into protoplasts in control buffer or that containing 0.5 M mannitol or 100 mM NaCl. GFP fluorescent (GFP) and bright filed (BF) images of protoplasts are shown (bar = 100 µM).
Poplar mesophyll protoplast transfection was used throughout the research as a first‐tier test of promoter activity in poplar. We fused native promoters of four of the representative genes mentioned above (water stress‐induced Potri.017G015700 and Potri.002G197900; salt stress‐induced Potri.002G039300 and Potri.006G148800) to GFP to test their inducibility in poplar leaf protoplasts (Figure 4c and Figure S2). We cloned promoters of significantly highest and lowest expressed gene under water cessation from the qRT‐PCR assay (Figure 4b). Whereas Potri.014G103000 appeared to have higher differential expression to drought and salt treatments (Figure 4b), the native promoter region in 717 1B4 poplar could not be PCR‐amplified. Thus, we replaced it with the Potri.017G015700 promoter, which was the highest responsive promoter. For the salt‐inducible native promoter assay, we included the promoters of Potri.002G039300 and Potri.006G148800, which were annotated as transcription factors (Figure 4b). All four native promoters drove GFP expression in poplar mesophyll protoplast under 500 mM mannitol or 100 mM NaCl treatments (Figure 4d). These results provided sufficient evidence to further explore the use of the SD and SS motifs for designing functional synthetic sequences in promoters to enable a strong gene induction in response to osmotic stress.
Synthetic promoter architecture and osmotic stress responses in poplar leaf mesophyll protoplasts
We developed a vector system that used two fluorescent protein genes for quantitative assessment of specific promoter activity in plant cells. Each stress‐inducible synthetic promoter was fused to TurboGFP as a reporter gene. A constitutively expressed 35S::TurboRFP internal control cassette was included in each construct in order to monitor transformation efficiency and relative expression strength (Figures 5a and 6a). A wide range of GFP inducibility was observed in poplar leaf‐derived protoplasts among the synthetic promoter variants (Figures 5d and 6d).
Figure 5.
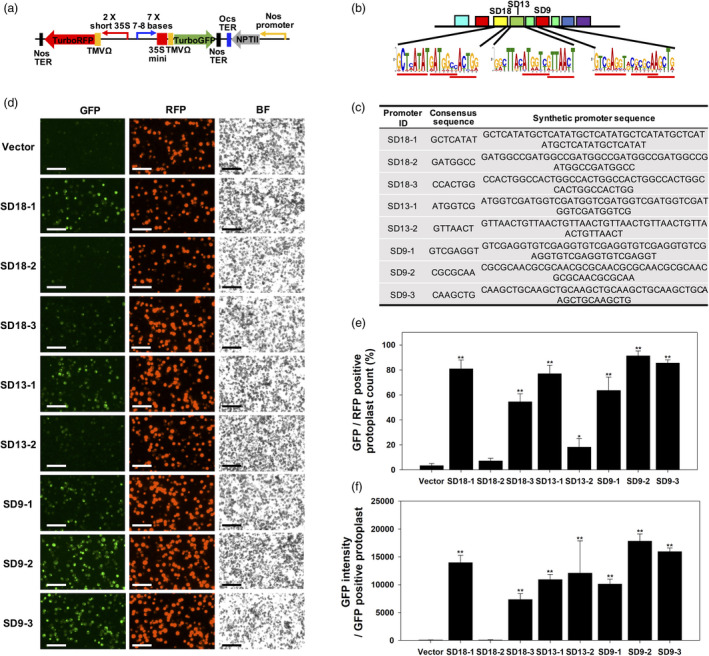
Stress induction analysis of synthetic promoters that were rationally designed from water stress‐inducible (SD) gene promoters under 0.5 M mannitol treatment in transfected poplar mesophyll protoplasts. (a) Binary vectors contained a series synthetic promoters (SD series) fused to GFP. The minimal CaMV 35S DNA core (−46 bp) with the TMV Ω 5’ UTR leader sequence was used for transcriptional and translational initiation. Vectors also contained a 35S::RFP reporter cassette that served as an internal positive control for constitutive expression to monitor transformation efficiency and ratio analysis for induced GFP expression. The synthetic promoters were composed of either 7 or 8 base pairs (SD motifs 18, 13 and 9) repeated 7 times. (b) The consensus location of motif 18, 13 and 9 in native promoters of candidate water stress‐induced poplar genes. The locations of core sequences for synthetic promoters are underlined. (c) The heptamerized DNA sequences of eight designed synthetic promoters from motif 18, 13 and 9. The most conserved sequences were selected for synthetic promoter design. (d) Representative GFP and RFP fluorescence images in transformed protoplasts (bar = 100 µm). Fluorescence microscopic images of synthetic promoter‐induced GFP and constitutively expressed RFP were taken at 2 day after protoplast transformation. (e) Percentage of GFP positive in RFP‐positive protoplast. Highly significant induced GFP fluorescence was observed in 6 of the synthetic promoters. (f) GFP intensity in GFP‐positive protoplast. All values in panels e and f are average of fluorescence intensity from three images of three different protoplast transformation experiments (n = 9). Error bars represent the SD Asterisks indicate significant differences of each value against fluorescence in vector‐transfected protoplast determined by t‐tests (*P < 0.05, **P < 0.01).
Figure 6.
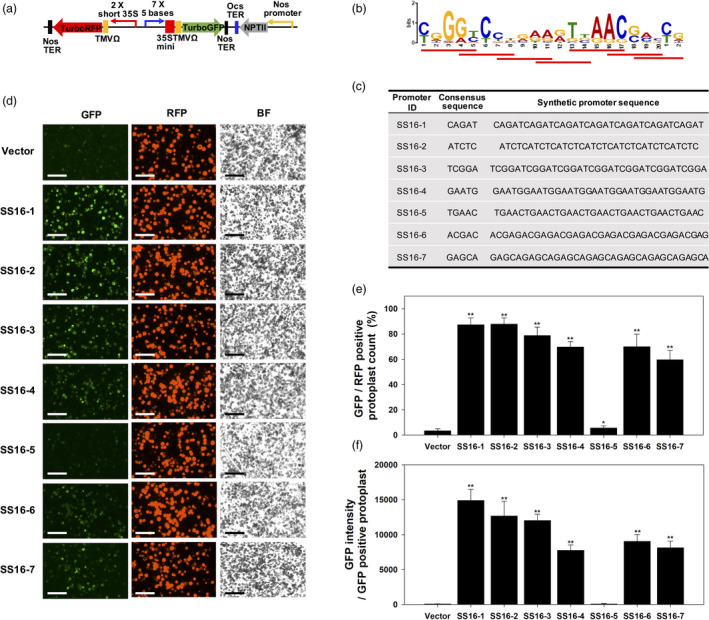
Stress induction analysis of synthetic promoters that were rationally designed from salt stress‐inducible (SS) gene promoters under 100 mM NaCl treatment in transfected poplar mesophyll protoplasts. (a) Binary vectors contained a series of synthetic promoters (SS‐motif series) fused to GFP as shown in SD synthetic promoter construct. (b) LOGO image of the SS16 motif. The synthetic promoters were composed of 5 base pairs of various subsets with the SS16 motif (red underlined). (c) The full sequences of heptamerized SS16 motifs. The 5 based core sequences were from the fixed sequence of SS16 motif on the promoter of Potri.002G039300. (d) Representative GFP and RFP fluorescence image in transformed protoplasts (bar = 100 µm). (e) Percentage of GFP positive in RFP‐positive protoplast. Induced GFP fluorescence was observed in 6 of the synthetic promoters. (f) GFP intensity in GFP‐positive protoplast. The value of panel e and f was calculated from three different images of three independent protoplast transformation (n = 9). The bar shows average, and error bar displays SD Asterisks indicate significant differences of synthetic promoter against vector transformation data determined by t‐tests (*P < 0.05, **P < 0.01).
Previous results reported that several copies of a length of 4 to 8 DNA sequence could induce reporter gene expression (Liu et al., 2014). Reflecting on this study, we selected repeated formats of 7‐8 and 5 bases to design synthetic promoter for water‐deficit and salt induction, respectively. Since the tandem version of synthetic promoter enhanced inducible gene expression, we optimized copy number of a core sequence by comparing GFP expression driven by a range of repeats from trimer to heptamer in transfected poplar protoplast, a logic suggested by Sharon et al. (2012). Increasing of the copy number of a core sequence led to increased GFP expression (Figure S3). Therefore, we employed the promoter format with heptamerized 7‐8 base or 5 base for SD or SS motif inducibility test, respectively.
Eight different heptamerized promoters of conserved 7–8 bases were synthetized from SD18, SD13 and SD9 motifs whose E‐values were higher than other predicted motifs (Figure 2a). Among the discovered sequences in SD18 and SD9 motifs were auxin and calmodulin‐binding elements (Table 1). In contrast, SD13 did not harbour any previously reported DNA elements. Based on the motif number designations, we denoted the derived synthetic promoters as SD18‐1, SD18‐2, SD18‐3, SD13‐1, SD13‐2, SD9‐1, SD9‐2 and SD9‐3 (Figure 5b,c). Protoplast transfection frequency ranged from 52.9% to 59.3%, which was calculated from scoring constitutively expressed RFP fluorescence in protoplasts (Figure 5d and Figure S4; black bars). GFP was induced in the range of 2.0%–46.6% of protoplasts, depending on the synthetic promoter; promoters exhibited various levels of induction (Figure 5d and Figure S4a; white bars). SD18‐1, SD13‐1, SD9‐2, and SD9‐3 exhibited the highest induction in the mannitol treatment (Figure 5e), whereas lower induction was observed in SD18‐3 and SD9‐1‐transfected cells. SD18‐2 and SD13‐2 induction was extremely low in the mannitol treatment. Transfection frequency was not different among promoter constructs tested (Figure S4a). Image analysis was used to gauge GFP intensity and protoplast counts, which appeared to positively correspond to one another, with the exception of SD13‐2 (Figure 5f).
For the purpose of discovering the minimal submotifs responsible for salt stress induction, we produced a permutational library using the SS16 motif on Potri. 002G039300’s promoter. The library contained heptamerized 5‐base‐long fragments (referred to as SS16‐1 through SS16‐7) (Figure 6a–c). Transfection frequency ranged from 52.3%–58.8% among the synthetic promoter constructs (Figure 6d and Figure S4b). Three synthetic promoters, SS16‐1, SS16‐2 and SS16‐3, were highly induced under salt treatments, whereas SS16‐5 displayed lower relative induction (Figure 6d,e). SS16‐4, SS16‐6 and SS16‐7 had a moderate level of salt induction (Figure 6d,e). As above, GFP intensity appeared to positively correspond to GFP protoplast frequency count (Figure 6f). Synthetic promoters SS16‐1, SS16‐2 and SS16‐3 were selected for further experimentation in other plant expression systems, which included agroinfiltrated Nicotiana benthamiana leaves and stably transformed Arabidopsis.
In summary, 12 of the 15 synthetic promoters we designed were stress‐inducible in transfected poplar leaf protoplasts (>50% of GFP/RFP‐positive protoplast count), 9 of which were deemed to have relatively high induction: six SD‐ and three SS‐based synthetic promoters.
Native and synthetic promoter inducibility under osmotic stress in transient agroinfiltrated Nicotiana benthamiana leaves
From the protoplast screens, six SD synthetic promoters and three SS synthetic promoters were selected for leaf agroinfiltration assays (Figures 5 and 6). In addition, one native water‐deficit‐inducible promoter of Potri.017G015700 and one native salt‐inducible promoter of Potri.002G039300 (Figure 4d) were included in agroinfiltration assays to compare the induction driven by native and synthetic promoters. Native and synthetic promoter constructs shared the same vector architecture (Figure 7a). Preliminary research (data not shown) was conducted to determine appropriate water‐deficit and salt stress treatments for N. benthamiana experiments. Treatments included 200 mM NaCl, 100 mM mannitol, and watering cessation for testing promoter activities (Figure 7b). These treatments were not lethal to N. benthamiana plants although leaf wilting was observed in water‐deficit conditions (Figure 7c). N. benthamiana leaves were undamaged in salt or mannitol treatment.
Figure 7.
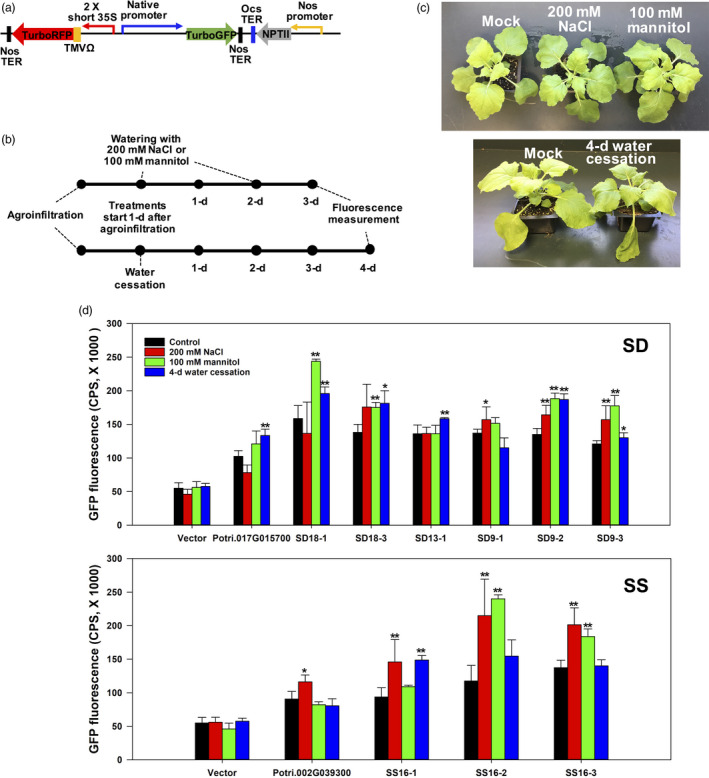
Transient expression analysis by leaf agroinfiltration of SD and SS synthetic promoter constructs in Nicotiana benthamiana leaves. (a) Binary vectors contained native promoters fused to GFP. Native promoters were represented by ca. 1 kb of the native promoter region of the water stress‐inducible Potri.017G015700 or the salt stress‐inducible Potri.002G039300. Vectors also contained a 35S::RFP reporter cassette that served as an internal positive control for constitutive expression to use for normalization of GFP fluorescence. (b) The experimental setup of stress treatments and fluorescence measurements on agroinfiltrated leaves. (c) A representation of plants from the agroinfiltration experiment. Mock and 100 mM mannitol or 200 mM NaCl‐treated plants were not damaged by 3‐day watering while halting watering resulted in wilt after 4 days. (d) GFP fluorescence in agroinfiltrated leaf under mock treatment, 200 mM NaCl, 100 mM mannitol and 4 days water cessation. GFP fluorescence and RFP fluorescence were measured at emission wavelength of 502 nm and 574 nm under a fixed excitation wavelength of 482 nm and 553 nm for GFP and RFP, respectively. The mock treatment (control) consisted of watering every 2 days. Bars represent count per second (CPS) value of normalized GFP fluorescence under stress versus and mock control. Values are the mean ± SD of three independent transformations (n = 3); asterisks denote significant differences via t‐tests: * P < 0.05, ** P < 0.01.
The 1 kb native water stress‐inducible (Potri.017G015700) promoter drove higher GFP expression than mock controls in the mannitol and water‐deficit treatments, respectively (Figure 7d), but was not induced in the salt treatment. Several synthetic promoters were highly induced under mannitol treatment, as measured by the increase in GFP fluorescence: SD18‐1, SD18‐3, SD9‐2 and SD9‐3 (Figure 7d). After 4 days from the last irrigation, we observed increased GFP fluorescence in leaves transformed with the following synthetic promoters: SD18‐1, SD18‐3, SD13‐1, SD9‐2 and SD9‐3 (Figure 7d). Synthetic promoters SD18‐1, SD18‐3, SD13‐1 and SD 9‐2 all controlled significantly higher induction than the native Potri.017G015700 promoter in the water‐deficit stress treatments (Figure 7d).
There was a variable response of the synthetic promoters designed to be salt stress‐inducible treatments. All three synthetic SS motif‐containing promoters (SS16‐1, SS16‐2 and SS16‐3) were induced in the salt treatments (Figure 7d). SS16‐2 and SS16‐3 were significantly induced by salt and mannitol (Figure 7d), while the native poplar promoter Potri.002G039300 was not induced in any treatments in the agroinfiltration experiment.
Stably transformed Arabidopsis as a synthetic promoter chassis
Five synthetic promoter constructs (SD18‐1, SD9‐2, SS16‐1, SS16‐2 and SS16‐3) were selected for stable transformation in Arabidopsis on the basis of the agroinfiltration experiments. T2 transgenic lines appeared to have no morphological differences in stress treatments (100 mM NaCl in tissue culture media over 10‐d water cessation) (Figure 8a). Each of the SS synthetic promoters was significantly induced under salt treatment with up to 279.8 and 345.0% increase in GFP gene expression driven by SS16‐1 and SS16‐3 promoters, respectively, whereas GFP was not induced in SD18‐1 and SD9‐2 (Figure 8b). Since synthetic promoters SD18‐1 and SD9‐2 responded to water‐deficit stress treatments in transiently transformed N. benthamiana leaves, transgenic plants expressing GFP under the control of SD18‐1 and SD9‐2 were tested under water‐deficit stress. Watering of 4‐week‐old Arabidopsis plants grown in soil was stopped for 10 days, until the rosette leaves displayed signs of wilting (Figure 8a). GFP fluorescence increased in both SD18‐1 and SD9‐2 plants relative to controls, which was commensurate with increases in GFP transcript abundance in the same plants (Figure 8c,d). Water‐use efficiency (WUE), which was measured to validate the water‐deficit condition, indeed increased after 10‐d water cessation (Figure 8e).
Figure 8.
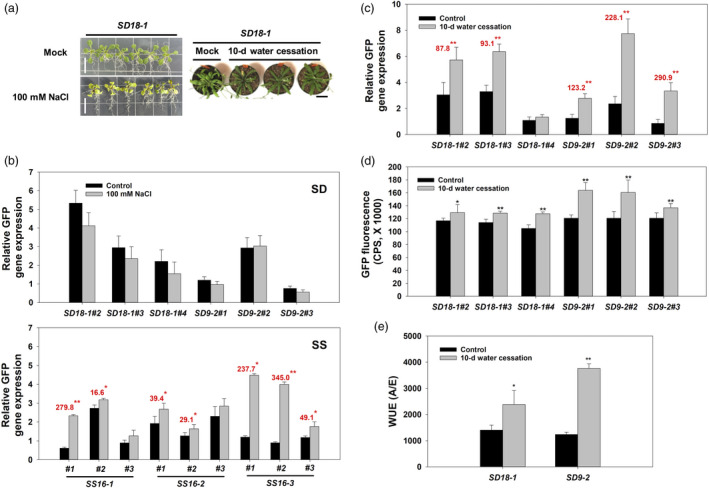
Stress induction analysis of SD and SS synthetic promoters in stable transgenic Arabidopsis. Stable transgenic Arabidopsis plants were generated via the floral dip method with binary vectors harbouring SD and SS synthetic promoters used in both protoplast transformation and agroinfiltration. (a) T2 SD18‐1 transgenic Arabidopsis plants grown on MS plates supplemented with 100 mM NaCl for 3 days, and in pots after watering was stopped for 10 days. No lethal effects were observed after stress treatment (bar = 1 cm). (b) GFP gene expression in transgenic Arabidopsis harbouring SD18‐1, SD9‐2, SS16‐1, SS16‐2 and SS16‐3 synthetic promoter constructs. Three T2 Arabidopsis seedlings (10‐day‐old) for each independent transgenic line were grown on each of three MS plates with 100 mM NaCl for 3 days. GFP gene expression in mock or treated samples by transgenic construct and event was calculated relative to the AtActin2 internal reference gene. The numerals in red font above each statistically significant bar denote reporter gene induction as a percentage over the control. (c) Relative GFP gene expression under the control of either SD18‐1 or SD9‐2 synthetic promoters in three independent T2 transgenic Arabidopsis lines under a 10‐day water stress (no watering). Transgenic control plants were watered every 2 days in the mock treatment (control). (d) GFP fluorescence in transgenic Arabidopsis harbouring either SD18‐1 or SD9‐2 promoter under water‐deficit stress. GFP fluorescence and RFP fluorescence were measured as described in Figure 7 at 10 days after water cessation, at which time GFP fluorescence between water stress and mock treatments was calculated. RFP values in mock and treated sample were used for GFP normalization. (e) WUE of SD18‐1 and SD9‐2 transgenic Arabidopsis 10 days after watering cessation. WUE was determined by calculating the ratio of carbon uptake (a) and the rate of gas exchange transpiration (e) measured as described in Methods section. In panels b, c and d, the bars represent means ± SD of 9 plants grown among three plates (n = 9). Each transgenic construct was represented by three independent transgenic T2 lines. qRT‐PCR data for each plant (single leaf) were based on three technical replicates. In panel e, one representative plant from each independent T2 line was measured to assess water stress (n = 3) for each construct. Statistical significance is shown via t‐tests between treated water cessation and mock (*P < 0.05, **P < 0.01).
Discussion
Synthetic biology will certainly revolutionize agriculture (Wurtzel et al., 2019). One of its important applications is in advanced biotechnology to improve crops and cropping systems (Goold et al., 2018). The most mature technologies in plant synthetic biology include gene editing, and synthetic promoters and transcription factors (Liu and Stewart, 2015; Liu and Stewart, 2016; Liu et al., 2013). Synthetic promoters have been employed to widely diversify the repertoire of tools for tuning gene expression (Hernandez‐Garcia and Finer, 2014; Kassaw et al., 2018). One of the significant challenges to design synthetic promoters is the identification of functional CREs (Roccaro et al., 2013). One end goal is to understand the regulation of plant gene expression, especially transcription regulation, which will enable parts (e.g. promoters and transcription factors) to be manufactured and installed in plant genomes to render high predictive functionality.
The combination of high‐throughput gene expression profiles and bioinformatic tools is a powerful approach to design synthetic promoters (Koschmann et al., 2012). Synthetic promoter design for predictable transcript inducibility largely rests on the interaction of induction conditions and critical CREs in native promoters. One route to discovery of CREs is via ‐omics data (e.g. Liu et al., 2014), which is now quite amenable in poplar. Poplar has a reference genome and deep transcriptomic data for abiotic stress traits. Indeed, co‐expressed gene data from microarray data have been used to construct inducible synthetic promoters (Koschmann et al., 2012; Liu et al., 2014), but only recently has sufficient RNA‐seq transcriptomic data been available for poplar, which we used exclusively in our study (Figures 2 and 3 and Figure S1).
Transient transfection of plant protoplasts has long been used to study gene expression (Sheen, 2001) and to screen synthetic promoter activity in a number of herbaceous plant species (Lehmeyer et al., 2016b; Roccaro et al., 2013; Schaumberg et al., 2016). Our study represents, to our knowledge, the first one using protoplasts for synthetic promoter screening in a woody plant species. The subsequent validation, through agroinfiltration assays on leaves of N. benthamiana and stable transgenic Arabidopsis, demonstrated orthologous responsiveness across eudicot species that are not closely related (Figures 5, 6, 7, 8).
A persistent challenge in synthetic promoter design has been false‐positive hits in de novo DNA motif discovery algorithms (Hu et al., 2005; Lihu and Holban, 2015; Tompa et al., 2005), and early studies achieved up to only 13% in sensitivity and 35% in precision (Hu et al., 2005). Although high‐quality sequence data are increasingly becoming available, motif discovery, synthetic promoter design and testing are still a hindrance to increase promoter precision (Lihu and Holban, 2015). One attempt to improve motif discovery has been explored through ensemble approaches, which combine different de novo motif‐finding tools, offering improved prediction over any single algorithm (Harbison et al., 2004; Hu et al., 2006; Liu et al., 2014; MacIsaac and Fraenkel, 2006). In the present study, we applied an ensemble approach to coordinate three motif‐finding tools coupled with high‐quality genome sequencing and transcriptomic data; 15 synthetic promoter candidates were built. Of these, 12 synthetic promoters appeared to be osmotic stress‐inducible in poplar protoplasts, 9 of which were confirmed to be inducible in agroinfiltration studies (Figure 7). We conclude that improved ‐omics data enabled more efficient and reliable CRE discovery, which led to better inducible synthetic promoter design. Nonetheless, we speculate that CRE selection is suboptimal by the current protocol that uses position weight matrix alignment of long DNA sequence based on co‐expressed gene clustering. Thus, improved bioinformatics algorithms may be needed to enable more predictive CRE selection. In addition, high‐throughput ChIP‐seq experiments using known stress‐responsive transcription factors may also be coupled with bioinformatics approaches to identify CREs.
Developing versatile synthetic promoters by constructing repeats of an appropriate short‐length motif(s) appears to be a powerful strategy with regard to abiotic stress induction. SD18‐1, SD18‐3, SD9‐2 and SD9‐3 were highly inducible under mannitol and water‐deficit stress treatments, whereas these promoters were weakly inducible to salt stress. On the other hand, the GFP expression driven by SS16‐1, SS16‐2 and SS16‐3, under salt stress was higher than the expression induced by mannitol and water‐deficit treatments. Our results were consistent with two previous studies showing a common response of synthetic promoters, consisting of DRE/C‐repeats (GCCGAC), to dehydration, high salinity, and cold stress in transgenic Arabidopsis (Hou et al., 2012; Kim et al., 2002). Therefore, repeated short‐length motifs in synthetic promoters appear to be a good strategy to endow promoter induction to abiotic stress. In contrast, native promoters tested in the present study were specifically induced either by water‐deficit or high salinity (Figure 7d), suggesting that different CREs may need to be synchronized with the designed synthetic promoters in order to acquire individual stress specificity. Furthermore, the present study was limited in scope. We focused on synthetic promoter activities in leaves only, while acknowledging the potential relevance in other tissues. Future research should combine research on additional tissues with synthetic promoters comprising tissue‐specific CREs for enhancing induced gene expression in space and time. Orthogonal regulatory systems expressing synthetic transcription factors to regulate synthetic promoters are a good strategy to further control gene expression in plants. A new plant orthogonal regulatory system was recently reported that leveraged a yeast transcriptional regulator to induce synthetic activators or repressors (Belcher et al., 2020). Such a system or a new synthetic Neurospora‐based 'Q‐system' (Persad et al., 2020) could be coupled with designed synthetic promoters to tune signal specificity and sensitivity in plants.
Synthetic promoter strength and specificity may be affected by combining multiple CRE sequences and altering their copy number and spacing (Liu and Stewart, 2016). To date, the basic gene construct design has largely included synthetic sequences upstream of a core promoter sequence such as the −46 35S promoter. Recently, a minimal synthetic promoter was designed for constitutive response by various arrangement of CREs at different positions to TATA box area (Cai et al., 2020). In our study, no downstream gene expression was observed in synthetic promoters including three copies of single CREs (Figure S3). However, 7–8 copies of the same short CRE induced GFP induction in our study. Therefore, optimizing copy number of CRE may be a pivotal factor to design useful synthetic promoters.
The present study used repeats of short motif without spacers between each motif. While our synthetic promoters were responsive to osmotic stress, we made no attempt to optimize spacing of repeats. Such optimization could be desirable. A correlation between the copy number of cis‐elements and productivity of synthetic promoters was observed in various plant species such as Arabidopsis (Sahoo et al., 2014), tobacco (Sawant et al., 2005) and rice (Wu et al., 1998), a pattern that was observed in our study. Abscisic acid and salicylic acid responsivity of two copies of ACGT was dependent on the spacing between the two copies, indicating that spacing is an important factor for synthetic promoter design (Mehrotra and Mehrotra, 2010). In addition to the spacing between multiple copies of motifs, the spacing between CREs and the core promoter is also important (Rushton, 2016); many unknowns remain about optimal spacing and repeats of CREs (Ali and Kim, 2019; Rushton, 2016).
In the past, well‐characterized CREs have been used to construct synthetic promoters inducible by abiotic stress. These cis‐elements include ABRE, DRE/C‐repeat and coupling factors derived from the cis‐regulatory region of wheat HVA21, Arabidopsis COR, RD29A and ERD1 constructed by classical promoter deletion assays (Shen et al., 1996; Simpson et al., 2003; Thomashow, 1999; Yamaguchi‐Shinozaki and Shinozaki, 1994). In one recent study, a 6 × ABRE CRE repeat was used to make a synthetic abscisic acid‐inducible promoter, which was tested in Arabidopsis (Wu et al., 2018). Although the present synthetic promoters were designed using motifs that are novel, from a prior characterization standpoint, their inducibility was similar to that of synthetic promoters using known promoter motifs, such as ABRE and DRE. Therefore, our present workflow is enabling to discover novel sequences with functional cis‐regulations to use in synthetic promoters.
Even though trees are subjected to various abiotic stresses throughout their lifetimes, there has been few studies that focus on abiotic stress‐inducible in poplar. Recently, five Populus trichocarpa C‐repeat binding factor (CBF)/DREB1 family proteins were found in 2263 annotated genes, but that study did not specifically address abiotic stress (Li et al., 2017). The promoter of Populus euphratica ascorbate peroxidase 2 (PeAPX2), a mediator of freezing tolerance through scavenging reactive oxygen species, is recognized by a cysteine‐2/histidine‐2 type zinc finger transcription factor (PeSTZ1) (He et al., 2019). That study points the way to a route for cold‐inducible synthetic promoter construction. Of interest is that our synthetic promoters SD18‐1, SD18‐3 and SD 9‐2 included auxin responsiveness (CATATG), ICE1‐binding (CANNTG) sequences, and the calmodulin‐binding sequence (ACGCGA), respectively. Therefore, other types of stress such as cold and freezing can be sensed by reconstructing expanded sequences into the present synthetic promoter construct architecture.
In our study, these synthetic promoters had relatively high induction under osmotic stress (Table 1, Figures 5, 6, 7, 8). The molecular/physiological function of SD and SS motifs as abiotic stress‐responsive CREs in poplar will be further characterized in upcoming field experiments.
Experimental procedures
Poplar plants
Female P. tremula × P. alba interspecific hybrid (717‐1B4) aspen clones were kindly donated by Dr. Steven Strauss (Oregon State University). Plants were grown on ProMix BK25 (Premier Tech, Quakertown, PA) in 4‐L pots in a greenhouse. Plants were watered once every 2 days. Poplar micropropagation via apical shoots from pot‐grown 2‐month‐old plants was conducted whereby apical shoots were sterilized by shaking them in 50% commercial bleach for 5 min followed by washing in 70% ethanol for another 5 min. Subsequently, cut shoots were washed five times with autoclaved water. Sterilized shoots were grown on rooting media (1 × MS salt with Gamborg's vitamin mixture (Caisson Labs, Smithfield, UT), 20 mM MES (Sigma, St. Louis, MO), 3% sucrose (Sigma), 0.5% activated charcoal (Sigma), 0.15% Gelrite (Sigma) and adjusted pH to 5.7) in Magenta GA7 boxes (Bioworld, Dublin, OH). Regenerated plants were propagated every 2 months.
Nicotiana and Arabidopsis plants
Nicotiana benthamiana and Arabidopsis thaliana (Col) were grown on ProMix BK25 soil in 1‐L pots after stratification at 4°C for 4 days in the sterile water without light. Plants were watered three times per week depending on conditions. All plants were grown under 16 h/8 h for light/dark cycles at constant 22°C with 150 μmol/m2s light intensity.
Bioinformatic analysis for selection of stress‐responsive motif
All promoter sequences for bioinformatic analysis were profiled from BioMart (Smedley et al., 2015) integrated in Phytozome v12.1.6 (www.phytozyme.org). Two kilobase upstream sequences flanking the ATG start codon were collected from P. trichocarpa v 3.0 genome annotation (Tuskan et al., 2006). We subjected these promoter sequences to command line application of MEME Suite (v5.0.5), MotifSampler (v3.2.2) and Weeder (v 2.0). DNA motifs up to 20 bases in length were predicted to design reliable core sequences for downstream experiments. To find cis‐elements over DNA motifs, we submitted them to the database of plant cis‐acting regulatory DNA elements (PLACE; https://www.dna.affrc.go.jp/PLACE/) with default analysis parameters.
Construction of fusion plasmids with fluorescent reporter genes
To screen inducible native or synthetic promoters against osmotic stress, we generated two different backbone genes harbouring GFP constructs with or without constitutive RFP expression gene constructs (Figures 4c, 5a, 6a and 7a). For native promoters used in protoplast transformation (Figure 4c), the backbone gene construct including TMV 5’ UTR (Ω) leader and TurboGFP was constructed with Gateway cloning sites via the Golden Gate modular cloning technique using the MoClo system (Engler et al., 2014; Figure 4c).
Through a BLAST search with the coding sequence of P. trichocarpa annotation of osmotic stress‐responsive genes to INRA 717‐1B4 genome assembly (Mader et al., 2016: http://urgi.versailles.inra.fr), about 1‐kb upstream promoter sequences from ATG were collected from the aspen genome. The native promoter was amplified from 717‐1B4 genomic DNA extracted following Kang’s method (Kang and Yang, 2004) using Phusion DNA polymerase with gene‐specific primers for Gateway cloning (Thermo Fisher Scientific, Waltham, MA, Table S7). The PCR was performed as described below; 1 cycle of 95°C for 2 min, 30 cycles repeating 95°C for 30 s, 57°C for 30 s and 72°C for 1 min. The insert was destined into the backbone construct by LR clonase (Invitrogen, Carlsbad, CA).
In the agroinfiltration test, native promoters fused with TurboGFP as described above were linked with TurboRFP driven by 2 × CaMV 35S promoter (Figure 7a). For synthetic promoter construct generation, a binary backbone plasmid constructing two different gene constructs was generated by the Golden Gate cloning technique (Figures 5a, 6a and 7a): i) a gene construct for synthetic promoter cloning site including BsaI restriction site, CaMV 35S minimal promoter (−46 bp) and TMV 5’ UTR (Ω) leader, fused with TurboGFP, and ii) a gene construct including 2 × CaMV 35S short promoter driving TurboRFP expression (Engler et al., 2014).
Synthetic promoter fragments were generated by reannealing two complementary single‐stranded oligonucleotides consisting of heptameric repeats of a DNA motif plus a 5ˊ overhangs of BsaI digestion sites on the backbone plasmid (Integrated DNA Technologies, Coralville, IA, Table S6). A pair of complementary strands was combined together and incubated at 95°C for 2 min followed by 65°C for 5 min, and then was cooled down gradually to 4°C using a thermal cycler. The annealed double strands were cloned into a BsaI‐digested final destination vector. All gene construct DNA sequences and ligation conditions were verified by Sanger sequencing analysis (UT Genomics Core, Knoxville, TN).
Poplar mesophyll protoplast transformation assay
Poplar mesophyll protoplasts were isolated from leaves of 1‐month‐old 717‐1B4 in vitro culture as described above. Cellulose enzyme treatment and recovery of extracted protoplasts were performed as described in Guo’s protocol (Guo et al., 2012). Ten micrograms of plasmid was transfected to 2 × 104 protoplasts. Since WI solution contains 0.5 M mannitol, osmotic condition was induced simultaneously with incubation for 2 days at room temperature. To find optimal NaCl concentration, we tested protoplast cell viability in 50 mM, 100 mM and 150 mM NaCl complemented with WI solution. Based on this result, the transformed protoplasts were incubated for 2 days at room temperature in 100 mM NaCl. The GFP and RFP fluorescent images were taken by EVOS M7000 (Thermo Fisher Scientific) and analysed by ImageJ software (Schneider et al., 2012). The fluorescent ratio was determined by counting fluorescent‐detected protoplast out of viable protoplasts in a bright field image. The relative fluorescence ratio was determined by calculating the number of GFP‐expressing protoplasts over the number of RFP‐expressing protoplasts.
Transient agroinfiltration assay in N. benthamiana leaves
The same plasmids for protoplast assay were transfected into Agrobacterium strain EHA105 by the liquid nitrogen transformation method (Höfgen and Willmitzer, 1988). For infiltration, Agrobacterium was cultured in LB medium containing 50 mg/L rifampicin and 50 mg/L kanamycin overnight shaking at 28°C. Resuspended cell culture was infiltrated into the abaxial side of the third and fourth leaves by pushing the syringe as previously described (Ma et al., 2012).
The GFP fluorescence and RFP fluorescence were measured by Fluorolog®‐3 (HORIBA, Kyoto, Japan) on the abaxial side of the agroinfiltrated leaf at the third day and the fourth day after salt or mannitol treatment or water cessation, respectively (Figure 7b). The fluorescence was measured at the emission wavelength of 502 and 574 nm under fixed excitation at 482 and 502 nm for TurboGFP and TurboRFP, respectively. GFP fluorescence increase was determined by calculating increase ratio of GFP fluorescence in stress‐treated leaves over that in mock. GFP intensity was normalized by RFP fluorescence ratio between stress‐treated and mock‐treated cells.
Stable transgenic Arabidopsis generation
To generate stable transgenic Arabidopsis, we used the floral dip technique on 1‐month‐old Arabidopsis plants (Clough and Bent, 1998). The same EHA105 Agrobacterium lines described above were transformed into Arabidopsis. Transgenic Arabidopsis plants were screened on 1/2 × MS plates including Gamborg’s vitamins (Caisson), 1% sucrose and 0.8% Phytoagar (Sigma) complementing 50 µg/ml kanamycin. For genotyping, genomic DNA was extracted from leaves of 3‐week‐old T2 generation Arabidopsis seedlings among multiple independent transgenic lines per construct (Kang and Yang, 2004). Two‐hundred and fifty nanograms of genomic DNA was subjected to DreamTaq Green PCR Master Mix (Thermo Scientific) including 200 nM of specific forward and reverse primers of GFP (Table S7). PCR was performed as 1 cycle of 95°C for 5 min, 30 cycles repeating a chain of 95°C for 30 sec, 60°C for 30 s, and 72°C for 30 s, and final extension at 72°C for 10 min. We selected three independent transgenic lines from each synthetic promoter (Figure S5).
Stress treatment for the osmotic inducibility experiment
To optimize osmotic stress conditions in present studies, 717‐1B4, N. benthamiana and Arabidopsis were first exposed various concentrations of mannitol or NaCl, or halting watering up to 10 days. We selected the chemical concentration or duration of water stress to detect enough reporter gene expression without severe damage in stress‐treated plants.
To verify endogenous poplar gene expression under water stress and high salinity (Figure 4b), we transplanted 717‐1B4 after 2 months of regeneration in Magenta GA7 boxes to 4‐L pots of potting mix. These plants were grown another 1 month with watering every 2 days. For drought condition, we ceased watering until leaves began to wilt (around 10 days after water cessation). The salt treatment consisted of watering plants with 200 mM NaCl solution every 3 days. Control poplar was watered with tap water.
In the agroinfiltration assay in N. benthamiana, the infiltrated plants were stressed by watering either with 200 mM NaCl or with 100 mM mannitol (Figure 7a). We ceased watering for 4 days after agroinfiltration until the leaf began to wilt.
To quantify GFP gene expression in transgenic Arabidopsis under osmotic stress (Figure 8), 10‐day‐old T2 seedlings from three different lines per each synthetic promoter were screened first on 1/2 × MS plates with 50 μg/ml kanamycin. The screened Arabidopsis plants were transferred to 1/2 × MS plates with 100 mM NaCl for 3 days. For water cessation treatment, screened T2 plants were transplanted and grown in pots watered every 2 days. After 4 weeks, watering was stopped for 10 days.
To determine water‐deficit stress on leaf surface, we measured CO2 uptake (A) and transpiration (E) by LI‐6800 portable photosynthesis system (LI‐COR Biosciences, Lincoln, NE). WUE (A/E) was used as a marker to assess drought stress (Yang et al., 2019).
Mock plants transformed with the same plasmids by the same process were watered every 2 days with deionized water.
qRT‐PCR
The qRT‐PCR process is summarized in Figure S6. Total RNA was extracted via Plant RNA Extraction reagent following the manufacturer’s manual (Invitrogen, Carlsbad, CA). One microgram of purified total RNA was used to perform the reverse transcription reaction with RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA). The five‐time diluted single‐strand cDNA was subjected into 20 μL of total volume with 1 × PowerUp SYBR Green Master Mix including gene‐specific primers as listed in Table S7 (Applied Biosystems, Foster City, CA). The qPCR profile was 50°C for 2 min, 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 30 s. The relative gene expression change was determined by the 2–ΔΔ Ct equation (Livak and Schmittgen, 2001).
Statistical analysis
Technical and biological repeat numbers are described in individual figure legends. Statistical analyses of all measurements were performed by Student's t‐test integrated in R (R core team, 2014).
Author contributions
YY and JHL equally contributed to designing and performing the experiments, analysing the data and writing the manuscript. MRP and YS assisted with the experiments and plant care. WL and SCL consulted to find DNA motif and construct working plasmid. AA, EB and CNS conceived of the study and its design and coordination, and assisted with interpretation of results and revisions to the manuscript. All authors read and approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Supporting information
Figure S1 The subset of published poplar RNA‐seq results of independent experiments used for the basis of promoter discovery in this study.
Figure S2 Cloned regions of four selected native promoters showing domains of interest.
Figure S3 Effect of copy number repeats of core sequences used in synthetic promoter constructs.
Figure S4 The ratio of GFP‐ and RFP‐positive transfected protoplasts in each respective experimental population.
Figure S5 PCR analysis of T2 transgenic Arabidopsis.
Figure S6 Scheme of qRT‐PCR process.
Table S1 Selected up‐regulated genes from four different RNA‐seq datasets: from leaves taken from drought‐treated poplar.
Table S2 Gene lists from two different RNA‐seq datasets from roots of salt‐stressed poplar.
Table S3 The predicted motifs in water‐stressed RNA‐seq promoter data (SD motifs) by de novo motif‐detecting software.
Table S4 The predicted motifs in salt‐stressed RNA‐seq promoter data (SS motifs) by de novo motif‐detecting software.
Table S5 Selected expressed genes from six different RNA‐seq datasets and included SD in A domain or SS motif on their promoters.
Table S6 DNA sequence for generating synthetic promoter fragment to construct binary plasmid using the reannealing method.
Table S7 List of qRT‐PCR and genomic DNA PCR primers.
Acknowledgements
We appreciate Cristiano Piasecki’s expertise and aid in the portable photosynthesis experiments. We also thank Reginald J. Millwood, Francisco Palacios and Tyler Newton for their assistance in plant care. This work was supported by funding from the Biological and Environmental Research in the U.S. Department of Energy Office of Science (DE‐SC0018347).
Yang, Y. , Lee, J. H. , Poindexter, M. R. , Shao, Y. , Liu, W. , Lenaghan, S. C. , Ahkami, A. H. , Blumwald, E. and Stewart, C. N. Jr (2021) Rational design and testing of abiotic stress‐inducible synthetic promoters from poplar cis‐regulatory elements. Plant Biotechnol. J., 10.1111/pbi.13550
References
- Ali, S. and Kim, W.‐C. (2019) A fruitful decade of using synthetic promoters in the improvement of transgenic plants. Front Plant Sci. 10, 1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, T.L. , Johnson, J. , Grant, C.E. and Noble, W.S. (2015) The MEME Suite. Nucleic Acids Res. 43, W39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcher, M.S. , Vuu, K.M. , Zhou, A. , Mansoori, N. , Ramos, A.A. , Thompson, M.G. , Scheller, H.V. et al.(2020) Design of orthogonal regulatory systems for modulating gene expression in plants. Nat. Chem. Biol. 16, 857–865. [DOI] [PubMed] [Google Scholar]
- Cai, Y. , Kallam, K. , Tidd, H. , Gendarini, G. , Salzman, A. and Patron, N.J. (2020) Rational design of minimal synthetic promoters for plants. Nucleic Acids Res. 48, 11845–11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J. and Bent, A.F. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cossu, R.M. , Giordani, T. , Cavallini, A. and Natali, L. (2014) High‐throughput analysis of transcriptome variation during water deficit in a poplar hybrid: a general overview. Tree Genet. Genomes, 10, 53–66. [Google Scholar]
- Das, M. , Chauhan, H. , Chhibbar, A. , Haq, Q.M.R. and Khurana, P. (2011) High‐efficiency transformation and selective tolerance against biotic and abiotic stress in mulberry, Morus indica cv. K2, by constitutive and inducible expression of tobacco osmotin. Transgenic. Res. 20, 231–246. [DOI] [PubMed] [Google Scholar]
- Datta, K. , Baisakh, N. , Ganguly, M. , Krishnan, S. , Yamaguchi Shinozaki, K. and Datta, S.K. (2012) Overexpression of Arabidopsis and Rice stress genes’ inducible transcription factor confers drought and salinity tolerance to rice. Plant Biotechnol. J. 10, 579–586. [DOI] [PubMed] [Google Scholar]
- Dey, N. , Sarkar, S. , Acharya, S. and Maiti, I.B. (2015) Synthetic promoters in planta. Planta, 242, 1077–1094. [DOI] [PubMed] [Google Scholar]
- Engler, C. , Youles, M. , Gruetzner, R. , Ehnert, T.M. , Werner, S. , Jones, J.D. , Patron, N.J. and et al. (2014) A golden gate modular cloning toolbox for plants. ACS Synth. Biol. 3, 839–843. [DOI] [PubMed] [Google Scholar]
- Filichkin, S.A. , Hamilton, M. , Dharmawardhana, P.D. , Singh, S.K. , Sullivan, C. , Ben‐Hur, A. , Reddy, A.S.N. and et al. (2018) Abiotic stresses modulate landscape of poplar transcriptome via alternative splicing, differential intron retention, and isoform ratio switching. Front. Plant Sci. 9, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold, H.D. , Wright, P. and Hailstones, D. (2018) Emerging opportunities for synthetic biology in agriculture. Genes, 9, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J. , Morrell‐Falvey, J.L. , Labbé, J.L. , Muchero, W. , Kalluri, U.C. , Tuskan, G.A. and Chen, J. (2012) Highly efficient isolation of Populus mesophyll protoplasts and its application in transient expression assays. PLoS One 7, e44908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin, C. (2005) Gene stacking in transgenic plants‐the challenge for 21st century plant biotechnology. Plant Biotechnol. J. 3, 141–155. [DOI] [PubMed] [Google Scholar]
- Han, K.H. , Meilan, R. , Ma, C. and Strauss, S.H. (2000) An Agrobacterium tumefaciens transformation protocol effective on a variety of cottonwood hybrids (genus Populus). Plant Cell Rep 19, 315–320. [DOI] [PubMed] [Google Scholar]
- Harbison, C.T. , Gordon, D.B. , Lee, T.I. , Rinaldi, N.J. , Macisaac, K.D. , Danford, T.W. , Hannett, N.M. et al. (2004) Transcriptional regulatory code of a eukaryotic genome. Nature, 431, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F. , Li, H. , Wang, J. , Wang, H. , Feng, C. , Yang, Y. , Niu, M. et al. (2019) PeSTZ1, a C2H2‐type zinc finger transcription factor from Populus euphratica, enhances freezing tolerance through modulation of ROS scavenging by directly regulating PeAPX2 . Plant Biotechol. J. 17, 2169–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez‐Garcia, C.M. and Finer, J.J. (2014) Identification and validation of promoters and cis‐acting regulatory elements. Plant Sci. 217–218, 109–119. [DOI] [PubMed] [Google Scholar]
- Higo, K. , Ugawa, Y. , Iwamoto, M. and Korenaga, T. (1999) Plant cis‐acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen, R. and Willmitzer, L. (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acid Res. 16, 9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, L. , Chen, L. , Wang, J. , Xu, D. , Dai, L. , Zhang, H. and Zhao, Y. (2012) Construction of stress responsive synthetic promoters and analysis of their activity in transgenic Arabidopsis thaliana . Plant Mol. Biol. Rep. 30, 1496–1506. [Google Scholar]
- Hu, J. , Li, B. and Kihara, D. (2005) Limitations and potentials of current motif discovery algorithms. Nucleic Acids Res. 33, 4899–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J. , Yang, Y.D. and Kihara, D. (2006) EMD: an ensemble algorithm for discovering regulatory motifs in DNA sequences. BMC Bioinformatics, 7, 342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, J. , Zhou, J. , Shi, W. , Cao, X. , Luo, J. , Polle, A. and Luo, Z.B. (2017) Comparative transcriptomic analysis reveals the roles of overlapping heat‐/drought‐responsive genes in poplars exposed to high temperature and drought. Sci. Rep. 7, 43215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, T.J. and Yang, M.S. (2004) Rapid and reliable extraction of genomic DNA from various wild‐type and transgenic plants. BMC Biotechnol. 4, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassaw, T.K. , Donayre‐Torres, A.J. , Antunes, M.S. , Morey, K.J. and Medford, J.I. (2018) Engineering synthetic regulatory circuits in plants. Plant Sci. 273, 13–22. [DOI] [PubMed] [Google Scholar]
- Kim, H.J. , Kim, Y.K. , Park, J.Y. and Kim, J. (2002) Light signalling mediated by phytochrome plays an important role in cold‐induced gene expression through the C‐repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana . Plant J. 29, 693–704. [DOI] [PubMed] [Google Scholar]
- Koschmann, J. , Machens, F. , Becker, M. , Niemeyer, J. , Schulze, J. , Bulow, L. , Stahl, D.J. and et al. (2012) Integration of bioinformatics and synthetic promoters leads to the discovery of novel elicitor‐responsive cis‐regulatory sequences in Arabidopsis . Plant Physiol. 160, 178–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.H. and Pijut, P.M. (2018) Optimization of Agrobacterium‐mediated genetic transformation of Fraxinus nigra and development of black ash for possible emerald ash borer resistance. Plant Cell Tiss Org. 134, 217–229. [Google Scholar]
- Lehmeyer, M. , Kanofsky, K. , Hanko, E.K. , Ahrendt, S. , Wehrs, M. , Machens, F. and Hehl, R. (2016b) Functional dissection of a strong and specific microbe‐associated molecular pattern‐responsive synthetic promoter. Plant Biotechnol. J. 14, 61–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Sima, W. , Ouyang, B. , Wang, T. , Ziaf, K. , Luo, Z. , Liu, L. et al. (2012) Tomato SlDREB gene restricts leaf expansion and internode elongation by downregulating key genes for gibberellin biosynthesis. J. Exp. Bot. 63, 6407–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Song, Y. , Xu, B. , Xie, J. , Zhang, D. and Cooke, J. (2017) Poplar CBF1 functions specifically in an integrated cold regulatory network. Tree Physiol. 37, 98–115. [DOI] [PubMed] [Google Scholar]
- Li, Xiong , Yang, Yunqiang , Sun, Xudong , Lin, Huaming , Chen, Jinhui , Ren, Jian , Hu, Xiangyang and et al. (2014) Comparative physiological and proteomic analyses of poplar (Populus yunnanensis) plantlets exposed to high temperature and drought. PLoS One, 9, e107605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lihu, A. and Holban, S. (2015) A review of ensemble methods for de novo motif discovery in ChIP‐Seq data. Brief Bioinform. 16, 964–973. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Mazarei, M. , Peng, Y. , Fethe, M.H. , Rudis, M.R. , Lin, J. , Millwood, R.J. et al. (2014) Computational discovery of soybean promoter cis‐regulatory elements for the construction of soybean cyst nematode‐inducible synthetic promoters. Plant Biotechnol. J. 12, 1015–1026. [DOI] [PubMed] [Google Scholar]
- Liu, W. and Stewart, C.N. Jr . (2015) Plant synthetic biology. Trends Plant Sci. 20, 309–317. [DOI] [PubMed] [Google Scholar]
- Liu, W. and Stewart, C.N. Jr . (2016) Plant synthetic promoters and transcription factors. Curr. Opin. Biotechnol. 37, 36–44. [DOI] [PubMed] [Google Scholar]
- Liu, W. , Yuan, J.S. and Stewart, C.N. Jr . (2013) Advanced genetic tools for plant biotechnology. Nat. Rev. Genet. 14, 781–793. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐delta delta C(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Ma, L. , Lukasik, E. , Gawehns, F. and Takken, F.L.W. (2012) The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. In Plant Fungal Pathogens Methods in Molecular Biology (Methods and Protocols), 835. ( Bolton, M. and Thomma, B. , eds), pp. 74–76. New York: Humana Press. [DOI] [PubMed] [Google Scholar]
- MacIsaac, K.D. and Fraenkel, E. (2006) Practical strategies for discovering regulatory DNA sequence motifs. PLoS Comput. Biol. 2, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader, M. , Le Paslier, M.C. , Bounon, R. , Berard, A. , Faivre Rampant, P. , Fladung, M. , Leple, J.C. and et al. (2016) Whole‐genome draft assembly of Populus tremula x Populus alba clone INRA 717–1B4. Silvae Genetica, 65, 74–79. [Google Scholar]
- Mehrotra, R. , Gupta, G. , Sethi, R. , Bhalothia, P. , Kumar, N. and Mehrotra, S. (2011) Designer promoter: an artwork of cis engineering. Plant Mol. Biol. 75, 527–536. [DOI] [PubMed] [Google Scholar]
- Mehrotra, R. and Mehrotra, S. (2010) Promoter activation by ACGT in response to salicylic and abscisic acids is differentially regulated by the spacing between two copies of the motif. J. Plant Physiol. 167, 1214–1218. [DOI] [PubMed] [Google Scholar]
- Ohta, Masaru , Sato, Aiko , Renhu, Na , Yamamoto, Tsuyoshi , Oka, Nodoka , Zhu, Jian‐Kang , Tada, Yasuomi et al. (2018) MYC‐type transcription factors, MYC67 and MYC70, interact with ICE1 and negatively regulate cold tolerance in Arabidopsis . Sci. Rep. 8, 11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura, A. , Shimada, A. , Yamasaki, S. , Horino, T. , Iwata, Y. , Koizumi, N. , Nishihara, M. and et al. (2016) CaMV‐35S promoter sequence‐specific DNA methylation in lettuce. Plant Cell Rep. 35, 43–51. [DOI] [PubMed] [Google Scholar]
- Pavesi, G. , Mereghetti, P. , Mauri, G. and Pesole, G. (2004) Weeder Web: discovery of transcription factor binding sites in a set of sequences from co‐regulated genes. Nucleic Acids Res. 32, W199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peramuna, A. , Bae, H. , Rasmussen, E.K. , Dueholm, B. , Waibel, T. , Critchley, J.H. , Brzezek, K. et al. (2018) Evaluation of synthetic promoters in Physcomitrella patens. Biochem. Biophys. Res. Commun. 500, 418–422. [DOI] [PubMed] [Google Scholar]
- Persad, R. , Reuter, D.N. , Dice, L.T. , Nguyen, M.‐A. , Rigoulot, S.B. , Layton, J.S. , Schmid, M.J. et al. (2020) The Q‐system as a synthetic transcriptional regulator in plants. Front Plant Sci. 11, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2014) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. http://www.R‐project.org/ [Google Scholar]
- Roberts, M.L. (2011) The use of functional genomics in synthetic promoter design. In: Compt. Biol. Appl. Bioinform. Intech ( Lopes, H.S. ed), pp 375–396.Available from: http://www.intechopen.com/books/computational‐biology‐and‐applied‐bioinformatics/the‐use‐offunctional‐genomics‐in‐synthetic‐promoter‐design
- Roccaro, M. , Ahmadinejad, N. , Colby, T. and Somssich, I.E. (2013) Identification of functional cis‐regulatory elements by sequential enrichment from a randomized synthetic DNA library. BMC Plant Biol. 13, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J. (2016) What have we learned about synthetic promoter construction? In Plant Synthetic Promoters Methods in Molecular Biology, 1482. ( Hehl, R. , ed), pp. 1–13. New York: Humana Press. [DOI] [PubMed] [Google Scholar]
- Sahoo, D.K. , Sarkar, S. , Raha, S. , Maiti, I.B. and Dey, N. (2014) Comparative analysis of synthetic DNA promoters for high‐level gene expression in plants. Planta, 240, 855–875. [DOI] [PubMed] [Google Scholar]
- Saint Pierre, C. , Crossa, J.L. , Bonnett, D. , Yamaguchi‐Shinozaki, K. and Reynolds, M.P. (2012) Phenotyping transgenic wheat for drought resistance. J. Exp. Bot. 63, 1799–1808. [DOI] [PubMed] [Google Scholar]
- Sannigrahi, P. , Ragauskas, A.J. and Tuskan, G.A. (2009) Poplar as a feedstock for biofuels: A review of compositional characteristics. Biofuels Bioprod. Bioref. 4, 209–226. [Google Scholar]
- Sawant, Samir V. , Kiran, Kanti , Mehrotra, Rajesh , Chaturvedi, Chandra Prakash , Ansari, Suraiya A. , Singh, Pratibha , Lodhi, Niraj and et al. (2005) A variety of synergistic and antagonistic interactions mediated by cis‐acting DNA motifs regulate gene expression in plant cells and modulate stability of the transcription complex formed on a basal promoter. J. Exp. Bot. 56, 2345–2353. [DOI] [PubMed] [Google Scholar]
- Schaumberg, K.A. , Antunes, M.S. , Kassaw, T.K. , Xu, W. , Zalewski, C.S. , Medford, J.I. and Prasad, A. (2016) Quantitative characterization of genetic parts and circuits for plant synthetic biology. Nat. Methods 13, 94–100. [DOI] [PubMed] [Google Scholar]
- Schneider, C.A. , Rasband, W.S. and Eliceiri, K.W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon, E. , Kalma, Y. , Sharp, A. , Raveh‐Sadka, T. , Levo, M. , Zeevi, D. , Keren, L. et al. (2012) Inferring gene regulatory logic from high‐throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 30, 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen, J. (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127, 1466–1475. [PMC free article] [PubMed] [Google Scholar]
- Shen, Q. , Zhang, P. and Ho, T.H. (1996) Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell, 8, 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, S. , Nakashima, K. , Narusaka, Y. , Seki, M. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2003) Two different novel cis‐acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark‐induced senescence. Plant J. 33, 259–270. [DOI] [PubMed] [Google Scholar]
- Smedley, Damian , Haider, Syed , Durinck, Steffen , Pandini, Luca , Provero, Paolo , Allen, James , Arnaiz, Olivier et al. (2015) The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 43, W589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, S. , Liang, H. , Yan, D. , Zhao, Y. , Han, X. , Carlson, J.E. , Xia, X. and et al. (2013) Populus euphratica: the transcriptomic response to drought stress. Plant Mol. Biol. 83, 539–557. [DOI] [PubMed] [Google Scholar]
- Thijs, G. , Marchal, K. , Lescot, M. , Rombauts, S. , De Moor, B. , Rouze, P. and Moreau, Y. (2002) A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J. Comput. Biol. 9, 447–464. [DOI] [PubMed] [Google Scholar]
- Thomashow, M.F. (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Tompa, Martin , Li, Nan , Bailey, Timothy L. , Church, George M. , De Moor, Bart , Eskin, Eleazar , Favorov, Alexander V. et al. (2005) Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 23, 137–144. [DOI] [PubMed] [Google Scholar]
- Tuskan, G.A. , Difazio, S. , Jansson, S. , Bohlmann, J. , Grigoriev, I. , Hellsten, U. , Putnam, N. et al. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science, 313, 1596–1604. [DOI] [PubMed] [Google Scholar]
- Wang, H. , Wang, H. , Shao, H. and Tang, X. (2016) Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 7, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, T. , Deng, K. , Zhang, Q. , Gao, Y. , Liu, Y. , Yang, M. , Zhang, L. et al. (2017) Modulating AtDREB1C expression improves drought tolerance in Salvia miltiorrhiza . Front. Plant Sci. 8, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, R. , Duan, L. , Pruneda‐Paz, J.L. , Oh, D.H. , Pound, M. , Kay, S. and Dinneny, J.R. (2018) The 6xABRE synthetic promoter enables the spatiotemporal analysis of ABA‐mediated transcriptional regulation. Plant Physiol. 177, 1650–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C.Y. , Suzuki, A. , Washida, H. and Takaiwa, F. (1998) The GCN4 motif in a rice glutelin gene is essential for endosperm‐specific gene expression and is activated by Opaque‐2 in transgenic rice plants. Plant J. 14, 673–683. [DOI] [PubMed] [Google Scholar]
- Wurtzel, E.T. , Vickers, C.E. , Hanson, A.D. , Millar, A.H. , Cooper, M. , Voss‐Fels, K.P. , Nikel, P.I. and et al. (2019) Revolutionizing agriculture with synthetic biology. Nat. Plants, 5, 1207–1210. [DOI] [PubMed] [Google Scholar]
- Yamaguchi‐Shinozaki, K. and Shinozaki, K. (1994) A novel cis‐acting element in an Arabidopsis gene is involved in responsiveness to drought, low‐temperature, or high‐salt stress. Plant Cell, 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki, Y. and Randall, S.K. (2016) Functionality of soybean CBF/DREB1 transcription factors. Plant Sci. 246, 80–90. [DOI] [PubMed] [Google Scholar]
- Yang, H. , Shukla, M. , Mao, X. , Kang, S. and Du, T. (2019) Interactive regimes of reduced irrigation and salt stress depressed tomato water use efficiency at leaf and plant scales by affecting leaf physiology and stem sap flow. Front. Plant Sci. 10, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L. , Meng, Y. , Ma, J. , Zhao, X. , Cheng, T. , Ji, J. , Chang, E. et al. (2015) Transcriptomic analysis reveals importance of ROS and phytohormones in response to short‐term salinity stress in Populus tomentosa . Front. Plant Sci. 6, 678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The subset of published poplar RNA‐seq results of independent experiments used for the basis of promoter discovery in this study.
Figure S2 Cloned regions of four selected native promoters showing domains of interest.
Figure S3 Effect of copy number repeats of core sequences used in synthetic promoter constructs.
Figure S4 The ratio of GFP‐ and RFP‐positive transfected protoplasts in each respective experimental population.
Figure S5 PCR analysis of T2 transgenic Arabidopsis.
Figure S6 Scheme of qRT‐PCR process.
Table S1 Selected up‐regulated genes from four different RNA‐seq datasets: from leaves taken from drought‐treated poplar.
Table S2 Gene lists from two different RNA‐seq datasets from roots of salt‐stressed poplar.
Table S3 The predicted motifs in water‐stressed RNA‐seq promoter data (SD motifs) by de novo motif‐detecting software.
Table S4 The predicted motifs in salt‐stressed RNA‐seq promoter data (SS motifs) by de novo motif‐detecting software.
Table S5 Selected expressed genes from six different RNA‐seq datasets and included SD in A domain or SS motif on their promoters.
Table S6 DNA sequence for generating synthetic promoter fragment to construct binary plasmid using the reannealing method.
Table S7 List of qRT‐PCR and genomic DNA PCR primers.


