Summary
Isopentenyltransferase (IPT) in plants regulates a rate‐limiting step of cytokinin (CTK) biosynthesis. IPTs are recognized as key regulators of CTK homeostasis and phytohormone crosstalk in both biotic and abiotic stress responses. Recent research has revealed the regulatory function of IPTs in gene expression and metabolite profiles including source‐sink modifications, energy metabolism, nutrient allocation and storage, stress defence and signalling pathways, protein synthesis and transport, and membrane transport. This suggests that IPTs play a crucial role in plant growth and adaptation. In planta studies of IPT‐driven modifications indicate that, at a physiological level, IPTs improve stay‐green characteristics, delay senescence, reduce stress‐induced oxidative damage and protect photosynthetic machinery. Subsequently, these improvements often manifest as enhanced or stabilized crop yields and this is especially apparent under environmental stress. These mechanisms merit consideration of the IPTs as ‘master regulators’ of core cellular metabolic pathways, thus adjusting plant homeostasis/adaptive responses to altered environmental stresses, to maximize yield potential. If their expression can be adequately controlled, both spatially and temporally, IPTs can be a key driver for seed yield. In this review, we give a comprehensive overview of recent findings on how IPTs influence plant stress physiology and yield, and we highlight areas for future research.
Keywords: IPT, plant yield, cytokinin, phytohormone, abiotic stress, biotic stress, stress response
Introduction
Multiple functions of CTKs in plant development and adaptation
Biotic and abiotic stresses negatively affect plant growth and development. The abiotic stresses which are major constraints in crop production include temperature stress (heat and cold), osmotic stress (drought and salinity), heavy metal exposure, irradiation, and oxidative stress. On the biotic side, attacks by pathogenic fungi, bacteria, viruses, and oomycetes are major stress factors, which impose losses in global agricultural production. During evolution, plants have developed a series of complex mechanisms to deal with environmental stressors. The communication from environmental sensing to an effective response is regulated by phytohormones. Cytokinins (CTKs) are adenine derivatives with isoprenoid or aromatic side chains; they represent a major phytohormone group and several of them act as key regulators for plant development and stress adaptation (Li et al., 2019; Wybouw and De Rybel, 2019). Cytokinins play pivotal roles in apical dominance, shoot meristem function, root expansion, source‐sink relationships, leaf senescence, reproductive development, and seed filling. Cytokinins are thus involved in many qualitative and quantitative components of yield. These regulatory functions make CTKs fascinating signalling molecules and there is much incentive to unravel their biosynthesis and signalling pathways in plants (Kieber and Schaller, 2014; Wybouw and De Rybel, 2019). Cytokinin action, induced by environmental stimuli, often invokes hormonal crosstalk. This, in turn, orchestrates the synthesis, transport, and function of other plant growth regulators, under both abiotic and biotic stresses. It is suggested that CTKs have evolved as a bridge in coordinating endogenous developmental processes and adaptive responses (Li et al., 2016, 2019).
Plant CTK homeostasis is complex and depends mainly on a number of key pathway enzymes in: biosynthesis [adenosine phosphate‐isopentenyltransferase (ATP/ADP‐IPTs) and tRNA‐isopentenyltransferase (tRNA‐IPT)]; side‐chain modification [cytokinin trans‐hydroxylase (CYP735A)]; O‐glucosylation [zeatin O‐glucosyltransferase (ZOG)]; hydrolysis of O‐glucosides [β‐glucosidase (GLU)]; N‐glucosylation [UDP‐glycosyltransferase (UGT)] primary activation [LONELY GUY (LOG)]; and degradation [cytokinin oxidase/dehydrogenase (CKX)] (Sakakibara, 2006). Cytokinins are known to occur in plants in at least 32 different forms (Kisiala et al., 2019). They are distributed and translocated among plant organs, and affect multiple biological processes, by way of their varying chemical properties and metabolic and signalling mechanisms (Durán‐Medina et al., 2017). An overview of CTK synthesis and translocation is summarized in Figure 1a and b. The initial steps in plant CTK biosynthesis are regulated by IPT gene family members (GFMs), which can be classified either into the adenylate ATP/ADP‐IPT GFMs or tRNA‐IPT GFMs (Kieber and Schaller, 2018). Generally, the ATP/ADP‐IPTs account for the biosynthesis of isopentenyladenine (iP)‐ and trans‐zeatin (tZ)‐type CTKs via using ATP or ADP as their preferred substrates, whereas tRNA‐IPTs are responsible for the synthesis of cis‐zeatin (cZ)‐type CTKs via transferring isopentenyl groups to the N6 atom of adenines in tRNAs (Figure 1a) (Sakakibara, 2006).
Figure 1.
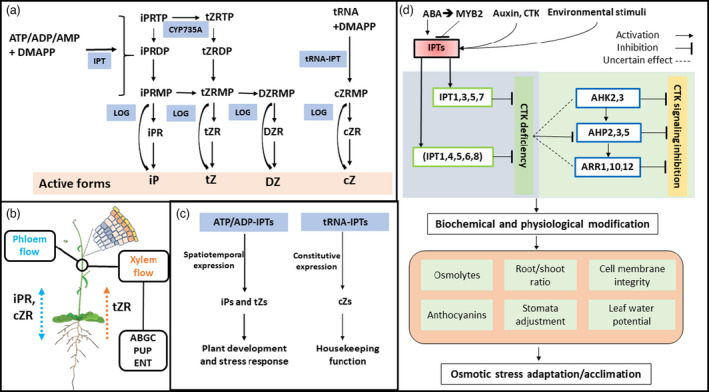
An overview of CTK biosynthesis (a); CTK translocation (b); general characteristics of two known CTKs biosynthesis pathways in angiosperms (c) and, the mode of action of IPTs in drought tolerance in Arabidopsis (d). (a) CTK types derived from two known pathways, adenylate (ATP/ADP‐IPTs and AMP‐IPTs) and tRNA types (adapted from Hirose et al. (2008), further details in Sakakibara (2006)). ADP: adenosine 5’‐diphosphate; ATP: adenosine 5’‐triphosphate; tRNA: Transfer ribonucleic acid; DMAPP: dimethylallyl diphosphate; IPT: isopentenyl transferase; CYP735A: cytochrome P450 monooxygenase, family 735, subfamily A (cytokinin trans‐hydroxylase); LOG: cytokinin phosphoribohydrolase ‘Lonely guy’; cZ: cis‐Zeatin, cZR: cZ riboside; cZRMP: cZ riboside 5’‐monophosphate, DZ: Dihydrozeatin, DZR: DZ riboside; DZRMP: DZ riboside 5’‐monophosphate; iP: isopentenyladenine; iPR: iP riboside; iPRMP: isopentenyladenosine‐5’–monophosphate; iPRDP: isopentenyladenosine‐5'‐diphosphate; iPRTP: isopentenyladenosine‐5'‐triphosphate; tZ: trans‐Zeatin, tZR: tZ riboside, tZRDP: tZ riboside 5’‐diphosphate; tZRMP: tZ riboside 5’‐monophosphate; tZRTP: tZ riboside 5’‐triphosphate. (b) Translocation of CTKs in plant. As multifunctional and mobile signalling molecules, active CTKs and their derivatives contribute to various developmental processes depending on CTK transport proteins across vascular tissues. Briefly, three kinds of CTK transporters have been systematically characterized: equilibrate nucleoside transporters (ENT), purine permeases (PUP), and G subfamily ATP‐binding cassette (ABCG) transporters (Durán‐Medina et al., 2017). tZR is the primary form of xylem transported CTKs, and iPR and cZR are major forms of phloem CTKs (Osugi and Sakakibara, 2015). Arabidopsis plant vector is adapted, with permission, from Figshare (Bouché, 2018). (c) General mode of action of two IPT GFMs (ATP/ADP‐IPTs and tRNA‐IPTs) in angiosperms (Köllmer et al 2014; Miyawaki et al 2006; Wang et al., 2020a). The investigation in Arabidopsis mutants showed that ATP/ADP‐IPT GFMs control biosynthesis of iP‐ and tZ‐type CTKs while tRNA‐type IPT genes regulate cZ‐type CTKs (Köllmer et al., 2014; Miyawaki et al., 2006). In the basal angiosperm Amborella trichopoda (Amborellaceae), and in Fragaria vesca (wild strawberry), eudicot woodland strawberry, transcriptomic analysis indicated that tRNA‐IPTs are constitutively expressed throughout the plant, whereas the expression of ATP/ADP‐IPTs is tissue‐specific and rapidly down‐regulated by abiotic stresses (Wang et al., 2020a). Generally, tRNA‐IPTs and the associated cZ‐type CTKs play a housekeeping role, whereas ATP/ADP‐IPTs and associated iP/tZ‐type CTKs play regulatory roles in organ development and stress responses in angiosperms (Köllmer et al., 2014; Miyawaki et al., 2004, 2006; Wang et al., 2020a). (d) The negative regulatory role of IPT‐repressed CTKs in osmotic stress tolerance in Arabidopsis. Arrowheads represent activation, and perpendicular bars indicate inhibition. Studies of drought stress tolerance using CTK‐deficient plants, such as the quadruple ATP/ADP‐ipt1,3,5,7 loss‐of‐function mutants, CTK signalling mutants [AHP mutants (Arabidopsis Histidine Phosphotransfer Proteins 2,3,5) and ARR mutants (type B Arabidopsis Response Regulators 1,10,12)], and down‐regulated expression of five ATP/ADP‐IPT genes (IPT1, IPT4, IPT5, IPT6, IPT8) found in the AtMYB2 (ABA‐dependent signalling pathway) overexpressor, have indicated that CTK‐depleted mutants have improved drought acclimation/adaptation (Guo and Gan, 2011; Nguyen et al., 2016; Nishiyama et al., 2011, 2013; Werner et al., 2010). Briefly, reducing endogenous CTK lowers the output of the CTK signalling cascades (i.e. quadruple ipt1,3,5,7 down‐regulates expression of AHP2, AHP3, AHP5) leading to drought acclimation/adaptation. CTK: cytokinin; AHK: Arabidopsis histidine kinase; AHP: Arabidopsis histidine phosphotransfer; ARR: Arabidopsis response regulator; ROS: reactive oxygen species.
Genome‐wide association studies for identification and annotation of novel IPT GFMs as well as altering the expression of IPTs have provided a new strategy for engineering CTKs for contemporary plant breeding programmes (Jameson and Song, 2016; Kieber and Schaller, 2018). For example, multiple Arabidopsis studies have indicated that manipulation of CTK levels via IPTs can enhance drought and salt stress tolerance (Li et al., 2016, 2019). Plants with modified expression of IPTs show improved growth and enhanced yield, opening new areas of exploration for future crop improvement technologies (Jameson and Song, 2016). Here, we critically review the current knowledge about IPTs, including their origins, evolution, induced‐stress sensing, and their roles in stress response and tolerance. We also discuss recent developments in elucidating the signalling pathways for plant stress physiology and field‐based achievements through in planta studies of IPTs. This review focuses on how IPTs are activated and connected to upstream sensing mechanisms and to downstream molecular cascades, which is attributed to changes in hormonal crosstalk, altered cellular metabolism, plant physiological responses, and plant yield.
A portrait of IPTs in the plant kingdom
There are three characteristics of plant IPT genes which point to their crucial role in plant development, yield formation, and adaptation processes: (i) IPTs are widespread in the plant kingdom; (ii) IPT expression occurs spatially throughout plant anatomy and temporally during all plant developmental stages; (iii) IPTs actively respond to environmental stresses including both abiotic and biotic factors, and to other phytohormones. These features make IPTs key developmental and adaptive regulators during plant evolution.
IPTs – ancient signalling molecules
For a long time, IPTs had been known to exist in various groups of bacteria and eukaryotes but not in Archaea (Lindner et al., 2014; Nishii et al., 2018). However, recently, putative IPT domains have been found in two archaeal species (belonging to the TACK superphylum), which have been proposed to be the direct ancestors of eukaryotes, suggesting that IPT homologs are also present in Archaea (Wang et al., 2020a). Thus, IPTs exist as pan‐kingdom signalling molecules and can be found in bacteria, slime moulds, animals, fungi, SAR, algae, prasinophytes, core chlorophytes, charophytes, liverworts, mosses, hornworts, lycophytes, monilophytes, gymnosperms, and angiosperms (Nishii et al., 2018).
Important efforts have been made to understand the complex history, origin, and classification of IPTs among kingdoms and within angiosperms. Frébort et al. (2011) classified IPT genes into five groups based on an unrooted gene tree reconstructed from full sequence lengths of IPT GFMs of two plant species, Arabidopsis and rice. To infer the deep origin and evolution of IPTs based on the matrix of a wide range of species, Lindner et al. (2014) performed a more comprehensive analysis using 30 species across different kingdoms. Nishii et al. (2018) focused their research on the conserved protein domain of the IPTs and found a deep history of IPTs that involved an interplay of possible horizontal gene transfers, gene duplication and loss events and functional diversification of IPT GFMs during evolution. Furthermore, through combined evolution and gene expression analyses of tRNA‐IPTs and ATP/ADP‐IPTs in land plants, it was determined that tRNA‐IPTs in angiosperms are associated with an initial endosymbiotic event, while ATP/ADP‐IPTs are derived from tRNA‐IPTs. Based on the combined unique transcriptional activities of these two known CTK biosynthesis pathways, together with knowledge from Arabidopsis mutant studies, a hypothesis was formed that tRNA‐IPT genes and the associated cZ‐type CTKs mainly play housekeeping roles, while ATP/ADP‐IPT genes and the associated iP/tZ‐type CTKs may be involved in regulating organ development and responses to environmental stresses (Köllmer et al., 2014; Miyawaki et al., 2004, 2006; Wang et al., 2020a) (Figure 1c). However, cZs are highly abundant in many plants (Schäfer et al., 2015), and they are the predominant CTKs in sweet potato (Hashizume et al., 1982), rice (Takagi et al., 1985), potato (Nicander et al., 1995), chickpea (Emery et al., 1998), maize (Veach et al., 2003), pea (Quesnelle and Emery, 2007) and moss (Lindner et al., 2014). They are also the predominant CTKs, at certain developmental stages and in response to environmental cues (Schäfer et al., 2015). A function of cZ in the regulation of plant response to abiotic stress, pathogen, and herbivore resistance has also been highlighted (Schäfer et al., 2015). cis‐Zeatin might also play a role as a competitor to the more active CTKs, since they bind and activate CTK receptors in the moss, P. patens (Gruhn et al., 2014), Arabidopsis (Spíchal et al., 2004), and a wide range of crops such as maize (Yonekura‐Sakakibara et al., 2004), rice (Choi et al., 2012), or apple (Daudu et al., 2017). However, in Arabidopsis, the trans‐isomers are by far the predominant forms at most growth stages (Gajdošová et al., 2011) and their CTK receptors have a lower affinity with cZs (Stolz et al., 2011). The situation is different for CHASE‐domain‐containing histidine kinases (CHKs) from crop plants, like rice or apple, for which the differences in binding affinities among tZ, iP, and cZ do not show the same clear bias against cZ (Choi et al., 2012; Daudu et al., 2017). Therefore, the functional predictions based on previous investigations of Arabidopsis (a low cZ‐containing plant), and studies on the evolution and gene expression of IPT GFMs, probably do not reflect the role of cZ‐type CTKs in planta (Schäfer et al., 2015). Many more experimental studies of tRNA‐IPT genes and the associated cZ‐type CTKs are needed.
IPTs in planta
Isopentenyltransferase GFMs are key enzymes in CTK biosynthesis, and they have been identified in a wide range of plant species including: nine IPT GFMs in Arabidopsis (Miyawaki et al., 2006; Takei et al., 2001); seven GFMs in hop (Sakano et al., 2004); nine GFMs in maize (Brugière et al., 2008); 10 GFMs in rice (Tsai et al., 2012); 17 GFMs in soybean (Le et al., 2012; Mens et al., 2018); 13 GFMs in Chinese cabbage (Liu et al., 2013b); six GFMs in pea (Dolgikh et al., 2017); seven GFMs in strawberry (Mi et al., 2017); 12 GFMs in apple (Tan et al., 2018); and nine GFMs in kiwifruit (Nardozza et al., 2020). In addition to identification of the IPT GFMs in various species, a systematic annotation of these putative genes has been conducted including characterization of their chromosome locus, enzyme domain features, and cis‐acting elements, among others. The diverse existence and unique expression of the IPT GFMs in crops emphasize the possibility for introducing IPT within or from one species to another to alter endogenous CTK levels for targeting desirable phenotypes (Figure 2). Upon exposure to the environmental changes, stress‐induced fluctuations in gene components of CTK metabolism can be associated with CTK action, including changes in IPT expression. Thus, it is important to understand IPT gene expression as an initial step to identifying stress response mechanisms and to develop new biotechnological strategies for enhancing plant performance under stress (Figure 2, Table 1).
Figure 2.
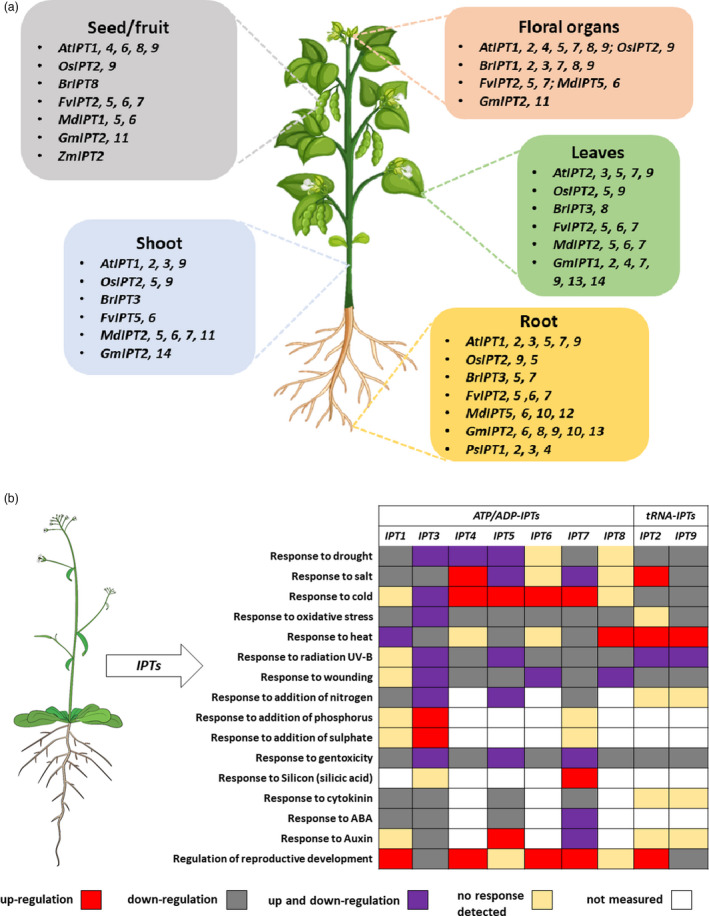
(a) Site‐specific expression of IPT GFMs in developmental tissues in planta, including Arabidopsis, rice, Chinese cabbage, strawberry, apple, soybean, pea, maize (Brugière et al., 2008; Dolgikh et al., 2017; Le et al., 2012; Liu et al., 2013b; Mi et al., 2017; Miyawaki et al., 2006; Takei et al., 2001; Tan et al., 2018; Tsai et al., 2012). The names of the polyploid BrIPT GFMs follow the Liu et al. (2013b) naming system. (b) IPT GFMs in Arabidopsis are involved in a variety of environmental responses and developmental processes (Ghosh et al., 2018; Hirose et al., 2008; Markovich et al., 2017; Miyawaki et al., 2004; Nishiyama et al., 2011; Takei et al., 2002; Woo et al., 2012). Color boxes indicate distinct transcriptional profiles of each IPT GFM in response to the noted stress factors or biological process Figure design is adapted from (Hallmark and Rashotte, 2019; Victor et al., 2019). The Arabidopsis plant vector is adapted, with permission, from Figshare (Bouché, 2018).
Table 1.
Impact of IPT genes on plant stress response and yield attributes. (↓) decrease, (↑) increase. Except for the loss of function study in Arabidopsis, all ectopic expressions of IPT genes were achieved using transformation with the Agrobacterium tumefaciens IPT gene
| Plant species | Genetic engineering approach | Promoter control | Physiological alteration | Stress tolerance | Plant component impacted | Reference |
|---|---|---|---|---|---|---|
| Rice (Oryza sativa) | stress/maturation‐induced overexpression | SARK | ↑ sink strength, photosynthesis; ↑ drought tolerance | Drought | ↑ grain yield | Peleg et al. (2011) |
| Peanut (Arachis hypogaea) | stress/maturation‐induced overexpression | SARK | ↑ photosynthesis; ↑ drought tolerance | Drought | ↑ grain yield | Qin et al. (2011) |
| Canola (Brassica napus) | maturation‐induced overexpression | AtMYB32 | ↑ chlorophyll; ↓ senescence; | Drought | ↑ grain yield | Kant et al. (2015) |
| Cotton (Gossypium hirsutum) | stress/maturation‐induced overexpression | SARK | ↓ senescence; ↑ biomass; ↑ photosynthesis | Drought | ↓ stress penalty | Kuppu et al. (2013) |
| stress/senescence‐induced overexpression | SAG12 | ↓ senescence; ↑ drought response genes | Salt | ↓ stress penalty | Liu et al. (2012); Zhao et al. (2013); Shan et al. (2019) | |
| Creeping bentgrass (Agrostis stolonifera) | stress/senescence‐induced overexpression | SAG12 | ↑ ROS‐scavenging systems; ↑ metabolites; ↑ drought response genes | Drought | ↓ stress penalty | Merewitz et al. (2010, 2012, 2016); Xu et al. (2016); Xu and Huang (2017) |
| HSP18.2 | ↑ ROS‐scavenging systems; ↑ metabolites; ↑ drought response genes | Drought | ↓ stress penalty | Merewitz et al. (2010, 2012, 2016); Xu et al. (2016); Xu and Huang (2017) | ||
| Wheat (Triticum aestivum) | maturation‐induced overexpression | Modified AtMYB32 | Drought | ↑ grain yield | Joshi et al. (2019) | |
| Sweet potato (Ipomoea batatas) | stress/maturation‐induced overexpression | SARK | ↓ senescence; ↑ photosynthesis; ↑ growth | Drought | ↑ yield | Nawiri et al. (2018) |
| Tobacco (Nicotiana tabacum) | stress/maturation‐induced overexpression | SARK | ↑ ABA and photosynthesis gene expression | Drought | ↓ stress penalty | Rivero et al. (2010) |
| Eggplant (Solanum melongena) | stress/senescence‐induced overexpression | SAG12 | ↑ ROS‐scavenging systems; ↓ senescence | Drought | ↓ stress penalty | Xiao et al. (2017) |
| Maize (Zea mays) | stress/maturation‐induced overexpression | SARK | ↑ relative water content; ↑ chlorophyll | Drought | ↑ grain yield | Bedada et al. (2016) |
| Broccoli (Brassica oleracea var. italica) | stress/senescence‐induced overexpression | SAG | ↓ post‐harvest senescence | ↓ senescence penalty | Liu et al. (2013a) | |
| Arabidopsis (Arabidopsis thaliana) | stress/senescence‐induced overexpression | SAG12 | ↑ ABA, SA, ET | Drought | ↓ stress penalty | Nishiyama et al. (2011) |
| loss of function (ipt1,3,5,7) | ↑ cell membrane integrity; ↑ ROS‐scavenging systems; ↑ drought response genes and metal‐binding encoded genes | Drought, salt, Selenium stress | ↓ stress penalty | Nishiyama et al. (2012); Jiang et al. (2019) | ||
| Tobacco (Nicotiana tabacum) | Dexamethasone‐induced overexpression | pOp/LhGR | ↑ necrotic lessons on leaves that prevent pathogen expansion | Biotic stress (pathogen attack) | ↓ biomass penalty | Novák et al. (2013) |
| Arabidopsis (Arabidopsis thaliana) | stress‐induced overexpression | 35S | ↑ callose deposition upon P. syringae pv. tomato DC3000 infection | Biotic stress (pathogen attack) | ↑ plant immunity | Choi et al. (2010) |
Spatiotemporal expression of IPTs
In general, the IPT genes are expressed widely and not restricted to specific vegetative or reproductive organs (Figure 2a). However, the expression patterns of the adenylate IPTs show more tissue‐ and stage‐specificity compared with the more ubiquitous tRNA‐IPTs (Ghosh et al., 2018; Liu et al., 2013b; Miyawaki et al., 2004). Arabidopsis has seven genes (AtIPT1 and AtIPT3–AtIPT8) that encode ATP/ADP‐IPTs while two others (AtIPT2 and AtIPT9) encode tRNA‐IPTs. Two Arabidopsis tRNA‐IPTs were ubiquitously expressed and showed the highest transcript levels within proliferating tissues (Ghosh et al., 2018; Miyawaki et al., 2004) while other AtIPTs exhibited specific, predominant expression patterns which varied among vegetative tissues such as phloem (AtIPT3), root primordia/cap (AtIPT5), siliques (AtIPT6), and other reproductive tissues such as immature seeds (AtIPT1, 4, 8), the chalazal endosperm (AtIPT4, 8) or pollen tubes (AtIPT7) (Miyawaki et al., 2004). Any IPT GFMs that specifically contribute to vegetative organs (root, shoot, and leaves) and reproductive organs (seeds/fruits) in plants can be strategically targeted candidates for crop development through molecular and genetic techniques (Figure 2a).
The involvement of IPTs in plant stress responses
Isopentenyltransferase expression is known to strongly respond to either abiotic or biotic stresses (Ghosh et al., 2018; Liu et al., 2013b). Endogenous CTK levels, associated with the reduced IPT expression, generally decrease during exposure to abiotic stress, depending on stress duration and intensity (Alvarez et al., 2008; Argueso et al., 2009; Hare et al., 1997), and this is opposite to trends observed for biotic stress. For example, CTK levels in infected plants often increase under infection with bacteria (Pertry et al., 2009; Radhika et al., 2015), fungi (Behr et al., 2012; Jiang et al., 2013; Morrison et al., 2017), or insects (Brütting et al., 2018; Schäfer et al., 2015). However, the increase in CTK levels in infected plants most likely originates from the invader (Brütting et al., 2018; Pertry et al., 2009; Trdá et al., 2017) and is not associated with any enhanced IPT expression.
In Arabidopsis, except for AtIPT5, gene expression of AtIPT1, AtIPT3, and AtIPT7 was repressed during dehydration treatment. AtIPT1 and AtIPT3 were repressed during salt stress. AtIPT5 and AtIPT7 were slightly induced after 1 and 2h of NaCl treatment and their expression returned to basal levels after 5h of treatment (Nishiyama et al., 2011). Transcriptional trajectories of all IPT GFMs in Arabidopsis, summarized in Figure 2b, have been thoroughly described before (Ghosh et al., 2018; Hirose et al., 2008; Markovich et al., 2017; Miyawaki et al., 2004; Nishiyama et al., 2011; Takei et al., 2002; Woo et al., 2012). These data suggest that IPTs not only coordinate developmental processes under favourable growth conditions but are also important regulators of plant stress response. Furthermore, IPTs can be either negative or positive regulators of growth processes under stress exposure. For example, in rice, salt stress up‐regulated only OsIPT5 levels and suppressed all other OsIPTs (Ghosh et al., 2018). OsIPT5 was up‐regulated in response to a range of other abiotic stresses and many toxic metals such as chromium, arsenic, lead, and cadmium. By contrast, OsIPT5 was down‐regulated in response to multiple pathogenic treatments, including attacks by an insect (Nilaparvata lugens), nematodes (Meloidogyne incognita and Meloidogyne graminicola), a fungus (Magnaporthe oryzae), and a parasitic plant (Striga hermonthica) (Ghosh et al., 2018). In soybean, CTK levels tend to decrease in leaves under drought conditions (Le et al., 2012). Among the soybean IPT GFMs, GmIPT08 and GmIPT10 were up‐regulated in response to drought conditions in the reproductive stage (Le et al., 2012). In Chinese cabbage, the majority of BrIPT GFMs were initially up‐regulated, before falling to basal levels during the extension of drought exposure, except for BrIPT7, for which transcription was continuously increased and maintained at high levels under severe drought conditions (Liu et al., 2013b).
As CTK biosynthesis genes, IPTs play important roles also in response to the other phytohormones involved in plant stress adaptation. The influence of auxin and ABA on IPT transcriptional activities are depicted in Figure 2b. Exogenous ABA showed an inhibitory effect on CTK levels by suppressing IPT genes (Nishiyama et al., 2011). The suppressed transcription of AtIPT1, AtIPT3, AtIPT5 (but not AtIPT7) under ABA treatment suggests that excess ABA inhibits CTK biosynthesis (Nishiyama et al., 2011). Besides ABA, treatment with other stress hormones, like jasmonic acid (JA) and salicylic acid (SA), could alter transcription of IPT genes. For example, in rice, OsIPT2 and OsIPT9 were down‐regulated by both JA and SA while OsIPT5 expression was induced by these two hormones (Ghosh et al., 2018). The occurrence of cis‐regulatory elements in IPT promoter regions also suggests there is involvement of IPTs in hormonal crosstalk. In particular, the analysis of OsIPT and AtIPT promoter sequences showed the presence of a variety of cis‐regulatory elements, including phytohormone‐sensitive, abiotic/biotic stress response, and development‐regulatory motifs and elements (Ghosh et al., 2018). The IPT gene expression can thus provide a foundation for in‐depth characterization, and development of CTK‐based selective markers for crop breeding programmes focused on high yield crops via manipulation of CTK levels.
As discussed above, IPTs are developmental and adaptive communicators, and recent efforts have mainly focused on plant functional studies to validate the role of IPTs and clarify their expression patterns at molecular, cellular, and physiological levels. In the next section, we map out how IPTs precisely modulate plant tolerance under environmental stress, thereby mitigating losses and stabilizing crop yields.
A path towards better‐engineered IPTs
IPTs in plant abiotic stress physiology
In Arabidopisis, mutations in two tRNA‐IPT genes (AtIPT2 and AtIPT9) do not affect the levels of iPs and tZs but they significantly reduce levels of cZ‐type CTKs, resulting in plant chlorosis (Miyawaki et al., 2006). CTK‐deficient plants with mutant ATP/ADP‐IPTs had reduced levels of iP‐type and tZ‐type CTKs but only slightly increased cZ content (Köllmer et al., 2014). A quadruple ATP/ADP‐IPT (ipt1,3,5,7) Arabidopsis mutant with very low levels of iPs and tZs, but slightly increased levels of cZs, displayed significantly enhanced salt and drought tolerance (Li et al., 2016; Nishiyama et al., 2011). Research in Arabidopsis suggests that the ABA‐AtMYB2 transcriptional factor (from the ABA‐dependent response pathway), down‐regulates several ATP/ADP‐IPT genes (IPT1, IPT4, IPT5, IPT6, and IPT8) leading to a CTK‐deficient phenotype and enhanced drought tolerance (Guo and Gan, 2011). The diverse physiological responses underlying increased abiotic stress tolerance in the CTK‐deficient Arabidopsis plants are listed in Figure 1d. These studies show that ATP/ADP‐IPTs and the associated iP/tZ‐type CTKs are negative regulators in plant responses to abiotic stress in Arabidopsis. The reduction in CTK content by quadruple loss of function of ipt1,3,5,7 reduces the action of CTK signalling components (e.g. AHP2, AHP3, AHP5) which results in improved stress response and acclimation (Cortleven et al., 2019; Li et al., 2016; Nishiyama et al., 2013). Suppression of CTK signalling (e.g. AHP2, AHP3, AHP5, ARR1, ARR10, ARR12) can lead to down‐regulation of many stress‐ and/or ABA‐responsive genes, and subsequently to drought‐tolerant phenotypes which exhibit improved cell membrane integrity, increased anthocyanin biosynthesis and accumulation of osmolytes, reduced stomatal aperture, enhanced leaf water potential, decreased shoot growth and increased root growth (reduced shoot/root ratio), resulting in higher survival rates (Figure 1d) (Nguyen et al., 2016; Nishiyama et al., 2011, 2013). Thus, studies using CTK‐deficient Arabidopsis lines, such as quadruple ipt1,3,5,7 mutants, or CTK signalling mutants (e.g. ahk2/3, ahp2/3/5, or arr1/10/12), have provided evidence that CTK metabolism and signalling components can act as negative regulators of plant drought adaptation, as the lowered CTK levels under unfavourable conditions result in reduced plant growth rates.
In contrast to what happens with IPT mutant lines of Arabidopsis where the chronically low CTK levels are inversely related to drought acclimation, increases of CTK, when more tightly controlled through transgenic manipulation, can show an opposite relationship. Dexamethasone spray‐controlled stimulation of the expression of a CTK biosynthetic gene (DEX::IPT) or the exogenous application of CTK [meta‐topolin (mT)] at the onset of stress, has resulted in a more rapid and vigorous plant recovery after drought (Prerostova et al., 2018). Also, transgenic crop plants with stress/senescence/maturation‐inducible promoters driving the expression of IPT genes, have shown improved plant tolerance to drought, heat, and other stresses. For example, CTK up‐regulation via overexpression of IPT as driven by a maturation‐inducible AtMYB32 promoter (AtMYB32::IPT), a stress‐inducible SARK promoter (SARK::IPT), or a senescence‐inducible SAG12 promoter (SAG::IPT), all resulted in adaptive responses under drought stress (Bedada et al., 2016; Joshi et al., 2019; Qin et al., 2011; Rivero et al., 2010; Xiao et al., 2017).
The growing number of in planta studies in Arabidopsis have shown the multifaceted nature of IPTs during drought stress, indicating IPT can be both, a positive, or a negative regulator of abiotic stress tolerance. These studies significantly advanced our understanding of the regulation of plant morphology, molecular genetics, biochemistry, and physiology by IPTs, and pointed the way to some remarkable features of IPTs that may be used to engineer crops species such as rice, maize, wheat, and sweet potato (Table 1). Supported by the improvements in genetic engineering/genome editing tools, the research on IPTs is now moving from discovery‐driven research towards more practical engineering and field crop‐based applications.
Functional specificity of IPT in response to abiotic stresses
The manipulations of CTKs in plants to cope with abiotic stresses, either by CTK exogenous application or by overexpressing CTK biosynthesis/signalling components, have brought promising outcomes (Chen et al., 2020; Hallmark and Rashotte, 2019; Wang et al., 2020b). Many findings have emerged, linking IPTs with various plant molecular and physiological responses under abiotic and biotic stresses (Cortleven et al., 2019; Jameson and Song, 2016). Besides the signalling function of phytohormones, their capacity to control leaf senescence, ROS detoxification, osmotic potential, and the protection of the photosynthetic apparatus, highlight IPTs as key molecules in cellular responses to cold, drought, salt, metal, and biotic stresses.
Osmotic and temperature stress regulation
The linkages between IPTs and hormonal crosstalk
Many phytohormones are involved in signal transduction networks of CTKs during plant responses to abiotic stress (Figure 3a). In Arabidopsis, IPTs have emerged as players in ABA‐mediated signalling pathways (Nishiyama et al., 2011). Generally, IPTs are suppressed upon ABA treatment, and thereby collaborate with ABA in drought stress alleviation (Liu et al., 2013b; Nishiyama et al., 2011).
Figure 3.
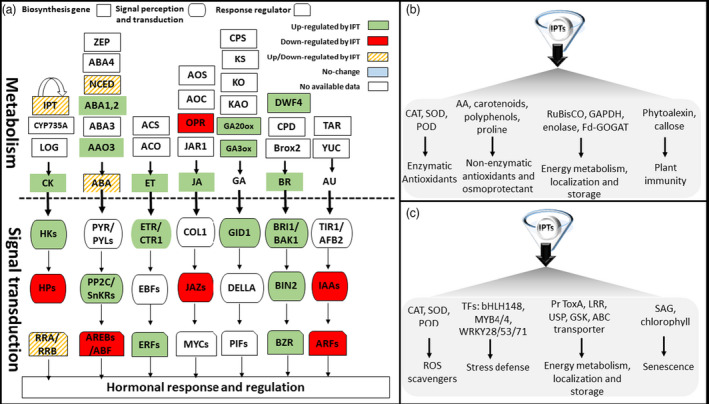
Functional specificities of IPT and their potential utilization for crop improvement. Manipulation of specific IPTs can help to obtain a certain trait, enhancing the feasibility of goal‐directed molecular design (Kuppu et al., 2013; Nishiyama et al., 2011; Peleg et al., 2011; Rivero et al., 2010; Skalák et al., 2016; Xu and Huang, 2017). (a) Schematic diagram representing contribution of IPTs in the regulation of CTKs with other plant hormone biosynthesis and signalling pathways. The results of in planta studies report that overexpression of IPTs changes endogenous CTK and other phytohormone levels. Manipulation of specific IPTs modifies transcriptional activities of hormone biosynthesis and signalling components. Cytokinin: CTK; IPT: isopentenyl transferase; CYP735A: cytochrome P450 monooxygenase, family 735, subfamily A (cytokinin trans‐hydroxylase); LOG: cytokinin phosphoribohydrolase ‘Lonely guy’; HK: histidine kinase; HP: histidine phosphotransfer; RRA/RRB: response regulator type A/B. Abscisic acid: ABA; ZEP: zeaxanthin epoxidase; ABA1,2: zeaxanthin epoxidase ABA deficient 1/2; ABA3: molybdenum cofactor (MoCo) synthase; ABA4: unidentified enzyme; AAO3: abscisic aldehyde oxidase 3; PYR: pyrabactin resistance; PYLs: pyrabactin 1‐Like; PP2Cs: protein phosphatase 2Cs; SnKRs: sucrose non‐fermenting1‐related protein kinase2; ABREs: ABA‐responsive elements; ABF: ABRE‐binding factors. Ethylene‐ET; ACS: 1‐aminocyclopropane‐1‐carboxylic acid (ACC) synthase; ACO: ACC oxidase; ETR: ET receptor; CTR1: constitutive triple response 1; EBFs: ET‐intensive (EIN3) binding F‐box proteins; ERF: ethylene response factor. Jasmonic acid: JA; AOS: Allene Oxide Synthase; AOC: Allene Oxide Cyclase; OPR: 12‐oxo‐phytodienoic acid (OPDA) reductase; JAR1: JA resistant 1; COL: constant‐like; JAZ: jasmonate ZIM‐domain; MYCs: basic helix–loop–helix (bHLH) transcription factors. Gibberellin: GA; CPS: ent‐copalyl diphosphate synthase; KS: ent‐kaurene synthase; KO: ent‐kaurene oxidase; KAO: ent‐kaurenoic acid oxidase; GA20ox: GA 20‐oxidases; GA3ox: GA 3‐oxidases; GID1: Gibberellin insensitive dwarf1 (GA receptor); DELLA: GA signalling repressors; PIFs: phytochrome interacting factors. Brassinosteroids: BR; DWF4: Dwaft4 (a cytochrome P450); CPD: BR biosynthetic cytochrome P450; BR6ox2: BR‐6‐oxidase2; BRI1: BR‐insensitive1; BAK1: BRI1‐associated‐kinase1; BIN2: BR insensitive2; BZR: Brassinazole‐resistant1. Auxin: AU; TAR: tryptophan aminotransferase‐related; YUC: YUCCA enzyme; TIR1: Transport inhibitor response1; AFB2: Auxin signalling F‐box2; IAAs: Indole‐3‐acetic acid transcriptional repressors; ARFs: Auxin response factors. Figure design adapted from (Nishiyama et al., 2018). (b) At the protein level, increasing endogenous CTK levels through the controlled expression of IPT GFMs can target various substrates to regulate specific functions. Arrowheads represent activation. AA: ascorbic acid; CAT: catalase, SOD: superoxide dismutase, POD: peroxidase; RuBisCO GAPDH: Ribulose‐1,5‐bisphosphate carboxylase/oxygenase; GAPDH: Glyceraldehyde‐3‐phosphate dehydrogenase; Fd‐GOGAT: Ferredoxin‐dependent glutamate synthase. (c) At the transcriptional level, IPTs act upstream of various genes to regulate specific functions. Arrowheads represent activation. ROS: reactive oxygen species; TFs: transcription factors; bHLH148: basic helix–loop–helix 148; MYB4/4: MYB domain protein 4; WRKY28/53/71: TFs containing highly conserved WRKY domain; Pr ToxA: proteinaceous host‐selective toxin; LRR: leucine‐rich repeat; USP: universal stress protein; GSK: glycogen synthase kinase; ABC: ATP‐binding cassette transporters; SAG: senescence‐associated gene.
Manipulation of CTKs in plants by targeting CTK metabolic genes affects ABA content. Transcriptionally, IPT genes are down‐regulated by exogenous ABA treatment suggesting that stress‐induced ABA might down‐regulate CTK levels and facilitate plant adaptation to adverse environmental conditions. Results of CTK metabolism and signalling studies suggest that IPTs contribute to the antagonistic actions between ABA and CTKs under water deficit conditions (Huang et al., 2018b; Li et al., 2016). Drought exposure increases ABA content, which activates the AtMYB2 transcriptional factor (TF) (Osakabe et al., 2014). The AtMYB2 TF subsequently down‐regulates several IPT genes, namely IPT1, 4, 5, 6, and 8, resulting in a reduction in endogenous CTK levels (Abe et al., 1997; Guo and Gan, 2011). Also consistent with an antagonistic CTK/ABA relationship, an eggplant IPT overexpressor had reduced levels of ABA under drought conditions which helped delay leaf senescence and induce abiotic stress tolerance (Xiao et al., 2017).
Research into the sophisticated and complex mechanisms of CTK‐ABA crosstalk in response to osmotic stress has been conducted to find out how ABA and CTKs antagonistically regulate drought stress response in plants (Huang et al., 2018b). In this regard, the reduction in CTK levels enables plants to cope with water deficit through a wide range of morphological and biochemical changes such as effective allocation of nutrient resources for root development, and enhanced ability to access water (Figure 1d). However, SAG12::IPT‐transgenic Arabidopsis had elevated ABA levels, which was opposite to ABA levels observed in DEX::IPT plants under drought stress (Prerostova et al., 2018). Similarly, an IPT‐transgenic tomato using the senescence/stress‐activated promoter, SARK, resulted in the induction of the carotenoid pathway leading to enhanced ABA biosynthesis (Rivero et al., 2010). Thus, constructs with differently‐driven promoters engineered with IPT genes can result in divergent profiles of hormone homeostasis and thereby induce dissimilar physiological responses in the transgenic plants (Li et al., 2019).
The expression of IPTs is affected by stress conditions and IPTs directly modify the content of CTKs and affect the content of other hormones through CTK action under abiotic stress (Figure 3a). In cotton, transcriptomic analyses of IPT overexpressors revealed an up‐regulation of ethylene (ET), brassinosteroids (BRs), JA, auxin, gibberellin (GA), and ABA‐related genes. In rice, SARK::IPT plants had increased expression of BR‐related genes and repressed expression of JA‐related genes (Peleg et al., 2011). In broccoli, both exogenous benzylaminopurine (BA) treatment and SAG::IPT‐induced elevation of CTKs resulted in reduced post‐harvest senescence via antagonist action with ET (Liu et al., 2013a). Ectopic expression of IPT in Arabidopsis displayed higher accumulations of JA, jasmonate‐isoleucine, and an ethylene precursor (aminocyclopropane carboxylic acid (ACC)) under drought conditions (Prerostova et al., 2018). Given the importance of IPTs in hormonal crosstalk, and their capacity to regulate various metabolic/signalling components in different phytohormone pathways (Figure 3a), there is a compelling case to undertake future research about how these modifications correspond to agronomic traits.
Delayed senescence
In addition to natural ageing factors, senescence can be triggered by abiotic stress. This phenomenon limits crop productivity via degradation of chloroplasts, reduced photosynthesis, massive degradation of organelles and macromolecules, and re‐allocation of nutrients (Zwack and Rashotte, 2013). Senescence is widely marked by a decline in the content of biologically active CTK forms in the leaf. As an inhibitor of senescence, CTK‐induction by IPTs can represent an effective strategy to reduce leaf senescence (Cortleven et al., 2019). There are already many examples of genetic manipulations of IPT that led to beneficial phenotypes via anti‐senescence effects of CTKs. For example, transgenic AtMYB2xs::IPT wheat demonstrated delayed leaf senescence and higher leaf water potential under both well‐watered and water stress conditions (Joshi et al., 2019). These features were also found in transgenic IPT maize, which had superior grain yields (Bedada et al., 2016). In eggplants, delayed senescence was found in IPT‐transgenic plants with enhanced cold/drought tolerance (Xiao et al., 2017). Overexpression of IPT in broccoli helped delay senescence and post‐harvest senescence (Liu et al., 2013a). Over the years, several molecular studies confirmed positive functions of inducible expression of IPTs which specifically delay senescence and thereby improve yield under drought stress conditions. This was achieved in petunia (Chang et al., 2003), tomato (Luo et al., 2005), cauliflower (Nguyen et al., 2008), creeping bentgrass (Merewitz et al., 2010), cotton (Kuppu et al., 2013), and canola (Kant et al., 2015). These results are critically important as they indicate that IPTs can be key targets for the development of transgenic crops with an enhanced ability to grow under reduced irrigation and without incurring yield penalties, ultimately contributing to savings in irrigation water. In general, active CTK levels decrease during leaf senescence. However, N7‐glucosides and O‐glucosides (in particular tZOG) are known to accumulate in senescing leaves (Šmehilová et al., 2016). Exogenously applied tZ and its glucoside derivatives (tZNGs: tZ7G, tZ9G) both delayed senescence in Arabidopsis and tZNGs altered plant transcriptome and proteome distinctly from the changes caused by tZ (Hallmark et al., 2020). A biological role in delaying leaf senescence through activation of CTK‐associated genes has been observed also for iP and its glucoside isopentenyladenine‐9‐glucoside (iP9G) (Hallmark and Rashotte, 2020). Manipulation of CTKs to increase plant yield either by targeting CTK metabolic genes or through exogenous hormone applications has provided promising outcomes (Gu et al., 2015; Holubová et al., 2018; Wang et al., 2020b; Zhao et al., 2015). The observed yield increases have been attributed mainly to delayed senescence and not to direct impact on mitotic cell division during endosperm development. However, this distinction remains quite uncertain and more effort is required to generate detailed data sets to complete a full inventory of IPTs involved in endosperm development (coenocyte, cellularization, cell division, expansion, differentiation, and maturation) at molecular, cellular, and tissue levels. For example, transcriptome analysis revealed the importance of CTK signalling during early endosperm development in Arabidopsis (Day et al., 2008).
Modification of major metabolic events and ROS detoxification pathways
Cellular events, such as alterations in carbohydrate and amino acid fluxes, are fundamental features in plant capacity to successfully cope with major osmotic stresses and involve a redirection of resources away from growth pathways and towards stress‐defensive responses. These events can be coordinated by IPTs, which induce major changes in active CTKs and cellular metabolism to affect the whole plant body which, in turn, induces stress acclimation/adaptation responses. For example, IPT overexpressing plants tolerate drought by maintaining normal accumulations of amino acids, sugars, and organic acids involved in the citric acid cycle (Merewitz et al., 2012). Also, overexpression of IPT regulates sink strength and coordinates regulation of carbon and nitrogen assimilation in rice under drought stress (Peleg et al., 2011; Reguera et al., 2013). In broccoli, SAG::IPT transgenes moderately increased levels of carbohydrate metabolism proteins and more strongly increased stress defence proteins, known as molecular chaperones (Liu et al., 2013a). Likewise, work with broccoli demonstrated that increasing CTK levels, either by overexpressing an IPT gene or exogenous hormone treatment, can regulate genes involved in sugar transport, and the metabolism of carbohydrates, amino acids, and lipids (Gapper et al., 2005; Liu et al., 2013a).
Reactive oxygen species (ROS) have partially reduced or excited states of molecular oxygen, and they are the unavoidable toxic by‐products of aerobic metabolism that have accompanied aerobic life forms since about 2.4–3.8 billion years ago (Mittler, 2017). Abiotic stress leads to excessive accumulation of ROS causing oxidative stress, leading to protein denaturation, lipid peroxidation, and nucleotide degradation. Eventually, this results in cellular damage and ultimately cell death (Choudhury et al., 2017). At the cellular level, ROS can be scavenged via non‐enzymatic systems (ascorbic acid, glutathione, tocopherols, carotenoids, phenols, etc.), enzymatic systems (CAT, SOD, POD, APX, etc.), and the osmolyte, proline (Das and Roychoudhury, 2014). IPTs were found to activate acclimation responses, as a result of alterations in the redox state of regulatory proteins, by way of transcription and translation, thereby mitigating effects of stress on metabolism and reducing metabolic ROS levels (Figure 3b) (Lai et al., 2007; Merewitz et al., 2012; Skalák et al., 2016; Thomas et al., 1995; Xu et al., 2016). For example, overexpressing IPT, under the control of SAG12, that was correlated with the elevation of the antioxidant enzyme activities, promoted cotton seed germination and seedling tolerance to salt stress (Liu et al., 2012; Shan et al., 2019). A similar construct in eggplant enhanced plant cold/drought tolerance and stimulated higher activities of ROS‐scavenging enzymes (Xiao et al., 2017). Enhancing CTK synthesis by overexpressing SAG12::IPT alleviated drought‐related inhibition of root growth and activated ROS‐scavenging systems in creeping bentgrass (Xu et al., 2016).
Protection of photosynthetic machinery
Photosynthesis is one of the most susceptible cellular processes that strongly respond to the effects of abiotic stress (Gururani et al., 2015). IPT‐induced CTKs, with their clear impact on the protection of the photosynthetic apparatus, can reduce the penalty on photosynthetic rate caused by drought stress, via regulation of stomatal conductance (Rivero et al., 2007) and chlorophyll biosynthesis (Xiao et al., 2017; Zhang et al., 2010). Transgenic peanut (SARK::IPT) was observed to have higher photosynthetic rates, stomatal conductance, transpiration, and yield under drought stress (Qin et al., 2011). Similarly, the enhanced CTK synthesis in tobacco expressing SARK::IPT prevented the degradation of photosynthetic protein complexes during drought (Rivero et al., 2010). Overexpression of IPT in canola was associated with higher chlorophyll levels, delay in leaf senescence, and enhanced yield under rainfed and irrigated conditions (Kant et al., 2015). To elucidate the effects of IPT‐altered CTKs on the proteome of the chloroplast and its subfractions (stroma and thylakoids), transgenic pSSU::IPT tobacco plants, that had high levels of CTKs, were analysed. Results revealed substantial quantitative differences in stroma proteins with significantly increased levels of CTKs in the transgenic plants but with no qualitative changes in the chloroplast proteomes between the transgenic and wild‐type plants (Cortleven et al., 2011). Paradoxically, such excessive amounts of CTKs do not result in any significant improvement in the chloroplast proteomes, and this emphasizes the important task of targeting the right hormone balance when employing any overexpression of IPTs for crop improvement. Overall, genetic engineering of IPTs in crops can ameliorate stress impacts and promote photosynthesis at different physiological and cellular organization levels under abiotic stress (stay‐green, leaf development, plastid function, protection of photosynthetic proteins, influence on photosynthetic genes, etc.) (Kant et al., 2015; Rivero et al., 2010).
Modulation of transcriptomic and proteomic responses
IPT is relevant to an array of abiotic stress responses, and its precise induction is associated with modified gene expression. Developments in plant biotechnology (the advances of transcriptomic approaches) flanked by parallel progress of in planta studies have enabled the elucidation of downstream transcriptional regulation of the IPTs involved in abiotic stress responses in plant genetic models and crop species. These diverse mechanisms include energy production, metabolic activities, stress defence, signalling, protein synthesis and transport, and membrane transport (Figure 3c).
In drought‐stressed creeping bentgrass, many downstream communicators were exposed by the use of a transgenically enhanced IPT (SAG12::IPT) and these included an oxygen‐evolving enhancer protein 3–1 and chloroplast precursor (OEE3) (metabolism), RuBisCo large subunit (energy production), leucine‐rich repeat (LRR) receptor‐like kinase (transmembrane receptor proteins), universal stress protein 5327 (stress defence) and CAT (ROS detoxification) (Merewitz et al., 2016). The analysis of differentially expressed genes (DEGs) in the transgenic IPT creeping bentgrass found that 170/250 DEGs related to energy production, metabolism, stress defence, signalling, protein synthesis and transport, and membrane transport under drought (Merewitz et al., 2016). Plant adaptive responses to abiotic stress are frequently activated by TFs, which are involved in the repression/activation of stress‐responsive genes (Colinas and Goossens, 2018; Ohama et al., 2017). Among 127 differentially expressing TFs in IPT‐transgenic creeping bentgrass, 65 exhibited up‐regulation and 62 were down‐regulated as compared to non‐transgenic plants. These up‐regulated gene expressions were involved in central hubs of bHLH148, MYB4/4‐like, and WRKY28/53/71 genes, which are associated with JA signalling and with down‐regulated, ABA signalling‐associated genes (Xu and Huang, 2017). By contrast, mutant Arabidopsis (quadruple ipt1,3,5,7 loss of function) displayed an up‐regulation of ABA marker genes (AIL1, COR47, RAB18, RD29B, SAG29), some TFs (NAC, DREB, and ZFHD), and several functional proteins that improved osmotic stress tolerance (Nishiyama et al., 2011, 2012).
Because IPTs regulate gene expression in many cellular metabolic pathways (Figure 3), regulation is also manifested at the level of proteomic responses under abiotic stress. Proteome analyses of transgenic plants (SAG12::IPT and HSP18::IPT) have revealed relatively high accumulation rates of leaf proteins (enolase, oxygen‐evolving enhancer protein 2, putative oxygen‐evolving complex, Rubisco small subunit, Hsp90, and glycolate oxidase) and root proteins (Fd‐GOGAT, nucleotide‐sugar dehydratase, NAD‐dependent isocitrate dehydrogenase, ferredoxin‐NADP reductase precursor, putative heterogeneous nuclear ribonucleoprotein A2, ascorbate peroxidase, dDTP‐glucose 4–6‐dehydratases‐like protein) as compared to non‐transgenic lines when exposed to heat stress (Xu et al., 2010). Drought tolerant SAG12‐IPT creeping bentgrass displayed an abundance of proteins involved in: photosynthesis and respiration [ribulose 1,5‐bisphosphate carboxylase (RuBisCO) and glyceraldehyde phosphate dehydrogenase (GAPDH)]; amino acid synthesis (methionine and glutamine); protein synthesis and destination [chloroplastic elongation factor (EF‐Tu) and protein disulphide isomerases (PDIs)] (Figure 3b) (Merewitz et al., 2011). These results indicate that IPT‐induced endogenous CTKs may directly and/or indirectly regulate functional proteins involved in the above‐mentioned cellular pathways, thereby reducing stress penalties. Researchers are getting closer to completing a full inventory of downstream regulatory mechanisms of IPTs which are involved in plant stress adaptation/acclimation, but they are still far from completing a comprehensive view of the signalling cascades that operate together to achieve final outputs at the molecular, cellular, and tissue level as well as long‐distance signalling under stress (e.g. CLAVATA3/EMBRYO‐SURROUNDING REGION‐RELATED 25 (CLE25) peptide) (Takahashi et al., 2018).
Metal stress response
Isopentenyltransferases are of particular importance for plant metal stress responses as their overexpression can result in improved heavy metal stress tolerance (Gomez Mansur et al., 2021; Thomas et al., 2005). Accordingly, IPT‐induced CTKs in transgenic tobacco enhanced copper stress tolerance, and this was explained by an increased expression of a metallothionein‐like gene (Thomas et al., 2005). Likewise, a transgenic wheat line, overexpressing IPT under the SARK::IPT promoter, showed a lower reduction in root growth under cadmium stress, and this was attributed to the activation of phenolic secondary metabolism, increased antioxidant defences, and cell wall reinforcement (Gomez Mansur et al., 2021). Another study, using Arabidopsis, reported an opposite relationship whereby, CTK depletion plants (ipt1,3,5,7 loss of function) gained selenium tolerance through the induced antioxidant enzyme activities and increased glutathione (GSH) content (Jiang et al., 2019). However, the antibody method used for CTK profiling was limited in its ability to discriminate among different CTK types and their identification and quantification should be interpreted with appropriate caution.
Biotic stress response
Along with SA and JA, CTKs are involved in plant responses to biotic stress factors (Akhtar et al., 2020; Ciura and Kruk, 2018). Therefore, manipulating genetic elements involved in CTK metabolism has potential to augment plant performance under biotic stress. In planta, overexpression of IPT genes has been demonstrated to promote plant immunity against bacteria, fungi, and insects (Choi et al., 2010; Großkinsky et al., 2011; Smigocki et al., 2000).
IPTs in bacterial/fungal pathogen and insect tolerance
While IPT activity and CTK accumulation are effective means by which plants deal with abiotic stresses they are also strong effectors for biotic stress tolerance. Increases in endogenous CTK level in tobacco, using a binary pOp‐ipt/LhGR system for dexamethasone‐inducible IPT expression (DEX::IPT), triggered rapid necrotic lesions on leaves that could act as a response to the plant detection of an ‘intruder’ attack, and limit the rate of the pathogen expansion (Novák et al., 2013). By overexpressing a bacterial IPT driven by a pathogen‐inducible synthetic promoter (4xJERE; 4x jasmonate and elicitor‐responsive expression), SAG12 (senescence‐associated gene 12), or TET (tetracycline‐dependent) promoter, transgenic tobacco was rendered significantly less susceptible to Pseudomonas syringae pv tabaci (Pst) while maintaining only mild symptoms of wildfire disease at infected sites (Großkinsky et al., 2011). This feature helps prevent the spread of bacteria as well as decreases the enlargement of the necrotic lesions. At the molecular level, IPT contributed to bactericidal activity of the transgenic tobacco through the expression of EAS and C4H, which encode for two antimicrobial phytoalexin compounds, scopoletin, and casidiol, respectively (Großkinsky et al., 2011). Individually overexpressing AtIPT1, 3, 5, or 7, driven by the 35S promoter, mitigated the damage caused by Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) in Arabidopsis by reducing pathogen growth (Choi et al., 2010). A 35S::IPT3 transgenic Arabidopsis displayed significantly stimulated callose deposition when treated with Pst DC3000 while there was no callose accumulation observed in wild‐type plants (Choi et al., 2010). Callose deposition is one of the primary defence responses that relates to plant cell wall reinforcement against pathogen attack, and it is often used as a parameter to evaluate plant immunity (Fan et al., 2020; Liu et al., 2020). Besides suppressing Pst DC3000 invasion, transgenic 35S::IPT3 Arabidopsis had improved resistance against a virulent necrotrophic fungus, Alternaria brassicicola (Choi et al., 2010). Reusche et al. (2013) showed that transgenic Arabidopsis overexpressing bacterial IPT under the regulation of the SAG12 promoter resulted in fewer chlorotic and necrotic leaves and less stunted growth compared with wild‐type plants upon exposure to infection by the fungus Verticillium longisporum. Additionally, V. longisporum‐infected Arabidopsis showed significant increases in expression of CKX1, CKX2, and CKX3, and this was consistent with a decrease in tZ level observed during fungal infection (Reusche et al., 2013). Transgenic IPT counteracted the CTK degradation normally prompted by infection of V. longisporum, producing an antifungal phenotype in host Arabidopsis.
Our understanding of the role of IPT genes in response to insect attack is quite limited compared with studies of pathogenic microbe infections and the few known examples suggest the existence of insect‐host plant‐specific mechanisms that regulate IPT involvement in plant defence reactions. Smigocki et al. (1993, 2000) had investigated an association between elevated CTK level and enhanced insecticidal effect in three transgenic plants that all carried PI‐II (Proteinase inhibitor‐II)‐IPT gene construct: Nicotiana plumbaginifolia, Nicotiana tabacum, and Lycopersicon esculentum (tomato). Both transgenic N. plumbaginifolia and transgenic tobacco exhibited robust tolerance against Manduca sexta with 50% to 70% less leaf consumption (Smigocki et al., 1993, 2000). Leaf extracts of transgenic N. plumbaginifolia had greater lethality to M. sexta second instar larvae, compared with less active suspension of the transgenic tobacco leaf (Smigocki et al., 2000) while anti‐insect effect on M. sexta was less consistent in the transgenic tomato since the reduction in larval weight gain could not be repeated in two independent experiments (Smigocki et al., 2000). On the other hand, analysis of the feeding habits of another insect herbivore, Tupiocoris notatus, indicated that the feeding damage on tobacco was more profound on the leaves with increased CTK levels (Brütting et al., 2018). Overall, the available data suggest that altering IPT gene expression can be used to modulate plant tolerance to insect attacks.
IPT involvement in crosstalk between CTKs and other hormones during biotic stress responses
Salicylic acid and JA are central factors in plant immune systems (Ávila et al., 2019; Zhang and Li, 2019). Previous study has elucidated the crosstalk between CTKs and SA signalling components in Arabidopsis via interactions between a type B ARR CTK transcription factor (ARR2) and the SA response factor (TGA3), the latter of which is required to induce the expression of SA‐related genes including defence marker gene PR1 (pathogenesis related 1) (Choi et al., 2010). Elevated endogenous CTK levels in transgenic 35S:IPT3 Arabidopsis plants significantly induced the expression of a PR1 gene upon the infection of P. syringae pv. tomato DC3000 (Choi et al., 2010), probably via the promotion of the CTK‐dependent phosphorylation of ARR2. By contrast, in tobacco, CTK‐induced immunity was reported to be SA‐independent, since there was no significant change in SA levels and only slight changes in transcription levels of SA signalling components, NPR1 and PR1, which mediate resistance against P. syringae (Großkinsky et al., 2011). Similarly, transgenic 4xJERE:IPT tobacco did not show any enhancement of JA levels, regardless of exposure to pathogenic infection (Großkinsky et al., 2011). In cotyledons of common bean, the accumulation of bactericidal phytoalexins was induced by exogenous SA (Durango et al., 2013) while in IPT‐transgenic tobacco, phytoalexin production was stimulated by elevated CTK levels, with SA levels remaining unaffected (Großkinsky et al., 2011). These observations suggest that synthesis of phytoalexins could be driven separately by IPT‐induced CTKs or by CTKs in concert with SA (Jeandet et al., 2013). However, more research is still needed in plant anti‐pathogen responses to clarify the involvement of IPT genes in interactions among CTKs and immune hormones, including SA and JA, as well as other hormones including auxin and ABA (Huang et al., 2018a; Shen et al., 2018), especially in important crop species such as rice, maize, wheat, or soybean.
Timing and design of IPT manipulations determine the extent of crop productivity and sustainability
Cytokinins exert many of their phenotypic effects by changing the nutrient source‐sink relationships among organs like seeds, pods, stems, leaves, and roots (Roitsch and Ehneß, 2000; Werner et al., 2008). The very nature of this dynamic means that spatial and temporal control of CTK production must be strategically and tightly controlled. Indeed, many early attempts at IPT transformation of crops for improved yields were thwarted by poorly controlled IPT expression with constitutive or leaky promoters (Atkins et al., 2011; Jameson and Song, 2016). This was often accompanied by systemic increases in CTKs, and off‐target growth changes such as hyperbranching, inhibited root production, and delayed senescence (Kuppu et al., 2013; Nawiri et al., 2018; Xiao et al., 2017). Even if expression is successfully localized within yield‐defining organs (i.e. in the flower or seed), excess CTKs can enter the vasculature and be translocated far from the point of synthesis (Atkins et al., 2011). Thus, the design of constructs and choice of promoter moieties will define if the IPT expression will be correctly targeted at the right tissue or organ and the right moment in development. In this regard, previously established IPT‐transgenic plants with strong constitutive promoters (35S promoter) and inducible heat shock‐responsive promoters (Phsp70 promoter), resulted in abnormally high endogenous CTK levels and exhibited the retardation phenotype (Loven et al, 1993; Smigocki and Owens, 1988). More narrowly responsive promoter constructs such as the senescence‐specific promoter, SAG12, a stress‐ and maturation‐induced promoter from the senescence‐associated receptor protein kinase (SARK) gene, and a developmental process‐related promoter from the AtMYB32 gene have been used to express IPT in a more controlled manner (Table 1). These have all been met with varying levels of success in improving seed yields, increasing canopy cover and shoot biomass, and improved water relations and drought tolerance (Chang et al., 2003; Gan and Amasino, 1995; Joshi et al., 2019; Kant et al., 2015). However, the success has not been absolute, and complications usually accompany the transgenic performance including morphological and physiological abnormalities such as delayed flowering, and nutrient deficiencies to name a few (Cowan et al., 2005; Jordi et al., 2000; McCabe et al., 2001). It is clear that more work on the strategic control of IPT expression needs to be done to refine CTK production with more precision.
One of the most potentially confounding issues of spatial IPT gene expression is the fact that CTKs can cause very different source‐sink effects between roots and shoots. In fact, not only does expression of various IPTs occur in different parts of the plant body, but also biosynthesis of different CTK forms is spatially diverse among plant organs (Durán‐Medina et al., 2017; Kiba et al., 2019). For example, tZ biosynthesis is mainly localized in the root tissues, while iP‐type CTKs are produced both in roots and in aerial plant parts (Ko et al., 2014). As long‐distance messengers, CTK ribosides are transported through the plant vascular system to their sites of action (Glanz‐Idan et al., 2020; Osugi et al., 2017). Cytokinins have contrasting effects on root and shoot development – they stimulate shoot growth, photosynthesis rates, and biomass accumulation; but they inhibit primary and lateral root elongation and branching (Ko et al., 2014; Ramireddy et al., 2018; Stenlid, 1982; Werner et al., 2010). Cytokinins regulate development of root vasculature, nodule formation, nutrient uptake, and allocation; however, stimulating CTK biosynthesis in the roots may alter inter‐organ source‐sink relationships and negatively affect shoot traits that contribute to yield. This emphasizes that manipulation of IPT expression must be spatially and tightly controlled to balance root and shoot phenotypes and to stabilize, or even enhance, crop yields.
Reprogramming endogenous CTK profiles for modification of plant architecture to improve resistance to stress can be achieved by two approaches. The first is by decreasing CTK content in roots by overexpression of CKX genes, which confers enhanced root fitness and remarkable phenotypic plasticity (Macková et al., 2013; Pospíšilová et al., 2016; Ramireddy et al., 2018; Werner et al., 2010). The second approach is realized by increasing endogenous CTK content via inducible expression of IPT genes to improve plant acclimatization/adaptation, or to delay senescence and minimize yield losses. This indirect mechanism could be used to limit damage caused by stress, by engineering stress‐ or senescence‐induced expression of IPT genes with specific promoters like the maturation‐inducible AtMYB32, the stress‐inducible SARK, or a senescence‐inducible SAG12, or a dexamethasone‐inducible pOp/LhGR (Table 1). Importantly, whenever CTK levels are elevated, via ectopic IPT expression or exogenous CTK treatment, increased transcriptional levels of CKX genes and/or CKX activity occurs (Panda et al., 2018; Prerostova et al., 2018). Positive correlations in gene expression of IPT and CKX GFMs were found in maize kernels (Brugière et al., 2008), rapid cycling field mustard (O’Keefe et al., 2011), wheat seed development (Nguyen et al., 2020; Song et al., 2012) and forage brassica (Song et al., 2015). Regulation of IPT and/or CKX genes in relationship to CTK metabolism and plant acclimation/adaptation might involve different stress tolerance pathways and crosstalk with other phytohormones (Figure 3). For example, CTKs regulate auxin‐efflux and influx carriers (Šimášková et al., 2015; Street et al., 2016) to control aspects of root development, such as root formation, emergence, elongation, and gravitropism (Inahashi et al., 2018). Understanding of these mechanisms and strategic promoter design can establish a scheme for the development of drought‐tolerant and high‐yielding crops through preprogrammed, via IPT, endogenous CTK levels.
The contribution of CTKs to crop yield determination has been thoroughly reviewed elsewhere (Chen et al., 2020; Jameson and Song, 2016). There are a number of studies linking seed yield in rice, soybean, barley and wheat with the increased CTK levels, and particularly with the higher levels of IPT expression/activities with various genetic modulation designs (Jameson and Song, 2016; Kambhampati et al., 2017; Powell et al., 2013). Regarding the specific function of IPTs in plant yield, one needs to focus on several research approaches implemented towards understanding IPTs and their role in controlling grain yield and enhancing crop production (Table 1).
In rice, IPT‐induced CTK synthesis maintained nitrogen (N) acquisition and reduced the environmental stress penalties on photosynthesis and yield (Reguera et al., 2013). Panda et al. (2018) suggested that overexpression of rice OsIPT9 can increase CTK level of the developing caryopses, leading to the enhanced grain filling in rice cultivars bearing large panicles with numerous spikelets, which subsequently improves yield. Many efforts have been undertaken to modify spatiotemporal expression of IPTs by strategically employing differently‐driven promoters to increase crop yield (Table 1). Transgenic wheat (IPT driven by promoter AtMYB2xs) had increased yield in well‐watered and water stress conditions (Joshi et al., 2019). In both glasshouse and field conditions, IPT‐transgenic maize (IPT driven by the SARK promoter) had higher grain yield (Bedada et al., 2016). Regulation of IPT via the AtMYB32 promoter improved yield under rainfed and irrigated conditions in canola (Kant et al., 2015). Transgenic peanut demonstrated higher photosynthetic rates and yield‐relevant traits (Qin et al., 2011). Interestingly, the timing of water deficit stress was critical for IPT‐transgenic cotton to enable its yield advantage over control plants. If water deficit was applied before flowering, the yield of IPT‐transgenic cotton was higher than non‐transgenic plants; however, when water stress was at, or after, flowering, there was no difference (Zhu et al., 2018). Overall, if their expression can be adequately controlled, both spatially and temporally, IPTs can be a key driver for seed yield, when considering multiple species that have shown improved productivity/yield under drought conditions, including: rice (Peleg et al., 2011), peanut (Qin et al., 2011), cotton (Kuppu et al., 2013), canola (Kant et al., 2015), tropical maize (Bedada et al., 2016), sweet potato (Nawiri et al., 2018), or wheat (Joshi et al., 2019).
Concluding remarks and future steps
A strong and constantly growing body of evidence highlights that IPTs play crucial roles in phytohormone crosstalk and stress‐responsive signalling pathways (Figures 1, 2, 3). These findings provide vital insights for crop breeding, especially for improving yield by increasing abiotic stress tolerance (Table 1). IPTs induce transcriptomic, proteomic, and metabolic responses, as well as physiological responses, enabling a more precise monitoring of, and acclimation to, abiotic stresses. We conclude that IPTs should be considered as master regulators of plant yield. As explained in this review, the mechanisms enhancing stress tolerance by the up‐ or down‐regulation of endogenous CTK levels might involve different pathways and crosstalk with other phytohormones (Figure 3). Seeking crops with an optimal balance of phytohormone homeostasis and/or precise responses to stress could be achieved by root‐specific or stress‐inducible promoters (Table 1).
Less clear is the contribution of IPT to the molecular response to metal and biotic stress resistance, as only a few cases involving CTK enhancement via IPTs have been shown to benefit the plant. Similarly, future research should be expanded to examine how IPTs are associated with signalling for other abiotic stresses and phytohormone pathways, such as metal and submergence stress, and strigolactone signalling.
Although current knowledge indicates that several IPT‐related genetic signalling components are required for nutrient allocation and transport, the mechanisms regarding IPT‐induced alterations of vascular cell differentiation relating to root/shoot fitness or the inter‐organ communication networks remain to be discovered. Further research on cell developmental and transcriptional trajectories using novel cellular imaging and single‐cell RNA sequencing techniques would enhance our understanding of these mechanisms.
Plants co‐exist with microorganisms in nature, and plant growth‐promoting microbiomes help plants resist stress via CTKs (Egamberdieva et al., 2017; Goh et al., 2019; Jorge et al., 2019). As CTKs are interkingdom signalling molecules, investigations into the possible function of IPTs, and how bacteria and fungi enhance plant stress resistance would increase our understanding of these beneficial interactions and could also provide novel technologies for crop stress management.
Conflict of interest
All authors agree to authorship and submission of the manuscript for peer review. The authors report no commercial or proprietary interest in any product or concept discussed in this article.
Author contributions
Hai Ngoc Nguyen involved in conceptualization, visualization, writing – original draft, and writing – review and editing. Nhan Lai Dinh contributed to writing – original draft; writing – review and editing. Anna Kisiala involved in writing – review and editing. R.J. Neil Emery contributed to funding acquisition, supervision, and writing – review and editing.
Acknowledgements
Financial support from the Natural Sciences and Engineering Council of Canada (RGPIN‐05436 and STPGP 521417 to RJNE) is gratefully acknowledged. HNN is supported by International Graduate Scholarship (IGS) from Environmental Life Sciences Graduate (EnLS) program, Trent University.
Nguyen, H. N. , Lai, N. , Kisiala, A. B. and Emery, R. J. N. (2021) Isopentenyltransferases as master regulators of crop performance: their function, manipulation, and genetic potential for stress adaptation and yield improvement. Plant Biotechnol. J, 10.1111/pbi.13603
References
- Abe, H. , Yamaguchi‐Shinozaki, K. , Urao, T. , Iwasaki, T. , Hosokawa, D. and Shinozaki, K. (1997) Role of Arabidopsis MYC and MYB homologs in drought‐and abscisic acid‐regulated gene expression. Plant Cell 9, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar, S.S. , Mekureyaw, M.F. , Pandey, C. and Roitsch, T. (2020) Role of cytokinins for interactions of plants with microbial pathogens and pest insects. Front. Plant Sc. 10, 1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, S. , Marsh, E.L. , Schroeder, S.G. and Schachtman, D.P. (2008) Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant, Cell Environ. 31, 325–340. [DOI] [PubMed] [Google Scholar]
- Argueso, C.T. , Ferreira, F.J. and Kieber, J.J. (2009) Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant, Cell Environ. 32, 1147–1160. [DOI] [PubMed] [Google Scholar]
- Atkins, C.A. , Emery, R.N. and Smith, P.M. (2011) Consequences of transforming narrow leafed lupin (Lupinus angustifolius [L.]) with an ipt gene under control of a flower‐specific promoter. Transgenic Res. 20, 1321–1332. [DOI] [PubMed] [Google Scholar]
- Ávila, A.C. , Ochoa, J. , Proaño, K. and Martínez, M.C. (2019) Jasmonic acid and nitric oxide protects naranjilla (Solanum quitoense) against infection by Fusarium oxysporum f. sp. quitoense by eliciting plant defense responses. Physiol. Mol. Plant Pathol. 106, 129–136. [Google Scholar]
- Bedada, L.T. , Seth, M.S. , Runo, S.M. , Teffera, W. , Mugoya, C. , Masiga, C.W. , Oduor, R.O. et al. (2016) Drought tolerant tropical maize (Zea mays L.) developed through genetic transformation with isopentenyltransferase gene. Afr. J. Biotech. 15, 2447–2464. [Google Scholar]
- Behr, M. , Motyka, V. , Weihmann, F. , Malbeck, J. , Deising, H.B. and Wirsel, S.G. (2012) Remodeling of cytokinin metabolism at infection sites of Colletotrichum graminicola on maize leaves. Mol. Plant Microbe Interact. 25, 1073–1082. [DOI] [PubMed] [Google Scholar]
- Bouché, F. (2018) 2018_Arabidopsis_flowering_plant. figshare. Figure. 10.6084/m9.figshare.7159937.v1 [DOI] [Google Scholar]
- Brugière, N. , Humbert, S. , Rizzo, N. , Bohn, J. and Habben, J.E. (2008) A member of the maize isopentenyl transferase gene family, Zea mays isopentenyl transferase 2 (ZmIPT2), encodes a cytokinin biosynthetic enzyme expressed during kernel development. Plant Mol. Biol. 67, 215–229. [DOI] [PubMed] [Google Scholar]
- Brütting, C. , Crava, C.M. , Schäfer, M. , Schuman, M.C. , Meldau, S. , Adam, N. and Baldwin, I.T. (2018) Cytokinin transfer by a free‐living mirid to Nicotiana attenuata recapitulates a strategy of endophytic insects. eLife 7, e36268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, H. , Jones, M.L. , Banowetz, G.M. and Clark, D.G. (2003) Overproduction of cytokinins in petunia flowers transformed with PSAG12‐IPT delays corolla senescence and decreases sensitivity to ethylene. Plant Physiol. 132, 2174–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Zhao, J. , Song, J. and Jameson, P.E. (2020) Cytokinin dehydrogenase: a genetic target for yield improvement in wheat. Plant Biotechnol. J. 18, 614–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. , Huh, S.U. , Kojima, M. , Sakakibara, H. , Paek, K.H. and Hwang, I. (2010) The cytokinin‐activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1‐dependent salicylic acid signaling in Arabidopsis. Dev. Cell 19, 284–295. [DOI] [PubMed] [Google Scholar]
- Choi, J. , Lee, J. , Kim, K. , Cho, M. , Ryu, H. , An, G. and Hwang, I. (2012) Functional identification of OsHk6 as a homotypic cytokinin receptor in rice with preferential affinity for iP. Plant Cell Physiol. 53, 1334–1343. [DOI] [PubMed] [Google Scholar]
- Choudhury, F.K. , Rivero, R.M. , Blumwald, E. and Mittler, R. (2017) Reactive oxygen species, abiotic stress and stress combination. Plant J. 90, 856–867. [DOI] [PubMed] [Google Scholar]
- Ciura, J. and Kruk, J. (2018) Phytohormones as targets for improving plant productivity and stress tolerance. J. Plant Physiol. 229, 32–40. [DOI] [PubMed] [Google Scholar]
- Colinas, M. and Goossens, A. (2018) Combinatorial transcriptional control of plant specialized metabolism. Trends Plant Sci. 23, 324–336. [DOI] [PubMed] [Google Scholar]
- Cortleven, A. , Leuendorf, J.E. , Frank, M. , Pezzetta, D. , Bolt, S. and Schmülling, T. (2019) Cytokinin action in response to abiotic and biotic stresses in plants. Plant, Cell Environ. 42, 998–1018. [DOI] [PubMed] [Google Scholar]
- Cortleven, A. , Noben, J.P. and Valcke, R. (2011) Analysis of the photosynthetic apparatus in transgenic tobacco plants with altered endogenous cytokinin content: a proteomic study. Proteome Sci. 9, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan, A.K. , Freeman, M. , Björkman, P.O. , Nicander, B. , Sitbon, F. and Tillberg, E. (2005) Effects of senescence‐induced alteration in cytokinin metabolism on source‐sink relationships and ontogenic and stress‐induced transitions in tobacco. Planta 221, 801–814. [DOI] [PubMed] [Google Scholar]
- Das, K. and Roychoudhury, A. (2014) Reactive oxygen species (ROS) and response of antioxidants as ROS‐scavengers during environmental stress in plants. Front. Environ. Sci. 2, 53. [Google Scholar]
- Daudu, D. , Allion, E. , Liesecke, F. , Papon, N. , Courdavault, V. , Dugé de Bernonville, T. , Mélin, C. et al. (2017) CHASE‐containing histidine kinase receptors in apple tree: from a common receptor structure to divergent cytokinin binding properties and specific functions. Front. Plant Sci. 8, 1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, R.C. , Herridge, R.P. , Ambrose, B.A. and Macknight, R.C. (2008) Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol. 148, 1964–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgikh, E.A. , Shaposhnikov, A.I. , Dolgikh, A.V. , Gribchenko, E.S. , Bodyagina, K.B. , Yuzhikhin, O.S. and Tikhonovich, I.A. (2017) Identification of Pisum sativum L. cytokinin and auxin metabolic and signaling genes, and an analysis of their role in symbiotic nodule development. Int. J. Plant Physiol. Biochem. 9, 22–35. [Google Scholar]
- Durango, D. , Pulgarin, N. , Echeverri, F. , Escobar, G. and Quiñones, W. (2013) Effect of salicylic acid and structurally related compounds in the accumulation of phytoalexins in cotyledons of common bean (Phaseolus vulgaris L.) cultivars. Molecules 18, 10609–10628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán‐Medina, Y. , Díaz‐Ramírez, D. and Marsch‐Martínez, N. (2017) Cytokinins on the move. Front. Plant Sci. 8, 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egamberdieva, D. , Wirth, S.J. , Alqarawi, A.A. , Abd_Allah, E.F. and Hashem, A. (2017) Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front. Microbiol. 8, 2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, R.J.N. , Leport, L. , Barton, J.E. , Turner, N.C. and Atkins, C.A. (1998) cis‐Isomers of cytokinins predominate in chickpea seeds throughout their development. Plant Physiol. 117, 1515–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, C. , Wang, G. , Wu, L. , Liu, P. , Huang, J. , Jin, X. , Zhang, G. et al. (2020) Distinct cellulose and callose accumulation for enhanced bioethanol production and biotic stress resistance in OsSUS3 transgenic rice. Carbohyd. Polym. 232, 115448. [DOI] [PubMed] [Google Scholar]
- Frébort, I. , Kowalska, M. , Hluska, T. , Frébortová, J. and Galuszka, P. (2011) Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 62, 2431–2452. [DOI] [PubMed] [Google Scholar]
- Gajdošová, S. , Spíchal, L. , Kamínek, M. , Hoyerová, K. , Novák, O. , Dobrev, P.I. , Galuszka, P. et al. (2011) Distribution, biological activities, metabolism, and the conceivable function of cis‐zeatin‐type cytokinins in plants. J. Exp. Bot. 62, 2827–2840. [DOI] [PubMed] [Google Scholar]
- Gan, S. and Amasino, R.M. (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270, 1986–1988. [DOI] [PubMed] [Google Scholar]
- Gapper, N.E. , Coupe, S.A. , McKenzie, M.J. , Sinclair, B.K. , Lill, R.E. and Jameson, P.E. (2005) Regulation of harvest‐induced senescence in broccoli (Brassica oleracea var. italica) by cytokinin, ethylene, and sucrose. J. Plant Growth Regul. 24, 153–165. [Google Scholar]
- Ghosh, A. , Shah, M.N.A. , Jui, Z.S. , Saha, S. , Fariha, K.A. and Islam, T. (2018) Evolutionary variation and expression profiling of Isopentenyl transferase gene family in Arabidopsis thaliana L. and Oryza sativa L. Plant Gene 15, 15–27. [Google Scholar]
- Glanz‐Idan, N. , Tarkowski, P. , Turečková, V. and Wolf, S. (2020) Root–shoot communication in tomato plants: cytokinin as a signal molecule modulating leaf photosynthetic activity. J. Exp. Bot. 71, 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, D.M. , Cosme, M. , Kisiala, A.B. , Mulholland, S. , Said, Z.M.F. , Spíchal, L. , Emery, R.J.N. et al. (2019) A stimulatory role for cytokinin in the arbuscular mycorrhizal symbiosis of pea. Front. Plant Sci. 10, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Mansur, N.M. , Pena, L.B. , Bossio, A.E. , Lewi, D.M. , Beznec, A.Y. , Blumwald, E. et al. (2021) An isopentenyl transferase transgenic wheat isoline exhibits less seminal root growth impairment and a differential metabolite profile under Cd stress. Physiol. Plant. 2021, 1–12. [DOI] [PubMed] [Google Scholar]
- Großkinsky, D.K. , Naseem, M. , Abdelmohsen, U.R. , Plickert, N. , Engelke, T. , Griebel, T. et al. (2011) Cytokinins mediate resistance against Pseudomonas syringae in tobacco through increased antimicrobial phytoalexin synthesis independent of salicylic acid signaling. Plant Physiol. 157, 815–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhn, N. , Halawa, M. , Snel, B. , Seidl, M.F. and Heyl, A. (2014) A subfamily of putative cytokinin receptors is revealed by an analysis of the evolution of the two‐component signaling system of plants. Plant Physiol. 165, 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, B. , Zhou, T. , Luo, J. , Liu, H. , Wang, Y. , Shangguan, Y. , Zhu, J. et al. (2015) An‐2 encodes a cytokinin synthesis enzyme that regulates awn length and grain production in rice. Mol. Plant 8, 1635–1650. [DOI] [PubMed] [Google Scholar]
- Guo, Y. and Gan, S. (2011) AtMYB2 regulates whole plant senescence by inhibiting cytokinin‐mediated branching at late stages of development in Arabidopsis . Plant Physiol. 156, 1612–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururani, M.A. , Venkatesh, J. and Tran, L.S.P. (2015) Regulation of photosynthesis during abiotic stress‐induced photoinhibition. Molecular Plant 8, 1304–1320. [DOI] [PubMed] [Google Scholar]
- Hallmark, H.T. , Černý, M. , Brzobohatý, B. and Rashotte, A.M. (2020) trans‐Zeatin‐N‐glucosides have biological activity in Arabidopsis thaliana . PLoS One 15, e0232762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmark, H.T. and Rashotte, A.M. (2019) Review–cytokinin response factors: responding to more than cytokinin. Plant Sci. 289, 110251. [DOI] [PubMed] [Google Scholar]
- Hallmark, H.T. and Rashotte, A.M. (2020) Cytokinin isopentenyladenine and its glucoside isopentenyladenine‐9G delay leaf senescence through activation of cytokinin‐associated genes. Plant Direct 4, e00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare, P.D. , Cress, W.A. and Van Staden, J. (1997) The involvement of cytokinins in plant responses to environmental stress. Plant Growth Regul. 23, 79–103. [Google Scholar]
- Hashizume, T. , Suye, S.I. and Sugiyama, T. (1982) Isolation and identification of cis‐zeatin riboside from tubers of sweet potato (Ipomoea batatas L.). Agric. Biol. Chem. 46, 663–665. [Google Scholar]
- Hirose, N. , Takei, K. , Kuroha, T. , Kamada‐Nobusada, T. , Hayashi, H. and Sakakibara, H. (2008) Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59, 75–83. [DOI] [PubMed] [Google Scholar]
- Holubová, K. , Hensel, G. , Vojta, P. , Tarkowski, P. , Bergougnoux, V. and Galuszka, P. (2018) Modification of barley plant productivity through regulation of cytokinin content by reverse‐genetics approaches. Front. Plant Sci. 9, 1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, R. , Li, Y. , Tang, G. , Hui, S. , Yang, Z. , Zhao, J. , Liu, H. et al. (2018a) Dynamic phytohormone profiling of rice upon rice black‐streaked dwarf virus invasion. J. Plant Physiol. 228, 92–100. [DOI] [PubMed] [Google Scholar]
- Huang, X. , Hou, L. , Meng, J. , You, H. , Li, Z. , Gong, Z. , Yang, S. et al. (2018b) The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis. Molecular Plant 11, 970–982. [DOI] [PubMed] [Google Scholar]
- Inahashi, H. , Shelley, I.J. , Yamauchi, T. , Nishiuchi, S. , Takahashi‐Nosaka, M. , Matsunami, M. et al. (2018) OsPIN2, which encodes a member of the auxin efflux carrier proteins, is involved in root elongation growth and lateral root formation patterns via the regulation of auxin distribution in rice. Physiol. Plant. 164, 216–225. [DOI] [PubMed] [Google Scholar]
- Jameson, P.E. and Song, J. (2016) Cytokinin: a key driver of seed yield. J. Exp. Bot. 67, 593–606. [DOI] [PubMed] [Google Scholar]
- Jeandet, P. , Clément, C. , Courot, E. and Cordelier, S. (2013) Modulation of phytoalexin biosynthesis in engineered plants for disease resistance. Int. J. Mol. Sci. 14, 14136–14170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C.‐J. , Shimono, M. , Sugano, S. , Kojima, M. , Liu, X. , Inoue, H. , Sakakibara, H. et al. (2013) Cytokinins act synergistically with salicylic acid to activate defense gene expression in rice. Mol. Plant Microbe Interact. 26, 287–296. [DOI] [PubMed] [Google Scholar]
- Jiang, L. , Liu, C. , Cao, H. , Chen, Z. , Yang, J. , Cao, S. and Wei, Z. (2019) The role of cytokinin in selenium stress response in Arabidopsis. Plant Sci. 281, 122–132. [DOI] [PubMed] [Google Scholar]
- Jordi, W. , Schapendonk, A.H.C.M. , Davelaar, E. , Stoopen, G.M. , Pot, C.S. , De Visser, R. et al. (2000) Increased cytokinin levels in transgenic PSAG12–IPT tobacco plants have large direct and indirect effects on leaf senescence, photosynthesis and N partitioning. Plant, Cell Environ. 23, 279–289. [Google Scholar]
- Jorge, G.L. , Kisiala, A. , Morrison, E. , Aoki, M. , Nogueira, A.P.O. and Emery, R.N. (2019) Endosymbiotic Methylobacterium oryzae mitigates the impact of limited water availability in lentil (Lens culinaris Medik.) by increasing plant cytokinin levels. Environ. Exp. Bot. 162, 525–540. [Google Scholar]
- Joshi, S. , Choukimath, A. , Isenegger, D. , Panozzo, J. , Spangenberg, G. and Kant, S. (2019) Improved wheat growth and yield by delayed leaf senescence using developmentally regulated expression of a cytokinin biosynthesis gene. Front. Plant Sci. 10, 1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambhampati, S. , Kurepin, L.V. , Kisiala, A.B. , Bruce, K.E. , Cober, E.R. , Morrison, M.J. and Emery, R.N. (2017) Yield associated traits correlate with cytokinin profiles in developing pods and seeds of field‐grown soybean cultivars. Field. Crop. Res. 214, 175–184. [Google Scholar]
- Kant, S. , Burch, D. , Badenhorst, P. , Palanisamy, R. , Mason, J. and Spangenberg, G. (2015) Regulated expression of a cytokinin biosynthesis gene IPT delays leaf senescence and improves yield under rainfed and irrigated conditions in canola (Brassica napus L.). PLoS One 10, e0116349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba, T. , Takebayashi, Y. , Kojima, M. and Sakakibara, H. (2019) Sugar‐induced de novo cytokinin biosynthesis contributes to Arabidopsis growth under elevated CO2. Sci. Rep. 9, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber, J.J. and Schaller, G.E. (2014) Cytokinins. Arabidopsis Book 12, e0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber, J.J. and Schaller, G.E. (2018) Cytokinin signaling in plant development. Development 145, dev149344. [DOI] [PubMed] [Google Scholar]
- Kisiala, A. , Kambhampati, S. , Stock, N.L. , Aoki, M. and Emery, R.N. (2019) Quantification of cytokinins using high‐resolution accurate‐mass orbitrap mass spectrometry and parallel reaction monitoring (PRM). Anal. Chem. 91, 15049–15056. [DOI] [PubMed] [Google Scholar]
- Ko, D. , Kang, J. , Kiba, T. , Park, J. , Kojima, M. , Do, J. , Kim, K.y. et al. (2014) Arabidopsis ABCG14 is essential for the root‐to‐shoot translocation of cytokinin. Proc. Natl Acad. Sci. USA 111, 7150–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllmer, I. , Novák, O. , Strnad, M. , Schmülling, T. and Werner, T. (2014) Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX 7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. 78, 359–371. [DOI] [PubMed] [Google Scholar]
- Kuppu, S. , Mishra, N. , Hu, R. , Sun, L.i. , Zhu, X. , Shen, G. , Blumwald, E. et al. (2013) Water‐deficit inducible expression of a cytokinin biosynthetic gene IPT improves drought tolerance in cotton. PLoS One 8, e64190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, Q.X. , Bao, Z.Y. , Zhu, Z.J. , Mao, B.Z. and Qian, Q.Q. (2007) Paraquat resistance in leaf discs of PSAG12‐IPT modified gerbera is related to the activities of superoxide dismutase, catalase, and dehydroascorbate reductase. Agric. Sci. China 6, 446–451. [Google Scholar]
- Le, D.T. , Nishiyama, R. , Watanabe, Y. , Vankova, R. , Tanaka, M. , Seki, M. , Ham, L.H. et al. (2012) Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels. PLoS One 7, e42411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Herrera‐Estrella, L. and Tran, L.S.P. (2016) The Yin‐Yang of cytokinin homeostasis and drought acclimation/adaptation. Trends Plant Sci. 21, 548–550. [DOI] [PubMed] [Google Scholar]
- Li, W. , Herrera‐Estrella, L. and Tran, L.S.P. (2019) Do cytokinins and strigolactones crosstalk during drought adaptation? Trends Plant Sci. 24, 669–672. [DOI] [PubMed] [Google Scholar]
- Lindner, A.‐C. , Lang, D. , Seifert, M. , Podlešáková, K. , Novák, O. , Strnad, M. , Reski, R. et al. (2014) Isopentenyltransferase‐1 (IPT1) knockout in Physcomitrella together with phylogenetic analyses of IPTs provide insights into evolution of plant cytokinin biosynthesis. J. Exp. Bot. 65, 2533–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M.S. , Li, H.C. , Lai, Y.M. , Lo, H.F. and Chen, L.F.O. (2013a) Proteomics and transcriptomics of broccoli subjected to exogenously supplied and transgenic senescence‐induced cytokinin for amelioration of postharvest yellowing. J. Proteomics. 93, 133–144. [DOI] [PubMed] [Google Scholar]
- Liu, N.‐J. , Zhang, T. , Liu, Z.‐H. , Chen, X. , Guo, H.‐S. , Ju, B.‐H. , Zhang, Y.‐Y. et al. (2020) Phytosphinganine affects plasmodesmata permeability via facilitating PDLP5‐stimulated callose accumulation in Arabidopsis. Molecular Plant 13, 128–143. [DOI] [PubMed] [Google Scholar]
- Liu, Y.D. , Yin, Z.J. , Yu, J.W. , Li, J. , Wei, H.L. , Han, X.L. and Shen, F.F. (2012) Improved salt tolerance and delayed leaf senescence in transgenic cotton expressing the Agrobacterium IPT gene. Biol. Plant. 56, 237–246. [Google Scholar]
- Liu, Z. , Lv, Y. , Zhang, M. , Liu, Y. , Kong, L. , Zou, M. , Lu, G. et al. (2013b) Identification, expression, and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 14, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven, K.V. , Beinsberger, S.E. , Valcke, R.L. , Van Onckelen, H.A. and Clisters, H.M. (1993) Morphometric analysis of the growth of P hsp70‐ipt transgenic tobacco plants. J. Exp. Bot. 44, 1671–1678. [Google Scholar]
- Luo, Y.Y. , Gianfagna, T.J. , Janes, H.W. , Huang, B. , Wang, Z. and Xing, J. (2005) Expression of the ipt gene with the AGPase S1 promoter in tomato results in unbranched roots and delayed leaf senescence. Plant Growth Regul. 47, 47–57. [Google Scholar]
- Macková, H. , Hronková, M. , Dobrá, J. , Turečková, V. , Novák, O. , Lubovská, Z. , Motyka, V. et al. (2013) Enhanced drought and heat stress tolerance of tobacco plants with ectopically enhanced cytokinin oxidase/dehydrogenase gene expression. J. Exp. Bot. 64, 2805–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovich, O. , Steiner, E. , Kouřil, Š. , Tarkowski, P. , Aharoni, A. and Elbaum, R. (2017) Silicon promotes cytokinin biosynthesis and delays senescence in Arabidopsis and Sorghum . Plant, Cell Environ. 40, 1189–1196. [DOI] [PubMed] [Google Scholar]
- McCabe, M.S. , Garratt, L.C. , Schepers, F. , Jordi, W.J. , Stoopen, G.M. , Davelaar, E. et al. (2001) Effects of PSAG12‐IPT gene expression on development and senescence in transgenic lettuce. Plant Physiol. 127, 505–516. [PMC free article] [PubMed] [Google Scholar]
- Mens, C. , Li, D. , Haaima, L.E. , Gresshoff, P.M. and Ferguson, B.J. (2018) Local and systemic effect of cytokinins on soybean nodulation and regulation of their isopentenyl transferase (IPT) biosynthesis genes following rhizobia inoculation. Front. Plant Sci. 9, 1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merewitz, E.B. , Du, H. , Yu, W. , Liu, Y. , Gianfagna, T. and Huang, B. (2012) Elevated cytokinin content in ipt transgenic creeping bentgrass promotes drought tolerance through regulating metabolite accumulation. J. Exp. Bot. 63, 1315–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merewitz, E.B. , Gianfagna, T. and Huang, B. (2010) Effects of SAG12‐ipt and HSP18.2‐ipt expression on cytokinin production, root growth, and leaf senescence in creeping bentgrass exposed to drought stress. J. Am. Soc. Hortic. Sci. 135, 230–239. [Google Scholar]
- Merewitz, E.B. , Gianfagna, T. and Huang, B. (2011) Protein accumulation in leaves and roots associated with improved drought tolerance in creeping bentgrass expressing an ipt gene for cytokinin synthesis. J. Exp. Bot. 62, 5311–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merewitz, E. , Xu, Y. and Huang, B. (2016) Differentially expressed genes associated with improved drought tolerance in creeping bentgrass overexpressing a gene for cytokinin biosynthesis. PLoS One 11, e0166676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, X. , Wang, X. , Wu, H. , Gan, L. , Ding, J. and Li, Y. (2017) Characterization and expression analysis of cytokinin biosynthesis genes in Fragaria vesca . Plant Growth Regul. 82, 139–149. [Google Scholar]
- Mittler, R. (2017) ROS are good. Trends Plant Sci. 22, 11–19. [DOI] [PubMed] [Google Scholar]
- Miyawaki, K. , Matsumoto‐Kitano, M. and Kakimoto, T. (2004) Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 37, 128–138. [DOI] [PubMed] [Google Scholar]
- Miyawaki, K. , Tarkowski, P. , Matsumoto‐Kitano, M. , Kato, T. , Sato, S. , Tarkowska, D. , Tabata, S. et al. (2006) Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl Acad. Sci. USA 103, 16598–16603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, E.N. , Emery, R.J.N. and Saville, B.J. (2017) Fungal derived cytokinins are necessary for normal Ustilago maydis infection of maize. Plant. Pathol. 66, 726–742. [Google Scholar]
- Nardozza, S. , Cooney, J. , Boldingh, H.L. , Hewitt, K.G. , Trower, T. , Jones, D. , Thrimawithana, A.H. et al. (2020) Phytohormone and transcriptomic analysis reveals endogenous cytokinins affect kiwifruit growth under restricted carbon supply. Metabolites 10, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawiri, S. , Oduor, R. and Mbinda, W. (2018) Isopentenyletransferase gene enhances drought tolerance in genetically engineered sweetpotato (Ipomoea batatas (L.) Lam). J. Plant Biochem. Physiol. 6, 221. 10.4172/2329-9029.1000221 [DOI] [Google Scholar]
- Nguyen, H.N. , Perry, L. , Kisiala, A. , Olechowski, H. and Emery, R.N. (2020) Cytokinin activity during early kernel development corresponds positively with yield potential and later stage ABA accumulation in field‐grown wheat (Triticum aestivum L.). Planta 252, 1–16. [DOI] [PubMed] [Google Scholar]
- Nguyen, K.‐H. , Jordi, W. , Van Dun, K. , Schepers, F. , Davelaar, E. , Stoopen, G. , Dix, P. et al. (2008) Delayed senescence in cauliflower transformed with an autoregulated isopentenyl transferase gene. Int. J. Plant Sci. 169, 339–347. [Google Scholar]
- Nguyen, K.H. , Van Ha, C. , Nishiyama, R. , Watanabe, Y. , Leyva‐González, M.A. , Fujita, Y. et al. (2016) Arabidopsis type B cytokinin response regulators ARR1, ARR10, and ARR12 negatively regulate plant responses to drought. Proc. Natl Acad. Sci. USA 113, 3090–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicander, B. , Bjorkman, P.O. and Tillberg, E. (1995) Identification of an N‐glucoside of cis‐zeatin from potato tuber sprouts. Plant Physiol. 109, 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishii, K. , Wright, F. , Chen, Y.Y. and Möller, M. (2018) Tangled history of a multigene family: The evolution of ISOPENTENYLTRANSFERASE genes. PLoS One 13, e0201198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, R. , Le, D.T. , Watanabe, Y. , Matsui, A. , Tanaka, M. , Seki, M. et al. (2012) Transcriptome analyses of a salt‐tolerant cytokinin‐deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS One 7, e32124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, R. , Watanabe, Y. , Fujita, Y. , Le, D.T. , Kojima, M. , Werner, T. , Vankova, R. et al. (2011) Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23, 2169–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, R. , Watanabe, Y. , Leyva‐Gonzalez, M.a. , Van Ha, C. , Fujita, Y. , Tanaka, M. , Seki, M. et al. (2013) Arabidopsis AHP2, AHP3, and AHP5 histidine phosphotransfer proteins function as redundant negative regulators of drought stress response. Proc. Natl Acad. Sci. USA 110, 4840–4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, T. , Sakayama, H. , de Vries, J. , Buschmann, H. , Saint‐Marcoux, D. , Ullrich, K.K. , Haas, F.B. et al. (2018) The Chara genome: secondary complexity and implications for plant terrestrialization. Cell 174, 448–464. [DOI] [PubMed] [Google Scholar]
- Novák, J. , Pavlů, J. , Novák, O. , Nožková‐Hlaváčková, V. , Špundová, M. , Hlavinka, J. , Koukalová, Š. et al. (2013) High cytokinin levels induce a hypersensitive‐like response in tobacco. Ann. Bot. 112, 41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe, D. , Song, J. and Jameson, P.E. (2011) Isopentenyl transferase and cytokinin oxidase/dehydrogenase gene family members are differentially expressed during pod and seed development in rapid‐cycling Brassica. J. Plant Growth Regul. 30, 92–99. [Google Scholar]
- Ohama, N. , Sato, H. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2017) Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22, 53–65. [DOI] [PubMed] [Google Scholar]
- Osakabe, Y. , Osakabe, K. , Shinozaki, K. and Tran, L.S.P. (2014) Response of plants to water stress. Front. Plant Sci. 5, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osugi, A. , Kojima, M. , Takebayashi, Y. , Ueda, N. , Kiba, T. and Sakakibara, H. (2017) Systemic transport of trans‐zeatin and its precursor have differing roles in Arabidopsis shoots. Nature Plants 3, 1–6. [DOI] [PubMed] [Google Scholar]
- Osugi, A. and Sakakibara, H. (2015) Q&A: How do plants respond to cytokinins and what is their importance? BMC Biol. 13, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda, B.B. , Sekhar, S. , Dash, S.K. , Behera, L. and Shaw, B.P. (2018) Biochemical and molecular characterisation of exogenous cytokinin application on grain filling in rice. BMC Plant Biol. 18, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg, Z. , Reguera, M. , Tumimbang, E. , Walia, H. and Blumwald, E. (2011) Cytokinin‐mediated source/sink modifications improve drought tolerance and increase grain yield in rice under water‐stress. Plant Biotechnol. J. 9, 747–758. [DOI] [PubMed] [Google Scholar]
- Pertry, I. , Václavíková, K. , Depuydt, S. , Galuszka, P. , Spíchal, L. , Temmerman, W. et al. (2009) Identification of Rhodococcus fascians cytokinins and their modus operandi to reshape the plant. Proc. Natl Acad. Sci. USA 106, 929–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospíšilová, H. , Jiskrová, E. , Vojta, P. , Mrízová, K. , Kokáš, F. , Čudejková, M.M. , Bergougnoux, V. et al. (2016) Transgenic barley overexpressing a cytokinin dehydrogenase gene shows greater tolerance to drought stress. New Biotechnol. 33, 692–705. [DOI] [PubMed] [Google Scholar]
- Powell, A.F. , Paleczny, A.R. , Olechowski, H. and Emery, R.N. (2013) Changes in cytokinin form and concentration in developing kernels correspond with variation in yield among field‐grown barley cultivars. Plant Physiol. Biochem. 64, 33–40. [DOI] [PubMed] [Google Scholar]
- Prerostova, S. , Dobrev, P.I. , Gaudinova, A. , Knirsch, V. , Körber, N. , Pieruschka, R. , Fiorani, F. et al. (2018) Cytokinins: their impact on molecular and growth responses to drought stress and recovery in Arabidopsis . Front. Plant Sci. 9, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, H. , Gu, Q. , Zhang, J. , Sun, L. , Kuppu, S. , Zhang, Y. , Burow, M. et al. (2011) Regulated expression of an isopentenyltransferase gene (IPT) in peanut significantly improves drought tolerance and increases yield under field conditions. Plant Cell Physiol. 52, 1904–1914. [DOI] [PubMed] [Google Scholar]
- Quesnelle, P.E. and Emery, R.N. (2007) cis‐Cytokinins that predominate in Pisum sativum during early embryogenesis will accelerate embryo growth in vitro. Botany 85, 91–103. [Google Scholar]
- Radhika, V. , Ueda, N. , Tsuboi, Y. , Kojima, M. , Kikuchi, J. , Kudo, T. and Sakakibara, H. (2015) Methylated cytokinins from the phytopathogen Rhodococcus fascians mimic plant hormone activity. Plant Physiol. 169, 1118–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramireddy, E. , Hosseini, S.A. , Eggert, K. , Gillandt, S. , Gnad, H. , von Wirén, N. and Schmülling, T. (2018) Root engineering in barley: increasing cytokinin degradation produces a larger root system, mineral enrichment in the shoot and improved drought tolerance. Plant Physiol. 177, 1078–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera, M. , Peleg, Z. , Abdel‐Tawab, Y.M. , Tumimbang, E.B. , Delatorre, C.A. and Blumwald, E. (2013) Stress‐induced cytokinin synthesis increases drought tolerance through the coordinated regulation of carbon and nitrogen assimilation in rice. Plant Physiol. 163, 1609–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusche, M. , Klásková, J. , Thole, K. , Truskina, J. , Novák, O. , Janz, D. et al. (2013) Stabilization of cytokinin levels enhances Arabidopsis resistance against Verticillium longisporum . Mol. Plant Microbe Interact. 26, 850–860. [DOI] [PubMed] [Google Scholar]
- Rivero, R.M. , Gimeno, J. , Van Deynze, A. , Walia, H. and Blumwald, E. (2010) Enhanced cytokinin synthesis in tobacco plants expressing PSARK: IPT prevents the degradation of photosynthetic protein complexes during drought. Plant Cell Physiol. 51, 1929–1941. [DOI] [PubMed] [Google Scholar]
- Rivero, R.M. , Kojima, M. , Gepstein, A. , Sakakibara, H. , Mittler, R. , Gepstein, S. and Blumwald, E. (2007) Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl Acad. Sci. USA 104, 19631–19636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch, T. and Ehneß, R. (2000) Regulation of source/sink relations by cytokinins. Plant Growth Regul. 32, 359–367. [Google Scholar]
- Sakakibara, H. (2006) Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449. [DOI] [PubMed] [Google Scholar]
- Sakano, Y. , Okada, Y. , Matsunaga, A. , Suwama, T. , Kaneko, T. , Ito, K. , Noguchi, H. et al. (2004) Molecular cloning, expression, and characterization of adenylate isopentenyltransferase from hop (Humulus lupulus L.). Phytochemistry 65, 2439–2446. [DOI] [PubMed] [Google Scholar]
- Schäfer, M. , Brütting, C. , Meza‐Canales, I.D. , Großkinsky, D.K. , Vankova, R. , Baldwin, I.T. and Meldau, S. (2015) The role of cis‐zeatin‐type cytokinins in plant growth regulation and mediating responses to environmental interactions. J. Exp. Bot. 66, 4873–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Y. , Zhao, P. , Liu, Z. , Li, F. and Tian, X. (2019) An isopentyl transferase gene driven by the senescence‐inducible SAG12 promoter improves salinity stress tolerance in cotton. J. Cotton Res. 2, 1–9. [Google Scholar]
- Shen, Q. , Liu, Y. and Naqvi, N.I. (2018) Fungal effectors at the crossroads of phytohormone signaling. Curr. Opin. Microbiol. 46, 1–6. [DOI] [PubMed] [Google Scholar]
- Šimášková, M. , O’Brien, J.A. , Khan, M. , Van Noorden, G. , Ötvös, K. , Vieten, A. , De Clercq, I. et al. (2015) Cytokinin response factors regulate PIN‐FORMED auxin transporters. Nat. Commun. 6, 1–11. [DOI] [PubMed] [Google Scholar]
- Skalák, J. , Černý, M. , Jedelský, P. , Dobrá, J. , Ge, E. , Novák, J. et al. (2016) Stimulation of ipt overexpression as a tool to elucidate the role of cytokinins in high temperature responses of Arabidopsis thaliana . J. Exp. Bot. 67, 2861–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmehilová, M. , Dobrůšková, J. , Novák, O. , Takáč, T. and Galuszka, P. (2016) Cytokinin‐specific glycosyltransferases possess different roles in cytokinin homeostasis maintenance. Front. Pant Sci. 7, 1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigocki, A. , Heu, S. and Buta, G. (2000) Analysis of insecticidal activity in transgenic plants carrying the ipt plant growth hormone gene. Acta Physiol. Plant. 22, 295–299. [Google Scholar]
- Smigocki, A. , Neal, J.W. , McCanna, I. and Douglass, L. (1993) Cytokinin‐mediated insect resistance in Nicotiana plants transformed with the ipt gene. Plant Mol. Biol. 23, 325–335. [DOI] [PubMed] [Google Scholar]
- Smigocki, A.C. and Owens, L.D. (1988) Cytokinin gene fused with a strong promoter enhances shoot organogenesis and zeatin levels in transformed plant cells. Proc. Natl Acad. Sci. USA 85, 5131–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J. , Jiang, L. and Jameson, P.E. (2012) Co‐ordinate regulation of cytokinin gene family members during flag leaf and reproductive development in wheat. BMC Plant Biol. 12, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J. , Jiang, L. and Jameson, P.E. (2015) Expression patterns of Brassica napus genes implicate IPT, CKX, sucrose transporter, cell wall invertase, and amino acid permease gene family members in leaf, flower, silique, and seed development. J. Exp. Bot. 66, 5067–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spíchal, L. , Rakova, N.Y. , Riefler, M. , Mizuno, T. , Romanov, G.A. , Strnad, M. and Schmülling, T. (2004) Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol. 45, 1299–1305. [DOI] [PubMed] [Google Scholar]
- Stenlid, G. (1982) Cytokinins as inhibitors of root growth. Physiol. Plant. 56, 500–506. [Google Scholar]
- Stolz, A. , Riefler, M. , Lomin, S.N. , Achazi, K. , Romanov, G.A. and Schmülling, T. (2011) The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J. 67, 157–168. [DOI] [PubMed] [Google Scholar]
- Street, I.H. , Mathews, D.E. , Yamburkenko, M.V. , Sorooshzadeh, A. , John, R.T. , Swarup, R. , Bennett, M.J. et al. (2016) Cytokinin acts through the auxin influx carrier AUX1 to regulate cell elongation in the root. Development 143, 3982–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi, M. , Yokota, T. , Murofushi, N. , Ota, Y. and Takahashi, N. (1985) Fluctuation of endogenous cytokinin contents in rice during its life cycle‐quantification of cytokinins by selected ion monitoring using deuterium‐labelled internal standards. Agric. Biol. Chem. 49, 3271–3277. [Google Scholar]
- Takahashi, F. , Suzuki, T. , Osakabe, Y. , Betsuyaku, S. , Kondo, Y. , Dohmae, N. , Fukuda, H. et al. (2018) A small peptide modulates stomatal control via abscisic acid in long‐distance signalling. Nature 556, 235–238. [DOI] [PubMed] [Google Scholar]
- Takei, K. , Sakakibara, H. and Sugiyama, T. (2001) Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana . J. Biol. Chem. 276, 26405–26410. [DOI] [PubMed] [Google Scholar]
- Takei, K. , Takahashi, T. , Sugiyama, T. , Yamaya, T. and Sakakibara, H. (2002) Multiple routes communicating nitrogen availability from roots to shoots: a signal transduction pathway mediated by cytokinin. J. Exp. Bot. 53, 971–977. [DOI] [PubMed] [Google Scholar]
- Tan, M. , Li, G. , Qi, S. , Liu, X. , Chen, X. , Ma, J. , Zhang, D. et al. (2018) Identification and expression analysis of the IPT and CKX gene families during axillary bud outgrowth in apple (Malus domestica Borkh.). Gene 651, 106–117. [DOI] [PubMed] [Google Scholar]
- Thomas, J.C. , Perron, M. , LaRosa, P.C. and Smigocki, A.C. (2005) Cytokinin and the regulation of a tobacco metallothionein‐like gene during copper stress. Physiol. Plant. 123, 262–271. [Google Scholar]
- Thomas, J.C. , Smigocki, A.C. and Bohnert, H.J. (1995) Light‐induced expression of ipt from Agrobacterium tumefaciens results in cytokinin accumulation and osmotic stress symptoms in transgenic tobacco. Plant Mol. Biol. 27, 225–235. [DOI] [PubMed] [Google Scholar]
- Trdá, L. , Barešová, M. , Šašek, V. , Nováková, M. , Zahajská, L. , Dobrev, P.I. , Motyka, V. et al. (2017) Cytokinin metabolism of pathogenic fungus Leptosphaeria maculans involves isopentenyltransferase, adenosine kinase and cytokinin oxidase/dehydrogenase. Front. Microbiol. 8, 1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, Y.‐C. , Weir, N.R. , Hill, K. , Zhang, W. , Kim, H.J. , Shiu, S.‐H. , Schaller, G.E. et al. (2012) Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 158, 1666–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veach, Y.K. , Martin, R.C. , Mok, D.W. , Malbeck, J. , Vankova, R. and Mok, M.C. (2003) O‐glucosylation of cis‐zeatin in maize. Characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol. 131, 1374–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor Roch, G. , Maharajan, T. , Ceasar, S.A. and Ignacimuthu, S. (2019) The role of PHT1 family transporters in the acquisition and redistribution of phosphorus in plants. Crit. Rev. Plant Sci. 38, 171–198. [Google Scholar]
- Wang, X. , Lin, S. , Liu, D. , Gan, L. , McAvoy, R. , Ding, J. and Li, Y. (2020a) Evolution and roles of cytokinin genes in angiosperms 1: Do ancient IPTs play housekeeping while non‐ancient IPTs play regulatory roles? Horticult. Res. 7, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X. , Liu, D. , Wei, M. and Man, J. (2020b) Spraying 6‐BA could alleviate the harmful impacts of waterlogging on dry matter accumulation and grain yield of wheat. PeerJ 8, e8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, T. , Holst, K. , Pörs, Y. , Guivarc'h, A. , Mustroph, A. , Chriqui, D. , Grimm, B. et al. (2008) Cytokinin deficiency causes distinct changes of sink and source parameters in tobacco shoots and roots. J. Exp. Bot. 59, 2659–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, T. , Nehnevajova, E. , Köllmer, I. , Novák, O. , Strnad, M. , Krämer, U. and Schmülling, T. (2010) Root‐specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 22, 3905–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, J. , MacPherson, C. , Liu, J. , Wang, H. , Kiba, T. , Hannah, M.A. , Wang, X.‐J. et al. (2012) The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Biol. 12, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wybouw, B. and De Rybel, B. (2019) Cytokinin–a developing story. Trends Plant Sci. 24, 177–185. [DOI] [PubMed] [Google Scholar]
- Xiao, X.O. , Zeng, Y.M. , Cao, B.H. , Lei, J.J. , Chen, Q.H. , Meng, C.M. and Cheng, Y.J. (2017) PSAG12‐IPT overexpression in eggplant delays leaf senescence and induces abiotic stress tolerance. J. Hortic. Sci. Biotechnol. 92, 349–357. [Google Scholar]
- Xu, Y. , Burgess, P. , Zhang, X. and Huang, B. (2016) Enhancing cytokinin synthesis by overexpressing ipt alleviated drought inhibition of root growth through activating ROS‐scavenging systems in Agrostis stolonifera . J. Exp. Bot. 67, 1979–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Gianfagna, T. and Huang, B. (2010) Proteomic changes associated with expression of a gene (ipt) controlling cytokinin synthesis for improving heat tolerance in a perennial grass species. J. Exp. Bot. 61, 3273–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. and Huang, B. (2017) Transcriptional factors for stress signaling, oxidative protection, and protein modification in ipt‐transgenic creeping bentgrass exposed to drought stress. Environ. Exp. Bot. 144, 49–60. [Google Scholar]
- Yonekura‐Sakakibara, K. , Kojima, M. , Yamaya, T. and Sakakibara, H. (2004) Molecular characterization of cytokinin‐responsive histidine kinases in maize. Differential ligand preferences and response to cis‐zeatin. Plant Physiol. 134, 1654–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P. , Wang, W.Q. , Zhang, G.L. , Kaminek, M. , Dobrev, P. , Xu, J. and Gruissem, W. (2010) Senescence‐inducible expression of isopentenyl transferase extends leaf life, increases drought stress resistance and alters cytokinin metabolism in cassava. J. Integr. Plant Biol. 52, 653–669. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. and Li, X. (2019) Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 50, 29–36. [DOI] [PubMed] [Google Scholar]
- Zhao, J. , Bai, W. , Zeng, Q. , Song, S. , Zhang, M.i. , Li, X. , Hou, L. et al. (2015) Moderately enhancing cytokinin level by down‐regulation of GhCKX expression in cotton concurrently increases fiber and seed yield. Mol. Breeding 35, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, P. , Zhang, N. , Yin, Z.J. , Liu, Y.D. and Shen, F.F. (2013) Analysis of differentially expressed genes in response to endogenous cytokinins during cotton leaf senescence. Biol. Plant. 57, 425–432. [Google Scholar]
- Zhu, X. , Sun, L.i. , Kuppu, S. , Hu, R. , Mishra, N. , Smith, J. , Esmaeili, N. et al. (2018) The yield difference between wild‐type cotton and transgenic cotton that expresses IPT depends on when water‐deficit stress is applied. Sci. Rep. 8, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwack, P.J. and Rashotte, A.M. (2013) Cytokinin inhibition of leaf senescence. Plant Signal. Behav. 8, e24737. [DOI] [PMC free article] [PubMed] [Google Scholar]


