Summary
Artemisinin, a sesquiterpene lactone widely used in malaria treatment, was discovered in the medicinal plant Artemisia annua. The biosynthesis of artemisinin is efficiently regulated by jasmonate (JA) and abscisic acid (ABA) via regulatory factors. However, the mechanisms linking JA and ABA signalling with artemisinin biosynthesis through an associated regulatory network of downstream transcription factors (TFs) remain enigmatic. Here we report AaTCP15, a JA and ABA dual‐responsive teosinte branched1/cycloidea/proliferating (TCP) TF, which is essential for JA and ABA‐induced artemisinin biosynthesis by directly binding to and activating the promoters of DBR2 and ALDH1, two genes encoding enzymes for artemisinin biosynthesis. Furthermore, AaORA, another positive regulator of artemisinin biosynthesis responds to JA and ABA, interacts with and enhances the transactivation activity of AaTCP15 and simultaneously activates AaTCP15 transcripts. Hence, they form an AaORA‐AaTCP15 module to synergistically activate DBR2, a crucial gene for artemisinin biosynthesis. More importantly, AaTCP15 expression is activated by the multiple reported JA and ABA‐responsive TFs that promote artemisinin biosynthesis. Among them, AaGSW1 acts at the nexus of JA and ABA signalling to activate the artemisinin biosynthetic pathway and directly binds to and activates the AaTCP15 promoter apart from the AaORA promoter, which further facilitates formation of the AaGSW1‐AaTCP15/AaORA regulatory module to integrate JA and ABA‐mediated artemisinin biosynthesis. Our results establish a multilayer regulatory network of the AaGSW1‐AaTCP15/AaORA module to regulate artemisinin biosynthesis through JA and ABA signalling, and provide an interesting avenue for future research exploring the special transcriptional regulation module of TCP genes associated with specialized metabolites in plants.
Keywords: Artemisia annua, jasmonate, abscisic acid, AaGSW1‐AaTCP15, AaORA transcriptional cascade, artemisinin biosynthesis
Introduction
Malaria, which is caused by Plasmodium species (Efferth et al., 2008), threatens humans in many countries and regions of the world every year. There were an estimated 228 million cases and 405 000 related deaths in 2018 (WHO, 2019). Artemisinin (AN), a sesquiterpene lactone, which is extracted from the medicinal plant Artemisia annua, is well‐known for its efficient anti‐malaria properties and broad pharmaceutical value. AN‐based combination therapies (ACTs) are the preferred treatment recommended by the World Health Organization (WHO). Chinese scientist Youyou Tu won the Lasker Award in 2011 and the Nobel Prize in 2015 for her discovery of AN in 1972. A. annua is the only natural source of AN, but its low content (0.01%–1.0% by dry weight) has prompted the semisynthetic biology approach in yeast to produce artemisinic acid (AA), the precursor of AN, using engineered Saccharomyces cerevisiae by Keasling’s group (Paddon et al., 2013; Ro et al., 2006). AA is then extracted from fermentation broth and chemically converted to AN. Although the improved engineered yeast was capable of producing 25 g artemisinic acid per litre (Paddon et al., 2013), the yield optimization and commercially relevant concentrations of AA still need to be increased for a viable industrial process, since a high concentration of AA is a prerequisite for the production of high concentrations of AN (Paddon and Keasling, 2014). Moreover, the limited production and high cost of the semisynthetic biology approach in yeast cannot meet worldwide demand and replace the agricultural production of AN at present (Peplow, 2016). Except the semisynthetic biology approach in yeast, a new synthetic biology approach was reported to produce AN using heterologous plant systems. For instance, tobacco plants are applied to produce AN by successfully introducing a core set of genes involved in the mevalonate and the AN biosynthetic pathway separately into the chloroplast and nuclear genomes at the same time (Malhotra et al., 2016), but the AN content 0.8 mg/g dry weight in engineered tobacco is less compared to A. annua. Hence, this finding lays a foundation for other alternative host plants except for A. annua to produce AN using compartmentalized metabolic engineering.
Substantial evidence suggests that A. annua possesses two kinds of trichomes including glandular trichomes (GSTs) and T‐shape trichomes (TSTs; Olofsson et al., 2012). Of these, AN is specifically synthesized in the GSTs and is transported to the epicuticular sac at the apex of GSTs (Olofsson et al., 2012; Wang et al., 2016). The AN biosynthetic pathway has almost been elucidated by several groups after years of effort (Figure S1; Bouwmeester et al., 1999; Chang et al., 2000; Paddon et al., 2013; Schramek et al., 2010; Teoh et al., 2009, 2006; Zhang et al., 2008). In summary, the cytosolic mevalonic acid (MVA) pathway and plastidial methylerythritol diphosphate (MEP) pathway‐derived isopentenyl diphosphate (IPP) and isomer dimethylallyl diphosphate (DMAPP) are catalysed by farnesyl diphosphate synthase (FPS) to produce farnesyl diphosphate (FPP), generating the common precursor of terpenoid biosynthesis (Schramek et al., 2010; Towler and Weathers, 2007).
The cyclization of FPP to amorpha‐4, 11‐diene by amorpha‐4, 11‐diene synthase (ADS) is considered as the preliminary step in the AN biosynthetic pathway (Bouwmeester et al., 1999). The next steps are two‐step oxidation of amorpha‐4, 11‐diene to artemisinic alcohol and artemisinic aldehyde by cytochrome P450‐dependent hydroxylase (CYP71AV1) along with NADPH: cytochrome P450 oxidoreductase (CPR) or alcohol dehydrogenase 1 (ADH1; Paddon et al., 2013; Ro et al., 2006; Teoh et al., 2006). The metabolic flux is then divided into two branches from artemisinic aldehyde: one branch involves artemisinic aldehyde being converted to dihydroartemisinic aldehyde via artemisinic aldehyde Δ11(13) reductase (a double‐bond reductase, DBR2) which is a crucial enzyme that efficiently promotes metabolic flux into the AN pathway (Zhang et al., 2008, see Figure S1). Then, dihydroartemisinic aldehyde is catalysed into dihydroartemisinic acid (DHAA) by aldehyde dehydrogenase 1 (ALDH1), and the second branch involves artemisinic aldehyde being converted to AA via ALDH1 (Teoh et al., 2009). Ultimately, DHAA and AA may be transported into the epicuticular sac of GSTs and subsequently converted to AN and arteannuin B (AB) via a light‐induced non‐enzymatic photochemical oxidation process (Brown and Sy, 2004, 2007; Czechowski et al., 2016).
The biosynthesis of specialized metabolites in plants is triggered by several indices such as environmental parameters and exogenous phytohormones. Jasmonic acid (JA) and abscisic acid (ABA) are essential in AN biosynthesis. Nevertheless, the mechanisms of their action are just beginning to be understood. A prior study pointed out that JA enhanced AN yield by activating the pathway structural genes (Maes et al., 2011). Later studies revealed that JA initially induced the expression of some transcription factors (TFs), which positively regulated AN biosynthesis through activating the transcription of AN biosynthetic genes. For instance, AaERF1/2 (ethylene response factor 1/2) and AaTAR1 (trichome and artemisinin regulator 1), which are induced by JA treatment, enhanced the transcripts of AN biosynthetic genes ADS and CYP71AV1 (Tan et al., 2015; Yu et al., 2012). Another JA‐responsive TF AaORA (octadecanoid‐derivative responsive AP2‐domain protein) enhanced the expression of four AN biosynthetic genes ADS, CYP71AV1, DBR2 and ALDH1 (Lu et al., 2013; Ma et al., 2018). In addition, AaWRKY1 and AabHLH1 TFs, which respond to JA treatment, also activated the expression of ADS and CYP71AV1 (Ji et al., 2014; Ma et al., 2009). Notably, elevated AN content by AaMYC2 (myelocytomatosis protein 2), a core activator of JA signalling, had a positive role in JA‐mediation of the specialized metabolites by binding to the CYP71AV1 and DBR2 promoters (Shen et al., 2016).
Besides JA, ABA is also reported by several studies to play a vital role in AN production through the activation of structural genes and downstream TFs (Jing et al., 2009; Zhang et al., 2015; Zhong et al., 2018). For example, ABA‐responsive TF AabZIP1 (Basic Leucine Zipper 1) increased ADS and CYP71AV1 expression (Zhang et al., 2015), and ABA‐induced TF AaABF3 activated the ALDH1 promoter (Zhong et al., 2018). Apart from the individual function of TFs in regulating structural genes, their combinatorial effect to form a transcriptional regulatory cascade is a delicate regulatory strategy at multiple layers. AaGSW1 (glandular trichome‐specific WRKY 1) is a JA and ABA dual‐responsive WRKY TF that activates the promoter regions of CYP71AV1 and AaORA, promoting AN biosynthesis (Chen et al., 2017). Moreover, its transcript is directly regulated by AaMYC2 and AabZIP1, two important regulators of JA and ABA signalling, thus forming AaMYC2/AabZIP1‐AaGSW1‐AaORA transcriptional cascades regulating AN accumulation (Chen et al., 2017), suggesting that the special transcriptional regulatory cascade acts at the nexus of JA and ABA signalling to control AN biosynthesis in A. annua. Other transcriptional regulatory modules involved in regulating AN biosynthesis through simultaneously linking JA and ABA signalling need to be identified.
TCP (teosinte branched1/cycloidea/proliferating cell factor) TFs are unique in the plant kingdom. They are divided into two subclasses (class I TCP and class II TCP) according to the TCP domain that is responsible for the specificity of protein‐DNA interactions (Cubas et al., 1999). In Arabidopsis, there are 13 class I and 11 class II TCP genes (Cubas et al., 1999). Subsequently, TCP family proteins have been systematically discovered in other plant species, including rice, wheat, tomato, and apple (Parapunova et al., 2014; Xu et al., 2014; Yao et al., 2007; Zhao et al., 2018). Over the past few years, substantial research uncovered diverse mechanisms adopted by TCP proteins to fulfil their broad function. The first mechanisms identified were related to controlling target gene expression by binding to conserved TCP binding sites (TBS; e.g. GGNCCCAC, GCCCR or G(T/C)GGNCCC; Aggarwal et al., 2010). Second, TCP proteins were found to interact with other TCP proteins (Danisman et al., 2012) or with various proteins including TFs (e.g. MYB, ERF, bZIP, NLP), ubiquitin receptors DA1 and DA1‐related proteins (DAR1 and DAR2), and MPK kinase 8 (MPK8; Guan et al., 2017; Nicolas and Cubas, 2016; Peng et al., 2015; Zhang et al., 2019), all of which were partially involved in regulating the transcriptional activities, or protein stability and phosphorylation in a context‐dependent fashion. Additionally, a few studies found that TCP activity was regulated by alternative splicing or microRNA319 (Bresso et al., 2018; Nicolas et al., 2015). However, our knowledge of the transcriptional regulatory factors that control TCP genes at the transcriptional level remains largely unknown.
Correspondingly, functional analyses also indicated that TCP genes have diverse roles in plant growth and development, seed germination, phytohormone signalling, biological clock and plant defence (Doebley et al., 1997; Pruneda‐Paz et al., 2009; Rueda‐Romero et al., 2012; Vadde et al., 2018; Weßling et al., 2014; Zhang et al., 2019, 2017, 2018). Furthermore, the same clade of TCP factors may have functional redundancy (Viola et al., 2016), but the different clades of TCP factors may have an antagonistic effect (Danisman et al., 2012). Moreover, several emerging lines of evidence uncovered the multifaceted role of TCP protein in plant specialized metabolism. In Arabidopsis, AtTCP3 promoted flavonoid biosynthesis by enhancing the transactivation activity of R2R3‐MYB proteins, which are components of the R2R3‐MYB/bHLH/WD40 (MBW) ternary complex (Li and Zachgo, 2013). AtTCP15 acted as a repressor in high light‐modulated anthocyanin biosynthesis (Viola et al., 2016). In apple, MdTCP46 promoted high light‐induced anthocyanin accumulation via interaction with MdMYB1 (An et al., 2020). In Lycium ruthenicum, LrTCP4 positively regulated kukoamine AN biosynthesis (Chahel et al., 2019). In A. annua, we previously found that AaTCP14 performed as a positive regulator in AN biosynthesis (Ma et al., 2018). However, the TCP‐related regulatory network associated with plant secondary metabolites has rarely been reported; thus, it is of great interest to further investigate the function of TCP genes in specialized plant metabolism such as biosynthesis of AN.
Here we report that AaTCP15 acts as a positive regulator of JA and ABA‐mediated AN biosynthesis by directly binding to and activating DBR2 and ALDH1 promoters. Furthermore, AaORA, a downstream component of JA and ABA signalling involved in promoting AN biosynthesis, exerts a double effect on AaTCP15, including enhancing the transactivation activity of AaTCP15 by direct interaction and elevating expression of AaTCP15, leading to the synergistic activation of DBR2 expression by AaORA‐AaTCP15 module. Strikingly, AaTCP15 expression is activated by the multiple JA‐ and ABA‐responsive TFs that specifically activate AN biosynthesis. Among them, AaGSW1 acts at the nexus of JA and ABA signalling and directly binds to and activates the AaTCP15 promoter apart from the reported AaORA promoter, and this facilitates formation of the AaGSW1‐AaTCP15/AaORA transcriptional cascade to integrate JA and ABA‐mediated AN biosynthesis. Thus, our data revealed a multilayer transcriptional regulatory cascade of AaGSW1‐AaTCP15/AaORA, which delicately modulates AN biosynthesis through linking the JA and ABA signalling pathways.
Results
AaTCP15 acts as a candidate regulator of AN biosynthesis in A. annua
Previous studies reported that the plant‐specific TCP TFs normally have similar functions or antagonistic effects (Danisman et al., 2013; Viola et al., 2016). Furthermore, another study showed that A. annua AaTCP14 is a positive regulator of AN biosynthesis (Ma et al., 2018), suggesting the possibility of other TCP TFs involved in AN biosynthesis. To identify all putative TCP family genes in A. annua, the hidden Markov model (HMM) profiles of the TCP domain (Pfam accession No.: PF03634) were used as queries against the local GST transcriptome database and annotated genome database (Shen et al., 2018) in the HMMER3.0 search program (Finn et al., 2011). Next, the obtained candidate sequences were used as queries against the conserved domain database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). After removing redundant sequences and sequences without the typical TCP domain, we obtained 38 full‐length TCP genes in A. annua.
To explore the evolutionary and phylogenetic relationships between A. annua TCP genes and other known TCPs, and also to characterize how the potential A. annua TCP genes functioned in AN biosynthesis, an unrooted neighbour‐joining (NJ) tree was constructed using 38 TCP genes from A. annua, 24 TCP genes from Arabidopsis, and 38 TCP genes from Gossypium raimondii (Figure S2). Based on the bootstrap values of clades and the topology of the tree, three candidate class I TCP proteins, AaTCP11, AaTCP15 and AaTCP16, were distributed into the same clade with AaTCP14, a pivotal regulator of AN biosynthesis (Figures 1a and S2). Among these, AaTCP15 clustered closest with AaTCP14 and contained a generally conserved TCP domain (Figure S3). Next, to screen one out of the three TCP genes that is mostly associated with AN biosynthesis, the heatmaps of expression levels of AaTCP11, AaTCP15, AaTCP16, AaTCP14 and AN biosynthetic genes ADS, CYP71AV1, DBR2 and ALDH1 were preliminarily analysed based on the early published transcriptome database from different A. annua tissues (Graham et al., 2010). Results revealed partial similarity with ADS, CYP71AV1, DBR2 and ALDH1, and expression of AaTCP15, AaTCP16 and AaTCP11 was also detected in trichomes (Figure 1b), where AN is mainly synthesized and accumulated in A. annua. In addition, quantitative RT‐PCR (qRT‐PCR) in the trichomes of A. annua was carried out to re‐evaluate the expression levels of AaTCP11, AaTCP15, AaTCP16, ADS, CYP71AV1, DBR2 and ALDH1. Results showed that, similar with the above heatmap result, all of them were expressed in trichomes, and the expression level of AaTCP15 was higher in trichomes compared with AaTCP11 and AaTCP16 (Figure 1c). Thus, according to the phylogenetic relationships and the results of the expression profiles of candidate TCP genes, AaTCP15 was chosen for further research due to its potential role in AN biosynthesis in A. annua.
Figure 1.
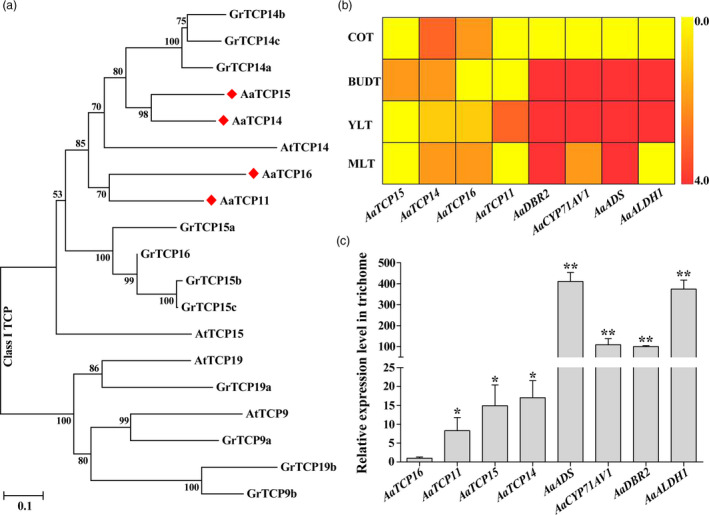
Identification of AaTCP15. (a) Phylogenetic tree showing the relationships of class I TCP transcription factors in Artemisia annua, Arabidopsis thaliana and Gossypium raimondii. TCP sequences were aligned with Clustal W, and a neighbour‐joining phylogenetic tree was constructed using MEGA. Bootstrap values are based on 2000 replicates. AaTCP14, AaTCP11, AaTCP15 and AaTCP16 are marked with red diamonds. The amino acid sequences are named based on the species name. (b) The heat map of transcriptome data includes indicated genes with differential expression in cotyledon (COT), flower bud trichomes (BUDT), young leaf trichomes (YLT) and mature leaf trichomes (MLT) of Artemisia annua. The colour scale at the right represents the value of transformed reads per kilobase per million mapped reads. (c) Relative expression levels of AaTCP16, AaTCP11, AaTCP15, AaTCP14, AaADS, AaCYP71AV1, AaDBR2 and AaALDH1 in trichomes were measured by quantitative real‐time PCR (qRT‐PCR). The expression level of AaTCP16 was set as 1. AaActin was used as an internal control. The data represent the means ± SD of three replicates from three independent A. annua plants. *P < 0.05, **P < 0.01, Student’s t‐test.
Expression profiles and nuclear localization of AaTCP15
To further investigate AaTCP15 expression, we examined the expression pattern of AaTCP15 in leaves at different positions and in different tissues by qRT‐PCR. The analysis showed that AaTCP15 was broadly expressed in different leaves, with the highest expression in leaf 5 followed by leaf 4, and relatively lower expression in leaf 2, leaf 6, leaf 7 and leaf 9 compared to leaf 1, whereas the expression level of AaTCP15 in leaf 3 and leaf 8 is comparable to leaf 1 (Figure 2a,b). Furthermore, the AaTCP15 transcript was also detected in different A. annua tissues, with the highest expression in trichomes (Figure 2c). To analyse the sites of AaTCP15 expression, we generated AaTCP15 promoter‐GUS fusion (1391Z‐ProTCP15‐GUS) transgenic A. annua plants and tested their GUS activity. Results revealed that GUS activity was observed in mesophyll cells in young leaves, and GUS expression was also observed in the two types of trichomes (TSTs and the two basal cells of GSTs; Figure 2d, iv, v and vi). No GUS activity was detected in A. annua plants transformed with the 1391Z‐GUS empty vector (control plants; Figure 2d, i, ii and iii). These results are in accordance with the previous expression pattern in trichomes (Figures 1c and 2c), indicating that AaTCP15 may function in the trichomes where the specialized metabolites are synthesized and stored.
Figure 2.
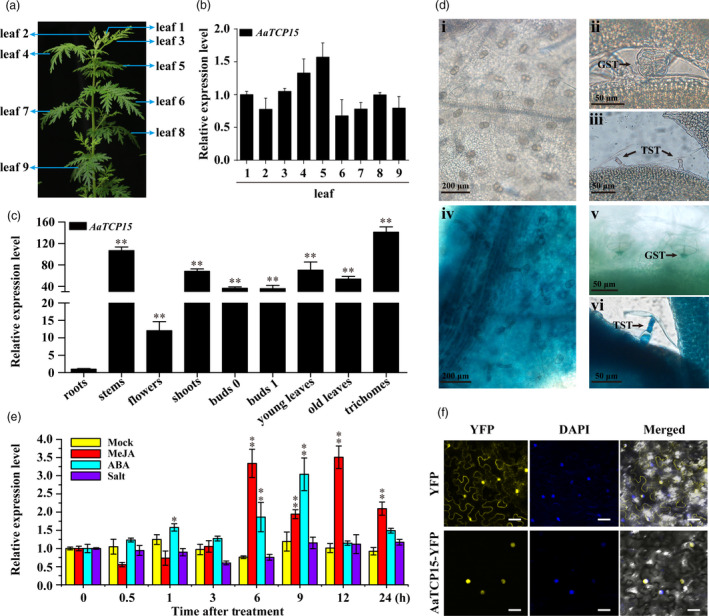
Expression pattern and subcellular localization of AaTCP15. (a) A schematic diagram of the labelled A. annua leaves used in quantitative real‐time PCR (qRT‐PCR) assays in (b). (b) Relative expression levels of AaTCP15 in leaves at different positions. The expression level of AaTCP15 in leaf 1 was set as 1. AaActin was used as an internal control. The data represent the means ± SD of three replicates from three independent A. annua plants. (c) Relative expression levels of AaTCP15 in roots, stems, flowers, shoots, buds (buds 0 and 1), leaves (young leaves and old leaves) and trichomes were measured by qRT‐PCR. The expression level of AaTCP15 in roots was set as 1. AaActin was used as an internal control. The data represent the means ± SD of three replicates from three independent A. annua plants. **P < 0.01, Student’s t‐test. (d) GUS expression (blue staining) in A. annua plants transformed with the 1391Z‐GUS empty vector (control plants) and 1391Z‐proTCP15‐GUS. (i, ii and iii), leaves of control plants. (iv, v and vi), leaves of 1391Z‐proTCP15‐GUS transgenic plants. TST, T‐shape trichome; GST, glandular trichome. Bars represent 200 μm in (i, iv) and 50 μm in (ii, iii, v, vi). (e) Relative expression levels of AaTCP15 in the leaves of A. annua plants treated with mock (0.1% ethanol), MeJA (100 μm), ABA (100 μm) and NaCl (150 mm) at the indicated times. AaActin was used as an internal control. The data represent the means ± SD of three replicates from three independent experiments. *P < 0.05, **P < 0.01, Student’s t‐test. (f) Subcellular localization of AaTCP15 in tobacco leaf epidermal cells. The nucleus was determined by 4′, 6‐diamidino‐2‐phenylindole (DAPI) staining. Yellow fluorescent protein (YFP) was used as a negative control. Three independent transfection experiments were performed. Scale bar = 20 μm.
Numerous reports have shown that methyl jasmonate (MeJA), ABA and salt stress are crucial mediators in AN biosynthesis by activating downstream regulatory genes (Jing et al., 2009; Li et al., 2014; Maes et al., 2011; Paul and Shakya, 2013; Zhou and Memelink, 2016). Hence, we examined whether AaTCP15 was regulated by these treatments. Based on qRT‐PCR analysis, the AaTCP15 transcript was induced by MeJA and ABA, respectively (Figure 2e). It is notable that the AaTCP15 transcript level displayed a delayed increment after prolonged MeJA exposure (6, 9, 12, and 24 h), that peaked at 12 h. In addition, AaTCP15 expression was increased at 1 h after ABA exposure and significantly increased and reached its highest level after 9 h, then returned to the original levels with prolonged ABA exposure (12 and 24 h). By contrast, the AaTCP15 transcript showed no differences under either mock or salt treatment (Figure 2e). These results suggested that AaTCP15 responds to JA and ABA signalling, which may be a potential downstream component of JA and ABA signalling in regulation of AN biosynthesis.
To determine the subcellular localization of AaTCP15, we transiently expressed an AaTCP15‐YFP (yellow fluorescent protein) fusion protein under the control of cauliflower mosaic virus (CaMV) 35S promoter in Nicotiana benthamiana leaf cells. As shown in Figure 2f, the recombinant AaTCP15‐YFP fusion protein was specifically localized to the nucleus in N. benthamiana leaf cells. This result showed that AaTCP15 is a nuclear‐localized protein, consistent with the role of AaTCP15 as a TF.
AaTCP15 enhances AN biosynthesis and is essential for JA‐ and ABA‐mediated AN biosynthesis in A. annua
To assess the biological role of AaTCP15 in controlling AN biosynthesis, stable AaTCP15‐overexpression (OE‐AaTCP15) and antisense (Anti‐AaTCP15) transgenic A. annua lines were generated. After testing the levels of AaTCP15 mRNA in these transgenic lines, we selected three independent overexpression lines (designated as OE‐AaTCP15‐2, 4, 9) or three antisense lines (designated as Anti‐AaTCP15‐6, 12, 29) for further characterization (Figure 3a,d). Results showed that, compared to the wild‐type (WT) and Vector controls (A. annua plants transformed with the empty vector), the expression levels of AN biosynthetic genes (ADS, CYP71AV1, DBR2 and ALDH1) and AN content followed the change of AaTCP15 expression, in that they were significantly up‐regulated in OE‐AaTCP15 (Figure 3a–c) and down‐regulated in Anti‐AaTCP15 A. annua lines (Figure 3d–f), suggesting a positive role of AaTCP15 in AN biosynthesis. However, we found that the dihydroartemisinic acid (DHAA) content was decreased in OE‐AaTCP15 and increased in anti‐AaTCP15 transgenic A. annua lines, compared to Vector controls (Figure S4a,c). This phenomenon may be attributed to the role of AaTCP15 in regulating some potential as‐yet unknown proteins involved in affecting the photo‐oxidation process from DHAA to AN. Moreover, no obvious morphological differences were observed in either the OE‐AaTCP15 or anti‐AaTCP15 transgenic plants compared with Vector controls (Figure S4b,d). Collectively, these results indicated that AaTCP15 enhanced AN biosynthesis through activating the AN biosynthetic genes.
Figure 3.
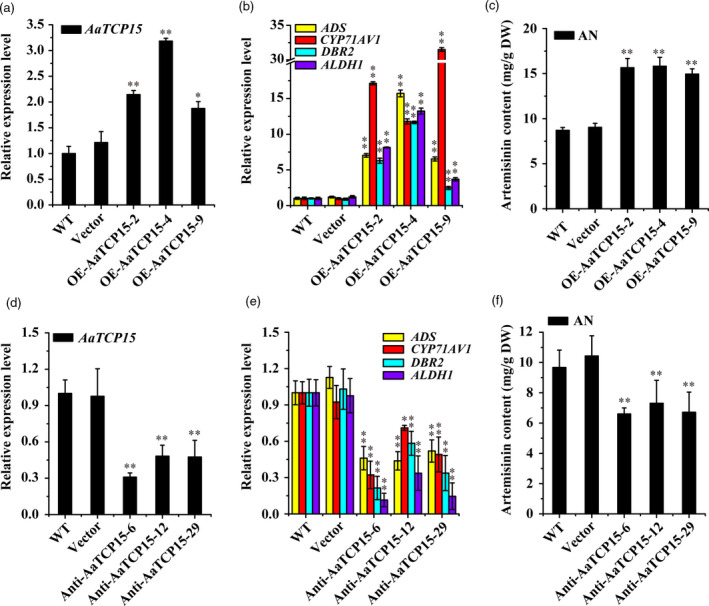
AaTCP15 transgenic plants modulate artemisinin biosynthesis. (a, d) Expression levels of AaTCP15 in the leaves of different A. annua AaTCP15‐overexpression (OE‐AaTCP15) (a) and AaTCP15‐antisense (Anti‐AaTCP15) lines (d), plants transformed with the empty vector (control plants, labelled as Vector) and wild‐type (WT) plants. AaActin was used as the internal standard. (b, e) Expression levels of ADS, CYP71AV1, DBR2 and ALDH1 in the leaves of different A. annua OE‐AaTCP15 (b) and Anti‐AaTCP15 lines (e), Vector control and WT plants. AaActin was used as the internal control. (c, f) HPLC analysis of artemisinin (AN) content in the leaves of different A. annua OE‐AaTCP15 (c) and Anti‐AaTCP15 lines (f), Vector control and WT plants. All data represent the means ± SD of three replicates from three cutting propagations. *P < 0.05, **P < 0.01, Student’s t‐test.
Subsequently, the function of AaTCP15 in JA‐ and ABA‐mediated AN biosynthesis was investigated. As expected, application of exogenous MeJA and ABA treatments increased AN accumulation significantly in the WT and Vector samples compared to Mock treatment (Figure S5), in accordance with a prior report (Maes et al., 2011). In addition, suppression of AaTCP15 significantly inhibited JA or ABA‐induced AN accumulation compared to the WT and Vector controls (Figure S5), further supporting that AaTCP15 was essential for JA‐ and ABA‐mediated AN accumulation in A. annua.
AaTCP15 enhances the transcription of both DBR2 and ALDH1 by binding to their promoters
To unravel the mechanism of how AaTCP15 regulated the expression of AN biosynthetic genes, we first carried out transient dual‐luciferase (Dual‐LUC) assays. The reporters (ADSpro/CYP71AV1pro/DBR2pro/ALDH1pro:LUC) along with the effectors (35S:GFP or 35S:AaTCP15, see Figure 4a) were transiently co‐expressed in the N. benthamiana leaf cells. As shown in Figure 4b, AaTCP15 only significantly activated the DBR2 or ALDH1 promoter compared to the GFP control (Figure 4b).
Figure 4.
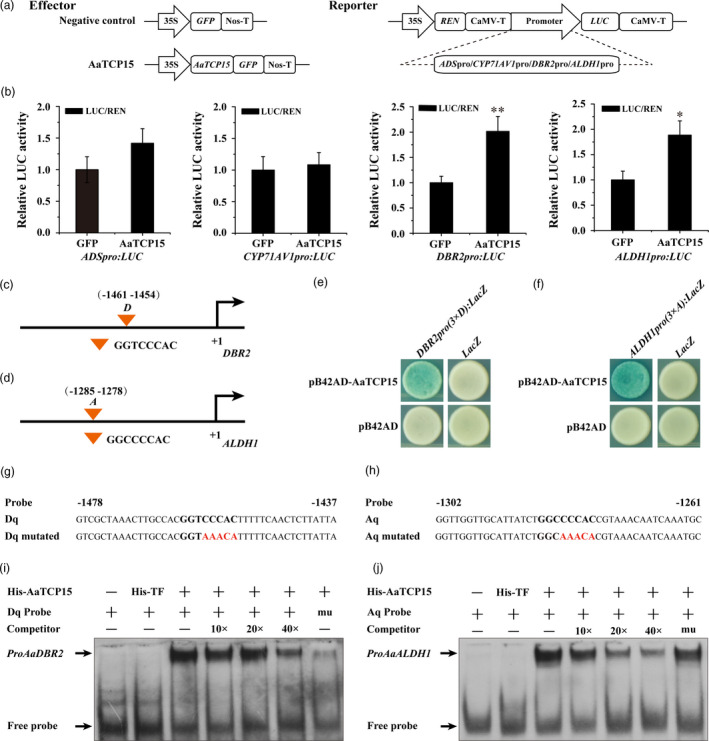
AaTCP15 is a transcriptional activator of DBR2 and ALDH1. (a) Schematic diagrams of the effector and reporter plasmids used in Dual‐LUC assays. REN, Renilla luciferase. LUC, firefly luciferase. (b) Dual‐LUC assay in N. benthamiana leaf cells using the constructs shown in (a). The GFP effector was used as a negative control, and the LUC/REN ratios of GFP were set as 1. Three independent transfection experiments were performed. The data represent the means ± SD of three replicates from three independent experiments. *P < 0.05, **P < 0.01, Student’s t‐test. (c, d) Schematic diagrams of the DBR2 (c) and ALDH1 (d) promoters. The positions of potential TBS (TCP binding site) DNA binding sites (D in DBR2 promoter and A in ALDH1 promoter) are shown as orange triangles and are numbered based on their distance from the translational start site (ATG), which is set as +1. (e, f) Y1H assays showing that AaTCP15 binds to the TBS motifs of DBR2 and ALDH1. Three tandem repeats of D or A motifs were used as baits. Transformed yeast cells were grown on selective medium SD/‐Trp/‐Ura containing 20 mg/L X‐gal, and pictures were taken after 4 days of incubation at 30 °C. Blue plaques indicate protein‐DNA interactions. The Y1H assays were repeated three times, and representative results are shown. (g, h) The sequences of wild‐type (Dq or Aq) and mutated probes (Dq mutated or Aq mutated) used for EMSAs corresponding to DBR2 or ALDH1 promoter. Class I TCPs binding motifs are shown in bold, and the mutated nucleotides are indicated by red. (i, j) EMSA assays showing that AaTCP15 binds to the Dq motif from DBR2 and the Aq motif from ALDH1. Unlabelled Dq, Aq and mutated Aq probes were used as cold competitors, and labelled mutated Dq probe was tested as a control. 10×, 20×, 40× indicate the fold excess of cold competitor relative to that of labelled probe. His‐TF protein was used as a negative control.
In plants, TCP TFs regulate their target genes by binding to the conserved promoter motifs, namely TBSs (Aggarwal et al., 2010). Our previous research uncovered that AaTCP14 could bind to DBR2 and ALDH1 promoters and activate their expression (Ma et al., 2018), and bioinformatics analysis revealed the presence of TBSs including D motif or A motif in the DBR2 or ALDH1 promoter (Figure 4c,d). Thus, we speculated that AaTCP15, which is closely homologous with AaTCP14 based on a phylogenetic analysis (Figure 1a), may also directly bind to the DBR2 or ALDH1 promoter. Yeast one‐hybrid assay showed that binding of the pB42AD‐AaTCP15 fusion protein, rather than pB42AD alone, to three tandem repeats of the D motif from DBR2 promoter or A motif from ALDH1 promoter, strongly activated the expression of the LacZ reporter gene (Figure 4e,f), indicating that AaTCP15 binds to the D or A motifs in the DBR2 or ALDH1 promoters, respectively.
Next, electrophoretic mobility shift assays (EMSAs) were conducted to verify AaTCP15 binding to DBR2 and ALDH1 promoters using His‐AaTCP15 fusion protein or His‐TF (trigger factor) and DBR2 or ALDH1 promoter probe (normal Dq or Aq probe, and mutated Dq or Aq probe, see Figure 4g,h). As shown in Figure 4i,j, a single shift band was detected in the presence of both His‐AaTCP15 and labelled Dq or Aq probe containing the D or A motif (Figure 4g,h), rather than the combination of His‐TF and labelled Dq or Aq probe; the intensity of this band decreased with increasing concentrations of cold competitor (unlabelled Dq or Aq probe; Figure 4i,j). Moreover, the His‐AaTCP15 protein was unable to bind to the labelled mutated Dq probe referred to as the DBR2 promoter (Figure 4i). Similarly, using the excess unlabelled mutated Aq probe as a cold competitor could not affect the binding of His‐AaTCP15 to the labelled Aq probe in the ALDH1 promoter (Figure 4j). Together, these results strongly supported the notion that AaTCP15 activated DBR2 and ALDH1 expression via specifically binding to the TBSs element (D or A motif) in their promoters.
AaTCP15 directly interacts with AaORA and synergistically activates DBR2 transcription by protein‐protein interaction
In Arabidopsis, homologous TCP proteins may act as common components of the same regulatory pathway (Peng et al., 2015). We previously reported that AaTCP14 together with AaORA formed a complex to regulate AN biosynthesis by their interaction (Ma et al., 2018). Therefore, we hypothesized that AaTCP15 might also interact with AaORA. Indeed, in bimolecular fluorescence complementation (BiFC) assays, the AaTCP15 or AaORA fused with the N‐ or C‐terminus of YFP were transiently co‐expressed in N. benthamiana leaf cells by infiltration. The reconstituted YFP fluorescence signal was obviously observed in the nucleus, and merged with the signal of DAPI, a nuclear stain, supporting the interaction between AaTCP15 and AaORA (Figure 5a). Next, the interaction of AaTCP15 with AaORA was further corroborated by a LUC complementation experiment. When Cluc‐AaTCP15 and AaORA‐Nluc fusion proteins were co‐expressed in N. benthamiana leaf cells, strong relative LUC activity was detected, whereas those expressing Cluc‐AaTCP15 or AaORA‐Nluc alone showed low LUC activity (Figure 5b). Taken together, these results suggest that AaTCP15 interacts with AaORA in plant cells.
Figure 5.
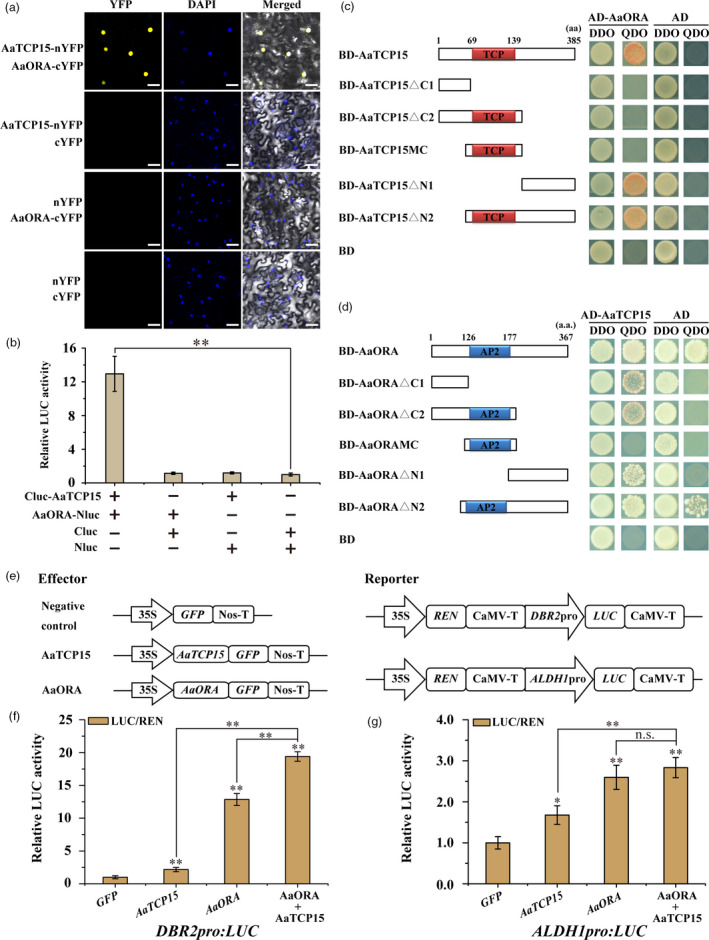
AaORA interacts with and enhances the transactivation activity of AaTCP15 on DBR2 promoter. (a) Bimolecular fluorescence complementation (BiFC) analysis of the interaction between AaTCP15 and AaORA in N. benthamiana leaf cells. AaTCP15 was fused to the N‐terminal fragment of YFP (AaTCP15‐nYFP), and AaORA was fused to the C‐terminal fragment of YFP (AaORA‐cYFP). The nucleus was indicated by DAPI staining. Three independent transfection experiments were performed. Yellow fluorescence was detected using a confocal laser‐scanning microscope. Scale bar = 20 μm. (b) LUC complementation assay to detect the interaction between AaTCP15 and AaORA. AaTCP15 was fused to the C‐terminal fragment of LUC (Cluc‐AaTCP15), and AaORA was fused to the N‐terminal fragment of LUC (AaORA‐Nluc). LUC activity of Nluc and Cluc was set to 1. Three independent transfection experiments were performed. The data represent the means ± SD of 3 independent experiments. **P < 0.01, Student’s t‐test. (c) Y2H assays showing the interactions between AaORA and truncated versions of AaTCP15. Left, schematic representations of the truncated AaTCP15 protein used in this experiment. Numbers indicate the amino acid positions of the truncated AaTCP15 variants. The TCP domains are indicated by red boxes. Right, Y2H assays of protein interactions between AD‐AaORA and truncated versions of BD‐AaTCP15. (d) Y2H assays showing the interactions between AaTCP15 and truncated versions of AaORA. Left, schematic representations of the truncated AaORA protein used in this experiment. Numbers indicate the amino acid positions of the truncated AaORA variants. The AP2 domains are indicated by blue boxes. Right, Y2H assays of protein interactions between AD‐AaTCP15 and truncated versions of BD‐AaORA. The data represent three independent experiments, and representative results are shown. (e) A schematic representation of the constructs used in Dual‐LUC assays. (f, g) Activation of the DBR2pro:LUC (f) and ALDH1pro:LUC (g) by indicated combinations of AaORA and AaTCP15 proteins in N. benthamiana leaf cells, respectively. The GFP effector served as a negative control, and the LUC/REN ratios of GFP were set as 1. Three independent transfection experiments were performed. The reporter strain harbouring DBR2pro:LUC or ALDH1pro:LUC was mixed with the effector strains harbouring 35Spro:AaTCP15 and 35Spro:AaORA at a ratio of 1 : 1 or 1 : 1 : 1. The data represent the means ± SD of three replicates from three independent experiments. *P < 0.05, **P < 0.01, Student’s t‐test, n.s., not significant.
To further identify which regions of AaTCP15 or AaORA may be responsible for their interactions, we generated yeast two‐hybrid (Y2H) constructs in which the different truncated AaTCP15 or AaORA variants were fused with the BD (binding domain) or AD (activating domain) of the pGBKT7 or pGADT7 vector, respectively. Consistent with the above results, AaTCP15 interacted with AaORA (Figure 5c). Furthermore, domain deletion analysis revealed that the C‐terminal region of AaTCP15 (AaTCP15ΔN1) alone was able to interact with AaORA, but the truncated versions of AaTCP15 that lacked the C‐terminal domain (AaTCP15ΔC1 and AaTCP15ΔC2) or TCP domain (AaTCP15MC) could not (Figure 5c). Additionally, we found that either the N‐terminal (AaORAΔC1) or C‐terminal (AaORAΔN1) domain of AaORA alone was able to interact with AaTCP15 (Figure 5d). Thus, these results suggested that the C‐terminal region of AaTCP15 and both of the N‐ and C‐terminal regions of AaORA are important for the AaTCP15‐AaORA interaction.
It was previously shown that the transactivation activity of TCP proteins was modulated by their interacting‐partner (Ma et al., 2018; Sun et al., 2019). Here, AaTCP15 and AaORA were involved in positively regulating the transcripts of AN biosynthetic genes. Thus, we next examined how the interaction between AaTCP15 and AaORA affected the transactivation activity of AaTCP15 on its target genes. We performed Dual‐LUC assays in N. benthamiana leaves using the effector constructs (35S:GFP, 35S:AaTCP15, and 35S:AaORA) and the reporter plasmid (DBR2pro:LUC or ALDH1pro:LUC, see Figure 5e). The co‐expression of AaTCP15 or AaORA alone with the reporter plasmid significantly increased LUC expression driven by DBR2 or ALDH1 promoters compared to the GFP control (Figure 5f,g). Most importantly, the LUC expression was further elevated when AaTCP15 and AaORA were simultaneously co‐expressed compared to those of AaTCP15 or AaORA expressed alone (Figure 5f,g). The activity of DBR2 promoter, rather than ALDH1 promoter, which was enhanced by co‐expressing AaTCP15 and AaORA, reached a significant level (P < 0.01) compared to AaTCP15 or AaORA expressed alone (Figure 5f,g). Taken together, these results suggested that AaORA cooperatively enhanced the transactivation activity of AaTCP15 in preferentially regulating DBR2 expression by their interaction.
The JA‐ and ABA‐responsive TF AaGSW1 directly activates AaTCP15 expression to regulate AN biosynthesis
Our current report demonstrated that the AaTCP15 transcript is induced after JA or ABA treatment (Figure 2e), and the suppression of AaTCP15 expression significantly reduced AN content and attenuated the JA‐ or ABA‐induced AN accumulation (Figures 3 and S5). These observations supported that AaTCP15 is a key positive regulator in AN biosynthesis, and JA and ABA promote AN biosynthesis by activating downstream AaTCP15 expression in A. annua. To better identify the upstream regulators that link JA or ABA signalling and lead to the activation of AaTCP15, we first analysed the cis‐acting regulatory elements in the promoter of AaTCP15 using PlantCARE tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Apart from the common light, hormonal (i.e. ABA and MeJA) and abiotic stress responsiveness elements (Figure S6), two or one conserved W‐box motif known to be bound by WRKY TFs (Chen et al., 2017) were also found in AaTCP15 or its homologous gene AaTCP14 promoter, respectively (Figure 6a). This suggested that AaTCP15 or AaTCP14 might be regulated by WRKY family genes.
Figure 6.
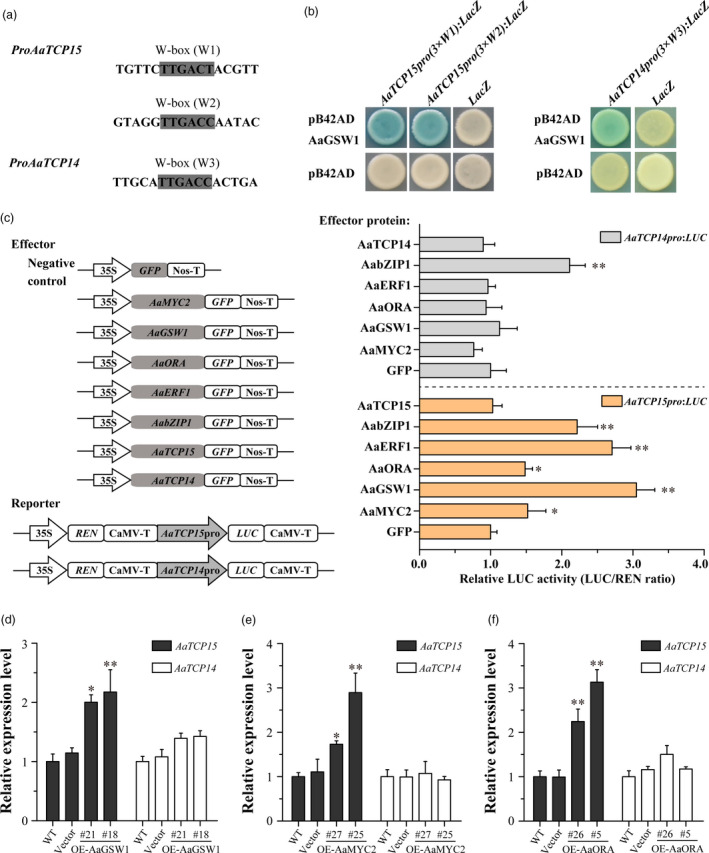
AaGSW1 directly and positively regulates the expression of AaTCP15 rather than AaTCP14. (a) The fragments of AaTCP15 and AaTCP14 promoters containing the intact W‐box. The W‐box motif sequences of W1, W2 and W3 are shown as grey boxes. (b) Yeast one‐hybrid (Y1H) assays showing that AaGSW1 binds to the W1 and W2 motif of AaTCP15 promoter, and W3 motif of the AaTCP14 promoter. Three tandem repeats of W1, W2 and W3 motifs were used as baits. Transformed yeast cells were grown on selective medium SD/‐Trp/‐Ura containing 20 mg/L X‐gal, and pictures were taken after 4 days of incubation at 30 °C. Blue plaques indicate protein‐DNA interactions. The Y1H assays were repeated three times, and representative results are shown. (c) Left, schematic diagrams of the effector and reporter plasmids used in Dual‐LUC assays. REN, Renilla luciferase. LUC, firefly luciferase. Right, Dual‐LUC assay in N. benthamiana leaf cells using the constructs shown at Left. The GFP effector was used as a negative control, and the LUC/REN ratios of GFP were set as 1. Three independent transfection experiments were performed. The data represent the means ± SD of three replicates from three independent experiments. *P < 0.05, **P < 0.01, Student’s t‐test. (d‐f) Expression levels of AaTCP15 and AaTCP14 in the leaves of different A. annua AaGSW1 (d), AaMYC2 (e) and AaORA (f) overexpression lines, and plants transformed with the empty vector (labelled as Vector) and WT. AaActin was used as the internal control. The data represent the means ± SD of three replicates from three cutting propagations. *P < 0.05, **P < 0.01, Student’s t‐test.
In this context, we tested whether the earlier reported JA and ABA dual‐responsive WRKY TF AaGSW1 (Chen et al., 2017), which acted at the nexus of JA and ABA signalling to positively regulate JA‐ and ABA‐induced AN biosynthesis, could bind to AaTCP15 or AaTCP14 promoters through Y1H assays. Results found that AaGSW1 directly bound to the W1 and W2 motifs in the AaTCP15 promoter or W3 motif in the AaTCP14 promoter (Figure 6a,b). Then, we investigated whether AaGSW1 could activate AaTCP15/14 expression by employing Dual‐LUC assays in N. benthamiana leaves and found that AaGSW1 significantly enhanced AaTCP15 but not AaTCP14 promoter activity (Figure 6c). To further verify this finding, we screened two independent AaGSW1 transgenic lines (OE‐AaGSW1‐18, 21) in which the expression of AaGSW1 and the AN content were significantly increased compared to Vector controls (Figure S7a,d). Expression of AaTCP15 rather than AaTCP14 was significantly increased in AaGSW1 transgenic lines (Figure 6d), which was in accordance with the Dual‐LUC assays (Figure 6c). These results revealed that JA and ABA promoted AaTCP15 but not AaTCP14 expression directly by JA and ABA dual‐responsive TF AaGSW1, and AaGSW1 along with AaTCP15 may form a JA and ABA stepwise responsive AaGSW1‐AaTCP15 transcriptional regulatory cascade to control AN biosynthesis.
In addition, our Dual‐LUC assays also showed that several JA‐responsive TFs, AaMYC2, AaORA, AaERF1 and ABA‐responsive TF AabZIP1, which positively promote AN biosynthesis by JA and ABA, enhanced the AaTCP15 promoter activity, whereas AaTCP15 itself had a negligible effect on its own promoter activity (Figure 6c). In a parallel assay, we found that although AaTCP14 is homologous with AaTCP15 (Figure 1a), only AabZIP1 but not AaMYC2, AaORA, AaERF1 or AaTCP14 itself could enhance the AaTCP14 promoter activity (Figure 6c). This finding was consistent with the result that expression of AaTCP14 was induced under ABA treatment (Figure S8), implying that JA and ABA signalling may regulate AaTCP15 or AaTCP14 expression through a distinct or partially similar upstream regulator in A. annua. Additionally, in accordance with the above results (Figure 6c), we found that the AaTCP15, but not the AaTCP14, transcript was significantly up‐regulated in AaMYC2 (OE‐AaMYC2‐27, 25) or AaORA (OE‐AaORA‐26, 5) overexpressed A. annua lines (Figures 6e,f and S7b,c), in which the AN content is significantly higher compared to WT or Vector controls (Figure S7e,f). Taken together, these results implied that apart from the JA and ABA dual‐responsive TF AaGSW1, the multiple JA or ABA‐responsive TFs could also activate AaTCP15 expression to control AN content in A. annua.
Discussion
Artemisinin (AN) is a sesquiterpene lactone endoperoxide derived from A. annua. AN is not only effective against malaria, but also has great application potential in treating lupus‐related nephritis, viral infections, schistosomiasis, tuberculosis, cancer and diabetes (Crespo‐Ortiz and Wei, 2012; Efferth et al., 2008; Li et al., 2017, 2006; Liu et al., 2011; Tin et al., 2012; Zheng et al., 2017). It has been recently reported that AN biosynthesis is controlled by external stimuli. Of these, JA and ABA have attracted extensive attention due to their vital roles in promoting AN biosynthesis (Jing et al., 2009; Maes et al., 2011). Notably, although previous studies have demonstrated that JA and ABA governed AN through activating downstream TFs including WRKY, bZIP, bHLH and ERF (Chen et al., 2017; Ji et al., 2014; Shen et al., 2016; Yu et al., 2012; Zhang et al., 2015), the specific molecular mechanism linking JA and ABA signalling with AN biosynthesis by activating the downstream TFs associated regulatory network as‐yet remains enigmatic. In this study, we found that AaTCP15 had an important role in positively promoting JA and ABA‐induced AN biosynthesis. Further, AaORA, an activator of AN biosynthesis, enhanced the transactivation activity of AaTCP15 on its target gene by direct interaction with AaTCP15, and simultaneously activated AaTCP15 expression. In addition, the AaTCP15 transcript was activated by JA and ABA through employing multiple downstream TFs involved in the JA‐ or ABA‐governed AN biosynthesis process. Of these, AaGSW1, a positive regulator that acts at the nexus of JA and ABA signalling to activate AN biosynthesis, activated AaTCP15 expression by directly binding to its promoter. Therefore, our study proposes an AaGSW1‐AaTCP15/AaORA transcriptional regulatory module that positively regulates AN biosynthesis through simultaneously responding to JA and ABA signalling in A. annua (Figure 7).
Figure 7.
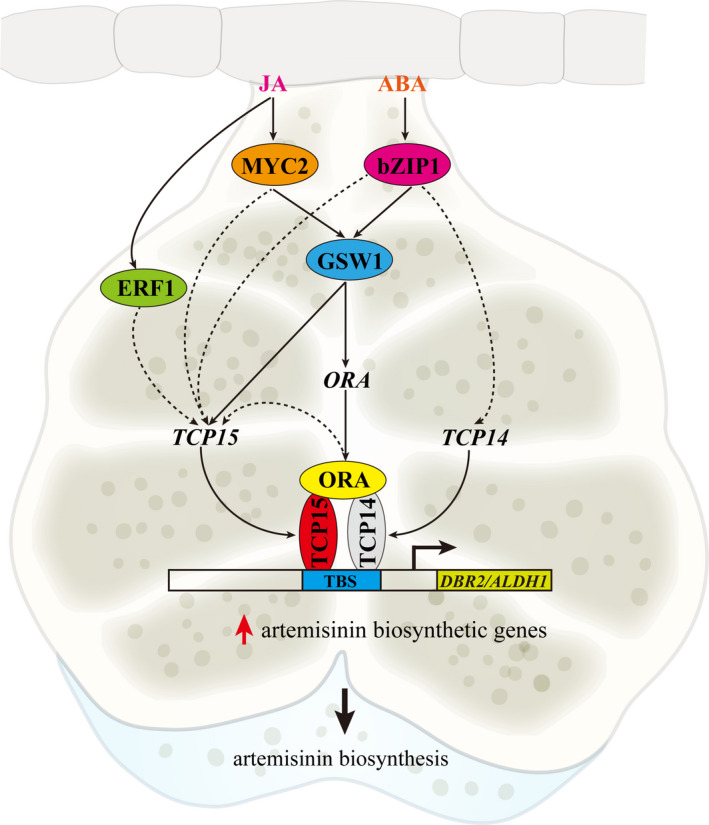
Proposed working model depicting how the AaTCP15‐related module regulates artemisinin biosynthesis in response to JA and ABA signalling. AaMYC2, AaERF1 and AabZIP1 transcription factors are positive regulators of artemisinin (AN) biosynthesis, which are positively activated by JA or ABA (Shen et al., 2016; Yu et al., 2012; Zhang et al., 2015). AaMYC2 and AabZIP1 directly activate the expression of their common target gene AaGSW1, a positive regulator of AN biosynthesis through linking JA and ABA signalling (Chen et al., 2017). AaGSW1 binds to and activates the AaTCP15 promoter except for the reported AaORA promoter, which may facilitate the formation of AaTCP15‐AaORA regulatory module by their interaction. In addition, AaMYC2, AabZIP1 and AaERF1 may be also involved in activating AaTCP15 expression. AaTCP15 enhances AN biosynthesis through directly binding to and activating the target genes DBR2 and ALDH1 promoters. AaORA interacts with AaTCP15 to enhance its transactivation activity on the AaTCP15 target gene and also promotes AaTCP15 expression, thereby leading to the synergistic activation of AN biosynthetic genes, and finally promoting AN biosynthesis through responding to upstream JA and ABA signalling. AabZIP1 is also involved in activating AaTCP14 expression, and the complex of AaTCP15‐AaORA and AaTCP14‐AaORA (Ma et al., 2018) may be involved in a common module to regulate artemisinin content. TBS, TCP binding site. Solid arrow, direct regulation; Red arrow, up‐regulation. Dashed line arrow, hypothetical direct links. The background image is the model of a glandular trichome of A. annua.
Up till now, only a few studies reported the upstream regulatory pathway of class II TCP factors. For instance, the C2H2 zinc finger gene RABBIT EARS (RBE) repressed the transcription of TCP4 during Arabidopsis petal development (Li et al., 2016). In Arabidopsis, miR319 targeted TCP2, TCP3, TCP4, TCP10 and TCP24 regulated leaf morphogenesis (Palatnik et al., 2003). However, upstream regulators of the class I TCP family are still unknown. Strikingly, we found that the expression level of AaTCP15 was induced by both JA and ABA treatments at later time points (Figure 2e), and AaTCP15‐antisense A. annua (Anti‐AaTCP15) had lower JA‐ or ABA‐induced AN accumulation (Figure S5). These results suggest that there might be a novel mechanism upstream of AaTCP15 involved in JA and ABA‐mediated AN biosynthesis. JA and ABA‐responsive AaMYC2, AaGSW1, AaERF1 and AabZIP1 are reported to positively regulate AN biosynthesis through activating structural gene promoters (Chen et al., 2017; Shen et al., 2016; Yu et al., 2012; Zhang et al., 2015). Interestingly, we found that these TFs all activated AaTCP15 promoter, and they might form a novel AaMYC2/AabZIP1‐AaGSW1‐AaTCP15 transcriptional cascade (Figure 7). It has been reported that AaORA activated all four structural genes (ADS, CYP71AV1, DBR2 and ALDH1) to modulate AN biosynthesis (Lu et al., 2013; Ma et al., 2018), and we found that AaORA modulated AN biosynthesis through AaTCP15 at multiple layers. On the one hand, AaORA increased the transcriptional activation activity of AaTCP15 on DBR2 promoter through forming an AaORA‐AaTCP15 complex at the protein level. On the other hand, AaORA activated the expression of AaTCP15 at the transcriptional level. Thus, AaORA is a key regulator in AN biosynthesis through various mechanisms. By contrast, JA‐responsive transcription factors AaMYC2, AaGSW1, AaERF1 and AaORA could not activate AaTCP14 promoter (Figure 6c). Although AaGSW1 could bind to AaTCP14 promoter in the Y1H assay (Figure 6b), it could not activate the expression of AaTCP14 (Figure 6c), suggesting that AaGSW1 may need another activator to coordinately modulate AaTCP14 expression. Hence, the AaGSW1‐AaTCP15/AaORA transcriptional cascade is specific to AaTCP15 but not AaTCP14, suggesting different molecular mechanisms between AaTCP15 and AaTCP14 in regulation of AN biosynthesis. These results broaden the network of JA and ABA regulation of specialized metabolites in plants. In addition, previous reports indicated that co‐expression of multiple genes than single gene in AN biosynthesis by metabolic engineering is an effective approach to increase AN content (Chen et al., 2013; Shi et al., 2017). In view of the components of AaGSW1‐AaTCP15/AaORA regulatory cascade could control the expression of different AN biosynthetic genes, co‐overexpression of AaGSW1, AaTCP15 and AaORA may be a potential and promising strategy to elevate the production of AN in the future in engineered A. annua plants.
AaTCP15 and AaTCP14 are homologous genes that belong to the class I TCP family, which can work synergistically or function alone. TCP15 and TCP14 interacted with SPINDLY to facilitate cytokinin responses in leaves and flowers in Arabidopsis (Steiner et al., 2012a), and they co‐regulated cytokinin‐induced branching and meristematic activity in tomato (Steiner et al., 2012b). Moreover, in Arabidopsis, TCP15 and TCP14 modulated internode length and leaf shape via cell division (Kieffer et al., 2011), regulated gynoecium development through balancing auxin and cytokinin responses (Lucero et al., 2015) and controlled endoreduplication together with DA1, DAR1 and DAR2 (Peng et al., 2015). Likewise, we found that AaTCP15 directly bound and activated DBR2 and ALDH1 promoters, similar to AaTCP14. Although AaTCP15 and AaTCP14 were both expressed in GSTs and TSTs, they showed different expression pattern in GSTs. AaTCP15 showed strong expression at the end of two cells of GST (Figure 2d), while AaTCP14 was expressed in all ten cells of GST (Ma et al., 2018), indicating that they may have different regulatory mechanisms in AN biosynthesis.
Interestingly, in OE‐AaTCP15 transgenic A. annua, the AN content increased but DHAA content decreased (Figures 3 and S4), whereas in OE‐AaTCP14 transgenic A. annua, both AN and DHAA content increased (Ma et al., 2018). This indicates that in OE‐AaTCP15 transgenic A. annua, the metabolic flux from artemisinic aldehyde to AN may remain constant, and the increased AN may be due to the enhanced efficiency from DHAA to AN, the last step in AN biosynthesis. However, in OE‐AaTCP14 transgenic A. annua, the metabolic flux from artemisinic aldehyde to AN may increase, thus increasing both AN and DHAA content. DHAA is considered the direct substrate of AN (Zhu et al., 2014) and is then converted to AN through an oxidation reaction, a process that requires species specific enzymes to obtain specialized metabolites. In rice, the short‐chain alcohol dehydrogenase/reductases including OsSDR110C‐MS2 and OsSDR110C‐MS1 (OsMAS) catalysed the final oxidation reaction in the biosynthesis of rice diterpenoid, momilactone A (Murphy and Zerbe, 2020). Moreover, in Mentha spicata, carveol dehydrogenase (CDH) catalysed the final oxidation reaction in the biosynthesis of carvone (Ahkami et al., 2015). The last step of AN biosynthesis is still mysterious due to the unidentified enzyme responsible for the conversion from DHAA to AN, thus AaTCP15 might be involved in the activation of the unknown enzyme activity, which finally leads to decreased or increased DHAA content in OE‐AaTCP15 or in Anti‐AaTCP15 transgenic A. annua, respectively. Correspondingly, elevated activity of this unknown enzyme might also be one of the reasons for the AN increase in OE‐AaTCP15 or decrease in Anti‐AaTCP15 transgenic A. annua.
Moreover, like AaTCP14, AaTCP15 also formed the AaTCP15‐AaORA complex by interaction (Figure 5), but unlike AaTCP14, which only interacted with the C‐terminus of AaORA (AaORAΔN1; Ma et al., 2018), AaTCP15 interacted with both the C‐terminus (AaORAΔN1) and N‐terminus of AaORA (AaORAΔC1; Figure 5d). Because of these different interaction regions, AaORA might have different synergistic activation effects between AaTCP15 and AaTCP14: the AaTCP15‐AaORA complex can only synergistically activate DBR2 promoter (Figure 5), but the AaTCP14‐AaORA complex can synergistically activate both DBR2 and ALDH1 promoters (Ma et al., 2018). Taken together, AaTCP15 and AaTCP14 may have some redundancy in regulating AN biosynthesis, but they have predominantly independent roles in AN biosynthesis. Therefore, it is of great significance to unravel the distinct roles of other TCP genes to modulate AN biosynthesis.
In conclusion, we found that AaTCP15 activates AN biosynthesis through JA and ABA signalling pathways; hence, AaTCP15 is a key point in these two pathways to modulate AN biosynthesis. In this study, the molecular network for JA and ABA regulation of AN biosynthesis was expanded, laying a firm foundation for cultivating varieties with high AN content by using transcriptional strategies and providing a reference for the regulation of specialized metabolites in other plants.
Methods
Plant materials and treatments
The high AN content A. annua, ‘Huhao 1’, which comes from Chongqing and was selected for several years in Shanghai, was used for all A. annua related assays (Shen et al., 2016). A. annua and N. benthamiana plants were grown in light chambers at 23 ± 2 °C under a 16‐h light/8‐h dark photoperiod.
For MeJA, ABA and salt treatments, 10‐day‐old A. annua seedlings were sprayed with 100 μm MeJA (Sigma, St. Louis, MO, USA), 100 μm ABA (Sigma) and 150 mm NaCl (Sigma). For the mock treatment, A. annua seedlings were sprayed with 0.1% ethanol. The leaves of A. annua seedlings in the same position were harvested at 0, 0.5, 1, 3, 6, 9, 12 and 24 h after treatment, respectively. To analyse AN content in AaTCP15‐antisense (Anti‐AaTCP15) plants after MeJA or ABA treatment, 2‐month‐old cutting propagations of Anti‐AaTCP15, wild‐type (WT) and Vector controls (A. annua plants transformed with the empty vector, labelled as Vector) were sprayed with 100 μm MeJA, 50 μm ABA or 0.05% ethanol (Mock treatment) and sampled at 72 h for AN extraction.
RNA extraction and quantitative real‐time PCR (qRT‐PCR)
Different leaves (leaf 1, leaf 2, leaf 3, leaf 4, leaf 5, leaf 6, leaf 7, leaf 8 and leaf 9, labelled in Figure 2a) of 3‐month‐old A. annua and different tissues (roots, stems, flowers, shoots, buds 0, buds 1, young leaves, old leaves and trichomes) of 4‐month‐old A. annua were collected to analyse the expression of the indicated genes. To detect the expression of AaTCP15, AaADS, AaCYP71AV1, AaDBR2 and AaALDH1 in AaTCP15 overexpression and antisense lines, and the expression of AaTCP15 or AaTCP14 in AaMYC2, AaGSW1 or AaORA overexpression lines, the leaves of 3‐month‐old WT plants, Vector controls and the aforementioned transgenic plants were collected. Total RNA of the above samples was extracted using the RNAprep pure Plant Kit (TianGen Biotech,Shanghai, China). cDNA was synthesized from 1.0 μg total RNA using the PrimeScript 1st Strand cDNA Synthesis Kit (Takara, Japan) according to the manufacturer’s instructions. The qRT‐PCR analysis was performed as previously reported (Lu et al., 2013) using the cDNA templates gained from the above indicated samples. All PCR reactions were conducted three times in independent experiments. The qRT‐PCR results were calculated by normalization to AaActin. Primers are listed in Table S1.
β‐glucuronidase (GUS) expression in 1391Z‐proTCP15‐GUS transgenic A. annua plants
The 2104 bp upstream region of the start codon of AaTCP15 was amplified with specific primers (Table S1) from the A. annua genomic DNA library and inserted into the pCAMBIA‐1391Z vector to construct 1391Z‐proTCP15‐GUS recombinant vector. Subsequently, the Agrobacterium tumefaciens strain EHA105 harbouring 1391Z‐proTCP15‐GUS or 1391Z‐GUS plasmids were introduced to A. annua leaves to obtain transgenic A. annua plants as previously reported (Zhang et al., 2009). Histochemical staining for GUS activity in transgenic plants was conducted as previously described (Ma et al., 2018). A. annua plants transformed with 1391Z‐GUS empty vector (control plants) were processed in parallel as negative controls.
Subcellular localization of AaTCP15
The GV3101 strains harbouring pHB‐AaTCP15‐YFP or pHB‐YFP vectors were transformed into 5‐week‐old N. benthamiana leaves for subcellular localization experiments. YFP fluorescence was observed at 60–72 h after infiltration by confocal laser microscopy (Leica TCS SP5‐II). Nuclei were stained with 4′, 6‐diamidino‐2‐phenylindole (DAPI, Sigma). Three biological repeats were performed to verify these results.
Plant transformation and phenotype analysis
The overexpression constructs pCAMBIA1300‐AaTCP15‐GFP, pHB‐AaMYC2 (Shen et al., 2016), pCYP‐AaGSW1 (Chen et al., 2017), pCAMBIA1300‐AaORA‐GFP, and the antisense construct pCAMBIA1300‐Anti‐AaTCP15 were transferred into A. tumefaciens strain EHA105 and then used to transform A. annua as previously described (Zhang et al., 2009). Briefly, the sterilized A. annua seeds were placed on MS0 medium and then cultured in a light chamber at 25 ± 1 °C under a 16‐h light/8‐h dark photoperiod. After 14 days, the leaves of germinated seedlings were collected and cut into 0.5 cm diameter discs and used as explants that were co‐cultivated with A. tumefaciens strain EHA105 containing the above construct at 25 °C for 3 days. After this, the transformed explants were selected in the selective medium MS1 (MS0 + 2.5 mg/L N6‐benzoyladenine + 0.3 mg/L naphthalene‐acetic acid) containing 50 mg/L hygromycin; the resistant plantlets were regenerated and sub‐cultured twice and then transferred into MS2 (½ MS0 + 250 mg/L carbenicillin) medium for root growth. After 1 month, the rooted plantlets were selected and planted in soil in a light chamber at 23 ± 2 °C under a 16‐h light/8‐h dark photoperiod. The phenotype of Vector control plants (labelled as Vector), AaTCP15‐overexpression (OE‐AaTCP15) and AaTCP15‐antisense (Anti‐AaTCP15) lines were observed at two and a half months in a greenhouse under normal conditions.
Measurement of artemisinin (AN) and DHAA content
Leaves of 3‐month‐old OE‐AaTCP15 and Anti‐AaTCP15 A. annua plants, Vector controls and WT plants grown in the greenhouse were harvested and dried at 50 °C for 2 days and then ground to powder. The extraction and measurement methods were conducted as previously described (Ma et al., 2018).
Dual‐luciferase assay
The full‐length coding sequences of AaTCP15, AaMYC2, AaGSW1, AaORA, AaERF1 and AabZIP1 were amplified and inserted into pCAMBIA1300‐GFP vectors (effectors), and the promoter regions upstream of the start codon of AaADS (~1.9 kb), AaCYP71AV1 (~1.2 kb), AaDBR2 (~2.3 kb), AaALDH1 (~2.3 kb), AaTCP15 (~2.1 kb) and AaTCP14 (~1.9 kb) were ligated into pGREENII0800‐LUC vector (reporters). The Renilla luciferase (REN) driven by the cauliflower mosaic virus (CaMV) 35S promoter in pGREENII0800‐LUC was used as an internal control. Empty pCAMBIA1300‐GFP was used as the negative control for the effector. Infiltration and detection were performed as described previously (Ma et al., 2018). The ratio of firefly luciferase to Renilla luciferase represents the relative activity of the promoter. All experiments were repeated three times for each combination. Primers are listed in Table S1.
Yeast one‐hybrid assay
The full‐length coding sequences of AaTCP15 and AaGSW1 were amplified and inserted into pB42AD vector. Three tandem copies of the D motif from DBR2 promoter, A motif from ALDH1 promoter, W1 and W2 motifs (W‐box, Figure 6a) from AaTCP15 promoter, and W3 motif (W‐box, Figure 6a) from AaTCP14 promoter were separately ligated into pLacZ vector. Y1H assays were conducted as described previously (Ma et al., 2018). All primers are listed in Table S1.
Electrophoretic mobility shift assay
For protein expression and purification, the full‐length coding sequence of AaTCP15 was cloned into pCold™‐TF (trigger factor) vector (Takara, Japan) to produce His‐tagged fusion protein. The expression and purification of His‐TF protein and His‐AaTCP15 fusion proteins were performed as described previously (Ma et al., 2018).
EMSA assays were conducted as described previously (Ma et al., 2018). The Dq, Dq‐mutated, Aq and Aq‐mutated DNA probes from the DBR2 or ALDH1 promoters were synthesized by Sangon (Shanghai, China). The mutated probes were designed in accordance with a previous report (Pruneda‐Paz et al., 2009). The primers and probes used in the EMSA assays are listed in Table S1.
Bimolecular fluorescence complementation assay
The amplicons of AaTCP15 or AaORA were ligated into pEarleyGate 201‐YN (N‐terminal of YFP) or pEarleyGate 202‐YC (C‐terminal of YFP), respectively. The resultant AaTCP15‐nYFP, AaORA‐cYFP vectors were transformed into Agrobacterium strains GV3101. The BiFC assays were conducted as previously described (Ma et al., 2018; Shen et al., 2016). Three independent experiments were conducted. The primers are listed in Table S1.
Luciferase complementation assay
The LUC complementation assays were performed as previously described (Ma et al., 2018). In brief, the amplicons of AaTCP15 and AaORA were inserted into pCAMBIA‐Nluc or pCAMBIA‐Cluc vector, respectively. The yield Cluc‐AaTCP15 and AaORA‐Nluc vectors were transformed into Agrobacterium strain GV3101. The indicated combinations were co‐transformed into 5‐week‐old N. benthamiana leaves. Leaf discs were harvested 3 days later and ground to powder in liquid nitrogen. Subsequently, the relative LUC activities were measured by the Commercial Luciferase Kit according to the manufacturers’ instructions (Promega, Durham, NC, USA). The relative LUC activities of Nluc and Cluc were set as 1. The primers are listed in Table S1.
Yeast two‐hybrid experiments
To test the physical interaction between AaTCP15 and AaORA, and map the regions responsible for this paired interaction, the full‐length coding sequences of AaTCP15 and AaORA were cloned into pGADT7 (AD) or pGBKT7 (BD) (Takara, Japan), respectively, and their truncated sequences including AaTCP15ΔC1, AaTCP15ΔC2, AaTCP15MC, AaTCP15ΔN1, AaTCP15ΔN2, AaORAΔC1, AaORAΔC2, AaORAMC, AaORAΔN1 and AaORAΔN2 were amplified using specific primers (Table S1) and inserted into the bait vector BD. Various combinations were co‐transformed into yeast strain AH109 according to the manufacturer’s instructions (Takara, Japan), and bait‐only or prey‐only was tested with empty AD or BD as negative controls. The Y2H assays were carried out as previously reported (Ma et al., 2018). All experiments were repeated three times with similar results.
Accession Numbers
The sequences of all genes mentioned in this article are available in the NCBI database as follows: AaTCP14 (MF770262), AaTCP15 (MK770359), AaORA (JQ797708), AaMYC2 (KP119607), AaGSW1 (KX465128), proADS (DQ448294), proCYP71AV1 (FJ870128), proDBR2 (KC118523.1) and proALDH1 (KC118525.1). All other data related to the conclusions in the paper can be found in the paper and/or the Supplemental Materials.
Conflict of interest
The authors declare that they have no competing interests.
Author contributions
K. X. T., Y. N. M. conceived of and supervised the research. Y. N. M., D. B. X. designed the experiments. Y. N. M., D. B. X., X. Y., Z. K. Y. W., and P. L. performed the experiments. S. I. K., X. Q. F., Q. S., Q. F. P., L. L., Z. Y. L., L. H. X., X. L. H., D. H., H. L. and X. F. S. analysed the data. Y. N. M., D. B. X. organized and wrote the manuscript. All authors read and approved the final manuscript.
Supporting information
Figure S1 A graphical representation of biosynthetic pathways and regulation of artemisinin in Artemisia annua.
Figure S2 Phylogenetic tree showing the relationship of TCP transcription factors in Artemisia annua, Arabidopsis and Gossypium raimondii.
Figure S3 Alignment of the protein sequences of TCP15 among Artemisia annua, Arabidopsis thaliana and Gossypium raimondii.
Figure S4 Characterization of the content of DHAA and phenotype of AaTCP15 transgenic plants.
Figure S5 Artemisinin content in AaTCP15‐antisense plants under MeJA and ABA treatment.
Figure S6 Analysis of the putative regulatory elements in the cloned AaTCP15 promoter nucleotide sequence.
Figure S7 Analysis of artemisinin contents in AaGSW1, AaMYC2, and AaORA overexpression A. annua plants.
Figure S8 The expression pattern of AaTCP14 under ABA treatment in A. annua.
Table S1 List of primers used in this study
Acknowledgements
This work was supported by National Key R&D Program of China (2018YFA0900600), the Bill & Melinda Gates Foundation (OPP1199872), SJTU Global Strategic Partnership Fund (2020 SJTU‐CORNELL) and SJTU Trans‐med Awards Research Program (20190104).
Ma, Y.‐N. , Xu, D.‐B. , Yan, X. , Wu, Z.‐K. , Kayani, S. I. , Shen, Q. , Fu, X.‐Q. , Xie, L.‐H. , Hao, X.‐L. , Hassani, D. , Li, L. , Liu, H. , Pan, Q.‐F. , Lv, Z.‐Y. , Liu, P. , Sun, X.‐F. and Tang, K.‐X. (2021) Jasmonate‐ and abscisic acid‐activated AaGSW1‐AaTCP15/AaORA transcriptional cascade promotes artemisinin biosynthesis in Artemisia annua . Plant Biotechnol. J., 10.1111/pbi.13561
References
- Aggarwal, P. , Gupta, M.D. , Joseph, A.P. , Chatterjee, N. , Srinivasan, N. and Nath, U. (2010) Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell, 22, 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahkami, A. , Johnson, S.R. , Srividya, N. and Lange, B.M. (2015) Multiple levels of regulation determine monoterpenoid essential oil compositional variation in the mint family. Mol. Plant, 8, 188–191. [DOI] [PubMed] [Google Scholar]
- An, J.P. , Liu, Y.J. , Zhang, X.W. , Bi, S.Q. , Wang, X.F. , You, C.X. and Hao, Y.J. (2020) Dynamic regulation of different light intensity‐modulated anthocyanin biosynthesis by BT2‐TCP46‐MYB1 in apple. J. Exp. Bot. 71, 3094–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester, H.J. , Wallaart, T.E. , Janssen, M.H. , van Loo, B. , Jansen, B.J. , Posthumus, M.A. , Schmidt, C.O. et al. (1999) Amorpha‐4, 11‐diene synthase catalyses the first probable step in artemisinin biosynthesis. Phytochemistry 52, 843–854. [DOI] [PubMed] [Google Scholar]
- Bresso, E.G. , Chorostecki, U. , Rodriguez, R.E. , Palatnik, J.F. and Schommer, C. (2018) Spatial control of gene expression by miR319‐regulated TCP transcription factors in leaf development. Plant Physiol. 176, 1694–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, G.D. and Sy, L.K. (2004) In vivo transformations of dihydroartemisinic acid in Artemisia annua plants. Tetrahedron 60, 1139–1159. [Google Scholar]
- Brown, G.D. and Sy, L.K. (2007) In vivo transformations of artemisinic acid in Artemisia annua plants. Tetrahedron 63, 9548–9566. [Google Scholar]
- Chahel, A.A. , Zeng, S.H. , Yousaf, Z. , Liao, Y.Y. , Yang, Z.Y. , Wei, X.Y. and Ying, W. (2019) Plant‐specific transcription factor LrTCP4 enhances secondary metabolite biosynthesis in Lycium ruthenicum hairy roots. Plant Cell Tissue Organ Cult. 136, 323–337. [Google Scholar]
- Chang, Y.J. , Song, S.H. , Park, S.H. and Kim, S.U. (2000) Amorpha‐4, 11‐diene synthase of Artemisia annua: cDNA isolation and bacterial expression of a terpene synthase involved in artemisinin biosynthesis. Arch. Biochem. Biophys. 383, 178–184. [DOI] [PubMed] [Google Scholar]
- Chen, Y.F. , Shen, Q. , Wang, Y.Y. , Wang, T. , Wu, S.Y. , Zhang, L. , Lu, X. et al. (2013) The stacked over‐expression of FPS, CYP71AV1 and CPR genes leads to the increase of artemisinin level in Artemisia annua L. Plant Biotechnol. Rep. 7, 287–295. [Google Scholar]
- Chen, M.H. , Yan, T.X. , Shen, Q. , Lu, X. , Pan, Q.F. , Huang, Y.R. , Tang, Y.L. et al. (2017) GLANDULAR TRICHOME‐SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua . New Phytol. 214, 304–316. [DOI] [PubMed] [Google Scholar]
- Crespo‐Ortiz, M.P. and Wei, M.Q. (2012) Antitumor activity of artemisinin and its derivatives: from a well‐known antimalarial agent to a potential anticancer drug. J. Biomed. Biotechnol. 2012, 247597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas, P. , Lauter, N. , Doebley, J. and Coen, E. (1999) The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. [DOI] [PubMed] [Google Scholar]
- Czechowski, T. , Larson, T.R. , Catania, T.M. , Harvey, D. , Brown, G.D. and Graham, I.A. (2016) Artemisia annua mutant impaired in artemisinin synthesis demonstrates importance of nonenzymatic conversion in terpenoid metabolism. Proc. Natl Acad. Sci. USA 113, 15150–15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman, S. , van Dijk, A.D.J. , Bimbo, A. , van der Wal, F. , Hennig, L. , de Folter, S. , Angenent, G.C. et al. (2013) Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 64, 5673–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danisman, S. , van der Wal, F. , Dhondt, S. , Waites, R. , de Folter, S. , Bimbo, A. , van Dijk, A. D.J. et al. (2012) Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J. , Stec, A. and Hubbard, L. (1997) The evolution of apical dominance in maize. Nature 386, 485–488. [DOI] [PubMed] [Google Scholar]
- Efferth, T. , Romero, M.R. , Wolf, D.G. , Stamminger, T. , Marin, J.J. and Marschall, M. (2008) The antiviral activities of artemisinin and artesunate. Clin. Infect. Dis. 47, 804–811. [DOI] [PubMed] [Google Scholar]
- Finn, R.D. , Clements, J. and Eddy, S.R. (2011) HMMER web server, interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, I. , Besser, K. , Blumer, S. , Branigan, C. , Czechowski, T. , Elias, L. , Guterman, I. et al. (2010) The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin. Science 327, 328–331. [DOI] [PubMed] [Google Scholar]
- Guan, P. , Ripoll, J.J. , Wang, R. , Vuong, L. , Bailey‐Steinitz, L.J. , Ye, D. and Crawford, N.M. (2017) Interacting TCP and NLP transcription factors control plant response to nitrate availability. Proc. Natl Acad. Sci. USA 114, 2419–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, Y.P. , Xiao, J.W. , Shen, Y.L. , Ma, D.M. , Li, Z.Q. , Pu, G.B. , Li, X. et al. (2014) Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua . Plant Cell Physiol. 55, 1592–1604. [DOI] [PubMed] [Google Scholar]
- Jing, F. , Zhang, L. , Li, M. , Tang, Y. , Wang, Y. , Wang, Y. , Wang, Q. et al. (2009) Abscisic acid (ABA) treatment increases artemisinin content in Artemisia annua by enhancing the expression of genes in artemisinin biosynthetic pathway. Biologia 64, 319–323. [Google Scholar]
- Kieffer, M. , Master, V. , Waites, R. and Davies, B. (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis . Plant Journal 68, 147–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Casteels, T. , Frogne, T. , Ingvorsen, C. , Honoré, C. , Courtney, M. , Huber, K.V.M. et al. (2017) Artemisinins target GABAA receptor signaling and impair alpha cell identity. Cell 168, 86–100.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.D. , Dong, Y.J. , Tu, Y.Y. and Lin, Z.B. (2006) Dihydroarteannuin ameliorates lupus symptom of BXSB mice by inhibiting production of TNF‐alpha and blocking the signaling pathway NF‐kappa B translocation. Int. Immunopharmacol. 6, 1243–1250. [DOI] [PubMed] [Google Scholar]
- Li, J. , Wang, Y. , Zhang, Y. , Wang, W. , Irish, V.F. and Huang, T. (2016) RABBIT EARS regulates the transcription of TCP4 during petal development in Arabidopsis . J. Exp. Bot. 67, 6473–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. and Zachgo, S. (2013) TCP3 interacts with R2R3‐MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana . Plant J. 76, 901–913. [DOI] [PubMed] [Google Scholar]
- Li, L. , Zhang, H.H. , Zhang, L. , Zhou, Y.H. , Yang, R.W. , Ding, C.B. and Wang, X.L. (2014) The physiological response of Artemisia annua L. to salt stress and salicylic acid treatment. Physiol. Mol. Biol. Plants 20, 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R. , Dong, H.F. , Guo, Y. , Zhao, Q.P. and Jiang, M.S. (2011) Efficacy of praziquantel and artemisinin derivatives for the treatment and prevention of human schistosomiasis: a systematic review and meta‐analysis. Parasites Vectors 4, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X. , Zhang, L. , Zhang, F. , Jiang, W. , Shen, Q. , Zhang, L. , Lv, Z. et al. (2013) AaORA, a trichome‐specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol. 198, 1191–1202. [DOI] [PubMed] [Google Scholar]
- Lucero, L.E. , Uberti‐Manassero, N.G. , Arce, A.L. , Colombatti, F. , Alemano, S.G. and Gonzalez, D.H. (2015) TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis . Plant J. 84, 267–282. [DOI] [PubMed] [Google Scholar]
- Ma, D. , Pu, G. , Lei, C. , Ma, L. , Wang, H. , Guo, Y. , Chen, J. et al. (2009) Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha‐4, 11‐diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 50, 2146–2161. [DOI] [PubMed] [Google Scholar]
- Ma, Y.N. , Xu, D.B. , Li, L. , Zhang, F. , Fu, X.Q. , Shen, Q. , Lyu, X.Y. et al. (2018) Jasmonate promotes artemisinin biosynthesis by activating the TCP14‐ORA complex in Artemisia annua . Sci. Adv. 4, eaas9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes, L. , Van Nieuwerburgh, F.C. , Zhang, Y. , Reed, D.W. , Pollier, J. , Vande Casteele, S.R. , Inzé, D. et al. (2011) Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytol. 189, 176–189. [DOI] [PubMed] [Google Scholar]
- Malhotra, K. , Subramaniyan, M. , Rawat, K. , Kalamuddin, M. , Qureshi, M.I. , Malhotra, P. , Mohmmed, A. et al. (2016) Compartmentalized metabolic engineering for artemisinin biosynthesis and effective malaria treatment by oral delivery of plant cells. Mol. Plant 9, 1464–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, K.M. and Zerbe, P. (2020) Specialized diterpenoid metabolism in monocot crops: Biosynthesis and chemical diversity. Phytochemistry 172, 112289. [DOI] [PubMed] [Google Scholar]
- Nicolas, M. and Cubas, P. (2016) TCP factors: new kids on the signaling block. Curr. Opin. Plant Biol. 33, 33–41. [DOI] [PubMed] [Google Scholar]
- Nicolas, M. , Rodríguez‐Buey, M. , Franco‐Zorrilla, J. and Cubas, P. (2015) A recently evolved alternative splice site in the BRANCHED1a gene controls potato plant architecture. Curr. Biol. 25, 1799–1809. [DOI] [PubMed] [Google Scholar]
- Olofsson, L. , Lundgren, A. and Brodelius, P.E. (2012) Trichome isolation with and without fixation using laser microdissection and pressure catapulting followed by RNA amplification: expression of genes of terpene metabolism in apical and sub‐apical trichome cells of Artemisia annua L. Plant Sci. 183, 9–13. [DOI] [PubMed] [Google Scholar]
- Paddon, C.J. and Keasling, J.D. (2014) Semi‐synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat. Rev. Microbiol. 12, 355–367. [DOI] [PubMed] [Google Scholar]
- Paddon, C. , Westfall, P. , Pitera, D. , Benjamin, K. , Fisher, K. , McPhee, D. , Leavell, M. et al. (2013) High‐level semi‐synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532. [DOI] [PubMed] [Google Scholar]
- Palatnik, J.F. , Allen, E. , Wu, X. , Schommer, C. , Schwab, R. , Carrington, J.C. and Weigel, D. (2003) Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Parapunova, V. , Busscher, M. , Busscher‐Lange, J. , Lammers, M. , Karlova, R. , Bovy, A.G. , Angenent, G.C. et al. (2014) Identification, cloning and characterization of the tomato TCP transcription fator family. BMC Plant Biol. 14, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, S. and Shakya, K. (2013) Arsenic, chromium and NaCl induced artemisinin biosynthesis in Artemisia annua L.: a valuable antimalarial plant. Ecotoxicol. Environ. Saf. 98, 59–65. [DOI] [PubMed] [Google Scholar]
- Peng, Y.C. , Chen, L.L. , Lu, Y.R. , Wu, Y.B. , Dumenil, J. , Zhu, Z.G. , Bevan, M.W. et al. (2015) The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis . Plant Cell 27, 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peplow, M. (2016) Synthetic biology’s first malaria drug meets market resistance. Nature 530, 389–390. [DOI] [PubMed] [Google Scholar]
- Pruneda‐Paz, J.L. , Breton, G. , Para, A. and Kay, S.A. (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323, 1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro, D.K. , Paradise, E.M. , Ouellet, M. , Fisher, K.J. , Newman, K.L. , Ndungu, J.M. , Ho, K.A. et al. (2006) Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943. [DOI] [PubMed] [Google Scholar]
- Rueda‐Romero, P. , Barrero‐Sicilia, C. , Gómez‐Cadenas, A. , Carbonero, P. and Oñate‐Sánchez, L. (2012) Arabidopsis thaliana DOF6 negatively affects germination in non‐after‐ripened seeds and interacts with TCP14. J. Exp. Bot. 63, 1937–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramek, N. , Wang, H. , Römisch‐Margl, W. , Keil, B. , Radykewicz, T. , Winzenhörlein, B. , Beerhues, L. et al. (2010) Artemisinin biosynthesis in growing plants of Artemisia annua. A 13CO2 study. Phytochemistry 71, 179–187. [DOI] [PubMed] [Google Scholar]
- Shen, Q. , Lu, X. , Yan, T. , Fu, X. , Lv, Z. , Zhang, F. , Pan, Q. et al. (2016) The jasmonate‐responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua . New Phytol. 210, 1269–1281. [DOI] [PubMed] [Google Scholar]
- Shen, Q. , Zhang, L. , Liao, Z. , Wang, S. , Yan, T. , Shi, P. , Liu, M. et al. (2018) The genome of Artemisia annua provides insight into the evolution of asteraceae family and artemisinin biosynthesis. Mol. Plant 11, 776–788. [DOI] [PubMed] [Google Scholar]
- Shi, P. , Fu, X.Q. , Liu, M. , Shen, Q. , Jiang, W.M. , Li, L. , Sun, X.F. et al. (2017) Promotion of artemisinin content in Artemisia annua by overexpression of multiple artemisinin biosynthetic pathway genes. Plant Cell Tissue Organ Cult. 129, 251–259. [Google Scholar]
- Steiner, E. , Efroni, I. , Gopalraj, M. , Saathoff, K. , Tseng, T.S. , Kieffer, M. , Eshed, Y. et al. (2012a) The Arabidopsis O‐linked N‐acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell 24, 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, E. , Yanai, O. , Efroni, I. , Ori, N. , Eshed, Y. and Weiss, D. (2012b) Class I TCPs modulate cytokinin‐induced branching and meristematic activity in tomato. Plant Signal. Behav. 7, 807–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T.H. , Zhou, F. , Huang, X.Q. , Chen, W.C. , Kong, M.J. , Zhou, C.F. , Zhuang, Z. et al. (2019) ORANGE represses chloroplast biogenesis in etiolated Arabidopsis cotyledons via interaction with TCP14. Plant Cell 31, 2996–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, H.X. , Xiao, L. , Gao, S.H. , Li, Q. , Chen, J.F. , Xiao, Y. , Ji, Q. et al. (2015) TRICHOME AND ARTEMISININ REGULATOR 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua . Mol. Plant 8, 1396–1411. [DOI] [PubMed] [Google Scholar]
- Teoh, K.H. , Polichuk, D.R. , Reed, D.W. and Covello, P.S. (2009) Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua . Botany 87, 635–642. [Google Scholar]
- Teoh, K.H. , Polichuk, D.R. , Reed, D.W. , Nowak, G. and Covello, P.S. (2006) Artemisia annua L. (Asteraceae) trichome‐specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 580, 1411–1416. [DOI] [PubMed] [Google Scholar]
- Tin, A.S. , Sundar, S.N. , Tran, K.Q. , Park, A.H. , Poindexter, K.M. and Firestone, G.L. (2012) Antiproliferative effects of artemisinin on human breast cancer cells requires the downregulated expression of the E2F1 transcription factor and loss of E2F1‐target cell cycle genes. Anticancer Drugs 23, 370–379. [DOI] [PubMed] [Google Scholar]
- Towler, M.J. and Weathers, P.J. (2007) Evidence of artemisinin production from IPP stemming from both the mevalonate and the nonmevalonate pathways. Plant Cell Rep. 26, 2129–2136. [DOI] [PubMed] [Google Scholar]
- Vadde, B.V.L. , Challa, K.R. and Nath, U. (2018) The TCP4 transcription factor regulates trichome cell differentiation by directly activating GLABROUS INFLORESCENCE STEMS in Arabidopsis thaliana . Plant J. 93, 259–269. [DOI] [PubMed] [Google Scholar]
- Viola, I.L. , Camoirano, A. and Gonzalez, D.H. (2016) Redox‐dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis . Plant Physiol. 170, 74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. , Kashkooli, A.B. , Sallets, A. , Ting, H.M. , de Ruijter, N.C.A. , Olofsson, L. , Brodelius, P. et al. (2016) Transient production of artemisinin in Nicotiana benthamiana is boosted by a specific lipid transfer protein from A. annua . Metab. Eng. 38, 159–169. [DOI] [PubMed] [Google Scholar]
- Weßling, R. , Epple, P. , Altmann, S. , He, Y. , Yang, L. , Henz, S.R , McDonald, N. et al. (2014) Convergent targeting of a common host protein‐network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16, 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . (2019) World Malaria Report 2019. Licence: CC BY‐NC‐SA 3.0 IGO. Geneva: World Health Organization. [Google Scholar]
- Xu, R. , Sun, P. , Jia, F. , Lu, L. , Li, Y. , Zhang, S. and Huang, J. (2014) Genome‐wide analysis of TCP transcription factor gene family in Malus domestica . J. Genet. 93, 733–746. [DOI] [PubMed] [Google Scholar]
- Yao, X. , Ma, H. , Wang, J. and Zhang, D. (2007) Genome‐wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thalianan and Oryza sativa . J. Integr. Plant Biol. 49, 885–897. [Google Scholar]
- Yu, Z.X. , Li, J.X. , Yang, C.Q. , Hu, W.L. , Wang, L.J. and Chen, X.Y. (2012) The jasmonate‐responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 5, 353–365. [DOI] [PubMed] [Google Scholar]
- Zhang, W. , Cochet, F. , Ponnaiah, M. , Lebreton, S. , Matheron, L. , Pionneau, C. , Boudsocq, M. et al. (2019) The MPK8‐TCP14 pathway promotes seed germination in Arabidopsis . Plant J. 100, 677–692. [DOI] [PubMed] [Google Scholar]
- Zhang, F. , Fu, X. , Lv, Z. , Lu, X. , Shen, Q. , Zhang, L. , Zhu, M. et al. (2015) A basic leucine zipper transcription factor, AabZIP1, connects abscisic acid signaling with artemisinin biosynthesis in Artemisia annua . Mol. Plant 8, 163–175. [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Jing, F. , Li, F. , Li, M. , Wang, Y. , Wang, G. , Sun, X. et al. (2009) Development of transgenic Artemisia annua (Chinese wormwood) plants with an enhanced content of artemisinin, an effective anti‐malarial drug, by hairpin‐RNA‐mediated gene silencing. Biotechnol. Appl. Biochem. 52, 199–207. [DOI] [PubMed] [Google Scholar]
- Zhang, T. , Qu, Y. , Wang, H. , Wang, J. , Song, A. , Hu, Y. , Chen, S. et al. (2017) The heterologous expression of a chrysanthemum TCP‐P transcription factor CmTCP14 suppresses organ size and delays senescence in Arabidopsis thaliana . Plant Physiol. Biochem. 115, 239–248. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Teoh, K.H. , Reed, D.W. , Maes, L. , Goossens, A. , Olson, D.J. , Ross, A.R. et al. (2008) The molecular cloning of artemisinic aldehyde Δ11 (13) reductase and its role in glandular trichome‐dependent biosynthesis of artemisinin in Artemisia annua . J. Biol. Chem. 283, 21501–21508. [DOI] [PubMed] [Google Scholar]
- Zhang, N. , Wang, Z.X. , Bao, Z.L. , Yang, L.Y. , Wu, D.X. , Shu, X.L. and Hua, J. (2018) MOS1 functions closely with TCP transcription factors to modulate immunity and cell cycle in Arabidopsis . Plant J. 93, 66–78. [DOI] [PubMed] [Google Scholar]
- Zhao, J. , Zhai, Z. , Li, Y. , Geng, S. , Song, G. , Guan, J. , Jia, M. et al. (2018) Genome‐wide identification and expression profiling of the TCP family genes in spike and grain development of wheat (Triticum aestivum L.). Front. Plant Sci. 9, 1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, H.Q. , Colvin, C.J. , Johnson, B.K. , Kirchhoff, P.D. , Wilson, M. , Jorgensen‐Muga, K. , Larsen, S.D. et al. (2017) Inhibitors of Mycobacterium tuberculosis DosRST signaling and persistence. Nat. Chem. Biol. 13, 218–225. [DOI] [PubMed] [Google Scholar]
- Zhong, Y. , Li, L. , Hao, X. , Fu, X. , Ma, Y. , Xie, L. , Shen, Q. et al. (2018) AaABF3, an abscisic acid‐responsive transcription factor, positively regulates artemisinin biosynthesis in Artemisia annua . Front. Plant Sci. 9, 1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, M. and Memelink, R. (2016) Jasmonate‐responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 34, 441–449. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Yang, J. , Zeng, Z. , Zhang, W. , Song, L. , Wen, W. and Yu, R. (2014) Inducing effect of dihydroartemisinic acid in the biosynthesis of artemisinins with cultured cells of Artemisia annua by enhancing the expression of genes. Sci. World J. 2014, 29319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 A graphical representation of biosynthetic pathways and regulation of artemisinin in Artemisia annua.
Figure S2 Phylogenetic tree showing the relationship of TCP transcription factors in Artemisia annua, Arabidopsis and Gossypium raimondii.
Figure S3 Alignment of the protein sequences of TCP15 among Artemisia annua, Arabidopsis thaliana and Gossypium raimondii.
Figure S4 Characterization of the content of DHAA and phenotype of AaTCP15 transgenic plants.
Figure S5 Artemisinin content in AaTCP15‐antisense plants under MeJA and ABA treatment.
Figure S6 Analysis of the putative regulatory elements in the cloned AaTCP15 promoter nucleotide sequence.
Figure S7 Analysis of artemisinin contents in AaGSW1, AaMYC2, and AaORA overexpression A. annua plants.
Figure S8 The expression pattern of AaTCP14 under ABA treatment in A. annua.
Table S1 List of primers used in this study


