Here a cycle of endophytic‐enteric‐soil‐endophytic microbes is proposed which has relevance for health and comprises the fate of animal faeces as natural microbial inoculants for plants that constitute bacterial sources for animal guts.

Summary
Health depends on the diet and a vegetal diet promotes health by providing fibres, vitamins and diverse metabolites. Remarkably, plants may also provide microbes. Fungi and bacteria that reside inside plant tissues (endophytes) seem better protected to survive digestion; thus, we investigated the reported evidence on the endophytic origin of some members of the gut microbiota in animals such as panda, koala, rabbits and tortoises and several herbivore insects. Data examined here showed that some members of the herbivore gut microbiota are common plant microbes, which derived to become stable microbiota in some cases. Endophytes may contribute to plant fibre or antimetabolite degradation and synthesis of metabolites with the plethora of enzymatic activities that they display; some may have practical applications, for example, Lactobacillus plantarum found in the intestinal tract, plants and in fermented food is used as a probiotic that may defend animals against bacterial and viral infections as other endophytic‐enteric bacteria do. Clostridium that is an endophyte and a gut bacterium has remarkable capabilities to degrade cellulose by having cellulosomes that may be considered the most efficient nanomachines. Cellulose degradation is a challenge in animal digestion and for biofuel production. Other endophytic‐enteric bacteria may have cellulases, pectinases, xylanases, tannases, proteases, nitrogenases and other enzymatic capabilities that may be attractive for biotechnological developments, indeed many endophytes are used to promote plant growth. Here, a cycle of endophytic‐enteric‐soil‐endophytic microbes is proposed which has relevance for health and comprises the fate of animal faeces as natural microbial inoculants for plants that constitute bacterial sources for animal guts.
Effects of vegetal diet and endophytes
Vegetal diet
Vegetables provide fibres, vitamins and metabolites that promote health (Cardona et al, 2013; Klinder et al, 2016; Makki et al, 2018), but their role as microbe providers is less known, unless these microbes are pathogens. When animals consume raw plants, they eat their associated bacteria. Eating an apple may provide hundred millions of bacteria (Wassermann et al, 2019), as does eating bananas, lettuce (Berg et al, 2014a) or other raw vegetables and non‐pasteurized juices. Even if vegetables are washed, peeled or disinfected, they still provide microbes because the endophytic bacteria or fungi reside in the plant interior protected from disinfectants. Herbivore guts had the largest diversity of bacteria, containing 14 phyla, while only six phyla were found in carnivores (Ley et al, 2008). Different species of Bifidobacterium and Lactobacillus were found in herbivores compared with carnivore or omnivore animals (Endo et al, 2010). The gut human microbiota has been extensively studied (reviewed in Thursby and Jurge, 2017; Rothschild et al, 2018) and depends on the diet (Muegge et al, 2011; David et al, 2014). Transient microbiota (called foreign microorganisms by David et al, 2014) may derive from food. Notably, the gut microbiome in humans is determined by the number of vegetables consumed (McDonald et al, 2018). Ingested bacteria may be metabolically active in human guts as revealed by gene transcripts from food‐bacteria in guts (David et al, 2014). Furthermore, plant‐borne pathogens provide an unfortunate example of human ingestion of plant bacteria (Berg et al, 2014a, 2014b; Rosenblueth and Martinez‐Romero, 2006). Certainly, plant bacteria have been in our diet for a long time (Berg et al, 2014a, 2014b) and for the whole evolutionary history of herbivores. The evolutionary history of insects is tightly dependent on plants as food (McKenna and Farell, 2006) and large radiations in insects followed plant diversification (Futuyma and Agrawal, 2009).
Besides containing bacteria, plants may modify gut bacterial composition and diversity due to their content of fibres, flavonoids, carotenoids, alkaloids, bioactive metabolites, antimetabolites or toxins (Cardona et al, 2013; Klinder et al, 2016; Makki et al, 2018; Baxter et al., 2019). Japanese that eat seaweeds have a peculiar microbiota (Hehemann et al, 2010).
Endophytes
All plants in nature and crops have associated microbes (Friesen et al, 2011) in apparently all organs and tissues. Microbes that colonize inner plant tissues are designated endophytes (Rosenblueth and Martinez‐Romero, 2006; Harrison and Griffin, 2020; Berg et al, 2014a, 2014b), as the Greek‐prefix ‘endo’ means inside or within and ‘phyton’ means plant. Endophytes are a selected group of plant‐associated microbes (Rosenblueth et al, 2004; Rosenblueth and Martinez‐Romero, 2006; Hardoim et al, 2015; Busby et al, 2016) in the sense that only particular microbial genotypes are capable of internally colonizing specific plants. Endophytes promote plant growth by different strategies, such as suppressing or out‐competing pathogens, fixing nitrogen, producing hormones that stimulate plant growth, protecting from stress or enhancing the availability of minerals (Rosenblueth and Martínez‐Romero, 2006).
Endophytes in animal guts
Gut microbiota may contain hundreds to thousands of bacterial species (McDonald et al, 2018; Rothschild et al, 2018) that may be acquired from or selected by the diet. Bacterial cultures from herbivore faeces showed bacteria that are evidently derived from plants, for example methylobacteria isolated in cultures from rhino (Rhinoceros sondaicus) and alpaca faeces (Jiang et al, 2013) and detected in cabbage white butterfly (Robinson et al, 2010b). Methylobacteria (commonly found in plants) use methanol, a sub‐product of plant cell wall biosynthesis and produce the plant hormones cytokinins (Lidstrom and Chistoserdova, 2002). Furthermore, cultured actinobacteria from herbivores showed that Streptomyces, Rhodococcus and Microbacterium were the dominant isolates from all six animal faeces tested, including an elephant (Jiang et al, 2013). These bacteria are common plant endophytes, for example, Microbacterium was isolated from legume nodules in arid regions (Zakhia et al, 2006) and from a halophyte in a salt‐marsh (Alves et al, 2014), Streptomyces was found as a seed endophyte in a Phaseolus vulgaris (common bean) cultivar with outstanding characteristics (López‐López et al 2010) and a Rhodococcus leaf endophyte enhanced resistance to pathogenic fungi in potato (Hong et al, 2016).
Endophytes that feed on plants would have fibre degrading capabilities that would help animals degrade plant polysaccharides in guts. Plant fibres containing cellulose and hemicellulose are not easily digested, and sets of large numbers and diverse degrading enzymes are needed, including glycoside hydrolases and polysaccharide lyases, which are not produced or rarely produced by mammals (El Kaoutari et al, 2013). Clostridium is found as an endophyte and in many animal guts (Figs 1 and 2). Clostridial cellulolytic activity is remarkable because these bacteria may contain cellulosomes (Bayer and Lamed, 1986; Schwarz, 2001), which are considered the most efficient natural nanomachines (Nunes, 2018) that degrade both cellulose and hemicellulose. Cellulosomes are protuberances on the bacterial cell wall (Bayer and Lamed, 1986) containing complex enzymatic systems. Enterobacter that is found in herbivore guts or faeces and in plants (Fig. 1) has diverse polysaccharide degrading enzymes such as cellobiosidase, endoglucanase, polygalacturonase, xylanase, β‐glucuronidase, pectinases and cellulases (Prem Anand et al, 2010; Naveed et al, 2014; Xia et al, 2017). Cellulases are found in many Proteobacteria, Firmicutes and Actinobacteria (Berlemont and Martiny, 2013).
Fig. 1.

Examples of animal gut bacteria (from cultures or identified in gut metagenomes) that are common plant endophytes.
Fig. 2.

16S rRNA gene phylogenies of gut bacteria and endophytes of selected genera. A. Clostridium . B. Weissella. C. Lactobacillus plantarum . 16S rRNA sequences from NCBI database were aligned using Clustal W and phylogenetic trees were constructed in MEGA X software using the maximum likelihood method and general time reversible model with 1000 bootstraps replicates. A total of 1369 positions were included for the phylogenetic analysis.
In contrast to fibre or wood‐chewing insects, sap‐sucking insects such as stinkbugs or cochineals may have bacteria with less fibre degrading capabilities. In sap‐sucking insects, protease activity was found in guts and it was suggested that ‘digestive proteolysis may be widespread in homoptera’ (Foissac et al, 2002). In the carmine cochineal, Dactylopiibacterium showed increased expression of protease and peptidase genes in a gut metatranscriptomic analysis suggesting a bacterial origin of proteases in guts (Bustamante‐Brito et al, 2019).
Some herbivores have a specialized diet, for example koalas eat eucalyptus, pandas eat bamboos, tortoises eat cactus, Monarch butterfly pupas eat Asclepias and maguey red worms eat Agave cactuses and their microbiota serves to digest some of the particular substances or antimetabolites in their host plants. Some of the antimetabolites may be degraded in guts by endophytes such as Pseudomonas, burkholderias and Enterobacter (Shanmuganandam et al, 2019). Tannins are the fourth more abundant plant molecules after cellulose, hemicellulose and lignin, with tannic acid as the most abundant reserve in plants (de Las Rivas et al, 2019). Tannases that degrade tannins found in eucalyptus, quercus and other tree leaves ingested by herbivores, are produced by many bacterial genera found in guts and plants, including Enterobacter, Weissella and Lactobacillus (de Las Rivas et al, 2019). Pinene, from pine trees that are ingested by different beetles, is degraded by some Pseudomonas using monooxygenases, lyases and aldehyde dehydrogenases (Linares et al, 2009).
The importance of endophytic fungi in plants and ecosystems is well appreciated (Harrison and Griffin, 2020). In turn, fungi may harbour bacterial communities that would also have an impact in plants (Bonfante et al, 2019). We suppose that from the outstanding diversity of fungi in plants (Harrison and Griffin, 2020) only a few fungi colonize animal guts. Some fungi are commonly found inside insects and plants (Pažoutová et al, 2013; Chen et al, 2018; Biederman and Vega, 2020) and may have cellulases and tannases (Martin, 1992; de Las Rivas et al, 2019). The most common contribution from fungi to their insect symbiont is the catalytic capacity to break down plant polysaccharides such as cellulose and pectin from their diet (Martin, 1992). Herrera et al (2011) found sequences 97% similar to root‐associated fungi in coprophilous fungal communities obtained from the dung of four species of mammalian herbivores. The effects of eating endophytic fungi were observed with sheep. Sheep were fed ryegrass with or without distinct fungal endophytes, and later sheep faeces were studied for their degradation rate. Faeces from animals that ate grass with fungal endophytes had the lowest faecal degradation rates (Cripps et al, 2013). This nice example showed that fungal endophytes from grasses that were consumed by sheep arrived into guts and exerted effects on faeces.
Insects among animals seem to have had unique associations with fungi for around 420 million years. These associations range from pathogenic with Cordyceps for example, to obligate mutualisms found in beetles, wasps and ants (Boucias et al, 2012a, 2012b; Gibson and Hunter, 2010). Remarkably, some of the insect‐associated fungal groups are well‐known endophytes within the Sordariomycetes and Agaromycetes (Naranjo‐Ortiz and Gabaldón, 2019).
Interestingly, some protists may inhibit cellulolytic activities in guts (Ray et al, 2012). Protists and archaea though they are associated with plants and guts, will not be revised here, neither the rumen or bee microbiota (Taxis et al, 2015; Engel et al, 2012; Zheng et al, 2019 Raymann et al, 2018; Motta et al, 2018. Powell et al, 2018; Raymann and Moran, 2018; Leonard et al, 2020). Previously, a deep analysis of anaerobic adaptations of bacteria in arthropod guts and the possible DNA contaminants in reagents were reported (Degli Esposti and Martinez‐Romero, 2017; Glassing et al, 2016). We show here examples of gut bacteria with a possible endophytic origin (Fig. 1), detected in pandas, koalas, rabbits, tortoises and several insects that are reviewed below.
Koala
The koala (Phascolarctos cinereus) has a diet based almost exclusively on Eucalyptus leaves (Moore and Foley, 2000). Secondary metabolites contained in Eucalyptus plants act as toxins and antimicrobial agents that could affect the koala and its microbiota (Moore et al, 2004; Brice et al, 2019). The koala gut microbiota is highly conserved and specialized in the digestion and detoxification of dietary components (Brice et al, 2019). Blyton et al (2019) demonstrated that oral‐faecal inoculation between wild koalas with different feeding habits allows them to feed on Eucalyptus species that were previously inedible. Similarly, gut microbiota are acquired by juvenile koalas when they feed on special maternal faeces called 'pap' (Osawa et al, 1993a, 1993b). The strict vegetarian diet of koalas leads to a constant supply of endophytic microbes, which may become resident or transitory inhabitants of their guts. The koala rectum is mainly colonized by Bacteroidetes, Firmicutes and Proteobacteria (Barker et al, 2013; Alfano et al, 2015). Staphylococcus, Bradyrhizobium and Acinetobacter have been found in koala guts and as Eucalyptus endophytes (Procópio et al, 2009; da Silva Fonseca et al, 2018). Likewise, tannin‐degrading strains of Streptococcus, Enterobacter and other Enterobacteriaceae have been isolated from koala faeces (Osawa, 1992), 'pap' (Osawa et al, 1993a, 1993b) and Eucalyptus leaves (Miguel et al, 2016). Cellulose‐degrading Pseudomonas and spore‐forming bacilli are other examples of microbiota shared between koala faeces and Eucalyptus tissues (Singh et al, 2015; Miguel et al, 2016).
Giant panda
The giant panda (Ailuropoda melanoleuca) consumes about 15 kg of leaves, stems and shoots of bamboo every day (Dierenfeld et al, 1982). Due to their carnivorous origin, pandas have a straight short gastrointestinal tract (Ley et al, 2008) whose microbiota is rich in Proteobacteria and Firmicutes (Fig. 1) (Tun et al, 2014; Xue et al, 2015; Guo et al, 2019). However, the gut microbiota is affected depending on the part and species of bamboo consumed. For instance, a high leaf consumption leads to an increase in Bacteroides and a decrease in Lactobacillus, but it does not affect Streptococcus and Clostridium populations (Williams et al, 2013). A phylogenetic tree showed Clostridium from plants and gut intermingled indicating that gut bacteria were recently acquired or constantly exchanged (Fig. 2A). Recent work by Jin et al (2020) showed that bacteria and fungi colonizing different bamboo species appeared as gut colonizers after consumed by giant pandas. Interestingly, the greater the microbial diversity of bamboo, the greater the diversity found in faecal samples.
The cellulolytic activity of Clostridium as well as the presence of bacterial genes that encode plant cell wall degrading enzymes (endocellulase, β‐glucosidase, xylan 1,4‐β‐xylosidase and endo‐1,4‐β‐xylanase) in panda guts highlight the microbial degradation of bamboo (Zhu et al, 2011; Guo et al, 2020) that could perhaps help to release endophytes during digestion.
Among the Proteobacteria that inhabit panda faeces, Pseudomonas, Klebsiella, Enterobacter and Pantoea are frequent endophytes of grasses and bamboo (Han et al, 2009b; Han et al, 2010; Liu et al, 2017). Most genes related to pathways for the plant secondary metabolite degradation have been associated with Pseudomonas from panda gut metagenomes (Zhu et al, 2018; Yao et al, 2019). Klebsiella and Enterobacter strains isolated from panda faeces carry genes involved in microbe‐plant interactions and cellulose degradation indicating their endophytic origin (Lu et al, 2015a, 2015b).
Weissellas from panda faeces are intermingled with plant weissellas in 16S rRNA gene phylogenetic trees, and this is the case with weissellas from human and bat faeces and from fish guts (Fig. 2B). Weissellas may contain tannases that would help to degrade tannins from plants (de Las Rivas et al, 2019).‐ Similarly, Cryptococcus, Ramichloridium, Shiraia, Ceramothyrium, Rhinocladiella and Cephalosporium are bamboo‐associated fungi detected in panda's faeces (Jin et al, 2020).
The gut microbiota of the giant panda resembles that of carnivorous and omnivorous bears because it has a different and lower diversity than other herbivores (Xue et al, 2015), but it also shows bacterial signatures that could result from being a bamboo specialist.
Rabbits
Rabbits (Oryctolagus cuniculus) eat grasses, tree leaves and faeces. They are considered caecotrophagic animals because they ingest their soft faecal pellets produced by digestion in the caecum. Within herbivorous mammals, rabbits have the shortest mean retention time to digest their food, while the ruminants have the longest (Uden, et al, 1982; Kararli, 1995; Crowley et al, 2017). The rabbit digestive tract is adapted to process large amounts of fibre‐rich feed. Microbial fermentation of the food takes place in the caecum (Mackie, 2002; Harcourt‐Brown, 2004). Bacteroidetes dominate the caecal population and may be associated with the high fibre content in the diet (Crowley et al, 2017).
Like koalas, newborn rabbits in wild habitats ingest faecal pellets excreted by their mothers (Kovacs et al, 2006). When rabbits have access to the faecal excreta, bacteria colonize the caeca and rabbits have a reduced mortality after weaning in comparison to rabbits not consuming faeces (Combes et al., 2014).
Cl ostridium, Anaerofustis, Blautia, Akkermansia and Bacteroides are abundantly found in caecal samples, and in faeces Oscillospira and Coprococcus (Velasco‐Galilea et al, 2018). Clostridium as well as Roseburia, Sutterella, Enterobacter and Desulfovibrio have been reported as endophytes and in rabbit faeces (Velasco‐Galilea et al, 2018; Shanmuganandam et al, 2019). Interestingly, the genome of a gut Enterobacter cloacae strain has genes for plant colonization revealing characteristics of an endophyte (Shastry et al, 2020). Roseburia can produce butyrate, which is a nutrient for enterocytes (Tamanai‐Shacoori et al, 2017). Butyrate was identified as a promotor of gut barrier formation (Beaumont et al 2020).
Herbivorous tortoise
Herbivory is not frequently found among reptiles (Yuan et al, 2015). However, there are herbivorous tortoises such as Gopherus berlandieri that is found in arid regions in Northeast Mexico and Southern United States (Judd and Rose, 1983). From the faeces of G. berlandieri healthy tortoises, we isolated Klebsiella variicola (Montes‐Grajales et al, 2019). K. variicola may be found as endophyte in maize, banana and rice plants (Rosenblueth et al, 2004) and was found as well in newborn baby faeces (Rosales‐Bravo et al, 2017). K. variicola has been proposed to be used as probiotic (Rosales‐Bravo et al, 2017) or crop inoculant. This bacterium is found in humans as an opportunistic pathogen (Martinez‐Romero et al, 2018).
From different G. berlandieri tortoises, distinct K. variicola isolates showed limited genetic diversity, suggesting that they are clones selected from a larger pool of these bacteria. Nitrogen‐fixing activity was detected by the acetylene‐reduction assay, both from faeces and K. variicola isolates. As tortoises are coprophagous, it seems possible that they acquire K. variicola tortoise‐borne strains directly from their mother or from the faeces of other tortoises. We surmise that K. variicola in tortoises were plant‐borne but we did not find them in their vegetal food; thus, they are not continuously ingested. Some K. variicola clones seem to have become stable microbiota, selectively maintained in tortoises by coprophagy. Clostridium was abundantly found in Gopherus flavormarginatus (Garcia‐De la Peña et al., 2019), Gopherus polyphemus (Yuan et al, 2015) and G. berlandieri faeces.
Stinkbugs
Some stinkbugs from the superfamilies Coreoidea, Lygaeoidea and Pyrrhocoroidea feed on sap of diverse plants that have burkholderia endophytes. Burkholderia were found in insect guts in specialized compartments with crypts that have restricted entry and high burkholderial densities (Kikuchi et al, 2007, 2011; Kim and Lee, 2015). Recently, two novel genera were named for burkholderia subclades, Paraburkholderia and Caballeronia (Sawana et al, 2014; Dobritsa and Samadpour, 2016), and these are differentially encountered in stinkbug families that have specialized diets (Takeshita et al, 2015; Takeshita and Kikuchi, 2020). The symbiosis of phytophagous stinkbugs with these bacteria seems to be ancient (Kikuchi et al, 2011; Takeshita et al, 2015). Symbionts are beneficial to the insects, as stinkbugs without them displayed developmental delays, impaired survival or reduced size (Kikuchi et al, 2007; Boucias et al, 2012a; Xu et al, 2016a). Bacteria may confer stinkbugs resistance to the insecticide fenitrothion (Kikuchi et al, 2012; Tago et al, 2015). A transcriptome analysis of midgut‐colonizing Burkholderia insecticola from the bean‐bug Riptortus pedestris showed that bacteria recycle the host nitrogen wastes allantoin and urea, provide B vitamins, especially B12 and supply methionine and tryptophan to the host (Ohbayashi et al, 2019). Burkholderias constitute nice examples of plant bacteria that become gut colonizers, which are acquired by each new insect generation (Kikuchi et al, 2007; Kikuchi et al, 2011). Consequently, reported phylogenies showed that insect‐gut and plant burkholderias are intermingled (Itoh et al, 2014; Tago et al, 2015; Xu et al, 2016b ).
Carmine cochineal
Carmine cochineals (Dactylopius spp.) are a group of hemipteran insects that have cultural and economic importance, as they produce a pigment called carminic acid that is used in industries like food, cosmetics and textiles. They originated in Mexico and South America (Chávez‐Moreno et al., 2009; Chávez‐Moreno et al., 2011; Mazzeo et al, 2019). Cochineals are sap‐suckers of Opuntia and other cactuses (Chávez‐Moreno et al, 2009). Our first microbial diversity study of different carmine cochineal species performed by Ramírez‐Puebla et al (2010) using PCR‐product sequencing, showed that the insect symbionts were related to plant endophytes such as Herbaspirillum, Acinetobacter and Mesorhizobium. Further metagenomic studies of D. coccus and D. opuntiae allowed us to obtain the genome of a betaproteobacterium (Dactylopiibacterium carminicum) related to the grass endophytes Uliginosibacterium and Azoarcus (Vera‐Ponce de León et al, 2017). Several characteristics of Dactylopiibacterium remind endophytes, like the ability to fix atmospheric nitrogen, to produce cellulases and pectinases, and to catabolize salicylic acid that is produced in plants (Vera‐Ponce de Leon et al 2017; Bustamante‐Brito et al, 2019). Our results suggested an endophytic origin of Dactylopiibacterium symbionts.
In the carmine cochineal guts, we found one species of endophytic fungi belonging to the genus Coniochaeta. Coniochaeta species are main endophytes of trees and grasses with some species well known for their production of a broad spectrum of antimycotics (Xie et al, 2015).
Bark beetles and weevils
Wood‐eating Dendroctonus beetle guts contain nitrogen‐fixing bacteria, for example Raoultella terrigena that is a common endophyte (Morales‐Jiménez et al, 2013). Endophytes that have been used as plant growth‐promoting bacteria were also isolated from Dendroctonus beetles. Among these, Serratia, Pseudomonas and Rhanella species were able to recycle uric acid (Morales‐Jiménez et al, 2013). An additional study using 16S rRNA gene identification of gut bacteria from the pine‐pest Monochamus alternatus (Coleoptera) showed Enterobacter, Raoultella, Serratia, Lactococcus and Pseudomonas, which are commonly found in plant tissues as well. Enterobacter was the most common in larval and Serratia in pupal intestines. These bacteria may help to degrade the terpene pinene found in pines (Chen et al 2020).
Weevils are important beetle pests of stored grain legumes that feed and reproduce on dried seeds (Tuda, 2007). The neotropical genus Acanthoscelides comprises a diverse group of weevils some specialized on Phaseolus seeds (Alvarez et al, 2005). We studied the gut microbiota of bean weevils, Acanthoscelides obtectus (Coleoptera: Chrysomelidae, Bruchinae). Weevils were collected from inside wild P. vulgaris seeds from vines growing in mountain fields in the state of Morelos, Mexico. 16S ribosomal RNA genes from midgut DNA or from isolates were sequenced to generate a census of bacterial communities. We identified bacteria related to Agrobacterium, Bacillus, Massilia, Gluconacetobacter, Propionibacterium, Asaia and Bradyrhizobium. Bacillus isolates were frequently identified in bean seeds (Lopez‐Lopez et al, 2010) and leaves as endophytes (de Oliveira Costa et al, 2012). We suppose that P. vulgaris endophytes are transferred to the guts of A. obtectus weevils when they feed on bean seeds. Weevil insects in turn may transfer these bacteria to other plants and seeds.
Maguey red worm
The maguey red worm (Comadia redtenbacheri) is edible and endemic to Mexico. Larvae are plant‐eating specialists of Agave inner plant tissues (Hernández‐Flores et al, 2015; Cárdenas‐Aquino et al, 2018). Enterococcus and Klebsiella that secrete indole‐acetic acid and solubilize phosphate were isolated as endophytes from leaf bases of agave plants (Martinez‐Rodriguez et al, 2019) and from the larva guts as well (our unpublished results). In the larvae from other Lepidoptera, Spodoptera littoralis an antimicrobial peptide was found secreted by Enterococcus located on the gut epithelium (Shao et al, 2017). Furthermore, microbiomes from distinct agave plants have been reported (Coleman‐Derr et al., 2016; Martinez‐Rodriguez et al., 2019; Flores‐Nuñez et al 2020) and several of the bacterial genera encountered therein were also identified in red worm guts (Hernández‐Flores et al, 2015) and from gut microbiomes in our laboratory.
Cellulosimicrobium found in cactus has an outstanding capability to degrade the plant cell wall using cellulases, xylanases and pectinases (Han et al, 2009a). Cellulosimicrobium was isolated from elephant and alpaca faeces (Jiang et al, 2013). We identified this bacterium from maguey worm microbiomes. Isolated strains from the larva guts of another lepidoptera, Plutella xylostella were capable of degrading plant phenolic compounds (Xia et al, 2017).
Monarch butterflies
Monarch butterflies have a specialized plant diet. Milkweeds of the genus Asclepias are the preferred food of monarch caterpillars and adult monarchs feed on the nectar and pollen of flowers that provide sugars and other nutrients. We analysed the overwintering microbiota from guts of adult monarch butterflies, Danaus plexippus (Lepidoptera: Nymphalidae, Danainae) by sequencing metagenomes and 16S rRNA genes from bacterial isolates. We identified 16S rRNA gene sequences for Asaia, Pseudomonas, Serratia, Enterococcus, Carnobacterium, Kinetoplastibacterium, Xylophilus, Polaromonas, Herbaspirillum and Lactococcus bacteria. Some of these bacteria such as Pseudomonas, Serratia, Enterococcus, and Herbaspirillum are well known endophytes of plants. In guts of adult monarch butterflies, the acetic acid bacterium Commensalibacter was the most abundant (Servín‐Garcidueñas et al., 2014; Servin‐Garcidueñas and Martínez‐Romero, 2014).
Cabbage white butterfly
The adult cabbage white butterflies (Pieris rapae) contain Enterobacter as well as Flavobacterium in their guts (Steinhaus, 1941); in the larvae, species from the genera Asaia, Acinetobacter, Methylobacterium, Enterobacter and Pantoea have been found (Robinson et al, 2010a, 2010b). Bacteria from these genera are common plant endophytes. Sinigrin, a glucosinolate found in Brussels sprouts, affects the bacterial community composition when fed to the cabbage white butterfly larvae. The bacteria found in the midgut may participate in sinigrin degradation (Robinson et al, 2010a, 2010b).
Drosophila and fruit flies
Lactobacillus plantarum is found in plants as endophyte (Minervini et al., 2018) and commonly in Drosophila gastrointestinal tracts (Siezen et al, 2010). L. plantarum from Drosophila is related to strains from the same species from human faeces and plants (Fig. 2C). Drosophila in natural habitats may feed on plants but mainly on yeast from fermented fruits (Becher et al, 2012). L. plantarum, considered a facultative symbiont, is continuously ingested and excreted by Drosophila (Storelli et al, 2018). It provides acetyl‐glutamine to the host, produces a hormone‐signalling control (Storelli et al, 2011) and stimulates the production of intestinal peptidases (Matos et al, 2017). Notably, an improvement of host beneficial effects was obtained by an experimental evolution assay of L. plantarum across 20 Drosophila generations (Martino et al, 2018).
Similarly, from the Mediterranean fruit fly several enterobacteria were isolated including Klebsiella that was found in all samples; nitrogen fixation was detected in the enterobacterial cultures (Behar et al, 2005). An additional study from our laboratory (using 16S rRNA gene identification of cultured gut bacteria) from wild larvae from the Mexican fruit fly Anastrepha ludens showed Bacillus, Lactobacillus, Pseudomonas, Enterobacter, Klebsiella, Acinetobacter, Leuconostoc and Weissella. All of them are reported endophytes. Nitrogen‐fixing activity was detected by the acetylene‐reduction assay with Klebsiella and Enterobacter. Pectinolytic activity was observed in Pseudomonas and Bacillus and uricolytic activity in Pseudomonas. Additionally, we isolated in culture Pichia and Hanseniaspora yeasts (that are known fungal endophytes) from A. ludens larvae. Pichia was present in the oranges where larvae were feeding. Both yeasts were found in Drosophila suzukii (Hamby et al, 2012). A. ludens is an important pest in Mexico and Central America that attacks several fruit species especially citrus and mangos. Larvae feed on the fruits causing great losses, and biological control with sterile males has been used to control this pest (Barker et al., 2013).
An endophytic‐enteric‐soil‐endophytic cycle
We showed data to support the endophytic origin of some gut bacteria, and we propose here the natural existence of an endophytic‐enteric‐soil‐endophytic microbiota cycle, designated hereafter as endophytic‐enteric cycle (Fig. 3) in which plant tissues could act as enteric ‘fibre capsules’ to protect plant endophytes from being digested in the stomach and allowing their later release in the intestine. If plant‐borne microbes from guts pass to the environment, then they may become soil bacteria and a natural biological inoculant of plants. By colonizing plants, these bacteria would complete a microbe cycle. Common ecological features in roots and gut (Ramírez‐Puebla et al, 2013) support their sharing bacteria. A shortcut in the endophytic‐enteric cycle would be faecal ingestion (coprophagy), which is a common practice among many animals, we surmise that animals were pioneers on faecal transplants. Fecal transplants are successfully used in modern Medicine by donating processed faeces from healthy humans to human patients with chronic gut diseases (Petrof and Khoruts, 2014).
Fig. 3.
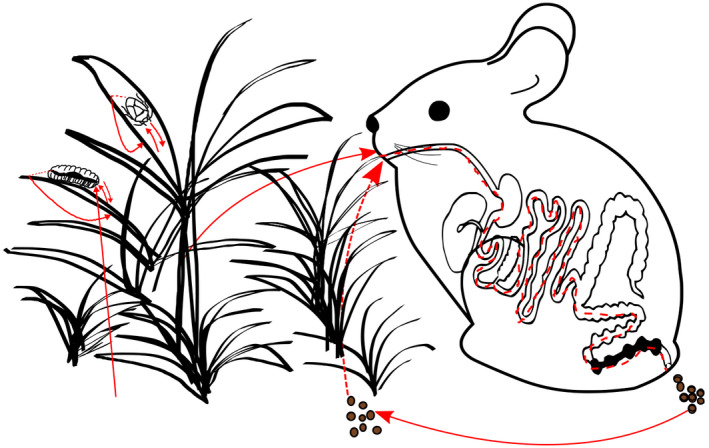
Schematic representation of the endophytic‐enteric microbiota cycle. Inside plant‐tissue endophytes may gain access to animal guts when animals eat plants, in turn animals produce faeces that may carry the ingested plant bacteria that would be available to colonize plants again. Coprophagy (faeces ingestion) is a shortcut in the cycle, as well as the transfer of insect bacteria to the plant during feeding or direct acquisition of bacteria from soil.
Endophytes, which may or may not reproduce in the gut, could pass in faeces to soil and water. Tracing bacteria from animal guts to the soil was made possible by identifying antibiotic‐resistant bacteria in soil that derived from manure obtained from chicken or cow faeces (Wichmann et al, 2014). Spread of enteric bacteria could lead to a flow of antibiotic resistance genes from sewage, manure or slurry to humans (Linton, 1986). Farmland soils that were fertilized with chicken manure had high levels of antibiotic‐resistant bacteria (Zhao et al, 2017), and the impact of the use of antibiotics in farm animals has been studied (Qian et al, 2018; Heuer et al., 2011). Manure would constitute not only a way to recycle large amounts of N to plants but a source of bacteria for plants, and this has been shown when plants got contaminated with pathogens from animal manures (Guan and Holley, 2003). It was reported that human health depends on plant health, which in turn is determined by soils (Hirt, 2020).
As soils are rich in bacteria, insect microbiota may derive directly from soil (Fig. 3) and indeed soils were the main source and not host plants for the leaf‐eating caterpillar microbiota (Hannula et al, 2019). This may be considered as another shortcut in the cycle and remains to be tested with other insects. Certainly, there is a very dynamic flux of microbes to the gut. In the reverse direction, phytophagous insects that harbour multiple gut bacteria in oral secretions may transfer microbes to plants during feeding (Fig. 3) (Chung et al, 2013) and this is how many insects are vectors of plant pathogens.
Endophytes normally feed on plant products, and their degradative capabilities could be of use in catabolizing plant‐derived nutrients or suppress pathogens and fix nitrogen in the gut, as they do in plants. Gut bacteria use plant fibres (dietary fibre) to produce short‐chain fatty acids (such as butyric acid) (Baxter et al., 2019) that are intestine cell (enterocyte) nutrients (Ríos‐Covián et al., 2016; Wang et al, 2019).
Not all plant bacteria would survive digestion and participate in the endophytic‐enteric cycle. Therein, spore‐producing bacteria may be particularly successful, as spores are resistant forms which could even become activated inside guts. Notably, spores may be an important constituent of gut microbiomes (Browne et al, 2016).
Critical issues
Even though there is clear evidence of plant‐derived bacteria in guts, a major question remains, are all small fragments of plant tissues effectively removed before macerating faecal samples? If not, they could be a source of DNA that would not reflect true gut bacteria. Work with panda faeces (Wei et al, 2007; Xue et al, 2015; Jin et al, 2020) addressed this issue, but it is not the case in all studies with faeces microbiota. Caution should be taken with metagenomic data, and microbial cultures from guts not to erroneously consider bacteria or fungi in plant fragments as bona fide gut microbiota. It is remarkable that most studies of gut microbiota are from faeces that may not well reflect gut bacteria (Zmora et al, 2018). Diet bacteria should be analysed concomitantly with gut microbiota and a comparison of the survival in the digestive tract of plant‐surface bacteria (Leff and Fierer, 2013) in relation to endophytes would be highly informative.
Seemingly, carnivores have endophytes in their guts as well. For example, Prevotella, which is known to be able to degrade plant derived carbohydrates, was the most abundant bacterium in the feline gut (Alessandri et al, 2020b). Endophytes in carnivore guts may derive from ingesting herbivore guts or faeces or soil‐contaminated meat. Notably, the gut microbiota of cats and dogs is determined as in humans by the diet and modified by overweight or inflammatory diseases (Alessandri et al, 2020a).
We suppose that plants may protect themselves from herbivory by possessing not only antimetabolites and toxins (in many cases produced by microbes) but also microbes that may cause disease or death to herbivorous animals when ingested. In contrast, plant bacteria may be useful, for example, Lactobacillus plantarum that is found in human and in omnivore gastrointestinal tracts (Siezen et al, 2010; Endo et al, 2010) is used as a probiotic (Panigrahi et al, 2017; Raveschot et al, 2020), which may help animals to better resist viral infections (Kumar et al, 2010; Kikuchi et al 2014), like other endophytic‐enteric bacteria do when used as probiotics (Chai et al, 2013; Mahooti et al 2020; Baud et al, 2020). However, probiotics constitute only transient members of the gut microbiota, remaining short periods in the gastrointestinal tract (Zmora et al, 2018) as we suppose some endophytes would do. Additionally, L. plantarum and Weissella are used in fermented food from plant products (Wacher‐Rodarte et al., 2015, Kavitake et al, 2016). Curiously, a metabolomic study showed that fermented food and stools were similar (Quinn et al, 2016).
Herbivores have most probably picked up their stable microbiota from plant‐associated bacteria. Particularly, the evolutionary history of insects is tightly dependent on plants as food (McKenna and Farell, 2006). When an herbivore has recently ingested an endophyte, gut strains would be identical or very similar to the plant isolates, as can be observed in phylogenies from Weissella from panda faeces, in L. plantarum and clostridia phylogenies (Fig. 2) or in burkholderias from the stinkbugs (Itoh et al, 2014; Tago et al, 2015; Xu et al, 2016b). On the other hand, when the endophytes coevolved with their animal hosts which maintain them by vertical transfer, then these bacteria would be divergent from plant bacteria, as observed in some insect symbionts such as Commensalibacter and Dactylopiibacterium (Servín‐Garcidueñas et al., 2014; Vera‐Ponce de León et al, 2017). Specialized insect endosymbionts in abdominal bacteriomes could have derived from gut bacteria, which in turn derived from endophytes that were found in plants that may not exist today. Flavobacterial endosymbionts of scale insects (Rosenblueth et al, 2012, 2018), which are around 200 million years old and provide essential amino acids to the insect, perhaps derived from plant flavobacteria which are found mainly in wild but not in domesticated plants (Pérez‐Jaramillo et al., 2018).
If we eat endophytes, we may excrete endophytes; this occurs with mealybug insects that eat and excrete honeydew containing Gluconacetobacter spp. (Ashbolt and Inkerman, 1990), a common nitrogen‐fixing endophyte from sugarcane (Caballero‐Mellado and Martinez‐Romero, 1994). Notably, the numbers of Gluconacetobacter bacteria diminish drastically in plants with nitrogen chemical fertilization (Fuentes‐Ramírez et al., 1999). Similarly, other agrochemicals may have strong effects on plant microbiota and thus on the endophytes which we ingest. Remarkably, fungal endophytes may vary depending on the habitat (Harrison and Griffin, 2020). Finally, inoculants used on plants should be considered not only in terms of their plant growth‐promoting capabilities but for their effects on human health as well. The use of cyanobacteria in plants should be carefully evaluated and avoided if possible. Cyanobacteria produce very harmful neurotoxins and hepatotoxins (Codd et al, 2005).
Conclusions
Certainly all herbivores eat endophytes, some of them may be digested or not liberated from plant tissues but others may be active in the gut and contribute to fibre or tannin digestion or to the synthesis of essential amino acids or vitamins, fix nitrogen or provide defence against pathogens. When studying herbivore gut microbiota, plant fragments in guts should be independently analysed.
While stable members of the gut microbiota may have derived from the vegetal diet, newly ingested endophytes may be quite variable. It would be desirable to eat the ones that contribute most to human health either as probiotics or with food. There is still lots to explore on this considering the variable responses in human individuals to probiotics, and the large effects of crop management on plant microbes. Ideally, plant microbial inoculants should benefit both plants and humans. Clearly, the endophyte‐enteric cycle has relevance to animal health. So, which endophytes would you like to eat?
Funding Information
This work was supported by grants from the National University of Mexico to EMR IN210021 and to LES‐G (PAPIIT‐DGAPA IA208019) to study the monarch microbiota.
Conflict of interest
None declared.
Acknowledgements
This work was supported by grants from the National University of Mexico to EMR IN210021 and to LES‐G (PAPIIT‐DGAPA IA208019) to study the monarch microbiota. We thank Rosendo Antonio Caro Gomez and Felipe Martinez at the Monarch Butterfly Biosphere Reserve (MBBR) in Mexico and Ministry of Environment and Natural Resources of Mexico for the permits for monarch samplings. We thank M. Dunn for critically reading the manuscript.
Microb. Biotechnol. (2021) 14(4), 1282–1299
References
- Alessandri, G. , Argentini, C. , Milani, C. , Turroni, F. , Cristina Ossiprandi, M. , van Sinderen, D. , and Ventura, M. (2020a) Catching a glimpse of the bacterial gut community of companion animals: a canine and feline perspective. Microb Biotechnol 13: 1708–1732. 10.1111/1751-7915.13656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessandri, G. , Milani, C. , Mancabelli, L. , Longhi, G. , Anzalone, R. , Lugli, G.A. , et al. (2020b) Deciphering the bifidobacterial populations within the canine and feline gut microbiota. Appl Environ Microbiol 86: e02875–19. 10.1128/AEM.02875-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano, N. , Courtiol, A. , Vielgrader, H. , Timms, P. , Roca, A.L. , and Greenwood, A.D. (2015) Variation in koala microbiomes within and between individuals: effect of body region and captivity status. Sci Rep 5: 10189. 10.1038/srep10189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez, N. , McKey, D. , Hossaert‐McKey, M. , Born, C. , Mercier, L. , and Benrey, B. (2005) Ancient and recent evolutionary history of the bruchid beetle, Acanthoscelides obtectus Say, a cosmopolitan pest of beans. Mol Ecol 14: 1015–1024. [DOI] [PubMed] [Google Scholar]
- Alves, A. , Correia, A. , Igual, J.M. , and Trujillo, M. E. (2014) Microbacterium endophyticum sp. nov. and Microbacterium halimionae sp. nov., endophytes isolated from the salt‐marsh plant Halimione portulacoides and emended description of the genus Microbacterium . Syst Appl Microbiol 37: 474–479. 10.1016/j.syapm.2014.08.004 [DOI] [PubMed] [Google Scholar]
- Ashbolt, N.J. , and Inkerman, P.A. (1990) Acetic acid bacterial biota of the pink sugar cane mealybug, Saccharococcus sacchari, and its environs. Appl Environ Microbiol 56: 707–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, C.J. , Gillett, A. , Polkinghorne, A. , and Timms, P. (2013) Investigation of the koala (Phascolarctos cinereus) hindgut microbiome via 16S pyrosequencing. Vet Microbiol 167: 554–564. [DOI] [PubMed] [Google Scholar]
- Baud, D. , Dimopoulou Agri, V. , Gibson, G.R. , Reid, G. , and Giannoni, E. (2020) Using probiotics to flatten the curve of coronavirus disease COVID‐2019 pandemic. Front Public Health 8: 186. 10.3389/fpubh.2020.00186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter, N.T. , Schmidt, A.W. , Venkataraman, A. , Kim, K.S. , Waldron, C. , and Schmidt, T.M. (2019) Dynamics of human gut microbiota and short‐chain fatty acids in response to dietary interventions with three fermentable fibers. mBio 10: e02566–18. 10.1128/mBio.02566-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer, E.A. , and Lamed, R. (1986) Ultrastructure of the cell surface cellulosome of Clostridium thermocellum and its interaction with cellulose. J Bacteriol 167: 828–836. 10.1128/jb.167.3.828-836.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont, M. , Paës, C. , Mussard, E. , Knudsen, C. , Cauquil, L. , Aymard, P. , and Gautier, R. (2020) Gut microbiota derived metabolites contribute to intestinal barrier maturation at the suckling‐to‐weaning transition. Gut Microbes 11: 1268–1286. 10.1080/19490976.2020.1747335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher, P.G. , Flick, G. , Rozpędowska, E. , Schmidt, A. , Hagman, A. , Lebreton, S. , et al. (2012) Yeast, not fruit volatiles mediate Drosophila melanogaster attraction, oviposition and development. Funct Ecol 26: 822–828. 10.1111/j.1365-2435.2012.02006.x [DOI] [Google Scholar]
- Behar, A. , Yuval, B. , and Jurkevitch, E. (2005) Enterobacteria‐mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata . Mol Ecol 14: 2637–2643. 10.1111/j.1365-294X.2005.02615.x [DOI] [PubMed] [Google Scholar]
- Berg, G. , Erlacher, A. , and Grube, M. (2014a) The edible plant microbiome: Importance and health issues. In Principles of Plant‐Microbe Interactions. Lugtenberg, B. (Ed.). Springer International Publishing Switzerland, B. Lugtenberg, Chapter 44. pp. 419–426. [Google Scholar]
- Berg, G. , Erlacher, A. , Smalla, K. , and Krause, R. (2014b) Vegetable microbiomes: is there a connection among opportunistic infections, human health and our 'gut feeling'? Microb Biotechnol 7: 487–95. 10.1111/1751-7915.12159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlemont, R. , and Martiny, A.C. (2013) Phylogenetic distribution of potential cellulases in bacteria. Appl Environ Microbiol 79: 1545–1554. 10.1128/AEM.03305-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann, P.H. , and Vega, F.E. (2020) Ecology and evolution of insect–fungus mutualisms. Annual Rev Entomol 65: 431–455. [DOI] [PubMed] [Google Scholar]
- Blyton, M.D. , Soo, R.M. , Whisson, D. , Marsh, K.J. , Pascoe, J. , Le Pla, M. , and Moore, B.D. (2019) Faecal inoculations alter the gastrointestinal microbiome and allow dietary expansion in a wild specialist herbivore, the koala. Animal Microbiome 1: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante, P. , Venice, F. , and Lanfranco, L. (2019) The mycobiota: fungi take their place between plants and bacteria. Curr Opin Microbiol 49: 18–25. 10.1016/j.mib.2019.08.004 [DOI] [PubMed] [Google Scholar]
- Boucias, D.G. , Garcia‐Maruniak, A. , Cherry, R. , Lu, H. , Maruniak, J.E. , and Lietze, V.U. (2012a) Detection and characterization of bacterial symbionts in the Heteropteran, Blissus insularis . FEMS Microbiol Ecol 82: 629–641. 10.1111/j.1574-6941.2012.01433.x [DOI] [PubMed] [Google Scholar]
- Boucias, D.G. , Lietze, V.U. , and Teal, P. (2012b) Chemical signals that mediate insect‐fungal interactions. In Biocommunication of Fungi. Springer, Dordrecht, pp. 305–336. [Google Scholar]
- Brice, K.L. , Trivedi, P. , Jeffries, T.C. , Blyton, M.D. , Mitchell, C. , Singh, B.K. , and Moore, B.D. (2019) The Koala (Phascolarctos cinereus) faecal microbiome differs with diet in a wild population. PeerJ 7: e6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne, H.P. , Forster, S.C. , Anonye, B.O. , Kumar, N. , Neville, B.A. , Stares, M.D. , et al. (2016) Culturing of 'unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature 533: 543–546. 10.1038/nature17645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busby, P.E. , Ridout, M. , and Newcombe, G. (2016) Fungal endophytes: modifiers of plant disease. Plant Mol Biol 90: 645–655. 10.1007/s11103-015-0412-0 [DOI] [PubMed] [Google Scholar]
- Bustamante‐Brito, R. , Vera‐Ponce de León, A. , Rosenblueth, M. , Martínez‐Romero, J.C. , and Martínez‐Romero, E. (2019) Metatranscriptomic analysis of the bacterial symbiont dactylopiibacterium carminicum from the carmine cochineal Dactylopius coccus (Hemiptera: Coccoidea: Dactylopiidae). Life 9: 4. 10.3390/life9010004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero‐Mellado, J. , and Martinez‐Romero, E. (1994) Limited genetic diversity in the endophytic sugarcane bacterium <em>Acetobacter diazotrophicus</em>. Appl Environ Microbiol 60: 1532–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas‐Aquino, M.D.R. , Alarcón‐Rodríguez, N.M. , Rivas‐Medrano, M. , González‐Hernández, H. , Vargas‐Hernández, M. , Sánchez‐Arroyo, H. , and Llanderal‐Cazares, C. (2018) Molecular delineation of the agave red worm Comadia redtenbacheri (Lepidoptera: Cossidae). Zootaxa 4375: 358–370. [DOI] [PubMed] [Google Scholar]
- Cardona, F. , Andrés‐Lacueva, C. , Tulipani, S. , Tinahones, F.J. , and Queipo‐Ortuño, M.I. (2013) Benefits of polyphenols on gut microbiota and implications in human health. J Nutritional Biochem 24: 1415–1422. [DOI] [PubMed] [Google Scholar]
- Chai, W. , Burwinkel, M. , Wang, Z. , Palissa, C. , Esch, B. , Twardziok, S. , et al. (2013) Antiviral effects of a probiotic Enterococcus faecium strain against transmissible gastroenteritis coronavirus. Arch Virol 158: 799–807. 10.1007/s00705-012-1543-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez‐Moreno, C.K. , Tecante, A. , and Casas, A. (2009) The Opuntia (Cactaceae) and Dactylopius (Hemiptera: Dactylopiidae) in Mexico: a historical perspective of use, interaction and distribution. Biodiversity Conserv 18: 3337. 10.1007/s10531-009-9647-x [DOI] [Google Scholar]
- Chávez‐Moreno, C.K. , Tecante, A. , Casas, A. , and Claps, L.E. (2011) Distribution and habitat in Mexico of Dactylopius Costa (Hemiptera: Dactylopiidae) and their cacti hosts (Cactaceae: Opuntioideae). Neotropical Entomol 40: 62–71. 10.1590/S1519-566X2011000100009 [DOI] [PubMed] [Google Scholar]
- Chen, B. , Du, K. , Sun, C. , Vimalanathan, A. , Liang, X. , Li, Y. , and Shao, Y. (2018) Gut bacterial and fungal communities of the domesticated silkworm (Bombyx mori) and wild mulberry‐feeding relatives. ISME J 12: 2252–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. , Hao, D. , Wei, Z. , Wang, L. , and Lin, T. (2020) Bacterial communities associated with the pine wilt disease insect vector Monochamus alternatus (Coleoptera: Cerambycidae) during the larvae and pupae stages. Insects 11(6): 376– 10.3390/insects11060376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, S.H. , Rosa, C. , Scully, E.D. , and Felton, G.W. (2013) Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc Natl Acad Sci USA 110: 15728–15733. 10.1073/pnas.1308867110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codd, G.A. , Morrison, L.F. , and Metcalf, J.S. (2005) Cyanobacterial toxins: risk management for health protection. Toxicol Appl Pharmacol 203: 264–272. 10.1016/j.taap.2004.02.016 [DOI] [PubMed] [Google Scholar]
- Coleman‐Derr, D. , Desgarennes, D. , Fonseca‐Garcia, C. , Gross, S. , Clingenpeel, S. , Woyke, T. , and Tringe, S.G. (2016) Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol 209: 798–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combes, S. , Gidenne, T. , Cauquil, L. , Bouchez, O. , and Fortun‐Lamothe, L. (2014) Coprophagous behavior of rabbit pups affects implantation of cecal microbiota and health status. J Anim Sci 92: 652–665. 10.2527/jas.2013-6394 [DOI] [PubMed] [Google Scholar]
- Cripps, M.G. , Edwards, G.R. , and McKenzie, S.L. (2013) Grass species and their fungal symbionts affect subsequent forage growth. Basic Appl Ecol 14: 225–234. [Google Scholar]
- Crowley, E.J. , King, J.M. , Wilkinson, T. , Worgan, H.J. , Huson, K.M. , Rose, M.T. , and McEwan, N.R. (2017) Comparison of the microbial population in rabbits and guinea pigs by next generation sequencing. PLoS One 12: e0165779. 10.1371/journal.pone.0165779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, L.A. , Maurice, C.F. , Carmody, R.N. , Gootenberg, D.B. , Button, J.E. , Wolfe, B.E. , et al. (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505: 559–63. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degli Esposti, M. , and Martinez Romero, E. (2017) The functional microbiome of arthropods. PLoS One 12: e0176573. 10.1371/journal.pone.0176573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierenfeld, E.S. , Hintz, H.F. , Robertson, J.B. , Van Soest, P.J. , and Oftedal, O.T. (1982) Utilization of bamboo by the giant panda. J Nutrition 112: 636–641. [DOI] [PubMed] [Google Scholar]
- Dobritsa, A.P. , and Samadpour, M. (2016) Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia . Int J Syst Evol Microbiol 66: 2836–2846. 10.1099/ijsem.0.001065 [DOI] [PubMed] [Google Scholar]
- Endo, A. , Futagawa‐Endo, Y. , and Dicks, L.M. (2010) Diversity of Lactobacillus and Bifidobacterium in feces of herbivores, omnivores and carnivores. Anaerobe 16: 590–596. 10.1016/j.anaerobe.2010.10.005 [DOI] [PubMed] [Google Scholar]
- Engel, P. , Martinson, V.G. , and Moran, N.A. (2012) Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci USA 109: 11002–11007. 10.1073/pnas.1202970109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores‐Núñez, V.M. , Fonseca‐García, C. , Desgarennes, D. , Eloe‐Fadrosh, E. , Woyke, T. , and Partida‐Martínez, L.P. (2020) Functional signatures of the epiphytic prokaryotic microbiome of agaves and cacti. Front Microbiol 10: 3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissac, X. , Edwards, M.G. , Du, J.P. , Gatehouse, A.M. , and Gatehouse, J.A. (2002) Putative protein digestion in a sap‐sucking homopteran plant pest (rice brown plant hopper; Nilaparvata lugens: Delphacidae) ‐ identification of trypsin‐like and cathepsin B‐like proteases. Insect Biochem Mol Biol 32: 967–978. 10.1016/s0965-1748(02)00033-4 [DOI] [PubMed] [Google Scholar]
- Friesen, M.L. , Porter, S.S. , Stark, S.C. , von Wettberg, E.J. , Sachs, J.L. , and Martinez‐Romero, E. (2011) Microbially mediated plant functional traits. Ann Rev Ecol Evol Systemat 42: 23–46. [Google Scholar]
- Fuentes‐Ramírez, L.E. , Caballero‐Mellado, J. , Sepúlveda, J. , and Martínez‐Romero, E. (1999) Colonization of sugarcane by Acetobacter diazotrophicus is inhibited by high N‐fertilization. FEMS Microbiol Ecol 29: 117–128. 10.1111/j.1574-6941.1999.tb00603.x [DOI] [Google Scholar]
- Futuyma, D.J. , and Agrawal, A.A. (2009) Macroevolution and the biological diversity of plants and herbivores. Proc Natl Acad Sci USA 106: 18054–18061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐De la Peña, C. , Garduño‐Nino, E. , Vaca‐Paniagua, F. , Diaz‐Velasquez, C. , Barrows, C.W. , Gomez‐Gil, B. , and Valenzuela‐Nunez, L.M. (2019) Comparison of the fecal bacterial microbiota composition between wild and captive bolson tortoises (Gopherus flavomarginatus). Herpetol Conservation Biol 14: 587–600. [Google Scholar]
- Gibson, C.M. , and Hunter, M.S. (2010) Extraordinarily widespread and fantastically complex: comparative biology of endosymbiotic bacterial and fungal mutualists of insects. Ecol Lett 13: 223–234. [DOI] [PubMed] [Google Scholar]
- Glassing, A. , Dowd, S. E. , Galandiuk, S. , Davis, B. , and Chiodini, R.J. (2016) Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathogens 8: 24. 10.1186/s13099-016-0103-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, T.Y. , and Holley, R.A. (2003) Pathogen survival in swine manure environments and transmission of human enteric illness–a review. J Environ Quality 32: 383–392. 10.2134/jeq2003.3830 [DOI] [PubMed] [Google Scholar]
- Guo, W. , Chen, Y. , Wang, C. , Ning, R. , Zeng, B. , Tang, J. , et al. (2020) The carnivorous digestive system and bamboo diet of giant pandas may shape their low gut bacterial diversity. Conserv Physiol 8: coz104. 10.1093/conphys/coz104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W. , Mishra, S. , Wang, C. , Zhang, H. , Ning, R. , Kong, F. , and Li, Y. (2019) Comparative study of gut microbiota in wild and captive giant pandas (Ailuropoda melanoleuca). Genes 10: 827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamby, K.A. , Hernández, A. , Boundy‐Mills, K. , and Zalom, F.G. (2012) Associations of yeasts with spotted‐wing Drosophila (Drosophila suzukii; Diptera: Drosophilidae) in cherries and raspberries. Appl Environ Microbiol 78: 4869–4873. 10.1128/AEM.00841-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, M.K. , Lee, J.S. , Oh, H.W. , Park, D.S. , Shin, D.H. , Bae, K.S. , and Park, H.Y. (2009a) Isolation and characterization of a cellulase‐free endo‐β‐1, 4‐xylanase produced by an invertebrate‐symbiotic bacterium, Cellulosimicrobium sp. HY‐13. Process Biochem 44: 1055–1059. [Google Scholar]
- Han, S. , Xia, D.L. , Li, L.B. , and Han, J.G. (2010) Diversity of the phosphate solubilizing bacteria isolated from the root of Phyllostachys pubescens . J Agric Univ Hebei 33: 26–31. [Google Scholar]
- Han, J. , Xia, D. , Li, L. , Sun, L. , Yang, K. , and Zhang, L. (2009b) Diversity of culturable bacteria isolated from root domains of moso bamboo (Phyllostachys edulis). Microb Ecol 58: 363–373. [DOI] [PubMed] [Google Scholar]
- Hannula, S.E. , Zhu, F. , Heinen, R. , and Bezemer, T.M. (2019) Foliar‐feeding insects acquire microbiomes from the soil rather than the host plant. Nat Commun 10: 1254. 10.1038/s41467-019-09284-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt‐Brown, F. (2004) Biological characteristic of domestic rabbit/ Digestive physiology, In: Textbook of Rabbit Medicine (Vol. 3). Elsevier Science, Oxford. [Google Scholar]
- Hardoim, P.R. , van Overbeek, L.S. , Berg, G. , Pirttilä, A.M. , Compant, S. , Campisano, A. , et al. (2015) The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev MMBR 79: 293–320. 10.1128/MMBR.00050-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, J.G. , and Griffin, E.A. (2020) The diversity and distribution of endophytes across biomes, plant phylogeny and host tissues: how far have we come and where do we go from here? Environ Microbiol 22: 2107–2123. 10.1111/1462-2920.14968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehemann, J.H. , Correc, G. , Barbeyron, T. , Helbert, W. , Czjzek, M. , and Michel, G. (2010) Transfer of carbohydrate‐active enzymes from marine bacteria to Japanese gut microbiota. Nature 464: 908–912. 10.1038/nature08937 [DOI] [PubMed] [Google Scholar]
- Hernández‐Flores, L. , Llanderal‐Cázares, C. , Guzmán‐Franco, A.W. , and Aranda‐Ocampo, S. (2015) Bacteria present in Comadia redtenbacheri larvae (Lepidoptera: Cossidae). J Med Entomol 52: 1150–1158. [DOI] [PubMed] [Google Scholar]
- Hernández‐García, J.A. , Briones‐Roblero, C.I. , Rivera‐Orduña, F.N. , and Zúñiga, G. (2017) Revealing the gut bacteriome of Dendroctonus bark beetles (Curculionidae: Scolytinae): diversity, core members and co‐evolutionary patterns. Sci Reports 7: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera, J. , Poudel, R. , and Khidir, H.H. (2011) Molecular characterization of coprophilous fungal communities reveals sequences related to root‐associated fungal endophytes. Microbial Ecol 61: 239–244. [DOI] [PubMed] [Google Scholar]
- Heuer, H. , Schmitt, H. , and Smalla, K. (2011) Antibiotic resistance gene spread due to manure application on agricultural fields. Curr Opin Microbiol 14: 236–243. 10.1016/j.mib.2011.04.009 [DOI] [PubMed] [Google Scholar]
- Hirt, H. (2020) Healthy soils for healthy plants for healthy humans: How beneficial microbes in the soil, food and gut are interconnected and how agriculture can contribute to human health. EMBO Rep 21: e51069. 10.15252/embr.202051069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, C.E. , Jeong, H. , Jo, S.H. , Jeong, J.C. , Kwon, S.Y. , An, D. , and Park, J.M. (2016) A leaf‐inhabiting endophytic bacterium, Rhodococcus sp. KB6, enhances sweet potato resistance to black rot disease caused by Ceratocystis fimbriata . J Microbiol Biotechnol 26: 488–492. 10.4014/jmb.1511.11039 [DOI] [PubMed] [Google Scholar]
- Itoh, H. , Aita, M. , Nagayama, A. , Meng, X.Y. , Kamagata, Y. , Navarro, R. , et al. (2014) Evidence of environmental and vertical transmission of Burkholderia symbionts in the oriental chinch bug, Cavelerius saccharivorus (Heteroptera: Blissidae). Appl Environ Microbiol 80: 5974–5983. 10.1128/AEM.01087-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y. , Chen, X. , Han, L. , Huang, X.S. , Qiu, S.M. , and Jiang, C.L. (2013) Diversity of cultivable actinomycetes in 6 species of herbivore feces. Int J Microbiol Res 1: 76–84. [Google Scholar]
- Jin, L. , Wu, D. , Li, C. , Zhang, A. , Xiong, Y. , Wei, R. , and Li, B. (2020) Bamboo nutrients and microbiome affect gut microbiome of giant panda. Symbiosis 80: 293–304. [Google Scholar]
- Judd, F.W. , and Rose, F.L. (1983) Population structure, density and movements of the Texas tortoise Gopherus berlandieri . Southwestern Naturalist 28: 387–398. [Google Scholar]
- El Kaoutari, A. , Armougom, F. , Leroy, Q. , Vialettes, B. , Million, M. , Raoult, D. , and Henrissat, B. (2013) Development and validation of a microarray for the investigation of the CAZymes encoded by the human gut microbiome. PLoS One 8: e84033. 10.1371/journal.pone.0084033 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kararli, T.T. (1995) Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharm Drug Dispos 16: 351–380. 10.1002/bdd.2510160502 [DOI] [PubMed] [Google Scholar]
- Kavitake, D. , Devi, P.B. , Singh, S.P. , and Shetty, P.H. (2016) Characterization of a novel galactan produced by Weissella confusa KR780676 from an acidic fermented food. Int J Biol Macromol 86: 681–689. 10.1016/j.ijbiomac.2016.01.099 [DOI] [PubMed] [Google Scholar]
- Kikuchi, Y. , Hayatsu, M. , Hosokawa, T. , Nagayama, A. , Tago, K. , and Fukatsu, T. (2012) Symbiont mediated insecticide resistance. Proc Nat. Acad Sci USA 109: 8618–8622. 10.1073/pnas.1200231109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, Y. , Hosokawa, T. , and Fukatsu, T. (2007) Insect‐microbe mutualism without vertical transmission. Environ Microbiol 73: 4308–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, Y. , Hosokawa, T. , and Fukatsu, T. (2011) An ancient but promiscuous host–symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J 5: 446–460. 10.1038/ismej.2010.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, Y. , Kunitoh‐Asari, A. , Hayakawa, K. , Imai, S. , Kasuya, K. , Abe, K. , et al. (2014) Oral administration of Lactobacillus plantarum strain AYA enhances IgA secretion and provides survival protection against influenza virus infection in mice. PLoS ONE 9: e86416– 10.1371/journal.pone.0086416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.K. , and Lee, B.B.L. (2015) Symbiotic factors in Burkholderia essential for establishing an association with the bean bug, Riptortus pedestris . Arch Insect Biochem Physiol 88: 4–17. 10.1002/arch.21218 [DOI] [PubMed] [Google Scholar]
- Klinder, A. , Shen, Q. , Heppel, S. , Lovegrove, J.A. , Rowland, I. , and Tuohy, K.M. (2016) Impact of increasing fruit and vegetables and flavonoid intake on the human gut microbiota. Food Function 7: 1788–1796. [DOI] [PubMed] [Google Scholar]
- Kovacs, M. , Szendro, Z. , Milisits, G. , Bota, B. , Biro‐Nemeth, E. , Radnai, I. , and Horn, P. (2006) Effect of nursing methods and faeces consumption on the development of the bacteroides, lactobacillus and coliform flora in the caecum of the newborn rabbits. Reprod Nutr Dev 46: 205–210. 10.1051/rnd:2006010 [DOI] [PubMed] [Google Scholar]
- Kumar, R. , Seo, B.J. , Mun, M.R. , Kim, C.J. , Lee, I. , Kim, H. , and Park, Y.H. (2010) Putative probiotic Lactobacillus spp. from porcine gastrointestinal tract inhibit transmissible gastroenteritis coronavirus and enteric bacterial pathogens. Trop Anim Health Prod 42: 1855–1860. 10.1007/s11250-010-9648-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Las Rivas, B. , Rodríguez, H. , Anguita, J. , and Muñoz, R. (2019) Bacterial tannases: classification and biochemical properties. Appl Microbiol Biotechnol. 103: 603–623. 10.1007/s00253-018-9519-y [DOI] [PubMed] [Google Scholar]
- Leff, J. W. , and Fierer, N. (2013) Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS One 8: e59310. 10.1371/journal.pone.0059310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard, S.P. , Powell, J.E. , Perutka, J. , Geng, P. , Heckmann, L.C. , Horak, R.D. , et al. (2020) Engineered symbionts activate honey bee immunity and limit pathogens. Science 367: 573–576. 10.1126/science.aax9039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley, R.E. , Hamady, M. , Lozupone, C. , Turnbaugh, P.J. , Ramey, R.R. , Bircher, J.S. , et al. (2008) Evolution of mammals and their gut microbes. Science 320: 1647–51. 10.1126/science.1155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidstrom, M.E. , and Chistoserdova, L. (2002) Plants in the pink: cytokinin production by Methylobacterium . J Bacteriol 184: 1818. 10.1128/jb.184.7.1818.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares, D. , Fontanille, P. , and Larroche, Ch (2009) Exploration of α‐pinene degradation pathway of Pseudomonas rhodesiae CIP 107491. Application to novalic acid production in a bioreactor. Food Res Int 42: 461–469. [Google Scholar]
- Linton, A.H. (1986) Flow of resistance genes in the environment and from animals to man. J Antimicrobial Chemotherapy 18: 189–197. 10.1093/jac/18.Supplement_C.189 [DOI] [PubMed] [Google Scholar]
- Liu, F. , Yuan, Z. , Zhang, X. , Zhang, G. , and Xie, B. (2017) Characteristics and diversity of endophytic bacteria in moso bamboo (Phyllostachys edulis) based on 16S rDNA sequencing. Arch Microbiol 199: 1259–1266. [DOI] [PubMed] [Google Scholar]
- Lopez‐Lopez, A. , Rogel, M.A. , Ormeno‐Orrillo, E. , Martinez‐Romero, J. , and Martinez‐Romero, E. (2010) Phaseolus vulgaris seed‐borne endophytic community with novel bacterial species such as Rhizobium endophyticum sp. nov. Syst Appl Microbiol 33: 322–327. [DOI] [PubMed] [Google Scholar]
- Lu, M.G. , Jiang, J. , Liu, L. , Ma, A.P. , and Leung, F C. (2015a) Complete genome Ssquence of Klebsiella variicola strain HKUOPLA, a cellulose‐degrading bacterium isolated from giant panda feces. Genome Announc 3(5): pii: e01200–15. 10.1128/genomeA.01200-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, M.G. , Jiang, J. , Liu, L. , Ma, A.P.‐Y. , and Leung, F.C.‐C. (2015b) Complete genome sequence of Klebsiella pneumoniae Strain HKUOPLC, a cellulose‐degrading bacterium isolated from giant panda feces. Genome Announc 3(6): pii: e01318–15. 10.1128/genomeA.01318-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie, R.I. (2002) Mutualistic fermentative digestion in the gastrointestinal tract: diversity and evolution. Integr Comp Biol 42: 319–326. 10.1093/icb/42.2.319 [DOI] [PubMed] [Google Scholar]
- Mahooti, M. , Miri, S. M. , Abdolalipour, E. , and Ghaemi, A. (2020) The immunomodulatory effects of probiotics on respiratory viral infections: a hint for COVID‐19 treatment? Microbial Pathogenesis 148: 104452. 10.1016/j.micpath.2020.104452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makki, K. , Deehan, E.C. , Walter, J. , and Bäckhed, F. (2018) The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe 23: 705–715. [DOI] [PubMed] [Google Scholar]
- Martin, M.M. (1992) The evolution of insect‐fungus associations: from contact to stable symbiosis. Am Zool 32: 593–605. [Google Scholar]
- Martinez‐Rodriguez, A. , Macedo‐Raygoza, G. , Huerta‐Robles, A. X. , Reyes‐Sepulveda, I. , Lozano‐Lopez, J. , García‐Ochoa, E. Y. , and Beltran‐Garcia, M. J. (2019) Agave seed endophytes: Ecology and impacts on root architecture, nutrient acquisition, and cold stress tolerance. In: Seed Endophytes. Springer, Cham, pp. 139–170. [Google Scholar]
- Martinez‐Romero, E. , Rodriguez‐Medina, N. , Beltran‐Rojel, M. , Toribio‐Jimenez, J. , and Garza‐Ramos, U. (2018) Klebsiella variicola and Klebsiella quasipneumoniae with capacity to adapt to clinical and plant settings. Salud Publica Mex 60: 29–40. 10.21149/8156 [DOI] [PubMed] [Google Scholar]
- Martino, M.E. , Joncour, P. , Leenay, R. , Gervais, H. , Shah, M. , Hughes, S. , et al. (2018) Bacterial adaptation to the host's diet is a key evolutionary force shaping Drosophila‐Lactobacillus symbiosis. Cell Host Microbe 24: 109–119.e6. 10.1016/j.chom.2018.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos, R.C. , Schwarzer, M. , Gervais, H. , Courtin, P. , Joncour, P. , Gillet, B. , et al. (2017) D‐Alanylation of teichoic acids contributes to Lactobacillus plantarum‐mediated Drosophila growth during chronic undernutrition. Nat Microbiol 2: 1635–1647. 10.1038/s41564-017-0038-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzeo, G. , Nucifora, S. , Russo, A. , and Suma, P. (2019) Dactylopius opuntiae, a new prickly pear cactus pest in the Mediterranean: an overview. Entomol Exp Appl 167: 59–72. 10.1111/eea.12756 [DOI] [Google Scholar]
- McDonald, D. , Hyde, E. , Debelius, J.W. , Morton, J.T. , Gonzalez, A. , Ackermann, G. , et al. (2018) Research. mSystems 3: e00031–18. 10.1128/mSystems.00031-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, D.D. , and Farrell, B.D. (2006) Tropical forests are both evolutionary cradles and museums of leaf beetle diversity. Proc Natl Acad Sci USA 103: 10947–10951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel, P.S.B. , de Oliveira, M.N.V. , Delvaux, J.C. , de Jesus, G.L. , Borges, A.C. , Tótola, M.R. , and Costa, M.D. (2016) Diversity and distribution of the endophytic bacterial community at different stages of Eucalyptus growth. Antonie Van Leeuwenhoek 109: 755–771. [DOI] [PubMed] [Google Scholar]
- Minervini, F. , Lattanzi, A. , Dinardo, F.R. , De Angelis, M. , and Gobbetti, M. (2018) Wheat endophytic lactobacilli drive the microbial and biochemical features of sourdoughs. Food Microbiol 70: 162–171. 10.1016/j.fm.2017.09.006 [DOI] [PubMed] [Google Scholar]
- Montes‐Grajales, D. , Jiménez, B. , Rogel, M.A. , Alagón, A. , Esturau‐Escofet, N. , Esquivel, B. , et al. (2019) Nitrogen‐fixing Klebsiella variicola in feces from herbivorous tortoises. BioRxiv Cold Spring Harbor. 666818. 10.1101/666818 [DOI] [Google Scholar]
- Moore, B.D. , and Foley, W.J. (2000) A review of feeding and diet selection in koalas (Phascolarctos cinereus). Australian J Zool 48: 317–333. [Google Scholar]
- Moore, B.D. , Wallis, I.R. , Palá‐Paúl, J. , Brophy, J.J. , Willis, R.H. , and Foley, W.J. (2004) Antiherbivore chemistry of Eucalyptus–cues and deterrents for marsupial folivores. J Chem Ecol 30: 1743–1769. [DOI] [PubMed] [Google Scholar]
- Morales‐Jiménez, J. , Vera‐Ponce de León, A. , García‐Domínguez, A. , Martínez‐Romero, Z.G. , and Hernández‐Rodríguez, C. (2013) Nitrogen‐fixing and uricolytic bacteria associated with the gut of Dendroctonus rhizophagus and Dendroctonus valens (Curculionidae: Scotytinae). Microbial Ecol 66: 200–210. 10.10007/s00248-013-0206-3 [DOI] [PubMed] [Google Scholar]
- Motta, E. , Raymann, K. , and Moran, N.A. (2018) Glyphosate perturbs the gut microbiota of honey bees. Proc Natl Acad Sci USA 115: 10305–10310. 10.1073/pnas.1803880115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge, B.D. , Kuczynski, J. , Knights, D. , Clemente, J.C. , González, A. , Fontana, L. , et al. (2011) Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332: 970–974. 10.1126/science.1198719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo‐Ortiz, M.A. , and Gabaldón, T. (2019) Fungal evolution: major ecological adaptations and evolutionary transitions. Biological Rev 94: 1443–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveed, M. , Mitter, B. , Yousaf, S. , Pastar, M. , Afzal, M. , and Sessitsch, A. (2014) The endophyte Enterobacter sp. FD17: a maize growth enhancer selected based on rigorous testing of plant beneficial traits and colonization characteristics. Biol Fertil Soils 50: 249–262. [Google Scholar]
- Nunes, C. S. (2018) Depolymerizating enzymes—cellulases. In: Enzymes in human and animal nutrition : principles and perspectives. Nunes, C. S. , and Kumar, V. (eds). Academic Press/Elsevier, London, UK, pp. 107–132. 10.1016/B978-0-12-805419-2.00006-X [DOI] [Google Scholar]
- Ohbayashi, T. , Futahashi, R. , Terashima, M. , Barrière, Q. , Lamouche, F. , Takeshita, K. , et al. (2019) Comparative cytology, physiology and transcriptomics of Burkholderia insecticola in symbiosis with the bean bug Riptortus pedestris and in culture. ISME J 13: 1469–1483. 10.1038/s41396-019-0361-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Costa, L.E. , de Queiroz, M.V. , Borges, A.C. , de Moraes, C.A. , and de Araújo, E.F. (2012) Isolation and characterization of endophytic bacteria isolated from the leaves of the common bean (Phaseolus vulgaris). Brazilian J Microbiol 43: 1562–1575. 10.1590/S1517-838220120004000041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa, R. (1992) Tannin‐protein complex‐degrading enterobacteria isolated from the alimentary tracts of koalas and a selective medium for their enumeration. Appl Environ Microbiol 58: 1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa, R. , Bird, P.S. , Harbrow, D.J. , Ogimoto, K. , and Seymour, G.J. (1993a) Microbiological studies of the intestinal microflora of the koala, Phascolarctos‐cinereus. 1. Colonization of the cecal wall by tannin‐protein‐complex‐degrading enterobacteria. Australian J Zool 41: 599–609. [Google Scholar]
- Osawa, R. , Blanshard, W.H. , and Ocallaghan, P.G. (1993b) Microbiological studies of the intestinal microflora of the koala, Phascolarctos‐cinereus. 2. Pap, a special maternal feces consumed by juvenile koalas. Australian J Zool 41: 611–620. [Google Scholar]
- Panigrahi, P. , Parida, S. , Nanda, N.C. , Satpathy, R. , Pradhan, L. , Chandel, D.S. , et al. (2017) A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 548: 407–412. 10.1038/nature23480 [DOI] [PubMed] [Google Scholar]
- Pažoutová, S. , Follert, S. , Bitzer, J. , Keck, M. , Surup, F. , Šrůtka, P. , and Stadler, M. (2013) A new endophytic insect‐associated Daldinia species, recognised from a comparison of secondary metabolite profiles and molecular phylogeny. Fungal Divers 60: 107–123. [Google Scholar]
- Pérez‐Jaramillo, J.E. , Carrión, V.J. , de Hollander, M. , and Raaijmakers, J.M. (2018) The wild side of plant microbiomes. Microbiome 6: 143. 10.1186/s40168-018-0519-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof, E.O. , and Khoruts, A. (2014) From stool transplants to next‐generation microbiota therapeutics. Gastroenterology 146: 1573–1582. 10.1053/j.gastro.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J.E. , Eiri, D. , Moran, N.A. , and Rangel, J. (2018) Modulation of the honey bee queen microbiota: effects of early social contact. PLoS One 13: e0200527. 10.1371/journal.pone.0200527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prem Anand, A.A. , Vennison, S.J. , Sankar, S.G. , Gilwax Prabhu, D.I. , Vasan, P.T. , Raghuraman, T. , and Vendan, S.E. (2010) Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J Insect Sci 10: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procópio, R.E.I. , Araújo, W.L. , Maccheroni, W. , and Azevedo, J.L. (2009) Characterization of an endophytic bacterial community associated with Eucalyptus spp. Genet Mol Res 8: 1408–1422. 10.4238/vol8-4gmr691 [DOI] [PubMed] [Google Scholar]
- Qian, X. , Gu, J. , Sun, W. , Wang, X.J. , Su, J.Q. , and Stedfeld, R. (2018) Diversity, abundance, and persistence of antibiotic resistance genes in various types of animal manure following industrial composting. J Hazard Mater 344: 716–722. 10.1016/j.jhazmat.2017.11.020 [DOI] [PubMed] [Google Scholar]
- Quinn, R.A. , Navas‐Molina, J.A. , Hyde, E.R. , Song, S.J. , Vázquez‐Baeza, Y. , Humphrey, G. , et al. (2016) From sample to Multi‐Omics conclusions in under 48 hours. mSystems 1: e00038–16. 10.1128/mSystems.00038-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez‐Puebla, S.T. , Rosenblueth, M. , Chávez‐Moreno, C.K. , de Lyra, M.C. , Tecante, A. , and Martínez‐Romero, E. (2010) Molecular phylogeny of the genus Dactylopius (Hemiptera: Dactylopiidae) and identification of the symbiotic bacteria. Environ Entomol 39: 1178–1183. 10.1603/EN10037 [DOI] [PubMed] [Google Scholar]
- Ramírez‐Puebla, S.T. , Servín‐Garcidueñas, L.E. , Jiménez‐Marín, B. , Bolaños, L.M. , Rosenblueth, M. , Martínez, J. , et al. (2013) Gut and root microbiota commonalities. Appl Environ Microbiol 79: 2–9. 10.1128/AEM.02553-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveschot, C. , Coutte, F. , Frémont, M. , Vaeremans, M. , Dugersuren, J. , Demberel, S. , et al. (2020) Probiotic Lactobacillus strains from Mongolia improve calcium transport and uptake by intestinal cells in vitro. Food Res Internat (Ottawa Ont.) 133: 109201. 10.1016/j.foodres.2020.109201 [DOI] [PubMed] [Google Scholar]
- Ray, A.K. , Ghosh, K. , and Ringø, E. (2012) Enzyme‐producing bacteria isolated from fish gut: a review. Aquac Nutr 18: 465–492. [Google Scholar]
- Raymann, K. , Coon, K.L. , Shaffer, Z. , Salisbury, S. , and Moran, N.A. (2018) Pathogenicity of Serratia marcescens strains in honey bees. mBio 9: e01649–18. 10.1128/mBio.01649-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymann, K. , and Moran, N.A. (2018) The role of the gut microbiome in health and disease of adult honey bee workers. Curr Op Insect Sci 26: 97–104. 10.1016/j.cois.2018.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos‐Covián, D. , Ruas‐Madiedo, P. , Margolles, A. , Gueimonde, M. , de Los Reyes‐Gavilán, C. G. , and Salazar, N. (2016) Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol 7: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, C.J. , Bohannan, B.J. , and Young, V.B. (2010a) From structure to function: the ecology of host‐associated microbial communities. Microbiol Mol Biol Rev 74: 453–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, C.J. , Schloss, P. , Ramos, Y. , Raffa, K. , and Handelsman, J. (2010b) Robustness of the bacterial community in the cabbage white butterfly larval midgut. Microb Ecol 59: 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales‐Bravo, H. , Morales‐Torres, H. C. , Vázquez‐Martínez, J. , Molina‐Torres, J. , Olalde‐Portugal, V. , and Partida‐Martínez, L.P. (2017) Novel consortium of Klebsiella variicola and Lactobacillus species enhances the functional potential of fermented dairy products by increasing the availability of branched‐chain amino acids and the amount of distinctive volatiles. J Appl Microbiol 123: 1237–1250. 10.1111/jam.13565 [DOI] [PubMed] [Google Scholar]
- Rosenblueth, M. , Martinez, J. , Ramirez‐Puebla, S. , Vera‐Ponce de León, A. , Rosas‐Perez, T. , Bustamante‐Brit, R. , et al. (2018) Endosymbiotic microorganisms of scale insects. TIP Revista Especializada en Ciencias Químico‐Biológicas 21: 53–69. 10.1016/j.recqb.2017.08.006 [DOI] [Google Scholar]
- Rosenblueth, M. , Martínez, L. , Silva, J. , and Martínez‐Romero, E. (2004) Klebsiella variicola, a novel species with clinical and plant‐associated isolates. Syst Appl Microbiol 27: 27–35. [DOI] [PubMed] [Google Scholar]
- Rosenblueth, M. , and Martínez‐Romero, E. (2006) Bacterial endophytes and their interactions with hosts. Mol Plant Microbe Interact 19: 827–837. [DOI] [PubMed] [Google Scholar]
- Rosenblueth, M. , Sayavedra, L. , Sámano‐Sánchez, H. , Roth, A. , and Martínez‐Romero, E. (2012) Evolutionary relationships of flavobacterial and enterobacterial endosymbionts with their scale insect hosts (Hemiptera: Coccoidea). J Evol Biol 25: 2357–2368. 10.1111/j.1420-9101.2012.02611.x [DOI] [PubMed] [Google Scholar]
- Rothschild, D. , Weissbrod, O. , Barkan, E. , Kurilshikov, A. , Korem, T. , Zeevi, D. , et al. (2018) Environment dominates over host genetics in shaping human gut microbiota. Nature 555: 210–215. 10.1038/nature25973 [DOI] [PubMed] [Google Scholar]
- Sawana, A. , Adeolu, M. , and Gupta, R.S. (2014) Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet 5: 429. 10.3389/fgene.2014.00429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, W.H. (2001) The cellulosome and cellulose degradation by anaerobic bacteria. Appl Microbiol Biotechnol 56: 634–649. 10.1007/s002530100710 [DOI] [PubMed] [Google Scholar]
- Servín‐Garcidueñas, L.E. , and Martínez‐Romero, E. (2014) Complete mitochondrial genome recovered from the gut metagenome of overwintering monarch butterflies, Danaus plexippus (L.) (Lepidoptera: Nymphalidae, Danainae). Mitochondrial DNA 25: 427–428. [DOI] [PubMed] [Google Scholar]
- Servín‐Garcidueñas, L.E. , Sánchez‐Quinto, A. , and Martínez‐Romero, E. (2014) Draft genome sequence of Commensalibacter papalotli MX01, a symbiont identified from the guts of overwintering monarch butterflies. Genome Announc 2: e00128–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevim, A. , Sevim, E. , Demirci, M. , and Sandallı, C. (2016) The internal bacterial diversity of stored product pests. Ann Microbiol 66: 749–764. [Google Scholar]
- Shanmuganandam, S. , Hu, Y. , Strive, T. , Schwessinger, B. , and Hall, R.N. (2019) Uncovering the microbiome of invasive sympatric European brown hares and European rabbits in Australia. BioRxiv, 8 e9564. 10.1101/832477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, Y. , Chen, B. , Sun, C. , Ishida, K. , Hertweck, C. , and Boland, W. (2017) Symbiont‐derived antimicrobials contribute to the control of the Lepidopteran gut microbiota. Cell Chem Biol 24: 66–75. [DOI] [PubMed] [Google Scholar]
- Shastry, R.P. , Welch, M. , Rai, V.R. , Ghate, S.D. , Sandeep, K. , and Rekha, P.D. (2020) The whole‐genome sequence analysis of Enterobacter cloacae strain Ghats1: insights into endophytic lifestyle‐associated genomic adaptations. Arch Microbiol 202: 1571–1579. 10.1007/s00203-020-01848-5 [DOI] [PubMed] [Google Scholar]
- Siezen, R.J. , Tzeneva, V.A. , Castioni, A. , Wels, M. , Phan, H.T. , Rademaker, J.L. , et al. (2010) Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ Microbiol 12: 758–773. 10.1111/j.1462-2920.2009.02119.x [DOI] [PubMed] [Google Scholar]
- da Silva Fonseca, E. , Peixoto, R.S. , Rosado, A.S. , de Carvalho Balieiro, F. , Tiedje, J.M. , and da Costa Rachid, C.T.C. (2018) The microbiome of Eucalyptus roots under different management conditions and its potential for biological nitrogen fixation. Microb Ecol 75: 183–191. [DOI] [PubMed] [Google Scholar]
- Singh, S. , Thavamani, P. , Megharaj, M. , and Naidu, R. (2015) Multifarious activities of cellulose degrading bacteria from Koala (Phascolarctos cinereus) faeces. J Animal Sci Technol 57: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhaus, E.A. (1941) A study of the bacteria associated with thirty species of insects. J Bacteriol 42: 757–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storelli, G. , Defaye, A. , Erkosar, B. , Hols, P. , Royet, J. , and Leulier, F. (2011) Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR‐dependent nutrient sensing. Cell Metab 14: 403–414. 10.1016/j.cmet.2011.07.012 [DOI] [PubMed] [Google Scholar]
- Storelli, G. , Strigini, M. , Grenier, T. , Bozonnet, L. , Schwarzer, M. , Daniel, C. , et al. (2018) Drosophila perpetuates nutritional mutualism by promoting the fitness of its intestinal symbiont Lactobacillus plantarum . Cell Metab 27: 362–377.e8. 10.1016/j.cmet.2017.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakaran, S. , Retz, F. , Kikuchi, Y. , Kost, C. , and Kaltenpoth, M. (2015) Evolutionary transition in symbiotic syndromes enabled diversification of phytophagous insects on an imbalanced diet. ISME J 9: 2587–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tago, K. , Okubo, T. , Itoh, H. , Kikuchi, Y. , Hori, T. , Sato, Y. , et al. (2015) Insecticide‐degrading Burkholderia symbionts of the stinkbug naturally occupy various environments of sugarcane fields in a Southeast island of Japan. Microbes Environ 30: 29–36. 10.1264/jsme2.ME14124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita, K. , and Kikuchi, Y. (2020) Genomic comparison of insect gut symbionts from divergent Burkholderia Subclades. Genes (Basel) 11: 744. 10.3390/genes11070744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita, K. , Matsuura, Y. , Itoh, H. , Navarro, R. , Hori, T. , Sone, T. , et al. (2015) Burkholderia of plant‐beneficial group are symbiotically associated with bordered plant bugs (Heteroptera: Pyrrhocoroidea: Largidae). Microbes Environ 30: 321–329. 10.1264/jsme2.ME15153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamanai‐Shacoori, Z. , Smida, I. , Bousarghin, L. , Loreal, O. , Meuric, V. , Fong, S.B. , et al. (2017) Roseburia spp.: a marker of health? Future Microbiol 12: 157–170. 10.2217/fmb-2016-0130 [DOI] [PubMed] [Google Scholar]
- Taxis, T.M. , Wolff, S. , Gregg, S.J. , Minton, N.O. , Zhang, C. , Dai, J. , et al. (2015) The players may change but the game remains: network analyses of ruminal microbiomes suggest taxonomic differences mask functional similarity. Nucleic Acids Res 43: 9600–9612. 10.1093/nar/gkv973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thursby, E. , and Juge, N. (2017) Introduction to the human gut microbiota. Biochem J 474: 1823–1836. 10.1042/BCJ20160510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuda, M. (2007) Applied evolutionary ecology of insects of the subfamily Bruchinae (Coleoptera: Chrysomelidae). Appl Entomol Zool 42: 337–346. [Google Scholar]
- Tun, H. M. , Mauroo, N. F. , San Yuen, C. , Ho, J. C. W. , Wong, M. T. , and Leung, F. C. C. (2014) Microbial diversity and evidence of novel homoacetogens in the gut of both geriatric and adult giant pandas (Ailuropoda melanoleuca). PLoS One 9: e79902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uden, P. , Rounsaville, T.R. , Wiggans, G.R. , and Van Soest, P.J. (1982) The measurement of liquid and solid digesta retention in ruminants, equines and rabbits given timothy (Phleum pratense) hay. Br J Nutr 48: 329–339. 10.1079/bjn19820117 [DOI] [PubMed] [Google Scholar]
- Velasco‐Galilea, M. , Piles, M. , Vinas, M. , Rafel, O. , Gonzalez‐Rodriguez, O. , Guivernau, M. , and Sanchez, J.P. (2018) Rabbit microbiota changes throughout the intestinal tract. Front Microbiol 9: 2144. 10.3389/fmicb.2018.02144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura, C. , Briones‐Roblero, C.I. , Hernández, E. , Rivera‐Orduña, F.N. , and Zúñiga, G. (2018) Comparative analysis of the gut bacterial community of four Anastrepha fruit flies (Diptera: Tephritidae) based on pyrosequencing. Curr Microbiol 75: 966–976. 10.1007/s00284-018-1473-5 [DOI] [PubMed] [Google Scholar]
- Vera‐Ponce de León, A. , Ormeño‐Orrillo, E. , Ramírez‐Puebla, S.T. , Rosenblueth, M. , Degli Esposti, M. , Martínez‐Romero, J. , and Martínez‐Romero, E. (2017) Candidatus Dactylopiibacterium carminicum, a nitrogen‐fixing symbiont of Dactylopius Cochineal insects (Hemiptera: Coccoidea: Dactylopiidae). Genome Biol Evol 9: 2237–2250. 10.1093/gbe/evx156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacher‐Rodarte, M.d.C. , Trejo‐Muñúzuri, T.P. , Montiel‐Aguirre, J.F. , Drago‐Serrano, M.E. , Gutiérrez‐Lucas, R.L. , Castañeda‐Sánchez, J.I. , and Sainz‐Espuñes, T. (2016) Antibiotic resistance and multidrug‐resistant efflux pumps expression in lactic acid bacteria isolated from pozol, a nonalcoholic Mayan maize fermented beverage. Food Sci Nutrition 4(3): 423–430. 10.1002/fsn3.304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Wichienchot, S. , He, X. , Fu, X. , and Zhang, B. (2019) In vitro colonic fermentation of dietary fibers: Fermentation rate, short‐chain fatty acid production and changes in microbiota. Trends Food Sci Technol 88: 1–9. [Google Scholar]
- Wassermann, B. , Müller, H. , and Berg, G. (2019) An apple a day: which bacteria do we eat with organic and conventional apples? Front Microbiol 10: 1629. 10.3389/fmicb.2019.01629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, G. , Lu, H. , Zhou, Z. , Xie, H. , Wang, A. , Nelson, K. , and Zhao, L. (2007) The microbial community in the feces of the giant panda (Ailuropoda melanoleuca) as determined by PCR‐TGGE profiling and clone library analysis. Microb Ecol 54: 194–202. [DOI] [PubMed] [Google Scholar]
- Wichmann, F. , Udikovic‐Kolic, N. , Andrew, S. , and Handelsman, J. (2014) Diverse antibiotic resistance genes in dairy cow manure. MBio 5: e01017. 10.1128/mBio.01017-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, C.L. , Willard, S. , Kouba, A. , Sparks, D. , Holmes, W. , Falcone, J. , and Brown, A. (2013) Dietary shifts affect the gastrointestinal microflora of the giant panda (Ailuropoda melanoleuca). J Animal Physiol Animal Nutrition 97: 577–585. [DOI] [PubMed] [Google Scholar]
- Xia, X. , Gurr, G.M. , Vasseur, L. , Zheng, D. , Zhong, H. , Qin, B. , and Lin, H. (2017) Metagenomic sequencing of diamondback moth gut microbiome unveils key holobiont adaptations for herbivory. Front Microbiol 8: 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, J. , Strobel, G.A. , Feng, T. , Ren, H. , Mends, M.T. , Zhou, Z. , and Geary, B. (2015) An endophytic Coniochaeta velutina producing broad spectrum antimycotics. J Microbiol 53: 390–397. [DOI] [PubMed] [Google Scholar]
- Xu, Y. , Buss, E.A. , and Boucias, D.G. (2016a) Environmental transmission of the gut symbiont Burkholderia to phloem‐feeding Blissus insularis . PLoS One 11: e0161699. 10.1371/journal.pone.0161699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y. , Buss, E.A. , and Boucias, D.G. (2016b) Impacts of antibiotic and bacteriophage treatments on the gut‐symbiont‐associated Blissus insularis (Hemiptera: Blissidae). Insects 7: 61. 10.3390/insects7040061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, Z. , Zhang, W. , Wang, L. , Hou, R. , Zhang, M. , Fei, L. , and Jiang, C. (2015) The bamboo‐eating giant panda harbors a carnivore‐like gut microbiota, with excessive seasonal variations. MBio 6: e00022–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, R. , Yang, Z. , Zhang, Z. , Hu, T. , Chen, H. , Huang, F. , and Zhu, L. (2019) Are the gut microbial systems of giant pandas unstable? Heliyon 5: e02480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, M.L. , Dean, S.H. , Longo, A.V. , Rothermel, B.B. , Tuberville, T.D. , and Zamudio, K.R. (2015) Kinship, inbreeding and fine‐scale spatial structure influence gut microbiota in a hindgut‐fermenting tortoise. Mol Ecol 24: 2521–2536. 10.1111/mec.13169 [DOI] [PubMed] [Google Scholar]
- Zakhia, F. , Jeder, H. , Willems, A. , Gillis, M. , Dreyfus, B. , and de Lajudie, P. (2006) Diverse bacteria associated with root nodules of spontaneous legumes in Tunisia and first report for nifH‐like gene within the genera Microbacterium and Starkeya . Microb Ecol 51: 375–393. 10.1007/s00248-006-9025-0 [DOI] [PubMed] [Google Scholar]
- Zhao, X. , Wang, J. , Zhu, L. , Ge, W. , and Wang, J. (2017) Environmental analysis of typical antibiotic‐resistant bacteria and ARGs in farmland soil chronically fertilized with chicken manure. Sci Total Environ 593–594: 10–17. 10.1016/j.scitotenv.2017.03.062 [DOI] [PubMed] [Google Scholar]
- Zheng, H. , Perreau, J. , Powell, J.E. , Han, B. , Zhang, Z. , Kwong, W.K. , et al. (2019) Division of labor in honey bee gut microbiota for plant polysaccharide digestion. Proc Natl Acad Sci USA 116: 25909–25916. 10.1073/pnas.1916224116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L. , Wu, Q. , Dai, J. , Zhang, S. , and Wei, F. (2011) Evidence of cellulose metabolism by the giant panda gut microbiome. Proc Natl Acad Sci USA 108: 17714–17719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L. , Yang, Z. , Yao, R. , Xu, L. , Chen, H. , Gu, X. , and Yang, X. (2018) Potential mechanism of detoxification of cyanide compounds by gut microbiomes of bamboo‐eating pandas. MSphere 3: e00229‐18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmora, N. , Zilberman‐Schapira, G. , Suez, J. , Mor, U. , Dori‐Bachash, M. , Bashiardes, S. , et al. (2018) Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 174: 1388–1405.e21. 10.1016/j.cell.2018.08.041 [DOI] [PubMed] [Google Scholar]


