Summary
Listeria monocytogenes is a highly pathogenic foodborne bacterium that is ubiquitous in the natural environment and capable of forming persistent biofilms in food processing environments. This species has a rich repertoire of surface structures that enable it to survive, adapt and persist in various environments and promote biofilm formation. We review current understanding and advances on how L. monocytogenes organizes its surface for biofilm formation on surfaces associated with food processing settings, because they may be an important target for development of novel antibiofilm compounds. A synthesis of the current knowledge on the role of Listeria surfactome, comprising peptidoglycan, teichoic acids and cell wall proteins, during biofilm formation on abiotic surfaces is provided. We consider indications gained from genome‐wide studies and discuss surfactome structures with established mechanistic aspects in biofilm formation. Additionally, we look at the analogies to the species L. innocua, which is closely related to L. monocytogenes and often used as its model (surrogate) organism.
Listeria monocytogenes has a rich repertoire of surface structures that enable it to survive, adapt, and persist in various environments, and promote biofilm formation. This review provides an overview of current understanding and advances on how the surfactome comprising peptidoglycan, teichoic acids and cell wall proteins of L. monocytogenes organizes for biofilm formation. Additionally, we look at the analogies to the closely related species L. innocua.
Introduction
Listeria monocytogenes is a foodborne pathogen found in soil, water, plants and animals. From these sources, it enters and prospers in agroecosystems, thus contaminating food surfaces (EFSA, 2018a). High‐risk products for contamination with L. monocytogenes are meat‐ and vegetable‐based ready‐to‐eat food products, dairy products and seafood (Desai et al., 2019). Although ingestion of contaminated food usually causes mild to severe gastroenteritis, the invasive form of the disease often develops in high‐risk individuals, the most vulnerable of which are the elderly, immunocompromised individuals, pregnant women and foetuses (Radoshevich and Cossart, 2018). The illness caused by L. monocytogenes is called listeriosis, and it is proportionally the leading cause of hospitalizations and death among zoonoses cases in the EU (EFSA, 2018b). Worldwide listeriosis is a relatively rare disease; however, according to the World Health Organization, the high death rate makes it a significant public health concern (CAC/GL61 2007). Strict food regulations regarding L. monocytogenes in ready‐to‐eat food products have been established in many countries, and all strains belonging to this species are currently considered virulent.
There are four evolutionary lineages of L. monocytogenes (I, II, III and IV) and 15 different serotypes (Liu, 2006; Orsi et al., 2011). Serotype 4b strains from lineage I are associated with the majority of human cases, whereas the serotypes 1/2a and 1/2c from lineage II are most frequently isolated from food (Lomonaco et al., 2015). On a genetic level, there are 156 different multilocus sequence typing sequence types, which are organized into 63 different clonal complexes (CC) and several singletons. CC1, CC2 and CC6 are associated with clinical cases, whereas CC9 and CC121 are hypovirulent food‐related isolates (Maury et al., 2016). This hypovirulence is correlated with the presence of a defective gene for internalin A (inlA), a cell wall protein that is present in many serotype 1/2a strains and is crucial for invading epithelial cells (Nightingale et al., 2005; Maury et al., 2016). The virulence of serotype 4b strains was found to be associated with full‐length inlA, pathogenicity island LIPI‐3 and gene clusters for teichoic acid synthesis; the hypervirulent lineage II CC4 additionally carries LIPI‐4, which encodes a putative cellobiose‐type phosphotransferase system (Maury et al., 2016).
Generally, L. monocytogenes strains survive extreme environmental stress that is often employed to inhibit or eradicate microbial loads in foods, including low pH, high osmolarity and low water activity (Conner et al., 1990; Nolan et al., 1992; Shabala et al., 2008). These strains grow in wide temperature ranges, from physiological to refrigeration temperatures and can even survive freezing at −18 °C (Walker et al., 1990; Miladi et al., 2008). Motility is an important temperature‐dependent phenotypic trait of L. monocytogenes, as major flagellin protein FlaA (Lmo0690) levels are high at temperatures below 30 °C and decrease at physiological temperatures of 37 °C (Way et al., 2004). Besides stress resistance, biofilms have been suggested to play an important role in surviving the food processing environment in which Listeria spp. persist for months or even years (Lappi et al., 2004). For the survival and adaptation under various environments, Listeria probably relies on its rich repertoire of surface structures (Bierne and Cossart, 2007). Their role has been well described in the pathogenicity of Listeria, but understanding of mechanistic aspects on how these structures affect the biofilm formation process is still limited. This review provides a synthesis of the current knowledge on the effect of individual surfactome components on the biofilm formation on abiotic surfaces. We use the term surfactome to describe the whole spectrum of molecules found at the surface of L. monocytogenes cell wall: peptidoglycan, teichoic acids and cell wall proteins. Because surfactome components have been proven to affect L. monocytogenes ability to form biofilm, they may serve as the novel antimicrobial targets affecting directly the biofilm formation as an important Listeria persistence mechanism. This knowledge can also pave the way for identification of novel antimicrobials in the fight against the increasing occurrence of antimicrobial resistance in gram‐positive bacteria.
We consider here also the analogies to the species L. innocua, which is closely related to L. monocytogenes, and they also coinhabit the food processing environments (Lappi et al., 2004; Milillo et al., 2012; Costa et al., 2018). Both species are tolerant to extreme environmental conditions and form biofilms, but unlike L. monocytogenes, L. innocua does not have virulence factors and is thus considered non‐pathogenic (Glaser et al., 2001; Costa et al., 2018). Therefore, L. innocua is being tested as a surrogate organism for L. monocytogenes, especially for determining the efficiency of antimicrobial and antibiofilm strategies against L. monocytogenes (Sommers et al., 2009; Mohan et al., 2019).
Listeria monocytogenes biofilms
Biofilm formation process starts by adhesion of planktonic bacteria to the abiotic surface followed by proliferation and formation of microcolonies (Renier et al., 2011). They grow and form mature biofilms being highly organized in L. monocytogenes, mostly in the form of a monolayer, but several strains were found to form three‐dimensional structures (Rieu et al., 2008). In vitro Listeria biofilms are not as extensive as observed for some other bacterial species, and the biofilm biomass quantified by crystal violet is usually congruent with the number of viable bacteria. The extracellular matrix of the biofilm is composed of DNA, proteins and teichoic acids (Colagiorgi et al., 2017). Listeria is able to adhere to various types of abiotic surfaces, and it attaches very firmly (Da Silva and De Martinis, 2013; Reis‐Teixeira et al., 2017). It is believed that Listeria is mostly present in complex biofilms in natural environments (Sasahara and Zottola, 1993). The biofilm‐forming ability of L. monocytogenes seems to differ between strains, which can be classified as weak, moderate and strong biofilm producers (Harvey et al., 2007). In a study on 98 clinical and food, L. monocytogenes isolates belonging to serotype groups 1/2a, 1/2b and 4b, the authors could not establish any correlation between the ability to form biofilms and serotype (Doijad et al., 2015). Additionally, Di Bonaventura et al. (2008) could not find significant differences in biofilm formation with regard to genetic lineage, source (environmental vs food) or food type (fish vs meat), regardless of the temperature and surface tested. This could also be due to differences in total fatty acid composition or hydrophobicity being strain‐specific properties correlating to the biofilm‐forming ability of the strain (Doijad et al., 2015). The known molecular determinants involved in Listeria biofilm formation include components of information pathways, signal transduction systems and surface structures (Renier et al., 2011).
The use of L. innocua as a surrogate organism in L. monocytogenes biofilm research has already been tested, but with conflicting results. Costa et al. (2018) found no difference between the biofilms of L. innocua and L. monocytogenes strains isolated from the same Gorgonzola factory, and these biofilms did not differ significantly in their susceptibility to the sanitizers tested. L. monocytogenes F6900 (serotype 1/2a) adhered to stainless steel to a larger extent than L. innocua ATCC 33090; however, after 72 h, no differences were found between the mature biofilms (Koo et al., 2014). And when grown as mixed biofilm on metal surfaces at 37 °C, L. innocua was shown to outgrow L. monocytogenes, implying they could be competitors in their natural environment (Koo et al., 2014).
Surface structures involved in biofilm development
Multiple indications on the potential molecular determinants affecting biofilm formation in Listeria provide results of four independent transposon mutant library screens (Chang et al., 2012; Ouyang et al., 2012; Alonso et al., 2014; Piercey et al., 2016). Background strains in these studies belonged to serotype 1/2a or 4b, and libraries were screened for their biofilm phenotype at 15, 32, 35 and 37 ºC (S1 Table). They infer that inactivation of proteins belonging to COGs (clusters of orthologous groups of proteins) cell envelope and cellular processes, information pathways and intermediary metabolism, affect biofilm formation. The enriched terms in the network of proteins affecting biofilm phenotype (S1 Table) are associated with intermediary metabolism (purine and other nucleobases metabolic processes, phosphorous associated metabolic processes) and carbohydrate derivative biosynthetic process. Description of interaction network analysis is provided in Appendix S1. Proteins associated with intermediary metabolism, for example metabolism of nucleotides and lipids, the citric acid cycle and membrane energetics were found to impair biofilm development only when a transposon mutant library was screened at 35 or 37 °C suggesting that these COGs may be more important at higher temperatures usually not relevant in food industry, though significant for Listeria virulence (Ouyang et al., 2012; Alonso et al., 2014). Eight proteins have been found to decrease biofilm formation regardless of the screening temperature or background strain serotype. They are functionally involved in peptidoglycan assembly (Lmo2229), lipoteichoic acid (LTA) synthesis (DltB, Lmo2554, Lmo0644) the flagellar apparatus (FlaA, FliP, Lmo0707), or have a yet unknown function (Lmo2056) (Fig. 1, Table 1) (Abachin et al., 2002; Bigot et al., 2005; Zawadzka‐Skomiał et al., 2006; Webb et al., 2009). Transposon mutant screens hence point to surface carbohydrate synthesis associated proteins as relevant to biofilm phenotype in Listeria as they are enriched in the network of potential molecular determinants of biofilm phenotype, and they affect biofilm phenotype regardless of the serotype and temperature. These findings, however, remain to be validated as described later. Protein Lmo2056 has not yet been characterized, but according to protein sequence analysis, it may be an extracellular protein and contains a signal peptide and cAMP receptor protein domain found in many secretory prokaryotic proteins (Table S1).
Fig. 1.
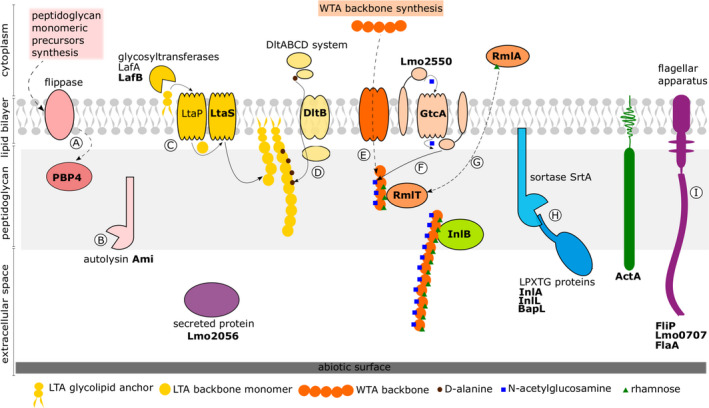
A schematic diagram of the Listeria monocytogenes surfactome and its structures affecting biofilm formation on abiotic surface. Surfactome is composed from carbohydrates (peptidoglycan, lipoteichoic acids – LTA and wall teichoic acids – WTA), cell wall proteins and part of the flagellar apparatus. These structures are located at the surface, but the metabolism of these components is taking place in the cytosol, at the membrane and on the surface, and when relevant we consider all enzymes affecting surface structures, directly or indirectly, by their activity. The structures in bold are involved in biofilm formation and were either validated for their role in biofilm formation or their role was indicated by two or more transposon mutagenesis library screens or both. The dashed line (‐ ‐ ‐ ‐) is used to indicate either multistep synthesis procedure that is not completely presented here or the synthesis steps are unknown.
The peptidoglycan synthesis (A) starts in the cytoplasm and continues at the cell surface after monomers are flipped across the membrane, the assembly of peptidoglycan is carried out by multimodular penicillin‐binding proteins (PBP). Peptidoglycan turnover (B) takes place directly at the cell wall by multiple autolysins. The glycolipid and the LTA backbone are synthesized by multiple enzyme system (C) and decorated by galactose (not shown) and d‐alanine (D) using DltABCD system. Currently, we have limited data on WTA backbone synthesis (E), but WTA glycosylation of serotype 1/2b by N‐acetylglucosamine (F) and rhamnose (G) is known and its absence negatively affects biofilm formation. The rhamnose on WTA is needed in serotype 1/2a for proper internalin B (InlB) positioning at the cell wall and its full function. Other internalins belong to the group of LPXTG proteins that are covalently bound to the peptidoglycan by sortase A via LPXTG motif (H). They are the most abundant proteins in the cell wall of Listeria. Flagellar apparatus (I) is the structure spanning the cell membrane and cell wall enabling Listeria motility at temperatures below 30 °C.
Table 1.
Surfactome components involved in Listeria monocytogenes biofilm development on abiotic surfaces.
| Protein affecting biofilm phenotype | Mechanism governing the biofilm phenotype | Primary biological role the protein plays | Homologues in L. innocua | Reference |
|---|---|---|---|---|
| PBP4 (Lmo2229) | unknown | peptidoglycan synthesis and turnover | Lin2331 |
Chang et al. (2012) Ouyang et al. (2012) |
| Ami (Lmo2558) | decreased adhesion to abiotic surface | peptidoglycan synthesis and turnover | Lin2703 | Kumar et al. (2009) |
| LafB (Lmo2554) | unknown | lipoteihoic acid synthesis | Lin2699 |
Chang et al. (2012) |
| LtaS (Lmo0644) | unknown | lipoteihoic acid synthesis | Lin0647 |
Chang et al. (2012) |
| DltB (Lmo0973) | unknown | lipoteihoic acid synthesis (D‐alanylation) | Lin0971 |
Alonso et al. (2014) Piercey et al. (2016) |
| Lmo2056 | unknown | secreted protein with unknown function | Lin2162 |
Chang et al. (2012) Ouyang et al. (2012) |
| Lmo2550 | decreased adhesion to abiotic surface | wall teichoic acid glycosylation | Lin2695 | Brauge et al. (2018) |
| GtcA (Lmo2549) | decreased adhesion to abiotic surface | wall teichoic acid glycosylation | Lin2694 | Brauge et al. (2018) |
| RmlA (Lmo1081) | decreased adhesion to abiotic surface | wall teichoic acid glycosylation | / | Hsu et al. (2020) |
| RmlT (Lmo1080) | decreased adhesion to abiotic surface | wall teichoic acid glycosylation | / | Hsu et al. (2020) |
| BapL (Lmo0435) | decreased adhesion to abiotic surface | LPXTG protein in cell envelope involved in colonization | Lin0457 | Jordan et al. (2008) |
| InlL (Lmo2026) | decreased adhesion to abiotic surface | LPXTG protein involved in colonization | Lin0740 | Popowska et al. (2017) |
| InlA (Lmo0433) a | decreased adhesion to abiotic surface | LPXTG protein virulence factor | / |
Chen et al. (2008) Travier et al. (2013) |
| InlB (Lmo0434) | decreased adhesion to abiotic surface |
GW protein virulence factor |
/ | Chen et al. (2008) |
| ActA (Lmo0204) | ActA‐ActA interaction between cells, enhancing aggregation |
cell envelope protein with hydrophobic tail virulence factor |
/ | Travier et al. (2013) |
| FliP (Lmo0676) | unknown | flagellar biosynthesis | Lin0684 |
Chang et al. (2012) Ouyang et al. (2012) |
| Lmo0707 | unknown | flagellar hook | Lin0715 |
Ouyang et al. (2012) Alonso et al. (2014) |
| FlaA (Lmo0690) a | unknown | flagellar protein | Lin0698 |
Chang et al. (2012) Alonso et al. (2014) Kumar et al. (2009) Piercey et al. (2016) |
The surfactome components presented in the table complement understanding of the schematic representation of L. monocytogenes surfactome by more detailed description of proteins, their role in biofilm development and presence of homologous genes in L. innocua.
Conflicting results among studies and the biofilm phenotype may be either enhanced or decreased.
Studies mostly focus on screening for impaired biofilm formation, whereas Piercey et al. (2016) reported also transposon mutants that exhibit enhanced biofilm phenotypes; these included surface proteins (muramidase MurA and flagellin A) and virulence factors (amidase Ami and internalins A, B and H). MurA is an autolysin, and its interruption results in formation of prolonged cells at 25 °C that sediment in planktonic culture leading to conclusion that the observed phenotype could be caused by excess biomass and not biofilm itself (Piercey et al., 2016). Mechanisms behind enhanced biofilm phenotype of the remaining transposon mutants are unknown, and may not be explained only by mediating interactions with abiotic surface or between cells like observed in Streptococcus gordonii. The deletion of the S. gordonii surface protein SspAB (involved in adhesion) enhanced the ability for biofilm formation because of inhibited histidine kinase signalling, which results in the upregulation of other adhesins, thus over‐compensating the lacking adhesin to maintain the ability to form biofilms (Hall et al., 2019).
Cell envelope carbohydrates
Surface carbohydrates are synthesized in pathways consisting of proteins located at the surface, at the membrane and in the cytosol. Here, we will discuss the enzymes from the whole synthesis pathways when relevant, because they affect the properties of the surfactome, directly or indirectly, by their activity. The most abundant structure in the cell wall of Listeria is peptidoglycan, which is composed of chains of alternating units of the disaccharides N‐acetylmuramic acid and N‐acetylglucosamine crosslinked by muropeptides. Peptidoglycan monomeric precursors are synthesized in the cytoplasm and are flipped across the membrane in the form of lipid‐anchored disaccharide–pentapeptide monomers (Fig 1A). These subunits are assembled by multimodular penicillin‐binding proteins (PBPs) at the cell surface or by the recently identified SEDS (shape, elongation, division and sporulation) protein family (Guinane et al., 2006; Meeske et al., 2016). Five different PBPs belonging to class A and B have been described in Listeria. The PBP4 from class A (lmo2229) has been indicated by transposon mutagenesis to play a role in biofilm formation (Zawadzka‐Skomiał et al., 2006; Chang et al., 2012; Ouyang et al., 2012). The interruption of lmo2229 led to decreased biofilm formation ability on polystyrene at 32 and 37 °C in L. monocytogenes ScottA (serotype 4b, CC2) and 10403S (serotype 1/2a, CC7) (Chang et al., 2012; Ouyang et al., 2012). But, this may not be due to the changes in peptidoglycan, because the phenotype of the lmo2229 mutant on the background strain EGD‐e (serotype 1/2b, CC9) does not differ in peptidoglycan composition compared with the wild type (Zawadzka‐Skomiał et al., 2006). PBP4 could be thus affecting biofilm phenotype by its presence at the cell wall (being part of surface proteome) mediating interactions with abiotic surface or between bacteria. Adaptations in peptidoglycan during growth and environmental changes are carried out by multiple hydrolytic enzymes (autolysins) (Fig 1B); they are non‐covalently bound to the cell wall by GW modules as described later. Autolysin amidase Ami associates to the cell wall by rhamnose in serotype 1/2 and is important also for virulence of L. monocytogenes (Carvalho et al., 2014; Carvalho et al., 2018). The mutant strain lacking the Ami protein did not adhere and form biofilm as efficiently on polystyrene at 30 °C as the wild‐type EGD‐e strain (Kumar et al., 2009). But, its overexpression caused by growth in medium with glucose and leucine, resulted in enhanced adhesion to stainless steel at 37 °C thus confirming its role in adhesion to abiotic surfaces by a yet unknown mechanism (Skovager et al., 2013). Autolysins amidases Atl were shown to play an important role in adhesion to abiotic surfaces also in S. aureus and S. epidermidis (Biswas et al., 2006). In S. epidermidis was also shown that Atl plays an active role in formation of exogenic DNA thereby increasing the initial adhesion and does not affect adhesion only by its presence (Qin et al., 2007). The other well‐known autolysin p60 (iap) of L. monocytogenes, which associated with the cell wall by a LysM domain, did not influence adhesion or biofilm formation on polystyrene at 30 °C (Kumar et al., 2009).
Cell envelope protein expression, including several genes for PBPs and proteins with putative cell wall‐hydrolysing activity, is regulated also by accessory gene regulator (agr) locus coding for peptide sensing (Zetzmann et al., 2019). The ΔagrD mutant that does not produce the autoinducing peptide responsible for autoregulation of the agr system exhibits decreased ability to form biofilm, and its motility and surface hydrophobicity are not different from wild type. But, its viability is significantly decreased following lysozyme digestion, inferring that agr inactivation leads to altered composition of the surfactome resulting in decreased adhesion to the abiotic surface or neighbouring bacteria (Zetzmann et al., 2019). The agr system is involved also in S. aureus biofilm formation, but its inactivation leads to enhanced biofilm phenotype possibly being consequence of decreased accumulation of extracellular proteases and increased expression of surface proteins such as fibronectin binding protein or protein A (Novick et al., 1993; Solano et al., 2014).
Peptidoglycan is decorated by teichoic acids that are divided into wall teichoic acids (WTAs), which are covalently bound to peptidoglycan, and LTAs, which are anchored to the cytoplasmic membrane by a glycolipid (Fig 1C and E). Because the teichoic acids are charged, they are considered to play an important role in initial adhesion to abiotic surfaces (Bos et al., 1999). Though transposon mutant library screens indicated importance of LTA and WTA in biofilm formation process, limited validation studies have been performed in Listeria to confirm these findings. LTA structure seems to be conserved among strains and consists of a polyglycerol phosphate backbone substituted by D‐alanine and galactose in L. monocytogenes (Nichterlein et al., 1997). L. monocytogenes synthesizes the LTA backbone by the enzymes LtaP (Lmo0644), a primase that produces glycerolphosphate glycolipids, and LtaS (Lmo0927), an LTA synthase that produces polyglycerolphosphate chains (Webb et al., 2009) (Fig 1C). Elongation of the LTA backbone is initiated on a glycolipid anchor synthesized by LafA (Lmo2555) and LafB (Lmo2554) (Fig 1C); mutants lacking these two enzymes produce only minute amounts of LTA (Webb et al., 2009). Transposon mutant library screens indicated that lmo0644 (ltaP) and lmo2554 (lafB) mutants exhibit deficient biofilm phenotype on background strains L. monocytogenes serotypes 4b and 1/2a at 32, 35 and 37 °C though these findings remain to be validated (Table 1) (Chang et al., 2012; Ouyang et al., 2012; Alonso et al., 2014). Enterococcus faecalis LTAs glycolipid anchor is also synthesized by orthologous two enzyme system similarly to Listeria and is named BgsA. Δbgs mutant contains longer LTA chains with less hydrophobic cells than the wild type and does not differ from the wild type regarding adherence, but differs significantly regarding biomass accumulation over prolonged incubation times, leading to negative biofilm phenotype (Theilacker et al., 2009). LTAs in L. monocytogenes are substituted by d‐alanine (Fig 1 D), and mutants lacking alanine esters exhibit increased electronegativity and more efficient binding of cationic compounds (e.g. colistin, nisin and polymyxin B) (Abachin et al., 2002). d‐alanylation of LTAs is carried out by the dlt operon in gram‐positive bacteria, and its activity determines biofilm formation ability in Staphylococcus aureus where its inactivation leads to biofilm inhibition on polystyrene and glass due to increased negative charge (Gross et al., 2001). Transposon mutants with interrupted dltB exhibit decreased ability to form biofilms on abiotic surfaces at different temperatures also on Listeria background strains with 1/2a serotype (Alonso et al., 2014; Piercey et al., 2016). Listeria changes LTA composition also in response to temperature changes, rendering LTAs less immunostimulatory and more hydrophobic (Dehus et al., 2011). This could lead to differences in biofilm formation ability at different temperatures, but biofilm phenotypes of LTA synthesis associated transposon mutants remained impaired between 15 and 37 ºC (Chang et al., 2012; Alonso et al., 2014; Piercey et al., 2016). Homologues of LTA synthesis genes involved in L. monocytogenes biofilm formation were also found in L. innocua CLIP 11262 (S1 Table); however, their role in biofilm formation is unknown. Furthermore, the structure of L. innocua LTAs has not yet been determined. Nevertheless, they exhibit similar potential to stimulate phagocytes to produce proinflammatory cytokines, implying large structural similarities with L. monocytogenes LTAs (Nichterlein et al., 1997).
Unlike LTAs, WTAs are more diverse and represent O‐antigens, which are the main determinants of serotypes along with flagellar proteins (H antigens). WTAs are composed of a variety of mono‐ and oligosaccharides that substitute hydroxyl groups on the 20 to 40 repeating ribitol‐phosphate units and are classified into type I and type II (Shen et al., 2017). WTA backbone synthesis is not well described in Listeria (Fig 1E); however, it has been established that the genes tagO1 (lmo0959) and tagO2 (lmo2519) are required in the initial steps of WTA synthesis and that only double mutants are devoid of WTA (Eugster and Loessner, 2012). The WTA backbone is further substituted by sugar moieties in Listeria, and these glycosylation patterns were found to be congruent with the current serotyping scheme (Shen et al., 2017). Listeria WTA type I are substituted by rhamnose and/or N‐acetylglucosamine (Fig 1F and G), and serotypes belonging to this group (1/2, 3, 7) differ only in small mutations in genes coding for rhamnosylation and/or GlcNAcylation causing serotype switch (Eugster et al., 2015). Absence of rhamnosylation caused by mutations in genes lmo1081 (rmlA) and lmo1080 (rmlT) in background strain EGD‐e (serotype 1/2a) was proven to have negative effect on adhesion and consequently biofilm formation on abiotic surface (Hsu et al., 2020). Serotype 1/2a decorates its LTA by galactose and WTAs by N‐acetylglucosamine using the same flippase GtcA in both pathways (Rismondo et al., 2020). GtcA in strain EGD, serotype 1/2a, is coded by lmo2549 and works together with the gene lmo2550 to decorate WTA with N‐acetylglucosamine (Fig 1G) (Eugster et al., 2011). Mutants Δlmo2549 and Δlmo2550 constructed by targeted mutagenesis had a more hydrophilic surface compared with wild type and adhered less to stainless steel at 30 °C; biofilm was growing in the form of individual bacterial clusters, and the mature biofilm exhibited architecture that was not carpet‐like in comparison with wild‐type EGD‐e biofilm (Brauge et al., 2018). The adhesion was similarly impaired in the strain carrying natural mutation in lmo2550, but there were no differences in morphology in mature biofilm compared with wildtype (Brauge et al., 2018).
Wall teichoic acids in the serotype 4b belong to type II WTAs where N‐acetylglucosamine is incorporated into the backbone and is decorated mostly by galactose or/and glucose, but the effect of glycosylation on biofilm remains unknown in this serotype (Sumrall et al., 2020a).
Wall teichoic acids were also found to be the only extracellular carbohydrates in biofilms formed by several different strains belonging to the serotypes 1/2a and 4b (Brauge et al., 2016). The structure of extracellular teichoic acid was the same as that of the WTAs found in the cell wall of the biofilm‐forming strain. The origin of WTAs in the extracellular matrix remains unknown; however, the origin is postulated to result from cell lysis or even active WTA export (Brauge et al., 2016).
Listeria innocua strains have been traditionally typed as serotype 6, and we can infer that their WTA structure is similar to serotypes 4a and 6b. An exception is L. innocua strains that exhibit serotype 4b antigens, because they harbour gtcA homologues; however, how this relates to their pathogenic potential and biofilm formation is unknown (Lan et al., 2000).
Cell envelope proteins
The L. monocytogenes EGD‐e genome harbours 133 putative surface proteins out of the total 2853 predicted proteins (Bierne and Cossart, 2007). These putative surface proteins were predicted based on the presence of specific domains or motifs that are known to mediate translocation across membranes and attachment to the cell envelope. They have been classified as follows: (i) proteins covalently attached to peptidoglycan moieties (LPXTG and NXXTX proteins), (ii) proteins non‐covalently associated with the surface (proteins with the GW, LysM or Wxl domain), and (iii) membrane‐bound proteins (proteins with hydrophobic tails and lipoproteins) (Cabanes et al., 2002). Surface proteins, including flagellar proteins, also determine the differences between the EGD (serotype 1/2a, CC9) and F2365 (serotype 4b) strains and may also account for differences in strains’ antigenicity and pathogenicity (Donaldson et al., 2009). The global proteome of these strains encompasses 1754 proteins (61.6% of the predicted proteome) for EGD and 1427 proteins (50.5% of the predicted proteome) for F2365. According to the function of the identified proteins, these strains were found to perform basic cellular processes similarly. Both strains also express the virulence‐related LPXTG protein internalin A and another uncharacterized internalin, whereas F2365 does not express internalin B (inlB) because its gene is truncated. However, five LPXTG proteins with unknown function at 37 °C were significantly more expressed in EGD cells, whereas a cell wall anchor protein was significantly more expressed in F2365 cells (Donaldson et al., 2009). Comparison of surface proteomes during biofilm growth mode is rarely performed because of the complexity of the extraction and analysis set‐up. The highly adherent strain 99‐38 from lineage I expresses 21 proteins exclusively during biofilm growth mode and not during growth in planktonic culture, including the surface proteins Lmo0275 (DNA uptake‐related protein), Lmo0394 (P60‐like protein), Lmo0204 (ActA), Lmo0434 (InlB) and Lmo2713 (internalin‐like protein) (Tiong et al., 2016). This study also identified moonlighting proteins, including glyceraldehyde‐3‐phosphate dehydrogenase (Lmo2459) with a yet unknown role at the surface (Tiong et al., 2016). Differences regarding surface proteomes have been found among studies that have often been attributed to different experimental temperatures (Calvo et al., 2005; Tiong et al., 2016). However, a comparative study of L. monocytogenes EGD‐e grown on stainless steel at 10, 25 or 37 °C has demonstrated that it is highly unlikely that a set of surface proteins could play a unique role in the adaptation to different temperatures, with the exception of motility‐related proteins (Santos et al., 2019).
The L. innocua CLIP 11262 genome also harbours several surface proteins, but not those associated with virulence in L. monocytogenes (e.g. InlA, InlB, Ami) (Glaser et al., 2001). Surfaceomes (i.e. all surface proteins) of L. monocytogenes EGD‐e and L. innocua CLIP11262 planktonic cultures grown at 37 °C revealed 19 and 11 proteins respectively (Calvo et al., 2005). The most abundant species were LPXTG proteins, followed by proteins involved in peptidoglycan synthesis (PBSs, P60 and P45‐like proteins) and NXXTG proteins (Calvo et al., 2005). In both species, orthologous genes were identified for proteins P60, P45 and NXXTG; however, several LPXTG proteins were absent in L. innocua, and some orthologous proteins could not be identified (Calvo et al., 2005).
Cell envelope proteins associated with L. monocytogenes pathogenicity
The most characterized virulence cell wall protein is internalin A (InlA) because it is crucial for the internalization of L. monocytogenes into epithelial cells (Lecuit et al., 1997). It belongs to the LPXTG protein group and has a sorting signal at the C‐terminal that is postulated to direct protein attachment to peptidoglycan by sortase SrtA, a membrane‐bound transpeptidase (Fig 1H). This type of proteins is also the most abundant among predicted cell wall proteins in Listeria. Inactivation of the inlA and inlB genes in L. monocytogenes EGD‐e was shown to decrease the strain’s ability to adhere to glass at 37 °C, and this effect was even stronger in the double mutant (Table 1) (Chen et al., 2008). Expression of full‐length InlA and InlB exhibited a positive correlation with the ability of L. monocytogenes to adhere to glass surfaces at 30 °C; this ability did not depend on the serotype (Chen et al., 2009). Unlike serotype 4b, many serotype 1/2c and 1/2a strains from lineage II express truncated versions of InlA (Franciosal et al., 2009). This truncated version lacks the cell wall‐binding domain and is secreted into the extracellular space where it undergoes proteolytic processing. The resulting peptides seem to enhance biofilm formation as they can be easily incorporated into the biofilm as extracellular matrix (Franciosal et al., 2009). On the other hand, transposon mutants with inactivated inlA, inlB and inlH led to an enhanced biofilm phenotype at 15 °C, but inlA and inlB are not expressed at lower temperatures, because their transcriptional activator PrfA is expressed only at temperatures above 30 °C (McGann et al., 2007; Piercey et al., 2016). Yet, Travier et al (2013) found that ΔinlA biofilm phenotype was enhanced compared to wild type and ΔinlB showed no change in biofilm formation at 37 °C on polyvinyl chloride. Conflicting observations could not be due to serotype related differences, because these studies were done on background strain serotype 1/2a, but could be associated with experimental set‐up or a yet unknown aspect of inlA and inlB expression regulation.
Another member of the internalin family with LPXTG motif, InlL, is also important for adhesion to polystyrene at 30 °C and as it binds secreted mucin type it could have also role in pathogenesis (Popowska et al., 2017). InlL is mainly present only in L. monocytogenes food isolates (CC9 and CC121), and hence, it might play an important role in the persistence of L. monocytogenes (Popowska et al., 2017). Another non‐core persistence‐associated gene with LPXTG motif, bapL (lmo0435), affects the adhesion of L. monocytogenes EGD‐e onto polystyrene and stainless steel at 37 °C (Jordan et al., 2008). Additionally, strains naturally lacking the bapL gene exhibit both stronger and weaker adhesion, suggesting that BapL is not the only determinant of adhesion (Jordan et al., 2008). These results also demonstrate that molecular determinants of biofilm phenotype can be isolate or serotype specific and thus results cannot be simply extrapolated to other strains.
InlB is an important virulence factor and crucial for invasion of non‐phagocytic human cells and plays a role in adhesion also to abiotic surfaces (Parida et al., 1998; Chen et al., 2008). InlB is associated with the cell wall by a GW motif‐containing domain to WTA via galactose (serotype 4b) or rhamnose (serotype 1/2) and in position‐dependent manner and is functional only when glycosylation pattern is complete (Carvalho et al., 2018; Sumrall et al., 2020a). This implies that glycosylation is responsible for increased virulence via InlB, and its mere expression does not alter strains pathogenic potential; that has been partially confirmed by expression of 4b serotype glycosylation gene gttA in serotype 4d causing increased invasiveness of this strain (Sumrall et al., 2020b). Even though this was a laboratory validation study, lateral transfer of glycosylation genes happens also naturally among Listeria species as demonstrated by L. innocua harbouring gtcA gene from L. monocytogenes leading to altered glycosylation in L. innocua (Lan et al., 2000). These events imply that surfactome is ever evolving property of the strain resulting in altered phenotype being more or less persistent or invasive.
An important part of Listeria surfaceome is autolysins (Fig. 1B), and some are involved in pathogenesis like Ami, and this group of proteins has been described in a previous section due to their role in peptidoglycan turnover.
Membrane protein ActA, responsible for actin polymerization promoting L. monocytogenes intracellular motility and cell‐to‐cell spread, is critical also for biofilm formation (Fig 1) (Travier et al., 2013). This protein mediates direct contact between bacteria by ActA‐ActA interaction resulting in aggregation, which is very important for biofilm formation. Active ActA enabled formation of larger clusters of bacteria thus promoting thicker biofilm biomass development on glass surface (Travier et al., 2013). Furthermore, the expression of L. monocytogenes ActA in L. innocua increased its ability to form biofilm thus confirming its direct role in biofilm formation (Travier et al., 2013).
The flagellar apparatus and motility
Listeria monocytogenes and L. innocua can swim in the extracellular environment by using four to six peritrichous flagella, and their motility is temperature‐dependent and to a lesser degree also strain‐dependent (Peel et al., 1988; Gründling et al., 2004). Motility is potent at temperatures below 30 °C because the main flagellin FlaA (Lmo0690) is highly expressed. FlaA expression decreases at physiological temperatures of 37 °C, and a small proportion of bacterial populations of certain strains may exceptionally still be flagellated at elevated temperatures (Way et al., 2004). In L. monocytogenes EGD‐e, flagellin synthesis is regulated by MogR (Lmo0674) and independently of PrfA (Lmo0200), a virulence transcription factor that is absent in L. innocua (Gründling et al., 2004). Flagella proteins of the L. monocytogenes serotypes 4b, 1/2c, 1/2a and 1/2b are additionally uniquely modified with O‐linked β‐N‐acetylglucosamine at up to six sites on each protein monomer (Schirm et al., 2004). All four genome‐wide studies identified at least one component of the flagellar apparatus (Fig 1I) that affects the ability of L. monocytogenes to form biofilms (Chang et al., 2012; Ouyang et al., 2012; Alonso et al., 2014; Piercey et al., 2016); however, their results are conflicting. Although the natural variability in the motility of L. monocytogenes is large, this property was only found to correlate with the ability to form biofilms on stainless steel where motile strains formed weaker biofilms (Di Bonaventura et al., 2008). L. monocytogenes 568 (serotype 1/2a) transposon mutant with interrupted lmo0690 (flaA) was able to form biofilms on polystyrene or stainless‐steel surfaces at 15 °C but not on polystyrene peg lids (Piercey et al., 2016). A FlaA transposon mutant on background strain 10403S (serotype 1/2a, CC7) exhibited a decreased ability to form biofilms on polystyrene at 35 °C (Alonso et al., 2014). This finding was confirmed by targeted mutagenesis in strain EGD‐e (serotype 1/2b, CC9) where ΔflaA formed less dense biofilm compared with the wild type at 30 °C detected by crystal violet staining and electron microscopy (Kumar et al., 2009). But, in both studies, biofilm screening temperatures were above 30 °C when motility decreases in Listeria, whereas at 15 °C, it is fully expressed, and we can hypothesize that biofilm did not form on the peg lids, because motility is required to swim towards the peg, whereas it is not required to form biofilm on the microplate bottom. Regulation of flagellin gene expression is extremely complex and may be affecting biofilm phenotypes by a yet unknown mechanism that would explain inhibited biofilm formation of ΔflaA at higher temperatures (Cossart, 2011). Discrepancies between studies could also be due to the variability of biofilm assays and sample preparations. Biofilm staining techniques mainly use rigorous washing of biofilms, leading to the detachment of weakly bound bacteria, resulting in possible falsely negative biofilm phenotype.
Antimicrobials targeting the surfactome
Understanding of L. monocytogenes surfactome components and associations between them is still limited, and only limited antimicrobials target them. The treatment of choice against listeriosis is the use of ampicillin or gentamicin belonging to the beta lactam group of antibiotics (Olaimat et al., 2018). They target the PBPs described above in the section ‘Cell envelope carbohydrates’ and play key roles in peptidoglycan biosynthesis. Penicillins usually work bacteriostatically and where shown to inhibit growth of planctonic bacteria and biofilm formation close to minimal inhibitory concentration (MIC), but enhanced biofilm formation at much lower concentrations (Nguyen et al., 2012). Several beta lactams were shown also to disperse mature biofilm at concentrations above MIC, but a small proportion of bacterial population survived treatment regardless of increasing antibiotic dose implying dispersal needs peptidoglycan turnover, which is absent in dormant bacteria (Nguyen et al., 2012).
Among alternative approaches, sortases have been suggested as antimicrobial targets for many gram‐positive pathogens because they are responsible for the anchoring of LPXTG proteins, many of which are virulence factors. The majority of LPXTG proteins in L. monocytogenes are substrates of sortase A (SrtA) (Bierne et al., 2002). The flavonoid chalcone, used in traditional Chinese medicine, was found to inhibit SrtA with an IC50 of 28.41 ± 5.34 µM by binding to the same active region as the substrate LPTTG (Li et al., 2016). At effective concentrations, chalcone reduces L. monocytogenes invasion of Caco‐2 cells (Li et al., 2016). Another chalcone compound, phloretin, which is naturally present in apples, was also shown to inhibit SrtA in in vitro assays (Wang et al., 2017). L. monocytogenes EGD‐e pre‐treated with phloretin exhibits a diminished ability to invade Caco‐2 cells, and the addition of phloretin after invasion reduces the multiplication of intracellular bacteria. Similar effects were also observed using baicalein, a flavonoid from Scutellaria baicalensis (Lu et al., 2019). The effect of these compounds on biofilm formation has not been evaluated to date.
Furthermore, bacteriophages also exploit surface structures as receptors and have gained a lot of interest as alternative antimicrobials. Phage P100 is commercially available in the United States for use against L. monocytogenes, L. innocua, and other Listeria spp. on foodstuff and has a status of ‘generally recognized as safe’ (GRAS). WTA glycosylation is directly affected by bacteriophage predation causing switches between L. monocytogenes serotypes with WTA type I (Eugster et al., 2015). Resistance mechanism against bacteriophages in serotype 4b was also found to be associated with altered WTA and LTA structures, which also abolish its ability to invade Caco‐2 and HepG2 cell lines (Sumrall et al., 2019). Altered glycosylation evidently affects bacterial surfactome and consequently interactions with environment as demonstrated by 4b serotype bacteriophage resistance and 1/2a serotype biofilm phenotypes, but it remains to be elucidated if and how it affects strain’s persistence.
Concluding remarks
Understanding the correlation between pathogenicity potential and key surface structures important for biofilm formation in L. monocytogenes, which enables long‐term persistence, will facilitate informed targeting of the strains that pose the highest risk to human and animal health. Insight into the molecular make‐up of bacterial surfaces can provide new ideas and directives for the development of novel antimicrobial strategies to prevent the development of bacterial resistance and extend our antimicrobial armoury.
Cell envelope carbohydrates present an important source of novel antimicrobial targets that are expected to expand as a result of the growing availability of glycobiology tools. They do not affect just overall properties of the surface like charge and hydrophobicity, but also direct localization of proteins composing the surface proteome. The latter has been characterized mainly in terms of pathogenic potential of Listeria, but as shown, these proteins play roles also in biofilm and possibly persistence. According to currently available knowledge, surfactome components contribute to adhesion to abiotic surfaces as expected, with exception of ActA mediating cohesion, the cell‐to‐cell interactions thus promoting aggregation and biofilm development. The role of LTA and some other components remains to be validated. Surfactome properties are further differentiated among serotypes, phylogenetic groups or clonal complexes as demonstrated by differences in WTA decoration and presence of LPXTG proteins.
Surfactome has been modulated using natural antimicrobials via sortase inhibition or bacteriophage treatment resulting in altered behaviour in cell line invasion models. Similar surfactome remodelling could lead to inhibition of biofilm process thereby negatively affecting the Listeria persistence. Based on the current research, the surface components of L. monocytogenes and L. innocua that promote biofilm formation differ and may thus cause differences in the responses to antimicrobials that target specific surface structures, but the classes of surfactome components are expected to be related to certain L. monocytogenes serotypes.
Conflict of interest
The authors declare that they have no competing interests.
Supporting information
Table S1. List of proteins affecting Listeria monocytogenes biofilm formation on abiotic surfaces.
Appendix S1. Interaction network analysis procedure and results.
Acknowledgements
This work was supported by Slovenian Research Agency with the grants no. P4‐0127, J4‐1771, N2‐0078, and P4‐0116. The work of Blaž Škrlj was funded by the Slovenian Research Agency through a young researcher grant.
Microb. Biotechnol. (2021) 14(4), 1269–1281
Funding information
This work was supported by Slovenian Research Agency with the grants no. P4‐0127, J4‐1771, N2‐0078, and P4‐0116.
References
- Abachin, E. , Poyart, C. , Pellegrini, E. , Milohanic, E. , Fiedler, F. , Berche, P. , and Trieu‐Cuot, P. (2002) Formation of D‐alanyl‐lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes . Mol Microbiol 43: 1–14. [DOI] [PubMed] [Google Scholar]
- Alonso, A.N. , Perry, K.J. , Regeimbal, J.M. , Regan, P.M. , and Higgins, D.E. (2014) Identification of Listeria monocytogenes determinants required for biofilm formation. PLoS One 9: e113696. 10.1371/journal.pone.0113696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne, H. , and Cossart, P. (2007) Listeria monocytogenes surface proteins: from genome predictions to function. Microbiol Mol Biol Rev 71: 377–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne, H. , Mazmanian, S.K. , Trost, M. , Pucciarelli, M.G. , Dehoux, P. , The European Listeria Genome Consortium , et al. (2002) Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol Microbiol 43: 869–881. [DOI] [PubMed] [Google Scholar]
- Bigot, A. , Pagniez, H. , Botton, E. , Fréhel, C. , Dubail, I. , Jacquet, C. , et al. (2005) Role of FliF and FliI of Listeria monocytogenes in flagellar assembly and pathogenicity. Infect Immun 73: 5530–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, R. , Voggu, L. , Simon, U.K. , Hentschel, P. , Thumm, G. , and Götz, F. (2006) Activity of the major staphylococcal autolysin Atl. FEMS Microbiol Lett 259: 260–268. 10.1111/j.1574-6968.2006.00281.x [DOI] [PubMed] [Google Scholar]
- Bos, R. , van der Mei, H.C. , and Busscher, H.J. (1999) Physico‐chemistry of initial microbial adhesive interactions–its mechanisms and methods for study. FEMS Microbiol Rev 23: 179–230. 10.1111/j.1574-6976.1999.tb00396.x [DOI] [PubMed] [Google Scholar]
- Brauge, T. , Faille, C. , Sadovskaya, I. , Charbit, A. , Benezech, T. , Shen, Y. , et al. (2018) The absence of N‐acetylglucosamine in wall teichoic acids of Listeria monocytogenes modifies biofilm architecture and tolerance to rinsing and cleaning procedures. PLoS One 13: e0190879. 10.1371/journal.pone.0190879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauge, T. , Sadovskaya, I. , Faille, C. , Benezech, T. , Maes, E. , Guerardel, Y. , and Midelet‐Bourdin, G. (2016) Teichoic acid is the major polysaccharide present in the Listeria monocytogenes biofilm matrix. FEMS Microbiol Lett 363: fnv229. 10.1093/femsle/fnv229 [DOI] [PubMed] [Google Scholar]
- Cabanes, D. , Dehoux, P. , Dussurget, O. , Frangeul, L. , and Cossart, P. (2002) Surface proteins and the pathogenic potential of Listeria monocytogenes . Trends Microbiol 10: 238–245. 10.1016/s0966-842x(02)02342-9 [DOI] [PubMed] [Google Scholar]
- Codex Alimentarius ‐ CAC/GL 61‐2007: Guidelines on the application of general principles of food hygiene to the control of Listeria monocytogenes in ready‐to‐eat foods. URL: http://www.codexalimentarius.net/download/standards/10740/CXG_061e.pdf
- Calvo, E. , Pucciarelli, M.G. , Bierne, H. , Cossart, P. , Albar, J.P. , and García‐Del Portillo, F. (2005) Analysis of the Listeria cell wall proteome by two‐dimensional nanoliquid chromatography coupled to mass spectrometry. Proteomics 5: 433–443. [DOI] [PubMed] [Google Scholar]
- Carvalho, F. , Sousa, S. , and Cabanes, D. (2014) How Listeria monocytogenes organizes its surface for virulence. Front Cell Infect Microbiol 4: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, F. , Sousa, S. , and Cabanes, D. (2018) l‐Rhamnosylation of wall teichoic acids promotes efficient surface association of Listeria monocytogenes virulence factors InlB and Ami through interaction with GW domains. Environ Microbiol 20: 3941–3951. [DOI] [PubMed] [Google Scholar]
- Chang, Y. , Gu, W. , Fischer, N. , and McLandsborough, L. (2012) Identification of genes involved in Listeria monocytogenes biofilm formation by mariner‐based transposon mutagenesis. Appl Microbiol Biotechnol 93: 2051–2062. [DOI] [PubMed] [Google Scholar]
- Chen, B.Y. , Kim, T.J. , Jung, Y.S. , and Silva, J.L. (2008) Attachment strength of Listeria monocytogenes and its internalin‐negative mutants. Food Biophys 3: 329–332. [Google Scholar]
- Chen, B.Y. , Kim, T.J. , Silva, J.L. , and Jung, Y.S. (2009) Positive correlation between the expression of inlA and inlB genes of Listeria monocytogenes and its attachment strength on glass surface. Food Biophys 4: 304–311. [Google Scholar]
- Colagiorgi, A. , Bruini, I. , Di Ciccio, P.A. , Zanardi, E. , Ghidini, S. , and Ianieri, A. (2017) Listeria monocytogenes biofilms in the wonderland of food industry. Pathogens 6: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner, D.E. , Scott, V.N. , and Bernard, D.T. (1990) Growth, inhibition, and survival of Listeria monocytogenes as affected by acidic conditions. J Food Prot 53: 652–655. [DOI] [PubMed] [Google Scholar]
- Cossart, P. (2011) Illuminating the landscape of host‐pathogen interactions with the bacterium Listeria monocytogenes . Proc Natl Acad Sci USA 108: 19484–19491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, A. , Lourenco, A. , Civera, T. , and Brito, L. (2018) Listeria innocua and Listeria monocytogenes strains from dairy plants behave similarly in biofilm sanitizer testing. LWT 92: 477–483. [Google Scholar]
- Da Silva, E.P. , and De Martinis, E.C.P. (2013) Current knowledge and perspectives on biofilm formation: The case of Listeria monocytogenes . Appl Microbiol Biotechnol 97: 957–968. [DOI] [PubMed] [Google Scholar]
- Dehus, O. , Pfitzenmaier, M. , Stuebs, G. , Fischer, N. , Schwaeble, W. , Morath, S. , et al. (2011) Growth temperature‐dependent expression of structural variants of Listeria monocytogenes lipoteichoic acid. Immunobiology 216: 24–31. [DOI] [PubMed] [Google Scholar]
- Desai, A.N. , Anyoha, A. , Madoff, L.C. , and Lassmann, B. (2019) Changing epidemiology of Listeria monocytogenes outbreaks, sporadic cases, and recalls globally: a review of ProMED reports from 1996 to 2018. Int J Infect Dis 84: 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bonaventura, G. , Piccolomini, R. , Paludi, D. , D’Orio, V. , Vergara, A. , Conter, M. , and Ianieri, A. (2008) Influence of temperature on biofilm formation by Listeria monocytogenes on various food‐contact surfaces: Relationship with motility and cell surface hydrophobicity. J Appl Microbiol 104: 1552–1561. [DOI] [PubMed] [Google Scholar]
- Doijad, S.P. , Barbuddhe, S.B. , Garg, S. , Poharkar, K.V. , Kalorey, D.R. , Kurkure, N.V. , et al. (2015) Biofilm‐forming abilities of Listeria monocytogenes serotypes isolated from different sources. PLoS One 10: e0137046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, J.R. , Nanduri, B. , Burgess, S.C. , and Lawrence, M.L. (2009) Comparative proteomic analysis of Listeria monocytogenes strains F2365 and EGD. Appl Environ Microbiol 75: 366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control) (2018) The European Union summary report on trends and sources of zoonoses, zoonotic agents and food‐borne outbreaks in 2017. EFSA Journal 16: 262. 10.2903/j.efsa.2018.5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EFSA Biohaz Panel (EFSA Panel on Biological Hazards) , Ricci, A. , Allende, A. , Bolton, D. , Chemaly, M. , Davies, R. , et al. (2018) Scientific opinion on the Listeria monocytogenes contamination of ready‐to‐eat foods and the risk for human health in the EU. EFSA Journal 16: 173. 10.2903/j.efsa.2018.5134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster, M.R. , Haug, M.C. , Huwiler, S.G. , and Loessner, M.J. (2011) The cell wall binding domain of Listeria bacteriophage endolysin PlyP35 recognizes terminal GlcNAc residues in cell wall teichoic acid. Mol Microbiol 81: 1419–1432. [DOI] [PubMed] [Google Scholar]
- Eugster, M.R. , and Loessner, M.J. (2012) Wall teichoic acids restrict access of bacteriophage endolysin Ply118, Ply511, and Plyp40 cell wall binding domains to the Listeria monocytogenes peptidoglycan. J Bacteriol 194: 6498–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster, M.R. , Morax, L.S. , Hüls, V.J. , Huwiler, S.G. , Leclercq, A. , Lecuit, M. , and Loessner, M.J. (2015) Bacteriophage predation promotes serovar diversification in Listeria monocytogenes . Mol Microbiol 97: 33–46. [DOI] [PubMed] [Google Scholar]
- Franciosal, G. , Mauglianp, A. , Scalfarol, C. , Floridf, F. , and Aurelp, P. (2009) Expression of internalin A and biofilm formation among Listeria monocytogenes clinical isolates. Int J Immunopathol Pharmacol 22: 183–193. [DOI] [PubMed] [Google Scholar]
- Glaser, P. , Frangeul, L. , Buchrieser, C. , Rusniok, C. , Amend, A. , Baquero, F. , et al. (2001) Comparative genomics of Listeria species. Science 294: 849–852. [DOI] [PubMed] [Google Scholar]
- Gross, M. , Cramton, S.E. , Götz, F. , and Peschel, A. (2001) Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect Immun 69: 3423–3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründling, A. , Burrack, L.S. , Bouwer, H.G.A. , and Higgins, D.E. (2004) Listeria monocytogenes regulates flagellar motility gene expression through MogR, a transcriptional repressor required for virulence. Proc Natl Acad Sci USA 101: 12318–12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinane, C.M. , Cotter, P.D. , Ross, R.P. , and Hill, C. (2006) Contribution of penicillin‐binding protein homologs to antibiotic resistance, cell morphology, and virulence of Listeria monocytogenes EGDe. Antimicrob Agents Chemother 50: 2824–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, J.W. , Lima, B.P. , Herbomel, G.G. , Gopinath, T. , McDonald, L.A. , Shyne, M.T. , et al. (2019) An intramembrane sensory circuit monitors sortase A–mediated processing of streptococcal adhesins. Sci Signal 12: eaas9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, J. , Keenan, K.P. , and Gilmour, A. (2007) Assessing biofilm formation by Listeria monocytogenes strains. Food Microbiol 24: 380–392. [DOI] [PubMed] [Google Scholar]
- Hsu, C.Y. , Cairns, L. , Hobley, L. , Abbott, J. , O’Byrne, C. , and Stanley‐Wall, N.R. (2020) Genomic differences between Listeria monocytogenes EGDe isolates reveal crucial roles for sigB and wall rhamnosylation in biofilm formation. J Bacteriol 202: e00692‐19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, S.J. , Perni, S. , Glenn, S. , Fernandes, I. , Barbosa, M. , Sol, M. , et al. (2008) Listeria monocytogenes biofilm‐associated protein (BapL) may contribute to surface attachment of L. monocytogenes but is absent from many field isolates. Appl Environ Microbiol 74: 5451–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo, O.K. , Ndahetuye, J.B. , O’Bryan, C.A. , Ricke, S.C. , and Crandall, P.G. (2014) Influence of Listeria innocua on the attachment of Listeria monocytogenes to stainless steel and aluminum surfaces. Food Control 39: 135–138. [Google Scholar]
- Kumar, S. , Parvathi, A. , George, J. , Krohne, G. , Karunasagar, I. , and Karunasagar, I. (2009) A study on the effects of some laboratory‐derived genetic mutations on biofilm formation by Listeria monocytogenes . World J Microbiol Biotechnol 25: 527–531. [Google Scholar]
- Lan, Z. , Fiedler, F. , and Kathariou, S. (2000) A sheep in wolf’s clothing: Listeria innocua strains with teichoic acid‐associated surface antigens and genes characteristic of Listeria monocytogenes serogroup 4. J Bacteriol 182: 6161–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappi, V.R. , Thimothe, J. , Walker, J. , Bell, J. , Gall, K. , Moody, M.W. , and Wiedmann, M. (2004) Impact of intervention strategies on Listeria contamination patterns in crawfish processing plants: A longitudinal study. J Food Prot 67: 1163–1169. [DOI] [PubMed] [Google Scholar]
- Lecuit, M. , Ohayon, H. , Braun, L. , Mengaud, J. , and Cossart, P. (1997) Internalin of Listeria monocytogenes with an intact leucine‐rich repeat region is sufficient to promote internalization. Infect Immun 65: 5309–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Chen, Y. , Zhang, B. , Niu, X. , Song, M. , Luo, Z. , et al. (2016) Inhibition of sortase A by chalcone prevents Listeria monocytogenes infection. Biochem Pharmacol 106: 19–29. [DOI] [PubMed] [Google Scholar]
- Liu, D. (2006) Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J Med Microbiol 55: 645–659. [DOI] [PubMed] [Google Scholar]
- Lomonaco, S. , Nucera, D. , and Filipello, V. (2015) The evolution and epidemiology of Listeria monocytogenes in Europe and the United States. Infect Genet Evol 35: 172–183. [DOI] [PubMed] [Google Scholar]
- Lu, G. , Xu, L. , Zhang, T. , Deng, X. , and Wang, J. (2019) A potential bio‐control agent from baical skullcap root against listeriosis via the inhibition of sortase A and listeriolysin O. J Cell Mol Med 23: 2042–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury, M.M. , Tsai, Y.‐H. , Charlier, C. , Touchon, M. , Chenal‐Francisque, V. , Leclercq, A. , et al. (2016) Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet 48: 308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGann, P. , Ivanek, R. , Wiedmann, M. , and Boor, K.J. (2007) Temperature‐dependent expression of Listeria monocytogenes internalin and internalin‐like genes suggests functional diversity of these proteins among the listeriae. Appl Environ Microbiol 73: 2806–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske, A.J. , Riley, E.P. , Robins, W.P. , Uehara, T. , Mekalanos, J.J. , Kahne, D. , et al. (2016) SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537: 634–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miladi, H. , Chaieb, K. , Bakhrouf, A. , Elmnasser, N. , and Ammar, E. (2008) Freezing effects on survival of Listeria monocytogenes in artificially contaminated cold fresh‐salmon. Ann Microbiol 58: 471–476. [Google Scholar]
- Milillo, S.R. , Friedly, E.C. , Saldivar, J.C. , Muthaiyan, A. , O’Bryan, C. , Crandall, P.G. , et al. (2012) A review of the ecology, genomics, and stress response of Listeria innocua and Listeria monocytogenes . Crit Rev Food Sci Nutr 52: 712–725. [DOI] [PubMed] [Google Scholar]
- Mohan, V. , Wibisono, R. , de Hoop, L. , Summers, G. , and Fletcher, G.C. (2019) Identifying suitable Listeria innocua strains as surrogates for Listeria monocytogenes for horticultural products. Front Microbiol 10: 2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, U.T. , Wenderska, I.B. , Chong, M.A. , Koteva, K. , Wright, G.D. , and Burrows, L.L. (2012) Small‐molecule modulators of Listeria monocytogenes biofilm development. Appl Environ Microbiol 78: 1454–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichterlein, T. , Kretschmar, M. , Ruhland, G.J. , Fiedler, F. , and Hof, H. (1997) Lipoteichoic acid fractions from pathogenic and apathogenic Listeria species and Staphylococcus aureus induce similar amounts of macrophage‐derived cytokines. Inttammopharmacology 5: 343–350. [DOI] [PubMed] [Google Scholar]
- Nightingale, K.K. , Windham, K. , and Wiedmann, M. (2005) Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J Bacteriol 187: 5537–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan, D.A. , Chamblin, D.C. , and Troller, J.A. (1992) Minimal water activity levels for growth and survival of Listeria monocytogenes and Listeria innocua . Int J Food Microbiol 16: 323–335. [DOI] [PubMed] [Google Scholar]
- Novick, R.P. , Ross, H.F. , Projan, S.J. , Kornblum, J. , Kreiswirth, B. , and Moghazeh, S. (1993) Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J 10: 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaimat, A.N. , Al‐Holy, M.A. , Shahbaz, H.M. , Al‐Nabulsi, A.A. , Abu Ghoush, M.H. , Osaili, T.M. , et al. (2018) Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: a comprehensive review. Compr Rev Food Sci Food Saf 17: 1277–1292. [DOI] [PubMed] [Google Scholar]
- Orsi, R.H. , de Bakker, H.C. , and Wiedmann, M. (2011) Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol 301: 79–96. [DOI] [PubMed] [Google Scholar]
- Ouyang, Y. , Li, J. , Dong, Y. , Blakely, L.V. , and Cao, M. (2012) Genome‐wide screening of genes required for Listeria monocytogenes biofilm formation. J Biotech Res 4: 13–25. [Google Scholar]
- Parida, S.K. , Domann, E. , Ronde, M. , Müller, S. , Darji, A. , Hain, T. , et al. (1998) Internalin B is essential for adhesion and mediates the invasion of Listera monocytognes into human endothelial cells. Mol Microbiol 28: 81–93. [DOI] [PubMed] [Google Scholar]
- Peel, M. , Donachie, W. , and Shaw, A. (1988) Temperature‐dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS‐PAGE and Western blotting. J Gen Microbiol 134: 2171–2178. [DOI] [PubMed] [Google Scholar]
- Piercey, M.J. , Hingston, P.A. , and Truelstrup Hansen, L. (2016) Genes involved in Listeria monocytogenes biofilm formation at a simulated food processing plant temperature of 15 °C. Int J Food Microbiol 223: 63–74. [DOI] [PubMed] [Google Scholar]
- Popowska, M. , Krawczyk‐Balska, A. , Ostrowski, R. , and Desvaux, M. (2017) InlL from Listeria monocytogenes is involved in biofilm formation and adhesion to mucin. Front Microbiol 8: 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, Z. , Ou, Y. , Yang, L. , Zhu, Y. , Tolker‐Nielsen, T. , Molin, S. , and Qu, D. (2007) Role of autolysin‐mediated DNA release in biofilm formation of Staphylococcus epidermidis . Microbiology 153: 2083–2092. [DOI] [PubMed] [Google Scholar]
- Radoshevich, L. , and Cossart, P. (2018) Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat Rev Microbiol 16: 32–46. [DOI] [PubMed] [Google Scholar]
- dos Reis‐Teixeira, F.B. , Alves, V.F. , and de Martinis, E.C.P. (2017) Growth, viability and architecture of biofilms of Listeria monocytogenes formed on abiotic surfaces. Brazilian J Microbiol 48: 587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier, S. , Hébraud, M. , and Desvaux, M. (2011) Molecular biology of surface colonization by Listeria monocytogenes: an additional facet of an opportunistic Gram‐positive foodborne pathogen. Environ Microbiol 13: 835–850. [DOI] [PubMed] [Google Scholar]
- Rieu, A. , Briandet, R. , Habimana, O. , Garmyn, D. , Guzzo, J. , and Piveteau, P. (2008) Listeria monocytogenes EGD‐e biofilms: no mushrooms but a network of knitted chains. Appl Environ Microbiol 74: 4491–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rismondo, J. , Haddad, T.F.M. , Shen, Y. , Loessner, M.J. , and Gründling, A. (2020) GtcA is required for LTA glycosylation in Listeria monocytogenes serovar 1/2a and Bacillus subtilis . Cell Surf 6: 100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, T. , Viala, D. , Chambon, C. , Esbelin, J. , and Hébraud, M. (2019) Listeria monocytogenes biofilm adaptation to different temperatures seen through shotgun proteomics. Front Nutr 6: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasahara, K.C. , and Zottola, E.A. (1993) Biofilm formation by Listeria monocytogenes utilizes a primary colonizing microorganism in flowing systems. J Food Prot 56: 1022–1028. [DOI] [PubMed] [Google Scholar]
- Schirm, M. , Kalmokoff, M. , Aubry, A. , Thibault, P. , Sandoz, M. , and Logan, S.M. (2004) Flagellin from Listeria monocytogenes is glycosylated with β‐O‐linked N‐acetylglucosamine. J Bacteriol 186: 6721–6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala, L. , Shih, H.L. , Cannesson, P. , and Ross, T. (2008) Acid and NaCl limits to growth of Listeria monocytogenes and influence of sequence of inimical acid and NaCl levels on inactivation kinetics. J Food Prot 71: 1169–1177. [DOI] [PubMed] [Google Scholar]
- Shen, Y. , Boulos, S. , Sumrall, E. , Gerber, B. , Julian‐Rodero, A. , Eugster, M.R. , et al. (2017) Structural and functional diversity in Listeria cell wall teichoic acids. J Biol Chem 292: 17832–17844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovager, A. , Larsen, M.H. , Castro‐Mejia, J.L. , Hecker, M. , Albrecht, D. , Gerth, U. , et al. (2013) Initial adhesion of Listeria monocytogenes to fine polished stainless steel under flow conditions is determined by prior growth conditions. Int J Food Microbiol 165: 35–42. [DOI] [PubMed] [Google Scholar]
- Solano, C. , Echeverz, M. , and Lasa, I. (2014) Biofilm dispersion and quorum sensing. Curr Opin Microbiol 18: 96–104. [DOI] [PubMed] [Google Scholar]
- Sommers, C.H. , Cooke, P.H. , Fan, X. , and Sites, J.E. (2009) Ultraviolet light (254 nm) inactivation of Listeria monocytogenes on frankfurters that contain potassium lactate and sodium diacetate. J Food Sci 74: 114–119. [DOI] [PubMed] [Google Scholar]
- Sumrall, E.T. , Keller, A.P. , Shen, Y. , and Loessner, M.J. (2020) Structure and function of Listeria teichoic acids and their implications. Mol Microbiol 113: 627–637. [DOI] [PubMed] [Google Scholar]
- Sumrall, E.T. , Schefer, C.R.E. , Rismondo, J. , Schneider, S.R. , Boulos, S. , Gründling, A. , et al. (2020) Galactosylated wall teichoic acid, but not lipoteichoic acid, retains InlB on the surface of serovar 4b Listeria monocytogenes . Mol Microbiol 113: 638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumrall, E.T. , Shen, Y. , Keller, A.P. , Rismondo, J. , Pavlou, M. , Eugster, M.R. , et al. (2019) Phage resistance at the cost of virulence: Listeria monocytogenes serovar 4b requires galactosylated teichoic acids for InlB‐mediated invasion. PLOS Pathog 15: e1008032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theilacker, C. , Sanchez‐Carballo, P. , Toma, I. , Fabretti, F. , Sava, I. , Kropec, A. , et al. (2009) Glycolipids are involved in biofilm accumulation and prolonged bacteraemia in Enterococcus faecalis . Mol Microbiol 71: 1055–1069. [DOI] [PubMed] [Google Scholar]
- Tiong, H.K. , Hartson, S.D. , and Muriana, P.M. (2016) Comparison of surface proteomes of adherence variants of Listeria monocytogenes using LC‐MS/MS for identification of potential surface adhesins. Pathogens 5: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travier, L. , Guadagnini, S. , Gouin, E. , Dufour, A. , Chenal‐Francisque, V. , Cossart, P. , et al. (2013) ActA promotes listeria monocytogenes aggregation, intestinal colonization and carriage. PLoS Pathog 9: e1003131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, S.J. , Archer, P. , and Banks, J.G. (1990) Growth of Listeria monocytogenes at refrigeration temperatures. J Appl Bacteriol 68: 157–162. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Liu, B. , Teng, Z. , Zhou, X. , Wang, X. , Zhang, B. , et al. (2017) Phloretin attenuates Listeria monocytogenes virulence both in vitro and in vivo by simultaneously targeting listeriolysin O and sortase A. Front Cell Infect Microbiol 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way, S.S. , Thompson, L.J. , Lopes, J.E. , Hajjar, A.M. , Kollmann, T.R. , Freitag, N.E. , and Wilson, C.B. (2004) Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell Microbiol 6: 235–242. [DOI] [PubMed] [Google Scholar]
- Webb, A.J. , Karatsa‐Dodgson, M. , and Gründling, A. (2009) Two‐enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes . Mol Microbiol 74: 299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawadzka‐Skomiał, J. , Markiewicz, Z. , Nguyen‐Distèche, M. , Devreese, B. , Frère, J.M. , and Terrak, M. (2006) Characterization of the bifunctional glycosyltransferase/acyltransferase penicillin‐binding protein 4 of Listeria monocytogenes . J Bacteriol 188: 1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetzmann, M. , Bucur, F.I. , Crauwels, P. , Borda, D. , Nicolau, A.I. , Grigore‐Gurgu, L. , et al. (2019) Characterization of the biofilm phenotype of a Listeria monocytogenes mutant deficient in agr peptide sensing. Microbiologyopen 8: e00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of proteins affecting Listeria monocytogenes biofilm formation on abiotic surfaces.
Appendix S1. Interaction network analysis procedure and results.


