Summary
Phi29 DNA polymerase (Phi29 Pol) has been successfully applied in DNA nanoball‐based sequencing, real‐time DNA sequencing from single polymerase molecules and nanopore sequencing employing the sequencing by synthesis (SBS) method. Among these, polymerase‐assisted nanopore sequencing technology analyses nucleotide sequences as a function of changes in electrical current. This ionic, current‐based sequencing technology requires polymerases to perform replication at high salt concentrations, for example 0.3 M KCl. Nonetheless, the salt tolerance of wild‐type Phi29 Pol is relatively low. Here, we fused helix–hairpin–helix (HhH)2 domains E‐L (eight repeats in total) of topoisomerase V (Topo V) from the hyperthermophile Methanopyrus kandleri to the Phi29 Pol COOH terminus, designated Phi29EL DNA polymerase (Phi29EL Pol). Domain fusion increased the overall enzyme replication efficiency by fourfold. Phi29EL Pol catalysed rolling circle replication in a broader range of salt concentrations than did Phi29 Pol, extending the KCl concentration range for activity up to 0.3 M. In addition, the mutation of Glu375 to Ser or Gln increased Phi29EL Pol activity in the presence of KCl. In this work, we produced a salt‐tolerant Phi29 Pol derivative by means of (HhH)2 domain insertion. The multiple advantages of this insertion make it a good substitute for Phi29 Pol, especially for use in nanopore sequencing or other circumstances that require high salt concentrations.
1. For ionic current based nanopore sequencing, polymerases, helicases or exonucleases were required to process DNA with relatively higher contents of salt (0.2–1 M KCl) which is necessary to attain sufficient ionic strength for nucleobase recognition and increase the signal to noise ratio. The delicate structure features endow Phi29 Pol intrinsic strand displacement capability, making it an ideal enzyme for various sequencing technologies, for example real‐time DNA sequencing from single polymerase molecules, DNA nanoball‐based sequencers and nanopore sequencing technology employing sequencing by synthesis method. 2. We have achieved a salt tolerant Phi29 Pol derivative working at 0.3 M KCl by means of helix‐hairpin‐helix domain insertion. The enzyme also showed improved rolling circle replication efficiency. 3. Besides salt tolerance, other properties of Phi29EL Pol such as the processivity, strand displacement and extension rate were not affected much by domain fusion. The combined advantages of Phi29EL Pol makes it a good substitute of Phi29 Pol, especially for nanopore sequencing or other circumstances depend on higher contents of salt.

Introduction
The discovery of DNA polymerase was a milestone in modern biotechnology (Bessman et al., 1958; Lehman et al., 1958). DNA polymerases are widespread in almost all organisms and are involved in cell division, genome replication and the maintenance of genetic integrity, in conjunction with other proteins (Johansson and Dixon, 2013). Generally, DNA polymerases (DNA pols) are classified into seven subfamilies: A, B, C, D, X, Y and RT, on the basis of conserved sequences and tertiary structures (Rothwell and Waksman, 2005). Numerous natural and mutant DNA polymerases have been characterized in terms of their conserved sequences, 3D structures, biochemical activities and enzymatic kinetics (Saiki et al., 1988; Yamagami et al., 2014; Wang et al., 2015), to identify ideal polymerases with high speed, fidelity, processivity, enhanced binding to DNA and dNTP substrates, and strong strand displacement activity (Kong et al., 1993; Cline et al., 1996). Many have been commercialized and successfully applied in scientific and industrial fields, including PCR, RT‐PCR, rolling circle replication (RCR), cDNA synthesis, DNA terminal modification and DNA sequencing (Saiki et al., 1988; Kong et al., 1993; Cline et al., 1996; Dean et al., 2001; Gardner and Jack, 2002; Yamagami et al., 2014; Wang et al., 2015; Baba et al., 2017).
Recently, the identification and development of salt‐tolerant DNA pols have been undertaken to resolve the difficulties associated with GC‐rich DNA sequencing and the new generation of nanopore sequencing technology (Bernick et al., 2012; Bruce et al., 2016; Fuller et al., 2016). For ionic‐current‐based nanopore sequencing, polymerases, helicases or exonucleases are required to process DNA in higher salt concentrations (0.2–1 M KCl) than in other procedures, necessary to attain sufficient ionic strength for nucleobase recognition and to increase the signal‐to‐noise ratio (Clarke et al., 2009; Bruce et al., 2016; Fuller et al., 2016). However, most enzymes lose activity in high‐salt conditions (Saiki et al., 1988; Kong et al., 1993; Cline et al., 1996; Dean et al., 2001; Gardner and Jack, 2002; Yamagami et al., 2014; Wang et al., 2015; Baba et al., 2017). To overcome such obstacles, several naturally halophilic DNA pols have been isolated from hypersaline ponds and deep‐sea brines of the Red Sea (Bernick et al., 2012; Bruce et al., 2016; Takahashi et al., 2018). We previously characterized a halotolerant DNA pol from the marine bacteriophage VpV262 that infects pathogenic Vibrio parahaemolyticus (Gao et al., 2020). Halophilic and halotolerant polymerases are typically enriched in acidic amino‐acid residues (Hardies et al., 2003; Bernick et al., 2012; Takahashi et al., 2018; Gao et al., 2020). For example, the dynamic structure of BR3 Pol, mediated by excess negatively charged residues and random coils, has been reported to contribute to the halophilic adaptation of BR3 Pol (Takahashi et al., 2018). Although polymerization by these enzymes could occur in the presence of chlorides (> 0.3 M), other properties, including low processivity or polymerization rate, make them unsuitable for direct use in sequencing (Hardies et al., 2003; Bernick et al., 2012; Gao et al., 2020). Cheng et al. harnessed the TBD (thioredoxin binding domain) from phage T7 to improve the processivity of DNAP I from Staphylococcus aureus, chosen because it can survive in tinned food at > 4 M NaCl, while Sau DNAP I was able to work at 0.16 M. Compared to wild‐type, chimeric Sau–TBD DNAP I was tolerant to KAc (0.18 M), with more than a twofold increase in processivity (Cheng et al., 2016).
Another approach to facilitate the salt resistance of DNA pols is to construct chimeric enzymes by coupling external domains to non‐halophilic DNA pols, which has been successful in several cases. The replacement of the exonuclease domain or thumb domain of KOD Pol (Thermococcus kodakarensis DNA polymerase) by those from halophilic BR3 Pol increased its salt tolerance from 0.2 to 0.3 M KCl (Takahashi et al., 2018). Gss‐polymerase derivatives fused with the DBD domain (DNA binding domain of DNA ligase from Pyrococcus abyssi) or Sto7d protein (from Sulfolobus tokodaii) maintain approximately 40% of polymerase activity in 0.5 M KCl, significantly higher than the < 10% of native enzymes (Oscorbin et al., 2017). Taq polymerase or Pfu polymerase fused with thermophilic Sso7d protein displayed a broader salt tolerance (from 0.01–0.05 to 0.01–0.1 M KCl) and increased amplification over wild‐type polymerases (Wang et al., 2004).
Pavlov et al. reported that helix–hairpin–helix motifs conferred salt tolerance to Taq Pol and Pfu Pol (Pavlov et al., 2002). There are 24 HhH repeats at the C‐terminus of Topo V from hyperthermophilic Methanopyrus kandleri, organized into 12 (HhH)2 domains: A, B, C, D, E, F, G, H, I, J, K and L, each consisting of two HhH motifs (Slesarev et al., 1993; Shao and Grishin, 2000; Belova et al., 2001). Among these domains, the E‐L domains are believed to function directly in the activity and processivity at high salt concentrations, based on biochemical analysis of truncated Topo‐61 (Belova et al., 2002). Topo‐61, which consists only of an N‐terminal topoisomerase domain and the A‐D domains, exhibited a maximum activity at 0.03 M KCl, compared to 0.45 M KCl for Topo V (Belova et al., 2002). Due to strong sequence‐nonspecific DNA binding, chimeric Taq Pol or Pfu Pol with 5‐8 (HhH)2 repeats, particularly for enzymes with E‐L cassettes, increased initial polymerization rates in the presence of either chlorides or potassium glutamate (Pavlov et al., 2002). Moreover, chimerism significantly increased processivity and thermostability. Importantly, this approach may be applicable to DNA pols of both families A and B (Pavlov et al., 2002).
Phi29 Pol, encoded by Phi29, a bacteriophage infecting Bacillus subtilis, is well known for its high fidelity and multiple isothermal displacement amplification at 30°C (Blanco et al., 1989). Although it is responsible for protein‐initiated genome replication in vivo, it also is used in DNA‐initiated replication experiments in vitro. It has 575 amino‐acid residues and belongs to the Pol B family (Blanco and Salas, 1984; Bernad et al., 1987). Crystallographic and biochemical studies have revealed that Phi29 Pol possesses five conserved domains: palm, finger, thumb, TPR1 and TPR2; together, they form one tunnel for incoming dNTP, allowing the upstream template/primer complex to leave, and another narrow tunnel that allows the displaced downstream strand to pass through (Blanco and Salas, 1996; Rodríguez et al., 2005; Berman et al., 2007). These delicate structural features endow Phi29 Pol with intrinsic strand displacement capability, making it an ideal enzyme for RCR and genome amplification without requiring thermal cycling. In recent years, Phi29 Pol has been exploited in multiple sequencing technologies, including real‐time DNA sequencing from single polymerase molecules (Korlach et al., 2010), DNA nanoball‐based sequencing (Drmanac et al., 2010) and nanopore sequencing employing the sequencing by synthesis (SBS) method (Fuller et al., 2016). However, previous research has shown that the DNA binding and salt tolerance of wild‐type Phi29 Pol are relatively low (Blanco et al., 1989; de Vega et al., 2010).
In this study, we followed a strategy described by Pavlov et al. (2002), selecting (HhH)2 domains as described above to create a chimeric Phi29 Pol with enhanced salt tolerance. The aim of this study was to obtain salt‐tolerant Phi29 Pol derivatives. Eight continuous (HhH)2 domains, used in the chimeras PfuC2 and TaqTopoC2 (Pavlov et al., 2002), were ligated to the C‐terminus of Phi29 Pol. An N‐terminal fusion was not attempted, as structural and functional analyses indicated that it may cause steric hindrance and interfere with the strand displacement capacity of wild‐type Phi29 Pol (Kamtekar et al., 2004; Rodríguez et al., 2005; de Vega et al., 2010). Phi29‐H Pol was also constructed as described previously (de Vega et al., 2010) for use in salt tolerance tests.
Results
Design and expression of chimeric Phi29 DNA polymerases
The E‐L domains of Topo V from M. kandleri were fused with Phi29 Pol (Fig. 1A), resulting in the chimeric protein Phi29EL Pol. Residues 518–964, representing the E‐L tandem repeats, were used to build a protein structural model. However, only residues 518–852, corresponding to E‐J domains, could be modelled, as the missing domains F and G in the crystal structure of Topo‐97 were predicted to be unstructured (Fig. S1) and suggested to be a hinge point (Rajan et al., 2016). Moreover, the other domains were superimposed well with each other (Fig. S1). Therefore, the modular structure of the chimeric polymerase was generated by combining the Phi29 Pol and Topo V‐EL models (Fig. 1B). With a short peptide connector linking the COOH‐termini of Phi29 Pol and Topo V‐EL, flexible (HhH)2 domains most likely lay at the end of dsDNA, extending away from the rigid polymerase module and participating in interactions with upstream template/primer strands (de Vega et al., 2010) in a similar manner to topoisomerase and other DNA processing enzymes with HhH motif(s) (Sawaya et al., 1997; Shao and Grishin, 2000; Newman et al., 2005).
Fig. 1.
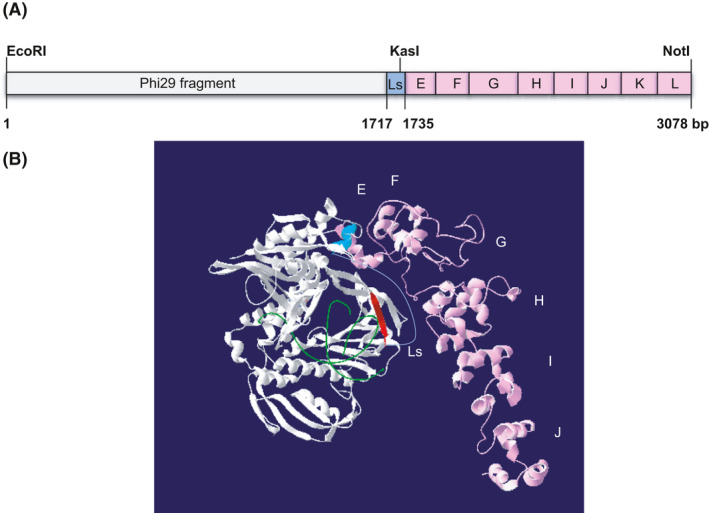
Diagram of chimeric Phi29EL Pol. (A) E‐L (HhH)2 repeats from Topo V (pink) were fused to the C‐terminal end of Phi29 Pol (white) with a linker sequence (GTGSGA) between them. Ls, linker sequence. (B) The 3D structure of Phi29EL Pol modelled by Swiss‐PdbViewer. Phi29 Pol structure (PDB: 2PYL) complexed with substrate DNA (green) was downloaded from PDB. The modular structure of E‐J domains was predicted using SWISS‐MODEL. Both domains are shown in the same colours as described for Panel A. The template/primer junction is shown in green. The COOH termini of Phi29 Pol and NH2 termini of (HhH)2 domains are shown in red and cyan, respectively. The linker sequence (Ls) is shown as a blue line. Except for the G motif and part of the F motif being disordered, all the other domains matched well with the crystal structure of the Topo‐97 fragment (PDB: 5HM5). The spatial arrangement of (HhH)2 domains is beneficial for DNA binding.
The recombinant protein was stable and active if stored in elution (Fig. S2A) or storage buffer (Fig. S2B, lane 1) with 0.5 M NaCl, in contrast to immediate precipitation (e.g. proteolysis) in common PBS with ~ 0.1 M NaCl (Fig. S2B, lane 2). This characteristic of Phi29EL is clearly different from that of Phi29 Pol, which is stable and active in low‐salt buffers, such as PBS (Fig. S3). The salt‐dependent stability of Phi29EL Pol is consistent with previous studies showing that Topo V required 0.45 M KCl or 0.3 M NaCl for maximal activity, with the (HhH)2 domains involved in DNA binding and processivity at high salt concentrations (Slesarev et al., 1994; Belova et al., 2002).
A high processivity is kept intact in the Phi29EL Pol chimera
Previous studies have reported that chimeric Phi29 polymerases produced by fusing one or two (HhH)2 domains increase DNA binding without decreasing processivity (de Vega et al., 2010). To examine the processivity of Phi29 wild‐type and chimeric DNA polymerases, we performed rolling circle replication (RCR) with singly primed M13mp18. We first performed the extension with Buffer 2 at 30°C (Figs S4 and S5).
After extension, a linear DNA fragment larger than 48 kb was produced as an elongated primer for both Phi29 Pol and Phi29EL Pol (Fig. 2A and B). Apart from DNA products larger than 48 kb, several weak bands appeared around the M13 templates of Phi29EL Pol (Fig. 2) and Phi29 Pol (Fig. S6) at multiple enzyme concentrations, indicating that they were not produced by dissociation. These bands possibly represented blocked extension products, or snap‐back products (after switching Pol to the displaced strand, which then became the template), which have also been detected before autoradiography (Blanco et al., 1989; de Vega et al., 2010). No shortened DNA product was obtained during the dilution of the corresponding enzymes, indicating that neither Phi29 Pol nor Phi29EL Pol had disassociated from the substrate DNA after DNA‒enzyme complex formation. The addition of (HhH)2 domains did not alter extension and processivity of the chimera.
Fig. 2.
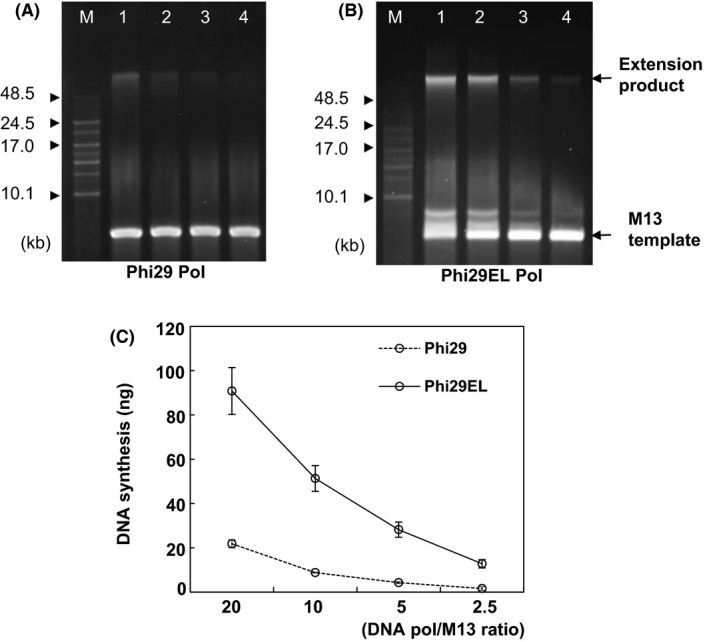
Processivity of Phi29 Pol and Phi29EL Pol with singly primed M13mp18 single‐strand DNA. Processivity was quantified by the gradual dilution of Phi29 Pol (A) and Phi29EL Pol (B). After incubating at 30°C for 30 min, samples were mixed with loading buffer for electrophoresis. The molar concentrations of enzymes were 20‐ (Lane 1), 10‐ (Lane 2), 5‐ (Lane 3) and 2.5‐fold (Lane 4) more than that of the M13 template. (C) Quantification of newly synthesized DNA from rolling circle replication. Data are shown as product concentrations versus molar ratios of DNA polymerase to M13 ssDNA. The dotted line and solid line represent Phi29 Pol and Phi29EL Pol, respectively. DNA length markers are labelled with arrowheads in A and B.
As shown in Fig. 2C, the extension product of Phi29EL Pol was approximately fourfold higher than that of Phi29 Pol when the molar ratio of the enzyme to the circular template was 20, indicating that chimeric DNA Pols exhibited better replication performance. As the ratio between the enzyme and template declined from 20 to 2.5, the total amount of replication product decreased for both enzymes. Additionally, Phi29EL Pol at a 2.5‐fold dilution produced as much DNA as Phi29 Pol at a 20‐fold dilution, demonstrating the high efficiency of the heterozyme in practical applications. The higher activity of Phi29EL Pol in the assay could be due either to an improved processivity or to a more efficient initial binding to the primed‐M13 template.
(HhH)2 domains increase polymerase resistance to potassium gluconate in hairpin extension
Using a 53‐nt hairpin template, we evaluated the activity of Phi29EL Pol with NaCl, KCl, potassium acetate (KAc), or potassium gluconate (KGlc) individually. As shown in Fig. 3A and B, Phi29EL Pol produced 31% more extension products than did Phi29 Pol without salt added, although higher salt concentrations all hindered the extension of hairpins to different extents.
Fig. 3.
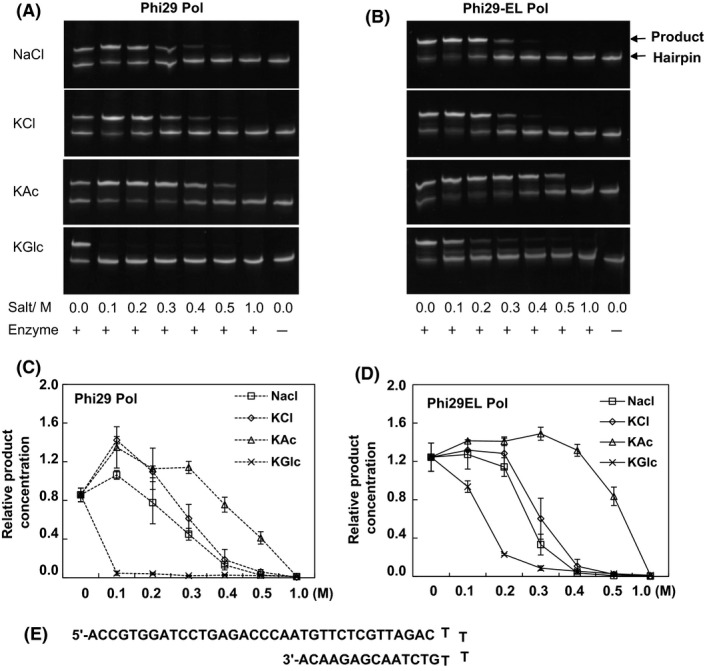
Salt tolerance of Phi29 Pol and Phi29EL Pol. Divergent salts ranging from 0 to 1 M were separately added into the hairpin extension reactions with Phi29 Pol (A) and Phi29EL Pol (B). TBE‐urea gels (20%) were used to separate the templates and extension products. Complete extension products in A and B were quantified by band intensity. The y‐axis in panels C and D indicates the product concentration relative to the total amount of template in the last lane of each gel. Data are presented as means ± SD. (E) The hairpin sequence was 53 nt, while the extension product was 74 nt.
The negative impact of NaCl or KCl on enzyme activity was between that of KAc and KGlc. Both enzymes exhibited the best performance in 0.1 M chloride (Fig. 3C and D). More than half of the total activities of both enzymes was prevented by 0.3 M chloride (Fig. 3C and D). A progressive increase in ionic strength (with NaCl and KCl) appeared to cause the same extent of inhibition for both Phi29 Pol and Phi29EL Pol. Although the wild‐type enzyme retained activity in the presence of higher salt concentration than Phi29EL Pol (0.4 vs 0.3 M for NaCl, 0.5 vs 0.4 M for KCl), Phi29EL Pol had a relatively small activity fluctuation among 0–0.2 M chloride (Fig. 3). Among the four salt solutions, KAc caused the least inhibition of the replication reaction for both enzymes. Phi29EL Pol generated more products than Phi29 Pol when treated with 0–0.5 M. More than 40% and 80% of the extension products were still obtained with 0.5 M KAc for Phi29 Pol and Phi29EL Pol, respectively. However, the assay was very saturated with most of the hairpin is extended at low salt with Phi29EL Pol, making it difficult to estimate the reduction produced by the increase in ionic strength. On the other hand, the activity of Phi29 Pol was inhibited at 0.1 M KGlc, while Phi29EL worked efficiently at 0.1 M KGlc and retained some activity at 0.3 M KGlc. In conclusion, the heterozyme Phi29EL Pol showed an increased halotolerance with the salt KGlc (Fig. 3).
Phi29EL DNA polymerase tolerates a broad salt concentration range in rolling circle replication
Compared to primer extension of hairpins less than 100 nt, the extension of singly primed M13mp18 requires more time to generate a much longer product progressively with circular templates. To test the salt tolerance, polymerization rate and strand displacement capability of Phi29EL Pol, four salts were added to the RCR reaction system separately. Higher concentrations of NaCl or KCl partially inhibited the activity of the two enzymes (Fig. 4A and B). Phi29EL Pol performance was significantly better than that of the wild‐type Phi29 Pol. We found that about 4 ng of DNA was synthesized by Phi29 Pol at 0.1 M KCl, but no product was detected when Phi29 Pol was incubated with 0.2 M NaCl or 0.3 M KCl for 30 min (Fig. 4A). However, weak bands appeared after 1 h of incubation with 0.3 M KCl for Phi29 Pol, and progressive accumulation of products was detected over time (Fig. S6). We found that 0.1 M NaCl or 0.2 M KCl almost halted the RCR activity of Phi29 Pol, whereas approximately 50% and 20% of the extension products, respectively, were obtained with Phi29EL Pol under the same conditions (Fig. 4C and D). We noticed that the product concentration of Phi29EL Pol with 0.3 M KCl was equivalent to that of Phi29 Pol without extra salt, demonstrating the efficient catalytic activity and higher salt tolerance of Phi29EL Pol (Fig. 4C and D).
Fig. 4.
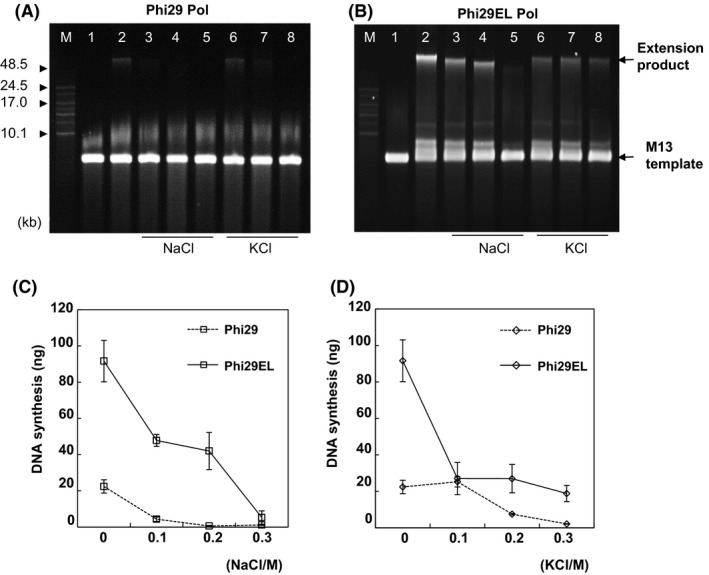
Salt tolerance of Phi29 Pol and Phi29EL Pol. Different concentrations of NaCl or KCl were added into the singly primed M13mp18 extension experiment. Products of Phi29 Pol (A) and Phi29EL Pol (B) were separated using 0.6% alkaline agarose gels. Lane 1, background reaction mixtures without enzyme; Lane 2, no added salt; Lanes 3–5, 0.1, 0.2 and 0.3 M NaCl; Lanes 6–8 with 0.1, 0.2 and 0.3 M KCl. The yielded DNA (> 48 kb) was further quantified in C (NaCl) and D (KCl). Data are shown as product amounts versus salt concentrations. Data are presented as means ± SD.
The influence of NaCl and KCl on polymerization by Phi29EL Pol differed according to the length of the synthesized products. Increasing concentrations of NaCl led to an abrupt reduction in both concentration and length of the extension product (Fig. 4A and B). KCl only affected concentration, while product length seemed to be identical with 0.1–0.3 M of KCl in the solution (Fig. 4A and B). The difference in product size with NaCl was not caused by the difference in the salt content of the reaction buffer. This is because only the mobility of the product band with NaCl was different, but not with other bands, such as M13 templates. Moreover, the same product loaded with various NaCl contents showed the same mobility (data not shown). In conclusion, mobility alteration was only related to product length (Fig. 4A and B).
Both Phi29 and Phi29EL Pol displayed the highest resistance to KAc compared to the other salts, consistent with the results of the hairpin extension experiments (Fig. 3). Both were able to replicate in 0.3 M KAc, although the amount of newly replicated DNA by Phi29 Pol was less than that of Phi29EL Pol (Fig. 5A and B). Conversely, KGlc caused the most serious damage to both proteins. We did not detect product DNA when Phi29 Pol was incubated with KGlc (0.1 M) (Fig. 5A). Phi29EL Pol generated a small amount of DNA with 0.1 M KGlc (Fig. 5B). Quantification of the extension products further confirmed these results (Fig. 5C and D). These results showed that Phi29EL Pol was more tolerant to a series of salts than was Phi29 Pol (Figs 4 and 5).
Fig. 5.
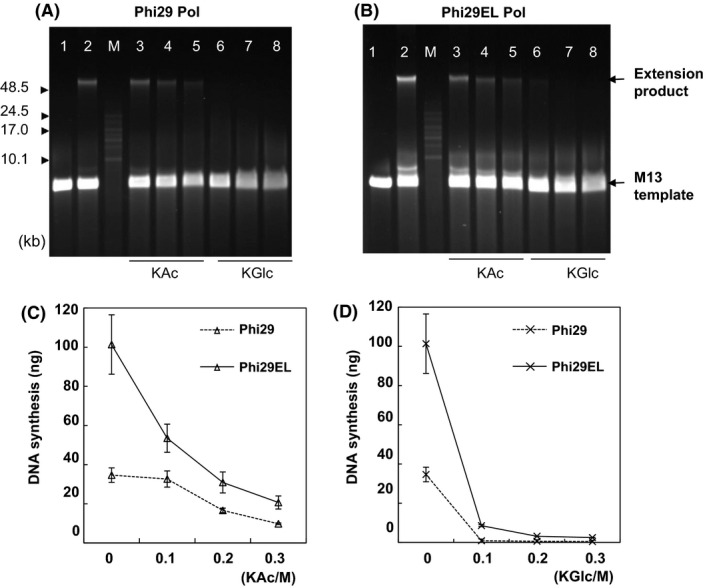
Salt tolerance of KAc and KGlc for Phi29 Pol (A) and Phi29EL Pol (B). KAc or KGlc at 0–0.3 M added to the reaction mixture separately with singly primed M13mp18 as templates. Lane 1, background reaction without enzyme; Lane 2, no added salt; Lanes 3–5, with 0.1, 0.2 or 0.3 M KAc; Lanes 6–8 with 0.1, 0.2 or 0.3 M KGlc. Quantification analysis of the synthesized DNA under increased concentrations of KAc (C) and KGlc (D). Compared to NaCl and KCl, both of the enzymes were much more resistant to KAc and sensitive to KGlc. Data are presented as means ± SD.
The addition of (HhH)2 motifs increases 3′‐5′ exonuclease activity
The 3′‐5′ proofreading exonuclease activity of Phi29 Pol contributes to fewer insertion errors and high‐fidelity replication. Since the (HhH)2 domains were flexible and organized into the C‐ terminal end of the enzyme, domain fusion may not significantly change the structure and activity of the exonuclease domains. To test this hypothesis, we synthesized three oligonucleotides to assess exonuclease activity (Guga and Koziołkiewicz, 2011; Gao et al., 2020). In this method, oligos with the two constitutive phosphorothioates at the 3′ end (Oligo B, Fig. 6A) were protected from degradation by 3′‐5′ exonuclease, while natural (Oligo A, Fig. 6A) or 5′‐modified oligos (Oligo C, Fig. 6A) were barely detected after incubation with 3′‐5′ exonuclease (Gao et al., 2020). As shown in Fig. 6B, Oligo A and 5′‐protected Oligo C were digested immediately after incubation with Phi29 Pol or Phi29EL Pol, deploying their 3′‐5′ exonuclease activity, while 3′‐protected Oligo C remained for both enzymes.
Fig. 6.
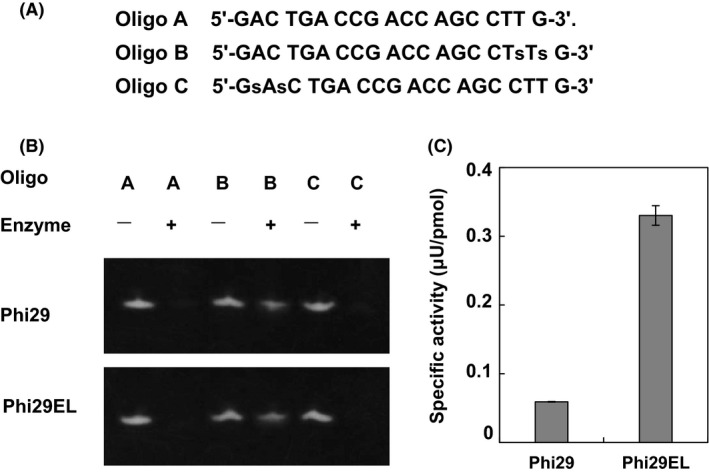
Exonuclease activity of Phi29 Pol and Phi29EL Pol.
A. Sequences and modifications of Oligos A, B and C. The lowercase ‘s’ denotes phosphorothioate.
B. Both polymerases (200 nM) were separately mixed with three oligos for 10 min. The reactions were analysed using a 20% TBE‐urea gel. 3′‐5′ proofreading exonuclease activity of both enzymes could degrade natural oligos (Oligo A) and 5′ phosphorothioate‐modified oligos (Oligo C) after a short time of incubation.
C. Specific activity assay. A 3′ to 5′ exonuclease activity assay was performed with Phi29 Pol or Phi29EL Pol according to the manufacturer’s instructions. Data are presented as means ± SD.
A 3′ to 5′ exonuclease activity assay was then performed (Fig. 6C). The specific activity of Phi29EL Pol (0.33 μU pmol−1) was approximately 5.5‐fold greater than that of Phi29 Pol (0.06 μU pmol−1) after monitoring the fluorescent signal in kinetic mode for 40 min. The sequence of the exonuclease domain in Phi29EL Pol did not change compared with Phi29 Pol, suggesting that the elevated exonuclease activity of Phi29EL Pol resulted from domain fusion and interaction.
Point mutations in the finger domain further enhance salt tolerance of the chimeras
Anions play a key role in the inhibition of DNA polymerase activity (Pavlov et al., 2002). Topo V (HhH)2 motifs have been reported to enhance DNA binding in a sequence‐nonspecific manner, especially at high salt concentrations (Belova et al., 2001; Belova et al., 2002). However, it is not known whether the incorporation of dNTPs mediated by the finger domain of Phi29EL Pol is affected by high salt concentrations. Thus, we focused our efforts on improving the salt tolerance of the finger domain. The non‐conserved residue glutamic acid 375 (E375) has been identified as anticipating the incorporation of nucleotide analogs with extra phosphate (Hanzel et al., 2017). We therefore generated a series of E375 mutants based on Phi29EL Pol for the salt tolerance assay, generating Phi29EL‐E375A/K/H/S/Q/W/Y (Fig. 7). RCR products of Phi29EL‐E375S/Q mutants were produced at a higher level than that of Phi29EL Pol with 0 or 0.3 M KCl. In contrast, the polymerization activities of Phi29EL‐E375A/K/H and Phi29EL Pol were comparable. The performance of Phi29EL‐E375W and Phi29EL‐E375Y was less efficient than that of the Phi29EL wild‐type. While W or Y mutations endowed Phi29 Pol with enhanced incorporation ability for tetraphosphate nucleotide analogs (Hanzel et al., 2017), the E375S/Q mutation conferred elevated salt tolerance of Phi29EL Pol. Changing residue E375 to A or Y in Phi29 Pol did not raise salt tolerance to the same extent as that of the Phi29EL mutants (Fig. S7). The combination of the (HhH)2 domain and site‐directed mutations contributed to the salt resistance of the Phi29EL Pol derivatives.
Fig. 7.
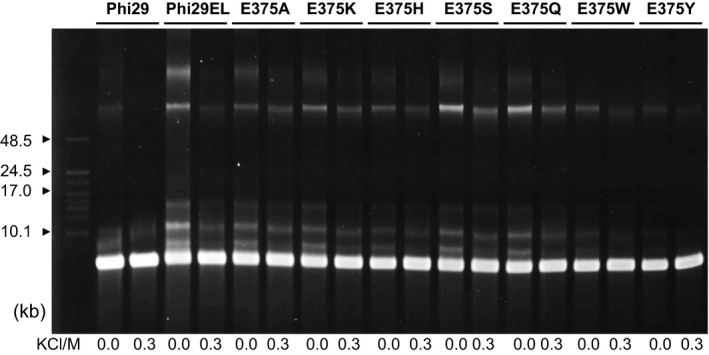
Salt tolerance of Phi29EL Pol mutants. KCl at 0 M or 0.3 M was separately added into the M13 extension system. The reaction time was 30 min. DNA length markers are labelled with arrowheads. Data are representative of several independent experiments.
Finally, the extension rate of the Phi29EL derivatives was lower than that of Phi29 Pol (Fig. S8), which is beneficial for sequencing because a slow reaction rate is essential for detecting each base in order. However, mutations in the finger domain, which is responsible for dNTP incorporation, of the Phi29EL variants mentioned above may impact the accuracy of dNTP incorporation. Further experiments are required to measure the fidelity of the chimeras.
Discussion
Roles of (HhH)2 repeats in sequence‐nonspecific DNA binding
Conferring DNA polymerase with salt tolerance is essential for polymerase‐assisted nanopore sequencing. Previous reports have shown that the salt tolerance of Phi29 Pol was relatively higher than that of the Klenow fragment from E. coli. While 0.05 M NaCl eliminated the activity of the Klenow fragment, Phi29 Pol generated DNA of the same size in both the absence and presence of 0.1 M NaCl (Blanco et al., 1989). These data emphasize that salt addition suppresses the polymerization activity of wild‐type Phi29 Pol. Salt stress may affect the structural integrity of the polymerase, hindering DNA anchoring on the polymerase, and the replication initiation.
The observation that chimeric Phi29EL Pol showed enhanced salt tolerance was in agreement with a previous study of domain tagging Taq Pol or Pfu Pol chimeras (Pavlov et al., 2002). Our work also confirmed the function of the last eight (HhH)2 repeats in sequence‐nonspecific DNA binding. While high salt concentration disrupts the protein–DNA interaction and hampers polymerization, flexible (HhH)2 repeats may stabilize the binary complex. Phi29‐H or Phi29‐HI with one or two (HhH)2 have been found to improve DNA binding and replication performance. Salt tolerance assays showed that Phi29EL Pol was most tolerant, followed by Phi29H and Phi29 Pol (Fig. S9). This suggests that salt tolerance is related to the length of (HhH)2 motifs and proportional to the number of (HhH)2 repeats, in accordance with a previous finding where the highest KCl concentration at which activity was detected was 0.55, 0.10 or 0.05 M for Topo V, Topo‐78 or Topo‐61, with 12, 8 or 4 (HhH)2 motifs, respectively (Pavlov et al., 2002). Multiple (HhH)2 repeats are necessary for DNA binding under hyperthermal or hypersaline conditions.
Specific structural features of the (HhH)2 domain contribute to its function in DNA binding. (HhH)2 domains contain highly conserved sequences and two HhH motifs connected by a short α‐helix (Pavlov et al., 2002; Rajan et al., 2016). This five‐helix domain forms a stabilized hydrophobic core for supporting DNA, whereas the pseudo‐2‐unit domain forms a symmetrical structure, allowing stronger binding with symmetrical double helix DNA (Shao and Grishin, 2000; Pavlov et al., 2002; Rajan et al., 2016). The GhG (h, a hydrophobic amino acid residue) element in the hairpin region is responsible for DNA binding via a hydrogen bond with the sugar‐phosphate backbone, rather than the base (Shao and Grishin, 2000; Newman et al., 2005). The unique structure of (HhH)2 shows that its interaction pattern is non‐specific and extremely strong. In fact, some DNA polymerases naturally harbour HhH motifs for strong DNA binding, such as DNA polymerase β (Sawaya et al., 1997; Newman et al., 2005; de Vega et al., 2010).
What is the mechanism of DNA binding for (HhH)2 at high salt concentrations? Considering that a characteristic feature of halophilic or halotolerant proteins is a high proportion of acidic residues (Graziano and Merlino, 2014), we analysed the amino‐acid composition of the (HhH)2 motifs. The E‐J domains of Topo V have equal numbers of acidic and basic residues (83 aa). The salt tolerance of Phi29EL Pol could be explained by the conserved protein sequence and intrinsic strong DNA binding of the (HhH)2 domains, rather than by halophilic properties or halophilic engineering (Graziano and Merlino, 2014; Ortega et al., 2015; Warden et al., 2015). However, a well‐defined interaction between multiple (HhH)2 domains and long, double‐stranded DNA needs to be confirmed to clearly explain the molecular mechanism of protein–DNA contacts in this instance.
The construction of a chimeric Phi29 Pol has been described earlier to improve DNA binding capacity and amplification efficiency with either circular or genomic DNA as templates (de Vega et al., 2010). The introduction of the H or HI repeats of Topo V to Phi29 Pol, producing Phi29‐H and Phi29‐HI, respectively, produced a fourfold increase in RCR efficiency (de Vega et al., 2010). In this study, we found that Phi29EL Pol could also produce fourfold to fivefold greater amounts of single‐stranded DNA than Phi29 Pol in the absence of salt, while product length was similar, as shown in Figs 2, 4 and 5. We also noticed that the total amount of newly synthesized DNA was comparable between Phi29H Pol and Phi29EL Pol after incubation for 30 min, suggesting that the amplification efficiency might have peaked in the reaction mixture, given that enzyme concentrations were 20‐times higher than the template/primer complex. DNA gel‐retardation assays demonstrated increased DNA binding of Phi29‐H and Phi29‐HI (de Vega et al., 2010). Thus, (HhH)2 domains may help polymerases anchor with greater affinity to DNA templates, especially when the template concentration is low (de Vega et al., 2010). In this sense, the outstanding replication performance of Phi29EL Pol could be attributed to strong DNA binding of multiple (HhH)2 domains.
Although both Phi29 Pol and Phi29EL Pol showed 3′‐5′ exonuclease activity, the specific activity of Phi29EL Pol was approximately sixfold higher than that of Phi29 Pol (Fig. 6C and D). However, the increased exonuclease activity was in accordance with the enhanced replication efficiency (Figs 2, 4, 5, 6) after domain fusion. The fact reveals that (HhH)2 domain has a positive effect on both polymerization and exonucleolysis. A more efficient initial binding to DNA or an improved processivity could play a critical role in the increased enzymatic activity. On the other hand, lying at the exit of upstream dsDNA product (Fig. 1), multiple (HhH)2 repeats may indirectly help stabilize DNA at the active sites of polymerization and 3′‐5′ exonuclease domain. Single substitutions in the conserved motif ‘YxGG/A’, which located between the 3′‐5′ exonuclease and polymerization domains of Phi29 Pol, showed high or low pol/exo ratio phenotypes (Truniger et al., 1996). The altered balance between synthesis and degradation of Phi29 Pol mutant derivatives was proposed to be related with defects in template/primer DNA binding at 3′‐5′ exonuclease or polymerization active sites (Truniger et al., 1996). We can propose that HhH motifs play an indirect role in the interaction between polymerase and DNA as well.
Acquisition of new properties by point mutations
Point mutation method has been proven efficient for regulation of the polymerase properties, including activity, fidelity of replication and the incorporation of nucleotide analogs (Truniger et al., 1996; Gardner and Jack, 1999; Gardner and Jack, 2002; Loh and Loeb, 2005; Hanzel et al., 2017). Analysis of Phi29EL mutant derivatives indicated that the acidic amino acid residue (E375) was not beneficial for substrate incorporation in high salt, as well as basic residues H, or K, apolar residues W, Y or A. Only polar residues S or Q with abilities to improve further the salt tolerance of Phi29EL Pol (Fig. 7). Negatively charged Glu375 belonging to the finger domain locates at the entrance of the pocket for incoming dNTP (Berman et al., 2007). Mutation of Glu375 to His, Lys, Trp, Tyr, Ala, Ser or Gln enhanced the incorporation of tetraphosphate nucleotide analogues, together with positively charged residues Lys371, Lys379 and Lys383 (Hanzel et al., 2017). However, halophilic proteins are characterized to have fewer basic or apolar residues (Fukuchi et al., 2003; Ortega et al., 2015). Thus, polar and neutral residues Ser or Gln appears to be preferred than the negatively charged Glu in high salt. Point mutation results provide indication of how to select residues with high incorporation efficiency at high salt concentrations.
Engineering of salt tolerant DNA polymerases
In this study, we generated a chimeric Phi29EL Pol with improved salt tolerance. In terms of salt resistance, DNA polymerases can be divided into two classes: salt‐tolerant and salt‐sensitive. Engineering non‐halophilic polymerases to become halophilic or salt tolerant is an effective, convenient and promising way to satisfy the requirements of nanopore sequencing technology. Unfortunately, the amplification efficiency of Phi29EL decreases with increasing salt concentration, with an optimum at 0 M KCl (Figs 4 and 5). To address this deficiency, new schemes for polymerase modifications, such as halophilic engineering of industrial enzymes via rational design of proteins with increasingly acidic residues, need to be developed (Warden et al., 2015; Wang et al., 2018; Zheng et al., 2019). Another potential future strategy is to introduce novel potential polymerases working optimally and naturally in high salt with the aid of genomic or metagenomic analysis of halophilic organisms.
Polymerase‐assisted nanopore sequencing at single‐nucleotide resolution should be more accurate than DNA passing through a pore constriction site directly because of the high fidelity of polymerase (Manrao et al., 2012; Heger, 2014; Fuller et al., 2016). An improved application of halophilic or halotolerant polymerases in sequencing will remove the ionic barriers for polymerases in this context. A higher ionic concentration would also help to provide a higher signal‐to‐noise ratio, allowing the use of low‐cost and easily synthesized substrates, such as PEG‐labelled nucleotides, which require high salt concentrations for analysis (Wang et al., 2014; Bruce et al., 2016; Fuller et al., 2016).
Experimental procedures
Plasmid preparation and protein expression
The coding sequences of Phi29 Pol and Topo V were synthesized and cloned into the pGEX‐6p‐1 and pET28‐a vectors by Generay (China). Sequence information was obtained from the NCBI database. DNA fragments encoding the H domain (residues 696–751) and E‐L domains (residues 518–964) of Topo V after PCR were tethered to the COOH terminus of Phi29 Pol using KasI (R0544S; NEB, USA). The sequence of the linker peptide was GTGSGA. The scheme for generating the chimeric gene was that described by de Vega et al. (2010), except that EcoRI and NotI were used for ligation with the vector. Mutants Phi29EL‐E375A, Phi29EL‐E375K, Phi29EL‐E375H, Phi29EL‐E375S, Phi29EL‐E375Q, Phi29EL‐E375W and Phi29EL‐E375Y were generated with the Fast mutagenesis system (FM111‐01; Transgene, Beijin, China). Primers used for mutagenesis are listed in Table S1.
The expression vectors pGEX6p‐1‐Phi29, pET28a‐Phi29H, pET28a‐Phi29EL and pET28a‐Phi29EL variants were transformed into chemically competent E. coli cells Transette DE3 (CD801‐02; Transgene, China) or BL21 (CD901‐02; Transgene, China) according to the manufacturer’s instructions. Overnight Express Autoinduction System 1 (71300‐4; Millipore, Billerica, MA, USA) was used for pGEX6p‐1‐Phi29 expression at 30°C for 16 h. Alternatively, expression was induced with IPTG at 16°C for 16 h. Then, GST‐tagged Phi29 Pol was purified with Glutathione Sepharose 4 B (17‐0756‐01; GE Healthcare, Uppsala, Sweden), and the GST tag cleaved using PreScission protease (27‐0843‐01; GE Healthcare, USA) overnight at 4°C. Histidine‐tagged recombinant Phi29H, Phi29EL Pol and Phi29EL mutants were purified using Ni Sepharose resin (17‐5318‐01; GE Healthcare, USA) following the manufacturer’s instructions. After purification, Phi29 Pol was exchanged with PBS buffer using an Amicon® Ultra‐15 Centrifugal Filter Unit (UFC905024; Millipore, USA). Other proteins were concentrated in the same way, but stored in 20 mM Tris‐HCl buffer (pH 8.0, 25°C) with 0.5 M. Purity and concentration of the purified proteins were analysed using a Qubit™ protein assay kit (Q33211; Thermo Fisher Scientific, Eugene, OR, USA) and 12% SDS‐PAGE. The Blue Plus® II Protein Marker (DM111‐01; Transgene, China) was used as a protein marker.
Hairpin extension
A 53 nt long hairpin (Generay, Shanghai, China) was designed as a template to generate product DNA (74 nt) for the extension experiment. The hairpin sequences are listed in Table S1. The reaction buffer, Phi29 buffer (or Buffer 2), was composed of 50 mM Tris‐HCl (pH 7.5), 10 mM MgCl2, 10 mM (NH4)2SO4, and 4 mM DTT. Other components included 200 μM dNTPs, 0.5 μM hairpin and 200 nM of the corresponding enzymes. To optimize pH value and other conditions, another three buffers with different pH values were tested as previously described (Gao et al., 2020). Buffer 1 consisted of 20 mM phosphate buffer, 10 mM MgCl2, 10 mM (NH4)2SO4 and 4 mM DTT, pH 5.8; Buffer 3 consisted of 10 mM Tris‐HCl, 50 mM KCl and 1.5 mM MgCl2, pH 8.3; and Buffer 4 consisted of 20 mM Tris‐HCl, 10 mM (NH4)2SO4, 0.1% Triton X‐100, 50 mM KCl and 2 mM MgSO4, pH 8.8. (Takahashi et al., 2018).
Reactions were incubated at 30°C for 10 min in a Mastercycler® nexus PCR thermocycler (Eppendorf, Germany). To stop the reactions, 0.5 M EDTA (R1021; Thermo Fisher Scientific, USA) was added. A total of 5 μl of each sample was loaded for 20% TBE‐urea gel electrophoresis. After staining with SYBR Gold (S11494; Thermo Fisher Scientific, USA), images were taken and analysed using the Azure Biosystem c300 (Azure Biosystems, Dublin, CA, USA). Band intensities of the complete extension products were used to evaluate polymerization activity. The relative product concentration was calculated using the total amount of template as a control.
Polymerase resistance to NaCl, KCl, potassium acetate (KAc) or potassium gluconate (KGlc) was tested separately. Concentrations of 0, 0.1, 0.2, 0.3, 0.4, 0.5 or 1 M of each salt were separately added into the extension mixture without changing the other components. For the thermal stability assay, proteins were incubated at 30, 37, 45, 55, 65, 75 and 85°C for 20 min prior to the hairpin extension experiments. The reaction temperature was 30°C. The reaction products were treated and analysed as described above. Each assay was repeated at least twice independently.
Rolling circle replication assay
Rolling circle replication (RCR) was performed using singly primed M13mp18 as a template. The M13mp18 template (N4040S; NEB, Ipswich, MA, USA) was hybridized with a 10‐fold higher amount of primers by heating at 95°C for 3 min, followed by holding at room temperature for 30 min (Kong et al., 1993). After annealing, 450 ng of M13mp18 was added to the 20‐μl reaction, which consisted of the same buffer for the hairpin extension and the indicated amounts of enzymes. In general, 200 nM enzyme was used for each assay. The reactions were incubated at 30°C for 30 min and then stopped by adding 0.5 M EDTA.
Each sample (5 μl) was then analysed by 0.6% alkaline agarose gel electrophoresis, performed according to a simplified protocol (Sambrook and Russell, 2002; Gao et al., 2020). After dissolving SeaKem GTG Agarose (50071; Lonza, Rockland, ME, USA) in ultrapure water by heating, 10× alkaline buffer was added after the solution was cooled to room temperature for 10–15 min. Samples were prepared in 10× alkaline buffer (NA0070; Leagene, Beijing, China) and 6× alkaline loading buffer (SL2212‐5 ml; Coolaber, Beijing, China). The GeneRuler High Range DNA Ladder (SM1351; Thermo Fisher Scientific, USA) including DNA from 10.1 kb–48.5 kb was treated in a similar manner to the samples. A flow rate peristaltic pump (BT100‐1L; Longerpump, Baoding, China) was set up at 12.15 ml min−1 to provide an appropriate flow rate of buffer through the chamber following the manufacturer’s standard protocols. After 2–3 h of electrophoresis performed at 3 V cm−1, the gel was stained with SYBR Gold in 1× TAE buffer for 40 min. The concentrations and molecular weights of the synthesized DNA were assessed according to the band migration using AzureSpot. For the processivity assay, the molar ratio of DNA polymerases to M13 was set to 20, 10, 5 or 2.5 (Blanco et al., 1989; de Vega et al., 2010). The dilution of enzymes would cause competition between different substrates if those with weak processivity disassociated from the DNA‒enzyme complex, leading to distributive DNA products of variable sizes. After extension for 30 min, the length and distribution of the products were detected by alkaline agarose gel electrophoresis and analysed using AzureSpot.
To analyse salt inhibition of RCR, 0–0.3 M of NaCl, KCl, KAc or KGlc were added into the RCR mix without changing other components or reaction conditions (Hardies et al., 2003; Wang et al., 2004; Oscorbin et al., 2017). The samples were subjected to the same procedure for detection. The decreased amounts of single‐stranded DNA generated by RCR represented salt inhibition.
To measure the replication rate of the Phi29 variants, the RCR reactions were stopped after extension for 10 min when the DNA fragment migrated to the resolving part of the gels (Blanco et al., 1989; de Vega et al., 2010). The concentrations and molecular weights of the synthesized DNA were assessed using the Azure system.
Exonuclease activity assay
Phosphorothioate (Thiol) modifications at the 3′ or 5′ end of oligonucleotides, which protect oligos from degradation of the 3′‐5′ or 5′‐3′ exonuclease, were used to analyse the exonuclease direction and activity in this methodology (Guga and Koziołkiewicz, 2011; Keith et al., 2013). Phi29 Pol and Phi29EL Pol were incubated with 1 μM natural or thiol‐modified oligonucleotides (Generay, China), respectively, in Phi29 buffer in a final volume of 10 μl (Gao et al., 2020). Briefly, natural oligos or oligos modified at the 5′ end were cleaved from the 3′ end by 3′‐5′ exonuclease activity of polymerase, whereas 3′ modified oligos were protected from exonuclease digestion by phosphorothioates at the 3′ end. After incubation for 10 min, the products were analysed using a 20% TBE‐urea polyacrylamide gel. The specific type and remaining concentration of the original oligos after exonuclease digestion were representative of the biochemical properties of the exonuclease domain.
Exonuclease activity was quantified using a 3′ to 5′ exonuclease activity assay kit (KP175; Biovision, Milpitas, CA, USA) using a Varioskan® Flash Multimode Reader (Thermo Fisher Scientific), according to the manufacturer’s instructions. To ensure that the kinetic curve fell within the range of the standard curve, 20 pmol of Phi29 Pol and 4 pmol of Phi29EL Pol were added to each well of the plate for each assay. Specific activities were calculated from three independent replicates.
Protein structure analysis
Structural data for Phi29 Pol (Berman et al., 2007) and Topo‐97 (Rajan et al., 2016) were downloaded from the Protein Data Bank (Berman et al., 2000). A structural model of Topo V‐EL (domains E‐L) was built from residues 518‐964 using SWISS‐MODEL (Waterhouse et al., 2018). The superimposition of the Topo‐97 crystal structure (PDB code: 5HM5) and Topo V‐EL model was carried out using UCSF Chimera v1.12 (Pettersen et al., 2004). The Phi29EL model was generated with Phi29 Pol (PDB code: 2PYL) and Topo V‐EL model by Swiss‐PdbViewer v4.1 software (Guex and Peitsch, 1997), as previously described for the modelling of Phi29 Pol or Taq Pol with the (HhH)2 domains (Pavlov et al., 2002; de Vega et al., 2010). The ProtParam tool was used to analyse physical and chemical parameters (http://web.expasy.org/protparam/).
Conflict of interests
The authors declare that they have no competing interests.
Author contributions
Y.P.G. and Y.H. designed the study. Y.P.G. carried out protein expression, salt tolerance assay and protein structure analysis. L.Y.C. performed site‐directed mutagenesis and protein expression analyses. Y.H. analysed the data. X.L. set up the instruments and prepared the resources. Y.P.G. wrote the manuscript, and all the authors commented on the manuscript. Y.H. and I.I. managed the project. H.T. and X.R.Y. supervised the project. H.T. and Y.P.G. conceived the study.
Supporting information
Fig. S1. The Topo V‐EJ model (cyan) containing E, F, G, H, I, and J domains superimposed onto the crystal structure of Topo‐97 (PDB ID: 5HM5, wheat). Part of the F domain and the entire G domain in Topo‐97 could not be modelled. The superposition was accomplished using UCSF Chimera software.
Fig. S2. Expression and purification of Phi29EL Pol. (A) Increasing concentrations of imidazole (0.02–0.5 M) were used to elute target protein with 6×His tag (Lanes 1–7). The molecular weight of Phi29EL Pol was 116.6 kDa. (B) Purified proteins were separately exchanged into two different storage buffers with 0.5 M (Lane 1) and 0.1 M NaCl (Lane 2). None of the target protein could be detected in Lane 2 after keeping the protein at low NaCl, while the other bands could be degradation products, as inferred by the gain in proportion of these bands when the storage buffer is 0.1 NaCl vs 0.5 M NaCl. Only Phi29EL Pol stored in buffers with 0.5 M NaCl was stable. Protein standards are labelled at the left in A and B.
Fig. S3. Expression of Phi29 Pol and Phi29EL Pol mutants. Lane 1, BSA; lane 2, Phi29 Pol (MW, 67.0 kD); lane 3, Phi29H Pol (72.9 kD); lane 4, Phi29EL Pol (116.6 kD); lanes 5–11, protein sample of Phi29EL‐E375A, Phi29EL‐E375K, Phi29EL‐E375H, Phi29EL‐E375S, Phi29EL‐E375Q, Phi29EL‐E375W and Phi29EL‐E375Y, respectively.
Fig. S4. Buffer optimization for Phi29 Pol and Phi29EL Pol. The hairpin extension experiment with Phi29 Pol and Phi29EL Pol was performed in four buffers with different pH values in the absence (left lanes) or presence (middle lanes) of NaCl. No enzymes were added to the control reactions in the rightmost lanes. All reactions were carried out at 30°C for 10 min. Both enzymes preferred basic or neutral buffers rather than acidic ones. Data are representative of several independent experiments.
Fig. S5. Thermal stability of Phi29 Pol and Phi29EL Pol. Proteins (10 µl) were held at 30, 37, 45, 55, 65, 75 and 85°C for 10 min, followed by hairpin extension experiments, as described before. Data are representative of several independent experiments.
Fig. S6. Salt tolerance of Phi29 Pol. RCR was carried out for two additional hours at multiple KCl concentrations, and stopped at defined times. After 20, 30, 60 and 120 min of incubation, each sample was loaded in a 0.6% alkaline agarose gel for analysis. Arrowheads on the left indicate DNA length markers.
Fig. S7. Salt tolerance of Phi29 Pol derivatives. Different concentrations of KCl were added into singly primed M13 extension reactions. After 30 min, products of Phi29‐E375A Pol and Phi29‐E375Y Pol were separated using a 0.6% alkaline agarose gel. Lane 1 and lane 5, reaction mixtures without salt; Lanes 2–4, with 0.1 M, 0.2 M or 0.3 M KCl; Lanes 6–8, with 0.1 M, 0.2 M or 0.3 M KCl.
Fig. S8. Extension rates of Phi29 Pol variants. After RCR with M13 templates for 10 min, replication was stopped by 0.5 M EDTA to measure the extension rate. Replication products (10 µl) were analysed on alkaline gels. The lengths of the extension products of Phi29 Pol were slightly larger than those of Phi29EL Pol variants. Arrowheads at the left indicate extension products (upper) and M13 templates (lower).
Fig. S9. Salt tolerance of Phi29H Pol in rolling circle replication. (A) Schematic of the Phi29H Pol. H domain (residues 696‐751) from Topo‐V fused to Phi29 Pol in the same manner as to Phi29 EL Pol. (B) Gel analysis of the products from rolling circle replication. Lane 1, without enzyme; Lane 2, without salt; Lanes 3–5, with 0.1, 0.2 and 0.3 M NaCl; Lanes 6‐8 with 0.1, 0.2 and 0.3 M KCl. Arrowheads at the left indicate DNA length markers. (C) Quantification of newly synthesized DNA larger than 48 kb in B.
Table S1. Oligos used in this study.
Acknowledgments
The authors thank Dr. Miaoqing Hu (Tsinghua University) for her assistance with the structural analysis of proteins. The authors also thank Dr. Suhua Deng (Axbio) and Dr. Vladimir Bashkirov (Axbio) for their experience with DNA polymerases. This work was supported by the Science, Technology, and Innovation Commission of Shenzhen Municipality (grant number 20160529234851639), and the Guangdong Basic and Applied Basic Research Foundation (grant number 2020A1515011582). Funding for open access charge was provided by the Guangdong Basic and Applied Basic Research Foundation.
Microb. Biotechnol. (2021) 14(4), 1642–1656
Funding Information
Grant no. 20160529234851639, Science, Technology, and Innovation Commission of Shenzhen Municipality. Grant no. 2020A1515011582, Guangdong Basic and Applied Basic Research Foundation.
Contributor Information
Yaping Gao, Email: gaoyp@tsinghua-sz.org.
Hui Tian, Email: tianhui@tsinghua-sz.org.
References
- Baba, M. , Kakue, R. , Leucht, C. , Rasor, P. , Walch, H. , Ladiges, D. , et al. (2017) Further increase in thermostability of Moloney murine leukemia virus reverse transcriptase by mutational combination. Protein Eng Des Sel 30: 551–557. [DOI] [PubMed] [Google Scholar]
- Belova, G.I. , Prasad, R. , Kozyavkin, S.A. , Lake, J.A. , Wilson, S.H. , and Slesarev, A.I. (2001) A type IB topoisomerase with DNA repair activities. Proc Natl Acad Sci USA 98: 6015–6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belova, G.I. , Prasad, R. , Nazimov, I.V. , Wilson, S.H. , and Slesarev, A.I. (2002) The domain organization and properties of individual domains of DNA topoisomerase V, a type 1B topoisomerase with DNA repair activities. J Biol Chem 277: 4959–4965. [DOI] [PubMed] [Google Scholar]
- Berman, A.J. , Kamtekar, S. , Goodman, J.L. , Lázaro, J.M. , de Vega, M. , Blanco, L. , et al. (2007) Structures of phi29 DNA polymerase complexed with substrate: the mechanism of translocation in B‐family polymerases. EMBO J 26: 3494–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H.M. , Westbrook, J. , Feng, Z. , Gilliland, G. , Bhat, T.N. , Weissig, H. , et al. (2000) The protein data bank. Nucleic Acids Res 28: 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad, A. , Zaballos, A. , Salas, M. , and Blanco, L. (1987) Structural and functional relationships between prokaryotic and eukaryotic DNA polymerases. EMBO J 6: 4219–4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernick, D. , Holmes, A. , and Nivala, J. (2012) Patent WO 2012/123905 A1; PCT, US/041802.
- Bessman, M.J. , Lehman, I.R. , Simms, E.S. , and Kornberg, A. (1958) Enzymatic synthesis of deoxyribonucleic acid. II. General properties of the reaction. J Biol Chem 233: 171–177. [PubMed] [Google Scholar]
- Blanco, L. , Bernad, A. , Lázaro, J.M. , Martín, G. , Garmendia, C. , and Salas, M. (1989) Highly efficient DNA synthesis by the phage 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem 264: 8935–8940. [PubMed] [Google Scholar]
- Blanco, L. , and Salas, M. (1984) Characterization and purification of a phage phi 29‐encoded DNA polymerase required for the initiation of replication. Proc Natl Acad Sci USA 81: 5325–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco, L. , and Salas, M. (1996) Relating structure to function in phi29 DNA polymerase. J Biol Chem 271: 8509–8512. [DOI] [PubMed] [Google Scholar]
- Bruce, M. , Heron, A.J. , Moysey, R. , Soeroes, S. , Wallace, E.J. , and White, J. (2016) Patent US20160257942 A1 PCT/GB2014/052736.
- Cheng, Q. , Zhai, B. , Chow, J. , Li, X.Z. , and Liu, G.X. (2016) Patent A1; PCT/CN2014/000285 US 2016/0369249.
- Clarke, J. , Wu, H.C. , Jayasinghe, L. , Patel, A. , Reid, S. , and Bayley, H. (2009) Continuous base identification for single‐molecule nanopore DNA sequencing. Nat Nanotechnol 4: 265–270. [DOI] [PubMed] [Google Scholar]
- Cline, J. , Braman, J.C. , and Hogrefe, H.H. (1996) PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res 24: 3546–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, F.B. , Nelson, J.R. , Giesler, T.L. , and Lasken, R.S. (2001) Rapid amplification of plasmid and phage DNA using Phi29 DNA polymerase and multiply‐primed rolling circle amplification. Genome Res 11: 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac, R. , Sparks, A.B. , Callow, M.J. , Halpern, A.L. , Burns, N.L. , Kermani, B.G. , et al. (2010) Human genome sequencing using unchained base reads on self‐assembling DNA nanoarrays. Science 327: 78–81. [DOI] [PubMed] [Google Scholar]
- Fukuchi, S. , Yoshimune, K. , Wakayama, M. , Moriguchi, M. , and Nishikawa, K. (2003) Unique amino acid composition of proteins in halophilic bacteria. J Mol Biol 32: 7347–7357. [DOI] [PubMed] [Google Scholar]
- Fuller, C.W. , Kumar, S. , Porel, M. , Chien, M. , Bibillo, A. , Stranges, P.B. , et al. (2016) Real‐time single‐molecule electronic DNA sequencing by synthesis using polymer‐tagged nucleotides on a nanopore array. Proc Natl Acad Sci USA 113: 5233–5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y.P. , He, Y. , Ivanov, I. , Yang, X.R. , Tian, H. , and Liu, X. (2020) Expression and functional study of VpV262 Pol, a moderately halophilic DNA polymerase from the Vibrio parahaemolyticus phage VpV262. Enzyme Microb Technol 139: 109588. [DOI] [PubMed] [Google Scholar]
- Gardner, A.F. , and Jack, W.E. (1999) Determinants of nucleotide sugar recognition in an archaeon DNA polymerase. Nucleic Acids Res 27: 2545–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, A.F. , and Jack, W.E. (2002) Acyclic and dideoxy terminator preferences denote divergent sugar recognition by archaeon and Taq DNA polymerases. Nucleic Acids Res 30: 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano, G. , and Merlino, A. (2014) Molecular bases of protein halotolerance. Biochim Biophys Acta 1844: 850–858. [DOI] [PubMed] [Google Scholar]
- Guex, N. , and Peitsch, M.C. (1997) SWISS‐MODEL and the Swiss‐PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- Guga, P. , and Koziołkiewicz, M. (2011) Phosphorothioate nucleotides and oligonucleotides‐ recent progress in synthesis and application. Chem Biodiv 8: 1642–1681. [DOI] [PubMed] [Google Scholar]
- Hanzel, D.K. , Otto, G. , Peluso, P. , Pham, T. , Rank, D.R. , Mitsis, P. , et al. (2017) Patent US 009556479 B2 PCT 14/538,653.
- Hardies, S.C. , Comeau, A.M. , Serwer, P. , and Suttle, C.A. (2003) The complete sequence of marine bacteriophage VpV262 infecting Vibrio parahaemolyticus indicates that an ancestral component of a T7 viral supergroup is widespread in the marine environment. Virology 310: 359–371. [DOI] [PubMed] [Google Scholar]
- Heger, M. (2014). URL http://www.genomeweb.com/sequencing/agbt‐first‐data‐oxford‐nanopore‐presented‐company‐issues‐early‐access‐invites.
- Johansson, E. , and Dixon, N. (2013) Replicative DNA polymerases. Cold Spring Harb Perspect Biol 5: a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamtekar, S. , Berman, A.J. , Wang, J. , Lázaro, J.M. , de Vega, M. , Blanco, L. , et al. (2004) Insights into strand displacement and processivity from the crystal structure of the protein‐primed DNA polymerase of bacteriophage phi29. Mol Cell 16: 609–618. [DOI] [PubMed] [Google Scholar]
- Keith, B.J. , Jozwiakowski, S.K. , and Connolly, B.A. (2013) A plasmid‐based lacZα gene assay for DNA polymerase fidelity measurement. Anal Biochem 433: 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, H. , Kucera, R.B. , and Jack, W.E. (1993) Characterization of a DNA polymerase from the hyperthermophile archaea Thermococcus litoralis. Vent DNA polymerase, steady state kinetics, thermal stability, processivity, strand displacement, and exonuclease activities. J Biol Chem 268: 1965–1975. [PubMed] [Google Scholar]
- Korlach, J. , Bjornson, K.P. , Chaudhuri, B.P. , Cicero, R.L. , Flusberg, B.A. , Gray, J.J. , et al. (2010) Real‐time DNA sequencing from single polymerase molecules. Methods Enzymol 472: 431–455. [DOI] [PubMed] [Google Scholar]
- Lehman, I.R. , Bessman, M.J. , Simms, E.S. , and Kornberg, A. (1958) Enzymatic synthesis of deoxyribonucleic acid. I. Preparation of substrates and partial purification of an enzyme from Escherichia coli . J Biol Chem 233: 163–170. [PubMed] [Google Scholar]
- Loh, E. , and Loeb, L.A. (2005) Mutability of DNA polymerase I: implications for the creation of mutant DNA polymerases. DNA Repair (Amst) 4: 1390–1398. [DOI] [PubMed] [Google Scholar]
- Manrao, E.A. , Derrington, I.M. , Laszlo, A.H. , Langford, K.W. , Hopper, M.K. , Gillgren, N. , et al. (2012) Reading DNA at single‐nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol 30: 349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, M. , Murray‐Rust, J. , Lally, J. , Rudolf, J. , Fadden, A. , Knowles, P.P. , et al. (2005) Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition. EMBO J 24: 895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega, G. , Diercks, T. , and Millet, O. (2015) Halophilic protein adaptation results from synergistic residue‐ion interactions in the folded and unfolded states. Chem Biol 22: 1597–1607. [DOI] [PubMed] [Google Scholar]
- Oscorbin, I.P. , Belousova, E.A. , Boyarskikh, U.A. , Zakabunin, A.I. , Khrapov, E.A. , and Filipenko, M.L. (2017) Derivatives of Bst‐like Gss‐polymerase with improved processivity and inhibitor tolerance. Nucleic Acids Res 45: 9595–9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov, A.R. , Belova, G.I. , Kozyavkin, S.A. , and Slesarev, A.I. (2002) Helix–hairpin–helix motifs confer salt resistance and processivity on chimeric DNA polymerases. Proc Natl Acad Sci USA 99: 13510–13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E.F. , Goddard, T.D. , Huang, C.C. , Couch, G.S. , Greenblatt, D.M. , Meng, E.C. , and Ferrin, T.E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Rajan, R. , Osterman, A. , and Mondragón, A. (2016) Methanopyrus kandleri topoisomerase contains three distinct AP lyase active sites in addition to the topoisomerase active site. Nucleic Acids Res 44: 3464–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez, I. , Lázaro, J.M. , Blanco, L. , Kamtekar, S. , Berman, A.J. , Wang, J. , et al. (2005) A specific subdomain in phi29 DNA polymerase confers both processivity and strand‐displacement capacity. Proc Natl Acad Sci USA 102: 6407–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell, P.J. , and Waksman, G. (2005) Structure and mechanism of DNA polymerases. Adv Protein Chem 71: 401–440. [DOI] [PubMed] [Google Scholar]
- Saiki, R.K. , Gelfand, D.H. , Stoffel, S. , Scharf, S.J. , Higuchi, R. , Horn, G.T. , et al. (1988) Primer‐directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239: 487–491. [DOI] [PubMed] [Google Scholar]
- Sambrook, J. , and Russell, D.W. (2002) Molecular Cloning, a Laboratory Manual (3rd edn). Cold Spring Harbor, NY: Cold Spring Harbor Press. [Google Scholar]
- Sawaya, M.R. , Prasad, R. , Wilson, S.H. , Kraut, J. , and Pelletier, H. (1997) Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry 36: 11205–11215. [DOI] [PubMed] [Google Scholar]
- Shao, X. , and Grishin, N.V. (2000) Common fold in helix–hairpin–helix proteins. Nucleic Acids Res 28: 2643–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slesarev, A.I. , Lake, J.A. , Stetter, K.O. , Gellert, M. , and Kozyavkin, S.A. (1994) Purification and characterization of DNA topoisomerase V. An enzyme from the hyperthermophilic prokaryote Methanopyrus kandleri that resembles eukaryotic topoisomerase I. J Biol Chem 269: 3295–3303. [PubMed] [Google Scholar]
- Slesarev, A.I. , Stetter, K.O. , Lake, J.A. , Gellert, M. , Krah, R. , and Kozyavkin, S.A. (1993) DNA topoisomerase V is a relative of eukaryotic topoisomerase I from a hyperthermophilic prokaryote. Nature 364: 735–737. [DOI] [PubMed] [Google Scholar]
- Takahashi, M. , Takahashi, E. , Joudeh, L.I. , Marini, M. , Das, G. , Elshenawy, M.M. , et al. (2018) Dynamic structure mediates halophilic adaptation of a DNA polymerase from the deep‐sea brines of the Red Sea. FASEB J 32: 3346–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truniger, V. , Lázaro, J.M. , Salas, M. , and Blanco, L. (1996) A DNA binding motif coordinating synthesis and degradation in proofreading DNA polymerases. EMBO J 15: 3430–3441. [PMC free article] [PubMed] [Google Scholar]
- de Vega, M. , Lázaro, J.M. , Mencía, M. , Blanco, L. , and Salas, M. (2010) Improvement of φ29 DNA polymerase amplification performance by fusion of DNA binding motifs. Proc Natl Acad Sci USA 107: 16506–16511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Li, S. , Zhao, H. , Bian, L. , Chen, L. , Zhang, Z. , et al. (2015) Expression and characterization of the RKOD DNA polymerase in Pichia pastoris . PLoS One 10: e0131757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.Z. , Zheng, C. , Cao, Y. , Tan, Z.T. , Liu, D. , Cheng, Z.P. , et al. (2018) Rational design of an efficient halotolerant enzymatic system for in vitro one‐pot synthesis of cytidine diphosphate choline. Biotechnol J 13: e1700577. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Prosen, D.E. , Mei, L. , Sullivan, J.C. , Finney, M. , and Vander Horn, P.B. (2004) A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Res 32: 1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Yang, Q. , and Wang, Z. (2014) The evolution of nanopore sequencing. Front Genet 5: 449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden, A.C. , Williams, M. , Peat, T.S. , Seabrook, S.A. , Newman, J. , Dojchinov, G. , and Haritos, V.S. (2015) Rational engineering of a mesohalophilic carbonic anhydrase to an extreme halotolerant biocatalyst. Nat Commun 6: 10278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, A. , Bertoni, M. , Bienert, S. , Studer, G. , Tauriello, G. , Gumienny, R. , et al. (2018) SWISS‐MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46: W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami, T. , Ishino, S. , Kawarabayasi, Y. , and Ishino, Y. (2014) Mutant Taq DNA polymerases with improved elongation ability as a useful reagent for genetic engineering. Front Microbiol 5: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, C. , Li, Z.J. , Yang, H.F. , Zhang, T.Y. , Niu, H.Q. , Liu, D. , et al. (2019) Computation‐aided rational design of a halophilic choline kinase for cytidine diphosphate choline production in high‐salt condition. J Biotechnol 290: 59–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The Topo V‐EJ model (cyan) containing E, F, G, H, I, and J domains superimposed onto the crystal structure of Topo‐97 (PDB ID: 5HM5, wheat). Part of the F domain and the entire G domain in Topo‐97 could not be modelled. The superposition was accomplished using UCSF Chimera software.
Fig. S2. Expression and purification of Phi29EL Pol. (A) Increasing concentrations of imidazole (0.02–0.5 M) were used to elute target protein with 6×His tag (Lanes 1–7). The molecular weight of Phi29EL Pol was 116.6 kDa. (B) Purified proteins were separately exchanged into two different storage buffers with 0.5 M (Lane 1) and 0.1 M NaCl (Lane 2). None of the target protein could be detected in Lane 2 after keeping the protein at low NaCl, while the other bands could be degradation products, as inferred by the gain in proportion of these bands when the storage buffer is 0.1 NaCl vs 0.5 M NaCl. Only Phi29EL Pol stored in buffers with 0.5 M NaCl was stable. Protein standards are labelled at the left in A and B.
Fig. S3. Expression of Phi29 Pol and Phi29EL Pol mutants. Lane 1, BSA; lane 2, Phi29 Pol (MW, 67.0 kD); lane 3, Phi29H Pol (72.9 kD); lane 4, Phi29EL Pol (116.6 kD); lanes 5–11, protein sample of Phi29EL‐E375A, Phi29EL‐E375K, Phi29EL‐E375H, Phi29EL‐E375S, Phi29EL‐E375Q, Phi29EL‐E375W and Phi29EL‐E375Y, respectively.
Fig. S4. Buffer optimization for Phi29 Pol and Phi29EL Pol. The hairpin extension experiment with Phi29 Pol and Phi29EL Pol was performed in four buffers with different pH values in the absence (left lanes) or presence (middle lanes) of NaCl. No enzymes were added to the control reactions in the rightmost lanes. All reactions were carried out at 30°C for 10 min. Both enzymes preferred basic or neutral buffers rather than acidic ones. Data are representative of several independent experiments.
Fig. S5. Thermal stability of Phi29 Pol and Phi29EL Pol. Proteins (10 µl) were held at 30, 37, 45, 55, 65, 75 and 85°C for 10 min, followed by hairpin extension experiments, as described before. Data are representative of several independent experiments.
Fig. S6. Salt tolerance of Phi29 Pol. RCR was carried out for two additional hours at multiple KCl concentrations, and stopped at defined times. After 20, 30, 60 and 120 min of incubation, each sample was loaded in a 0.6% alkaline agarose gel for analysis. Arrowheads on the left indicate DNA length markers.
Fig. S7. Salt tolerance of Phi29 Pol derivatives. Different concentrations of KCl were added into singly primed M13 extension reactions. After 30 min, products of Phi29‐E375A Pol and Phi29‐E375Y Pol were separated using a 0.6% alkaline agarose gel. Lane 1 and lane 5, reaction mixtures without salt; Lanes 2–4, with 0.1 M, 0.2 M or 0.3 M KCl; Lanes 6–8, with 0.1 M, 0.2 M or 0.3 M KCl.
Fig. S8. Extension rates of Phi29 Pol variants. After RCR with M13 templates for 10 min, replication was stopped by 0.5 M EDTA to measure the extension rate. Replication products (10 µl) were analysed on alkaline gels. The lengths of the extension products of Phi29 Pol were slightly larger than those of Phi29EL Pol variants. Arrowheads at the left indicate extension products (upper) and M13 templates (lower).
Fig. S9. Salt tolerance of Phi29H Pol in rolling circle replication. (A) Schematic of the Phi29H Pol. H domain (residues 696‐751) from Topo‐V fused to Phi29 Pol in the same manner as to Phi29 EL Pol. (B) Gel analysis of the products from rolling circle replication. Lane 1, without enzyme; Lane 2, without salt; Lanes 3–5, with 0.1, 0.2 and 0.3 M NaCl; Lanes 6‐8 with 0.1, 0.2 and 0.3 M KCl. Arrowheads at the left indicate DNA length markers. (C) Quantification of newly synthesized DNA larger than 48 kb in B.
Table S1. Oligos used in this study.


