We demonstrate that medium‐chain fatty acids mimic the quorum‐sensing molecule farnesol. Six sets of evidence, that is, (i) structural similarities, (ii) inhibition of hyphal growth, (iii) inhibition of biofilm formation, (iv) inhibition of farnesol production, (v) inhibition of sterol production, and (vi) effects on gene expressions, which showed medium‐chain fatty acids interfere with farnesol signaling and that their antibiofilm and antivirulence activities are superior to that of farnesol. Our findings show that fatty acids found in prokaryotes and eukaryotes might interfere with fungal communication and that medium‐chain fatty acids might be used as developmental starting points for the design of potent antibiofilm and antifungal agents against fungal Candida species.
Summary
Candida biofilms are tolerant to conventional antifungal therapeutics and the host immune system. The transition of yeast cells to hyphae is considered a key step in C. albicans biofilm development, and this transition is inhibited by the quorum‐sensing molecule farnesol. We hypothesized that fatty acids mimicking farnesol might influence hyphal and biofilm formation by C. albicans. Among 31 saturated and unsaturated fatty acids, six medium‐chain saturated fatty acids, that is, heptanoic acid, octanoic acid, nonanoic acid, decanoic acid, undecanoic acid and lauric acid, effectively inhibited C. albicans biofilm formation by more than 75% at 2 µg ml−1 with MICs in the range 100–200 µg ml−1. These six fatty acids at 2 µg ml−1 and farnesol at 100 µg ml−1 inhibited hyphal growth and cell aggregation. The addition of fatty acids to C. albicans cultures decreased the productions of farnesol and sterols. Furthermore, down‐regulation of several hyphal and biofilm‐related genes caused by heptanoic or nonanoic acid closely resembled the changes caused by farnesol. In addition, nonanoic acid, the most effective compound diminished C. albicans virulence in a Caenorhabditis elegans model. Our results suggest that medium‐chain fatty acids inhibit more effectively hyphal growth and biofilm formation than farnesol.
Introduction
Biofilms are communities of bacteria, fungi or yeasts that can form on diverse biotic or abiotic surfaces including those of inert materials, synthetic polymers and indwelling medical devices. Biofilms protect microbial communities from nutrient limitations, host defence systems and antimicrobial agents, and thus, are often associated with persistent infections (Donlan, 2002; Hall‐Stoodley et al., 2004). Hence, novel strategies are required to control biofilm formation by pathogens. However, traditional antibiotics and antifungal agents were primarily designed to inhibit cell growth, which often results in drug resistance. Therefore, it has become important that non‐toxic biofilm inhibitors be identified that do not inhibit planktonic cell growth, and thus, reduce the risk of drug resistance (Wright, 2015; Defoirdt, 2018).
Candida albicans is found in mucosal surfaces and in the gastrointestinal and genitourinary tracts and is the most common cause of systemic and invasive infections. The fungus easily colonizes host tissues and indwelling medical devices (Ramage et al., 2005), such as urinary catheters, dental materials, artificial heart valves, joint prostheses, penile implants and intrauterine devices (Sardi et al., 2013; de Oliveira et al., 2019; Handorf et al., 2019). Furthermore, biofilms are often tolerant to conventional antifungal therapeutics and the host immune system (Nobile et al., 2006). Candida biofilms contain yeast cells, pseudohyphae and hyphae. The transition of yeast cells to hyphae (filamentation) is required for stable biofilm formation, and thus, hyphal transition is considered a crucial virulence factor of Candida infections (Carradori et al., 2016). Various genes, including transcription factors, cell wall‐related proteins, and others, are involved in hyphae formation and biofilm development by C. albicans (Finkel and Mitchell, 2011; Araujo et al., 2017; Pandin et al., 2017; Song et al., 2020). Also, it has been well‐reported that the quorum‐sensing (QS) molecule, farnesol, inhibits filamentation and biofilm formation by C. albicans (Ramage et al., 2002; Polke et al., 2018) and Candida dubliniensis (Jabra‐Rizk et al., 2006). In addition, several natural and synthetic farnesol analogs (Shchepin et al., 2003) inhibit C. albicans filamentation, although their modes of action remains undetermined (Polke et al., 2018).
Fatty acids are widespread in all forms of life. More than 70 naturally occurring fatty acids have been identified, and there are literally thousands of natural sources (Kenar et al., 2017). Fatty acids are important cellular structural components and important energy sources for animals (Desbois and Smith, 2010) and have been suggested to be potential alternative antimicrobial agents (Desbois and Smith, 2010; Yoon et al., 2018). Recently, several studies reported that fatty acids exhibit anti‐hyphal and antibiofilm activities at concentrations less than their MICs (Kumar et al., 2020). For example, several fatty acids have been shown to selectively disrupt or inhibit biofilm formation by various microbial pathogens, such as Staphylococcus aureus (Davies and Marques, 2009; Kim et al., 2018), Pseudomonas aeruginosa (Inoue et al., 2008; Wenderska et al., 2011), Candida albicans (Murzyn et al., 2010; Muthamil et al., 2018; Prasath et al., 2019), and others (Wenderska et al., 2011; Santhakumari et al., 2017; Ramanathan et al., 2018; Cui et al., 2019). More specifically, capric acid (10:0) and lauric acid (12:0) inhibit the growth of planktonic Candida cells (Bergsson et al., 2001), and butanoic acid (4:0) inhibit hyphal formation by C. albicans (Noverr and Huffnagle, 2004). Stearidonic acid (18:4 n‐3), eicosapentaenoic acid (20:5), docosapentaenoic acid (22:5) (Thibane et al., 2010) and conjugated linoleic acid inhibit hyphal growth by C. albicans (Shareck et al., 2011), and trans‐2‐decenoic acid (10:Δ2) (Vilchez et al., 2010), undecenoic acid (10:0) (Shi et al., 2016; Muthamil et al., 2018) and myristic acid (14:0) (Prasath et al., 2019) inhibit hyphal growth and biofilm formation. However, long‐chain unsaturated fatty acids such as arachidonic acid, oleic acid, linolenic acid or γ‐linolenic acid did not influence hyphal development (Noverr and Huffnagle, 2004). Most recently, it was reported that oleic acid (18:1) (Muthamil et al., 2020) and linoleic acid (18:2) (Kim et al., 2020) inhibited filamentation and biofilm formation by C. albicans without affecting planktonic cell growth.
The QS molecule farnesol (3,7,11‐trimethyl‐2,6,10‐dodecatriene‐1‐ol) is structurally similar to several saturated and unsaturated fatty acids. Hence, we hypothesized that some fatty acids might mimic farnesol and inhibit filamentation and biofilm formation by C. albicans. In the present study, 31 natural fatty acids (2 short‐chain, 7 medium‐chain, 16 long‐chain and 6 very long‐chain) were initially screened for their antifungal and antibiofilm activities against C. albicans. To understand how fatty acids control hyphal and biofilm development, we used confocal laser scanning microscopy and scanning electron microscopy to investigate hyphal growth, cell aggregation and biofilm formation. Furthermore, the molecular basis of fatty acid induced alterations to the physiology of C. albicans was investigated using transcriptomic assays and by assaying farnesol and sterol production. In addition, an in vivo Caenorhabditis elegans model was used to confirm the anti‐hyphal and antibiofilm efficacies of fatty acids and their non‐cytotoxic natures. Farnesol was used as a control throughout this study.
Results
Antifungal and antibiofilm activities of fatty acids
To investigate the antibiofilm activities of fatty acids against C. albicans, 31 fatty acids (17 saturated fatty acids, and 14 unsaturated fatty acids) were initially screened in 96‐well plates at a concentration of 10 µg ml−1. Several of them were found to inhibit biofilm formation by C. albicans, but with widely different efficacies. Detailed information on biofilm formation by two C. albicans strains DAY185 and ATCC 10231 in the presence of fatty acids is provided in Table S1. Most notably, six medium‐chain saturated fatty acids (7:0, 8:0, 9:0, 10:0, 11:0 and 12:0) at 10 µg ml−1 significantly inhibited C. albicans biofilm formation by more than 85%. The fatty acids 14:0, 14:1, 16:1, 18:2, 18:3 and 20:4 also appreciably inhibited biofilm formation by both strains (Table S1). The antibiofilm efficacies of most fatty acids were similar for the two strains, and thus, we focused on the well‐studied DAY185 strain during further study. These results matched those of previous reports, which found that undecylenic acid (11:0) (Shi et al., 2016; Muthamil et al., 2018), myristic acid (14:0) (Prasath et al., 2019), oleic acid (18:1) (Muthamil et al., 2020) and linoleic acid (18:2) (Kim et al., 2020) inhibited biofilm formation by C. albicans. However, the present study is to report that fatty acids 7:0, 8:0, 9:0, 10:0 and 12:0 at low concentrations have antibiofilm activities comparable to that of 11:0.
Minimum inhibitory concentrations (MICs) of the 24 fatty acids are shown in Table S1. The MICs of most fatty acids were > 500 µg ml−1, but those of five medium‐chain saturated fatty acids (7:0, 8:0, 9:0, 10:0 and 11:0) ranged from 100 to 200 µg ml−1. MICs of other 7 fatty acids could not be determined due to low solubility (< 100 µg ml−1) in aqueous phase. The antifungal activities of capric acid (10:0 or called decanoic acid) and lauric acid (12:0) have been previously reported (Bergsson et al., 2001), and concur with our results. Importantly, six medium‐chain saturated fatty acids (7:0, 8:0, 9:0, 10:0, 11:0 and 12:0) at a sub‐inhibitory concentration (10 µg ml−1) were found to potently inhibit biofilm formation by two C. albicans strains (Table S1).
The medium‐chain saturated fatty acids inhibited C. albicans biofilm formation
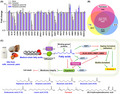
A detailed biofilm study showed the six medium‐chain saturated fatty acids (7:0, 8:0, 9:0, 10:0, 11:0 and 12:0) dose‐dependently inhibited C. albicans biofilm formation in 96‐well polystyrene plates (Fig. 1A). In particular, octanoic acid (8:0), nonanoic acid (9:0), decanoic acid (10:0) and undecanoic acid (11:0) inhibited biofilm formation by more than 75% at 2 µg ml−1. Nonanoic acid (9:0) appeared to be the most active fatty acid. In addition, farnesol at 100 µg ml−1 also significantly inhibited C. albicans biofilm formation (Fig. 1B), as has been previously reported (Ramage et al., 2002). However, the antibiofilm activities of the six medium‐chain fatty acids were much greater than that of farnesol. Notably, the antibiofilm concentrations (1–2 µg ml−1) of these fatty acids were about 100 times lower than their MICs (100–200 µg ml−1), which means that at low concentrations they effectively inhibit C. albicans biofilm formation without having a fungicidal effect. Since heptanoic acid (7:0) and nonanoic acid (9:0) exhibited most antibiofilm activity at low concentrations and had MICs of ˜ 100 µg ml−1, we focused on these in subsequent studies along with the less active lauric acid (12:0) and farnesol (the positive control).
Fig. 1.
Antibiofilm activities of medium‐chain saturated fatty acids against C. albicans. Biofilm formations by C. albicans DAY185 were quantified in the presence of each fatty acid, heptanoic acid (7:0), octanoic acid (8:0), nonanoic acid (9:0), decanoic acid (10:0), undecanoic acid (11:0) or lauric acid (12:0) (A) or farnesol (B) after incubation for 24 h in 96‐well plates. Error bars indicate standard deviations. *P < 0.05 vs. non‐treated controls. Biofilm formations by C. albicans on polystyrene plates were observed in the presence of each fatty acid (2 µg ml−1) or farnesol (50 or 100 µg ml−1) by confocal laser microscopy (C) and COMSTAT analysis (D).
Biofilm inhibition was further analysed by a confocal laser scanning microscope. In non‐treated controls after 24 h culture, C. albicans formed dense biofilms (thickness > 40 µm and achieved almost 100% surface coverage), whereas the presence of 7:0, 9:0 or 12:0 at 2 µg ml−1 dramatically reduced biofilm densities and thicknesses. On the other hand, farnesol at 50 µg ml−1 less appreciably inhibited and farnesol at 100 µg ml−1 clearly inhibited (Fig. 1C). Biofilm reduction was also quantified using COMSTAT biofilm software (Heydorn et al., 2000), and results showed 7:0, 9:0 and 12:0 significantly reduced biofilm biomass, average thickness and substrate coverage. Specifically, biofilm biomass, thickness and substrate coverage were reduced by 7:0, 9:0 and 12:0 by > 95% versus untreated controls (Fig. 1D).
Medium‐chain fatty acids inhibited hyphal growth and cell aggregation
The dimorphic switch of yeast cells to hyphal cells and cell aggregation are prerequisites of biofilm maturation by C. albicans (Chandra et al., 2001). To study the effect of fatty acids on C. albicans morphology, we monitored C. albicans colony formation on potato dextrose agar, performed cell aggregation assay, and used scanning electron microscopy (SEM) to assess hyphal growth.
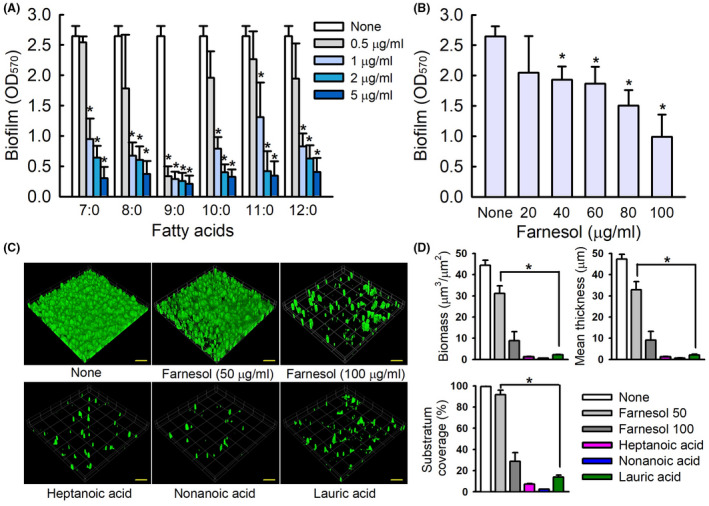
In untreated C. albicans, marked hyphal protrusions from colonies were observed after 6 days of incubation, whereas the presence of 7:0, 9:0 or 12:0 at 2 µg ml−1 completely suppressed these protrusions after 6 days. Farnesol at 100 µg ml−1 partially reduced hyphal protrusion (Fig. 2A). After incubation for 24 h in PDB medium, mostly hyphae and large cell aggregations entangled by hyphae were observed in non‐treated controls. However, treatments with 7:0, 9:0 or 12:0 much reduced cell aggregations and farnesol at 100 µg ml−1 slightly reduced aggregation (Fig. 2B). SEM analysis confirmed that 7:0, 9:0 and 12:0 all substantially suppressed hyphal formation. Non‐treated control biofilms consisted predominately of hyphae and few pseudohyphae, but biofilms grown in the presence of 7:0, 9:0 or 12:0 had much shorter hyphae and were predominantly composed of yeast and pseudohyphae cells (Fig. 2C). Notably, 7:0, 9:0 and 12:0 at 2 µg ml−1 more effectively suppressed hyphal protrusions, cell aggregation and hyphal growth than farnesol at 100 μg ml−1, and hyphal growth and cell aggregation results were in‐line with the observed antibiofilm activities of these fatty acids. Taken together, these results show medium‐chain fatty acids potently inhibit C. albicans hyphal formation, cell aggregation and biofilm formation.
Fig. 2.
Inhibition of hyphal filamentation and aggregation by medium‐chain fatty acids. C. albicans morphology on solid media (A). C. albicans was streaked on PDA solid plates in the absence or presence of heptanoic acid (7:0), nonanoic acid (9:0) or lauric acid (12:0) at 2 µg ml−1 or farnesol at 100 µg ml−1. Colony morphologies were observed during incubation for 6 days at 37°C. Inhibition of filamentation and of cell aggregation in PDB medium (B). Hyphae were visualized after incubation for 24 h. The scale bars in panels A and B represent 100 µm. None; non‐treated control. SEM observation of hyphal inhibition in C. albicans biofilms grown in PDB medium by fatty acids (C). The scale bars represent 15 µm.
Medium‐chain fatty acids decreased the productions of farnesol and sterols
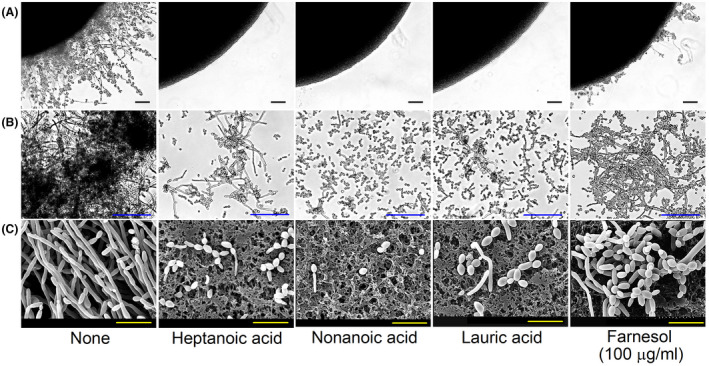
In Candida species, farnesol is a well‐known QS molecule that controls various virulence traits, such as biofilm formation, morphogenic transition and even cell death (Ramage et al., 2002; Polke et al., 2018). We hypothesized that fatty acids structurally similar to farnesol interfere with farnesol associated QS, and thus, we investigated their effects on the productions of farnesol and sterols. Interestingly, the addition of either of the three medium‐chain fatty acids (7:0, 9:0 or 12:0) significantly reduced the production of farnesol, while the less active antibiofilm compound 16:0 (palmitic acid) slightly decreased farnesol production (Fig. 3A). For example, in the absence of a fatty acid, C. albicans DAY185 produced 15.6 ± 0.5 µM farnesol in PDB medium, which concurs with previous reports (Hornby et al., 2001; Weber et al., 2008), whereas in the presence of 7:0, 9:0, 12:0 or 16:0 at 10 μg ml−1 farnesol production decreased by 85 ± 3%, 91 ± 1%, 66 ± 15% and 13 ± 9%, respectively (Fig. 3A), while culture biomasses in the present of 7:0 or 9:0 at 10 µg ml−1 were decreased by only 5 and 13% compared to the untreated control (3.9 g l−1). These results show that these medium‐chain fatty acids, but not long fatty acids, negatively influenced the synthesis of farnesol.
Fig. 3.
Impacts of medium‐chain fatty acids on the productions of farnesol and sterols. Farnesol levels were measured by HPLC after culturing C. albicans DAY185 in PDB medium for 24 h (A). Heptanoic acid (7:0), nonanoic acid (9:0), lauric acid (12:0) or palmitic acid (16:0) at 10 µg ml−1 or farensol at 50 or 100 µg ml‐1 were added at culture start. Sterol production was assessed by filipin staining after culturing C. albicans in PDB medium for 60 min (B). The scale bars represent 5 µm.
Farnesol is a precursor of the synthesis of sterols in C. albicans (Hornby et al., 2001; Nickerson et al., 2013). Ergosterol (ergosta‐5,7,22‐trien‐3β‐ol) is a sterol found in fungi and is an essential component of fungal cell membranes (Weete et al., 2010). We investigated the effect of fatty acids on the production of sterols by staining for filipin, which is often used to study sterol distribution in C. albicans. In the absence of fatty acids, sterols were localized at the tips of pseudohyphae (Fig. 3B). Treatment with 7:0, 9:0 or 12:0 at 10 μg ml−1 markedly inhibited sterol production, while treatment with 16:0 had little inhibitory effect. Farnesol at 50 μg ml−1 slightly inhibited and at 100 μg ml−1 further inhibited sterol production (Fig. 3B). It appears that sterol inhibition by 7:0, 9:0 or 12:0 is probably due to the inhibition of farnesol synthesis. It was reported that sterol inhibition by farnesol at high concentrations probably led to the apoptotic activity of farnesol in C. albicans (Shirtliff et al., 2009). Therefore, it appears that 7:0, 9:0 and 12:0 at high concentrations could damage the cell membrane of C. albicans via sterol inhibition.
Differential expressions of genes by heptanoic acid (7:0), nonanoic acid (9:0) or farnesol in C. albicans
qRT‐PCR was used to investigate the effects of heptanoic acid (7:0) and nonanoic acid (9:0) on the expressions of 35 biofilm‐ and hypha‐related genes associated with inhibitions of biofilm formation and hyphal growth. Overall, expressional changes were similar after treatment with 7:0 at 5 μg ml−1, 9:0 at 5 μg ml−1 or farnesol at 50 μg ml−1 (Fig. 4). Notably, four key biofilm‐ and hypha‐related genes, that is, ALS3 (agglutinin‐like protein 3), ECE1 (hypha‐specific protein, also known as HWP2), HWP1 (hyphal cell wall protein, also known as ECE2), and UME6 (filament‐specific regulator), were repressed by 7:0, 9:0 and by farnesol. For example, 9:0 (the most active) down‐regulated ALS3 by 25.5‐fold, ECE1 by 93‐fold, HWP1 by 17.2‐fold and UME6 by 7.9‐fold, while farnesol down‐regulated these genes by 3.5, 11.4, 3.0 and 2.7‐fold, respectively. As was observed for biofilm inhibition (Fig. 1), the impact of 9:0 at 5 μg ml−1 on gene expression was significantly greater than that of farnesol at 50 μg ml−1 (Fig. 4A). On the other hand, the expression of UCF1 was up‐regulated by 7:0, 9:0 or farnesol (Fig. 4A) while UCF1 was positively up‐regulated in filamentous growth (Bahn et al., 2007).
Fig. 4.
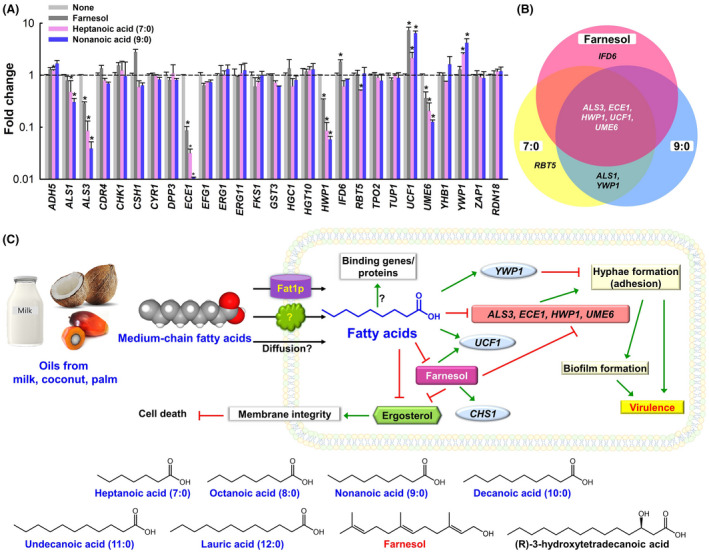
Relative transcriptional profiles of C. albicans cells treated with or without fatty acids or farnesol and summary of fatty acid associated processes in C. albicans. C. albicans was incubated with or without farnesol at 50 μg ml−1, heptanoic acid (7:0) at 5 μg ml−1, or nonanoic acid (9:0) at 5 μg ml−1 for 6 h without shaking. Transcriptional profiles were obtained by qRT‐PCR (A). Fold changes represent changes in the transcriptions of treated versus untreated C. albicans. RDN18 was a housekeeping gene. Venn diagram of qRT‐PCR results (B). The experiment was performed in duplicate (six qRT‐PCR reactions were performed per gene). *P < 0.05 vs. non‐treated controls (None). → indicates induction of gene expression or stimulation of a phenotype and ⊥ indicates repression of gene expression or repression of a phenotype. Chemical structures of medium‐chain fatty acids, farnesol and farnesol‐like compounds that exhibit antibiofilm activity against C. albicans (C).
In addition, the expression of ALS1 (agglutinin‐like protein 1) was slightly inhibited by 7:0 and 9:0 but unaffected by farnesol, and the expression of YWP1 (yeast‐form wall protein 1) was up‐regulated by 7:0 and 9:0. Also, the expression of CSH1 (cell‐surface hydrophobicity protein 1) was up‐regulated 2.8‐fold by farnesol but unaffected by 7:0 or 9:0. The expressions of other biofilm and hyphae‐related genes (ADH5, CDR4, CHK1, CSH1, CYR1, DPP3, EFG1, ERG1, ERG11, FKS1, GST3, HGC1, HGT10, IFD6, RBT5, TPO2, TUP1, YHB1 and ZAP1), DPP3 (farnesol pyrophosphatase gene) and of CYR1 (a possible farnesol binding protein) were unaffected by 7:0, 9:0 or farnesol. The expression of nine more ERG genes in the sterol biosynthesis pathway such as ERG2, ERG3, ERG4, ERG5, ERG6, ERG9, ERG10, ERG20 and ERG24, was not affected by 9:0 at this condition (Fig. S2). Taken together, qRT‐PCR showed 7:0, 9:0 and farnesol significantly down‐regulated biofilm‐ and hyphae‐related genes (i.e. ALS3, ECE1, HWP1 and UME6) and up‐regulated the biofilm‐related genes UCF1 and YWP1 (Fig. 4B).
Nonanoic acid (9:0) reduced C. albicans virulence in a C. elegans nematode with minimal cytotoxicity
We also examined whether nonanoic acid (9:0) affected C. albicans virulence in a C. elegans nematode model, an accepted alternative to mammalian models (Tampakakis et al., 2008). The hyphal form of C. albicans lethally pierces the nematode’s cuticle (Pukkila‐Worley et al., 2009). In this model, C. albicans caused 85 ‐ 95% fatality (5–15% survival) at 5 and 7 days post‐infection, but the presence of nonanoic acid at 1, 2 or 5 µg ml−1 protected C. elegans (50–80% survival) (Fig. S1A). In addition, in non‐infected C. elegans, nonanoic acid was non‐toxic at concentrations up to 100 µg ml−1 (Fig. S1B), while nonanoic acid rescued the C. elegans survival by inhibiting hyphal and biofilm growth of C. albicans (Fig. S1C).
Discussion
Biofilms confer resistance to conventional antifungals or antibiotics, and thus, represent a serious threat to human health. Although fatty acids have been known for some time to have non‐specific broad‐range antimicrobial activities at high concentrations (Desbois and Smith, 2010; Yoon et al., 2018), their antibiofilm and antivirulence effects at concentrations well below their MICs have been seemingly overlooked (Kumar et al., 2020). This study demonstrates for the first time that six medium‐chain saturated fatty acids at concentrations of few µg ml−1 suppress C. albicans biofilm formation by inhibiting hyphal growth and cell aggregation, and reducing fungal virulence. We found that medium‐chain fatty acids structurally similar to the QS molecule farnesol interfere with farnesol and sterol production in C. albicans, and thus, cause physiological changes of fungal dimorphism, biofilm formation and even cell death.
The antifungal activities of several fatty acids such as capric acid (10:0), lauric acid (12:0) (Bergsson et al., 2001), stearidonic acid (18:4), eicosapentaenoic acid (20:5) and docosapentaenoic acid (22:5) (Thibane et al., 2010), have been previously reported at concentrations above 1 mM. The present study shows that all six medium‐chain saturated fatty acids examined (7:0, 8:0, 9:0, 10:0, 11:0 and 12:0) exhibited antifungal activity with MICs in the range 100–200 µg ml−1, while other shorter or longer‐chain fatty acids had MICs of above 500 µg ml−1. It has been reported 10:0 caused cytoplasm disorganization and that this was probably due to changes in intracellular hydrostatic turgor pressure (Bergsson et al., 2001). In another study, 18:4, 20:5 and 22:5 inhibited mitochondrial metabolism probably by increasing oxidative stress (Thibane et al., 2010). The current result indicates that the antifungal effects of medium‐chain fatty acids at high concentrations (Table S1) are probably due to the inhibition of sterol production (Fig. 3B), which could impede cell wall synthesis.
It has been reported farnesol induces apoptosis in C. albicans via ROS (reactive oxygen species) accumulation, mitochondrial degradation and caspase activation (Shirtliff et al., 2009). Despite intensive research on farnesol extending for more than a decade, it is not clear how C. albicans cells sense farnesol or how farnesol exerts its biological effects (Polke et al., 2018). Our results (Fig. 3B) suggest farnesol at high concentrations (> 100 µg ml−1) inhibits the production of ergosterol and essential cell membrane component, which led to fungal cell death. Interestingly, the inhibition of sterol production by medium‐chain fatty acids was reminiscent of the action mechanism of commercial antifungal azoles that inhibit 4‐α‐sterol demethylase (encoded by the ERG11 gene), which is required for the biosynthesis of ergosterol. While azole‐resistant Candida species have been developed due to enzyme modification, sterol uptake and genetic mutation (Whaley et al., 2016), medium‐chain fatty acids inhibited the virulence characteristics of C. albicans biofilm at sub‐inhibitory concentrations of only a few µg ml−1.
Interference of QS signalling using QS‐degrading enzymes or QS inhibitors has been widely proposed as a means of controlling microbial infections and biofilm formation (Zhang and Dong, 2004; Kalia, 2013; Grandclement et al., 2016). For example, halogenated furanones produced by the marine red alga Delisea pulchra disrupt the N‐acylated homoserine lactone (AHL) regulatory system in several Gram‐negative bacteria (Rasmussen et al., 2000). Plants such as rice, tomato, soybean and Medicago truncatula can also produce substances that mimic the activities of AHL (Teplitski et al., 2000; Koh et al., 2013). Several QS interfering compounds have also been reported in C. albicans. For example, quercetin (a flavonoid) was found to inhibit farnesol‐dependent biofilm formation, probably by inhibiting adenylate cyclase activity (Singh et al., 2015). In one study, four azole antifungals increased farnesol production (Hornby and Nickerson, 2004), and in another, various natural and synthetic farnesol analogs were synthesized and found to have the activity of farnesol (Shchepin et al., 2003). Interestingly, (R)‐3‐hydroxytetradecanoic acid, a metabolite of linoleic acid, was able to reverse the farnesol‐induced inhibition of biofilm formation in C. albicans (Nigam et al., 2011), which suggests fatty acids and fatty acid‐like compounds structurally similar to farnesol might interfere with farnesol‐induced QS signalling (Fig. 4C).
The present study suggests that medium‐chain fatty acids mimic the QS molecule farnesol (Fig. 4C), as indicated by six lines of evidence, (i) structural similarities, (ii) inhibition of hyphal growth, (iii) inhibition of biofilm formation, (iv) inhibition of farnesol production, (v) inhibition of sterol production and (vi) effects on gene expressions, which show the activities of medium‐chain fatty acids and farnesol are similar.
The chain lengths of medium‐chain fatty acids and farnesol are similar except three branches and three double bonds from farnesol structure (Fig. 4C). It appears that carbon chains containing 8, 9 or 10 carbon atoms are optimal for antibiofilm activity since shorter‐ or longer‐chain fatty acids with double bonds had weaker antibiofilm effects (Table S1). Since several farnesol‐like compounds, such as geraniol (Dalleau et al., 2008), citral (Silva Cde et al., 2008) and nerolidol (Curvelo et al., 2014), which all have 10 carbon atoms, and linanool (Hsu et al., 2013), which has 15, have been shown to possess antibiofilm activity against C. albicans, it would appear that studies on the effects of structural modifications of medium‐chain fatty acids might result in the identification of molecules with much enhanced antifungal and antibiofilm activities.
Interestingly, qRT‐PCR studies showed that the expressions of several hypha‐ and biofilm‐related genes (ALS3, ECE1, HWP1, UCF1 and UME6) were simultaneously altered in C. albicans cells by 7:0, 9:0 or farnesol (Fig. 4A). Specifically, ALS3, ECE1, HWP1 and UME6 were significantly down‐regulated, and UCF1 was significantly up‐regulated (Fig. 4A). ALS3 is a multifunctional adhesion (Phan et al., 2007; Liu and Filler, 2011) and ECE1 (also called HWP2) and HWP1 (also called ECE2) are essential for hyphal development and their expressions have been shown to be correlated with cell elongation, biofilm formation (Nobile et al., 2006a, 2006b,2006a, 2006b) and intercellular adhesion (Orsi et al., 2014). UME6 is a filament‐specific regulator of C. albicans hyphal extension (Banerjee et al., 2008) and enhances biofilm formation (Banerjee et al., 2013). Hence, it appears the suppression of hyphal growth and biofilm formation by 7:0, 9:0 or farnesol may be explained, at least in part, by the down‐regulations of these hypha‐specific genes. Although UCF1 was reported to be up‐regulated by cAMP (Bahn et al., 2007) and was up‐regulated by 7:0, 9:0 or farnesol in the present study (Fig. 4A and B), treatment of C. albicans with cAMP at concentrations up to 10 mM did not complement hyphal growth or biofilm formation (data not shown).
The observed inhibition of farnesol production by medium‐chain fatty acids (7:0, 9:0 or 12:0), but not by 16:0, was somewhat unexpected (Fig. 3A). The mechanism responsible for this effect is not clear as the expressions of farnesol‐regulatory genes (CYR1, DPP3, EFG1, ERG1, ERG2, ERG3, ERG4, ERG5, ERG6, ERG9, ERG10, ERG11, ERG20 and ERG24) were not directly affected by these three fatty acids (Fig. 4A and Fig. S2). We speculate, C. albicans might confuse these fatty acids with farnesol or that medium‐chain fatty acids interfere with QS. It has been previously reported that C. albicans contains fatty acyl‐CoA synthetase (CaFaa4p), which can convert long‐chain fatty acids into CoA esters (Black and DiRusso, 2003). Additional studies are required to determine how fungal cells sense medium‐chain fatty acids and regulate its gene expression. In addition, our transcriptome results broadly match expressional changes induced by oleic acid (18:1) and linoleic acid (18:2) in C. albicans cells. Oleic acid (18:1) was found to down‐regulated the expressions of ALS1, ALS3, ERG11, SAP2, HWP1, CST20 and RAS1 by more than twofold (Muthamil et al., 2020), whereas linoleic acid (18:2) down‐regulated CHT2, ECE1, HWP1, RAS1, RBT1 and UME6 (Kim et al., 2020). Our results also support a previous report that expressional changes induced by farnesol suppress the expression of hypha‐specific HWP1 (Ramage et al., 2002). Also, linalool (C10H18O), a plant metabolite structurally similar to medium‐chain fatty acids, inhibited hyphal growth and biofilm formation by C. albicans by down‐regulating the expressions of ALS3, HWP1, UME6, HGC1 and EED1 (Hsu et al., 2013), which is similar to the gene expression changes induced by 7:0, 9:0 or farnesol observed in the present study (Fig. 4A). Therefore, it appears plants and animals may both utilize fatty acids and fatty acid‐like compounds to control C. albicans hyphal growth and biofilm formation and diminish its virulence.
Medium‐chain fatty acids are found in mammalian milk, palm kernel oil and coconut oil and are widely used in foods, drugs and cosmetics (Fig. 4C) (Traul et al., 2000). Toxicity rankings of fatty acids conducted in two human leukaemic cell lines showed that short‐ and medium‐chain saturated fatty acids are much less toxic than long‐chain fatty acids (Lima et al., 2002). Several authors have reported fatty acids can exert antimicrobial activity at high concentrations and antibiofilm activity at sub‐inhibitory concentrations (Kumar et al., 2020). To enhance the antifungal efficacy, combinatorial therapy and fatty acids can be simultaneously used in tandem or combination. We speculate that fatty acids alone or in combination delivered using liposomes or novel nanocarrier systems might protect eukaryotes from fungal infections (Kumar et al., 2020).
In conclusion, the rapid emergence of drug resistant microorganisms has driven the development of novel antifungals and antibiotics. This study shows that six medium‐chain fatty acids mimic the QS molecule farnesol and have antibiofilm and anti‐hyphal activities in C. albicans. Fatty acids are widespread in most organisms and our findings suggest they may be utilized for defence purposes against C. albicans and interfere with its quorum‐sensing system. Nonanoic acid (9:0), which was the most active fatty acid, also reduced C. albicans virulence effectively in vivo in our Caenorhabditis elegans (Fig. S1) model and exhibited only minimal cytotoxicity. A variety of biotechnological applications of the current study is possibility to control pathogenic biofilms, such as coating medical devices with fatty acids, synthesizing composites of polymer‐ or nanoparticle‐fatty acids, producing liposomes or emulsion with fatty acids, and developing fatty acids as alternatives of antibiotics or antibiotic adjuvants. As regards medical applications, our findings indicate that medium‐chain fatty acids offer a basis for the design of potent antibiofilm and anti‐hyphal forming agents against Candida species.
Experimental procedures
Strains, chemicals and culture materials
The fluconazole‐resistant C. albicans strains DAY185 and ATCC 10231 were used in the present study and cultured in potato dextrose agar (PDA) and potato dextrose broth (PDB). All experiments were performed at 37°C. Twenty four fatty acids, namely butanoic acid (4:0), pentanoic acid (5:0), hexanoic acid (6:0), heptanoic acid (7:0), octanoic acid (8:0), nonanoic acid (9:0), decanoic acid (10:0), undecanoic acid (11:0), lauric acid (12:0), myristic acid (14:0), myristoleic acid (14:1), palmitic acid (16:0), palmitoleic acid (16:1), heptadecanoic acid (17:0), stearic acid (18:0), oleic acid (18:1), elaidic acid (18:1), petroselinic acid (18:1), linoleic acid (18:2), conjugated linoleic acid (18:2), linolenic acid (18:3), arachidonic acid (20:4), eicosapentaenoic acid (20:5), behenic acid (22:0), erucic acid (22:1), docosahexaenoic acid (22:6), tricosanoic acid (23:0), hexacosanoic acid (26:0) and octacosanoic acid (C28:0) were purchased from either Sigma‐Aldrich (St. Louis, USA) or TCI (Tokyo, Japan). Dimethyl sulfoxide (DMSO, Sigma‐Aldrich, St. Louis, MO) was used as solvent to dissolve all the fatty acids, except butanoic acid, which was dissolved in water. DMSO (0.1% v/v) was used as the negative control and at < 0.1% did not affect bacterial growth or biofilm formation.
Planktonic cell growths and turbidities were measured using an Optizen 2120UV spectrophotometer (Mecasys, Daejeon, Korea) at 600 nm. MIC was defined as the lowest concentration that inhibited planktonic cell growth by 80% and also confirmed by colony counting.
Biofilm assay using crystal violet
Biofilm formation by C. albicans was produced on 96‐well polystyrene plates (SPL Life Sciences, Korea), as previously described (Lee et al., 2011). Briefly, a two day old single colony was inoculated into 25 ml of PDB medium and incubated overnight at 37°C. Overnight cultures at an initial turbidity of 0.1 at OD600 nm (˜ 105 CFU ml−1) were inoculated into PDB (final volume 300 μl) with or without fatty acids in 96‐well polystyrene plates and incubated for 24 h without shaking at 37°C. Biofilm cells that adhered to 96‐well plates were stained with 0.1% crystal violet Sigma‐Aldrich (St. Louis, USA) for 20 min after washing planktonic cells with distilled water three times, then washed repeatedly with distilled water three times, and resuspended in 95% ethanol. Plates were read at 570 nm to measure biofilm formation and results are presented as the means of at least six repetitions. The percentage of inhibition ratio represents the relative biofilm formation (100× biofilm formation with chemical/biofilm formation of untreated control).
Biofilm observations by confocal microscopy
Biofilm formation by C. albicans was developed on 96‐well polystyrene plates with or without fatty acids or farnesol without shaking for 24 h. Planktonic cells were then removed by washing with distilled water three times, and biofilms were stained with carboxyfluorescein diacetate succinimidyl ester (Invitrogen, Molecular Probes, Eugene, OR, USA). Bottoms of plate were then visualized using a 488 nm Ar laser (emission 500 to 550 nm) under a confocal laser microscope (Nikon Eclipse Ti, Tokyo, Japan). To quantify biofilm structures, COMSTAT software (Heydorn et al., 2000) was used to determine biovolumes (μm3 μm−2), mean biofilm thicknesses (μm) and substratum coverages (%). Two independent cultures were performed under each experimental condition and at least 10 random positions were assayed.
C. albicans colony morphologies on solid media
To examine the colony morphology of C. albicans on solid agar plates, a freshly prepared glycerol stock of C. albicans was streaked onto PDA plates supplemented with or without fatty acids (2 μg ml−1) or farnesol (100 μg ml−1). Plates were then incubated for 6 days at 37°C and changes in colony morphologies were observed using an iRiS™ Digital Cell Imaging System (Logos Bio Systems, Anyang, Korea).
Hyphae and cell aggregation in liquid media
To investigate hyphal growth and cell aggregation, C. albicans cells were inoculated into 2 ml of PDB medium at a density of 105 CFU ml−1 in 14 ml test tubes with or without fatty acids (2 μg ml−1) or farnesol (100 μg ml−1) and incubated at 37°C for 24 h without shaking. After incubation for 24 h, aggregated cells and hyphal growths were visualized in bright field using the iRiSTM Digital Cell Imaging System (Logos Bio Systems) at magnifications of 4x and 10x. At least, four independent experiments were conducted.
Analysis of biofilm and hyphal formation by scanning electron microscopy (SEM)
To observe biofilm and hyphal formation, SEM was used as previously described (Kim et al., 2016). Briefly, a nylon membrane was cut into 0.4 × 0.4 cm pieces, placed in 96‐well plates containing C. albicans grown with or without fatty acids (2 μg ml−1) or farnesol (100 μg ml−1), and incubated for 24 h at 37°C. Cells that adhered to membranes were fixed with a glutaraldehyde (2.5%) and formaldehyde (2%) for 24 h, post‐fixed using OsO4, and dehydrated using an ethanol series (50, 70, 80, 90, 95 and 100%) and isoamyl acetate. After critical‐point drying, cells on filters were sputter‐coated with palladium/gold and imaged under a S‐4100 scanning electron microscope (Hitachi, Tokyo, Japan) at a voltage of 15kV.
Farnesol assay by high‐performance liquid chromatography (HPLC)
Farnesol production in C. albicans was measured as previously described (Hornby et al., 2001) using a HPLC unit (YL9100HPLC, Young Lin, Anyang, Korea) equipped with a reverse‐phase HPLC column (4.6 × 250 mm; Agilent ZORBAX Eclipse XDB‐C18). The mobile phase used was water containing methanol (20:80, v/v) and the flow rate was 1.0 ml min‐1. Eluates were monitored at 210 nm for farnesol (retention time 17 min). The fungus was cultured in PDB with and without fatty acid for 24 h at 37°C with shaking at 250 rpm. Farnesol was extracted from 20 ml of C. albicans culture by vortexing for 5 min with 10 ml of n‐hexane. The hexane fraction was then dried and dissolved in 0.5 ml of methanol. Trans, trans‐farnesol purchased from Sigma‐Aldrich (St. Louis, USA) was used as the standard.
Sterol assay with filipin
Sterol production in C. albicans was investigated as previously described (Liu et al., 2017). Briefly, cells (˜ 105 CFU ml−1) were inoculated into PDB broth in 2 ml test tubes and incubated at 37°C for 1 h without agitation in the presence or absence of fatty acids (10 μg ml−1) or farnesol (50 or 100 μg ml−1). After incubation, cells were stained with filipin (Sigma‐Aldrich, St. Louis, USA, 25 µg ml−1 final concentration) for 30 min at room temperature. Cells were analysed by optical microscopy (iRiS™ Digital Cell Imaging System).
RNA isolation for transcriptomic studies
For transcriptomic analyses, 10 ml of C. albicans at an initial turbidity of 0.1 at OD600 (˜ 105 CFU ml−1) was inoculated into PDB broth in 250 ml Erlenmeyer flasks and incubated for 6 h at 37°C without agitation in the presence or absence of fatty acids (5 μg ml−1) or farnesol (20 μg ml−1). To prevent RNA degradation, RNase inhibitor (RNAlater, Ambion, TX, USA) was added to cells immediately after incubation. Total RNA was isolated using a hot acidic phenol method (Amin‐ul Mannan et al., 2009), and RNA was purified using a Qiagen RNeasy mini Kit (Valencia, CA, USA).
Quantitative Real‐Time PCR (qRT‐PCR)
To determine the expressions of hyphae‐related genes (ADH5, ALS1, ALS3, CDR4, CHK1, CSH1, CYR1, DPP3, ECE1, EFG1, ERG1, ERG2, ERG3, ERG4, ERG5, ERG6, ERG9, ERG10, ERG11, ERG20, ERG24, FKS1, GST3, HGC1, HGT10, HWP1, IFD6, RBT5, TPO2, TUP1, UCF1, UME6, YHB1, YWP1 and ZAP1), qRT‐PCR was performed. The specific primers and housekeeping gene (RDN18) used for qRT‐PCR are listed in Table S2. The expression of RDN18 was not affected by fatty acids or farnesol. The qRT‐PCR method used was as described by Kim et al, 2016 (Kim et al., 2016) and was performed using SYBR Green master mix (Applied Biosystems, Foster City, USA) and an ABI StepOne Real‐Time PCR System (Applied Biosystems). At least two independent cultures were used.
Antivirulence and toxicity assays in the nematode model
To investigate the effects of fatty acids on the virulence of C. albicans, we used C. elegans strain fer‐15(b26); fem‐1(hc17), as previously described (Lee et al., 2018). Briefly, synchronized adult nematodes were washed with M9 buffer before starting experiments and approximately 30 worms were added into each well of 96‐well plates containing PDB:M9 (20:80) medium (200 µl) with or without nonanoic acid (0, 1, 2 and 5 µg ml−1). Also, the untreated control and fatty acid‐treated C. albicans cells (˜ 105 CFU ml−1) were added to into wells containing worms. Plates were then incubated for 5 days at 25°C without shaking. For chemical toxicity assays, about 30 non‐infected worms were pipetted into each well of a 96‐well plate containing M9 buffer and nonanoic acid was added to final concentrations of 0, 10, 20, 50 or 100 µg ml−1 without C. albicans. Plates were then incubated for 7 days at 25°C without shaking. Three independent experiments were performed in triplicate. Results are expressed as percentages of live worms (survival), as determined by responses to platinum wire touching after incubation for 5 and 7 days. Observations were made using an iRiS™ Digital Cell Imaging System (Logos Bio Systems, Anyang, Korea).
Statistical analysis
Replication numbers for assays are provided above and results are expressed as means ± standard deviations. The statistical analysis was performed by one‐way ANOVA followed by Dunnett’s test using spss version 23 (SPSS, Chicago, IL, USA). P values of < 0.05 were considered significant and asterisks indicate significant differences between treated and untreated samples.
Conflict of interests
None declared.
Supporting information
Table S1. Inhibitory effects of various fatty acids on C. albicans biofilm formation and planktonic growth. The antibiofilm activity of fatty acids against two C. albicans strains (DAY185 and ATCC 10231) in PDB medium was determined after culture for 24 h in 96‐well plates. For the biofilm assay, each fatty acid was used at 10 μg ml‐1. The MICs of each fatty acid against planktonic cells of C. albicans DAY185 are shown. Active fatty acids with low MIC are indicated in blue.
Table S2. Primer sequences used for qRT‐PCR.
Fig. S1. Effects of nonanoic acid (9:0) on C. albicans infected C. elegans. Nematode survival after exposure to C. albicans for 5 or 7 days in the presence of nonanoic acid (A). The toxicity of nonanoic acid was investigated by treating non‐infected nematodes for 5 or 7 days (B). Nonanoic acid rescued the C. elegans survival by inhibiting hyphal growth of C. albicans (C). None indicates non‐treated controls. Worm survival was determined based on movement. *P < 0.05 vs. non‐treated controls.
Fig. S2. Relative transcriptional profiles of ergosterol biosynthesis related genes in C. albicans cells treated with or without nonanoic acid (9:0) at 5 µg ml−1. C. albicans was incubated with or without nonanoic acid (9:0) at 5 μg ml−1 for 6 h without shaking. Transcriptional profiles were obtained by qRT‐PCR. RDN18 was a housekeeping gene.
Acknowledgements
This research was supported by grants from the Basic Science Research Program through the NRF funded by the Ministry of Education (2018R1D1A3B07040699 to J.‐H. Lee, 2019R1C1C1008329 to Y.‐G. Kim), by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Innovational Food Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (119034‐3), and by a grant from the Priority Research Centers Program through the NRF funded by the Ministry of Education (2014R1A6A1031189).
Microbial Biotechnology (2021) 14(4), 1353–1366
Funding information This research was supported by grants from the Basic Science Research Program through the NRF funded by the Ministry of Education (2018R1D1A3B07040699 to J.‐H. Lee, 2019R1C1C1008329 to Y.‐G. Kim), by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Innovational Food Technology Development Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (119034‐3), and by a grant from the Priority Research Centers Program through the NRF funded by the Ministry of Education (2014R1A6A1031189).
References
- Amin‐ul Mannan, M. , Sharma, S. , and Ganesan, K. (2009) Total RNA isolation from recalcitrant yeast cells. Anal Biochem 389: 77–79. [DOI] [PubMed] [Google Scholar]
- Araujo, D. , Henriques, M. , and Silva, S. (2017) Portrait of Candida species biofilm regulatory network genes. Trends Microbiol 25: 62–75. [DOI] [PubMed] [Google Scholar]
- Bahn, Y.S. , Molenda, M. , Staab, J.F. , Lyman, C.A. , Gordon, L.J. , and Sundstrom, P. (2007) Genome‐wide transcriptional profiling of the cyclic AMP‐Dependent signaling pathway during morphogenic transitions of Candida albicans . Eukaryot Cell 6: 2376–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, M. , Thompson, D.S. , Lazzell, A. , Carlisle, P.L. , Pierce, C. , Monteagudo, C. , et al. (2008) UME6, a novel filament‐specific regulator of Candida albicans hyphal extension and virulence. Mol Biol Cell 19: 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, M. , Uppuluri, P. , Zhao, X.R. , Carlisle, P.L. , Vipulanandan, G. , Villar, C.C. , et al. (2013) Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1‐and Sun41‐dependent mechanisms. Eukaryot Cell 12: 224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsson, G. , Arnfinnsson, J. , Steingrimsson, O. , and Thormar, H. (2001) In vitro killing of Candida albicans by fatty acids and monoglycerides. Antimicrob Agents Chemother 45: 3209–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, P.N. , and DiRusso, C.C. (2003) Transmembrane movement of exogenous long‐chain fatty acids: proteins, enzymes, and vectorial esterification. Microbiol Mol Biol Rev 67: 454–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carradori, S. , Chimenti, P. , Fazzari, M. , Granese, A. , and Angiolella, L. (2016) Antimicrobial activity, synergism and inhibition of germ tube formation by Crocus sativus‐derived compounds against Candida spp. J Enzyme Inhib Med Chem 31: 189–193. [DOI] [PubMed] [Google Scholar]
- Chandra, J. , Kuhn, D.M. , Mukherjee, P.K. , Hoyer, L.L. , McCormick, T. , and Ghannoum, M.A. (2001) Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol 183: 5385–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, C. , Song, S. , Yang, C. , Sun, X. , Huang, Y. , Li, K. , et al. (2019) Disruption of quorum sensing and virulence in Burkholderia cenocepacia by a structural analogue of the cis‐2‐dodecenoic acid signal. Appl Environ Microbiol 85: e00105‐00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curvelo, J.A. , Marques, A.M. , Barreto, A.L. , Romanos, M.T. , Portela, M.B. , Kaplan, M.A. , and Soares, R.M. (2014) A novel nerolidol‐rich essential oil from Piper claussenianum modulates Candida albicans biofilm. J Med Microbiol 63: 697–702. [DOI] [PubMed] [Google Scholar]
- Dalleau, S. , Cateau, E. , Berges, T. , Berjeaud, J.M. , and Imbert, C. (2008) In vitro activity of terpenes against Candida biofilms. Int J Antimicrob Agents 31: 572–576. [DOI] [PubMed] [Google Scholar]
- Davies, D.G. , and Marques, C.N.H. (2009) A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol 191: 1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defoirdt, T. (2018) Quorum‐sensing systems as targets for antivirulence therapy. Trends Microbiol 26: 313–328. [DOI] [PubMed] [Google Scholar]
- Desbois, A.P. , and Smith, V.J. (2010) Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl Microbiol Biotechnol 85: 1629–1642. [DOI] [PubMed] [Google Scholar]
- Donlan, R.M. (2002) Biofilms: microbial life on surfaces. Emerg Infect Dis 8: 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel, J.S. , and Mitchell, A.P. (2011) Genetic control of Candida albicans biofilm development. Nat Rev Microbiol 9: 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandclement, C. , Tannieres, M. , Morera, S. , Dessaux, Y. , and Faure, D. (2016) Quorum quenching: role in nature and applied developments. FEMS Microbiol Rev 40: 86–116. [DOI] [PubMed] [Google Scholar]
- Hall‐Stoodley, L. , Costerton, J.W. , and Stoodley, P. (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2: 95–108. [DOI] [PubMed] [Google Scholar]
- Handorf, O. , Schnabel, U. , Bosel, A. , Weihe, T. , Bekeschus, S. , Graf, A.C. , et al. (2019) Antimicrobial effects of microwave‐induced plasma torch (MiniMIP) treatment on Candida albicans biofilms. Microb Biotechnol 12: 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn, A. , Nielsen, A.T. , Hentzer, M. , Sternberg, C. , Givskov, M. , Ersboll, B.K. , and Molin, S. (2000) Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146: 2395–2407. [DOI] [PubMed] [Google Scholar]
- Hornby, J.M. , Jensen, E.C. , Lisec, A.D. , Tasto, J.J. , Jahnke, B. , Shoemaker, R. , et al. (2001) Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol 67: 2982–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby, J.M. , and Nickerson, K.W. (2004) Enhanced production of farnesol by Candida albicans treated with four azoles. Antimicrob Agents Chemother 48: 2305–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, C.C. , Lai, W.L. , Chuang, K.C. , Lee, M.H. , and Tsai, Y.C. (2013) The inhibitory activity of linalool against the filamentous growth and biofilm formation in Candida albicans . Med Mycol 51: 473–482. [DOI] [PubMed] [Google Scholar]
- Inoue, T. , Shingaki, R. , and Fukui, K. (2008) Inhibition of swarming motility of Pseudomonas aeruginosa by branched‐chain fatty acids. FEMS Microbiol Lett 281: 81–86. [DOI] [PubMed] [Google Scholar]
- Jabra‐Rizk, M.A. , Meiller, T.F. , James, C.E. , and Shirtliff, M.E. (2006) Effect of farnesol on Staphylococcus aureus biofilm formation and antimicrobial susceptibility. Antimicrob Agents Chemother 50: 1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia, V.C. (2013) Quorum sensing inhibitors: an overview. Biotechnol Adv 31: 224–245. [DOI] [PubMed] [Google Scholar]
- Kenar, J.A. , Moser, B.R. , and List, G.R. (2017) Chapter 2 ‐ Naturally occurring fatty acids: source, chemistry, and uses. In Fatty Acids. Ahmad, M.U. (ed). AOCS Press, pp. 23–82. [Google Scholar]
- Kim, Y.‐G. , Lee, J.‐H. , Gwon, G. , Kim, S.‐I. , Park, J.G. , and Lee, J. (2016) Essential oils and eugenols inhibit biofilm formation and the virulence of Escherichia coli O157:H7. Sci Rep 6: 36377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.‐G. , Lee, J.‐H. , Park, J.G. , and Lee, J. (2020) Inhibition of Candida albicans and Staphylococcus aureus biofilms by centipede oil and linoleic acid. Biofouling 36: 126–137. [DOI] [PubMed] [Google Scholar]
- Kim, Y.‐G. , Lee, J.‐H. , Raorane, C.J. , Oh, S.T. , Park, J.G. , and Lee, J. (2018) Herring oil and omega fatty acids inhibit Staphylococcus aureus biofilm formation and virulence. Front Microbiol 9: 1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, C.L. , Sam, C.K. , Yin, W.F. , Tan, L.Y. , Krishnan, T. , Chong, Y.M. , and Chan, K.G. (2013) Plant‐derived natural products as sources of anti‐quorum sensing compounds. Sensors 13: 6217–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, P. , Lee, J.‐H. , Beyenal, H. , and Lee, J. (2020) Fatty acids as antibiofilm and antivirulence agents. Trends Microbiol 28: 753–768. [DOI] [PubMed] [Google Scholar]
- Lee, J.‐H. , Cho, M.H. , and Lee, J. (2011) 3‐Indolylacetonitrile decreases Escherichia coli O157:H7 biofilm formation and Pseudomonas aeruginosa virulence. Environ Microbiol 13: 62–73. [DOI] [PubMed] [Google Scholar]
- Lee, J.‐H. , Kim, Y.‐G. , Choi, P. , Ham, J. , Park, J.G. , and Lee, J. (2018) Antibiofilm and antivirulence activities of 6‐gingerol and 6‐shogaol against Candida albicans due to hyphal inhibition. Front Cell Infect Microbiol 8: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima, T.M. , Kanunfre, C.C. , Pompeia, C. , Verlengia, R. , and Curi, R. (2002) Ranking the toxicity of fatty acids on Jurkat and Raji cells by flow cytometric analysis. Toxicol In Vitro 16: 741–747. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , and Filler, S.G. (2011) Candida albicans als3, a multifunctional adhesin and invasin. Eukaryot Cell 10: 168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, R.H. , Shang, Z.C. , Li, T.X. , Yang, M.H. , and Kong, L.Y. (2017) In vitro antibiofilm activity of eucarobustol E against Candida albicans . Antimicrob Agents Chemother 61: e02707–02716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzyn, A. , Krasowska, A. , Stefanowicz, P. , Dziadkowiec, D. , and Łukaszewicz, M. (2010) Capric acid secreted by S. boulardii inhibits C. albicans filamentous growth, adhesion and biofilm formation. PLoS One 5: e12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthamil, S. , Balasubramaniam, B. , Balamurugan, K. , and Pandian, S.K. (2018) Synergistic effect of quinic acid derived from syzygium cumini and undecanoic acid against Candida spp. biofilm and virulence. Front Microbiol 9: 2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthamil, S. , Prasath, K.G. , Priya, A. , Precilla, P. , and Pandian, S.K. (2020) Global proteomic analysis deciphers the mechanism of action of plant derived oleic acid against Candida albicans virulence and biofilm formation. Sci Rep 10: 5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, K.W. , Atkin, A.L. , Hargarten, J.C. , Pathirana, R. , and Hasim, S. (2013) Thoughts on quorum sensing and fungal dimorphism. In Biocommunication of Fungi. Dordrecht, Netherlands: Springer, pp. 189–204. [Google Scholar]
- Nigam, S. , Ciccoli, R. , Ivanov, I. , Sczepanski, M. , and Deva, R. (2011) On mechanism of quorum sensing in Candida albicans by 3(R)‐hydroxy‐tetradecaenoic acid. Curr Microbiol 62: 55–63. [DOI] [PubMed] [Google Scholar]
- Nobile, C.J. , Andes, D.R. , Nett, J.E. , Smith, F.J. , Yue, F. , Phan, Q.T. , et al. (2006a) Critical role of Bcr1‐dependent adhesins in C. albicans biofilm formation in vitro and in vivo . PLoS Pathog 2: e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile, C.J. , Nett, J.E. , Andes, D.R. , and Mitchell, A.P. (2006b) Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell 5: 1604–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr, M.C. , and Huffnagle, G.B. (2004) Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect Immun 72: 6206–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira, D.B.C. , Silva, L.B. , da Silva, B.V. , Borges, T.C. , Marques, B.C. , Dos Santos, M.B. , et al. (2019) A new acridone with antifungal properties against Candida spp. and dermatophytes, and antibiofilm activity against C. albicans . J Appl Microbiol 127: 1362–1372. [DOI] [PubMed] [Google Scholar]
- Orsi, C.F. , Borghi, E. , Colombari, B. , Neglia, R.G. , Quaglino, D. , Ardizzoni, A. , et al. (2014) Impact of Candida albicans hyphal wall protein 1 (HWP1) genotype on biofilm production and fungal susceptibility to microglial cells. Microb Pathog 69–70: 20–27. [DOI] [PubMed] [Google Scholar]
- Pandin, C. , Le Coq, D. , Canette, A. , Aymerich, S. , and Briandet, R. (2017) Should the biofilm mode of life be taken into consideration for microbial biocontrol agents? Microb Biotechnol 10: 719–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan, Q.T. , Myers, C.L. , Fu, Y. , Sheppard, D.C. , Yeaman, M.R. , Welch, W.H. , et al. (2007) Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol 5: 543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polke, M. , Leonhardt, I. , Kurzai, O. , and Jacobsen, I.D. (2018) Farnesol signalling in Candida albicans ‐ more than just communication. Crit Rev Microbiol 44: 230–243. [DOI] [PubMed] [Google Scholar]
- Prasath, K.G. , Sethupathy, S. , and Pandian, S.K. (2019) Proteomic analysis uncovers the modulation of ergosterol, sphingolipid and oxidative stress pathway by myristic acid impeding biofilm and virulence in Candida albicans . J Proteomics 208: 103503. [DOI] [PubMed] [Google Scholar]
- Pukkila‐Worley, R. , Peleg, A.Y. , Tampakakis, E. , and Mylonakis, E. (2009) Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot Cell 8: 1750–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage, G. , Saville, S.P. , Thomas, D.P. , and Lopez‐Ribot, J.L. (2005) Candida biofilms: an update. Eukaryot Cell 4: 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage, G. , Saville, S.P. , Wickes, B.L. , and Lopez‐Ribot, J.L. (2002) Inhibition of Candida albicans biofilm formation by farnesol, a quorum‐sensing molecule. Appl Environ Microbiol 68: 5459–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan, S. , Ravindran, D. , Arunachalam, K. , and Arumugam, V.R. (2018) Inhibition of quorum sensing‐dependent biofilm and virulence genes expression in environmental pathogen Serratia marcescens by petroselinic acid. Antonie Van Leeuwenhoek 111: 501–515. [DOI] [PubMed] [Google Scholar]
- Rasmussen, T.B. , Manefield, M. , Andersen, J.B. , Eberl, L. , Anthoni, U. , Christophersen, C. , et al. (2000) How Delisea pulchra furanones affect quorum sensing and swarming motility in Serratia liquefaciens MG1. Microbiology 146: 3237–3244. [DOI] [PubMed] [Google Scholar]
- Santhakumari, S. , Nilofernisha, N.M. , Ponraj, J.G. , Pandian, S.K. , and Ravi, A.V. (2017) In vitro and in vivo exploration of palmitic acid from Synechococcus elongatus as an antibiofilm agent on the survival of Artemia franciscana against virulent vibrios. J Invertebr Pathol 150: 21–31. [DOI] [PubMed] [Google Scholar]
- Sardi, J.C. , Scorzoni, L. , Bernardi, T. , Fusco‐Almeida, A.M. , and Mendes Giannini, M.J. (2013) Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J Med Microbiol 62: 10–24. [DOI] [PubMed] [Google Scholar]
- Shareck, J. , Nantel, A. , and Belhumeur, P. (2011) Conjugated linoleic acid inhibits hyphal growth in Candida albicans by modulating Ras1p cellular levels and downregulating TEC1 expression. Eukaryot Cell 10: 565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchepin, R. , Hornby, J.M. , Burger, E. , Niessen, T. , Dussault, P. , and Nickerson, K.W. (2003) Quorum sensing in Candida albicans: Probing farnesol's mode of action with 40 natural and synthetic farnesol analogs. Chem Biol 10: 743–750. [DOI] [PubMed] [Google Scholar]
- Shi, D. , Zhao, Y. , Yan, H. , Fu, H. , Shen, Y. , Lu, G. , et al. (2016) Antifungal effects of undecylenic acid on the biofilm formation of Candida albicans . Int J Clin Pharmacol Ther 54: 343–353. [DOI] [PubMed] [Google Scholar]
- Shirtliff, M.E. , Krom, B.P. , Meijering, R.A.M. , Peters, B.M. , Zhu, J. , Scheper, M.A. , et al. (2009) Farnesol‐induced apoptosis in Candida albicans . Antimicrob Agents Chemother 53: 2392–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Cde, B. , Guterres, S.S. , Weisheimer, V. , and Schapoval, E.E. (2008) Antifungal activity of the lemongrass oil and citral against Candida spp. Braz J Infect Dis 12: 63–66. [DOI] [PubMed] [Google Scholar]
- Singh, B.N. , Upreti, D.K. , Singh, B.R. , Pandey, G. , Verma, S. , Roy, S. , et al. (2015) Quercetin sensitizes fluconazole‐resistant Candida albicans to induce apoptotic cell death by modulating quorum sensing. Antimicrob Agents Chemother 59: 2153–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S.H. , Sun, X.Y. , Meng, L.L. , Wu, Q.H. , Wang, K. , and Deng, Y.Y. (2020) Antifungal activity of hypocrellin compounds and their synergistic effects with antimicrobial agents against Candida albicans . Microb Biotechnol, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tampakakis, E. , Okoli, I. , and Mylonakis, E. (2008) A C. elegans‐based, whole animal, in vivo screen for the identification of antifungal compounds. Nat Protoc 3: 1925–1931. [DOI] [PubMed] [Google Scholar]
- Teplitski, M. , Robinson, J.B. , and Bauer, W.D. (2000) Plants secrete substances that mimic bacterial N‐acyl homoserine lactone signal activities and affect population density‐dependent behaviors in associated bacteria. Mol Plant Microbe In 13: 637–648. [DOI] [PubMed] [Google Scholar]
- Thibane, V.S. , Kock, J.L.F. , Ells, R. , van Wyk, P.W.J. , and Pohl, C.H. (2010) Effect of marine polyunsaturated fatty acids on biofilm formation of Candida albicans and Candida dubliniensis . Mar Drugs 8: 2597–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traul, K.A. , Driedger, A. , Ingle, D.L. , and Nakhasi, D. (2000) Review of the toxicologic properties of medium‐chain triglycerides. Food Chem Toxicol 38: 79–98. [DOI] [PubMed] [Google Scholar]
- Vílchez, R. , Lemme, A. , Ballhausen, B. , Thiel, V. , Schulz, S. , Jansen, R. , et al. (2010) Streptococcus mutans inhibits Candida albicans hyphal formation by the fatty acid signaling molecule trans‐2‐decenoic acid (SDSF). ChemBioChem 11: 1552–1562. [DOI] [PubMed] [Google Scholar]
- Weber, K. , Sohr, R. , Schulz, B. , Fleischhacker, M. , and Ruhnke, M. (2008) Secretion of E, E‐farnesol and biofilm formation in eight different Candid a species. Antimicrob Agents Chemother 52: 1859–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weete, J.D. , Abril, M. , and Blackwell, M. (2010) Phylogenetic distribution of fungal sterols. PLoS One 5: e10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenderska, I.B. , Chong, M. , McNulty, J. , Wright, G.D. , and Burrows, L.L. (2011) Palmitoyl‐DL‐carnitine is a multitarget inhibitor of Pseudomonas aeruginosa biofilm development. ChemBioChem 12: 2759–2766. [DOI] [PubMed] [Google Scholar]
- Whaley, S.G. , Berkow, E.L. , Rybak, J.M. , Nishimoto, A.T. , Barker, K.S. , and Rogers, P.D. (2016) Azole antifungal resistance in Candida albicans and emerging non‐albicans Candida species. Front Microbiol 7: 2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, G.D. (2015) Solving the antibiotic crisis. ACS Infect Dis 1: 80–84. [DOI] [PubMed] [Google Scholar]
- Yoon, B.K. , Jackman, J.A. , Valle‐Gonzalez, E.R. , and Cho, N.J. (2018) Antibacterial free fatty acids and monoglycerides: Biological activities, experimental testing, and therapeutic applications. Int J Mol Sci 19: 1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L.H. , and Dong, Y.H. (2004) Quorum sensing and signal interference: diverse implications. Mol Microbiol 53: 1563–1571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Inhibitory effects of various fatty acids on C. albicans biofilm formation and planktonic growth. The antibiofilm activity of fatty acids against two C. albicans strains (DAY185 and ATCC 10231) in PDB medium was determined after culture for 24 h in 96‐well plates. For the biofilm assay, each fatty acid was used at 10 μg ml‐1. The MICs of each fatty acid against planktonic cells of C. albicans DAY185 are shown. Active fatty acids with low MIC are indicated in blue.
Table S2. Primer sequences used for qRT‐PCR.
Fig. S1. Effects of nonanoic acid (9:0) on C. albicans infected C. elegans. Nematode survival after exposure to C. albicans for 5 or 7 days in the presence of nonanoic acid (A). The toxicity of nonanoic acid was investigated by treating non‐infected nematodes for 5 or 7 days (B). Nonanoic acid rescued the C. elegans survival by inhibiting hyphal growth of C. albicans (C). None indicates non‐treated controls. Worm survival was determined based on movement. *P < 0.05 vs. non‐treated controls.
Fig. S2. Relative transcriptional profiles of ergosterol biosynthesis related genes in C. albicans cells treated with or without nonanoic acid (9:0) at 5 µg ml−1. C. albicans was incubated with or without nonanoic acid (9:0) at 5 μg ml−1 for 6 h without shaking. Transcriptional profiles were obtained by qRT‐PCR. RDN18 was a housekeeping gene.


