Mowat et al. show that anti-tumor immunity in dMMR CRCs relies on recruitment and retention of systemic CD8+ T cells. This is driven by extensive genomic instability that upregulates CCL5 and CXCL10 in the CRC cells by endogenously activating cGAS/STING and type I IFN signaling.
Abstract
Colorectal cancers (CRCs) deficient in DNA mismatch repair (dMMR) contain abundant CD8+ tumor-infiltrating lymphocytes (TILs) responding to the abundant neoantigens from their unstable genomes. Priming of such tumor-targeted TILs first requires recruitment of CD8+ T cells into the tumors, implying that this is an essential prerequisite of successful dMMR anti-tumor immunity. We have discovered that selective recruitment and activation of systemic CD8+ T cells into dMMR CRCs strictly depend on overexpression of CCL5 and CXCL10 due to endogenous activation of cGAS/STING and type I IFN signaling by damaged DNA. TIL infiltration into orthotopic dMMR CRCs is neoantigen-independent and followed by induction of a resident memory-like phenotype key to the anti-tumor response. CCL5 and CXCL10 could be up-regulated by common chemotherapies in all CRCs, indicating that facilitating CD8+ T cell recruitment underlies their efficacy. Induction of CCL5 and CXCL10 thus represents a tractable therapeutic strategy to induce TIL recruitment into CRCs, where local priming can be maximized even in neoantigen-poor CRCs.
Introduction
Colorectal cancer (CRC) is a prevalent and often fatal disease initiated by loss of either the tumor suppressor APC or the DNA mismatch repair gene MLH1 (Kloor and von Knebel Doeberitz, 2016; Picard et al., 2020). Whereas APC inactivation promotes large-scale DNA damage leading to chromosomal instability (CIN), MLH1 inactivation promotes hypermutability in the form of genome-wide point mutations and microsatellite instability (MSI) from deficient mismatch repair (dMMR). Although accounting for only 10–15% of CRCs, dMMR CRCs are of particular interest due to their potent immune-stimulating capacity and overall favorable patient outcome (Guinney et al., 2015). These positive features are commonly attributed to the high tumor mutation burden (TMB) induced by their hypermutability, which generates abundant neoantigens that can stimulate tumor-infiltrating lymphocytes (TILs) to attack (Sahan et al., 2018). Considerable effort is being invested in identifying immunogenic neoantigens in the genomes of dMMR CRCs, and there is no doubt that a high TMB contributes to immune activation by these CRCs (Chae et al., 2019; Maby et al., 2015; Spranger et al., 2016). However, identifying MSI CRC neoantigens is of limited therapeutic value to CIN CRC patients since there is little overlap in the neoantigens of these CRC subtypes due to their different underlying mutational signatures (Bonneville et al., 2017; Lim et al., 2017). Fortunately, the observation that dMMR tumors in other tissues do not have higher immunogenicity or a better prognosis indicates that a high TMB is neither necessary nor sufficient to explain these features in dMMR CRCs (Bonneville et al., 2017; Kim et al., 2020). There is thus a significant benefit to understanding the antigen-independent mechanisms of immune activation that are orchestrated in the colon microenvironment by dMMR CRCs and could be developed into therapies for a wider patient population.
Abundant TIL infiltration directly into the tumor epithelium is an independent, positive predictive factor in many cancers, including dMMR CRCs (Bindea et al., 2013; Galon and Bruni, 2020). While the clinical significance of this pattern of TIL distribution within a tumor is clear, the processes regulating it are poorly understood. Critical outstanding questions relate to the source of these TILs, their activation or exhaustion level, their specificity for tumor neoantigens, and the method of their recruitment. While systemic CD8+ T cell activation can occur in cancer, there is also substantial evidence that CD8+ TILs harboring a phenotype similar to resident memory T (TRM) cells are critical mediators of anti-tumor immunity (Low et al., 2020; Park et al., 2019; Wu et al., 2020). In addition to their localization within the diseased tissue and their inherently preprimed state, TRM cells express the appropriate surface receptors, such as CD103, for interacting with local epithelial cells (Low et al., 2020; Park et al., 2019). TILs in dMMR CRCs could thus arise from either an expansion of the resident intestinal intraepithelial lymphocyte population or recruitment of systemic T cells into the tumor. This is an important unresolved question since the former are inherently tolerized by their exposure to the suppressive mucosal immune environment and have a much higher activation threshold than CD8+ T cells recruited from systemic circulation (Cario, 2008; MacDonald et al., 2011). Furthermore, the mechanisms for recruiting these two immune populations will differ substantially.
Immune cell recruitment is regulated through a complex network of chemokines that guide cell trafficking throughout the body (Kistner et al., 2017; Nagarsheth et al., 2017). T cells in particular are responsive to a set of chemokines whose production is controlled by IFN signaling (Hubel et al., 2019; Schneider et al., 2014). In most cases, this is initiated by viral infection of a cell and detection of pathogenic nucleic acids by intracellular DNA- and RNA-sensing systems such as cGAS/STING, AIM2, and TLR3 (Schlee and Hartmann, 2016). Numerous reports indicate that dMMR CRCs up-regulate IFN-stimulated genes (ISGs), yet what role this plays in TIL infiltration remains unknown (Llosa et al., 2015; Vanpouille-Box et al., 2018; Lu et al., 2020; Guan et al., 2021). Additionally, many of these reports have drawn their conclusions from gene expression analysis of whole tissue transcripts without correcting for the disproportionately higher amount of immune cells in the dMMR CRCs (Boissière-Michot et al., 2014; Russo et al., 2019). Since immune cells express high levels of IFNs and ISGs, it is unclear if the dMMR CRC cells themselves or the infiltrated immune cells are the primary source of high IFN signaling in the unique in vivo setting of the intestine.
In the current study, we used an orthotopic model with isogenic CRC cells differing in their MMR capacity to study how loss of the DNA mismatch repair gene MLH1 regulates antigen-independent activation and recruitment of CD8+ T cells. This was driven by overexpression of the IFN-dependent chemokines CCL5 and CXCL10, which led to preferential recruitment and retention of systemic, not local, CD8+ T cells into the tumor epithelium. While activation of this immunogenic gene signature is endogenous in dMMR CRCs, it can be exogenously induced in CIN CRCs via treatment with type I IFN or DNA-damaging chemo- and radiotherapies. These findings identify potential therapeutic targets for inducing T cell infiltration into CIN CRCs. Given that immune recruitment is necessary for subsequent antigen-specific T cell–mediated killing, this enhanced T cell infiltration will increase the likelihood of TIL activation even by tumors with few neoantigens.
Results
Mismatch repair deficiency in CRC induces a chemokine signature associated with CD8+ T cell recruitment
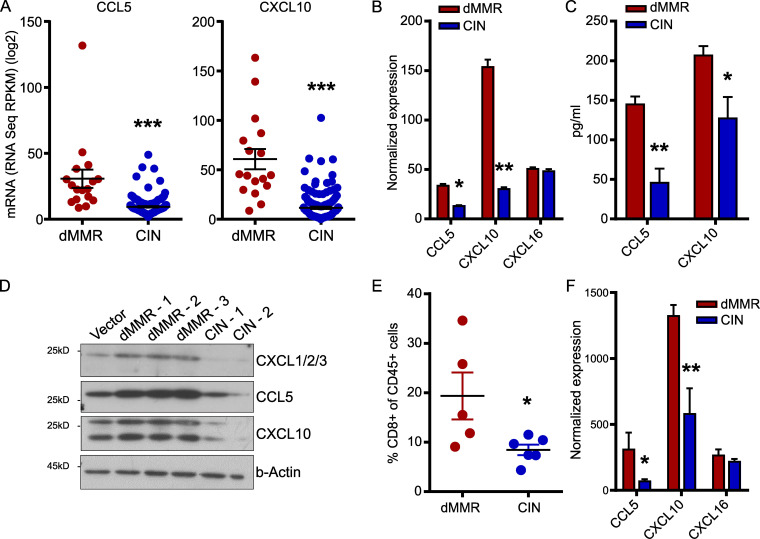
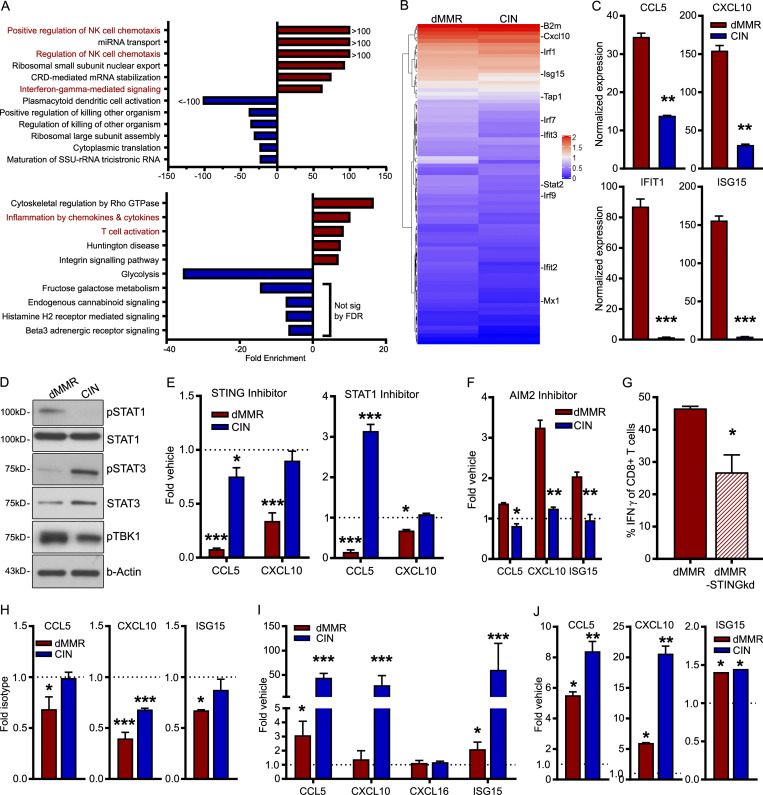
Immune-associated genes are well-known to be overexpressed in dMMR CRC, but few studies have investigated the functional significance of these genes for the anti-tumor immune response (Guinney et al., 2015; Picard et al., 2020). Furthermore, most reports are based on whole-tissue transcript analysis, making it difficult to determine the relative contribution of the CRC cells themselves to the expression of immune genes in the tissue (Russo et al., 2019). To better understand the factors governing TIL recruitment into dMMR CRCs, we examined expression of various chemokines in data obtained from The Cancer Genome Atlas (TCGA) PanCancer Atlas dataset (Hoadley et al., 2018). We noted that two chemokines known to regulate CD8+ T cell trafficking, CCL5 and CXCL10, were more highly expressed in dMMR CRCs than their CIN counterparts (Fig. 1 A; Boissière-Michot et al., 2014; Zumwalt et al., 2015). To determine if the dMMR CRC cells themselves were the primary source of these chemokines, we generated dMMR and CIN variants of the murine MC38 CRC cell line by mutating Mlh1 or the CIN-associated gene Kras, respectively (Fig. S1, A and B). This led to the consistent up-regulation of chemokines CCL5 and CXCL10 specifically in the dMMR CRC cells (Fig. 1, B–D). To ensure that the dMMR and CIN MC38 cells accurately modeled their human CRC counterparts, we used fluorescence-based reporter plasmids to verify that the dMMR cells specifically gained instability in repetitive microsatellites but did not develop more double-strand breaks (Fig. S1, C and D). We then subcutaneously injected the cells into the flanks of immunocompetent WT C57BL/6 mice to examine the immune profile of the resulting tumors. Consistent with human CRC types, dMMR tumors contained greater numbers of CD8+ T cells than CIN tumors (Fig. 1 E). Furthermore, whole tissue transcript analysis confirmed that the dMMR MC38 CRC tumors expressed greater amounts of CCL5 and CXCL10 compared with the CIN MC38 CRC tumors (Fig. 1 F). In contrast, dMMR CRCs did not express more CXCL16, which is not associated with CD8+ T cell recruitment (Fig. 1, B and F). These data indicate that MMR loss in CRC cells up-regulates their expression of CCL5 and CXCL10, both of which can attract CD8+ T cells, and suggest that directly regulating TIL recruitment is an important component of dMMR CRC immunogenicity.
Figure 1.
Mismatch repair deficiency in CRC induces a chemokine signature associated with CD8+ T cell recruitment.(A) Chemokine expression in dMMR CRCs and CIN CRCs from the TCGA PanCancer Atlas. (B) dMMR and CIN models of the MC38 mouse CRC cell line were created by mutating Mlh1 and Kras, respectively. Chemokine expression in the cell lines was assessed by qPCR. n = 3 repeats. (C and D) Chemokine secretion was analyzed by cytokine bead array of the cell supernatants (C) or Western blotting (D) of lysates from three different clones of dMMR MC38 cells and two different clones of CIN MC38 CRC cells. Clones 1 were used in all subsequent experiments. n = 3 repeats each. (E) Infiltration of CD8+CD3+ T cells into subcutaneously injected dMMR and CIN MC38 CRC tumors was assessed by flow cytometry. n ≥ 4 mice per group, four repeats. (F) Chemokine expression was assessed by qPCR on whole tissue transcripts isolated from subcutaneous dMMR and CIN CRCs. n ≥ 4 mice per group, four repeats. dMMR versus CIN: *, P ≤ 0.05; **, P ≤ 0.01; *** P, ≤ 0.005. RPKM, reads per kilobase million.
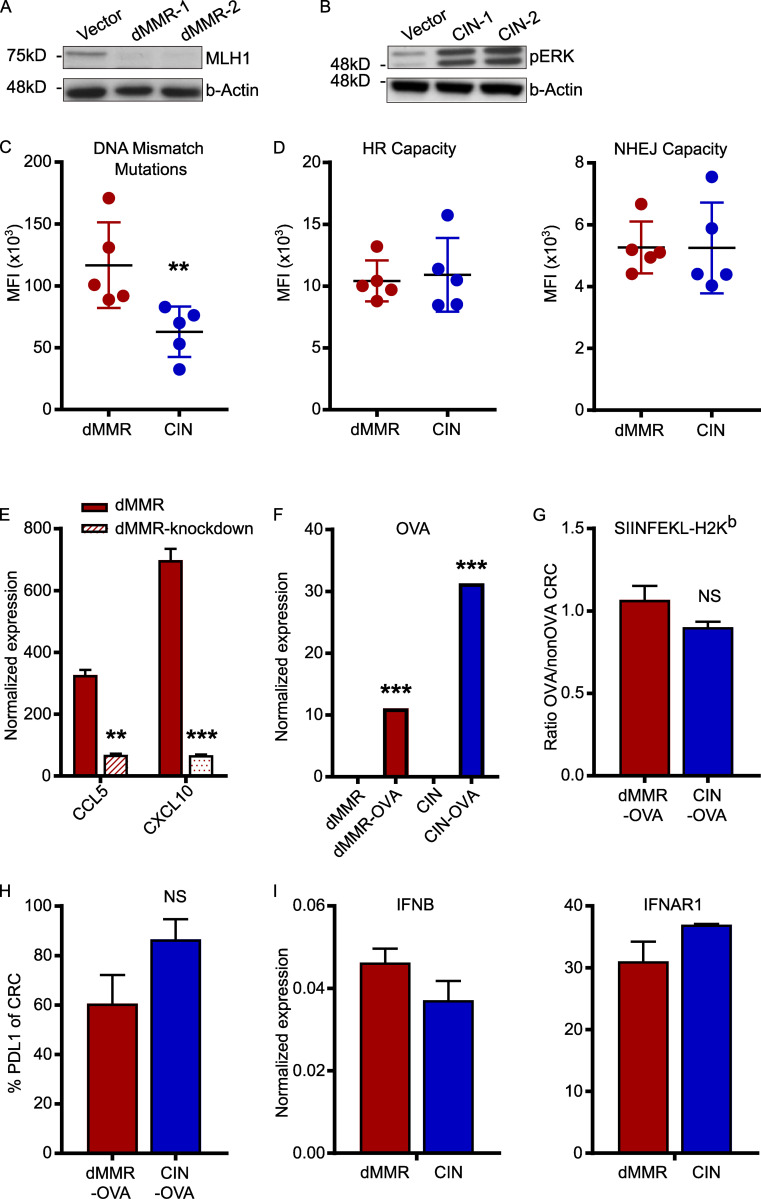
Figure S1.
Confirmation of successful generation of dMMR and CIN MC38 CRC cell line variants. dMMR and CIN models of the MC38 mouse CRC cell line were created by mutating Mlh1 and Kras, respectively. (A and B) Protein expression was analyzed by Western blotting lysates from two different clones of dMMR MC38 cells and two different clones of CIN MC38 CRC cells. Clones 1 were used in all subsequent experiments. n = 3 repeats. (C) Microsatellite instability was measured using a reporter plasmid where uncorrected deletion and insertion mutations in microsatellite regions of a promoter enable expression of β-galactosidase. Cells were transiently transfected with the reporter plasmid, and β-galactosidase expression was measured after 24 h by flow cytometry using a fluorescence-based readout. Increasing fluorescence corresponds to increased microsatellite instability due to the failure of DNA mismatch repair to correct insertions and deletions. (D) HR and NHEJ were measured using a fluorescent reporter assay. Cells were transiently transfected with the reporter plasmids and the amount of HR and NHEJ were quantified by flow cytometry after 24 h. Increasing fluorescence corresponds to increased DNA repair by each mechanism. (E) Depletion of CCL5 and CXCL10 in dMMR-CCL5kd and CXCL10kd cells following stable transduction with shRNA plasmids was verified by qPCR. (F) Expression of OVA was verified in stable clones of dMMR-OVA and CIN-OVA using qPCR. (G) Surface expression of MHC-I SIINFEKL-H-2Kb was verified in dMMR-OVA and CIN-OVA CRC cells using flow cytometry. (H) Surface expression of PDL1 on dMMR and CIN CRC was measured with flow cytometry. (I) Expression of IFNB and IFNAR1 in untreated dMMR and CIN CRC cells using qPCR. All panels: representative data from n = 3–5 repeats. dMMR versus indicated bar. **, P ≤ 0.01; ***, P ≤ 0.001. MFI, mean fluorescence intensity.
Overexpression of CCL5 and CXCL10 by dMMR CRC cells is essential for recruitment and activation of systemic CD8+ T cells but not for neoantigen-targeted cytotoxicity
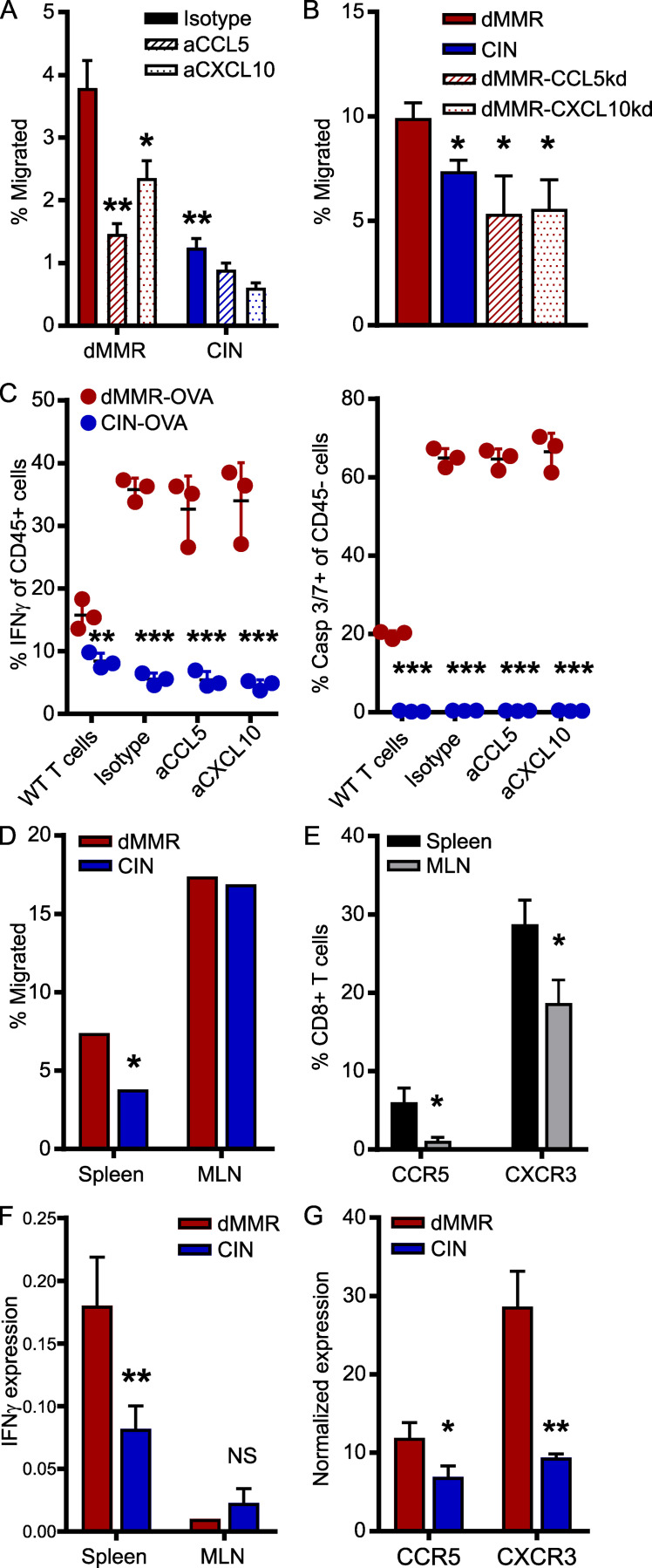
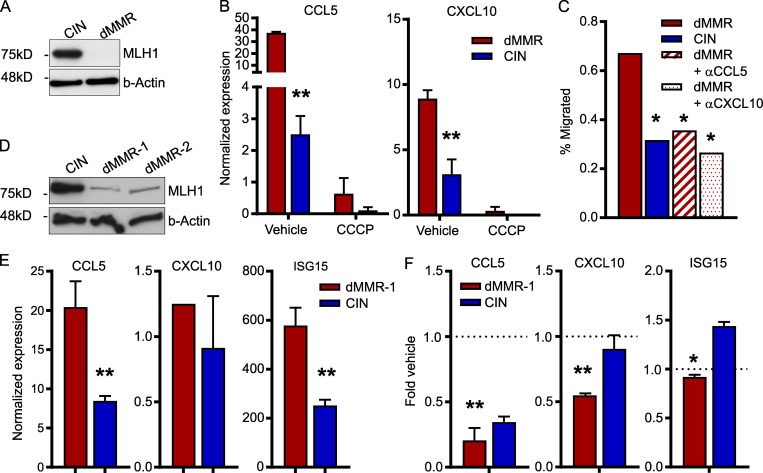
Although abundant infiltration of the dMMR CRC tumor epithelium by CD8+ TILs is a defining feature of these CRCs, the origin of these TILs and their mechanism of recruitment remain unknown (Baker et al., 2007; Kloor and von Knebel Doeberitz, 2016). Of particular importance is determining if they originate from the systemic pool of CD8+ T cells or from the resident pool of immunologically tolerized intestinal T cells. In addition to their different activation states, these cells have fundamentally different recruitment mechanisms (Low et al., 2020; Park et al., 2019; Wu et al., 2020). We first determined if the CCL5 and CXCL10 produced by dMMR CRCs could recruit systemic CD8+ T cells using Transwell migration assays. Greater numbers of CD8+ T cells migrated toward supernatant conditioned by dMMR CRC cells than by CIN CRC cells, and this was significantly inhibited with blocking antibodies against either CCL5 or CXCL10 (Fig. 2 A). We confirmed the importance of CCL5 and CXCL10 in CD8+ T cell recruitment by knocking down CCL5 and CXCL10 in the dMMR MC38 cells. Supernatant from these chemokine-deficient cells attracted significantly fewer CD8+ T cells than supernatant from the original dMMR CRC cells (Fig. 2 B and Fig. S1 E). However, cytotoxicity assays with OTI T cells and OVA-transfected dMMR and CIN CRCs revealed no role for CCL5 or CXCL10 in T cell activation or antigen-specific tumor cell killing (Fig. 2 C). Notably, these experiments did confirm that dMMR CRCs induce both greater antigen-dependent and antigen-independent CD8+ T cell activation since both OTI and WT T cells were preferentially activated by dMMR CRC cells despite the CRC subtypes expressing equivalent total OVA and surface SIINFEKL-H-2Kb MHC-I complexes (Fig. S1, F–H). To further explore the origin site and activation mechanism of TILs by dMMR CRCs, we investigated how effectively dMMR and CIN MC38 CRC cells could induce migration of CD8+ T cells from different sources across a Transwell membrane. Whereas splenic CD8+ T cells were more attracted by conditioned media from dMMR CRC cells than media conditioned by CIN CRC cells, CD8+ T cells derived from the mesenteric lymph nodes (MLNs), which are the source of intestinal CD8+ T cells, were equally attracted by both tumor types (Fig. 2 D). This is consistent with our finding that splenic CD8+ T cells express more CCR5 and CXCR3, the receptors for CCL5 and CXCL10, respectively, than MLN CD8+ T cells (Fig. 2 E). Since both our in vivo experiments and patient data indicated that TILs in dMMR CRCs are more activated than those in CIN CRCs, we investigated if dMMR CRCs are superior at stimulating CD8+ T cells from each of these locations. After coculturing splenic or MLN CD8+ T cells with the CRC cells for 24 h, only the splenic CD8+ T cells were preferentially activated by dMMR CRC cells (Fig. 2 F). We further noted that dMMR CRCs can directly induce expression of CCR5 and CXCR3 on CD8+ T cells, which may facilitate both their initial recruitment and subsequent retention (Fig. 2 G). Collectively, these data suggest that the abundant TILs in dMMR CRCs result from selective recruitment and activation of systemic CD8+ T cells in a CCL5- and CXCL10-dependent manner. This finding highlights the fact that dMMR CRCs use both neoantigen-dependent and -independent mechanisms to activate CD8+ T cell–mediated anti-tumor immunity.
Figure 2.
CCL5 and CXCL10 expression is essential for the recruitment and activation of systemic CD8+ T cells by dMMR CRCs.(A) CD8+ T cell migration through a Matrigel-coated 5.0-µm Transwell insert toward supernatant conditioned for 24 h by dMMR or CIN MC38 CRCs. Anti-CCL5, CXCL10, or isotype control antibodies were added at 2.0 µg/ml 30 min before T cells were added. Migrated cells were quantified after 2 h by flow cytometry. n = 5 repeats. (B) CD8+ T cell migration toward supernatant conditioned by dMMR, CIN, or dMMR cells deficient in either CCL5 or CXCL10. n = 3 repeats. (C) OTI CD8+ T cells were cultured with OVA-transfected variants of dMMR and CIN CRC cells at a 5:1 ratio for 48 h. T cell activation was measured by intracellular IFN-γ staining, and cytotoxicity was measured by caspase (Casp) 3/7 cleavage. Anti-CCL5, CXCL10, or isotype control was added at 2.0 µg/ml 30 min before T cells were added. WT CD8+ T cells isolated from C57BL/6 mice were used as controls. Representative data from n = 3 repeats. (D) Migration of CD8+ T cells isolated from the spleen or MLNs toward supernatant conditioned by dMMR or CIN CRCs. Representative data from n = 4 repeats. (E) Baseline chemokine receptor expression on CD8+ T cells isolated from the spleen or MLN was assessed by flow cytometry directly after isolation. n ≥ 3 mice per group, four repeats. Spleen versus MLN: *, P ≤ 0.05. (F) Activation of CD8+ T cells isolated from the spleen or MLN and co-cultured at a 5:1 ratio with dMMR or CIN CRC cells for 24 h. CD8+ cells were isolated, and IFN-γ expression was quantified by qPCR. n = 3 repeats. (G) Expression of CCR5 and CXCR3 on CD8+ T cells co-cultured with dMMR or CIN CRC cells for 24 h. CD8+ cells were isolated, and IFN-γ expression was quantified by qPCR. n = 3 repeats. (A–E and G) dMMR versus indicated bar. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
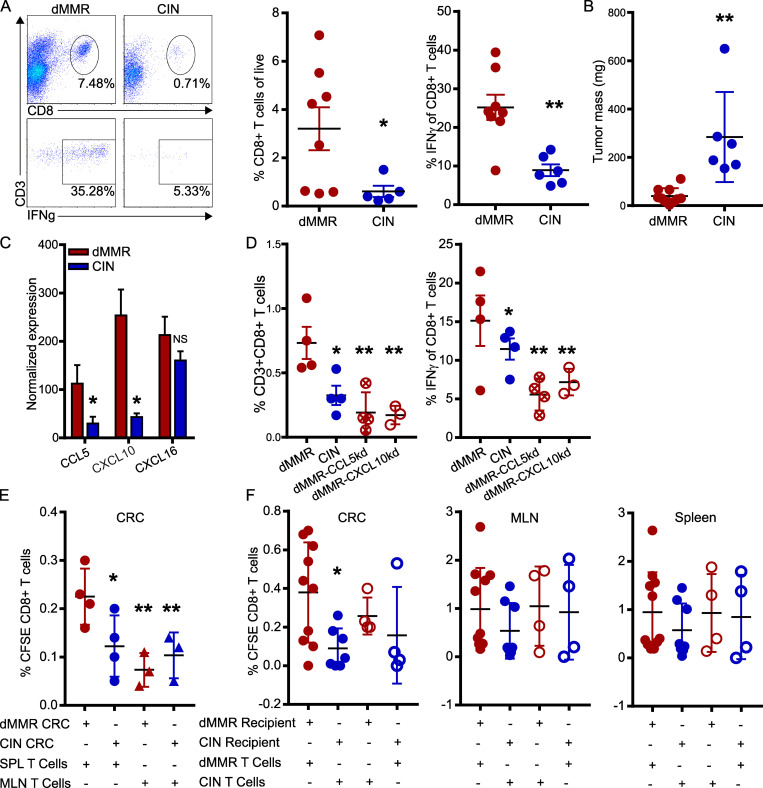
Orthotopic dMMR CRCs use CCL5 and CXCL10 to efficiently recruit systemic CD8+ TILs
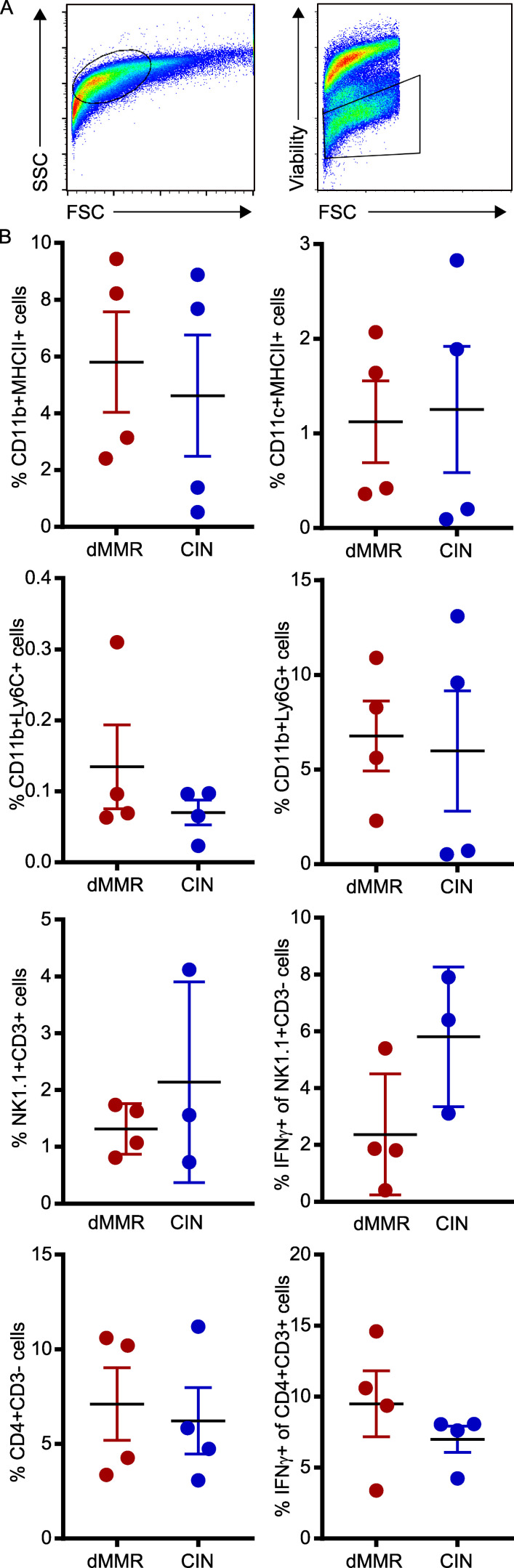
T cell migration is a complex process involving tissue homing factors, adhesion to extracellular matrices of varying composition and cell–cell interactions. The only way to study such a complex process is using in vivo models where tumors grow in their endogenous tissue environment. We thus orthotopically implanted the dMMR and CIN MC38 CRC cells directly into the colonic wall of WT C57BL/6 mice via noninvasive endoscopic injection in order to observe immune cell infiltration in tumors growing in their native environment (Roper et al., 2018; Zhao et al., 2017). dMMR CRC tumors were consistently smaller than CIN CRCs and contained both more total CD8+ TILs and more IFN-γ+ activated TILs than their CIN counterparts (Fig. 3, A and B). As reported for CRC patient tissues, transcript analysis of whole tumor tissue indicated that dMMR orthotopic CRCs expressed more CCL5 and CXCL10, but not CXCL16, than did their CIN counterparts (Fig. 3 C). We did not observe differences in tumor infiltration by, or activation of, macrophages, granulocytes, natural killer cells, or CD4+ T cells, suggesting that differential recruitment by dMMR CRC was limited to CD8+ T cells and could be orchestrated by the tumor cells themselves (Fig. S2). We furthermore confirmed the importance of CCL5 and CXCL10 for recruiting CD8+ TILs into dMMR CRCs by orthotopically injecting CCL5- and CXCL10-deficient dMMR CRC cells. These chemokine-deficient tumors contained significantly fewer CD8+ T cells, and far fewer of these expressed IFN-γ, indicating a lower activation potential in the absence of these chemokines (Fig. 3 D). To gauge the relative importance of TIL recruitment versus TIL activation in dMMR-mediated anti-tumor immunity, we isolated CD8+ T cells from the spleens or MLNs of mice bearing either dMMR or CIN orthotopic CRCs, labeled them with CFSE, and adoptively transferred them into recipient mice with orthotopic dMMR or CIN CRCs. 48 h later, tumors in dMMR CRC-bearing recipients had recruited more spleen-derived, but not MLN-derived, TILs regardless of whether they were primed in dMMR or CIN donor mice (Fig. 3 E). This clearly highlights the superior ability of dMMR CRCs to recruit systemic CD8+ T cells regardless of their antigen specificity. Although it failed to reach statistical significance, we consistently observed that more TILs from dMMR than from CIN donors were recruited into dMMR CRCs, indicating that systemic CD8+ T cells primed by dMMR tumors also have a superior ability to home to CRCs than do those primed by CIN CRCs (Fig. 3 F).
Figure 3.
Orthotopically implanted dMMR CRCs use CCL5 and CXCL10 to efficiently recruit systemic CD8+ TILs.(A) CD8+ T cell infiltration and activation in orthotopic tumors 14–21 d after injecting 1.5 × 105 CRC cells in 50 µl PBS into the colon wall of WT mice using an endoscope. n ≥ 4 mice per group, five repeats. (B) Mass of orthotopic dMMR and CIN CRCs. (C) Chemokine expression in whole-tissue transcripts of orthotopic dMMR and CIN CRCs was analyzed by qPCR. (D) Infiltration and activation of CD8+ T cells in orthotopic dMMR, CIN, or dMMR cells deficient in either CCL5 or CXCL10. n = 4 mice per group, four repeats. (E and F) Mice with orthotopic CRCs were adoptively transferred i.v. with 2 × 106 CFSE-stained CD8+ T cells from the MLN (E) or spleens (SPL; E and F) of tumor-bearing donors. 48 h later, T cell infiltration was assessed by flow cytometry in the CRC (E and F), MLN, and spleens (F) of recipients. Pooled data from n = 3 repeats. All panels, dMMR versus indicated bar. *, P ≤ 0.05; **, P ≤ 0.01.
Figure S2.
Immune cell subset frequencies in orthotopically implanted dMMR and CIN CRC tumors. 1.5 × 105 CRC cells in 50 µl were nonsurgically injected into the colonic wall using an endoscope. n ≥ 4 mice per group, five repeats. (A) Initial gating strategy. (B) Immune cell subsets infiltrating the tumors were quantified by flow cytometry using the indicated markers. FSC, forward scatter; SSC, side scatter.
dMMR CRCs up-regulate TRM markers on infiltrating TILs
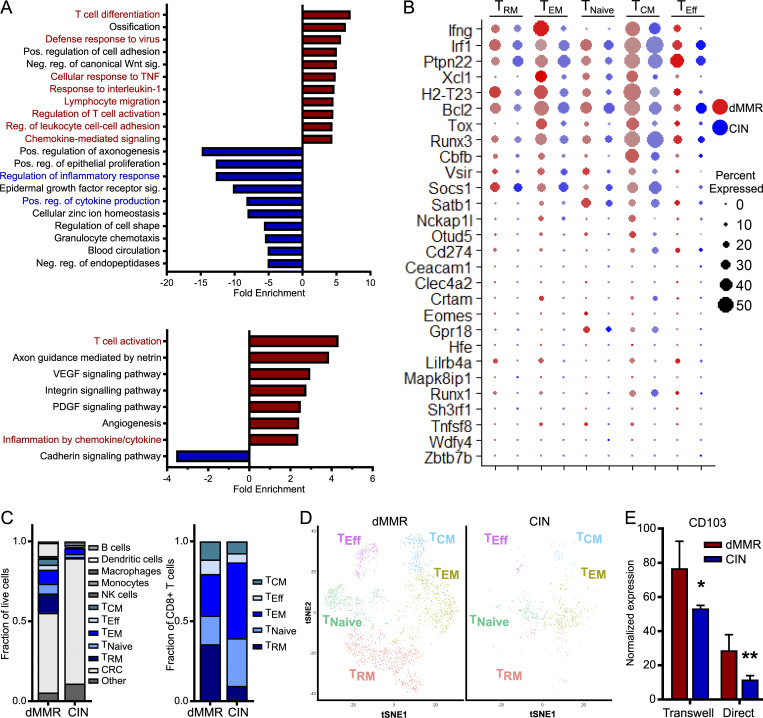
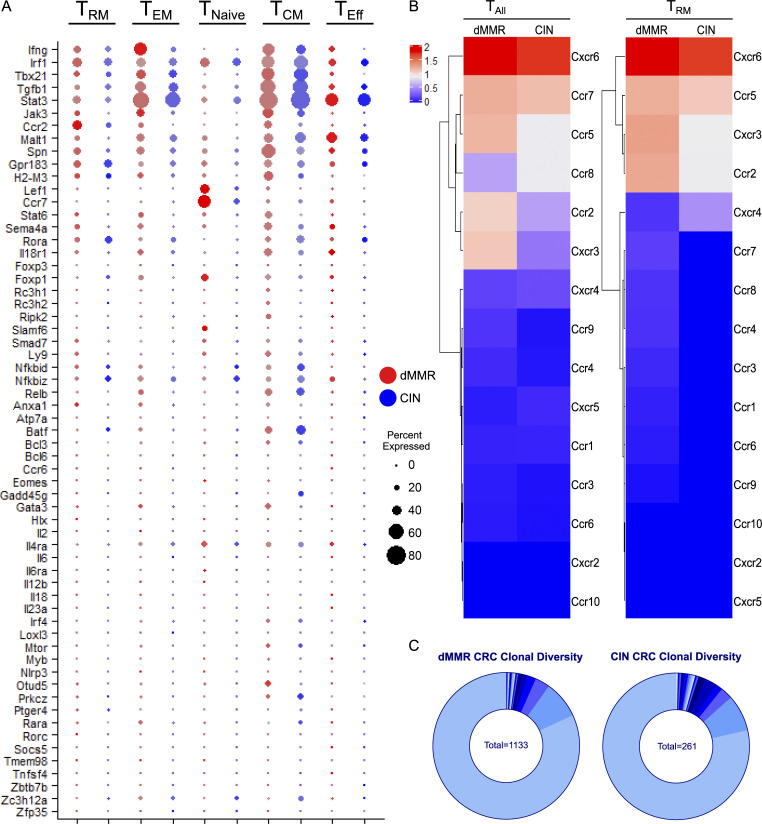
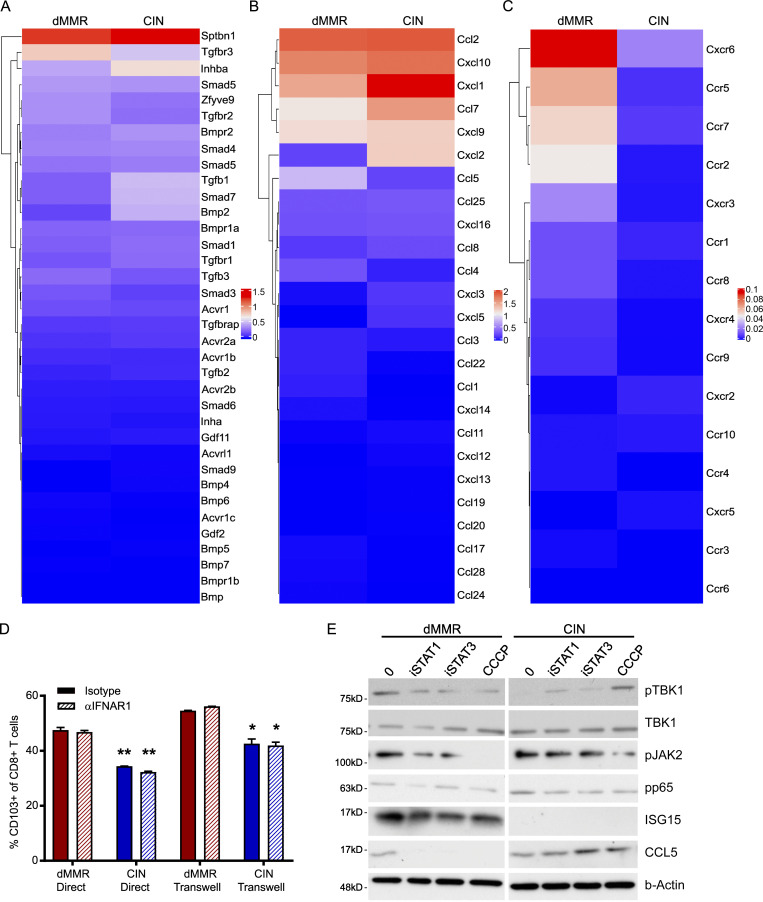
To better understand the complex interplay between dMMR CRCs and their immune microenvironment, we performed single-cell RNA sequencing (scRNAseq) on orthotopically grown dMMR and CIN CRCs. Gene ontology (GO) enrichment analysis indicated that CD8+ TILs within the dMMR CRCs were enriched in genes associated with T cell activation and chemokine-related signaling pathways in addition to several biological processes regulating chemotaxis (Fig. 4 A). Specifically, more CD8+ TILs in dMMR CRCs expressed genes in the GO terms lists for “CD8+ T cell activation” and “T cell activation” and often at a higher level than TILs in CIN CRCs (Fig. 4 B and Fig. S3 A). Significant differences were also noted in expression of chemokine receptors, notably more CCR5 and CXCR3, on CD8+ TILs in dMMR compared with CIN CRCs, which is consistent with our in vitro co-cultures (Fig. S3 B and Fig. 2 G). In addition to containing significantly more CD8+ TILs, dMMR tumors contained different proportions of TILs expressing markers associated with canonical T cell subsets (Fig. 4, C and D). Specifically, more TILs in dMMR CRCs expressed the CD103+CD69+ markers characteristic of TRM cells (Low et al., 2020; Park et al., 2019). Given that there was no difference in clonal diversity of the TILs in dMMR and CIN CRCs, these TILs did not arise through expansion of an oligoclonal pool of intestinal intraepithelial lymphocytes but rather through up-regulation of CD103 and CD69 following infiltration of systemic CD8+ T cells into the dMMR CRCs (Fig. S3 C). We confirmed the superior ability of dMMR CRCs to up-regulate CD103 on systemic CD8+ T cells by coculturing these two cell populations either in direct contact or separated by a 0.4-µm-pore-size Transwell membrane (Fig. 4 E). CD103 up-regulation did not require direct cell contact and appeared independent of TGFβ, the canonical inducer of CD103, since dMMR CRC cells did not express more TGFβ-associated genes (Fig. S4 A). Furthermore, blocking studies indicated that this up-regulation was independent of type I IFN signaling or production of CCL5 and CXCL10 by the dMMR CRCs (Fig. S4, B–D). Although further study is clearly needed to identify the underlying mechanisms, our results indicate that in addition to recruitment, retention of systemic CD8+ T cells by local up-regulation of TRM-associated genes is crucial to successful anti-tumor immunity in dMMR CRCs.
Figure 4.
Orthotopic dMMR CRCs up-regulate TRM markers on infiltrating systemic CD8+ T cells. scRNAseq was performed on orthotopically grown dMMR and CIN CRCs. Five mice were pooled from each CRC type. (A) GO enrichment analysis of the most enriched biological processes (top) and signaling pathways (bottom) for CD8+ T cells in orthotopic dMMR compared with CIN CRCs. (B) Expression of genes in the “CD8 T Cell Activation” GO term list. Size of the dot represents percent of cells and color intensity represents expression level. (C) Proportion of cell types identified by scRNAseq within the tumor microenvironment of orthotopic dMMR and CIN CRC (left) and within the CD8+ T cell subsets (right). TNaive, naive T cells; TEM, effector memory T cells; TEff, effector T cells; TCM, central memory T cells. (D) tSNE plots of CD8+ T cell subsets identified by scRNAseq in orthotopic dMMR and CIN CRCs. (E) Induction of CD103 in CD8+ T cells by dMMR and CIN CRCs was measured by qPCR after coculturing cells directly or separated by a 0.4-µm Transwell filter for 24 h. Representative data from n = 3 repeats. dMMR versus CIN: *, P ≤ 0.05; **, P ≤ 0.01. Neg., negative; Pos., positive; Reg., regulation; Sig., signaling.
Figure S3.
dMMR CRCs contain more activated TILs than CIN CRCs but are equally clonally diverse.(A) Expression of genes in the “T Cell Activation” GO term list for different subsets of CD8+ TILs in orthotopic dMMR and CIN MC38 tumors analyzed by scRNAseq. Size of the dot represents percent of CD8+ T cells and color intensity represents expression level. TNaive, naive T cells; TEM, effector memory T cells; TEff, effector T cells; TCM, central memory T cells. (B) Expression of chemokine receptors on tumor infiltrating CD8+ T cells in orthotopic CRCs. TAll, All CD8+ T cells. (C) TCR frequency in the CD8+ TILs from orthotopic tumors analyzed by scRNAseq. Each section of the arc represents the number of cells with the indicated frequency. No significant difference exists between the frequency distribution of dMMR and CIN CRC TILs.
Figure S4.
dMMR CRCs express more of select chemokines and chemokine receptors than CIN CRCs but similar levels of TGFβ-associated genes. (A–C) Expression of genes associated with the “TGFβ signaling” GO term gene (A), chemokines (B), and chemokine receptors (C) in orthotopic CRC cells. (D) Induction of CD103 on CD8+ T cells by dMMR and CIN CRCs was measured by flow cytometry after coculturing cells directly or separated by a 0.4-µm Transwell filter for 24 h. CRC cells were pretreated for 24 h by 10 µg/ml anti-IFNAR1 or isotype control. Representative data from n = 3 repeats. dMMR versus CIN of same treatment condition: *, P ≤ 0.05; **, P ≤ 0.01. (E) Dependence of chemokine endogenous signaling on cGAS/STING, STAT1, or STAT3 was assessed by treatment of CRC cells for 1 h with 10 µM CCCP, 10 µM fludarabine (iSTAT1), or 50 µM S3I-201 (iSTAT3), respectively. n = 3 repeats.
CCL5 and CXCL10 overexpression is driven by active endogenous type I IFN signaling in dMMR CRCs but can be exogenously induced in CIN CRCs
Chemokines CCL5 and CXCL10 are well-known members of the ISGs family, a group of genes induced by type I IFN and cGAS/STING signaling, and are most strongly expressed in immune cells (Hubel et al., 2019; Schneider et al., 2014). GO analysis from scRNAseq analysis of orthotopic CRCs, however, revealed that dMMR CRC cells themselves are enriched in genes involved in chemokine-related signaling pathways and the biological processes of lymphocyte migration and T cell activation (Fig. 5 A; Mi et al., 2019). dMMR CRC cells also differentially expressed IFNA pathway–associated genes (Fig. 5 B), a finding that we confirmed by quantitative PCR (qPCR) was inherent to the CRC cells and was accompanied by high endogenous expression of ISGs (Fig. 5 C). This indicated that knocking out DNA mismatch repair increased either the endogenous activation of type I IFN signaling pathways within the cancer cells or the sensitivity of the cells to exogenous IFN. Given that we had not noticed consistently different expression of type I IFN between our dMMR and CIN CRC cells (Fig. S1 I) and that there was not a significant source of exogenous IFN in the in vitro culture where differential CCL5 and CXCL10 gene expression were first noticed, we investigated the latter possibility first.
Figure 5.
CCL5 and CXCL10 production results from active endogenous type I IFN signaling in dMMR CRCs but can be exogenously induced in CIN CRCs.(A) GO enrichment analysis of the most enriched biological processes (top) and signaling pathways (bottom) in dMMR compared with CIN orthotopic CRC cells analyzed by scRNAseq. (B) Expression of genes associated with “IFNA Signaling” GO term gene list in orthotopically grown CRC cells. (C) Baseline expression of key ISGs was assessed by qPCR in unstimulated dMMR and CIN CRC MC38 cells grown in vitro. n = 4 repeats. dMMR versus CIN: **, P ≤ 0.01; ***, P ≤ 0.005. (D) Baseline activation of proteins in the IFN and cGAS/STING signaling pathways in dMMR and CIN CRC cells grown in vitro. n = 3 repeats. (E and F) Dependence of chemokine expression on cGAS/STING and STAT1 (E) or Aim2 (F) signaling was assessed by treatment of CRC cells for 24 h with 2 µM H-151, 10 µM fludarabine, or 9 µg/ml phosphorothioate oligo, respectively. n = 3 repeats. (G) Dependence of CD8+ T cell activation on STING signaling in dMMR CRC. OTI CD8+ T cells were co-cultured with SIINFEKL-pulsed dMMR STING knockdown (STINGkd) or control dMMR cells for 48 h, and IFN-γ expression was measured by flow cytometry. n = 3 repeats. dMMR versus STINGkd: *, P ≤ 0.05. (H) Dependence on endogenous type I IFN signaling was assessed following 24 h treatment with 10 µg/ml anti-IFNAR1-blocking antibody. n = 4 repeats. Isotype versus indicated bar. *, P ≤ 0.05; **, P ≤ 0.01. (I and J) Sensitivity to exogenous induction of type I IFN signaling was assessed by qPCR following treatment of cells with 9 µg/ml cGAMP (I) or 1,000 U/ml IFNB (J) for 24 h. n = 3 repeats. (E and H–J) Vehicle versus treatment: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005. FDR, false discovery rate; NK, natural killer; sig, signaling.
We observed greater baseline activation of TBK1 and STAT1, but not STAT3, in dMMR compared with CIN CRC cells (Fig. 5 D). TBK1 is a primary mediator of the cGAS/STING signaling pathway that drives innate type I IFN signaling, and STAT1 is a downstream responder to activation of this pathway (Vanpouille-Box et al., 2018). Treating MC38 cells with inhibitors of STAT1 (fludarabine) or STING (H-151) down-regulated CCL5 and CXCL10 selectively in dMMR but not CIN cells (Fig. 5 E; Haag et al., 2018). We further observed down-regulation in the associated signaling pathways (Fig. S4 E). Since we did not observe decreased chemokine production upon blocking the AIM2 nucleic acid–sensing pathway, this does not seem to represent a generalized increase in cytoplasmic DNA sensing but rather specific activation of the cGAS/STING pathway (Fig. 5 F). We further confirmed the dependence of dMMR CCL5 and CXCL10 production on STING by knocking down its gene expression in dMMR MC38 CRC cells, which significantly reduced their ability to activate CD8+ T cells (Fig. 5 G). While we did not detect significantly more production of IFNA/B by dMMR CRC cells (Fig. S1 I), blocking IFNAR1 did significantly decrease CCL5, CXCL10, and ISG15 in dMMR CRC cells (Fig. 5 H). This confirms an important role for endogenously produced IFNA/B in the anti-tumor immune response mediated by dMMR CRC cells despite our inability to detect high levels of production. Given our desire to find dMMR CRC–mediated mechanisms of TIL activation that could be therapeutically translated to other CRC subsets, we tested whether or not either of these pathways was fundamentally defective in the CIN CRCs since this would preclude their therapeutic targeting. Stimulating both dMMR and CIN CRC cells with either the cGAS ligand cGAMP or IFNB significantly up-regulated ISGs including CCL5 and CXCL10, eliminating defective cGAS/STING or type I IFN signaling as the main mechanism for failed anti-tumor immunity in CIN CRCs (Fig. 5, I and J). This set of findings confirms that loss of MLH1 activates endogenous IFN-related signaling in dMMR CRC cells and supports our hypothesis that the resulting IFN-associated chemokine secretion transforms these CRC cells into central architects of the tumor TIL microenvironment.
Loss of DNA mismatch repair alters the stimulatory capacity of cytoplasmic DNA generated by baseline or treatment-induced genetic instability
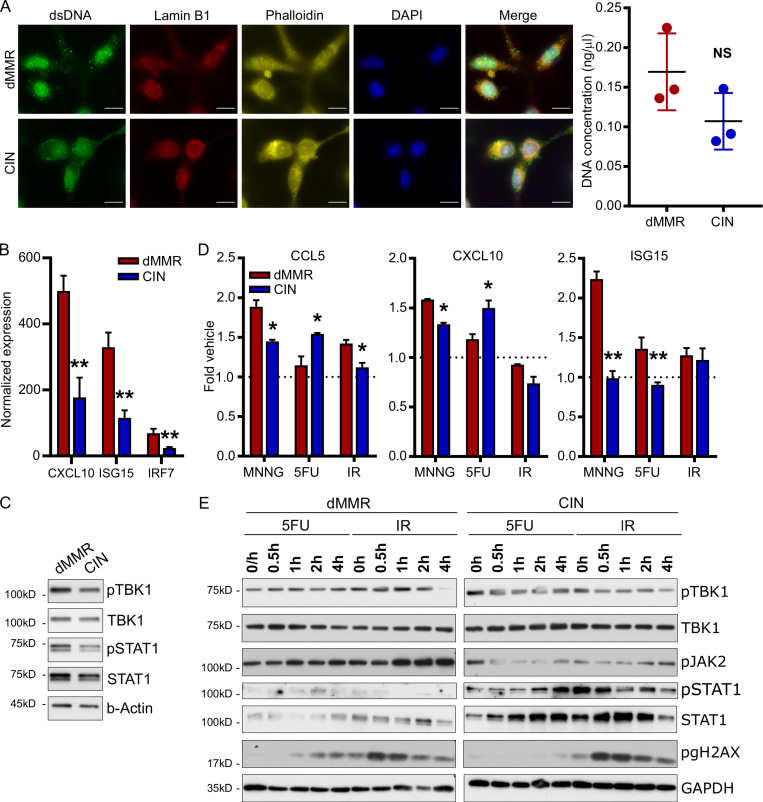
Loss of the DNA mismatch repair function in dMMR CRCs confers widespread genomic instability that renders the tumors hypermutable. A notable consequence of extensive genomic instability in cancer cells is the release of damaged DNA into the cytoplasm in the form of either free DNA or micronuclei (Vanpouille-Box et al., 2018). Since cytoplasmic DNA can activate nucleic acid sensing pathways in the cells, a plausible explanation for high endogenous activation of cGAS/STING and type I IFN signaling in dMMR CRC cells is that these cells have alterations in the amount or composition of their cytoplasmic DNA due to extreme underlying genomic instability. Immunofluorescence staining with an anti–double-stranded DNA antibody confirmed that our cells did contain significant quantities of cytoplasmic DNA although we did not observe any difference in quantity between dMMR and CIN CRCs (Fig. 6 A). We thus reasoned that loss of mismatch repair could alter the composition of cytoplasmic DNA in a way that more strongly activated endogenous type I IFN signaling. To test this, we isolated cytoplasmic DNA from dMMR and CIN CRC cells and used equal amounts to stimulate bone marrow–derived dendritic cells (BMDCs). Cytoplasmic DNA from dMMR CRCs up-regulated more ISGs and induced greater IFN signaling than did that from CIN CRCs, indicating that the nature of cytoplasmic DNA in dMMR CRCs is particularly stimulatory for cGAS/STING (Fig. 6, B and C). To investigate if this is specifically related to DNA damage associated with MMR as opposed to other DNA repair pathways, we made use of the compound N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) to induce DNA damage that is corrected via the mismatch repair pathway (Jascur et al., 2011). MNNG treatment increased production of ISGs including CCL5 and CXCL10 in both dMMR and CIN CRCs to a greater extent than treatment with 5-fluorouracil (5FU) or ionizing radiation (IR), which induce DNA damage corrected by base excision repair or homologous recombination (HR), respectively (Fig. 6 D; Li et al., 2016). Notably, MNNG-induced up-regulation was highest in dMMR CRC cells, which cannot repair the damage. In contrast, 5FU induced greater up-regulation of CCL5 and CXCL10 in CIN CRC cells than dMMR CRC cells. This is consistent with the longstanding observation that dMMR CRCs respond differently, and often less effectively, than CIN CRCs to many standard-of-care DNA-damaging radio- and chemotherapies, especially 5FU (Helleday et al., 2008). To better understand this differential response, we looked at the signaling pathways induced by 5FU and IR in dMMR and CIN CRC cells (Fig. 6 E). We noticed similar activation patterns for TBK1, STAT1, and JAK2, indicating that a pathway other than cGAS/STING may account for the different response patterns of these two CRC subtypes. Collectively, these findings confirm that increased genomic damage generally increases ISG and chemokine expression via cGAS/STING and type I IFN signaling but that genetic damage resulting from defective mismatch repair is the most effective at doing so and likely accounts for high endogenous expression of TIL-recruiting CCL5 and CXCL10 in dMMR CRCs.
Figure 6.
Loss of DNA mismatch repair alters the stimulatory capacity of cytoplasmic DNA generated by baseline or treatment-induced genetic instability.(A) Cytoplasmic DNA was visualized by staining with an anti–double-stranded DNA antibody (left). Cytoplasmic DNA was isolated from untreated dMMR and CIN MC38 cells and quantified by Qubit (right). n = 4 repeats. Scale bar, 10 µm. (B) BMDCs were stimulated for 3 h with 500 ng of cytoplasmic DNA isolated from dMMR or CIN CRC cells. Gene expression was evaluated by qPCR. n = 3 repeats. (C) BMDCs were treated as in B for 20 min and analyzed for signaling activation. n = 3 repeats. (D) Expression of IFN-associated genes in dMMR and CIN MC38 CRC cells treated with genotoxic agents (10 µM MNNG, 1 µM 5FU, 10 Gy IR) for 24 h. Expression was assessed by qPCR. n = 4 repeats. (E) Activation of STING signaling in cells treated as in D. n = 3 repeats. For all panels, dMMR versus CIN: *, P ≤ 0.05; **, P ≤ 0.01.
Loss of DNA mismatch repair in CRC patient organoids up-regulates chemokine production and type I IFN signaling
Cell line studies can be limited by artifacts from long-term culture. To determine if depletion of Mlh1 in primary cells also up-regulated ISGs, we generated organoids from CRCs induced in WT C57BL/6 mice through repeated doses of azoxymethane (Meunier et al., 2010). Stably knocking down Mlh1 in these organoids to model dMMR CRC indeed up-regulated CCL5 and CXCL10 (Fig. 7, A and B). Treatment of dMMR organoids with the STING inhibitor carbonyl cyanide 3-chlorophenylhydrazone (CCCP) decreased chemokine expression, confirming that chemokine induction in these primary cultures was also due to endogenous activation of the cGAS/STING signaling pathway (Fig. 7 B). Migration of CD8+ T cells toward supernatant conditioned by dMMR organoids was also suppressed to the same level as the control organoids by blocking antibodies against CCL5 and CXCL10, confirming the importance of these chemokines for T cell infiltration in the setting of primary dMMR CRC cells (Fig. 7 C).
Figure 7.
Loss of DNA mismatch repair in primary mouse and CRC patient organoids up-regulates chemokine production and type I IFN signaling.(A) Baseline protein expression from primary mouse CRC organoids. CRC was induced by 10 weekly injections of 10 µg/kg azoxymethane followed by another 10 wk without treatment. Mlh1 was stably knocked down by lentiviral transduction with shRNA and hygromycin selection to establish dMMR organoids. Scrambled shRNA-transduced organoids were used as CIN controls. n = 4 repeats. (B) Reduced expression of ISGs following 24 h incubation of the dMMR and CIN organoids with 10 µM of the cGAS/STING inhibitor CCCP. n = 3 repeats. dMMR versus CIN: **, P ≤ 0.01. (C) CD8+ T cell migration toward supernatant conditioned for 24 h by established dMMR or CIN organoids. dMMR versus indicated bar. *, P ≤ 0.05. (D and E) Baseline protein (D) and RNA (E) expression in organoids established from CRC patients. dMMR organoids were generated by stably knocking down MLH1 as in A with the appropriate shRNA and Scramble CIN control. dMMR versus CIN: **, P ≤ 0.01. (F) Inhibition of chemokine and ISG expression by treatment of human CRC organoids for 24 h with 10 µM CCCP. n = 2 patients, three repeats each. Vehicle versus *, P ≤ 0.05; **, P ≤ 0.01.
To demonstrate the relevance of our findings to human CRCs, we generated organoids from resected CRC samples and stably knocked down MLH1 in these organoids using lentiviral transduction (Fig. 7 D). This led to increased expression of the ISGs CCL5, CXCL10, and ISG15, indicating that these chemokines are likely a significant contributor to TIL recruitment into dMMR tumors in CRC patients (Fig. 7 E). Treatment of the dMMR organoids with CCCP confirmed the importance of the cGAS/STING pathway to chemokine up-regulation (Fig. 7 F). These data confirm that induction of CCL5 and CXCL10 via activated cGAS/STING signaling in dMMR CRCs is a direct result of loss of MLH1 and that activation of this pathway in CIN CRCs represents a promising therapeutic approach for increasing TIL infiltration into this normally immune-resistant subset of CRCs.
Discussion
Successful anti-tumor immunity requires the coordination of many processes; chief among these are effective recruitment and retention of TILs in the tumor epithelium. We show here that chemokine-mediated recruitment of systemic CD8+ T cells is an essential component of successful anti-tumor immunity in dMMR CRCs. Specifically, genomic instability induced by defective DNA mismatch repair endogenously stimulates cGAS/STING and innate type I IFN signaling in dMMR CRCs, leading to up-regulated expression of ISGs, including the chemokines CCL5 and CXCL10. Blocking either STING or type I IFN sensing down-regulates CCL5 and CXCL10 production by dMMR CRC cells, while genetic instability induced by DNA-damaging agents, especially those requiring MMR, increases CCL5 and CXCL10 production. Neutralization of CCL5 and CXCL10 blocks migration of CD8+ T cells toward dMMR CRC cells, thereby precluding effective neoantigen-specific killing by TILs. Critically, cGAS/STING and type I IFN signaling remains functional in CIN CRCs despite its low baseline activation, suggesting that therapeutically targeting this pathway could increase T cell trafficking into these normally immunologically silent tumors.
Abundant evidence links high TMB to tumor immunogenicity and activation of neoantigen-specific T cell–mediated killing. Paradoxically, some high TMB tumors fail to stimulate an endogenous anti-tumor immune response and remain refractory to checkpoint inhibition therapy (Bonneville et al., 2017; Kim et al., 2020). This includes dMMR tumors at other body sites as well as tumors rendered hypermutable by other mechanisms. One possible explanation for this is that mutational signatures caused by some processes are more immunogenic than others (Lim et al., 2017). Alternatively, hypermutation caused by different defects could selectively affect different gene families depending on their sequence or structure. This is consistent with the observation that, despite their mutator phenotype, specific genes are consistently mutated in dMMR CRCs (Hause et al., 2016). The fact that only some of these are also consistently mutated in dMMR endometrial cancers implies that site-specific selection factors may work in tandem with each mutational process to select for persistence of specific mutations from among a large body of randomly occurring passenger mutations. A final possible explanation is that mutations induced by certain forms of DNA repair defects could produce immunogenic DNA byproducts that activate endogenous DNA-sensing antiviral machinery (Raeker and Carethers, 2020; van Wietmarschen et al., 2020). Our findings suggest that dMMR-induced hypermutability affects genes that regulate the production of chemokines such as CCL5 and CXCL10 and that this, in turn, generates strongly immunogenic tumors that not only have a high TMB but are also proficient at recruiting systemic CD8+ T cells into the tumors, where they can affect neoantigen-specific killing. Notably, our work implies that therapeutically inducing the production of CCL5 and CXCL10 by CIN CRC cells could increase T cell recruitment into the tumor microenvironment. Even low levels of targeted killing of these neoantigen-poor tumors could initiate development of effective anti-tumor immunity via processes such as epitope spreading (Menares et al., 2019). The lack of prognostic benefit seen in dMMR tumors in nonintestinal tissues may be due to the need for a different set of non–ISG-dependent chemokines for T cell homing to those sites. Our evidence that both dMMR and CIN CRCs remain sensitive to exogenous type I IFN further indicates that initial recruitment of CD8+ T cells, which are potent IFN producers, could establish a positive feedback loop where TILs induce tumor secretion of CCL5 and CXCL10, leading in turn to more TIL recruitment and activation.
Recruiting more CD8+ T cells into the tumor microenvironment can promote anti-tumor immunity only if the infiltrating T cells are competent at tumor cell killing. Our work indicates that effective immunity in CRC requires recruitment of systemic CD8+ T cells rather than expansion of the local pool of resident cells. This is consistent with many studies showing systemic immune activation by localized tumors (Pauken et al., 2021; Wu et al., 2020). However, we also found that following recruitment, TILs in dMMR CRCs up-regulate the tissue resident marker CD103, which is a ligand for E-cadherin on tumor cells that facilitates direct interactions between the TILs and CRC cells (Low et al., 2020; Park et al., 2019). Numerous studies have shown that CD103+CD8+ T cells are particularly effective at cytotoxic tumor cell killing, but in most of these studies the origin of the CD103+ T cells was not identified (Duhen et al., 2018; Okła et al., 2021; Webb et al., 2014). Indeed, TILs in dMMR CRCs were discovered long ago to express CD103, leading to the assumption that these derived from expansion of local resident intraepithelial lymphocytes (Dolcetti et al., 1999; Guidoboni et al., 2001). Given the tolerized nature of such resident intestinal cells, this created a paradox for how they could effectively mediate tumor cell killing. Our findings place these earlier observations into context by revealing the systemic source of these CD103+ TILs and identifying the mechanism by which they are recruited into the tumors.
Chemokines are potent but pleiotropic chemoattractants. Chemokine receptors often recognize multiple ligands and are broadly expressed on immune cells of different types (Nagarsheth et al., 2017). Our data indicate that both CCL5 and CXCL10 are selectively up-regulated in dMMR CRCs. We consistently failed to find differences in expression of other chemokines such as CXCL16 that are not regulated by IFN. CXCL10 expression by cancers is consistently associated with effective anti-tumor immunity and CD8+ T cell regulation. In contrast, CCL5 expression has been linked to both pro- and anti-tumor immune responses. In particular, CCL5 can recruit tumor-associated macrophages that can drive tumor-promoting inflammation (Halama et al., 2016; Zumwalt et al., 2015). However, we did not find any increase in tumor-associated macrophages in orthotopically grown dMMR MC38 CRCs compared with CIN CRCs, which is consistent with clinical data. In addition, cancer cells themselves can up-regulate CCR5, the CCL5 receptor, and respond to autocrine or paracrine secretion of CCL5. Indeed, blocking CCR5 with small molecule inhibitors such as maraviroc has shown some promise in clinical trials as an anti-cancer agent (Jiao et al., 2019). Whether these compounds also inhibit anti-tumor immunity or TIL recruitment into tumors will be important to determine in the future since this could compromise efficacy of the drug. In CRC, the majority of reports indicate that CCL5 expression is predictive of a positive patient outcome and productive anti-tumor immunity and that CCR5+CD8+ T cells express more IFN-γ and lead to a good prognosis (Boissière-Michot et al., 2014; Musha et al., 2005; Zumwalt et al., 2015). In light of our data showing differential expression between dMMR and CIN CRCs, whatever discrepancy does exist between studies could be explained by the fact that none of the reports examined dMMR and CIN CRCs separately. Our blocking experiments indicate that CCL5 and CXCL10 additively or synergistically regulate CD8+ T cell trafficking and activation. This indicates that expression of both of these chemokines could be predictive of patient prognosis and immunotherapy response even in CIN CRCs and particularly in the context of chemo- and radiotherapies that induce genomic instability.
The inherent ability of dMMR CRCs to stimulate anti-tumor immunity provides an important opportunity to learn the mechanisms necessary for this process. While there is an undeniable role for the high TMB content of these cancers, the lack of antigenic overlap with CIN CRCs means that identifying neoantigen-independent immune-stimulatory processes used by dMMR CRCs could provide more tractable therapeutic opportunities for treating other CRCs subsets. We show here a that a critical component of the successful anti-tumor immune program of dMMR CRCs is effective recruitment of systemic CD8+ T cells by high endogenous production of the chemokines CCL5 and CXCL10. This recruitment process is independent of neoantigen production by the tumor cells, thereby establishing it as an achievable therapeutic target for improving immune killing of CIN CRCs with few neoantigens. Since TIL infiltration is a prerequisite for effective adaptive immune killing of tumor cells, our work not only provides a potential new screening tool for identifying patients likely to respond to immunotherapies but also establishes the basal criteria by which the immunogenic potential of other therapies, particularly DNA damaging agents, can be evaluated.
Materials and methods
Cell line generation and stimulation
MC38 murine CRC cells were purchased from Kerafast. Cells were grown in high-glucose DMEM supplemented with 10% FBS, 1% penicillin-streptomycin, and 1% Hepes at 37°C with 5% CO2. Cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) with the pSpCas9(BB)-2A-puro (px459) V2.0 plasmid (Addgene; #62988) containing CRISPR guide RNAs (Table S1) targeting the Mlh1 gene to model dMMR CRCs or the Kras gene to model CIN CRCs (Ran et al., 2013). Transfectants were selected with 2 µg/ml puromycin, and mutations were confirmed either by sequencing or Western blot. shRNA-mediated knockdown of CCL5, CXCL10, and STING in the dMMR MC38 cells was achieved using the pLKO.1 system (Addgene; #10878) and containing the shRNA sequences in Table S1 or a scrambled sequence as a control (Moffat et al., 2006). Stably knocked down cells were selected with 250 µg/ml hygromycin, and knockdown was confirmed by Western blot. OVA-expressing cells were made by transfection with the pCI-neo-cOVA plasmid (Addgene; #25097) and selection with 200 µg/ml G418 (Yang et al., 2010). OVA expression was confirmed by qPCR using the primers in Table S1.
For verification of MSI, cells were transiently cotransfected with the pCAR-OF reporter plasmid (Addgene; #16627) and the pmCherry-C1 plasmid (Clontech; Nicolaides et al., 1998). 24 h later, expression of a β-galactosidase reporter gene (which is only expressed if a frame-restoring deletion or insertion mutation in the promoter occurs) was measured by flow cytometry using the FluoReporter lacZ/Galactosidase Quantitation Kit (Thermo Fisher Scientific) and gating on mCherry+ cells (Fig. S1 C). Increasing fluorescence indicates increased MSI. Verification of double-strand break repair capacity via HR or nonhomologous end joining (NHEJ) was measured using the Traffic Light reporter system by transiently cotransfecting cells with the pCVL Traffic Light Reporter 1.1 (Sce target) Ef1a BFP (Addgene; 31481) and pCVL SFFV d14GFP EF1s HA.NLS.Sce(opt) (Addgene; #31476) plasmids (Fig. S1 D; Certo et al., 2011; Chroma et al., 2016; Mateos-Gomez et al., 2015). 24 h later, cells were analyzed via flow cytometry by gating on BFP+ cells. Increased GFP fluorescence indicates increased HR capacity, and increased mCherry fluorescence indicates increased NHEJ capacity.
For sequencing, genomic DNA was isolated from cell pellets using the Quick Genomic DNA Extraction kit (Truin Science). Primers possessing EcoRI and BamHI cut sites at their ends were used (Table S1) with the Q5 High-Fidelity PCR Kit (New England Biolabs) to amplify CRISPR target site regions. PCR products were purified using a GeneJet Gel Extraction Kit (Thermo Fisher Scientific), and then the ends were digested using EcoRI-HF and BamHI-HF (New England Biolabs). PCR products were subcloned into the pUC19 plasmid (Addgene; #50005) and sequenced with the M13 primer.
Cancer cell stimulations
MC38 CRC cells were seeded into plates 24 h before treatments as indicated in the figure legends. The following treatments were used: 10 µM fludarabine, 50 µM STAT3 Inhibitor VI (S3I-201), 10 µM CCCP, 9 µg/ml 2’,3′-cGAMP, 1 µM 5FU, 10 µM MNNG, 2 µM H-151 (all from Sigma-Aldrich), phosphorothioate oligo (IDT), 1,000 U/ml IFNb1 (Thermo Fisher Scientific), and 10 Gy irradiation (60Co source). After the indicated incubation time, cells supernatant or cells were harvested as indicated below.
Mouse CRC experiments
C57BL/6 WT mice originally purchased from Charles River were bred and maintained in the Cross Cancer Institute vivarium. OTI mice were purchased from The Jackson Laboratory. Mixed groups of male and female littermates 6–20 wk old were used for experiments. All animal work was approved by the Cross Cancer Institute’s Animal Care Committee.
Subcutaneous CRC experiments were performed by injecting 5 × 105 MC38 CRC cells in 100 µl PBS into the hind flank. Tumors were harvested after 2–3 wk. Resected tumors were minced and digested in enzyme cocktail (RPMI containing 0.5 mg/ml collagenase IV [Sigma-Aldrich], 10 µg/ml DNaseI [Sigma-Aldrich], 10% FBS, 1% penicillin-streptomycin, and 1% Hepes buffer) for 30 min at 37°C in a shaking incubator at 200 rpm (Baker et al., 2013). Fragments were rigorously pipetted to dissociate, filtered through a 100-µm cell strainer, and washed. To separate the cancer and immune cells in the tumors, dissociated cells were resuspended in 40% Percoll (GE Healthcare), overlaid onto 80% Percoll, and centrifuged at 500 g (with brake off) for 30 min at room temperature. The top layer (tumor cells) and the interface (immune cells) were collected into separate tubes, washed, and processed for RNA isolation as below.
Orthotopic CRC experiments were performed by injecting 1.5 × 105 MC38 CRC cells in 50 µl PBS into the wall of the descending colon using a flexible needle (Hamilton) inserted through the working channel of a Wolfe endoscope and visualized via the ColoView imaging system (Storz; Roper et al., 2018). Orthotopic tumor growth was monitored by endoscopy, and tumors were harvested after 14–21 d and dissociated as above before flow staining. Adoptive transfer experiments were performed by isolating CD8+ T cells from the spleens or MLNs using the EasySep Mouse CD8+ T Cell Isolation Kit (StemCell Technologies) from donor mice harboring orthotopic CRC tumors. CD8+ T cells were stained with 2 µM CFSE, and 2 × 106 cells were injected i.v. into recipient mice bearing orthotopic CRC tumors. 48 h later, spleens, MLNs, and tumors were harvested, dissociated, and analyzed by flow cytometry.
Mouse and human CRC-derived organoids
Murine organoids from either the normal colonic epithelium or colorectal tumors induced by repeated doses of azoxymethane (10 weekly doses of 10 mg/kg azoxymethane) were generated as described previously and cultured as below (Meunier et al., 2010; Sato et al., 2011).
Resected human CRC tumors were collected in HBSS within 10 min of devitalization. Tumors were processed as described previously (Sato et al., 2011) In brief, tumors were dissociated for 1 h in DMEM with 2.5% FBS, 75 U/ml collagenase XI (Sigma-Aldrich), and 125 µg/ml dispase II (Sigma-Aldrich). Following filtration, cells were plated at 500–1,000 per well in growth factor–reduced Matrigel (Corning) and cultured in basal crypt media (Advanced DMEM/F12 containing 10% FBS, 2 mM glutamine, 10 mM Hepes, 1 mM N-acetylcystein, 1X N2 supplement, 1X B27 supplement, 10 mM nicotinamide, 500 nM A83-01, 10 µM SB202190, and 50 ng/ml epidermal growth factor; Thermo Fisher Scientific) mixed 1:1 with conditioned supernatant from L-cells expressing Wnt3a, R-spondin, and noggin (ATCC #CRL-3276; Miyoshi and Stappenbeck, 2013). All work with human samples was approved by the Health Research Ethics Board of Alberta Cancer Committee and performed after obtaining informed patient consent.
Primary dMMR mouse or human organoids were generated using lentiviral transduction as described previously (Koo et al., 2011; Roper et al., 2018). Lentivirus was prepared as previously described using the pLKO.1 system (Addgene; #10878) and containing the shRNA sequences in Table S1 (Moffat et al., 2006). Lentivirus was concentrated 100× by ultracentrifugation. Organoids were pretreated for 4–5 d with 10 mM nicotinamide to enrich for stem cells. Organoids were dislodged from the plate by pipetting and then treated for 5 min at 37°C with TrypLE Express (Life Technologies). Organoids were mixed with lentivirus along with 8 µg/ml polybrene and 10 µM Y27632 (Sigma-Aldrich) and seeded into a 96-well plate. The plate was centrifuged for 60 min at 600 g at 32°C and then incubated at 37°C for 6 h. The organoids were then embedded in Matrigel and cultured in media containing 50–100 µg/ml hygromycin to select for successful transduction. Gene knockdown was verified by Western blot.
For stimulations, equal numbers of organoids were plated in Matrigel, cultured for 3 d, and then treated as indicated in the figure legends. To harvest, organoids were resuspended in ice-cold Cell Recovery Solution (Corning) and incubated for 10 min on ice to dissolve Matrigel. Cell Recovery Solution was diluted fourfold and then spun down. Pellets were processed for RNA or protein isolation as below.
Flow cytometry and cytokine bead arrays
Flow cytometry staining was performed using the antibodies in Table S2 in addition to the Zombie Aqua viability stain (BioLegend) and the Foxp3 Transcription Factor Staining Buffer Set (eBioscience). Flow cytometry was performed on a CytoFlex S cytometer (Beckman Coulter) with subsequent analysis using FlowJo (BD Biosciences).
Secretion of CCL5 and CXCL10 by dMMR and CIN CRCs into cell supernatants was analyzed using a custom LegendPlex cytokine bead array (BioLegend). Data were acquired on a CytoFlex S cytometer and analyzed in FlowJo.
T cell activation, migration, and cytotoxicity assays
CD8+ T cells were isolated from the spleens or MLNs using the EasySep Mouse CD8+ T Cell Isolation Kit (StemCell Technologies). Where indicated, T cells were first expanded by incubation at a 5:1 ratio with BMDCs that had been pulsed with tumor lysates (100 µg/ml with 106 BMDCs) for 30 min and then irradiated (20 Gy). For direct co-culture experiments, tumor cells were first incubated with the indicated treatment, and this was removed before adding CD8+ T cells at a 5:1 T cell:tumor cell ratio. Where indicated, cytotoxicity was assessed after 24 h or 48 h using the CellEvent Caspase-3/7 Green Detection Reagent (Thermo Fisher Scientific) at 1.0 µM and gating on CD45-negative cells. To assess T cell activation by STING knockdown or scramble controls, CRC cells were first pulsed with 1 µg/ml of the SIINFEKL peptide for 30 min. Cells were washed twice before addition of OTI CD8+ T cells as above. For chemokine blocking experiments, the antibodies (Table S2) were present throughout the co-culture at 2 µg/ml.
For migration assays, conditioned supernatants (DMEM plus 10% FBS) were collected from CRC cells treated as indicated in figure legends. Tissue culture inserts with 5.0-µm pores (Sarstedt) were precoated with 0.8 mg/ml Matrigel for 2 h at 37°C and rinsed twice with warm media. 105 CFSE-labeled CD8+ T cells were added to the upper portion of the insert in 100 µl media containing 2% FBS. 500 µl of conditioned supernatants was added to the bottom well. For chemokine blocking experiments, antibodies were added to the media in the lower chamber 30 min before T cell addition and were present during the entire assay. T cells were allowed to migrate for 2 h, and then cells in the upper insert and bottom well were collected and quantified by flow cytometry. Percentage migrated cells were calculated as follows: percentage migrated cells = (number migrated cells) / (number input cells + number migrated cells).
BMDC stimulation with cytoplasmic DNA
Cytoplasmic DNA was isolated from dMMR and CIN CRCs as described previously (Baghirova et al., 2015; Härtlova et al., 2015). Briefly, cells were lysed in cytoplasmic extraction buffer (150 mM NaCl, 50 mM Hepes, 200 µg/ml digitonin, and 1 M hexylene glycol) for 10 min on ice and spun 10 min at 2,000 g. 1 mg of protein from the supernatant was treated with 1 mg/ml proteinase K for 1 h at 55°C. DNA was purified with phenol/chloroform/isoamyl alcohol, and then RNA was removed by digestion with 500 µg/ml RNaseA for 1 h at 37°C. DNA was purified as above and quantified by nanodrop.
BMDCs were derived from WT C57BL/6 mice for 7 d using 20 ng/ml GM-CSF, as described previously (Baker et al., 2011). 5 × 105 BMDCs were stimulated with 500 ng of cytoplasmic DNA that had been premixed at a 0.5:1 Lipofectamine 2000:DNA ratio. After the indicated times, RNA or protein was extracted for analysis.
Immunofluorescence staining
Cells were seeded onto poly-L-lysine–coated coverslips and allowed to adhere overnight at 37°C. Cells were fixed with methanol and blocked (5% normal serum and 0.3% Triton X-100 in PBS). Cells were stained with the indicated primary antibodies for 1 h at room temperature. After washing, cells were stained with anti-mouse–Alexa 488 or anti-rabbit–Alexa 647 secondary antibodies (Thermo Fisher Scientific) for 1 h at room temperature before staining with phalloidin Alexa 546 (Thermo Fisher Scientific) for 20 min. Nuclei were stained with DAPI, and then coverslips were mounted on slides. Cells were imaged on a Zeiss Axioscope 2 microscope. Post-image processing was performed using ImageJ (Schneider et al., 2012).
Western blotting
Protein was isolated in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 50 mM sodium pyrophosphate, 1 mM EDTA, 0.5% NP-40, and 1% Triton X-100) containing 1 mM sodium orthovanadate and 1× protease inhibitor (Sigma-Aldrich). Protein was quantified using a BCA protein assay kit (Thermo Fisher Scientific). 5 or 10 µg of protein was loaded per lane of SDS-PAGE gels and transferred to nitrocellulose membranes. The antibodies used are listed in Table S2. Bands were visualized using the ECL Prime Western Blotting Detection Reagent (GE Healthcare Amersham).
RNA and qPCR
RNA was extracted using TRIzol and reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). qPCR reactions were set up using the primers indicated in Table S1 and POWRUP SYBR Master Mix (Thermo Fisher Scientific). qPCR was performed on the QuantStudio6 real-time PCR system (Applied Biosystems).
scRNAseq
Live cells were isolated from orthotopically grown dMMR and CIN MC38 CRC tumors. Each sample represents pooled cells from five mice per group. Viable CD45+ cells were enriched using magnetic selection (StemCell Technologies) and submitted to the University of Alberta High Content Analysis Core for processing with a Single Cell Immune Profiling kit (10X Genomics). The data were processed using the Seurat package for R (v3.0) as described previously (Stuart et al., 2019; Wang et al., 2019). Briefly, cells with either a unique gene count >2,500 or <200 as well as genes found in fewer than three cells were filtered out. Principal component analysis was used to visualize and explore these datasets, and we used the default settings of the RunTSNE function to summarize the principal component analysis with tSNE (t-distributed stochastic neighbor embedding) dimensionality. Cell clusters in the two-dimensional representations were annotated to known cell types using FindAllMarkers for all clusters. Differential expression analysis was performed using the DESeq2 package (Love et al., 2014). GO enrichment analysis was performed using the fgsea package and mouse versions of the MSigDB genesets from http://bioinf.wehi.edu.au (Mi et al., 2019). ISGs were defined as previously (Liao et al., 2019). Other genesets were taken from MSigDB. CD8+ T cell subsets were defined using the following canonical markers: naïve CD8+ T cell = Cd62HiCd44Low, effector CD8+ T cell = Cd62LowCd44HiKlrgHiTbetLowBlimpHiCxcr3Low, effector memory CD8+ T cell = Cd62LowCd44HiKlrgLowTbetHiBlimpHiCxcr3Hi, TRM = ItgaeHiCd69HiIl7rHiCxcr3Hi, and central memory CD8+ T cell = Ccr7HiIl7rHiCxcr3Hi (Low et al., 2020; Park et al., 2019).
Data from TCGA analysis
Human RNA sequencing data and DNA sequencing data (Illumina HiSeq RNASeqV2) from the Colorectal Adenocarcinoma dataset from the TCGA Nature 2012 and TCGA PanCancer Atlas from TCGA were downloaded from cBioPortal for Cancer Genomics (https://www.cbioportal.org/; Cancer Genome Atlas Network, 2012; Hoadley et al., 2018; Sanchez-Vega et al., 2018). Chemokine expression analysis was performed using the DESeq2 package in R (v3.0; Love et al., 2014).
Statistical analysis
Prism (GraphPad Software) was used for statistical analysis. Gene expression analysis was processed by log2 transformation, and the resulting data were evaluated for Gaussian distribution. Comparison of two unpaired groups was made by two-tailed Student’s t test for normal data, or Mann–Whitney for nonparametric tests. For three or more groups with two parameters, two-way ANOVA or multiple t test procedures were used as appropriate, for data with Gaussian distribution. Responses to multiple stimuli of a single cell type or of cells from a single donor were analyzed using paired tests. Post hoc analysis to correct for multiple comparisons and detect differences between groups was by the two-stage linear step-up procedure of Benjamin, Krieger, and Yekutieli with false discovery rate <0.05. A two-sided probability (P) of α error <0.05 defined significance.
Online supplemental material
Fig. S1 provides data validating the phenotype and genotype of all MC38 dMMR and CIN cell line variants used. Fig. S2 demonstrates the quantity and activation state of immune cell types other than CD8+ T cells that infiltrate orthotopically grown dMMR and CIN MC38 CRC tumors. Fig. S3 shows expression of GO list “T Cell Activation” as well as chemokines and chemokine receptors genes in TILs of orthotopically grown dMMR and CIN tumors. It also shows the TCR clonal diversity of TILs in the orthotopic tumors. Fig. S4 shows expression of GO list “TGFβ Signaling” as well as chemokines and chemokine receptor genes in dMMR and CIN CRC cells grown as orthotopic tumors. It also shows the relative capacity for dMMR and CIN CRCs to induce CD103 expression on systemic CD8+ T cells and the dependence of intracellular signaling on the cGAS/STING and STAT pathways. Table S1 provides information on all of the DNA oligos and primers used. Table S2 provides information on all antibodies used.
Supplementary Material
provides information on all of the DNA oligos and primers used.
provides information on all antibodies used.
Acknowledgments
The authors thank Rose-Marie Cornand, Dan McGinn, Cheryl Santos, Daming Li, Joaquín López-Orozco, Sharmin Sultana Sumi, Geraldine Barron, Dr. Xuejun Sun, Dr. Anne Galloway, and Dr. Aja Rieger as well as the Faculty of Medicine and Dentistry flow cytometry and high throughput cores at the University of Alberta for technical support. Dr. Jatin Roper (Duke University) provided expert advice on orthotopic CRC protocols.
This work was supported by funding from the Canadian Institutes of Health Research (grant 407882), the Natural Sciences and Engineering Research Council of Canada (grant RGPIN-2016-05152), the Canadian Foundation for Innovation (grant 37832), the Cancer Research Society (grant 22277), and the University Hospital Foundation (K. Baker).
Author contributions: C. Mowat, S.R. Mosley, and A. Namdar designed and performed experiments, analyzed data, and wrote the manuscript. D. Schiller provided human CRC tissue samples. K. Baker conceived of the project, designed and performed experiments, analyzed data, wrote the manuscript, and secured funding.
Data availability
Data from scRNAseq have been deposited in GEO under accession no. GSE178706.
References
- Baghirova, S., Hughes B.G., Hendzel M.J., and Schulz R.. 2015. Sequential fractionation and isolation of subcellular proteins from tissue or cultured cells. MethodsX. 2:440–445. 10.1016/j.mex.2015.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, K., Zlobec I., Tornillo L., Terracciano L., Jass J.R., and Lugli A.. 2007. Differential significance of tumour infiltrating lymphocytes in sporadic mismatch repair deficient versus proficient colorectal cancers: a potential role for dysregulation of the transforming growth factor-beta pathway. Eur. J. Cancer. 43:624–631. 10.1016/j.ejca.2006.11.012 [DOI] [PubMed] [Google Scholar]
- Baker, K., Qiao S.W., Kuo T.T., Aveson V.G., Platzer B., Andersen J.T., Sandlie I., Chen Z., de Haar C., Lencer W.I., et al. 2011. Neonatal Fc receptor for IgG (FcRn) regulates cross-presentation of IgG immune complexes by CD8-CD11b+ dendritic cells. Proc. Natl. Acad. Sci. USA. 108:9927–9932. 10.1073/pnas.1019037108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, K., Rath T., Flak M.B., Arthur J.C., Chen Z., Glickman J.N., Zlobec I., Karamitopoulou E., Stachler M.D., Odze R.D., et al. 2013. Neonatal Fc receptor expression in dendritic cells mediates protective immunity against colorectal cancer. Immunity. 39:1095–1107. 10.1016/j.immuni.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea, G., Mlecnik B., Tosolini M., Kirilovsky A., Waldner M., Obenauf A.C., Angell H., Fredriksen T., Lafontaine L., Berger A., et al. 2013. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 39:782–795. 10.1016/j.immuni.2013.10.003 [DOI] [PubMed] [Google Scholar]
- Boissière-Michot, F., Lazennec G., Frugier H., Jarlier M., Roca L., Duffour J., Du Paty E., Laune D., Blanchard F., Le Pessot F., et al. 2014. Characterization of an adaptive immune response in microsatellite-instable colorectal cancer. OncoImmunology. 3:e29256. 10.4161/onci.29256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneville, R., Krook M.A., Kautto E.A., Miya J., Wing M.R., Chen H.Z., Reeser J.W., Yu L., and Roychowdhury S.. 2017. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis. Oncol. 2017:1–15. 10.1200/PO.17.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network . 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 487:330–337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cario, E.2008. Innate immune signalling at intestinal mucosal surfaces: a fine line between host protection and destruction. Curr. Opin. Gastroenterol. 24:725–732. 10.1097/MOG.0b013e32830c4341 [DOI] [PubMed] [Google Scholar]
- Certo, M.T., Ryu B.Y., Annis J.E., Garibov M., Jarjour J., Rawlings D.J., and Scharenberg A.M.. 2011. Tracking genome engineering outcome at individual DNA breakpoints. Nat. Methods. 8:671–676. 10.1038/nmeth.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae, Y.K., Anker J.F., Oh M.S., Bais P., Namburi S., Agte S., Giles F.J., and Chuang J.H.. 2019. Mutations in DNA repair genes are associated with increased neoantigen burden and a distinct immunophenotype in lung squamous cell carcinoma. Sci. Rep. 9:3235. 10.1038/s41598-019-39594-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chroma, K., Mistrik M., Moudry P., Gursky J., Liptay M., Strauss R., Skrott Z., Vrtel R., Bartkova J., Kramara J., and Bartek J.. 2016. Tumors overexpressing RNF168 show altered DNA repair and responses to genotoxic treatments, genomic instability and resistance to proteotoxic stress. Oncogene. 36:2405–2422. 10.1038/onc.2016.392 [DOI] [PubMed] [Google Scholar]
- Dolcetti, R., Viel A., Doglioni C., Russo A., Guidoboni M., Capozzi E., Vecchiato N., Macrì E., Fornasarig M., and Boiocchi M.. 1999. High prevalence of activated intraepithelial cytotoxic T lymphocytes and increased neoplastic cell apoptosis in colorectal carcinomas with microsatellite instability. Am. J. Pathol. 154:1805–1813. 10.1016/S0002-9440(10)65436-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhen, T., Duhen R., Montler R., Moses J., Moudgil T., de Miranda N.F., Goodall C.P., Blair T.C., Fox B.A., McDermott J.E., et al. 2018. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat. Commun. 9:2724. 10.1038/s41467-018-05072-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon, J., and Bruni D.. 2020. Tumor Immunology and Tumor Evolution: Intertwined Histories. Immunity. 52:55–81. 10.1016/j.immuni.2019.12.018 [DOI] [PubMed] [Google Scholar]
- Guan, J., Lu C., Jin Q., Lu H., Chen X., Tian L., Zhang Y., Ortega J., Zhang J., Siteni S., et al. 2021. MLH1 Deficiency-Triggered DNA Hyperexcision by Exonuclease 1 Activates the cGAS-STING Pathway. Cancer Cell. 39:109–121.e5. 10.1016/j.ccell.2020.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidoboni, M., Gafà R., Viel A., Doglioni C., Russo A., Santini A., Del Tin L., Macrì E., Lanza G., Boiocchi M., and Dolcetti R.. 2001. Microsatellite instability and high content of activated cytotoxic lymphocytes identify colon cancer patients with a favorable prognosis. Am. J. Pathol. 159:297–304. 10.1016/S0002-9440(10)61695-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinney, J., Dienstmann R., Wang X., de Reyniès A., Schlicker A., Soneson C., Marisa L., Roepman P., Nyamundanda G., Angelino P., et al. 2015. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21:1350–1356. 10.1038/nm.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag, S.M., Gulen M.F., Reymond L., Gibelin A., Abrami L., Decout A., Heymann M., van der Goot F.G., Turcatti G., Behrendt R., and Ablasser A.. 2018. Targeting STING with covalent small-molecule inhibitors. Nature. 559:269–273. 10.1038/s41586-018-0287-8 [DOI] [PubMed] [Google Scholar]
- Halama, N., Zoernig I., Berthel A., Kahlert C., Klupp F., Suarez-Carmona M., Suetterlin T., Brand K., Krauss J., Lasitschka F., et al. 2016. Tumoral Immune Cell Exploitation in Colorectal Cancer Metastases Can Be Targeted Effectively by Anti-CCR5 Therapy in Cancer Patients. Cancer Cell. 29:587–601. 10.1016/j.ccell.2016.03.005 [DOI] [PubMed] [Google Scholar]
- Härtlova, A., Erttmann S.F., Raffi F.A., Schmalz A.M., Resch U., Anugula S., Lienenklaus S., Nilsson L.M., Kröger A., Nilsson J.A., et al. 2015. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 42:332–343. 10.1016/j.immuni.2015.01.012 [DOI] [PubMed] [Google Scholar]
- Hause, R.J., Pritchard C.C., Shendure J., and Salipante S.J.. 2016. Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med. 22:1342–1350. 10.1038/nm.4191 [DOI] [PubMed] [Google Scholar]
- Helleday, T., Petermann E., Lundin C., Hodgson B., and Sharma R.A.. 2008. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer. 8:193–204. 10.1038/nrc2342 [DOI] [PubMed] [Google Scholar]
- Hoadley, K.A., Yau C., Hinoue T., Wolf D.M., Lazar A.J., Drill E., Shen R., Taylor A.M., Cherniack A.D., Thorsson V., et al. Cancer Genome Atlas Network . 2018. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 173:291–304.e6. 10.1016/j.cell.2018.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubel, P., Urban C., Bergant V., Schneider W.M., Knauer B., Stukalov A., Scaturro P., Mann A., Brunotte L., Hoffmann H.H., et al. 2019. A protein-interaction network of interferon-stimulated genes extends the innate immune system landscape. Nat. Immunol. 20:493–502. 10.1038/s41590-019-0323-3 [DOI] [PubMed] [Google Scholar]
- Jascur, T., Fotedar R., Greene S., Hotchkiss E., and Boland C.R.. 2011. N-methyl-N'-nitro-N-nitrosoguanidine (MNNG) triggers MSH2 and Cdt2 protein-dependent degradation of the cell cycle and mismatch repair (MMR) inhibitor protein p21Waf1/Cip1. J. Biol. Chem. 286:29531–29539. 10.1074/jbc.M111.221341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, X., Nawab O., Patel T., Kossenkov A.V., Halama N., Jaeger D., and Pestell R.G.. 2019. Recent Advances Targeting CCR5 for Cancer and Its Role in Immuno-Oncology. Cancer Res. 79:4801–4807. 10.1158/0008-5472.CAN-19-1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.R., Pina A., Albert A., McAlpine J.N., Wolber R., Gilks B., Carey M.S., and Kwon J.S.. 2020. Mismatch repair deficiency and prognostic significance in patients with low-risk endometrioid endometrial cancers. Int. J. Gynecol. Cancer. 30:783–788. 10.1136/ijgc-2019-000910 [DOI] [PubMed] [Google Scholar]
- Kistner, L., Doll D., Holtorf A., Nitsche U., and Janssen K.P.. 2017. Interferon-inducible CXC-chemokines are crucial immune modulators and survival predictors in colorectal cancer. Oncotarget. 8:89998–90012. 10.18632/oncotarget.21286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloor, M., and von Knebel Doeberitz M.. 2016. The Immune Biology of Microsatellite-Unstable Cancer. Trends Cancer. 2:121–133. 10.1016/j.trecan.2016.02.004 [DOI] [PubMed] [Google Scholar]
- Koo, B.K., Stange D.E., Sato T., Karthaus W., Farin H.F., Huch M., van Es J.H., and Clevers H.. 2011. Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods. 9:81–83. 10.1038/nmeth.1802 [DOI] [PubMed] [Google Scholar]
- Li, Z., Pearlman A.H., and Hsieh P.. 2016. DNA mismatch repair and the DNA damage response. DNA Repair (Amst.). 38:94–101. 10.1016/j.dnarep.2015.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, W., Overman M.J., Boutin A.T., Shang X., Zhao D., Dey P., Li J., Wang G., Lan Z., Li J., et al. 2019. KRAS-IRF2 Axis Drives Immune Suppression and Immune Therapy Resistance in Colorectal Cancer. Cancer Cell. 35:559–572.e7. 10.1016/j.ccell.2019.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, B., Mun J., Kim Y.S., and Kim S.Y.. 2017. Variability in Chromatin Architecture and Associated DNA Repair at Genomic Positions Containing Somatic Mutations. Cancer Res. 77:2822–2833. 10.1158/0008-5472.CAN-16-3033 [DOI] [PubMed] [Google Scholar]
- Llosa, N.J., Cruise M., Tam A., Wicks E.C., Hechenbleikner E.M., Taube J.M., Blosser R.L., Fan H., Wang H., Luber B.S., et al. 2015. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 5:43–51. 10.1158/2159-8290.CD-14-0863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low, J.S., Farsakoglu Y., Amezcua Vesely M.C., Sefik E., Kelly J.B., Harman C.C.D., Jackson R., Shyer J.A., Jiang X., Cauley L.S., et al. 2020. Tissue-resident memory T cell reactivation by diverse antigen-presenting cells imparts distinct functional responses. J. Exp. Med. 217:e20192291. 10.1084/jem.20192291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., Guan J., Lu S., Jin Q., Rousseau B., Lu T., Stephens D., Zhang H., Zhu J., Yang M., et al. 2020. DNA Sensing in Mismatch Repair-Deficient Tumor Cells Is Essential for Anti-tumor Immunity. Cancer Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maby, P., Tougeron D., Hamieh M., Mlecnik B., Kora H., Bindea G., Angell H.K., Fredriksen T., Elie N., Fauquembergue E., et al. 2015. Correlation between Density of CD8+ T-cell Infiltrate in Microsatellite Unstable Colorectal Cancers and Frameshift Mutations: A Rationale for Personalized Immunotherapy. Cancer Res. 75:3446–3455. 10.1158/0008-5472.CAN-14-3051 [DOI] [PubMed] [Google Scholar]
- MacDonald, T.T., Monteleone I., Fantini M.C., and Monteleone G.. 2011. Regulation of homeostasis and inflammation in the intestine. Gastroenterology. 140:1768–1775. 10.1053/j.gastro.2011.02.047 [DOI] [PubMed] [Google Scholar]
- Mateos-Gomez, P.A., Gong F., Nair N., Miller K.M., Lazzerini-Denchi E., and Sfeir A.. 2015. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature. 518:254–257. 10.1038/nature14157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menares, E., Gálvez-Cancino F., Cáceres-Morgado P., Ghorani E., López E., Díaz X., Saavedra-Almarza J., Figueroa D.A., Roa E., Quezada S.A., and Lladser A.. 2019. Tissue-resident memory CD8+ T cells amplify anti-tumor immunity by triggering antigen spreading through dendritic cells. Nat. Commun. 10:4401. 10.1038/s41467-019-12319-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier, C., Cai J., Fortin A., Kwan T., Marquis J.F., Turbide C., Van Der Kraak L., Jothy S., Beauchemin N., and Gros P.. 2010. Characterization of a major colon cancer susceptibility locus (Ccs3) on mouse chromosome 3. Oncogene. 29:647–661. 10.1038/onc.2009.369 [DOI] [PubMed] [Google Scholar]
- Mi, H., Muruganujan A., Ebert D., Huang X., and Thomas P.D.. 2019. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47(D1):D419–D426. 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi, H., and Stappenbeck T.S.. 2013. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 8:2471–2482. 10.1038/nprot.2013.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat, J., Grueneberg D.A., Yang X., Kim S.Y., Kloepfer A.M., Hinkle G., Piqani B., Eisenhaure T.M., Luo B., Grenier J.K., et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 124:1283–1298. 10.1016/j.cell.2006.01.040 [DOI] [PubMed] [Google Scholar]
- Musha, H., Ohtani H., Mizoi T., Kinouchi M., Nakayama T., Shiiba K., Miyagawa K., Nagura H., Yoshie O., and Sasaki I.. 2005. Selective infiltration of CCR5(+)CXCR3(+) T lymphocytes in human colorectal carcinoma. Int. J. Cancer. 116:949–956. 10.1002/ijc.21135 [DOI] [PubMed] [Google Scholar]
- Nagarsheth, N., Wicha M.S., and Zou W.. 2017. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 17:559–572. 10.1038/nri.2017.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaides, N.C., Littman S.J., Modrich P., Kinzler K.W., and Vogelstein B.. 1998. A naturally occurring hPMS2 mutation can confer a dominant negative mutator phenotype. Mol. Cell. Biol. 18:1635–1641. 10.1128/MCB.18.3.1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okła, K., Farber D.L., and Zou W.. 2021. Tissue-resident memory T cells in tumor immunity and immunotherapy. J. Exp. Med. 218:e20201605. 10.1084/jem.20201605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.L., Gebhardt T., and Mackay L.K.. 2019. Tissue-Resident Memory T Cells in Cancer Immunosurveillance. Trends Immunol. 40:735–747. 10.1016/j.it.2019.06.002 [DOI] [PubMed] [Google Scholar]
- Pauken, K.E., Shahid O., Lagattuta K.A., Mahuron K.M., Luber J.M., Lowe M.M., Huang L., Delaney C., Long J.M., Fung M.E., et al. 2021. Single-cell analyses identify circulating anti-tumor CD8 T cells and markers for their enrichment. J. Exp. Med. 218:e20200920. 10.1084/jem.20200920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard, E., Verschoor C.P., Ma G.W., and Pawelec G.. 2020. Relationships Between Immune Landscapes, Genetic Subtypes and Responses to Immunotherapy in Colorectal Cancer. Front. Immunol. 11:369. 10.3389/fimmu.2020.00369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeker, M.O., and Carethers J.M.. 2020. Immunological Features with DNA Microsatellite Alterations in Patients with Colorectal Cancer. J. Cancer Immunol. (Wilmington). 2:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper, J., Tammela T., Akkad A., Almeqdadi M., Santos S.B., Jacks T., and Yilmaz O.H.. 2018. Colonoscopy-based colorectal cancer modeling in mice with CRISPR-Cas9 genome editing and organoid transplantation. Nat. Protoc. 13:217–234. 10.1038/nprot.2017.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, M., Crisafulli G., Sogari A., Reilly N.M., Arena S., Lamba S., Bartolini A., Amodio V., Magrì A., Novara L., et al. 2019. Adaptive mutability of colorectal cancers in response to targeted therapies. Science. 366:1473–1480. 10.1126/science.aav4474 [DOI] [PubMed] [Google Scholar]
- Sahan, A.Z., Hazra T.K., and Das S.. 2018. The Pivotal Role of DNA Repair in Infection Mediated-Inflammation and Cancer. Front. Microbiol. 9:663. 10.3389/fmicb.2018.00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vega, F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., Dimitriadoy S., Liu D.L., Kantheti H.S., Saghafinia S., et al. Cancer Genome Atlas Research Network . 2018. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell. 173:321–337.e10. 10.1016/j.cell.2018.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., and Clevers H.. 2011. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 141:1762–1772. 10.1053/j.gastro.2011.07.050 [DOI] [PubMed] [Google Scholar]
- Schlee, M., and Hartmann G.. 2016. Discriminating self from non-self in nucleic acid sensing. Nat. Rev. Immunol. 16:566–580. 10.1038/nri.2016.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, W.M., Chevillotte M.D., and Rice C.M.. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32:513–545. 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger, S., Luke J.J., Bao R., Zha Y., Hernandez K.M., Li Y., Gajewski A.P., Andrade J., and Gajewski T.F.. 2016. Density of immunogenic antigens does not explain the presence or absence of the T-cell-inflamed tumor microenvironment in melanoma. Proc. Natl. Acad. Sci. USA. 113:E7759–E7768. 10.1073/pnas.1609376113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive Integration of Single-Cell Data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wietmarschen, N., Sridharan S., Nathan W.J., Tubbs A., Chan E.M., Callen E., Wu W., Belinky F., Tripathi V., Wong N., et al. 2020. Repeat expansions confer WRN dependence in microsatellite-unstable cancers. Nature. 586:292–298. 10.1038/s41586-020-2769-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanpouille-Box, C., Demaria S., Formenti S.C., and Galluzzi L.. 2018. Cytosolic DNA Sensing in Organismal Tumor Control. Cancer Cell. 34:361–378. 10.1016/j.ccell.2018.05.013 [DOI] [PubMed] [Google Scholar]
- Wang, H., Xiang D., Liu B., He A., Randle H.J., Zhang K.X., Dongre A., Sachs N., Clark A.P., Tao L., et al. 2019. Inadequate DNA Damage Repair Promotes Mammary Transdifferentiation, Leading to BRCA1 Breast Cancer. Cell. 178:135–151.e19. 10.1016/j.cell.2019.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, J.R., Milne K., Watson P., Deleeuw R.J., and Nelson B.H.. 2014. Tumor-infiltrating lymphocytes expressing the tissue resident memory marker CD103 are associated with increased survival in high-grade serous ovarian cancer. Clin. Cancer Res. 20:434–444. 10.1158/1078-0432.CCR-13-1877 [DOI] [PubMed] [Google Scholar]
- Wu, T.D., Madireddi S., de Almeida P.E., Banchereau R., Chen Y.-J.J., Chitre A.S., Chiang E.Y., Iftikhar H., O’Gorman W.E., Au-Yeung A., et al. 2020. Peripheral T cell expansion predicts tumour infiltration and clinical response. Nature. 579:274–278. 10.1038/s41586-020-2056-8 [DOI] [PubMed] [Google Scholar]
- Yang, J., Sanderson N.S., Wawrowsky K., Puntel M., Castro M.G., and Lowenstein P.R.. 2010. Kupfer-type immunological synapse characteristics do not predict anti-brain tumor cytolytic T-cell function in vivo. Proc. Natl. Acad. Sci. USA. 107:4716–4721. 10.1073/pnas.0911587107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., Li L., Starr T.K., and Subramanian S.. 2017. Tumor location impacts immune response in mouse models of colon cancer. Oncotarget. 8:54775–54787. 10.18632/oncotarget.18423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumwalt, T.J., Arnold M., Goel A., and Boland C.R.. 2015. Active secretion of CXCL10 and CCL5 from colorectal cancer microenvironments associates with GranzymeB+ CD8+ T-cell infiltration. Oncotarget. 6:2981–2991. 10.18632/oncotarget.3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
provides information on all of the DNA oligos and primers used.
provides information on all antibodies used.
Data Availability Statement
Data from scRNAseq have been deposited in GEO under accession no. GSE178706.