Abstract
Coronavirus disease, which comes up in China at the end of 2019 and showed different symptoms in people infected, affected millions of people. Computer-aided expert systems are needed due to the inadequacy of the reverse transcription-polymerase chain reaction kit, which is widely used in the diagnosis of this disease. Undoubtedly, expert systems that provide effective solutions to many problems will be very useful in the detection of Covid-19 disease, especially when unskilled personnel and financial deficiencies in underdeveloped countries are taken into consideration. In the literature, there are numerous machine learning approaches built with different classifiers in the detection of this disease. This paper proposes an approach based on deep learning which detects Covid-19 and no-finding cases using chest X-ray images. Here, the classification performance of the Bi-LSTM network on the deep features was compared with the Deep Neural Network within the frame of the fivefold cross-validation technique. Accuracy, sensitivity, specificity and precision metrics were used to evaluate the classification performance of the trained models. Bi-LSTM network presented better performance compare to DNN with 97.6% value of high accuracy despite the few numbers of Covid-19 images in the dataset. In addition, it is understood that concatenated deep features more meaningful than deep features obtained with pre-trained networks by one by, as well. Consequently, it is thought that the proposed study based on the Bi-LSTM network and concatenated deep features will be noteworthy in the design of highly sensitive automated Covid-19 monitoring systems.
Keywords: Covid-19, Artifcial intelligence, Deep learning, Concatenated deep features, Bi-LSTM, X-ray imaging
Introduction
Covid-19 is a new enrolee of the coronavirus family of acute respiratory syndromes and has been denominated SARS-CoV-2 [1]. The virus caused an epidemic and affected the whole world economy. The global impact of this disease, which can be seen in both humans and animals, is not yet known, and its spread rate is quite high [2]. The coronavirus disease, which shows up in China in late 2019 and showed many different symptoms, affected millions of people [3]. The virus has spread to many countries through cross-country transport. The full symptoms of Covid-19 in an infected person can appear in about 14 days. Currently, there is no complete treatment method or medicine to get rid of this disease [4]. Early diagnosis of the symptoms of SARS-CoV-2 disease are difficult [5]. A kit called reverse transcription-polymerase chain reaction is used to diagnose this disease [6]. Inadequacy of this kit has been one of the biggest threats in preventing the rapid spread of the coronavirus [7]. During this time, people with the virus may have transmitted the virus to other people. To prevent this, a solution must be found that will detect the presence of the disease more quickly in people with Covid-19 [8]. Consequently, the quick spread of this disease and the negatives on people necessitate early diagnosis of this disease [9].
Undoubtedly, computer-aided decision support systems, which offer effective solutions to many problems, will be very useful for detecting Covid-19 disease, especially considering the unskilled personnel and financial deficiencies in underdeveloped countries. In this context, considering the time and financial costs required of laboratory kits used to fight this epidemic to diagnose, artificial intelligence-based solutions have been launched to support doctors who aim to treat patients and fight the disease [10]. Artificial intelligence-based studies are carried out effectively and successfully in the medical field. Deep features obtained from pre-trained Convolutional Neural Networks (CNN) layers are widely used in the computer vision. Chest X-ray and computed tomography (CT) medical images are widely used to detect Covid-19 cases [2, 4, 11, 12].
Field specialists make evaluations on chest X-ray or CT images to monitor the development of Covid-19 case. But, using CT images in the diagnosis of the Covid-19 has disadvantages such as the fact that many hospitals do not have the necessary equipment to obtain the CT image and the cost of tomographic imaging is high. Disinfection time between patients is approximately 15 min. Different from CT imaging, chest X-ray imaging is widely used for detecting Covid-19 as it requires both short imaging time and low cost [13]. This technique minimizes the possibility of spreading the virus and also exposes the patient to lower doses of ionizing radiation. Moreover, this technique costs less than a CT scan. However, detecting Covid-19 from these images is often quite a complex task, as it has similarities with other types of viral pneumonia [14].
Big-scale visual examination of X-ray images needs a long time and could lead to misdiagnosis of physicians due to a lack of prior knowledge of infected areas [15]. Moreover, due to the uncertainty of Covid-19 findings in most cases, expert radiologists can only identify 65% of those who are Covid-19 positive [16]. Therefore, there is a great need for quick and accurate Covid-19 automatic diagnosis. In this context, artificial intelligence approaches including deep learning architectures are mostly used to decrease the workload of radiologists [15]. The financial costs of diagnostic laboratory kits are an important problem in fighting the disease especially for developing and underdeveloped countries [17]. Clinicians and field experts can use deep learning-based expert systems for early detection of Covid-19 [14]. The recent popularity of deep learning architectures that offer high performance has given the opportunity to use models based on these architectures effectively. Deep learning methods, as used in many other fields, can be successfully applied in the fixation and classification of various diseases in the medical field without the need for the handy craft features required for machine learning.
Deep learning-based models offer solutions for problems in different areas have become very popular in this field by showing successful performance for both image-based classification applications and prediction problems. Studies of coronary artery disease [18], eye disease [19, 20], breast cancer [21–26], brain tumor [27–34], Alzheimer disease [35–42], prediction [43–45] are some of them in literature.
Recently, deep learning models including different approaches with high success have been implemented for the detection of Covid-19 disease using CT and CXR images. For example, Nigam et al. used five well-liked deep learning architectures to diagnose the Covid-19. They obtained X-ray images which are not including text and other unnecessary information using YOLO in their study. They accomplished the highest recognition accuracy of 93.48% with the EfficientNet model [2]. Shah et al. examined the performances of different deep learning techniques to accurate and impetuous Covid-19 detecting. In their study, the VGG-19 outperformed others with an accuracy of 94.52% [46]. Pathak et al. conducted experimental studies by applying a deep transfer learning approach and tenfold cross-validation to detect the Covid-19 disease. They obtained efficient results with 93.0189% value of accuracy by using the ResNet-50 network [47]. Loey et al. reported that ResNet50 is superior to other models to diagnose the Covid-19 with a testing accuracy of 82.91% on the chest CT radiograph digital images [48]. Sethy and Behera extracted deep features from chest X-ray images using different pre-trained CNN architectures. In their experimental study, the authors achieved the highest accuracy of 95.38% with the Support Vector Machine on deep features extracted by ResNet50 architecture [49]. Narin et al. detected Covid-19 and others with 96.1% classification accuracy using fivefold cross-validation [50]. Turkoglu extracted deep features from CT scan images with the transfer learning deep learning approach. Then, the author voted the classification results obtained by different activation methods in the proposed model called Multiple Kernels-ELM-based Deep Neural Network (MKs-ELM-DNN) for detecting Covid-19 disease. According to the results obtained, the MKs-ELM-DNN model presented 98.36% accuracy [12]. Lastly, Nayak performed a contrastive analysis of eight pre-trained networks. Besides, the author performed more analysis considering various factors such as type of optimization techniques, batch size, learning rate, epoch number, misclassification rate of the models. According to the results, the ResNet-34 model with 98.33% accuracy outperformed other models [4].
The main aim of this study is to present an effective and distinct deep learning-based model that shows a successful performance in Covid-19 disease which has adversely affects humanity since 2020. Here, the performances of two deep learning architecture on the deep features which are obtained by pre-trained models by one by and concatenated of them for Covid-19 and no-finding (normal) cases are discussed. In this context, the main contribution of this study is as follows:
The effectiveness of the Deep Neural Networks (DNN) and Bidirectional Long Short Term Memory (Bi-LSTM) deep learning models were compared comprehensively.
The performances of the models within the framework of the fivefold cross-validation technique were verified on the validation datasets.
Finally, a highly efficient deep learning model was derived for detecting Covid-19 disease.
The rest of the paper is structured as follows. Section 2 gives information about the dataset and used methods. Section 3 presents the experiments in detail. Section 4 discusses the results with comprehensive comparisons. Finally, Sect. 5 concludes the study with final remarks.
Materials and Method
This section summarizes the methods used in experimental studies for Covid-19 detection as well as explanations about the dataset used to evaluate the success of constructed models.
Dataset
To assess the proposed approach in this study, a dataset was comprised of chest X-ray images from two different public sources. This dataset consists of two sub-datasets with a total of 625 images. The first one consists of 125 Covid-19 cases randomly selected by Muhammed Talo from the dataset prepared by Cohen et al. [51]. The dataset prepared by Cohen [51] including chest X-ray images labeled as Covid-19 has received widespread attention since its publication [4]. The second one consists of 500 no finding cases randomly selected by Muhammed Talo from the dataset prepared by Wang et al. [52]. Each X-ray image was rescaled to a size of 224 × 224 in this study. Examples of chest X-ray images are shown in Fig. 1.
Fig. 1.

Sample images; a Covid-19 case, b no-finding case
Proposed Methodology
Recently, several deep learning architectures have been used effectively in computer vision and medical imaging classification studies. The main aim of this study is to build a model that performs positive and negative classification robustly and efficiently. With this aim, deep features extracted from the pre-trained deep learning architectures were sent as input to two different deep learning models built from scratch and the performances of these models were compared. Experimental studies include the validation of these models within the framework of the fivefold cross-validation technique. The description of each step was explained in detail in the next sub-sections. Figure 2 shows the general blog diagram of the proposed study. The deep learning model proposed in this study includes three main processes which are as follows:
Training and validation of DNN and Bi-LSTM were conducted using deep features within the framework of fivefold cross-validation.
Finally, the class prediction results of the models for test images were examined and evaluated.
Fig. 2.
The general block diagram of the proposed model
Both models were compiled using the Adam (learning rate = 0.001) optimization method [53]. Model training was carried out by 100 epochs with 32 batch sizes. The last layer with softmax activation function in the models was set to two neurons that represent Covid-19 and no-finding classes. To enhance the classification accuracy, the best models with the best weights in both deep neural network models were saved by utilizing the model checkpoint method. Then, the performance of the models for both DNN and Bi-LSTM was evaluated on the test datasets.
Transfer Learning
Transfer learning is a knowledge transferring strategy using a pre-trained CNN model instead of training a CNN from scratch to solve the classification problem in new images in different populations [54]. In this strategy, the deep features set representing images are sent as input to a network that will perform the classification job. This method is often used to overcome the computational costs that will become with a deep-network model that will be built from the scratch or to use ancillary feature extractors belong to trained models in the first step [10].
In this strategy, knowledge of a deep learning network trained on a big dataset is used to perform a task on a small dataset. This eliminates the need for a big dataset required for training and also long-term training of deep learning methods [55, 56]. Transfer learning provides the models trained on one million images in the Imagenet dataset and their weights to be used on other datasets. There are many studies in the literature using this technique. While some of these studies directly use the pre-trained models, some of them send the deep features extracted from CNN layers to different machine learning algorithms as input data.
Experiments
Experimental Setup
The training, validation, and test procedures of the models were carried out in a 64-bit Windows 10 operating system having 8 GB RAM and Intel 1.80 GHz CPU. All codes were implemented with Python language using Keras 2.3.1 framework.
Model Training
The number of X-ray images in each class was summarized in Table 1. 20% of the dataset which is a different part in each fold was reserved as a test set to measure the model performance within the frame of fivefold cross-validation [57]. The remaining 80% of the chest X-ray images were reserved as the train set in each fold. 20% of the training set was reserved for model validation. So, the tests of the models were performed five times on the different test sets in each fold. Figure 3 shows the flowchart of the data split and model validation processes.
Table 1.
Division of images into train, validation and test sets on the datasets
| Original dataset | Training | Validation | Testing | Total |
|---|---|---|---|---|
| Classes | ||||
| No-finding cases | 320 | 80 | 100 | 500 |
| Covid-19 cases | 80 | 20 | 25 | 125 |
| Total | 400 | 100 | 125 | 625 |
Fig. 3.

Experiments performed with fivefold cross-validation technique
As given in Table 1, training, validation, and test sets were reserved for each fold with values of 400,100 and 125, respectively. The training set consists of 80 images which having Covid-19 cases and 320 images which having no-finding cases. The validation set consists of 20 images which having Covid-19 cases and 80 images which having no-finding cases. The validation set was used to prevent the overfitting of the model and to obtain the most suitable model. Besides, the test set consists of 25 images which having Covid-19 cases and 100 images which having no-finding cases.
In this study, firstly fine-tuning was applied to VGG-16, DenseNet-121, and ResNet-50 deep learning architectures trained on a large-scale dataset called ImageNet [58] for chest X-ray images. Two deep neural network models built from scratch were used to classify the deep features extracted by the pre-trained models. DNN used in this study has four hidden layers with batch size = 32. Bi-LSTM network was set with the parameters of rnn_width = 16, dropOut = 0.1 and recurent DropOut = 0.1. While the dropout layers were added to prevent overfitting for both deep neural networks, the adaptive moment estimation algorithm (Adam) was used in the training process with a 0.001 learning rate value. The model checkpoint method was applied to save the best weights of the model. Therefore, the network parameters were load at the point of the maximum validation accuracy. A maximum of 100 training epochs was performed during the training of the DNN and Bi-LSTM. To evaluate the success of the models, accuracy (Acc), sensitivity (Sen), specificity (Spe), and precision (Pre) measures shown in the following equations were used.
| 1 |
| 2 |
| 3 |
| 4 |
where TP true positive, TN true negative, FP false positive, FN false negative.
Results
This section presents experimental results performed with two deep learning models on several deep features. The performance of the DNN model built from scratch on the feature sets extracted by transfer learning from the image set was examined (Scenario A). The performances of the Bi-LSTM model built from scratch on these feature sets were discussed (Scenario B). Also, the performances of the Bi-LSTM and DNN on the concatenated deep features were also examined (Scenario C). With these scenarios, it is aimed to determine a reliable and successful model by comparing the performances of different deep learning models.
Table 2 presents the average Acc, Sen, Spe and Pre values achieved by the models built in this study. By examining the performances of the DNN and Bi-LSTM models used in the classification of deep features extracted from each pre-trained model in Table 2, it was seen that the Bi-LSTM model presented the best performance on the concatenated deep features. While DNN model achieved 95.84% value of average accuracy, Bi-LSTM achieved 97.6% value of average accuracy on the concatenated deep features which presented the best accuracy. Bi-LSTM model presented high accuracy, high sensitivity, and high precision. It is seen that the Sen value is slightly low in both Bi-LSTM and DNN compare to other metrics. High Acc means that the ratio of the sum of samples correctly classified in each class to the total of all samples is high. The high Sen value means that the ratio of correctly classified Covid-19 images to all Covid-19 images is high. When the Sen values presented by the models are examined, it is stand out that the Bi-LSTM network detected Covid-19 cases on the concatenated deep features more successfully than other deep features. The high Spe value means that the ratio of correctly classified no-finding cases to all no-finding cases is high. When the Spe values presented by the models are examined, it also takes attention that the Bi-LSTM network detected no-finding cases on the concatenated deep features more successfully than others. High Pre means that the ratio of correctly classified Covid-19 images to all images which are classified as Covid-19 is high. Finally, when the Pre values presented by the models are examined, it is also strikeout that the Bi-LSTM network distinguished Covid-19 cases and no-finding cases on the concatenated deep features more successfully than other deep features. Also, this model also has a small standard deviation showing that it is effective and stable on the concatenated deep features.
Table 2.
Average results obtained by developed models on the deep features
| Deep features/deep learning model | Acc (%) | Sen (%) | Spe (%) | Pre (%) | |
|---|---|---|---|---|---|
| VGG-16 | DNN | 91.84 ± 2.23 | 69.6 ± 13.05 | 97.4 ± 2.8 | 89.21 ± 9.51 |
| Bi-LSTM | 95.68 ± 1.93 | 88.0 ± 7.16 | 97.6 ± 2.33 | 91.02 ± 7.31 | |
| ResNet-50 | DNN | 92.64 ± 1.38 | 70.4 ± 8.24 | 98.2 ± 0.75 | 90.93 ± 3.02 |
| Bi-LSTM | 93.28 ± 1.09 | 76.0 ± 5.66 | 97.6 ± 1.85 | 89.58 ± 6.28 | |
| DenseNet-121 | DNN | 88.32 ± 0.64 | 50.4 ± 6.5 | 98.0 ± 2.1 | 88.89 ± 10.69 |
| Bi-LSTM | 89.76 ± 1.48 | 60.0 ± 8.0 | 97.4 ± 1.5 | 86.27 ± 8.02 | |
| Concatenated deep features | DNN | 95.84 ± 1.06 | 82.4 ± 7.42 | 99.2 ± 0.75 | 96.59 ± 3.14 |
| Bi-LSTM | 97.6 ± 0.88 | 91.2 ± 1.6 | 99.2 ± 1.17 | 96.86 ± 4.5 |
The best result is shown in bold font
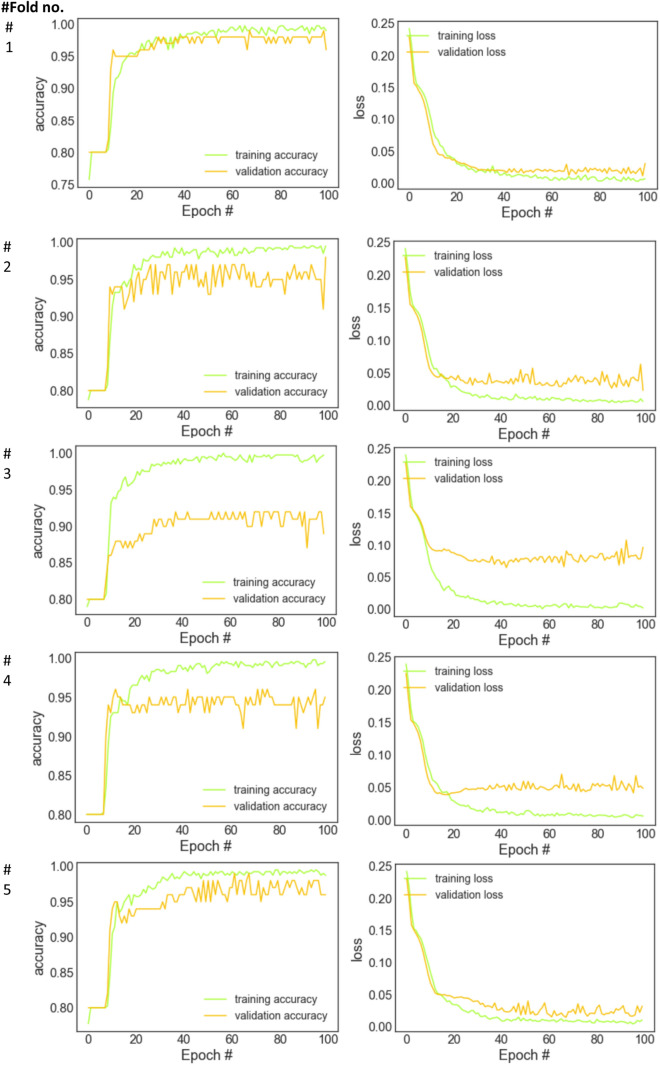
Figure 4 shows the plots of epoch vs accuracy and epoch vs loss obtained for each fold with the Bi-LSTM model on the concatenated deep features. It was observed that the loss value of the model decreased as the epoch value increased along with slight differences in general. Besides, the accuracy of the model increased as the number of epoch increased along with slight differences.
Fig. 4.
The accuracy/loss graphs of the training and validation process of the Bi-LSTM network on the concatenated deep features
Figure 5 shows overlapped confusion matrices obtained by the trained DNN and Bi-LSTM trained on the deep features extracted from the images in test sets. The classification results are seen as TP, FN, TN, and FP in the confusion matrices. With following the matrices, for example, row #3, The Bi-LSTM model classified 11 out of 125 images of the Covid-19 class as no-finding cases by using concatenated deep features. Besides, the model incorrectly classified 4 out of the 500 images of the no-finding case. For the same deep features, the DNN model classified 22 out of 125 images of the Covid-19 cases as no-finding cases. The model incorrectly classified 4 out of the 500 images of the no-finding case. The Bi-LSTM model is very successful with low false numbers which are 11 FN and 4 FP. According to these results, the Bi-LSTM effectively distinguishes Covid-19 cases from no-finding cases compared to DNN.
Fig. 5.
Overlapped confusion matrices obtained from the test datasets
Receiver operating characteristic curves which present the performance of the models built in this study on test data are presented in Fig. 6. It could be seen that the Bi-LSTM model has good classification performance on the test sets including the concatenated deep features for each fold. Moreover, the highest ROC AUC scores with Bi-LSTM was also stand out on these deep features set.
Fig. 6.
ROC curves of each deep features and classifier model
Discussion
In this section, the results and inferences in detecting the Covid-19 disease were discussed. As can be seen by examining the overlapped confusion matrix results, false negatives presented by DNN quite high compare to the Bi-LSTM network for all deep features extracted by pre-trained models. But, false positives presented by Bi-LSTM slightly high than DNN for deep features extracted by ResNet-50 and DenseNet-121 models. In the literature, there are many models that used different classifiers or for detecting Covid-19 disease. The best model obtained by the Bi-LSTM network with an acceptable classification performance was compared with other studies within the frame of two-class classification (Covid-19 and no-finding cases). The Bi-LSTM network achieved 97.6% average accuracy in classifying the concatenated deep features. Table 3 presents the comparison of the proposed study and others in the literature. Specifically, the proposed method achieves good performance on the no-finding cases compared to Covid-19 cases. In spite of the classification accuracy of the proposed model slightly low from the studies of Nayak et. [4] and Türkoğlu [12], it is successful compared to the studies of Sethy and Behera [49]. Nayak et al. [4] demonstrated higher success compared to the model proposed in this study, but they used hold-out technique, which is called a simple technique, in their study. Moreover, the proposed model presented a significant performance improvement compared to the studies performed on the CT images except for the study of Turkoğlu [12]. In addition, some of the limitations of this study may be clear in future research. In particular, many more samples of Covid-19 cases are needed for a more in-depth analysis. The low number of Covid-19 images limits model performance and may result in a biased result.
Table 3.
Comparison of the proposed model and related studies
| Study | Method | Data source | Split technique | Acc (%) |
|---|---|---|---|---|
| Shah et al. [46] | VGG-19 deep learning model | CT | Hold-out | 94.52 |
| Pathak et al. [47] | Deep transfer learning | CT | Tenfold cv | 93.01 |
| Loey et al. [48] | ResNet-50 deep learning model with data augmentation | CT | Hold-out | 82.91 |
| Turkoğlu [12] | Transfer learning and MKs-ELM-DNN model | CT | Tenfold cv | 98.36 |
| Sethy and Behera [49] | ResNet50 + support vector machine classifier | Chest X-ray | Hold-out | 95.38 |
| Narin et al. [50] | Transfer learning (ResNet-50) | Chest X-ray | Fivefold cv | 96.1 |
| Nayak et al. [4] | Transfer learning (ResNet-34) | Chest X-ray | Hold-out | 98.33 |
| This study | Concatenated deep features and Bi-LSTM | Chest X-ray | Fivefold cv | 97.6 |
The result of the proposed study is in bold
acv: cross-validation
Conclusion
Covid-19 disease caused an epidemic and affected the world economy. The spreading ratio of this virus, which the global effect is still unknown, is very high and can be seen in both humans and animals. Deep learning models were proven to achieve superior results in many medical image processing applications. The role of early warning systems in classifying Covid-19 and undetected cases is considered crucial today and will continue to be important in the future.
In this study, it was aimed to predict Covid-19 cases by sending deep features extracted from X-ray images into two deep neural network models namely DNN and Bi-LSTM developed from scratch. Experiments involving the comparison of DNN and Bi-LSTM models on deep features in this study differ from the approaches in the literature and therefore it is thought to contain a unique approach. In experiments, firstly, three different pre-trained models were used to compare their distinctive characteristics. According to the experiments, in terms of machine learning, the concatenated deep features are more meaningful than the deep features obtained individually by pre-trained networks. As seen from overlapped matrices, the concatenated deep features are an encouraging result, with optimal results with the lowest number of FN (11) and FP (4) compared to other pre-trained architectures. Moreover, the results indicate that the Bi-LSTM model outperformed DNN on the concatenated deep features with 97.6% value of Acc, 91.2% value of Sen, 99.2% value of Spe, and 96.86% value of Pre. Considering the high classification accuracy obtained, the model proposed could be used in the development of expert systems to be used in real clinical environments. Such an expert system to be developed using a low-cost based on deep learning architecture will help field experts to diagnose Covid-19 disease quickly and also down the errors arising from the subjective opinions of experts. In summary, the proposed study indicates that the Bi-LSTM is able to classify Covid-19 and no-finding cases, even with a little number of training images and a small number of Covid19 images. The proposed model can help radiologists or field experts for accurate and robust diagnosis of Covid-19 infection. Besides, the performance of this system can be increased by using different deep learning architectures and also by applying data augmentation techniques. The approach proposed in this study can be used as a low-cost tool that can provide faster and more accurate recognition of the virus to prevent its spread.
Particularly, it is thought that combine of clinical studies with expert systems is the next milestone for the solution of this problem. In the future, it is aimed to make a study by extracting the features based on activation maps. Moreover, a model with higher generalization capacity using different Covid-19 image sources is planned for future work. Also, the performances of the ensemble learning methods namely stacking or snapshot for detecting the Covid-19 will be examined. Finally, future work will involve applying different deep learning architectures to an automated detection system that will serve as the backbone of Covid-19 monitoring and alerting systems.
Acknowledgements
We especially thank Cohen et al and Wang et al because they prepared the images. Also, we thank Mohammed Talo because he prepared the sub-datasets by selecting the images randomly.
Declaration
Conflict of interest
Authors declare that they have no conflict of interest.
Contributor Information
Kemal Akyol, Email: kakyol@kastamonu.edu.tr.
Baha Şen, Email: bsen@ybu.edu.tr.
References
- 1.Lai CC, Shih TP, Ko WC, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nigam B, Nigam A, Jain R, et al. COVID-19: automatic detection from X-ray images by utilizing deep learning methods. Expert Syst Appl. 2021 doi: 10.1016/j.eswa.2021.114883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salman S, Salem ML. Routine childhood immunization may protect against COVID-19. Med Hypotheses. 2020;140:109689. doi: 10.1016/j.mehy.2020.109689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nayak SR, Nayak DR, Sinha U, et al. Application of deep learning techniques for detection of COVID-19 cases using chest X-ray images: a comprehensive study. Biomed Signal Process Control. 2021;64:102365. doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pan F, Ye T, Sun P, et al. Time course of lung changes at chest CT during recovery from Coronavirus disease 2019 (COVID-19) Radiology. 2020;295:715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang Y, Zhang H, Xie J, et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Deng X, Fu Q, et al. A weakly-supervised framework for COVID-19 classification and lesion localization from chest CT. IEEE Trans Med Imaging. 2020;39:2615–2625. doi: 10.1109/TMI.2020.2995965. [DOI] [PubMed] [Google Scholar]
- 8.Kanne JP, Little BP, Chung JH, et al. Essentials for radiologists on COVID-19: an update-radiology scientific expert panel. Radiology. 2020;296:E113–E114. doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie M, Chen Q. Insight into 2019 novel coronavirus—an updated interim review and lessons from SARS-CoV and MERS-CoV. Int J Infect Dis. 2020;94:119–124. doi: 10.1016/j.ijid.2020.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong D, Tang Z, Wang S, et al. The role of imaging in the detection and management of COVID-19: a review. IEEE Rev Biomed Eng. 2021;14:16–29. doi: 10.1109/RBME.2020.2990959. [DOI] [PubMed] [Google Scholar]
- 12.Turkoglu M. COVID-19 detection system using chest CT images and multiple Kernels-extreme learning machine based on deep neural network. IRBM. 2021 doi: 10.1016/j.irbm.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh Y, Park S, Ye JC. Deep learning COVID-19 features on CXR using limited training data sets. IEEE Trans Med Imaging. 2020;39:2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 14.López-Cabrera JD, Orozco-Morales R, Portal-Diaz JA, et al. Current limitations to identify COVID-19 using artificial intelligence with chest X-ray imaging. Health Technol (Berl) 2021;1:3. doi: 10.1007/s12553-021-00520-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi F, Wang J, Shi J, et al. Review of artificial intelligence techniques in imaging data acquisition, segmentation, and diagnosis for COVID-19. IEEE Rev Biomed Eng. 2021;14:4–15. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 16.Castiglioni I, Ippolito D, Interlenghi M, et al. Artificial intelligence applied on chest X-ray can aid in the diagnosis of COVID-19 infection: a first experience from Lombardy, Italy. medRxiv. 2020 doi: 10.1101/2020.04.08.20040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekeroglu B, Ozsahin I. Detection of COVID-19 from chest X-ray images using convolutional neural networks. SLAS Technol. 2020;25:553–565. doi: 10.1177/2472630320958376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.JayaSree M, Koteswara Rao L. Survey on—identification of coronary artery disease using deep learning. Mater Today Proc. 2020 doi: 10.1016/j.matpr.2020.09.526. [DOI] [Google Scholar]
- 19.Badar M, Haris M, Fatima A. Application of deep learning for retinal image analysis: a review. Comput Sci Rev. 2020;35:100203. doi: 10.1016/j.cosrev.2019.100203. [DOI] [Google Scholar]
- 20.Perdomo O, Rios H, Rodríguez FJ, et al. Classification of diabetes-related retinal diseases using a deep learning approach in optical coherence tomography. Comput Methods Programs Biomed. 2019;178:181–189. doi: 10.1016/j.cmpb.2019.06.016. [DOI] [PubMed] [Google Scholar]
- 21.Liu T, Huang J, Liao T, et al. A hybrid deep learning model for predicting molecular subtypes of human breast cancer using multimodal data. IRBM. 2021 doi: 10.1016/j.irbm.2020.12.002. [DOI] [Google Scholar]
- 22.Prasath Alias Surendhar S, Vasuki R. Breast cancers detection using deep learning algorithm. Mater Today Proc. 2021 doi: 10.1016/j.matpr.2020.11.600. [DOI] [Google Scholar]
- 23.Gangaputra S, Lovato JF, Hubbard L, et al. Comparison of standardized clinical classification with fundus photograph grading for the assessment of diabetic retinopathy and diabetic macular edema severıty. Retina. 2013;33:1393–1399. doi: 10.1097/IAE.0b013e318286c952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang T, Wong JHD, Ng WL, Chan CS. Deep learning radiomics in breast cancer with different modalities: overview and future. Expert Syst Appl. 2020;158:113501. doi: 10.1016/j.eswa.2020.113501. [DOI] [Google Scholar]
- 25.Singla C, Sarangi PK, Sahoo AK, Singh PK. Deep learning enhancement on mammogram images for breast cancer detection. Mater Today Proc. 2020 doi: 10.1016/j.matpr.2020.10.951. [DOI] [Google Scholar]
- 26.Houssein EH, Emam MM, Ali AA, Suganthan PN. Deep and machine learning techniques for medical imaging-based breast cancer: a comprehensive review. Expert Syst Appl. 2020;167:114161. doi: 10.1016/j.eswa.2020.114161. [DOI] [Google Scholar]
- 27.Saranya C, Priya JG, Jayalakshmi P, Pavithra EH. Brain tumor identification using deep learning. Mater Today Proc. 2021 doi: 10.1016/j.matpr.2020.11.555. [DOI] [Google Scholar]
- 28.Mehrotra R, Ansari MA, Agrawal R, Anand RS. A transfer learning approach for AI-based classification of brain tumors. Mach Learn with Appl. 2020;2:100003. doi: 10.1016/j.mlwa.2020.100003. [DOI] [Google Scholar]
- 29.Mohsen H, El-Dahshan E-SA, El-Horbaty E-SM, Salem A-BM. Classification using deep learning neural networks for brain tumors. Futur Comput Informatics J. 2018;3:68–71. doi: 10.1016/j.fcij.2017.12.001. [DOI] [Google Scholar]
- 30.Saba T, Sameh Mohamed A, El-Affendi M, et al. Brain tumor detection using fusion of hand crafted and deep learning features. Cogn Syst Res. 2020;59:221–230. doi: 10.1016/j.cogsys.2019.09.007. [DOI] [Google Scholar]
- 31.Deepak S, Ameer PM. Brain tumor classification using deep CNN features via transfer learning. Comput Biol Med. 2019;111:103345. doi: 10.1016/j.compbiomed.2019.103345. [DOI] [PubMed] [Google Scholar]
- 32.Hashemzehi R, Mahdavi SJS, Kheirabadi M, Kamel SR. Detection of brain tumors from MRI images base on deep learning using hybrid model CNN and NADE. Biocybern Biomed Eng. 2020;40:1225–1232. doi: 10.1016/j.bbe.2020.06.001. [DOI] [Google Scholar]
- 33.Gupta A, Verma A, Kaushik D, Garg M. Applying deep learning approach for brain tumor detection. Mater Today Proc. 2020 doi: 10.1016/j.matpr.2020.10.063. [DOI] [Google Scholar]
- 34.Srikanth B, Venkata Suryanarayana S. Multi-class classification of brain tumor images using data augmentation with deep neural network. Mater Today Proc. 2021 doi: 10.1016/j.matpr.2021.01.601. [DOI] [Google Scholar]
- 35.Li H, Habes M, Wolk DA, Fan Y. A deep learning model for early prediction of Alzheimer’s disease dementia based on hippocampal magnetic resonance imaging data. Alzheimer’s Dement. 2019;15:1059–1070. doi: 10.1016/j.jalz.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abuhmed T, El-Sappagh S, Alonso JM. Robust hybrid deep learning models for Alzheimer’s progression detection. Knowledge-Based Syst. 2021;213:106688. doi: 10.1016/j.knosys.2020.106688. [DOI] [Google Scholar]
- 37.Bringas S, Salomón S, Duque R, et al. Alzheimer’s disease stage identification using deep learning models. J Biomed Inform. 2020;109:103514. doi: 10.1016/j.jbi.2020.103514. [DOI] [PubMed] [Google Scholar]
- 38.Puente-Castro A, Fernandez-Blanco E, Pazos A, Munteanu CR. Automatic assessment of Alzheimer’s disease diagnosis based on deep learning techniques. Comput Biol Med. 2020;120:103764. doi: 10.1016/j.compbiomed.2020.103764. [DOI] [PubMed] [Google Scholar]
- 39.Raza M, Awais M, Ellahi W, et al. Diagnosis and monitoring of Alzheimer’s patients using classical and deep learning techniques. Expert Syst Appl. 2019;136:353–364. doi: 10.1016/j.eswa.2019.06.038. [DOI] [Google Scholar]
- 40.Zhou Y, Lu Y, Pei Z. Intelligent diagnosis of Alzheimer’s disease based on internet of things monitoring system and deep learning classification method. Microprocess Microsyst. 2021;83:104007. doi: 10.1016/j.micpro.2021.104007. [DOI] [Google Scholar]
- 41.Bi X, Li S, Xiao B, et al. Computer aided Alzheimer’s disease diagnosis by an unsupervised deep learning technology. Neurocomputing. 2020;392:296–304. doi: 10.1016/j.neucom.2018.11.111. [DOI] [Google Scholar]
- 42.An N, Ding H, Yang J, et al. Deep ensemble learning for Alzheimer’s disease classification. J Biomed Inform. 2020;105:103411. doi: 10.1016/j.jbi.2020.103411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ingle DV, Deshmukh DS. Ensemble deep learning framework for stock market data prediction (EDLF-DP) Glob Transitions Proc. 2021 doi: 10.1016/j.gltp.2021.01.008. [DOI] [Google Scholar]
- 44.Xin Z, Jun S, Yan T, et al. A deep learning based regression method on hyperspectral data for rapid prediction of cadmium residue in lettuce leaves. Chemom Intell Lab Syst. 2020;200:103996. doi: 10.1016/j.chemolab.2020.103996. [DOI] [Google Scholar]
- 45.da Silva BLS, Inaba FK, Salles EOT, Ciarelli PM. Fast deep stacked networks based on extreme learning machine applied to regression problems. Neural Netw. 2020;131:14–28. doi: 10.1016/j.neunet.2020.07.018. [DOI] [PubMed] [Google Scholar]
- 46.Shah V, Keniya R, Shridharani A, et al. Diagnosis of COVID-19 using CT scan images and deep learning techniques. Emerg Radiol. 2021 doi: 10.1007/s10140-020-01886-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pathak Y, Shukla PK, Tiwari A, et al. Deep transfer learning based classification model for COVID-19 disease. IRBM. 2020 doi: 10.1016/j.irbm.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loey M, Manogaran G, Khalifa NEM. A deep transfer learning model with classical data augmentation and CGAN to detect COVID-19 from chest CT radiography digital images. Neural Comput Appl. 2020 doi: 10.1007/s00521-020-05437-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar Sethy P, Kumari Behera S (2020) Detection of coronavirus disease (COVID-19) based on deep features, preprints. 10.20944/preprints202003.0300.v1
- 50.Narin A, Kaya C, Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Anal Applic. 2021 doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cohen JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M (2020) COVID-19 image data collection: prospective predictions are the future. ArXiv: 2006.11988. https://github.com/ieee8023/covid-chestxray-dataset.
- 52.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) ChestX-ray8: hospital scale chest X-Ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases, in Proc. IEEE Conf. Comput. Vis. Pattern Recognit.
- 53.Hawkins DM. The problem of overfitting. J Chem Inf Comput Sci. 2004;44:1–12. doi: 10.1021/ci0342472. [DOI] [PubMed] [Google Scholar]
- 54.Weiss K, Khoshgoftaar TM, Wang D. A survey of transfer learning. J Big Data. 2016;3:9. doi: 10.1186/s40537-016-0043-6. [DOI] [Google Scholar]
- 55.Shin HC, Roth HR, Gao M, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35:1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nayak DR, Dash R, Majhi B. Automated diagnosis of multi-class brain abnormalities using MRI images: a deep convolutional neural network based method. Pattern Recognit Lett. 2020;138:385–391. doi: 10.1016/j.patrec.2020.04.018. [DOI] [Google Scholar]
- 57.Refaeilzadeh P, Tang L, Liu H (2009) Cross-validation. In: Liu L, Özsu MT (eds) Encyclopedia of database systems, pp 532–538. Springer US, New York. 10.1007/978-0-387-39940-9_565
- 58.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60(6):84–90. doi: 10.1145/3065386. [DOI] [Google Scholar]