Abstract
The emergence of the COVID-19 epidemic in the United States (U.S.) went largely undetected due to inadequate testing. New Orleans experienced one of the earliest and fastest accelerating outbreaks, coinciding with Mardi Gras. To gain insight into the emergence of SARS-CoV-2 in the U.S. and how large-scale events accelerate transmission, we sequenced SARS-CoV-2 genomes during the first wave of the COVID-19 epidemic in Louisiana. We show that SARS-CoV-2 in Louisiana had limited diversity compared to other U.S. states and that one introduction of SARS-CoV-2 led to almost all of the early transmission in Louisiana. By analyzing mobility and genomic data, we show that SARS-CoV-2 was already present in New Orleans before Mardi Gras, and the festival dramatically accelerated transmission. Our study provides an understanding of how superspreading during large-scale events played a key role during the early outbreak in the U.S. and can greatly accelerate epidemics.
Keywords: SARS-CoV-2, genomic epidemiology, phylogenetics, mobility, viral emergence, viral sequencing
Graphical abstract

Genomic and epidemiological analyses provide a clearer picture of one of the earliest SARS-CoV-2 superspreader events in the United States in accelerating transmission, with a single introduction of the virus being responsible for most cases during this period.
Introduction
In December 2019, SARS-CoV-2 was first identified in cases of unknown pneumonia in Wuhan, China (Wu et al., 2020; Zhou et al., 2020). Initially, community transmission was confined to China, but in late February 2020, large-scale outbreaks were increasingly detected in Europe, the Middle East, and elsewhere (World Health Organization, 2020a, 2020b). Although SARS-CoV-2 was first detected in the United States (U.S.) in January 2020 (Centers for Disease Control, 2020a), the majority of early COVID-19 cases were associated with travel from high-risk countries or close contact with travelers (Centers for Disease Control, 2020b).
By late February, widespread community transmission of SARS-CoV-2 in the U.S. was identified in Washington state (Worobey et al., 2020), New York City (Maurano et al., 2020), and Santa Clara County in California (Deng et al., 2020), but it is estimated that local transmission in the U.S. started earlier and was more widespread than recognized at the time (Davis et al., 2020; Perkins et al., 2020). Elsewhere, outside of these early virus “hotspots” in the U.S., transmission of SARS-CoV-2 occurred mostly silently due to lack of testing until the second week of March (Jorden et al., 2020; Davis et al., 2020; Lu et al., 2020). In contrast to the emergence of inherently more transmissible virus variants in the fall of 2020 and beyond (Davies et al., 2021; Faria et al., 2021), in the early phase of the epidemic transmission was mainly driven by favorable epidemiological circumstances. It seems likely that large-scale events in this period dramatically accelerated early SARS-CoV-2 transmission and that subsequent interstate seeding amplified the COVID-19 epidemic in the U.S.
More than one million people from all over the U.S. were drawn to the Mardi Gras parades in New Orleans starting on February 14th and culminating on February 25th, 2020 (Mardi Gras day, or “Fat Tuesday”). The timing and the scale of this event, as well as the absence of any meaningful mitigation efforts (in agreement with official guidelines at the time), provides a unique opportunity to investigate how large-scale events can accelerate SARS-CoV-2 transmission and amplify local outbreaks during the ongoing pandemic. To investigate this, we sequenced SARS-CoV-2 from cases in New Orleans and other locations in Louisiana and compared them with SARS-CoV-2 genomes from the U.S. and globally to reconstruct the timing, origin, and emergence of the virus in Louisiana. By integrating genomic, epidemiological, and mobility data, we show that SARS-CoV-2 overdispersion during Mardi Gras greatly accelerated the early outbreak in New Orleans, comparable to the emergence of more transmissible SARS-CoV-2 variants in the winter of 2020, and seeded the virus to other parts of Louisiana and nearby states. Our findings suggest that large-scale events in the beginning of 2020 may have contributed significantly to SARS-CoV-2 transmission early in the COVID-19 epidemic in the U.S., which is in contrast to epidemic waves later in the epidemic that were also fueled by inherently more transmissible lineages. Without widespread availability of vaccination and testing, large gatherings of people without strict control efforts will continue to amplify the COVID-19 pandemic.
Results
SARS-CoV-2 was likely introduced into Louisiana via domestic travel
To understand the early emergence of SARS-CoV-2 in Louisiana, we investigated epidemiological, genomic, and travel data of SARS-CoV-2 during the first wave of the epidemic (March 9th–May 15th). We found that SARS-CoV-2 in Louisiana displayed little genetic diversity compared to other states and was likely introduced from a domestic source.
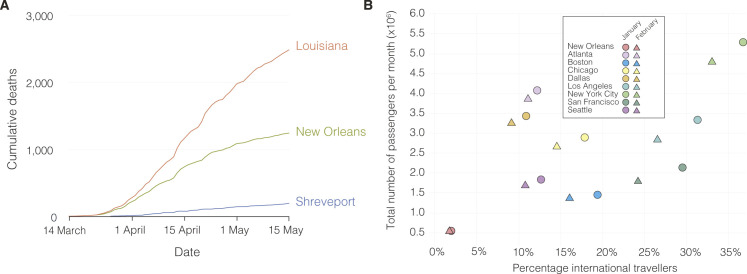
Using aggregated parish-level COVID-19 case data (Outbreak.info, 2021a), we analyzed reported cases and deaths during the first wave of the epidemic in Louisiana. The first reported case of COVID-19 in Louisiana was detected on March 9th, 2020, and the epidemic rapidly increased with reported cases reaching a peak on April 4th (Figure 1 A). While COVID-19 cases were reported throughout Louisiana during the first wave, the New Orleans-Metairie metropolitan statistical area (MSA; henceforth referred to as New Orleans) accounted for more than 54.9% of all deaths in the period up until May 1st (Figure S1, Figure S10 ) and was the focal point of the epidemic in Louisiana.
Figure 1.
SARS-CoV-2 epidemiology in Louisiana
(A) Epidemiological curve and number of sequenced samples in New Orleans, Shreveport and other parishes in Louisiana.
(B) Sampling location of sequenced SARS-CoV-2 samples in Louisiana: New Orleans metro area (blue), Shreveport metro area (green), and other parishes in Louisiana (orange).
(C) Maximum clade credibility tree of whole genome SARS-CoV-2 sequences sampled from Louisiana, U.S., and outside the U.S. The black circles show the strength of the posterior support for each node.
(D) Domestic and international air travel passenger volumes to Louisiana in February and March.
(E) Relative NextStrain clade prevalence per U.S. state up until May 15th (bottom). Number of sequences per U.S. state up until May 15th (top).
(F) Shannon evenness of NextStrain clades per U.S. state in relation to available SARS-CoV-2 sequences.
Figure S1.
Number of COVID-19 deaths and international arrivals in New Orleans in Louisiana, related to Figure 1
(A) Cumulative COVID-19 deaths during the first wave of the SARS-CoV-2 epidemic in Louisiana. (B) International arrivals for New Orleans and other major airports in the U.S. in January and February.
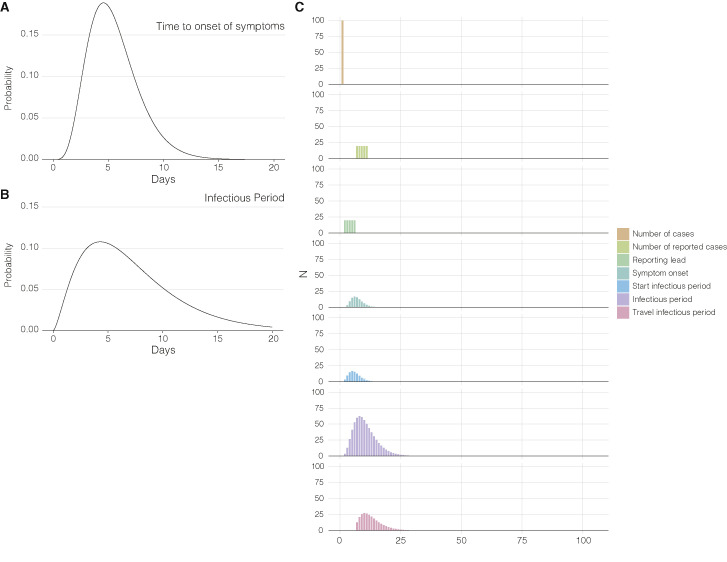
Figure S10.
Underlying distributions to infer import and export risk, related to Figures 4 and 5 and STAR Methods
(A) Gamma distribution of the time to onset of symptoms used to infer the number of infectious travelers. (B) Gamma distribution of the infectious period used to infer the number of infectious travelers. (C) Illustration of how the number of infectious travelers is derived from the number of cases. The number of infectious travelersa is used to calculate SARS-CoV-2 import risk. The panel shows how a 100 cases at day 1 result in a distribution of the infectious travelers several days later given heterogeneity in symptom onset and reporting, and assuming cases won’t travel after having received a positive SARS-CoV-2 test.
Early SARS-CoV-2 epidemics in New York and the West Coast were seeded by international introductions from Europe and Asia, respectively (Worobey et al., 2020). However, the source of many other local epidemics in the U.S., including the one in Louisiana, is unknown. To determine whether the emergence of SARS-CoV-2 in Louisiana originated from a domestic or international source, we sequenced 235 SARS-CoV-2 virus genomes collected from COVID-19 patients in New Orleans, Shreveport (Shreveport-Bossier City, LA MSA), and other parishes in Louisiana (Figures 1A and 1B). We reconstructed phylogenetic trees together with 1,263 whole-genome sequences that were representative of the global SARS-CoV-2 sequence diversity between January and May 2020. We found that the lineages responsible for the first wave in Louisiana all closely resembled SARS-CoV-2 sequences sampled within the U.S., suggesting that the epidemic in Louisiana was seeded from a domestic source (Figure 1C).
To further investigate the origin of the SARS-CoV-2 introduction into Louisiana, we investigated domestic and foreign air travel into Louisiana and found that in February, 360,000 passengers arrived from within the U.S., while only 40,000 international travelers were reported (Figure 1D). In particular, we found that travel from Europe and Asia, where the majority of SARS-CoV-2 transmissions occurred in February, accounted for less than 5% of all travel movements to Louisiana (Figures 1D and Figure S1, Figure S10). Consistent with our phylogenetic analysis, the travel data strongly suggest that the COVID-19 epidemic in Louisiana was due to seeding from domestic sources of SARS-CoV-2, and, unlike New York (Maurano et al., 2020) and Washington (Worobey et al., 2020), not the result of importations from Europe, Asia, or other foreign regions.
Early SARS-CoV-2 transmission in Louisiana predominantly originated from a single introduction
Unrestricted domestic travel in the U.S. in February 2020 and associated large travel volumes likely facilitated the emergence of SARS-CoV-2 in Louisiana. To investigate how many times SARS-CoV-2 was introduced into Louisiana, we first conducted a high-level genomic analysis by comparing NextStrain clade distributions of all available SARS-CoV-2 sequences from the continental U.S. up until May 15th, 2020. We found that SARS-CoV-2 sequences from Louisiana almost exclusively belonged to a single clade, 20C (Figure 1E). In other U.S. states with more than 10 sequences available, including neighboring states of Louisiana, we observed the co-circulation of multiple clades at more equal frequencies than in Louisiana (Figures 1E and 1F). In fact, we found that the genetic diversity of SARS-CoV-2 in Louisiana strongly resembled outbreaks on cruise ships (Figures 1E and 1F). These findings suggest that, like on the Diamond Princess and Grand Princess cruise ships (Deng et al., 2020; Sekizuka et al., 2020), SARS-CoV-2 in Louisiana most likely originated from a single source.
To further support these findings, we reconstructed a maximum likelihood tree of our SARS-CoV-2 genomes from Louisiana together with a representative selection of 1,399 clade 20C sequences collected across the U.S. (Figure 2 A). We found that within clade 20C, the majority of SARS-CoV-2 sequences in Louisiana belonged to a single cluster (“Louisiana clade”; Figures 2A and 2B), which is characterized by a single defining nucleotide mutation (C27964T; Figure 2A). Within the Louisiana clade, we identified three additional subclades supported by single nucleotide mutations, but the Louisiana clade was otherwise strongly dominated by polytomies, consistent with rapid local transmission (Figure 2A). Outside the main Louisiana clade, we found ten singleton sequences, but these either resulted in very limited or no onward transmission and likely did not contribute substantially to the overall SARS-CoV-2 transmission during the first wave (Figure 2A). The clustering of SARS-CoV-2 sequences within a single well-supported Louisiana clade strongly suggests that a single introduction was responsible for the vast majority of transmission events during the first wave of the epidemic in Louisiana.
Figure 2.
Phylogenetic analysis of SARS-CoV-2 in Louisiana
(A) Maximum likelihood tree of SARS-CoV-2 genomes sequenced from other parts of the U.S. and Louisiana. U.S. states that are not color-coded are indicated in gray. Arrows indicate clades.
(B) Illustration of maximum clade credibility tree. Gradients are used to illustrate uncertainty in the topology and node heights. Numbered arrows are nodes with a relatively high posterior support and correspond to the arrows in panel A. The red colored arrow indicates the most recent common ancestor of SARS-CoV-2 in Louisiana and represents the start of local transmission in Louisiana.
(C) Posterior distribution of the first emergence into New Orleans (blue) and Shreveport (green). The time of the first location transition (Markov jump) to New Orleans and Shreveport along the phylogenetic tree of each posterior sample was computed, and the posterior distribution was learned by summarizing across all the posterior samples.
SARS-CoV-2 likely emerged in Louisiana prior to the Mardi Gras festival
Both the timing and the onset of the COVID-19 epidemic in New Orleans as well as media reports (Table S1) suggest that Mardi Gras, which culminated in large parades on Mardi Gras day on February 25th, 2020, may have played a role in the spread or emergence of SARS-CoV-2 in Louisiana. It is unclear, however, if SARS-CoV-2 was introduced during Mardi Gras or if local transmission was already ongoing prior to the festival. To evaluate when SARS-CoV-2 started circulating in Louisiana, we created time-aware phylogenies to estimate the median time to the most recent common ancestor (TMRCA) for the Louisiana clade, which indicates the likely start of sustained local transmission (Grubaugh et al., 2019a; Suchard et al., 2018). We found that the posterior median TMRCA of the Louisiana clade was February 13th (95% highest posterior density [HPD] interval: January 24th, 2020–February 27th, 2020), suggesting that low levels of local SARS-CoV-2 transmission within Louisiana were likely already ongoing prior to Mardi Gras (Figure 2B).
To further investigate potential local transmission prior to Mardi Gras, we determined the emergence of SARS-CoV-2 in Louisiana by inferring the timing of the first introduction (location transition), often called a Markov jump (Minin and Suchard, 2008), into New Orleans and Shreveport across our full model posterior distribution that includes uncertainty on the tree and model parameters. We estimated that SARS-CoV-2 lineages belonging to the Louisiana clade emerged in New Orleans with a median time of February 11th, 2020, which is two weeks before Mardi Gras day (Pr[introduction < February 25th] = 97.9%), and, in confirmation, two days before our TMRCA estimates of sustained local transmission on February 13th (Figures 2B and 2C). In Shreveport, we found that SARS-CoV-2 emerged noticeably later than in New Orleans, after Mardi Gras on March 17th (Pr[introduction > February 25th] = 95.5%; Figure 2C). Combined, our phylodynamic analyses suggest that SARS-CoV-2 emerged and spread locally in New Orleans a couple of weeks prior to Mardi Gras day.
Favorable epidemiological circumstances resulted in superspreading during Mardi Gras
Although we found that SARS-CoV-2 likely began spreading in New Orleans mid-February 2020, the first official COVID-19 case was not reported until March 9th. This suggests that SARS-CoV-2 was likely spreading undetected and unmitigated during the large-scale gathering of people during Mardi Gras. To determine whether the festival may have accelerated the early COVID-19 epidemic in Louisiana, we modeled the number of likely daily cases using reported deaths (Figure 3 A) and compared these with a forward simulation of case numbers using a negative binomial branching process model starting from the onset of local transmission on February 13th, 2020 (Figures 2C, 3B, and S2). We found that the number of infections inferred based on observed death counts was substantially higher than the expected number of infections, suggesting superspreading during Mardi Gras (Figure 3C). In addition, we show that superspreading during Mardi Gras likely resulted in increased transmission in New Orleans in the immediate period after Mardi Gras (Figure 3D) and that it was caused by favorable epidemiological circumstances rather than virus genetics (Figure 3E).
Figure 3.
Acceleration of SARS-CoV-2 transmission during Mardi Gras
(A) Modeled incidence of SARS-CoV-2 in New Orleans based on registered COVID-19 deaths as inferred using Epidemia. The inset shows SARS-CoV-2 incidence in February and the hashed area indicates the cumulative number of COVID-19 cases up until Mardi Gras day (February 25th, 2020).
(B) Forward simulation of the cumulative number of infections between the TMRCA (February 13th) and the end of Mardi Gras using a negative binomial branching process model. The red dotted lines indicate the estimated median number of infections.
(C) Probability density curve of the number of COVID-19 cases required on Mardi Gras day to recapitulate the epi curve in New Orleans (random sampling of the probability distributions of A and B, see Figure S2 for additional details). The red dotted line indicates the median number of cases. The hashed area is the probability that no increased transmission occurred during Mardi Gras. The black lines indicate the probability of accelerated transmission by 100, 200, 300, 400, and 500 COVID-19 cases.
(D) SARS-CoV-2 incidence inferred from reported COVID-19 deaths between Mardi Gras day and the statewide stay at home order in Louisiana for New Orleans, Shreveport, and 52 metro areas with a population of more than 1 million.
(E) Lineage growth rate and normalized genetic distance of Pango lineages across counties in the United States. Lineage growth rate was calculated based on a 10-day interval after at least 5 sequences per week were reported. Variants of concern are outlined in red, whereas lineages that emerged during the first pandemic wave are outlined in black.
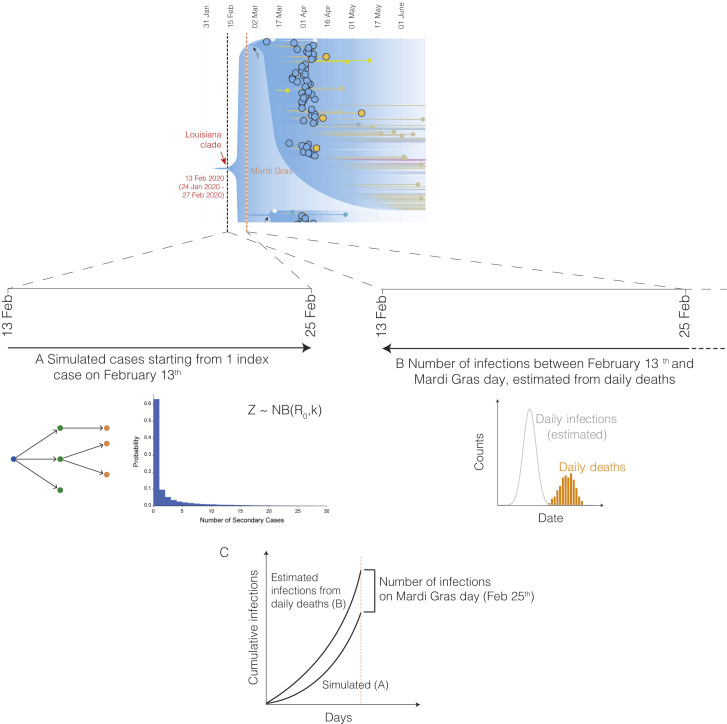
Figure S2.
Overview of forward and backward simulation to determine the number of infections on Mardi Gras day, related to Figure 3 and STAR Methods
(A) Forward simulation of cases starting with a single introduction on February 13th using a negative binomial branching process model (B). Estimated number of infections using the Epidemia model based on daily reported COVID-19 deaths (C). The number of infections on Mardi Gras day (February 25th) is determined by estimating the difference between the forward and backward simulated infections on Mardi Gras day.
To estimate daily COVID-19 case numbers in the absence of reporting during February 2020, we reconstructed the number of likely infections based on the number of reported deaths using a Bayesian regression model (Flaxman et al., 2020). Since our model was not able to accommodate sudden increases in transmission that are typically associated with superspreading events (Flaxman et al., 2020), we estimated the number of cases between February 11th and Mardi Gras day on February 25th, 2020. We found that by Mardi Gras day, 793 (95% HPD: 400–1,497) cumulative cases would have been required to align our model with the estimated daily number of SARS-CoV-2 infections during the first wave of the COVID-19 epidemic in New Orleans (Figure 3A). To estimate the likely number of infections in New Orleans between February 13th (start of local transmission of the Louisiana clade; Figure 2B) and the end of Mardi Gras (February 25th), assuming a constant reproduction number and an epidemic initiated from a single individual, we simulated the number of cases using a negative binomial branching process model (Lloyd-Smith et al., 2005). We estimated a total of 42 (95% confidence interval [CI]: 5-491) infections occurred between February 13th and Mardi Gras day (Figure 3B), which is substantially lower than the estimated 793 infections that would have been required to recapitulate the number of cases seen later in March (Figure 3A).
To estimate the number of likely SARS-CoV-2 infections during Mardi Gras, we calculated the median difference between our previously estimated number of infections up until Mardi Gras day inferred from observed deaths (793; Figure 3A) and the number of cases that were expected based on the start of local transmission from a single individual on February 13th (42; Figure 3B). We estimated that a median of 713 infections would have been required during Mardi Gras to recapitulate our modeled epidemiological curve (Figure 3C), with only a 0.9% probability that no transmission occurred at all during the festival. To better understand the magnitude of SARS-CoV-2 transmission during Mardi Gras, we randomly sampled the probability distribution of the inferred (from deaths) and simulated (via branching process model started on February 13th) cases and calculated the probability of various transmission scenarios ranging from 100 to 500 additional infections during the festival. We found that at least 100 infections occurred during Mardi Gras with a 98.4% probability, and that at least 500 occurred with a 79.9% probability (Figure 3C). These findings suggest that superspreading very likely occurred during the festival resulting in hundreds of SARS-CoV-2 infections.
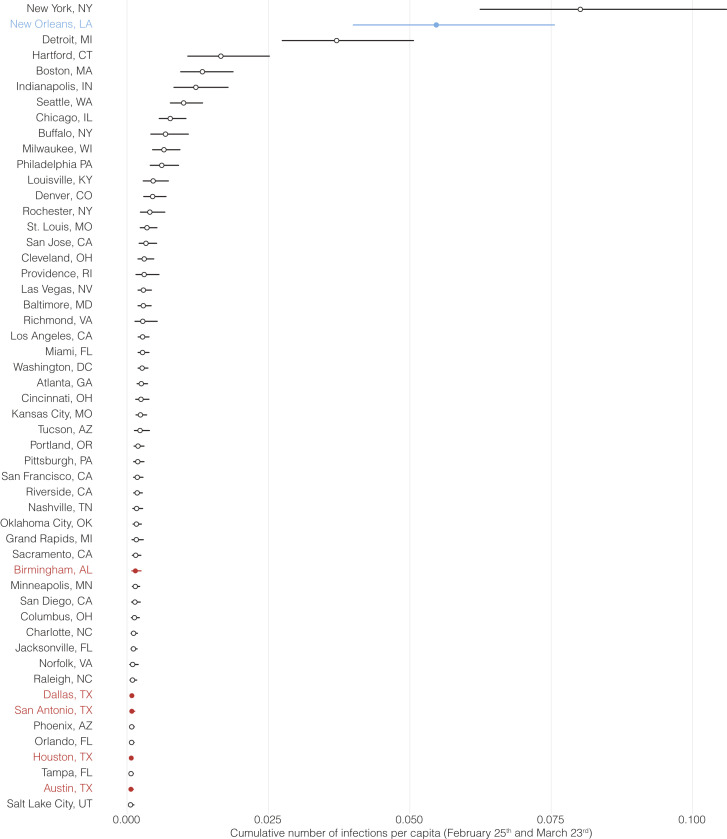
We hypothesized that superspreading during Mardi Gras should have resulted in a more rapid increase of early COVID-19 cases in New Orleans compared to other U.S. cities. To investigate this, we used a Bayesian regression model to estimate daily case numbers in New Orleans and other large population centers in the weeks after Mardi Gras until the statewide stay-at-home order in Louisiana on March 23rd, 2020 (Flaxman et al., 2020). We found that infection rates were substantially higher in New Orleans than in other large population centers, including cities with the next eight highest infection rates in the U.S. (Detroit, Boston, New York, Indianapolis, Chicago, Seattle, Buffalo, and Hartford; Figures 3D and S3 ). Since all of these population centers were located in the north or the west of the U.S., we also compared New Orleans to regional population centers in the South (Houston, Dallas, Birmingham, and Shreveport). We found 3.7- to 73-fold higher infection rates in New Orleans compared with these regional cities, indicating that infection rates in New Orleans were uniquely high in the Southern U.S. (Figures 3D and S3). The increased rate of COVID-19 cases in New Orleans in the weeks immediately after Mardi Gras suggests that superspreading occurred during the festival, and is in agreement with our previous analyses (Figures 3A, 3B, and 3C).
Figure S3.
Cumulative number of SARS-CoV-2 infections, related to Figure 3
Median estimates for the number of SARS-CoV-2 infections and their 95% HPD between February 25th and March 23rd in 52 metro areas with a population of more than 1 million. New Orleans is indicated in blue, and regional metro areas closest to New Orleans are indicated in red.
To understand whether the first COVID-19 wave in Louisiana was unique or representative of SARS-CoV-2 transmission observed elsewhere in the U.S. during the early epidemic, we compared the growth rate of individual lineages across counties in the U.S. (Figure 3E). We found that SARS-CoV-2 lineages B.1 and B.1.595 in New Orleans (using the Pango naming scheme [Rambaut et al., 2020]; both fall in the Louisiana clade; Figure 2A) showed a unique combination of high lineage growth rate and low genetic diversity, indicating a uniquely rapidly expanding virus population in Louisiana during the first wave (Figure 3E). In fact, we found that except for New York, all other counties in the U.S. had much slower growth rates during the first wave of the pandemic than the Louisiana clade (Figure 3E), suggesting that virus transmission in New Orleans was unusually high at the beginning of the first wave.
To investigate to what extent rapid transmission during Mardi Gras was the result of favorable epidemiological circumstances or potential virus genetics, we also compared the growth rates of SARS-CoV-2 lineages across the U.S. with variants of concern that emerged in the winter of 2020 (Washington et al., 2021). We found that the lineage growth rates in New Orleans were only slightly lower compared to the emergence of B.1.1.7 in the UK but were more than 50% higher than other variants of concern, such as B.1.427, B.1.351, and B.1.526 (Figure 3E). Since B.1.1.7 is inherently more transmissible than other SARS-CoV-2 lineages (Davies et al., 2021), this suggests that favorable epidemiological circumstances alone can be sufficient to achieve growth rates similar to much more transmissible SARS-CoV-2 variants.
SARS-CoV-2 in Louisiana was highly similar to SARS-CoV-2 lineages circulating in Texas
Our analyses showed that SARS-CoV-2 was most likely introduced into Louisiana via domestic travel (Figure 1C). To more precisely determine the likely source of SARS-CoV-2 into Louisiana, we performed Bayesian phylogeographic analyses and analyzed mobility data from across the U.S. and found that SARS-CoV-2 in Louisiana may have originated in Texas. Prior to Mardi Gras, our analyses demonstrated that Texas is more than twice as likely as the next most probable state to be the source of SARS-CoV-2 lineages in New Orleans, while SARS-CoV-2 in Shreveport likely originated from New Orleans itself (Figures 4A and 4B).
Figure 4.
Origin of SARS-CoV-2 emergence in Louisiana
(A) Relative distribution of location transitions inferred by phylogeographic analysis, by origin state. Only location transitions that occurred before Mardi Gras day (February 25th) were included.
(B) Estimated number of location transitions into New Orleans (left) and Shreveport (right).
(C) Estimated number of travelers from states with the highest travel volumes to New Orleans, Shreveport, and other parishes in Louisiana.
(D) Import risk to New Orleans. Import risk was estimated based on the number of infectious travelers relative to the population size and the total number of travelers at the origin (see Figure S10 for more details). Large Southern U.S. states and U.S. states that had early outbreaks of SARS-CoV-2 are color-coded. Other U.S. states that were included in the phylogenetic analysis are shown in gray.
(E) Relative import risk into New Orleans. Gray area represents other U.S. states that were included in the phylogenetic analysis.
Although these analyses point to Texas as a likely source of the Louisiana clade, our phylogeographic inference is limited by geographic and temporal sampling (Bloomquist et al., 2010). Therefore, we also investigated movement between New Orleans, Shreveport, and other U.S. states by analyzing human mobility patterns. To determine the number of travelers into Louisiana from states in the U.S. that were represented in our phylogenetic analysis, we used weekly mobility data generated by SafeGraph (SafeGraph, 2020). We found that travel movements in the week of February 13th into Louisiana were strongly dominated by Texas, which accounted for 13% of travel to New Orleans, and 35% of travel to Shreveport (Figure 4C). These findings suggest that Texas and other regions of Louisiana were the main origins of travel into New Orleans and Shreveport during February 2020.
To investigate the SARS-CoV-2 importation risk into New Orleans during February 2020, we estimated the import risk based on the number of incoming travelers and the SARS-CoV-2 incidence rate at likely U.S. states of origin. We found that although the overall import risk into New Orleans was small, during the week of the likely initial introduction (February 13th; Week 7; Figure 4D), Florida and Texas represented 29% and 24% of the total import risk, respectively, whereas we estimated a lower proportion of import risk from more distant states, including California (3%), Washington (20%), and New York (0.2%; Figure 4E). These results are in agreement with the findings from our phylogenetic and mobility analyses, suggesting that the Louisiana clade may have originated via an introduction of SARS-CoV-2 from Texas.
Exportation of SARS-CoV-2 from New Orleans may have caused localized outbreaks in nearby states
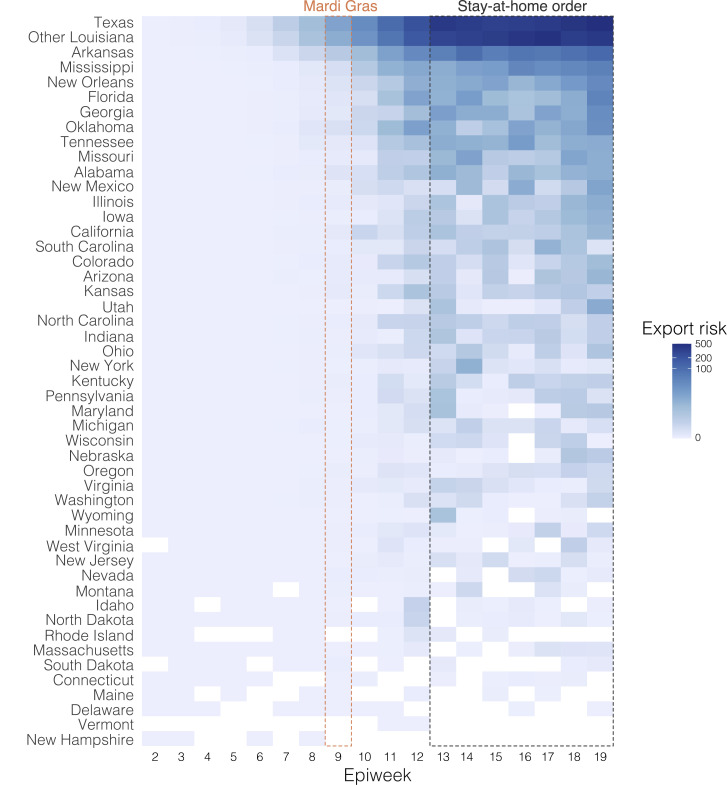
Our observation that superspreading during Mardi Gras likely led to increased transmission rates within New Orleans prompted us to investigate if this could also have resulted in spread to other U.S. states. We analyzed SARS-CoV-2 exports from New Orleans using mobility and genomic data in the four weeks after Mardi Gras until the stay-at-home order on March 23rd, which resulted in a large decline of travel and incidence. We found that the export from New Orleans was highest for nearby states and regions, in particular other parts of Louisiana, Mississippi, Alabama, and Texas (Figure 5 ).
Figure 5.
SARS-CoV-2 export risk from Louisiana
(A) Estimated number of location transitions inferred by phylogeographic analysis from New Orleans (left) and Shreveport (right). On the right of each graph the number of sequences in the dataset belonging to clade 20C and the Louisiana clade is shown. The strength of a connection between a particular location and New Orleans/Shreveport is relative to the difference between the number of location transitions and the number of sequences in clade 20C.
(B) Estimated number of infected travelers from New Orleans per week. The number of infected travelers was estimated based on local incidence and the total number of travelers between New Orleans and the destination.
(C) Percentage of import risk in the lower 48 U.S. states that can be attributed to New Orleans in the four epidemiological weeks after Mardi Gras. Import risk was estimated based on the number of infectious travelers relative to the population size and the total number of travelers at the origin (see Figure S10 for more details). Inset shows local relative import risk from New Orleans within Louisiana.
To determine to what extent increased transmission following superspreading during Mardi Gras could have resulted in SARS-CoV-2 infections in other states, we analyzed location transitions from New Orleans to regions in Louisiana and states across the U.S. We found that SARS-CoV-2 from New Orleans may have primarily spread to nearby regions, in particular Texas and Louisiana (Figure 5A). In contrast, transmission in Shreveport, where we did not observe increased transmission following Mardi Gras, did not show large amounts of spread to other locations other than New Orleans (Figure 5A). However, since we found that location transitions from New Orleans following Mardi Gras exclusively occurred within the Louisiana clade, we compared the number of transitions to the number of genomes in the Louisiana clade for each location. We found that the majority of all SARS-CoV-2 jumps into Mississippi and Alabama can be traced back to New Orleans (Figure 5A), suggesting that SARS-CoV-2 transmission in New Orleans may have resulted in regional spreading of COVID-19.
To further investigate to what extent increased transmission in New Orleans may have acted as a source for seeding SARS-CoV-2 to other U.S. states, we estimated the export risk from New Orleans by analyzing travel movements between New Orleans and U.S. states. We found that the export risk from New Orleans was highest to nearby regions and states, in particular to other parts of Louisiana, Mississippi, and Texas (Figure 5B). In the four weeks between the end of Mardi Gras and the stay-at-home order, these accounted for 60% of all exported risk from New Orleans, increasing to 70% of all risk in the subsequent weeks when air travel was highly restricted (Figure 5B). In line with our phylogenetic analyses, we found that SARS-CoV-2 exports from Shreveport were substantially lower than from New Orleans (Figure S4 ).
Figure S4.
Export risk from Shreveport per epiweek, related to Figure 5
Mardi Gras and the stay-at-home-order are indicated by the dotted lines.
As export risk from New Orleans was strongly driven by travel movements, our estimates were inherently biased toward states with larger populations. Therefore, to determine the impact of SARS-CoV-2 exports from New Orleans on local SARS-CoV-2 transmission in each U.S. state, we estimated the relative import risk from New Orleans by calculating the percentage of total SARS-CoV-2 import risk for each state that could be attributed to New Orleans. We found that the relative import risk from New Orleans was highest in neighboring U.S. states or regions (Figure 5C). In particular, for Mississippi and other parts of Louisiana, we found that the majority of the SARS-CoV-2 imports may have come from New Orleans (Figure 5C). Although the relative import risk from New Orleans declined everywhere after the statewide stay-at-home order, the decline was less pronounced for Mississippi and Louisiana, which both consistently had the highest relative import risks from New Orleans throughout the entire first wave of the COVID-19 epidemic in Louisiana (Videos S1 and S2). Taken together, both our phylogenetic and mobility analysis suggest that the early COVID-19 epidemic in New Orleans was amplified by superspreading during Mardi Gras and may have helped seed local outbreaks in neighboring U.S. states and regions.
Frequent reintroductions largely determine the lineage prevalence in later epidemic waves
Since the superspreading we observed during Mardi Gras resulted in the early dominance of a single SARS-CoV-2 lineage (Figure 2A, the “Louisiana clade”), we next investigated how first wave events may influence the prevalence of lineages in later epidemic waves. By reconstructing SARS-CoV-2 lineage dynamics during multiple consecutive COVID-19 waves, we found that new waves are largely characterized by reintroductions of new lineages and not by resurgence of lingering low-level transmission of preexisting lineages.
The COVID-19 epidemic in Louisiana during 2020 and early 2021 had three distinct epidemic waves, each interrupted by troughs of low transmission (Figure 6 A). To investigate SARS-CoV-2 lineage dynamics, we constructed a maximum likelihood phylogenetic tree containing all available SARS-CoV-2 sequences (n = 3,196) from Louisiana spanning March 2020 to March 2021 and found that SARS-CoV-2 was strongly temporally clustered into different lineages (Figure 6A). To estimate the turnover of the Louisiana clade, which was dominant during the first wave (Figure 2A) through all successive waves, we calculated the prevalence of this clade in each epidemic phase. We found that the Louisiana clade rapidly declined between the first trough and second epidemic wave, followed by a more gradual decline in subsequent epidemic phases (Figure 6B), resulting in less than 5% of all COVID-19 cases by February 2021 (Figure 6B). These findings suggest that the statewide stay-at-home order that was in effect between March and May 2020 (Figure 5) resulted in a rapid decline of the Louisiana clade that extinguished the first wave, only to be later replaced by different lineages during later waves via domestic reintroductions of SARS-CoV-2.
Figure 6.
Lineage and clade persistence of SARS-CoV-2 in Louisiana
(A) Maximum likelihood tree of SARS-CoV-2 showing sequences collected throughout three consecutive epidemic waves in Louisiana. Sequences from Louisiana are annotated according to their epidemic phase, as shown in the epicurve inset.
(B) Evolution of Louisiana clade prevalence over time. Sequences belonging to the Louisiana clade are indicated in (A) in blue.
(C) Pango lineage distribution of SARS-CoV-2 sequences from Louisiana per epidemic phase. The total number of sequences in each phase is shown next to the graph.
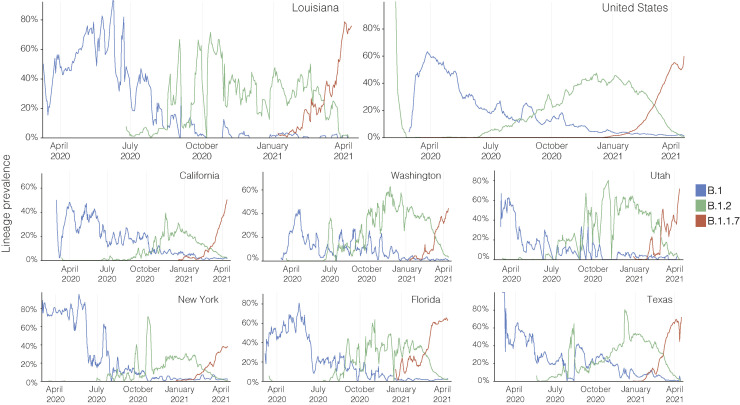
To investigate how often lineage replacement occurred in Louisiana over the course of the pandemic, we determined the lineage distribution during each epidemic phase (Figure 6C). We found a frequent lineage turnover, and lineage B.1.2 and B.1.596 (green) replaced the initially dominant B.1 lineages (blue) after the second wave, after which B.1.1 and descending lineages (red) largely replaced B.1.2 and B.1.596 after the third wave (Figure 6C). We found that this frequent lineage turnover followed a larger national trend in the U.S. with similar shifts in lineage dominance observed in other U.S. states such as Texas, California, Florida, and New York (Figure S5 ) (Outbreak.info, 2021b). The rapid replacement of the Louisiana clade after the first wave suggests that reintroductions of SARS-CoV-2 largely shape later epidemic waves, especially during periods of low local transmission in between epidemic waves.
Figure S5.
Lineage prevalence during the epidemic in the United States, related to Figure 6
Lineage prevalence of B.1, B.1.2, and B.1.1.7 in the United States, Louisiana and other U.S states from March 2020 until April 2021.
Discussion
In this study, we show that domestic travel likely introduced SARS-CoV-2 into Louisiana and that a single introduction directly led to the vast majority of transmission during the first wave. Furthermore, we present several lines of evidence showing that it is likely that the Mardi Gras festival in New Orleans was a superspreading event: (1) an unusual lack of genetic diversity of SARS-CoV-2 in Louisiana, which is in sharp contrast with what has been seen in other large U.S. cities and more similar to what has been observed during cruise ship outbreaks; (2) although our analyses suggest that SARS-CoV-2 was likely transmitting locally before Mardi Gras, we found that it is unlikely that the observed epidemiological curve in New Orleans could have been recapitulated without superspreading during Mardi Gras; (3) infection rates in New Orleans in the weeks immediately following Mardi Gras were substantially higher than in other major cities throughout the U.S.; and (4) the growth rate of lineages falling within the Louisiana clade was close to the highly transmissible B.1.1.7 variant, suggesting highly favorable epidemiological circumstances.
The rapid nature of the early COVID-19 epidemic in New Orleans likely resulted in thousands of additional cases, which is supported by seroprevalence studies showing exposure rates of close to ten percent by May 15th, 2020 in New Orleans (Feehan et al., 2020). Compared to neighboring states that did not experience the same explosive first waves as Louisiana, the CDC’s Nationwide Commercial Laboratory Seroprevalence Survey estimated that the seroprevalence in Louisiana was 35%–134% higher than in other states in the Southern U.S. (Centers for Disease Control, 2020c).
SARS-CoV-2 superspreading events can rapidly change the course of local outbreaks. Previously, superspreading during a biotech conference in Boston in early 2020 (Lemieux et al., 2021) and a motorcycle rally in Sturgeon, South Dakota in August, 2020 (Dave et al., 2020) have been estimated to have resulted in more than 250,000 SARS-CoV-2 infections. Although we did not attempt to estimate the exact magnitude of the Mardi Gras superspreading event, given the lack of genetic diversity of SARS-CoV-2 within Louisiana, it seems likely that the majority of the ∼50,000 confirmed COVID-19 cases during the first wave (Outbreak.info, 2020) can be traced back to Mardi Gras. However, we show here that subsequent epidemic waves are not defined by previous ones, indicating that effective non-pharmaceutical interventions can effectively cancel the effect of previous superspreading events.
We used a combination of genomic and mobility data to investigate the import and export of SARS-CoV-2 into and out of Louisiana. Our phylogenetic analyses show that SARS-CoV-2 in Louisiana most likely originated from Texas (Figure 4). However, most of the Louisiana clade consists of sequences from various U.S. states that either share the basal node of the Louisiana clade or belong to unresolved polytomies originating from this node. This makes accurate phylogeographic inference challenging, particularly in situations with rapid spread between different locations (Villabona-Arenas et al., 2020). Previous genomic epidemiology studies investigating the emergence of SARS-CoV-2 in San Francisco (Deng et al., 2020), Boston (Lemieux et al., 2020), and New York (Maurano et al., 2020) showed that determining the source of introduction during the early stages of the COVID-19 pandemic can be challenging. A particularly illustrative example is the (re-)emergence of SARS-CoV-2 in Washington state in January and February 2020. The first case in Washington was linked to recent travel from China (Bedford et al., 2020), and when six weeks later other, genetically similar cases were detected, it was initially thought to be the result of community transmission in the context of inadequate testing (Bedford et al., 2020). Only after a reanalysis with related SARS-CoV-2 genomes from nearby British Columbia, Canada could prolonged local transmission be excluded in favor of a more likely explanation of additional virus introduction(s) into the state (Worobey et al., 2020). In this study, we supplemented our phylogenetic analyses with large-scale analyses of travel and mobility patterns to gain more confidence in our finding that the SARS-CoV-2 in Louisiana may have been introduced via travel from Texas. However, our estimates remain unsure and much more extensive sequencing of SARS-CoV-2 from early in the U.S. epidemic would be required to obtain more conclusive answers.
We showed that lineage growth rates can vary considerably depending on either epidemiological or virus genetic factors. Soon after the first wave, we observed that newly imported lineages replaced the lineages falling within the Louisiana clade (Figure 6), indicating that these lineages are not inherently more transmissible due to virus genetic factors. This shows that epidemiological factors alone can increase the growth rate of lineages that are not inherently more transmissible to a level that is similar to those of highly transmissible variants, like B.1.1.7 (Davies et al., 2021; Washington et al., 2021). However, epidemiological factors and genetic factors can also amplify each other, as is the case in a recent outbreak in India, where large-scale gatherings and the emergence of SARS-CoV-2 variants resulted in the largest COVID-19 outbreak to date (Outbreak.info, 2021a).
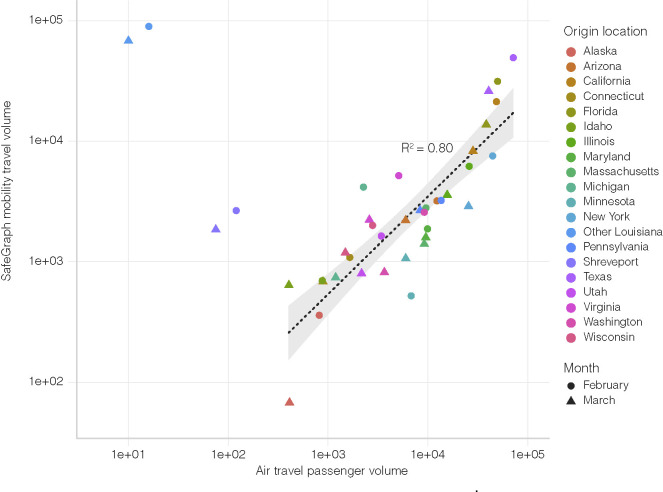
We used mobility data to determine human movement between U.S. states. Such movement, however, changed dramatically over the course of the pandemic, particularly air travel (Transport Security Agency, 2020). In addition, we found that air travel, as expected, can be a poor indicator of short-distance movement (Figure S6 ). To capture human movements of short distances, we therefore used weekly SafeGraph mobility data, which is based on cell phone tracking (SafeGraph, 2021). Cell phone tracking data has been shown to capture human movements on various distance scales (Chang et al., 2021; Kraemer et al., 2020). To further increase the accuracy of our mobility analysis and mitigate large swings in human movements due to government intervention, we only analyzed travel until mid-March, before Louisiana and many other states adopted stay-at-home orders and travel substantially decreased.
Figure S6.
Correlation between travel datasets, related to STAR Methods
Air travel passenger volumes and SafeGraph mobility travel volumes from various U.S. states into New Orleans. Spearman rank correlation does not include Shreveport and Other Louisiana, since air travel is not the dominant mode of transport to New Orleans for these locations.
Our phylogenetic analyses indicate that SARS-CoV-2 was introduced into New Orleans multiple times but that only one main clade (the “Louisiana clade”) was eventually successful in establishing widespread community transmission. We estimated that the emergence of the Louisiana clade in New Orleans occurred in mid-February, just prior to Mardi Gras. However, estimating an accurate introduction date with limited genetic diversity can be challenging (Grubaugh et al., 2019a). We therefore investigated timing by estimating both the time of introduction by analyzing location transitions and the start of local transmission by determining the TMRCA of the Louisiana clade. We found that both analyses suggest that the Louisiana clade was likely present in New Orleans prior to Mardi Gras.
With the recent emergence of more transmissible SARS-CoV-2 variants in the U.S. (Galloway et al., 2021) and elsewhere (Volz et al., 2021), robust virus genomic surveillance systems and analysis frameworks will be critical to provide insights into the ongoing spread and evolution of SARS-CoV-2. We show that a single introduction of SARS-CoV-2 can rapidly find its way through an unprotected population and cause large-scale epidemics in the absence of adequate testing and control efforts. Our study provides a key example of how a large-scale event played an important role during the early epidemic in the U.S. and how such events may continue to play a role in amplifying local outbreaks if SARS-CoV-2 is left unchecked.
Limitations of the study
In this study we analyze genetic and epidemiological data to show that Mardi Gras was most likely a superspreading event in the early phase of the COVID-19 pandemic in the U.S. Our phylodynamic and phylogeographic analyses are biased by uneven collection and sequencing of SARS-CoV-2 samples in New Orleans and Shreveport, Louisiana as well as other U.S. states. Due to the lack of testing in February and early March 2020, we relied on modeling the number of cases based on the number of COVID-19 deaths to estimate early COVID-19 prevalence in the U.S.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| Omega BioTek MagBind Viral DNA/RNA Kit | Omega Biotek | Cat#M6246-03 |
| QIAmp Viral RNA Mini Kit | QIAGEN | Cat#52904 |
| Quick-RNA Viral Kit | Zymo Research | Cat#R1034 |
| SuperScript IV VILO Master Mix | ThermoFisher Scientific | Cat#11756500 |
| AMPure XP | Beckman Coulter | Cat#A63882 |
| Maxima H Minus First Strand cDNA Synthesis Kit | ThermoFisher Scientific | Cat#K1652 |
| Nextera Flex for Enrichment Library Preparation kit | Illumina | Cat#20025524 |
| Nextera XT | Illumina | Cat#FC-131-1096 |
| Illumina MiSeq with MiSeq reagent kit V3. | Illumina | Cat#MS-102-3003 |
| Illumina NextSeq with 500/550 Mid Output Kit v2.5 | Illumina | Cat#20024908 |
| KingFisher Flex Purification System | ThermoFisher Scientific | Cat#5400630 |
| Q5 Hot Start High-Fidelity DNA Polymerase Kit | NEB | Cat #0493L |
| NEBNext Ultra II DNA Library Kit for Illumina | NEB | Cat#E7645L |
| Deposited data | ||
| SARS-CoV-2 reference genome | NCBI | NCBI: NC_045512.2 |
| SARS-CoV-2 consensus sequences | GISAID | Table S2 |
| SARS-CoV-2 raw data | NCBI | BioProject accession ID: PRJNA643574, PRJNA681020, PRJNA643575, and PRJNA612578 |
| BEAST XML and log files | This paper | https://github.com/andersen-lab/paper_2020_new-orleans-hcov-genomics |
| Epidemiological data | Outbreak.info | https://outbreak.info/ |
| Oligonucleotides | ||
| ARTIC Network n-CoV-19 V3 primers | ARTIC Network | https://github.com/artic-network/artic-ncov2019/tree/master/primer_schemes/nCoV-2019/V3 |
| Software and algorithms | ||
| Pangolin v2.0 | Rambaut et al., 2020 | https://github.com/cov-lineages/pangolin |
| NextClade v0.12.0 | Hadfield et al., 2018 | https://github.com/nextstrain/nextclade |
| IQtree2 | Minh et al., 2020 | https://github.com/iqtree/iqtree2 |
| BEASTv1.10.5pre | Suchard et al., 2018 | https://github.com/beast-dev/beast-mcmc/tree/v1.10.5pre_thorney_v0.1.0 |
| BEAGLE | Ayres et al., 2019 | https://faculty.washington.edu/browning/beagle/beagle.html#download |
| Baltic | GitHub | https://github.com/evogytis/baltic |
| Snakemake | Köster and Rahmann, 2012 | https://snakemake.readthedocs.io/en/stable/ |
| BWA-mem | Li, 2013 | https://github.com/lh3/bwa |
| BreSeq v.0.34.1 | Deatherage and Barrick, 2014 | https://github.com/barricklab/breseq |
| iVar v1.2.2 | Grubaugh et al., 2019b | https://github.com/andersen-lab/ivar/releases/tag/v1.2.2 |
Resource availability
Lead contact
Further information and requests for reagents may be directed to the lead contact Kristian Andersen (andersen@scripps.edu).
Materials availability
This study did not generate new unique reagents, but raw data and code generated as part of this research can be found in the supplemental files, as well as on public resources as specified in the Data and code availability section below.
Experimental model and subject details
Ethical Statement
Sample collection, RNA extraction, and viral sequencing was evaluated by the Institutional Review Boards (IRBs) at Tulane University (IRB# 2020-396), Louisiana State University Health System (LSUHS) (IRB# STUDY00001445) and Ochsner Health (IRB# 2019.334). All samples were de-identified before receipt by the study investigators.
Method Details
Sample Collection and RNA extraction
Nasopharyngeal swabs from Tulane Medical Center were collected March-April 2020 from 1) hospitalized COVID-19 patients consenting to participate in viral isolation and sequencing studies and 2) left-over clinical samples from individuals presenting to the Emergency Department (ED) with COVID-19 symptoms. Nasopharyngeal swabs from LSUHS and Ochsner health were left-over clinical samples from either outpatient or hospitalized individuals.
Viral RNA was extracted using the QIAmp Viral RNA Mini Kit (QIAGEN), Quick-RNA Viral Kit (Zymo Research) or Mag-Bind Viral DNA/RNA kit (Omega Bio-tek) according to the manufacturer’s instructions. RNA extracts from samples collected at Tulane Medical Center were screened for presence of SARS-CoV-2 Nucleocapsid gene according to the 2019-nCoV Real Time rRT-PCR Panel protocol (Centers for Disease Control, 2020d) on the QuantStudio 3 (Applied Biosciences); only the N1 Primer/Probe Mix was used(F: 5'-GACCCCAAAATCAGCGAAAT-3', R: 5'-TCTGGTTACTGCCAGTTGAATCTG-3', Probe: 5'- FAM-ACCCCGCAT/ZEN/TACGTTTGGTGGACC-3IABkFQ-3'). Samples with a Ct < 30 (correlating to ∼500 copies of virus/mL) were selected for amplicon sequencing and viral RNA was shipped to Scripps Research Institute. RNA extracts from samples collected at LSUHS were screened with an EUA diagnostic RT-qPCR at the LSUHS emerging viral threat laboratory and shipped for sequencing to the Microbial Genome Sequencing Center (MiGS) in Pittsburgh, PA.
SARS-CoV-2 Amplicon Sequencing
SARS-CoV-2 was sequenced using PrimalSeq-Nextera XT. This protocol is based on the ARTIC PrimalSeq protocol and adapted for Illumina Nextera XT library preparation (Quick et al., 2017). The ARTIC network nCoV-2019 V3 primer scheme uses two multiplexed primer pools to create overlapping 400 bp amplicon fragments in two PCR reactions. Instead of ligating Illumina adapters, Nextera XT is used to circumvent the 2x250 or 2x300 read length requirement. A detailed version of this protocol can be found here: https://andersen-lab.com/secrets/protocols/. Briefly, SARS-CoV-2 RNA (2 mL) was reverse transcribed with SuperScript IV VILO (ThermoFisher Scientific). The virus cDNA was amplified in two multiplexed PCR reactions (one reaction per ARTIC network primer pool) using Q5 DNA High-fidelity Polymerase (New England Biolabs). Following an AMPureXP bead (Beckman Coulter) purification of the combined PCR products, the amplicons were diluted and libraries were prepared using Nextera XT (Illumina) or NEBNext Ultra II DNA Library Prep Kits (New England Biolabs). The libraries were purified with AMPureXP beads and quantified using the Qubit High Sensitivity DNA assay kit (Invitrogen) and Tapestation D5000 tape (Agilent). The individual libraries were normalized and pooled in equimolar amounts at 2 nM. The 2 nM library pool was sequenced on an Illumina NextSeq using a 500/550 Mid Output Kit v2.5 (300 Cycles). A subset of samples from Ochsner Health were processed without tagmentation and sequenced on a Illumina MiSeq using a MiSeq reagent kit V3 (600 cycles). Raw reads were deposited under BioProject accession ID’s PRJNA643575 and PRJNA612578.
Consensus sequences were assembled using an inhouse Snakemake (Köster and Rahmann, 2012) pipeline with bwa-mem (Li, 2013) and iVar v1.2.2 (Grubaugh et al., 2019b; Li, 2013).
SARS-CoV-2 metagenomic Sequencing
For samples that were collected at Ochsner Health we used the following metagenomic sequencing protocol: RNA isolated from VTM was converted to double stranded cDNA and sequencing libraries prepared using TruSeq Stranded RNA Library Preparation Kit (Illumina) according to the manufacturer’s instructions. The sequencing libraries were evaluated using high sensitivity D5000 ScreenTape in the 4200 TapeStation system (Agilent) and quantified using Library Quantitation Kit (Roche). The libraries normalized and pooled, and subsequently sequenced using the NextSeq and 500/550 2x150 MID Output format (Illumina). Raw reads were deposited under BioProject accession ID PRJNA643574.
For samples that were collected at LSUHS we used the following metagenomic sequencing protocol: For each sample, 13μL of extracted RNA was reverse transcribed using the Maxima H-minus ds cDNA kits (ThermoFisher Scientific). Libraries were enriched using a Nextera Flex for Enrichment Library Preparation kit with a Respiratory Virus Oligo Set v1 (Illumina), with samples being pooled in 12-plex enrichment reactions. The resulting pools were quantified and grouped in sets of no more than 48 samples and run on a NextSeq 550 using a 150cyc High Output Flow Cell (Illumina). We used BreSeq v.0.34.1 (Deatherage and Barrick, 2014) to map reads to Wuhan-Hu-1 SARS-CoV-2 (NC_045512) or 2019-nCoV WIV04 (EPI_ISL_402124) (Zhou et al., 2020) and call the consensus sequence. All predicted mutations were reported for isolates exceeding mean 40x coverage. Raw reads were deposited under BioProject accession ID PRJNA681020.
Phylogenetic Analysis
We used the global SARS-CoV-2 phylogeny provided by Rob Lanfear (Lanfear and Mansfield, 2020) as of Oct 21st from GISAID (Table S2) and narrowed it down to 1,171 full-length genomes representing the genetic diversity from 19 different states in the USA and 228 sequences from outside the USA. The number of genomes from each state are shown in Table S3. We also masked sites in the alignment that were homoplastic as shown in Table S4. We used this dataset to estimate a starting tree using a HKY (Hasegawa et al., 1985) nucleotide substitution model, with a strict clock model using a non-informative continuous-time Markov chain (CTMC) reference prior (Ferreira and Suchard, 2008) and an exponential population prior implemented in BEAST v1.10.5pre (Suchard et al., 2018). We used the maximum clade credibility tree from this analysis as a starting tree to estimate the movement of the virus between geographic locations under a flexible discrete-state phylogeographic framework (Lemey et al., 2009) using BEAST v1.10.5pre (Suchard et al., 2018). We used a HKY nucleotide substitution model under an uncorrelated relaxed clock model (Drummond et al., 2006), an exponential population prior and a symmetric discrete-state substitution model. We included a Markov jump counting procedure (Minin and Suchard, 2008) to estimate the number of specific transitions between locations while simultaneously accounting for the large uncertainty in phylogenetic reconstruction. Specifically, to characterize the proportion of introductions from each discrete state into New Orleans and Shreveport, we first compute the relative number of the earliest Markov jump from each discrete state to New Orleans or Shreveport along the phylogenetic tree for each posterior sample. We then summarize these proportions over all samples to learn their posterior distributions. We simulated two independent MCMC chains for 100 million steps each and discarded the first 10 million steps as burnin in each. Effective sample sizes for scientifically relevant model parameters were all above 200. The BEAST XML and log files are available at https://github.com/andersen-lab/paper_2020_new-orleans-hcov-genomics.
Travel data
We calculated travel between counties using the weekly patterns data from SafeGraph (SafeGraph, 2020) a data company that aggregates anonymized location data from numerous applications in order to provide insights about physical places, via the Placekey (Placekey, 2020) Community. To enhance privacy, SafeGraph excludes census block group information if fewer than five devices visited an establishment in a month from a given census block group. We estimated the true number of travelers for a given week, w, between a source census block group (which is determined by monitoring the nighttime location over a period of 6 weeks), cbgs and a destination census block group, cbgd (Vw,cbgs,cbgd) using the raw number of visitor counts for week, w, identified from points of interest in cbgd from cbgs (Cw,cbgs,cbgd), the total number of visitors with a known source census block group in census block group, cbgd, and the population of cbgd, , according to,
We also obtained monthly air travel passenger data between the 19 U.S. states from the International Air Transportation Association. We used Apache Spark v2.4.6 and PySpark v2.4.6 to preprocess data from SafeGraph to estimate the travel between states.
There was a strong correlation in travel trends between mobility data and air travel passenger counts, but unlike SafeGraph mobility data, air travel data was unable to capture travel over short distances (R2 = 0.80; Figure S6). The code used to estimate movement between states using mobility data is available at https://github.com/andersen-lab/paper_2020_new-orleans-hcov-genomics.
Incidence
We used the R package Epidemia (Flaxman et al., 2020) to estimate the number of infections over time for each state and metro area, independently, using the number of deaths. Epidemia estimates a time-varying reproduction number, from the observed number of deaths, informed by an infection-to-death distribution and infection fatality rate (IFR) estimate. We assigned the IFR a normal prior with a mean of 0.01 and a standard deviation of 0.0001. We assumed the same infection-to-death distribution as described in Flaxman et al., 2020 (Flaxman et al., 2020), informed from data in Europe. Briefly, we assumed a gamma-distributed infection-to-onset time period with mean 5.1 days and a coefficient of variation of 0.86, a gamma-distributed symptom onset-to-death time period with a mean of 17.8 days and a coefficient of variation 0.45. Thus, the infection-to-death distribution was given by: π ∼ Gamma (5.1, 0.86) + Gamma (17.8, 0.45). Epidemia allows users to model as a log-linear function of a set of predictors. To estimate the effects of a lockdown, we used a “lockdown” predictor for each location which is set to 0 if the date was before the institution of a lockdown and set to 1 if the date was after. We used a normal prior with a mean of 0 and a standard deviation of 1 on the estimated parameters. We observed a reduction of ∼80% in Rt with a lockdown which was consistent with previously estimated Rt reductions due to lockdowns in Europe (Flaxman et al., 2020). We obtained the number of deaths for each location through the outbreak.info R package (R-outbreak.info, 2020), which aggregates epidemiological data from the COVID-19 data repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (Dong et al., 2020) and the COVID-19 data repository by the New York Times (https://github.com/nytimes/covid-19-data). The code used to estimate the number of infections is available at https://github.com/andersen-lab/paper_2020_new-orleans-hcov-genomics.
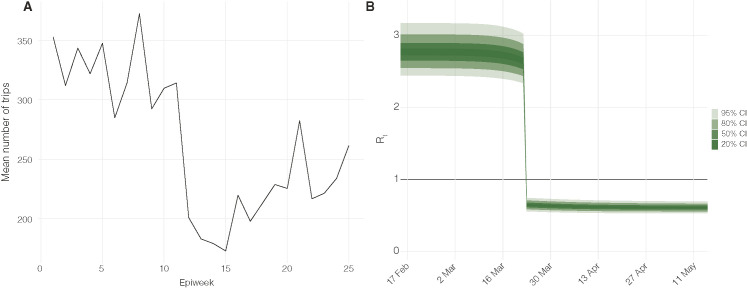
We created a predictor based on SafeGraph mobility data and used that to model the increase in in New Orleans on Mardi Gras (February 25th) using the Epidemia package. We analyzed the number of trips made during each week within New Orleans based on the mobility data obtained from SafeGraph (SafeGraph, 2020) (Figure S7 ), but we found only a slight increase in mobility during the week of Mardi Gras (Week 7), and hence, the SafeGraph mobility data was not representative for the increase in travel during Mardi Gras which drew over one million visitors to New Orleans. In addition, Testing, delayed reporting, and the variation in time-to-death among individual cases biases the accurate reporting of COVID-19 deaths. Due to these limitations, we did not include mobility as a predictor to assess the increase in using the framework provided by Epidemia. Instead, to quantify the number of infections that would have occurred on February 25th, we relied on case estimates from two separate models (Figure S2): (1) the cumulative number of infections until February 25th from daily deaths estimated using Epidemia (median: 713 (95% HPD: [174, 1426])), and (2) the cumulative number of infections until February 25th starting with 1 index case on February 13th.
Figure S7.
Estimates of mobility and epidemiological parameters, related to STAR Methods
(A) Mean number of trips over each epiweek made within New Orleans, Louisiana. (B) Daily Rt estimated from daily deaths using Epidemia.
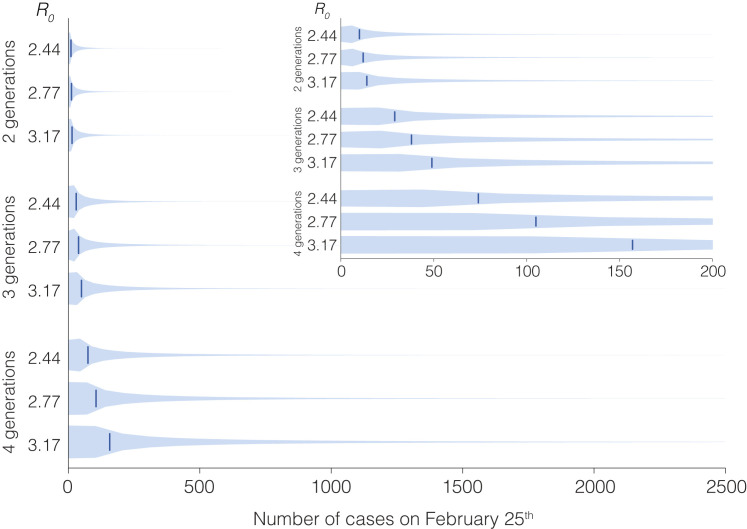
We calculated the number of infections that resulted from one index case on February 13th (Figure 2B) until February 25th based on 100,000 simulations of a negative binomial branching process model. Following Lloyd-Smith et al. (Lloyd-Smith et al., 2005), we assumed that secondary infections from a single infection would follow a negative binomial distribution described by and the overdispersion parameter, . We estimated a median of 2.77 (95% HPD: [2.44, 3.17]) in New Orleans based on the daily deaths using Epidemia before February 25th (Figure S7). Based on this, we assumed an of 2.77 and based on Althouse et al., 2020 (Althouse et al., 2020), of 0.16. In addition, we assumed that there was sustained local transmission in New Orleans that started with a single introduction of the virus on February 13th (median TMRCA of Louisiana clade) and 3 generations between February 13th and February 25th. We varied (2.77, 2.44, and 3.17) and the number of generations (2, 3, and 4 generations) independently (Figure S8 ), and found that even with an of 3.17 and 4 generations, the median cumulative number of infections (162 (95% CI [8, 2213])) was still below the median cumulative number of infections of 713 (95% HPD: [174, 1426]) as estimated from daily deaths using Epidemia. Hence, showing that a majority of 713 infections probably occurred on February 25th (Mardi Gras day) itself. The code to run the branching process model is available at https://github.com/andersen-lab/paper_2020_new-orleans-hcov-genomics.
Figure S8.
Sensitivity analysis for two parameters of the negative binomial branching process model, related to STAR Methods
The total number of generations (between February 13th and 25th) and the R0 were varied independently.
Mean growth rate, prevalence and normalized genetic distance of lineages
In order to calculate the mean growth rate over the first 10 days of the detection of a lineage we applied the methodology from Davies et al. (Davies et al., 2021). We pulled the number of sequences per day for each lineage from every county in the U.S., with at least 1000 sequences from Jan, 2020 to March, 2021 from https://outbreak.info/ which is enabled by genomic data provided by GISAID (GISAID - Initiative, 2021). In addition, we pulled the lineage counts for the B.1.1.7 and B.1.1.177 lineage in the United Kingdom, and the B.1.351 lineage in South Africa. We took the 7-day rolling average of these counts for each lineage and estimated the time-varying exponential growth rates of cases of each lineage, r(i, t), using a negative binomial state-space model correcting for day-of-week effects whose dispersion parameter was optimized for each strain by marginal likelihood maximization. We defined the relativized growth rate of a lineage i at time t as , where is the average growth rate of all circulating strains at time t and is the standard deviation of growth rates across all lineages at time t. We start estimating the growth rate of a lineage starting with the first week with at least 5 sequences and we average the growth rate over the first 10 days from this initial date. We selected a window of 10 days since, based on the first week of at least 5 sequences of B.1 in New Orleans on March 23rd and the peak of the B.1 lineage on April 3rd. The prevalence of each lineage was estimated based on the fraction of sequences within this 10 day window that were classified as the given lineage.
To estimate a normalized genetic distance for each lineage during the 10 day window, we used the global phylogeny provided by Rob Lanfear (Lanfear and Mansfield, 2020) from GISAID and identified sequences from each lineage that were used to calculate the mean growth rate as explained above. We then calculated the genetic distance of these sequences from the most recent common ancestor (MRCA) for each lineage. We normalized this genetic sequence by the number of sequences to account for sampling biases, according to .
Import/export risk
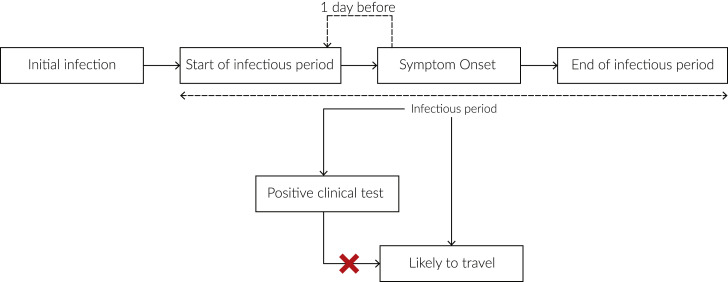
We estimated the number of infectious individuals likely to travel for a given location (Figure S9 ), and used weekly travel between two locations estimated using the same methodology as described above (see “Travel data” section), to determine the risk of import or export of the virus for two locations. For any given location on a given day, , we estimated the median number of infections, , from the daily reported deaths using Epidemia as previously described in the “Incidence” section. We assumed a gamma distributed incubation period with shape 5.807 and rate 1.055 (mean = 5.504; standard deviation = 2.284) (Lauer et al., 2020) (Figure S10). We estimated the number of cases that started showing symptoms using,
Figure S9.
Schematic showing when infectious cases would be likely to travel, related to STAR Methods
Infectious cases are unlikely to travel after receiving a positive clinical test.
where is the probability distribution function of the incubation period and is the estimated number of infections on a given day, .
We assumed that cases were infectious one day before symptom onset (Fauver et al., 2020) and a gamma distributed infectious period with shape, 2.5 and rate, 0.35 (mean = 7.143; standard deviation = 4.518) (Jung et al., 2020) (Figure S10). As per Fauver et al. (Fauver et al., 2020), we assumed that cases would not travel after receiving a positive clinical test. We pulled the number of confirmed cases as reported by state and local health departments using the outbreak.info R package. We assumed a uniform ascertainment period of 5 days for the reported cases and hence, excluded the reported cases on day, , from the cases that started showing symptoms on day, . We estimated the number of infectious cases that could travel on a given day, , using
where is the cumulative distribution function of the infectious period and is the number of cases that start showing symptoms on a given day, , and is the number of reported cases on day, . We show a schematic of how we estimated the number of infectious cases likely to travel in Figure S10.
We estimated the number of infectious travelers coming into a destination, , from a source, , on a given day, t, using
where is the population at the source, is the number of infectious cases likely to travel at the source and is the number of travelers from the source to the destination. We used this estimate to compare importation and exportation risk. The code to estimate the import and export risk is available at https://github.com/andersen-lab/paper_2020_new-orleans-hcov-genomics.
Acknowledgments
We thank the administrators of the GISAID database for supporting rapid and transparent sharing of genomic data during the COVID-19 pandemic and all our colleagues sharing data on GISAID. A full list acknowledging the authors submitting genome sequence data used in this study can be found in Table S2. The research leading to these results has received funding from the National Institutes of Health (grants U19AI135995, 3U19AI135995-03S2, UL1TR002550, U01AI151812, U01AI124302, R01AI153044, and R01HG006139), the CDC BAA contract 75D30120C09795, the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 725422-ReservoirDOCS), and from the European Union’s Horizon 2020 project MOOD (grant agreement no. 874850). The Artic Network receives funding from the Wellcome Trust through project 206298/Z/17/Z. P.L. acknowledges support by the Research Foundation—Flanders (“Fonds voor Wetenschappelijk Onderzoek—Vlaanderen”; G066215N, G0D5117N, and G0B9317N). S.L.L. acknowledges support by the National Science Foundation Small Business Innovation Research grants 2027424 and 1830867. We also gratefully acknowledge support from NVIDIA Corporation and Advanced Micro Devices, Inc., with the donation of parallel computing resources used for this research.
Author contributions
Conceptualization, M.Z., K.G., C.A., P.L., J.P.K., S.L.L., L.G., R.F.G., M.A.S., and K.G.A.; Methodology, M.Z., K.G., M.A.S., and K.G.A.; Software Programming, K.G., P.L., and M.A.S.; Formal Analysis, M.Z., K.G., D.J.S., M.A., M. Marshall, R.R., V.S.C., P.L., and M.A.S.; Investigation, M.Z., C.A., A.R.S., D.J.N., G.S.-S., A.R.B.-K., P.S., L.I.M., K.J.G., K.C., D.J.S., R.R.-S., and R.K.; Resources, A.R.S., J.A.V., R.S.S., J.G.-D., A.K.F., A.C.D., D.N.F., and J.P.K.; Data Curation, M.Z., K.G., C.A., D.J.S., M.A., M. Marshall, R.R., V.S.C., and M.A.S.; Writing – Original Draft, M.Z., K.G., M.A.S., and K.G.A.; Writing – Review & Editing, M.Z., K.G., C.A., P.L., J.P.K., S.L.L., L.G., R.F.G., M.A.S., and K.G.A; Visualization Preparation, M.Z., K.G., and P.L.; Supervision, P.L., J.P.K., S.L.L., L.G., R.F.G., M.A.S., and K.G.A.; Project Administration, M.Z., C.A., M. McGraw, S.T., E.S., and L.N.; Funding Acquisition, J.P.K., S.L.L., L.G., R.F.G., M.A.S., and K.G.A.; All authors contributed to interpreting and reviewing the manuscript.
Declaration of interests
M.A.S. reports grants from the National Institutes of Health, European Research Council, and Wellcome Trust during the conduct of this research and grants and contracts from the Bill & Melinda Gates Foundation, Janssen Research and Development, Private Health Management, IQVIA, and the U.S. Department of Veterans Affairs outside the submitted work. S.L.L., R.R., and D.J.N. are employed by BioInfoexperts LLC. R.F.G. reports grants from the National Institutes of Health, the Coalition for Epidemic Preparedness Innovations, the Burroughs Wellcome Fund, the Wellcome Trust, the Center for Disease Prevention and Control, and the European & Developing Countries Clinical Trials Partnership. He is the co-founder and Chief Scientific Advisor of Zalgen Labs, a biotechnology company developing countermeasures to emerging viruses, including SARS-CoV-2. K.G.A. has received consulting fees and compensated expert testimony on SARS-CoV-2 and the COVID-19 pandemic.
Published: July 27, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.07.030.
Supplemental information
Data and code availability
Genomes used in this analysis can be downloaded from GISAID.
References
- Althouse B.M., Wenger E.A., Miller J.C., Scarpino S.V., Allard A., Hébert-Dufresne L., Hu H. Superspreading events in the transmission dynamics of SARS-CoV-2: Opportunities for interventions and control. PLoS Biol. 2020;18:e3000897. doi: 10.1371/journal.pbio.3000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres D.L., Cummings M.P., Baele G., Darling A.E., Lewis P.O., Swofford D.L., Huelsenbeck J.P., Lemey P., Rambaut A., Suchard M.A. BEAGLE 3: Improved Performance, Scaling, and Usability for a High-Performance Computing Library for Statistical Phylogenetics. Syst. Biol. 2019;68:1052–1061. doi: 10.1093/sysbio/syz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford T., Greninger A.L., Roychoudhury P., Starita L.M., Famulare M., Huang M.-L., Nalla A., Pepper G., Reinhardt A., Xie H., et al. Seattle Flu Study Investigators Cryptic transmission of SARS-CoV-2 in Washington state. Science. 2020;370:571–575. doi: 10.1126/science.abc0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomquist E.W., Lemey P., Suchard M.A. Three roads diverged? Routes to phylogeographic inference. Trends Ecol. Evol. 2010;25:626–632. doi: 10.1016/j.tree.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control (2020a). https://www.cdc.gov/media/releases/2020/p0121-novel-coronavirus-travel-case.html (Centers for Disease Control).
- Centers for Disease Control (2020b). https://www.cdc.gov/media/releases/2020/p0130-coronavirus-spread.html.
- Centers for Disease Control (2020c). https://covid.cdc.gov/covid-data-tracker.
- Centers for Disease Control (2020d). https://www.fda.gov/media/134922/download (Centers for Disease Control).
- Chang S., Pierson E., Koh P.W., Gerardin J., Redbird B., Grusky D., Leskovec J. Mobility network models of COVID-19 explain inequities and inform reopening. Nature. 2021;589:82–87. doi: 10.1038/s41586-020-2923-3. [DOI] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. CMMID COVID-19 Working Group. COVID-19 Genomics UK (COG-UK) Consortium Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J.T., Chinazzi M., Perra N., Mu K., Piontti A.P.y., Ajelli M., Dean N.E., Gioannini C., Litvinova M., Merler S., et al. Estimating the establishment of local transmission and the cryptic phase of the COVID-19 pandemic in the USA. medRxiv. 2020 2020.07.06.20140285. [Google Scholar]
- Deatherage D.E., Barrick J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Gu W., Federman S., du Plessis L., Pybus O.G., Faria N.R., Wang C., Yu G., Bushnell B., Pan C.-Y., et al. Genomic surveillance reveals multiple introductions of SARS-CoV-2 into Northern California. Science. 2020;369:582–587. doi: 10.1126/science.abb9263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave D., Friedson A.I., McNichols D., Sabia J.J. The Contagion Externality of a Superspreading Event: The Sturgis Motorcycle Rally and COVID-19. Southern Economic Journal. 2020;87:769–807. doi: 10.1002/soej.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A.J., Ho S.Y.W., Phillips M.J., Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauver J.R., Petrone M.E., Hodcroft E.B., Shioda K., Ehrlich H.Y., Watts A.G., Vogels C.B.F., Brito A.F., Alpert T., Muyombwe A., et al. Coast-to-Coast Spread of SARS-CoV-2 during the Early Epidemic in the United States. Cell. 2020;181:990–996.e5. doi: 10.1016/j.cell.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feehan A.K., Fort D., Garcia-Diaz J., Price-Haywood E.G., Velasco C., Sapp E., Pevey D., Seoane L. Seroprevalence of SARS-CoV-2 and Infection Fatality Ratio, Orleans and Jefferson Parishes, Louisiana, USA, May 2020. Emerg. Infect. Dis. 2020;26:2766–2769. doi: 10.3201/eid2611.203029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M.A.R., Suchard M.A. Bayesian analysis of elapsed times in continuous-time Markov chains. Can. J. Stat. 2008;36:355–368. [Google Scholar]
- Flaxman S., Mishra S., Gandy A., Unwin H.J.T., Mellan T.A., Coupland H., Whittaker C., Zhu H., Berah T., Eaton J.W., et al. Imperial College COVID-19 Response Team Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature. 2020;584:257–261. doi: 10.1038/s41586-020-2405-7. [DOI] [PubMed] [Google Scholar]
- Galloway S.E., Paul P., MacCannell D.R., Johansson M.A., Brooks J.T., MacNeil A., Slayton R.B., Tong S., Silk B.J., Armstrong G.L., et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:95–99. doi: 10.15585/mmwr.mm7003e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GISAID - Initiative (2021). https://www.gisaid.org/.
- Grubaugh N.D., Ladner J.T., Lemey P., Pybus O.G., Rambaut A., Holmes E.C., Andersen K.G. Tracking virus outbreaks in the twenty-first century. Nat. Microbiol. 2019;4:10–19. doi: 10.1038/s41564-018-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N.D., Gangavarapu K., Quick J., Matteson N.L., De Jesus J.G., Main B.J., Tan A.L., Paul L.M., Brackney D.E., Grewal S., et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20:8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Jorden M.A., Rudman S.L., Villarino E., Hoferka S., Patel M.T., Bemis K., Simmons C.R., Jespersen M., Iberg Johnson J., Mytty E., et al. CDC COVID-19 Response Team Evidence for Limited Early Spread of COVID-19 Within the United States, January-February 2020. MMWR Morb. Mortal. Wkly. Rep. 2020;69:680–684. doi: 10.15585/mmwr.mm6922e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S.-M., Akhmetzhanov A.R., Hayashi K., Linton N.M., Yang Y., Yuan B., Kobayashi T., Kinoshita R., Nishiura H. Real-Time Estimation of the Risk of Death from Novel Coronavirus (COVID-19) Infection: Inference Using Exported Cases. Journal of Clinical Medicine. 2020;9:523. doi: 10.3390/jcm9020523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster J., Rahmann S. Snakemake--a scalable bioinformatics workflow engine. Bioinformatics. 2012;28:2520–2522. doi: 10.1093/bioinformatics/bts480. [DOI] [PubMed] [Google Scholar]
- Kraemer M.U.G., Yang C.-H., Gutierrez B., Wu C.-H., Klein B., Pigott D.M., du Plessis L., Faria N.R., Li R., Hanage W.P., et al. Open COVID-19 Data Working Group The effect of human mobility and control measures on the COVID-19 epidemic in China. Science. 2020;368:493–497. doi: 10.1126/science.abb4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear, R., and Mansfield, R. (2020). https://zenodo.org/record/4289383.
- Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P., Rambaut A., Drummond A.J., Suchard M.A. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux J., Siddle K.J., Shaw B.M., Loreth C., Schaffner S., Gladden-Young A., Adams G., Fink T., Tomkins-Tinch C.H., Krasilnikova L.A., et al. Phylogenetic analysis of SARS-CoV-2 in the Boston area highlights the role of recurrent importation and superspreading events. medRxiv. 2020 doi: 10.1101/2020.08.23.20178236. [DOI] [Google Scholar]
- Lemieux J.E., Siddle K.J., Shaw B.M., Loreth C., Schaffner S.F., Gladden-Young A., Adams G., Fink T., Tomkins-Tinch C.H., Krasilnikova L.A., et al. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science. 2021;371:eabe3261. doi: 10.1126/science.abe3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM.https://arxiv.org/abs/1303.3997 [Google Scholar]
- Lloyd-Smith J.O., Schreiber S.J., Kopp P.E., Getz W.M. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F.S., Nguyen A.T., Link N.B., Lipsitch M., Santillana M. Estimating the Early Outbreak Cumulative Incidence of COVID-19 in the United States: Three Complementary Approaches. medRxiv. 2020 doi: 10.1101/2020.04.18.20070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano M.T., Ramaswami S., Zappile P., Dimartino D., Boytard L., Ribeiro-Dos-Santos A.M., Vulpescu N.A., Westby G., Shen G., Feng X., et al. Sequencing identifies multiple early introductions of SARS-CoV-2 to the New York City region. Genome Res. 2020;30:1781–1788. doi: 10.1101/gr.266676.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Molecular Biology and Evolution. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minin V.N., Suchard M.A. Counting labeled transitions in continuous-time Markov models of evolution. J. Math. Biol. 2008;56:391–412. doi: 10.1007/s00285-007-0120-8. [DOI] [PubMed] [Google Scholar]
- Outbreak.info (2020). https://outbreak.info/epidemiology?location=USA_US-LA&log=false&variable=confirmed&xVariable=date&fixedY=false&percapita=false.
- Outbreak.info (2021a). https://outbreak.info/.
- Outbreak.info (2021b). https://outbreak.info/location-reports?loc=USA.
- Perkins A., Cavany S.M., Moore S.M., Oidtman R.J., Lerch A., Poterek M. Estimating unobserved SARS-CoV-2 infections in the United States. Proc. Natl. Acad. Sci. USA. 2020;117:22597–22602. doi: 10.1073/pnas.2005476117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Placekey (2020). https://placekey.io/.
- Quick J., Grubaugh N.D., Pullan S.T., Claro I.M., Smith A.D., Gangavarapu K., Oliveira G., Robles-Sikisaka R., Rogers T.F., Beutler N.A., et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017;12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R-outbreak.info (2020). https://github.com/outbreak-info/R-outbreak-info.
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SafeGraph (2020). https://www.safegraph.com/.
- SafeGraph (2021). https://docs.safegraph.com/docs/weekly-patterns.
- Sekizuka T., Itokawa K., Kageyama T., Saito S., Takayama I., Asanuma H., Nao N., Tanaka R., Hashino M., Takahashi T., et al. Haplotype networks of SARS-CoV-2 infections in the Diamond Princess cruise ship outbreak. Proc. Natl. Acad. Sci. USA. 2020;117:20198–20201. doi: 10.1073/pnas.2006824117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard M.A., Lemey P., Baele G., Ayres D.L., Drummond A.J., Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Transport Security Agency 2020. https://www.tsa.gov/coronavirus/passenger-throughput
- Villabona-Arenas C.J., Hanage W.P., Tully D.C. Phylogenetic interpretation during outbreaks requires caution. Nat. Microbiol. 2020;5:876–877. doi: 10.1038/s41564-020-0738-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á., et al. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. medRxiv. 2021 doi: 10.1101/2020.12.30.20249034. [DOI] [Google Scholar]
- Washington N.L., Gangavarapu K., Zeller M., Bolze A., Cirulli E.T., Schiabor Barrett K.M., Larsen B.B., Anderson C., White S., Cassens T., et al. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell. 2021;184:2587–2594.e7. doi: 10.1016/j.cell.2021.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2020a). https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200123-sitrep-3-2019-ncov.pdf?sfvrsn=d6d23643_8.
- World Health Organization (2020b). https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200229-sitrep-40-covid-19.pdf?sfvrsn=849d0665_2.
- Worobey M., Pekar J., Larsen B.B., Nelson M.I., Hill V., Joy J.B., Rambaut A., Suchard M.A., Wertheim J.O., Lemey P. The emergence of SARS-CoV-2 in Europe and North America. Science. 2020;370:564–570. doi: 10.1126/science.abc8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomes used in this analysis can be downloaded from GISAID.