Abstract
Since its first appearance in 1996, H9N2 avian influenza virus (AIV) of the Y439 lineage persisted in Korean live bird markets (LBMs) until the last documented occurrence in 2018. However, in June 2020, the avian influenza surveillance program detected a novel H9N2 AIV belonging to the Y280 lineage, which has zoonotic potential, in a Korean native chicken (KNC) from a LBM. In this study, we infected KNCs and ducks (the 2 major species held at LBMs), as well as SPF chickens, with Y280-lineage H9N2 AIV LBM261/20 and Y439-equivalent LBM294/18 to compare pathogenicity and transmissibility. In SPF chickens, LBM261/20 replicated mostly in the respiratory tract and spread rapidly among birds. By contrast, LBM294/18 replicated preferentially in the gastrointestinal tract and transmitted more slowly than LBM261/20. LBM261/20 replicated for a longer time in KNCs than in SPF chickens, and only in the respiratory tract; by contrast, LBM294/18 was detected in the oropharynx and cloaca. Ducks did not shed either virus or seroconvert. Taken together, the data suggest that the scheme used to monitor the newly introduced H9N2 AIV of the Y280 lineage needs to be modified to place emphasis on oropharyngeal sampling. Such changes will facilitate better disease control and protect public health.
Key words: H9N2, influenza A virus, live bird market, pathogenicity
INTRODUCTION
The H9N2 avian influenza virus (AIV) was isolated for the first time in 1966 from turkey flocks in Wisconsin, United States (Homme and Easterday, 1970). Since then, H9N2 AIVs have been detected worldwide in wild birds, domestic poultry, and mammals (Kawaoka et al., 1988; Peiris et al., 2001; Choi et al., 2004; Butt et al., 2005; Lee et al., 2007; Yu et al., 2013). H9N2 AIVs can be broadly categorized into 2 major lineages: Eurasian and American. American lineage H9N2 AIVS are found mostly in wild birds, whereas Eurasian lineage H9N2 AIVs are classified genetically into several sublineages: the G1 lineage (represented by A/quail/Hong Kong/G1/1997), the Y280 lineage (represented by A/duck/Hong Kong/Y280/1997), and the Y439 lineage (represented by A/duck/Hong Kong/Y439/1997) (Guan et al., 1999; Guo et al., 2000). Notably, H9N2 AIVs belonging to the G1 and Y280 lineages have zoonotic potential; indeed, as of June 2019, 59 cases have been reported in China, Hong Kong, Bangladesh, Egypt, Pakistan, and Oman (Zhang et al., 2014; Pu et al., 2015; Bi et al., 2016; Carnaccini and Perez, 2020). These viruses have donated their internal genes, leading to emergence of novel zoonotic AIVs such as H7N9, H10N8, and H5N6 (Liu et al., 2013; Chen et al., 2014; Yang et al., 2015). H9N2 AIVs of the Y439 lineage have been identified primarily in wild aquatic birds, with sporadic outbreaks in poultry, throughout Eurasia. (Peacock et al., 2019).
In 1996, an H9N2 AIV was isolated for the first time on a Korean native chicken (KNC) farm; the virus was H9N2 of the Y439 lineage. Since 1999, H9N2 AIVs of the Y439 lineage have been circulating predominantly in chickens, forming a Korean sublineage within the Y439 lineage (Lee et al., 2012; Youk et al., 2020). In 2007, a comprehensive vaccination policy was adopted to overcome the endemic, aided by biosecurity campaigns. Since then, H9N2 AI outbreaks have become less common in layers and broiler breeders, which are the major pillars of the poultry industry. However, H9N2 AIVs are circulating continuously, mainly among KNCs in live bird markets (LBMs); these viruses evolve via reassortment with Eurasian aquatic bird viruses, resulting in altered pathogenicity (Lee et al., 2007; Kim et al., 2010; Moon et al., 2010). Amid the massive H5N6 HPAI outbreaks of 2016–2017, enhanced control measures (including closure of LBMs) were implemented, contributing to a marked reduction in detection of H9N2 AIVs. Since the identification of a single case of H9N2 avian influenza of the Y439 lineage in 2018, no further cases were reported.
However, in June 2020, a novel H9N2 AIV was detected in a KNC during nationwide surveillance of LBMs. Phylogenetic analysis revealed that the H9N2 AIV belonged to the Y280 lineage, which has not been detected previously in Korea. Therefore, we conducted experiments in SPF chickens, KNCs, and ducks to compare the pathogenicity and transmissibility of the novel H9N2 AIV with those of the most recent Y439-like virus isolated in 2018.
MATERIALS AND METHODS
Animals
Experiments were conducted using mixed-sex, 5-wk-old SPF chickens, 6-wk-old KNCs, and 2-wk-old domestic ducks. SPF chickens were purchased from a commercial provider (Namduk, Korea). KNCs and ducks were acquired from breeding farms. All birds were tested for influenza A virus infection by serological assay with a commercial competitive ELISA kit (Bionote, Korea). All birds were negative. All birds were housed in a self-contained isolation unit placed in a biosafety level 3 (BSL3) facility at the Animal and Plant Quarantine Agency. All procedures were approved by the institutional animal care and use committee (IACUC) (No. 2020-550). The feeding and care of all birds was done in accordance with the approved guidelines, and feed and water were available ad libitum.
Viruses
A/chicken/Korea/LBM261/2020(H9N2) (abbreviated to LBM261/20) and A/chicken/Korea/LBM294/2018(H9N2) (abbreviated to LBM294/18) were used for the sequence analysis and animal experiments. The full genome sequences of both viruses are deposited in the Global Initiative on Sharing Avian Influenza Data (GISAID) under isolate IDs EPI_ISL492107 (for LBM261/20) and EPI_ISL492106 (for LBM294/18). The 50% egg infectious dose (EID50) of the virus stocks was calculated using the Reed and Muench method (Reed and Muench, 1938) based on serial 10-fold dilution in PBS and culture in 9- to 11-day-old embryonated chicken eggs. In addition, 6 Korean H9N2 AIVs isolated in 2020 were subjected to phylogenetic analysis of the HA gene: A/Chicken/Korea/20LBM314/2020(H9N2), A/Duck/Korea/20H49/2020(H9N2), A/Chicken/Korea/20H101/2020(H9N2), A/Duck/Korea/H112/2020, A/Chicken/Korea/20H195/2020(H9N2), and A/Chicken/Korea/20H90/2020(H9N2).
Sequence Analysis
For subtyping, the hemagglutinin (HA) and neuraminidase (NA) genes of the viruses were amplified using gene-specific primers (Hoffmann et al., 2001) and the One-Step RT-PCR kit (cat. no.: 210212, Qiagen, Valencia, CA); amplicons were confirmed by sequencing. The whole genomes were sequenced using the Miseq NGS platform (Illumina Inc., San Diego, CA) and a de novo assembly program (CLC genomics workbench 8.1), as previously described (Zhou et al., 2017). For phylogenetic analysis, the sequences of the HA gene of seven H9N2 AIVs isolated in 2020 and LBM294/18 were compared with available sequences in the GISAID and GenBank databases, and the most closely related sequences were identified from a BLAST research. A neighbor-joining tree was constructed using the maximum composite likelihood model in MEGA (Molecular Evolutionary Genetics Analysis, version 6.05), with a gamma distribution of among-site rate variation. The reliability of the tree was measured using 1,000 bootstrap trials.
Experimental Design
To compare pathogenicity and transmissibility in different bird species, 8 KNCs, 8 SPF chickens, and 8 ducks were inoculated intranasally with 0.1 mL of each virus (each inoculum contained 106.0 EID50). Ten hours later, 3 contact birds were co-housed with each inoculation group of the same bird species. Finally, 15 control birds (5 per species) were inoculated with PBS via the intranasal route. All birds were monitored daily for 14 days postinfection (dpi) to assess clinical symptoms.
Oropharyngeal (OP) and cloacal (CL) swabs were collected in 1 mL of PBS at 1, 3, 5, 7, 10, and 14 dpi to detect viral shedding. To investigate viral replication in tissues, 3 birds from each inoculation group plus 1 control bird species were euthanized at 3 dpi, and 6 organs (lung, brain, thymus, proventriculus, spleen, and pancreas) were harvested and processed for viral analysis. Briefly, the tissue samples were homogenized in maintenance medium containing antibiotics to yield a wt/vol ratio of 10%. Samples were then centrifuged at 3,500 rpm for 5 min, and 0.1 mL of supernatant was inoculated into two 9- to 11-day-old embryonated chicken eggs to verify the presence of virus. Blood samples were collected from the surviving birds at 14 dpi to determine seroconversion. The birds were sacrificed by cervical dislocation at 14 dpi.
Detection and Quantification of Viruses
To assess viral shedding, the OP and CL swabs collected at 1, 3, 5, 7, 10, and 14 dpi were suspended in 1 mL PBS. Viral titers in the tissues from H9N2 AIV-infected birds were measured using the centrifuged tissue homogenates (10%, w/v). Briefly, 200 μL of the suspension (supernatant) was subjected to RNA extraction using the Maxwell RSC simplyRNA Tissue kit and a Maxwell RSC 48 instrument (Promega, Germany). The cycle threshold (Ct) value was used to calculate the viral load after real-time reverse transcriptase-PCR (rRT-PCR) of the M-gene (Spackman et al., 2003).
To convert the Ct values to infectious units, quantitative viral standards (ranging from 106.0 to 100.0 EID50/0.1 mL) of each virus were prepared in egg allantoic fluid. Viral RNA was extracted from these standards and quantified by rRT-PCR. The resulting standard curves showed a high correlation (r2 > 0.99) and were used to convert Ct values to EID50 equivalents/0.1 mL. The detection limit for each virus was 101.0 EID50/0.1 mL, with Ct values of 39 for LBM261/20 and 38 for LBM284/18 (Supplementary Figure 1). If the Ct value was >35, swabs were retested by isolating virus as described below.
Virus Isolation
Virus isolation was performed by inoculation into the allantoic cavity of 9- to 11-day-old embryonated chicken eggs in accordance with the World Animal Health (OIE) protocol (OIE, 2019). The harvested chorio-allantoic fluid was tested for hemagglutination and subjected to rRT-PCR as previously described.
Serological Analysis
To confirm seroconversion, serum samples were collected at 14 dpi. Serum samples from KNCs and ducks were treated with receptor-destroying enzyme and tested for hemagglutination inhibition (HI) antibodies. Briefly, to detect homologous anti-H9 antibodies, serum samples were tested for HI antibodies using each homologous antigen and 1% chicken red blood cells, as specified in the OIE Terrestrial Manuals (OIE, 2019). A commercial multispecies ELISA kit (Bionote, Korea) was used to detect anti-influenza A NP-specific antibodies to identify influenza A virus infection before study entry. The ELISA was performed in accordance with the manufacturer's instructions.
Statistical Analysis
The viral titers in the swab samples were compared using a two-tailed unpaired t test (Prism 5; GraphPad Software, La Jolla, CA). A P value of < 0.05 was considered statistically significant.
RESULTS
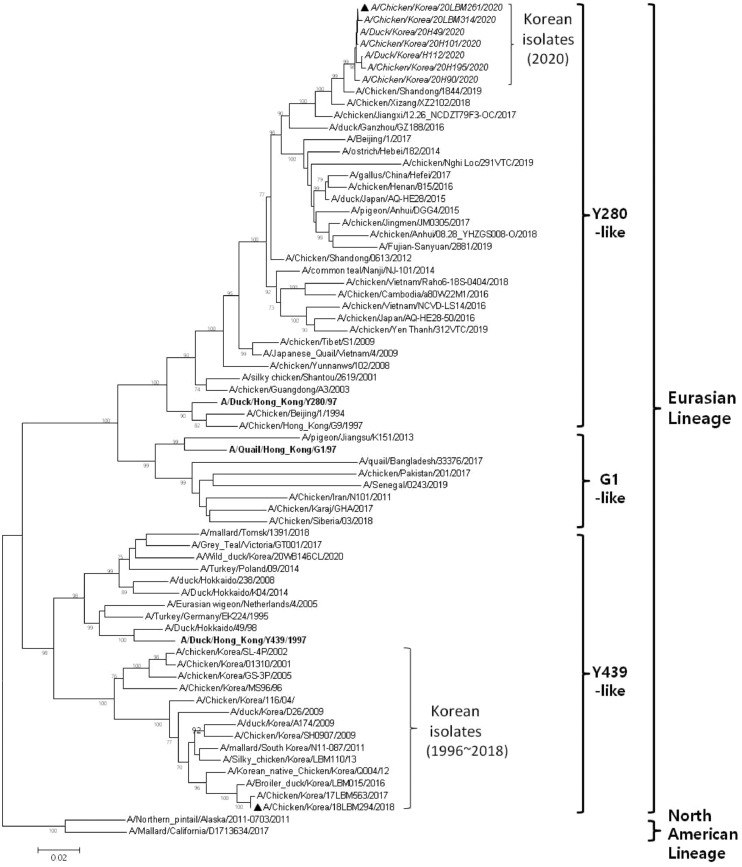
Phylogenetic analysis of the HA gene sequences revealed differences between H9N2 AIVs collected in 2020 and those circulating in 2018. LBM261/20 of the Y280 lineage clustered with other 6 H9N2 Korean AIVs identified in 2020, along with H9N2 AIVs isolated in China (Figure 1). The HA sequence of LBM261/20 showed highest homology (98.9%) with that of a 2019 Chinese H9N2 strain (A/chicken/Shandong/1844/2019(H9N2)). The HA sequences of Korean H9N2 AIVs isolated from 1996 to 2018 fell into the Y439 lineage, clustering with previously circulating H9N2 AIVs. Seven Korean H9N2 AIVs collected in 2020 were genetically very closely related (nucleotide identity: 99.2–100.0%), whereas the HA genes from these isolates were <80% homologous with those of Korean H9N2 AIVs belonging to the Y439 lineage, which had been circulating from 1996 to 2018. The HA gene of all Korean H9N2 AIVs belonging to the Y280 lineage contained the substitutions (H)183(N) and (Q)226(L) (H3 numbering) (data not shown), which confers high binding affinity for the α2,6-linked sialic acid receptors that are predominant in humans (Matrosovich et al., 2001). To determine the pathogenicity of Korean H9N2 AIVs of different lineages, we selected 2 representative H9N2 viruses. Specifically, LBM261/20 was from the Y280 lineage and LBM294/18 was from the Y439 lineage. Both viruses were tested in 3 poultry species: SPF chickens, KNCs, and ducks.
Figure 1.
Phylogenetic tree for the hemagglutinin gene of H9N2 influenza viruses. The tree was generated using the neighbor-joining method in MEGA 6.0, with 1,000 bootstrap replicates. Bootstrap values > 70% are shown on the branches. The tree was rooted by outgroup (North American lineage). The isolates subjected to genetic analysis in this study are indicated in italics. The isolates used for the pathogenicity investigation are marked with a black triangle.
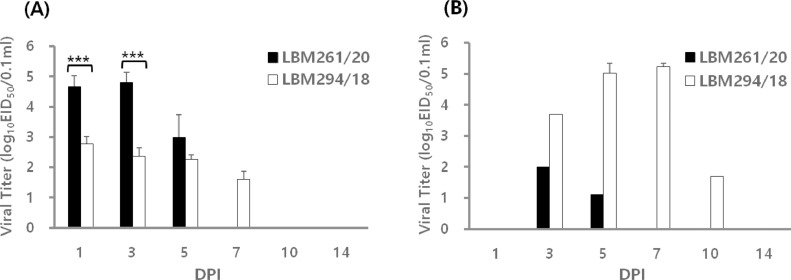
In the experiment using SPF chickens, no clinical symptoms were observed during the 14-d experimental period. All inoculated birds shed viruses via the OP route, with titers ranging from 103.0 to 104.8 EID50/0.1 mL for LBM261/20 and 101.6 to 102.8 EID50/0.1 mL for LBM294/18 (Table 1). It is noteworthy that replication of LBM261/20 was significantly higher (by approximately 100-fold; P < 0.001) than that of LBM294/18 during the early phase (Table 1 and Figure 2A). Only a limited number of the birds in each inoculation group shed virus via the CL route. LBM294/18 showed clear shedding in the cloaca, with titers up to 1,000 times higher than those of LBM261/20 (Table 1 and Figure 2B). All birds in the contact group shed virus via the OP route (Table 1). Overall, birds inoculated with LBM261/20 shed virus for a shorter period than those exposed to LBM294/18. Birds infected with LBM261/20 shed virus from 1 to 5 dpi, whereas contact birds shed virus from 3 to 7 dpi (Table 1). Birds infected with LBM294/18 shed virus from 1 to 10 dpi, whereas contact birds shed virus from 5 to 14 dpi. Based on these findings, it is reasonable to infer that virus replication at the oropharyngeal site might lead to more rapid transmission of LBM261/20 than LBM294/18, which also replicated in the gastrointestinal tract. All tested birds seroconverted to homologous influenza A viruses by 14 dpi, with HI titers ranging from 6.0 to 6.7 log2 (Table 1).
Table 1.
Detection of H9N2 viruses in SPF chicken swabs1.
| Virus titer (log10 EID50/0.1 mL, mean ± SD) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Group | Route | 1 dpi | 3 dpi | 5 dpi | 7 dpi | 10 dpi | 14 dpi | HI titer (log2, mean ± SD) |
| A/chicken/Korea/LBM261/2020 (H9N2) | Inoculated | OP | 4.7 ± 0.3 (8/8) | 4.8 ± 0.3 (8/8) | 3.0 ± 0.7 (5/5) | - (0/5) | - (0/5) | - (0/5) | 6.0 ± 0.7 (5/5) |
| CL | - (0/8) | 2.0 (1/8) | 1.1 (1/5) | - (0/5) | - (0/5) | - (0/5) | |||
| Contact | OP | - (0/3) | 4.5 ± 0.5 (3/3) | 4.5 ± 0.5 (3/3) | 2.5 ± 0.7 (3/3) | - (0/3) | - (0/3) | 6.0 ± 1.0 (3/3) | |
| CL | - (0/3) | - (0/3) | - (0/3) | - (0/3) | - (0/3) | - (0/3) | |||
| A/chicken/Korea/LBM294/2018 (H9N2) | Inoculated | OP | 2.8 ± 0.2 (8/8) | 2.4 ± 0.3 (7/8) | 2.3 ± 0.1 (3/5) | 1.6 ± 0.2 (4/5) | - (0/5) | - (0/5)- | 6.4 ± 0.9 (5/5) |
| CL | - (0/8) | 3.7 (1/8) | 5.0 ± 0.3 (2/5) | 5.2 ± 0.1 (2/5) | 1.7 (1/5) | - (0/5) | |||
| Contact | OP | - (0/3) | - (0/3) | 1.1 (1/3) | 3.4 ± 0.4 (3/3) | 3.2 ± 0.8 (3/3) | - (0/3) | 6.7 ± 0.6 (3/3) | |
| CL | - (0/3) | - (0/3) | - (0/3) | 1.4 ± 0.1 (2/3) | 5.1 ± 0.4 (2/3) | 2.1 (1/3) | |||
Abbreviations: CL, cloacal swab sample; DPI, days postinfection; EID50, 50% egg infective dose; OP, oropharyngeal swab sample.
The number of affected birds/birds per group is indicated in parenthesis. Eight birds were inoculated intranasally with 106.0 EID50/0.1 mL of each virus and 3 birds (contact group) were co-housed with the infected birds. Three birds inoculated with each of the viruses were euthanized for autopsy on d 3 postinfection. Seroconversion was measured in a hemagglutinin-inhibition assay at the end of the experimental period.
Figure 2.
Patterns of viral shedding via the oropharyngeal (OP) (A) and cloacal (CL) (B) routes in SPF chickens. Groups of eight SPF chickens were inoculated intranasally with 106.0 EID50 of each tested virus (LBM261/20 or LBM294/18). Three inoculated birds were euthanized at 3 dpi. At 1, 3, 5, 7, 10, and 14 dpi, OP and CL swabs were taken and subjected to rRT-PCR. The Ct value was converted to viral load using standard curves. Each bar represents the mean and standard deviation of 2–8 birds. ***; P < 0.0005, unpaired t test.
Similar to SPF chickens, KNCs showed no signs of clinical disease. Viral shedding was detected at 1 dpi in both inoculation groups (Table 2). All birds inoculated with LBM261/20 shed virus via the OP route, with titers ranging from 103.7 to 104.7 EID50/0.1 mL, whereas 4 out of 5 birds inoculated with LBM294/18 shed virus, with titers ranging from 101.5 to 102.5 EID50/0.1 mL (3 of 8 birds inoculated with LBM294/18 were lost at 1 dpi due to a mechanical disorder in the isolator). Of interest, as in SPF chickens, titers of LBM261/20 were approximately 100 times higher than those of LBM294/18. With respect to the CL route, LBM294/18 was detected at high titers (103.2 to 104.3 EID50/0.1 mL), whereas LBM261/20 was not detected at all. All birds in the LBM294/18 contact group shed virus via the OP route (102.2 to 102.8 EID50/0.1 mL) and CL route (104.1 to 104.5 EID50/0.1 mL), whereas no birds in the LBM261/20 group shed virus via the CL route. In general, LBM261/20 in KNCs replicated for a longer time (10 d) than LBM294/18; this is in contrast to the observation in SPF chickens (5 d). In addition, virus was shed via the CL route by all KNCs in the LBM294/18 contact group, while only a few birds in the corresponding group of SPF chickens shed virus via this route. All tested KNCs showed positive response in an HI assay, with titers of 5.0 to 7.3 log2 by 14 dpi; titers of LBM294/18 were higher than those of LBM261/20 (Table 2).
Table 2.
Detection of H9N2 viruses in Korean native chicken swabs1.
| Virus titer (log10 EID50/0.1 mL, mean ± SD) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Group | Route | 1 dpi | 3 dpi | 5 dpi | 7 dpi | 10 dpi | 14 dpi | HI titer (log2, mean ± SD) |
| A/chicken/Korea/LBM261/2020 (H9N2) | Inoculated | OP | 4.6 ± 0.5 (8/8) | 4.7 ± 0.7 (8/8) | 3.9 ± 0.6 (5/5) | 3.8 ± 1.8 (3/5) | 3.7 (1/5) | - (0/5) | 5.0 ± 1.2 (5/5) |
| CL | - (0/8) | - (0/8) | - (0/5) | - (0/5) | - (0/5) | - (0/5) | |||
| Contact | OP | - (0/3) | 4.5 ± 0.3 (3/3) | 4.2 ± 0.3 (3/3) | 3.9 ± 0.7 (3/3) | 1.9 (1/3) | - (0/3) | 5.3 ± 0.6 (3/3) | |
| CL | - (0/3) | - (0/3) | - (0/3) | - (0/3) | - (0/3) | - (0/3) | |||
| A/chicken/Korea/LBM294/2018 (H9N2) | Inoculated | OP | 1.9 ± 0.2 (2/5)a | 2.3 ± 0.3 (3/5) | 2.5 ± 0.4 (2/2) | 1.5 ± 0.4 (2/2) | - (0/2) | - (0/2) | 6.0 ± 0.0 (2/2) |
| CL | - (0/5)a | 3.2 (1/5) | 4.3 ± 0.9 (2/2) | 4.1 ± 2.0 (2/2) | - (0/2) | - (0/2) | |||
| Contact | OP | - (0/3) | - (0/3) | 2.6 (1/3) | 2.8 ± 0.8 (3/3) | 2.2 ± 0.5 (3/3) | - (0/3) | 7.3 ± 0.6 (3/3) | |
| CL | - (0/3) | - (0/3) | - (0/3) | 4.1 ± 2.2 (3/3) | 4.5 ± 0.7 (3/3) | - (0/3) | |||
Abbreviations: CL, cloacal swab sample; DPI, days postinfection; EID50, 50% egg infective dose; OP, oropharyngeal swab sample.
The number of affected birds/birds per group is indicated in parenthesis. Eight birds were inoculated intranasally with 106.0 EID50/0.1 mL of each virus and three birds (contact group) were co-housed with the infected birds. Three birds inoculated with each virus were euthanized for autopsy on d 3 postinfection. Seroconversion was measured in an hemagglutinin-inhibition assay at the end of the experimental period.
There were three accidental deaths (due to mechanical failure of the isolator on d 1 postinfection) among eight inoculated birds.
To assess virus replication in tissues, we harvested lung, brain, thymus, proventriculi, spleen, and pancreas at 3 dpi. LBM261/20 was recovered from all lung samples from each chicken species, with viral titers of 2.4 ± 0.3 log10EID50/0.1 mL for SPF chickens and 4.8 ± 0.3 log10EID50/0.1 mL for KNCs, whereas LBM294/18 was detected in 1 out of 3 lung samples from SPF chickens (titer = 2.3log10 EID50/0.1 mL) (Table 3). None of the lung samples from KNCs showed evidence of LBM294/18 replication. This observation was consistent with our finding that LBM261/20 replicated more efficiently in the respiratory tract than LBM294/18. LBM261/20 replicated in the brain of each chicken species, with titers of 2.1 ± 0.4log10EID50/0.1 mL in SPF chickens and 2.2 ± 0.4log10EID50/0.1 mL in KNCs. LBM261/20 was also detected in other extrapulmonary organs, including thymus, proventriculi, and spleen (Table 3).
Table 3.
Virus titers in different tissues taken from chicken species inoculated with LBM261/20 and LBM294/18.1
| Tissue (log10 EID50/0.1 mL, mean ± SD) |
|||||||
|---|---|---|---|---|---|---|---|
| Viruses | Species | Lung | Bra | Thy | Pro | Spl | Pan |
| A/chicken/Korea/LBM261/2020(H9N2) | SPF chicken | 2.4 ± 0.3 (3/3)a | 2.1 ± 0.4 (2/3) | 2.1 (1/3) | 3.0 ± 2.3 (2/3) | - (0/3) | - (0/3) |
| KNC | 4.8 ± 0.3 (3/3) | 2.2 ± 0.4 (2/3) | 1.6 ± 1.0 (2/3) | 4.6 ± 0.1 (2/3) | 1.1 (1/3) | - (0/3) | |
| A/chicken/Korea/LBM294/2018(H9N2) | SPF chicken | 2.3 (1/3) | - (0/3) | 1.1 (1/3) | - (0/3) | - (0/3) | - (0/3) |
| KNC | - (0/3) | - (0/3) | 0.4b (1/3) | 3.5 (1/3) | - (0/3) | - (0/3) | |
Abbreviations: Bra, brain; KNC, Korean native chicken; Pan, pancreas; Pro, proventriculus; Spl, spleen; Thy, thymus.
At 3 d postinfection, tissue samples were collected from three birds experimentally infected with LBM261/20 or LBM294/18.
The number of birds that recovered versus the number of birds subjected to autopsy.
Although quantification of <1.0 log10 EID50/0.1 mL may be inaccurate, the rRT-PCR showed evidence of viral replication.
During the 14-d experiment period, none of the ducks tested at 2 wk old, or control birds of each species, showed obvious clinical signs, and no virus was detected in OP and CL swabs (data not shown). In addition, neither virus replicated in any of the tissues tested. Consequently, no seroconversion occurred in ducks or any of the surviving control birds of each species.
DISCUSSION
Since nationwide surveillance was implemented in 2008, H9N2 AIV of the Y439 lineage accounted for the vast majority of AIVs detected in LBMs in Korea (Lee et al., 2017). In June 2020, a H9N2 AIV of the Y280 lineage was detected for the first time in a Korean LBM (Heo et al., 2021), followed by a series of reported cases (mainly among KNCs) as of December 9, 2020. Since Korean LBMs hold mainly KNCs and ducks, we performed animal experiments to characterize this newly emerged H9N2 virus in both species. We also used SPF chickens. We compared the pathogenicity of the newly introduced H9N2 virus of the Y280 lineage (herein LBM261/20) with that of the most recent H9N2 virus (herein named LBM294/18) of the Y439 lineage, which was isolated in 2018.
In terms of viral shedding, all SPF chickens inoculated with LBM261/20 shed virus at high titers via the OP route; however, LBM294/18 was detected in high titers only via the CL route. The preferential replication in the respiratory tract of SPF chickens might enable LBM261/20 to transmit earlier than LBM294/18, possibly via the oral-to-oral route or the respiratory route; similar results were obtained in experiments using KNCs. According to previous studies of the pathogenicity of H9N2 AIVs of the Y280 lineage, the titers of 12 test viruses were higher via the OP route than the CL route (Pu et al., 2015; Song et al., 2019). In addition, Song et al. reported that 3 H9N2 AIVs of the Y280 lineage, which were isolated in 2011, were detected in the contact group at 1 or 3 dpi (Song et al., 2019), suggesting rapid transmission; this pattern of transmission is similar to that of LBM261/20 reported herein.
Here, we found that LBM261/20 replicated in the extrapulmonary organs, including the brain. Replication of low pathogenic AIVs is largely confined to trypsin-expressing epithelial cells (Koshikawa et al., 1998) lining the respiratory and gastrointestinal tracts (Swayne, 2007). Therefore, our result suggests that further study of the pathogenesis of H9N2 AIVs is necessary to examine their replication in extrapulmonary tissues. Despite replication of LBM261/20 in extrapulmonary tissues, no clinical symptoms were observed. Song et al. (2019) reported that H9N2 AIVs of the Y280 lineage replicate in brain tissue without causing clinical symptoms, with low titers ranging from 101.8 to 102.3 EID50/0.1 mL. In this study, a similar replication pattern was observed; the virus titers in brain tissue ranged from 101.8 to 102.4 EID50/0.1 mL for SPF chickens and from 102.0 to 102.5 EID50/0.1 mL in KNCs. Based on these findings, we can infer that the relatively low titers in the brain may result in the absence of disease symptoms. The finding that LBM261/20 replicates in extrapulmonary tissues implies that LBM261/20 has the potential to be more harmful to poultry flocks than LBM294/18.
There was no sign of disease in either SPF chickens or KNCs during the 14-d experimental period. However, there was a difference in the level of viral replication between species. KNCs inoculated with LBM261/20 shed virus for longer than SPF chickens. All the KNCs inoculated with LBM294/18 shed virus via the CL route, while a small number of inoculated SPF chickens shed virus via this route. These observations suggest that KNCs sustain H9N2 AIV replication and shedding for a longer duration, resulting in a higher risk of transmission. In this respect, KNCs might be a reservoir for H9N2 AIVs, mainly in LBMs and on small-scale KNC farms in Korea (Lee and Song, 2013; Youk et al., 2020). The Korean poultry industry suffered from outbreaks of infection caused by H9N2 AIVs of the Y439 lineage for 22 yr; these results suggest that the virus persisted in KNCs. Therefore, it cannot be ruled out that the newly introduced H9N2 AIV of the Y280 lineage may become another endemic H9N2 AIV, unless proper counteractive measures are taken in a timely manner.
Domestic ducks are considered to be a link between wild aquatic birds and terrestrial poultry; as such, they play an important role in viral transmission and evolution of AIVs (Chen et al., 2004; Hulse-Post et al., 2005; Sturm-Ramirez et al., 2005; Bi et al., 2016). In this study, we found that ducks were not susceptible to infection by either H9N2 AIV. H9N2 AIVs belonging to the Y280 lineage were distributed extensively in China and detected mostly in chickens (Xu et al., 2007). H9N2 AIVs belonging to the Y280 lineage did not replicate at all in Pekin ducks (Wang et al., 2019). Likewise, we found no evidence of LBM261/20 replication in Pekin ducks in this study, which is in line with the high adaption of H9N2 Y280 lineage AIVs to chickens. H9N2 AIVs belonging to the Y439 lineage have been found in poultry, as well as in wild birds. Several studies showed that H9N2 AIVs belonging to the Y439 lineage, especially the Korean sublineage, replicated less efficiently in Pekin ducks than in chickens (Guo et al., 2000; Park et al., 2011). In this study, LBM294/18 failed to replicate in Pekin ducks. Considering that H9N2 AIVs belonging to the Y439 lineage had been circulating in Korean poultry from 1996 to 2018, it is feasible that these viruses had evolved and better adapted to chickens than to duck species.
Since the first detection in an LBM in June 2020, most Korean H9N2 AIVs of the Y280 lineage have been detected in chickens, mostly KNCs (data not shown). Ducks were resistant to infection by either H9N2 virus in the experimental setting. In poultry flocks, however, 2 cases were confirmed in ducks. Epidemiological investigation revealed that these ducks were kept together with infected KNCs in the same LBM stores. Contextually, a possible explanation may be that the prolonged and repeated exposure to the source of infection, namely infected KNCs shedding virus, might have led to establishment of infection in ducks. This explanation is supported by the findings that the inoculated KNCs shed LBM261/20 in relatively high titers, ranging from 103.7 to 104.7 EID50/0.1 mL from 1 to 10 dpi. Also, it cannot be ruled out that samples from ducks might have been contaminated during sampling, considering the management conditions in the LBM stores. Given that the likelihood of spillover of the chicken-adapted H9N2 AIVs into aquatic birds is low, it is even less likely that H9N2 AIV of the Y280 lineage was introduced into Korea by wild migratory birds from other H9N2-endemic regions.
Phylogenetic analysis revealed that major human isolates belonged to the Y280/G1 lineage (Carnaccini and Perez, 2020; Song and Qin, 2020). As of June 2019, there have been 59 laboratory-confirmed cases of H9 in humans, in which the majority of the infections were caused by contact with live poultry in China, Hong Kong, Bangladesh, Egypt, and Pakistan (Peacock et al., 2019; Carnaccini and Perez, 2020). To date, there has been no human case of H9N2 AIV infection in Korea. However, it is noteworthy that LBM261/20 isolated from an LBM contained leucine (L) amino acid residues at position 226 in the HA protein (Heo et al., 2021); this finding increases concerns related to public health. LBMs are ideal sites for viral reassortment and interspecies transmission due to the wide variety of potential hosts (Shortridge, 1992; Liu et al., 2003a,b). Therefore, it is necessary to examine the suitability of the on-going surveillance strategy for accurately monitoring the prevalence of H9N2 AIV of the Y280 lineage in LBMs. This study suggests that the veterinary authority should tailor the sampling scheme to focus on the OP route, thereby reducing the risk of spillover to humans. Such carefully planned surveillance would be highly effective in controlling the disease, particularly when accompanied by implementation of increased biosecurity and hygiene practices.
In this study, we found that replication of the currently circulating LBM261/20 of the Y280 lineage, isolated from an LBM, is superior to that of the Y480 lineage isolated in 2018. LBMs are a recognized source of AIVs that pose a great hazard to the poultry industry. With respect to zoonotic potential, LBM261/20 of the Y280 lineage raises public health concerns. Thus, it is important to undertake well-designed surveillance measures fit for LBM261/20 of the Y280 lineage to better understand its prevalence in LBMs, along with its intrinsic zoonotic risk. Such an effort would enable us to develop measures to properly counteract the newly introduced H9N2 AIV of the Y280 lineage.
ACKNOWLEDGMENTS
This work was supported by the Animal and Plant Quarantine Agency (APQA) R&D project (no. N-1543418-2019-27-01).
DISCLOSURES
The authors declare no conflicts of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2021.101318.
Appendix. Supplementary materials
REFERENCES
- Bi Y., Chen Q., Wang Q., Chen J., Jin T., Wong G., Quan C., Liu J., Wu J., Yin R., Zhao L., Li M., Ding Z., Zou R., Xu W., Li H., Wang H., Tian K., Fu G., Huang Y., Shestopalov A., Li S., Xu B., Yu H., Luo T., Lu L., Xu X., Luo Y., Liu Y., Shi W., Liu D., Gao G.F. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe. 2016;20:810–821. doi: 10.1016/j.chom.2016.10.022. [DOI] [PubMed] [Google Scholar]
- Butt K.M., Smith G.J., Chen H., Zhang L.J., Leung Y.H., Xu K.M., Lim W., Webster R.G., Yuen K.Y., Peiris J.S., Guan Y. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J. Clin. Microbiol. 2005;43:5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnaccini S., Perez D.R. H9 influenza viruses: An emerging challenge. Cold Spring Harb Perspect. Med. 2020;10 doi: 10.1101/cshperspect.a038588. (6):a038588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Deng G., Li Z., Tian G., Li Y., Jiao P., Zhang L., Liu Z., Webster R.G., Yu K. The evolution of H5N1 influenza viruses in ducks in southern China. PNAS. 2004;101:10452–10457. doi: 10.1073/pnas.0403212101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Yuan H., Gao R., Zhang J., Wang D., Xiong Y., Fan G., Yang F., Li X., Zhou J., Zou S., Yang L., Chen T., Dong L., Bo H., Zhao X., Zhang Y., Lan Y., Bai T., Dong J., Li Q., Wang S., Zhang Y., Li H., Gong T., Shi Y., Ni X., Li J., Zhou J., Fan J., Wu J., Zhou X., Hu M., Wan J., Yang W., Li D., Wu G., Feng Z., Gao G.F., Wang Y., Jin Q., Liu M., Shu Y. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- Choi Y.K., Ozaki H., Webby R.J., Webster R.G., Peiris J.S., Poon L., Butt C., Leung Y.H., Guan Y. Continuing evolution of H9N2 influenza viruses in Southeastern China. J. Virol. 2004;78:8609–8614. doi: 10.1128/JVI.78.16.8609-8614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.J., Krauss S., Senne D.A., Mo I.P., Lo K.S., Xiong S.P., Norwood M., Shortridge K.F., Webster R.G., Guan Y. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- Guan, Y., K. F. Shortridge, S. Krauss, and R. G. Webster. 1999. Molecular characterization of H9N2 influenza viruses: were they the donors of the "internal" genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. U. S. A. 96:9363-9367. [DOI] [PMC free article] [PubMed]
- Heo G.B., Kye S.J., Sagong M., Lee E.K., Lee K.N., Lee Y.N., Choi K.S., Lee M.H., Lee Y.J. Genetic characterization of H9N2 avian influenza virus previously unrecognized in Korea. J. Vet. Sci. 2021;22:e21. doi: 10.4142/jvs.2021.22.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E., Stech J., Guan Y., Webster R.G., Perez D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- Homme P.J., Easterday B.C. Avian influenza virus infections. I. Characteristics of influenza A-turkey-Wisconsin-1966 virus. Avian Dis. 1970;14:66–74. [PubMed] [Google Scholar]
- Hulse-Post D.J., Sturm-Ramirez K.M., Humberd J., Seiler P., Govorkova E.A., Krauss S., Scholtissek C., Puthavathana P., Buranathai C., Nguyen T.D., Long H.T., Naipospos T.S., Chen H., Ellis T.M., Guan Y., Peiris J.S., Webster R.G. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. PNAS. 2005;102:10682–10687. doi: 10.1073/pnas.0504662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y., Chambers T.M., Sladen W.L., Webster R.G. Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? Virology. 1988;163:247–250. doi: 10.1016/0042-6822(88)90260-7. [DOI] [PubMed] [Google Scholar]
- Kim H.R., Park C.K., Oem J.K., Bae Y.C., Choi J.G., Lee O.S., Lee Y.J. Characterization of H5N2 influenza viruses isolated in South Korea and their influence on the emergence of a novel H9N2 influenza virus. J. Gen. Virol. 2010;91:1978–1983. doi: 10.1099/vir.0.021238-0. [DOI] [PubMed] [Google Scholar]
- Koshikawa N., Hasegawa S., Nagashima Y., Mitsuhashi K., Tsubota Y., Miyata S., Miyagi Y., Yasumitsu H., Miyazaki K. Expression of trypsin by epithelial cells of various tissues, leukocytes, and neurons in human and mouse. Am. J. Pathol. 1998;153:937–944. doi: 10.1016/S0002-9440(10)65635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Song C.S. H9N2 avian influenza virus in Korea: evolution and vaccination. Clin. Exp. Vaccine Res. 2013;2:26–33. doi: 10.7774/cevr.2013.2.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E.K., Kang H.M., Song B.M., Lee Y.N., Heo G.B., Lee H.S., Lee Y.J., Kim J.H. Surveillance of avian influenza viruses in South Korea between 2012 and 2014. Virol. J. 2017;14:54. doi: 10.1186/s12985-017-0711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.J., Lee D.H., Lee Y.N., Kwon J.S., Lee Y.J., Lee J.B., Park S.Y., Choi I.S., Song C.S. Generation of reassortant influenza viruses within the non-industrial poultry system. Infect. Genet. Evol. 2012;12:933–946. doi: 10.1016/j.meegid.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Lee Y.J., Shin J.Y., Song M.S., Lee Y.M., Choi J.G., Lee E.K., Jeong O.M., Sung H.W., Kim J.H., Kwon Y.K., Kwon J.H., Kim C.J., Webby R.J., Webster R.G., Choi Y.K. Continuing evolution of H9 influenza viruses in Korean poultry. Virology. 2007;359:313–323. doi: 10.1016/j.virol.2006.09.025. [DOI] [PubMed] [Google Scholar]
- Liu D., Shi W., Shi Y., Wang D., Xiao H., Li W., Bi Y., Wu Y., Li X., Yan J., Liu W., Zhao G., Yang W., Wang Y., Ma J., Shu Y., Lei F., Gao G.F. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381:1926–1932. doi: 10.1016/S0140-6736(13)60938-1. [DOI] [PubMed] [Google Scholar]
- Liu H., Liu X., Cheng J., Peng D., Jia L., Huang Y. Phylogenetic analysis of the hemagglutinin genes of twenty-six avian influenza viruses of subtype H9N2 isolated from chickens in China during 1996-2001. Avian Dis. 2003;47:116–127. doi: 10.1637/0005-2086(2003)047[0116:PAOTHG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Liu M., He S., Walker D., Zhou N., Perez D.R., Mo B., Li F., Huang X., Webster R.G., Webby R.J. The influenza virus gene pool in a poultry market in South central china. Virology. 2003;305:267–275. doi: 10.1006/viro.2002.1762. [DOI] [PubMed] [Google Scholar]
- Matrosovich M.N., Krauss S., Webster R.G. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001;281:156–162. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- Moon H.J., Song M.S., Cruz D.J., Park K.J., Pascua P.N., Lee J.H., Baek Y.H., Choi D.H., Choi Y.K., Kim C.J. Active reassortment of H9 influenza viruses between wild birds and live-poultry markets in Korea. Arch. Virol. 2010;155:229–241. doi: 10.1007/s00705-009-0577-4. [DOI] [PubMed] [Google Scholar]
- OIE(World Organization for Animal Health), Paris, France, 2019. Chapter 3.3.4.-Avian influenza (including infection with high pathogenicity avian influenza viruses). Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.03.04_AI.pdf
- Park K.J., Kwon H.I., Song M.S., Pascua P.N., Baek Y.H., Lee J.H., Jang H.L., Lim J.Y., Mo I.P., Moon H.J., Kim C.J., Choi Y.K. Rapid evolution of low-pathogenic H9N2 avian influenza viruses following poultry vaccination programmes. J. Gen. Virol. 2011;92:36–50. doi: 10.1099/vir.0.024992-0. [DOI] [PubMed] [Google Scholar]
- Peacock T.H.P., James J., Sealy J.E., Iqbal M. A global perspective on H9N2 avian influenza virus. Viruses. 2019;11 doi: 10.3390/v11070620. 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S., Guan Y., Markwell D., Ghose P., Webster R.G., Shortridge K.F. Cocirculation of avian H9N2 and contemporary "human" H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 2001;75:9679–9686. doi: 10.1128/JVI.75.20.9679-9686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Wang S., Yin Y., Zhang G., Carter R.A., Wang J., Xu G., Sun H., Wang M., Wen C., Wei Y., Wang D., Zhu B., Lemmon G., Jiao Y., Duan S., Wang Q., Du Q., Sun M., Bao J., Sun Y., Zhao J., Zhang H., Wu G., Liu J., Webster R.G. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. PNAS. 2015;112:548–553. doi: 10.1073/pnas.1422456112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L.J., Muench H.A. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- Shortridge K.F. Pandemic influenza: a zoonosis? Semin. Respir. Infect. 1992;7:11–25. [PubMed] [Google Scholar]
- Song W., Qin K. Human-infecting influenza A (H9N2) virus: a forgotten potential pandemic strain? Zoonoses Public Health. 2020;67:203–212. doi: 10.1111/zph.12685. [DOI] [PubMed] [Google Scholar]
- Song Y., Zhang Y., Chen L., Zhang B., Zhang M., Wang J., Jiang Y., Yang C., Jiang T. Genetic characteristics and pathogenicity analysis in chickens and mice of Three H9N2 avian influenza viruses. Viruses. 2019;11 doi: 10.3390/v11121127. (12):1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E, Senne D.A., Bulaga L.L., Myers T.J., Perdue M.L., Garber L.P., Lohman K., Daum L.T., Suarez D.L. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J. Virol. 2003;47:1079–1082. doi: 10.1637/0005-2086-47.s3.1079. [DOI] [Google Scholar]
- Sturm-Ramirez K.M., Hulse-Post D.J., Govorkova E.A., Humberd J., Seiler P., Puthavathana P., Buranathai C., Nguyen T.D., Chaisingh A., Long H.T., Naipospos T.S., Chen H., Ellis T.M., Guan Y., Peiris J.S., Webster R.G. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J. Virol. 2005;79:11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swayne D.E. Understanding the complex pathobiology of high pathogenicity avian influenza viruses in birds. Avian Dis. 2007;51:242–249. doi: 10.1637/7763-110706-REGR.1. [DOI] [PubMed] [Google Scholar]
- Wang C., Wang Z., Ren X., Wang L., Li C., Sun Y., Wang M., Tong Q., Sun H., Pu J. Infection of chicken H9N2 influenza viruses in different species of domestic ducks. Vet. Microbiol. 2019;233:1–4. doi: 10.1016/j.vetmic.2019.04.018. [DOI] [PubMed] [Google Scholar]
- Xu K.M., Smith G.J., Bahl J., Duan L., Tai H., Vijaykrishna D., Wang J., Zhang J.X., Li K.S., Fan X.H., Webster R.G., Chen H., Peiris J.S., Guan Y. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J. Virol. 2007;81:10389–10401. doi: 10.1128/JVI.00979-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.F., Mok C.K., Peiris J.S., Zhong N.S. Human infection with a novel avian influenza A(H5N6) virus. N. Engl. J. Med. 2015;373:487–489. doi: 10.1056/NEJMc1502983. [DOI] [PubMed] [Google Scholar]
- Youk S.S., Lee D.H., Jeong J.H., Pantin-Jackwood M.J., Song C.S., Swayne D.E. Live bird markets as evolutionary epicentres of H9N2 low pathogenicity avian influenza viruses in Korea. Emerg. Microbes Infect. 2020;9:616–627. doi: 10.1080/22221751.2020.1738903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Liu L., Pu J., Zhao J., Sun Y., Shen G., Wei H., Zhu J., Zheng R., Xiong D., Liu X., Liu J. Risk perceptions for avian influenza virus infection among poultry workers, China. Emerg. Infect. Dis. 2013;19:313–316. doi: 10.3201/eid1902.120251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Bi Y., Tian H., Li X., Liu D., Wu Y., Jin T., Wang Y., Chen Q., Chen Z., Chang J., Gao G.F., Xu B. Human infection with influenza virus A(H10N8) from live poultry markets, China, 2014. Emerg. Infect. Dis. 2014;20:2076–2079. doi: 10.3201/eid2012.140911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Deng Y.M., Barnes J.R., Sessions O.M., Chou T.W., Wilson M., Stark T.J., Volk M., Spirason N., Halpin R.A., Kamaraj U.S., Ding T., Stockwell T.B., Salvatore M., Ghedin E., Barr I.G., Wentworth D.E. Multiplex reverse transcription-PCR for simultaneous surveillance of influenza A and B viruses. J. Clin. Microbiol. 2017;55:3492–3501. doi: 10.1128/JCM.00957-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.