Abstract
Cryptosporidium is an intracellular coccidian parasite causing gastrointestinal disturbances resulting in diarrhoea in humans and animals. It is more frequently detected in calves and early childhood, and one of the major causes of mortality in low-income countries. National estimates of Cryptosporidium infection rate in cattle and humans are lacking in Ethiopia. Therefore, this systematic review and meta-analysis estimated the prevalence and assess the risk factors of Cryptosporidium infection in cattle and humans over 20 years. Article searches were made using PubMed, HINARI, Research Gates, AJOLs and Google Scholar databases. Studies that met the inclusion criteria under the preferred reporting items for systematic reviews and meta-analyses (PRISMA) checklist were used. Random effects models and Inverse Variance Index were used to calculate the pooled prevalence of cryptosporidiosis and heterogeneity among studies, respectively. A total of 23 eligible studies published between 2000 and 2020 were selected for this study. The estimated pooled prevalence of cryptosporidiosis was found to be 16.2% and 11% in cattle and humans, respectively. Ten Cryptosporidium species were documented with cattle and human-based studies. C. andersoni, C. parvum, C. bovis and C. ryanae were the reported species in cattle. Similarly, in humans, seven types of Cryptosporidium species (such as C. parvum, C. hominis, C. viatorum, C. felis, C. meleagridis, C. canis and C. xiaoi) were recorded. C. parvum and C. hominis were the dominant and responsible species for human illness. Using gp60 gene locus analysis, various zoonotic C. parvum subgenotypes were determined in humans; but it was limited in anthroponotic C. hominis. In conclusion, the overall prevalence of Cryptosporidium infection in cattle and humans was high and linked with several risk factors. Thus, there is a need for further epidemiological and genetic diversity studies, and awareness of creations on the disease to provide strategies that mitigate the disease in cattle and humans.
Keywords: Cattle, Cryptosporidiosis, Ethiopia, Humans, Prevalence, Meta-analysis, Systematic review
1. Introduction
Cryptosporidium is an extracytoplasmic intracellular parasite causing gastrointestinal disturbances resulting in diarrhoea in young animals and immunocompromised individuals (Swai and Schoonman, 2010). In cattle, it causes an acute or chronic gastrointestinal disturbance, which results in anorexia, loss of weight gain, lower growth rate, reduced milk production and mortality (Smith et al., 2006; Brook et al., 2008). The parasite has a homoxenous life cycle and a large capacity to reproduce and disseminate in various animal hosts (Wegayehu et al., 2013). It can develop and multiply in the brush border of gastrointestinal cells of infected animals and humans (Shafieyan et al., 2016).
To date, about seven species and two Cryptosporidium genotypes have been identified in cattle (Hunter et al., 2004). The parasite in the form of viable sporulated oocysts can be transmitted via the faecal-oral route from a human to human or animal to animal (anthroponotic transmission) or from animal to human (zoonotic transmission) with contaminated food or water (Abu-Madi et al., 2011). Its infection is found to be self-limiting in immunocompetent hosts, but it leads to life-threatening acute and chronic diarrhoea in young and immunocompromised animals (Snelling et al., 2007).
The occurrence of Cryptosporidium infection in cattle can be associated with age, bed depth and environmental sanitation (Brook et al., 2008). Bovine cryptosporidiosis has been noted as an important cause of neonatal diarrhoea and economic losses in dairy farms (Goma et al., 2007). This disease can be characterized by anorexia, diarrhoea, poor growth rate and death. Clinically, the severity of its infection can be attributed to animals' age, immune and nutritional status (Urquhart et al., 1996; Nasir et al., 2009).
Similarly, human cryptosporidiosis is most prevalent during early childhood and it is one of the major causes of mortality from an infectious disease in children under 24 months in low-income countries and is associated with an increased risk of death in toddlers aged 12–23 months. Even in children who survive, there is growing evidence of a link between cryptosporidiosis, childhood malnutrition and stunting (Kotloff et al., 2012).
Reports have indicated that the prevalence of cryptosporidiosis ranges from 6.25 to 39.65% in cattle in different parts of the world (Azami, 2007; Venu et al., 2013; Joute et al., 2016). Similarly, in humans, it ranges from 0.1% to 73.3% (Berahmat et al., 2017; Odeniran and Ademola, 2019; Mohebali et al., 2020). Various factors have been identified to affect the prevalence of Cryptosporidium infection in different hosts. These include age, hygiene, absence of toilets, colostrum feeding, season, contact with infected person or animals, socioeconomic status, overcrowded living style, feed and water sources, diarrhoea and climate (Berahmat et al., 2017; Ogendo et al., 2017; Ayele et al., 2018; Odeniran and Ademola, 2019; Mohebali et al., 2020).
Despite the public and economic importance of this pathogen, no national survey addressed the epidemiological aspects (temporal and spatial distribution and the risk factors associated with the occurrence of cryptosporidiosis in Ethiopia, where more than 56 million cattle and 108 million people are living within various agroecological zones). Besides, no national data show multiple disorders imposed on humans and the impacts on cattle production. Therefore, the objectives of this systematic review and meta-analysis were aimed to estimate the prevalence and assess the potential risk factors of Cryptosporidium infection in cattle and humans in Ethiopia. It also assesses the Cryptosporidium species involved in cattle and humans infection.
2. Methodology
The study was conducted based on the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analysis) checklist (Moher et al., 2010).
2.1. Search strategy
A comprehensive and exhaustive search strategy was made to identify all relevant studies. Databases such as Web of Science, PubMed, Google Scholar, HINARI, AJOLs and Research Gates were used for literature searches to identify relevant papers. Condition, Context and Population (CoCoPop) format were used to identify the required searching terms. The search strategy included Medical Subject Heading (MeSH) terms and a range of relevant keywords. Epidemiologically, cryptosporidiosis is a worldwide infection, which infects animals and humans. The research question was “what is the epidemiology of Cryptosporidium infection and associated risk factors in cattle and humans in Ethiopia?” Searching terms used were (Cryptosporidiosis OR Cryptosporidium infection OR Cryptosporidium) AND (epidemiology OR prevalence OR infection rate) AND (child OR children OR human infected individuals with HIV OR immunocompromised people OR calves OR cattle) AND (Ethiopia). Moreover, unpublished thesis manuscripts were also accessed from Ethiopian universities. All identified studies were imported to EndNote software to remove duplicates.
2.2. Study eligibility and application of inclusion and exclusion criteria
Upon the compilation of study reports from the different databases, duplicates were removed. After that, studies were considered for systematic review if they were conducted in Ethiopia and reported the prevalence and risk factors of Cryptosporidium infection in cattle and humans. After preliminary screening of all titles obtained from our searches, abstracts were then assessed for relevance and eligibility based on the inclusion criteria. Finally, studies that met all of the following criteria were included in this systematic review. The criteria were cross-sectional population-based studies, studies concerning a parasite other than Cryptosporidium species, studies done on the prevalence and risk factors in calf/cattle and children/human, studies that provided a definite total sample size and positive cases, diagnostic methods, sample size greater than 20 for data analysis, studies conducted in different parts of Ethiopia, duplicated data and study year. To provide contemporaneous and representative estimates, studies were excluded if they presented data collected before 2000 G.C and published other than the English language.
2.3. Study selection and data extraction process
A standardized data collection format was prepared to extract necessary data from the articles. The data included were the title of the study, the first author's last name, the study population where the study was conducted (calf/cattle or children/HIV infected individuals/human), study design, year of publication, sample size, main findings and quality assessment tools. Some duplicates were addressed manually due to variation in reference styles across sources. After that, the authors have screened the title and abstracts with predefined inclusion criteria. The author also collected full texts and evaluated their eligibility for final inclusion. Any data discrepancy was resolved by referring back to the original study.
2.4. Study quality assessment
The included papers qualities (risk of biases) were evaluated by two independent investigators following the quality assessment checklist (standard strengthening the Reporting of Observational Studies in Epidemiology checklist (STROBE) (von Elm et al., 2008). This checklist includes 22 items constituting various sections of the articles such as title, abstract, introduction, methods, results, and discussion. The checklist focuses on the objectives, different components of the methods (e.g., study design, sample size, study population, bias, statistical methods), results, limitations, and funding of the studies. The assigned scores were determined from 0 to 44. Based on the checklist, searched articles were categorized into 3 groups: low-quality score (<15.5), moderate quality (15.5–29.5), and high quality (30.0–44.0).
2.5. Data synthesis and statistical analysis
Extracted data were entered and kept into a Microsoft Excel spreadsheet and exported into Stata version 16 (Stata Corp., College Station, TX, USA) for analysis. The inverse variance index (I2) was calculated to determine the heterogeneity among studies. Similarly, the I2 values of 25, 50, and 75% were considered low, medium and high heterogeneity, respectively (Higgins and Thompson, 2002). In pooled prevalence analysis and 95% confidence intervals (95% CIs), the random-effects model with the DerSimonian-Laird method of selection (Moher et al., 2015) was used. A Forest plot diagram was employed to present the variations among studies, outcomes of meta-analysis that display estimates of the prevalence, and their corresponding CIs of all included studies with the pooled effect size. Similarly, subgroup analyses for the prevalence of Cryptosporidium infection in cattle and humans were conducted with sample size and study year. Small study effects and publication bias presence were then visualized using funnel plot diagrams and, Egger's test (Borenstein et al., 2009). Figures and tables were used to show the summarized and descriptive results.
3. Results
3.1. Search result
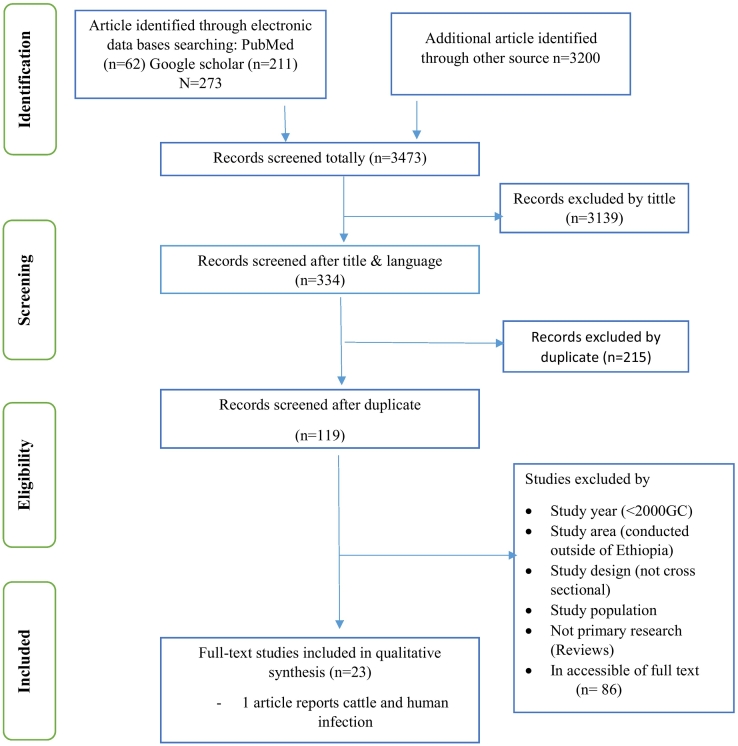
This review included both published and unpublished (thesis) reports of Cryptosporidium infection in cattle and humans in Ethiopia. The literature search period was started from November 2020 to February 2021. A total of 3473 articles were identified using the English language and cattle and human domain restrictions. Of these, 3139 were rejected based on the title confirmed non-relevance to this review. The remaining 334 articles were further screened, and subsequently, 215 were considered duplicates or inappropriate. The abstracts of 119 full-text articles were accessed and assessed for eligibility based on the pre-set criteria, which resulted in the further exclusion of 86 articles primarily due to the study area, study year (<2000 G.C.), study design (not cross-sectional or cohort/retrospective), not primary research (Reviews) and inaccessible of the full text. Ultimately, 23 studies met the eligibility criteria and were included in the final systematic review. (See Table 1) The PRISMA flow diagram is used to present stages of the review process (Fig. 1).
Table 1.
List of included studies in a meta-analysis on human and cattle.
| Author | Study year | Geog. location | Region | Sampling method | Source | Diag. test | Sample Size | Events | Event rate (AP) | LCI | UCI |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Adamu et al. (2010) | 2007–2008 | S.A. Ethiopia | SA Ethiopia | Purposive | Human | MZN | 1038 | 79 | 0.076 | 0.06 | 0.092 |
| Adamu et al. (2014) | 2009–2011 | Central Ethiopia | Addis Ababa | Purposive | HIV + | PCR | 520 | 140 | 0.269 | 0.23 | 0.307 |
| Ayalew et al. (2008) | 2005–2006 | Eastern Ethiopia | Dire Dawa | Simple R | Children | MZN | 655 | 80 | 0.122 | 0.097 | 0.147 |
| de Lucio et al. (2016) | 2013 | N. west Ethiopia | Amhara | Simple R | Children | PCR | 393 | 18 | 0.046 | 0.025 | 0.067 |
| Flecha et al. (2015) | 2014 | Southern Ethiopia | Oromia | Purposive | Human | PCR | 92 | 1 | 0.011 | −0.010 | 0.032 |
| Kibru and Mekete (2000) | 1997 | S. west Ethiopia | Oromia | Purposive | Children | MZN | 150 | 5 | 0.033 | 0.004 | 0.062 |
| Tigabu et al. (2010) | 2006 | N. west Ethiopia | Ben. Gumuz | Simple R | Children | MZN | 384 | 31 | 0.081 | 0.054 | 0.108 |
| Girma et al. (2014) | 2007 | Southern Ethiopia | SNNRP | Purposive | HIV + | MZN | 268 | 92 | 0.343 | 0.286 | 0.4 |
| Getaneh et al. (2010) | 2007 | Southern Ethiopia | SNNRP | Purposive | HIV +/− | MZN | 384 | 52 | 0.135 | 0.101 | 0.17 |
| Shimelis et al. (2016) | 2013–2014 | Southern Ethiopia | SNNRP | Purposive | HIV + | MZN | 491 | 65 | 0.132 | 0.102 | 0.162 |
| Gebre et al. (2019) | 2016–2017 | Southern Ethiopia | SNNRP | Purposive | HIV+ children | MZN | 384 | 37 | 0.096 | 0.067 | 0.126 |
| Wegayehu et al. (2013) | 2009 | Central Ethiopia | Oromia | Simple R | Children | MZN | 384 | 28 | 0.073 | 0.047 | 0.099 |
| Ayalew (2006) | 2005–2006 | Eastern Ethiopia | Dire Dawa | Simple R | Children | MZN | 1259 | 145 | 0.115 | 0.098 | 0.133 |
| Abebe et al. (2008) | 2004–2005 | Central Ethiopia | SA Ethiopia | Cluster R | Calf | MZN | 580 | 102 | 0.176 | 0.145 | 0.207 |
| Ayele et al. (2018) | 2014–2015 | N. west Ethiopia | Amhara | Simple R | Calf | MZN | 360 | 67 | 0.186 | 0.146 | 0.226 |
| Birhanu et al. (2017) | 2014–2015 | S. east Ethiopia | Oromia | Simple R | Calf | MZN | 384 | 92 | 0.24 | 0.197 | 0.283 |
| Ayana and Alemu (2015) | 2014–2015 | Central Ethiopia | Oromia | Systematic R | Calf | MZN | 214 | 29 | 0.136 | 0.090 | 0.182 |
| Manyazewal et al. (2017) | 2014–2015 | Central Ethiopia | Oromia | Purposive | Calf | PCR | 270 | 40 | 0.148 | 0.106 | 0.19 |
| Manyazewal et al. (2018) | 2014–2015 | Central Ethiopia | SA Ethiopia | Stratified R | Calf | PCR | 392 | 73 | 0.186 | 0.148 | 0.225 |
| Regassa et al. (2013) | 2010–2011 | Eastern Ethiopia | Oromia | Purposive | Calf | MZN | 133 | 37 | 0.278 | 0.202 | 0.354 |
| Wegayehu et al. (2013) | 2009 | Central Ethiopia | Oromia | Simple R | Calf | MZN | 384 | 30 | 0.078 | 0.051 | 0.105 |
| Wegayehu et al. (2016) | 2014 | Central Ethiopia | Oromia | Simple R | Calf | PCR | 449 | 71 | 0.158 | 0.124 | 0.192 |
| Gashaw et al. (2020) | 2017–2018 | Central Ethiopia | Oromia | Systematic R | Cattle | MZN | 378 | 41 | 0.109 | 0.077 | 0.140 |
| Hailu et al. (2020) | 2017–2018 | Southern Ethiopia | SA Ethiopia | Simple R | Calf | Serology | 330 | 43 | 0.1303 | 0.094 | 0.167 |
Fig. 1.
Flowchart of study selection for a systematic review of the prevalence of Cryptosporidium infection in cattle and humans in Ethiopia.
3.2. Study characteristics
All the twenty-three included studies were conducted using a cross-sectional study design with convenient/purposive, stratified, systematic, cluster, multistage, and simple random sampling procedures and published between 2000 and 2020. The studies were undertaken from some regions of Ethiopia: Addis Ababa, Amhara, Benshangulgumuz, Dire Dawa, Oromia and Southern Nations and Nationalities of Peoples. No studies were reported from Afar, Gambela, Harari, Somali and Tigray regions. Of the 23studies, 15 were performed using Modified Ziehl–Neelsen acid-fast staining (MZN)/direct microscope, 7 with PCR (five in combined with MZN/direct microscopy or Immunochromatographic (IC) test) and one with a serological diagnostic test. Out of the thirteen studies done in a human, five reported HIV coinfection and six on child infection. The identified studies were included 10,256 (3874 cattle and 6382 humans) study participants to determine the overall pooled prevalence of Cryptosporidium infection. The sample size of the studies ranged from 92 to 1239. From the total sample size, 1398 were found to be positive for Cryptosporidium species infection. The overall apparent infection rate in cattle and human was 16.1% and 12.1%, respectively. Table 1 illustrates the lists and characteristics of the included studies.
3.3. Meta-analysis and bias assessment
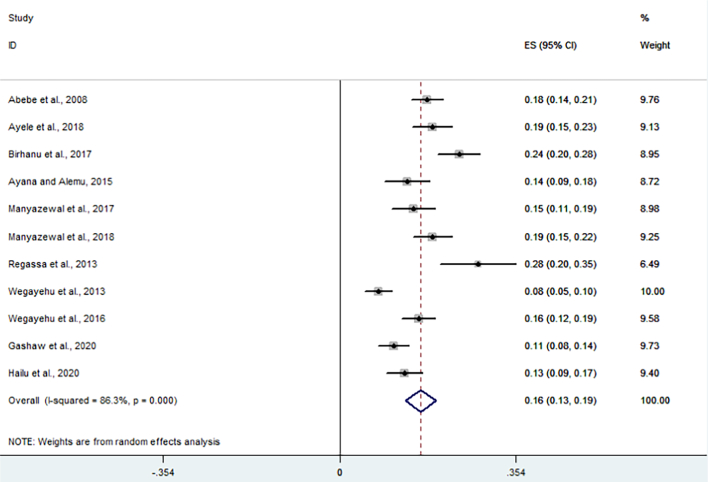
All 11 studies were included for meta-analysis to determine the overall pooled prevalence of Cryptosporidium infections in cattle. Thus, the random-effects model with DerSimonian-Laird selection model and inverse-variance procedures showed an overall pooled prevalence of 16.2% (95% CI: 13.1%–19.2%) Cryptosporidium infection in cattle (Fig. 2). The apparent overall prevalence ranges from 7.8% (Tigabu et al., 2010; Wegayehu et al., 2013) to 27.8% (Regassa et al., 2013). A substantial heterogeneity/variability was observed across the included studies on Cryptosporidium infection rate in cattle (I2 = 86.3%, Q-test = 72.92, T2 = 0.0023, df = 10, p < 0.001). The Galbraith plot assessment among studies on the infection rate in cattle (Fig. 3) also revealed that about 36.4% of the included reports are laid outside the 95% confidence limit and proved the relative variability presence between reports. Using subgroup analysis, the heterogeneity of studies was due to study location, sampling method and the publication year (p < 0.001).
Fig. 2.
Forest plot for the pooled prevalence of Cryptosporidium infection in cattle in Ethiopia.
Fig. 3.
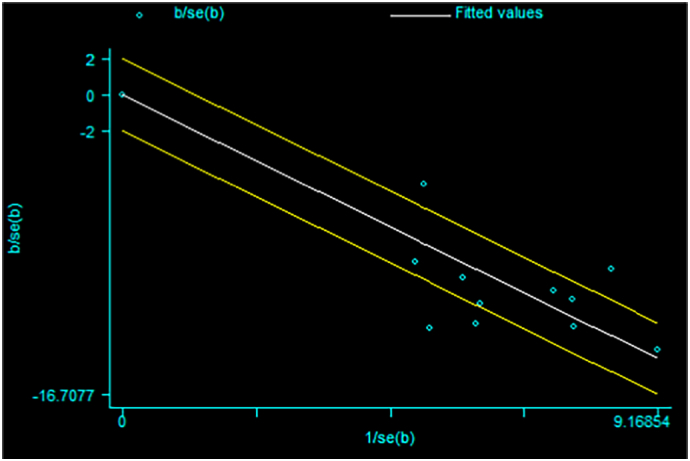
Galbraith plot for the prevalence of Cryptosporidium infection in cattle in Ethiopia.
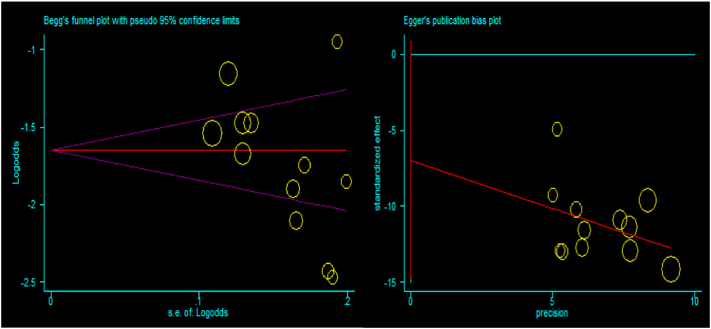
Besides, funnel plot asymmetry observation and bias coefficients for the studies published on Cryptosporidium infection in cattle were conducted to prove the presence of publication bias and small-study effects. The result of the funnel plot showed that there was no asymmetrical distribution of articles (Fig. 4), which depicts that smaller studies do not tend to be missed to be reported for the scientific community. Likewise, the results of Egger's and Begg's test plot showed that there was no statistically significant publication bias in estimating the prevalence of Cryptosporidium infection in cattle (Egger's test: b = −5.53, 95% CI: −13.65%–2.60%, p = 0.158 and Begg's test: p = 0.161) as indicated in Fig. 5.
Fig. 4.
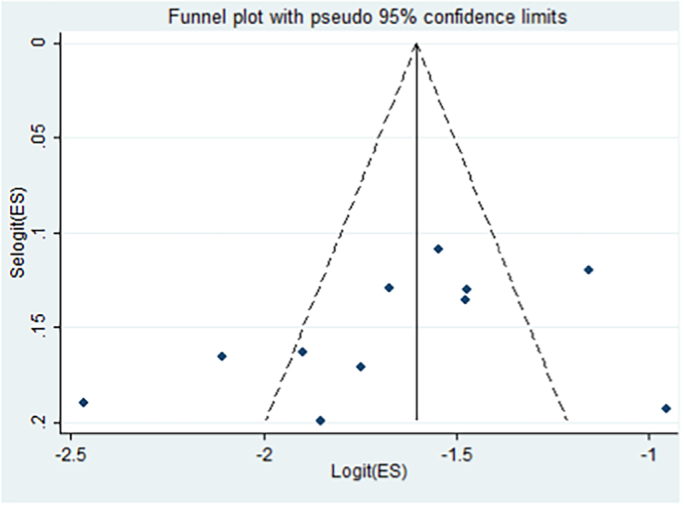
Funnel plot with 95% confidence limits the pooled prevalence of Cryptosporidium infection in cattle in Ethiopia.
Fig. 5.
Egger's publication bias plot (left) and Begg's funnel plot (right) reports for cattle studies.
Similarly, thirteen studies were compiled to determine the overall pooled prevalence of Cryptosporidium infections in humans. Therefore, the random-effects model with inverse-variance procedures showed an overall pooled prevalence of 11% (95% CI: 8%–15%) Cryptosporidium infection in humans (Fig. 6). Individual study prevalence estimates ranged from 1% (Flecha et al., 2015) to 34.3% (Girma et al., 2014). Considerable heterogeneity/variability was observed among the included studies in humans (I2 = 95.7%, Q-test = 281.9, T2 = 0.0037, df = 12, p < 0.001). The Galbraith plot assessment among studies on Cryptosporidium infection rate in humans (Figure 7:) also revealed that the majority (69%) of the included reports are laid outside the 95% confidence limit and provided clear evidence of the variability of reports. Based on subgroup analysis, the variability among studies was attributed to study location, publication year and sample size of the studies (p < 0.001).
Fig. 6.
Forest plot of the pooled prevalence of Cryptosporidium infection in humans in Ethiopia.
Fig. 7.
Galbraith plot for the prevalence of Cryptosporidium infection in humans in Ethiopia.
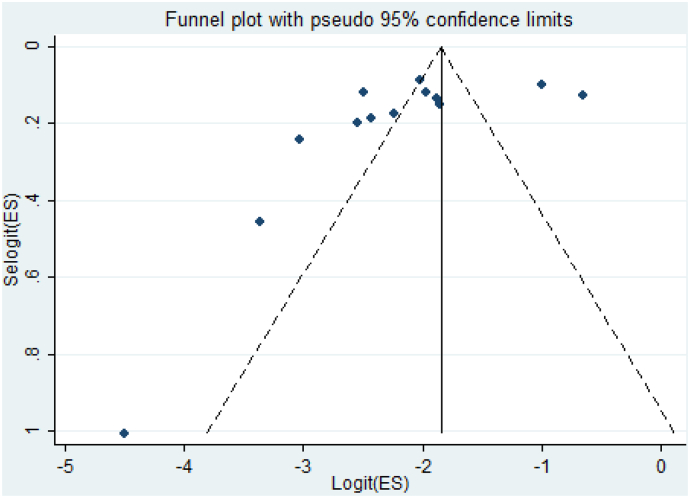
Further, publication bias was assessed using funnel plot observation and bias coefficient tests (Egger's and Begg's tests) for small-study effects of prevalence of Cryptosporidium infection in humans in Ethiopia. The result of funnel plot observation depicts an asymmetrical distribution of articles (Fig. 8). However, the result of small-study effects showed that there was statistically insignificant evidence of publication bias (small study bias) (Egger's test: b = −5.15, 95% CI: −11.8%–1.48%, p = 0.115 and Begg's test: p = 0.127) on studies reporting the prevalence of Cryptosporidium infection in humans in Ethiopia (Fig. 9).
Fig. 8.
Funnel plot with 95% confidence limits the pooled prevalence of Cryptosporidium infection in humans in Ethiopia.
Fig. 9.
Egger's publication bias plot (left) and Begg's funnel plot (right) reports for human studies.
3.4. Cryptosporidium genotypes and subgenotypes reported so far in Ethiopia
Studies to characterize Cryptosporidium species infecting animals and humans in Ethiopia are limited. However, to date, seven studies had been conducted to identify and subtype the parasite infecting cattle and humans. The studies were conducted using nested PCR to amplify the Cryptosporidium oocyst wall protein (COWP) gene, ssu-rRNA gene or a gene encoding 60-kDa glycoprotein (GP60) gene fragments. Restriction fragmented length polymorphism (RFLP) analysis was also used to determine the species type or subtypes using nested PCR (secondary) products of 18 s-rRNA targeting gene by digesting the isolated DNA template with endonucleases AseI, RsaI, SspI and VspI or MboII restriction enzymes. Thus, 10 Cryptosporidium species were documented with cattle and human-based studies. The reported species/genotypes in cattle were Cryptosporidium andersoni, C. parvum, C. bovis and C. ryanae. Of these, C. andersoni and C. parvum were the dominant and more frequently reported species responsible for cattle cases. Similarly, in humans, seven types of Cryptosporidium species (such as C. parvum, C. hominis, C. viatorum, C. felis, C. meleagridis, C. canis and C. xiaoi) were recorded (Table 2). C. parvum and C. hominis were the dominant and responsible species for human illness. Reports using sequence analysis of the gp60 or ssu-rRNA (18S rRNA) genes revealed a high genetic diversity of C. parvum and C. hominis. The subtypes of C. parvum identified in humans are the zoonotic subtype families (IIa and IId) and anthroponotic subtype families (IIc, IIb, IIe and If-like). Besides, C. parvum subtype originated from cattle include bovine type IIa & IId subtypes. Similarly, reports have documented various C. hominis subtype families, including Id, Ie and Ib subtypes. Further, with gp60 gene locus analysis, few studies have noted various zoonotic C. parvum subgenotypes from human subjects, including IIaA13G2R1, IIaA14G2R1, IIaA15G2R1, IIaA16G2R1, IIaA17G2R1, IIaA18G2R1, IIaA16G3R1, IIaA16G1R1, IIaA19G1R1, IIbA12, IIcA5G3a, IIdA17G1, IIdA19G1, IIdA22G1, IIdA24G1, IIeA12G1, and If-like. Likewise, limited anthroponotic C. hominis subgenotypes (IdA20, IdA24, IdA26, IeA11G3T3, IbA10G2, and IbA9G3) had also been reported so far (Table 2). But, no Cryptosporidium subgenotype studies were conducted in the cattle population.
Table 2.
Composition of Cryptosporidium genotypes reported so far in cattle and humans in Ethiopia.
| Species type | Genotype family | Subgenotype using gp60 gene locus analysis | Target group | References |
|---|---|---|---|---|
| C. andersoni | Ia, IIa & IIIa subtypes | Cattle (calf, heifer, adult) | Abebe et al. (2008), Wegayehu et al. (2016), Manyazewal et al., 2017, Manyazewal et al., 2018 | |
| C. parvum | Bovine type IIa & IId subtypes | No report | Cattle (calf) | Manyazewal et al., 2017, Manyazewal et al., 2018 |
| C. bovis | Ib & IIb subtypes | Cattle (calf) | Wegayehu et al. (2016) | |
| C. ryanae | I, II & III subtypes | Cattle (calf) | Wegayehu et al. (2016) | |
| C. parvum | Zoonotic subtypes (llaa & lld) and anthroponotic subtypes (llc, llb, lle & lf-like) | IIaA13G2R1, IIaA14G2R1, IIaA15G2R1a, IIaA16G2R1a, IIaA17G2R1, IIaA18G2R1, IIaA16G3R1, IIaA16G1R1a, IIaA19G1R1, IIbA12, IIcA5G3a, IIdA17G1, IIdA19G1, IIdA22G1, IIdA24G1, IIeA12G1, If-like | Human (HIV/AIDS), patients with diarrhoeaa | Adamu et al., 2010, Adamu et al., 2014; de Lucio et al. (2016)b |
| C. hominis | Subtype: ld, le & lba | IdA20, IdA24, IdA26, IeA11G3T3, IbA10G2, IbA9G3a | Human (HIV/AIDS), patients with GIT symptoms/diarrhoeaa | Adamu et al., 2010, Adamu et al., 2014, Flecha et al. (2015)a, de Lucio et al. (2016)b |
| C. viatorum | Human (HIV/AIDS) | Adamu et al. (2014), de Lucio et al. (2016) | ||
| C. felis | Human (HIV/AIDS) | Adamu et al. (2014) | ||
| C. meleagridis | Human (HIV/AIDS) | Adamu et al. (2014) | ||
| C. canis | Human (HIV/AIDS) | Adamu et al. (2014) | ||
| C. xiaoi | Human (HIV/AIDS) | Adamu et al. (2014) |
Designates similar findings with different authors.
Designates failing to determine subgenotypes.
3.5. Risk factors
Several potential risk factors have been documented in previous reports in Ethiopia and are presented in Table 3, Table 4 & The risk factors considered to affect the occurrence of Cryptosporidium infection in cattle are altitude, age, sex, body condition score (poor, medium or good), genetic blood level (indigenous, cross, or exotic breed), parity of the dam, faecal consistency, farm management practices (intensive, semi-intensive or extensive), farm location (urban/rural), feeding system, hygiene of calf-rearing houses, herd size and composition, farm type (smallholder/commercial), house floor type, preweaning house, presence of calving pen/bedding, method of colostrum feeding, weaning age (<6 or ≥ 6 months), source of drinking water (a pipe, well or river/stream), disposal of farm wastewater, pen cleanness (clean/poor), pen-type (individual/group pen), the occurrence of other diseases in the farm and presence of contact with other domestic animals). Among the factors, intensive farm management system, urban farms, group penning, poor condition score, presence of other cattle diseases in the farm, colostrum feeding through suckling, highland agroclimate, lower age groups, early weaning age, cross/exotic breed, number of parity, ground-feeding system, poor/fair house hygiene, river/stream drinking water sources, tethering of young animals in cow-barn, and diarrheic/soft faecal consistency were noted to have a significant association with increased infection rate in cattle. Similarly, in humans, several factors were considered to have an association with the occurrence of Cryptosporidium infection. Thus, being HIV-positive, urban residence, poor hygiene drinking water, having animal contact and diarrhoea influenced the infection rate in humans.
Table 3.
Risk factors identified in cattle so far in Ethiopia.
Table 4.
Risk factors identified in humans so far in Ethiopia.
| Variables | Risks (p < 0.05) | Not associated (p > 0.05) | Num. of studies |
|---|---|---|---|
| Age | Gebre et al., 2019; Shimelis et al., 2016; Girma et al., 2014; Getaneh et al., 2010; Adamu et al., 2014; de Lucio et al., 2016; Wegayehu et al., 2013 | 7 | |
| Sex | Gebre et al., 2019; Girma et al., 2014; Getaneh et al., 2010; Adamu et al., 2014; de Lucio et al., 2016; Wegayehu et al., 2013; Tigabu et al., 2010 | 7 | |
| HIV status | Positive & stage 1–4 (Getaneh et al., 2010) | 1 | |
| Mother's occupation | Gebre et al., 2019 | 1 | |
| Mother's education | Gebre et al., 2019 | 1 | |
| Residence | Urban (Getaneh et al., 2010) | Gebre et al., 2019 | 2 |
| Drinking water source | Tigabu et al., 2010 | Gebre et al., 2019; Shimelis et al., 2016; Ayalew et al., 2008 | 4 |
| Animal contact | Adamu et al., 2014 | Gebre et al., 2019; Shimelis et al., 2016; Wegayehu et al., 2013 | 4 |
| Diarrhoea status | Adamu et al., 2014 | Gebre et al., 2019 | 2 |
| CD4 level | Gebre et al., 2019; Girma et al., 2014; Adamu et al., 2014 | 3 |
4. Discussion
Cryptosporidium is one of the important zoonotic diseases that cause life-threatening diarrhoea in young, immunodeficient and malnourished hosts. It has been reported in animals and humans across the world in more than 106 countries and mainly in developing countries (Odeniran and Ademola, 2019; Gebre et al., 2019; Haghi et al., 2020; Mohebali et al., 2020). Nowadays, various epidemiological reports on the prevalence and associated risk factors of Cryptosporidium infection in animals and humans are available. Based on the reports, the prevalence of infection varied from country to country and from host to host. However, the infection rate is higher in low-income countries, young animals and immunodeficient individuals (Berahmat et al., 2017).
Ethiopia has diverse agroclimatic environments favouring the survival of different parasites (like Cryptosporidium species) affecting domestic animals and humans. On the other hand, Ethiopian communities are categorized by low resources, including poor hygienic practices and poor drinking water supply, coupled with different domestic animals (cats, dogs, goats, sheep, cattle, camels, and equines) potentially increase the risk of Cryptosporidium infection. Considering the above situations, synthesizing information using retrospective data is crucial to depict the pooled prevalence and associated risk factors of Cryptosporidium infection. Estimating country-level and regional pooled prevalence with associated risk factors of Cryptosporidium infection playing a central role in developing suitable strategies for Cryptosporidium infection diagnosis, prevention, treatment, and control in Ethiopia. As far as the author's knowledge, this study is the first to determine the pooled prevalence and assess the risk factors of Cryptosporidium infection in cattle and humans of Ethiopia. Importantly, Cryptosporidium is widespread in Ethiopia among domestic animals (cattle/calf) and humans (children/HIV infected individuals), and the high prevalence indicates a significant risk of clinical cryptosporidiosis. But, there is a lack of comprehensive and systematically organized data on overall prevalence and associated risk factors contributing to Cryptosporidium infection in cattle and humans in Ethiopia.
4.1. Prevalence of Cryptosporidium infection in cattle and humans
This meta-analysis evaluated the overall prevalence of Cryptosporidium infection among humans and animals. Thus, the weighted prevalence of Cryptosporidium infection was estimated to be 16.2% in cattle. This suggests that the probability of a high risk of clinical cryptosporidiosis among young and immunodeficient cattle in Ethiopia. This report agrees with the findings of Abebe et al. (2008), Manyazewal et al., 2017, Manyazewal et al., 2018, Ayele et al. (2018) and Jokar et al. (2021). The finding is also comparable with the pooled prevalence of 14.4% (Haghi et al., 2020) from Iran and 17.0% (Cai et al., 2019). However, a higher prevalence (28%) of cryptosporidiosis was reported in Ethiopia (Regassa et al., 2013). Odeniran and Ademola (2019) also documented an overall pooled prevalence (26.1%) of Cryptosporidium infection in cattle from Nigeria. Moreover, higher prevalence in cattle also confirmed earlier in various countries (Almeida et al., 2010; Ayinmode et al., 2010; Paudyal et al., 2013; García-Romo et al., 2014; Ghoneim et al., 2017; Diaz et al., 2018; Lombardelli et al., 2019). In contrast, lower infection rates in cattle were reported from Iran at 4.7% (Firoozi et al., 2019), China at 5.09% (Zhang et al., 2015) and Ethiopia at 13% (Hailu et al., 2020). Similarly, Gong et al. (2017) was also noted a lower prevalence (11.9%) in cattle in China. Mallinath et al. (2009) and Abdullah et al. (2019) also documented a lower prevalence of 5.71 and 12.5% in India and Malaysia, respectively.
The pooled prevalence of humans in this study was found to be 11.5%, which is comparable with reports of 15% from Nigeria (Odeniran and Ademola, 2019) and 11.2% in global HIV-positive people (Ahmadpour et al., 2020). Comparable reports were also confirmed in human elsewhere previously by Tombang et al. (2019), Krumkamp et al. (2020) and Chacín-Bonilla et al. (2008). The pooled prevalence of Cryptosporidium infection from studies administered to people living with HIV/AIDS in Ethiopia has noted 11% (Mohebali et al., 2020). However, a significantly higher overall prevalence (34%) was recorded from infected people with HIV (Girma et al., 2014) and 27% from patients with HIV/AIDS (Adamu et al., 2014). Likewise, the higher prevalence was also recorded in humans (Ghoneim et al., 2017; Paudyal et al., 2013). In contrast, the significantly lower pooled prevalence in the general population of humans was noted (Dong et al., 2020; Liu et al., 2020; Bouzid et al., 2018). Chen et al. (2017) also reported a lower infection rate of 6.9% from hospitalized children for diarrhoea in China.
The variation in the prevalence reports among studies might be attributed to the difference in local climatic situations, which determine the survival of oocysts and favours the dissemination of oocysts, sample size, sampling method, study location, diagnostic methods, nutritional habit, the status of public health and sanitary services, personal/environmental hygiene, season, sources of drinking water, socio-cultural differences, residence, livestock management/production system, colostrum-feeding practices, herd size, composition and breed, co-infection and levels of close contact with domestic animals. Further, the infection rate reported being influenced by education level, socioeconomic status and age groups.
4.2. Cryptosporidium species/genotypes reported in cattle and human
Molecular information regarding the diversity and frequency of Cryptosporidium species/genotypes circulating in Africa, in particular, Ethiopia is scarce. However, with limited studies, ten genotypes were recorded from cattle and human study subjects in Ethiopia. Four species/genotypes including C. andersoni, C. parvum, C. bovis and C. ryanae were reported in cattle. Of these, C. andersoni and C. parvum were the dominant and more frequently reported species responsible for cattle cases. This agrees with the findings of Taha et al. (2017), Lombardelli et al. (2019) and, Odeniran and Ademola (2019) from Sudan, Argentina and Nigeria, respectively. Zhang et al. (2015) and Gong et al. (2017) also reported similar genotypes in China. Similarly, in humans, seven types of Cryptosporidium species (such as C. hominis, C. canis, C. felis, C. meleagridis, C. parvum, C. viatorum, and C. xiaoi) were recorded. C. hominis and C. parvum were the dominant and responsible species for human illness. This supports various previous reports (Brankston et al., 2018; Robertson et al., 2020; Thomson et al., 2017). Reports using sequence analysis of the gp60 or ssu-rRNA (18S rRNA) genes revealed a high genetic diversity of C. hominis and C. parvum. The subtype families of C. hominis identified in human studies are Id, Ie and Ib. Similarly, the subtypes of C. parvum identified in humans are the zoonotic subtype families (IIa and IId) and anthroponotic subtype families (IIc, IIb, IIe and If-like). Besides, C. parvum subtype originated from cattle include bovine type IIa & IId subtypes. This is consistent with the findings of Taha et al. (2017) and Odeniran and Ademola (2019). The presence of C. parvum genotypes in cattle and humans demonstrate the zoonotic potential of the pathogen for farmers (attendants/producers) and cattle product users in Ethiopia. Since asymptomatic cattle infected with C. parvum can be the main reservoir of the pathogen, we should pay attention to the control and prevention of this infection both in cattle and humans to avoid mutual transmission between animals and humans.
4.3. Risk factors associated with Cryptosporidium infection in cattle and humans
Several risk factors have been reported to have a significant association with the occurrence of Cryptosporidium infection in the cattle population of Ethiopia. These include management system, altitude, farm location, age of animals, weaning period, the blood level of cattle, parity of dam, feeding system, feed source, calf house/pen hygiene, drinking water sources, herd size, preweaning house and faecal consistency. Similarly, various risk factors were reported in human study subjects for the occurrence of human cryptosporidiosis, including HIV status, residence site, drinking water sources, presence of contact with domestic animals, and diarrhoea status. Manyazewal et al. (2018) indicated that the association of herd size, farm location (urban), grouping pen, suckling method of colostral feeding, intensive farm management, exotic blood level (>75%), fair/poor hygiene and river/stream water source, with the prevalence of Cryptosporidium infection. Abebe et al. (2008) also showed a statistically significant (p < 0.05) association of Cryptosporidium infection rate and altitude of rearing environment (highland area), feeding on the ground, poor hygiene of calf house, tethering of calves in cow-barn. Similarly, Manyazewal et al. (2017) found statistically significant interaction (p < 0.05) between pre-weaning age and prevalence of Cryptosporidium infection. Furthermore, studies have shown the effect of early age (Ayele et al., 2018; Gashaw et al., 2020) and breed (exotic blood level) (Birhanu et al., 2017; Wegayehu et al., 2016) on the exposure rate of cattle with Cryptosporidium infection. In recent studies by Ayele et al. (2018), Gashaw et al. (2020) and Hailu et al. (2020), significant prevalence variation was observed with faecal consistency. Similarly, the study by Abebe et al. (2008) and Manyazewal et al. (2017) reported a similar effect. Several investigations revealed age as an important risk factor associated with Cryptosporidium infection in cattle (Wegayehu et al., 2016; Gashaw et al., 2020; Manyazewal et al., 2018; Regassa et al., 2013). In a study by Ayele et al. (2018), the odds of Cryptosporidium infection occurrence was 2.12 times more in the young calf (<6 months old) compared to cattle with a higher age category (>6 months). Manyazewal et al. (2017) also indicated as the prevalence of Cryptosporidium infection decreases as the age of animal increases.
In humans, the epidemiology of cryptosporidiosis is complex, involving direct (person-to-person and animal-to-person) and indirect (through water, food and fomites contaminated with infectious oocysts) transmission routes (Cacciò and Chalmers, 2016; Odeniran and Ademola, 2019). Drinking untreated/river or stream water, consuming water during recreational water activities, toileting children, touching another person with diarrhoea and engaging in contact with farm animals increase the risk of becoming infected with Cryptosporidium species (Cacciò and Chalmers, 2016). Likewise, in this study, the occurrence of Cryptosporidium infection in humans is greatly associated with immunodeficiency (HIV status) (Getaneh et al., 2010). Getaneh et al. (2010) also noted that the prevalence of cryptosporidiosis in humans is higher in urban residency than in rural. The prevalence is also influenced by having river/stream or open well water sources for drinking (Tigabu et al., 2010). Moreover, Adamu et al. (2014) reported direct animal contact and diarrhoeic stool increase the exposure of humans to cryptosporidiosis.
In conclusion, the prolonged and severe diarrhoea caused by Cryptosporidium is associated with significant morbidity and mortality, especially in young animals and immunodeficient individuals. This study is the first systematic review and meta-analysis providing a comprehensive view of the prevalence of cryptosporidiosis in cattle and humans, genotypes and related risk factors in Ethiopia. The results of this meta-analysis revealed that the pooled prevalence estimates of the disease in the country are 16.2% and 11% in cattle's and humans, respectively. Cryptosporidium parvum has been reported to infect both cattle and humans in Ethiopia. This study has certain limitations such as the lack of studies in some regions of the country, limited molecular studies reported genotypes/subgenotypes, and the findings were heterogeneous. Thus, further studies on molecular characterization and risk factor analysis are recommended. Study on C. parvum transmission dynamics between livestock and humans requires attention. Besides, awareness of creations is required about the disease to provide strategies that mitigate the disease in cattle's and humans.
Declaration of competing interest
We have no conflict of interest with this manuscript or report.
References
- Abdullah D.A., Ola-Fadunsin S.D., Ruviniyia K., Gimba F.I., Chandrawathani P., Lim Y., Jesse F., Sharma R. Molecular detection and epidemiological risk factors associated with Cryptosporidium infection among cattle in Peninsular Malaysia. Food Waterborne Parasitol. 2019;14 doi: 10.1016/j.fawpar.2019.e00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abebe R., Wossene A., Kumsa B. An epidemiological study of Cryptosporidium infection in dairy calves on selected dairy farms of central Ethiopia. Rev. Med. Vet. 2008;159(2):107. [Google Scholar]
- Abu-Madi M.A., Behnke J.M., Ismail A., Al-Olaqi N., Al-Zaher K., El-Ibrahim R. Comparison of intestinal parasitic infection in newly arrived and resident workers in Qatar. Parasit. Vectors. 2011;4:211. doi: 10.1186/1756-3305-4-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamu H., Petros B., Hailu A., Petry F. Molecular characterization of Cryptosporidium isolates from humans in Ethiopia. Acta Trop. 2010;115(1–2):77–83. doi: 10.1016/j.actatropica.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Adamu H., Petros B., Zhang G., Kassa H., Amer S., Ye J., Feng Y., Xiao L. Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PLoS Negl. Trop. Dis. 2014;8(4) doi: 10.1371/journal.pntd.0002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadpour E., Safarpour H., Xiao L., Zarean M., Hatam-Nahavandi K., Barac A., Picot S., Rahimi M.T., Rubino S., Mahami-Oskouei M., Spotin A., Nami S., Baghi H.B. Cryptosporidiosis in HIV-positive patients and related risk factors: A systematic review and meta-analysis. Parasite (Paris, France) 2020;27:27. doi: 10.1051/parasite/2020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida A., Moreira M.J., Soares S., Delgado M., Figueiredo J., Silva E., Castro A., Cosa J.M. Presence of Cryptosporidium spp. and Giardia duodenalis in drinking water samples in the north of Portugal. Kor. J. Parasitol. 2010;48(1):43–48. doi: 10.3347/kjp.2010.48.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew D. Addis Ababa University; 2006. Association of Cryptosporidium parvum, Giardia Lamblia and Entamoeba Histolytica/Dispar Infection with Drinking Water Sources among Children in Rural Part of Dire-Dawa, Eastern Ethiopia. MSc thesis. [Google Scholar]
- Ayalew D., Boelee E., Endeshaw T., Petros B. Cryptosporidium and Giardia infection and drinking water sources among children in Lege Dini, Ethiopia. Trop. Med. Int. Health TM & IH. 2008;13(4):472–475. doi: 10.1111/j.1365-3156.2008.02024.x. [DOI] [PubMed] [Google Scholar]
- Ayana D., Alemu B. Cryptosporidiosis in calves, lambs and goat kids in Bishoftu, Oromia regional state, Ethiopia. Afr. J. Basic Appl. Sci. 2015;7(5):233–239. doi: 10.5829/idosi.ajbas.2015.7.5.95160. [DOI] [Google Scholar]
- Ayele A., Seyoum Z., Leta S. Cryptosporidium infection in bovine calves: prevalence and potential risk factors in northwest Ethiopia. BMC Res. Notes. 2018;11(1):105. doi: 10.1186/s13104-018-3219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayinmode A.B., Olakunle F.B., Xiao L. Molecular characterization of Cryptosporidium spp. in native calves in Nigeria. Parasitol. Res. 2010;107(4):1019–1021. doi: 10.1007/s00436-010-1972-1. [DOI] [PubMed] [Google Scholar]
- Azami M. Prevalence of Cryptosporidium infection in cattle in Isfahan, Iran. J. Eukaryot. Microbiol. 2007;54(1):100–102. doi: 10.1111/j.1550-7408.2006.00236.x. [DOI] [PubMed] [Google Scholar]
- Berahmat R., Spotin A., Ahmadpour E., Mahami-Oskouei M., Rezamand A., Aminisani N., Ghojazadeh M., Ghoyounchi R., Mikaeili-Galeh T. Human cryptosporidiosis in Iran: a systematic review and meta-analysis. Parasitol. Res. 2017;116(4):1111–1128. doi: 10.1007/s00436-017-5376-3. [DOI] [PubMed] [Google Scholar]
- Birhanu F., Lemma D., Eticha E., Abera B., Adem A. Prevalence and risk factors of cryptosporidiosis in dairy calves in Asella town, south eastern, Ethiopia. Acta Parasitol. Globalis. 2017;8(1):50–57. doi: 10.5829/idosi.apg.2017.50.57. [DOI] [Google Scholar]
- Borenstein M., Hedges L.V., Higgins J.P.T., Rothstein H.R. 1st Ed. John Wiley and Sons, Ltd., Chichester, U.K; Chichester, U.K: 2009. Introduction to Meta-Analysis; pp. 277–291. [Google Scholar]
- Bouzid M., Kintz E., Hunter P.R. Risk factors for Cryptosporidium infection in low and middle-income countries: a systematic review and meta-analysis. PLoS Negl. Trop. Dis. 2018;12(6) doi: 10.1371/journal.pntd.0006553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brankston G., Boughen C., Ng V., Fisman D.N., Sargeant J.M., Greer A.L. Assessing the impact of environmental exposures and Cryptosporidium infection in cattle on human incidence of cryptosporidiosis in southwestern Ontario, Canada. PLoS One. 2018;13(4) doi: 10.1371/journal.pone.0196573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook E., Hart C.A., French N., Christley R. Prevalence and risk factors for Cryptosporidium spp. infection in young calves. Vet. Parasitol. 2008;152(1–2):46–52. doi: 10.1016/j.vetpar.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Cacciò S.M., Chalmers R.M. Human cryptosporidiosis in Europe. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2016;22(6):471–480. doi: 10.1016/j.cmi.2016.04.021. [DOI] [PubMed] [Google Scholar]
- Cai Y., Zhang N.Z., Gong Q.L., Zhao Q., Zhang X.X. Prevalence of Cryptosporidium in dairy cattle in China during 2008-2018: A systematic review and meta-analysis. Microb. Pathog. 2019;132:193–200. doi: 10.1016/j.micpath.2019.05.006. [DOI] [PubMed] [Google Scholar]
- Chacín-Bonilla L., Barrios F., Sanchez Y. Environmental risk factors for Cryptosporidium infection in an island from Western Venezuela. Memorias do Instituto Oswaldo Cruz. 2008;103(1):45–49. doi: 10.1590/s0074-02762008005000007. [DOI] [PubMed] [Google Scholar]
- Chen S., Atwill E.R., Zhong F., Wei Y., Hou S. Prevalence and risk factors of Cryptosporidium infection in children hospitalized for diarrhea in Guangzhou, China. J. Bacteriol. Parasitol. 2017;8:308. doi: 10.4172/2155-9597.1000308. [DOI] [Google Scholar]
- de Lucio A., Amor-Aramendía A., Bailo B., Saugar J.M., Anegagrie M., Arroyo A., López-Quintana B., Zewdie D., Ayehubizu Z., Yizengaw E., Abera B., Yimer M., Mulu W., Hailu T., Herrador Z., Fuentes I., Carmena D. Prevalence and genetic diversity of Giardia duodenalis and Cryptosporidium spp. among school children in a rural area of the Amhara region, north-West Ethiopia. PLoS One. 2016;11(7) doi: 10.1371/journal.pone.0159992. e0159992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz P., Varcasia A., Pipia A.P., Tamponi C., Sanna G., Prieto A., Ruiu A., Spissu P., Díez-Baños P., Morrondo P., Scala A. Molecular characterisation and risk factor analysis of Cryptosporidium spp. in calves from Italy. Parasitology research. 2018;117(10):3081–3090. doi: 10.1007/s00436-018-6000-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S., Yang Y., Wang Y., Yang D., Yang Y., Shi Y., Li C., Li L., Chen Y., Jiang Q., Zhou Y. Prevalence of Cryptosporidium infection in the global population: A systematic review and meta-analysis. Acta Parasitol. 2020;65(4):882–889. doi: 10.2478/s11686-020-00230-1. [DOI] [PubMed] [Google Scholar]
- Firoozi Z., Sazmand A., Zahedi A., Astani A., Fattahi-Bafghi A., Kiani-Salmi N., Akrami-Mohajeri F. Prevalence and genotyping identification of Cryptosporidium in adult ruminants in Central Iran. Parasit. Vectors. 2019;12(1):510. doi: 10.1186/s13071-019-3759-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flecha M.J., Benavides C.M., Tissiano G., Tesfamariam A., Cuadros J., de Lucio A., Bailo B., Cano L., Fuentes I., Carmena D. Detection and molecular characterisation of Giardia duodenalis, Cryptosporidium spp. and Entamoeba spp. among patients with gastrointestinal symptoms in Gambo Hospital, Oromia region, southern Ethiopia. Trop. Med. Int. health: TM & IH. 2015;20(9):1213–1222. doi: 10.1111/tmi.12535. [DOI] [PubMed] [Google Scholar]
- García-Romo D., Cruz Vázquez C., Quezada Tristán T., Silva Peña E., Valdivia Flores A., Vázquez Flores S., Ramos Parra M. Prevalence and risk factors associated with infection by Cryptosporidium spp. in suckling calves in Aguascalientes, Mexico. Veterinaria México OA. 2014;1(1) [Google Scholar]
- Gashaw M., Welde N., Ayana D., Waktole H., Addis Study on eimeria and Cryptosporidium infection in dairy cattle farms of Holeta, West Shoa Zone, Oromia, Ethiopia. J. Am. Sci. 2020;16(8):44–60. doi: 10.7537/marsjas160820.06. [DOI] [Google Scholar]
- Gebre B., Alemayehu T., Girma M., Ayalew F., Tadesse B.T., Shemelis T. Cryptosporidiosis and other intestinal parasitic infections and concomitant threats among HIV-infected children in southern Ethiopia receiving first-line antiretroviral therapy. HIV/AIDS (Auckland, N.Z.) 2019;11:299–306. doi: 10.2147/HIV.S215417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getaneh A., Medhin G., Shimelis T. Cryptosporidium and Strongyloides stercoralis infections among people with and without HIV infection and efficiency of diagnostic methods for Strongyloides in Yirgalem Hospital, southern Ethiopia. BMC Res. Notes. 2010;3:90. doi: 10.1186/1756-0500-3-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoneim N.H., Hassanain M.A., Hamza D.A., Shaapan R.M., Draz S.H. Prevalence and molecular epidemiology of Cryptosporidium infection in calves and hospitalized children in Egypt. Res. J. Parasitol. 2017;12:19–26. doi: 10.3923/jp.2017.19.26. [DOI] [Google Scholar]
- Girma M., Teshome W., Petros B., Endeshaw T. Cryptosporidiosis and Isosporiasis among HIV-positive individuals in south Ethiopia: a cross sectional study. BMC Infect. Dis. 2014;14:100. doi: 10.1186/1471-2334-14-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goma F.Y., Geurden T., Siwila J., Phiri I.G.K., Gabriël S., Claerebout E., Vercruysse J. The prevalence and molecular characterisation of Cryptosporidium spp. in small ruminants in Zambia. Small Rumin. Res. 2007;72(1):77–80. [Google Scholar]
- Gong C., Cao X.F., Deng L., Li W., Huang X.M., Lan J.C., Xiao Q.C., Zhong Z.J., Feng F., Zhang Y., Wang W.B., Guo P., Wu K.J., Peng G.N. Epidemiology of Cryptosporidium infection in cattle in China: a review. Épidémiologie de l’infection à Cryptosporidium chez les bovins en Chine: une synthèse. Parasite (Paris, France) 2017;24:1. doi: 10.1051/parasite/2017001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghi M.M., Khorshidvand Z., Khazaei S., Foroughi-Parvar F., Sarmadian H., Barati N., Etemadifar F., Ghasemikhah R. Cryptosporidium animal species in Iran: a systematic review and meta-analysis. Trop. Med. Health. 2020;48(1):97. doi: 10.1186/s41182-020-00278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailu M., Asmare K., Gebremedhin E.Z., Sheferaw D., Gizaw D., Di Marco V., Vitale M. Cryptosporidium and Giardia infections in dairy calves in southern Ethiopia. Parasite Epidemiol. Control. 2020;10 doi: 10.1016/j.parepi.2020.e00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J.P.T., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Hunter P.R., Hughes S., Woodhouse S., Syed Q., Verlander N.Q., Chalmers R.M., Morgan K., Nichols G., Beeching N., Osborn K. Sporadic cryptosporidiosis case-control study with genotyping. Emerg. Infect. Dis. 2004;10(7):1241–1249. doi: 10.3201/eid1007.030582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokar M., Rabiee M.H., Bokaie S., Rahmanian V., Dehesh P., Hasannejad H., Hushmandi K., Keshipour H. Prevalence of cryptosporidiosis in animals in Iran: a systematic review and metaanalysis. Asian Pac J Trop Med. 2021;14(3):99–112. doi: 10.4103/1995-7645.307532. [DOI] [Google Scholar]
- Joute J.R., Gill J.P., Singh B.B. Prevalence and molecular epidemiology of Cryptosporidium parvum in dairy calves in Punjab (India) J. Parasitic Dis. Off. Organ Ind. Soc. Parasitol. 2016;40(3):745–749. doi: 10.1007/s12639-014-0571-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibru G., Mekete G. Prevalence of Cryptosporidium infection at the pediatrics clinic of Jimma Hospital, Southwest Ethiopia. Ethiop. J. Health Sci. 2000;10(2) [Google Scholar]
- Kotloff K.L., Blackwelder W.C., Nasrin D., Nataro J.P., Farag T.H., van Eijk A., Adegbola R.A., Alonso P.L., Breiman R.F., Faruque A.S., Saha D., Sow S.O., Sur D., Zaidi A.K., Biswas K., Panchalingam S., Clemens J.D., Cohen D., Glass R.I., Mintz E.D.…Levine M.M. The global enteric multicenter study (GEMS) of diarrheal disease in infants and young children in developing countries: epidemiologic and clinical methods of the case/control study. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2012;55(Suppl. 4):S232–S245. doi: 10.1093/cid/cis753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumkamp R., Aldrich C., Maiga-Ascofare O., Mbwana J., Rakotozandrindrainy N., Borrmann S., Caccio S.M., Rakotozandrindrainy R., Adegnika A.A., Lusingu J., Amuasi J. Transmission of Cryptosporidium spp. among human and animal local contact networks in sub-Saharan Africa: a multi-country study. Clin. Infect. Dis. 2020:ciaa223. doi: 10.1093/cid/ciaa223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., Gong B., Liu X., Shen Y., Wu Y., Zhang W., Cao J. A retrospective epidemiological analysis of human Cryptosporidium infection in China during the past three decades (1987-2018) PLoS Negl. Trop. Dis. 2020;14(3) doi: 10.1371/journal.pntd.0008146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardelli J.A., Tomazic M.L., Schnittger L., Tiranti K.I. Prevalence of Cryptosporidium parvum in dairy calves and GP60 subtyping of diarrheic calves in central Argentina. Parasitol. Res. 2019;118(7):2079–2086. doi: 10.1007/s00436-019-06366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallinath H.K., Chikkachowdappa G., Javare Gowda A., D’Souza P. Studies on the prevalence of cryptosporidiosis in bovines in organized dairy farms in and around Bangalore, South India. Veterinarski arhiv. 2009;79(5):461–470. https://hrcak.srce.hr/45606 [Google Scholar]
- Manyazewal A., Francesca S., Gezahegn M., Getachew T. Cryptosporidium infection in dairy cattle calves and its public health significance in Central Ethiopia. J. Adv. Vet. Res. 2017;7(2):59–65. [Google Scholar]
- Manyazewal A., Francesca S., Pal M., Gezahegn M., Tesfaye M., Lucy M., Teklu W., Getachew T. Prevalence, risk factors and molecular characterization of Cryptosporidium infection in cattle in Addis Ababa and its environs, Ethiopia. Vet. Parasitol. Region. Stud. Rep. 2018;13:79–84. doi: 10.1016/j.vprsr.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohebali M., Yimam Y., Woreta A. Cryptosporidium infection among people living with HIV/AIDS in Ethiopia: a systematic review and meta-analysis. Pathog. Global Health. 2020;114(4):183–193. doi: 10.1080/20477724.2020.1746888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D., Liberati A., Tetzlaff J., Altman D.G., PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int. J. Surg. (London, England) 2010;8(5):336–341. doi: 10.1016/j.ijsu.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Moher D., Shamseer L., Clarke M., Ghersi D., Liberati A., Petticrew M., Shekelle P., Stewart L.A., PRISMA-P Group Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst. Rev. 2015;4(1):1. doi: 10.1186/2046-4053-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir A., Avais M., Khan M.S., Ahmad N. Prevalence of Cryptosporidium parvum infection in Lahore (Pakistan) and its association with diarrhea in dairy calves. Int. J. Agric. Biol. 2009;11(2):221–224. [Google Scholar]
- Odeniran P.O., Ademola I.O. Epidemiology of Cryptosporidium infection in different hosts in Nigeria: a meta-analysis. Parasitol. Int. 2019;71:194–206. doi: 10.1016/j.parint.2019.04.007. [DOI] [PubMed] [Google Scholar]
- Ogendo A., Obonyo M., Wasswa P., Bitek A., Mbugua A., Thumbi S.M. Cryptosporidium infection in calves and the environment in Asembo, Western Kenya: 2015. Pan Afric. Med. J. 2017;28(Suppl. 1):9. doi: 10.11604/pamj.supp.2017.28.1.9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paudyal S., Karna S., Khatiwada S., Joshi L., Tiwari A., Shrestha S. Study on the prevalence of Cryptosporidium in calves and HIV infected humans in the periphery of river basins of Kathmandu valley. Int. J. Infect. Microbiol. 2013;2(1) [Google Scholar]
- Regassa A., Gizaw O., Abunna F., Abebe R., Beyene D., Megersa B., Debela E., Asmare K., Skierve E. Cryptosporidium in calves, lambs and kids at Haramaya, eastern Ethiopia. Ethiop. Vet. J. 2013;17(1):81–94. doi: 10.4314/evj.v17i1.7. [DOI] [Google Scholar]
- Robertson L.J., Johansen Ø.H., Kifleyohannes T., Efunshile A.M., Terefe G. Cryptosporidium infections in Africa-how important is zoonotic transmission? A review of the evidence. Front. Vet. Sci. 2020;7:575881. doi: 10.3389/fvets.2020.575881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafieyan H., Alborzi A., Hamidinejat H., Tabandeh M.R., Hajikolaei M.R. Prevalence of Cryptosporidium spp. in ruminants of Lorestan province, Iran. J. Parasitic Dis. Off. Organ Indian Soc. Parasitol. 2016;40(4):1165–1169. doi: 10.1007/s12639-014-0642-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimelis T., Tassachew Y., Lambiyo T. Cryptosporidium and other intestinal parasitic infections among HIV patients in southern Ethiopia: significance of improved HIV-related care. Parasit. Vectors. 2016;9(1):270. doi: 10.1186/s13071-016-1554-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H.V., Cacciò S.M., Tait A., McLauchlin J., Thompson R.C. Tools for investigating the environmental transmission of Cryptosporidium and Giardia infections in humans. Trends Parasitol. 2006;22(4):160–167. doi: 10.1016/j.pt.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Snelling W.J., Xiao L., Ortega-Pierres G., Lowery C.J., Moore J.E., Rao J.R., Smyth S., Millar B.C., Rooney P.J., Matsuda M., Kenny F., Xu J., Dooley J.S. Cryptosporidiosis in developing countries. J. Infect. Dev. Countries. 2007;1(3):242–256. [PubMed] [Google Scholar]
- Swai E.S., Schoonman L. Investigation into the prevalence of Cryptosporidium infection in calves among small-holder dairy and traditional herds in Tanzania. Vet. Med. Int. 2010;2010:676451. doi: 10.4061/2010/676451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha S., Elmalik K., Bangoura B., Lendner M., Mossaad E., Daugschies A. Molecular characterization of bovine Cryptosporidium isolated from diarrheic calves in the Sudan. Parasitol. Res. 2017;116(11):2971–2979. doi: 10.1007/s00436-017-5606-8. [DOI] [PubMed] [Google Scholar]
- Thomson S., Hamilton C.A., Hope J.C., Katzer F., Mabbott N.A., Morrison L.J., Innes E.A. Bovine cryptosporidiosis: impact, host-parasite interaction and control strategies. Vet. Res. 2017;48(1):42. doi: 10.1186/s13567-017-0447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigabu E., Petros B., Endeshaw T. Prevalence of giardiasis and cryptosporidiosis among children in relation to water sources in selected village of Pawi special district in Benishangul-Gumuz region, northwestern Ethiopia. Ethiop. J. Health Dev. 2010;24(3) [Google Scholar]
- Tombang A.N., Ambe N.F., Bobga T.P. Prevalence and risk factors associated with cryptosporidiosis among children within the ages 0–5 years attending the Limbe regional hospital, southwest region, Cameroon. BMC Public Health. 2019;19(1144) doi: 10.1186/s12889-019-7484-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquhart G.M., Armour J., Duncan J.L., Dunn A.M., Jennings F.W. 2nd ed. Blackwell Science; London: 1996. Veterinary parasitology; pp. 226–227. [Google Scholar]
- Venu R., Latha B.R., Basith S.A., Sreekumar C., Raj G.D., Raman M. Factors influencing on prevalence of Cryptosporidium infection in south Indian dairy calves. J. Parasitic Dis. Off. Organ Indian Soc. Parasitol. 2013;37(2):168–172. doi: 10.1007/s12639-012-0153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Elm E., Altman D.G., Egger M., Pocock S.J., Gøtzsche P.C., Vandenbroucke J.P., STROBE Initiative The strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. J. Clin. Epidemiol. 2008;61(4):344–349. doi: 10.1016/j.jclinepi.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Wegayehu T., Adamu H., Petros B. Prevalence of Giardia duodenalis and Cryptosporidium species infections among children and cattle in north Shewa zone, Ethiopia. BMC Infect. Dis. 2013;13:419. doi: 10.1186/1471-2334-13-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegayehu T., Karim R., Anberber M., Adamu H., Erko B., Zhang L., Tilahun G. Prevalence and genetic characterization of Cryptosporidium species in dairy calves in Central Ethiopia. PLoS One. 2016;11(5) doi: 10.1371/journal.pone.0154647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.X., Tan Q.D., Zhou D.H., Ni X.T., Liu G.X., Yang Y.C., Zhu X.Q. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle, northwest China. Parasitol. Res. 2015;114(7):2781–2787. doi: 10.1007/s00436-015-4537-5. [DOI] [PubMed] [Google Scholar]