Summary
Peroxynitrite is a reactive nitrogen species (RNS) that plays critical roles in signal transduction, stress response, and numerous human diseases. Advanced molecular tools that permit the selective, sensitive, and non-invasive detection of peroxynitrite is essential for understanding its pathophysiological functions. Here, we present pnGFP-Ultra, a high performance, reaction-based, genetically encodable biosensor for imaging peroxynitrite in live cells. pnGFP-Ultra features a p-boronophenylalanine-modified chromophore as the sensing moiety and exhibits a remarkable ~110-fold fluorescence turn-on response towards peroxynitrite while displaying virtually no cross-reaction with other reactive oxygen/nitrogen species. To facilitate the expression of pnGFP-Ultra in mammalian cells, we engineered an efficient noncanonical amino acid (ncAA) expression system that is broadly applicable to the mammalian expression of ncAA-containing proteins. pnGFP-Ultra robustly detected peroxynitrite production in activated macrophages and primary glial cells. pnGFP-Ultra fills an important technical gap and represents a valuable addition to the molecular toolbox for probing RNS biology.
Keywords: peroxynitrite, reactive nitrogen species, biosensor, fluorescent probes, genetic code expansion, noncanonical amino acids, live cell imaging, protein engineering, peroxynitrite production, fluorescence microscopy
eTOC Blurb
Advanced molecular tools that permit the selective, sensitive, and non-invasive detection of peroxynitrite is essential for understanding its pathophysiological functions. Chen et al. report pnGFP-Ultra, a high-performance fluorescent turn-on biosensor for minimally invasive and selective imaging of peroxynitrite production in live cells.
Graphical Abstract
Introduction
Reactive oxygen species (ROS) have been studied intensively for decades, but our understanding of their cousins, reactive nitrogen species (RNS), lagged far behind. Peroxynitrite (ONOO−) is a RNS formed from a diffusion-controlled reaction between nitric oxide (•NO) and superoxide anion (O2•−) (Ischiropoulos et al., 1992; Radi, 2013a), and has been implicated in numerous pathophysiological processes, including cardiac (Mungrue et al., 2002; Ronson et al., 1999), vascular (Förstermann and Münzel, 2006), circulatory (Szabó, 1996), inflammatory (Van Der Veen et al., 1997), diabetic (Zou et al., 2004), and neurodegenerative (Smith et al., 1997; Torreilles et al., 1999) diseases. Although no enzyme dedicated for direct ONOO− production has been identified, enzymes responsible for the biogenesis of the parental species of peroxynitrite are widely known: •NO (Beckman and Koppenol, 1996) and O2•− (Fridovich, 1997) can be generated by nitric oxide synthetase (NOS) and NADPH oxidase (NOX) (Brewer et al., 2015), respectively (Figure 1A). In addition, the mitochondrial electron transport chain has been recognized as an important source of O2•− (Brand, 2016). These primary species present limited cellular toxicity but can react with each other and/or transition metals to produce highly reactive secondary species (e.g. hypochlorous acid, hydroxyl radical, peroxynitrite) that could have deleterious consequences. For example, the direct toxicity of •NO is modest but can be greatly enhanced by reacting with O2•− to produce ONOO−. As such, many nitrosative stresses historically attributed to •NO have been gradually reassigned to ONOO− (Pacher et al., 2007), a far more potent RNS capable of mediating radicals (•OH, CO3•− and •NO2) generation, DNA damage, lipid peroxidation, protein nitration, and cell death (apoptosis or necrosis) (Szabó et al., 2007). In particular, ONOO−-mediated post-translational protein tyrosine nitration plays important roles in cell signal transduction (Liaudet et al., 2009). In macrophages, ONOO− protects cells against invading pathogens by acting as a strong oxidant (Allen et al., 2012). Thus, like many reactive oxygen/nitrogen species (ROS/RNS), peroxynitrite displays a sophisticated signal/stress dichotomy that is inextricably linked to its complex biology (Pacher et al., 2007).
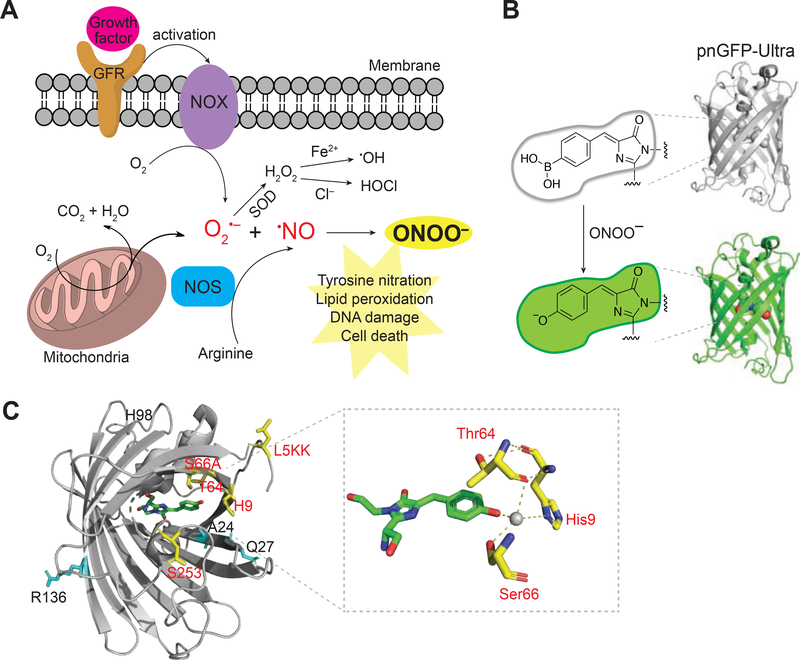
Figure 1. Peroxynitrite Biogenesis and Sensing Mechanism of pnGFP-Ultra.
(A) Illustration of the major peroxynitrite (ONOO–) biogenesis pathway. ONOO– is generated from the diffusion-controlled reaction between superoxide (O2•–) and nitric oxide (•NO). O2•– could be produced during mitochondrial respiration (oxidative phosphorylation) or by activated NADPH oxidase (NOX). NOX can be activated during cell signaling such as a growth factor binding to a growth factor receptor (GFR). Superoxide dismutase (SOD) can convert O2•– to hydrogen peroxide (H2O2), which can mediate secondary radical (e.g. •OH or –OCl) production. •NO is produced by nitric oxide synthetase (NOS), which is coupled to arginine metabolism. ONOO– is a highly reactive nitrogen species that plays crucial roles in tyrosine nitration, lipid peroxidation, DNA damage, and cell death.
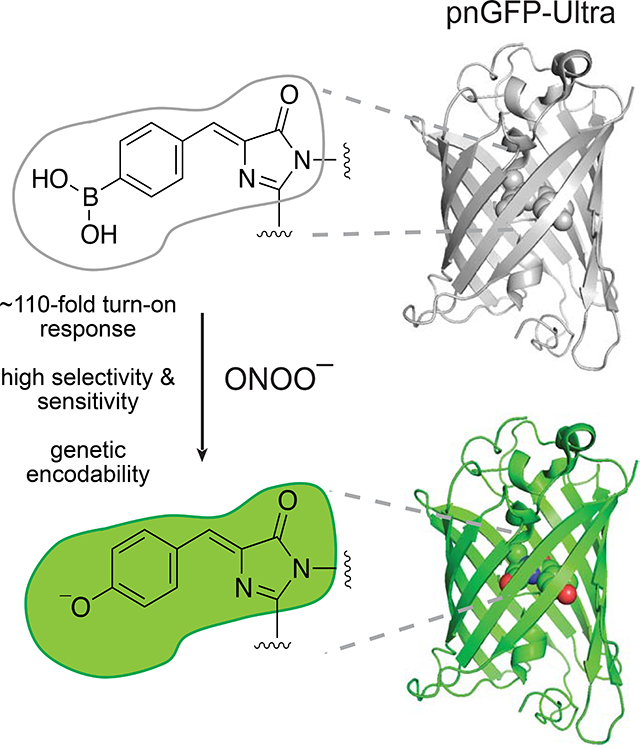
(B) Sensing mechanism of pnGFP-Ultra. pnGFP-Ultra has a boronic acid-modified chromophore that can be converted to a phenolate chromophore upon reaction with ONOO–, resulting in a large fluorescence turn-on response.
(C) Cartoon representation of pnGFP1.5-Y.Cro (PDB 5F9G) and key residues targeted during pnGFP-Ultra development. Residues identified during directed evolution that increase the brightness and folding of cpsGFP are highlighted as cyan sticks. Residues that tune of the chemoselectivity of pnGFP-Ultra are highlighted as yellow sticks. The chromophore is show in green. The inset box on the right shows the chromophore and its interacting residues. The grey sphere denotes a water molecule. The structure was prepared using Pymol.
Elucidating the various roles ONOO− plays, whether in the context of normal cell signaling or pathogenesis, is pivotal for developing targeted therapeutics (Beckman, 2009; Szabó et al., 2007). Efforts to study ONOO− encounter similar hurdles to that of studying other ROS/RNS, such as a very short half-life, permeability through and reactions within membrane, multiplex reaction pathways, and signal/stress dichotomy (Ferrer-Sueta and Radi, 2009; Hardy et al., 2018). Traditionally, detection of ONOO− relied primarily on immunostaining of 3-nitrotyrosine footprints, but this method suffers from poor antibody specificity, a requisite for cell lysis, and interference from other nitration sources (Radi, 2013b). More recently, fluorescent probes for ONOO− (Chen et al., 2013; Li et al., 2015; Lin et al., 2013; Peng and Yang, 2010; Peng et al., 2014; Sikora et al., 2009, 2020; Sun et al., 2014, 2015, 2009; Tian et al., 2011; Ueno et al., 2006; Xu et al., 2011; Yang et al., 2006; Yu et al., 2011, 2013; Zhang et al., 2012; Zielonka et al., 2010) have emerged as powerful tools for ONOO− detection, due to their high responsiveness, signal-to-background ratio, spatial-temporal resolution, and compatibility with well-established fluorescence microscopy platforms (Nadler and Schultz, 2013; Ueno and Nagano, 2011). Deployment of these tools in pathophysiological relevant settings such as in activated macrophage cells (Weber et al., 2020), in live smooth muscles from a mouse model of atherosclerosis (Peng et al., 2014), and in the cerebral vasculature of live mice (Li et al., 2015), has revealed unprecedentedly rich information on peroxynitrite production and pathogenesis. Despite the progress, developing probes with both high responsiveness and high selectivity for ONOO− remains a fundamental challenge (Chen et al., 2016a; Hardy et al., 2018). Of all the peroxynitrite probes reported thus far, none have shown absolute selectivity towards ONOO− over other ROS/RNS such as H2O2, •OH and ClO–. For example, HKGreen-4, one of the most advanced probe for ONOO−, shows reactivity toward •OH and ClO– at 1 equivalent of the probe concentration (1 μM) (Peng et al., 2014); NP3, a probe with high ONOO− reactivity cross-reacts (albeit mildly) with H2O2 and other species at 2 equivalents of the probe concentration (10 μM) (Li et al., 2015); Probe 1–D-fructose shows substantial cross-reaction with ClO– (Sun et al., 2014). In particular, H2O2, a common cellular ROS that can be generated at markedly higher concentrations in certain pathophysiological conditions (Brewer et al., 2015), represents a major species competing for probe reactivity in a complex cellular milieu, confounding the unambiguous detection of ONOO−. Finally, existing probes—predominately in the form of small molecule dye derivatives—often require organic synthesis and lack the capability to be genetically encoded, a feature that would greatly lower the barrier to the widespread adoption of these probes by other laboratories. A genetically encodable sensor would also facilitate cell-/tissue-specific expression for in vivo studies, as exemplified by the popular genetically encoded calcium sensors (e.g. GCaMP series) (Chen et al., 2017; Lin and Schnitzer, 2016).
We previously developed pnGFP (Chen et al., 2013), a genetically encoded fluorescent probe for the selective detection of peroxynitrite, by site-specific incorporation of p-boronophenylalanine (pBoF) into the chromophore of a circularly permuted superfolder green fluorescent protein (cpsGFP). This is achieved via genetic code expansion (Chin, 2017; Liu and Schultz, 2010), a revolutionary technique that uses an orthogonal aminoacyl-tRNA synthetase (aaRS)–tRNA pair to direct the incorporation of a ncAA into proteins in response to a nonsense codon (often the amber UAG codon). Interestingly, we serendipitously discovered that the boronic acid-based chromophore of pnGFP shows unusual high selectivity for ONOO−—essentially nonresponsive to even 1 mM H2O2 (> 2000 equivalents of the probe concentration). Mechanistic studies using site-directed mutagenesis, X-ray crystallography, 11B NMR, and computational simulation collectively revealed that the boron atom in pnGFP is converted to an sp3-hybridized form by a nearby histidine residue via a polarized water bridge (Chen et al., 2016b). The unique pathway by which pnGFP gains high selectivity suggests that the protein scaffold of genetically encoded probes can be engineered to tune chemoselectivity, which remains challenging to obtain from small molecule probes.
Here, using directed evolution, rational mutagenesis, and targeted reactivity screening guided by the crystal structure of pnGFP1.5-Y.Cro (Chen et al., 2016b), we engineered a high-performance peroxynitrite probe, pnGFP-Ultra, which show low detection limit and ultra-high responsiveness and selectivity for ONOO−. Compared to pnGFP, pnGFP-Ultra has better chromophore maturation, brighter fluorescence, and a 6-fold enhancement on both reactivity and selectivity, making it the most advanced genetically encoded biosensor for peroxynitrite. To facilitate the use of pnGFP-Ultra in mammalian cells, we systematically optimized the plasmid-based expression system for ncAA incorporation, leading to a 13.3-fold higher pBoF-incorporated protein expression in HEK 293T cells. We demonstrate that pnGFP-Ultra can be used to robustly detect physiological peroxynitrite production in activated macrophages and primary mouse glia. Together, these developments make pnGFP-Ultra a powerful tool for imaging peroxynitrite, especially in experiments where genetic encodability and high selectivity are of primary interests.
Results
Design and Engineering of pnGFP1.5
pnGFP was engineered by replacing the chromophore tyrosine residue of a circularly permutated superfolder green fluorescent protein (cpsGFP) with a ncAA, pBoF (Chen et al., 2013). Upon reaction with ONOO−, the boronic acid-based chromophore (dark) is converted into its phenolate form (bright), leading to potent fluorescence turn-on response (Figure 1B). Despite the conceptual success, the broad utility of pnGFP is stymied by several of its drawbacks, including poor protein folding/chromophore maturation, low mammalian cell expression, and dim fluorescence before and after ONOO− conversion. Given the sensing mechanism of pnGFP, we aimed to first improve the folding, expression, and brightness of cpsGFP (Chen and Ai, 2014), the parental template of pnGFP that has a tyrosine derived chromophore (Figure S1). We carried out multiple rounds of directed protein evolution followed by colony-based screening in Escherichia coli (E. coli) cells (Figures S1A), leading to a brighter mutant, cpsGFP2 (cpsGFP-V24A K27Q Y98H S136R) (Figure S1B). Next, we substituted the chromophore tyrosine residue of cpsGFP2 with pBoF (Y174B, where B = pBoF) through genetic code expansion. To screen for mutants with high reactivity and selectivity toward ONOO−, we performed saturation mutagenesis on threonine 5 (T5) and threonine 253 (T253), two terminal residues that have previously been shown to influence the reactivity of ncAA-containing fluorescent biosensors (Chen and Ai, 2014; Chen et al., 2012, 2013, 2016b). Library screening coupled with fluorescence-based in vitro assays (Figure S1) resulted in pnGFP1.5 (cpsGFP2-T5L Y174B T253S) (Figure 2), which exhibited a remarkable 132-fold fluorescence turn-on response towards 100 μM ONOO− (Figure 3A). pnGFP1.5 has a higher protein expression level in E. coli as compared to pnGFP, presumably because of enhanced protein folding and chromophore maturation.
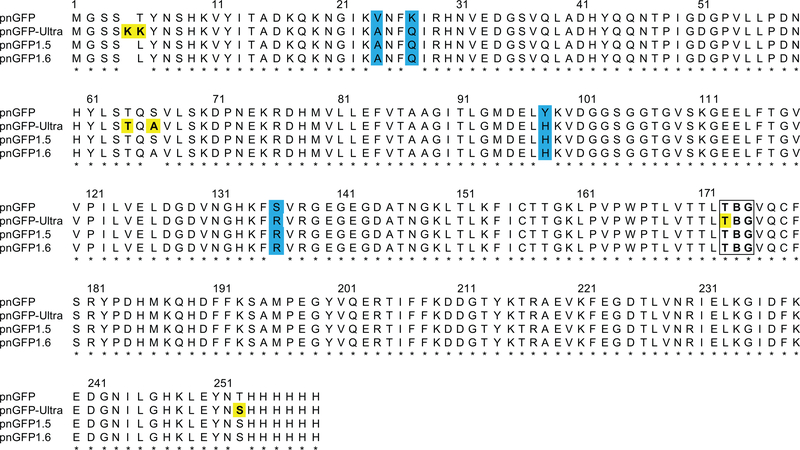
Figure 2. Sequence Alignment of pnGFP-Ultra and Related Proteins.
Mutations identified during directed evolution are highlighted in cyan. Residues subjected to rational mutagenesis are yellow-colored. The chromophore-forming residues (B denoting pBoF) are highlighted in a box. Residues are numbered according to the numbering of pnGFP1.5-Y.Cro (PDB 5F9G).
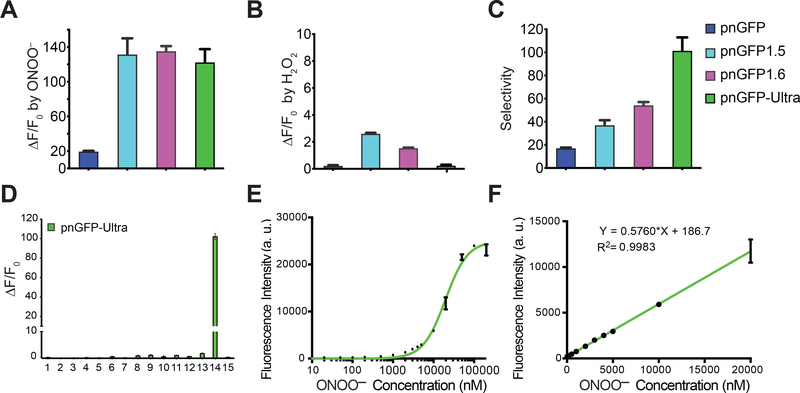
Figure 3. In vitro Characterization of pnGFP-Ultra.
(A) Dynamic range (expressed as ΔF/F0) of pnGFP-Ultra and relevant mutants towards ONOO–. F0 is the initial fluorescence intensity. ΔF is the final fluorescent intensity (after treatment) minus F0. Samples were treated with 100 μM ONOO− for 1 hour.
(B) Dynamic range (ΔF/F0) of pnGFP-Ultra and relevant mutants towards H2O2. Samples were treated with 100 μM H2O2 for 1 hour.
(C) Selectivity of pnGFP-Ultra and its mutants. Arbitrarily defined selectivity is calculated as the fold of fluorescence enhancement by ONOO− / fold of fluorescence enhancement by H2O2.
(D) Response of pnGFP-Ultra post 1-h incubation with a panel of redox-active chemicals: (1) •OtBu (1 mM Fe2+ and 100 μM HOOtBu), (2) 100 μM HOOtBu, (3) •OH (1 mM Fe2+ and 100 μM H2O2), (4) 5 mM oxidized glutathione, (5) 100 μM O2•−, (6) 100 μM NOC-7 (•NO donor), (7) 100 μM HOCl, (8) 100 μM NaHS (H2S donor), (9) 5 mM L-cysteine, (10) 1 mM DL-homocysteine, (11) 1 mM vitamin C, (12) 100 μM H2O2, (13) 1 mM H2O2, (14) 100 μM ONOO−, (15) Tris buffer.
(E) Fluorescence responses of pnGFP-Ultra (0.2 μM) to nanomolar to high micromolar concentrations of ONOO−. The half maximal response concentration (EC50) was determined to be 19.5 μM.
(F) Linear fluorescence responses of pnGFP-Ultra (0.2 μM) to nanomolar to low micromolar concentrations of ONOO−. The limit of detection was determined to be 122 nM with a signal-to-noise ratio (S/N) of 3.
Data in all panels represent the mean ± SD of triplicates.
Structure-Guided Engineering and in Vitro Characterization of pnGFP-Ultra
Compared to pnGFP, pnGFP1.5 showed a 6-fold enhancement in the dynamic range towards ONOO− (Figure 3A), but also an undesirable ~3-fold increase in the dynamic range towards H2O2 (Figure 3B). The overall selectivity (arbitrarily defined as fold of fluorescence enhancement by ONOO− / fold of fluorescence enhancement by H2O2) of pnGFP1.5 remains higher than that of pnGFP (Figure 3C). We next sought to tune the selectivity of pnGFP1.5 by rational mutagenesis. In our previous attempt to decipher the unusual chemoselectivity of pnGFP, we solved the X-ray crystal structure of pnGFP1.5-Y.Cro (pnGFP1.5-B174Y, PDB 5F9G), which contains a tyrosine-derived chromophore (Chen et al., 2016b). The structure indicates that three residues—namely Thr64, Ser66, His9—are in close proximity to the phenolate oxygen of the chromophore (Figure 1C). We speculate that these residues may interact with the pBoF-derived chromophore in pnGFP1.5 to fine tune the chromophore environment (electrostatics, hydrophobicity, and solvent accessibility). In addition, the first chromophore-forming residue (Thr173) can also affect electron distributions within the conjugate chromophore structure. Thus, these four residues are next subjected to site-directed mutagenesis (Figure 2 and S1). Based on our prior experiences with pnGFP, we created a panel of pnGFP1.5 mutants and assayed their selectivity in vitro (Figure S2). From this screening, we identified pnGFP1.6 (pnGFP1.5-S66A), which showed much lower reactivity towards H2O2, but maintained the same amplitude of response towards ONOO−. All other mutants exhibited lower selectivity than pnGFP1.5, either due to low fluorescence intensity after ONOO− conversion or high reactivity towards H2O2 (Figure S2). We next focused on improving pnGFP1.6 (Figure S1B).
pnGFP gains selectivity, in part, by leveraging the nucleophilic attack efficiency differences between ONOO− and HOO− (Chen et al., 2016b). Due to pKa differences (11.6 for H2O2 and 6.8 for ONOOH), the portion of HOO− at neutral pH is much lower than that of ONOO−, thereby disfavoring the reaction of pnGFP with H2O2. However, this difference alone is not sufficient to explain the remarkable selectivity of pnGFP, suggesting that the protein scaffold may further modulate reactivity and specificity. To this end, we substituted threonine 5—which gates an opening towards the chromophore—with two randomized residues by using degenerate NNK codons. Screening of the resultant library against both H2O2 and ONOO− led to pnGFP-Ultra (pnGFP1.6-T5KK, Figure 2 and S1B), which showed virtually no response to 100 μM H2O2 even after 1-hour incubation (Figure 3B). While this came at a price of a slightly reduced reactivity to ONOO− (Figure 3A), the selectivity of pnGFP-Ultra is 1.9- and 6-fold that of pnGFP1.6 and pnGFP, respectively (Figure 3C). Interestingly, in pnGFP-Ultra, threonine 5 is replaced with two consecutive lysine residues (Figure 2), which possibly abolish the reactivity of pnGFP-Ultra towards H2O2 by channeling a positively charged entry route that diminishes local availability of H2O2 near the pBoF-derived chromophore. This unexpected screening result further highlights the unique scaffolding effect of protein-based sensors, which could be harnessed to achieve unusual chemoselectivity.
We tested in vitro-purified pnGFP-Ultra against a panel of redox-active molecules, including ROS/RNS and thiols that are commonly present in a complex cellular milieu (Figure 3D). Except for ONOO−, none of the species surveyed triggered appreciable fluorescent changes, confirming that pnGFP-Ultra is highly selective towards ONOO− (Figure 3D). Immediately (less than 10 seconds) upon incubation with 100 μM ONOO−, pnGFP-Ultra displayed a robust ~110-fold fluorescence enhancement. By contrast, 1-hour incubation of pnGFP-Ultra with 100 μM and 1 mM H2O2 triggered only 0.31- and 1.46-fold fluorescence increase (ΔF/F0), respectively (Figure 3D). Given the low basal fluorescence intensity of pnGFP-Ultra (essentially dark), these small fluorescence increases are negligible in most applications. pnGFP-Ultra responded to high nanomolar to high micromolar concentrations of ONOO− and the half maximal response concentration (EC50) was determined to be 19.5 μM (Figure 3E). Furthermore, the fluorescence intensity of 0.2 μM pnGFP-Ultra increased linearly from high nanomolar to ~20 μM ONOO− concentrations, with the limit of detection (LOD) determined to be 122 nM (Figure 3F). To note, the EC50 and LOD determined from these assays depend on sensor concentrations. Indeed, when a higher sensor concentration (1 μM) was used, the EC50 and LOD increased (Figure S3AB). Compared to pnGFP, pnGFP-Ultra exhibits an overall 6-fold improvement in both the dynamic range and selectivity (Figure 3A and 3C). We further compared the time-lapse responses of pnGFP-Ultra with those of coumarin boronic acid (CBA), a small-molecule boronate-based probe (Zielonka et al., 2010, 2012a). The fluorescence of CBA increased drastically in response to both H2O2 and SIN-1 (a ONOO− donor), while pnGFP-Ultra showed excellent selectivity although the response magnitude is lower and kinetics are slower than those of CBA (Figure S3C). This phenomenon is expected given the solvent-exposed boronic acid group (i.e. high accessibility to both H2O2 and ONOO−) in CBA. Taken together, these results strongly demonstrate that pnGFP-Ultra is a high performance RNS biosensor that is highly responsive and selective for peroxynitrite.
Development of an Enhanced ncAA Incorporation System in Mammalian Cells
Efficient co-translational ncAA incorporation in response to the amber TAG codon is crucial for the success of pnGFP-Ultra and other applications that require ncAA incorporation. Indeed, one of the fundamental limitations of pnGFP and other ncAA-based biosensors is their inadequate expression in mammalian cells. While ncAA incorporation in E. coli. has been thoroughly optimized to an extent that ncAA incorporation can reach a level comparable to that of canonical amino acids, mammalian incorporation of ncAA has proven to be far more challenging, presumably due to a more complicated ribosomal translation machinery.
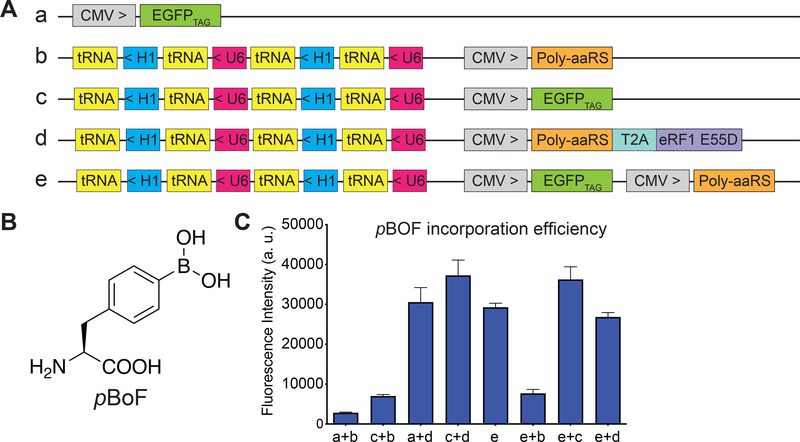
Existing plasmid-based systems for ncAA incorporation (Chen et al., 2009; Hino et al., 2012; Liu et al., 2007; Schmied et al., 2014; Xiao et al., 2013) vary in the type and copy number of the engineered orthogonal tRNA-aminoacyl tRNA synthetase pair, and the choices of vectors and promoters, making it difficult to cross compare the efficiencies of these systems. Several strategies for enhancing amber codon suppression efficiency such as increasing the copy number of tRNAs (Liu et al., 2007), use of type-3 RNA polymerase III promoters to express the engineered tRNAs (Wang et al., 2007), and ectopic expression of a dominant-negative eukaryotic release factor 1 mutant (eRF1-E55D) (Schmied et al., 2014) have proven to be generalizable. To systematically optimize ncAA incorporation into proteins in mammalian cells, we constructed a series of expression plasmids (plasmids a-e, Figure 4A) and tested ncAA incorporation in HEK293T cells using an EGFPTAG fluorescent reporter gene that bears an amber TAG codon at position 39 of EGFP. Incorporation of a ncAA in response to the amber TAG codon (amber suppression) would generate the full length EGFP while failure to do so would result in a truncated EGFP (non-fluorescent). Previously, pBoF was incorporated into pnGFP by co-transfecting the CMV promoter-driven pnGFP reporter plasmid with the suppressor plasmid, which harbors one copy of CMV promoter-driven poly-specific aminoacyl tRNA synthetase (Poly-aaRS) and four copies of U6/H1 promoter-driven suppressor tRNAs (Figure 5A). Since orthogonal tRNA expression is a limiting factor in ncAA incorporation efficiency, we cloned four copies of tRNAs on the reporter plasmid c as well (Figure 4A). Compared to our previous expression method using plasmids a + b, addition of extra copies of tRNA on the reporter plasmid doubled the incorporation efficiency of pBoF (Figure 4B) into the EGFPTAG reporter (Figure 4C, plasmids c + b). To test whether ectopic expression of eRF1-E55D further enhances ncAA incorporation efficiency, eRF1-E55D was bicistronically expressed with Poly-aaRS via a self-cleaving T2A peptide linker (Figure 4A, plasmid d). Co-expression of eRF1-E55D-containing suppressor plasmid d, in lieu of b, with the original reporter plasmid a resulted in a 10.9-fold enhancement in pBoF incorporation (Figure 4C). Remarkably, we observed further improvements in incorporation efficiency using plasmids c + d, suggesting that the benefit from expression of extra copies of tRNA and the eRF1-E55D mutant are synergistic (Figure 4C). To simplify the expression system, we integrated all genetic components necessary for ncAA incorporation—including the reporter gene, Poly-aaRS, and orthogonal tRNAs—into a single plasmid (Figure 4A, plasmid e). Expression of plasmid e alone gave rise to comparable incorporation efficiency to that of using a + d. Co-transfection of plasmid e with other plasmids led to various degrees of expression efficiency. Nevertheless, all strategies tested surpass our original method of using plasmids a + b for ncAA incorporation, with plasmids c + d and c + e having the highest incorporation efficiency for pBoF (13.3-fold enhancement) (Figure 4C). We achieved similar enhancement in incorporation efficiency for p-azido-phenylalanine (pAzF) (Figure S4), suggesting that this plasmid-based system is quite general and can be used to incorporate other ncAAs.
Figure 4. Engineering of a Highly Efficient Plasmid-based ncAAs Incorporation System in Mammalian Cells.
(A) Schematics of various expression plasmids. Poly-aaRS is the poly-specific aminoacyl tRNA synthetase, eRF1-E55D is the mutant eRF1 gene, T2A is a self-cleaving 2A peptide, tRNA is the orthogonal tRNA, U6 indicates the U6 promoter, H1 indicates the H1 promoter, CMV is the CMV promoter, EGFPTAG is the EGFP reporter gene with a TAG codon at position 39. > or < denotes the direction of the gene cassettes.
(B) Chemical structure of p-Boronophenylalanine (pBoF).
(C) Quantification of pBoF (1 mM) incorporation into the EGFPTAG reporter gene measured in a cell-based fluorescence assay. The indicated constructs were transiently expressed in HEK 293T cells and the cell lysate green fluorescence was quantified in a plate reader at 515 nm emission with excitation at 490 nm. Data represent the mean ± SD of triplicates.
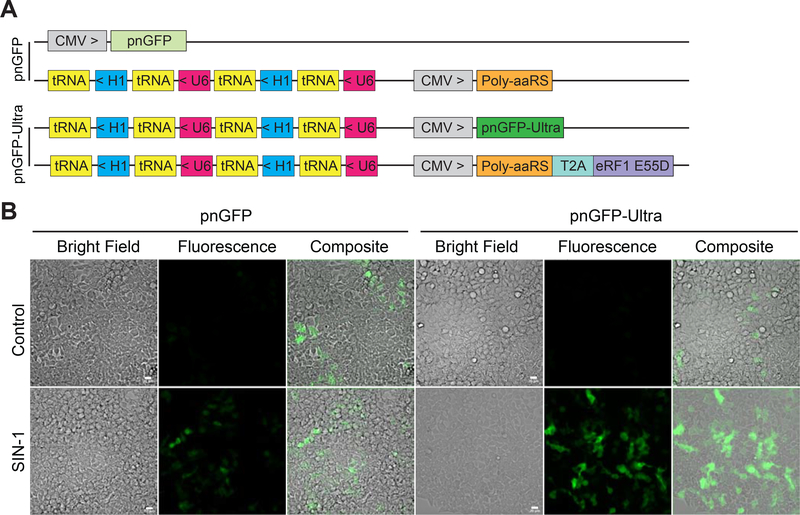
Figure 5. Live-cell imaging of peroxynitrite in HEK 293T cells.
(A) Constructs used for expressing pnGFP (top two plasmids) and pnGFP-Ultra (bottom two plasmids).
(B) Green fluorescent and bright-field images of cells expressing pnGFP (left) or pnGFP-Ultra (right). In the bottom row, HEK 293T cells in HBSS were treated with 100 μM SIN-1 for 90 min before imaging. Experiments were repeated three times with independent cultures and similar results were obtained. Scale bar, 20 μm.
Imaging of Peroxynitrite in Live Cells using pnGFP-Ultra
Next, we constructed a mammalian expression plasmid by replacing the EGFPTAG gene with pnGFP-Ultra gene in plasmid c (Figure 5A). We co-expressed the resultant plasmid with plasmid d, as this combination gave rise to the highest pBoF incorporation efficiency in the above EGFPTAG reporter assay (Figure 4C). To compare with the first-generation biosensor, we expressed pnGFP in parallel by utilizing our previous method (Figure 5A). After treatment with SIN-1, the fluorescence intensities from pnGFP-Ultra-expressing HEK 293T cells were much higher than those from pnGFP-expressing cells (Figure 5B). It is also evident (from the number of fluorescent cells) that the expression efficiency of pnGFP-Ultra is much higher than that of pnGFP. Both pnGFP and pnGFP-Ultra are selective towards ONOO−, with no obvious response toward H2O2 (1 mM) or HOCl (100 μM) (Figure S5AB). We further expressed pnGFP-Ultra-Y.Cro, which contains a tyrosine-derived chromophore, in HEK 293T cells. Treating these cells with SIN-1 did not change the fluorescence (Figure S5CD), confirming that the pBoF-derived chromophore in pnGFP-Ultra is responsible for ONOO− sensing. Moreover, in the presence of 100 μM urate or minocycline, both of which are well-established ONOO− scavengers (Schildknecht et al., 2011), the fluorescence response of pnGFP-Ultra in HEK 293T cells to SIN-1 was largely attenuated (Figure S6AB). Because ONOO− was not enzymatically synthesized under this treatment condition, 1400W, a selective inhibitor of inducible nitric oxide synthase (iNOS) (Garvey et al., 1997), was unable to attenuate the fluorescence turn-on caused by SIN-1 (Figure S6AB). Furthermore, the SIN-1 treatment significantly reduced the viability of HEK 293T cells, while urate or minocycline, but not 1400W, rescued SIN-1-induced cell death (Figure S6C). Taken together, pnGFP-Ultra is a high-performance biosensor for imaging ONOO− in mammalian cells. We attribute the superior performance of pnGFP-Ultra to both its large dynamic range (~ 110-fold fluorescence enhancement) and the substantially improved pBoF incorporation efficiency.
Visualization of Peroxynitrite Production in Activated Macrophages
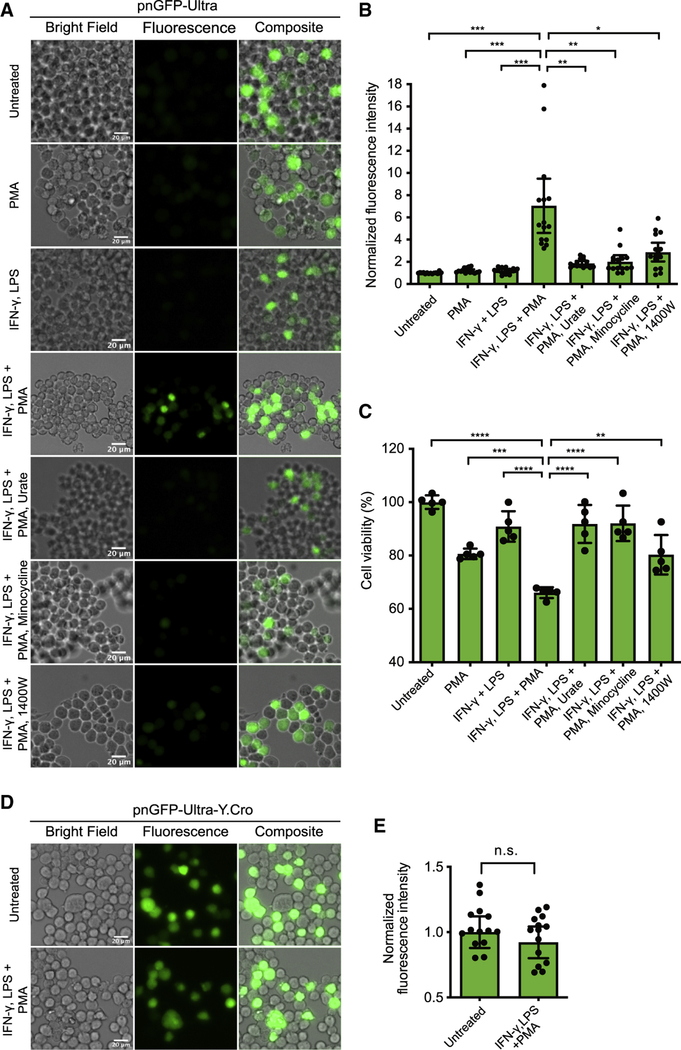
To further demonstrate the utility of pnGFP-Ultra in imaging physiological ONOO−, we expressed pnGFP-Ultra in mouse RAW264.7 macrophage immune cells, which are known to produce ONOO− from nitric oxide and superoxide under immune stimulation (Allen et al., 2012; Ischiropoulos et al., 1992; Peng et al., 2014; Xia and Zweier, 1997). We first pre-stimulated the cells for 10 h using bacterial endotoxin lipopolysaccharide (LPS) and pro-inflammatory cytokine interferon-gamma (IFN-γ) to increase iNOS expression, and next, incubated the cells with phorbol myristate acetate (PMA) for 1 h to activate NADPH oxidase (Zielonka et al., 2012b). We observed strong fluorescence signal in pnGFP-Ultra expressing cells (Figure 6A). The mean fluorescence of cells in the treated group was ~ 7.1-fold higher than that of untreated cells (Figure 6B), while IFN-γ and LPS, or PMA alone only minimally affected the fluorescence. Furthermore, in the presence of ONOO− scavengers (100 μM urate or minocycline) or the iNOS inhibitor (100 μM 1400W), the fluorescence increase of pnGFP-Ultra was attenuated (Figure 6B). We next evaluated the impact of these conditions on the viability of RAW264.7 cells. As expected, treatment with LPS, IFN-γ and PMA induced cell death; either urate, or minocycline, or 1400W partially rescued cell viability (Figure 6C). The observed fluorescence and cell viability changes are aligned with our further validation, which used SIN-1 to deliver ONOO− to RAW264.7 macrophage cells. SIN-1 activated the fluorescence of pnGFP-Ultra and induced the death of RAW264.7, both of which can be largely reverted by supplementing the ONOO− scavengers (Figure S7). As a control, we expressed pnGFP-Ultra-Y.Cro in RAW264.7 cells and no fluorescence change was observed post-treatment of cells with LPS, IFN-γ and PMA (Figure 6DE). Collectively, these results demonstrate that pnGFP-Ultra can reliably detect physiological ONOO− production.
Figure 6. Live-cell Imaging of Peroxynitrite Production in Activated Macrophages.
(A) Mouse RAW264.7 cells were transfected with pnGFP-Ultra. Cells were next pretreated with IFN-γ (50 U/mL) and LPS (0.5 μg/mL) for 10 h. After LPS and IFN-γ were removed, cells were further treated with PMA (200 ng/mL) in HBSS for 1 h before imaging. Several control groups were used. In the “untreated” group were cells untreated with either of LPS, IFN-γ and PMA. In the “PMA” group were cells without pretreatment but directly treated with PMA for 1 h before imaging. In the “IFN-γ, LPS” group were cells pretreated with LPS and IFN-γ but without additional PMA treatment. In addition, cells in other control groups were co-treated with ONOO− scavengers (urate or minocycline) or an iNOS inhibitor (1400W) when the PMA treatment was implemented. Representative bright field (left), fluorescence (middle) and overlay (right) images of each group were shown. Experiments were repeated three times with independent cultures and similar results were obtained. Scale bar, 20 μm.
(B) Quantification of fluorescence intensities of cells in each treatment group in panel A. Data are presented as mean ± 95% confidential intervals and black dots indicate fluorescent intensities of single cells (n = 15 individual cells from 3 independent replicates in each group). Statistical test was performed using Brown-Forsythe and Welch’s ANOVA followed by Dunnett’s T3 multiple comparisons test (***P < 0.001, **P < 0.01 and *P < 0.05).
(C) MTT cell viability assay of RAW264.7 cells in each treatment group. Raw data were normalized to pnGFP-Ultra-expressing cells untreated with either of LPS, IFN-γ and PMA. Data are presented as mean ± SD and black dots indicate the cell viability of individual replicates (n = 5 wells for each group). Statistical test was performed using ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test (****P < 0.0001, ***P < 0.001, and **P < 0.01).
(D) Mouse RAW264.7 cells transiently expressing pnGFP-Ultra-Y.Cro (which contains a tyrosine-derived chromophore) were untreated or treated with LPS, IFN-γ, and PMA. Experiments were repeated three times with independent cultures and similar results were obtained. Scale bar, 20 μm.
(E) Quantification of fluorescence intensities of cells in each group in panel D. Data are presented as mean ± 95% confidential intervals and black dots indicate fluorescent intensities of single cells (n = 15 individual cells from 3 independent replicates in each group). Comparison was made using an unpaired two-tailed t-test (n.s., not significant, P > 0.05).
Detection of Peroxynitrite Production in Amyloid β-Activated Primary Mouse Glia
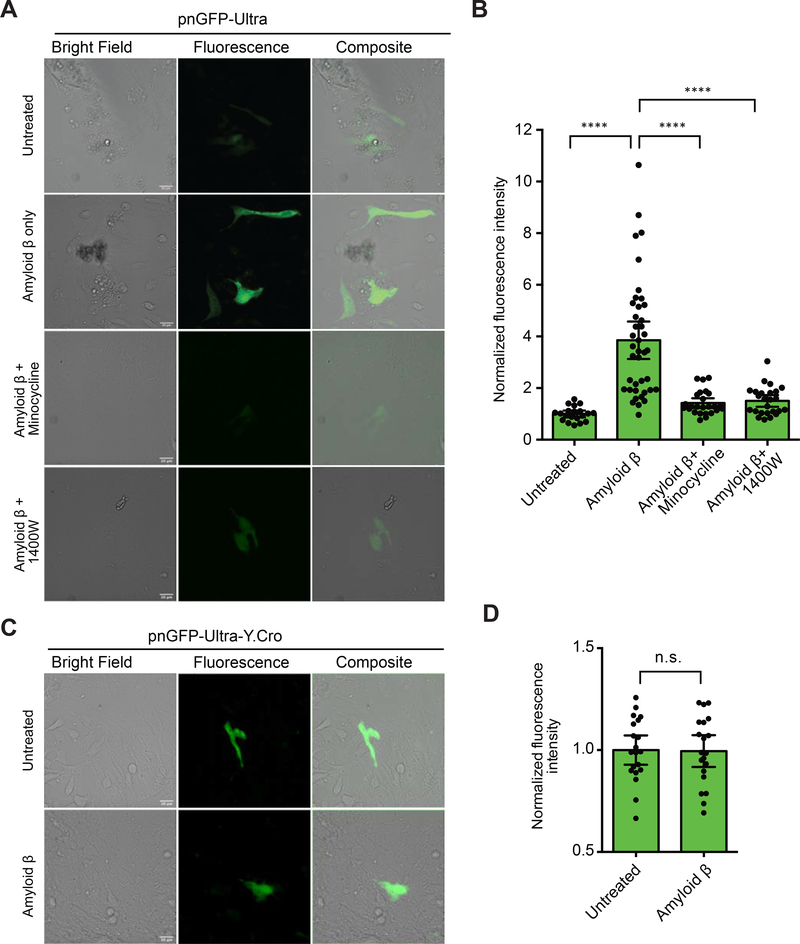
Despite the continuous improvement in life expectancy, Alzheimer’s disease (AD) and other neurodegenerative disorders have become a global health and social challenge (Alzheimer’s Association, 2020). ONOO− is known to play important roles in numerous neurodegenerative diseases. For example, ONOO−-mediated damage has been broadly observed in AD (Smith et al., 1997) and the amyloid β peptide, whose accumulation has been considered to be the hallmark of AD, is known to induce ONOO− production in glial cells (González-Reyes et al., 2017; Xie et al., 2002) and subsequently mediate neurotoxicity (Xie et al., 2002). To test whether pnGFP-Ultra can detect ONOO− production in neurological systems, we expressed pnGFP-Ultra in cultured primary mouse glia cells. Indeed, we observed dramatically enhanced fluorescence signal (~3.9-fold compared with untreated cells) from glia activated with amyloid β (Figure 7AB). Either the ONOO− scavenger, minocycline, or the iNOS inhibitor, 1400W, attenuated the fluorescence increase of pnGFP-Ultra (Figure 7AB). In addition, treating glia cells expressing pnGFP-Ultra-Y.Cro with amyloid β caused no fluorescence enhancement (Figure 7CD). Thus, pnGFP-Ultra represents a valuable tool enabling the imaging of ONOO− production in neurological systems.
Figure 7. Live-cell Imaging of Amyloid β-Induced Peroxynitrite in Primary Glial Cells.
(A) Primary mouse glia cells were isolated and transfected with pnGFP-Ultra. After 24 hours, the samples were untreated or treated with amyloid β (5 μM) in the absence or presence of an ONOO− scavenger (minocycline) or an iNOS inhibitor (1400W) for 42 hours before imaging. Representative bright field (left), fluorescence (middle) and overlay (right) images of each group were shown. Experiments were repeated three times with independent cultures and similar results were obtained. Scale bar, 20 μm.
(B) Quantification of fluorescence intensities of cells in each treatment group in panel A. Data are presented as mean ± 95% confidential intervals and black dots indicate fluorescent intensities of single cells (n = 20, 40, 25, and 25 individual cells from 3 independent cultures for untreated, amyloid β-treated, amyloid β and minocycline-treated, and amyloid β and 1400W-treated groups, respectively). Statistical test was performed using Brown-Forsythe and Welch’s ANOVA followed by Dunnett’s T3 multiple comparisons test (****P < 0.0001).
(C) Primary mouse glia cells expressing pnGFP-Ultra-Y.Cro (which contains a tyrosine-derived chromophore) untreated or treated with amyloid β (5 μM) for 42 hours before imaging. Experiments were repeated three times with independent cultures and similar results were obtained. Scale bar, 20 μm.
(D) Quantification of fluorescence intensities in panel C. Data are presented as mean ± 95% confidential intervals and black dots indicate fluorescent intensities of single cells (n = 20 individual cells from 3 cultures for each group). Comparison was made using an unpaired two-tailed t-test (n.s., not significant, P > 0.05).
Discussion
In summary, we have developed pnGFP-Ultra—a highly responsive and selective fluorescent ONOO− biosensor—by directed evolution, structure-guided rational design, and reactivity-based library screening. Compared to the first-generation ONOO− probe pnGFP, pnGFP-Ultra has much better chromophore maturation and a 6-fold larger ONOO−-induced fluorescence response, while retaining the high chemoselectivity against H2O2 and other ROS/RNS. An apparent hurdle of applying ncAA-based biosensors to cell imaging studies has been the inefficiency with which ncAA can be incorporated into proteins in mammalian cells. Here, we systematically optimized and increased pBoF incorporation efficiency in HEK 293T cells by 13.3-fold. These developments make pnGFP-Ultra a high-performance biosensor capable of imaging physiological peroxynitrite, as we have demonstrated in activated macrophages and primary glial cells. pnGFP-Ultra thus represents an important advancement enabling the practical implementation of a genetically encoded biosensor for live-cell detection of peroxynitrite. We expect pnGFP-Ultra to find broad utility in probing RNS biology in diverse pathophysiological settings.
pnGFP-Ultra, in the purified form and at the concentration of 0.2 μM, detected as low as 122 nM ONOO−, and responded up to ~100 μM ONOO− with a linear response range below 20 μM ONOO−. We applied a bolus addition of ONOO− in these measurements and because ONOO− degrades in neutral aqueous solutions and biological systems within seconds and milliseconds, respectively (Radi, 2018; Szabó et al., 2007), it is difficult to directly translate the in vitro responses of pnGFP-Ultra to its performance in live cells. Nevertheless, given the irreversibility and 1:1 stoichiometry of the reaction, we estimate that only a small portion of ONOO− molecules reacted with pnGFP-Ultra, presumably due to the highly unstable and transient nature of ONOO− and its relatively restricted accessibility to the chromophore. These results should further minimize concerns of perturbing and scavenging of endogenous ONOO− when the biosensor is expressed in cells.
Previous studies estimated that the steady-state concentrations of ONOO− in biological systems are likely in the nanomolar range, but its formation rate could be as high as ~5 μM·s−1 (Radi, 2018; Szabó et al., 2007). Owning to the facts that cellular production of ONOO− is often sustained and that pnGFP-Ultra is an irreversible biosensor, we indeed successfully used pnGFP-Ultra to detect physiological ONOO− in both activated macrophages and primary glial cells. Since pnGFP-Ultra was responsive to a large concentration range of ONOO−, it may be compatible with diverse cell types and microenvironments. Moreover, the large intensiometric response of pnGFP-Ultra will facilitate its future conversion into a genetically encoded ratiometric biosensor for in situ measurement and calibration of ONOO− concentrations by fusing pnGFP-Ultra to a FRET donor (such as EBFP2 (Ai et al., 2007)), as we have recently demonstrated with a hydrogen sulfide biosensor (Youssef et al., 2019).
pnGFP-Ultra not only fills an important technological gap in the field of RNS, but also conveys innovations that will inspire the development of future biosensors for other bioanalytical targets. First, the chromophore (and other residues) of circularly permutated fluorescent proteins can be modified into a hub of chemical sensing, binding, reactivity, and catalysis, thereby greatly expanding the use of fluorescent proteins as reporters. Second, direct evolution and rational mutagenesis performed on ncAA-modified proteins can lead to new and sometimes, surprising functions, as we have demonstrated here via the engineering of exquisite chemoselectivity of pnGFP-Ultra. While this paper was under review, the genetic introduction of thyronine (Thy), another ncAA, into sfGFP and cpsGFP was reported and the resultant proteins showed early promise for the detection of ONOO− (Li et al., 2020). Since aryloxyphenols also react with other highly reactive ROS such as hydroxyl radical (•OH) (Price et al., 2009), we envision that the method we describe here may be used to further engineer Thy-based biosensors specifically responsive to either ONOO− or other highly oxidizing species.
We compared the responses of pnGFP-Ultra with a synthetic boronate-based probe, CBA, using activity-based assays. Although the selectivity is of course a function of the incubation time, pnGFP-Ultra is convincingly a much more selective ONOO− biosensor than CBA. The increased chemoselectivity of pnGFP-Ultra is accompanied with reduced reactivity, as the sensory moiety is shielded by the protein scaffold. To date, significant efforts have been devoted into modulating the chemoselectivity of various small-molecule ONOO− biosensors (Sikora et al., 2020), and our study provides a unique example which uses a protein scaffold to modulate reactivity and specificity. The engineered protein scaffold may directly scavenger highly reactive ROS, such as HOCl. More generally, the protein scaffold provides a special environment—which affects the reactivity and accessibility of various ROS and RNS—and can serve as a widely adaptable strategy for developing chemoselective biosensors. Furthermore, pnGFP-Ultra still has residual activity with H2O2. With extended incubation, it is difficult to completely rule out the contribution of H2O2 to the fluorescence increase of pnGFP-Ultra. We thus recommend the use of complementary tools, such as peroxynitrite scavengers, •NO synthesis inhibitors, and additional nitrotyrosine detection methods (Teixeira et al., 2016), for further confirmation.
Compared to existing small-molecule based ONOO− probes, pnGFP-Ultra has another advantage of being genetically encodable. This feature permits long-term in vivo imaging and organelle-/cell-/tissue-specific targeting, making it possible to apply pnGFP-Ultra to a variety of biological/disease models. More importantly, because of the convenient fluorescence readout and robust off-on response of pnGFP-Ultra, cell lines stably expressing this reporter can be used in high-throughput drug screening assays. Such capability will aid the development of drugs that protect neurons or other cell types against ONOO−-mediated cell toxicity/death (Szabó et al., 2007). pnGFP-Ultra may also be used in genome-wide CRISPR screens (Shalem et al., 2015) to uncover new genes involved in •NO, O2•–, or ONOO− production pathways. Since pnGFP-Ultra can be widely distributed in the form of plasmids, it represents a valuable catalyst to future breakthroughs in the field of RNS.
In addition, our optimized plasmid system will greatly facilitate the site-specific incorporation of various ncAAs into proteins in mammalian cells. It can be adapted to overexpress other engineered orthogonal aaRS-tRNA pairs. Also, because the Poly-aaRS can cross-react with an array of para-substituted phenylalanine analogs including pBoF, pAzF, p-acetylphenylalanine, p-iodophenylalanine, p-bromophenylalanine, p-cyanophenylalanine, p-nitrophenylalanine, p-benzoyl-phenylalanine, 4,4′-biphenylalanine, O-methyl-tyrosine, O-propargyl-tyrosine and O-allyl-tyrosine (Chatterjee et al., 2013; Chen et al., 2013), the plasmids we describe here may directly be used to express proteins harboring these diverse functional groups, thereby easing and enabling many new studies.
STAR★Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Huiwang Ai (huiwang.ai@virginia.edu).
Materials Availability
All reagents created in this study (see Key Resources Table) are available on request. Key plasmids generated have been deposited to Addgene (http://www.addgene.org/Huiwang_Ai/) and the accession numbers for these plasmids are presented in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli DH10B | Thermo Fisher Scientific | Cat# 18297010 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| p-borono-DL-phenylalanine | Synthonix | Cat#: B5001; CAS:90580-64-6 |
| p-azido-phenylalanine | Bachem | Cat#: F-4305; CAS:1241681-80-0 |
| sodium peroxynitrite | Cayman Chemical | Cat#: 81565; CAS:14042-01-4 |
| 3-morpholinosydnonimine (SIN-1) | Sigma Aldrich | Cat#: M5793; CAS:16142-27-1 |
| urate | Alfa Aesar | Cat# A13346-14; CAS: 69-93-2 |
| minocycline | TCI | Cat# M2288; CAS: 13614-98-7 |
| 1400W | Enzo Life Sciences | Cat# ALX-270-073-M025; CAS: 214358-33-5 |
| human Interferon-γ Protein, recombinant | Sigma Aldrich | Cat#: IF002 |
| lipopolysaccharides (LPS) from Salmonella typhosa | Sigma Aldrich | Cat#: L7895 |
| phorbol myristate acetate (PMA) | Sigma Aldrich | Cat#: P8139 CAS: 16561-29-8 |
| coumarin boronic acid pinacolate ester (CBA) | Cayman Chemical | Cat#: 10818; CAS: 190788-61-5 |
| L-tyrosine | Spectrum Chemical | Cat#: T1150; CAS: 60-18-4 |
| β-amyloid (1–42), human | GenScript | Cat#: RP10017 |
| TRI reagent | Thermo Fisher Scientific | Cat#: AM9738 |
| β-mercaptoethanol | MP Biomedicals | Cat# 773648 CAS: 60-24-2 |
| Viromer | ORIGENE | Cat# TT100312 |
| lipofectamine 2000 | Thermo Fisher Scientific | Cat# 11668027 |
| SuperScript® IV Reverse Transcriptase | Thermo Fisher Scientific | Cat#: 18090010 |
| RIPA Lysis and Extraction Buffer | Thermo Fisher Scientific | Cat# 89900 |
| Critical Commercial Assays | ||
| B-PER | Thermo Fisher Scientific | Cat#: 78243 |
| Cell Proliferation Kit I (MTT) | Roche Diagnostics | Cat# 11465007001 |
| Deposited Data | ||
| Structure of pnGFP1.5-Y.Cro (PDB ID: 5F9G) | (Chen et al., 2016b) | PDB ID: 5F9G |
| Experimental Models: Cell Lines | ||
| Human HEK 293T cells | ATCC | CRL-3216™ |
| RAW264.7 Macrophage cells | ATCC | TIB-71™ |
| Experimental Models: Organisms/Strains | ||
| BALB/C mice | The Jackson Laboratory | JAX: 000651 |
| Oligonucleotides | ||
| Primers for molecular cloning, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| Plasmid pEvol-BoF | (Young et al., 2010) | N/A |
| Plasmid pBAD-cpsGFP2 | This paper | N/A |
| Plasmid pcDNA3-EGFPTAG | This paper, plasmid a in Figure 4A | N/A |
| Plasmid pMAH-EGFPTAG | This paper, plasmid c in Figure 4A | N/A |
| Plasmid pMAH-EGFPTAG-POLY | This paper, plasmid e in Figure 4A | N/A |
| Plasmid pUltra | (Lou et al., 2012) | Addgene # 24129 |
| Plasmid pMAH-POLY | (Chen et al., 2013), plasmid b in Figure 4A | Addgene # 64915 |
| Plasmid pcDNA3-pnGFP | (Chen et al., 2013) | Addgene # 64913 |
| Plasmid pBAD-pnGFP-Ultra | This paper | Addgene #157923 |
| Plasmid pMAH-POLY-eRF1E55D | This paper, plasmid d in Figure 4A | Addgene #157925 |
| Plasmid pMAH-pnGFP-Ultra | This paper | Addgene #157924 |
| Software and Algorithms | ||
| Fiji | (Schindelin et al., 2012) | https://fiji.sc |
| PyMOL | Schrödinger | https://pymol.org/2/ |
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Data and Code Availability
Raw source data are available upon reasonable request. The sequence information for key plasmids is available from Addgene and the accession numbers for these plasmids are presented in the Key Resources Table.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HEK 293T cells (human, female, embryonic kidney) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) FBS at 37°C and 5% CO2 atmosphere. RAW264.7 cells (mouse, male, macrophage) were grown in DMEM supplemented with 10% FBS, 100 μg/mL penicillin, 100 μg/mL streptomycin, and 58 μM β-mercaptoethanol at 37°C and 5% CO2 atmosphere. Primary glial cells were isolated from the brain of newborn (0–2 days postnatal) BALB/C mice (both male and female) and cultured in DMEM (4.5 mg/L glucose) supplemented with 10% FBS, 5 mM glutamine and 100 U/ml penicillin and streptomycin at 37°C and 5% CO2 atmosphere. Mice were maintained under standard housing conditions (12h/12h light-dark cycle, 22–25°C ambient temperature, and standard chow diet). All animal procedures were conducted in accordance with the protocol approved by the Animal Care and Use Committee at the University of Virginia.
METHODS DETAILS
Materials and General Methods
Synthetic DNA oligonucleotides (Table S1) were purchased from Integrated DNA Technologies (San Diego, CA). Restriction endonucleases were purchased from Thermo Fisher Scientific (Waltham, MA). PCR and restriction digestion products were purified by gel electrophoresis and extracted using the Syd Labs Gel Extraction kit (Malden, MA). Plasmid DNA was purified using the Syd Labs Miniprep kit (Malden, MA). DNA sequencing was analyzed by Retrogen (San Diego, CA). Mammalian cell imaging was performed on a Leica DMi8 microscope equipped with a Photometrics Prime 95B sCMOS camera. Other materials and general procedures were acquired or performed as previously described (Chen et al., 2012, 2013, 2016b).
Engineering and Screening of pnGFP1.5
To improve the brightness and folding of pnGFP, several rounds of error prone PCR (EP-PCR) based directed evolution were performed to improve its template—cpsGFP (Chen and Ai, 2014), which contains a tyrosine-derived chromophore. Briefly, oligos pBAD-FP and pBAD-RP were used to amplify the cpsGFP gene from the pBAD-cpsGFP plasmid in an EP-PCR reaction condition. The mutated PCR products were digested with XhoI and HindIII and ligated into a predigested compatible pBAD vector. The ligated product was used to transform Escherichia coli DH10B competent cells by electroporation. Cells were grown on Luria-Bertani (LB) broth agar plates supplemented with 100 μg/mL ampicillin and 0.02% L-arabinose. LB agar plates were incubated at 37°C overnight. Bacterial colonies were illuminated under a laboratory-built colony fluorescence imaging system and examined using a pair of forensic yellow goggles with a cutoff wavelength of ~490 nm. Plasmid DNA from the brightest colonies of each round were extracted and combined to serve as the template for the next round of EP-PCR. The fluorescence of the B-PER (Pierce, Rockford, IL) extracted cell lysate from the brightest colonies of the final round was quantitatively compared on the Synergy Mx Microplate Reader. The best mutant was sequenced and renamed as cpsGFP2.
To screen for pnGFP1.5, the codon for the chromophore tyrosine (Tyr174) of cpsGFP2 were mutated to TAG to allow for pBoF incorporation using the pEvol-BoF-based amber suppression system (Chen et al., 2013). Next, threonine 5 (Thr5) and threonine 253 (Thr253) of cpsGFP2-Tyr174TAG were randomized using NNK degenerate codons to construct a gene library. The library was used to screen for mutants with high reactivity to ONOO−. The best mutant from the library was sequenced and designated as pnGFP1.5.
Development and Characterization of pnGFP-Ultra
To create the various pnGFP1.5 mutants, site-directed mutagenesis of pnGFP1.5 was performed using an overlap extension PCR based strategy (Chen et al., 2016b), and the mutagenesis primers can be found in Supplementary Information Table S1. Proteins for each mutant were purified, buffer-exchanged and concentrated as detailed below for pnGFP-Ultra. To test the reactivity and selectivity of the pnGFP1.5 mutants, 0.2 μM purified protein was incubated with Tris buffer (150 mM Tris-HCl, 150 mM NaCl, pH 7.4), 100 μM ONOO− or 100 μM H2O2 for 1 hour at room temperature in Tris buffer. Fluorescence intensities at 510 nm with a 490 nm excitation were quantified and represented as means ± SD from three independent measurements. Fluorescence enhancements of pnGFP1.5 and relevant mutants towards ONOO– or H2O2 were calculated as “dynamic range” expressed as ΔF/F0, where F0 is the initial fluorescence intensity (before treatment) and ΔF is the final fluorescent intensity (after treatment) minus F0. The selectivity of each mutant was defined as the ratio of the fluorescence enhancement from ONOO−-treated group over that from H2O2 -treated group. The mutant with the highest selectivity, namely pnGFP1.5-S66A (pnGFP1.6), was chosen for further engineering. To screen for mutants with further enhanced selectivity, the threonine 5 (Thr5) codon of pnGFP1.6 was mutated into two consecutive degenerative codons (NNKNNK) using primers pnGFP1.6-F-NNK2 and pBAD-RP. The gene library was screened and constructed as described before. The mutant with the highest selectivity from this library was sequenced and designated as pnGFP-Ultra.
Characterization protocols for pnGFP-Ultra including the purification, reactivity measurements, selectivity tests over other reactive chemical species, and determination of the limits of detection (LOD) were the same as those for pnGFP (Chen et al., 2013), as detailed below.
To express pnGFP-Ultra, the pBAD plasmid harboring pnGFP-Ultra was used to co-transform DH10B E. coli cells with pEvol-pBoF plasmid. Cells were grown on LB agar plates containing 100 μg/mL ampicillin and 50 μg/ml chloramphenicol at 37°C overnight. A single colony from the co-transformation was grown overnight at 37°C in 5 ml LB media supplemented with appropriate antibiotics. Saturated overnight cultures were then diluted by 100-fold and grown in 2YT media to OD600 = 0.6. L-arabinose (final conc. 0.2%) and pBoF (final conc. 1 mM) were added to express the full-length proteins. Growth continued with vigorous shaking at 37°C for 24 hours, and next, at room temperature for additional 24 hours. Cells were harvested and lysed by sonication. The His-tagged protein was purified using Ni-NTA agarose beads (Qiagen) according to the manufacturer’s instruction. The protein was next concentrated in Tris buffer (30 mM Tris-Cl, pH 7.4) using Amicon Ultra Centrifugal Filter Units (3,000 Da cutoff). Bradford Assays were performed to determined protein concentrations by comparing with a set of BSA standards.
Reactivity measurements were carried out on a monochromator-based Synergy Mx Microplate Reader (BioTek, Winooski, VT). Excitation at 490 nm and emission at 510 nm were used for single-point measurements, and a blank solution was used to subtract the background. Reactivity was reported as either arbitrary fluorescence emission intensity or dynamic range (ΔF/F0 = (final intensity – initial intensity)/initial intensity).
To test protein responses to various redox-active chemicals, concentrated protein stock solutions were diluted into 1x PBS to gain a final concentration of 200 nM (for data in Figure 3) or 1 μM (for data in Figure S3). The following methods were utilized to supply individual redox-active species. They were added into proteins, followed with incubation at room temperature. Excitation at 490 nm and emission at 510 nm were used for measurement on a monochromator-based Synergy Mx Microplate Reader.
Nitric oxide (NO•): 1-Hydroxy-2-oxo-3-(N-methyl-3-aminopropyl)-3-methyl-1-triazene (NOC-7) stock solution (100 mM) was prepared in 0.3 M NaOH. An appropriate amount of the stock solution was added to generate NO•. The final mixtures were confirmed to have no notably pH change from 7.4.
Hydrogen sulfide (H2S/HS−): Solid NaHS was dissolved in PBS to afford a 100 mM stock solution, whose pH was adjusted to 7.4. An appropriate amount of a freshly prepared stock solution was added. At physiological pH, H2S and HS− are in equilibrium.
tert-butyl hydroperoxide (HOOtBu): 70% solution in H2O was purchased from Sigma-Aldrich (St. Louis, MO). It was diluted with PBS to gain a 100 mM stock solution. An appropriate amount of a freshly prepared stock solution was used.
Hydrogen Peroxide (H2O2): 30% solution in H2O was purchased from Fisher (Hampton, NH). It was diluted with PBS to gain a 100 mM stock solution. An appropriate amount of a freshly prepared stock solution was added.
Hyperchlorite (ClO−/HOCl): 5% NaOCl solution was purchased from Fisher (Hampton, NH). It was diluted with PBS to gain a 100 mM stock solution. The pH was adjusted to 7.4. An appropriate amount of a freshly prepared stock solution was added. At physiological pH, ClO− and HOCl are in equilibrium.
Superoxide (O2•−): solid KO2 was suspended in DMSO by sonication. An appropriate amount was directly added to a protein in aqueous buffer. Please note that superoxide decomposes quickly to form H2O2 in aqueous solution.
Peroxynitrite (ONOO−/ONOOH): Purchased as a solution in 0.3 M NaOH (stored at −80°C). Concentration was determined before use by measuring absorbance at 302 nm. A 100 mM fresh stock solution was made by dilution using cold 0.3 M NaOH. An appropriate amount was utilized to provide ONOO−. The final mixtures were confirmed to have no notably pH change from 7.4. At physiological pH, ONOOH and ONOO− are in equilibrium.
Hydroxy Radical (•OH): Generated by Fenton reaction between Fe2+ and H2O2. Fresh FeCl2 and H2O2 stock solutions were prepared for dilution.
tert-butoxy radical (•OtBu): Generated by Fenton reaction between Fe2+ and HOOtBu. Fresh FeCl2 and HOOtBu stock solutions were prepared for dilution.
Construction of Mammalian Expression Plasmids
Plasmids a and b (also known as pcDNA3-EGFPTAG and pMAH-POLY, respectively) were previously reported (Chen et al., 2013). To construct plasmid c (pMAH-EGFPTAG), the EGFPTAG gene was amplified from plasmid a with oligos EGFP-F and EGFP-R. The PCR product was digested with HindIII and ApaI, and ligated into plasmid b, which was predigested with the same enzymes. To construct plasmid d (pMAH-POLY-eRF1E55D), the eRF1 gene was first cloned from the total cDNA of HEK 293T cells with oligos eRF1-F and eRF1-R. To prepare the cDNA, total mRNA from HEK293T cell was isolated with the TRI Reagent (Sigma, St. Louis, MO) and reverse-transcribed with the SuperScript® IV Reverse Transcriptase (Thermo Fisher Scientific, Carlsbad, CA). To introduce the E55D point mutation within eRF1, oligos eRF1-NheI-F and E55D-R, E55D-F and eRF1-EcoRI-R were used to amplify the two overlapping fragments flanking E55 position of eRF1, respectively. Next, the two fragments were joined by overlap extension PCR with oligos eRF1-NheI-F and eRF1-EcoRI-R, and sub-cloned into the pUltra vector (Addgene Plasmid #24129) between restriction sites NheI and EcoRI to generate pUltra-eRF1-E55D, which contains the T2A-eRF1-E55D-WPRE cassette. This cassette was amplified with oligos eRF1-GB-F and eRF1-GB-R and ligated into ApaI-linearized plasmid b using Gibson assembly cloning (Gibson et al., 2009). To construct plasmid e (pMAH-EGFPTAG-POLY), the CMV- EGFPTAG cassette was amplified from plasmid a with oligos CMV-39TAG-XhoI-F and CMV-39TAG-XhoI-R. The PCR product was digested with XhoI and ligated into XhoI digested, DpnI dephosphorylated plasmid b. To construct the mammalian expression plasmid for pnGFP-Ultra, the pnGFP-Ultra gene was amplified from pBAD-pnGFP-Ultra with oligos pnGFP-Ultra-HindIII-F and pnGFP-Ultra-ApaI-R, and sub-cloned into plasmid c between restriction sites Hind III and ApaI.
Mammalian Cell Culture, Transfection and Imaging
Human Embryonic Kidney (HEK) 293T cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Cells were incubated at 37°C with 5% CO2 in humidified air. To test the transfection and non-canonical amino acid (ncAA) incorporation efficiency of the various plasmids, cells were split and seeded into 24-well plates 16 hours before transfection. Cells were transfected with 600 ng DNA and 2 μg PEI (polyethyleneimine, linear, M.W. 25 kDs) per well for three hours, and then cultured in complete medium supplemented with 2 mM DL-pBoF for 48 hours. Cells were lysed with 150 μL RIPA buffer and centrifuged at 13,000 rpm, 4°C for 5 mins. The fluorescence of the EGFP-containing supernatant (100 μL) was measured on the microplate reader with excitation and emission wavelength set at 490 nm and 515 nm, respectively. Transfections were triplicated and data were represented as mean ± SD.
To express pnGFP or pnGFP-Ultra, HEK 293T cells in each 35-mm plastic culture dish were co-transfected with 1.5 μg pnGFP-containing plasmid a and 1.5 μg plasmid b or 1.5 μg pnGFP-Ultra-containing plasmid c and 1.5 μg plasmid d, in the presence of 10 μg PEI. Other procedures were the same as those for pnGFP (Chen et al., 2013). Cells were imaged under a Leica DMi8 microscope equipped with a Photometrics Prime 95B sCMOS camera. A GFP filter cube with a 470/40 nm bandpass excitation filter and a 525/50 nm bandpass emission filter was used for cell imaging. To test the effect of ONOO− scavengers, transfected HEK 293T cells in Hank’s Balanced Salt Solution (HBSS) were treated with 100 μM SIN-1 in the absence or presence of 100 μM urate or 100 μM minocycline. Incubation lasted for 90 min at 37°C under 5% CO2 before cells were imaged. HEK 293T cells expressing pnGFP-Ultra-Y.Cro were prepared using the same procedure for pnGFP-Ultra, except for that 5 mM additional L-tyrosine (but no DL-pBoF) was supplemented into the complete culture medium.
RAW264.7 Macrophage Cell Culture, Transfection and Activation
RAW264.7 cells were obtained from ATCC and grown at 37°C under 5% CO2 in DMEM supplemented with 10% FBS, 100 μg/mL penicillin, 100 μg/mL streptomycin, and 58 μM β-mercaptoethanol. Cells were passaged 18 h prior to transfection, and next, transfected with 1.5 μg pnGFP-Ultra-containing plasmid c and 1.5 μg plasmid d in the presence of Viromer according to the manufacturer’s instruction. After transfection, cells were cultured in the aforementioned complete medium supplemented with 2 mM DL-pBoF for 24 h. Next, the medium was replaced with fresh complete medium without DL-pBoF and cells were cultured for another 24 h. IFN-γ (50 U/ml) and LPS (0.5 μg/mL) were added into the cell culture media, and the incubation lasted for another 10 h. After LPS and IFN-γ were removed, cells were rinsed with HBSS and further treated with PMA (200 ng/mL) in HBSS for 1 h before imaging. Additionally, cells in some groups were co-treated with 100 μM urate, or 10 μM minocycline, or 100 μM 1400W when PMA (200 ng/mL) was used for stimulation. Cells were imaged under the same condition as used for imaging HEK 293T. In addition, the procedure to treat RAW264.7 cells with SIN-1 and ONOO− scavengers is identical to that used for treating HEK 293T. RAW264.7 cells expressing pnGFP-Ultra-Y.Cro were prepared by supplementing 5 mM additional L-tyrosine into the complete culture medium.
Cell Viability Assay
MTT assays were performed using the Roche Cell Proliferation Kit I (MTT) according to the manufacturer’s instruction. Briefly, HEK 293T or RAW264.7 cells that transiently expressing pnGFP-Ultra were cultured at a density of 1,000 cells/well in a 96-well glass-bottom plate (Cellvis, Cat. # P96–1.5H-N). Cells in individual wells were subjected to various chemical treatments. To determine cell viability, 10 μl of the MTT labeling reagent was added into each well. Cells were incubated at 37°C and 5% CO2 for additional 4 h. 100 μl of the Solubilization solution was then added to the cells and incubated overnight at 37°C and 5% CO2. Absorbance for each well was determined at 550 nm using a monochromator-based BioTek Synergy Mx plate reader. Baseline absorbance at 690 nm was also measured and used to subtract the absorbance at 550 nm. The absorbance differences (A550 nm – A690 nm) were normalized to the values of the control groups that were untreated by chemicals.
Isolation and Culture of Primary Glial Cells
Primary glial cells were isolated from the brain of newborn (0–2 days postnatal) wild-type BALB/C mice and cultured as previously described (Redlich et al., 2013; Seele et al., 2016). Briefly, meninges of mouse brains were removed and whole brains were cut into small pieces and treated with 0.25% trypsin (5 mL/brain) for 15 min at 37°C. After centrifugation (1000 × g, 1 min), trypsin was removed and DMEM (4.5 g/L glucose, 3 mL/brain) was added into the cell precipitation, which was further minced by pipetting up and down with a 1 mL pipette. After another centrifugation (750 × g, 8 min), the supernatant, which contained the needed cells, was filtered with a 40-μm sterile cell strainer (Nylon mesh, Fisherbrand, Cat#: 22363547). Cells were next seeded onto 35 mm cell culture dishes with poly-D-lysine coated glass bottoms and cultured in DMEM (4.5 mg/L glucose) supplemented with 10% FBS, 5 mM glutamine and 100 U/ml penicillin and streptomycin at 37°C, 5% CO2 for 8–10 days before transfection. This well-established procedure is known to derive mixed glial cells composed of astrocytes and microglia (Redlich et al., 2013; Seele et al., 2016).
Transfection and Activation of Primary Glial Cells
Primary glial cells were transfected with Lipofectamine 2000 by following the manufacturer’s instruction. After transfection, cells were cultured in the complete culture media supplemented with 2 mM DL-pBoF. β-amyloid (1–42, human) was added at a final concentration of 5 μM at 24 h post transfection. The incubation lasted for another 24 h and the culture media were replaced with fresh complete media supplemented with 5 μM β-amyloid but without DL-pBoF. The treatment lasted for another 18 h before microscopic images were acquired under the same condition as used for imaging HEK 293T. Cells in the control groups were untreated with β-amyloid, or co-treated with 10 μM minocycline or 100 μM 1400W when 5 μM β-amyloid was used for stimulation. To express pnGFP-Ultra-Y.Cro in primary glial cells, after transfection, cells were cultured in DMEM supplemented with 10% FBS and 5 mM additional L-tyrosine.
QUANTIFICATION AND STATISTICAL ANALYSIS
Microscopic images were analyzed with Fiji (ImageJ). Imaging background was subtracted by setting the rolling ball radius to 300 pixels, and the fluorescence intensities of each selected cells were next determined using Fiji (ImageJ). GraphPad Prism 8 was used for data and statistical analysis. Statistical methods used to determine P values are presented in the figure legends. Sample size, the number of replications for experiments, and dispersion and precision measures (individual data points, mean and SD or 95% confidential intervals) are also presented in figures or figure legends. For Students’ t-tests, F-tests were pre-performed to compare the variability of the two samples. For AVOVA tests, Brown-Forsythe tests were pre-performed to compare the variability of the samples. For all comparisons, P values of less than 0.05 were considered statistically significant.
Supplementary Material
Significance.
pnGFP-Ultra, a high performance, reaction-based, genetically encodable biosensor for imaging ONOO− is presented. pnGFP-Ultra uses a pBoF-modified chromophore as the sensing moiety and an engineered circularly permuted fluorescent protein scaffold for further modulation of reactivity and specificity. pnGFP-Ultra exhibits a remarkable ~110-fold fluorescence turn-on response towards ONOO− while displaying virtually no cross-reaction with other reactive oxygen/nitrogen species. We developed an optimized plasmid system to facilitate the expression of pnGFP-Ultra in mammalian cells and demonstrated that pnGFP-Ultra could reliably detect physiological ONOO− in various cell culture models such as activated macrophages and primary glial cells. pnGFP-Ultra thus represents an important advancement enabling the practical implementation of a genetically encoded biosensor for live-cell detection of ONOO−. In addition to live-cell imaging, we envision high-throughput drug/genetic screening to be an important avenue of application where pnGFP-Ultra shall prove to be an enabling tool. Moreover, our unique sensor design and development strategy may be widely adaptable and will inspire the development of chemoselective biosensors for other bioanalytical targets. Furthermore, the optimized plasmid system presented here will facilitate the site-specific incorporation of diverse ncAAs into proteins in mammalian cells.
Highlights.
pnGFP-Ultra is a genetically encoded biosensor with a ~110-fold turn-on response
pnGFP-Ultra exhibits high selectivity and sensitivity toward peroxynitrite
An optimized plasmid-based system increases noncanonical amino acid incorporation
pnGFP-Ultra robustly detects peroxynitrite in macrophages and primary glial cells
Acknowledgements
Research reported in this publication was supported in part by the National Science Foundation under Award CHE-1750660 and the National Institute of General Medical Science of National Institutes of Health under Awards R01GM118675 and R01GM129291 (including the Supplement Award 3R01GM129291–02S2 from the National Institute on Aging). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Supplemental Information
Supplemental Information includes a supplementary table of oligonucleotides sequences used in this study and seven supplementary figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ai HW, Shaner NC, Cheng Z, Tsien RY, and Campbell RE (2007). Exploration of new chromophore structures leads to the identification of improved blue fluorescent proteins. Biochemistry 46, 5904–5910. [DOI] [PubMed] [Google Scholar]
- Allen RG, Lafuse WP, Powell ND, Marketon JIW, Stiner-Jones LM, Sheridan JF, and Bailey MT (2012). Stressor-induced increase in microbicidal activity of splenic macrophages is dependent upon peroxynitrite production. Infect. Immun. 80, 3429–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer’s Association (2020). 2020 Alzheimer’s disease facts and figures. Alzheimer’s Dement. 16, 391–460. [Google Scholar]
- Beckman JS (2009). Understanding peroxynitrite biochemistry and its potential for treating human diseases. Arch. Biochem. Biophys. 484, 114–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman JS, and Koppenol WH (1996). Nitric oxide, superoxide, and peroxynitrite: The good, the bad, and the ugly. Am. J. Physiol. - Cell Physiol. 271. [DOI] [PubMed] [Google Scholar]
- Brand MD (2016). Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic. Biol. Med. 100, 14–31. [DOI] [PubMed] [Google Scholar]
- Brewer TF, Garcia FJ, Onak CS, Carroll KS, and Chang CJ (2015). Chemical Approaches to Discovery and Study of Sources and Targets of Hydrogen Peroxide Redox Signaling Through NADPH Oxidase Proteins. Annu. Rev. Biochem. 84, 765–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A, Xiao H, Bollong M, Ai HW, and Schultz PG (2013). Efficient viral delivery system for unnatural amino acid mutagenesis in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 110, 11803–11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, and Ai HW (2014). A Highly Responsive and Selective Fluorescent Probe for Imaging Physiological Hydrogen Sulfide. Biochemistry 53, 5966–5974. [DOI] [PubMed] [Google Scholar]
- Chen PR, Groff D, Guo J, Ou W, Cellitti S, Geierstanger BH, and Schultz PG (2009). A facile system for encoding unnatural amino acids in mammalian cells. Angew. Chemie - Int. Ed. 48, 4052–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Chen Z, Ren W, and Ai HW (2012). Reaction-based genetically encoded fluorescent hydrogen sulfide sensors. J. Am. Chem. Soc. 134, 9589–9592. [DOI] [PubMed] [Google Scholar]
- Chen Z, Truong TM, and Ai HW (2016a). Chapter 10: Development of fluorescent probes for the detection of peroxynitrite, in Peroxynitrite Detection in Biological Media: Challenges and Advances, Royal Society of Chemistry, pp. 186–207. [Google Scholar]
- Chen Z, Truong T, and Ai HW (2017). Illuminating Brain Activities with Fluorescent Protein-Based Biosensors. Chemosensors 5, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Ren W, Wright QE, and Ai HW (2013). Genetically encoded fluorescent probe for the selective detection of peroxynitrite. J. Am. Chem. Soc. 135, 14940–14943. [DOI] [PubMed] [Google Scholar]
- Chen Z, Tian Z, Kallio K, Oleson AL, Ji A, Borchardt D, Jiang DE, Remington SJ, and Ai HW (2016b). The N-B Interaction through a Water Bridge: Understanding the Chemoselectivity of a Fluorescent Protein Based Probe for Peroxynitrite. J. Am. Chem. Soc. 138, 4900–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW (2017). Expanding and reprogramming the genetic code. Nature 550, 53–60. [DOI] [PubMed] [Google Scholar]
- Ferrer-Sueta G, and Radi R (2009). Chemical biology of peroxynitrite: Kinetics, diffusion, and radicals. ACS Chem. Biol. 4, 161–177. [DOI] [PubMed] [Google Scholar]
- Förstermann U, and Münzel T (2006). Endothelial nitric oxide synthase in vascular disease: From marvel to menace. Circulation 113, 1708–1714. [DOI] [PubMed] [Google Scholar]
- Fridovich I (1997). Superoxide anion radical (O2/-·), superoxide dismutases, and related matters. J. Biol. Chem. 272, 18515–18517. [DOI] [PubMed] [Google Scholar]
- Garvey EP, Oplinger JA, Furfine ES, Kiff RJ, Laszlo F, Whittle BJR, and Knowles RG (1997). 1400W is a slow, tight binding, and highly selective inhibitor of inducible nitric-oxide synthase in vitro and in vivo. J. Biol. Chem. 272, 4959–4963. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, and Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. [DOI] [PubMed] [Google Scholar]
- González-Reyes RE, Nava-Mesa MO, Vargas-Sánchez K, Ariza-Salamanca D, and Mora-Muñoz L (2017). Involvement of astrocytes in Alzheimer’s disease from a neuroinflammatory and oxidative stress perspective. Front. Mol. Neurosci. 10, 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy M, Zielonka J, Karoui H, Sikora A, Michalski R, Podsiadły R, Lopez M, Vasquez-Vivar J, Kalyanaraman B, and Ouari O (2018). Detection and Characterization of Reactive Oxygen and Nitrogen Species in Biological Systems by Monitoring Species-Specific Products. Antioxidants Redox Signal. 28, 1416–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino N, Sakamoto K, and Yokoyama S (2012). Site-specific incorporation of unnatural amino acids into proteins in mammalian cells. Methods Mol. Biol. 794, 215–228. [DOI] [PubMed] [Google Scholar]
- Ischiropoulos H, Zhu L, and Beckman JS (1992). Peroxynitrite formation from macrophage-derived nitric oxide. Arch. Biochem. Biophys. 298, 446–451. [DOI] [PubMed] [Google Scholar]
- Li S, Yang B, Kobayashi T, Yu B, Liu J, and Wang L (2020). Genetically encoding thyronine for fluorescent detection of peroxynitrite. Bioorganic Med. Chem. 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tao RR, Hong LJ, Cheng J, Jiang Q, Lu YM, Liao MH, Ye WF, Lu NN, Han F, et al. (2015). Visualizing Peroxynitrite Fluxes in Endothelial Cells Reveals the Dynamic Progression of Brain Vascular Injury. J. Am. Chem. Soc. 137, 12296–12303. [DOI] [PubMed] [Google Scholar]
- Liaudet L, Vassalli G, and Pacher P (2009). Role of peroxynitrite in the redox regulation of cell signal transduction pathways. Front. Biosci. 14, 4809–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MZ, and Schnitzer MJ (2016). Genetically encoded indicators of neuronal activity. Nat. Neurosci. 19, 1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KK, Wu SC, Hsu KM, Hung CH, Liaw WF, and Wang YM (2013). A N-(2-aminophenyl)-5-(dimethylamino)-1-naphthalenesulfonic amide (Ds-DAB) based fluorescent chemosensor for peroxynitrite. Org. Lett. 15, 4242–4245. [DOI] [PubMed] [Google Scholar]
- Liu CC, and Schultz PG (2010). Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem. 79, 413–444. [DOI] [PubMed] [Google Scholar]
- Liu W, Brock A, Chen S, Chen S, and Schultz PG (2007). Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat. Methods 4, 239–244. [DOI] [PubMed] [Google Scholar]
- Lou E, Fujisawa S, Morozov A, Barlas A, Romin Y, Dogan Y, Gholami S, Moreira AL, Manova-Todorova K, and Moore MAS (2012). Tunneling nanotubes provide a unique conduit for intercellular transfer of cellular contents in human malignant pleural mesothelioma. PLoS One 7, e33093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungrue IN, Gros R, You X, Pirani A, Azad A, Csont T, Schulz R, Butany J, Stewart DJ, and Husain M (2002). Cardiomyocyte overexpression of iNOS in mice results in peroxynitrite generation, heart block, and sudden death. J. Clin. Invest. 109, 735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler A, and Schultz C (2013). The power of fluorogenic probes. Angew. Chemie - Int. Ed. 52, 2408–2410. [DOI] [PubMed] [Google Scholar]
- Pacher P, Beckman JS, and Liaudet L (2007). Nitric oxide and peroxynitrite in health and disease. Physiol. Rev. 87, 315–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, and Yang D (2010). HKGreen-3: A rhodol-based fluorescent probe for peroxynitrite. Org. Lett. 12, 4932–4935. [DOI] [PubMed] [Google Scholar]
- Peng T, Wong NK, Chen X, Chan YK, Ho DHH, Sun Z, Hu JJ, Shen J, El-Nezami H, and Yang D (2014). Molecular imaging of peroxynitrite with HKGreen-4 in live cells and tissues. J. Am. Chem. Soc. 136, 11728–11734. [DOI] [PubMed] [Google Scholar]
- Price M, Reiners JJ, Santiago AM, and Kessel D (2009). Monitoring singlet oxygen and hydroxyl radical formation with fluorescent probes during photodynamic therapy. Photochem. Photobiol. 85, 1177–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radi R (2013a). Peroxynitrite, a stealthy biological oxidant. J. Biol. Chem. 288, 26464–26472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radi R (2013b). Protein tyrosine nitration: Biochemical mechanisms and structural basis of functional effects. Acc. Chem. Res. 46, 550–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radi R (2018). Oxygen radicals, nitric oxide, and peroxynitrite: Redox pathways in molecular medicine. Proc. Natl. Acad. Sci. U. S. A. 115, 5839–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redlich S, Ribes S, Schütze S, Eiffert H, and Nau R (2013). Toll-like receptor stimulation increases phagocytosis of Cryptococcus neoformans by microglial cells. J. Neuroinflammation 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronson RS, Nakamura M, and Vinten-Johansen J (1999). The cardiovascular effects and implications of peroxynitrite. Cardiovasc. Res. 44, 47–59. [DOI] [PubMed] [Google Scholar]
- Schildknecht S, Pape R, Müller N, Robotta M, Marquardt A, Bürkle A, Drescher M, and Leist M (2011). Neuroprotection by minocycline caused by direct and specific scavenging of peroxynitrite. J. Biol. Chem. 286, 4991–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmied WH, Elsässer SJ, Uttamapinant C, and Chin JW (2014). Efficient multisite unnatural amino acid incorporation in mammalian cells via optimized pyrrolysyl tRNA synthetase/tRNA expression and engineered eRF1. J. Am. Chem. Soc. 136, 15577–15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seele J, Nau R, Prajeeth CK, Stangel M, Valentin-Weigand P, and Seitz M (2016). Astrocytes enhance streptococcus suis-glial cell interaction in primary astrocyte-microglial cell co-cultures. Pathogens 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, and Zhang F (2015). High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 16, 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora A, Zielonka J, Lopez M, Joseph J, and Kalyanaraman B (2009). Direct oxidation of boronates by peroxynitrite: Mechanism and implications in fluorescence imaging of peroxynitrite. Free Radic. Biol. Med. 47, 1401–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora A, Zielonka J, Dębowska K, Michalski R, Smulik-Izydorczyk R, Pięta J, Podsiadły R, Artelska A, Pierzchała K, and Kalyanaraman B (2020). Boronate-Based Probes for Biological Oxidants: A Novel Class of Molecular Tools for Redox Biology. Front. Chem. 8, 580899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Richey Harris PL, Sayre LM, Beckman JS, and Perry G (1997). Widespread peroxynitrite-mediated damage in Alzheimer’s disease. J. Neurosci. 17, 2653–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Xu Q, Kim G, Flower SE, Lowe JP, Yoon J, Fossey JS, Qian X, Bull SD, and James TD (2014). A water-soluble boronate-based fluorescent probe for the selective detection of peroxynitrite and imaging in living cells. Chem. Sci. 5, 3368–3373. [Google Scholar]
- Sun X, Lacina K, Ramsamy EC, Flower SE, Fossey JS, Qian X, Anslyn EV, Bull SD, and James TD (2015). Reaction-based Indicator displacement Assay (RIA) for the selective colorimetric and fluorometric detection of peroxynitrite. Chem. Sci. 6, 2963–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZN, Wang HL, Liu FQ, Chen Y, Tam PKH, and Yang D (2009). BODIPY-based fluorescent probe for peroxynitrite detection and imaging in living cells. Org. Lett. 11, 1887–1890. [DOI] [PubMed] [Google Scholar]
- Szabó C (1996). The pathophysiological role of peroxynitrite in shock, inflammation, and ischemia-reperfusion injury. Shock 6, 79–88. [DOI] [PubMed] [Google Scholar]
- Szabó C, Ischiropoulos H, and Radi R (2007). Peroxynitrite: Biochemistry, pathophysiology and development of therapeutics. Nat. Rev. Drug Discov. 6, 662–680. [DOI] [PubMed] [Google Scholar]
- Teixeira D, Fernandes R, Prudêncio C, and Vieira M (2016). 3-Nitrotyrosine quantification methods: Current concepts and future challenges. Biochimie 125, 1–11. [DOI] [PubMed] [Google Scholar]
- Tian J, Chen H, Zhuo L, Xie Y, Li N, and Tang B (2011). A highly selective, cell-permeable fluorescent nanoprobe for ratiometric detection and imaging of peroxynitrite in living cells. Chem. - A Eur. J. 17, 6626–6634. [DOI] [PubMed] [Google Scholar]
- Torreilles F, Salman-Tabcheh S, Guérin MC, and Torreilles J (1999). Neurodegenerative disorders: The role of peroxynitrite. Brain Res. Rev. 30, 153–163. [DOI] [PubMed] [Google Scholar]
- Ueno T, and Nagano T (2011). Fluorescent probes for sensing and imaging. Nat. Methods 8, 642–645. [DOI] [PubMed] [Google Scholar]
- Ueno T, Urano Y, Kojima H, and Nagano T (2006). Mechanism-based molecular design of highly selective fluorescence probes for nitrative stress. J. Am. Chem. Soc. 128, 10640–10641. [DOI] [PubMed] [Google Scholar]
- Van Der Veen RC, Hinton DR, Incardonna F, and Hofman FM (1997). Extensive peroxynitrite activity during progressive stages of central nervous system inflammation. J. Neuroimmunol. 77, 1–7. [DOI] [PubMed] [Google Scholar]
- Wang W, Takimoto JK, Louie GV, Baiga TJ, Noel JP, Lee KF, Slesinger PA, and Wang L (2007). Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat. Neurosci. 10, 1063–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Yamada N, Tian X, Bull SD, Minoshima M, Kikuchi K, Mackenzie AB, and James TD (2020). Sensing Peroxynitrite in Different Organelles of Murine RAW264.7 Macrophages With Coumarin-Based Fluorescent Probes. Front. Chem. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, and Zweier JL (1997). Superoxide and peroxynitrite generation from inducible nitric oxide synthase in macrophages. Proc. Natl. Acad. Sci. U. S. A. 94, 6954–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Chatterjee A, Choi SH, Bajjuri KM, Sinha SC, and Schultz PG (2013). Genetic incorporation of multiple unnatural amino acids into proteins in mammalian cells. Angew. Chemie - Int. Ed. 52, 14080–14083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Wei M, Morgan TE, Fabrizio P, Han D, Finch CE, and Longo VD (2002). Peroxynitrite Mediates Neurotoxicity of Amyloid β-Peptide 1–42- and Lipopolysaccharide-Activated Microglia. J. Neurosci. 22, 3484–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Chen H, Tian J, Ding B, Xie Y, Qiang M, and Tang B (2011). A near-infrared reversible fluorescent probe for peroxynitrite and imaging of redox cycles in living cells. Chem. Commun. 47, 9468–9470. [DOI] [PubMed] [Google Scholar]
- Yang D, Wang HL, Sun ZN, Chung NW, and Shen JG (2006). A highly selective fluorescent probe for the detection and imaging of peroxynitrite in living cells. J. Am. Chem. Soc. 128, 6004–6005. [DOI] [PubMed] [Google Scholar]
- Young TS, Ahmad I, Yin JA, and Schultz PG (2010). An Enhanced System for Unnatural Amino Acid Mutagenesis in E. coli. J. Mol. Biol. 395, 361–374. [DOI] [PubMed] [Google Scholar]
- Youssef S, Zhang S, and Ai HW (2019). A Genetically Encoded, Ratiometric Fluorescent Biosensor for Hydrogen Sulfide. ACS Sensors 4, 1626–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Li P, Li G, Zhao G, Chu T, and Han K (2011). A near-IR reversible fluorescent probe modulated by selenium for monitoring peroxynitrite and imaging in living cells. J. Am. Chem. Soc. 133, 11030–11033. [DOI] [PubMed] [Google Scholar]
- Yu F, Li P, Wang B, and Han K (2013). Reversible near-infrared fluorescent probe introducing tellurium to mimetic glutathione peroxidase for monitoring the redox cycles between peroxynitrite and glutathione in vivo. J. Am. Chem. Soc. 135, 7674–7680. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhu Z, Zheng Y, Cheng J, Zhang N, Long YT, Zheng J, Qian X, and Yang Y (2012). A three-channel fluorescent probe that distinguishes peroxynitrite from hypochlorite. J. Am. Chem. Soc. 134, 18479–18482. [DOI] [PubMed] [Google Scholar]
- Zielonka J, Sikora A, Joseph J, and Kalyanaraman B (2010). Peroxynitrite is the major species formed from different flux ratios of co-generated nitric oxide and superoxide: Direct reaction with boronate-based fluorescent probe. J. Biol. Chem. 285, 14210–14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielonka J, Sikora A, Hardy M, Joseph J, Dranka BP, and Kalyanaraman B (2012a). Boronate probes as diagnostic tools for real time monitoring of peroxynitrite and hydroperoxides. Chem. Res. Toxicol. 25, 1793–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielonka J, Zielonka M, Sikora A, Adamus J, Joseph J, Hardy M, Ouari O, Dranka BP, and Kalyanaraman B (2012b). Global profiling of reactive oxygen and nitrogen species in biological systems: High-throughput real-time analyses. J. Biol. Chem. 287, 2984–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou MH, Cohen RA, and Ullrich V (2004). Peroxynitrite and vascular endothelial dysfunction in diabetes mellitus. Endothel. J. Endothel. Cell Res. 11, 89–97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw source data are available upon reasonable request. The sequence information for key plasmids is available from Addgene and the accession numbers for these plasmids are presented in the Key Resources Table.