Abstract
Background
Bronchoscopy serves as direct visualisation of the airway. Virtual bronchoscopy provides similar visual information using a non-invasive imaging procedure(s). Early and accurate image-guided diagnosis requires the possible highest performance, which might be approximated by combining anatomical and functional imaging. This communication describes an advanced functional virtual bronchoscopic (fVB) method based on the registration of PET images to high-resolution diagnostic CT images instead of low-dose CT images of lower resolution obtained from PET/CT scans. PET/CT and diagnostic CT data were collected from 22 oncological patients to develop a computer-aided high-precision fVB. Registration of segmented images was performed using elastix.
Results
For virtual bronchoscopy, we used an in-house developed segmentation method. The quality of low- and high-dose CT image registrations was characterised by expert’s scoring the spatial distance of manually paired corresponding points and by eight voxel intensity-based (dis)similarity parameters. The distribution of (dis)similarity parameter correlating best with anatomic scoring was bootstrapped, and 95% confidence intervals were calculated separately for acceptable and insufficient registrations. We showed that mutual information (MI) of the eight investigated (dis)similarity parameters displayed the closest correlation with the anatomy-based distance metrics used to characterise the quality of image registrations. The 95% confidence intervals of the bootstrapped MI distribution were [0.15, 0.22] and [0.28, 0.37] for insufficient and acceptable registrations, respectively. In case of any new patient, a calculated MI value of registered low- and high-dose CT image pair within the [0.28, 0.37] or the [0.15, 0.22] interval would suggest acceptance or rejection, respectively, serving as an aid for the radiologist.
Conclusion
A computer-aided solution was proposed in order to reduce reliance on radiologist’s contribution for the approval of acceptable image registrations.
Keywords: Computed tomography, Diagnostics, Image registration, Image segmentation, Image-guided bronchoscopy
Background
There is a growing interest in increasing the diagnostic yield in investigating peripheral pulmonary lesions and malignant tracheal tumours [1–4]. Visualisation of the structure of the thorax and lung and combining it with taking tissue samples is of primary importance. For this purpose, bronchoscopy is commonly used; it provides an optical image of the inner surface of the bronchi using fibre optics and thus guides sampling.
The disadvantage of bronchoscopic examination is that it provides visual information on details within the bronchus only, but not the outside extension of tumour tissue (suggesting how to orient the sampling needle), and it is invasive. Virtual bronchoscopy (VB) is a non-invasive procedure and offers visualisation of the airway from the in- and outside along with its neighbouring blood vessels and lymph nodes, derived from computed tomography (CT) and/or positron emission tomography (PET) [3, 5–7]. Additionally, there are further imaging modalities capable of providing three-dimensional images of the bronchial tree and lung, including, among others, ultrasound [8, 9].
The PET method excels from others due to its ability to give information about the tissue biochemistry. The fluorodeoxyglucose (FDG) accumulation map displays the intensity distribution of carbohydrate metabolism; thus, reports on tumour suspect localisation(s). FDG-PET studies have been proven to be useful in the selection of benign from malignant lesions [10–13] and staging [14–17].
Acquisition of simultaneous information about the structure and function implies a presumable improvement in the diagnostic yield. The 3D rendering of 18 F-FDG PET/CT images was first used for virtual bronchoscopy in 2006 [18]. Unfortunately, the majority, if not all of the few publications reporting on the use of PET imaging in VB, applied low-resolution ldCT images obtained from PET/CT scan. Thus, this kind of VB using both CT and PET lacked the high resolution offered by the diagnostic CT images. The high resolution in this combined type of VB can be assured by substituting ldCT with hdCT, but it involves complex registration problems.
Registration of PET images to ldCT images is remarkably more straightforward than that to hdCT because of the identical geometry [19]. We decided to work out an image processing pipeline based on high-resolution hdCT images to perform precise registration of PET images to diagnostic CT images for functional virtual bronchoscopy (fVB). Thus, a radiologist expert was asked to validate and decide on the accept/reject issue in the initial phase of the project. To spare the contribution of the radiologist expert, we developed a computer-aided method to evaluate the hdCT image registration to the ldCT image allowing the performance of high-resolution CT-PET-based fVB.
Methods
Patients
PET/CT and hdCT scans were performed on 22 oncological patients (11 females and 11 males) aged between 42 and 81 years (mean 63 years, median 63 years, SD 9 years). The Regional and Institutional Ethics Committee at our university approved this clinical study, which was then carried out under the relevant guidelines and regulations. Clinical data of the patients were collected from medical records and entered into a patient database.
Imaging protocols
The hdCT images were constructed by a Siemens Emotion 16 camera (Siemens Healthcare GmbH, Erlangen, Germany). These investigations were carried out with 110 kVp voltage and pitch 0.8. Other hospitals also sent hdCT data obtained by Philips Brilliance 10 (Koninklijke Philips N.V., Amsterdam, Netherlands) with 120 kVp and pitch 0.9, or Siemens Emotion Duo (Siemens Healthcare GmbH, Erlangen, Germany) with 110 kVp and pitch 0.8. The ldCT and PET scans were performed by a PET/CT camera Philips GEMINI TF TOF 64 (Koninklijke Philips N.V., Amsterdam, Netherlands) applying 120 kVp and pitch 0.829. Scans were performed using automatic exposure control (AEC).
The ldCT and hdCT imaging was carried out with less than 1-year time difference. Scan conditions were not standardised; hands of the subjects were along the trunk during the ldCT scan, while in case of hdCT they were over the head, making ldCT–hdCT registration more difficult. Also, there was no air retention constraint during the ldCT scan (PET/CT) as opposed to hdCT. Scan time and the imaged region of the body were not uniform either.
Registration and segmentation
Registration
Performing VB, functional information on the living tissues can be anatomically localised by projecting the adequate radioactivity accumulation to the surface of hdCT segmented organs. To transfer functional information to the space of high-performance hdCT (reference frame), the hdCT and ldCT images must be anatomically fitted by image registration as PET, and ldCT images are inherently fitted due to the identical geometry of the appropriate scans.
For image registration, we used the elastix algorithm [20, 21] based on Insight Toolkit [22], which supports both rigid and non-rigid body transformations. Having completed the registration of the ldCT and hdCT images of the patients, a radiologist expert categorised the registration of the image pairs as good or bad in the initial phase of the project.
Segmentation
The aim was to establish a dedicated fVB protocol with the simultaneous visualisation of both anatomical and functional information. To have a more structured image of the airway, one should work with the hdCT image. Figure 1 shows the remarkable difference between the surface model of the segmented airway images obtained from low-dose and high-dose CT scans of the chest.
Fig. 1.
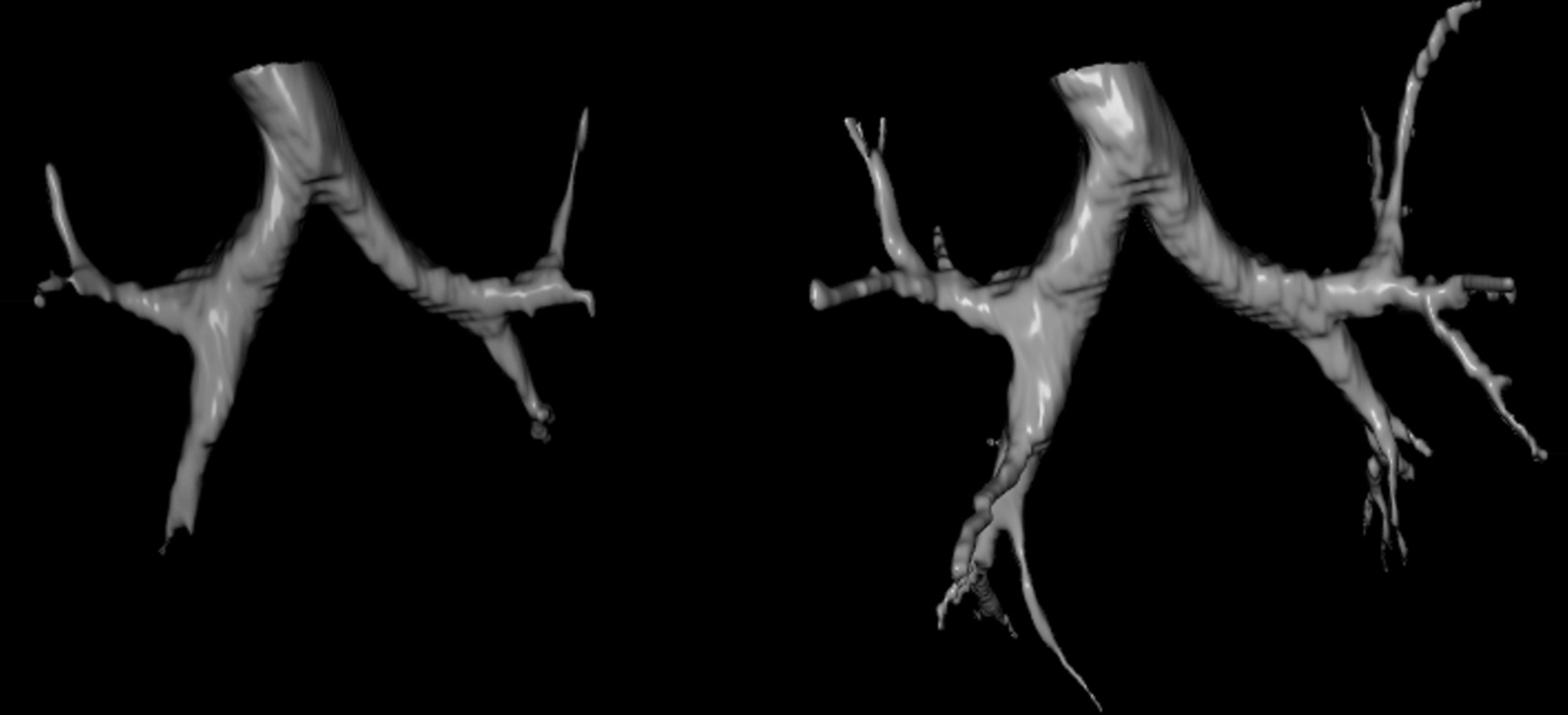
Surface model of airway images segmented from ldCT (left) and hdCT (right) images
The high-dose CT image of the airway offers more detailed structural data for VB with higher-generation branches. Thus, high-quality fVB necessitates the registration of PET image to hdCT. As previously explained, this registration can be performed using the transformation matrix of the ldCT to hdCT registration. To obtain precise registration of ldCT and hdCT images, only the essential part of the data should be used excluding irrelevant details, e.g. bed and ribs. The relevant data can be selected by constructing and using an appropriate lung mask. A segmented hdCT lung image was obtained with the help of the M-SEGM (see “Appendix”) algorithm, used after selecting voxels with intensities between − 1000 and − 400 HU representing bronchial airway and parenchyma. As a first step, the intensity value of the non-segmented voxels of the lung image was set to zero, while that of the segmented ones was set to one, adding up to a lung-mask (referred to as the “lung-mask”). This procedure was followed by constructing lung-masked ldCT and hdCT images with voxel intensities set to the product of the “lung-mask” intensity values and that of the ldCT or hdCT. The registration of the ldCT and hdCT images was performed using the lung-masked images.
Sometimes, the rib cage may be well aligned to the detriment of vessel structures near the border of the lung [20, 21], occasionally causing the relocation of the lung apex to the abdomen region in the hdCT image. Successful registration of lung-masked ldCT and hdCT images using elastix was not possible because the software inevitably stopped. We implemented a specific application to get a functioning lung-mask, eliminating all major discontinuities and adding the mediastinal voxels to the lung-mask. The attachment of the mediastinal region was necessary because a part of the lung-masked ldCT image frequently overlapped the hdCT mediastinum, which was eliminated from the lung-masked hdCT image. The obtained mask is referred to as “functioning lung-mask.” Using this application elastix worked satisfactorily with the default input parameters [20, 21].
The bronchial tree segmentation from the ldCT and hdCT was performed using modified GeoS 2.2 Microsoft software [23], or alternately our region-growing segmentation (multi-SEGMentation: M-SEGM a part of M3I framework [24]) method, a voxel intensity-based iterative procedure (to be published elsewhere). In addition to the application in the current project, M-SEGM can be used to solve similar tasks. Our segmentation procedure also has the advantage that the necessary modification can be easily solved for the actual local needs. M-SEGM allows automatic segmentation having specified a parameter (seed point) and the homogeneity criterion for the tissue to be segmented (airway, blood vessel or parenchyma, etc.). The application of M-SEGM enabled the segmentation of the hdCT tracheal tree up to the origin of the sub-segment bronchi.
Construction of airway tree and surface model
The ensemble of the segmented airway voxels can be used to generate their centre of gravity slice by slice (centre points are located in the air compartment). The total of the centre of gravity points will be referred to as airway tree or skeleton (dotted line in Fig. 2). The individual points of the skeleton are characterised by a set of indices reporting on the ordinal number (i) and the coordinates of the points (x, y, z) and the ordinal number of the first adjacent point(s) along the new branch(es) (i1, i2, i3…) also indicating the number of branches at the particular point (the format of an element in the skeleton file: (i, x, y, z, i1, i2, i3…). After its creation, the skeleton was cleansed by erasing short branches containing less than six voxels unless they comprise a new branch before the sixth point.
Fig. 2.
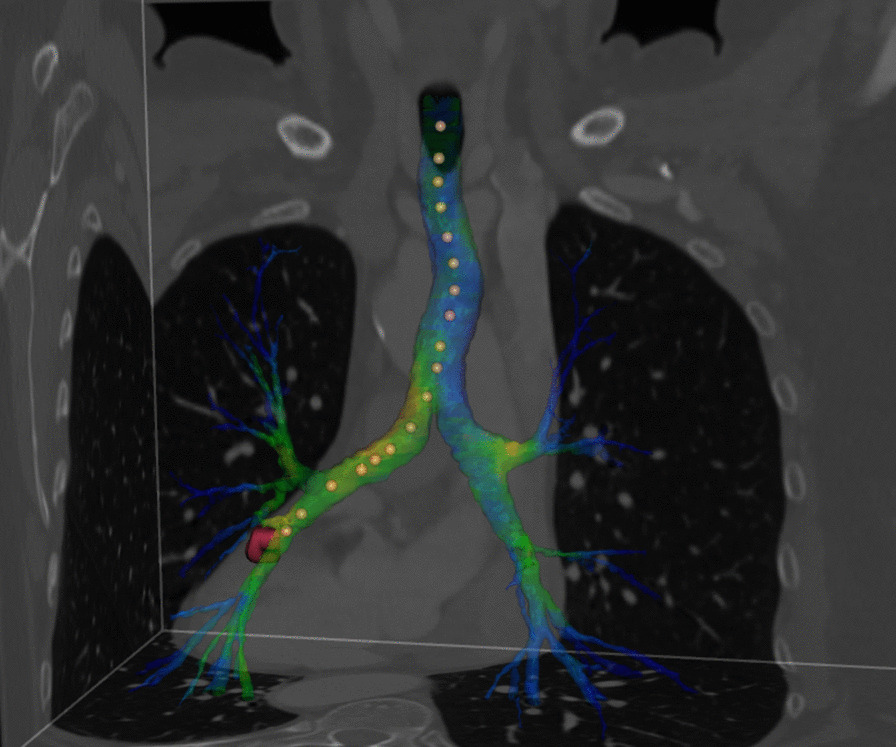
Parts of the three orthogonal sections of the thorax and the segmented bronchial tree comprising a possible traversal (indicated by dots) in the 3D surface model
The bronchial skeleton allows easy traversal of the bronchial tree by a virtual camera to observe the structure and function of the airway or to compare these items with those on the bronchoscope images. Figure 3 displays a snapshot of such a traversal (see a detailed discussion of the figure below). The number of branching points as precisely defined anatomical locations can also be easily counted with the help of this bronchial skeleton. The output of the bronchial tree segmentation served as input for the marching cube algorithm [25] generating a three-dimensional surface model (comprising a high number of triangles) along the sheath of the airway. The number of triangles on this surface model was reduced by a mesh simplification algorithm [26]. Touring along the bronchial tree, the clinician can navigate in space within the luminal structure. Thus, the surface model allows performing VB using the bronchial tree generated by the segmentation algorithm. As mentioned above, the anatomic information was completed with functional data. FDG accumulation reporting on tissue metabolism was projected perpendicularly to the surface elements of the 3D surface model. Accumulation data were summed up from voxels 10 mm from or closer to the surface located outside of the airway. The 10-mm characteristic distance was chosen according to the range of transbronchial needle. Figure 2 shows the 3D surface model of the airway with the superimposed specific functional information derived from the PET image. This metabolic information in Fig. 2 is localised exclusively within the airway contour. Visualisation occurred with an appropriate blending value to make CT voxel intensities negligible.
Fig. 3.
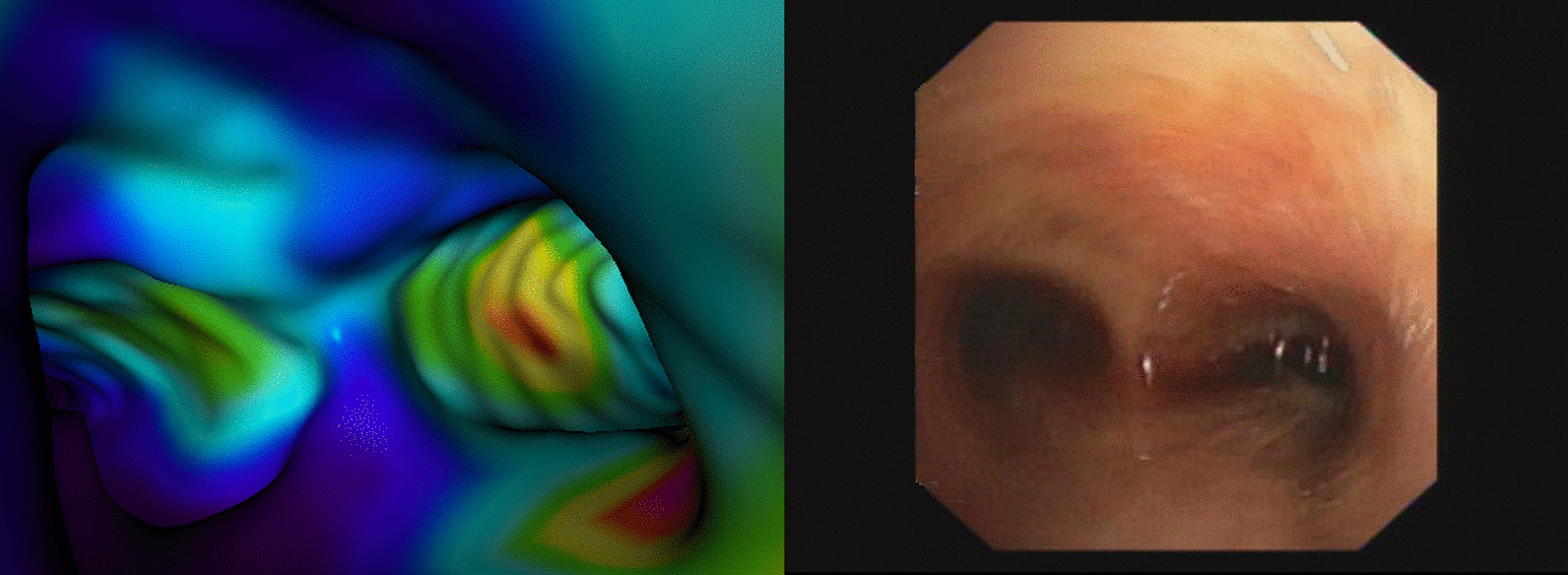
Visualisation of the bifurcation point, as it can be seen from the trachea. The segmented trachea defines the geometry, while the texture originates from the radioactivity accumulated in pixels located 10 mm or closer to the outer surface of the trachea (panel A). Real bronchoscopic photograph of the same carinal region (panel B)
Visualisation of the 3D surface model can be performed from both inside (Fig. 3) and outside. Outside visualisation also allows the display of regional blood vessels. To do this, we also segmented the appropriate blood vessels in the same way as described for the airway. Figure 4 shows the final result of visualised structural hdCT segmentations (bronchial tree and blood vessels) and functional data (PET).
Fig. 4.
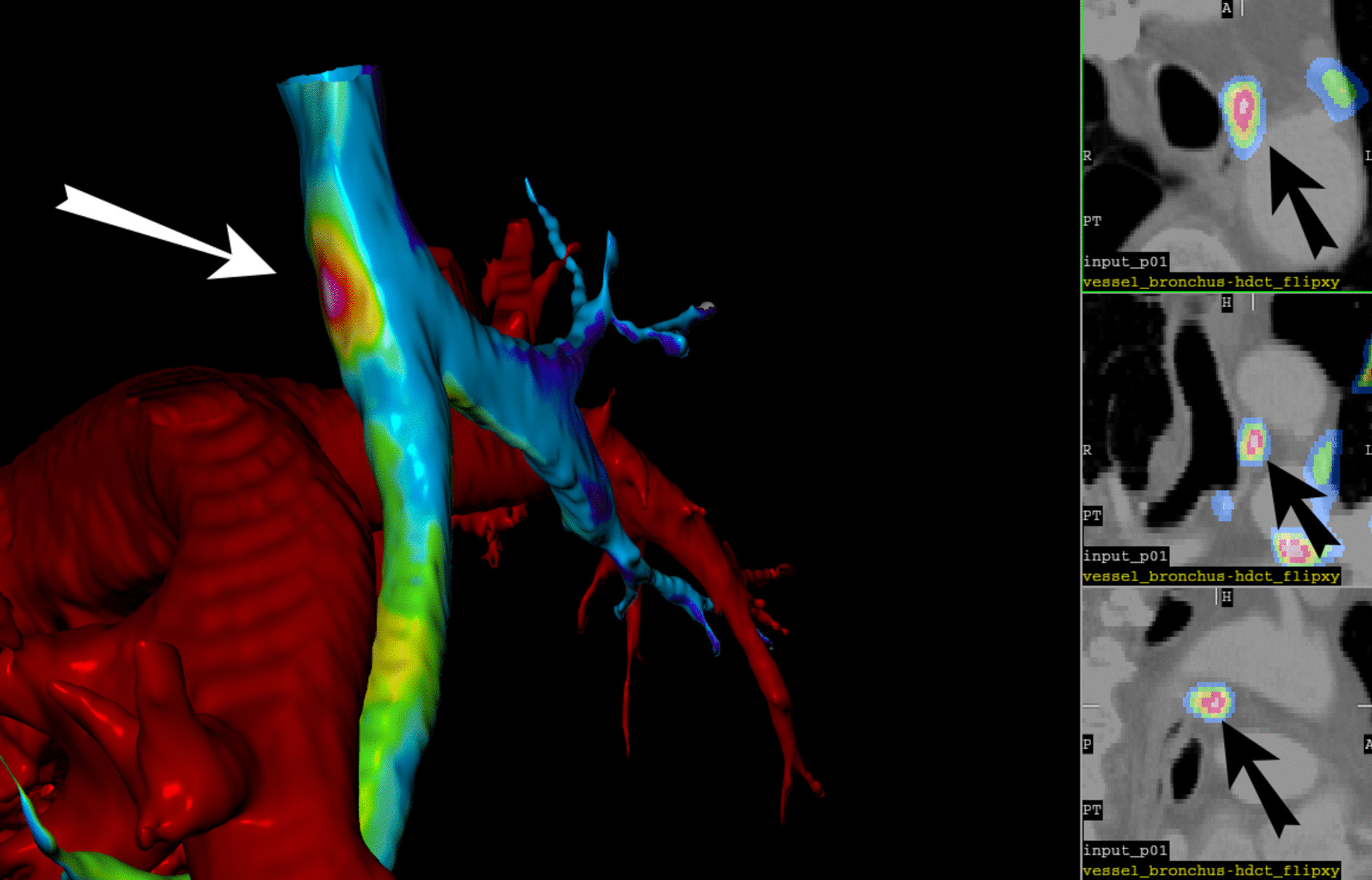
Visualisation of the airway tree and blood vessels from the outside (left) and orthogonal sections in the vicinity of the lesion indicated by the arrow (right). The projected FDG accumulation in the lesion is displayed in spectral scale
Registration validation method
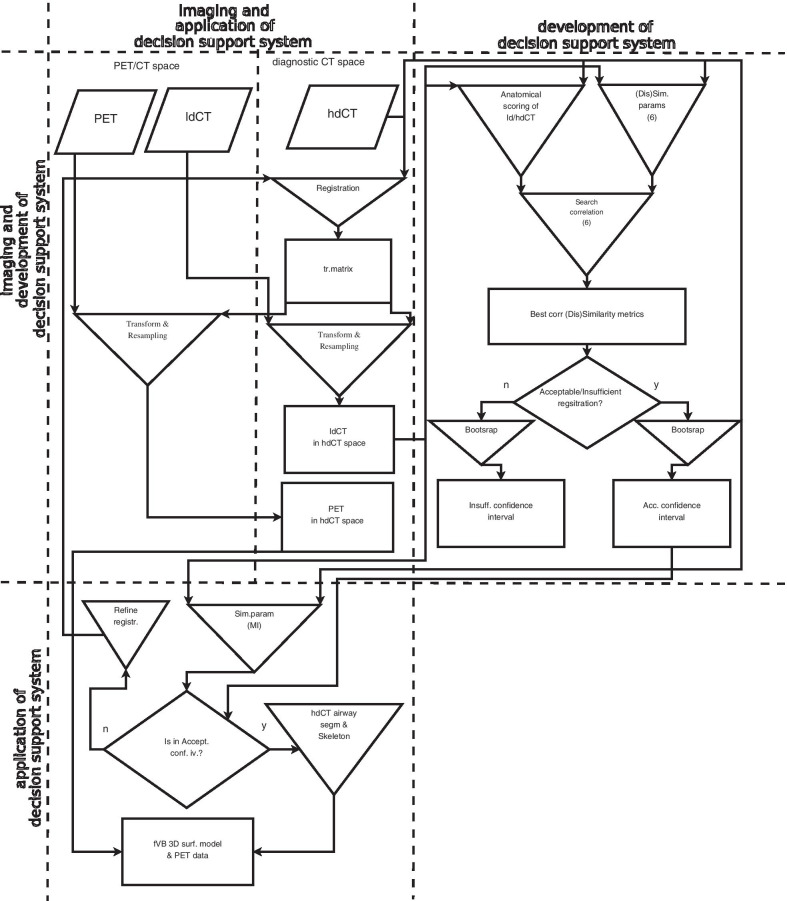
The high-precision fVB requires high-quality steric and functional information, in other words; both hdCT and PET data are required the latter having been transformed into the hdCT space. The necessary transformation matrix is identical to that applied for the registration of the ldCT image to the hdCT image (Fig. 5). During the realignment processes, we used trilinear interpolation to evaluate registered ldCT and PET images with the same matrix and voxel size as the hdCT was acquired. For this registration, both rigid and non-rigid body transformations were applied due to the eventually different conditions (see “Imaging protocols” section) of the chest CT scannings. Because of the low quality of the ldCT, some registrations of the two CT images were insufficient.
Fig. 5.
Flowchart of transforming ldCT and PET into diagnostic hdCT space, calculation of intensity based (dis)similarity parameters, distance-based registration’s quality-judging metrics as well as calculation of correlation between intensity- and distance-based parameters. The flowchart also displays the steps necessary to determine the confidence intervals of MI (see detailed explanation in the text)
These registrations are unsuitable to proceed towards fVB, while acceptable registrations allow construction of the 3D airway surface model from the hdCT and visualisation of the PET accumulation on the surface model as detailed in “Construction of airway tree and surface model” section. Distinction between acceptable and insufficient registration was made by a radiologist; however, this decision requires careful, meticulous and time-consuming work and is subjective.
We replaced this procedure with an objective method. A numeric parameter, D (score) was introduced to characterise the registration quality, derived from 25 distance-type quantities calculated from real distances measured in mm or compared to real measure of anatomical structures (Table 1). A medical imaging specialist physician visually reviewed the hdCT images and identified various parts of the tracheobronchial tree, including the tracheal bifurcation, the origins of the main, lobar and segmental bronchi. The expert marked all the luminal centres in hdCT and checked the localisation of these points on the appropriate registered ldCT image. In case of larger structures (trachea, main bronchi), the distance between the centre of the lumens on the registered hdCT and ldCT image pairs was measured in mm. For each smaller bronchus (columns 5–26 in Table 1), the location of the marked point on the hdCT image was investigated as to whether it was inside the corresponding ldCT bronchial lumen. If so, the distance-type quantity was set to one, while in all other cases to zero.
Table 1.
Distances, measured in mm, and distance-type quantities of corresponding anatomical localisations (see detailed explanation in the text). Segmental bronchi were labelled according to Boyden’s nomenclaturea
| Pat. | TB [mm] | RMB [mm] | LMB [mm] | RULB | RMLB | RLLB | RB1 | RB2 | RB3 | RB4 | RB5 | RB6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p01 | 1.7 | 0.7 | 0.9 | 1 | 1 | 1 | NA | 1 | 1 | 0 | 1 | 1 |
| p03 | 0 | 0 | 1.1 | 1 | 1 | 1 | NA | 1 | 1 | 1 | 1 | 1 |
| p04 | 0 | 0.5 | 0.8 | 1 | 1 | 1 | NA | 1 | 1 | 0 | 1 | 1 |
| p05 | 2 | 1.1 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 |
| p06 | 1.9 | 0 | 0.4 | 1 | 1 | 1 | NA | 1 | 1 | 1 | 1 | 1 |
| p07 | 1.2 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p08 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p09 | 2.6 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p10 | 1.9 | 0 | 0 | 1 | NULL | NULL | 1 | 0 | 0 | NULL | NULL | NULL |
| p11 | 2.4 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p12 | 1.1 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p13 | 1.5 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p14 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| p15 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| p16 | 2.5 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p17 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p18 | 0 | 0 | 0 | NULL | 0 | 0 | NULL | NULL | NULL | 1 | 1 | 1 |
| p20 | 2.8 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p21 | 0 | 0 | 0 | 1 | 1 | 1 | NA | 1 | 1 | 1 | 1 | NA |
| p22 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p23 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p25 | 2.3 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Pat. | RB8 | RB9 | RB10 | LULB | LLLB | LB1/2 | LB3 | LB4 | LB5 | LB6 | LB8 | LB9 | LB10 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p01 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | NA | NA | 1 | 1 | 1 | 1 |
| p03 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p04 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 |
| p05 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p06 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| p07 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p08 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | NULL | NULL | 1 | 1 | 1 | 1 |
| p09 | NA | 1 | 1 | 1 | 1 | 1 | 1 | NULL | NULL | 1 | 1 | 1 | 1 |
| p10 | NULL | NULL | NULL | 1 | 1 | 1 | 0 | NA | NA | 1 | 1 | 1 | 1 |
| p11 | 0 | NA | 1 | 1 | 1 | 1 | NA | NA | NA | 1 | 1 | 1 | 1 |
| p12 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p13 | 1 | NA | 1 | 1 | 1 | 1 | 1 | NA | NA | 1 | 1 | 1 | 1 |
| p14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| p15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| p16 | NA | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p17 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 |
| p18 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p20 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p21 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p22 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p23 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p25 | 1 | 1 | 1 | 1 | 1 | NA | NA | NA | NA | 1 | NA | 1 | 1 |
aTB tracheal bifurcation; main bronchi: RMB right main, LMB left main; right lobar bronchi: RULB upper lobe, RMLB middle lobe, RLLB lower lobe; Right segmental bronchi: RB1 apical, RB2 posterior, RB3 anterior, RB4 lateral, RB5 medial, RB6 superior, RB8 anterior basal, RB9 lateral basal, RB10 posterior basal; Left lobar bronchi: LULB upper lobe, LLLB lower lobe; left segmental bronchi: LB1/2 apicoposterior, LB3 anterior, LB4 superior lingular, LB5 inferior lingular, LB6 superior, LB8 anterior basal, LB9 lateral basal, LB10 posterior basal; NA not identifiable in the ldCT (not visually found by an expert); NULL cannot be identified in hdCT (because the bronchus is absent (after surgery) or obscured by pathology)
We aimed to identify the (dis)similarity of registered ldCT and hdCT image pairs for both acceptable and insufficient registrations by calculating the numerical value (Fig. 5) of six parameters (Pearson correlation coefficient, mutual information (MI), normalised mutual information, Kullback–Leibler divergence, L1norm, L2norm2 [27]). For the objective characterisation of the quality of the registrations [28], absolute distances were determined between corresponding point pairs of the registered images. We investigated how the (dis)similarity parameters and the measure of the registration's quality of the coherent ldCT and hdCT image pairs correlate. Close and low correlations refer to acceptable and insufficient registration, respectively (see the rhomboid-shaped decision symbol in “development of decision support system” part of Fig. 5).
Results
Calculation of (dis)similarity parameters
The numerical value of the voxel-based (dis)similarity parameters can be calculated without any spatial limitation using all the voxels (method α—see below) or with spatial confinement using only a reduced number of voxels (method β and γ—see below). For method γ, this constraint is more significant.
Parameter values were calculated in three different ways:
(α) masks were not used at all in both ldCT and hdCT images (“no mask”),
(β) the same “functioning lung-mask” was used to both type of the CT images,
(γ) modification of the segmented airway was used as a mask to extend the original airway beyond its boundary up to 10 mm thickness. The modification was completed separately for the ldCT and hdCT images.
Distance metrics
The parameter values in Table 1 measured in mm were converted (see below) to obtain distance data of the same type. Converted data for the trachea and the two main bronchi (columns 2–4 in Table 1) were set to:
1 if the distance between the bronchial centres in hdCT and ldCT was less than 10% of the bronchial diameter (≤ 1.8 mm for trachea, ≤ 1.22 mm for main bronchi).
0.5 if this distance was between 10 and 20% of the bronchial diameter ([1.8, 3.6] mm for trachea, [1.22, 2.44] mm for main bronchi).
0 if this distance was above 20% of the bronchial diameter (> 3.6 mm for trachea, > 2.44 mm for main bronchi).
The quality D of the hdCT–ldCT registration was defined as the weighted sum (score) of the distance-type quantities of the same type. We set the weight factor for the trachea, the main bronchi, the lobular bronchi, the segment bronchi and the sub-segment ones to 16, 7, 3, 1 and 1, respectively (Table 2, first line). Table 2 displays the products of the weight factors and the value of the distance-type quantities recorded in Table 1. Applied weight factors are nearly proportional to the cross section of the appropriate bronchus [29] with scaling based on the idea that a good registration algorithm should unequivocally register large structures acceptably. Thus, D provides a score characterising the distance relation of the corresponding anatomical points as a measure of registration’s quality.
Table 2.
Weighted values of distance-type quantities and their sum (score); we used Boyden’s nomenclaturea for abbreviations
| Pat. | TB | RMB | LMB | RULB | RMLB | RLLB | RB1 | RB2 | RB3 | RB4 | RB5 | RB6 | RB8 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Weight | 16 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p01 | 16 | 7 | 7 | 3 | 3 | 3 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| p03 | 16 | 7 | 7 | 3 | 3 | 3 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| p04 | 16 | 7 | 7 | 3 | 3 | 3 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| p05 | 8 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 0 | 1 | 0 | 1 | 1 |
| p06 | 8 | 7 | 7 | 3 | 3 | 3 | 0 | 1 | 1 | 1 | 1 | 1 | 1 |
| p07 | 16 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p08 | 16 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p09 | 8 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| p10 | 8 | 7 | 7 | 3 | NA | NA | 1 | 0 | 0 | NA | NA | NA | NA |
| p11 | 8 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| p12 | 16 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p13 | 16 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p14 | 16 | 7 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| p15 | 16 | 7 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| p16 | 8 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| p17 | 16 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 0 |
| p18 | 16 | 7 | 7 | NA | 0 | 0 | NA | NA | NA | 1 | 1 | 1 | 1 |
| p20 | 8 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p21 | 16 | 7 | 7 | 3 | 3 | 3 | 0 | 1 | 1 | 1 | 1 | 0 | 1 |
| p22 | 16 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p23 | 16 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| p25 | 8 | 7 | 7 | 3 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Pat. | RB9 | RB10 | LULB | LLLB | LB1/2 | LB3 | LB4 | LB5 | LB6 | LB8 | LB9 | LB10 | Score (D) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Weight | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 62 |
| p01 | 0 | 0 | 3 | 3 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 54 |
| p03 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 61 |
| p04 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 59 |
| p05 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 52 |
| p06 | 1 | 1 | 3 | 3 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 52 |
| p07 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 62 |
| p08 | 1 | 1 | 3 | 3 | 1 | 1 | NA | NA | 1 | 1 | 1 | 1 | 60 |
| p09 | 1 | 1 | 3 | 3 | 1 | 1 | NA | NA | 1 | 1 | 1 | 1 | 51 |
| p10 | NA | NA | 3 | 3 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 37 |
| p11 | 0 | 1 | 3 | 3 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 49 |
| p12 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 62 |
| p13 | 0 | 1 | 3 | 3 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 59 |
| p14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 30 |
| p15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 30 |
| p16 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 53 |
| p17 | 1 | 1 | 3 | 3 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 1 | 58 |
| p18 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 50 |
| p20 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 54 |
| p21 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 60 |
| p22 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 62 |
| p23 | 1 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 62 |
| p25 | 1 | 1 | 3 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 49 |
aTB tracheal bifurcation; Main bronchi: RMB right main, LMB left main; right lobar bronchi: RULB upper lobe, RMLB middle lobe, RLLB lower lobe; right segmental bronchi: RB1 apical, RB2 posterior, RB3 anterior, RB4 lateral, RB5 medial, RB6 superior, RB8 anterior basal, RB9 lateral basal, RB10 posterior basal; Left lobar bronchi: LULB upper lobe, LLLB lower lobe; Left segmental bronchi: LB1/2 apicoposterior, LB3 anterior, LB4 superior lingular, LB5 inferior lingular, LB6 superior, LB8 anterior basal, LB9 lateral basal, LB10 posterior basal. The last column (score-D) is the weighted sum of the displayed distance-type data
Correlation calculation
Linear model-based regression calculations (MASS R-package [30]) were performed to determine whether there is any (dis)similarity parameter, correlating with the value of D. The ϱ correlation coefficients are listed in Table 3. Numerical values of D and (dis)similarity parameters are displayed in Table 4. (Dis)similarity parameters giving significant correlation (p < 0.05) with the score values are shown in Table 5 in case of γ masking technique.
Table 3.
Correlation coefficients (ϱ) between the distance metrics and the intensity-based (dis)similarity parameters (calculated in three different manners: α, β, γ; see details in the text)
| Masking | (dis)similarity parameter | |||||
|---|---|---|---|---|---|---|
| Pearson (s) | MI (s) | NMI (s) | HKL (d) | L1norm (d) | L2norm2 (d) | |
| α | 0.15 | 0.02 | 0.02 | − 0.07 | 0.25 | 0.22 |
| β | 0.38 | 0.56 | 0.5 | 0.45 | − 0.04 | − 0.09 |
| γ | 0.4 | 0.6 | 0.51 | − 0.02 | 0.07 | 0.09 |
MI mutual information, NMI normalised mutual information, HKL Kullback–Leibler divergence, L1Norm L1 norm or Manhattan norm, L2norm2 square L2 norm or square Euclidean distance, (s) similarity parameter, (d) dissimilarity parameter (data are rounded to two significant digits, the highest correlation coefficient is emphasized in bold text)
Table 4.
Calculated values of the quality of registration (score) and the (dis)similarity parameters of corresponding ldCT and hdCT image pairs in case of γ masking technique (data are rounded to two significant digits)
| Pat | Score | (dis)similarity parameter | |||||
|---|---|---|---|---|---|---|---|
| Pearson | MI | NMI | HKL | L1norm * 1e−7 | L2norm2 * 1e−10 | ||
| p01 | 54 | 0.86 | 0.44 | 1.21 | 0.44 | 20.55 | 13.61 |
| p03 | 61 | 0.25 | 0.25 | 1.1 | 0.74 | 4.1 | 2.07 |
| p04 | 59 | 0.74 | 0.44 | 1.21 | 0.62 | 15.1 | 10.4 |
| p05 | 52 | 0.32 | 0.32 | 1.11 | 0.55 | 8.18 | 5.42 |
| p06 | 52 | 0.31 | 0.31 | 1.11 | 0.74 | 5.89 | 3.45 |
| p07 | 62 | 0.24 | 0.25 | 1.09 | 0.78 | 4.36 | 2.34 |
| p08 | 60 | 0.28 | 0.28 | 1.1 | 0.8 | 4.71 | 2.45 |
| p09 | 51 | 0.19 | 0.19 | 1.08 | 0.85 | 3.11 | 1.41 |
| p10 | 37 | 0.19 | 0.2 | 1.09 | 0.76 | 4.75 | 2.5 |
| p11 | 49 | 0.24 | 0.22 | 1.1 | 0.79 | 6.8 | 4.1 |
| p12 | 62 | 0.47 | 0.47 | 1.17 | 0.79 | 5.35 | 2.88 |
| p13 | 59 | 0.17 | 0.18 | 1.07 | 0.68 | 6.47 | 4.21 |
| p14 | 30 | 0.15 | 0.15 | 1.06 | 0.75 | 7.03 | 4.15 |
| p15 | 30 | 0.1 | 0.1 | 1.04 | 0.75 | 4.67 | 2.34 |
| p16 | 53 | 0.41 | 0.31 | 1.13 | 0.69 | 10.52 | 6.87 |
| p17 | 58 | 0.34 | 0.34 | 1.12 | 0.69 | 7.12 | 4.62 |
| p18 | 50 | 0.2 | 0.2 | 1.08 | 0.84 | 3.83 | 1.58 |
| p20 | 54 | 0.21 | 0.21 | 1.08 | 0.7 | 8.24 | 5.2 |
| p21 | 60 | 0.43 | 0.43 | 1.17 | 0.81 | 2.47 | 1.08 |
| p22 | 62 | 0.41 | 0.41 | 1.19 | 0.8 | 3.97 | 1.75 |
| p23 | 62 | 0.22 | 0.22 | 1.08 | 0.62 | 10.57 | 7.32 |
| p25 | 49 | 0.26 | 0.26 | 1.09 | 0.44 | 7.8 | 5.08 |
Table 5.
Values of the p, estimated correlation of score and similarity parameters, and the lower and upper limits of confidence intervals (with γ masking technique)
| (dis)similarity parameter | p | Corr | CI_lower | CI_upper |
|---|---|---|---|---|
| Pearson | 0.0673 | 0.3971 | − 0.0295 | 0.7013 |
| MI | 0.0033 | 0.5981 | 0.2360 | 0.8143 |
| NMI | 0.0150 | 0.5115 | 0.1146 | 0.7676 |
Data are displayed for similarity parameters giving significant correlation with the score values (p < 0.05)
The analysis revealed that the MI displayed the closest correlation with the anatomy-based distance metrics (ϱ = 0.5981, p = 0.0033).
Confidence intervals
We aimed to establish whether the numerical value of the MI can suggest acceptable vs insufficient ldCT–hdCT lung image pair registration classification is applicable to any patient. To answer this question, one should draw a large number of samples from the population of interest. However, the time required to gather this vast amount of data is extensive, and the procedure is labour intensive and too expensive. Instead, we used the frequently applied bootstrap statistical method [31, 32] providing a bootstrapped distribution of the MI statistics, which approximates the MI distribution in the whole population. For this purpose, we divided our 22 cases into two parts according to the quality of the registration as measured by score value (the higher the better) and constructed the bootstrapped MI distribution separately for the acceptable (15 subjects) and insufficient (7 subjects) subsets assigned to the 22-member scanned population (Fig. 6).
Fig. 6.
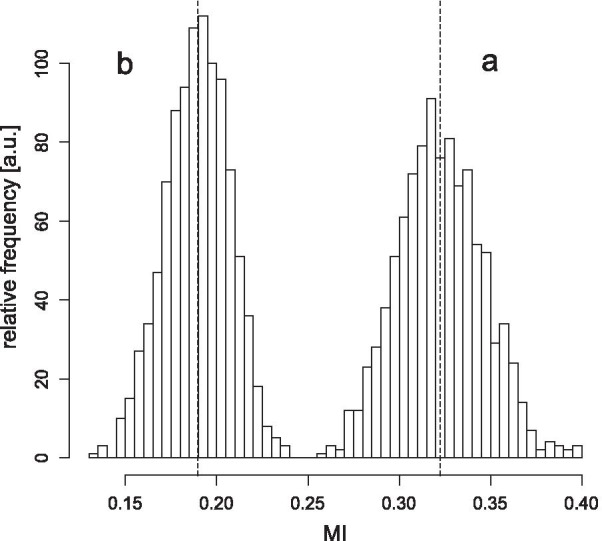
MI distribution calculated from the bootstrapped samples (a: for the accepted registrations; b: for the rejected registrations). A vertical dashed line indicates the mean of MI values for both parts of the distribution (0.19 and 0.324)
The numerical value of MI is image pair dependent, and its value fluctuates from sample to sample. The fluctuation can be characterised by confidence intervals. If one trims off a small percentage (e.g. 2.5%) from both the lower and upper end of the distribution, the remaining interval includes 95% of all the cases.
The distribution displayed in Fig. 6 indicates that the registrations can be separated into two groups based on MI values. We also examined the confidence intervals of the bootstrapped MI for insufficient and acceptable registrations. The 95% confidence intervals were [0.15, 0.22], [0.28, 0.37], respectively.
For the subpopulation acceptable and insufficient registration, we separately examined how the mean of their bootstrapped distributions (Fig. 6) differed from the mean of the appropriate scanned populations (bias). Values obtained:
Consequently, in case of any new registration, a calculated MI value within the [0.28, 0.37] (mean: 0.324) or the [0.15, 0.22] (mean: 0.19) interval would suggest acceptance or rejection, respectively, with 95% confidence serving as an aid for the radiologist. In case of insufficient registration, the registration has to be repeated with slightly different conditions. MI values between the two confidence intervals (values between 0.22 and 0.28) also indicate the necessity of a new registration.
Discussion
VB is frequently used as a potent diagnostic modality. Application of fVB is even more advantageous because along with the morphological details, it also visualises the areas of malignancy and tumour growth and the tumorous tissue both in- and outside the bronchus. The functional information enables early diagnosis as the metabolic change within the infiltrated region precedes the development of anatomical changes. Complementing anatomical information with functional data would assist the tissue sampling from the right location. The glucose accumulation map delivered by PET scan can also help in the assessment of the tumour aggressiveness, therapeutic response and recurrences. The incorporation of functional information into the practice of virtual diagnostics is generally done by registering the PET image to the bronchoscope video image. Thus, the physician does not have to solve the “merging” of a pre-viewed, digitally generated hdCT–PET image and a video image viewed during tissue sampling. Fortunately, there are working algorithms [33, 34] that display the two 3D images (hdCT–PET and digitised bronchoscope video image) in a single merged image.
The quality of the fVB is improved if we use the higher-resolution hdCT instead of ldCT images (Fig. 1). The use of hdCT is justified because it can map structures not resolved by ldCT. For high-quality fVB, the registration of the ldCT and hdCT images is indispensable, because the transformation matrix of this registration is necessary to fit the PET and hdCT images anatomically correctly.
In the initial phase of our project, a radiologist decided whether ldCT–hdCT registration was acceptable or not. However, the judgement of a radiologist expert is subjective and difficult to scale. Therefore, we were looking for a solution characterising registration quality objectively and independently of the radiologist. Numeric value of (dis)similarity parameters of registered image pairs can be calculated easily, so it is worth looking at whether there are any of them correlating with the measure of the registration quality (scoring of the appropriate structures’ fit by an expert). In case of a close correlation, a simple software procedure can substitute the radiologist’s categorisation of any new ldCT–hdCT registration. Rohlfing has shown that distance-type errors should be used for reporting reliable registration errors [28]. As D provides a distance-type characterisation of the point pair's set of the registered images, a parameter correlated with D maintains approximately the same characterisation. Application of this software procedure is advantageous due to its simplicity compared to the labour-intensive calculation of D. At the same time, it might give a somewhat looser characterisation of registration quality.
For software judgement of the registration’s quality, we used parameters to show the similarity (the larger, the better) and the dissimilarity (the less, the better) in the voxel intensity data of the fitted images. For this purpose, the easily calculable Pearson correlation coefficient, mutual information, normalised mutual information, Kullback–Leibler divergence, L1norm and L2norm2 were chosen. Out of these parameters, those providing a close correlation with the distance-based D registration quality can be used to classify the registration as acceptable or insufficient. Contrary to expectations, we found a close correlation in only one case, suggesting that the (dis)similarity parameters define the concept of similarity and dissimilarity in different ways.
The categorisation can be performed using confidence intervals of the (dis)similarity parameters. Registrations with MI parameter value falling within the confidence interval “a” and “b” of the distributions (Fig. 6) are considered acceptable and insufficient, respectively. Parameter values falling within the “b” confidence interval indicate that the registrations are to be repeated under more tightly controlled conditions.
Substitution of ldCT images with diagnostic hdCT images will further strengthen the successful applicability of the fVB and improve its clinical achievements. A disadvantageous feature of the developed procedure is that it has to be reconstructed (the confidence intervals have to be recalculated) in case of any change in the imaging equipment, segmentation or registration protocols.
Conclusion
Patients undergoing bronchoscopy have previously undergone a series of examinations, including in the majority of cases CT and PET examinations. The latter investigation is generally performed in the form of PET/ldCT. Many PET centres usually do not even perform hdCT testing because the indication is usually tumour search. If the whole investigation process indicates that bronchoscopy is required, a prospective hdCT test either alone or with a concomitant PET scan would involve an extra dose of radiation, which would increase the likelihood of developing a second tumour.
An automatic pipeline was worked out for virtual bronchoscopic examinations based on both PET and hdCT data using retrospective scans to avoid additional prospective HRCT study with the extra radiation dose involved. Accurate manual verification of the quality of ldCT–hdCT registration, which is a critical element of the procedure, would traditionally require time-consuming radiologist expert work, for the replacement of which we have developed an automated procedure.
The proposed procedure will likely increase the diagnostic yield of fVB as the added value of HRCT and PET methods is difficult to question. The precise visualisation of functional information can decrease the number of biopsies taken from inadequate locations. Displaying the blood vessel system (Fig. 4) can reduce the risk of artery or vein perforation [35].
Results of fVB examinations supported by PET and hdCT imaging have not been published so far to our best knowledge; thus, numerical data on how our method improves the current workflow can only be obtained by performing a large number of studies.
Acknowledgements
The authors thank Emma Emri (East Sussex Healthcare NHS Trust) for proofreading.
Abbreviations
- CT
Computed tomography
- PET
Positron emission tomography
- FDG
Fluorodeoxyglucose
- VB
Virtual bronchoscopy
- hdCT
High-dose CT
- ldCT
Low-dose CT
- fVB
Functional virtual bronchoscopy
- HRCT
High-resolution CT
- kVp
Kilovolt peak
- AEC
Automatic exposure control
- M-SEGM
Multi-SEGMentation
- M3I
Multi-modal medical imaging
- Pearson
Pearson correlation coefficient
- MI
Mutual information
- NMI
Normalised mutual information
- HKL
Kullback–Leibler divergence
- L1norm
Manhattan distance
- L2norm2
Euclidean distance
- ANOVA
Analysis of variance
Appendix
Our M-SEGM segmentation algorithm is semi-automatic (Multi-SEGMentation: M-SEGM). The user has to specify the placement of the starting point (seed point) of the segmentation within the desired region (e.g. inside the trachea) and the intensity range with low and high limits as the homogeneity criterion. This algorithm attaches voxels one by one to the previous voxel set performing iterations, starting from the seed point. The intensity of the last attached voxel is in the range specified by the homogeneity criterion (airway: [− 1000, − 950], parenchyma: [− 800, − 400], contrast-enhanced blood vessel: [200,400]). Voxel addition continues until the last voxel is not yet tested, followed by the stop of the process and the generation of the segmented mask.
Authors' contributions
LG and GO were involved in conceptualisation; GO, MN, ZB, CA and DS were involved in methodology; GO, ME, LB, AM and IV were involved in formal analysis and investigation; LT, GO and ME were involved in writing—original draft preparation; LT and GO were involved in writing—review and editing; ME and LB were involved in funding acquisition; and GO, DS, ME and CA contributed to software. All authors read and approved the final manuscript.
Funding
This work was partially supported by VKSZ 14-1-2015-0072, SCOPIA: Development of diagnostic tools based on endoscope technology supported by the European Union, co-financed by the European Social Fund.
Availability of data and materials
The datasets analysed during the current study are not publicly available due to containing information that could compromise research participant privacy but are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
The Regional and Institutional Ethics Committee, Clinical Center at the University of Debrecen, approved this clinical study, which was then carried out in accordance with the relevant guidelines and regulations. As our study involves retrospective analysis of previously acquired data, informed consent was not required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gábor Opposits, Email: opposits.gabor@med.unideb.hu.
Marianna Nagy, Email: nagy.marianna@med.unideb.hu.
Zoltán Barta, Email: bartaz.md@gmail.com.
Csaba Aranyi, Email: aranyics11@gmail.com.
Dániel Szabó, Email: szabo.daniel.andras@med.unideb.hu.
Attila Makai, Email: makai.attila@med.unideb.hu.
Imre Varga, Email: varga.imre@med.unideb.hu.
László Galuska, Email: galuska.laszlo@med.unideb.hu.
Lajos Trón, Email: tron.lajos@med.unideb.hu.
László Balkay, Email: balkay.laszlo@med.unideb.hu.
Miklós Emri, Email: emri.miklos@med.unideb.hu.
References
- 1.Leong S, Ju H, Marshall H, Bowman R, Yang I, Ree AM, et al. Electromagnetic navigation bronchoscopy: a descriptive analysis. J Thorac Dis. 2012;4(2):173–185. doi: 10.3978/j.issn.2072-1439.2012.03.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stevic R, Milenkovic B. Tracheobronchial tumors. J Thorac Dis. 2016;8(11):3401–3413. doi: 10.21037/jtd.2016.11.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shinagawa N. A review of existing and new methods of bronchoscopic diagnosis of lung cancer. Respir Investig. 2019;57(1):3–8. doi: 10.1016/j.resinv.7018.08.004. [DOI] [PubMed] [Google Scholar]
- 4.McLean AEB, Barnes DJ, Troy LK. Diagnosing lung cancer: the complexities of obtaining a tissue diagnosis in the era of minimally invasive and personalised medicine. J Clin Med. 2018;7(7):163. doi: 10.3390/jcm7070163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferguson JS, McLennan G. Virtual bronchoscopy. Proc Am Thorac Soc. 2005;2(6):488–491. doi: 10.1513/pats.200508-082DS. [DOI] [PubMed] [Google Scholar]
- 6.Fernández PG, Sánchez ER, García VAM, Luna AA, Ceballos VJ, Delgado BRC, et al. SEOM–SERAM–SEMNIM guidelines on the use of functional and molecular imaging techniques in advanced non-small-cell lung cancer. Radiologia. 2018;60(4):332–346. doi: 10.1016/j.rx.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Fledelius J, Winther-Larsen A, Khalil AA, Bylov CM, Hjorthaug K, Bertelsen A, et al. 18F-FDG PET/CT for very early response evaluation predicts CT response in erlotinib-treated non-small cell lung cancer patients: a comparison of assessment methods. J Nucl Med. 2017;58(12):1931–1937. doi: 10.2967/jnumed.117.193003. [DOI] [PubMed] [Google Scholar]
- 8.Huang YH, Chen KC, Chen JS. Ultrasound for intraoperative localization of lung nodules during thoracoscopic surgery. Ann Transl Med. 2019;7(2):37. doi: 10.21037/atm.2019.01.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomos I, Tziolos N, Raptakis T, Kavatha D. Thoracic ultrasound for the detection of rib metastases of non-small cell lung cancer. Adv Respir Med. 2018;86(2):101–102. doi: 10.5603/ARM.2018.0014. [DOI] [PubMed] [Google Scholar]
- 10.Cho A, Hur J, Kang WJ, Cho HJ, Lee J, Yun M, Lee JD. Usefulness of FDG PET/CT in determining benign from malignant endobronchial obstruction. Eur Radiol. 2010;21(5):1077–1087. doi: 10.1007/s00330-010-2006-1. [DOI] [PubMed] [Google Scholar]
- 11.Park CM, Goo JM, Lee HJ, Kim MA, Lee CH, Kang MJ. Tumors in the tracheobronchial tree: CT and FDG PET features. Radiographics. 2009;29(1):55–71. doi: 10.1148/rg.291085126. [DOI] [PubMed] [Google Scholar]
- 12.Lim CH, Seok HY, Hyun SH, Moon SH, Cho YS, Lee KH, et al. Evaluation of a diagnostic 18F-FDG PET/CT strategy for differentiating benign from malignant retroperitoneal soft-tissue masses. Clin Radiol. 2019;74(3):207–215. doi: 10.1016/j.crad.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 13.Ciftci E, Turgut B, Cakmakcilar A, Erturk SA. Diagnostic importance of 18F-FDG PET/CT parameters and total lesion glycolysis in differentiating between benign and malignant adrenal lesions. Nucl Med Commun. 2017;38(9):788–794. doi: 10.1097/MNM.0000000000000712. [DOI] [PubMed] [Google Scholar]
- 14.De Wever W, Ceyssens S, Mortelmans L, Stroobants S, Marchal G, Bogaert J, Verschakelen JA. Additional value of PET-CT in the staging of lung cancer: comparison with CT alone, PET alone and visual correlation of PET and CT. Eur Radiol. 2006;17(1):23–32. doi: 10.1007/s00330-006-0284-4. [DOI] [PubMed] [Google Scholar]
- 15.Buchbender C, Herbrik M, Treffert J, Forsting M, Bockisch A, Antoch G, Heusner TA. Virtual 18F-FDG PET/CT bronchoscopy for lymph node staging in non-small-cell lung cancer patients: present and future applications. Expert Rev Med Devices. 2012;9(3):241–247. doi: 10.1586/erd.12.9. [DOI] [PubMed] [Google Scholar]
- 16.Voigt W. Advanced PET imaging in oncology: status and developments with current and future relevance to lung cancer care. Curr Opin Oncol. 2017;29:1–7. doi: 10.1097/CCO.0000000000000430. [DOI] [PubMed] [Google Scholar]
- 17.Shroff GS, Carter BW, Viswanathan C, Benveniste MF, Wu CC, Marom EM, et al. Challenges in interpretation of staging PET/CT in thoracic malignancies. Curr Probl Diagn Radiol. 2017;46(4):330–341. doi: 10.1067/j.cpradiol.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 18.Quon A, Napel S, Beaulieu CF, Gambhir SS. "Flying through" and "flying around" a PET/CT scan: pilot study and development of 3D integrated 18F-FDG PET/CT for virtual bronchoscopy and colonoscopy. J Nucl Med. 2006;47(7):1081–1087. [PubMed] [Google Scholar]
- 19.Baluwala HY, Risser L, Schnabel JA, Saddi KA. Toward physiologically motivated registration of diagnostic CT and PET/CT of lung volumes. Med Phys. 2013;40(2):021903-1–021903-13. doi: 10.1118/1.4771682. [DOI] [PubMed] [Google Scholar]
- 20.Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW. elastix: a toolbox for intensity based medical image registration. IEEE Trans Med Imaging. 2010;29(1):196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 21.Shamonin DP, Bron EE, Lelieveldt BPF, Smits M, Klein S, Staring M. Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer’s disease. Front Neuroinform. 2014;7(50):1–15. doi: 10.3389/fninf.2013.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Insight Segmentation and Registration Toolkit. http://www.itk.org, 2019. Accessed 31 Dec 2020.
- 23.Criminisi A, Sharp T, Blake A, GeoS: Geodesic image segmentation. In Proceedings of European conference on computer vision (ECCV), vol 5302. p. 99–112; 2008. https://www.microsoft.com/en-us/research/publication/geos-geodesic-image-segmentation/. Accessed 31 Dec 2020.
- 24.Multimodal medical imaging tools (M3I), 2003–2021. Available at: https://pet.dote.hu/minipetct/. Accessed 21 May 2020.
- 25.Lorensen WE, Cline HE. Marching cubes: a high resolution 3D surface construction algorithm. ACM Comput Graph. 1987;21(4):163–169. doi: 10.1145/37402.37422. [DOI] [Google Scholar]
- 26.Hamann B. A data reduction scheme for triangulated surfaces. Comput Aided Geom Des. 1994;11(2):197–214. doi: 10.1016/0167-8396(94)90032-9. [DOI] [Google Scholar]
- 27.Opposits G, Kis SA, Trón L, Berényi E, Takács E, Dobai JG, Bognár L, Szűcs B, Emri M. Population based ranking of frameless CT-MRI registration methods. Z Med Phys. 2015;25(4):353–367. doi: 10.1016/j.zemedi.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Rohlfing T. Image similarity and tissue overlaps as surrogates for image registration accuracy: widely used but unreliable. IEEE Trans Med Imaging. 2012;31(2):153–163. doi: 10.1109/TMI.2011.2163944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chovancová M, Elcner J. The pressure gradient in the human respiratory tract. EPJ Web Conf. 2014;67:02047. doi: 10.1051/epjconf/20146702047. [DOI] [Google Scholar]
- 30.Venables WN, Ripley BD. Modern applied statistics with S. 4th edn. New York: Springer. 2002. ISBN 0-387-95457-0, http://www.stats.ox.ac.uk/pub/MASS4. Accessed 31 Dec 2020.
- 31.Bickel PJ, Freedman D. Some asymptotic theory for the bootstrap. Ann Stat. 1981;9:1196–1217. [Google Scholar]
- 32.Singh K. On asymptotic accuracy of Efron’s bootstrap. Ann Stat. 1981;9:1187–1195. [Google Scholar]
- 33.Kaufman A, Wang J. 3D Surface reconstruction from endoscopic videos. In: Linsen L, Hagen H, Hamann B, editors. Visualisation in medicine and life sciences. Mathematics and visualisation. Berlin: Springer; 2008. ISBN 978-3-540-72629-6. 10.1007/978-3-540-72630-2_4.
- 34.Winter C, Scholz I, Rupp S, Wittenberg T. Reconstruction of tubes from monocular fiberscopic images—application and first results. Vision Model Vis. 2005;20:57–64. [Google Scholar]
- 35.Bauer TL, Steiner KV. Virtual bronchoscopy: clinical applications and limitations. Surg Oncol Clin N Am. 2007;16(2):323–328. doi: 10.1016/j.soc.2007.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analysed during the current study are not publicly available due to containing information that could compromise research participant privacy but are available from the corresponding author on reasonable request.