Abstract
The airborne transmission path for SARS-CoV-2 is of primary scientific and health-related interest as it could actually involve management, accessibility, use and functionality of many activities, including hospitals), schools, workplaces, factories, transport, sport venues and outdoor environment. It is necessary to develop a sampling and analytical method for virus-laden bioaerosol that could be considered reliable and validated according to ISO/IEC 17025 requirements.
The present paper defines sample pretreatments aiming at recover SARS-CoV-2 from glass-fiber and PTFE filters employed by low and high-volume air samplers. Recovery test results focused on the sample concentration step carried out by means of ultracentrifugation are reported as well. Human coronavirus strain OC43 (a surrogate β-coronavirus with the same SARS-CoV-2 particle structure) was used to validate each step of the recovery tests.
We found that the elution efficiency of coronavirus OC43 from glass-fiber and quartz filters could be strongly enhanced by using an elution buffer containing up to 40% of fetal calf serum. Moreover, the recovery from PTFE filters is much higher and easier than from glass-fiber filters: for glass-fiber filters a 3 h-shaking phase, followed by a 30 s-vortexing step, are necessary to elute viral infective particles; for PTFE, 60 min-shaking is enough. The effect of suction time on filters could be resumed as follows: sampling durations up to 20 min at a flow rate of 500 L/min do not affect recovery efficiencies from 10 cm glass-fiber filters, whereas the recovery efficiency of infectious virions from 4.7 cm PTFE filters decreases of a factor 2 after 3 h of sampling at a flow rate of 20 L/min. The recovery efficiency of ultracentrifugation turns out to be around 57%. The effect of storage temperature of filters immersed in a transport medium on coronavirus infectivity is assessed as well.
Based on the sampling techniques and the analytical methods developed as described in the present study, many field tests were carried out reporting virus concentrations up to 50 genomic copies per cubic meter of air in domestic environment with poor ventilation condition, whereas in hospital wards the detectable concentrations of SARS-CoV-2 were generally lower than 10 genomic copies per cubic meter of air.
Keywords: SARS-CoV-2, Airborne transmission, PTFE filter, Glass-fiber filter, Bioaerosol, High-volume sampler
1. Introduction
SARS-CoV-2 is a pathogenic virus belonging to the Coronaviridae family. The infection causes a syndrome called COVID-19 that, in the most severe cases, leads to pneumonia and loss of respiratory function. Unlike other members of Coronaviridae family that can cause severe pathology in the lower respiratory airways (namely SARS-CoV and MERS-CoV), SARS-CoV-2 apparently fits the human host better (Platto S et al., 2020), as it is characterized by a high transmissibility that led to a pandemic spread. On January 30, 2020, the World Health Organization (WHO) declared SARS-CoV-2 outbreak as a public health emergency of international concern (PHEIC), its highest level of alarm.
SARS-CoV-2 infection is transmitted by different routes, directly (1), through respiratory droplets emitted by infected people, while sneezing, coughing or talking, even if they are asymptomatic (Morawska and Cao, 2020), indirectly (2), through contact of a susceptible person with a contaminated object or surface (fomite transmission), and through airborne transmission (3), defined as the spread of an infectious agent caused by the dissemination of aerosols that remain infectious when suspended in air over long distances and time and could be inhaled (WHO, 2020). These small particles of pathogen-containing respiratory secretions expelled into the air can remain airborne for long periods (Atkinson et al., 2009), carrying their contents away from where they were originated (Siegel et al., 2007), therefore raising the possibility of transmission also without aerosol-generating procedures. Notably, on April 5, 2021, the ASHRAE Epidemic Task Force released an updated statement on the airborne transmission of SARS-CoV-2, acknowledging its risk in indoor environment: “Airborne transmission of SARS-CoV-2 is significant and should be controlled. Changes to building operations, including the operation of heating, ventilating, and air-conditioning systems, can reduce airborne exposures” (ASHRAE, 2021) This statement replaces the April 2020 one (ASHRAE, 2020), that referred to airborne transmission as “sufficiently likely”. On the contrary, again on April 5, 2021, the Center for Disease Control and Prevention of the USA reports that “the risk of SARS-CoV-2 infection via the fomite transmission route is low, and generally less than 1 in 10,000, which means that each contact with a contaminated surface has less than a 1 in 10,000 chance of causing an infection” (CDC, 2021a). Even more recently, the World Health Organization (WHO, 2021) on April 30, 2021 and the Center for Disease Control and Prevention of the USA (CDC, 2021b) on May 7, 2021 have clearly reported the importance of airborne transmission.
Although close contact with contagious people, up to now, has been improperly considered the main way the pandemic spreads, several obstacles prevent a complete clarification of the actual epidemiological role played by airborne transmission. Firstly, most part of the studies that address this topic are focused only on the detection of viral genome in the air, and do not correlate these molecular biology data with a solid assessment of viral particles infectivity. While indirect and analogy-based evidences have been provided by comparing the infectivity of airborne SARS-CoV and MERS-CoV with SARS-CoV-2 one (da Silva PG et al., 2021), to date only few published studies provided information on SARS-CoV-2 viability in air, focused on COVID-19 hospital wards (Binder et al., 2020; Lednicky et al., 2020; Santarpia et al., 2020; van Doremalen et al., 2020).
Interestingly, the findings of these studies provided useful hints about SARS-CoV-2 spread, supporting the position that aerosol transmission can occur early in the course of disease, even before the onset of symptoms, concluding that aerosol transmission should be assessed closely after the infection (Binder et al., 2020).
The lack of more studies aiming at monitoring the presence of SARS-CoV-2 infective particles in the air represents a remarkable knowledge gap that requires urgent attention. The described aspect is due to the technical limits that hinder the development of reliable techniques to capture and isolate infective particles in the air: in other words, in order to perform an adequate monitoring of the presence of SARS-CoV-2 in indoor and outdoor air, it is necessary to develop a bioaerosol sampling method capable of maintaining the infectivity of sampled viral particles.
Various sampling devices can be used to capture bioaerosol containing viruses (Verrault et al., 2008; Pan et al., 2019). The most common ones are solid impactors, liquid impactors and filters, whose sampling technology is based on a few principles such as inertia, Brownian motions, adhesion properties, mainly depending on the aerodynamic diameter of the airborne particles. It is important to highlight that bioaerosol originates from small droplets that tend to evaporate their water content before they settle down; droplets smaller than 5 μm, which correspond to a fraction of less than 5% of exhaled particles, do not settle at all (Xie et al., 2007; ASHRAE Board of Directors, 2020). The particle size of primary interest for human expiratory activities is around 1 μm.
Solid impactors, such as Andersen samplers, slit samplers and cyclone samplers, are usually more efficient at capturing large particles. Despite recent advancements, cut-off sizes (particle diameter with 50% collection efficiency) for cyclones (mostly > 1 μm) cannot meet the sampling requirements for small sized virus-containing particles. Moreover, the cyclone action can damage and deactivate viruses, resulting in an underestimate of the infectious viruses collected.
Liquid impactors are liquid impingers using different liquid solutions such as sterile distilled water, physiological saline, phosphate-buffered saline, nutrient broth, peptone water, or mineral oil to collect particles. All-Glass Impingers (AGI) samplers, characterized by a critical flow orifice, accelerating the air passing through it to sonic velocity, are the most often used samplers for the capture of airborne viruses; the formation of air bubbles in the liquid phase can improve the collection of small particles through diffusion but part of the sample could be lost because of transport of droplets towards the extraction pump or particles re-aerosolization, thus limiting sampling durations. Nevertheless, collection through liquid media prevents desiccation and allows the preservation of the infectivity of the sample. A “swirling aerosol collector” with three tangential sonic nozzles causing a swirling motion of the liquid phase has been developed to make the sampling procedure less violent and destructive than with traditional AGI sampler. The tangential components of the aerosol jets produce swirling air motion inside the device that rotates the liquid and swirls it up the inner wall thus wetting the region where the aerosol jets meet the inner wall. This way, swirling aerosol collectors can improve collection efficiency of particles smaller than 1 μm up to 80% at 0.3 μm, as highlighted by Willeke et al. (1998); moreover, this kind of sampler is suitable to high-viscosity non-evaporating collection fluid, such as white mineral oil, thus permitting longer sampling times, up to 8 h (Lin et al., 1999).
On the other hand, filters are the most effective device to capture micrometric (or smaller) particles; they could be made of cellulose, polycarbonate, glass-fibers, quartz fibers, polytetrafluoroethylene (PTFE). The above-cited literature reports that 0.3 μm PTFE filters appear to be the best option for long-term sampling of 10–900 nm diameter virus-laden particles.
Filters are very efficient, but literature reports that they can cause desiccation of the sample, likely compromising virus infectivity (Verrault et al., 2008). Modern molecular biology techniques can detect both viral genome and antigens, so they do not require viral particle integrity to be maintained to detect viruses; however, prolonged filtration could also damage and reduce the amount of detectable genetic material, making its final concentration assessment less reliable. Moreover, in order to be further analyzed, the genetic material captured by filters must be eluted by a suitable solution.
A partial solution could be represented by gelatine filters, because they are very efficient while they do not appear to significantly affect viral infectivity: they can be dissolved into liquid for molecular or virus enumeration in cell cultures without significantly affecting the viability of many viruses. Some authors report that low humidity can cause them to dry and break, while high humidity can cause dissolution of the filter; therefore, the sampling duration is usually quite short, around 15 min (Pan et al., 2019). On the contrary, studies of some manufacturer demonstrated that gelatine membranes could be qualified for continuous air monitoring in industrial pharmaceutical production environments covering a whole 8 h work shift without the need for human intervention (Scherwing et al., 2019).
Among the described bioaerosol sampling technologies, the most suitable should be chosen to match the following minimal requirements, defined also in a previous study produced by our research group (Robotto et al., 2021):
-
1.
Large volumes of air should be sampled in consideration of the expected dilution in environmental media;
-
2.
High bioaerosol capture efficiency, for fractions around or smaller than 1 μm;
-
3.
Preservation of sample infectivity, to allow viral replication in vitro on susceptible cell cultures;
-
4.
Temperatures should be not hostile to the pathogen throughout the sampling process and the sample transport and manipulation;
-
5.
Low degradation or re-aerosolization, bounce, inlet and wall losses in samplers;
-
6.
Standardized procedures and methods for sampling airborne viruses and enable measurement of the detection limit of the measurement should be established;
-
7.
Use of optimal media for suspension or collection;
-
8.
Sample pre-treatment should be optimized (elution from filters, concentration of transport medium, RNA extraction).
Therefore, a conceptual approach like the one described by Robotto et al. (2021) should be developed to allow the choice of suitable collection materials, sampling duration, transport medium and sample pre-treatments with a reliable and reproducible plan. Recovery tests represent a necessary step to validate an analytical method, assuring the right quality to environmental sampling and analysis according to ISO/IEC 17025 standard.
To address these points, in the present study we provide evidence of the feasibility and reproducibility of the approaches described in our previous paper.
2. Materials and methods
2.1. Sampling devices and filters
In the last 10 months our research group formed by the Environmental Protection Agency of Piedmont (Arpa Piemonte) and Università degli Studi di Torino, Dept. of Clinical and Biological Science, Azienda Ospedaliero-Universitaria San Luigi Gonzaga, focused on the use of 3 different sampling devices:
-
1)
a low-volume sampler for air filtration on PTFE filters, with a flow rate of 20 L/min;
-
2)
a high-volume sampler for air filtration on glass-fiber or quartz filters, with a flow rate of 500 L/min;
-
3)
a swirling aerosol collector (BioSampler, SKC Inc., Eighty-Four, PA, USA) accelerating the flow of aspirated air to sonic speed, minimizing losses due to evaporation, maintaining the infectivity and integrity of the viral particles by transferring them directly to a suitable transport solution (phosphate buffered saline (PBS), Dulbecco's Modified Eagle Medium (DMEM, Sigma, St. Louis, MO, USA) or non-aqueous fluids), sampling a flow rate of 12.5 L/min.
Concerning air filtration, we employed:
-
•
PTFE filters (0.2-μm pore size, diameter of 4.7 cm, Sartorius AG, Göttingen, Germany)
-
•
glass-fiber filters (grade MG G, 1.5-μm pore size, diameter of 4.7 cm and 10 cm, Munktell Filter AB, Falun, Sweden)
2.2. Cell lines and viruses
Human lung fibroblast MRC-5 (ATCC® CCL-171) were propagated in DMEM (Sigma, St. Louis, MO, USA) supplemented with 1% (v/v) penicillin/streptomycin solution (Euroclone, Milan, Italy) and heat inactivated, 10% (v/v) fetal bovine serum (Sigma). Human coronavirus strain OC43 (HCoV-OC43) (ATCC® VR-1558) was purchased from ATCC (American Type Culture Collection, Rockville, MD, USA) and propagated in MRC-5 cells, at 34 °C, in a humidified 5% CO2 incubator. When the full cytopathic effect (CPE) developed, cells and supernatants were harvested, pooled, frozen, and thawed three times, then clarified and aliquoted. The virus was stored at -80 °C. HCoV-OC43 titers were determined by the indirect immunoperoxidase staining procedure. Briefly, MRC-5 cells were seeded 2 days before infection in 96-well plates, reaching 60%–70% confluence at the time of infection. The viral suspension was serially diluted in DMEM supplemented with 2% fetal bovine serum and inoculated; the infected wells were incubated at 34 °C for 24 h, allowing viruses to enter the cells and replicate. After this time, cells were washed with medium, and fixed with cold acetone-methanol (50:50). Cells were then permeabilized with Triton X-100 0.1% in PBS, and incubated with an OC43-specific monoclonal antibody (MAB9013; Merck Life Science Srl, Milan, Italy). After three quick washes with PBS, the secondary antibody peroxidase-conjugated AffiniPure F(ab’)2 Fragment Goat Anti-Mouse IgG (H + L) (Jackson ImmunoResearch Laboratories Inc., 872 W. Baltimore Pike, West Grove, PA, 19390) was added. Finally, after three more washes with PBS, 3, 3′ diaminobenzidine tetrahydrochloride (DAB Substrate; 11,718,096,001, Merck Life Science Srl, Milan, Italy) was added. It is one of the major sensitive substrates for HRP (horseradish peroxidase): the reaction product is brown in color and insoluble, thereby precipitating over the antigen-antibody site and staining infected cells. Viral titers are expressed as focus-forming unit (FFU) per ml.
2.3. Virus-filter elution procedures
2.3.1. Glass-fiber filters
A fixed inoculum of HCoV-OC43 suspension (50 μl) was spotted on glass-fiber filters, alternatively of 4.7 cm or 10 cm diameter. Different procedures were tested to develop a method to elute viral particles collected on the filter. Three different approaches were explored, namely (A) a washing procedure, (B) a vortexing procedure, and (C) a shaking procedure.
Concerning the washing procedure, 4 ml of DMEM supplemented with 2%, 10%, or 20% fetal bovine serum was added to the filter “spotted” and placed in a Petri dish. Alternatively, PBS at pH 3.3, 7.4, or 10 were used as eluent buffer. The Petri dish was then placed on a tilting plate (therefore, under stirring) for 30 min at room temperature. The residual volume of eluent (i.e., not absorbed by the filter) was recovered from the plate, and the contained viral titer was evaluated as described above. Moreover, in order to extract the elution buffer still absorbed in the filter at the end of the process, the procedure was integrated with a further step: after having recovered the residual volume of buffer, the filter was taken with sterile forceps, placed in a 50 ml sterile syringe and subjected to pressure, so as to release the volume of liquid still adsorbed in the filter. This was added to the elution buffer collected from the plate and the viral titer was evaluated.
Regarding the vortexing procedure, the spotted filter was transferred to a sterile 50 ml tube and 4 ml of DMEM supplemented with 2%, 10%, or 20% fetal bovine serum was added. The tube was vortexed for 30 s at room temperature, and finally centrifuged at 1200 rpm at room temperature for 10 min. The residual volume of eluent was then collected and the viral titer was assessed as described above.
As far as the shaking procedure is concerned, the spotted filter was transferred to a sterile 50 ml tube and 8 ml of DMEM supplemented with 10%, 20%, or 40% fetal bovine serum was added. The test tube was placed on an orbital shaker and stirred for 60 min or 180 min at about 150 revolutions per minute. The residual volume of eluent was then collected and the viral titer contained was evaluated as described above. Alternatively, a combination of the shaking and vortexing procedure was tested, by vortexing the shaken sample for 30 s, then spinning it at 1200 rpm at room temperature for 10 min and recovering the obtained supernatant.
For each single experiment, the viral inoculum itself was titrated and percentages of infective particles recovery were calculated according to the following formula: % of recovery = (FFU recovered x 100)/FFU inoculated.
Tests were also carried out to optimize storage times and temperatures (room temperature, 4 °C, -20 °C, -80 °C) of the spotted filters before they were subjected to elution techniques.
2.3.2. PTFE filters
A fixed inoculum of HCoV-OC43 suspension (50 μl) was spotted on PTFE filters of 4.7 cm diameter. The shaking procedure was selected to elute the viral particles entrapped in the filters, with some modifications: briefly, the spotted filter was transferred to a sterile 50 ml tube and 8 ml of DMEM supplemented with 10% fetal bovine serum was added. The test tube was placed on an orbital shaker and stirred for 60 min at about 150 revolutions per minute. The residual volume of eluent was then collected, and the viral titer contained was evaluated as described above. For each single experiment, the viral inoculum itself was titrated and percentages of infective particles recovery were calculated according to the following formula: % of recovery = (FFU recovered x 100)/FFU inoculated.
Tests were also carried out to optimize storage times and temperatures (room temperature, 4 °C, -20 °C, -80 °C) of the spotted filters before they were subjected to elution techniques.
2.4. Recovery tests with aspiration
2.4.1. Glass-fiber filters
A fixed inoculum of HCoV-OC43 suspension (50 μl) was spotted on glass-fiber filters of 10 cm diameter and placed on a sampling head (the laboratory setup is showed in Fig. 1 ). The spotted filters were aspirated for different times (namely, 1 min, 5 min, 10 min, 15 min, and 20 min) at a flow rate of 500 L of air per minute. The shaking procedure integrated with the vortexing step was selected to elute the viral particles entrapped in the filters. The residual volume of eluent was then collected, and the viral titer contained was evaluated as described above.
Fig. 1.
Experimental setting for recovery tests with aspiration in biosafety level 2 (BSL-2) conditions. A fixed inoculum of HCoV-OC43 suspension (50 μl) was spotted on glass-fiber filters of 10 cm diameter and placed on a sampling head (A), and a high-volume sampler for air filtration was used (B).
2.4.2. PTFE filters
A fixed inoculum of HCoV-OC43 suspension (50 μl) was spotted on PTFE filters of 4.7 cm diameter and placed on a sampling head. The spotted filters were aspirated for different times (namely, 5 min, 15 min, 60 min, 180 min, and 900 min) at a flow rate of 20 L of air per minute. The shaking procedure was selected to elute the viral particles entrapped in the filters, with some modifications: briefly, the spotted filter was transferred to a sterile 50 ml tube and 8 ml of DMEM supplemented with 10% fetal bovine serum was added. The test tube was placed on an orbital shaker and stirred for 60 min at about 150 revolutions per minute. The residual volume of eluent was then collected, and the viral titer contained was evaluated as described above.
2.5. Virus inactivation assays
In order to evaluate the possible effect of filter materials or their components eventually released during the elution process, viral inactivation assays were performed. Briefly, both glass-fiber filters and PTFE filters were subjected to the shaking protocol or the combined shaking and vortexing protocol as described above. Subsequently, the residual volume of eluent was inoculated with approximately 105 FFU of HCoV-OC43 and incubated at room temperature for 3 h or 1 h, respectively for glass-fiber filters and PTFE filters. Control samples were prepared by inoculating the same inoculum of fresh eluent. After the incubation, the samples were titrated, and the residual viral infectivity was determined by indirect immunostaining as described above.
2.6. SARS-CoV-2 reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The samples are extracted in single with the MagMAX ™ Viral/Pathogen Nucleic Acid Isolation Kit (ThermoFisher) protocol. 200 μl of each sample were resuspended in 265 μl of inactivating solution (binding solution), then the magnetic beads and proteinase K are added. The extraction procedure is carried out automatically using the King Fisher Flex instrumentation. At the end of the extraction process, the RNA extracted from the samples is resuspended in 50 μl of elution solution.
The eluate obtained from the previous step are analyzed in duplicate by multiplex PCR using the SARS-CoV-2 ELITe MGB Kit (ElitechGroup). The targets are RdRp and ORF8 genes specific for SARS-CoV-2 and RNase P gene as endogenous Internal Control. The volume of sample loaded into PCR is 10 μl. The instrument used for PCR analysis is QuantStudio 5 Real-Time PCR (Applied Biosystems).
Data processing is carried out following the instructions of the PCR kit, setting the Thresholds for each individual gene and evaluating the presence of suitable PCR curves.
The results are expressed with the CT values (Cycle Threshold) for each detected target. In case of absence of amplification (absence of the desired target) the result is reported as Ct > 40. RT-qPCR assay limit of detection (LoD) can be conservatively defined as 3 genome copies per reaction, that is around 75 RNA copies per mL of transport medium.
The quantification curves have been determined by means of a SARS-CoV-2 RNA standard (LGC Standards): the RNA concentration of the transport medium (copies/mL) can be calculated, on average, as:
-
•
for ORF8 gene and
-
•
for RdRp gene.
2.7. HCoV-OC43 reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The samples are extracted in single with the MagMAX ™ Viral/Pathogen Nucleic Acid Isolation Kit (ThermoFisher) protocol. 200 μl of each sample were resuspended in 265 μl of the inactivating solution (binding solution), then the magnetic beads and proteinase K are added. The extraction procedure is carried out automatically using the King Fisher Flex instrumentation. At the end of the extraction process, the RNA extracted from the samples is resuspended in 50 μl of elution solution.
The eluate obtained from the previous step is analyzed in single by multiplex PCR using the VIASURE Coronavirus 229E, NL63, OC43 & HKU1 Real Time PCR Detection Kit (VIASURE). The target is a specific sequence for OC43. The volume of sample loaded into PCR is 5 μl. The CFX96 Touch Real-Time PCR (Bio-Rad) instrument was used for PCR analysis. Data processing was carried out by setting the Threshold above the non-specific background noise and evaluating the presence of suitable PCR curves.
2.8. Sampling tests in real-life setting
Based on the described sampling techniques and analytical methods, many field tests have been carried out involving indoor environments such as private houses, hospitals and means of transport. In particular we generally applied the biosampler device sampling 12.5 L/min of air from 30 to 90 min, a low volume pump sampling 20 L/min on 4.7 cm PTFE filters for 60 min and a high-volume device sampling 500 L/min on 10 cm glass-fiber filters for 20 min. These sampling techniques have been applied both singularly and contemporarily.
After the sampling phase, filters are immersed in 10 ml DMEM and transported to the laboratory at a temperature about 4 °C. The transport normally takes less than 2 h since the end of the sampling.
Once arrived at the laboratory, the transport medium is supplemented with a volume of FBS from 10 to 40% of the final volume. Samples are then subjected to the combined shaking-vortexing protocol as described above. The eluate (13 ml) is ultracentrifuged by using an ultracentrifuge Optima LE-80K (Beckman Coulter Life Science) for 1 h at 150000g; then, the supernatant is discarded and the pelleted virus is concentrated in 0.3 ml of autoclave-sterilized PBS. The presence of SARS-CoV-2 genomic RNA is then assessed by PCR.
Proof-of-concept experiments were performed to validate this approach by measuring the recovery efficiency and the capacity of ultracentrifugation to concentrate the eluted coronavirus. Briefly, 500,000 FFU of HCoV-OC43 were inoculated in 13 ml of DMEM supplemented with 40% FBS, then subjected to ultracentrifuge and titrated as previously described. For each single experiment, the viral inoculum itself was titrated and percentage of infective particles recovered after ultracentrifugation was calculated according to the following formula: % of recovery = (FFU recovered x 100)/FFU inoculated.
2.9. Statistical analysis
Where possible, descriptive statistic values (i.e. mean, median, and 95% confidence interval (CI)) were calculated with GraphPad PRISM 7 software (GraphPad Software, San Diego, CA, USA). Where appropriate, regression analysis was performed to assess a correlation between detected CT resulting from Real Time PCR and FFU/mL obtained by in vitro titration, using GraphPad PRISM 7. The square of the correlation coefficient (R2) was used to evaluate the linear regression. One-way ANOVA, followed by Bonferroni test, was used to assess the statistical significance of the differences between treated and untreated samples, where appropriate. Significance threshold was set at 95% level.
3. Results
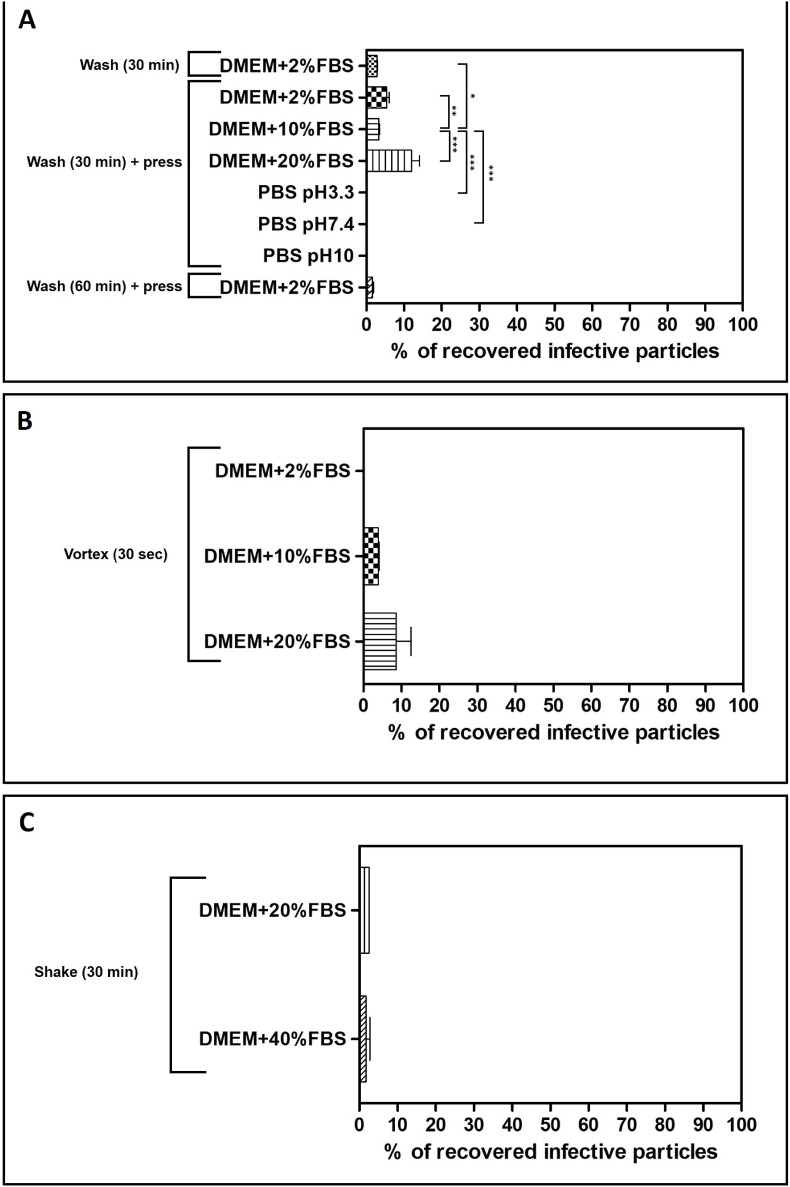
The results of the washing elution protocol on glass-fiber filters are presented in Fig. 2 A. DMEM supplemented with 2% of FBS was firstly chosen as the eluent since it is the medium canonically used to culture HCoV-OC43 on MRC-5 cells. For this setting, the percentage of recovery of viral particles was very modest, about 2.7%. Since glass-fiber filters are highly absorbent, the protocol was integrated with a further step to recover the eluent soaked in the filter by pressing it inside a sterile syringe. The addition of this step in the procedure partially increased the percentage of virus recovered (5.4%), yet not statistically significant (pANOVA > 0.05). Extending the incubation time of the filter with the eluent up to 60 min resulted in a low recovery (1.6%), probably because of a loss of viral titer due to excessive incubation of the virus at room temperature. As depicted, the use of PBS at different pHs as eluent buffer did not allow the recovery of infectious viral particles. On the contrary, the percentage of recovered viral particles was increased to 12%, when DMEM supplemented with 20% FBS was used as eluent buffer, a result significantly different if compared to the ones obtained with lower percentages of serum (0.01<pANOVA < 0.05) or PBS (pANOVA < 0.001).
Fig. 2.
Evaluation of the extraction efficiency of infective HCoV-OC43 particles from 4.7 cm glass-fiber filters, accordingly to the wash procedure (panel A), vortexing procedure (panel B), or the shaking procedure (C). The titers of filter-recovered HCoV-OC43 are expressed as a percentage of the titer of the inoculated virus (% of recovery = (FFU recovered x 100)/FFU inoculated). Error bars represent the standard error of the mean (SEM) of independent experiments. *pANOVA < 0.05; **pANOVA < 0.01; ***pANOVA < 0.001.
In order to evaluate the effect of a mechanical stimulus on the adhesiveness of viral particles trapped in the glass-fiber filters, two different approaches were tested: a first procedure based on vortexing the filter together with the eluent, and a second one based on shaking them on an orbital shaker. The results of the “vortex” protocol are depicted in Fig. 2B: while they are generally similar to the ones of the “wash” protocol, they clearly indicate that the increase of FBS in the eluent improves the release of the entrapped infective HCoV-OC43 particles (ranging from 3.6% to 8.6%, respectively for DMEM+10% FBS and DMEM+20% FBS). The shaking protocol resulted in even lower recovery efficiencies (Fig. 2C), even increasing the percentage of FBS supplemented in the eluent (2.6% of recovery and 1.8% of recovery respectively for DMEM+20% FBS and DMEM+40% FBS).
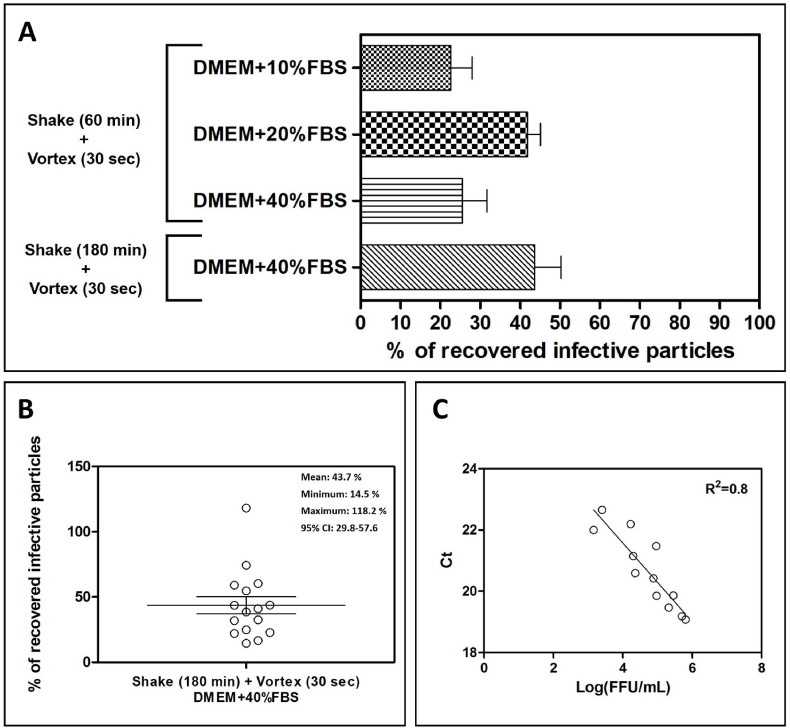
Therefore, we tested a combined protocol where a shaking step of 60 min was followed by a vortexing step of 30 s. The results (depicted in Fig. 3 A) show a relevant improvement if compared with the previously tested protocols, resulting in HCoV-OC43 infective particles recovery of 22.7%, 41.9%, and 25.5%, respectively for DMEM+10% FBS, DMEM+20% FBS, and DMEM+40% FBS. A further improvement was obtained by increasing the shaking time of the glass-fiber filter together with the eluent to 180 min (eluent: DMEM+40% FBS): according to the statistical analysis of the results obtained in each single independent experiment (n = 16; Fig. 3B), an average recovery of 43.7% can be obtained with this protocol, with a 95% CI between 29.8% and 57.6%. Each sample eluted following this protocol, in addition to the titration, was tested by means of PCR to determine the presence of the genetic material of HCoV-OC43; a linear regression analysis was performed in order to test the correlation between the two endpoints. As depicted in Fig. 2C, the titer of eluted infectious HCoV-OC43 particles correlates by direct proportionality (R2 = 0.8) with the Threshold Cycle measured by real time PCR.
Fig. 3.
Evaluation of the extraction efficiency of infective HCoV-OC43 particles from 4.7 cm glass-fiber filters when using a combined shaking and vortexing procedure. In panel A, the titers of filter-recovered HCoV-OC43 are expressed as a percentage of the titer of the inoculated virus (% of recovery = (FFU recovered x 100)/FFU inoculated). Error bars represent the SEM of independent experiments. In panel B, the dispersion of the different percentages of recovery obtained with the shaking-vortexing and using DMEM + 40%FBS are depicted; the horizontal line represents the mean value of 16 independent experiments, while the error bars represent the SEM of independent experiments. In panel C, a linear regression analysis correlates for each eluate obtained with the combined shaking and vortexing procedure (eluent: DMEM+40%FBS), the FFU recovered with the respective threshold cycles (Ct) measured with the quantitative real-time RT-PCR. 95% CI: 95% confidence interval; R2 = square of the correlation coefficient.
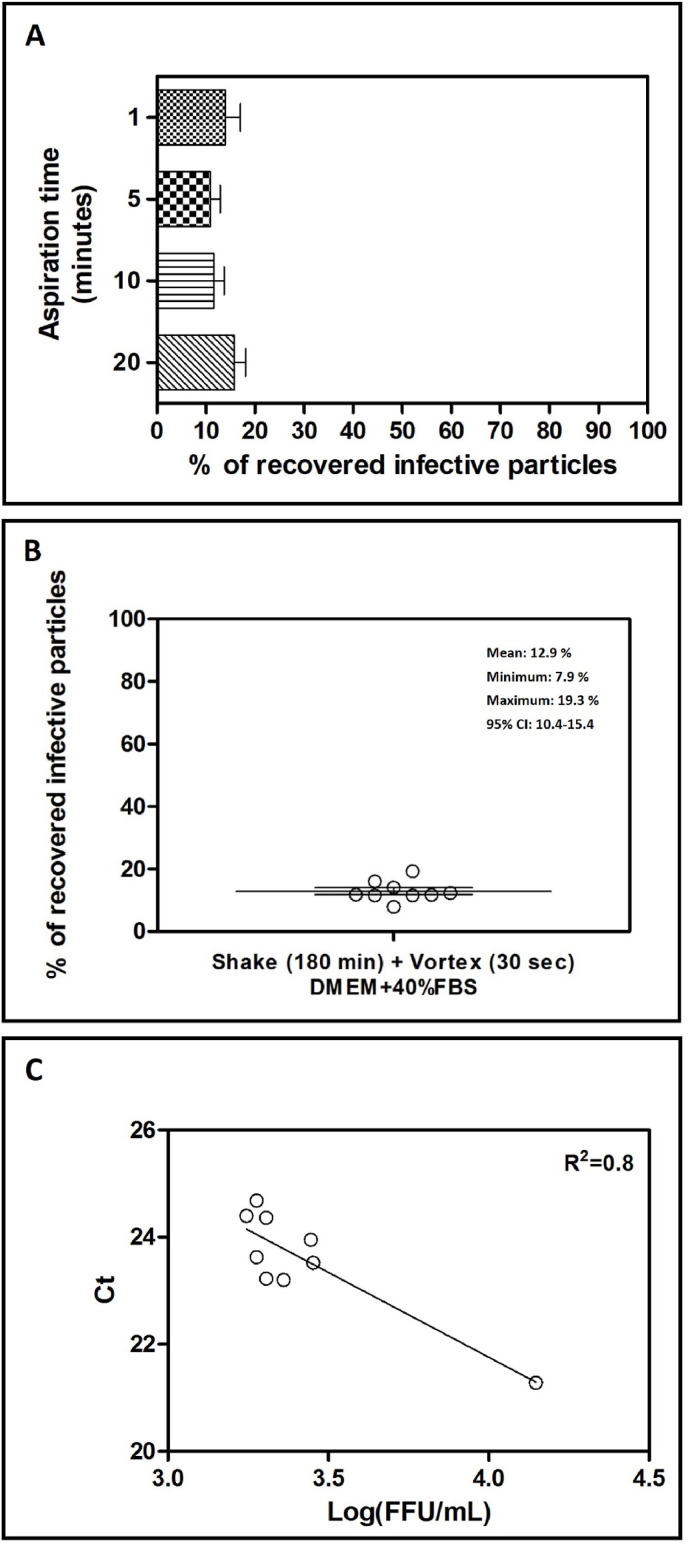
Once a reliable elution method had been developed for glass-fiber filters, we tested the effect of aspiration on the viral titer recovered from filters. This set of experiments was performed both on glass-fiber filters and PTFE filters. As depicted in Fig. 4 A, recovery from glass-fiber filters ranges from a minimum of 4.7% to a maximum of 19.3%, independently from the time of aspiration, thus excluding a virucidal effect on the population of particles trapped in the filter, for example due to desiccation. According to the statistical analysis of the percentages obtained in each single independent experiment (n = 18; Fig. 4B), an average recovery of 12.9% can be obtained with this protocol, with a 95% CI between 10.4% and 15.4%. Moreover, as shown in Fig. 4C, real time PCR detected the presence of HCoV-OC43 genome in each of the tested sample, showing a direct correlation of Threshold Cycles assessed and viral titers measured.
Fig. 4.
Evaluation of the extraction efficiency of infective HCoV-OC43 particles from 10 cm (panel A) glass-fiber filters when using a combined shaking and vortexing procedure. In both panels, the titers of filter-recovered HCoV-OC43 are expressed as a percentage of the titer of the inoculated virus (% of recovery = (FFU recovered × 100)/FFU inoculated), and error bars represent the SEM of independent experiments. In panel B, the dispersion of the different percentages of recovery obtained with the shaking-vortexing and using DMEM + 40%FBS (10 cm glass-fiber filters) are depicted; the horizontal line represents the mean value of 16 independent experiments, while the error bars represent the SEM of independent experiments. In panel C, a linear regression analysis correlates for each eluate obtained with the combined shaking and vortexing procedure (10 cm glass-fiber filter; eluent: DMEM + 40%FBS), the FFU recovered with the respective threshold cycles (Ct) measured with the RT-PCR. 95% CI: 95% confidence interval; R2 = square of the correlation coefficient.
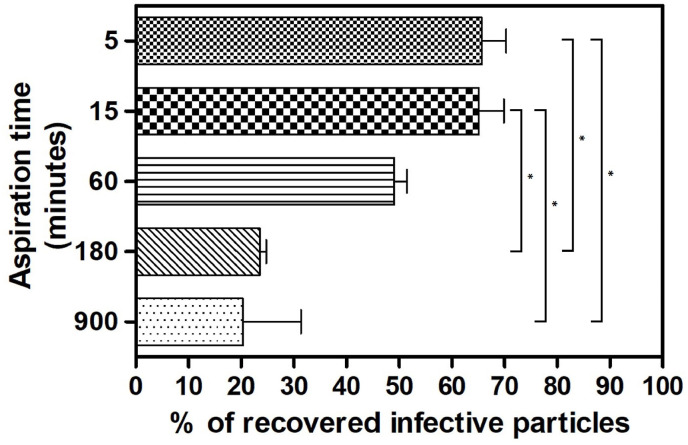
The shaking protocol, with appropriate modifications (60 min at 150 revolutions per minute), was used to elute viral particles also from PTFE filters; in this set of experiments the effect of longer aspiration times on entrapped infective particles was tested. As shown in Fig. 5 , the percentage of eluted HCoV-OC43 particles is inversely related to the aspiration time, ranging from a maximum of 65.7% at 5 min of aspiration to a minimum of 20.4% at 900 min of aspiration.
Fig. 5.
Evaluation of the extraction efficiency of infective HCoV-OC43 particles from 4.7 cm PTFE filters when using the shaking procedure. The titers of filter-recovered HCoV-OC43 are expressed as a percentage of the titer of the inoculated virus (% of recovery = (FFU recovered × 100)/FFU inoculated), and error bars represent the SEM of independent experiments. *pANOVA < 0.05.
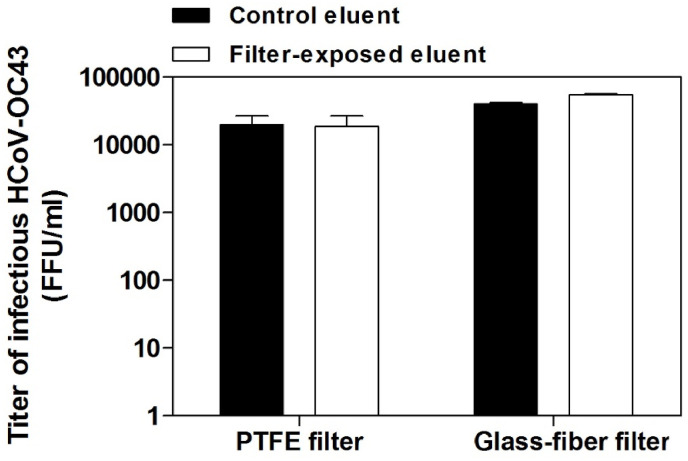
We performed viral inactivation assays to exclude that the filters subjected to elution processes released into the eluent substances that could inactivate HCoV-OC43 particles. The results shown in Fig. 6 demonstrate that HCoV-OC43 does not lose infectivity when it is inoculated in a known volume of eluent, which was previously put in contact with the glass-fiber or PTFE filters and subsequently subjected to all the steps of the elution process.
Fig. 6.
Virus inactivation assay. The white bars refer to eluent samples inoculated with HCoV-OC43 and previously exposed to PTFE or glass-fiber filters and subjected to the shake-vortex procedure. Control samples (black bars) were prepared by inoculating the same inoculum of fresh eluent. On Y axis, the titers of HCoV-OC43 are expressed as FFU/ml. Error bars represent the SEM of independent experiments.
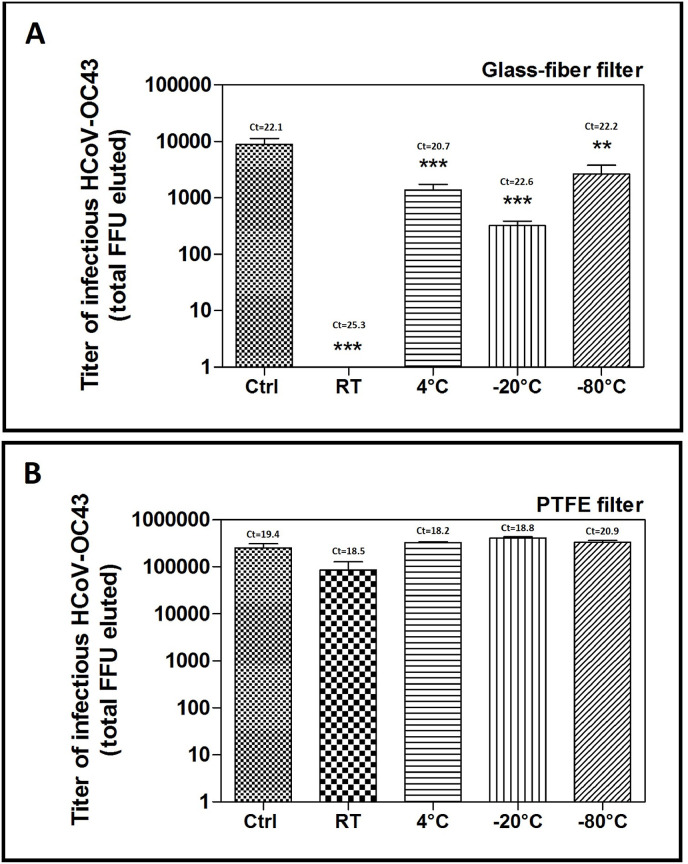
Furthermore, we assessed the effect of the storage temperature of the filters immersed in the transport medium on the infectivity of HCoV-OC43. Briefly, we inoculated the glass-fiber filters or PTFE filters with HCoV-OC43, then immersed them in the elution buffer and stored them for 24 h at room temperature, 4 °C, -20 °C, or -80 °C. As shown in Fig. 7 , storing the glass-fiber filters (panel A) at room temperature significantly (pANOVA < 0.001) reduced HCoV-OC43 infectivity to undetectable levels. On the contrary, the viral genome was still detectable in all samples, as assessed by PCR, regardless of the storage temperature. We obtained similar results with the PTFE filters (Fig. 7B), although the decrease in viral titer in samples stored at room temperature was considerably lower and not significant (pANOVA > 0.05).
Fig. 7.
Effect of filter storage temperature on the extraction efficiency of infective HCoV-OC43 particles from 4.7 cm glass-fiber (A) or PTFE (B) filters. On Y axis, the titers of eluted HCoV-OC43 are expressed as total extracted FFU when using the combined shaking-vortexing procedure, and error bars represent the SEM of independent experiments. At the top of each bar, threshold cycle (Ct) values measured with RT-PCR are indicated for each sample.
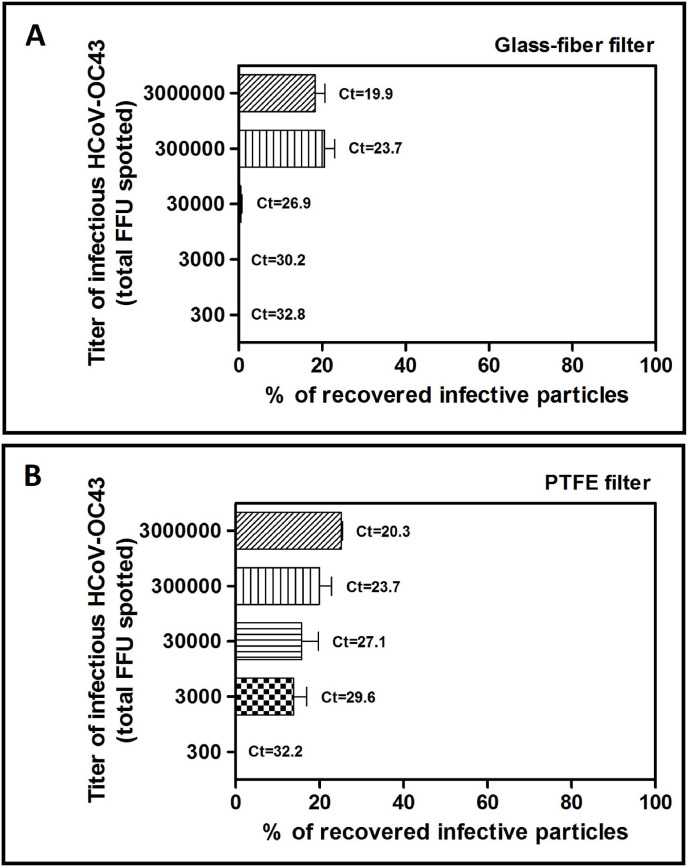
Finally, we evaluated whether the titer of the inoculum spotted on the glass-fiber or PTFE filters affected the recovery percentage. As shown in Fig. 8 A, the recovery for glass-fiber filters ranges between 18% and 21%, respectively for 3,000,000 or 300,000 spotted FFU, it decreases considerably up to 0.5% if the inoculum is equal to 30,000 FFU, while it is equal to 0% for inocula equal to 3000 or 300 FFU. We obtained similar results with PTFE filters (Fig. 8B), demonstrating that the recovery decreases with direct proportionality with respect to the viral inoculum, from a maximum of 25% (inoculum: 3,000,000 FFU) to a minimum of 14% (inoculum: 3000 FFU); in the sample inoculated with 300 FFU the recovery was equal to 0%. In addition, the eluate samples were subjected to PCR and we demonstrated that the viral genetic material was detectable even in samples where the infecting units of the virus (i.e., the FFUs) were not detectable.
Fig. 8.
Evaluation of the extraction efficiency of infective HCoV-OC43 particles from 4.7 cm glass-fiber (A) or PTFE filters (B) as a function of viral inoculum. On Y axis, the titers of infective HCoV-OC43 inoculated on filters are expressed as total FFU. The titers of filter-recovered HCoV-OC43 are expressed as a percentage of the titer of the inoculated virus (% of recovery = (FFU recovered x 100)/FFU inoculated), and error bars represent the SEM of independent experiments. At the top of each bar, threshold cycle (Ct) values measured with RT-PCR are indicated for each sample.
According to the proof-of-concept experiments performed with HCoV-OC43, the applied ultracentrifugation step can concentrate the eluted viral suspension to maxima of 15 times (data not shown), while the recovery efficiency of ultracentrifugation (calculated by comparing the number of total FFU diluted in the initial volume before ultracentrifugation and the number of FFU recovered in the pellet after ultracentrifugation) is 57% (data not shown). These results are particularly promising if compared with recovery efficiencies reported by Ahmed et al. (2020), who tested the efficiency of seven wastewater virus concentration methods (adsorption-extraction with three different pre-treatment options, centrifugal filter device methods with two different devices, polyethylene glycol (PEG 8000) precipitation, and ultracentrifugation) on murine hepatitis virus (MHV), finding mean recoveries ranging from 26.7 to 65.7%.
As far as sampling tests are concerned, we reported in Table 1 the results of 20 samples taken in a private house where a positive person has been isolated for more than 3 weeks. The table highlights that in the first phase of the disease indoor concentration with poor ventilation conditions could be higher than 40 RNA copies/m3 of air; three different sampling devices, namely a biosampler run for 90 min at 12.5 L/min, a PTFE sampler run for 60 min at 20 L/min and a glass-fiber filter high volume sampler run for 20 min at 500 L/min, could give comparable results.
Table 1.
SARS-CoV-2 air concentration, expressed as RNA copies per m3 of air, measured in a private house where a positive person has been isolated.
| days from the 1st test | Sampling methods (sampling duration, air volume sampled) |
Sampling place (room, volume, n° of patients) | ||
|---|---|---|---|---|
| PTFE (60min, 1 m3) |
Glass fiber (20min, 10 m3) |
Biosampler (90 min, 1.1 m3) |
||
| Copies/m3, mean values (n° of samples) | Copies/m3, mean values (n° of samples) | Copies/m3, mean values (n° of samples) | ||
| 3 | 31.1 (3) | 42.1 (1) | – | Office, 33 m3, 1 patient inside |
| 6 | – | – | 39.0 (1) | Bedroom, 55 m3, 1 patient inside |
| 8 | – | 17.5 (5) | – | Office, 33 m3, 1 patient inside |
| 9 | – | 4.9 (2) | – | Bedroom, 55 m3, 1 patient inside |
| 20 | Not detectable (1) | 8.1 (2) | Not detectable (1) | Bedroom, 55 m3, 1 patient inside |
4. Discussion
The aim of this study is to develop a specific and repeatable method for the detection of coronavirus infectious particles in ambient air, by exploiting the devices and techniques currently used for sampling air in the external environment as long as indoor. In particular, among the various devices that can be used to sample bioaerosol containing viruses, we focused on the use of filters because they appear to be the most effective device both to capture micrometric (or smaller) particles and collect large air volumes at the same time.
The use of air filtration allows the recovery by aspiration of the viral particles present in the air, and their consequent trapping inside the filters, thus requiring a subsequent elution step to extract the virus from the solid matrix in which it is sequestered. To this end, we evaluated different types of eluents; based on our data, the addition of FBS to the elution buffer is essential to trigger a more conspicuous release of viral particles. We showed that HCoV-OC43 infective particles can be recovered both from PTFE and glass-fiber filters, and that the use of these materials does not inactivate viral particles. We identified DMEM supplemented with 40% FBS as the best eluent for glass-fiber filters, and DMEM supplemented with 10% FBS as the most suitable one for PTFE filters.
While PTFE filters grant the highest viral recovery with the less complex procedures, glass-fiber filters are more suitable for sampling procedures in a real-life setting because they enable the recovery of a much higher volume of air if compared to PTFE filters: for example, the use of a high-volume sampler filtering on glass-fiber filters allows the sampling of 10 cubic meter of air in 20 min, whereas 1 h of sampling with PTFE filters collects 1 cubic meter of air.
Regarding glass-fiber filters, we found that a two-step procedure is necessary to elute viral infective particles: a 3 h-shaking step, followed by a 30 s-vortexing step. For PTFE filters, 60 min shaking procedure is adequate.
We have shown that a simple mechanical stimulus (pressure or agitation) is not sufficient to release the viral particles trapped in the glass-fiber filters but that a specific combination of mechanical stimuli and a suitable eluent is required to facilitate their release. While, on the one hand, this aspect limits the amount of HCoV-OC43 particles that can be recovered, on the other hand it demonstrates that virions show a tenacious adhesiveness towards fiberglass: that is to say that these filters are particularly suitable for capturing and trapping infectious particles sampled from the air.
Moreover, we demonstrated that the use of filters to capture bioaerosol is very efficient and, as far as the sampling duration is lower than 60 min, the expected inactivation of coronavirus is not drastic.
Table 1 reports SARS-CoV-2 air concentration trends inside a private house, along with the development of the disease of the owner (more or less, 3 weeks, from the first positive test). These data highlight a clear decreasing trend of air concentration since the first days of the disease, that means that airborne virus tends to reflect viral load trends of patients. At the same time, it is possible to hypothesize that sampling on glass-fiber filters at very high flow rates make analytical sensitivity higher than other sampling techniques.
It is important to underline that PTFE filters are made up of a porous membrane with a complex structure with tortuous routes through the filter material, while glass-fiber filters are fibrous filters consisting of a deep mesh of fibers with random orientations.
As a matter of facts, aerosol filtration is far more complex than a simple “sieve” model as it is based on 5 different mechanism, namely interception, impaction, diffusion (Brownian motion), electrostatic attraction, and sedimentation. Therefore, aerosol filters can efficiently collect particles much smaller than would be expected based on the pore size of the filter.
PTFE filters could reach very high collection efficiency, but they are generally characterized by high pressure drops (only low flow rates can be realized). On the contrary, the high porosity of glass-fiber filters allows very high flow rates to be aspired and good collection efficiencies at the same time.
It should be remembered that recovery efficiency of the genetic materials coming from collected SARS-CoV-2 particles would be surely higher that the reported data on infectious virus.
4.1. Real-life sampling test results
On the basis of the sampling techniques and the analytical methods developed as described in the previous chapters, many field tests have been carried out involving COVID-19 hospital wards, private houses of infected families, outdoor air in the center of a 1 million people city.
We started our sampling activities focusing on hospital rooms where 2 to 3 COVID-19 patients were present: in these spaces, with generally high mechanical ventilation rates (6–12 air changes per hour), the detectable concentrations of SARS-CoV-2 were very low, lower than 10 genomic copies per cubic meter, even when patients with high viral loads (i.e. Cycle Thresholds <20) were hospitalized or aerosol-generating procedures (intubation, noninvasive positive pressure ventilation, sputum induction, high-flow oxygen) take place. Air samplings have been carried out within Intensive Care Units as well, in order to maximize places at disposal without diminishing safety levels for healthcare operators. When air exchange rates respect the standard requirements (in Italy 2 to 6 air changes per hour are recommended for hospital wards) and air inlet/outlet openings are correctly designed, realized and operated, SARS-CoV-2 concentrations are often below the LoD.
In domestic environment, on the contrary, where single patients or entire families are COVID-19 positive, virus concentrations were found to be more consistent, up to 40 ÷ 50 genomic copies per cubic meter of air. These values are strongly influenced by air changes frequencies and the number of positive subjects present in the house, as well as by the development of the most common symptoms of the disease (dry cough), the stage of the disease, the initial viral load.
Air sampling campaigns applying the validated methods involved also nursing homes and schools, means of transport (subways and trains) and supermarkets: according to our elaborations, only traces of SARS-CoV-2 were rarely found in these spaces.
As far as external environment is concerned, air quality quartz filters sampling 55 cubic meter in a day as well as glass-fiber filters employed by high-volume pump sampling 10 cubic meter of air in 20 min, referring to different seasons and different areas of the same one million people city, have been analyzed. SARS-CoV-2 has never been found in these samples. The reported results refer to external areas away from aggregated people, queues or crowded and narrow street and can be considered valid exclusively in the described conditions.
The described results confirm those reported by some studies published by Liu et al. (2020) and Stern et al. (2021), showing concentrations in the range 5 ÷ 50 copies/m3 of air within hospital wards, even though these results were not supported by the use of validated analytical methods such as those described in the present paper.
The described sampling and analytical methods are very interesting in order to compare different sampling techniques as well, for example different filtering materials (PTFE or gelatine filters vs fiber-glass filters), different capture mechanisms (filtering vs impaction) and different sampling duration, by carrying out air sampling with different devices in the same room, simultaneously.
At the same time, the availability of a reliable sampling and analytical method allows to thoroughly investigate the influence of environmental characteristics, such as temperature and humidity, on transport and dispersion mechanisms of virus-containing bioaerosols.
Moreover, the role of air exchange in indoor environments operated by natural and/or forced ventilation, as well as the effectiveness of air sanification systems, as preventive agents for infection, represent essential research targets that could be fully achieved only if reliable air monitoring could validate models and modeling outputs and be applied for tests.
In this regard, targeted experiments could contribute decisively to the definition of an emission model of viral particles exhaled by COVID-19 patients during different respiratory and metabolic activities; this kind of model is of fundamental importance to calculate the probability of infection and the individual risk, in particular for indoor spaces.
5. Conclusions
In the present paper we described the development of reliable sampling and pretreatment methods in order to quantify the presence of SARS-CoV-2 in air as bioaerosol.
The study defines samples pretreatment aiming at optimizing virus recovery from glass-fiber and PTFE filters. Recovery test results focused on the sample concentration step carried out by means of ultracentrifugation are also reported.
On the basis of the described approach, we can conclude that:
-
•
the recovery of HCoV-OC43 from PTFE filters is much higher and easier than from glass-fiber filters or quartz filter; regarding glass-fiber filters, we found that a two-step procedure is necessary to elute viral infective particles: a 3 h-shaking step followed by a 30 s-shaking step. On the contrary, 60 min-shaking is enough to elute viral particles from PTFE filters.
-
•
the recovery efficiency from glass-fiber filters and quartz filters could be strongly enhanced by using an elution buffer containing up to 40% of fetal calf serum;
-
•
the effect of suction time on filters could be resumed as follows: concerning 10 cm glass-fiber filters, sampling durations up to 20 min at a flow rate of 500 L per minute do not affect recovery efficiencies. On the contrary, the recovery efficiency of infectious virions from 4.7 cm PTFE filters decreases of a factor 2 after 3 h of sampling at a flow rate of 20 L per minute;
-
•
the applied ultracentrifugation step can concentrate the eluted viral suspension to maxima of 15 times, while the recovery efficiency of ultracentrifugation is 57%.
The developed methods, aiming at providing the community with reliable determinations about the presence of SARS-CoV-2 and other airborne pathogens in air, proves essential for the development, during the pandemic, of a coherent management of places, in particular crowded ones such as schools, means of transport, stations, gyms, theaters, cinemas. In this regard, the studies described here provide a solid basis for developing reliable methods for quantifying the viral load in the air, allowing, in perspective, to assess the health risk in a specific environment. They also represent an important acquisition of technical knowledge that can be readily applied in the future in the case of the emergence of new epidemics or pandemics.
Funding sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT author statement
Angelo Robotto: conceptualization, project administration, supervision, David Lembo: conceptualization, methodology, supervision, validation, Andrea Civra: methodology, investigation, Writing- Reviewing and Editing, Enrico Brizio: methodology, investigation, Writing- Reviewing and Editing, Paola Quaglino: methodology, project administration, Denis Polato: methodology, validation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors acknowledge the financial support received from Assicurazioni Generali and Intesa San Paolo to realize the Environmental Virology laboratory run by Arpa Piemonte (Environmental Protection Agency of Piedmont) in La Loggia, Torino, Italy. The authors are grateful to the Molecular Virology Laboratory at Dept. of Clinical and Biological Science, Università degli Studi di Torino, for the technical support received.
References
- CDC (Center for Disease Control and Prevention, USA) 2021. Science Brief: SARS-CoV-2 and Surface (Fomite) Transmission for Indoor Community Environments. [PubMed] [Google Scholar]
- Ahmed W., Bertsch P., Bivins A., Bibby K., Farkas K., Gathercole A., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739(2020) doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ASHRAE EPIDEMIC TASK FORCE . 2021. Core Recommendations for Reducing Airborne Infectious Aerosol Exposure. [Google Scholar]
- ASHRAE Position Document on Infectious Aerosols Approved by ASHRAE Board of Directors April 14, 2020.
- Atkinson J., Chartier Y., Lúcia Pessoa-Silva C., Jensen P., Li Y., Seto W.-H. WHO; Geneva: 2009. Natural Ventilation for Infection Control in Health-Care Settings. [PubMed] [Google Scholar]
- Binder R.A., Alarja N.A., Robie E.R., Kochek K.E., Xiu L., Rocha-Melogno L., Abdelgadir A., Goli S.V., Farrell A.S., Coleman K.K., Turner A.L., Lautredou C.C., Lednicky J.A., Lee M.J., Polage C.R., Simmons R.A., Deshusses M.A., Anderson B.D., Gray G.C. Environmental and aerosolized severe acute respiratory syndrome coronavirus 2 among hospitalized coronavirus disease 2019 patients. J. Infect. Dis. 2020;222(11):1798–1806. doi: 10.1093/infdis/jiaa575. 2020 Nov 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (Center for Disease Control and Prevention, USA) 2021. Scientific Brief: SARS-CoV-2 Transmission. [Google Scholar]
- Da Silva P.G., Nascimento M.S.J., Soares R.R.G., Sousa S.I.V., Mesquita J.R. Airborne spread of infectious SARS-CoV-2: moving forward using lessons from SARS-CoV and MERS-CoV. Sci. Total Environ. 2021;764 doi: 10.1016/j.scitotenv.2020.142802. 2021 Apr 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lednicky J.A., Lauzardo M., Fan Z.H., Jutla A., Tilly T.B., Gangwar M., Usmani M., Shankar S.N., Mohamed K., Eiguren-Fernandez A., Stephenson C.J., Alam M.M., Elbadry M.A., Loeb J.C., Subramaniam K., Waltzek T.B., Cherabuddi K., Morris J.G., Jr., Wu C.Y. Viable SARS-CoV-2 in the air of a hospital room with COVID-19 patients. Int. J. Infect. Dis. 2020;100:476–482. doi: 10.1016/j.ijid.2020.09.025. 2020 Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X., Reponene T.A., Willeke K., Grinshpun S.A., Foarde K.K., Ensor D.S. Long-term sampling of airborne bacteria and fungi into a non-evaporating liquid. Atmos. Environ. 1999;33(1999):4291–4298. [Google Scholar]
- Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., et al. Aerodynamic analysis of SARS-CoV-2 in Two Wuhan Hospitals. BioRxiv. 2020 doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Morawska L., Cao J. Airborne transmission of SARS-CoV-2: the world should face the reality. Environ. Int. 2020;139 doi: 10.1016/j.envint.2020.105730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M., Lednicky J.A., Wu C.-Y. Collection, particle sizing and detection of airborne viruses. Journal of Applied Microbiology ISSN. 2019:1364–5072. doi: 10.1111/jam.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platto S., Xue T., Carafoli E. COVID19: an announced pandemic. Cell Death Dis. 2020;11(9) doi: 10.1038/s41419-020-02995-9. 2020 Sep. 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robotto A., Quaglino P., Lembo D., Morello M., Brizio E., Bardi L., Civra A. SARS-CoV-2 and indoor/outdoor air samples: a methodological approach to have consistent and comparable results. Environ. Res. 2021;195(2021) doi: 10.1016/j.envres.2021.110847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santarpia J.L., Rivera D.N., Herrera V.L., Morwitzer M.J., Creager H.M., Santarpia G.W., Crown K.K., Brett-Major D.M., Schnaubelt E.R., Broadhurst M.J., Lawler J.V., Reid S.P., Lowe J.J. Aerosol and surface contamination of SARS-CoV-2 observed in quarantine and isolation care. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-69286-3. 2020 Jul 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherwing C., Bunke J. Sartorius application note; 2019. Continuous Microbial Air Monitoring in Clean Room Environments. [Google Scholar]
- Siegel J.D., Rhinehart E., Jackson M., Chiarello L. 2007 guideline for isolation precautions: preventing transmission of infectious agents in healthcare settings (updated July 2019) Centers Dis. Control Prev. 2007:1–232. doi: 10.1016/j.ajic.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern R.A., Koutrakis P., Martins M.A.G., Lemos B., Dowd S.E., Sunderland E.M., Garshick E. Characterization of hospital airborne SARS-CoV-2. Respir. Res. 2021;22(2021):73. doi: 10.1186/s12931-021-01637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Morris D.H., Holbrook M.G., Gamble A., Williamson B.N., Tamin A., Harcourt J.L., Thornburg N.J., Gerber S.I., Lloyd-Smith J.O., de Wit E., Munster V.J. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N. Engl. J. Med. 2020;382(16):1564–1567. doi: 10.1056/NEJMc2004973. 2020 Apr 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault D., Moineau S., Duchaine C. Methods for sampling of airborne viruses. Microbiol. Mol. Biol. Rev. 2008;72:413–444. doi: 10.1128/MMBR.00002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; 2020. Transmission of SARS-CoV-2 – Implications for Infection Prevention Precautions: Scientific Brief.https://www.who.int/news-room/commentaries/detail/transmission-of-sars-cov-2-implications-for-infection-prevention-precautions [WWW Document] [Google Scholar]
- WHO 2021. https://www.who.int/news-room/q-a-detail/coronavirus-disease-covid-19-how-is-it-transmitted (update 30th April 2021)
- Willeke K., Lin X., Grinshpun S.A. Improved aerosol collection by combined impaction and centrifugal motion. Aerosol. Sci. Technol. 1998;28:439–456. (1998) © 1998 American Association for Aerosol Research. [Google Scholar]
- Xie X., Li Y. How far droplets can move in indoor environments—revisiting the Wells evaporation-falling curve. Indoor Air. 2007;17(3):211–225. doi: 10.1111/j.1600-0668.2007.00469.x. [DOI] [PubMed] [Google Scholar]