Abstract
ESX-1 is a major virulence factor of Mycobacterium tuberculosis, a secretion machinery directly involved in the survival of the microorganism from the immune system defence. It disrupts the phagosome membrane of the host cell through a contact-dependent mechanism. Recently, the structure of the inner-membrane core complex of the homologous ESX-3 and ESX-5 was resolved; however, the elements involved in the secretion through the outer membrane or those acting on the host cell membrane are unknown. Protein substrates might form this missing element. Here, we describe the oligomerisation process of the ESX-1 substrate EspB, which occurs upon cleavage of its C-terminal region and is favoured by an acidic environment. Cryo-electron microscopy data shows that quaternary structure of EspB is conserved across slow growing species, but not in the fast growing M. smegmatis. EspB assembles into a channel with dimensions and characteristics suitable for the transit of ESX-1 substrates, as shown by the presence of another EspB trapped within. Our results provide insight into the structure and assembly of EspB, and suggests a possible function as a structural element of ESX-1.
Keywords: Cryo-EM, EspB, ESX-1, Mycobacteria, Preferential orientation
Graphical abstract
Highlights
-
•
Acidic environment and removal of C-terminal end primes EspB oligomerisation.
-
•
EspB from slow-growing species oligomerise while EspB from M. smegmatis does not.
-
•
New structure of EspB from M. marinum presented.
-
•
Residues within the C-terminal region interact with hydrophobic surfaces.
-
•
Monomer inside the channel supports the idea that EspB is part of ESX-1 machinery.
1. Introduction
Tuberculosis (TB) is an infectious disease caused by the bacillus Mycobacterium tuberculosis. In 2018, 10 million people developed the disease from which 0.5 million were caused by multidrug-resistant strains. Even though TB is curable, 1.5 million people succumb to it every year (World Health Organization, 2019). The current treatment is long and with serious side effects, often driving the patient to terminate the therapy before its conclusion (Schaberg et al., 1996). This has contributed to an increase in the number of patients suffering from multidrug- and extensively drug-resistant TB. While treatment is available for some of these resistant strains, the regimen is usually longer, more expensive and sometimes more toxic. For this reason, research on mycobacterial pathogenesis is vital to find a proper target in order to develop more effective therapeutics and vaccines.
The high incidence of TB relates to the ability of M. tuberculosis to evade the host immune system (Ferluga et al., 2020). This ability is related to multiple factors, one of which is a complex cell envelope of low permeability that plays a crucial role in drug resistance and in survival under harsh conditions (Brennan and Nikaido, 1995). Likewise, pathogenic mycobacteria secrete virulence factors that manipulate the environment and compromise the host immune response. Mycobacteria have up to five specialised secretion machineries that carry out this process, named ESX-1 to −5 (together known as the type VII secretion system or T7SS). The core components of the inner-membrane part of the T7SS have been identified (Pym et al., 2003). Nevertheless, it remains unknown whether the translocation of substrates through the inner and outer membranes is functionally coupled or not, and if it deploys a specific outer-membrane complex to do so (Bunduc et al., 2021). Proteins from the PE/PPE family, characterised by Pro-Glu and Pro-Pro-Glu motifs and secreted by T7SS, are often associated with the outer most layer of the mycobacterial cell envelope, and have been suggested to play a role in the membrane channel formation (Burggraaf et al., 2019; Cascioferro et al., 2007; Wang et al., 2020). Recently, the intake of nutrients by M. tuberculosis was shown to be dependent on PE/PPE proteins, suggesting that these form small molecule–selective porins that allow the bacterium to take up nutrients over an otherwise impermeable barrier (Wang et al., 2020).
ESX-1 to -5 are paralogue protein complexes with specialised functions and substrates, unable to complement each other (Abdallah et al., 2007; Phan et al., 2017). ESX-1 is essential for the virulence of M. tuberculosis. It has been implicated in phagosomal escape, cellular inflammation, host cell death, and dissemination of the bacteria to neighbouring cells (Abdallah et al., 2011; Houben et al., 2012a; Simeone et al., 2012; Stanley et al., 2007; van der Wel et al., 2007). Our knowledge about the structure of the machinery as well as the mechanism of secretion and regulation remains limited. ESX-3 is involved in iron homeostasis (Siegrist et al., 2009), and only recently the molecular architecture of its inner-membrane core has been determined (Famelis et al., 2019; Poweleit et al., 2019). The complex consists of a dimer of protomers, made of four proteins: the ESX-conserved component (Ecc)-B, C, D (×2), and E. Despite the resolution achieved in both studies, there was no obvious channel through which the proteins substrates can traverse. Rosenberg and collaborators have described that one of the elements of the secretion system (EccC) forms dimers upon substrate binding, which then forms higher-order oligomers (Rosenberg et al., 2015). This is in agreement with observations that ESX-5, involved in nutrient uptake (Ates et al., 2015) and host cell death (Abdallah et al., 2011), forms a hexamer (Beckham et al., 2021; Bunduc et al., 2021; Houben et al., 2012b). A recent structure of an ESX-5 trimer of dimers shows that is it stabilised by a mycosin protease (MycP) positioned in the periplasm on top of EccB5 (Bunduc et al., 2021). ESX-2 and ESX-4 are the least characterised, where ESX-4 is involved in DNA transfer (Gray et al., 2016) and is seen as the ancestor of the five ESX-systems (Gey Van Pittius et al., 2001).
Located in different positions in the genome, the esx loci contain the genes that code for the Ecc proteins, MycP, a heterodimer of EsxA/B-like proteins, and one or more PE-PPE pairs. With high sequence similarity and conservation between paralogues (Poweleit et al., 2019; van Winden et al., 2016), one could expect the inner-membrane core of the different systems to share a similar architecture. So what makes each one of them unique? Experimental data suggest that the answer lies with the substrates (Lou et al., 2017). The esx-1 locus encodes for more than ten unique proteins that are known to be secreted (Sani et al., 2010), termed the ESX-1 secretion-associated proteins (Esp) (Bitter et al., 2009). Amongst those is EspC, a protein present in pathogenic mycobacteria that was described to form filamentous structures in vitro and to localise on the bacterial surface in vivo (Lou et al., 2017). Due to the similarities between EspC and the needle protein of the type III secretion system, Lou et al. hypothesised that ESX-1 could be an injectosome system with EspC as its needle. This is of particular importance because, compared to the other secretion systems, ESX-1 function has been described to take place through a contact-dependent mechanism (Conrad et al., 2017), which makes the discovery of an outer-membrane complex essential for understanding the system. Other proteins, like EspE that has been localised on the cell wall (Carlsson et al., 2009; Phan et al., 2018; Sani et al., 2010), are of interest as possible elements of such outer-membrane complex. The protein EspB has been the focus of attention due to its ability to oligomerise upon secretion (Korotkova et al., 2015; Solomonson et al., 2015), making it a strong candidate as a structural component of the machinery (Piton et al., 2020).
EspB belongs to the PE/PPE family, but unlike other family members that form heterodimers in mycobacteria, EspB consists of a single poly-peptide chain fusing the PE and PPE domains (Korotkova et al., 2015). EspB is a 48-kDa protein that matures during secretion: Its largely unstructured C-terminal region is cleaved in the periplasm by the protease MycP1, leaving a mature 38-kDa isoform (Ohol et al., 2010; Solomonson et al., 2013; Xu et al., 2007). The purpose of this maturation is not yet clear but it was shown that inactivation of MycP1, and thus cleavage of EspB, deregulates the secretion of proteins by ESX-1 (Ohol et al., 2010). Chen et al. observed specific binding of EspB to phosphatidylserine and phosphatidic acid after cleavage (Chen et al., 2013), suggesting that the C-terminal processing of EspB is important for its function, possibly involving lipid binding. The crystal structure of the monomeric N-terminal region of EspB from M. tuberculosis and M. smegmatis has been determined: It forms a four-helix bundle with high structural homology between species (Korotkova et al., 2015; Solomonson et al., 2015). During the preparation of this work for publication, the structure of an EspB oligomer from M. tuberculosis was published by Piton et al., showing features of a pore-like transport protein (Piton et al., 2020).
EspB is the only member of the PE/PPE family described to date to form higher-order oligomers. In this work, we studied the oligomerisation ability and structures of EspB from M. tuberculosis, M. marinum, M. haemophilum and M. smegmatis. We show that truncation of EspB at the MycP1 cleavage site and an acidic environment promote the oligomerisation of EspB from the three slow-growing species but not from fast-growing M. smegmatis. Oligomerisation is mediated by intermolecular hydrogen bonds and amide bridges between residues highly conserved in the slow-growing species, but absent in M. smegmatis. The structures of oligomeric EspB consist of two domains: an N-terminal region that forms a cylinder-like structure with a tunnel large enough to accommodate a folded PE-PPE pair, and a partly hydrophobic C-terminal region that interacts with hydrophobic surfaces. The oligomer has similar inner-pore dimensions as was described for the pore within the periplasmic region of ESX-5 (Bunduc et al., 2021). Visualisation of a trapped EspB monomer within the channel supports the idea that it could transit secreted proteins through its tunnel. Overall, in this work we describe factors that prime the oligomerisation of EspB, and provide insight into its potential role in the ESX-1 machinery.
2. Materials and methods
2.1. Cloning, expression and purification of EspB constructs
Different constructs used in this study are listed in Table S1. DNA fragments were PCR-amplified with KOD Hot Start Master Mix (Novagen®) from genomic DNA of M. tuberculosis H37Rv, M. marinum or M. smegmatis [BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID)], and cloned in a modified pRSET backbone (Invitrogen™) using NsiI and HindIII restriction sites. Constructs included an N-terminal 6 × His-tag followed by a tobacco etch virus (TEV) protease cleavage site. EspB mutants and construct EspB2-358 and EspB2-287 were generated using KOD-Plus- Mutagenesis kit (Toyobo Co., Ltd.) from the plasmid encoding the full-length protein. All plasmids were sequenced to verify absence of inadvertent mutations. M. haemophilum and PE25-PPE41 construct were synthesised and codon optimised for expression in Escherichia coli (Eurofins Genomics).
For the non–codon optimised constructs, proteins were expressed in Rosetta™ 2(DE3) E. coli cells in Overnight Express™ Instant LB Medium (EMD Millipore) supplemented with 100 μg/mL of carbenicillin and 25 μg/mL of chloramphenicol for 50 h at 25 °C. In the case of codon optimisation, the protein was expressed in C41 (DE3) E. coli cells in the same conditions with the respective antibiotic. Prior to protein purification, cells were resuspended in buffer containing 20 mM Tris-HCl (pH 8.0), 300 mM NaCl, 40 mM imidazole, 1 mM PMSF, and 25 U/mL benzonase, and were lysed using an EmulsiFlex-C3 homogenizer (Avestin). Proteins were purified with HisPur™ Ni-NTA Resin (ThermoFisher) equilibrated in the lysis buffer and eluted in the same buffer supplemented with 400 mM imidazole. The 6 × His-tag was cleaved using TEV protease followed by a second Ni-NTA purification to remove the free 6 × His-tag, uncleaved protein and the His-tagged protease (Kapust et al., 2001). In case higher purity was needed, proteins were purified on a size-exclusion Superdex® 200 Increase 10/300 GL column (GE Healthcare) in buffer containing 20 mM Tris-HCl (pH 8.0), 300 mM NaCl. Protein was stored at −80 °C until further use.
2.2. Analytical size exclusion chromatography (SEC)
Samples were dialysed overnight in the corresponding buffers and different concentrations of protein were loaded onto a size-exclusion Superdex® 200 Increase 3.2/300 column (GE Healthcare Life Science) at a flow rate of 50 μL/min. Basic buffer comprised 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, while the acidic buffer was 20 mM acetate buffer (pH 5.5), 150 mM NaCl.
2.3. Native mass spectrometry
Native mass spectrometry was used to obtain high resolution mass information of the samples. M. tuberculosis EspB2-348 (5 mg/mL) was buffer exchanged with 100 mM NH4CH3CO2 (at pH 5.5 and 8.5) using 3-kDa molecular weight cut-off dialysis membrane overnight followed by an extra hour buffer exchange with a fresh NH4CH3CO2 solution at 4 °C. The buffer exchange of fragments produced by limited proteolysis (2 mg/mL) was performed using SEC on a Superdex® 200 Increase 3.2/300 column (GE Healthcare Life Science) with 100 mM NH4CH3CO2 at pH 6.8. CH3COOH and NH4OH were used to adjust the pH of NH4CH3CO2 solution. The mass spectrometry measurements were performed in positive ion mode on an ultra-high mass range (UHMR) Q-Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific) with a static nano-electrospray ionization (nESI) source. In-house pulled, gold-coated borosilicate capillaries (Mathew et al., 2021) were used for the sample introduction to the mass spectrometer, and a voltage of 1.2 kV was applied. Mass spectral resolution was set at 4375 to 8750 (at m/z = 200) and an injection time of 100–200 ms was used. For each spectrum, 10 scans were combined, containing 5 to 10 microscans. The inlet capillary temperature was kept at 320 °C. Parameters such as in-source trapping, transfer m/z, detector m/z, trapping gas pressure and mass range were optimised for each analyte separately. All mass spectra were analysed using Thermo Scientific Xcalibur software and spectral deconvolutions were performed with the UniDec software (Marty et al., 2015).
2.4. Cryo-EM sample preparation, data acquisition and image processing
Samples, in 20 mM acetate buffer (pH 5.5), 150 mM NaCl, were diluted to the respective concentrations (Table S1). A volume of 2.5 μL of each sample was applied on glow-discharged UltrAuFoil Au300 R1.2/1.3 grids (Quantifoil), and excess liquid was removed by blotting for 3 s (blot force 5) using filter paper followed by plunge freezing in liquid ethane using a FEI Vitrobot Mark IV at 100% humidity and 4 °C. For PE25-PPE41, an acetate buffer (pH 6.5), 150 mM NaCl was used, due to precipitation of the protein at lower pH.
Cryo-EM single particle analysis (SPA) data were collected using untilted and tilted schemes (Tan et al., 2017). For EspB2-287 from M. tuberculosis and EspB2-286 from M. marinum, untilted images were recorded on a Titan Krios (Thermo Fischer Scientific) at 300 kV with a K3 detector operated in super-resolution counting mode. Tilted SPA data were collected for EspB2-348 from M. tuberculosis on a 200-kV Tecnai Arctica TEM using SerialEM (Mastronarde, 2005), with a Falcon III detector in counting mode. Table S2 shows all specifications and statistics for the data sets. Individual micrographs of EspB2-287 from M. haemophilum, EspB2-407 from M. smegmatis as well as PE25-PPE41 from M. tuberculosis were collected on the 200-kV Arctica.
Data were processed using the RELION-3 pipeline (Zivanov et al., 2018). Movie stacks were corrected for drift (5 × 5 patches) and dose-weighted using MotionCor2 (Zheng et al., 2017). The local contrast transfer function (CTF) parameters were determined for the drift-corrected micrographs using Gctf (Zhang, 2016). The EspB2-348 data set was collected at two angles of the stage: 0° and 40°. For each tilt angle, a first set of 2D references was generated from manually picked particles in RELION (Scheres, 2012) and this was used for subsequent automatic particle picking. Table S2 lists the number of particles in the final data set after particle picking, 2D and 3D classification. The 3D classification was run without imposing symmetry and used to select the heptameric particles. Local CTF parameters were iteratively refined (Zivanov et al., 2018), which was particularly important for the tilted data set; beam-tilt parameters were estimated, and particles were polished. Particle subtraction followed by focused classification was used to characterise densities other than those described by the refined model described below. Due to the extreme preferred orientation of EspB2-348 heptamers, automatic masking and automatic B-factor estimation in post-processing were hampered by missing wedge artefacts. For this data set, parameters were manually optimised by visual inspection of the resulting maps. Density within the heptameric pore was obtained by a combination of 2D and 3D classification. The initial density map of a loaded complex was generated by symmetry expansion of a C7 3D-refined particle list, followed by 3D classification in C1 without further image alignment. Later iterations employed 45,671 unique particles and 3D refinement in C1 while imposing local symmetry for the heptamer. The resulting 5.3 Å map was used to identify a total of eight EspB monomers (heptamer plus one in the middle), and local symmetry averaged. The final resolution of the heptamer maps, listed in Table S2, varied between 2.3 and 3.4 Å, using the gold-standard FSC = 0.143 criterion (Scheres and Chen, 2012).
2.5. Structure determination and refinement
The PDB model 4XXX (Korotkova et al., 2015) was used as a starting model in Coot (Emsley and Cowtan, 2004) for manual docking and building into the tilted-scheme SPA data set of EspB2-348 of M. tuberculosis. The final model was refined against the high-resolution sharpened map of EspB2-287 of M. tuberculosis. This model was later used as reference for M. marinum model. Models were refined iteratively through rounds of manual adjustment in Coot (Emsley et al., 2010), real space refinement in Phenix (Afonine et al., 2018) and structure validation using MolProbity (Williams et al., 2018).
2.6. Cryo-electron tomography
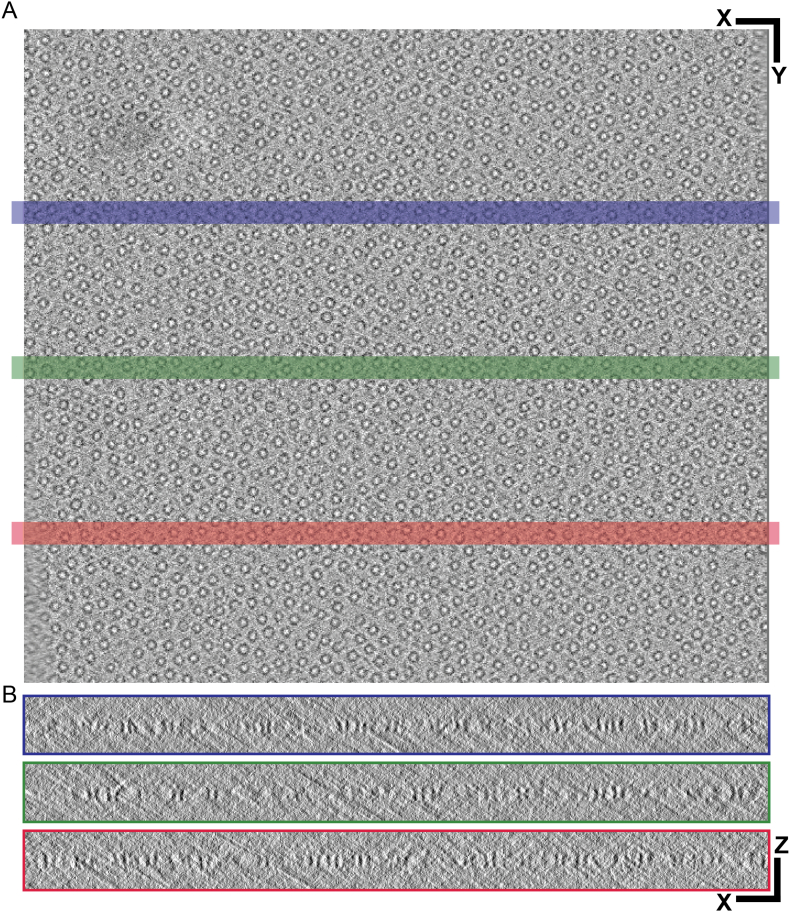
EspB2-348 from M. tuberculosis, in 20 mM acetate buffer (pH 5.5), 150 mM NaCl, was diluted to 0.5 mg/mL. A volume of 2.5 μL was applied on glow-discharged UltrAuFoil Au200 R2/2 grids (Quantifoil), and excess liquid was removed by blotting for 3 s (blot force 5) using filter paper followed by plunge freezing in liquid ethane using a FEI Vitrobot Mark IV at 100% humidity at 4 °C.
Tomography data was acquired with a Titan Krios (Thermo Fisher Scientific) equipped with a K3 direct electron detector (Gatan) in electron counting mode and pixel size of 1.37 Å. Movies were acquired in SerialEM (Mastronarde, 2005) using a stage tilt scheme of −60°–60° in increments of 3° through a total fluence of 80 e−/Å2 and a defocus target range of −3 to −6 μm. Recorded movie fractions were corrected for beam-induced sample motion and processed into dose weighted stacks with MotionCor2 (Zheng et al., 2017). Tilt series were aligned, 4 × binned and reconstructed with IMOD (Mastronarde and Held, 2017) using patch tracking and SIRT, respectively.
2.7. Limited proteolysis and Edman sequencing
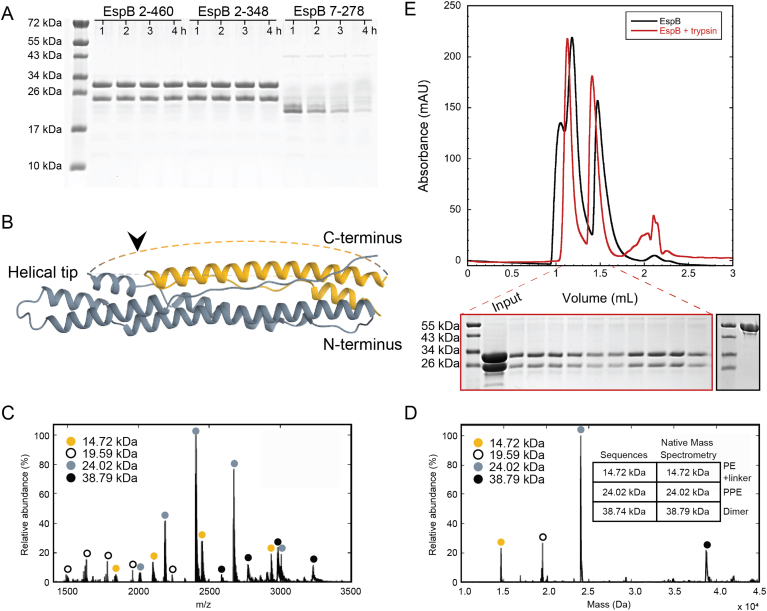
Samples were incubated with trypsin for different length of time at a molar ratio of 1:6 (enzyme:substrate) following the Proti-Ace™ Kit (Hampton Research) recommendation. The reaction was stopped by adding SDS-PAGE loading buffer (63 mM Tris-HCl, 2% SDS, 10% glycerol, 0.1% bromophenol blue) and samples were resolved on a 12% polyacrylamide gel. Bands were transferred from the SDS-PAGE gel to a PVDF membrane and stained with 0.1% Coomassie Brilliant Blue R-250, 40% methanol, and 10% acetic acid until bands were visible. The membrane was then washed with water and dried, and EspB cleavage products were cut out. The first ten amino acids were determined by Edman sequencing at the Plateforme Protéomique PISSARO IRIB at the Université de Rouen.
2.8. Circular dichroism (CD) spectroscopy
The CD spectra of 5 μM EspB279-460 were recorded either in 50 mM phosphate (pH 8.0), 50 mM NaCl or 10 mM acetate (pH 5.5), 50 mM NaCl at 25 °C in the far-UV region using a Jasco J-1500 CD spectropolarimeter (JASCO Analytical Instruments) on a 0.1 cm path-length cell. Spectra correspond to the average of five repetitive scans acquired every 1 nm with 5-s average time per point and 1-nm band pass. Temperature was regulated with a Peltier temperature-controlled cell holder. Data were corrected by subtracting the CD signal of the buffer over the same wavelength region. The effect of 2,2,2-trifluoroethanol (TFE) was recorded using the aforementioned phosphate buffer. Temperature dependence of ellipticity was followed at 222 nm from 20 to 80 °C, a bandwidth of 1 nm, a time response of 16 s and a temperature gradient of 1 °C/min. The data was fitted to a Boltzmann distribution describing a standard two-state transition model to obtain the melting temperature.
3. Results
3.1. Oligomerisation is favoured by an acidic pH and maturation of EspB
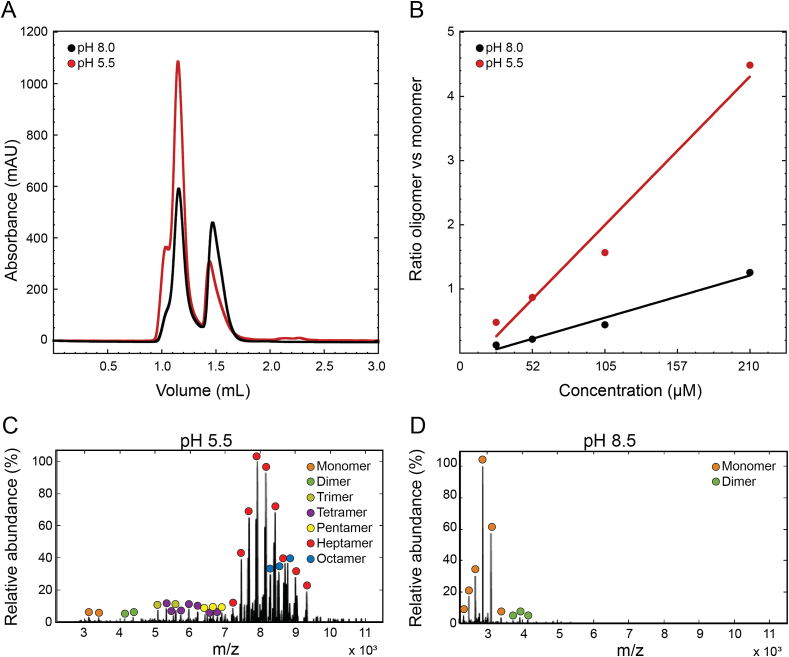
Previously, it has been described that oligomerisation of EspB occurs after secretion (Korotkova et al., 2015). In the infection context, this secretion would lead the protein to the phagosomal lumen of a macrophage, an organelle known to have pH acidification as a functional mechanism. To evaluate the putative role of pH in the oligomerisation process, the mature form of M. tuberculosis EspB (residues 2–358) was incubated at different pH values and analysed by size exclusion chromatography (SEC). Results showed that the equilibrium is favoured towards an oligomer form at pH 5.5 compared to pH 8.0 (Fig. 1A), as observed by a higher oligomer/monomer ratio at any protein concentration (Fig. 1B). Native mass spectrometry experiments confirmed this behaviour and could identify different oligomeric states of EspB, with the heptamer being the most predominant. Intermediate states were observed (dimer to pentamer) and even higher oligomeric states (octamer) but in lower abundance compared to the heptamer (Fig. 1C and D).
Fig. 1.
Oligomerisation of EspB is promoted by an acidic environment. (A) Size exclusion chromatography profiles of M. tuberculosis EspB2-358 at 210 μM in 20 mM acetate buffer (pH 5.5), 150 mM NaCl and 20 mM Tris (pH 8.0), 150 mM NaCl. Void volume corresponds to 0.8 mL elution volume. (B) Oligomer/monomer ratios at different protein concentrations in conditions from panel (A). The absorbance values of the oligomer were taken at 1.14 mL while the monomer values were at 1.48 mL. (C–D) Presence of the different oligomer species from M. tuberculosis EspB2-348 at pH 5.5 and pH 8.5 obtained by native mass spectrometry.
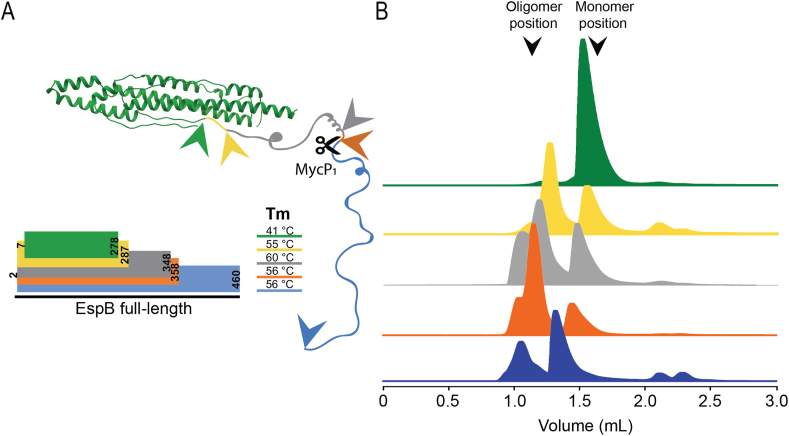
Because EspB undergoes proteolytic processing of its C-terminus during secretion, we investigated the effect of this cleavage at pH 5.5 on the quaternary structure of different M. tuberculosis EspB constructs that vary in their C-terminus length (Fig. 2). With the exception of EspB7-278 that did not oligomerise, we observed that oligomerisation was favoured for all other constructs at pH 5.5 (Fig. 2B). The full-length EspB2-460 (Fig. 2B, blue trace) presented the lowest amounts of complex formation compared to the other constructs tested, while the highest was observed for the mature isoform, EspB2-358 (Fig. 2B, orange trace). Shorter versions showed oligomerisation, though not to the extent of EspB2-358. These results suggest that MycP1 cleaves EspB to allow oligomerisation, and that the remaining residues of the unstructured C-terminal region could aid complex stabilisation. Melting temperatures (Tm) calculated by circular dichroism (CD) highlighted EspB7-278 as the least stable construct of all those tested, showing a 14 °C difference or more (Fig. 2A), which could explain the negative impact on the oligomerisation.
Fig. 2.
Impact of EspB C-terminus processing on oligomerisation. (A) Scheme of the different constructs used in this work, where EspB2-460 is in blue, EspB2-358 in orange (MycP1 cleavage site), EspB2-348 in grey, EspB2-287 in yellow and EspB7-278 in green. Structural model from PDB ID 4XXX, while the C-terminal region is a representation of an unfolded protein. Arrows represent the end of each construct. Melting temperatures were obtained following the dependency of the circular dichroism signal as a function of temperature. (B) Size exclusion chromatograms of each construct corresponding to the colours in panel (A), resulting from 50 μL sample injection at 220 μM eluted in 20 mM acetate buffer (pH 5.5), 150 mM NaCl. Void volume corresponds to 0.8 mL elution volume.
3.2. EspB from M.smegmatis is unable to oligomerise
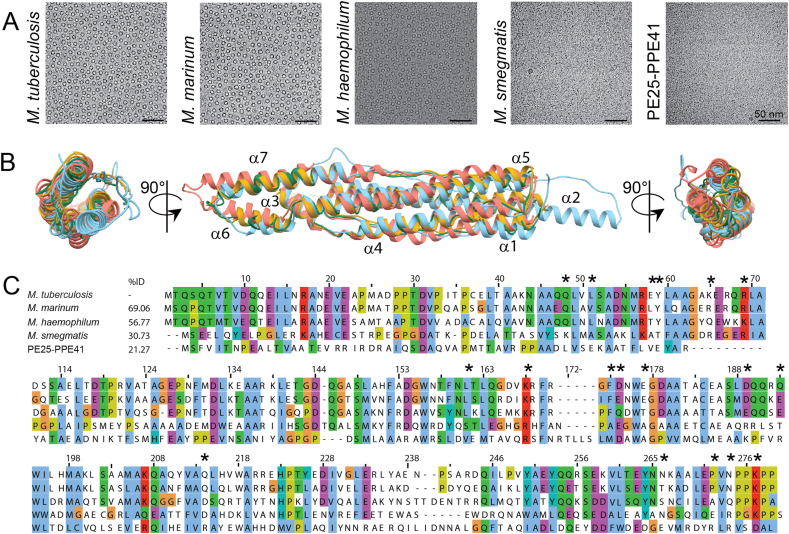
To determine whether the oligomerisation ability is conserved across species, we performed cryo-EM analysis on different orthologues of the mature EspB. We selected M. marinum due to its wide use as model microorganism to study TB, M. haemophilum as an evolutionary distant specie from M. tuberculosis, and M. smegmatis representing a fast-growing mycobacterium and a common model organism used in research. Proteins from slow-growing species M. tuberculosis, M. marinum and M. haemophilum were able to oligomerise into ring-like structures while the fast-growing M. smegmatis did not, as seen by the lack of visible particles (Fig. 3A); the structured region of an EspB monomer (30 kDa) has a signal-to-noise ratio too low to be visualised within these micrographs (Henderson, 1995; Zhang et al., 2020). Interestingly, comparison of the tertiary structure from the slow-growing species studied here with the published crystallographic model of EspB from M. smegmatis did not show substantial differences (RMSD Cα′s 0.979–1.128 Å), apart from an extended α-helix 2 (Fig. 3B), absent in our oligomeric structures. To determine whether the differences in oligomerisation ability between M. smegmatis and the slow-growing species were due to variations in their primary structure, we performed sequence alignment of multiple EspB orthologues. The species able to oligomerise have high sequence identity, together with other slow-growing mycobacteria species, whereas M. smegmatis has a low sequence identity, similar to other fast-growing species (Fig. 3C and Fig. S1).
Fig. 3.
Oligomerisation differences between EspB orthologues despite sharing similar tertiary structure. (A) Evaluation of the oligomerisation of EspB orthologues and PE25-PPE41 by cryo-electron microscopy. Scale bars represent 50 nm. (B) Different views of structural alignment of EspBMtb (yellow – this work), EspBMmar (green – this work), EspBMsmeg (light blue – PDB ID 4WJ1), and PE25-PPE41 (orange – PDB ID 4W4K). (C) Multi-alignment of amino acid sequences of different species from the Mycobacterium genus, as well as the protein pair PE25-PPE41. Numbering and sequence identity is based on the sequence of M. tuberculosis. Asterisks denote residues making inter-subunit contact in EspB from M. tuberculosis and M. marinum. Alignment was generated using ClustalW server, and figure was created using software Jalview (Waterhouse et al., 2009). The colour scheme of ClustalX is used (Larkin et al., 2007).
Because EspB belongs to the PE/PPE family, we included in the analysis a PE-PPE pair with a structure already published (Ekiert and Cox, 2014). PE25-PPE41 did not oligomerise (Fig. 3), despite sharing a similar tertiary structure (RMSD Cα′s 1.134 Å). With a low percentage identity (21.3%), it confirms the importance of specific amino acids sequence for the conservation of the quaternary structure.
3.3. High-resolution cryo-EM structures of EspB oligomers
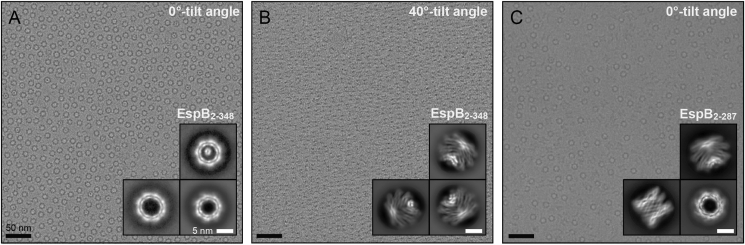
Next, we aimed to solve the high-resolution structure of EspB oligomers by cryo-EM. Initial experiments were performed with EspB2-460 and EspB2-348 from M. tuberculosis, which displayed an extreme preferential orientation where only “top views” could be seen (Fig. 4A). Cryo-electron tomography revealed these molecules to be attached to the air-water interface (Fig. S2). Different oligomers were found: hexamers, heptamers, rings with an extra density in the middle, and octamers, with the heptameric ensemble being the predominant one (Fig. 4A), in agreement with the results obtained in solution (Fig. 1C). Preliminary 3D reconstructions could be obtained from data collected at one or more tilt angles (Fig. 4B). Data processing resulted in 3–4 Å resolution maps from which the first heptamer models were built. Removal of C-terminus to residue 287 led to a different distribution of particles on the cryo-EM grids, now with random orientations (Fig. 4C), denoting an interaction between this region and the air-water interface on the EM grid (Noble et al., 2018).
Fig. 4.
Loss of the EspB preferential orientation by removal of its C-terminal residues. (A–B) Representative micrograph of EspB2-348 with preferential orientation at 0°-tilt angle or 40°-tilt angle. (C) Representative micrograph of EspB2-287 with random orientation taken at 0°-tilt angle. Insets correspond to the respective 2D classes. Scale bars in A–C represent 50 nm; scale bars in insets represent 5 nm.
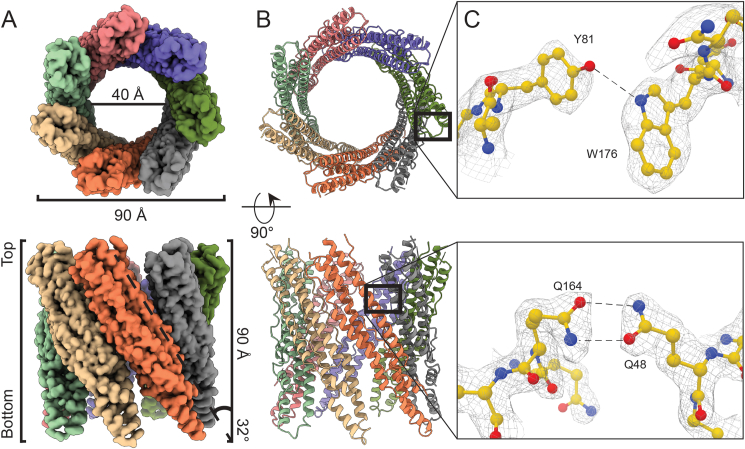
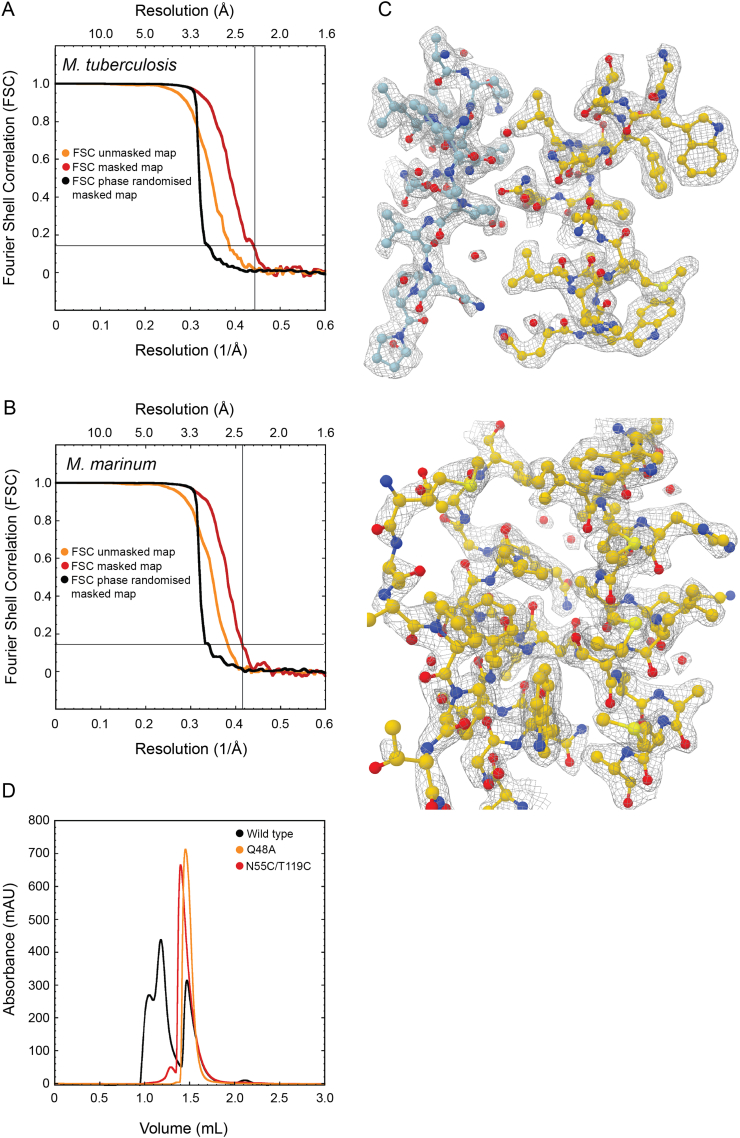
Experiments were repeated for constructs EspB2-287 from M. tuberculosis, and the equivalent construct from M. marinum (at 0°-stage tilt), leading to high-resolution EM maps of 2.3 Å and 2.5 Å average resolution, respectively (Fig. 5A and B, and Fig. SA–C). We observed high structural conservation between the two structures. Both displayed a four-helix bundle, like the EsxA-B complex and PE25-PPE41 complex, with the WxG and YxxD located on one end of the elongated molecule, referred to the top hereafter, making an H-bond interaction between the nitrogen of W176 with the oxygen of Y81, as was observed in the crystal structure (Fig. 5C) (Korotkova et al., 2015). The helical tip is located on the opposite end, referred to as the bottom, for both EspB and PE25-PPE41 (Korotkova et al., 2015; Solomonson et al., 2015). The C-terminal region starts near the top end of the elongated molecule. The overall structure shows seven copies tilted 32° with respect to the symmetry-axis forming a cylinder-like oligomer with a width and a height of 90 Å (Fig. 5A and B).
Fig. 5.
Cryo-EM reconstruction of EspB2-287 heptamer complex. (A–B) Density map and structural model made with ChimeraX (Goddard et al., 2018), showing each monomer in different colours. For (A) and (B), the upper panels show the top views and the bottom panels show the side views. Representation of the smallest inner-diameter calculated by HOLE (Smart et al., 1993). (C) Model and densities of intramolecular interaction at W176–Y81 and intermolecular interaction at Q48-Q164. Colours follow the conventional colouring code for chemical elements.
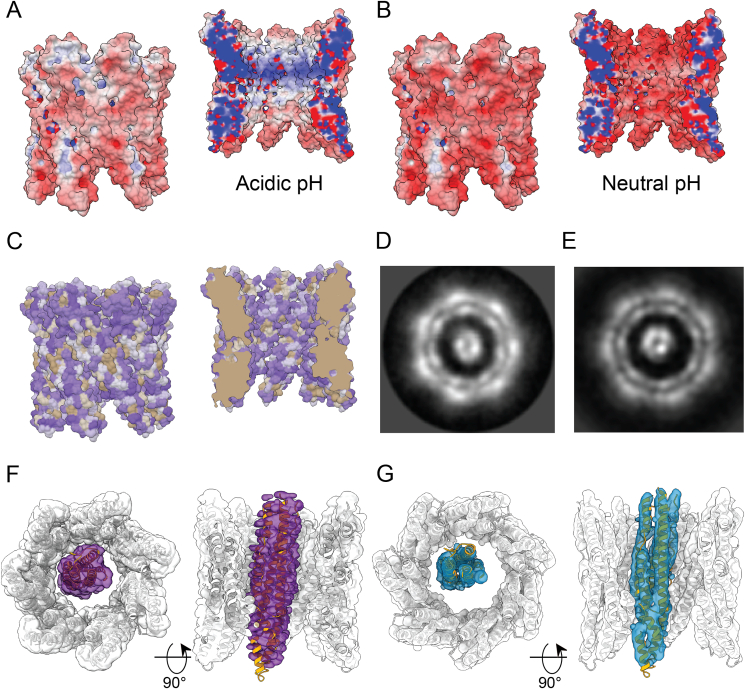
The single-particle analysis (SPA) map from M. tuberculosis EspB revealed several interaction pairs between monomers (Table S3), involving residues highly conserved in slow-growing mycobacteria (Fig. 5C and Fig. S1). Mutation in one of these residues (Q48A) in the M. tuberculosis orthologue resulted in the disruption of the oligomer (Fig. S4D, yellow trace) as evidenced by the absence of a high molecular weight peak in the size exclusion chromatogram. Q48 forms an amide bridge with Q164 (Fig. 5C), an uncommon interaction seen within the structures present in the Protein Data bank (PDB) (Joosten et al., 2009). However, they are stronger interactions than typical hydrogen bonds and less affected by pH changes than salt bridges, another strong interaction (Xie et al., 2015). According to the theoretical pKa of amino acids, Histidines would be the only residue that would change its protonation state in the pH interval 5.5–8.0; however, no Histidines were found in the oligomerisation interface. Alterations of the pKa can result from the influence of neighbouring residue in a 3D structure. Prediction by PROPKA did not show a significant change in other ionisable groups, suggesting that these residues mainly play a role in the pH-dependent overall charge distribution of the monomers (Fig. 6A and B).
Fig. 6.
Evaluation of EspB pore. (A–B) Electrostatic potential of EspB oligomer at pH 5.4 and (B) neutral pH 6.7 (−10 kT/e − red, 10 kT/e − blue). The protonation state was assigned by PROPKA (Olsson et al., 2011) and electrostatic calculations were generated by APBS (Baker et al., 2001) and PDBPQR (Dolinsky et al., 2004). (C) Surface representation of amino acid hydrophobicity according to the Kyte-Doolittle scale (polar residues – purple, non-polar residues – gold). (D) High-resolution 2D class of EspB heptamer with extra density in the middle. (E) Projection of the 3D map obtained for the 7 + 1 EspB oligomer. (F) C1 3D map of 7 + 1 EspB oligomer with local symmetry applied to the heptamer ring. (G) C1 3D map of the 7 + 1 EspB oligomer with 8-fold local symmetry applied and models fitted to the map.
The high-resolution EspB oligomer maps did not reveal a continuous density for the PE-PPE linker. The proposed location of the linker within the crystal structure (Korotkova et al., 2015) overlaps with the oligomerisation interface, and would need to adopt a different position upon oligomerisation. Particle subtraction followed by focused classification showed partial densities for the linker at the periphery of the structure. To confirm that the PE-PPE linker adopts a different conformation upon oligomerisation, we created a double mutation, N55C (in the core of the monomer) and T119C (in the PE-PPE linker), that would lock the linker in its crystal position, thus preventing it from adopting a different conformation needed for the oligomerisation. This double mutant abolished oligomerisation of EspB, suggesting that such intramolecular disulfide bond was formed (Fig. S3D, red trace).
3.4. EspB, a possible transport channel for T7SS proteins
The EspB cylinder-like structure has an internal pore diameter of 40 Å (Fig. 5A), large enough to accommodate folded T7SS substrates such as EsxA/EsxB (diameter 35 Å), PE25-PPE41 (diameter 27 Å) or an EspB monomer itself (diameter 28 Å). Analysis of the degree of hydrophobicity in the structure showed that the internal surface of the oligomer is mainly hydrophilic (Fig. 6C), allowing other hydrophilic molecules to pass.
During the cryo-EM data processing, additional densities were consistently found within the EspB heptamer of all the different constructs, including constructs that lack the C-terminal region. Fig. 6D shows a high-resolution 2D class of EspB2-348 with a well-defined density inside the channel from a subset of the data collected at 0°-stage tilt. This 2D class was found in ~7% of the particles recorded. The 2D classes obtained at 40°-stage tilt could not be unambiguously manually assigned to specific oligomerisation forms. Instead, 3D classification in RELION (Scheres, 2012) was used to identify one class with solely C7 symmetry and one class with an extra density within the heptameric channel. Local symmetry averaging of the heptamer model while processing the overall map in C1 map revealed an extra density spanning the entire channel, in which an EspB monomer model can be fitted (Fig. 6E–G).
3.5. Integrity of the PE-PPE linker is not essential for the oligomerisation of EspB
To determine if the PE-PPE linker absent in our model was essential for oligomerisation, we performed limited proteolysis analysis on the M. tuberculosis constructs. Incubation with trypsin fully digested EspB7-278, perhaps due to a lower stability (Fig. 2A), but resulted in two major fragments for the constructs EspB2-460 and EspB2-348, as shown by SDS-PAGE (Fig. S4A). N-terminal sequencing and mass spectrometry analysis revealed that the larger fragment corresponded to a section of the protein comprising residues V122 to R343 (corresponding to the PPE domain), while the smaller fragment included the N-terminal end of the protein sequence, with a few residues from the affinity tag, up to residue R121 (PE domain and linker) (Figs. S4B−D). Despite being split within the PE-PPE linker into two fragments, EspB2-348 behaved in SEC as a single entity with the capacity to form oligomers (Figs. S4E and F), confirming that the integrity of this region is not necessary for the complex to form. It is noteworthy that trypsin did not cut before R343, even though there is a cleavage site in the allegedly unfolded C-terminal region, raising the question whether this region is truly fully unstructured.
3.6. Properties of the EspB C-terminal region
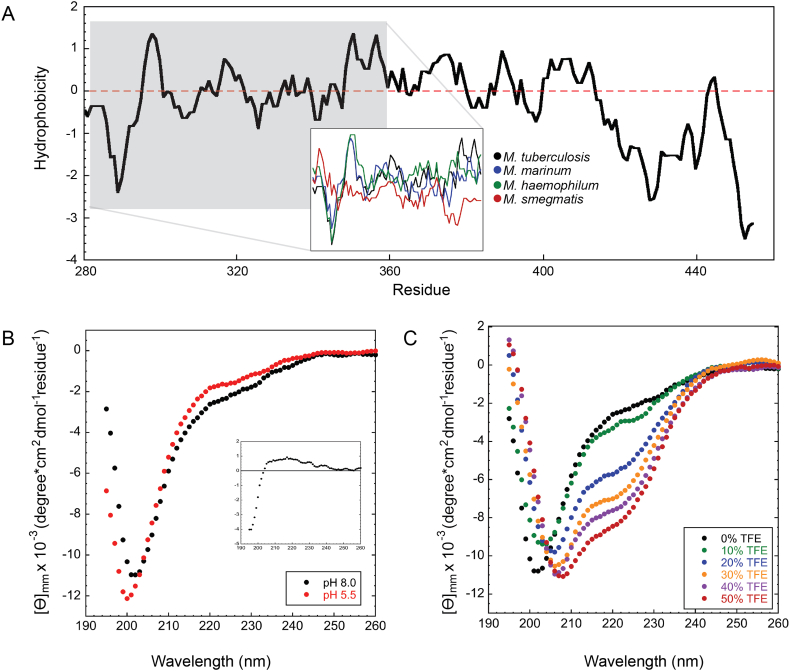
The function of the C-terminal region has puzzled the scientific community for a long time, partly because EspB is the only substrate known to date of the MycP1 protease. Here, we described its processing as an important factor for the oligomerisation of the N-terminal region (1–287); however, the cleavage leaves ~70 residues for no-obvious reason. To gain insight in the properties of the C-terminal region that could hint for its function, and based on the preferential orientation-effect seen in cryo-EM, we performed a hydrophobicity analysis of this region on the different EspB orthologues. Analysis evidenced the presence of hydrophobic patches in the slow-growing species, and absent in EspB from M. smegmatis (Fig. S5A). Some of these patches are present in all the constructs with preferential orientation, leading to speculate that residues in the 297–324 region interact with the air-water interface of the cryo-EM grid (Noble et al., 2018).
To understand whether this effect is related to a structural change or a particular characteristic in the C-terminal region of the protein, we expressed a construct corresponding to residues 279–460 and carried out circular dichroism (CD) studies on it. Far UV CD spectra analysis of this region showed a negative band around 198 nm (Fig. S5B), characteristic of random coil structures. This result is in line with the high fraction (54%) of “disorder-promoting” residues within this region (lysine, glutamine, serine, glutamic acid, proline, and glycine: amino acids commonly found in intrinsically disordered protein regions). Interestingly, its proline content is 2.5 times higher than that observed for proteins in the PBD (Theillet et al., 2013; Uversky, 2013). Analysis of the CD difference spectra obtained at two pH [ΔƟ (pH 5.5 – pH 8.0)] revealed a positive signal close to 220 nm and a negative signal near 200 nm (Fig. S5B inset), showing that this region is able to adopt extended left-handed helical conformations [poly-L-proline type II or PPII (Rucker and Creamer, 2002)]. CD analysis of the C-terminal region in the presence of different concentrations of 2,2,2-trifluoroethanol (TFE) showed that this region has an intrinsic ability to attain helicity based on the decrease in the ellipticity signal at 222 nm (Fig. S5C) (Luo and Baldwin, 1997). The lack of a single isodichroic point at 200 nm suggests that the conformational changes elicited by TFE do not comply with a two-state model; most probably, the transition is accompanied by an intermediate, e.g., the presence of more than one α-helix.
4. Discussion
In the present study, we describe different factors that facilitate oligomerisation of EspB: an acidic environment, the truncation of its C-terminal region, a flexible PE-PPE linker and the residues involved in the interaction. Our findings are in agreement with previous observations that EspB oligomerises upon secretion (Korotkova et al., 2015). Based on these results, the C-terminus of the full-length protein could prevent premature oligomerisation in the cytosol of mycobacteria, possibly through steric hindrance. However, this region is also likely to have other functions. Deletion of EspB C-terminus does not affect its own secretion (McLaughlin et al., 2007; Xu et al., 2007) but rather the secretion of EsxA/EsxB, possibly by loss of interaction with the last residues of EspB (Xu et al., 2007). The sequence of the C-terminal end is highly conserved (Fig. S1), which makes it possible that this region interacts with other molecules in the cytoplasm of the bacterium.
The ability of EspB to oligomerise seems to be conserved across slow-growing species, in contrast to fast-growing mycobacteria, like M. smegmatis. This microorganism is a non-pathogenic species that uses ESX-1 system for horizontal DNA transfer (Flint et al., 2004). The exact mechanism of this transfer is unknown; however, evidence suggests that ESX-1 is not the DNA conduit but rather secretes proteins that act like pheromones, which in turn induce the expression of esx-4 genes resulting in mating-pair interactions (Gray et al., 2016). The ESX-1 substrate EsxA was shown to undergo a structural change that allows membrane insertion in M. tuberculosis when exposed to an acidic environment, however, this effect does not occur in its M. smegmatis orthologue (De Leon et al., 2012; Ma et al., 2015). Taking the aforementioned antecedents and the oligomerisation differences between EspB proteins observed in this work, it is plausible that the mechanism of action of ESX-1 is distinct between these two species. High degree of sequence conservation in slow-growing species compared to the fast-growing ones suggests that EspB gained the ability to oligomerise during evolution. Slow-growth is usually a characteristic of pathogenic species; however, pathogenesis is a complex process involving more elements than the ESX-1 secretion system (Abdallah et al., 2007). Follow-up experiments are needed to fully understand the role of EspB in mycobacterial pathogenesis.
EspB interacts with the lipids phosphatidylserine and phosphatidic acid (Chen et al., 2013). It was suggested that EspB could transport phosphatidic acid (Piton et al., 2020), but the complex's interior is mainly hydrophilic, making this scenario less plausible. Despite the presence of lipids in the crystallisation setup, Korotkova et al. (2015) could not find lipids within the crystal structure of EspB7-278, which lacks the C-terminus. Our results show that the C-terminus of EspB contributes to the protein's preferred orientation on an EM grid caused by interaction with the hydrophobic air-water interface (Noble et al., 2018), analogous to what could happen on a lipid membrane. With a PPII helix at the end of the channel followed by hydrophobic patches at the C-terminus, we hypothesise that this secondary structure interacts with the head group of the lipids, as it has been described for other PPII helices (Franz et al., 2016), allowing the hydrophobic residues to insert into bilayer membranes. Based on the chemical properties of the channel and supported by the evidence of an extra EspB monomer observed within the oligomer, we propose that EspB could be a structural element of ESX-1, allowing other substrates to transit through the channel.
The combined data presented in our work leads us to hypothesise three models of the role of the EspB oligomer. EspB within the cytosol is likely to be monomeric (Korotkova et al., 2015), either free or chaperoned by EspK (McLaughlin et al., 2007), similar to the role of EspG and PE/PPE pairs (Korotkova et al., 2014; Williamson et al., 2020). The binding of a chaperone to the helical tip of EspB would place the WxG and YxxD bipartite secretion signal exposed on the top of EspB, ready to interact with the T7SS machinery. Upon exiting ESX-1 inner-membrane pore, the pre-protein EspB will be cleaved within the periplasm by MycP1. Analogous to ESX-5 (Bunduc et al., 2021), we expect MycP1 to cap the central periplasmic dome-like chamber formed by EccB1, and to have its proteolytic site facing towards its central pore. The cleavage of the C-terminal region at A358 (Solomonson et al., 2013) will remove the most hydrophilic part of the C-terminus, leaving a hydrophobic tail (Fig. S5A).
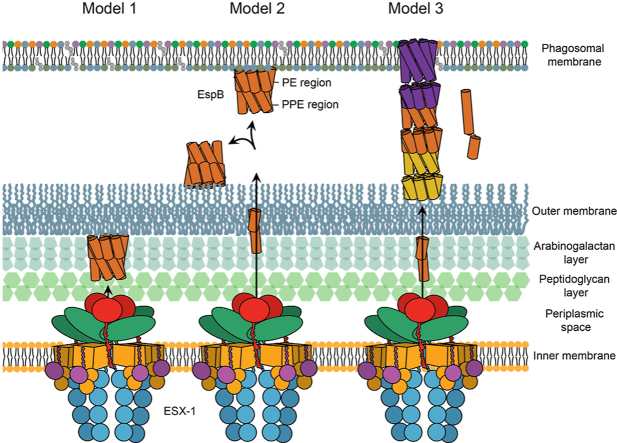
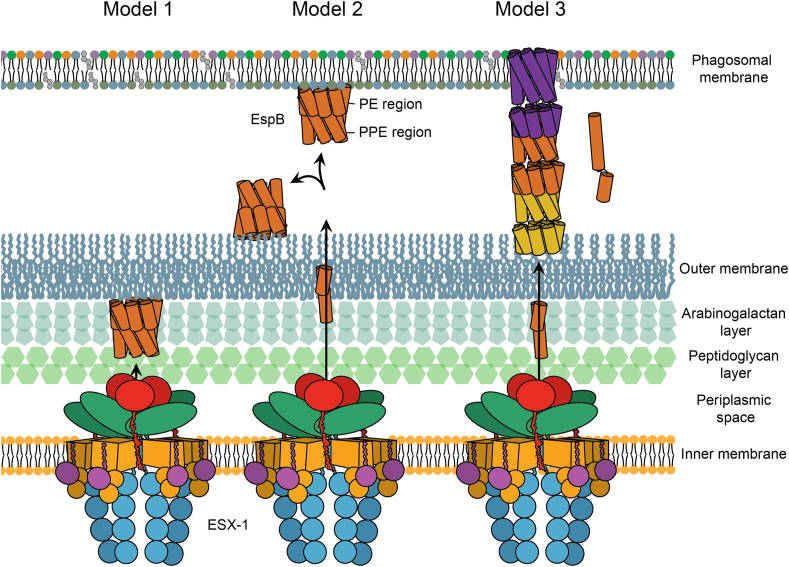
From here, we propose three possible pathways for the oligomerisation of EspB. In one scenario, after processing the C-terminus, EspB binds the outer membrane of mycobacteria, decreasing its critical concentration to form an oligomer (Fig. 7 model 1). As suggested above, it can be assumed that EspB monomers would transit through the inner-membrane pore with the top first, where the C-terminus, as well as the WxG and YxxD motifs, are located. In this position, the monomers would already be properly oriented to form an oligomer on the outer membrane inner leaflet just like EspB2-358 attaches to the air-water interface of a cryo-EM grid (Fig. 4A). The inner pore of the EspB heptamer has similar dimensions compared to that proposed by Beckham et al. for the ESX-5 hexameric structure (Beckham et al., 2017), albeit they later published a higher resolution structure with a more constricted pore in a close state (Beckham et al., 2021). The space between the inner and outer-membrane has been reported to be 20–24 nm wide (Dulberger et al., 2020; Sani et al., 2010; Zuber et al., 2008), which could accommodate the 9-nm long EspB heptamer. It was postulated (Piton et al., 2020) that the positively charged interior of the EspB channel could play a role in the transfer of negatively charged substrates such as DNA or phospholipids. However, in analogy to the negative lumen of a bacteriophage tail that is used to transfer DNA (Zinke et al., 2020), we propose that the positively charged interior space of the EspB oligomer would channel substrates of the same charge, as negatively charged substrates would most likely bind and get trapped. Since the heptameric structure presented here lacks any trans-membrane domains and is highly soluble, it is unlikely to be embedded within the outer membrane. However, it could be anchored by its C-terminus forming part of a larger machinery that completes the ESX-1 core complex. EspB is well known to be secreted to the culture medium of mycobacteria (Lodes et al., 2001); thus, in this model, EspB will help in its own secretion by forming a channel through which additional substrates could travel. This proposal is based on the fact that EspB is cleaved right after crossing the inner membrane of the bacterium. This cleavage is one of the factors that prime oligomerisation.
Fig. 7.
Putative pathways for the oligomerisation of EspB. In Model 1, EspB is cleaved in its C-terminus by the protease MycP1 in the periplasm of mycobacteria leaving hydrophobic residues to insert into the outer membrane; an increase in the local concentration on the membrane leads to oligomerisation of EspB. In model 2, secretion of EspB across the double membrane after MyP1 cleavage allows the protein to bind to either the phagosomal membrane or the external part of the outer membrane. In model 3, after cleavage in the periplasm and secretion to the exterior of the bacterium, EspB goes through a conformational change dissociating the PE and PPE domains and exposing hydrophobic residues that would allow the insertion into the membrane; while the PPE gets embedded into the membrane in an oligomeric form, the respective PE is able to interact with the PPE of a second molecule forming a tubular structure. Different colours are used for each heptamer-subunit. Regardless of what oligomerisation pathway EspB follows, oligomerised EspB is hypothesised to form part of the larger machinery that completes the inner-membrane complex of ESX-1.
In a second scenario, when EspB is secreted outside the bacterium, it could interact with either the phagosomal membrane or the external face of the outer membrane (Fig. 7 model 2). The aforementioned hypothesis of how EspB is secreted (C-terminus and WxG/YxxD motifs first) would favour interaction with the phagosomal membrane; however, there is also some evidence EspB being extracted from the outer-most layer of the bacterium (Sani et al., 2010). From different experiments (Fig. 1), it is expected that for soluble oligomers, a high amount of protein would be needed before the complex is formed. Interaction with the secretion machinery or a membrane would increase the local concentration, potentially reducing the amount of protein required and making the system more efficient.
In the third more speculative model, EspB would undergo a conformational change, as observed for some pore-forming proteins such as the amphitropic gasdermins (Liu and Lieberman, 2020). Upon proteolysis, a pre-pore ring could assemble prior to membrane insertion (Ruan et al., 2018). Recently, it was suggested that the PE/PPE family of proteins could form small molecule-selective channels analogous to outer-membrane porins, allowing M. tuberculosis to take up nutrients through its almost impermeable cell wall (Wang et al., 2020). Despite the evidence, it remains a mystery how such soluble heterodimers would insert into a membrane. We hypothesise that, analogous to the heterodimer EsxA/EsxB where EsxA alone can insert into a membrane in acidic conditions (De Leon et al., 2012), the amphiphilic helices of either PE or PPE alone might insert into the membrane. EspB is fundamentally different from PE/PPE pairs in the sense that its PE and PPE parts are fused into a single protein, joined by one long flexible linker able to adopt multiple conformations (Piton et al., 2020). Unlike the EsxA-EsxB heterodimer, where EsxA would act independently from EsxB upon membrane insertion, the PE moiety of EspB would still be linked to its PPE counterpart even if the latter would insert itself into a membrane. We speculate that such a linker could allow EspB to form tubular-like structures while exchanging PE and PPE domains between different molecules (Fig. 7 model 3). As described for EspC (Lou et al., 2017) and occasionally also found in our data (Fig. S6), such higher-order oligomers could be a component of the secretion apparatus.
Our hypothesis that EspB acts as a scaffold or structural component of the secretion apparatus is supported by earlier findings. As ESX-1 works through a contact-dependent mechanism and not by secretion of toxins (Conrad et al., 2017), it is possible that the cytotoxic effects on macrophages observed by Chen et al. (2013) for EspB was the result of an increment in the machinery activity. Most of the work described here favours model 1 or 2. More evidence needs to be gathered to verify or falsify any of the models (Table S4). Techniques like in situ cryo-electron tomography of infected immune cells (Berger et al., 2021) could be used to provide more relevant insight.
In summary, this study reveals factors that prime the oligomerisation of EspB and presents evidence that supports the hypothesis that EspB is a structural element of ESX-1 secretion system, possibly acting on a lipid membrane. ESX-1 is essential for the virulence of infectious mycobacteria such as M. tuberculosis. However, after decades of arduous research, our understanding on the structure and the mechanism of action of this system remains limited. Here we provide a structural and possibly functional understanding of an ESX-1 element that could be used to guide structure-based drug and vaccine design in order to tackle the global health threat that tuberculosis is.
Data availability
The final global and local B-factor sharpened maps as well as the half-maps and masks have been deposited in EMPIAR for M. tuberculosis and M. marinum under the access number EMD-13154 and EMD-13153, respectively. Likewise, refined models were deposited in the Protein Data Bank with the access code 7P13 and 7P0Z, respectively. Deposition of heptamer model contaning a monomer inside (7 + 1) has been linked to the high-resolution heptamer model access code.
CRediT authorship contribution statement
Abril Gijsbers: Conceptualization, Methodology, Formal analysis, Investigation, Visualization, Writing – original draft, Writing – review & editing. Vanesa Vinciauskaite: Methodology, Investigation. Axel Siroy: Methodology, Investigation, Writing – review & editing. Ye Gao: Methodology. Giancarlo Tria: Methodology, Formal analysis. Anjusha Mathew: Methodology, Formal analysis. Nuria Sánchez-Puig: Methodology, Formal analysis, Writing – review & editing. Carmen López-Iglesias: Supervision, Writing – review & editing. Peter J. Peters: Conceptualization, Supervision, Funding acquisition, Writing – review & editing. Raimond B.G. Ravelli: Conceptualization, Formal analysis, Investigation, Supervision, Funding acquisition, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Paul van Schayck (UM) for indispensable SerialEM and IT support; the Microscopy CORE Lab (UM) for their technical and scientific support; Yue Zhang (UM) for model refinement support; Chris Lewis (UM) for tomogram reconstruction support; Florence Pojer and Stewart Cole (Global Health Institute, Lausanne, Switzerland) for initial sample aliquots and preliminary studies; Ron Heeren and Shane Ellis (UM) for native mass spectrometry support; and Hang Nguyen (UM) for critical reading of the manuscript. Nuria Sánchez-Puig acknowledges the support from the PASPA-DGAPA programme from UNAM and CONACYT 283909. This research received funding from the Netherlands Organisation for Scientific Research (NWO) in the framework of the Fund New Chemical Innovations, numbers 731.016.407 and 184.034.014, from the European Union's Horizon 2020 Research and Innovation Programme under Grant Agreement No 766970 Q-SORT. This research is also part of the M4I research programme supported by the Dutch Province of Limburg through the LINK programme.
Handling editor: Natalie Strynadka
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crstbi.2021.06.001.
Contributor Information
Peter J. Peters, Email: pj.peters@maastrichtuniversity.nl.
Raimond B.G. Ravelli, Email: rbg.ravelli@maastrichtuniversity.nl.
Appendix A. Supplementary data
The following is the supplementary data to this article:
Fig S1.
Fig S2.
Fig S3.
Fig S4.
Fig S5.
Fig S6.
References
- Abdallah A.M., Bestebroer J., Savage N.D., de Punder K., van Zon M., Wilson L., Korbee C.J., van der Sar A.M., Ottenhoff T.H., van der Wel N.N. Mycobacterial secretion systems ESX-1 and ESX-5 play distinct roles in host cell death and inflammasome activation. J. Immunol. 2011;187:4744–4753. doi: 10.4049/jimmunol.1101457. [DOI] [PubMed] [Google Scholar]
- Abdallah A.M., Gey van Pittius N.C., Champion P.A., Cox J., Luirink J., Vandenbroucke-Grauls C.M., Appelmelk B.J., Bitter W. Type VII secretion--mycobacteria show the way. Nat. Rev. Microbiol. 2007;5:883–891. doi: 10.1038/nrmicro1773. [DOI] [PubMed] [Google Scholar]
- Afonine P.V., Klaholz B.P., Moriarty N.W., Poon B.K., Sobolev O.V., Terwilliger T.C., Adams P.D., Urzhumtsev A. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D. 2018;74:814–840. doi: 10.1107/S2059798318009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ates L.S., Ummels R., Commandeur S., van de Weerd R., Sparrius M., Weerdenburg E., Alber M., Kalscheuer R., Piersma S.R., Abdallah A.M. Essential role of the ESX-5 secretion system in outer membrane permeability of pathogenic mycobacteria. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham K.S., Ciccarelli L., Bunduc C.M., Mertens H.D., Ummels R., Lugmayr W., Mayr J., Rettel M., Savitski M.M., Svergun D.I. Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis. Nat Microbiol. 2017;2:17047. doi: 10.1038/nmicrobiol.2017.47. [DOI] [PubMed] [Google Scholar]
- Beckham K.S., Ritter C., Chojnowski G., Ziemianowicz D.S., Mullapudi E., Rettel M., Savitski M.M., Mortensen S.A., Kosinski J., Wilmanns M. Structure of the mycobacterial ESX-5 Type VII secretion system pore complex. Science Advances. 2021;7(26) doi: 10.1126/sciadv.abg9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C., Ravelli R.B.G., Lopez-Iglesias C., Kudryashev M., Diepold A., Peters P.J. Structure of the Yersinia injectisome in intracellular host cell phagosomes revealed by cryo FIB electron tomography. J. Struct. Biol. 2021;213:107701. doi: 10.1016/j.jsb.2021.107701. [DOI] [PubMed] [Google Scholar]
- Bitter W., Houben E.N., Bottai D., Brodin P., Brown E.J., Cox J.S., Derbyshire K., Fortune S.M., Gao L.Y., Liu J. Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan P.J., Nikaido H. The envelope of mycobacteria. Annu. Rev. Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- Bunduc C.M., Fahrenkamp D., Wald J., Ummels R., Bitter W., Houben E.N.G., Marlovits T.C. Structure and dynamics of a mycobacterial type VII secretion system. Nature. 2021;593:445–448. doi: 10.1038/s41586-021-03517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggraaf M.J., Ates L.S., Speer A., van der Kuij K., Kuijl C., Bitter W. Optimization of secretion and surface localization of heterologous OVA protein in mycobacteria by using LipY as a carrier. Microb. Cell Factories. 2019;18:44. doi: 10.1186/s12934-019-1093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson F., Joshi S.A., Rangell L., Brown E.J. Polar localization of virulence-related Esx-1 secretion in mycobacteria. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascioferro A., Delogu G., Colone M., Sali M., Stringaro A., Arancia G., Fadda G., Palu G., Manganelli R. PE is a functional domain responsible for protein translocation and localization on mycobacterial cell wall. Mol. Microbiol. 2007;66:1536–1547. doi: 10.1111/j.1365-2958.2007.06023.x. [DOI] [PubMed] [Google Scholar]
- Chen J.M., Zhang M., Rybniker J., Boy-Rottger S., Dhar N., Pojer F., Cole S.T. Mycobacterium tuberculosis EspB binds phospholipids and mediates EsxA-independent virulence. Mol. Microbiol. 2013;89:1154–1166. doi: 10.1111/mmi.12336. [DOI] [PubMed] [Google Scholar]
- Conrad W.H., Osman M.M., Shanahan J.K., Chu F., Takaki K.K., Cameron J., Hopkinson-Woolley D., Brosch R., Ramakrishnan L. Mycobacterial ESX-1 secretion system mediates host cell lysis through bacterium contact-dependent gross membrane disruptions. Proc. Natl. Acad. Sci. U. S. A. 2017;114:1371–1376. doi: 10.1073/pnas.1620133114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon J., Jiang G., Ma Y., Rubin E., Fortune S., Sun J. Mycobacterium tuberculosis ESAT-6 exhibits a unique membrane-interacting activity that is not found in its ortholog from non-pathogenic Mycobacterium smegmatis. J. Biol. Chem. 2012;287:44184–44191. doi: 10.1074/jbc.M112.420869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky T.J., Nielsen J.E., McCammon J.A., Baker N.A. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulberger C.L., Rubin E.J., Boutte C.C. The mycobacterial cell envelope - a moving target. Nat. Rev. Microbiol. 2020;18:47–59. doi: 10.1038/s41579-019-0273-7. [DOI] [PubMed] [Google Scholar]
- Ekiert D.C., Cox J.S. Structure of a PE-PPE-EspG complex from Mycobacterium tuberculosis reveals molecular specificity of ESX protein secretion. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14758–14763. doi: 10.1073/pnas.1409345111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famelis N., Rivera-Calzada A., Degliesposti G., Wingender M., Mietrach N., Skehel J.M., Fernandez-Leiro R., Bottcher B., Schlosser A., Llorca O. Architecture of the mycobacterial type VII secretion system. Nature. 2019 doi: 10.1038/s41586-019-1633-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferluga J., Yasmin H., Al-Ahdal M.N., Bhakta S., Kishore U. Natural and trained innate immunity against Mycobacterium tuberculosis. Immunobiology. 2020;225:151951. doi: 10.1016/j.imbio.2020.151951. [DOI] [PubMed] [Google Scholar]
- Flint J.L., Kowalski J.C., Karnati P.K., Derbyshire K.M. The RD1 virulence locus of Mycobacterium tuberculosis regulates DNA transfer in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12598–12603. doi: 10.1073/pnas.0404892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz J., Lelle M., Peneva K., Bonn M., Weidner T. SAP(E) - a cell-penetrating polyproline helix at lipid interfaces. Biochim. Biophys. Acta. 2016;1858:2028–2034. doi: 10.1016/j.bbamem.2016.05.021. [DOI] [PubMed] [Google Scholar]
- Gey Van Pittius N.C., Gamieldien J., Hide W., Brown G.D., Siezen R.J., Beyers A.D. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-10-research0044. RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T.A., Clark R.R., Boucher N., Lapierre P., Smith C., Derbyshire K.M. Intercellular communication and conjugation are mediated by ESX secretion systems in mycobacteria. Science. 2016;354:347–350. doi: 10.1126/science.aag0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q. Rev. Biophys. 1995;28:171–193. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- Houben D., Demangel C., van Ingen J., Perez J., Baldeon L., Abdallah A.M., Caleechurn L., Bottai D., van Zon M., de Punder K. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol. 2012;14:1287–1298. doi: 10.1111/j.1462-5822.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- Houben E.N., Bestebroer J., Ummels R., Wilson L., Piersma S.R., Jimenez C.R., Ottenhoff T.H., Luirink J., Bitter W. Composition of the type VII secretion system membrane complex. Mol. Microbiol. 2012;86:472–484. doi: 10.1111/j.1365-2958.2012.08206.x. [DOI] [PubMed] [Google Scholar]
- Joosten R.P., Salzemann J., Bloch V., Stockinger H., Berglund A.C., Blanchet C., Bongcam-Rudloff E., Combet C., Da Costa A.L., Deleage G. PDB_REDO: automated re-refinement of X-ray structure models in the PDB. J. Appl. Crystallogr. 2009;42:376–384. doi: 10.1107/S0021889809008784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapust R.B., Tozser J., Fox J.D., Anderson D.E., Cherry S., Copeland T.D., Waugh D.S. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- Korotkova N., Freire D., Phan T.H., Ummels R., Creekmore C.C., Evans T.J., Wilmanns M., Bitter W., Parret A.H., Houben E.N. Structure of the Mycobacterium tuberculosis type VII secretion system chaperone EspG5 in complex with PE25-PPE41 dimer. Mol. Microbiol. 2014;94:367–382. doi: 10.1111/mmi.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkova N., Piton J., Wagner J.M., Boy-Rottger S., Japaridze A., Evans T.J., Cole S.T., Pojer F., Korotkov K.V. Structure of EspB, a secreted substrate of the ESX-1 secretion system of Mycobacterium tuberculosis. J. Struct. Biol. 2015;191:236–244. doi: 10.1016/j.jsb.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Liu X., Lieberman J. Knocking 'em dead: pore-forming proteins in immune defense. Annu. Rev. Immunol. 2020;38:455–485. doi: 10.1146/annurev-immunol-111319-023800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes M.J., Dillon D.C., Mohamath R., Day C.H., Benson D.R., Reynolds L.D., McNeill P., Sampaio D.P., Skeiky Y.A., Badaro R. Serological expression cloning and immunological evaluation of MTB48, a novel Mycobacterium tuberculosis antigen. J. Clin. Microbiol. 2001;39:2485–2493. doi: 10.1128/JCM.39.7.2485-2493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y., Rybniker J., Sala C., Cole S.T. EspC forms a filamentous structure in the cell envelope of Mycobacterium tuberculosis and impacts ESX-1 secretion. Mol. Microbiol. 2017;103:26–38. doi: 10.1111/mmi.13575. [DOI] [PubMed] [Google Scholar]
- Luo P., Baldwin R.L. Mechanism of helix induction by trifluoroethanol: a framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- Ma Y., Keil V., Sun J. Characterization of Mycobacterium tuberculosis EsxA membrane insertion: roles of N- and C-terminal flexible arms and central helix-turn-helix motif. J. Biol. Chem. 2015;290:7314–7322. doi: 10.1074/jbc.M114.622076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty M.T., Baldwin A.J., Marklund E.G., Hochberg G.K., Benesch J.L., Robinson C.V. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem. 2015;87:4370–4376. doi: 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N., Held S.R. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 2017;197:102–113. doi: 10.1016/j.jsb.2016.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A., Buijs R., Eijkel G.B., Giskes F., Dyachenko A., van der Horst J., Byelov D., Spaanderman D.J., Heck A.J.R., Porta Siegel T., Ellis S.R., Heeren R.M.A. Ion Imaging of Native Protein Complexes Using Orthogonal Time-of-Flight Mass Spectrometry and a Timepix Detector. J. Am. Soc. Mass Spectrom. 2021;32(2):569–580. doi: 10.1021/jasms.0c00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin B., Chon J.S., MacGurn J.A., Carlsson F., Cheng T.L., Cox J.S., Brown E.J. A mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog. 2007;3:e105. doi: 10.1371/journal.ppat.0030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble A.J., Wei H., Dandey V.P., Zhang Z., Tan Y.Z., Potter C.S., Carragher B. Reducing effects of particle adsorption to the air-water interface in cryo-EM. Nat. Methods. 2018;15:793–795. doi: 10.1038/s41592-018-0139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohol Y.M., Goetz D.H., Chan K., Shiloh M.U., Craik C.S., Cox J.S. Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe. 2010;7:210–220. doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M.H., Sondergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theor. Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Phan T.H., Ummels R., Bitter W., Houben E.N. Identification of a substrate domain that determines system specificity in mycobacterial type VII secretion systems. Sci. Rep. 2017;7:42704. doi: 10.1038/srep42704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan T.H., van Leeuwen L.M., Kuijl C., Ummels R., van Stempvoort G., Rubio-Canalejas A., Piersma S.R., Jimenez C.R., van der Sar A.M., Houben E.N.G. EspH is a hypervirulence factor for Mycobacterium marinum and essential for the secretion of the ESX-1 substrates EspE and EspF. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton J., Pojer F., Wakatsuki S., Gati C., Cole S.T. High resolution CryoEM structure of the ring-shaped virulence factor EspB from Mycobacterium tuberculosis. J. Struct. Biol. X. 2020;4 doi: 10.1016/j.yjsbx.2020.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poweleit N., Czudnochowski N., Nakagawa R., Trinidad D.D., Murphy K.C., Sassetti C.M., Rosenberg O.S. The structure of the endogenous ESX-3 secretion system. Elife. 2019;8 doi: 10.7554/eLife.52983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pym A.S., Brodin P., Majlessi L., Brosch R., Demangel C., Williams A., Griffiths K.E., Marchal G., Leclerc C., Cole S.T. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat. Med. 2003;9:533–539. doi: 10.1038/nm859. [DOI] [PubMed] [Google Scholar]
- Rosenberg O.S., Dovala D., Li X., Connolly L., Bendebury A., Finer-Moore J., Holton J., Cheng Y., Stroud R.M., Cox J.S. Substrates control multimerization and activation of the multi-domain ATPase motor of type VII secretion. Cell. 2015;161:501–512. doi: 10.1016/j.cell.2015.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J., Xia S., Liu X., Lieberman J., Wu H. Cryo-EM structure of the gasdermin A3 membrane pore. Nature. 2018;557:62–67. doi: 10.1038/s41586-018-0058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucker A.L., Creamer T.P. Polyproline II helical structure in protein unfolded states: lysine peptides revisited. Protein Sci. 2002;11:980–985. doi: 10.1110/ps.4550102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani M., Houben E.N., Geurtsen J., Pierson J., de Punder K., van Zon M., Wever B., Piersma S.R., Jimenez C.R., Daffe M. Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaberg T., Rebhan K., Lode H. Risk factors for side-effects of isoniazid, rifampin and pyrazinamide in patients hospitalized for pulmonary tuberculosis. Eur. Respir. J. 1996;9:2026–2030. doi: 10.1183/09031936.96.09102026. [DOI] [PubMed] [Google Scholar]
- Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H., Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist M.S., Unnikrishnan M., McConnell M.J., Borowsky M., Cheng T.Y., Siddiqi N., Fortune S.M., Moody D.B., Rubin E.J. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone R., Bobard A., Lippmann J., Bitter W., Majlessi L., Brosch R., Enninga J. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart O.S., Goodfellow J.M., Wallace B.A. The pore dimensions of gramicidin A. Biophys. J. 1993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomonson M., Huesgen P.F., Wasney G.A., Watanabe N., Gruninger R.J., Prehna G., Overall C.M., Strynadka N.C. Structure of the mycosin-1 protease from the mycobacterial ESX-1 protein type VII secretion system. J. Biol. Chem. 2013;288:17782–17790. doi: 10.1074/jbc.M113.462036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomonson M., Setiaputra D., Makepeace K.A.T., Lameignere E., Petrotchenko E.V., Conrady D.G., Bergeron J.R., Vuckovic M., DiMaio F., Borchers C.H. Structure of EspB from the ESX-1 type VII secretion system and insights into its export mechanism. Structure. 2015;23:571–583. doi: 10.1016/j.str.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Stanley S.A., Johndrow J.E., Manzanillo P., Cox J.S. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J. Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- Tan Y.Z., Baldwin P.R., Davis J.H., Williamson J.R., Potter C.S., Carragher B., Lyumkis D. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods. 2017;14:793–796. doi: 10.1038/nmeth.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theillet F.X., Kalmar L., Tompa P., Han K.H., Selenko P., Dunker A.K., Daughdrill G.W., Uversky V.N. The alphabet of intrinsic disorder: I. Act like a Pro: on the abundance and roles of proline residues in intrinsically disordered proteins. Intrinsically Disord. Proteins. 2013;1 doi: 10.4161/idp.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V.N. The alphabet of intrinsic disorder: II. Various roles of glutamic acid in ordered and intrinsically disordered proteins. Intrinsically Disord. Proteins. 2013;1 doi: 10.4161/idp.24684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel N., Hava D., Houben D., Fluitsma D., van Zon M., Pierson J., Brenner M., Peters P.J. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- van Winden V.J., Ummels R., Piersma S.R., Jimenez C.R., Korotkov K.V., Bitter W., Houben E.N. Mycosins are required for the stabilization of the ESX-1 and ESX-5 type VII secretion membrane complexes. mBio. 2016;7 doi: 10.1128/mBio.01471-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Boshoff H.I.M., Harrison J.R., Ray P.C., Green S.R., Wyatt P.G., Barry C.E., 3rd PE/PPE proteins mediate nutrient transport across the outer membrane of Mycobacterium tuberculosis. Science. 2020;367:1147–1151. doi: 10.1126/science.aav5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.J., Headd J.J., Moriarty N.W., Prisant M.G., Videau L.L., Deis L.N., Verma V., Keedy D.A., Hintze B.J., Chen V.B. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson Z.A., Chaton C.T., Ciocca W.A., Korotkova N., Korotkov K.V. PE5-PPE4-EspG3 heterotrimer structure from mycobacterial ESX-3 secretion system gives insight into cognate substrate recognition by ESX systems. J. Biol. Chem. 2020 doi: 10.1074/jbc.RA120.012698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; Geneva: 2019. Global Tuberculosis Report. [Google Scholar]
- Xie N.Z., Du Q.S., Li J.X., Huang R.B. Exploring strong interactions in proteins with quantum chemistry and examples of their applications in drug design. PloS One. 2015;10 doi: 10.1371/journal.pone.0137113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Laine O., Masciocchi M., Manoranjan J., Smith J., Du S.J., Edwards N., Zhu X., Fenselau C., Gao L.Y. A unique Mycobacterium ESX-1 protein co-secretes with CFP-10/ESAT-6 and is necessary for inhibiting phagosome maturation. Mol. Microbiol. 2007;66:787–800. doi: 10.1111/j.1365-2958.2007.05959.x. [DOI] [PubMed] [Google Scholar]
- Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tammaro R., Peters P.J., Ravelli R.B.G. Could egg white lysozyme be solved by single particle cryo-EM? J. Chem. Inf. Model. 2020;60:2605–2613. doi: 10.1021/acs.jcim.9b01176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke M., Sachowsky K.A.A., Oster C., Zinn-Justin S., Ravelli R., Schroder G.F., Habeck M., Lange A. Architecture of the flexible tail tube of bacteriophage SPP1. Nat. Commun. 2020;11:5759. doi: 10.1038/s41467-020-19611-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber B., Chami M., Houssin C., Dubochet J., Griffiths G., Daffe M. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 2008;190:5672–5680. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The final global and local B-factor sharpened maps as well as the half-maps and masks have been deposited in EMPIAR for M. tuberculosis and M. marinum under the access number EMD-13154 and EMD-13153, respectively. Likewise, refined models were deposited in the Protein Data Bank with the access code 7P13 and 7P0Z, respectively. Deposition of heptamer model contaning a monomer inside (7 + 1) has been linked to the high-resolution heptamer model access code.