Abstract
Background
The coronavirus disease 2019 (COVID-19) affects billions of lives around the world and has a significant impact on public healthcare. For quantitative assessment and disease monitoring medical imaging like computed tomography offers great potential as alternative to RT-PCR methods. For this reason, automated image segmentation is highly desired as clinical decision support. However, publicly available COVID-19 imaging data is limited which leads to overfitting of traditional approaches.
Methods
To address this problem, we propose an innovative automated segmentation pipeline for COVID-19 infected regions, which is able to handle small datasets by utilization as variant databases. Our method focuses on on-the-fly generation of unique and random image patches for training by performing several preprocessing methods and exploiting extensive data augmentation. For further reduction of the overfitting risk, we implemented a standard 3D U-Net architecture instead of new or computational complex neural network architectures.
Results
Through a k-fold cross-validation on 20 CT scans as training and validation of COVID-19, we were able to develop a highly accurate as well as robust segmentation model for lungs and COVID-19 infected regions without overfitting on limited data. We performed an in-detail analysis and discussion on the robustness of our pipeline through a sensitivity analysis based on the cross-validation and impact on model generalizability of applied preprocessing techniques. Our method achieved Dice similarity coefficients for COVID-19 infection between predicted and annotated segmentation from radiologists of 0.804 on validation and 0.661 on a separate testing set consisting of 100 patients.
Conclusions
We demonstrated that the proposed method outperforms related approaches, advances the state-of-the-art for COVID-19 segmentation and improves robust medical image analysis based on limited data.
Keywords: COVID-19, Segmentation, Limited data, Computed tomography, Deep learning, Artificial intelligence
Abbreviations
- CNN
Convolutional neural network
- CT
Computed tomography
- COVID-19
coronavirus disease 2019
- DSC
Dice Similarity Coefficient
- FP
False positive rate
- FN
False negative rate
- GGO
Ground-glass opacity
- HU
Hounsfield units
- IoU
Intersection-over-Union
- MIS
Medical image segmentation
- ROI
Regions of interest
- RT-PCR
Reverse transcription polymerase chain reaction
- TN
True negative rate
- TP
True positive rate
1. Introduction
The ongoing coronavirus pandemic has currently (May 18, 2021) spread to 220 countries in the world [1]. The World Health Organization (WHO) declared the outbreak as a “Public Health Emergency of International Concern” on the January 30, 2020 and as a pandemic on the March 11, 2020 [2,3]. Because of the rapid spread of severe respiratory syndrome coronavirus 2 (SARS-CoV-2), billions of lives around the world were changed. A SARS-CoV-2 infection can lead to a severe pneumonia with potentially fatal outcome [[3], [4], [5]]. Until now, there are 163,714,589 confirmed cases in total resulting in 3,392,649 deaths [1]. Through a combined international effort, multiple vaccines were rapidly developed, and various countries already began large vaccine campaigns. However, there is still no effective treatment in case of an infection [3,4,6,7]. Additionally, the rapid increase of confirmed cases and the resulting estimated basic reproduction numbers show that SARS-CoV-2 is highly contagious [4,6,8]. The WHO named this new disease “coronavirus disease 2019”, short form: COVID-19.
An alternative solution to the established reverse transcription polymerase chain reaction (RT-PCR) as standard approach for COVID-19 screening or monitoring is medical imaging like X-ray or computed tomography (CT). The medical imaging technology has made significant progress in recent years and is now a commonly used method for diagnosis, as well for quantification assessment of numerous diseases [[9], [10], [11]]. Particularly, chest CT screening has emerged as a routine diagnostic tool for pneumonia. Therefore, chest CT imaging has also been strongly recommended for COVID-19 diagnosis and follow-up [12]. In addition, CT imaging is playing an important role in COVID-19 quantification assessment, as well as disease monitoring. COVID-19 infected areas are distinguishable on CT images by ground-glass opacity (GGO) in the early infection stage and by pulmonary consolidation in the late infection stage [6,12,13]. An illustration of COVID-19 infected regions on a CT scan can be seen in Fig. 1 . In comparison to RT-PCR, several studies showed that CT is more sensitive and effective for COVID-19 screening, and that chest CT imaging is more sensitive for COVID-19 testing even without the occurrence of clinical symptoms [10,[12], [13], [14]]. Notably, a large clinical study with 1014 patients in Wuhan (China) [12] determined that chest CT analysis can achieve 0.97 sensitivity, 0.25 specificity and 0.68 accuracy for COVID-19 detection.
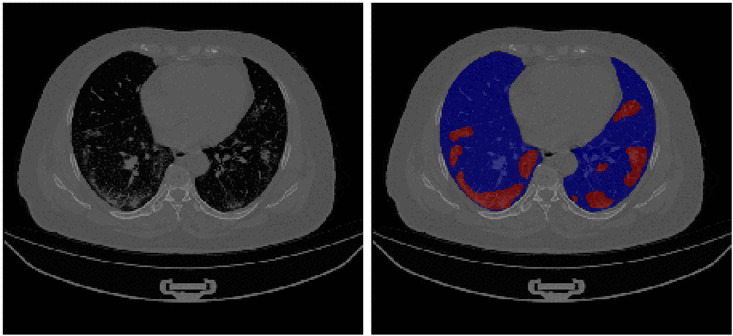
Fig. 1.
Visualization of COVID-19 infected regions in a chest CT. The left image is the unsegmented CT scan, whereas the right image shows segmentation of lungs (blue) and infection (red). The infected regions are distinguishable by GGOs and pulmonary consolidation in the lung regions. The image was obtained from the analyzed CT dataset [45]. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Still, evaluation of medical images is a manual, tedious and time-consuming process performed by radiologists. Even though increasing CT scan resolution and number of slices resulted in higher sensitivity and accuracy, these improvements also increased the workload. Additionally, annotations of medical images are often highly influenced by clinical experience [15,16].
A solution for these challenges could be clinical decision support systems based on automated medical image analysis. In recent years, artificial intelligence has seen a rapid growth with deep learning models, whereas image segmentation is a popular sub-field [9,17,18]. The aim of medical image segmentation (MIS) is the automated identification and labeling of regions of interest (ROI) e.g. organs like lungs or medical abnormalities like cancer and lesions. In recent studies, medical image segmentation models based on neural networks proved powerful prediction capabilities and achieved similar results as radiologists regarding the performance [9,19]. It would be a helpful tool to implement such an automatic segmentation for COVID-19 infected regions as clinical decision support for physicians. By automatic highlighting abnormal features and ROIs, image segmentation is able to aid radiologists in diagnosis, disease course monitoring, reduction of time-consuming inspection processes and improvement of accuracy [9,10,20]. Nevertheless, training accurate and robust models requires sufficient annotated medical imaging data. Because manual annotation is labor-intensive, time-consuming and requires experienced radiologists, it is common that publicly available data is limited [9,10,16]. This lack of data often results in an overfitting of the traditional data-hungry models. Especially for COVID-19, large enough medical imaging datasets are currently unavailable [10,16].
In this work, we push towards creating an accurate and state-of-the-art MIS pipeline for COVID-19 lung infection segmentation, which is capable of being trained on small datasets consisting of 3D CT volumes. In order to avoid overfitting, we exploit extensive on-the-fly data augmentation, as well as diverse preprocessing methods. In order to further reduce the risk of overfitting, we implement the standard U-Net architecture instead of other more computational complex variants, like the residual architecture of the U-Net. Furthermore, we use a sensitivity analysis with k-fold cross-validation for reliable performance evaluation.
Our manuscript is organized as follows: Section 1 introduces the current challenges, our research question and related work on COVID-19 image analysis research. In Section 2, we describe our proposed pipeline including the datasets, preprocessing methods, proposed neural network and evaluation techniques. In Section 3, we report the experimental results, and discuss these in detail in Section 4. In Section 5, we conclude our paper and give insights on future work. The Appendix contains further information on the availability of our trained models, all result data and the code used in this research.
1.1. Related work
Since the breakthrough of convolutional neural network (CNN) architectures for computer vision, neural networks became one of the most accurate and popular machine learning algorithms for automated medical image analysis [9,17,21]. Two of the major tasks in this field are classification and segmentation. Whereas medical image classification aims to label a complete image to predefined classes (e.g. to a diagnosis), medical image segmentation aims to label each pixel in order to identify ROIs (e.g. organs or medical abnormalities). Popular deep learning architectures, which achieved performance equivalent to humans, are Inception-v3 [22], ResNet [23], as well as DenseNet [24] for classification and VB-Net [25], U-Net [26] and various variants of the U-Net for segmentation [10,27].
For measuring the performance of image segmentation models, it is important to select suited metrics for reliable evaluation. Especially, in medical image segmentation, images reveal a large class imbalance between a small but important ROI and the large number of remaining pixels defined as background. The ideal metric should heavily focus on the correct predictability for the ROI, which is usually less than 5 % of pixels of the total image. Taha et al. [28] discussed the behavior and requirements of 3D medical image segmentation metrics in detail and demonstrated that metric behavior can have advantages as well as disadvantages. The disadvantage lays in the restrictiveness of the segmentation patterns. Even if a ROI is correctly identified, small annotation differences, which can arise from not computational refined annotations, can lead to drastic scoring variances due to the large class imbalance in medical imaging. Still, the advantage as well as the necessity of using false negative focused metrics lays in the class imbalance, too. Other common metrics like accuracy are not suited for medical image segmentation due to the true negative influence. Therefore, the scientific community, strongly favors F-score based metrics like the Dice similarity coefficient (1), also called F-1, or the Intersection-over-Union (2), also called F-0 or Jaccard index. Due to their reliable capability of handling class imbalance by focusing on false positive and false negative predictions, the two are the most widespread metrics in computer vision. All related studies, referenced later for medical image segmentation, are using either one or both of the two metrics for evaluation. In contrast, the sensitivity (3) and specificity (4) are one of the most popular metrics in medical fields. All metrics are based on the confusion matrix for binary classification, where TP, FP, TN and FN represent the true positive, false positive, true negative and false negative rate, respectively.
| (1) |
| (2) |
| (3) |
| (4) |
In reaction to the rapid spread of the coronavirus, many scientists quickly reacted and developed various approaches based on deep learning to contribute to the efforts against COVID-19. Furthermore, the scientific community focused their efforts on the development of models for COVID-19 classification, because X-ray and CT images of infected patients could be collected without further required annotations [10,20]. These classification algorithms can be categorized through their objectives: 1) Classification of COVID-19 from non-COVID-19 (healthy) patients, which resulted into models achieving a sensitivity of 94.1 %, specificity of 95.5 %, and AUC of 0.979 by Jin et al. [29]. 2) Classification of COVID-19 from other pneumonia, which resulted in models achieving a sensitivity of 100.0 %, specificity of 85.18 %, and AUC of 0.97 by Abbas et al. [30]. 3) Severity assessment of COVID-19, which resulted in a model achieving a true positive rate of 91.0 %, true negative rate of 85.8 %, and accuracy of 89.0 % by Tang et al. [31].
In the middle of the year 2020, clinicians started to publish COVID-19 CT images with annotated ROIs, which allowed the training of segmentation models. Automated segmentation is highly desired as COVID-19 application [10,32]. The segmentation of lung, lung lobes and lung infection provide accurate quantification data for progression assessment in follow-up, comprehensive prediction of severity in the enrollment and visualization of lesion distribution using percentage of infection (POI) [10]. Still, the limited amount of annotated imaging data causes a challenging task for detecting the variety of shapes, textures and localizations of lesions or nodules. Nonetheless, multiple approaches try to solve these problems with different methods. The most popular network models for COVID-19 segmentation are variants of the U-Net which achieved reasonable performance on sufficiently sized 2D datasets [5,10,[33], [34], [35], [36], [37], [38], [39], [40]]. In order to compensate limited dataset sizes, more attention has been drawn to semi-supervised learning pipelines [10,41,42]. These methods optimize a supervised training on labeled data along with an unsupervised training on unlabeled data. Another approach is the development of special neural network architectures for handling limited dataset sizes. Frequently, attention mechanisms are built into the classic U-Net architecture like the Inf-Net from Fan et al. [41] or the MiniSeg from Qiu et al. [43]. Wang et al. [44] utilized transfer learning strategies based on models trained on non-COVID-19 related conditions. Particularly worth mentioning is the development of a benchmark model with a 3D U-Net from Ma et al. [16,45], because the authors also provide high reproducibility through a publicly available dataset.
2. Methods
This pipeline was based on MIScnn [46], which is an in-house developed open-source framework to setup complete medical image segmentation pipelines with convolutional neural networks and deep learning models on top of Tensorflow/Keras [47]. MIScnn supports extensive preprocessing, data augmentation, state-of-the-art deep learning models and diverse evaluation techniques. The implemented medical image segmentation pipeline is illustrated in Fig. 2 .
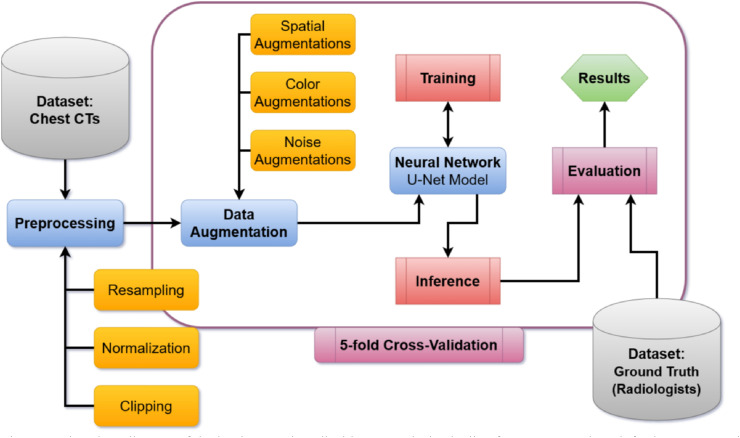
Fig. 2.
Flowchart diagram of the implemented medical image analysis pipeline for COVID-19 lung infection segmentation. The workflow is starting with the COVID-19 dataset and ending with the computed evaluation results for each fold in the cross-validation.
2.1. Datasets of COVID-19 chest CTs
In this study, we used two public datasets: Ma et al. [45] as limited dataset for model training as well as validation, and An et al. [48] as a larger hold-out dataset for additional testing purpose.
The Ma et al. dataset consists of 20 annotated COVID-19 chest CT volumes [16,45]. All cases were confirmed COVID-19 infections with a lung infection proportion ranging from 0.01 % to 59 % [16]. This dataset was one of the first publicly available 3D volume sets with annotated COVID-19 infection segmentation [16]. The CT scans were collected from the Coronacases Initiative and Radiopaedia and were licensed under CC BY-NC-SA. Each CT volume was first labeled by junior annotators, then refined by two radiologists with 5 years of experience and afterwards the annotations verified by senior radiologists with more than 10 years of experience [16]. Despite the fact that the sample size is rather small, the annotation process led to an excellent high-quality dataset. The volumes had a resolution of 512x512 (Coronacases Initiative) or 630x630 (Radiopaedia) with a number of slices of about 176 by mean (200 by median). The CT images were labeled into four classes: Background, lung left, lung right and COVID-19 infection.
The An et al. dataset consists of unenhanced chest CT volumes from 632 patients with COVID-19 infections and is one of the largest publicly available COVID-19 CT datasets [48]. The CT scans were collected through the outbreak settings from patients with a combination of symptoms, exposure to an infected patient or travel history to an outbreak region [48,49]. All patients had a positive RT-PCR for SARS-CoV-2 from a sample obtained within 1 day of the initial CT [48,49]. The annotation of the dataset was made possible through the joint work of Children's National Hospital, NVIDIA and National Institutes of Health for the COVID-19-20 Lung CT Lesion Segmentation Grand Challenge [50]. The challenge authors were able to annotate a subset of 295 patients through American board certified radiologists [50]. Through the characteristic as a challenge, not all volumes had publicly available annotations. Nevertheless, we were able to obtain a subset of 100 patients as additional testing set. The volumes had a resolution of 512x512 with a number of slices of about 75 by mean (65 by median). The CT images were labeled into two classes: Background and COVID-19 infection.
2.2. Preprocessing
In order to simplify the pattern finding and fitting process for the model, we applied several preprocessing methods on the dataset.
We exploited the Hounsfield units (HU) scale by clipping the pixel intensity values of the images to −1250 as minimum and +250 as maximum, because we were interested in infected regions (+50 to +100 HU) and lung regions (−1000 to −700 HU) [51]. It was possible to apply the clipping approach on the Coronacases Initiative and An et al. CTs, because the Radiopaedia volumes were already normalized to a grayscale range between 0 and 255.
Varying signal intensity ranges of images can drastically influence the fitting process and the resulting performance of segmentation models [52]. For achieving dynamic signal intensity range consistency, it is recommended to scale and standardize imaging data. Therefore, we normalized the remaining CT volumes likewise to grayscale range. Afterwards, all samples were standardized via z-score.
Medical imaging volumes have commonly inhomogeneous voxel spacings. The interpretation of diverse voxel spacings is a challenging task for deep neural networks. Therefore, it is possible to drastically reduce complexity by resampling volumes in an imaging dataset to homogeneous voxel spacing, which is also called target spacing. Resampling voxel spacings also directly resizes the volume shape and determines the contextual information, which the neural network model is able to capture. As a result, the target spacing has a huge impact on the final model performance. We decided to resample all CT volumes to a target spacing of 1.58x1.58x2.70, resulting in a median volume shape of 267x254x104.
2.3. Data augmentation
The aim of data augmentation is to create more data of reasonable variations of the desired pattern and, thus, artificially increase the number of training images. This technique results into improved model performance and robustness [[53], [54], [55]]. In order to compensate the small dataset size, we performed extensive data augmentation by using the batchgenerators interface within MIScnn. The batchgenerators package [56] is an API for state-of-the-art data augmentation on medical images from the Division of Medical Image Computing at the German Cancer Research Center. We implemented three types of augmentations: Spatial augmentation by mirroring, elastic deformations, rotations and scaling. Color augmentations by brightness, contrast and gamma alterations. Noise augmentations by adding Gaussian noise. Furthermore, each augmentation method had a random probability of 15 % to be applied on the current image with random intensity or parameters (e.g. random angle for rotation) [56,57].
Instead of traditional upsampling approaches, we performed on-the-fly data augmentation on each image before it was forwarded into the neural network model. The innovative one-the-fly augmentation technique is defined as the creation of novel and unique images in each iteration of the training process instead of generating once a fixed number of augmented images beforehand. Through this technique, the probability that the model encounters the exact same image twice during the training process decreases significantly, which proved to reduce the risk of overfitting drastically [57].
2.4. Patch-wise analysis
In image analysis there are three popular methods: The analysis of full images, the slice-wise analysis for 3D data or patch-wise by slicing the volume into smaller cuboid patches [9]. We selected the patch-wise approach in order to exploit random cropping for the fitting process. Through random forwarding only a single cropped patch from the image to the fitting process, another type of data augmentation is induced, and the risk of overfitting additionally decreased. Furthermore, full image analysis requires unnecessary resolution reduction of the 3D volumes in order to handle the enormous GPU memory requirements. By slicing the volumes into patches with a shape of 160x160x80, we were able to utilize high-resolution data. All slicing processes were done via manual image matrix slicing.
For inference, the volumes were sliced into patches according to a grid. Between the patches, we introduced an overlap of half the patch size (80x80x40) to increase prediction performance. After the inference of each patch, they were reassembled into the original volume shape, whereas overlapping regions were averaged.
2.5. Neural network model
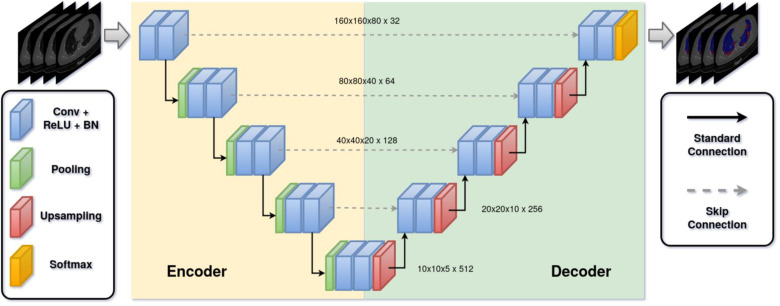
The neural network architecture and its hyper parameters are one of the key parts in a medical image segmentation pipeline. The current landscape of deep learning architectures for semantic segmentation accommodates a variety of variants which distinguish by efficiency, robustness or performance. Nevertheless, the U-Net is currently the most popular and promising architecture in terms of the interaction between performance and variability [[57], [58], [59], [60]]. In this work, we implemented the standard 3D U-Net as architecture without any custom modification in order to avoid unnecessary parameter increase by more complex architectures like the residual variant of the 3D U-Net [26,61,62]. The input of our architecture was a 160x160x80 patch with a single channel consisting of normalized HUs. The output layer of our architecture normalized the class probabilities through a softmax function (normalized exponential function) and returned the 160x160x80 mask with 4 channels representing the probability for each class (background, lung left, lung right and COVID-19 infection). Upsampling was achieved via transposed convolution and downsampling via maximum pooling. The architecture used 32 feature maps at its highest resolution and 512 at its lowest. All convolutions were applied with a kernel size of 3 × 3 × 3 in a stride of 1 × 1 × 1, except for up- and downsampling convolutions which were applied with a kernel size of 2 × 2 × 2 in a stride of 2 × 2 × 2. After each convolutional block, batch normalization was applied. The architecture can be seen in Fig. 3 .
Fig. 3.
The architecture of the standard 3D U-Net. The network takes a 3D patch (cuboid) and outputs the segmentation of lungs and infected regions by COVID-19. Skip connections were implemented with concatenation layers. Conv: Convultional layer; ReLU: Rectified linear unit layer; BN: Batch normalization.
In medical image segmentation, it is common that semantic annotation includes a strong bias in class distribution towards the background class. Our dataset revealed a class distribution of 89 % for background, 9 % for lungs and 1 % for infection. In order to compensate this class bias, we utilized the sum of the Tversky index [63] and the categorical cross-entropy as loss function for model fitting (5).
| (5) |
| (6) |
| (7) |
We implemented a multi-class adaptation for the Tversky index (6), which is an asymmetric similarity index to measure the overlap of the segmented region with the ground truth. It allows for flexibility in balancing the false positive rate (FP) and false negative (FN) rate. The cross-entropy (7) is a commonly used loss function in machine learning and calculates the total entropy between the predicted and true distribution. The multi-class adaptation for multiple categories (categorical cross-entropy) is represented through the sum of the binary cross-entropy for each class c, whereas y o,c is the binary indicator whether the class label c is the correct classification for observation o. The variable p o,c is the predicted probability that observation o is of class c.
For model fitting, an Adam optimization [64] was used with the initial weight decay of 1e-3. We utilized a dynamic learning rate which reduced the learning rate by a factor of 0.1 in case the training loss did not decrease for 15 epochs. The minimal learning rate was set to 1e-5. In order to further reduce the risk of overfitting, we exploited the early stopping technique for training, in which the training process stopped without a fitting loss decrease after 100 epochs. The neural network model was trained for a maximum of 1000 epochs. Instead of the common epoch definition as a single iteration over the dataset, we defined an epoch as the iteration over 150 training batches. This allowed for an improved fitting process for randomly generated batches in which the dataset acts as a variation database. According to our available GPU VRAM, we selected a batch size of 2.
2.6. Sensitivity analysis with cross-validation
For reliable robustness evaluation, we performed a sensitivity analysis to estimate the generalizability and sensitivity of our pipeline. Thus, we performed multiple k-fold cross-validations on the Ma et al. dataset to obtain various models based on limited training data as well as different validation subsets.
As k-fold multitude, we used a range from 2 up to 5 for the sensitivity analysis resulting in to 4 separate cross-validation analyses with in total 14 models. Each model was created through a training process on k-1 folds and validated through the leftover fold in each cross-validation sampling. Training and validation were performed on the small Ma et al. dataset, whereas the An et al. dataset was used as additional testing set to further ensure a robust evaluation. As example, this technique resulted in the following sampling for a 5-fold cross-validation: 16 samples as training dataset (Ma et al.), 4 samples as validation dataset (Ma et al.) and 100 samples as testing dataset (An et al.).
Furthermore, we analyzed the impact of the preprocessing and data augmentation techniques on model performances for the 5-fold cross-validation. We did not configure any hyper parameters afterwards on basis of validation results and did not perform any validation monitoring based training techniques, which allowed us to utilize our validation results for hold-out evaluation, as well.
2.7. Evaluation metrics
During the fitting process, we computed the segmentation performance for each epoch on randomly cropped and data augmented patches from the validation dataset. This allowed for an evaluation of the overfitting on the training data.
After the training, we used mainly four widely popular evaluation metrics in the community for medical image analysis to do the inference performance measurement on the validation and testing set: Dice similarity coefficient, Intersection-over-Union, sensitivity, and specificity. Furthermore, we computed the accuracy and precision as supplementary metrics for the Appendix. The performance measurement was based on the segmentation overlap between prediction and ground truth, which was manually annotated through the consensus of multiple radiologists, as described in the dataset section. For the Ma et al. dataset, the two lung classes (‘lung left’ and ‘lung right’) were averaged by mean into a single class (‘lungs’) during the evaluation.
2.8. Code reproducibility
In order to ensure full reproducibility and to create a base for further research, the complete code of this project, including extensive documentation, is available in a public Git repository which is referenced in the Appendix.
3. Results
The sequential training of the complete cross-validation on 2 NVIDIA QUADRO RTX 6000 with 24 GB VRAM, an Intel Xeon Gold 5220R using 4 CPUs and 20 GB RAM took around 182 h. All models did not require the entire 1000 epochs for training and instead were early stopped after an average of 312 epochs.
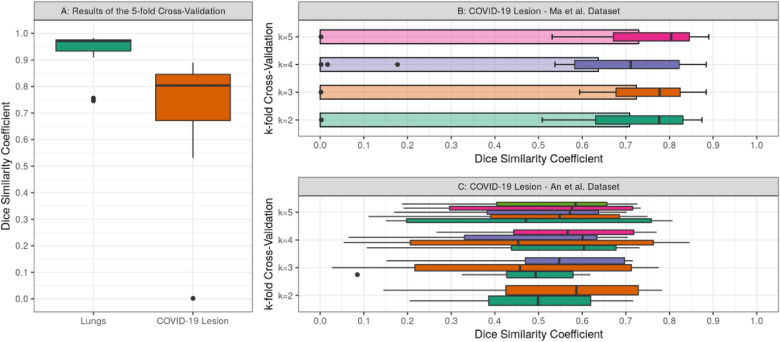
After the training, the inference revealed a strong segmentation performance for lungs and COVID-19 infected regions. Overall, the k-fold cross-validation models achieved a DSC and IoU of around 0.971 and 0.944 for lungs, as well as 0.804 and 0.672 for COVID-19 infection segmentation on the Ma et al. dataset, respectively. On the additional testing set from An et al. the models achieved a DSC of around 0.661 and an IoU of around 0.494 for COVID-19 infection segmentation. Furthermore, the models obtained a sensitivity and specificity of 0.778 and 0.999 on the validation set, as well as 0.580 and 0.999 on the testing set for COVID-19 infection, respectively. More details on inference performance are listed in Table 1 and visualized in Fig. 4 .
Table 1.
Achieved results showing the median Dice similarity coefficient (DSC), the Intersection-over-Union (IoU) the sensitivity (Sens) and specificity (Spec) on Lung and COVID-19 infection segmentation for each k-fold cross-validation of the sensitivity analysis for the Ma et al. and An et al. dataset. Standard deviation is included for DSC and IoU.
| Dataset: Ma et al. |
Dataset: An et al. |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lungs |
COVID-19 Lesion |
COVID-19 Lesion |
||||||||||
| k-fold CV | DSC | IoU | Sens. | Spec. | DSC | IoU | Sens. | Spec. | DSC | IoU | Sens. | Spec. |
| k=2 | 0.960 ± 0.06 | 0.923 ± 0.10 | 0.970 | 0.998 | 0.775 ± 0.20 | 0.635 ± 0.19 | 0.747 | 0.999 | 0.555 ± 0.07 | 0.386 ± 0.07 | 0.485 | 0.998 |
| k=3 | 0.966 ± 0.07 | 0.934 ± 0.10 | 0.968 | 0.999 | 0.778 ± 0.19 | 0.636 ± 0.18 | 0.730 | 0.999 | 0.598 ± 0.10 | 0.426 ± 0.11 | 0.580 | 0.999 |
| k=4 | 0.951 ± 0.22 | 0.907 ± 0.29 | 0.948 | 0.999 | 0.711 ± 0.27 | 0.552 ± 0.25 | 0.731 | 0.999 | 0.661 ± 0.07 | 0.494 ± 0.09 | 0.561 | 0.999 |
| k=5 | 0.971 ± 0.07 | 0.944 ± 0.11 | 0.971 | 0.999 | 0.804 ± 0.20 | 0.672 ± 0.19 | 0.778 | 0.999 | 0.623 ± 0.04 | 0.453 ± 0.04 | 0.513 | 0.998 |
Fig. 4.
Summaries showing the Dice similarity coefficient distributions from validation and testing on the Ma et al. and An et al. datasets. A: Boxplot showing the results of the 5-fold cross-validation on the Ma et al. dataset. B: Boxplots and bar plots showing the average Dice similarity coefficient for each k-fold cross-validation run on the Ma et al. dataset. C: Boxplots for each model of the k-fold cross-validation on the An et al. testing dataset.
For the sensitivity analysis, average evaluation metrics were calculated for each k-fold cross-validation (Table 1) as well as for each data augmentation and preprocessing configuration (Table 2 ). The 5-fold cross-validation revealed the best performance on all evaluation metrics on the validation set, whereas the 4-fold cross-validation was superior on the testing set. The Dice similarity coefficient difference between the best k-fold cross-validation and the worst is 0.093 on validation and 0.106 on testing for COVID-19 lesion segmentation. The inclusion of data augmentation and preprocessing increased the pipeline performance on average by 0.647 for lung and by 0.630 for COVID-19 lesion segmentation based on the Dice similarity coefficient, which is summarized in Table 2.
Table 2.
The segmentation pipeline was applied four times with in-/excluded preprocessing and data augmentation in order to evaluate their performance influence on the model. Achieved results showing the median Dice similarity coefficient (DSC) on Lung and COVID-19 infection segmentation for each CV fold of the 5-fold cross-validation and the global average (AVG) based on the Ma et al. dataset.
| Data Augmentation:Excluded Preprocessing: Excluded |
Data Augmentation: Included Preprocessing: Excluded |
Data Augmentation: Excluded Preprocessing: Included |
Data Augmentation: Included Preprocessing: Included |
|||||
|---|---|---|---|---|---|---|---|---|
| Fold | Lungs | COVID-19 | Lungs | COVID-19 | Lungs | COVID-19 | Lungs | COVID-19 |
| 1 | 0.711 | 0.031 | 0.397 | 0.166 | 0.867 | 0.530 | 0.907 | 0.556 |
| 2 | 0.046 | 0.186 | 0.275 | 0.050 | 0.979 | 0.819 | 0.977 | 0.801 |
| 3 | 0.190 | 0.241 | 0.168 | 0.057 | 0.951 | 0.814 | 0.952 | 0.829 |
| 4 | 0.080 | 0.005 | 0.175 | 0.114 | 0.979 | 0.819 | 0.979 | 0.853 |
| 5 | 0.520 | 0.194 | 0.360 | 0.201 | 0.964 | 0.798 | 0.967 | 0.765 |
| AVG | 0.309 | 0.131 | 0.275 | 0.118 | 0.948 | 0.756 | 0.956 | 0.761 |
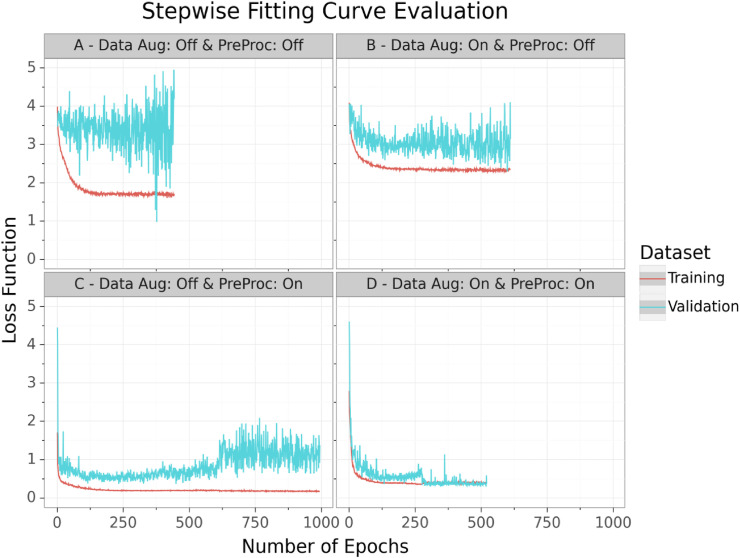
Through validation monitoring, no overfitting was observed. The training and validation loss function revealed no significant distinction from each other, which can be seen in Fig. 5 . During the fitting, the performance settled down at a loss of around 0.383 for the 5-fold cross-validation (Fig. 5-D) which is a generalized DSC (average of all class-wise DSCs) of around 0.919. Because of this robust training process without any signs of overfitting, we concluded that fitting on randomly generated patches via extensive data augmentation and random cropping from a variant database, is highly efficient for limited imaging data.
Fig. 5.
The illustration showing the loss course during the training process for training (red) and validation (cyan) data for the 5-fold cross-validation from four pipeline runs including (‘on’) or excluding (‘off’) data augmentation (Data Aug) and preprocessing (PreProc) techniques. The lines were computed via Gaussian Process Regression and represent the average loss across all folds for each 5-fold cross-validation pipeline run. The final pipeline fitting curve is illustrated in the bottom-right corner (D). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
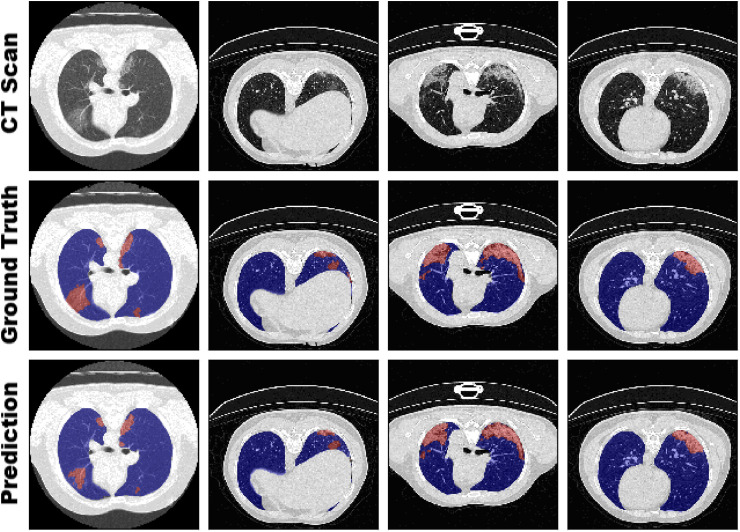
Exemplary for model performance of the 5-fold cross-validation, 4 samples with annotated ground truth and predicted segmentation are visualized in Fig. 6 . The performance evaluation of our sensitivity analysis revealed that there is only a marginal but notable difference between the k-fold cross-validations. As example, the 3-fold cross-validation with a training dataset size of only 13 samples achieved accurate segmentation results on the validation as well as testing set. Interestingly, the 4-fold cross-validation (15 training samples) obtained the best DSC and IoU and the 3-fold cross-validation the best sensitivity on the larger testing set. This demonstrated that generalizability is one of the most important hallmarks of a model, especially if trained on a limited dataset. If all important visual features for the medical condition are present in the training set, a low number of samples can be sufficient by using extensive image augmentation and preprocessing techniques as our pipeline for creating a powerful model. However, if too many samples share similar morphological features without any variation, the risk of overfitting or generating a less generalized model is still present.
Fig. 6.
Visual comparison of the segmentation between ground truth from radiologist annotations and our model (5-fold cross-validation) on four slices from different CT scans of the Ma et al. dataset. Visualization for all samples for both datasets is provided in the appendix.
4. Discussion
From a medical perspective, detection of COVID-19 infection is a challenging task and one of the reasons for the weaker segmentation accuracy in contrast to the lung segmentation. The reason for this is the variety of GGO and pulmonary consolidation morphology. In contrast to the specificity, the dice similarity coefficient as well as the sensitivity are showing a lower but more reliable performance evaluation comparable with the visualized segmentation correctness. The reason for this is that false negative predictions have a strong impact on these two metrics. Especially, in medical image segmentation, in which ROIs are quite small compared to the remaining image, a few incorrect predicted pixels have a large impact on the resulting score. Such strict metrics are required in order to compensate the class unbalance between mostly background and small ROIs in medical imaging. Nevertheless, our medical image segmentation pipeline allowed fitting a model which is able to segment COVID-19 infection with state-of-the-art accuracy that is comparable to models trained on large datasets.
In order to provide further insights on the influence of our methodology on the achieved performance, we run and analyzed our pipeline through a sensitivity analysis based on cross-validation and variable data augmentation as well as applied preprocessing configuration. All other configurations as well as the neural network architecture remained the same as described in the methods section. Thus, this experiment resulted into 30 models (14 models from cross-validation ranging from k-fold 2 up to 5 and 15 models from three 5-fold cross-validation runs with variable data augmentation as well as preprocessing configuration).
The fitting process of the different runs revealed that extensive data augmentation plays an important role for avoiding overfitting and to improve model robustness, as it can be seen in the fitting curves of Fig. 5. Therefrom, the model overfitted on the training data. The on-the-fly data augmentation helped the model to learn a more generalized pattern for recognizing the lungs and infected regions instead of just memorizing the training data. In contrast, the preprocessing methods increased the overall performance of the model by simplifying the computer vision task. The applied methods like resampling or clipping led to a search space reduction which increased the chances of the model to identify patterns in the imaging data. This advantage was also shown in the resulting performances, which can be seen in Table 2. As expected, the pipeline run with no data augmentation as well as no preprocessing appeared to be the worst model. In contrast, the preprocessing techniques demonstrated the highest performance increase on the testing data of the 5-fold cross-validation. Therefore, the final pipeline build combined data augmentation, for improving robustness, and preprocessing techniques, for increasing performance, in order for optimizing inference quality.
4.1. Comparison with prior work
For further evaluation, we compared our pipeline to other available COVID-19 segmentation approaches based on CT scans. Information and further details of related work was structured and summarized in Table 3 . The authors (Ma et al.), who also provided the dataset we used for our analysis, implemented a 3D U-Net approach as a baseline for benchmarking [16]. They were able to achieve a DSC of 0.70355 and 0.6078 for lungs and COVID-19 infection, respectively. With our model, we were able to outperform this baseline. It is important to mention that the authors of this baseline trained with a 5-fold cross-validation sampling of 20 % training and 80 % validation, whereas we used the inverted distribution for our k-fold cross-validations (k-1 folds for training and the k's fold for validation). Based on the Ma et al. dataset, Wang et al. [44] gathered more samples, expanded the dataset and also applied a 3D U-Net which resulted in a DSC of 0.704. Another approach from Yan et al. [65] developed a novel neural network architecture (COVID-SegNet) specifically designed for COVID-19 infection segmentation with limited data. The authors tested their architecture on a limited dataset consisting of ten COVID-19 cases from Brainlab Co. Ltd (Germany) and were able to achieve a DSC of 0.987 and 0.726 for lungs and infection, respectively. Hence, COVID-SegNet as well as our approach achieved similar results. This raises the question, if it is possible to further increase our performance by switching from the standard U-Net of our pipeline to an architecture specifically designed for COVID-19 infection segmentation like COVID-SegNet. Further approaches, with the aim to utilize specifically designed architectures, were Inf-Net (Fan et al.) [41] and MiniSeg (Qiu et al.) [43]. Both were trained on 2D CT scans and achieved for COVID-19 infection segmentation DSCs of 0.764 and 0.773, respectively. Although diverse datasets were used for training, which leads to incomparability of the results, it is highly impressive that they achieved similar performance as approaches based on 3D imaging data. The 3D transformation of these architectures and the integration into our pipeline would be an interesting experiment to evaluate improvement possibilities. Other high-performance 2D approaches like Saood et al. [37] and Pei et al. [38] were difficult to compare due to these models are purely trained and evaluated on 2D slices with COVID-19 presence [66].
Table 3.
Related work overview for COVID-19 segmentation and comparison of resulting segmentation performances. The table categories the related work in terms of model architecture, training dataset information for comparability like source, dimension (Dim.), sample size as well as the presence of non-COVID-19 slices (Control) and their performance on a validation/testing set.
| Related Work |
Training Dataset |
Validation/Testing Performance |
|||||
|---|---|---|---|---|---|---|---|
| Author | Model Architecture | Source | Dim. | Sample Size | Control | DSC – COVID-19 | Sample Size |
| Amyar et al. [5] | U-Net (Standard) | Amyar et al. [5] | 2D | 1219 | Yes | 0.78 | 150 |
| Fan et al. [41] | Inf-Net (Attention U-Net) | Fan et al. [41] | 2D | 1650 | Yes | 0.764 | 50 |
| Qiu et al. [43] | MiniSeg (Attention U-Net) | Qiu et al. [43] | 2D | 3558 | Yes | 0.773 | 3558 |
| Saood et al. [37] | U-Net (Standard) | SIRM [66] | 2D | 80 | No | 0.733 | 20 |
| Saood et al. [37] | SegNet | SIRM [66] | 2D | 80 | No | 0.749 | 20 |
| Pei et al. [38] | MPS-Net (Supervision U-Net) | SIRM [66] | 2D | 300 | No | 0.833 | 68 |
| Zheng et al. [39] | MSD-Net | Zheng et al. [39] | 2D | 3824 | Yes | 0.785 | 956 |
| Wang et al. [40] | COPLE-Net (enhanced U-Net) | Wang et al. [40] | 2D | 59,045 | Yes | 0.803 | 17,205 |
| Ma et al. [16] | U-Net (Standard) | Ma et al. [16] | 3D | 20 | Yes | 0.608 | 20 |
| Ma et al. [16,57] | nnU-Net | Ma et al. [16] | 3D | 20 | Yes | 0.673 | 20 |
| Wang et al. [44] | U-Net (Standard) | Wang et al. [44] | 3D | 211 | Yes | 0.704 | 211 |
| Yan et al. [65] | COVID-SegNet | Yan et al. [65] | 3D | 731 | Yes | 0.726 | 130 |
| He et al. [42] | M2UNet (Segmentation only) | He et al. [42] | 3D | 666 | Yes | 0.759 | 666 |
| Our Pipeline | U-Net (Standard) | Ma et al. [16] | 3D | 20 | Yes | 0.804/0.661 | 20/100 |
4.2. Limitations
However, it is important to note that the majority of current segmentation approaches in research are not suited for clinical usage. The bias of current models is that the majority are only trained with COVID-19 related images. Therefore, it is not certain how good the models can differentiate between COVID-19 lesions and other pneumonia, or entirely unrelated medical conditions like cancer. Furthermore, identical to COVID-19 classification, the models reveal huge differences depending on which dataset they were trained on. Segmentation models purely based on COVID-19 scans are often not able to segment accurately in the presence of other medical conditions [16]. Additionally, there is a high potential for false positive segmentation of pneumonia lesions that are not caused by COVID-19. This demonstrates that these models could be biased and are not suitable for COVID-19 screening. Nevertheless, current infection segmentation models are already highly accurate for confirmed COVID-19 imaging. This offers the opportunity for quantitative assessment and disease monitoring as applications in clinical studies.
Despite that our model and those of others, which are based on limited data, are capable for accurate segmentation, it is essential to discuss their robustness. Currently, there are only a handful annotated imaging datasets publicly available for COVID-19 segmentation. More imaging data with especially more variance (different COVID-19 states, other pneumonia, healthy control samples, etc.) need to be collected, annotated, and published for researchers. Similar to Ma et al. [16,45], community accepted benchmark datasets have to be established in order to fully ensure robustness as well as comparability of models.
5. Conclusions
Even so, neural networks are capable of accurate decision support, their robustness is highly dependent on dataset size for training. Various medical conditions like rare or novel diseases lack available data for model training which decreases generalizability and increases the risk of overfitting. In this paper, we developed and evaluated an approach for automated as well as robust segmentation of COVID-19 infected regions in CT volumes based on a limited dataset. Our method focuses on on-the-fly generation of unique and random image patches for training by performing several preprocessing methods and exploiting extensive data augmentation. Thus, it is possible to handle limited dataset sizes which act as variant database. Instead of novel and complex neural network architectures, we utilized the standard 3D U-Net. We proved that our medical image segmentation pipeline is able to successfully train accurate and robust models without overfitting on limited data. Furthermore, we were able to outperform current state-of-the-art semantic segmentation approaches for COVID-19 infected regions. Our work has great potential to be applied as a clinical decision support system for COVID-19 quantitative assessment and disease monitoring in a clinical environment. As further research, we are planning to integrate ensemble learning techniques in our pipeline to combine the predictive strengths of the k-fold cross-validation models. Additional, clinical studies are needed for robust validation on clinical performance and generalizability of models based on limited data. Also, we are going expand our testing data and evaluation by adding cases with non-COVID-19 conditions like bacterial pneumonia or lung cancer.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We want to thank Bernhard Bauer and Fabian Rabe for sharing their GPU hardware (Nvidia Quadro P6000) with us which was used for this work. We also want to thank Dennis Klonnek, Jana Glöckler, Johann Frei, Florian Auer, Peter Parys, Zaynab Hammoud and Edmund Müller for their useful comments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2021.100681.
Authors' contributions
Dr. Frank Kramer and Dr. Iñaki Soto-Rey were in charge of coordination, review, and correction of the manuscript. Dominik Müller contributed to the conception and design of this work, its data analysis and interpretation, and was in charge for draft and revise the manuscript. All the authors are accountable for the integrity of this work. All authors read and approved the final manuscript.
Funding
This work is a part of the DIFUTURE project funded by the German Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung, BMBF) grant FKZ01ZZ1804E.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
The COVID-19 CT dataset analyzed during this study is available in the Zenodo repository, https://doi.org/10.5281/zenodo.3757476.
All data generated and analyzed during this study is available in the Zenodo repository, https://doi.org/10.5281/zenodo.3902293.
The code for this article was implemented in Python (platform independent) and is available under the GPL-3.0 License at the following GitHub repository:
https://github.com/frankkramer-lab/covid19.MIScnn.
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.W. H. O. Coronavirus . 2020. Disease (COVID-19) pandemic.https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [Google Scholar]
- 3.Sohrabi C., Alsafi Z., O'Neill N., Khan M., Kerwan A., Al-Jabir A., et al. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020;76 doi: 10.1016/j.ijsu.2020.02.034. February:71–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rki - coronavirus SARS-CoV-2 - SARS-CoV-2 Steckbrief zur Coronavirus-Krankheit-2019 (COVID-19). https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Steckbrief.html. Accessed 24 May 2020.
- 5.Amyar A., Modzelewski R., Ruan S. Multi-task deep learning based CT imaging analysis for COVID-19: classification and segmentation. medRxiv. 2020 doi: 10.1101/2020.04.16.20064709. 2020.04.16.20064709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez-Morales A.J., Cardona-Ospina J.A., Gutiérrez-Ocampo E., Villamizar-Peña R., Holguin-Rivera Y., Escalera-Antezana J.P., et al. Clinical, laboratory and imaging features of COVID-19: a systematic review and meta-analysis. Trav Med Infect Dis. 2020;34(February) doi: 10.1016/j.tmaid.2020.101623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. Am J Roentgenol. 2020;215:87–93. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 9.Litjens G., Kooi T., Bejnordi B.E., Setio A.A.A., Ciompi F., Ghafoorian M., et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. December 2012. [DOI] [PubMed] [Google Scholar]
- 10.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., et al. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. IEEE Rev Biomed Eng. 2020 doi: 10.1109/RBME.2020.2987975. 1–1. [DOI] [PubMed] [Google Scholar]
- 11.Rubin G.D., Ryerson C.J., Haramati L.B., Sverzellati N., Kanne J.P., Raoof S., et al. The role of chest imaging in patient management during the COVID-19 pandemic. Radiology. 2020;158:106–116. doi: 10.1148/radiol.2020201365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., et al. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020:23. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng M.-Y., Lee E.Y., Yang J., Yang F., Li X., Wang H., et al. Imaging profile of the COVID-19 infection: radiologic findings and literature review authors. Radiol Cardiothorac Imaging. 2020;2 doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020:8. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manning D., Ethell S., Donovan T., Crawford T. How do radiologists do it? The influence of experience and training on searching for chest nodules. Radiography. 2006;12:134–142. [Google Scholar]
- 16.Ma J., Wang Y., An X., Ge C., Yu Z., Chen J., et al. 2020. Towards efficient covid-19 CT annotation: a benchmark for lung and infection segmentation; pp. 1–7.http://arxiv.org/abs/2004.12537 [Google Scholar]
- 17.Wang G. A perspective on deep imaging. IEEE Access. 2016;4:8914–8924. [Google Scholar]
- 18.Aggarwal P., Vig R., Bhadoria S., Dethe A C.G. Role of segmentation in medical imaging: a comparative study. Int J Comput Appl. 2011;29:54–61. [Google Scholar]
- 19.Lee K., Zung J., Li P., Jain V., Seung H.S. 2017. Superhuman accuracy on the SNEMI3D connectomics challenge; pp. 1–11.http://arxiv.org/abs/1706.00120 Nips. [Google Scholar]
- 20.Bullock J., Luccioni A., Pham K.H., Lam C.S.N., Luengo-Oroz M. 2020. Mapping the landscape of artificial intelligence applications against COVID-19; pp. 1–32.http://arxiv.org/abs/2003.11336 [Google Scholar]
- 21.Shen D., Wu G., Suk H.-I. Deep learning in medical image analysis. Annu Rev Biomed Eng. 2017;19:221–248. doi: 10.1146/annurev-bioeng-071516-044442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE computer society conference on computer vision and pattern recognition. IEEE Computer Society; 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [DOI] [Google Scholar]
- 23.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE computer society conference on computer vision and pattern recognition. IEEE Computer Society; 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- 24.Huang G., Liu Z., van der Maaten L., Weinberger K.Q. Proc - 30th IEEE conf comput vis pattern recognition, CVPR 2017. 2016. Densely connected convolutional networks.http://arxiv.org/abs/1608.06993 2017-January:2261–9. Accessed. [Google Scholar]
- 25.Mu G, Lin Z, Han M, Yao G, Gao Y. Segmentation of kidney tumor by multi-resolution VB-nets. :1–5.
- 26.Ronneberger O., Fischer Philipp, Brox T. U-net: convolutional networks for biomedical image segmentation. Lect Notes Comput Sci. 2015;9351:234–241. [Google Scholar]
- 27.Jin S., Wang B., Xu H., Luo C., Wei L., Zhao W., et al. AI-assisted CT imaging analysis for COVID-19 screening: building and deploying a medical AI system in four weeks. medRxiv. 2020 doi: 10.1101/2020.03.19.20039354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taha A.A., Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imag. 2015;15:29. doi: 10.1186/s12880-015-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin C., Chen W., Cao Y., Xu Z., Tan Z., Zhang X., et al. Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat Commun. 2020;11:1–14. doi: 10.1038/s41467-020-18685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abbas A., Abdelsamea M.M., Gaber M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intell. 2020 doi: 10.1007/s10489-020-01829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Z., Zhao W., Xie X., Zhong Z., Shi F., Ma T., et al. Severity assessment of COVID-19 using CT image features and laboratory indices. Phys Med Biol. 2021;66 doi: 10.1088/1361-6560/abbf9e. [DOI] [PubMed] [Google Scholar]
- 32.Gozes O., Frid-Adar M., Greenspan H., Browning P.D., Bernheim A., Siegel E. 2020. Rapid AI development cycle for the coronavirus (COVID-19) pandemic: initial results for automated detection & patient monitoring using deep learning CT image analysis.http://arxiv.org/abs/2003.05037 [Google Scholar]
- 33.Chen X., Yao L., Zhang Y. vol. 14. 2020. Residual attention U-net for automated multi-class segmentation of COVID-19 chest CT images; pp. 1–7.http://arxiv.org/abs/2004.05645 [Google Scholar]
- 34.Gozes O., Frid-Adar M., Sagie N., Zhang H., Ji W., Greenspan H. 2020. Coronavirus detection and analysis on chest CT with deep learning; pp. 1–8.http://arxiv.org/abs/2004.02640 [Google Scholar]
- 35.Gaál G., Maga B., Lukács A. 2020. Attention U-net based adversarial architectures for chest X-ray lung segmentation; pp. 1–7.http://arxiv.org/abs/2003.10304 [Google Scholar]
- 36.Zhou T., Canu S., Ruan S. 2020. An automatic COVID-19 CT segmentation based on U-Net with attention mechanism; pp. 1–14.http://arxiv.org/abs/2004.06673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saood A., Hatem I. COVID-19 lung CT image segmentation using deep learning methods: U-Net versus SegNet. BMC Med Imag. 2021;21:19. doi: 10.1186/s12880-020-00529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pei H.Y., Yang D., Liu G.R., Lu T. MPS-net: multi-point supervised network for ct image segmentation of covid-19. IEEE Access. 2021;9:47144–47153. doi: 10.1109/ACCESS.2021.3067047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng B., Liu Y., Zhu Y., Yu F., Jiang T., Yang D., et al. Msd-net: multi-scale discriminative network for covid-19 lung infection segmentation on CT. IEEE Access. 2020;8:185786–185795. doi: 10.1109/ACCESS.2020.3027738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang G., Liu X., Li C., Xu Z., Ruan J., Zhu H., et al. A noise-robust framework for automatic segmentation of COVID-19 pneumonia lesions from CT images. IEEE Trans Med Imag. 2020;39:2653–2663. doi: 10.1109/TMI.2020.3000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan D.-P., Zhou T., Ji G.-P., Zhou Y., Chen G., Fu H., et al. Inf-Net: automatic COVID-19 lung infection segmentation from CT scans. IEEE Trans Med Imag. 2020:1–11. doi: 10.1109/tmi.2020.2996645. 2019. [DOI] [PubMed] [Google Scholar]
- 42.He K., Zhao W., Xie X., Ji W., Liu M., Tang Z., et al. 2020. Synergistic learning of lung lobe segmentation and hierarchical multi-instance classification for automated severity assessment of COVID-19 in CT images.http://arxiv.org/abs/2005 03832. Accessed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu Y., Liu Y., Xu J. 2020. MiniSeg: an extremely minimum network for efficient COVID-19 segmentation; pp. 1–10.http://arxiv.org/abs/2004.09750 [DOI] [PubMed] [Google Scholar]
- 44.Wang Y., Zhang Y., Liu Y., Tian J., Zhong C., Shi Z., et al. Does non-COVID-19 lung lesion help? investigating transferability in COVID-19 CT image segmentation. Comput Methods Progr Biomed. 2021;202:106004. doi: 10.1016/j.cmpb.2021.106004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jun M., Cheng G., Yixin W., Xingle A., Jiantao G., Ziqi Y., et al. 2020. COVID-19 CT lung and infection segmentation dataset. [DOI] [Google Scholar]
- 46.Müller D., Kramer F. 2021. MIScnn: a framework for medical image segmentation with convolutional neural networks and deep learning. arXiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abadi M., Agarwal A., Barham P., Brevdo E., Chen Z., Citro C., et al. 2015. TensorFlow: large-scale machine learning on heterogeneous systems.https://www.tensorflow.org/ [Google Scholar]
- 48.An P., Xu S., Harmon S., Turkbey E., Sanford T., Amalou A., et al. TCIA); 2020. CT images in COVID-19 - the cancer imaging archive. [DOI] [Google Scholar]
- 49.Harmon S.A., Sanford T.H., Xu S., Turkbey E.B., Roth H., Xu Z., et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat Commun. 2020;11:1–7. doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.COVID-19 lung CT lesion segmentation challenge. 2020. https://covid-segmentation.grand-challenge.org/COVID-19-20/ Grand Challenge. [Google Scholar]
- 51.Toennies K.D. Guide to medical image analysis. Springer London; 2012. The analysis of medical images; pp. 1–19. [Google Scholar]
- 52.Roy S., Carass A., Prince J.L. Proceedings - international symposium on biomedical imaging. 2013. Patch based intensity normalization of brain MR images. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shorten C., Khoshgoftaar T.M. A survey on image data augmentation for deep learning. J Big Data. 2019;6 doi: 10.1186/s40537-019-0197-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez L., Wang J. 2017. The effectiveness of data augmentation in image classification using deep learning.http://arxiv.org/abs/1712.04621 Accessed. [Google Scholar]
- 55.Taylor L., Nitschke G. 2017. Improving deep learning using generic data augmentation.http://arxiv.org/abs/1708.06020 Accessed. [Google Scholar]
- 56.Isensee F., Jäger P., Wasserthal J., Zimmerer D., Petersen J., Kohl S., et al. 2020. Batchgenerators - a python framework for data augmentation. [DOI] [Google Scholar]
- 57.Isensee F., Jaeger P.F., Kohl S.A.A., Petersen J., Klaus H. 2020. Automated design of deep learning methods for biomedical image segmentation; pp. 1–55.https://arxiv.org/abs/1904.08128 [Google Scholar]
- 58.Minaee S., Boykov Y., Porikli F., Plaza A., Kehtarnavaz N., Terzopoulos D. 2020. Image segmentation using deep learning: a survey.http://arxiv.org/abs/2001.05566 Accessed. [DOI] [PubMed] [Google Scholar]
- 59.Asgari Taghanaki S., Abhishek K., Cohen J.P., Cohen-Adad J., Hamarneh G. Deep semantic segmentation of natural and medical images: a review. Artif Intell Rev. 2020:1–42. doi: 10.1007/s10462-020-09854-1. [DOI] [Google Scholar]
- 60.Zahra E., Ali B., Siddique W. 2020. Medical image segmentation using a U-net type of architecture.http://arxiv.org/abs/2005 05218. Accessed. [Google Scholar]
- 61.Çiçek Ö., Abdulkadir A., Lienkamp S.S., Brox T., Ronneberger O. 3D U-net: learning dense volumetric segmentation from sparse annotation. Lect Notes Comput Sci. 2016:424–432. 9901 LNCS. [Google Scholar]
- 62.Zhang Z., Liu Q., Wang Y. Road extraction by deep residual U-net. Geosci Rem Sens Lett IEEE. 2018 [Google Scholar]
- 63.Seyed S.S.M., Erdogmus D., Gholipour A., Salehi S.S.M., Erdogmus D., Gholipour A. Lecture notes in computer science. Springer Verlag; 2017. Tversky loss function for image segmentation using 3D fully convolutional deep networks; pp. 379–387. [DOI] [Google Scholar]
- 64.Kingma D.P., Lei Ba J. 2014. Adam: a method for stochastic optimization.https://arxiv.org/abs/1412.6980 [Google Scholar]
- 65.Yan Q., Wang B., Gong D., Luo C., Zhao W., Shen J., et al. 2020. COVID-19 chest CT image segmentation -- A deep convolutional neural network solution; pp. 1–10.http://arxiv.org/abs/2004.10987 [Google Scholar]
- 66.Italian Society of Medical and Interventional Radiology . 2020. COVID-19 - medical segmentation.http://medicalsegmentation.com/covid19/ Accessed. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.