ABSTRACT
Signaling through the platelet-derived growth factor receptor alpha (PDGFRα) is crucial for mammalian craniofacial development, although the mechanisms by which the activity of downstream intracellular effectors is regulated to mediate gene expression changes have not been defined. We find that the RNA-binding protein Srsf3 is phosphorylated at Akt consensus sites downstream of PI3K-mediated PDGFRα signaling in mouse palatal mesenchyme cells, leading to its nuclear translocation. We further demonstrate that ablation of Srsf3 in the mouse neural crest lineage leads to facial clefting due to defective cranial neural crest cell proliferation and survival. Finally, we show that Srsf3 regulates the alternative RNA splicing of transcripts encoding protein kinases in the mouse facial process mesenchyme to regulate PDGFRα-dependent intracellular signaling. Collectively, our findings reveal that alternative RNA splicing is an important mechanism of gene expression regulation downstream of PI3K/Akt-mediated PDGFRα signaling in the facial mesenchyme and identify Srsf3 as a critical regulator of craniofacial development.
KEY WORDS: Srsf3, PDGFRα, Palate, Neural crest, Facial clefting, Alternative RNA splicing
Summary: PDGFRα signaling regulates gene expression through alternative RNA splicing; ablation in the murine neural crest lineage of a downstream effector of PDGFRα signaling, the RNA-binding protein Srsf3, results in midline facial clefting.
INTRODUCTION
Craniofacial development is a complex morphogenetic process that, upon disruption, results in several of the most prevalent birth defects in humans (Mai et al., 2019). Signaling through the receptor tyrosine kinase platelet-derived growth factor receptor alpha (PDGFRα) is crucial for this process in both humans and mice. Missense mutations in the PDGFRA coding region and single base-pair substitutions in the 3′ untranslated region are associated with nonsyndromic cleft palate (Rattanasopha et al., 2012). Similarly, Pdgfra mutant mouse models display phenotypes ranging from a cleft palate to complete facial clefting (Klinghoffer et al., 2002; Soriano, 1997; Tallquist and Soriano, 2003). Phosphatidylinositol 3-kinase (PI3K) has been identified as the primary effector of PDGFRα signaling during skeletal development in the mouse (Klinghoffer et al., 2002). Activated PI3K signaling results in the recruitment of the serine/threonine kinase Akt to the cell membrane. Once phosphorylated, active Akt dissociates from the membrane and phosphorylates hundreds of target proteins with roles in diverse cellular processes (Manning and Cantley, 2007). To identify which proteins are phosphorylated by Akt downstream of PI3K-mediated PDGFRα signaling, we previously performed a phosphoproteomic screen using primary mouse embryonic palatal mesenchyme (MEPM) cells, ultimately identifying 56 proteins that were differentially phosphorylated upon PDGF-AA ligand treatment (Fantauzzo and Soriano, 2014). A gene ontology (GO) analysis of these proteins indicated that the most significant terms for biological process were mRNA processing (14/56 target proteins, P=1.4×10−8) and RNA splicing (12/56 target proteins, P=2.5×10−7) (Fantauzzo and Soriano, 2014).
Alternative RNA splicing (AS), which is characterized by differential inclusion of exons in a mature RNA transcript, is an important mechanism used to regulate gene expression and increase the diversity of protein isoforms (Licatalosi and Darnell, 2010; Wang et al., 2008). Approximately 95% of multi-exon human genes are subject to AS, often in a tissue-specific manner (Pan et al., 2008; Wang et al., 2008). Dysregulation of AS has been shown to cause numerous diseases, stemming from mutations in precursor RNA sequence elements that regulate splicing, mutations in core spliceosome components and/or mutations in auxiliary RNA-binding proteins (RBPs) (Scotti and Swanson, 2016). These RBPs are trans-acting factors that bind splicing regulatory elements in introns and/or exons of pre-mRNA to promote exon inclusion or skipping (Fu and Ares, 2014; Licatalosi and Darnell, 2010). AS mediated through RBPs is an essential process in the developing mouse face, as demonstrated by the midline facial clefting and craniofacial bone hypoplasia phenotypes resulting from global and/or tissue-specific ablation of Esrp1, Esrp2 and Rbfox2 (Bebee et al., 2015; Cibi et al., 2019; Lee et al., 2020). Despite these recent studies, the crucial process of AS remains understudied in neural crest cells (NCCs) and their derivatives in the facial mesenchyme. Furthermore, the question of how the activity of RBPs is regulated to affect the AS of subsets of transcripts in a tissue-specific and spatiotemporal manner has not been explored in the context of craniofacial development.
Here, we focused on one of the RBPs detected in our phosphoproteomic screen, serine/arginine-rich splicing factor 3 (Srsf3). Srsf3 has been implicated in NCC development in non-mouse models, as overexpression of srsf3 mRNA in Xenopus led to a change in the shape and location of the expression domain of the NCC marker slug (Dichmann et al., 2008). Srsf3 belongs to the highly conserved and widely expressed family of serine/arginine-rich (SR) proteins, which typically bind exonic splicing enhancer elements to promote exon inclusion (Fu and Ares, 2014; Licatalosi and Darnell, 2010). Srsf3 specifically was shown to bind pyrimidine-rich motifs in both exons and introns in murine embryonic carcinoma cells, with a preference for the former (Änkö et al., 2012). Importantly, RBPs can be regulated by post-translational modifications, including phosphorylation, that can alter several aspects of their function, such as subcellular localization, RNA binding and/or sequence specificity (Stamm, 2008). Phosphorylation of Akt consensus sites within the C-terminal arginine/serine-rich (RS) domain of Srsf3 has been shown to drive its translocation to the nucleus (Bavelloni et al., 2014; Long et al., 2019), although the upstream inputs that stimulate these modifications are incompletely understood.
In this work, we demonstrated that PI3K/Akt-mediated PDGFRα signaling regulates the expression of genes involved in palatal shelf morphogenesis through AS, in part through the phosphorylation and subsequent nuclear translocation of Srsf3. We further showed that ablation of Srsf3 in the NCC lineage leads to a severe midline facial clefting phenotype due to defective NCC proliferation and survival, and the mis-splicing of multiple protein kinases. Taken together, our results provide significant insight into the mechanisms underlying gene expression regulation during mammalian craniofacial development.
RESULTS
Differential alternative splicing of select transcripts in response to PI3K-mediated PDGFRα signaling
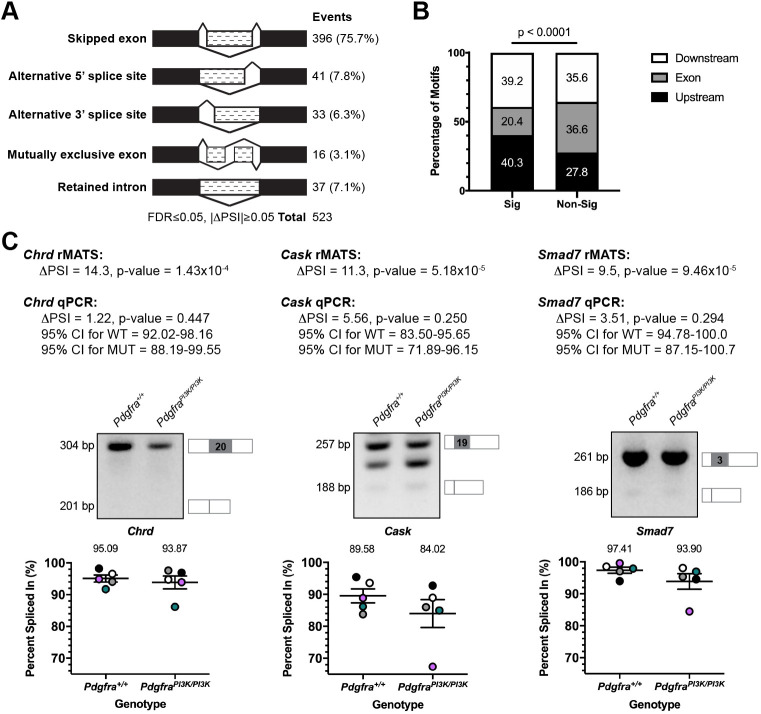
To determine whether disrupted PI3K-mediated PDGFRα signaling in the secondary palatal shelves (PS) leads to changes in AS, we harvested and sequenced PS mesenchyme RNA from three biological replicates of embryonic day 13.5 (E13.5) Pdgfra+/+ versus PdgfraPI3K/PI3K embryos in which PDGFRα is unable to bind PI3K (Klinghoffer et al., 2002) (Table S1). rMATS (Shen et al., 2014) was used to detect AS events, identifying 523 events that were significantly different between genotypes, with the majority of events (75.7%) involving skipped exons (SE) (Fig. 1A; Table S2). A GO analysis of the 348 genes represented in the SE class using the MGI Mammalian Phenotype Level 4 2019 library of the Enrichr gene list enrichment analysis tool (Chen et al., 2013; Kuleshov et al., 2016) revealed a number of significant terms corresponding to phenotypes observed in PdgfraPI3K/PI3K embryos, including abnormal secondary palate development (P=0.03).
Fig. 1.
Differential alternative splicing of select transcripts in response to PI3K-mediated PDGFRα signaling. (A) Summary of differential AS events detected by rMATS analysis of RNA-seq data from E13.5 Pdgfra+/+ versus PdgfraPI3K/PI3K PS mesenchyme. n=3 biological replicates per genotype. FDR, false discovery rate; ΔPSI, difference in percent spliced in. (B) Bar graph depicting percentage of Srsf3 motifs detected by RBPmap within or flanking exons between significant (Sig) and non-significant (Non-Sig) skipped exon events. Statistical analyses were performed using a χ2 test. (C) ΔPSI values for three differentially alternatively spliced transcripts between E13.5 Pdgfra+/+ versus PdgfraPI3K/PI3K PS mesenchyme as assessed by rMATS analysis of RNA-seq data and qPCR. Representative qPCR gels with depictions of differentially alternatively spliced exon (gray), and upstream and downstream sequences that were assessed by qPCR, as well as scatter dot plots depicting percent spliced in, as assessed by qPCR, are presented. Data are mean±s.e.m. Statistical analyses were performed using a paired t-test. Colored circles correspond to independent experiments. n=5 biological replicates per genotype. CI, confidence interval; WT, Pdgfra+/+; MUT, PdgfraPI3K/PI3K.
We then used RBPmap (Paz et al., 2014) to examine annotated Srsf3 motifs in the 396 SE plus 250 bp flanking each end, revealing Srsf3 motifs in 381 (96.2%) of these regions of interest (ROIs) (Table S2). Although the density of these motifs was similar between significant (4.4 motifs per ROI) versus non-significant (4.6 motifs per ROI) SE events in our dataset (P=0.2), Srsf3 motifs were enriched upstream and downstream of the SE in the significant SE events (P<0.0001) (Fig. 1B). Among the SE events with one or more Srsf3 motifs in the ROI, significantly more events had a positive percent spliced in (PSI; 260 out of 381), in which the differentially alternatively spliced exon was included more often in Pdgfra+/+ versus PdgfraPI3K/PI3K samples. As SR proteins promote exon inclusion (Fu and Ares, 2014; Licatalosi and Darnell, 2010), this finding suggests that the ability of Srsf3 to regulate AS is partially disrupted in PdgfraPI3K/PI3K embryos. Among the genes represented in the SE class that had one or more Srsf3 motifs in the ROI, 22 have a corresponding mouse model with a craniofacial phenotype, eight of which have a cleft secondary palate (Table S3). We examined the differential AS of three of these eight transcripts, Chrd, Cask and Smad7 (Atasoy et al., 2007; Bachiller et al., 2003; Papangeli and Scambler, 2013), between Pdgfra+/+ and PdgfraPI3K/PI3K E13.5 PS mesenchyme samples by qPCR using primers within constitutively expressed exons flanking the alternatively spliced exon, confirming the trends observed in the rMATS analysis in four out of five biological replicates for each transcript (Fig. 1C). We next employed SPLINTER (https://github.com/dianalow/SPLINTER/) to predict outcomes stemming from the AS of the 25 events in Table S3, revealing that 10 (40%) are predicted to result in a truncated protein, six (24%) in nonsense-mediated decay and potential downregulation of expression, and two (8%) in an alternative protein (Table S3). All 10 truncated protein outcomes, including those for Cask and Smad7, and five of six nonsense-mediated decay outcomes, including that for Chrd, are predicted to occur more often in PdgfraPI3K/PI3K as opposed to wild-type samples. Identifying how the differential AS of such transcripts ultimately affects protein function and contributes to the PdgfraPI3K/PI3K palatal clefting phenotype will be the topic of future research studies.
Next, differential gene expression was assessed in the above RNA-seq samples via DESeq2 (Love et al., 2014). Surprisingly, this analysis identified only 13 genes with significant differences in expression between the two genotypes (Table S4), none of which were differentially alternatively spliced. Among these 13 genes, three (Foxp2, Aldh1a2 and Pdgfra) have a corresponding mouse model with a craniofacial phenotype, with only Pdgfra models exhibiting a cleft secondary palate (Klinghoffer et al., 2002; Niederreither et al., 1999; Shu et al., 2005; Soriano, 1997). Collectively, these findings demonstrate that AS is an important mechanism of gene expression regulation downstream of PI3K/Akt-mediated PDGFRα signaling in the mid-gestation PS.
Srsf3 expression is enriched in the facial processes
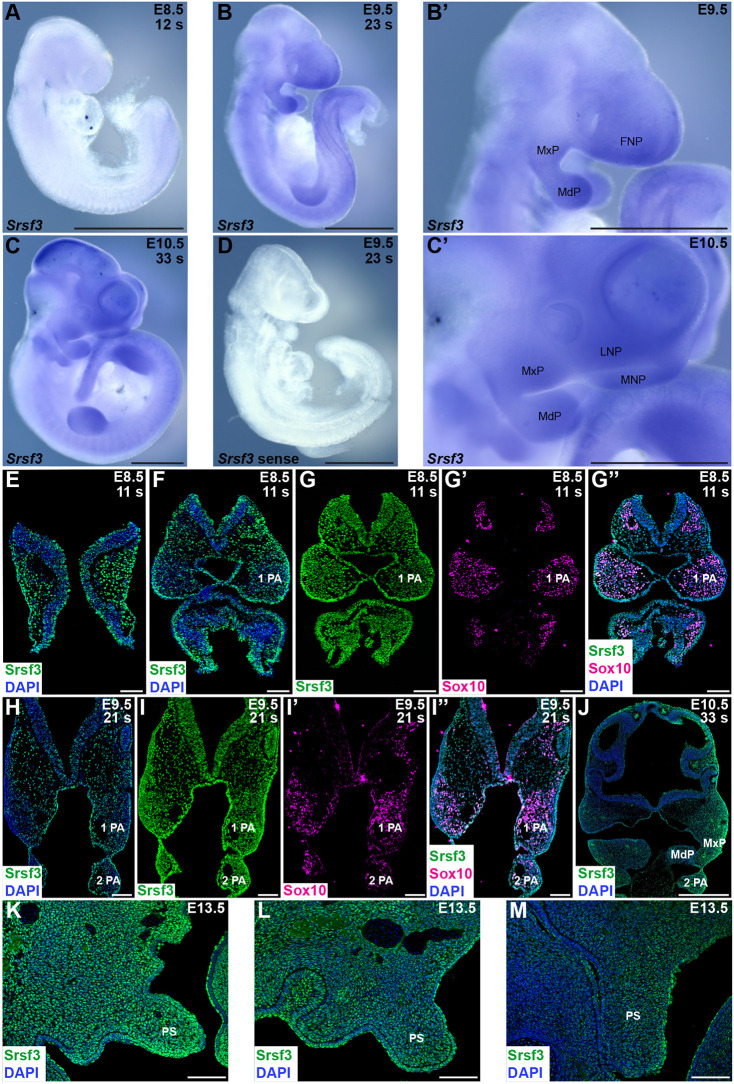
To assess where Srsf3 is expressed during development, we performed whole-mount RNA in situ hybridization from E8.5 to E10.5. We saw an enrichment of transcripts in the head region at all timepoints (Fig. 2A-D), and particularly in the facial processes at E9.5-E10.5 (Fig. 2B′,C′), including the maxillary processes (MxP) from which the PS will extend. We then performed immunofluorescence analysis to examine expression of Srsf3 protein during development. From E8.5 to E10.5, we observed predominantly nuclear Srsf3 expression in the pharyngeal arch (PA) and facial process mesenchyme, as well as the overlying ectoderm (Fig. 2E,F,H,J). Srsf3 was expressed in Sox10-positive NCCs migrating away from the cranial neural folds and in PAs 1 and 2 from E8.5 to E9.5 (Fig. 2G-G″,I-I″). At E13.5, Srsf3 expression was detected along the anterior-posterior axis of the PS, with noticeably increased expression in the anterior PS (Fig. 2K-M).
Fig. 2.
Srsf3 expression is enriched in the facial processes. (A-C′) Srsf3 expression as assessed by whole-mount in situ hybridization analysis at E8.5 (A), E9.5 (B,B′) and E10.5 (C,C′). s, somite pairs; FNP, frontonasal prominence; MxP, maxillary process; MdP, mandibular process; LNP, lateral nasal process; MNP, medial nasal process. (D) No signal was detected with a control sense probe. n≥two biological replicates per timepoint. Scale bars: 1 mm. (E-M) Srsf3 (green) and/or Sox10 expression (magenta), as assessed by immunofluorescence analysis, on sections of cranial neural folds and PAs at E8.5 (E-G″) and E9.5 (H-I″), facial processes at E10.5 (J), and anterior (K), middle (L) and posterior (M) PS at E13.5. Nuclei were stained with DAPI (blue). n=1 biological replicate at E8.5-10.5; n=4 biological replicates at E13.5. 1 PA, first pharyngeal arch; 2 PA, second pharyngeal arch; PS, secondary palatal shelves. Scale bars: 100 µm in E-I″,K-M; 500 µm in J.
Phosphorylation of Srsf3 at Akt consensus sites in response to PDGF ligand stimulation leads to nuclear translocation
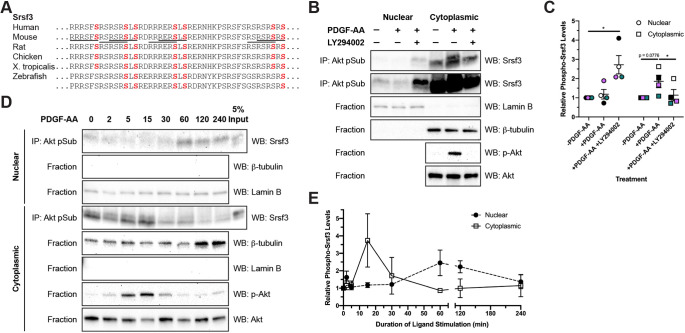
The full complement of Akt consensus sites (RxRxxS/T) (Alessi et al., 1996) found in the mouse Srsf3 RS domain are conserved from human to Xenopus, with five of the seven sites conserved in zebrafish (Fig. 3A), demonstrating a high level of conservation among vertebrate species. To validate the PI3K-mediated phosphorylation of Srsf3 upon PDGF-AA ligand treatment detected in our previous mass spectrometry analysis (Fantauzzo and Soriano, 2014), we immunoprecipitated phosphorylated Akt substrates from nuclear and cytoplasmic fractions of immortalized MEPM (iMEPM) cells that were unstimulated or stimulated for 20 min with PDGF-AA ligand in the absence or presence of the PI3K inhibitor LY294002 (Vlahos et al., 1994) with an anti-Akt phosphosubstrate antibody (Zhang et al., 2002) followed by western blotting with an anti-Srsf3 antibody. This analysis revealed increased band intensities over baseline levels in response to PDGF-AA ligand treatment in the cytoplasmic fractions (1.851±0.3222-fold induction over unstimulated levels), indicative of increased phospho-Srsf3 levels, and a return to band intensities near baseline levels upon treatment with LY294002 (1.136±0.2930-fold induction over unstimulated levels) (Fig. 3B,C).
Fig. 3.
Phosphorylation of Srsf3 at Akt consensus sites in response to PDGF ligand stimulation leads to nuclear translocation. (A) Akt consensus sequence conservation in Srsf3 across vertebrate species. Full consensus sequences are underlined in mouse, with terminal serine residues highlighted in red. (B) Immunoprecipitation of Akt phosphorylation targets from nuclear and cytoplasmic fractions of iMEPM cells that were untreated, treated for 20 min with PDGF-AA ligand or pre-treated with PI3K inhibitor LY294002 with an anti-Akt phosphosubstrate antibody followed by western blotting with an anti-Srsf3 antibody. Top two panels are the same blot imaged at different exposures to reveal the expression of phospho-Srsf3 in cytoplasmic (top) and nuclear (bottom) fractions. n=4 biological replicates per condition. IP, immunoprecipitation; WB, western blot. (C) Scatter dot plot depicting quantification of band intensities from four independent experiments as in B. Data are mean±s.e.m. Statistical analyses were performed using a paired t-test. *P<0.05. Colored circles and squares correspond to independent experiments. (D) Similar biochemical experiments as in B, with a time course analysis of PDGF-AA ligand treatments. n=3 biological replicates per condition. (E) Line graph depicting quantification of band intensities from three independent experiments as in D. Data are mean±s.e.m.
To further investigate the effects of PI3K/Akt-mediated phosphorylation of the above sites on the subcellular localization of Srsf3, iMEPM cells were left unstimulated or stimulated with PDGF-AA ligand from 2 min to 4 h. Immunoprecipitation of phosphorylated Akt substrates from the fractionated lysates using the anti-Akt phosphosubstrate antibody followed by western blotting with the anti-Srsf3 antibody revealed that while phosphorylated Srsf3 was detected in both fractions at all timepoints, the cytoplasmic and nuclear fractions had the greatest amount of phosphorylated Srsf3 at 15 min (3.741±1.527-fold induction over unstimulated levels) and 60 min (2.455±0.7326-fold induction over unstimulated levels), respectively (Fig. 3D,E). Significantly, the changes observed in phosphorylated Srsf3 levels in the nucleus over time in response to PDGF-AA ligand stimulation do not result from changes in total Srsf3 protein levels in this compartment following 2 h of ligand stimulation (Fig. S1A,B). These results suggest that phosphorylation of Srsf3 at Akt consensus sites downstream of PDGFRα signaling drives translocation of phosphorylated Srsf3 into the nucleus, where alternative splicing takes place.
Ablation of Srsf3 in the neural crest lineage results in a midline facial clefting phenotype
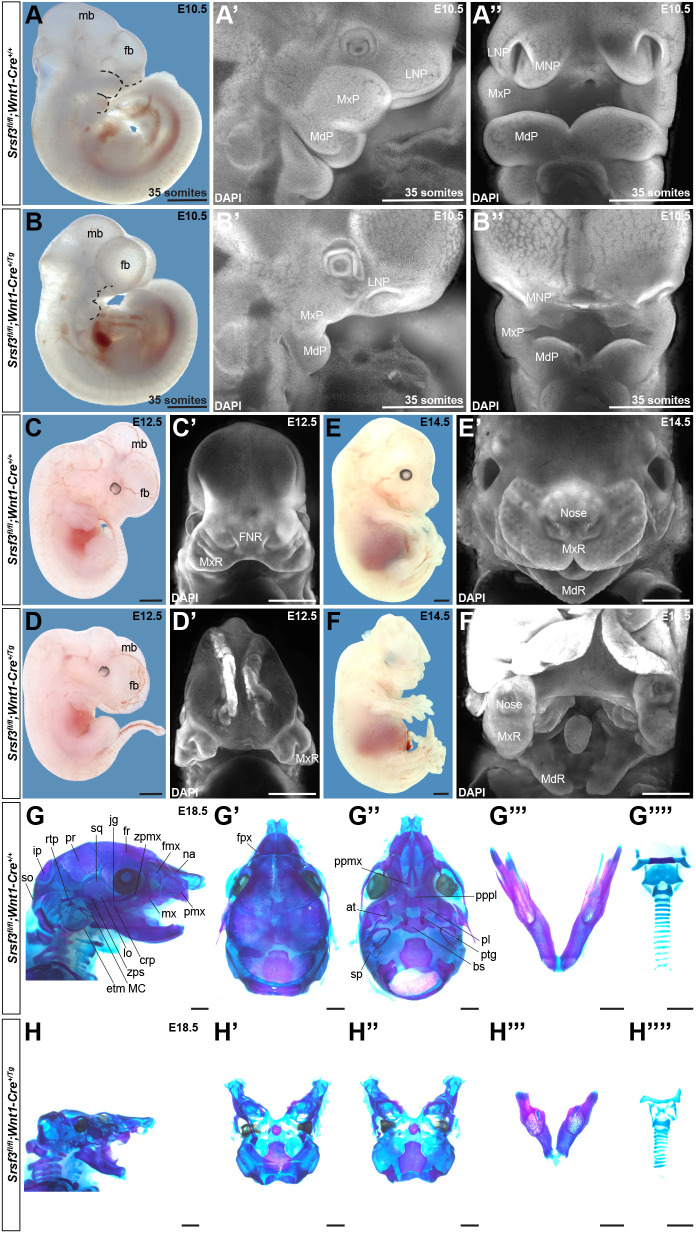
To characterize the role of Srsf3 during craniofacial development in vivo, we combined a Srsf3 conditional allele (Jumaa et al., 1999) with the Wnt1-Cre transgene (Danielian et al., 1998) to ablate Srsf3 in the NCC lineage. Quantitative PCR, whole-mount in situ hybridization and western blotting confirmed deletion of Srsf3 exons 2 and 3 in the facial processes and loss of Srsf3 protein expression in MxP mesenchyme lysates from cKO embryos (Fig. S2A-F). We crossed Srsf3fl/fl males with Srsf3+/fl;Wnt1-Cre+/Tg females and initially harvested progeny at birth. Genotyping revealed that cKO pups were recovered well below Mendelian frequency at this timepoint (two cannibalized pups versus 14 expected pups out of 56 total, P=0.0002), with no live pups present at birth (Table S5). Additional harvests at embryonic timepoints indicated that the majority of cKO embryos died after mid-gestation (Table S5). We first assessed the craniofacial phenotypes of cKO embryos by examining gross morphology during development. At E10.5, cKO embryos had hypoplastic facial processes, a widening of the space between the nasal pits, an enlarged forebrain and a reduced midbrain (Fig. 4B-B″, Fig. S3B) compared with their control littermates (Fig. 4A-A″, Fig. S3A). At E12.5, cKO embryos exhibited facial clefting, wherein the medial nasal processes had failed to fuse at the midline (Fig. 4C′,D′). Moreover, the PS, tongue and midbrain were hypoplastic, and the forebrain was misshapen (Fig. 4C-D′, Fig. S3C′-D‴). Sectioning of cKO embryos at this timepoint revealed that the lateral ventricles had formed, although they were enlarged compared with those of control littermates, indicating that the anterior neural tube had successfully closed (Fig. S3C-D‴). By E14.5, the facial clefting phenotype in cKO embryos was more pronounced, with a striking cleft at the midline of the upper jaw and a subtler cleft in the mandible (Fig. 4E′,F′). The tongue remained severely hypoplastic at this timepoint (Fig. 4F′), and continued forebrain overgrowth and exencephaly were observed (Fig. 4E-F′). A subset of cKO embryos additionally exhibited facial subepidermal blebbing (43% at E12.5, n=7), facial hemorrhaging (33% at E10.5, n=33) and a wavy neural tube (19% at E11.5, n=32). Importantly, the defects above were not due to a general delay in cKO embryo development or global growth reduction, as structures such as the limb buds were indistinguishable from those of control embryos (Fig. 4E,F).
Fig. 4.
Ablation of Srsf3 in the neural crest lineage results in a midline facial clefting phenotype. (A-F′) Gross morphology of E10.5 (A-B″), E12.5 (C-D′) and E14.5 (E-F′) Srsf3fl/fl;Wnt1-Cre+/+ control embryos (top) and Srsf3fl/fl;Wnt1-Cre+/Tg cKO embryos (bottom) viewed either laterally before (A-F) and after (A′,B′) DAPI staining (white) or frontally after DAPI staining (white) (A″,B″,C′-F′). Dotted black lines (A,B) outline nasal pits and PA 1. fb, forebrain; mb, midbrain; LNP, lateral nasal process; MxP, maxillary process; MdP, mandibular process; MNP, medial nasal process; FNR, frontonasal region; MxR, maxillary region; MdR, mandibular region. (G-H″″) Lateral (G,H), dorsal (G′,H′) and ventral (G″,H″) views of craniofacial skeletal preparations, dissected mandibles (G‴,H‴), and dissected hyoid bones and thyroid, cricoid and tracheal cartilages (G″″,H″″) generated from E18.5 control embryos (G-G″″) and cKO embryos (H-H″″). n≥3 biological replicates per genotype per timepoint. na, nasal; fmx, frontal process of maxilla; zpmx, zygomatic process of maxilla; fr, frontal; jg, jugal; sq, squamosal; pr, parietal; rtp, retrotympanic process; ip, interparietal; so, supraoccipital; pmx, premaxilla; mx, maxilla; crp, coronoid process; lo, lamina obturans; zps, zygomatic process of squamosal; MC, Meckel's cartilage; etm, ectotympanic; fpx, frontal process of premaxilla; ppmx, palatal process of maxilla; pppl, palatal process of palatine; pl, palatine; ptg, pterygoid; at, ala temporalis; bs, basisphenoid; sp, processus styloidius. Scale bars: 1 mm.
We next evaluated the craniofacial phenotypes of three cKO embryos that survived to E18.5 by generating skeletal preparations. Generally, cKO embryos exhibited a smaller head than their littermate controls (Fig. 4G,H). The anterior part of the cKO face was clefted, and those bones and cartilages tended to be hypoplastic. These elements included the premaxilla, frontal process of premaxilla, maxilla, frontal process of maxilla and zygomatic process of maxilla bones as well as the nasal cartilage. The nasal and frontal bones were missing entirely. The middle of the face had elements that were more severely affected and in several cases absent. These hypoplastic structures included the jugal, zygomatic process of squamosal, squamosal, parietal, basisphenoid, pterygoid and retrotympanic process bones as well as the cartilages of the middle ear. The palatal process of the palatine, palatine, ala temporalis, lamina obturans and ectotympanic bones were absent from the middle of the face as was the processus styloidius cartilage. The palatal process of the maxilla was absent in two of three cKO embryos and hypoplastic in the remaining embryo. The posterior part of the skull derived from the paraxial mesoderm was relatively unaffected, although the interparietal and supraoccipital bones were clefted and hypoplastic (Fig. 4G′-H″). The mandible, coronoid process and Meckel's cartilage of cKO embryos were hypoplastic (Fig. 4G‴,H‴), with clefting of the mandible observed in one embryo. The hyoid was present but not ossified in two cKO embryos and severely hypoplastic in the third embryo, while the lesser and greater horns of the hyoid were hypoplastic (Fig. 4G″″,H″″). Finally, the thyroid and cricoid cartilages were hypoplastic and the tracheal cartilage rings were misshapen and occasionally fused (Fig. 4G″″,H″″).
Cranial neural crest cell proliferation and survival are defective in Srsf3 conditional knockout embryos
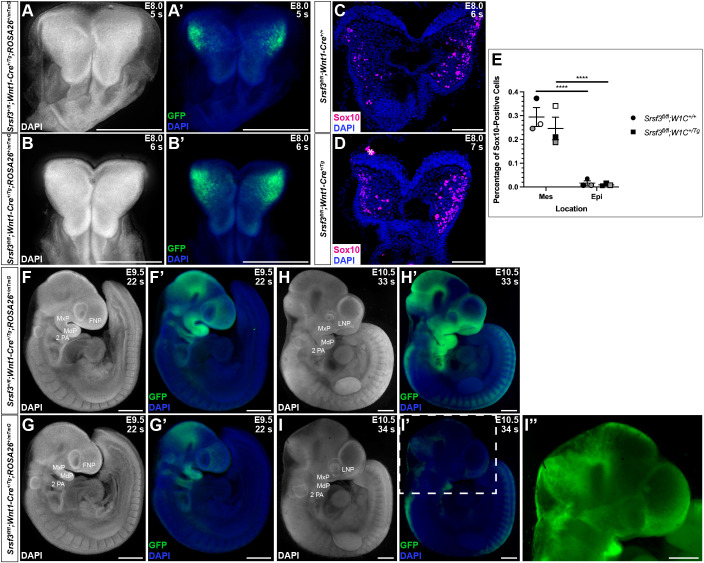
We next introduced the ROSA26mTmG double-fluorescent Cre reporter allele (Muzumdar et al., 2007) through the male germline in the above crosses to examine NCC distribution. At E8.0, Srsf3+/fl;Wnt1-Cre+/Tg;ROSA26+/mTmG control embryos and Srsf3fl/fl;Wnt1-Cre+/Tg;ROSA26+/mTmG cKO littermates had bright, GFP-positive cells in the cranial neural folds (Fig. 5A-B′). Consistently, sections through embryos at this same timepoint revealed that the percentage of Sox10-positive NCCs was not significantly different in the cranial neural folds or underlying mesenchyme between genotypes (Fig. 5C-E), suggesting that cKO embryos do not have defects in NCC specification or epithelial-to-mesenchymal transition. At E9.5, Srsf3fl/fl;Wnt1-Cre+/Tg;ROSA26+/mTmG cKO embryos exhibited reduced GFP intensity in the frontonasal prominence and PA 1 (Fig. 5G,G′) compared with Srsf3+/fl;Wnt1-Cre+/Tg;ROSA26+/mTmG control embryos (Fig. 5F,F′). At E10.5, whereas control embryos had bright GFP-positive cells throughout the facial processes and clearly delineated NCC streams entering PAs 3 and 4 (Fig. 5H,H′), cKO littermates had noticeably fewer and less intense GFP-positive cells in the facial processes and no obvious NCC streams entering the PAs (Fig. 5I-I″).
Fig. 5.
Cranial neural crest cell specification, epithelial-to-mesenchymal transition and migration are normal in Srsf3 conditional knockout embryos. (A-B′,F-I″) Dorsal (A-B′) and lateral (F-I″) whole-mount fluorescence images of DAPI (white/blue) (A-B′,F-I″) and GFP (green) (A′,B′,F′-I″) expression in Srsf3+/fl;Wnt1-Cre+/Tg;ROSA26+/mTmG control embryos (top) and Srsf3fl/fl;Wnt1-Cre+/Tg;ROSA26+/mTmG cKO embryos (bottom) at E8.0 (A-B′), E9.5 (F-G′) and E10.5 (H-I″). Image in I″ is the same embryo as in I′ with increased GFP exposure. n≥3 biological replicates per genotype per timepoint. s, somite pairs; FNP, frontonasal process; MxP, maxillary process; MdP, mandibular process; 2 PA, second pharyngeal arch; LNP, lateral nasal process. Scale bars: 500 µm. (C,D) Sox10 staining (magenta) on sections of cranial neural folds in E8.0 Srsf3fl/fl;Wnt1-Cre+/+ control embryos (C) and Srsf3fl/fl;Wnt1-Cre+/Tg cKO embryos (D). Nuclei were stained with DAPI (blue). White asterisk denotes non-specific staining. n=3 biological replicates per genotype. Scale bars: 100 µm. (E) Scatter dot plot depicting average percentage of Sox10-positive cells per embryo in cranial neural folds (Epi) and underlying mesenchyme (Mes) at E8.0. Data are mean±s.e.m. Statistical analyses were performed using a two-tailed, unpaired t-test with Welch's correction for comparisons between genotypes and a paired t-test for comparisons within a given genotype. ****P<0.0001. Shaded circles and squares correspond to independent experiments. n=3 biological replicates per genotype.
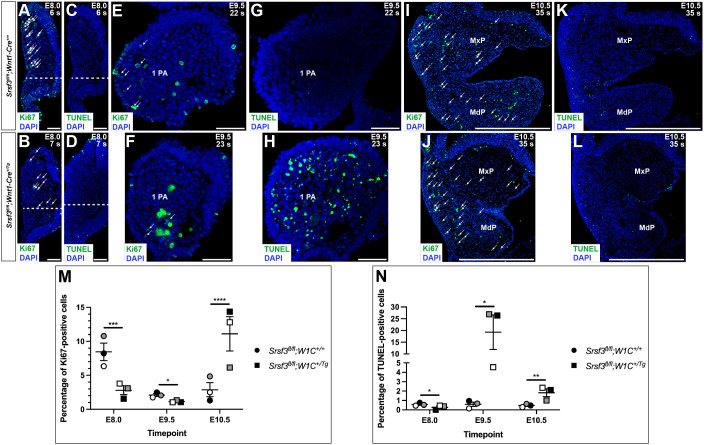
We next assessed both cell proliferation and cell death via Ki67 immunofluorescence analysis and terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL), respectively, in the mesenchyme underlying the cranial neural folds at E8.0 and the mesenchyme of PA 1 at E9.5 and E10.5 (including the maxillary and mandibular processes) in Srsf3fl/fl;Wnt1-Cre+/+ control versus Srsf3fl/fl;Wnt1-Cre+/Tg cKO embryos. Consistent with the above findings, cKO embryos had a considerable reduction in the percentage of Ki67-positive cells (P=0.0002) and a modest decrease in TUNEL-positive cells (P=0.03) compared with control embryos at E8.0 (Fig. 6A-D,M,N). At E9.5, cKO embryos had a slight reduction in proliferation (P=0.03) and a greater than 30-fold increase in cell death (P=0.01) relative to control embryos (Fig. 6E-H,M,N). By E10.5, cKO embryos had an approximately fourfold increase in the percentage of Ki67-positive (P<0.0001) and TUNEL-positive cells (P=0.007) compared with control embryos (Fig. 6I-N). Together, these results reveal an early requirement for Srsf3 in promoting proliferation of cranial NCCs that have recently undergone an epithelial-to-mesenchymal transition and a later requirement for Srsf3 in promoting survival of the NCC-derived craniofacial mesenchyme.
Fig. 6.
Cranial neural crest cell proliferation and survival are defective in Srsf3 conditional knockout embryos. (A-L) Ki67 (A,B,E,F,I,J) and TUNEL (C,D,G,H,K,L) staining (green) on sections of E8.0 (A-D), E9.5 (E-H) and E10.5 (I-L) Srsf3fl/fl;Wnt1-Cre+/+ control embryos (top) and Srsf3fl/fl;Wnt1-Cre+/Tg cKO embryos (bottom). Nuclei were stained with DAPI (blue). White arrows distinguish Ki67 signal from auto-fluorescent red blood cells. Ki67-positive and TUNEL-positive cells were counted above the dashed white lines. n=3 biological replicates per genotype. s, somite pairs; 1 PA, first pharyngeal arch; MxP, maxillary process; MdP, mandibular process. Scale bars: 50 µm in A-H; 500 µm in I-L. (M,N) Scatter dot plots depicting average percentage of Ki67-positive (M) and TUNEL-positive (N) cells per embryo in mesenchyme underlying cranial neural folds at E8.0 and mesenchyme of PA 1 at E9.5 and E10.5. Data are mean±s.e.m. Statistical analyses were performed using a two-tailed, unpaired t-test with Welch's correction. *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. Shaded circles and squares correspond to independent experiments. n=3 biological replicates per genotype.
Srsf3 regulates the alternative RNA splicing of transcripts encoding protein kinases in the facial process mesenchyme
Given the severity of the facial phenotype in cKO embryos, we next examined whether AS remained intact in this setting. Fgfr2 is subject to AS that produces tissue-specific isoforms depending on the inclusion of exon 8 (epithelial ‘b’ isoforms) or exon 9 (mesenchymal ‘c’ isoforms). Both isoforms are required for proper craniofacial development (De Moerlooze et al., 2000; Eswarakumar et al., 2002). Analysis of Fgfr2 expression from RNA isolated from the facial processes of E12.5 control and cKO embryos revealed that both epithelia- and mesenchyme-specific splice isoforms were observed in each genotype (Fig. S2G), indicating that Srsf3 likely regulates the AS of a specific subset of transcripts in the facial mesenchyme. To identify those transcripts, we harvested and sequenced MxP mesenchyme RNA from three biological replicates of E11.5 control versus cKO embryos (Table S1). We chose this timepoint in order to profile AS and gene expression changes between control and cKO embryos immediately before the onset of PS development and to harvest tissue close to the E13.5 timepoint of the RNA-seq experiment above to better compare the results from the two sequencing experiments. Consistent with the qPCR results above (Fig. S2C), reads from control embryos at the Srsf3 locus included exons 2 and 3, whereas reads from cKO embryos often skipped these exons (Fig. S4).
Differential gene expression between control and cKO embryos was assessed via DESeq2 as above. This analysis identified 423 genes with significant differences in expression between the two genotypes (Fig. S5A; Table S6). A GO analysis of these 423 genes using various Enrichr libraries indicated that the top terms related to focal adhesions, the extracellular matrix and cellular signaling, particularly the PI3K/Akt and MAPK/Erk pathways (Fig. S5B). We examined the differential expression of representative genes from these terms with associated mouse craniofacial phenotypes, including Pdgfc, Col2a1 and Kdr (Aszódi et al., 2001; Ding et al., 2004; Sandell et al., 2011), between control and cKO E11.5 MxP mesenchyme samples by qRT-PCR. This analysis confirmed the trends observed in the DESeq2 analysis for all three genes, with significantly increased levels of Col2a1 in the cKO samples compared with the control samples (P=0.04) (Fig. S5C). Among the 423 differentially expressed genes, 65 have mouse models with craniofacial phenotypes, 21 of which exhibit a cleft secondary palate (Table S7). A GO analysis of these 65 genes using the WikiPathways 2019 Mouse library of Enrichr revealed that the second most significant term was neural crest differentiation (P=1.9×10−5). We examined the differential expression of three genes from this term, Foxd3, Col2a1 and Zic5, and one additional gene involved in mouse craniofacial development, Pou3f4 (Aszódi et al., 2001; Inoue et al., 2004; Phippard et al., 1999; Teng et al., 2008), between control and cKO E11.5 MxP mesenchyme samples by qRT-PCR, confirming a significant increase in Zic5 and Pou3f4 expression in cKO samples compared with the control samples (P=0.04) (Fig. S5C).
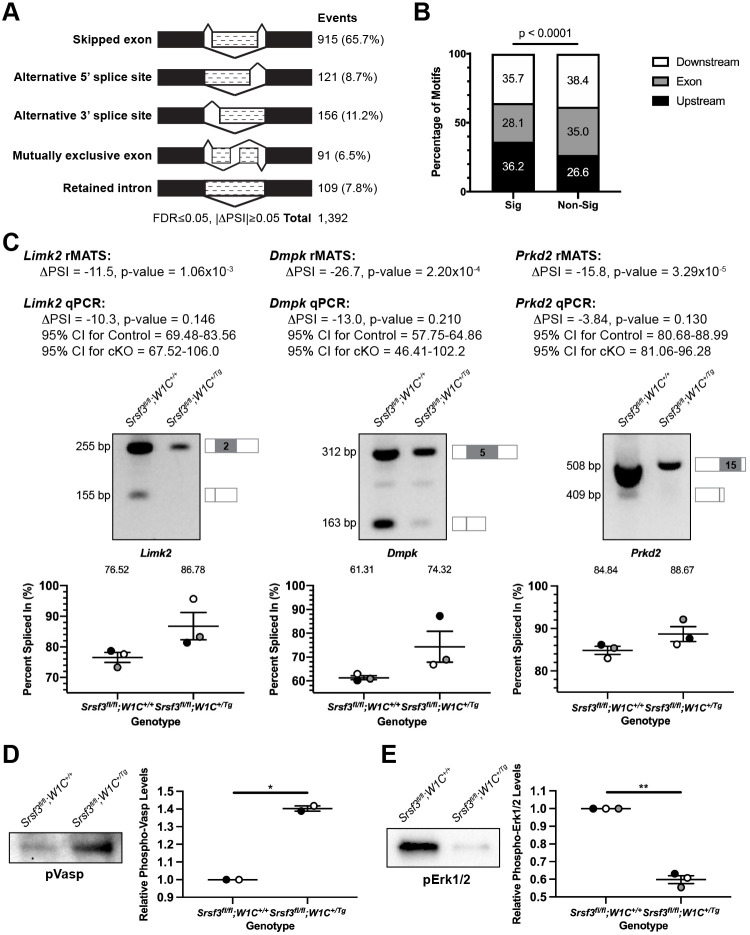
rMATS was again used to detect AS events in this same dataset, revealing 1392 events that were significantly different between genotypes, with the majority of events (65.7%) involving SE (Fig. 7A; Table S8). Only 30 genes (2.9% of all rMATs events) were detected in both the DESeq2 and rMATS analyses (Table S6, bold). We used RBPmap as above to examine annotated Srsf3 motifs in the 915 SE plus 250 bp flanking each end, revealing Srsf3 motifs in 895 (97.8%) of these ROIs (Table S8). The density of these motifs was greater in significant (5.1 motifs per ROI) versus non-significant (4.5 motifs per ROI) SE events in our dataset (P=1.3×10−4). Again, Srsf3 motifs were enriched flanking the SE in the significant SE events (P<0.0001) (Fig. 7B). Among the 728 genes represented in the SE class that had one or more Srsf3 motifs in the ROI, 45 have a corresponding mouse model with a craniofacial phenotype, 14 of which have a cleft or arched secondary palate (Table S9). We again employed SPLINTER to predict outcomes stemming from the AS of the SE events (Table S8). Of the 51 events in Table S9, 14 (26%) are predicted to result in nonsense-mediated decay and potential downregulation of expression, nine (17%) in a truncated protein, four (8%) in no termination codon and two (4%) in an alternative protein (Table S9). Half of the nonsense-mediated decay and alternative protein outcomes, and the majority of truncated protein and no termination codon outcomes, are predicted to occur more often in Srsf3 cKO as opposed to control samples.
Fig. 7.
Srsf3 regulates the alternative RNA splicing of transcripts encoding protein kinases in the facial process mesenchyme. (A) Summary of differential alternative splicing events detected by rMATS analysis of RNA-seq data from E11.5 Srsf3fl/fl;Wnt1-Cre+/+ control versus Srsf3fl/fl;Wnt1-Cre+/Tg cKO MxP mesenchyme. n=3 biological replicates per genotype. FDR, false discovery rate; ΔPSI, difference in percent spliced in. (B) Bar graph depicting percentage of Srsf3 motifs detected by RBPmap within or flanking exons between significant (Sig) and non-significant (Non-Sig) skipped exon events. Statistical analyses were performed using a χ2 test. (C) ΔPSI values for three differentially alternatively spliced transcripts between E11.5 control versus cKO MxP mesenchyme as assessed by rMATS analysis of RNA-seq data and qPCR. Representative qPCR gels with depictions of differentially alternatively spliced exon (gray), and upstream and downstream sequences that were assessed by qPCR, as well as scatter dot plots depicting the percent spliced in, as assessed by qPCR, are presented. Data are mean±s.e.m. Statistical analyses were performed using a paired t-test. Shaded circles correspond to independent experiments. n=3 biological replicates per genotype. CI, confidence interval; control, Srsf3fl/fl;Wnt1-Cre+/+; cKO, Srsf3fl/fl;Wnt1-Cre+/Tg. (D,E) Representative western blots and scatter dot plots depicting relative phospho-Vasp (D) and phospho-Erk1/2 (E) levels between E11.5 Srsf3fl/fl;Wnt1-Cre+/+ control versus Srsf3fl/fl;Wnt1-Cre+/Tg cKO MxP mesenchyme. Data are mean±s.e.m. Statistical analyses were performed using a paired t-test. *P<0.05; **P<0.01. Shaded circles correspond to independent experiments. n≥2 biological replicates per genotype.
A GO analysis of the genes represented in the SE class using the GO Biological Process 2018, GO Molecular Function 2018 and InterPro Domains 2019 libraries of Enrichr revealed that the top terms were phosphorylation (P=1.3×10−4), protein serine/threonine kinase activity (P=5.7×10−5) and protein kinase domain (P=6.4×10−4), respectively. We examined the differential AS of Limk2, Dmpk and Prkd2, each of which was included in these three GO terms, between control and cKO E11.5 MxP mesenchyme samples by qPCR using primers within constitutively expressed exons flanking the alternatively spliced exon. These analyses confirmed the trends observed in the rMATS analysis in all three biological replicates for each transcript (Fig. 7C). Among these 21 protein serine/threonine kinases, Wnk1, Map3k12, Prkaa1 and Irak1 are demonstrated Akt substrates (Chen et al., 2002; Heathcote et al., 2016; Vitari et al., 2004; Wu et al., 2015), as are several of their phosphorylation substrates. Of interest, we detected increased phosphorylation of the protein serine/threonine kinase Prkd2 and its substrate Vasp upon PDGF-AA ligand treatment of primary MEPM cells in our previous phosphoproteomic screen (Fantauzzo and Soriano, 2014), and have confirmed that phospho-Vasp levels are decreased in PdgfraPI3K/PI3K E13.5 PS mesenchyme lysates (Fig. S6A). Vasp was also found to be differentially alternatively spliced in our E13.5 PS mesenchyme rMATS analysis (Table S2), a finding that we confirmed by qPCR (Fig. S6B), resulting in skipping of exon 9 and loss of amino acids containing the Akt consensus motif and the final Prkd2 consensus motif more often in PdgfraPI3K/PI3K samples, indicating an additional mechanism of Vasp regulation downstream of PI3K/Akt-mediated PDGFRα signaling. SPLINTER predicted that AS of Prkd2 would result in a truncated protein in Srsf3fl/fl;Wnt1-Cre+/+ control embryos that affected amino acids that fall within the protein kinase domain and include the proton acceptor. Alternatively, Srsf3 cKO embryos were predicted to express more full-length Prkd2 protein containing the protein kinase domain. Consistent with these predictions, phosphorylation of the Prkd2 substrate Vasp was significantly increased in pooled Srsf3 cKO E11.5 MxP mesenchyme lysates compared with those of control embryos (P=0.02) (Fig. 7D).
Finally, a separate GO analysis of the 21 genes encoding the above protein serine/threonine kinases using the WikiPathways 2019 Mouse library of Enrichr revealed that the second and fourth most significant terms were MAPK signaling pathway (P=2.1×10−5) and focal adhesion-PI3K-Akt-mTOR-signaling pathway (P=3.3×10−4), respectively. In line with changes in MAPK signaling between genotypes, phospho-Erk1/2 levels were significantly decreased in pooled Srsf3 cKO E11.5 MxP mesenchyme lysates compared with those of control embryos (P=0.003) (Fig. 7E). Taken together, these results indicate that Srsf3 serves to regulate PDGFRα-dependent intracellular signaling through the AS of protein serine/threonine kinases.
DISCUSSION
Despite the breadth of studies on PDGF signaling, the intracellular effectors that bridge the receptor-bound signaling molecules and gene expression changes downstream of PDGFRα activation remain largely unknown. Only two studies have shed light on the transcriptional changes downstream of this signaling pathway in the context of craniofacial development, both using primary MEPM cells. PDGFRα signaling was shown to specifically upregulate the expression of 41 genes bound by the SRF transcription factor, a subset of which were enriched for MRTF co-factor binding sites and associated with the cytoskeleton (Vasudevan and Soriano, 2014). Furthermore, PDGFRα signaling was demonstrated to principally depend on PI3K activity and ultimately promote osteoblast differentiation through the upregulation of 24 transcripts associated with cell differentiation (Vasudevan et al., 2015). Here, we have revealed that the PI3K/Akt-mediated PDGFRα signaling axis regulates gene expression through AS in vivo in the mid-gestation PS and have discovered 523 transcripts that are differentially alternatively spliced when this pathway is disrupted. This study thus highlights a novel role for PDGFRα signaling in regulating RNA processing and identifies the phosphorylation of RBPs downstream of this pathway as a mechanism that contributes to this response.
Transcripts encoding each of the 11 SR proteins expressed in mouse were detected in both RNA-seq analyses presented here. In the E13.5 PS mesenchyme, Srsf1, Srsf2 and Srsf6 had average normalized read counts in the control samples on the same order of magnitude as Srsf3. Furthermore, each of the murine SR proteins contains at least one Akt consensus sequence and as many as 22 in the case of Srsf4. In addition to human SRSF3 (Bavelloni et al., 2014), human SRSF1 and SRSF7 have previously been shown to be phosphorylated by Akt in in vitro kinase assays (Blaustein et al., 2005). It is therefore interesting that Srsf3 was the only SR protein detected in our previous mass spectrometry screen as an Akt phosphorylation target downstream of PI3K-mediated PDGFRα signaling in the palatal mesenchyme (Fantauzzo and Soriano, 2014). Further confirming this finding, the severe facial clefting phenotype of Srsf3 cKO embryos indicates that other SR proteins expressed in the facial mesenchyme are unable to compensate for the loss of Srsf3 in this setting. This idea is consistent with previous results demonstrating that two human SR proteins, SRSF3 and SRSF4, have distinct consensus binding motifs and mostly unique target transcripts in the P19 mouse embryonic carcinoma cell line (Änkö et al., 2012). Furthermore, none of the genes encoding SR proteins were differentially expressed in either DESeq2 analysis here, nor were the transcripts differentially alternatively spliced in the Pdgfra+/+ versus PdgfraPI3K/PI3K rMATS analysis. Only Srsf3 was found to be differentially alternatively spliced in the Srsf3fl/fl;Wnt1-Cre+/+ versus Srsf3fl/fl;Wnt1-Cre+/Tg rMATS analysis. These results demonstrate that, unlike in P19 cells (Änkö et al., 2012), Srsf3 is not a master regulator of SR protein-encoding transcript splicing in the facial mesenchyme.
Interestingly, the Srsf3 western blot bands present in the absence of PDGF-AA ligand stimulation following Akt phosphosubstrate immunoprecipitation further confirm that the majority of Akt phosphorylation targets in MEPM cells are phosphorylated at baseline levels (Fantauzzo and Soriano, 2014). This finding suggests that input(s) other than PDGF-AA-stimulated PDGFRα signaling contributes to Srsf3 phosphorylation at Akt consensus sites. SR proteins have been shown to be phosphorylated in response to multiple treatments, such as downstream of hepatocyte, keratinocyte and epidermal growth factor stimulation and all-trans retinoic acid treatment (Bavelloni et al., 2014; Blaustein et al., 2005; Zhou et al., 2012). As these growth factors, metabolites and/or their receptors have demonstrated roles in regulating the activity of NCCs and the development of their derivatives in the mouse (Fantauzzo and Soriano, 2015; Williams and Bohnsack, 2019), it is likely that one or more of these pathways, or an as yet undetermined signaling pathway, contributes to Srsf3 phosphorylation in the palatal mesenchyme. Although we have examined the effect of Srsf3 phosphorylation in response to PDGF-AA ligand stimulation on its subcellular localization, future studies will determine whether these same post-translational modifications downstream of PI3K/Akt-mediated PDGFRα signaling also affect Srsf3 RNA binding and/or sequence specificity.
Our study has revealed a novel requirement for an SR protein in NCCs and the facial mesenchyme. Upon ablation of Srsf3 in the NCC lineage, cKO cells underlying the cranial neural folds exhibit a proliferation defect. These cranial NCCs are able to migrate into the facial processes, although fewer of these cells reach their target site. By E9.5, cKO embryos exhibit significantly increased cell death in the facial process mesenchyme, leading to hypoplastic facial processes. However, this cell death is partially offset by increased cell proliferation at E10.5, thereby allowing derivatives of these processes to form in embryos that survive beyond mid-gestation. In accordance with Srsf3 acting as an effector downstream of PDGFRα signaling, Srsf3 cKO embryos have a number of overlapping phenotypes with mutant mouse models of Pdgfra and/or one of its ligands, including midline facial clefting, PS hypoplasia, facial subepidermal blebbing, facial hemorrhaging, brain malformations and a wavy neural tube (Andrae et al., 2016; Ding et al., 2004; Fantauzzo and Soriano, 2014; Fredriksson et al., 2012; Klinghoffer et al., 2002; Soriano, 1997). However, the craniofacial skeletal phenotypes in Srsf3 cKO embryos are more severe than those found upon ablation of Pdgfra in the NCC lineage (He and Soriano, 2013; Tallquist and Soriano, 2003), again indicating that Srsf3 acts to mediate more than PDGFRα signaling in this setting.
Comparing the two RNA-seq experiments performed here, there were 38 AS events commonly detected in both rMATS analyses. Among these, only 12 (32%) were differentially alternatively spliced in the same direction (included or excluded) in PdgfraPI3K/PI3K and Srsf3 cKO embryos compared with their respective control embryos. A GO analysis of the genes represented by the 38 common AS events using the WikiPathways 2019 Mouse and KEGG 2019 Mouse libraries of Enrichr demonstrated that the top terms were focal adhesion-PI3K-Akt-mTOR-signaling pathway (P=0.006) and PI3K-Akt signaling pathway (P=0.008), respectively. For the DESeq2 analyses, only two genes were commonly detected: Aldh1a2 and Rragd. The relatively low extent of overlap between identified genes is not surprising given that these RNA-seq experiments were performed in different, but related, tissues across a 48 h timeframe. In fact, AS changes in the developing murine face from E10.5 to E12.5 are more significant across age than across facial prominence location (Hooper et al., 2020). Furthermore, there was little to no overlap between the genes detected in the rMATS and DESeq2 analyses within either RNA-seq experiment, similar to previous analyses of differential AS and differential gene expression upon ablation of the RBPs Esrp1 and Esrp2 in the embryonic murine epidermis (Bebee et al., 2015). The majority of genes identified in the control versus Srsf3 cKO MxP mesenchyme DESeq2 analysis encode transcripts that are likely regulated by Srsf3 through mechanisms other than AS, such as transcription, export, translation and degradation (Howard and Sanford, 2015). Notably, however, the rMATS and DESeq2 results from the control versus Srsf3 cKO MxP mesenchyme RNA-seq analysis were commonly enriched for genes involved in focal adhesion, PI3K/Akt signaling and MAPK/Erk signaling, similar to results from a xenograft glioblastoma model in the presence or absence of SRSF3 (Fuentes-Fayos et al., 2020). These findings indicate that Srsf3 activity may influence each of these processes through multiple mechanisms of gene expression regulation. Relatedly, we found that phospho-Erk1/2 levels were significantly reduced in Srsf3 cKO embryos. As the MAPK/Erk pathway has demonstrated roles in murine facial midline development and has been shown to function downstream from PDGFRα activation in this setting (Parada et al., 2015; Vasudevan et al., 2015), we propose that dysregulation of this signaling axis in Pdgfra and Srsf3 mutant mouse models may underlie, at least in part, the common facial clefting phenotypes. Going forward, it will be crucial to identify transcripts that are directly bound by Srsf3 in the facial mesenchyme through eCLIP analyses. Of interest, of the genes represented by the differentially alternatively spliced transcripts identified in our control versus Srsf3 cKO MxP mesenchyme rMATS analysis, transcripts from 63.0%, 48.8% and 13.5% were shown to be bound by human SRSF3 in mouse embryonic stem cells, HeLa cells and P19 cells, respectively (Änkö et al., 2012; Ratnadiwakara et al., 2018; Xiao et al., 2016).
Taken together, our findings significantly expand our understanding of the molecular mechanisms by which PDGFRα signaling controls gene expression and identify Srsf3 as a novel intracellular effector of this pathway during mammalian midface development. Future studies will seek to establish an AS ‘code’ in the facial mesenchyme to address how RBPs might function in a cooperative or antagonistic fashion in this context and determine how the activity of these proteins is affected by post-translational modification downstream of receptor tyrosine kinase activity.
MATERIALS AND METHODS
Mouse strains and husbandry
All animal experimentation was approved by the Institutional Animal Care and Use Committee of the University of Colorado Anschutz Medical Campus. Pdgfratm5Sor mice (Klinghoffer et al., 2002), referred to in the text as PdgfraPI3K mice; Srsf3tm1Pjln mice (Jumaa et al., 1999), referred to in the text as Srsf3fl mice; H2afvTg(Wnt1-cre)11Rth mice (Danielian et al., 1998), referred to in the text as Wnt1-CreTg mice; and Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo mice (Muzumdar et al., 2007), referred to in the text as ROSA26mTmG mice, were maintained on a 129S4 co-isogenic genetic background and housed at a sub-thermoneutral temperature of 21-23°C. Embryos were obtained from intercrosses of Srsf3fl/fl males with Srsf3+/fl;Wnt1-Cre+/Tg females or intercrosses of Srsf3fl/fl;ROSA26+/mTmG males with Srsf3+/fl;Wnt1-Cre+/Tg females. Mice were euthanized by inhalation of carbon dioxide from compressed gas. Cervical dislocation was used as a secondary verification of death. Both male and female embryos were analyzed in this study and no differences in phenotype were detected between these two groups. Developmental stages are described in the Results section for individual experiments. Control and experimental embryos were harvested from the same litter and embryos were age-matched to the greatest extent possible by somite counting and/or digit volume measurement. Statistical analyses of Mendelian inheritance among mice were performed with the GraphPad QuickCalcs data analysis resource using a χ2 test.
RNA-seq and bioinformatics analyses
Secondary palatal shelves (PS) were dissected on ice from three independent biological replicates each of E13.5 Pdgfra+/+ and PdgfraPI3K/PI3K embryos, and the overlying ectoderm was removed by digestion with 20 mg/ml trypsin for 20 min at 4°C. Maxillary processes (MxP) were dissected on ice from three independent biological replicates each of E11.5 Srsf3fl/fl;Wnt1-Cre+/+ and Srsf3fl/fl;Wnt1-Cre+/Tg embryos, and the overlying ectoderm was removed by digestion with 20 mg/ml trypsin for 15-20 min at 4°C. PS and MxP were subsequently rinsed in 10% fetal bovine serum (FBS) (Hyclone Laboratories) for 1 min followed by 1×phosphate buffered saline (PBS). Excess PBS was removed and PS and MxP were submerged in 200 μl RNAlater stabilization solution (Invitrogen) for overnight storage at 4°C. The following day excess RNAlater was removed and the samples were stored at −80°C. Total RNA was simultaneously isolated from all samples using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. RNA was forwarded to the University of Colorado Anschutz Medical Campus Genomics and Microarray Core for quality control, library preparation and sequencing. RNA integrity was assessed on an Agilent 4200 TapeStation using TapeStation Analysis Software A.02.02, revealing RINe values of 9.6-10.0 for the PS samples and 9.5-10.0 for the MxP samples. RNA concentration was measured with an Infinite M200 PRO Microplate Reader (Tecan Group). Total RNA (100 ng) was used for input into the Universal Plus mRNA-Seq with NuQuant kit (NuGEN). Dual index, stranded libraries were prepared and sequenced on a NovaSeq 6000 Sequencing System using an S4 Flow Cell (Illumina) to an average depth of ∼44 million read pairs (2×151 bp reads).
BBDuk (from the BBmap v35.85 tool suite) (Bushnell, 2015) was used to perform adapter sequence contamination removal and light quality trimming. For STAR (Dobin et al., 2013) mapping and rMATS (Shen et al., 2014) analysis, read length was standardized to 125 bp to satisfy the input requirements for rMATS. After quality trimming and adapter contamination removal, reads shorter than 125 bp were discarded, and reads longer than 125 bp were cropped to 125 bp. For discovery of AS events, sequence alignment was performed with STAR (v2.7.0) two-pass mapping, using combined splice junctions reported in the first alignment round. AS events detected via rMATS (v4.0.2, default parameters plus --readLength 125 --libType fr-secondstrand) with false discovery rate≤0.05, a difference in PSI (|ΔPSI|)≥0.05 and an average count of 10 in either population are reported. For differential expression analysis, RNA-sequencing data were quantified at the transcript level via Salmon (v1.1.0) (Patro et al., 2017) using GENCODE (Frankish et al., 2019) vM19 annotation, summarized to gene-level estimated counts via the R package tximport (Soneson et al., 2016) and analyzed for differential expression via DESeq2 (Love et al., 2014). Significant changes in gene-level expression are reported for cases with adjusted P≤0.05. Raw read pairs, trimmed read pairs (125 bp) for STAR input, STAR unique mapping rate, trimmed read pairs for Salmon input and Salmon mapping rate per sample can be found in Table S1. For exons identified to have a skipped exon event, a region of interest (ROI) was generated to include the skipped exon plus 250 bp flanking both ends. ROIs were specific to the strand of the skipped exon. ROIs were analyzed by RBPmap (Paz et al., 2014) to search for Srsf3 (both ‘cuckucy’ and ‘wcwwc’) motifs using default parameters and the mm10 genome assembly. RBPmap output was parsed and filtered using a custom R script and only exact motif matches were kept. Motifs were summarized by ‘n’ per ROI. Motif location was determined by scoring each motif hit location as upstream, within or downstream of the exon. Statistical analyses of motif density between significant versus non-significant skipped exon events were performed using a Wilcoxon rank sum test with continuity correction and statistical analyses of motif location were performed using a χ2 test. To predict potential outcomes of AS events, the R package SPLINTER (https://github.com/dianalow/SPLINTER/) was run with default parameters using the skipped exon ‘JCEC’ rMATS output file. A transcript and coding sequence database was prepared from the GTF file used above for STAR alignment. For each AS event identified by rMATS, SPLINTER identified compatible transcripts and compared the compatible transcripts before and after removal of the exon. One outcome was predicted per compatible transcript. Gene ontology analysis was performed with various libraries from the Enrichr gene list enrichment analysis tool (Chen et al., 2013; Kuleshov et al., 2016) and terms with P<0.05 were considered significant.
qPCR
Total RNA was isolated using the RNeasy mini kit (Qiagen) according to the manufacturer's instructions. First-strand cDNA was synthesized using a ratio of 2:1 random primers: oligo (dT) primer and SuperScript II RT (Invitrogen) according to the manufacturer's instructions. All reactions were performed with 1× ThermoPol buffer [0.02 M Tris (pH 8.8), 0.01 M KCl, 0.01 M (NH4)2SO4, 2 mM MgSO4 and 0.1% Triton X-100], 200 µM dNTPs, 200 nM primers (Integrated DNA Technologies), 0.6 U Taq polymerase and 1 µg cDNA in a 25 µl reaction volume. The primers used can be found in Table S10. The following PCR protocol was used for Chrd, Cask and Smad7: step 1, 3 min at 94°C; step 2, 30 s at 94°C; step 3, 30 s at 50°C; step 4, 30 s at 72°C; repeat steps 2-4 for 34 cycles; and step 5, 2 min at 72°C. The following PCR protocol was used for Limk2: step 1, 3 min at 94°C; step 2, 30 s at 94°C; step 3, 30 s at 51°C; step 4, 30 s at 72°C; repeat steps 2-4 for 34 cycles; and step 5, 2 min at 72°C. The following PCR protocol was used for Dmpk: step 1, 3 min at 94°C; step 2, 30 s at 94°C; step 3, 30 s at 55°C; step 4, 30 s at 72°C; repeat steps 2-4 for 34 cycles; and step 5, 2 min at 72°C. The following PCR protocol was used for Prkd2: step 1, 3 min at 94°C; step 2, 30 s at 94°C; step 3, 30 s at 49.2°C; step 4, 30 s at 72°C; repeat steps 2-4 for 34 cycles; and step 5, 2 min at 72°C. Two-thirds of total PCR products were electrophoresed on a 2% agarose/TBE gel containing ethidium bromide and photographed on an Aplegen Omega Fluor Gel Documentation System. Quantifications of band intensity were performed with ImageJ software (version 1.51m9, National Institutes of Health). The PSI was calculated independently for each sample as the percentage of the larger isoform divided by the total abundance of all isoforms within the given gel lane. Statistical analyses were performed with Prism 8 (GraphPad Software) using a paired t-test. qPCR reactions were performed using samples from at least three embryos per genotype.
In situ hybridization
Template for the Srsf3 probe spanning exon 7 was amplified by PCR from E13.5 head cDNA using the following primers: 5′-GACTCTAGAGATAAGGGTAGGAACCACAC-3′ and 5′-GAGAAGCTTGGAATGTTTTACCTGGACTTG-3′. PCR product was cloned into the pBluescript II KS(+) dual promoter (T7 and T3) phagemid vector (Agilent Technologies) and standard procedures were followed for the preparation of DIG-labeled cRNA (Roche Diagnostics) anti-sense (AS) and control sense (S) probes. Whole-mount in situ hybridization was performed based on a previously published protocol (Wilkinson, 1998) using 1 µg/ml probe and anti-DIG-AP Fab fragment primary antibody (1:2000; 11093274910; Roche Diagnostics). Stained embryos were photographed using an Axiocam 105 color digital camera (Carl Zeiss) fitted onto a Stemi 508 stereo microscope (Carl Zeiss). In situ hybridization experiments were performed simultaneously using antisense and sense probes across at least two independent experiments per timepoint.
Immunofluorescence analyses
Embryos were fixed in 4% paraformaldehyde (PFA) in 1× PBS and infiltrated with 30% sucrose in PBS before being mounted in OCT compound (Sakura Finetek USA). Sections (8-10 µm) were deposited on glass slides. Sections were fixed in 4% PFA in PBS with 0.1% Triton X-100 for 10 min and washed in PBS with 0.1% Triton-X 100. Sections were blocked for 1 h in 5% normal donkey serum (Jackson ImmunoResearch Laboratories) in PBS and incubated overnight at 4°C in primary antibody in 1% normal donkey serum in PBS. After washing in PBS, sections were incubated in Alexa Fluor 488-conjugated donkey anti-rabbit secondary antibody (1:1000; A21206; Invitrogen) or Alexa Fluor 546-conjugated donkey anti-goat secondary antibody (1:800; A11056; Invitrogen) diluted in 1% normal donkey serum in PBS with 2 μg/ml 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) for 1 h. Sections were mounted in Aqua Poly/Mount mounting medium (Polysciences) and photographed using an Axiocam 506 mono digital camera (Carl Zeiss) fitted onto an Axio Observer 7 fluorescence microscope (Carl Zeiss). The following antibodies were used for immunofluorescence analysis: Srsf3 (1:500; ab73891; Abcam), previously validated by Sen et al. (2013); Sox10 (1:2000; AF2864; R&D Systems); Ki67 (1:300; PA1-38032; Invitrogen). Srsf3 immunofluorescence analysis was performed on multiple sections from individual E8.5-E10.5 embryos and from four E13.5 embryos. Srsf3 and Sox10 double immunofluorescence analysis was performed on multiple sections from individual E8.5-E9.5 embryos. All Sox10- and Ki67-positive signals were confirmed by DAPI staining. The percentage of Sox10-positive or Ki67-positive cells was determined in three embryos per genotype, with up to five sections analyzed per individual embryo. Analyzed sections within a given embryo were 5-10 sections apart, representing a distance of 40-100 µm. Graphed data represent averages from three independent embryos. Statistical analyses were performed on values from individual sections with Prism 8 (GraphPad Software) using a two-tailed, unpaired t-test with Welch's correction for comparisons between genotypes and a paired t-test for comparisons within a given genotype.
Immunoprecipitations and western blotting
Immortalized mouse embryonic palatal mesenchyme (iMEPM) cells were derived from a male Cdkn2a−/− embryo as previously described (Fantauzzo and Soriano, 2017). iMEPM cells were cultured in medium [Dulbecco's modified Eagle's medium (Gibco, Invitrogen) supplemented with 50 U/ml penicillin (Gibco), 50 µg/ml streptomycin (Gibco) and 2 mM L-glutamine (Gibco) containing 10% FBS (Hyclone Laboratories)] and grown at 37°C in 5% carbon dioxide. To induce PDGFRα signaling, passage 23-29 iMEPM cells at ∼70% confluence were serum starved for 24 h in medium containing 0.1% FBS and stimulated with 10 ng/ml PDGF-AA ligand (R&D Systems) for the indicated length of time. When applicable, cells were pretreated with 50 µM LY294002 (Sigma-Aldrich) 1 h before PDGF-AA ligand stimulation. Subcellular fractions were generated by harvesting cells in ice-cold PBS, resuspending cells in hypotonic buffer [20 mM Tris-HCl (pH 7.4), 10 mM NaCl, 3 mM MgCl2 and 0.5% Nonidet P-40] and collecting cytoplasmic lysates by centrifugation at 800 g at 4°C for 10 min. The nuclear pellet was resuspended in cell extraction buffer [100 mM Tris-HCl (pH 7.4), 100 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA, 0.1% SDS, 1×complete Mini protease inhibitor cocktail (Roche Diagnostics), 1 mM PMSF, 1 mM NaF, 2 mM Na3VO4, 0.5% deoxycholate and 20 mM Na4P2O7] and nuclear lysates collected by centrifugation at 15,800 g at 4°C for 30 min. For immunoprecipitations, cell lysates were incubated with magnetic bead-conjugated phospho-Akt substrate primary antibody (1:30; 110B7E; 8050S; Cell Signaling Technology) overnight at 4°C. Beads were washed with lysis buffer five times and the precipitated proteins were eluted with Laemmli buffer containing 10% β-mercaptoethanol, heated for 5 min at 100°C and separated by SDS-PAGE. Maxillary processes (MxP) were dissected on ice from multiple E11.5 Srsf3fl/fl;Wnt1-Cre+/+ and Srsf3fl/fl;Wnt1-Cre+/Tg embryos, and the overlying ectoderm was removed by digestion with 20 mg/ml trypsin for 15-20 min at 4°C. MxP were subsequently rinsed in 10% FBS (Hyclone Laboratories) for 1 min followed by 1×PBS. Excess PBS was removed and MxP were snap frozen in 100% ethanol on dry ice and stored at −80°C. MxP from three embryos per genotype were thawed on ice and pooled to form biological replicates. Protein lysates were generated by resuspending tissues in ice-cold NP-40 lysis buffer [20 mM Tris-HCl (pH 8), 150 mM NaCl, 10% glycerol, 1% Nonidet P-40, 2 mM EDTA, 1× complete Mini protease inhibitor cocktail (Roche Diagnostics), 1 mM PMSF, 10 mM NaF, 1 mM Na3VO4 and 25 mM β-glycerophosphate] and collecting cleared lysates by centrifugation at 13,400 g at 4°C for 20 min. Laemmli buffer containing 10% β-mercaptoethanol was added to the lysates, which were heated for 5 min at 100°C. Proteins were subsequently separated by SDS-PAGE. Western blot analysis was performed according to standard protocols using horseradish peroxidase-conjugated secondary antibodies. The following antibodies were used for western blotting: Srsf3 (1:1000; ab73891; Abcam), previously validated by Sen et al. (2013); lamin B1 (1:1000; D4Q4Z; 12586S; Cell Signaling Technology); β-tubulin (1:1000; E7; E7; Developmental Studies Hybridoma Bank); phospho-Akt (1:1000; Ser473; 9271S; Cell Signaling Technology); Akt (1:1000; 9272S; Cell Signaling Technology); phospho-VASP (1:1000; Ser157; 3111; Cell Signaling Technology); VASP (1:1000; 9A2; 3132; Cell Signaling Technology); phospho-Erk1/2 (1:1000; Thr202/Thr204; 9101; Cell Signaling Technology); Erk1/2 (1:1000; 9102; Cell Signaling Technology); horseradish peroxidase-conjugated goat anti-rabbit IgG (1:10,000; 111035003; Jackson ImmunoResearch Laboratories); horseradish peroxidase-conjugated goat anti-mouse IgG (1:10,000; 115035003; Jackson ImmunoResearch Laboratories). Quantifications of signal intensity were performed with ImageJ software (version 1.51m9, National Institutes of Health). Relative phospho-Srsf3 levels were determined by normalizing to lamin B1 and β-tubulin for the nuclear and cytoplasmic fractions, respectively. Relative phospho-Vasp levels were determined by normalizing to total Vasp levels. Relative phospho-Erk1/2 levels were determined by normalizing to total Erk1/2 levels. When applicable, statistical analyses were performed with Prism 8 (GraphPad Software) using a paired t-test. Immunoprecipitation and western blotting experiments were performed across at least three independent experiments, with the exception of the phospho-Vasp and Vasp western blots, which were performed across two independent experiments, each consisting of pooled lysates from three embryos per genotype.
Morphological analyses
Embryos were dissected at multiple timepoints (day of plug considered 0.5 days) in 1×PBS and fixed overnight at 4°C in 4% PFA in PBS. Embryos were photographed using an Axiocam 105 color digital camera (Carl Zeiss) fitted onto a Stemi 508 stereo microscope (Carl Zeiss). One representative embryo per genotype per timepoint was photographed for morphological analysis.
Whole-mount DAPI staining
Whole-mount DAPI staining was performed according to a previously published protocol (Sandell et al., 2012), with the exception that staining was performed with 10 µg/ml DAPI (Sigma-Aldrich) for 1 h at room temperature. Embryos were photographed using an Axiocam 506 mono digital camera (Carl Zeiss) fitted onto an Axio Observer 7 fluorescence microscope (Carl Zeiss). Extended Depth of Focus was applied to z-stacks using ZEN Blue software (Carl Zeiss) to generate images with the maximum depth of field. One representative embryo per genotype per timepoint not possessing the ROSA26mTmG allele was photographed for E10.5-E14.5 embryos. At least three embryos per genotype per timepoint possessing the ROSA26mTmG allele were photographed for E8.0-E10.5 embryos.
Skeletal preparations
E18.5 embryos were skinned, eviscerated, fixed in 95% ethanol and stained in 0.015% Alcian Blue, 0.005% Alizarin Red and 5% glacial acetic acid in 70% ethanol at 37°C. Embryos were then cleared in 1% KOH and transferred to solutions of decreasing KOH concentration and increasing glycerol concentration. Skeletons were photographed using an Axiocam 105 color digital camera (Carl Zeiss) fitted onto a Stemi 508 stereo microscope (Carl Zeiss). At least three embryos per genotype were assayed.
TUNEL assay
Sections (8-10 µm) of PFA-fixed, sucrose-infiltrated, OCT-mounted embryos were deposited on glass slides. Apoptotic cells were identified using the In Situ Cell Death Detection Kit, Fluorescein (Sigma-Aldrich) according to the manufacturer's instructions for the treatment of cryopreserved tissue sections. Sections were mounted in VECTASHIELD Antifade Mounting Medium with DAPI (Vector Laboratories) and photographed using an Axiocam 506 mono digital camera (Carl Zeiss) fitted onto an Axio Observer 7 fluorescence microscope (Carl Zeiss). All positive signals were confirmed by DAPI staining. The percentage of TUNEL-positive cells was determined in three embryos per genotype, with up to four sections analyzed per individual embryo. Analyzed sections within a given embryo were 5-10 sections apart, representing a distance of 40-100 µm. Graphed data represent averages from three independent embryos. Statistical analyses were performed on values from individual sections with Prism 8 (GraphPad Software) using a two-tailed, unpaired t-test with Welch's correction.
Supplementary Material
Acknowledgements
We thank Damian Garno, Jessica Patrick and Robert Long for technical assistance. RNA-seq experiments were performed at the Genomics and Microarray Core at the University of Colorado Anschutz Medical Campus. Srsf3fl mice were a gift from Dr Nicholas Webster at the University of California San Diego. We are grateful to Drs David Clouthier and Julie Siegenthaler at the University of Colorado Anschutz Medical Campus for assistance with analysis of Srsf3fl/fl;Wnt1-Cre+/Tg phenotypes. We thank members of the Fantauzzo laboratory, and Drs Matthew Taliaferro, David Clouthier and Philippe Soriano for their critical comments on the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: K.A.F.; Methodology: B.J.C.D., E.D.L., K.A.F.; Formal analysis: B.J.C.D., E.D.L., R.F., K.A.F.; Investigation: B.J.C.D., J.M., K.A.F.; Writing - original draft: K.A.F.; Writing - review & editing: B.J.C.D., E.D.L., R.F., J.M., K.A.F.; Visualization: B.J.C.D., K.A.F.; Supervision: K.A.F.; Funding acquisition: B.J.C.D., K.A.F.
Funding
This work was supported by the National Institutes of Health (R01DE027689 to K.A.F., K02DE028572 to K.A.F. and F31DE029364 to B.J.C.D.) and the RNA Bioscience Initiative at the University of Colorado Anschutz Medical Campus (Graduate Scholar Award to B.J.C.D. and RNA-seq grant to K.A.F.). Deposited in PMC for release after 12 months.
Data availability
The RNA-sequencing datasets generated during this study have been deposited in GEO under accession number GSE161510.
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/article-lookup/doi/10.1242/dev.199448
References
- Alessi, D. R., Caudwell, F. B., Andjelkovic, M., Hemmings, B. A. and Cohen, P. (1996). Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 399, 333-338. 10.1016/S0014-5793(96)01370-1 [DOI] [PubMed] [Google Scholar]
- Andrae, J., Gouveia, L., Gallini, R., He, L., Fredriksson, L., Nilsson, I., Johansson, B. R., Eriksson, U. and Betsholtz, C. (2016). A role for PDGF-C/PDGFRα signaling in the formation of the meningeal basement membranes surrounding the cerebral cortex. Biol. Open 5, 461-474. 10.1242/bio.017368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Änkö, M.-L., Müller-McNicoll, M., Brandl, H., Curk, T., Gorup, C., Henry, I., Ule, J. and Neugebauer, K. M. (2012). The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 13, R17. 10.1186/gb-2012-13-3-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aszódi, A., Hunziker, E. B., Olsen, B. R. and Fässler, R. (2001). The role of collagen II and cartilage fibril-associated molecules in skeletal development. Osteoarthr. Cartil. 9, S150-S159. [PubMed] [Google Scholar]
- Atasoy, D., Schoch, S., Ho, A., Nadasy, K. A., Liu, X., Zhang, W., Mukherjee, K., Nosyreva, E. D., Fernandez-Chacon, R., Missler, M.et al. (2007). Deletion of CASK in mice is lethal and impairs synaptic function. Proc. Natl. Acad. Sci. USA 104, 2525-2530. 10.1073/pnas.0611003104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachiller, D., Klingensmith, J., Shneyder, N., Tran, U., Anderson, R., Rossant, J. and De Robertis, E. M. (2003). The role of chordin/Bmp signals in mammalian pharyngeal development and DiGeorge syndrome. Development 130, 3567-3578. 10.1242/dev.00581 [DOI] [PubMed] [Google Scholar]
- Bavelloni, A., Piazzi, M., Faenza, I., Raffini, M., D'Angelo, A., Cattini, L., Cocco, L. and Blalock, W. L. (2014). Prohibitin 2 represents a novel nuclear AKT substrate during all-trans retinoic acid-induced differentiation of acute promyelocytic leukemia cells. FASEB J. 28, 2009-2019. 10.1096/fj.13-244368 [DOI] [PubMed] [Google Scholar]
- Bebee, T. W., Park, J. W., Sheridan, K. I., Warzecha, C. C., Cieply, B. W., Rohacek, A. M., Xing, Y. and Carstens, R. P. (2015). The splicing regulators Esrp1 and Esrp2 direct an epithelial splicing program essential for mammalian development. eLife 4, e08954. 10.7554/eLife.08954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaustein, M., Pelisch, F., Tanos, T., Muñoz, M. J., Wengier, D., Quadrana, L., Sanford, J. R., Muschietti, J. P., Kornblihtt, A. R., Cáceres, J. F.et al. (2005). Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat. Struct. Mol. Biol. 12, 1037-1044. 10.1038/nsmb1020 [DOI] [PubMed] [Google Scholar]
- Bushnell, B. (2015). BBMap. https://sourceforge.net/projects/bbmap/.
- Chen, B.-C., Wu, W.-T., Ho, F.-M. and Lin, W.-W. (2002). Inhibition of interleukin-1β-induced NF-κB activation by calcium/calmodulin-dependent protein kinase kinase occurs through Akt activation associated with interleukin-1 receptor-associated kinase phosphorylation and uncoupling of MyD88. J. Biol. Chem. 277, 24169-24179. 10.1074/jbc.M106014200 [DOI] [PubMed] [Google Scholar]
- Chen, E. Y., Tan, C. M., Kou, Y., Duan, Q., Wang, Z., Meirelles, G. V., Clark, N. R. and Ma'ayan, A. (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibi, D. M., Mia, M. M., Guna Shekeran, S., Yun, L. S., Sandireddy, R., Gupta, P., Hota, M., Sun, L., Ghosh, S. and Singh, M. K. (2019). Neural crest-specific deletion of Rbfox2 in mice leads to craniofacial abnormalities including cleft palate. eLife 8, e45418. 10.7554/eLife.45418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian, P. S., Muccino, D., Rowitch, D. H., Michael, S. K. and McMahon, A. P. (1998). Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, 1323-1326. 10.1016/S0960-9822(07)00562-3 [DOI] [PubMed] [Google Scholar]
- De Moerlooze, L., Spencer-Dene, B., Revest, J., Hajihosseini, M., Rosewell, I. and Dickson, C. (2000). An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127, 483-492. 10.1242/dev.127.3.483 [DOI] [PubMed] [Google Scholar]
- Dichmann, D. S., Fletcher, R. B. and Harland, R. M. (2008). Expression cloning in Xenopus identifies RNA-binding proteins as regulators of embryogenesis and Rbmx as necessary for neural and muscle development. Dev. Dyn. 237, 1755-1766. 10.1002/dvdy.21590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, H., Wu, X., Boström, H., Kim, I., Wong, N., Tsoi, B., O'Rourke, M., Koh, G. Y., Soriano, P., Betsholtz, C.et al. (2004). A specific requirement for PDGF-C in palate formation and PDGFR-α signaling. Nat. Genet. 36, 1111-1116. 10.1038/ng1415 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., Batut, P., Chaisson, M. and Gingeras, T. R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswarakumar, V. P., Monsonego-Ornan, E., Pines, M., Antonopoulou, I., Morriss-Kay, G. M. and Lonai, P. (2002). The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development 129, 3783-3793. 10.1242/dev.129.16.3783 [DOI] [PubMed] [Google Scholar]
- Fantauzzo, K. A. and Soriano, P. (2014). PI3K-mediated PDGFRα signaling regulates survival and proliferation in skeletal development through p53-dependent intracellular pathways. Genes Dev. 28, 1005-1017. 10.1101/gad.238709.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantauzzo, K. A. and Soriano, P. (2015). Receptor tyrosine kinase signaling: regulating neural crest development one phosphate at a time. Curr. Top. Dev. Biol. 111, 135-182. 10.1016/bs.ctdb.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantauzzo, K. A. and Soriano, P. (2017). Generation of an immortalized mouse embryonic palatal mesenchyme cell line. PLoS ONE 12, e0179078. 10.1371/journal.pone.0179078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish, A., Diekhans, M., Ferreira, A.-M., Johnson, R., Jungreis, I., Loveland, J., Mudge, J. M., Sisu, C., Wright, J., Armstrong, J.et al. (2019). GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47, D766-D763. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson, L., Nilsson, I., Su, E. J., Andrae, J., Ding, H., Betsholtz, C., Eriksson, U. and Lawrence, D. A. (2012). Platelet-derived growth factor C deficiency in C57BL/6 mice leads to abnormal cerebral vascularization, loss of neuroependymal integrity, and ventricular abnormalities. Am. J. Pathol. 180, 1136-1144. 10.1016/j.ajpath.2011.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X.-D. and Ares, M. (2014). Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15, 689-701. 10.1038/nrg3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Fayos, A. C., Vázquez-Borrego, M. C., Jiménez-Vacas, J. M., Bejarano, L., Pedraza-Arévalo, S., L.-López, F., Blanco-Acevedo, C., Sánchez-Sánchez, R., Reyes, O., Ventura, S.et al. (2020). Splicing machinery dysregulation drives glioblastoma development/aggressiveness: oncogenic role of SRSF3. Brain 143, 3273-3293. 10.1093/brain/awaa273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F. and Soriano, P. (2013). A critical role for PDGFRα signaling in medial nasal process development. PLoS Genet. 9, e1003851. 10.1371/journal.pgen.1003851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heathcote, H. R., Mancini, S. J., Strembitska, A., Jamal, K., Reihill, J. A., Palmer, T. M., Gould, G. W. and Salt, I. P. (2016). Protein kinase C phosphorylates AMP-Activated protein kinase α1 Ser487. Biochem. J. 473, 4681-4697. 10.1042/BCJ20160211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper, J. E., Jones, K. L., Smith, F. J., Williams, T. and Li, H. (2020). An alternative splicing program for mouse craniofacial development. Front. Physiol. 11, 1099. 10.3389/fphys.2020.01099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, J. M. and Sanford, J. R. (2015). The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip. Rev. RNA 6, 93-110. 10.1002/wrna.1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, T., Hatayama, M., Tohmonda, T., Itohara, S., Aruga, J. and Mikoshiba, K. (2004). Mouse Zic5 deficiency results in neural tube defects and hypoplasia of cephalic neural crest derivatives. Dev. Biol. 270, 146-162. 10.1016/j.ydbio.2004.02.017 [DOI] [PubMed] [Google Scholar]
- Jumaa, H., Wei, G. and Nielsen, P. J. (1999). Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr. Biol. 9, 899-902. 10.1016/S0960-9822(99)80394-7 [DOI] [PubMed] [Google Scholar]
- Klinghoffer, R. A., Hamilton, T. G., Hoch, R. and Soriano, P. (2002). An allelic series at the PDGFαR locus indicates unequal contributions of distinct signaling pathways during development. Dev. Cell 2, 103-113. 10.1016/S1534-5807(01)00103-4 [DOI] [PubMed] [Google Scholar]
- Kuleshov, M. V., Jones, M. R., Rouillard, A. D., Fernandez, N. F., Duan, Q., Wang, Z., Koplev, S., Jenkins, S. L., Jagodnik, K. M., Lachmann, A.et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90-W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. K., Sears, M. J., Zhang, Z., Li, H., Salhab, I., Krebs, P., Xing, Y., Nah, H.-D., Williams, T. and Carstens, R. P. (2020). Cleft lip and cleft palate in Esrp1 knockout mice is associated with alterations in epithelial-mesenchymal crosstalk. Development 147, dev187369. 10.1242/dev.187369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi, D. D. and Darnell, R. B. (2010). RNA processing and its regulation: Global insights into biological networks. Nat. Rev. Genet. 11, 75-87. 10.1038/nrg2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, Y., Sou, W. H., Yung, K. W. Y., Liu, H., Wan, S. W. C., Li, Q., Zeng, C., Law, C. O. K., Chan, G. H. C., Lau, T. C. K.et al. (2019). Distinct mechanisms govern the phosphorylation of different SR protein splicing factors. J. Biol. Chem. 294, 1312-1327. 10.1074/jbc.RA118.003392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M. I., Huber, W. and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai, C. T., Isenburg, J. L., Canfield, M. A., Meyer, R. E., Correa, A., Alverson, C. J., Lupo, P. J., Riehle-Colarusso, T., Cho, S. J., Aggarwal, D.et al. (2019). National population-based estimates for major birth defects, 2010–2014. Birth Defects Res. 111, 1420-1435. 10.1002/bdr2.1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, B. D. and Cantley, L. C. (2007). AKT/PKB signaling: navigating downstream. Cell 129, 1261-1274. 10.1016/j.cell.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. and Luo, L. (2007). A global double-fluorescent cre reporter mouse. Genesis 45, 593-605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Niederreither, K., Subbarayan, V., Dollé, P. and Chambon, P. (1999). Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat. Genet. 21, 444-448. 10.1038/7788 [DOI] [PubMed] [Google Scholar]
- Pan, Q., Shai, O., Lee, L. J., Frey, B. J. and Blencowe, B. J. (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 40, 1413-1415. 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- Papangeli, I. and Scambler, P. J. (2013). Tbx1 genetically interacts with the transforming growth factor-β/bone morphogenetic protein inhibitor Smad7 during great vessel remodeling. Circ. Res. 112, 90-102. 10.1161/CIRCRESAHA.112.270223 [DOI] [PubMed] [Google Scholar]
- Parada, C., Han, D., Grimaldi, A., Sarrión, P., Park, S. S., Pelikan, R., Sanchez-Lara, P. A. and Chai, Y. (2015). Disruption of the ERK/MAPK pathway in neural crest cells as a potential cause of Pierre Robin sequence. Development 142, 3734-3745. 10.1242/dev.125328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. and Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417-419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz, I., Kosti, I., Ares, M., Cline, M. and Mandel-Gutfreund, Y. (2014). RBPmap: A web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 42, W361-W367. 10.1093/nar/gku406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phippard, D., Lu, L., Lee, D., Saunders, J. C. and Crenshaw, E. B. (1999). Targeted mutagenesis of the POU-domain gene Brn4/Pou3f4 causes developmental defects in the inner ear. J. Neurosci. 19, 5980-5989. 10.1523/JNEUROSCI.19-14-05980.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnadiwakara, M., Archer, S. K., Dent, C. I., Ruiz De Los Mozos, I., Beilharz, T. H., Knaupp, A. S., Nefzger, C. M., Polo, J. M. and Anko, M.-L. (2018). SRSF3 promotes pluripotency through nanog mRNA export and coordination of the pluripotency gene expression program. eLife 7, e37419. 10.7554/eLife.37419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattanasopha, S., Tongkobpetch, S., Srichomthong, C., Siriwan, P., Suphapeetiporn, K. and Shotelersuk, V. (2012). PDGFRa mutations in humans with isolated cleft palate. Eur. J. Hum. Genet. 20, 1058-1062. 10.1038/ejhg.2012.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell, L. L., Iulianella, A., Melton, K. R., Lynn, M., Walker, M., Inman, K. E., Bhatt, S., Leroux-Berger, M., Crawford, M., Jones, N. C.et al. (2011). A phenotype-driven ENU mutagenesis screen identifies novel alleles with functional roles in early mouse craniofacial development. Genesis 49, 342-359. 10.1002/dvg.20727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell, L. L., Kurosaka, H. and Trainor, P. A. (2012). Whole mount nuclear fluorescent imaging: Convenient documentation of embryo morphology. Genesis 50, 844-850. 10.1002/dvg.22344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti, M. M. and Swanson, M. S. (2016). RNA mis-splicing in disease. Nat. Rev. Genet. 17, 19-32. 10.1038/nrg.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, S., Jumaa, H. and Webster, N. J. G. (2013). Splicing factor SRSF3 is crucial for hepatocyte differentiation and metabolic function. Nat. Commun. 4, 1336. 10.1038/ncomms2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, S., Park, J. W., Lu, Z.-X., Lin, L., Henry, M. D., Wu, Y. N., Zhou, Q. and Xing, Y. (2014). rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 111, E5593-E5601. 10.1073/pnas.1419161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu, W., Cho, J. Y., Jiang, Y., Zhang, M., Weisz, D., Elder, G. A., Schmeidler, J., De Gasperi, R., Sosa, M. A. G., Rabidou, D.et al. (2005). Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc. Natl. Acad. Sci. USA 102, 9643-9648. 10.1073/pnas.0503739102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson, C., Love, M. I. and Robinson, M. D. (2016). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences [version 2; referees: 2 approved]. F1000Research 4, 1521. 10.12688/f1000research.7563.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano, P. (1997). The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development 124, 2691-2700. 10.1242/dev.124.14.2691 [DOI] [PubMed] [Google Scholar]
- Stamm, S. (2008). Regulation of alternative splicing by reversible protein phosphorylation. J. Biol. Chem. 283, 1223-1227. 10.1074/jbc.R700034200 [DOI] [PubMed] [Google Scholar]
- Tallquist, M. D. and Soriano, P. (2003). Cell autonomous requirement for PDGFRα in populations of cranial and cardiac neural crest cells. Development 130, 507-518. 10.1242/dev.00241 [DOI] [PubMed] [Google Scholar]
- Teng, L., Mundell, N. A., Frist, A. Y., Wang, Q. and Labosky, P. A. (2008). Requirement for Foxd3 in the maintenance of neural crest progenitors. Development 135, 1615-1624. 10.1242/dev.012179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan, H. N. and Soriano, P. (2014). SRF regulates craniofacial development through selective recruitment of MRTF Cofactors by PDGF signaling. Dev. Cell 31, 332-344. 10.1016/j.devcel.2014.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan, H. N., Mazot, P., He, F. and Soriano, P. (2015). Receptor tyrosine kinases modulate distinct transcriptional programs by differential usage of intracellular pathways. eLife 4, e07186. 10.7554/eLife.07186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitari, A. C., Deak, M., Collins, B. J., Morrice, N., Prescott, A. R., Phelan, A., Humphreys, S. and Alessi, D. R. (2004). WNK1, the kinase mutated in an inherited high-blood-pressure syndrome, is a novel PKB (protein kinase B)/Akt substrate. Biochem. J. 378, 257-268. 10.1042/bj20031692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahos, C. J., Matter, W. F., Hui, K. Y. and Brown, R. F. (1994). A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)- 8-phenyl-4H-1-benzopyran-4-one (LY294002). J. Biol. Chem. 269, 5241-5248. 10.1016/S0021-9258(17)37680-9 [DOI] [PubMed] [Google Scholar]
- Wang, E. T., Sandberg, R., Luo, S., Khrebtukova, I., Zhang, L., Mayr, C., Kingsmore, S. F., Schroth, G. P. and Burge, C. B. (2008). Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470-476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, D. G. (1998). In Situ Hybridization: A Practical Approach, 2nd Edn. Oxford University Press. [Google Scholar]
- Williams, A. L. and Bohnsack, B. L. (2019). What's retinoic acid got to do with it? Retinoic acid regulation of the neural crest in craniofacial and ocular development. Genesis 57, e23308. 10.1002/dvg.23308 [DOI] [PubMed] [Google Scholar]
- Wu, C.-C., Wu, H.-J., Wang, C.-H., Lin, C.-H., Hsu, S.-C., Chen, Y.-R., Hsiao, M., Schuyler, S. C., Lu, F. L., Ma, N.et al. (2015). Akt suppresses DLK for maintaining self-renewal of mouse embryonic stem cells. Cell Cycle 14, 1207-1217. 10.1080/15384101.2015.1014144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, W., Adhikari, S., Dahal, U., Chen, Y.-S., Hao, Y.-J., Sun, B.-F., Sun, H.-Y., Li, A., Ping, X.-L., Lai, W.-Y.et al. (2016). Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 61, 507-519. 10.1016/j.molcel.2016.01.012 [DOI] [PubMed] [Google Scholar]
- Zhang, H., Zha, X., Tan, Y., Hornbeck, P. V., Mastrangelo, A. J., Alessi, D. R., Polakiewicz, R. D. and Comb, M. J. (2002). Phosphoprotein analysis using antibodies broadly reactive against phosphorylated motifs. J. Biol. Chem. 277, 39379-39387. 10.1074/jbc.M206399200 [DOI] [PubMed] [Google Scholar]
- Zhou, Z., Qiu, J., Liu, W., Zhou, Y., Plocinik, R. M., Li, H., Hu, Q., Ghosh, G., Adams, J. A., Rosenfeld, M. G.et al. (2012). The Akt-SRPK-SR Axis Constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol. Cell 47, 422-433. 10.1016/j.molcel.2012.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.