Abstract
Vibrio natriegens is emerging as a promising host for biotechnology which is basically due to the remarkable intrinsic properties such as the exceptionally high growth and substrate consumption rates. The facultatively anaerobic marine bacterium possesses a versatile metabolism, is able to utilize a variety of substrates as carbon and energy sources and is easy to handle in the lab. These features initiated the rapid development of genetic tools and resulted in extensive engineering of production strains in the past years. Although recent examples illustrate the potential of V. natriegens for biotechnology, a comprehensive understanding of the metabolism and its regulation is still lacking but essential to exploit the full potential of this bacterium. In this review, we summarize the current knowledge on the physiological traits and the genomic organization, provide an overview of the available genetic engineering tools and recent advances in metabolic engineering of V. natriegens. Finally, we discuss the obstacles which have to be overcome in order to establish V. natriegens as industrial production host.
Keywords: genetic engineering, industrial biotechnology, metabolic engineering, physiology, synthetic biology, Vibrio natriegens
Introduction
Currently, most biotechnological production processes are carbohydrate-based (e.g. glucose or sucrose) [1] applying industrially established microbial systems such as Escherichia coli or Corynebacterium glutamicum. Although sugar-based fermentation approaches are generally state of the art, various developed processes do not reach commercialization level since the overall costs cannot compete with existing chemical approaches. The key performance indicators (KPIs) to achieve industrial relevance are closely related to the desired product. For bulk chemicals particularly high titers (approx. 100 g l−1), yields (80–90% of the theoretical maximum) and volumetric productivities (approx. 2.5 g l−1 h−1) are often considered as requirements for the economic feasibility [2–4]. Vibrio natriegens has recently been investigated as novel host for biotechnology due to its promising intrinsic properties, namely, the high growth rate (µ) and biomass-specific substrate consumption rate (qS) [5–7]. These characteristics bear the potential to outreach the mentioned KPIs.
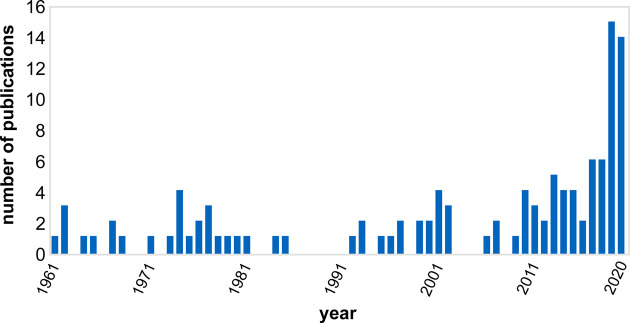
Although, the fast growing V. natriegens has already been discovered in 1958 [8], it took almost 60 years until several research groups independently recognized its potential and started to utilize this bacterium for biotechnology [5–7]. These publications served as a primer for steadily increasing research efforts until today (Figure 1), that endeavor to use V. natriegens as novel host for molecular biology, (cell-free) protein synthesis and as platform strain for small molecule production. In this review, we summarize the current knowledge on physiological properties, genomic organization, genetic engineering tools as well as metabolic engineering approaches to illustrate the status quo of developing V. natriegens into a novel production platform for biotechnology.
Figure 1. Record of PubMed-indexed publications on V. natriegens.
(Query = ‘Vibrio natriegens’ or ‘Beneckea natriegens’ or ‘Pseudomonas natriegens’; https://pubmed.ncbi.nlm.nih.gov/; accessed: 19/01/2021).
Physiological properties
V. natriegens was isolated in 1958 from salt marsh mud of Sapelo Island (Georgia, U.S.A.) [8]. The Gram-negative γ-proteobacterium was formerly classified within the genus Pseudomonas and Beneckea. Although various strains have been isolated [6,9,10], V. natriegens ATCC 14048 (DSM 759, NCBR 15636) is commercially available, the most commonly used wildtype strain so far, and referred to in the following. The facultatively anaerobic bacterium is non-pathogenic and belongs to the biosafety level 1 [7,11]. V. natriegens is rod-shaped with a single polar flagellum (Figure 2) and prototrophic, however, requires sodium ions (Na+) for cell proliferation, although it retains metabolic activity in the absence of Na+ [9,12,13]. The genome features genes for at least one Na+-extruding oxaloacetate decarboxylase (PN96_01205-10), Na+-transporting NADH:ubiquinone oxidoreductase (PN96_02115-40) and Na+-translocating ferredoxin:NAD+ oxidoreductase (RNF complex, PN96_03245-70). The fact, that all genes belonging to the latter two complexes are essential [14], indicates the relevance of Na+ for cellular energetics comparable with the relatives Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus [15]. The biomass composition, the cell volume, and weight as well as basic information about the energetics were determined and are summarized in Table 1 [16–18].
Figure 2. Transmission electron microscopy of V. natriegens with one polar flagellum per cell.
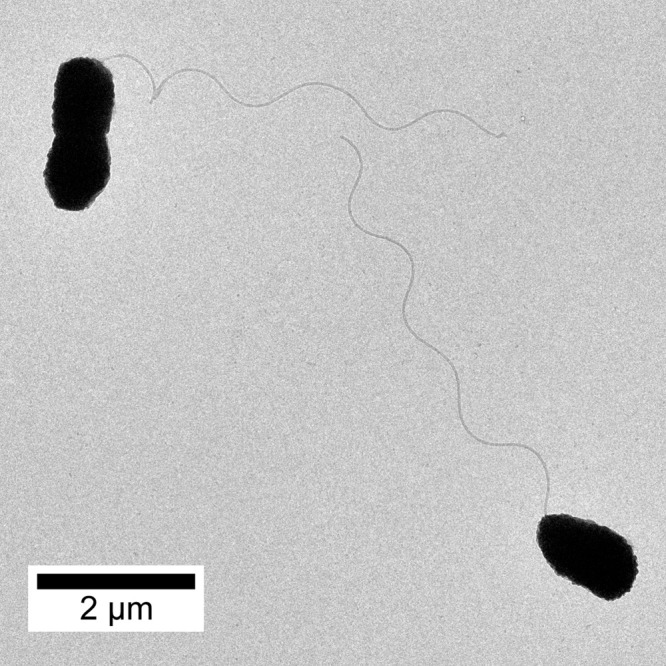
Bar at the left bottom corner represents the scale of 2 µm.
Table 1. Cell composition, cell volume, weight, and cellular energetics of V. natriegens.
| Elemental composition | C1.000H1.770O0.607N0.153P0.015S0.003 | [16] |
| Carbon content per biomass | 450 gC kgCDW−1 | [16] |
|
Cell volume Exponentially growing cells Stationary phase cells |
3.50 µm3 cell−1 0.93 µm3 cell−1 |
[17] |
|
Cell weight Exponentially growing cells Stationary phase cells |
850 fg cell−1 145 fg cell−1 |
[17] |
|
Biomass composition
Protein RNA Lipids Glycogen |
(During exponential growth) 47% 29% 7% 3% |
[18] |
| Cellular energeticsIntracellular ATP poolAdenylate energy charge (AEC) | (During exponential growth)5–8 µmol gCDW−1≥0.9 | [22] |
| Ribosomes per cell (at µ= 2.78 h−1) | 115 000 | [55] |
Providing sufficient sodium ions, V. natriegens grows quickly on standard complex media such as lysogeny broth (LB) and brain heart infusion (BHI) medium [5,6,19]. It is easy to handle in the lab, however, storage at 4°C for a prolonged time negatively impacts the viability [6]. Under optimum cultivation conditions (aerobic growth in BHI medium containing 15 g sea salts or NaCl l−1, 37°C, pH 7.5) minimum doubling times of 9.4–9.8 min (µ = 4.24–4.42 h−1) were shown, which have not been reported for any other non-pathogenic bacterium to date [5,19]. In aerobic cultivations (batch mode) with defined minimal medium containing glucose as sole carbon and energy source, V. natriegens grows at µ = 1.48–1.70 h−1 and exhibits a qS of 3.5–3.9 gGlc gCDW−1 h−1 (Table 2) which are at least two times higher compared with established microbial hosts [5,18]. Along with the high µ, V. natriegens possesses a high biomass-specific oxygen uptake rate (qO2) of 28 mmolO2 gCDW−1 h−1 and directs approx. 50% of carbon to biomass and 25% to CO2. Due to the overflow metabolism, another 25% of carbon is secreted as acetate (Table 2) [5,18] similar to E. coli. Occasional pyruvate formation in shaking flasks indicates that the high oxygen demand is not properly satisfied in this cultivation system [20,21]. In the presence of oxygen, glucose is mainly metabolized via the Embden–Meyerhof–Parnas (EMP) pathway (80–92%). Only 8–18% of the carbon flux is directed into the oxidative pentose phosphate pathway (PPP), which is at least 33% lower compared with E. coli [18,21]. Although the Entner–Doudoroff pathway is practically not operative in glucose-grown cells, it becomes the major catabolic pathway for growth on gluconate and approx. 80% of the carbon is routed through it [18,21]. Interestingly, the nicotinamide adenine dinucleotide phosphate (NADPH) gap caused by the comparably low PPP flux during growth on glucose, is apparently compensated by transhydrogenase activity (catalyzing the conversion of NADH into NADPH) which is almost three-times higher than in E. coli lysates [18]. As a consequence, 20% of the generated NADH (50% via glycolysis, 40% via citric acid cycle) is converted into NADPH by this reaction. The remaining 80% of the NADH is finally used for adenosine triphosphate (ATP) generation via the respiratory chain [18]. Exponentially growing V. natriegens cells from non-limited and nitrogen-limited batch cultivations using either glucose or succinate as sole carbon source exhibit an intracellular pool of 5–8 µmol ATP per gram of dry biomass and an adenylate energy charge of ≥0.9 (Table 1), which is in the upper range of actively growing bacteria [22].
Table 2. Key performance parameters of V. natriegens in bioreactor cultivations (batch) under aerobic and anaerobic conditions and with anaerobic resting cells using minimal medium with glucose as carbon and energy source.
| Aerobically growing cells | ||
|---|---|---|
|
Growth rate µ | h−1 |
1.48–1.70 | [5,18] |
|
Biomass yield YX/S | gCDW gGlc−1 |
0.38–0.44 | [5,18] |
|
Byproduct yield acetate YAc/Glc | molAc molGlc−1 carbon dioxide YCO2/Glc | molCO2 molGlc−1 |
0.5–0.8 1.5 |
[5,18] [18] |
|
Biomass-specific substrate uptake rate qS | gGlc gCDW−1 h−1 |
3.50–3.90 | [5,18] |
|
Biomass-specific oxygen uptake rate qO2 | mmolO2 gCDW−1 h−1 |
28 | [18] |
| Anaerobically growing cells | ||
|---|---|---|
|
Growth rate µ | h−1 |
0.92 | [5] |
|
Biomass yield YX/S | gCDW gGlc−1 |
0.12 | [5] |
|
Biomass-specific substrate uptake rate qS | gGlc gCDW−1 h−1 |
7.81 | [5] |
| Anaerobically resting cells | ||
|---|---|---|
|
Biomass-specific substrate uptake rate qS | gGlc gCDW−1 h−1 |
1.00 | [5] |
The facultatively anaerobic metabolism allows V. natriegens to grow in the absence of oxygen at a µ of 0.92 h−1 by fermenting glucose to the main products acetate, succinate, formate, lactate, and ethanol as well as minor amounts of amino acids. As observed for E. coli, the qS of V. natriegens is approx. twice as high (7.81 gGlc gCDW−1 h−1) compared with aerobic conditions (Table 2) [5,23]. Under anaerobic conditions non-growing cells remain metabolically active, show a qS of 1.0 gGlc gCDW−1 h−1 and additionally secrete considerable amounts of alanine (Table 2) [5] which is probably synthesized through reductive amination of pyruvate catalyzed by the alanine dehydrogenase. Moreover, V. natriegens is capable of reducing alternative electron acceptors, i.e. nitrate or Fe(III) citrate, under oxygen-deprived conditions by nitrate respiration [9] or extracellular electron transfer [24], respectively.
V. natriegens exhibits a remarkable nutritional flexibility with regard to the carbon source and turned out as the most versatile member of its genus (Beneckea by that time) [9]. More than 60 different carbon sources (including various of industrial relevance) can be utilized by V. natriegens as sole source of carbon and energy under aerobic conditions and at least d-gluconate, d-glucose, d-mannitol, and d-ribose support biomass generation in anaerobic cultivations [5,9,16,21]. Further studies suggest moreover, that various substrates do not support growth but are metabolized by V. natriegens as detected by respiratory activity of the cells [10,25] or reduction of alternative terminal electron acceptors like Fe(III) citrate [24]. Table 3 provides an overview of all carbohydrates and polymers, amino and carboxylic acids, alcohols and polyamines, that promote growth of V. natriegens under aerobic and anaerobic conditions or that are metabolized without supporting growth [5,9,10,16,21,24,25]. Although the genome features at least two chitinase genes and V. natriegens grows rapidly on the chitin monomers glucosamine and N-acetylglucosamine [5], it was shown to be negative in the chitinase test [9].
Table 3. Substrates utilized by V. natriegens.
| Substrates that served as sole carbon and energy sources (under aerobic conditions) [5,9,16,21] | ||
|---|---|---|
| Carbohydrates | ||
| N-Acetylglucosamine | Gluconate | Maltose |
| l-Arabinose | Glucosamine | l-Rhamnose |
| Cellobiose | d-Glucose | d-Ribose |
| d-Fructose | Glycerol | Sucrose |
| d-Galactose | d-Mannitol | Trehalose |
| Polymers/molasses | ||
| Soluble starch | Sugar beet molasses | |
| Amino acids | ||
| β-Alanine | l-Arginine | l-Ornithine |
| d-α-Alanine | l-Citrulline | l-Proline |
| l-α-Alanine | l-Glutamate | Sarcosine |
| δ-Aminovalerate | Glycine | l-Serine |
| γ-Aminobutyrate | l-Histidine | l-Threonine |
| Carboxylic acids | ||
| Acetate | d-Glucuronate | dl-Lactate |
| Aconitate | Glutarate | dl-Malate |
| Butyrate | dl-Glycerate | Malonate |
| Caprate | Heptanoate | Pelargonate |
| Caprylate | Hippurate | Propionate |
| Citrate | dl-β-Hydroxybutyrate | Pyruvate |
| Fumarate | Isobutyrate | Succinate |
| d-Gluconate | α-Ketoglutarate | Valerate |
| Aromatic acids | ||
| Benzoate | p-Hydroxybenzoate | Quinate |
| Di-/polyamines | ||
| Putrescine | Spermine | |
| Alcohols | ||
| Ethanol | Inositol | Propanol |
| Others | ||
| Betaine | Salicin | |
| Substrates that served as sole carbon and energy sources (anaerobic conditions) [9] | ||
|---|---|---|
| d-Gluconate | d-Mannitol | |
| d-Glucose | d-Ribose | |
| Substrates that served as electron donor in extracellular electron transfer (anaerobic conditions) [24] | ||
|---|---|---|
| Formate | Gluconate | Malate |
| Fructose | Glucosamine | Pyruvate |
| Galactose | Glycerol | Ribose |
| Additional substrates for that metabolic acitivity were detected (by cellular respiration); only substrates are listed, which have not been assigned to any category before [10,25] | ||
|---|---|---|
| Carbohydrates | ||
| Adenosine | Gentiobiose | Maltotriose |
| d-Arabitol | Glycogen | β-Methyl-d-glucoside |
| Arbutin | Inosine | Thymidine |
| 2′-Deoxyadenosine | l-Lyxose | Uridine |
| Polymers | ||
| γ-Cyclodextrin | Laminarin | |
| Dextrin | Pectin | |
| Amino acids | ||
| Ala-Gly | Gly-Asp | Hydroxy-l-proline |
| l-Asparagine | Gly-Glu | l-Pyroglutamic acid |
| l-Glutamine | Gly-Pro | d-Serine |
| Carboxylic acids | ||
| α-Ketobutyric acid | 5-Keto-d-gluconic acid | Oxalomalic acid |
Genomic organization
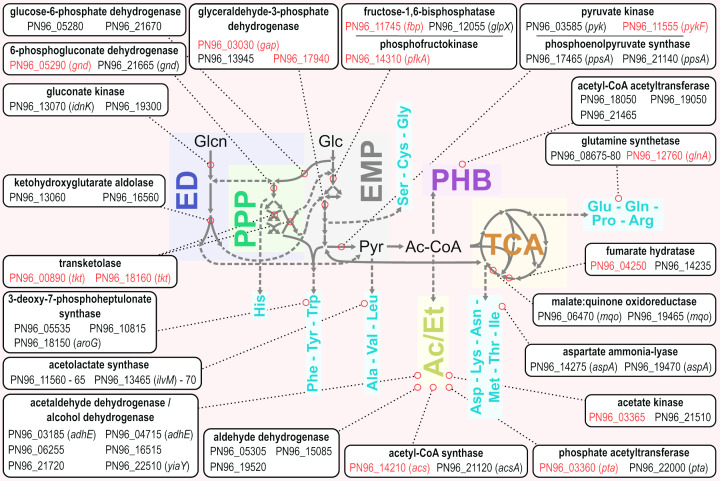
The annotated sequence of the V. natriegens WT (ATCC 14048) genome was published in 2013 in two independent studies [26,27] and re-sequenced in 2015 to obtain a closed genome map [7]. The genome of V. natriegens is distributed on two circular chromosomes of approx. 3.24 Mb (chromosome 1; chr 1) and 1.92 Mb (chromosome 2; chr 2) with a total GC content of 43.1–46.4% (Table 4) as typically found in members of the fast replicating Vibrionaceae family [11]. The native codon usage was uncovered and, thus, affords optimization of heterologous genes to enhance expression [7]. Lee et al. identified 4578 coding sequences (CDSs) on both chromosomes, determined transcriptome abundances, and developed and applied a clustered regularly interspaced short palindromic repeats interference (CRISPRi) screen to identify putative essential and growth supporting genes for proliferation in LB3 complex medium and M9 medium with glucose or sucrose [7,14]. Interestingly, 96% of these 587 genes (defined as core genes) are apparently encoded on chr 1 and many of them find a growth-neutral duplicate on chr 2. However, harboring another 16 essential genes, chr 2 is indispensable nonetheless [14]. Figure 3 illustrates the metabolic pathways of the central carbon metabolism of V. natriegens with redundantly annotated genes, that might represent prominent targets for future metabolic engineering approaches. Confusion that might arise from the many different gene and protein identifiers, resulting from various genome sequencing approaches, can be clarified by using some handy tools for bidirectional translation, that are accessible on https://github.com/citizenlee/vnat_glib [14]. For this manuscript, we stick to the identifier of the locus tags derived from the assembly deposited under Biosample SAMN03178087 (i.e. PN96_12345) to denote genes unambiguously [7]. This identifier was acquired by the KEGG database (kegg.jp) and, thus, provides a link of genomic equipment and illustrated metabolic pathways.
Table 4. Basic information about the genome of V. natriegens.
Figure 3. Schematic representation of the central carbon metabolic pathways in V. natriegens.
Locus tags of prominent genes are depicted if redundantly annotated and core genes (under the given test conditions) are highlighted in red [14]. Amino acid synthesis pathways are clustered in families and only key branching points are depicted. Amino acids are abbreviated with the universal three-letter code. Abbreviations: Ac, acetate; Ac-CoA, acetyl-coenzyme A; ED, Entner–Doudoroff pathway; Et, ethanol; Glc, glucose; Glcn, gluconate; PHB, polyhydroxybutyrate biosynthesis pathway; Pyr, pyruvate; TCA, tricaboxylic acid cycle.
Genetic engineering tools
Since 2016, when Weinstock et al. and Lee et al. made V. natriegens accessible for genetic manipulation [6,7] and proposed to utilize this fast-growing bacterium as novel host for routine applications in molecular biology, a wealth of tools to manipulate its metabolism were established within less than 5 years.
Plasmid DNA can be introduced into V. natriegens by heat-shock transformation, electroporation, and conjugation [5–7,28]. V. natriegens is also capable of taking up naked linear DNA fragments from its environment, when the endogenous tfoX gene (PN96_07370) or the homolog from V. cholerae encoding an activator of natural competence is expressed [29]. The suitability of this approach to introduce chromosomal mutations by homologous recombination in parallel (multiplexed genome editing by natural transformation – MuGENT) was demonstrated by transforming tfoX expressing cells with a number of different PCR fragments containing sufficiently long homologous arms (best with 3 kilobase (kb) size).
Commercially available standard plasmids can stably replicate in V. natriegens as many origins of replication are recognized, that function in E. coli (e.g. RSF1010, p15A, pMB1, RK2, ColE1, pBR322, pBBR1, pUC, pES213, pSC101, pRO1600) [6,7,28]. Differences were noted with regard to their transformability and stability in non-selective media, as derivatives of the ColE1/pBR322 origin of replication (ori) were only maintained in 60% of the cells after 24 h [28]. Plasmids containing the R6K ori, which requires the initiation protein π (encoded by the pir gene), cannot replicate and, consequently, were applied in suicide vector systems to introduce chromosomal modifications in V. natriegens [5–7,20]. A number of commonly used antibiotics (ampicillin/carbenicillin, chloramphenicol, gentamicin, kanamycin, rifampicin, spectinomycin, streptomycin, tetracycline) have been tested for positive selection and the range of working concentrations was reviewed recently [11]. Levansucrase encoded by the sacB gene from Bacillus subtilis as well as the inducible toxin-based ccdB system were successfully employed for counter-selection [5,6,20]. In addition, deletion of the dns locus (PN96_00865) was demonstrated by recombination with the Cre-loxP system [6]. The disruption of the same gene locus by inserting the spectinomycin resistance gene was also achieved through a recombineering approach based on the SXT (λ-Red homologous) system originating from V. cholera [30]. Genome-wide screening systems to address gene function were established by generating a genome-wide transposon library as well as by the construction of a CRISPRi system based on the nuclease-deficient variant dCas9 [7,14].
To drive adjusted gene expression in V. natriegens, synthetic promoter libraries have been constructed and characterized [28,31]. Permutation of promoter elements (−35, −10, and UP element), spacer sequences, ribosomal-binding sites, and terminator sequences yielded a library which varies up to five orders of magnitude in gfp expression. Also various constitutive promoters of the Anderson library have been proven to function in V. natriegens and covered an expression range over two to three orders of magnitude [28]. However, strongest expression was obtained from the endogenous rrnA P1 promoters of V. natriegens [28]. Tunable gene expression was achieved by the use of promoter systems, that respond to chemical induction (e.g. with isopropyl β-d-1-thiogalactopyranoside (IPTG), arabinose, anhydrotetracycline) or by physical stimuli (light and temperature) as outlined in Table 5 [6,7,28,29]. Protein levels were reduced by the factor 2–5 by the fusion of N-terminal ssrA degradation tags, that might have directed proteins to the endogenous Clp protease of V. natriegens [28].
Table 5. Overview of inducible promoters suitable for V. natriegens.
Metabolic engineering approaches
By exploiting the above-described genetic engineering tools a number of production and platform strains with improved properties have been generated successfully. Genome reduction of V. natriegens by the sequential deletion of 194 kb (3.5% reduction in the genome size), integration of the inducible T7-RNA polymerase [6] and potentially further modifications resulted in the commercially available Vmax™ express (Synthetic Genomics Inc., La Jolla, CA, U.S.A.) with improved properties for protein expression. Pfeifer et al. (2019) showed that the genome of V. natriegens harbors two prophage regions (VNP1 [PN96_04290–04520] and VNP2 [PN96_06880 – 07090]) which are induced under stress but also under standard conditions [20]. Consequently, the authors constructed V. natriegens ∆vnp1-2 which represents a genetically more stable platform for future metabolic engineering studies (Pfeifer et al. (2019) [20]).
Metabolic engineering of the native metabolism was demonstrated by re-direction of carbon flux towards the intracellular storage compound polyhydroxybutyrate (PHB) which is an interesting precursor of bioplastics [29]. Within less than one week the authors applied the combinatorial MuGENT approach to construct a strain which produced about 100 times more PHB than the WT strain. The substitution of the native promoter of the PHB biosynthetic genes phaBAC with the strong tac promoter (Ptac) had, apparently, the strongest impact on PHB formation. The titer could be further increased to 4% PHB per cell dry weight (CDW) by the inactivation of the pta and gltA genes encoding phosphate acetyltransferase and citrate synthase, respectively [29]. Considering that V. natriegens and close relatives naturally accumulate PHB under growth limiting conditions [9,32,33], it might be interesting to validate the performance of the engineered PHB-producing strain under optimized process conditions. For the anaerobic alanine production, the lldh and dldh genes (PN96_16800 and PN96_16785) encoding l- and d-lactate dehydrogenase, the pfl gene (PN96_08455) coding for the pyruvate formate lyase and the malate dehydrogenase encoding gene mdh (PN96_11695) were deleted, which avoided/reduced by-product formation and improved pyruvate availability. In an anaerobic resting cell approach, the resulting strain V. natriegens ∆lldh ∆dldh ∆pfl ∆mdh produced about 17 g alanine l−1 with a yield of 0.73 g alanine g−1 glucose within 30 min, corresponding to a volumetric productivity of 34 g alanine l−1 h−1, which outcompetes previously reported values for engineered E. coli and C. glutamicum strains by the factor of 9 and 13, respectively [5]. This work demonstrated that the high metabolic activity of V. natriegens can be translated into high space time yields. Recently, Erian et al. (2020) engineered V. natriegens for 2,3-butanediol (2,3-BDO) production by introduction of a pUC-derived plasmid harboring the 2,3-BDO biosynthesis genes budBACfrom Enterobacter cloacae subsp. dissolvens. Within 12 h a titer of 36 g of 2,3-BDO l−1 was generated from glucose in microaerobic fed-batch cultivations supplemented with yeast extract. Fine-tuning the oxygen availability in chemostat cultivations increased the combined diol yield (2,3-BDO and acetoin) to 70% of the maximum theoretical value [16]. Remarkably, this was achieved without further engineering of competing pathways and without adapting the codon-usage of the plasmid-encoded genes for expression in V. natriegens. Recently, Lim et al. (2019) isolated Vibrio sp. dhg which is a close relative of V. natriegens ATCC 14048 but in contrast to the latter is able to utilize the macroalgae constituent alginate as substrate. The authors sequenced the genome and established a genetic engineering toolbox which has been applied to construct Vibrio sp. dhg strains for the production of 2,3-BDO, ethanol, and lycopene from macroalgae sugars [34].
Overproduction of the natural compound β-carotene was achieved in V. natriegens by expression of the heterologous mevalonate pathway from Lactobacillus acidophilus and four genes from Vibrio campbellii that encode β-carotene forming enzymes [25]. In the same study, violacein was produced by expression of five heterolous genes from Chromobacterium violaceum. Highest titers of both substances (3 mg β-carotene l−1 and 13 mg violacein l−1), that are particularly challenging with regard to precursor and redox cofactor requirements, were obtained in test tubes containing complex media. But only minor amounts (<1 mg β-carotene L−1 and <4 mg violacein l−1) could be generated from various substrates in minimal medium [25]. When the tyrosinase gene tyr1 from Bacillus megaterium was overexpressed in V. natriegens, melanin production from l-tyrosine proceeded at least three times faster than in previously reported systems [35]. Since V. natriegens has an exceptionally high tolerance towards selenium, Fernandez-Llamosa et al. (2017) utilized this bacterium for efficient production of selenium nanoparticles.
In addition to this, various studies have proven V. natriegens as a suitable producer of different proteins. Examples include several reporter constructs (e.g. GFP, sfGFP, mCherry, mSCFP3, and LacZ, worked nicely, whereas functionality of RFP, YFP, and eBFP2 was poor) as well as proteins, that are largely insoluble, typically yielded at low concentrations in E. coli, isotopically labeled proteins or even multimeric membrane protein complexes [6,7,20,28,31,36–38].
Reflecting the high growth rate, V. natriegens possesses a well equipped translational maschinery with 11-12 rRNA operons, up to 129 genes for tRNAs and up to 115,000 ribosomes per cell (Table 1 and 4). In order to utilize the potentially more active extracts of V. natriegens for cell-free protein synthesis (CFPS), several groups optimized the procedure for extract preparation and the reaction conditions with plasmid DNA as template [39–41]. Recently, Zhu et. al (2020) established CFPS from linear expression templates. Addition of the double-stranded DNA binding protein Cro protected the linear DNA in the CFPS against degradation and improved sfGFP yields 18-fold with V. natriegens extracts [42].
Conclusion and outlook
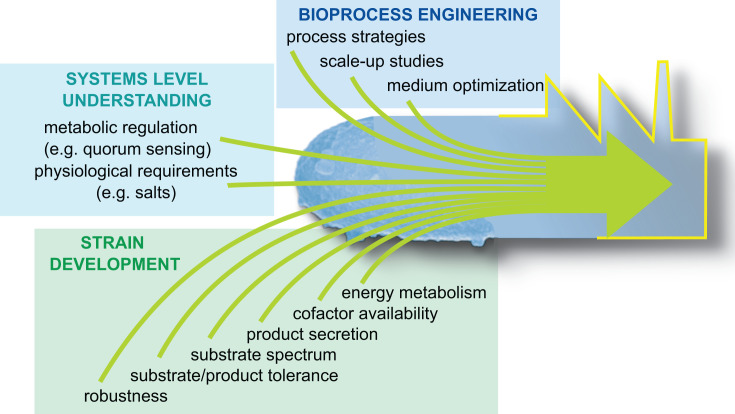
The exceptionally fast cell proliferation is a remarkable property of V. natriegens which represents a promising basis not only to understand the molecular fundamentals of rapid microbial growth but also to spur biotechnology. Constantly repeating workflows of molecular biology and design-built-test-learn cycles of synthetic biology [7,43] might directly benefit from the application of V. natriegens. As proven by first metabolic engineering studies, the high substrate consumption rates of V. natriegens encourage the development of superior production processes for industrial biotechnology. In the following, we discuss some aspects, that need to be be addressed in future studies in order to fully exploit the potential of this bacterium (Figure 4).
Figure 4. Anticipated steps in systems metabolic engineering to transform V. natriegens into a biotechnological platform strain.
Recent systems level analysis of the metabolism provided first insights into the genome, transcriptome, proteome, and metabolome of V. natriegens [6,7,14,18]. However, a comprehensive understanding of the metabolism and its regulation is lacking and predictions rely mainly on the knowledge of better studied relatives such as Vibrio fisheri or Vibrio cholera. The marine habitat represents a special environment with regard to salinity which apparently impacts the transport and energy metabolism of the sodium dependent V. natriegens [44]. Moreover, the genome compromises several elements of a complex quorum sensing apparatus as found in other Vibrionaceae such as V. harveyi (Waters und Bassler, 2005). Thus, a holistic understanding of the metabolic repertoire and underlying regulatory mechanisms is indispensable to realize sophisticated (systems) metabolic engineering strategies with V. natriegens.
Through strain development carbon fluxes of V. natriegens were successfully re-directed towards small molecule production using mainly glucose as substrate. Since the future bioeconomy relies on the circular use of biogenic resources and waste streams such as lignocellulosic hydrolysates, biorefinery side streams and non-sugar carbon sources such as organic acids [1,45–47], the (parallel) consumption of such substrates by V. natriegens, potentially impacted by catabolite repression, has to be investigated and might be optimized by substrate utilization engineering [48]. For such applications the tolerance of V. natriegens towards alternative carbon sources, (lignocellulosic) inhibitors and the envisioned products has to be analyzed and might be increased to levels of industrial relevance by methods such as adaptive laboratory evolution [49]. Both approaches might benefit from the short generation time of V. natriegens. As stated above, glucose-grown V. natriegens exhibits a comparably low carbon flux in the PPP but provides high transhydrogenase activity [18] which is of particular interest with regard to optimizing the cofactor availability (cofactor engineering) in future metabolic engineering studies. ATP-demanding products might require sophisticated energy engineering approaches to tightly adjust the energy state of the cell for efficient production [50]. We anticipate that such an approach might face the intricate entanglement with sodium-dependent energetics. In general, to ensure efficient metabolic engineering procedures tailoring V. natriegens for the production of natural and non-natural products, in silico metabolic modeling approaches have to be developed (Figure 4).
Eventually, successfully developed lab-scale processes have to be transferred to industrial scale. Thus, scale up studies need to provide a comprehensive understanding of the metabolism and its regulation under fluctuating process conditions such as substrate, oxygen and CO2/HCO3− gradients as typically found in large-scale bioreactors (Figure 3; [51–53]). To satisfy the demand of V. natriegens for sodium ions, most cultivation media are supplemented with at least 15 g NaCl l−1. Since chloride promotes corrosion processes on metal surfaces of a bioreactor, Hoffart et al. (2017) designed a defined medium containing only traces of this halogen but sufficient sodium ions to maintain the high growth rate of V. natriegens [5]. However, a significant reduction of the overall salt load would be desirable for future bioprocesses with this bacterium. The high growth rate of V. natriegens comes along with a high oxygen uptake rate which can quickly exceed the mass transfer capacity of typical large-scale bioreactors. As a consequence, unlimited aerobic growth will hardly be applicable and sophisticated bioprocesses have to be developed to utilize the full potential of this bacterium. Noteworthy, V. natriegens shows also under anaerobic conditions exceptionally high substrate consumption rates, which might be utilized in zero-growth processes scenarios [54] in the large-scale environment.
Summary
V. natriegens possesses a versatile metabolism and promising intrinsic properties for biotechnolocical processes.
A genetic engineering toolbox to manipulate the metabolism of V. natriegens is readily available and constantly expanded.
Production strains performing with high yield and productivity have already been engineered which emphasizes the high potential of V. natriegens to become a novel host for biotechnology.
Fundamental systems level knowledge about the metabolism and its regulation is essentially lacking but required for future sophisticated metabolic engineering strategies.
To utilize the full potential in the industrial environment, intensive biochemical engineering with V. natriegens is indispensable.
Acknowledgements
We thank Sebastian Grenz (University of Stuttgart, Institute of Biochemical Engineering) and Michael Schweikert (University of Stuttgart, Institute of Biomaterials and Biomolecular Systems) for support in preparing the transmission electron micrographs of V. natriegens.
Abbreviations
- 2,3-BDO
2,3-butanediol
- ATP
adenosine triphosphate
- BHI
brain heart infusion
- CDW
cell dry weight
- CFPS
cell-free protein synthesis
- chr 1/2
chromosome 1 (larger)/2 (smaller)
- CRISPRi
clustered regularly interspaced short palindromic repeats (interference)
- Fe(III)
ferric iron (Fe3+)
- gC
gram of carbon
- Glc
glucose
- kb
kilobase
- KPI
key performance indicator
- MuGENT
multiplexed genome editing by natural transformation
- Na+
sodium ion
- NAD+/NADH
nicotinamide adenine dinucleotide (oxidized/reduced)
- NADPH
nicotinamide adenine dinucleotide phosphate
- ori
origin of replication
- PHB
polyhydroxybutyrate
- PPP
pentose phosphate pathway
- qO2
oxygen consumption rate in mmolO2 gCDW h−1
- qS
biomass-specific substrate consumption rate in gsubstrate gCDW h−1
- WT
wildtype (i.e. ATCC 14048)
- µ
growth rate in h−1
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft [grant number BL1408/2-1].
Author Contribution
F.T. and B.B. conceived and wrote the manuscript.
References
- 1.Kiefer D., Merkel M., Lilge L., Henkel M. and Hausmann R. (2020) From acetate to bio-based products: underexploited potential for industrial biotechnology. Trends Biotechnol. 39, 397–411 10.1016/j.tibtech.2020.09.004 [DOI] [PubMed] [Google Scholar]
- 2.Upadhyaya B.P., DeVeaux L.C. and Christopher L.P. (2014) Metabolic engineering as a tool for enhanced lactic acid production. Trends Biotechnol. 32, 637–644 10.1016/j.tibtech.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 3.Werpy T. and Petersen G. (2004) Top value added chemicals from biomass: volume I – results of screening for potential candidates from sugars and synthesis gas. U.S. NREL. Golden, CO (United States), 10.2172/15008859 [DOI] [Google Scholar]
- 4.Dien B.S., Cotta M.A. and Jeffries T.W. (2003) Bacteria engineered for fuel ethanol production: current status. Appl. Microbiol. Biotechnol. 63, 258–266 10.1007/s00253-003-1444-y [DOI] [PubMed] [Google Scholar]
- 5.Hoffart E., Grenz S., Lange J., Nitschel R., Müller F., Schwentner A.et al. (2017) High substrate uptake rates empower Vibrio natriegens as production host for industrial biotechnology. Appl. Environ. Microbiol. 83, 1–10 10.1128/AEM.01614-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinstock M.T., Hesek E.D., Wilson C.M. and Gibson D.G. (2016) Vibrio natriegens as a fast-growing host for molecular biology. Nat. Methods 13, 849–851 10.1038/nmeth.3970 [DOI] [PubMed] [Google Scholar]
- 7.Lee H.H., Ostrov N., Wong B.G., Gold M.A., Khalil A.S. and Church G.M. (2016) Vibrio natriegens, a new genomic powerhouse. bioRxiv 10.1101/058487 [DOI] [Google Scholar]
- 8.Payne W.J. (1958) Studies on bacterial utilization of uronic acids III. Induction of oxidative enzymes in a marine isolate. J. Bacteriol. 76, 301–307 10.1128/JB.76.3.301-307.1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baumann P., Baumann L., Mandel M. and Allen R.D. (1971) Taxonomy of marine bacteria: the genus Beneckea. J. Bacteriol 108, 1380–1383 10.1128/JB.107.1.268-294.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng Y., Han X., Xu P. and Tao F. (2020) Next-generation microbial workhorses: comparative genomic analysis of fast-growing Vibrio strains reveals their biotechnological potential. Biotechnol. J. 15, 1–8 10.1002/biot.201900499 [DOI] [PubMed] [Google Scholar]
- 11.Hoff J., Daniel B., Stukenberg D., Thuronyi B.W., Waldminghaus T. and Fritz G. (2020) Vibrio natriegens: an ultrafast‐growing marine bacterium as emerging synthetic biology chassis. Environ. Microbiol. 22, 4394–4408 10.1111/1462-2920.15128 [DOI] [PubMed] [Google Scholar]
- 12.Payne W.J. (1960) Effects of sodium and potassium ions on growth and substrate penetration of a marine pseudomonad. J. Bacteriol. 80, 696–700 10.1128/JB.80.5.696-700.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Webb C.D. and Payne W.J. (1971) Influence of Na+ on synthesis of macromolecules by a marine bacterium. Appl. Microbiol. 21, 1080–1088 10.1128/AM.21.6.1080-1088.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee H.H., Ostrov N., Wong B.G., Gold M.A., Khalil A.S. and Church G.M. (2019) Functional genomics of the rapidly replicating bacterium Vibrio natriegens by CRISPRi. Nat. Microbiol. 4, 1105–1113 10.1038/s41564-019-0423-8 [DOI] [PubMed] [Google Scholar]
- 15.Mulkidjanian A.Y., Dibrov P. and Galperin M.Y. (2008) The past and present of sodium energetics: May the sodium-motive force be with you. Biochim. Biophys. Acta Bioenerg. 1777, 985–992 10.1016/j.bbabio.2008.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erian A.M., Freitag P., Gibisch M. and Stefan P. (2020) High rate 2,3-butanediol production with Vibrio natriegens. Bioresour. Technol. Rep. 10, 100408 10.1016/j.biteb.2020.100408 [DOI] [Google Scholar]
- 17.Fagerbakke K., Heldal M. and Norland S. (1996) Content of carbon, nitrogen, oxygen, sulfur and phosphorus in native aquatic and cultured bacteria. Aquat. Microb. Ecol. 10, 15–27 10.3354/ame010015 [DOI] [Google Scholar]
- 18.Long C.P., Gonzalez J.E., Cipolla R.M. and Antoniewicz M.R. (2017) Metabolism of the fast-growing bacetrium Vibrio natriegens elucidated by 13C metabolic flux analysis. Metab. Eng. 44, 191–197 10.1016/j.ymben.2017.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eagon R.G. (1962) Pseudomonas natriegens, a marine bacterium with a generation time of less than 10 minutes. J. Bacteriol. 83, 736–737 10.1128/JB.83.4.736-737.1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeifer E., Michniewski S., Gätgens C., Müller F., Polen T., Millard A.et al. (2019) Generation of a prophage-free variant of the fast-growing bacterium Vibrio natriegens. Appl. Environ. Microbiol. 85, 1–17 10.1128/AEM.00853-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eagon R.G. and Wang C.H. (1962) Dissimilation of glucose and gluconic acid by Pseudomonas natriegens. J. Bacteriol. 83, 879–886 10.1128/JB.83.4.879-886.1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niven D.F., Collins P.A. and Knowles C.J. (1977) Adenylate energy charge during batch culture of Beneckea natriegens. J. Gen. Microbiol. 98, 95–108 10.1099/00221287-98-1-95 [DOI] [PubMed] [Google Scholar]
- 23.Varma A. and Palsson B.O. (1994) Metabolic flux balancing: basic concepts, scientific and practical use. Nat. Biotechnol. 12, 994–998 10.1038/nbt1094-994 [DOI] [Google Scholar]
- 24.Conley B.E., Weinstock M.T., Bond D.R. and Gralnick J.A. (2020) A hybrid extracellular electron transfer pathway enhances the survival of Vibrio natriegens. Appl. Environ. Microbiol. 86, 1–18 10.1128/AEM.01253-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis G.A., Tschirhart T., Spangler J., Walper S.A., Medintz I.L. and Vora G.J. (2019) Exploiting the feedstock flexibility of the emergent synthetic biology chassis Vibrio natriegens for engineered natural product production. Mar. Drugs 17, 1–21 10.3390/md17120679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maida I., Bosi E., Perrin E., Papaleo M.C., Orlandini V., Fondi M.et al. (2013) Draft genome sequence of the fast-growing bacterium Vibrio natriegens strain DSMZ 759. Genome Announc. 1, e00648–e00713 10.1128/genomeA.00648-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z., Lin B., Hervey W.J. and Vora G.J. (2013) Draft genome sequence of the fast-growing marine bacterium Vibrio natriegens strain ATCC 14048. Genome Announc. 1, e00589–e00613 10.1128/genomeA.00589-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tschirhart T., Shukla V., Kelly E.E., Schultzhaus Z., Newringeisen E., Erickson J.S.et al. (2019) Synthetic biology tools for the fast-growing marine bacterium Vibrio natriegens. ACS Synth. Biol. 8, 2069–2079 10.1021/acssynbio.9b00176 [DOI] [PubMed] [Google Scholar]
- 29.Dalia T.N., Hayes C.A., Stolyar S., Marx C.J., McKinlay J.B. and Dalia A.B. (2017) Multiplex genome editing by natural transformation (MuGENT) for synthetic biology in Vibrio natriegens. ACS Synth. Biol. 6, 1650–1655 10.1021/acssynbio.7b00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee H.H., Ostrov N., Gold M.A. and Church G.M. (2017) Recombineering in Vibrio natriegens. bioRxiv 10.1101/130088 [DOI] [Google Scholar]
- 31.Wu F., Chen W., Peng Y., Tu R., Lin Y., Xing J.et al. (2020) Design and reconstruction of regulatory parts for fast-frowing Vibrio natriegens synthetic biology. ACS Synth. Biol. 9, 2399–2409 10.1021/acssynbio.0c00158 [DOI] [PubMed] [Google Scholar]
- 32.Cho H.W. and Eagon R.G. (1967) Factors affecting the pathways of glucose catabolism and the tricarboxylic acid cycle in Pseudomonas natriegens. J. Bacteriol. 93, 866–873 10.1128/JB.93.3.866-873.1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chien C.C., Chen C.C., Choi M.H., Kung S.S. and Wei Y.H. (2007) Production of poly-β-hydroxybutyrate (PHB) by Vibrio spp. isolated from marine environment. J. Biotechnol. 132, 259–263 10.1016/j.jbiotec.2007.03.002 [DOI] [PubMed] [Google Scholar]
- 34.Lim H.G., Kwak D.H., Park S., Woo S., Yang J.S., Kang C.W.et al. (2019) Vibrio sp. dhg as a platform for the biorefinery of brown macroalgae. Nat. Commun. 10, 2486 10.1038/s41467-019-10371-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z., Tschirhart T., Schultzhaus Z., Kelly E.E., Chen A., Oh E.et al. (2020) Melanin produced by the fast-growing marine bacterium Vibrio natriegens through heterologous biosynthesis: Characterization and application. Appl. Environ. Microbiol. 86, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eichmann J., Oberpaul M., Weidner T., Gerlach D. and Czermak P. (2019) Selection of high producers from combinatorial libraries for the production of recombinant proteins in Escherichia coli and Vibrio natriegens. Front. Bioeng. Biotechnol. 7, 254 10.3389/fbioe.2019.00254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schleicher L., Muras V., Claussen B., Pfannstiel J., Blombach B., Dibrov P.et al. (2018) Vibrio natriegens as host for expression of multisubunit membrane protein complexes. Front. Microbiol. 9, 2537 10.3389/fmicb.2018.02537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becker W., Wimberger F. and Zangger K. (2019) Vibrio natriegens: an alternative expression system for the high-yield production of isotopically labeled proteins. Biochemistry 58, 2799–2803 10.1021/acs.biochem.9b00403 [DOI] [PubMed] [Google Scholar]
- 39.Failmezger J., Scholz S., Blombach B. and Siemann-Herzberg M. (2018) Cell-free protein synthesis from fast-growing Vibrio natriegens. Front. Microbiol. 9, 1146 10.3389/fmicb.2018.01146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiegand D.J., Lee H.H., Ostrov N. and Church G.M. (2018) Establishing a cell-free Vibrio natriegens expression system. ACS Synth. Biol. 7, 2475–2479 10.1021/acssynbio.8b00222 [DOI] [PubMed] [Google Scholar]
- 41.Des Soye B.J., Davidson S.R., Weinstock M.T., Gibson D.G. and Jewett M.C. (2018) Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens. ACS Synth. Biol. 7, 2245–2255 10.1021/acssynbio.8b00252 [DOI] [PubMed] [Google Scholar]
- 42.Zhu F., Gan R., Cabezas M.D., Kojima T., Nicol R., Jewett M.et al. (2020) Increasing cell-free gene expression yields from linear templates in Escherichia coli and Vibrio natriegens extracts by using DNA-binding proteins. Biotechnol. Bioeng. 117, 3849–3857 10.1002/bit.27538 [DOI] [PubMed] [Google Scholar]
- 43.Hillson N., Caddick M., Cai Y., Carrasco J.A., Chang M.W., Curach N.C.et al. (2019) Building a global alliance of biofoundries. Nat. Commun. 10, 1038–1041 10.1038/s41467-019-10079-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khanna G., DeVoe L., Brown L., Niven D.F. and MacLeod R.A. (1984) Relationship between ion requirements for respiration and membrane transport in a marine bacterium. J. Bacteriol. 157, 59–63 10.1128/JB.157.1.59-63.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kutscha R. and Pflügl S. (2020) Microbial upgrading of acetate into value-added products—examining microbial diversity, bioenergetic constraints and metabolic engineering approaches. Int. J. Mol. Sci. 21, 1–30 10.3390/ijms21228777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lange J., Müller F., Bernecker K., Dahmen N., Takors R. and Blombach B. (2017) Valorization of pyrolysis water: a biorefinery side stream, for 1,2-propanediol production with engineered Corynebacterium glutamicum. Biotechnol. Biofuels 10, 277 10.1186/s13068-017-0969-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lange J., Müller F., Takors R. and Blombach B. (2018) Harnessing novel chromosomal integration loci to utilize an organosolv-derived hemicellulose fraction for isobutanol production with engineered Corynebacterium glutamicum. Microb. Biotechnol. 11, 257–263 10.1111/1751-7915.12879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee J.W., Na D., Park J.M., Lee J., Choi S. and Lee S.Y. (2012) Systems metabolic engineering of microorganisms for natural and non-natural chemicals. Nat. Chem. Biol. 8, 536–546 10.1038/nchembio.970 [DOI] [PubMed] [Google Scholar]
- 49.Portnoy V.A., Bezdan D. and Zengler K. (2011) Adaptive laboratory evolution-harnessing the power of biology for metabolic engineering. Curr. Opin. Biotechnol. 22, 590–594 10.1016/j.copbio.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 50.Schwentner A., Feith A., Münch E., Stiefelmaier J., Lauer I., Favilli L.et al. (2019) Modular systems metabolic engineering enables balancing of relevant pathways for l-histidine production with Corynebacterium glutamicum. Biotechnol. Biofuels 12, 1–21 10.1186/s13068-019-1410-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blombach B. and Takors R. (2015) CO2 - Intrinsic product, essential substrate, and regulatory trigger of microbial and mammalian production processes. Front. Bioeng. Biotechnol. 3, 108 10.3389/fbioe.2015.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuschel M. and Takors R. (2020) Simulated oxygen and glucose gradients as a prerequisite for predicting industrial scale performance a priori. Biotechnol. Bioeng. 117, 2760–2770 10.1002/bit.27457 [DOI] [PubMed] [Google Scholar]
- 53.Lara A.R., Galindo E., Ramírez O.T. and Palomares L.A. (2006) Living with heterogeneities in bioreactors: Understanding the effects of environmental gradients on cells. Mol. Biotechnol. 34, 355–381 10.1385/MB:34:3:355 [DOI] [PubMed] [Google Scholar]
- 54.Lange J., Takors R. and Blombach B. (2016) Zero-growth bioprocesses - a challenge for microbial production strains and bioprocess engineering. Eng. Life Sci. 17, 27–35 10.1002/elsc.201600108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aiyar S.E., Gaal T. and Gourse R.L. (2002) rRNA promoter activity in the fast-growing bacterium Vibrio natriegens. J. Bacteriol. 184, 1349–1358 10.1128/JB.184.5.1349-1358.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]