Abstract
Allergies to food and environmental antigens have steeply grown to epidemic proportions. IgE antibodies are key mediators of allergic disease, including life-threatening anaphylaxis. There is now compelling evidence that one of the hallmarks of anaphylaxis-inducing IgE molecules is their high affinity for allergen, and the cellular pathway to high-affinity IgE is typically through sequential switching of IgG B cells. Further, in contrast to the previously held paradigm that a subset of CD4+ T cells called Th2 cells promotes IgE responses, recent studies suggest that T follicular helper cells are crucial for inducing anaphylactic IgE. Here we discuss recent studies that have enabled us to understand the nature, induction, and regulation of this enigmatic antibody isotype in allergic sensitization.
Keywords: Allergy, Anaphylaxis, IgE, Tfh cells, Th2 cells
1. INTRODUCTION
The humoral arm of the adaptive immune response is mediated through antibodies secreted by B lymphocytes. Antibodies (also called immunoglobulins) can be classified into distinct classes, or isotypes, based on the differences in their heavy chain constant regions, allowing them to perform distinct biologic functions ranging from beneficial, such as neutralizing pathogens and maintaining gut homeostasis, to harmful, such as involvement in autoimmune disorders and allergies. IgE remains one of the most enigmatic of all antibody classes. Whereas IgE confers protection against toxins in venom and in certain models of helminth infections, it is instead pathogenic in allergy and asthma.1–4 Despite being one of the least abundant antibody isotypes, IgE can have devastating consequences when it triggers anaphylaxis, a severe, life-threatening allergic reaction.5
One of the key features of the adaptive humoral immune response is clonal selection of B cells, a process typically dependent on CD4 T cell help, which leads to the generation of high-affinity antibodies.6 Based on the cytokines they provide, T cells also influence the isotype of the immunoglobulin (IgG subtypes, IgE, or IgA) that is made during an immune response.7 It has long been known that antigen-specific IgE induction is also reliant on CD4T cell help.8 The specific CD4T cell subset that regulates IgE production in allergic and helminth responses has been an area of active investigation. A subset of CD4 T cells called Th2 cells were initially thought to be the key cells that help B cells make antibodies including IgE, partly because they made the cytokine IL-4, which is a B cell growth factor.9 However, the discovery of a dedicated B cell helper cell subset, known as T follicular helper cells (Tfh cells), changed the paradigm of Th2-dependent antibody responses.9,10 Tfh cells help promote not only the generation of high-affinity antibodies but also influence the class of antibody that the B cells make. In this review, we discuss recent research highlighting the importance of high-affinity IgE in allergic disease and the nature of the Tfh cells that promote anaphylactic IgE to allergens.
2. HIGH-AFFINITY IGE IN ALLERGIES AND ANAPHYLAXIS
Allergen binding to IgE molecules on mast cells leads to cross-linking of the FcεRI, resulting in mast cell degranulation and, in the most severe cases, systemic anaphylaxis. To better understand the instigators and mechanisms of anaphylaxis, the field has been using rodent models. In mice, there are two distinct pathways of anaphylaxis. One is typically called “classical,” which is the IgE-mediated pathway. The other is the IgG-mediated “alternative” pathway. The IgE-mediated pathway involves mast cell degranulation and release of mediators such as histamine, chymase, and mast cell proteases including MMCP-1.11,12 The IgG-mediated pathway involves macrophages and platelet-activating factor. Symptoms such as temperature drop and increased hematocrit are common to both pathways, but certain markers of mast cell degranulation including the presence of particular proteases in the blood are specific to the mast cell/IgE-dependent pathway.11–13 The IgE-mediated pathway requires almost 100-fold less antigen compared to IgG-mediated pathways.11 Whether the IgG-mediated alternative pathway occurs in human anaphylaxis is currently unclear.14 Further, as anaphylaxis can be induced by small allergen doses in humans, focus has centered on IgE- and mast cell-mediated mechanisms of anaphylaxis. One key determinant of IgE-mediated clinical symptoms is the affinity of the IgE antibody for a given allergen.8,15,16 Affinity refers to the strength with which a single region of an antibody binds to an antigen. Affinity differs from avidity, which is the collective strength arising from multiple binding interactions.
2.1. High-affinity but not low-affinity IgE causes anaphylaxis
Various methods are used to evaluate the affinity of IgE to an allergen; these include competitive inhibition assays, surface plasmon resonance, and sequencing of BCRs for somatic hypermutations. Irrespective of the method used and the type of allergies, clinical data as well as experimental evidence from mouse models indicate that IgE to allergens are typically high-affinity.8,17–21 Mita et al. studied lowand high-affinity IgE reactive to Der p 2, an antigen found in house dust mite, from allergic patients.22 They estimated the affinity to range from 5.6 to 177.8pM (Kd value of IgE antibody to Der p 2). Through an in vitro histamine release assay, they found a strong correlation between the affinity of a given Der p 2-specific IgE antibody and the degree of histamine release. In a study by Christensen et al., murine and human Der p 2-specific IgE antibodies were cloned and their affinity was measured with surface plasmon resonance to classify the antibodies as high-, medium-, and low-affinity binders.23 By testing IgE antibodies of varying affinities in isolation or in combination, they found that when equimolar concentration of low- or high-affinity antibody was tested on basophils, the degranulation efficacy was poor with low-affinity IgE, even when using 1000 times more antigen. Thus, high-affinity allergen-specific IgE seems to be superior at inducing degranulation, suggesting a qualitatively distinct downstream signaling process. In yet another study, one group found that the affinity of milk-specific IgE positively correlated with the severity of the allergic symptoms of the patient from whom the IgE antibodies were derived.18 Further, Croote et al. found a convergent pattern of mutations in the IgE antibodies from unrelated peanut-allergic patients, suggesting that a common stepwise mutational pathway leads to the generation of high-affinity IgE in allergic individuals.21 Observations from patients are also supported by findings in murine models of allergic sensitization in which only high-affinity IgE induces anaphylaxis.8,15,16,24
Why is high-affinity IgE more adept at inducing anaphylaxis? Elegant signaling experiments have demonstrated that even though all IgE antibodies bind by their Fc region to the high-affinity IgE receptor, FcεRI, the signaling downstream of FcεRI differs depending on whether cross-linking is mediated by high-affinity versus low-affinity antigen-binding fragment (Fab) of IgE, leading to qualitatively distinct outcomes.25 The FcεRI-IgE-antigen signaling complex recruits Src kinases Lyn and Fyn. Lyn phosphorylates ITAMson FcεRI, leading to the recruitment of the tyrosine kinase Syk, which subsequently phosphorylates adapters LAT1 and LAT2 that nucleate the FcεRI signalosome. LAT1 and LAT2 play antagonistic roles in FcεRI-mediated signaling.25 Lyn-Syk-LAT1 signaling predominantly occurs downstream of high-affinity IgE interactions, whereas LAT2 signaling is favored downstream of low-affinity interactions.26 This leads to degranulation and the release of β-hexosaminidase and leukotrienes downstream of high-affinity interactions, whereas low-affinity interactions instead promote chemokine (CCL2,3,4) production.26,27 The Lyn-Syk-LAT1 axis is integral for degranulation by high-affinity IgE as a single amino acid change in Syk (Y130E) impaired LAT1 phosphorylation and thus degranulation.28
Helminth infections are the other major immunologic insult that provokes IgE responses. In contrast to allergens, helminth infections are not thought to elicit high affinity or anaphylactic IgE responses.8,16,29 Antigen-specific IgE or IgE with mutations have been reported in some helminth infections, and the number of mutations in IgE antibodies has been shown to increase upon secondary infection.30,31 However, it is unclear whether these IgE molecules are of high affinity or can persist for a long duration. Our recent work showed that mice co-immunized with helminths and haptenated ovalbumin developed only low- but not high-affinity anti-hapten IgE antibodies.15 McCoy et al. have shown that the IgE antibodies induced to the natural mouse parasite H. polygyrus are nonspecific and polyclonal and that loss of this IgE does not impact worm clearance during either primary or secondary infection.32 Patients with recurrent worm infections do not show evidence of antigen-driven selection or mutations in IgE as compared to other isotypes.33
One tenet of the hygiene hypothesis draws upon findings that people from helminth-endemic areas or mice infected with helminths are protected from severe anaphylaxis to allergens. One mechanism that might partly explain this association includes the production of nonspecific or low-affinity IgE that can occupy the IgE receptor on mast cells and thereby block the ability of allergen-reactive IgE to degranulate those mast cells.29,34,35 Intriguingly, some studies have shown that IgE induced by helminths can be cross-reactive to allergens, presumably due to molecular mimicry.35 Though cross-reactive, such IgE was found to have poor biologic activity, especially in its ability to induce degranulation.29 Further evidence comes from a study of IgE cross-reactivity and function among school children in Ghana.36 The authors found that infection with the helminth Schistosoma haematobium was associated with increased levels of peanut-specific IgE that could bind cross-reactive carbohydrate determinants. Such cross-reactive IgE had poor biologic activity by skin prick test and basophil histamine release assay.36
Aside from affinity, another possible reason for poorly anaphylactic IgE in helminth infections could be differences in IgE glycosylation. A recent study by Shade et al. has shown an absolute requirement for IgE glycosylation in allergic reactions.37 The asparagine residues in the constant domain 3 (Cε3) of IgE (at asparagine-394 [N394] in human and N384 in mouse) must be glycosylated in order for the IgE to be anaphylactic. Thus, an open question in the field that remains unanswered is whether glycosylation of IgE differs in allergic sensitization versus helminth infections.
2.2. High-affinity IgE is typically generated via sequential switching of IgG1
Affinity maturation is the process by which B cell clones that begin with low-affinity antibodies for a given antigen ultimately generate antibodies with up to a 5000-fold increase in affinity during the course of an immune response.38 The process of affinity maturation typically takes place in organized structures within secondary lymphoid organs called germinal centers (GCs), where antigen-experienced B cells mutate their BCRs and compete to capture and present antigens to T cells. B cells with higher-affinity BCRs have an advantage over lower-affinity B cells because they can efficiently acquire antigens from follicular dendritic cells in the GC that act as antigen depots. Thus, high-affinity B cells are more likely to receive CD40 signaling and cytokine help from T cells present in GCs to proliferate and differentiate into memory or plasma cells (PCs).39 Somatic hypermutation is a process that enables these B cells to mutate their BCRs through the induction of the enzyme activation-induced cytidine deaminase (AID). Typically, the higher the rate of mutation in a BCR, the higher the affinity it develops for a given antigen.38
Much of the evidence for the GC selection-based models for affinity maturation comes from studying IgG.6,39 Whether the GC-dependent selection model holds true for IgE has been an area of active investigation during the past decade. One of the main bottlenecks for studying IgE B cells has been their rarity in vivo.40 An earlier attempt to study the origin of IgE B cells in vivo during helminth infection concluded that although IgE switching appeared to occur within GCs, IgE GC B cells were transient, and most IgE B cells differentiated into PCs and exited the GCs.40 The generation of IgE reporter mice by three independent groups was a major technical advancement for the field.41–43 Using type 2 immunization models such as the adjuvant aluminum hydroxide (alum) or helminth infections, studies in these reporter mice concluded that IgE GC B cells were not sustained like IgG1 GC B cells; IgE GC B cells instead quickly differentiated into PCs.41–43 Several reasons have been proposed for the compromised presence of IgE B cells within GCs. These include higher basal level signals of IgE BCR compared to IgG1 BCR (which likely predisposes them to PC differentiation), lower expression of IgE BCR in GC B cells due to differences in polyadenylation, poor ability to capture antigen and present to T cells, and greater tendency for apoptosis.44–48
How then is affinity-matured IgE made? One mechanism that has been demonstrated is sequential switching to IgE via IgG.8,16,24 When IgM B cells switch to IgE B cells, the process is called direct switching. When instead the switching occurs via an intermediary IgG step (IgM to IgG to IgE), the process is called sequential switching. Classic experiments reveal that IgE antibodies from both mice and humans show evidence of sequential switching, given the presence of IgG switch remnant sequences.17,49–51 In an elegant study in mice by Xiong et al., it was shown that IgM to IgE direct switching typically leads to low-affinity IgE production and that sequential switching was essential to generate high-affinity IgE.16 They used a mouse model in which the mice could not make IgG1 antibodies, so any IgE made would be a result of direct switching from IgM (IgM to IgE). Immunization with the hapten (4-hydroxy-3-nitrophenyl) acetyl (NP) conjugated to model protein antigens such as ovalbumin allows the measurement of affinity of the antibody made against the hapten NP. When immunized with NP-conjugated antigen in alum, the IgG1-deficient mice made comparable levels of total and low-affinity IgE to wild-type mice. However, they did not make high-affinity IgE, suggesting that high-affinity IgE predominantly arise from IgG1 through the sequential switching pathway.16 Further, the IgE generated in these mice did not cause an anaphylactic response but, at high molar ratios, prevented anaphylaxis triggered by high-affinity IgE. Clinical studies in patients provide further support for sequential switching of IgE; analysis of IgE sequences or clonal relationships of IgE B cells in allergic patients have indicated that IgE are sequentially switched from IgG B cells.8,17,19–21
2.3. The IgE response has memory but not memory B cells
Humoral memory is maintained by long-lived PCs in the bone marrow and memory B cells that can be reactivated to differentiate into antibody-secreting cells. Whereas the existence of class switched memory B cells are well established for isotypes including IgG and IgA, many studies suggest a dearth of IgE memory B cells. Studies examining the presence of memory IgE B cells using reporter mice have concluded that, in contrast to IgG1 memory B cells, IgE memory B cells rarely form.41–43 This may be in part due to poor development of IgE GC B cells, as memory B cells typically arise from GC responses.52 Therefore, the maintenance of humoral IgE responses relies on IgE PCs. However, recent studies suggest that IgE PCs only persist for a few months in the absence of antigen re-exposure.53–55 Employing a peanut allergen sensitization model, Jordana and colleagues estimated that, in contrast to IgG1 PCs with a half-life of more than 200 d, IgE PCs have a half-life of only about 60 d. After serum levels of IgE were no longer detectable, IgE bound to mast cells and basophils persisted for only an additional 2 mo.55 How then are life-long humoral IgE responses maintained? Recent data suggest that IgG1 memory B cells, upon re-exposure to antigen, can differentiate into IgE PCs to maintain humoral IgE.24,55 Thus, for both affinity maturation and maintenance of the IgE response, IgG B cells are a key intermediary.
3. TFH CELLS AS DRIVERS OF IGE RESPONSES
3.1. The old paradigm—Th2 cells drive IgE responses
One of the etymologic reasons for naming CD4 T cells as T helper cells was their requirement for generating strong antibody responses to immunogens.9 Classic experiments showed that transfer of B cells alone into irradiated mice did not result in antibody response to immunizations but required co-transfer of thymus-derived T cells.56,57 Another significant breakthrough came about two decades later, with the functional classification of CD4 T cells into distinct helper subsets based on their cytokine profiles: the Th1-Th2 paradigm.58,59 The distinction of CD4 T cells into Th1 and Th2 cells based on the cytokines and distinctive transcription factors provided key mechanistic understanding of how polarized immune responses occur in vivo.60,61 The basic flavor of immune responses was therefore divided into type 1 and type 2 based on the Th1-Th2 paradigm. The cytokines secreted by polarized T cell subsets dictated the particular class-switched antibody produced by B cells, suggesting that they may profoundly influence the outcome of the humoral response.62,63 Because IgE is induced during type 2 responses and is dependent on the canonical Th2 cytokine IL-4, Th2 cells were thought to be the main drivers of IgE in vivo.64–66 However, as IL-4 was described as a B cell growth factor, Th2 cells were thought to be the principal subset that helped B cell responses in general, whereas Th1 cells drove cell-mediated immunity. This notion became dogma in the 1990s but could not account for inconsistencies between the model and observations, such as IgG antibody production during a Th1 type response, and Th2-driven cellular responses.9
3.2. The new paradigm—Tfh cells are required for IgE responses in vivo
The Th1-Th2 paradigm for B cell help saw a major change in the 2000s when a dedicated subset of Th cells for B cell help, Tfh cells, were described.9 These cells were defined by their expression of the surface markers PD-1 and CXCR5 and the cytokines IL-21 and IL-4.9,67 Identification of BCL6 as a lineage-specific transcription factor for Tfh cells allowed the interrogation of their role in shaping antibody responses, which established these cells as a distinct and dedicated subset of CD4 T cells that help B cells.68–70 Tfh cells were shown to be important in two key processes of the adaptive humoral response: affinity maturation of B cells and isotype switching.10 Tfh cells could produce Th1 or Th2 cytokines such as IFNγ and IL-4 and to induce B cell switching to IgG2 and IgG1 isotypes, respectively. In a seminal study by Reinhardt et al. using reporters for IFNγ and IL-4 cytokine expression, it was observed that murine B cells that interacted with IFNγ+ Tfh cells made IgG2 transcripts, whereas B cells that interacted with IL-4+ Tfh cells made IgG1 transcripts during L. major infection.71 Recent experiments using mice with conditional loss of Bcl6 or IL-6 receptor signaling in T cells, which abrogates Tfh cell development, showed that Tfh cells are critical for inducing IgE in multiple type 2 responses including to allergens.15,72–74 Further, bone marrow chimera experiments that isolated IL-4 deficiency to Tfh cells also demonstrated severely reduced IgE responses, suggesting that Tfh cell-derived IL-4 is the main driver of IgE in vivo.75 Supporting these findings of the importance of Tfh cells in directing IgE responses, a functional dichotomy between Th2 and Tfh cells in regulating peripheral inflammation and IgE, respectively, has begun to emerge. Adoptive transfer of Th2 cells led to allergic airway inflammation as evidenced by lung eosinophilia but weak IgE responses, whereas transfer of Tfh cells led to poor airway inflammation but robust IgE responses.71,72 Further, deletion of Tfh cells does not impair type 2 cellular immune responses, such as lung infiltration of eosinophils in models of allergic airway disease.15,72
Intriguingly, a recent study suggested that most class switching to IgG occurs at extrafollicular sites prior to GC formation in protein antigen immunizations.76 If the same is true for other isotypes (e.g., IgE and IgA) remains to be determined. But an important question to be answered now is where Tfh cells and their derived cytokines regulate class switching during different types of immune insults such as infections or allergic responses.
4. NATURE OF THE CELLS THAT DRIVE IGE RESPONSE TO HELMINTHS AND ALLERGENS
Based on the recent evidence, the model that emerges for IgE induction is that IL-4-producing Tfh cells drive IgE responses.10,75 However, allergens typically induce high-affinity anaphylactic IgE responses, which are rarely induced during helminth infections as previously discussed. As Tfh cells are critical to induce antigen-specific IgE response, does the nature of Tfh cells induced during high-affinity IgE responses such as allergic sensitization differ from helminth infection? IL-4-producing Tfh cells, typically called Tfh2 cells, are present in both conditions. Using IL-4 reporter mice or IL-4 intracellular staining, multiple groups have demonstrated the presence of IL-4+ Tfh cells in the spleen and lymph nodes of mice infected with helminths.15,77,78 IL-4+ Tfh cells have also been identified in various inhalational as well as oral allergen sensitization models.15,72,74 However, IL-4 is also considered a canonical Tfh cytokine and as such, IL-4-expressing Tfh cells have also been observed in models of type 1 immune responses including viral infection.67,79–81 Altogether, these findings suggest that IL-4 is necessary but not sufficient for high-affinity IgE induction. Our group recently described a unique population of IL-4+ IL-13+ “Tfh13” cells that were induced to allergens but not helminth infection or during type 1 immune responses and were required for induction of anaphylactic IgE during allergic sensitization.15 Tfh13 cells were induced to aero- and food allergens in both mouse models and allergic patients, produced IL-13 along with IL-4, and expressed the Th2 transcription factor GATA3 in addition to the canonical Tfh cell transcription factor BCL6. Loss of these cells in mouse models resulted in poor induction of high-affinity anaphylactic IgE to allergens and impaired passive cutaneous anaphylaxis (PCA) upon serum transfer without impairing total or low-affinity IgE titers. These IL-13-producing Tfh13 cells were also present in a mouse model of dedicator of cytokinesis 8 (DOCK8) deficiency, an immunodeficiency syndrome that paradoxically presents with aberrant hyper-IgE production and allergic responses. Indeed, isolated loss of DOCK8 in T cells in mice immunized with a type 1 adjuvant to mimic a bacterial infection lead to induction of anaphylaxis-inducing IgE, systemic release of MMCP-1 and multi-tissue edema upon antigenic challenge. Based on these data we propose that IL-4+ Tfh2 cells drive low-affinity IgE production (likely during both helminth and allergen type 2 immune responses), whereas Tfh13 cells, which are uniquely induced in allergic conditions, drive high-affinity IgE.15
4.1. Role of IL-13 in IgE production
IL-13 is well known for its function in the effector T cell arm of type 2 immunity; however, its role in inducing IgE antibodies has been less clear. Whereas it was shown that IL-13 could act on human B cells to promote IgE production, it was thought that IL-13 did not act on murine B cells based on an early review.82 However, later studies demonstrated that murine B cells respond in vitro and in vivo to IL-13.83 Complete IL-13 or IL-13 receptor knockout mice demonstrated partial reduction of IgE responses in some studies but not others.78,84–86 Importantly, many of these studies examined total IgE levels rather than affinity-matured or anaphylactic IgE. Additionally, mice lacking IL-4 or IL-13 alone fail to generate WT levels of IgE supporting the conclusion that IL-4 and IL-13 have nonredundant roles that cannot fully compensate for the loss of the other.85,87 Although some studies have observed IL-13 production by Tfh cells,74,88 the functional significance of Tfh cell-derived IL-13 was previously unknown; we found that the isolated loss of IL-13 production by Tfh cells impaired the production of high-affinity IgE to allergens.15 We and others have found that IL-13Rα1 is up-regulated on activated B cells, especially GC B cells.15,89,90 Intriguingly, IgG1 and IgE GC B cells but not IgM GC B cells have high expression of IL-13Rα1.15 Thus, IL-13Rα1hi IgG1 GC B cells may be involved in sequential switching to IgE, presumably through their interaction with Tfh13 cells, whereas direct switching to IgE may proceed via IL-13Rα1lo IgM GC B cells, likely guided by IL-4 (Fig. 1).
FIGURE 1. A model for IgE induction during helminth infections and allergen exposure.
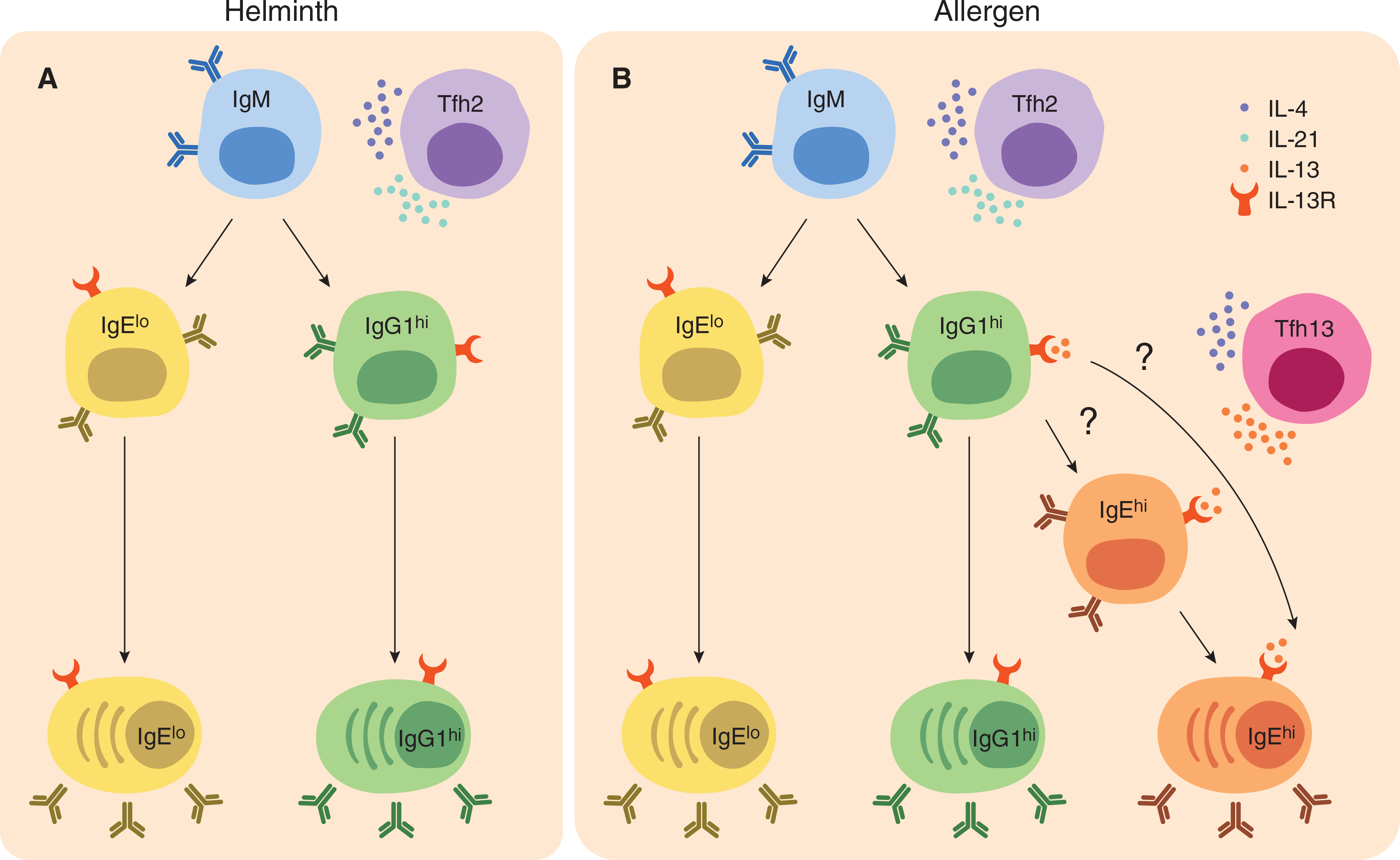
In both helminth infections and allergic sensitization, Tfh2 cells that make IL-4 and IL-21 are induced. Tfh2 cells help B cells make high-affinity IgG and low-affinity IgE through direct switching of IgM B cells. Following activation, these B cells up-regulate the receptor for IL-13, yet little Tfh cell-derived IL-13 should be present. In contrast, during allergic sensitization, distinct populations of IL-4- and IL-13-producing Tfh13 cells are induced in addition to Tfh2 cells. Through their production of IL-13 and IL-4, Tfh13 cells promote sequential switching of high-affinity IgG1 to high-affinity IgE-producing plasma cells (PCs). It is unclear whether this switching occurs through a high-affinity IgE B cell intermediate, or whether high-affinity IgE-producing PCs directly differentiate from high-affinity IgG1 B cells (see question marks). Further, isotype-switched B cells and PCs during both helminth infection and allergen sensitization express IL-13 receptor (IL-13R); however, only during allergen sensitization, but not helminth infection, is IL-13 available to promote high-affinity IgE
As previously discussed, one of the distinctive features of IgE responses is the lack of sustained IgEGCB cells.91 This is thought to be a critical limitation for inducing antigen-specific IgE. IL-13 may overcome this limitation, as it has been shown to promote anti-apoptotic factor Bcl-xL in cultured human B cells.92 Further, there are distinct differences between IL-4 and IL-13 signaling that could account for the requirement of IL-13 from a Tfh cell already producing IL-4. IL-4 can signal through both the type I receptor, composed of IL-4Rα and the common gamma chain (γC), and the type II receptor, composed of IL-4Rα and IL-13Rα1. In contrast, IL-13 primarily uses the type II receptor.93 IL-4-bound type I receptor complex results in both STAT6 activation, which is critical for IgE induction, and PI3K signaling, which negatively regulates IgE induction.93–95 As IL-13-bound type II receptor complex exclusively activates STAT6 and is also more stable (Kd = 20 nM) than the IL-4 bound type I (Kd = 559 nM) or type II (Kd = 487 nM) complexes, IL-13 may lead to more sustained STAT6 activation and promote IgE production.93,96,97
4.2. Transcription factors that drive IgE-promoting cytokines in Tfh cells
One of the main drivers of IgE-promoting cytokines IL-4 and IL-13 production in T cells is the transcription factor GATA3. However, Tfh cells are thought not to express high levels of GATA3. Overexpression of the Tfh transcription factor BCL6 was shown to inhibit GATA3 expression in T cells.69,70,98 Further, characterization of Tfh cells induced in vivo during viral or helminth infections also showed that they poorly express Gata3, suggesting a GATA3-independent mechanism of Il4 induction in Tfh cells in contrast to Th2 cells.78,80 Supporting distinct mechanisms of Il4 transcriptional activation in these two cell types, studies of Il4 enhancers show a divergent requirement for these regulatory elements in Th2 cells versus Tfh cells. Namely, Th2 cells utilize hypersensitivity site 2 (HS2) and Rad50 hypersensitive site 6 (RHS6) for IL-4 production.99,100 Tfh cells instead require hypersensitivity site V (HS V), also known as conserved noncoding sequence 2 (CNS2), for Il4 expression.101,102 In turn, the enhancers active in Th2 cells are bound by GATA3, whereas those active in Tfh cells are bound by BATF.99,103,104 These data led to a model in which Th2 cells may selectively utilize GATA3 and thereby produce IL-4, −5, and −13, whereas Tfh cells rely on BATF for isolated IL-4 production. However, BATF has been shown more recently to regulate IL-4 production in Th2 cells, demonstrating that its Il4 regulatory activity is not specific to Tfh cells as previously argued.105,106
Although not considered canonical Tfh TFs, multiple studies have since observed that Tfh cells can co-express other lineage TFs such as T-bet and GATA3 along with BCL6 and make cytokines such as IFNγ or IL-13 during particular type 1 or 2 immune responses.15,88,107–110
Our recent analysis of single-cell RNA-sequencing of Tfh cells induced to the allergen Alternaria alternata showed that IL-13+ IL-4+ Tfh13 cells expressed high levels of Gata3 whereas the IL-4+ Tfh2 cells did not. Therefore, Gata3 up-regulation could be a key determinant in inducing IL-5 and −13 in addition to IL-4 production in Tfh13 cells. Taken together, these studies indicate that GATA3 and BATF may not be dichotomously utilized by Th2 cells and Tfh cells, respectively, for IL-4 induction as previously suggested104,111; rather, BATF regulates IL-4 production in both Th2 and Tfh cells, whereas GATA3 is utilized by Th2 cells and, under certain conditions (namely allergic sensitization), Tfh cells as well.
5. DEVELOPMENT OF IGE-PROMOTING TFH CELLS—PROGRESSIVE DIFFERENTIATION VERSUS PLASTICITY
How do Tfh cells differentiate into these subsets to induce specific classes of antibodies and thus tailor the humoral response? One model to explain the occurrence of type 2 cytokines in Tfh cells is the progressive differentiation model (Fig. 2A). An elegant study by Weinstein et al. showed that Tfh cells start off as IL-21 single-producing cells but then gradually differentiate into IL-4 single-producing cells.77 This pathway seems to be unidirectional as IL-4 single producers or IL-21/IL-4 double producers cannot revert back to an IL-21 single producer state. Whether this model applies to the Tfh13 cells in allergic sensitization and, if so, what signals drive this process of stepwise differentiation are unknown. Alternatively, Tfh13 cells could directly differentiate from early Tfh cells, in parallel with Tfh2 cells (Fig. 2B). Another model that could explain the occurrence of type 2 cytokines in Tfh cells is the conversion of Th2 cells into Tfh cells (Fig. 2C). Adoptively transferred IL-4-expressing Th2 cells were found to convert to Tfh cells during helminth antigen immunization.88 On the other hand, Tfh cells have been shown to convert into Th2 cells in a house dust mite model of allergic sensitization.112 Thus, it appears that the expression of GATA3 and the Th2 cytokine profile could be acquired by Tfh-Th2 cell interconversion. Whether a Tfh-Th2 plastic pathway is mutually exclusive to, or can occur in parallel with a progressive differentiation pathway and how stable GATA3+ Tfh cells are open questions. As more detailed lineage tracing and high-throughput studies on T helper cells emerge, subset heterogeneity and plasticity seem to be the norm rather than the exception.113,114 Therefore, a unidimensional model of subsetting T cells based on lineage-specifying transcription factors is proving insufficient; instead, a revised paradigm incorporating function, heterogeneity, and plasticity is required.
FIGURE 2. Progressive differentiation and plasticity models of Tfh13 cell induction.

One possible pathway for Tfh13 cell induction is that Tfh13 cells progressively differentiate from IL-21+ Tfh cells (A, B). IL-21+ Tfh cells are induced early during an immune response, and they differentiate into IL-4-producing Tfh2 cells by up-regulating BATF, which induces IL-4 but not IL-5 or IL-13 production. These Tfh2 cells could in turn differentiate into Tfh13 cells through GATA3 upregulation (A). Alternatively, Tfh13 cells may also arise directly from IL-21+ Tfh cells (B). In contrast to progressive differentiation, a plasticity model suggests that Tfh13 cells and Tfh2 cells differentiate from Th2 cells (C). This pathway may be bi-directional as Tfh cells could also acquire a Th2 phenotype and migrate to the periphery as effector cells
6. CONCLUSIONS AND FUTURE DIRECTIONS
Anaphylaxis and its principal causative factor, the IgE antibody, were described several decades ago. Research over the last two decades has given phenomenal insights into the nature of the IgE molecules induced in allergic responses, multiple regulatory constraints for the induction and long-term maintenance of IgE responses by the immune system, and the nature of T cells that help in this process. Conclusions that emerge from recent studies are that (i) high-affinity, but not low-affinity IgE causes anaphylaxis; (ii) the pathway to high-affinity IgE is typically through sequential switching of IgG molecules; and (iii) the T cell subset that drives the induction of high-affinity anaphylactic IgE is not Th2 cells but Tfh cells, in particular Tfh13 cells. However, several aspects of IgE induction during allergic sensitization remain to be explored. One of the key questions that remain to be addressed is why Tfh13 cells are induced to allergens but not to helminths. Unique features of allergens, such as protease activity or other intrinsic properties, may promote the immune system to generate Tfh13 cells. Cytokines play a key role in skewing the immune response to type 1 versus type 2 immunity. Although many studies have defined crucial roles for cytokines such as IL-25, IL-33, TSLP, and IL-1 in promoting Th2 differentiation, little is known about how they may influence the differentiation of Tfh cell subsets.74,114–116 Other key areas that require further investigation are understanding the precise mechanisms by which Tfh cells instruct B cells to make high-affinity IgE and why only certain individuals develop life-threatening allergic reactions to allergens whereas others do not.
IgE-mediated allergic responses to food and environmental allergens have been on the rise, which indicates that IgE regulatory mechanisms often go awry in our current industrialized environments. Understanding the nuances of IgE regulation is paramount to designing clinical interventions to reverse this trend. Identification of cellular players and key checkpoints in the pathway that result in anaphylactic IgE to allergens will help identify biomarkers and new potential therapeutic targets for patients with allergies.
ACKNOWLEDGMENTS
We thank Adam Williams (The Jackson Laboratory for Genomic Medicine, Farmington, CT, USA), Jake Gertie (Yale University, New Haven, CT, USA) for their comments and critical reading of the manuscript, and members of the Eisenbarth Laboratory for helpful discussions. This work was supported by Food Allergy Research and Education Ira and Diana Riklis Family Research Award in Food Allergy, R01 AI136942, and R01 AI108829.
Abbreviations:
- BATF
Basic leucine zipper transcription factor, ATF-like
- BCL6
B-cell lymphoma 6 protein
- BCR
B cell receptor
- FcεRI
FC epsilon receptor 1
- GATA3
GATA binding protein 3
- IgE
Immunoglobulin E
- IgG
Immunoglobulin G
- IL-4
Interleukin 4
- IL-13
Interleukin 13
- ITAM
Immunoreceptor Tyrosine-based Activation Motif
- MMCP1
Mouse Mast Cell Protease 1
- PI3K
Phosphatidylinositol 3-kinase
- STAT
Signal Transducer and Activator of Transcription
- Tfh cell
T follicular helper cell
- WT
Wild type
REFERENCES
- 1.Marichal T, Starkl P, Reber LL, et al. A beneficial role for immunoglobulin E in host defense against honeybee venom. Immunity. 2013;39:963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galli SJ, Starkl P, Marichal T, Tsai M. Mast cells and IgE in defense against venoms: possible “good side” of allergy? Allergol Int. 2016;65:3–15. [DOI] [PubMed] [Google Scholar]
- 3.Mukai K, Tsai M, Starkl P, Marichal T, Galli SJ. IgE and mast cells in host defense against parasites and venoms. Semin Immunopathol. 2016;38:581–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palm NW, Rosenstein RK, Yu S, Schenten DD, Florsheim E, Medzhitov R. Bee venom phospholipase A2 induces a primary type 2 response that is dependent on the receptor ST2 and confers protective immunity. Immunity. 2013;39:976–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finkelman FD, Rothenberg ME, Brandt EB, Morris SC, Strait RT. Molecular mechanisms of anaphylaxis: lessons from studies with murine models. J Allergy Clin Immunol. 2005;115:449–457. quiz 458. [DOI] [PubMed] [Google Scholar]
- 6.Mesin L, Ersching J, Victora GD. Germinal center B cell dynamics. Immunity. 2016;45:471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gowthaman U, Chodisetti SB, Agrewala JN. T cell help to B cells in germinal centers: putting the jigsaw together. Int Rev Immunol. 2010;29:403–420. [DOI] [PubMed] [Google Scholar]
- 8.He JS, Narayanan S, Subramaniam S, Ho WQ, Lafaille JJ, Curotto de Lafaille MA. Biology of IgE production: IgE cell differentiation and the memory of IgE responses. Curr Top Microbiol Immunol. 2015;388:1–19. [DOI] [PubMed] [Google Scholar]
- 9.Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol. 2015;15:185–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crotty S. T follicular helper cell biology: a decade of discovery and diseases. Immunity. 2019;50:1132–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkelman FD. Anaphylaxis: lessons from mouse models. J Allergy Clin Immunol. 2007;120:506–515. quiz 516–7. [DOI] [PubMed] [Google Scholar]
- 12.Reber LL, Hernandez JD, Galli SJ. The pathophysiology of anaphylaxis. J Allergy Clin Immunol. 2017;140:335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khodoun MV, Strait R, Armstrong L, Yanase N, Finkelman FD. Identification of markers that distinguish IgE- from IgG-mediated anaphylaxis. PNAS. 2011;108:12413–12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castells M. Diagnosis and management of anaphylaxis in precision medicine. J Allergy Clin Immunol. 2017;140:321–333. [DOI] [PubMed] [Google Scholar]
- 15.Gowthaman U, Chen JS, Zhang B, et al. Identification of a T follicular helper cell subset that drives anaphylactic IgE. Science. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong H, Dolpady J, Wabl M, Curotto de Lafaille MA, Lafaille JJ. Sequential class switching is required for the generation of high affinity IgE antibodies. J Exp Med. 2012;209:353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janezic A, Chapman CJ, Snow RE, Hourihane JO, Warner JO, Stevenson FK. Immunogenetic analysis of the heavy chain variable regions of IgE from patients allergic to peanuts. J Allergy Clin Immunol. 1998;101:391–396. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, Lin J, Bardina L, et al. Correlation of IgE/IgG4 milk epitopes and affinity of milk-specific IgE antibodies with different phenotypes of clinical milk allergy. J Allergy Clin Immunol. 2010;125:695–702. 702 e1–702 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoh RA, Joshi SA, Liu Y, et al. Single B-cell deconvolution of peanut-specific antibody responses in allergic patients. J Allergy Clin Immunol. 2016;137:157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Looney TJ, Lee JY, Roskin KM, et al. Human B-cell isotype switching origins of IgE. J Allergy Clin Immunol. 2016;137:579–586. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croote D, Darmanis S, Nadeau KC, Quake SR. High-affinity allergen-specific human antibodies cloned from single IgE B cell transcriptomes. Science. 2018;362:1306–1309. [DOI] [PubMed] [Google Scholar]
- 22.Mita H, Yasueda H, Akiyama K. Affinity of IgE antibody to antigen influences allergen-induced histamine release. Clin Exp Allergy. 2000;30:1583–1589. [DOI] [PubMed] [Google Scholar]
- 23.Christensen LH, Holm J, Lund G, Riise E, Lund K. Several distinct properties of the IgE repertoire determine effector cell degranulation in response to allergen challenge. J Allergy Clin Immunol. 2008;122: 298–304. [DOI] [PubMed] [Google Scholar]
- 24.He JS, Subramaniam S, Narang V, et al. IgG1 memory B cells keep the memory of IgE responses. Nat Commun. 2017;8:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki R, Scheffel J, Rivera J. New insights on the signaling and function of the high-affinity receptor for IgE. Curr Top Microbiol Immunol. 2015;388:63–90. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki R, Leach S, Liu W, et al. Molecular editing of cellular responses by the high-affinity receptor for IgE. Science. 2014;343:1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu ZJ, Haleem-Smith H, Chen H, Metzger H. Unexpected signals in a system subject to kinetic proofreading. PNAS. 2001;98:7289–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz SL, Cleyrat C, Olah MJ, et al. Differential mast cell outcomes are sensitive to FcepsilonRI-Syk binding kinetics. Mol Biol Cell. 2017;28:3397–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamid F, Amoah AS, van Ree R, Yazdanbakhsh M. Helminth-induced IgE and protection against allergic disorders. Curr Top Microbiol Immunol. 2015;388:91–108. [DOI] [PubMed] [Google Scholar]
- 30.Pochanke V, Koller S, Dayer R, et al. Identification and characterization of a novel antigen from the nematode Nippostrongylus brasiliensis recognized by specific IgE. Eur J Immunol. 2007;37:1275–1284. [DOI] [PubMed] [Google Scholar]
- 31.Turqueti-Neves A, Otte M, Schwartz C, et al. The extracellular domains of IgG1 and T cell-derived IL-4/IL-13 are critical for the polyclonal memory IgE response in vivo. PLoS Biol. 2015;13:e1002290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy KD, Stoel M, Stettler R, et al. Polyclonal and specific antibodies mediate protective immunity against enteric helminth infection. Cell Host Microbe. 2008;4:362–373. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Jackson KJ, Chen Z, et al. IgE sequences in individuals living in an area of endemic parasitism show little mutational evidence of antigen selection. Scand J Immunol. 2011;73:496–504. [DOI] [PubMed] [Google Scholar]
- 34.Yazdanbakhsh M, Kremsner PG, van Ree R. Allergy, parasites, and the hygiene hypothesis. Science. 2002;296:490–494. [DOI] [PubMed] [Google Scholar]
- 35.Fitzsimmons CM, Falcone FH, Dunne DW. Helminth allergens, parasite-specific IgE, and its protective role in human immunity. Front Immunol. 2014;5:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amoah AS, Obeng BB, Larbi IA, et al. Peanut-specific IgE antibodies in asymptomatic Ghanaian children possibly caused by carbohydrate determinant cross-reactivity. J Allergy Clin Immunol. 2013;132: 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shade KT, Platzer B, Washburn N, et al. A single glycan on IgE is indispensable for initiation of anaphylaxis. J Exp Med. 2015;212:457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mishra AK, Mariuzza RA. Insights into the structural basis of antibody affinity maturation from next-generation sequencing. Front Immunol. 2018;9:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cyster JG, Allen CDC. B cell responses: cell interaction dynamics and decisions. Cell. 2019;177:524–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erazo A, Kutchukhidze N, Leung M, et al. Unique maturation program of the IgE response in vivo. Immunity. 2007;26:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Talay O, Yan D, Brightbill HD, et al. IgE(+) memory B cells and plasma cells generated through a germinal-center pathway. Nat Immunol. 2012;13:396–404. [DOI] [PubMed] [Google Scholar]
- 42.Yang Z, Sullivan BM, Allen CD. Fluorescent in vivo detection reveals that IgE(+) B cells are restrained by an intrinsic cell fate predisposition. Immunity. 2012;36:857–872. [DOI] [PubMed] [Google Scholar]
- 43.He JS, Meyer-Hermann M, Xiangying D, et al. The distinctive germinal center phase of IgE+ B lymphocytes limits their contribution to the classical memory response. J Exp Med. 2013;210:2755–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haniuda K, Fukao S, Kodama T, Hasegawa H, Kitamura D. Autonomous membrane IgE signaling prevents IgE-memory formation. Nat Immunol. 2016;17:1109–1117. [DOI] [PubMed] [Google Scholar]
- 45.Vanshylla K, Opazo F, Gronke K, Wienands J, Engels N. The extracellular membrane-proximal domain of membrane-bound IgE restricts B cell activation by limiting B cell antigen receptor surface expression. Eur J Immunol. 2018;48:441–453. [DOI] [PubMed] [Google Scholar]
- 46.Laffleur B, Duchez S, Tarte K, et al. Self-restrained B cells arise following membrane IgE expression. Cell Rep. 2015;10:900–909. [DOI] [PubMed] [Google Scholar]
- 47.Tong P, Granato A, Zuo T, et al. IgH isotype-specific B cell receptor expression influences B cell fate. PNAS. 2017;114:E8411–E8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z, Robinson MJ, Chen X, et al. Regulation of B cell fate by chronic activity of the IgE B cell receptor. eLife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mandler R, Finkelman FD, Levine AD, Snapper CM. IL-4 induction of IgE class switching by lipopolysaccharide-activated murine B cells occurs predominantly through sequential switching. J Immunol. 1993;150:407–418. [PubMed] [Google Scholar]
- 50.Siebenkotten G, Esser C, Wabl M, Radbruch A. The murine IgG1/IgE class switch program. Eur J Immunol. 1992;22:1827–1834. [DOI] [PubMed] [Google Scholar]
- 51.Jabara HH, Loh R, Ramesh N, Vercelli D, Geha RS. Sequential switching from mu to epsilon via gamma 4 in human B cells stimulated with IL-4 and hydrocortisone. J Immunol. 1993;151: 4528–4533. [PubMed] [Google Scholar]
- 52.Saunders SP, Ma EGM, Aranda CJ, Curotto de Lafaille MA. Non-classical B cell memory of allergic IgE responses. Front Immunol. 2019;10:715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luger EO, Fokuhl V, Wegmann M, et al. Induction of long-lived allergen-specific plasma cells by mucosal allergen challenge. J Allergy Clin Immunol. 2009;124:819–826. e4. [DOI] [PubMed] [Google Scholar]
- 54.Moutsoglou DM, Dreskin SC. B cells establish, but do not maintain, long-lived murine anti-peanut IgE(a). Clin Exp Allergy. 2016;46: 640–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jimenez-Saiz R, Chu DK, Mandur TS, et al. Lifelong memory responses perpetuate humoral TH2 immunity and anaphylaxis in food allergy. J Allergy Clin Immunol. 2017;140:1604–1615. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller JF, Mitchell GF. Cell to cell interaction in the immune response. I. Hemolysin-forming cells in neonatally thymectomized mice reconstituted with thymus or thoracic duct lymphocytes. J Exp Med. 1968;128:801–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchell GF, Miller JF. Cell to cell interaction in the immune response. II. The source of hemolysin-forming cells in irradiated mice given bone marrow and thymus or thoracic duct lymphocytes. J Exp Med. 1968;128:821–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 59.Woods A, West J, Rasmussen R, Bottomly K. Granulocyte-macrophage colony stimulating factor produced by cloned L3T4a+, class II-restricted T cells induces HT-2 cells to proliferate. J Immunol. 1987;138:4293–4297. [PubMed] [Google Scholar]
- 60.Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–173. [DOI] [PubMed] [Google Scholar]
- 61.Fang D, Zhu J. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J Exp Med. 2017;214:1861–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Snapper CM, Paul WE. Interferon-gamma and B cell stimulatory factor-1 reciprocally regulate Ig isotype production. Science. 1987;236:944–947. [DOI] [PubMed] [Google Scholar]
- 63.Stevens TL, Bossie A, Sanders VM, et al. Regulation of antibody isotype secretion by subsets of antigen-specific helper T cells. Nature. 1988;334:255–258. [DOI] [PubMed] [Google Scholar]
- 64.Finkelman FD, Katona IM, Urban JF, et al. IL-4 is required to generate and sustain in vivo IgE responses. J Immunol. 1988;141: 2335–2341. [PubMed] [Google Scholar]
- 65.Katona IM, Urban JF Jr, Finkelman FD. The role of L3T4+ and Lyt-2+ T cells in the IgE response and immunity to Nippostrongylus brasiliensis. J Immunol. 1988;140:3206–3211. [PubMed] [Google Scholar]
- 66.Kopf M, Le Gros G, Bachmann M, Lamers MC, Bluethmann H, Kohler G. Disruption of the murine IL-4 gene blocks Th2 cytokine responses. Nature. 1993;362:245–248. [DOI] [PubMed] [Google Scholar]
- 67.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Johnston RJ, Poholek AC, DiToro D, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nurieva RI, Chung Y, Martinez GJ, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu D, Rao S, Tsai LM, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31: 457–468. [DOI] [PubMed] [Google Scholar]
- 71.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol. 2009;10: 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kobayashi T, Iijima K, Dent AL, Kita H. Follicular helper T cells mediate IgE antibody response to airborne allergens. J Allergy Clin Immunol. 2017;139:300–313. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Noble A, Zhao J. Follicular helper T cells are responsible for IgE responses to Der p 1 following house dust mite sensitization in mice. Clin Exp Allergy. 2016;46:1075–1082. [DOI] [PubMed] [Google Scholar]
- 74.Dolence JJ, Kobayashi T, Iijima K, et al. Airway exposure initiates peanut allergy by involving the IL-1 pathway and T follicular helper cells in mice. J Allergy Clin Immunol. 2018;142:1144–1158. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meli AP, Fontes G, Leung Soo C, King IL. T follicular helper cell-derived IL-4 is required for IgE production during intestinal helminth infection. J Immunol. 2017. [DOI] [PubMed] [Google Scholar]
- 76.Roco JA, Mesin L, Binder SC, et al. Class-switch recombination occurs infrequently in germinal centers. Immunity. 2019;51:337–350. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weinstein JS, Herman EI, Lainez B, et al. TFH cells progressively differentiate to regulate the germinal center response. Nat Immunol. 2016;17:1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liang HE, Reinhardt RL, Bando JK, Sullivan BM, Ho IC, Locksley RM. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat Immunol. 2012;13:58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu X, Yan X, Zhong B, et al. Bcl6 expression specifies the T follicular helper cell program in vivo. J Exp Med. 2012;209: 1841–1852: S1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yusuf I, Kageyama R, Monticelli L, et al. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J Immunol. 2010;185: 190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Luthje K, Kallies A, Shimohakamada Y, et al. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat Immunol. 2012;13:491–498. [DOI] [PubMed] [Google Scholar]
- 82.Zurawski G, de Vries JE. Interleukin 13, an interleukin 4-like cytokine that acts on monocytes and B cells, but not on T cells. Immunol Today. 1994;15:19–26. [DOI] [PubMed] [Google Scholar]
- 83.Lai YH, Mosmann TR. Mouse IL-13 enhances antibody production in vivo and acts directly on B cells in vitro to increase survival and hence antibody production. J Immunol. 1999;162:78–87. [PubMed] [Google Scholar]
- 84.McKenzie GJ, Emson CL, Bell SE, et al. Impaired development of Th2 cells in IL-13-deficient mice. Immunity. 1998;9:423–432. [DOI] [PubMed] [Google Scholar]
- 85.McKenzie GJ, Fallon PG, Emson CL, Grencis RK, McKenzie AN. Simultaneous disruption of interleukin (IL)-4 and IL-13 defines individual roles in T helper cell type 2-mediated responses. J Exp Med. 1999;189:1565–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramalingam TR, Pesce JT, Sheikh F, et al. Unique functions of the type II interleukin 4 receptor identified in mice lacking the interleukin 13 receptor alpha1 chain. Nat Immunol. 2008;9:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Emson CL, Bell SE, Jones A, Wisden W, McKenzie AN. Interleukin(IL)-4-independent induction of immunoglobulin (Ig)E, and perturbation of T cell development in transgenic mice expressing IL-13. J Exp Med. 1998;188:399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Glatman Zaretsky A, Taylor JJ, King IL, Marshall FA, Mohrs M, Pearce EJ. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. J Exp Med. 2009;206:991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Graber P, Gretener D, Herren S, et al. The distribution of IL-13 receptor alpha1 expression on B cells, T cells and monocytes and its regulation by IL-13 and IL-4. Eur J Immunol. 1998;28:4286–4298. [DOI] [PubMed] [Google Scholar]
- 90.Poudrier J, Graber P, Herren S, et al. A novel monoclonal antibody, C41, reveals IL-13Ralpha1 expression by murine germinal center B cells and follicular dendritic cells. Eur J Immunol. 2000;30:3157–3164. [DOI] [PubMed] [Google Scholar]
- 91.Yang Z, Robinson MJ, Allen CD. Regulatory constraints in the generation and differentiation of IgE-expressing B cells. Curr Opin Immunol. 2014;28:64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lomo J, Blomhoff HK, Jacobsen SE, Krajewski S, Reed JC, Smeland EB. Interleukin-13 in combination with CD40 ligand potently inhibits apoptosis in human B lymphocytes: upregulation of Bcl-xL and Mcl-1. Blood. 1997;89:4415–4424. [PubMed] [Google Scholar]
- 93.McCormick SM, Heller NM. Commentary: IL-4 and IL-13 receptors and signaling. Cytokine. 2015;75:38–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Doi T, Obayashi K, Kadowaki T, Fujii H, Koyasu S. PI3K is a negative regulator of IgE production. Int Immunol. 2008;20:499–508. [DOI] [PubMed] [Google Scholar]
- 95.Linehan LA, Warren WD, Thompson PA, Grusby MJ, Berton MT. STAT6 is required for IL-4-induced germline Ig gene transcription and switch recombination. J Immunol. 1998;161:302–310. [PubMed] [Google Scholar]
- 96.LaPorte SL, Juo ZS, Vaclavikova J, et al. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell. 2008;132:259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Junttila IS. Tuning the cytokine responses: an update on interleukin (IL)-4 and IL-13 receptor complexes. Front Immunol. 2018;9:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kusam S, Toney LM, Sato H, Dent AL. Inhibition of Th2 differentiation and GATA-3 expression by BCL-6. J Immunol. 2003;170:2435–2441. [DOI] [PubMed] [Google Scholar]
- 99.Tanaka S, Motomura Y, Suzuki Y, et al. The enhancer HS2 critically regulates GATA-3-mediated Il4 transcription in T(H)2 cells. Nat Immunol. 2011;12:77–85. [DOI] [PubMed] [Google Scholar]
- 100.Williams A, Lee GR, Spilianakis CG, Hwang SS, Eisenbarth SC, Flavell RA. Hypersensitive site 6 of the Th2 locus control region is essential for Th2 cytokine expression. PNAS. 2013;110:6955–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Harada Y, Tanaka S, Motomura Y, et al. The 3’ enhancer CNS2 is a critical ical regulator of interleukin-4-mediated humoral immunity in follicular helper T cells. Immunity. 2012;36:188–200. [DOI] [PubMed] [Google Scholar]
- 102.Vijayanand P, Seumois G, Simpson LJ, et al. Interleukin-4 production by follicular helper T cells requires the conserved Il4 enhancer hypersensitivity site V. Immunity. 2012;36:175–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sahoo A, Alekseev A, Tanaka K, et al. Batf is important for IL-4 expression in T follicular helper cells. Nat Commun. 2015;6:7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sahoo A, Wali S, Nurieva R. T helper 2 and T follicular helper cells: regulation and function of interleukin-4. Cytokine Growth Factor Rev. 2016;30:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bao K, Carr T, Wu J, et al. BATF modulates the Th2 locus control region and regulates CD4+ T cell fate during antihelminth immunity. J Immunol. 2016;197:4371–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kuwahara M, Ise W, Ochi M, et al. Bach2-Batf interactions control Th2-type immune response by regulating the IL-4 amplification loop. Nat Commun. 2016;7:12596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Morita R, Schmitt N, Bentebibel SE, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim CJ, Lee CG, Jung JY, et al. The transcription factor Ets1 suppresses T follicular helper type 2 Cell differentiation to halt the onset of systemic lupus erythematosus. Immunity. 2018;49:1034–1048. e8. [DOI] [PubMed] [Google Scholar]
- 109.Weinstein JS, Laidlaw BJ, Lu Y, et al. STAT4 and T-bet control follicular helper T cell development in viral infections. J Exp Med. 2018;215:337–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fang D, Cui K, Mao K, et al. Transient T-bet expression functionally specifies a distinct T follicular helper subset. J Exp Med. 2018;215:2705–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kubo M. T follicular helper and TH2 cells in allergic responses. Allergol Int. 2017;66:377–381. [DOI] [PubMed] [Google Scholar]
- 112.Ballesteros-Tato A, Randall TD, Lund FE, Spolski R, Leonard WJ, Leon B. T follicular helper cell plasticity shapes pathogenic T helper 2 cell-mediated immunity to inhaled house dust mite. Immunity. 2016;44:259–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Walker JA, McKenzie ANJ. TH2 cell development and function. Nat Rev Immunol. 2018;18:121–133. [DOI] [PubMed] [Google Scholar]
- 115.Guo L, Wei G, Zhu J, et al. IL-1 family members and STAT activators induce cytokine production by Th2, Th17, and Th1 cells. PNAS. 2009;106:13463–13468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhu J. T helper 2 (Th2) cell differentiation, type 2 innate lymphoid cell (ILC2) development and regulation of interleukin-4 (IL-4) and IL-13 production. Cytokine. 2015;75:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]


