Abstract
Background
Although imaging, endoscopy, and inflammatory biomarkers are associated with future Crohn disease (CD) outcomes, common laboratory studies may also provide prognostic opportunities. We evaluated machine learning models incorporating routinely collected laboratory studies to predict surgical outcomes in U.S. Veterans with CD.
Methods
Adults with CD from a Veterans Health Administration, Veterans Integrated Service Networks (VISN) 10 cohort examined between 2001 and 2015 were used for analysis. Patient demographics, medication use, and longitudinal laboratory values were used to model future surgical outcomes within 1 year. Specifically, data at the time of prediction combined with historical laboratory data characteristics, described as slope, distribution statistics, fluctuation, and linear trend of laboratory values, were considered and principal component analysis transformations were performed to reduce the dimensionality. Lasso regularized logistic regression was used to select features and construct prediction models, with performance assessed by area under the receiver operating characteristic using 10-fold cross-validation.
Results
We included 4950 observations from 2809 unique patients, among whom 256 had surgery, for modeling. Our optimized model achieved a mean area under the receiver operating characteristic of 0.78 (SD, 0.002). Anti-tumor necrosis factor use was associated with a lower probability of surgery within 1 year and was the most influential predictor in the model, and corticosteroid use was associated with a higher probability of surgery. Among the laboratory variables, high platelet counts, high mean cell hemoglobin concentrations, low albumin levels, and low blood urea nitrogen values were identified as having an elevated influence and association with future surgery.
Conclusions
Using machine learning methods that incorporate current and historical data can predict the future risk of CD surgery.
Keywords: prediction models, Lasso, Crohn disease, complications
INTRODUCTION
Crohn disease (CD) is a chronic inflammatory bowel disease (IBD) affecting nearly 1 million patients in the United States, an estimated 30,000 of whom are veterans.1,2 The disease exhibits symptoms with a range of severity but may not always produce symptoms that alert the patient to take action. Regardless of these preceding symptoms, when chronic intestinal inflammation is inadequately controlled, progressive and irreversible bowel damage will occur. Potential complications of progressive damage are serious and include bowel obstruction from fibrotic strictures, intra-abdominal abscesses, fistulas, and bowel perforation.3-5 Unfortunately, these severe CD complications remain common and result in approximately 50% of all patients with CD eventually undergoing surgery within 10 years of diagnosis despite attempts at medication rescue.6 Beyond the need for relieving debilitating patient symptoms, caregivers are often charged with long-term prevention of these irreversible severe complications.
The goal of modern CD management is to employ sufficient medical therapy, early enough in the disease course, to both control symptoms and prevent severe structural complications that result in hospitalization, disability, and surgery.7 Alternatively, patients remaining symptomatic despite using substantial medical therapy may be best served by timely surgery rather than empiric cycling through alternative medical regimens that may be ultimately unsuccessful and only prolong symptoms. However, the benefits of surgery must be balanced against the noncurative aspect of surgery and the reduction of the intestinal absorptive surface area that becomes significant with repeated bowel resections. In addition, decision-making is also influenced by potential surgical complications including anastomotic leak and major wound infection, which occur in approximately 20% of patients with CD undergoing ileocecal resection.8 Predicting which patients will require or benefit most from the intensification of medical therapy compared to immediate surgery remains challenging.
When considering population health management strategies to improve CD care, identifying patients who are at risk of future surgery and severe complications is needed. Objective features of disease activity including laboratory biomarkers of inflammation, endoscopy, and imaging have been shown to be superior predictors of clinical outcomes compared to self-reported symptoms.9, 10 Biomarkers indicating persistent disease activity are associated with clinical outcomes of corticosteroid use, hospitalization, and surgery.11 However, unlike in research settings where laboratory values, imaging, and endoscopy are scheduled and frequently performed, in usual clinical care the frequency and timing of data collection are less uniform. Many tests, especially cross-sectional imaging and endoscopy, are often performed as a reaction to clinical symptoms.
Alternatively, laboratory studies are more frequently and routinely collected in patients with IBD as part of usual care. Though attention is often focused on C-reactive protein (CRP), erythrocyte sedimentation rate, and other biomarkers of inflammation, common laboratory tests including hemoglobin, platelets, and albumin can provide information regarding disease status and prognosis.11 Our aim was to examine the ability of readily available longitudinal clinical and laboratory data to yield personalized predictions of future complications requiring surgery in veterans with CD. We tested machine learning methods to handle unbalanced time-sensitive data with a goal of improving CD population health management using readily available clinical information collected in the course of usual care.
METHODS
Patient Selection
Using a cohort of Veterans with IBD from the 12 facilities comprising the Veterans Health Administration Veterans Integrated Service Networks (VISN) 10 (formerly VISNs 10 and 11) geographic region, patients between ages 18 and 89 years with CD were selected for analysis between January 1, 2001, through December 31, 2015. A CD diagnosis was verified by the presence of >2 CD administrative codes on 2 separate encounters and 1 or more IBD visits.12 Patients were required to have information on 18 different laboratory tests (detailed in the next section) for inclusion. The primary outcome was a CD-related bowel surgery, as defined by administrative current procedural terminology codes and detailed in Supplementary Data Content 1.
Preparation of Training Data
Clinical variables
Patient demographics collected included sex, race, and ethnicity. Medication use history was classified as current or prior use of corticosteroids, immunomodulators, aminosalicylates, immunomodulators (methotrexate and thiopurines), and anti-tumor necrosis factor (TNF) therapies. Medication combinations (eg, anti-TNFs and immunomodulators) were explored as interaction terms.
Laboratory data and variable preprocessing
Laboratory variables utilized in the models included the components of complete blood counts (white blood cell count; hemoglobin, hematocrit, and platelet count; lymphocytes; neutrophils; monocytes; eosinophils; basophils) and comprehensive metabolic panels (electrolytes, albumin, alanine aminotransferase, aspartate aminotransferase, bilirubin, alkaline phosphatase, blood urea nitrogen, creatinine). The inflammatory biomarkers CRP and erythrocyte sedimentation rate were not available for 59% and 43% of patients, respectively, which was common during the time period studied at these were not routinely ordered.
Laboratory variables were described in 2 temporal contexts: (1) the last laboratory value before the prediction of surgery and (2) historical laboratory values describing prior laboratory value behavior. Each laboratory variable’s historical behavior was described using summary statistics (median, minimum, maximum, mean) and fluctuation measures (slope between 2 consecutive measurements, slope of linear regression of observed history, normalized total variation). Principal component analysis (PCA) was performed on the historical laboratory value statistics as a preprocessing step before model-building. In general, PCA retains much of the variation while reducing the high dimensionality and collinearity between historical laboratory values. We performed PCA on each laboratory’s historical information, keeping the components that captured 90% of the variation for each laboratory test history. The PCA transformations of historical laboratory data were used in both training and testing data.
Time-sensitive prediction model architecture
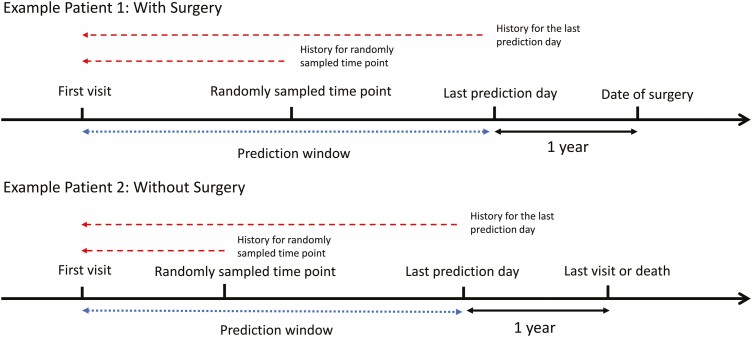
To make the model practical and reflective of clinical scenarios, we aimed to predict the risk of surgery in 1 year from a given clinic visit using the demographic, medication, and laboratory data collected. A prediction window for each patient was defined as the time between the first visit and 1 year before surgery or 1 year before the last day of available clinical data in those not undergoing surgery, as illustrated in Fig. 1. Two time points for prediction were selected for each patient: (1) the last day of the prediction window using the laboratory value closest to but before the last visit, and (2) a randomly selected clinic visit date within the prediction window. The purpose of including a randomly sampled time point was to reduce the bias of the model toward the patients who had surgery and to reflect the clinical scenarios encountered in clinical practice, where the time between visits is uneven. Therefore, each patient had 2 observations in the constructed dataset. Patients missing more than 25% of variables were excluded from the final dataset, with the missing data in the remaining patients imputed using random-forest imputation.13-15 Both the randomly sampled time point within the prediction window together with the last day of the prediction window were used to construct the model for predicting future CD-related surgery in 1 year, using all the variables described above.
FIGURE 1.
Illustration of prediction window at 2 separate time points used to model future CD-related surgery. The prediction window is composed of the time from the first clinic visit until 1 year before the surgery outcome or the date of the last visit. Two distinct time points were used to model outcome prediction: the last prediction day (1 year before surgery or last visit) and a random time point within the prediction window. In addition to single time points, the historical summary statistics of laboratory values before time points were also calculated and included in the models of surgery.
Model development and statistical analysis
Performance evaluations for all models used 10-fold cross-validation by patient identifier. We evaluated both lasso-regularized logistic regression and random-forest methods. The lasso-regularized logistic regression model was built using the following steps: (1) an initial 5-fold cross-validation within the subgroup of training data to identify the best tuning parameters, (2) optimization of model fit using the selected tuning parameter on the training data, and (3) calculation of the area under the receiver operating characteristic (AuROC) on the testing data. This procedure was repeated 10 times within each fold. The final AuROC was the average over the 10 repetitions performed, with the AuROC mean and SD reported. To evaluate the effect of the sampling step that randomly sampled time points on the prediction performance, the above procedure was repeated 30 times, each time constructing a dataset using a different random time point seed. In addition to cross-validation, we also tested our approach on 50 replications of random splits where 90% of the data were used as training and 10% were used as testing in a randomly chosen dataset from the 30 datasets constructed. To compare the prediction performances of the different models, we evaluated the AuROC on the testing data and counted the number of times that the AuROC of one model was greater than the other. These findings were then compared to the AuROC curves of the different models using a 1-sided paired Delong test.16 Finally, we compared model Brier scores, a global measure of model accuracy, using the multiple sampling approached described above.
RESULTS
Study Cohort Characteristics
Of 6092 observations constructed from 3046 patients meeting our definition of CD and including data from both randomly sampled visits and the last day of the prediction window, roughly 1100 observations (18.1%) were excluded because they were missing 25% or more of laboratory values. The final cohort contained 4950 observations from 2809 unique patients, of whom 93.4% were male with a median age of 59 years (interquartile ratio, 48-68 years; Table 1; full laboratory components in Supplementary Data Content 2). Of those patients included, laboratory values were imputed in 1.5% of the dataset. In this patient cohort, the median follow-up time was 8.5 years (interquartile ratio, 4.3-13.2 years), during which 256 (9.1%) patients underwent CD-related surgery. Corticosteroid exposure occurred in 41.7% of patients, 10.1% of patients in the cohort were exposed to anti-TNF therapy, and 4.6% used combination therapy.
TABLE 1.
Patient Characteristics Summary With Selected Laboratory Values
| Variables | No Surgery (n = 2553) | Surgery (n = 256) | ||
|---|---|---|---|---|
| Sex (M) | 2384 (93.4%) | 238 (93.0%) | ||
| Race | White | 1977 (77.4%) | 199 (77.7%) | |
| Black | 225 (8.8%) | 27 (10.5%) | ||
| Asian/Pacific islander | 34 (1.3%) | 2 (0.78%) | ||
| Unknown | 317 (12.4%) | 28 (10.9%) | ||
| Ethnicity | Hispanic | 15 (0.6%) | 1 (0.4%) | |
| Non-Hispanic | 2251 (88.1%) | 239 (93.4%) | ||
| Unknown | 287 (11.2%) | 16 (6.3%) | ||
| Medication History | 5-aminosalicylates | 1649 (64.6%) | 181 (70.7%) | |
| Steroids | 1023 (40.0%) | 149 (58.2%) | ||
| Anti-TNF | 244 (9.6%) | 40 (15.6%) | ||
| Thiopurine or methotrexate | 702 (27.5%) | 116 (45.3%) | ||
| Combination therapy | 112 (4.4%) | 17 (6.6%) | ||
| Laboratory | HCT | Last measurement | 40.38 (5.12) | 40.12 (4.75) |
| Historical mean | 40.43 (4.05) | 39.81(4.19) | ||
| WBC count | Last measurement | 7.58 (2.81) | 8.25 (3.07) | |
| Historical mean | 7.68 (2.10) | 8.46 (2.51) | ||
| PLATE | Last measurement | 226.0 (184.0-277.0) | 257.0 (207.8-330.2) | |
| Historical mean | 239.8 (199.5-284.8) | 277.1 (231.7-338.0) | ||
| ALB | Last measurement | 3.71 (0.55) | 3.68 (0.58) | |
| Historical mean | 3.86 (0.41) | 3.71 (0.50) | ||
| BUN | Last measurement | 16.0 (12.0-21.0) | 13.0 (10.0-16.0) | |
| Historical mean | 15.3 (12.4-19.4) | 12.8 (9.8-16.0) | ||
| MCHC | Last measurement | 33.19 (1.26) | 33.66 (1.15) | |
| Historical mean | 33.50 (0.90) | 33.61 (0.94) | ||
| Days from last visit to Surgery | N/A | 411.5 (383.0-464.2) |
The summary statistics are presented as count (%), mean (SD), or median (first quartile to third quartile).
ALB indicates albumin; BUN, blood urea nitrogen; HCT, hematocrit; MCHC, mean corpuscular hemoglobin concentration; PLATE, platelets; WBC, white blood cell.
Model Prediction Performance
The results of the 5 models to predict CD-related surgery are presented in Table 2. Models predicting surgery using demographic, medication use, and laboratory values to predict a CD-related surgery had a mean AuROC of 0.78 (SD, 0.002) and a sensitivity and specificity of 0.74 and 0.73, respectively. The combination of both a singular laboratory value time point and historical laboratory data (model 1) summary improved the performance compared to the model without historical data (model 2). The sensitivity and specificity were determined by using the cutoff value closest to the top-left corner of the ROC curve, corresponding to minimizing the quantity (1–sensitivity)2+ (1–specificity)2. Model comparison indicated that the models combining laboratory test data, demographic information, and medication history data (models 1 and 2) performed significantly better than the models using only demographic information and medication history (model 3) or laboratory test data only (model 4), or the random-forest model (model 5).
TABLE 2.
Comparison of Models Containing Different Data Elements for Predicting Future Surgery in CD
| Model | Sensitivity | Specificity | AuROC | Brier | AuROC* | Brier* |
|---|---|---|---|---|---|---|
| 1. Best model demographic + medication + last laboratory measurement + historical laboratory summary | 0.735 (0.013) | 0.726 (0.013) | 0.782 (0.0019) | 0.0451 (0.0002) | 0.775 (0.0447) | 0.0465 (0.0018) |
| 2. Demographic + medication + last laboratory measurement | 0.722 (0.011) | 0.714 (0.010) | 0.761 (0.0014) | 0.0455 (0.0002) | 0.761 (0.0446) | 0.0466 (0.0018) |
| 3. Demographic + medication | 0.631 (0.103) | 0.702 (0.012) | 0.714 (0.0016) | 0.0473 (0.0002) | 0.715 (0.0473) | 0.0482 (0.0012) |
| 4. Last laboratory measurement alone | 0.690 (0.009) | 0.670 (0.009) | 0.691 (0.0021) | 0.0477 (0.0002) | 0.673 (0.0494) | 0.0489 (0.0010) |
| 5. Random-forest method for all variables | 0.673 (0.017) | 0.652 (0.016) | 0.686 (0.0049) | 0.0488 (0.0002) | 0.675 (0.0526) | 0.0500 (0.0016) |
The first 4 columns of the table are based on cross-validation, reporting mean measures collected over 30 constructed datasets.
*The last 2 columns of the table are based on random splitting, reporting the mean value over 50 replications from a randomly chosen dataset.
Optimized Model Component Elements
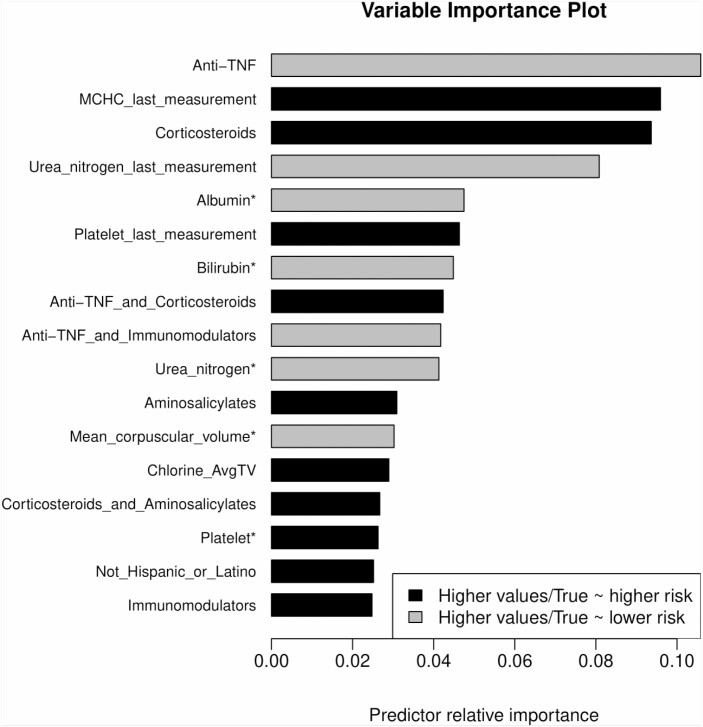
The final model selected roughly 40 features with an impact on predicting surgery. The model variable importance plot showed that anti-TNF use was associated with a lower probability of surgery within 1 year (log odds, 0.50) compared to patients without anti-TNF exposure (log odds, 0.73) in this cohort (Fig. 2). Further, anti-TNF use exhibited the strongest influence in this model for predicting surgery at 1 year from any time point. Although combination anti-TNF and immunomodulator use had less influence on the overall model relative to anti-TNF monotherapy, it was also associated with avoidance of surgery in 1 year. Interestingly, corticosteroid use had a strong influence on the model prediction of surgery after 1 year (log odds, 1.67) in that it was associated with an increased risk of surgery. Immunomodulator monotherapy and 5-aminosalicylate use had little effect on the likelihood of surgery. High platelet values, high mean cell hemoglobin concentration values, low albumin values, and low blood urea nitrogen values were also influential components in models predicting future surgery.
FIGURE 2.
Variable importance plot for time-dependent variables used to predict future surgery in CD. Anti-TNF use was associated with a lower probability of surgery within 1 year. Combination anti-TNF and immunomodulator use was also associated with avoidance of surgery at 1 year. Corticosteroid use had a strong influence on model predictions of surgery in 1 year. Immunomodulator monotherapy and 5-aminosalicylate use had little effect on the likelihood of surgery. High platelet values, high mean cell hemoglobin concentration values, low albumin values, and low blood urea nitrogen values were influential model components predicting future surgery. * First principal component.
DISCUSSION
We found that the application of machine learning methods to routinely available clinical and laboratory data can aid in the identification of patients at risk of undergoing future surgery for CD with a sensitivity and specificity of approximately 73% and an AuROC of 0.78. Unsurprisingly, anti-TNF use was protective against and corticosteroid use was a risk factor for future surgery in this CD cohort. In addition to the interpretation of data at a single point in time, we showed that incorporating a summarization of historical laboratory value behavior can improve outcome prediction model performance. Several analytic features of prior laboratory values, such as slope, PCA, and other trend-based behavior over time (eg, albumin levels) had a greater influence in the models compared to the single absolute laboratory value 1 year before surgery. Beyond improving outcome prediction models in CD, these results highlight the relevance and importance of incorporating historical data trends and behaviors into disease assessments and prognostic tools.
Although endoscopy, histology, and imaging are key biomarkers, they are performed infrequently and are often collected in the setting of acute symptomatic disease rather than on a scheduled protocol. Alternatively, traditional laboratory panels of complete blood counts and comprehensive metabolic panels, although not specifically tailored to IBD, are more frequently collected in consideration of drug toxicity monitoring, routine health assessment, and their low cost and availability. Routine laboratory parameters have been shown to be important surrogates of both IBD disease activity and prognosis.17 Platelets have been shown to be a good surrogate marker for subacute inflammation, in some instances reflecting inflammatory disease activity better than CRP.18, 19 Serum albumin is recognized as a biomarker of chronic bowel injury and reflects multifactorial malnutrition associated with increasingly severe CD.7, 20 Interestingly, in our study, the historical behavior of albumin (captured using the albumin PCA) had more importance for predicting surgical outcomes than single time point measurements, suggesting that the temporal context of albumin values over time is an important consideration. Although the need to account for laboratory value trends is intuitive, studies infrequently consider these historical laboratory behaviors in population-based analyses owing to the challenges of doing so using traditional statistical approaches.
Machine learning approaches to modeling real-world data can also provide insight into the relationship of medication use behavior and clinical outcomes and the relative importance of variables. Unsurprisingly, anti-TNF use was protective against surgical outcomes, although monotherapy seemed to be more protective than combination therapy with an immunomodulator. We hypothesize that the improved avoidance of surgery conferred by anti-TNF monotherapy vs combination therapy was not a function of efficacy but instead reflected the relatively late use of combination therapy in the CD course, by which time excessive bowel damage can occur.10, 21, 22 Similarly, the need for corticosteroid use in the setting of existing anti-TNF use may be a marker of excessive bowel damage and a lower probability of medication responsiveness rather than a corticosteroid-related effect. Interestingly, relative to the predictive utility of medication use, laboratory values reflective of nutritional status, including blood urea nitrogen and serum albumin, still have a meaningful impact on the future probability of surgery. Beyond quantifying the variable importance of readily available information, these results may identify medication use behaviors and opportunities to improve the selection of patients for early CD interventions.
Often, questions arise regarding the utility of different machine learning methodologies compared to traditional statistical methods. Although logistic regression is widely used for binary classification problems and is easy to interpret, it does not perform well when the number of predictors is large and variable relevance is unknown. Machine learning methods better handle large numbers of known and unknown predictors and define relationships between and within variables and the outcome of interest. Lasso-regularized logistic regression can identify linear relationships, whereas ensemble methods such as random-forest methods can model the nonlinear relations between the predictors and the outcome. Random-forest methods offer flexibility in the structure of predictors and require less preprocessing compared to the lasso-penalized logistic regression but at the cost of needing a larger sample size to achieve good performance. In general, when the sample size is moderate and the relation between the predictors is close to linear, the lasso-penalized logistic regression may be a better choice.
This work is subject to several limitations impacting the results. Accurate phenotyping information was not available on individual patients in this cohort. Intestinal stricturing, fistulas, disease distribution, and severity covariates can directly impact predictions of surgical outcomes but could not be retrieved in this dataset. In addition, we used an aggregate of all abdominal surgeries associated with IBD administrative codes as a single common surgical outcome event. However, the proposed models may perform differently based on the type or location of surgery performed. Further, prior CD-related surgeries may have been performed for included patients before their entry into the Veterans Health Administration, although we expect this to be infrequent among veterans utilizing the Veterans Health Administration. Finally, lasso methodologies may discard variables that clinicians believe are relevant or include variables where the biological cause of the laboratory variable’s influence on outcomes is difficult to determine, impacting model interpretability.
CONCLUSIONS
Machine learning analysis of routinely collected clinical and laboratory information can aid in predicting surgical outcomes in CD. Although new diagnostics and biological assays will be needed, maximizing the benefit of readily available and low-cost laboratory tests for predicting the disease course offers high value. In addition, this work reveals the potential for machine learning prediction models using “big data” analytics that are deployable in lower-resource areas. Finally, this work provides an example of machine learning analytics offering new insights into clinically relevant data patterns that are not readily apparent using traditional statistics.
Supplementary Material
Author contributions: RWS: study design, data analysis, data acquisition, data interpretation, drafting of manuscript, critical review of manuscript, study supervision. YL: data analysis, data interpretation, drafting of manuscript, review of manuscript. BE: data analysis, data acquisition, data interpretation, drafting of manuscript, critical review of manuscript. TV: data analysis, data acquisition, data interpretation, drafting of manuscript, critical review of manuscript. VK: data acquisition, drafting of manuscript, critical review of manuscript, study supervision. GLS: data interpretation, data analysis, critical review of manuscript. JZ: study design, data analysis, data interpretation, drafting of manuscript, critical review of manuscript, study supervision. AKW: study design, data analysis, data acquisition, data interpretation, drafting of manuscript, critical review of manuscript, study supervision.
Supported by: Work for this study was supported by the U.S. Department of Defense (CDMRP-PR151614; to RWS/AKW) and the National Institutes of Health (K23-DK101687; to RWS).
Conflicts of interest: RWS has served as a consultant for AbbVie, Janssen, Merck, and Takeda.
REFERENCES
- 1. Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2018;390:2769–2778. [DOI] [PubMed] [Google Scholar]
- 2. Hou JK, Kramer JR, Richardson P, et al. The incidence and prevalence of inflammatory bowel disease among U.S. veterans: a national cohort study. Inflamm Bowel Dis. 2013;19:1059–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kotze PG, Shen B, Lightner A, et al. Modern management of perianal fistulas in Crohn’s disease: future directions. Gut. 2018;67:1181–1194. [DOI] [PubMed] [Google Scholar]
- 4. Safar B, Sands D. Perianal Crohn’s disease. Clin Colon Rectal Surg. 2007;20:282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schwartz DA, Loftus EV Jr, Tremaine WJ, et al. The natural history of fistulizing Crohn’s disease in Olmsted County, Minnesota. Gastroenterology. 2002;122:875–880. [DOI] [PubMed] [Google Scholar]
- 6. Frolkis AD, Dykeman J, Negrón ME, et al. Risk of surgery for inflammatory bowel diseases has decreased over time: a systematic review and meta-analysis of population-based studies. Gastroenterology. 2013;145:996–1006. [DOI] [PubMed] [Google Scholar]
- 7. Stidham RW, Guentner AS, Ruma JL, et al. Intestinal dilation and platelet:albumin ratio are predictors of surgery in stricturing small bowel Crohn’s disease. Clin Gastroenterol Hepatol. 2016;14:1112–1119.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fumery M, Seksik P, Auzolle C, et al. ; REMIND study group investigators . Postoperative complications after ileocecal resection in Crohn’s disease: a prospective study from the REMIND group. Am J Gastroenterol. 2017;112:337–345. [DOI] [PubMed] [Google Scholar]
- 9. Colombel JF, Panaccione R, Bossuyt P, et al. Effect of tight control management on Crohn’s disease (CALM): a multicentre, randomised, controlled phase 3 trial. Lancet. 2018;390:2779–2789. [DOI] [PubMed] [Google Scholar]
- 10. Colombel JF, Sandborn WJ, Reinisch W, et al. ; SONIC Study Group . Infliximab, azathioprine, or combination therapy for Crohn’s disease. N Engl J Med. 2010;362:1383–1395. [DOI] [PubMed] [Google Scholar]
- 11. Peyrin-Biroulet L, Reinisch W, Colombel JF, et al. Clinical disease activity, C-reactive protein normalisation and mucosal healing in Crohn’s disease in the SONIC trial. Gut. 2014;63:88–95. [DOI] [PubMed] [Google Scholar]
- 12. Hou JK, Tan M, Stidham RW, et al. Accuracy of diagnostic codes for identifying patients with ulcerative colitis and Crohn’s disease in the Veterans Affairs Health Care System. Dig Dis Sci. 2014;59:2406–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Waljee AK, Mukherjee A, Singal AG, et al. Comparison of imputation methods for missing laboratory data in medicine. BMJ Open 2013;3:e002847. doi: 10.1136/bmjopen-2013-002847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Friedman J, Hastie T, Tibshirani R.. The Elements of Statistical Learning. Vol. 1. Spinger Series in Statistics. New York, NY: Springer; 2001. [Google Scholar]
- 15. Stekhoven DJ, Bühlmann P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28:112–118. [DOI] [PubMed] [Google Scholar]
- 16. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 17. Waljee AK, Lipson R, Wiitala WL, et al. Predicting hospitalization and outpatient corticosteroid use in inflammatory bowel disease patients using machine learning. Inflamm Bowel Dis. 2017;24:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shen EX, Lord A, Doecke JD, et al. A validated risk stratification tool for detecting high-risk small bowel Crohn’s disease. Aliment Pharmacol Ther. 2020;51:281–290. [DOI] [PubMed] [Google Scholar]
- 19. Takeyama H, Mizushima T, Iijima H, et al. Platelet activation markers are associated with Crohn’s disease activity in patients with low C-reactive protein. Dig Dis Sci. 2015;60:3418–3423. [DOI] [PubMed] [Google Scholar]
- 20. Hyams J, Markowitz J, Otley A, et al. ; Pediatric Inflammatory Bowel Disease Collaborative Research Group . Evaluation of the pediatric crohn disease activity index: a prospective multicenter experience. J Pediatr Gastroenterol Nutr. 2005;41:416–421. [DOI] [PubMed] [Google Scholar]
- 21. Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc2011;73:273–282. [Google Scholar]
- 22. Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.