Abstract
Mammalian oocytes and embryos rely exclusively on maternal mRNAs to accomplish early developmental processes. Since oocytes and early embryos are transcriptionally silent after meiotic resumption, most of the synthesised maternal mRNA does not undergo immediate translation but is instead stored in the oocyte. Quantitative RT-PCR is commonly used to quantify mRNA levels, and correct quantification relies on reverse transcription and the choice of reference genes. Different methods for reverse transcription may affect gene expression determination in oocytes. In this study, we examined the suitability of either random or oligo(dT) primers for reverse transcription to be used for quantitative RT-PCR. We further looked for changes in poly(A) length of the maternal mRNAs during oocyte maturation. Our data indicate that depending on the method of reverse transcription, the optimal combination of reference genes for normalisation differed. Surprisingly, we observed a shortening of the poly(A) tail lengths of maternal mRNA as oocytes progressed from germinal vesicle to metaphase II. Overall, our findings suggest dynamic maternal regulation of mRNA structure and gene expression during oocyte maturation and early embryo development.
Keywords: maternal mRNA /, bovine embryo /, qRT-PCR /, reverse transcription /, gene expression /, poly(A) tail
Introduction
Early embryonic development is primarily controlled by maternal mRNAs, which are produced by the oocyte during oogenesis (Vastenhouw et al., 2019). Unlike in somatic cells, most mRNA synthesised in an oocyte is not immediately translated but is stored for future events (Hamatani et al., 2004). After breakdown of the germinal vesicle (GV), the chromatin in an oocyte becomes condensed, resulting in transcriptional silencing of meiotic oocytes (Tan et al., 2009; Dumdie et al., 2018). This period of reduced transcription lasts until the maternal to zygotic transition (MZT), when the zygotic genome is activated and maternal mRNA is largely degraded. Before the MZT, oocytes and embryos therefore rely on maternal mRNA to accomplish dynamic events accompanying final oocyte maturation, fertilisation and early embryo development (Winata and Korzh, 2018). In short, precise regulation of maternal mRNA dynamics plays an essential role during maturation of the oocyte and early embryo development. Precise regulation of maternal mRNA expression ensures that dynamic events required for successful oocyte maturation and early embryo development can be accomplished; quantitative reverse transcription PCR (qRT-PCR) is a useful method for monitoring the regulation of (maternal) mRNA abundance or expression. It is routinely used in life science research because of its sensitivity and cost-effectiveness and has proven to be a useful method to quantify gene expression levels in sparse or limited RNA samples such as that from oocytes and embryos.
One of the key steps in qRT-PCR is the synthesis of complementary DNA (cDNA) by reverse transcription, which should result in a cDNA pool that quantitatively reflects the original mRNA copy number (Stangegaard et al., 2006). There are two major priming strategies for reverse transcription used in qRT-PCR, namely random primers and oligo(dT) primers. Random primers with randomly ordered base sequences can potentially anneal to any RNA species, at any position (from 5′ to 3′). Reverse transcription based on oligo(dT) primers can only anneal to the 3′ poly(A) tail of RNA. Maternal mRNA is, however, usually stored in a short poly(A) tail state (Mendez and Richter, 2001; Stangegaard et al., 2006). It is therefore possible that, depending on the method of synthesis, cDNA may not quantitatively reflect the population of maternal mRNA available for translation.
Normalisation is an essential step when analysing gene expression to account for factors such as the total amount of mRNA recovered from different samples (Evans et al., 2018). Even though selection of reference genes for normalisation in preimplantation embryos has been documented in various mammalian species (Goossens et al., 2005; Kuijk et al., 2007; Mamo et al., 2007), the possible effect of different reverse transcription priming strategies has not been analysed in detail.
Translational control of maternal mRNA is achieved mainly by polyadenylation and deadenylation, and in particular by regulating the length of the poly(A) tail (Eichhorn et al., 2016; Winata and Korzh, 2018). The regulation of maternal mRNA availability has been investigated primarily in Drosophila, Xenopus and the mouse (Sallés et al., 1994; Mendez and Richter, 2001; Richter, 2007; Morgan et al., 2017). It has been reported that, in immature oocytes, maternal mRNA is kept in a relatively short poly(A) tail state, stored in cytoplasmic granules (Anderson and Kedersha, 2009; Winata and Korzh, 2018). Since the oocyte is transcriptionally silent during and after its nuclear maturation, it has been hypothesised that stored maternal mRNA is released from RNA granules when translation is required (Anderson and Kedersha, 2009; Kotani et al., 2013; Winata and Korzh, 2018).
Oocyte progression through meiosis is highly dependent on the activity of maturation-promoting factor (MPF), a complex of the regulatory subunit cyclin B, coded for by the CCNB gene and the catalytic subunit cyclin-dependent kinase 1 (CDK1). An increase in MPF activity from germinal vesicle breakdown (GVBD) to the metaphase I (MI) and metaphase II (MII) stages is achieved by temporally controlled synthesis of cyclin B from stored maternal mRNA (Ihara et al., 1998; Mendez and Richter, 2001; Nakahata et al., 2003; Nagahama and Yamashita, 2008). Recently, it has been reported that a high CCNB mRNA translation rate is associated with elongation of the CCNB poly(A) tail in mouse oocytes (Kotani et al., 2013; Daldello et al., 2019). Cyclin A (coded by CCNA) can also bind to CDK1 and regulate the activity of MPF during oocyte maturation (Li et al., 2019). Moreover, a recent study demonstrated that cyclin A1 expression prevents segregation of chromosomes and anaphase entry (Radonova et al., 2020).
We hypothesised that different priming strategies for reverse transcription may result in different fidelities of cDNA generation when using samples from oocytes or embryos. We chose cyclin genes to study, because they are highly regulated during oocyte maturation, which might lead to different gene expression patterns if different reverse transcription strategies were performed. We also focused on CDK2 and EIF4A3, since they play important role during oocyte maturation and early embryo development. We therefore extracted mRNA from bovine oocytes and embryos, and synthesised cDNA using random primers and oligo(dT) primers. We used qRT-PCR to examine the gene expression patterns of cyclin genes and examined poly(A) tail length of various genes in oocytes from the GV to the MII stage (Supplementary Fig. S1). Our data uncovered differences in poly(A)tail length of mRNA during oocyte maturation and early embryo development. It is concluded that one should be critical in deciding which primer-type to use for reverse transcription when gene expression levels are examined in oocytes and pre-MZT embryos.
Materials and methods
Bovine in vitro embryo culture and sample collection
Bovine ovaries were collected from a local slaughterhouse, rinsed with water and kept in 0.9% NaCl (B. Braun, Melsungen, Germany) supplemented with penicillin/streptomycin (100 µg/ml) (Gibco, Paisley, UK) at 30°C during processing. Cumulus–oocyte complexes (COCs) were aspirated from follicles with a diameter of 2–8 mm and identified using a stereomicroscope. The COCs were matured in vitro as described previously (Brinkhof et al., 2015) and for subsequent analysis, GV oocytes were collected immediately after COC recovery while GVBD, MI and MII stage oocytes were collected at 6, 12 and 23 h of invitro maturation, respectively; in all cases, cumulus cells were removed by vortexing. Invitro fertilisation was done as described (Brinkhof et al., 2015). In short, after 23 h maturation, COCs were transferred to fertilisation medium. Motile sperm cells were introduced into the fertilisation medium at a final concentration of 1 × 106 per ml, and this was considered as day 0. After incubation with sperm for 20–22 h, presumptive zygotes were denuded of their cumulus cells by vortexing for 3 min, and then cultured further in synthetic oviductal fluid (SOF) (Brinkhof et al., 2017) in a humidified atmosphere containing 5% CO2 and 7% O2 at 39°C. At day 5 after fertilisation, developing embryos were transferred to fresh SOF for further culture until day 8.
RNA extraction and cDNA generation
Oocytes or embryos in pools of 50 were rinsed in PBS and stored in 200 μl RLT lysis buffer (Qiagen, Valencia, CA, USA) at −80°C until RNA extraction. Total RNA isolation was performed using the RNeasy Micro Kit (Qiagen) according to the manufacturer’s instruction. Reverse transcription was carried out directly after RNA isolation, using two different priming strategies. Reverse transcription (RT) mixtures were prepared from 10 µl of the RNA sample, 4 µl of 5 × RT buffer (Invitrogen, Breda, the Netherlands), 10 mM DTT (Invitrogen), 0.5 mM dNTP (Promega, Leiden, the Netherlands), 8 units RNAsin/RNAse inhibitor (Promega) and 150 units Superscript III reverse transcriptase (Invitrogen) in a total volume of 20 µl, supplemented with 1.8 units per ml random primers (Invitrogen) or 2.5 μM oligo(dT) primers (Invitrogen) respectively. Minus-RT controls were made up of the same reagents as above, using primers but without the reverse transcriptase. The mixtures were incubated at 70°C for 5 min, followed by 1 h at 50°C and 5 min at 80°C. Samples were subsequently stored at −20°C until further analysis.
Quantitative PCR
All amplification reactions were performed on three independent cDNA samples or -RT blanks in duplicate, following the manufacturer’s protocol of the CFX detection system (Bio-Rad, Hercules, CA, USA). The reaction mixture contained 1 µl cDNA, 9 µl RNAse- and DNAse-free water (Invitrogen) and 10 µl iQ SYBR Green supermix (Biorad) with a final primer concentration of 500 nM. Initial denaturation took place for 3 min at 95°C, followed by 40 cycles of 95°C for 20 s, the primer specific annealing temperature (Supplementary Table S1) for 20 s, and elongation at 72°C for 20 s. To determine the quantitative amplification efficiency, standard curves for each primer pair were made by four-fold dilution series of cDNA from 400 oocytes or 100 blastocysts.
Poly(A) tail assay
Poly(A) tail assays were performed as described (Rassa et al., 2000) with a few modifications. Total RNA was extracted from 50 GV or MII oocytes as described above. After denaturation at 70°C, the mRNA was ligated with 50 pmol of primer GB-135 (5′-P-GGTCACCTTGATCTGAAGC-NH2-3′) (Eurogentec, Maastricht, the Netherlands) at 37°C for 1 h in a total volume of 20 µl using T4 RNA ligase (New England Biolabs, Ipswich, MA, USA). GB-135 contained a 3′ amino modification to block ligation at this end. To inactivate RNA ligase, samples were boiled at 100°C for 5 min and cooled on ice. Reverse transcription was performed as described above using 50 pmol of primer GB-136 (5′-GCTTCAGATCAAGGTGACCTTTTT-3′) (Eurogentec), and the anchored cDNA was used for amplification. First round amplification was performed using a gene-specific primer (P1) (Supplementary Table S2) and GB-136. The product of the first round amplification was used as template for the second round amplification, using a gene-specific primer starting after the 3′ site of P1 (P2) (Supplementary Table S2) and GB-136. The PCR was performed as described above with 40 cycles for first round amplification and 20 cycles for second round amplification. Samples were electrophoresed on 1.0% agarose (Eurogentec) gels and visualised with ethidium bromide (Invitrogen).
Transformation and DNA sequencing
The PCR products resulting from the second round amplification as described above were inserted into the pGEM-T Easy vector (Promega) using T4 DNA ligase (New England Biolabs) and transformed into Escherichiacoli JM109 competent cells (Promega) by heat-shock transformation according to the manufacturer’s instructions. Transformants were selected on LB/Ampicillin/IPTG/X-Gal plates according to the manufacturer’s instructions. At least 12 clones from each reaction were examined by digestion with Eco52I (Thermo Fisher Scientific, Eindhoven, the Netherlands) restriction followed by sequencing in case of the correct insert size.
DNA sequencing reactions were conducted using T7 primer (5′-TAATACGACTCACTATAGGG-3′) (Eurogentec) according to the manufacturer’s instructions for the BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Beverly, MA, USA), and determined using a 3500XL Genetic Analyser (Applied Biosystems). DNA sequencing results were analysed using Sequencing Analysis Software v6.0 (Thermo Fisher Scientific) and aligned by cluster W method using DNASTAR Lasergene 14 (DNASTAR, Madison, WI, USA).
Statistical analysis
Stability analysis for the reference genes was performed using geNorm (Vandesompele et al., 2002). Data from the PCRs were tabulated in Microsoft Excel and statistical differences were examined using GraphPad Prism 7 (https://www. graphpad.com/scientific-software/prism/). Pools of embryos from three biological replicates were analysed for gene expression. Normal distributions of data sets were determined by the Shapiro–Wilk tests. Differences between two groups were analysed by the Mann–Whitney test or, in the case of multiple groups, by one-way ANOVA followed by a posthoc Tukey test. In bar graphs, differences between groups are indicated with different letters above the bars, with statistical significance set as P < 0.05. Asterisks indicate levels of significance (*P < 0.05, **P < 0.005, ***P < 0.0005 and ****P < 0.0001). Error bars indicate standard deviation.
Results
Stability of potential reference genes during oocyte maturation
As different priming strategies for reverse transcription in oocytes may result in different pools of cDNA, we hypothesised that suitable reference genes for normalisation are different between cDNA synthesised using random primers and cDNA generated using oligo(dT) primers. To determine whether the optimal set of stably expressed reference genes differs in differently generated cDNA samples from GV, GVBD, MI and MII oocytes, expression of six commonly used reference genes (ACTB, GAPDH, HPRT1, RPL15, SDHA and YWHAZ) was analysed using qRT-PCR and the software packages of geNorm (Vandesompele et al., 2002).
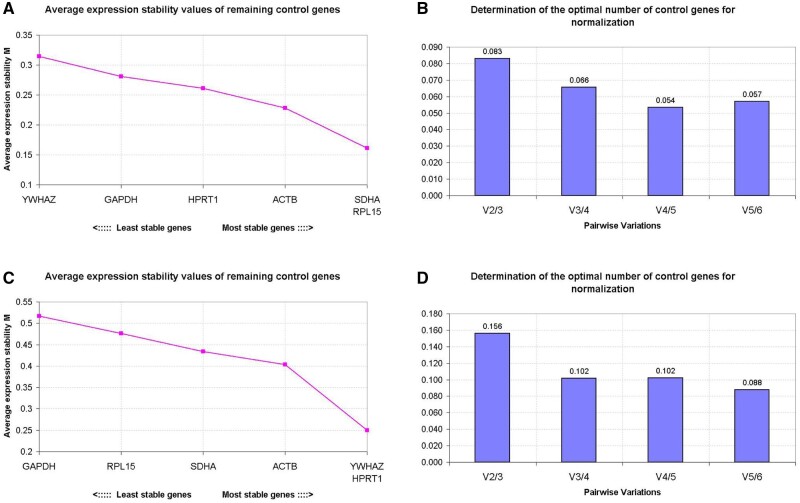
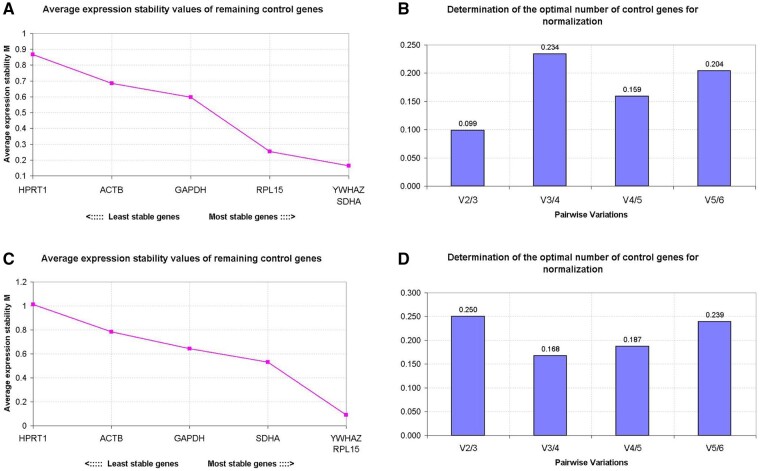
Using geNorm, gene expression stability can be evaluated by the average expression stability (M value), with a low M value indicating high stability of expression. In cDNA samples synthesised using random primers, RPL15 and SDHA were the most stably expressed, followed by ACTB, HPRT1, GAPDH and YWHAZ (Fig. 1A). The highest stability of gene expression in cDNA samples synthesised using oligo(dT) primers was recorded for HRPT1 and YWHAZ, followed by ACTB, SDHA, RPL15 and GAPDH (Fig. 1C).
Figure 1.
Stability of potential reference genes during oocyte maturation. The calculation of the average expression stability (M value) of candidate reference genes as determined by quantitative RT-PCR. The y-axis presents the M value and the x-axis presents the ranking of reference genes in order of increasing stability (from left to right); cDNA samples were synthesised with random primers (A) or oligo(dT) primers (C). The optimal number of reference genes for normalisation was determined by Pairwise variation (V) between the normalisation factors (Vn and Vn + 1). The optimal number of reference genes is shown for cDNA samples synthesised with random primers (B) or oligo(dT) primers (D). Y-axis represents the v-value indicating the pairwise variation between two sequential normalisation factors. Samples were derived from pools of 50 oocytes with three biological replicates.
To determine the optimal number of reference genes for accurate normalisation, the pairwise variation (V) was calculated between two sequential normalisation factors. In cDNA samples synthesised with either random primers and oligo(dT) primers, the inclusion of a third gene improved normalisation (high V2/3 value), but the inclusion of a fourth gene had little effect (low V3/4 value) (Fig. 1B and D). Overall, the optimal combination of reference genes for normalisation in cDNA samples differed between cDNA synthesised using random primers and oligo(dT) primers. With random primers, the combination of RPL15, SDHA and ACTB was favoured, whereas in cDNA samples synthesised using oligo(dT) primers, it was HPRT1, YWHAZ and ACTB.
Gene expression patterns during oocyte maturation
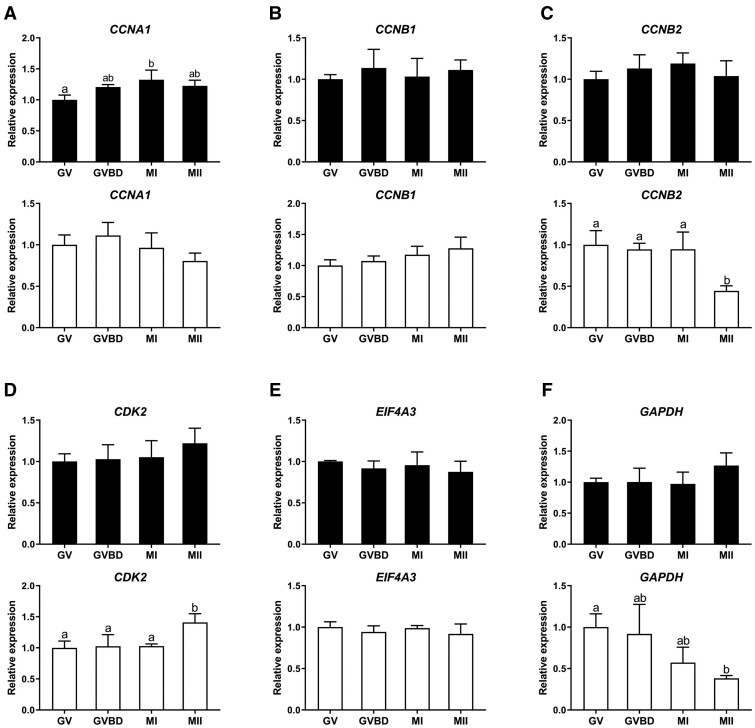
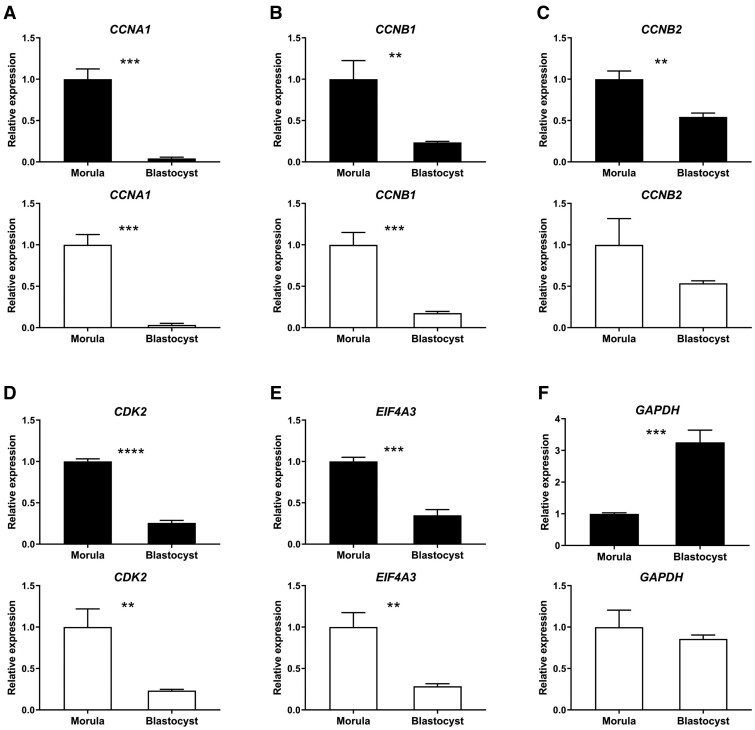
We next focused on the expression patterns of CCNA1, CCNB1, CCNB2, CDK2 and EIF4A3 in oocyte cDNA samples synthesised using random primers and oligo(dT) primers, since these genes play important roles during oocyte and early embryo development (Mendez and Richter, 2001; Jansova et al., 2018). In cDNA samples synthesised with random primers, the expression of CCNA1, CCNB1, CCNB2, CDK2 and EIF4A3 was relatively stable throughout maturation from GV to MII (Fig. 2A–E). Even though expression of CCNA1 increased significantly from the GV to the MI stage, the GV/MI ratio of CCNA1 expression was only 1.32 (Fig. 2A). In cDNA samples synthesised using oligo(dT) primers, the expression level patterns for CCNA1, CCNB1, CDK2 and EIF4A3 were similar to those for cDNA synthesised using random primers (Fig. 2A–C and E). By contrast, the expression of CCNB2 decreased significantly from the MI to the MII stage (Fig. 2C).
Figure 2.
Gene expression patterns during oocyte maturation. The relative expression of genes in cDNA samples synthesised using random (black bars) or oligo(dT) primers (white bars) from bovine GV to MII oocytes. (A) CCNA1, (B) CCNB1, (C) CCNB2, (D) CDK2, (E) EIF4A3, (F) GAPDH. GV, GVBD, MI and MII, respectively refer to germinal vesicle, germinal vesicle breakdown, metaphase I and metaphase II stages. Relative expression in GV oocytes was set as 1. Significant differences between groups are indicated by different letters above the bars (P < 0.05) and error bars represent standard deviation. ACTB, RPL15 and SDHA were used for normalisation in cDNA samples synthesised using random primers; ACTB, HPRT1, and YWHAZ were used for normalisation in cDNA samples synthesised using oligo(dT) primers. Samples were collected from pools of 50 oocytes with three biological replicates.
We then examined the relative expression of GAPDH in oocytes from GV to MII, since GAPDH expression was not used for normalisation. The patterns of apparent oocyte GAPDH expression were similar from GV to MII in cDNA samples synthesised with random primers (Fig. 2F). However, to our surprise, GAPDH expression levels decreased stepwise from the GV to MII stage in cDNA samples synthesised using oligo(dT) primers (Fig. 2F).
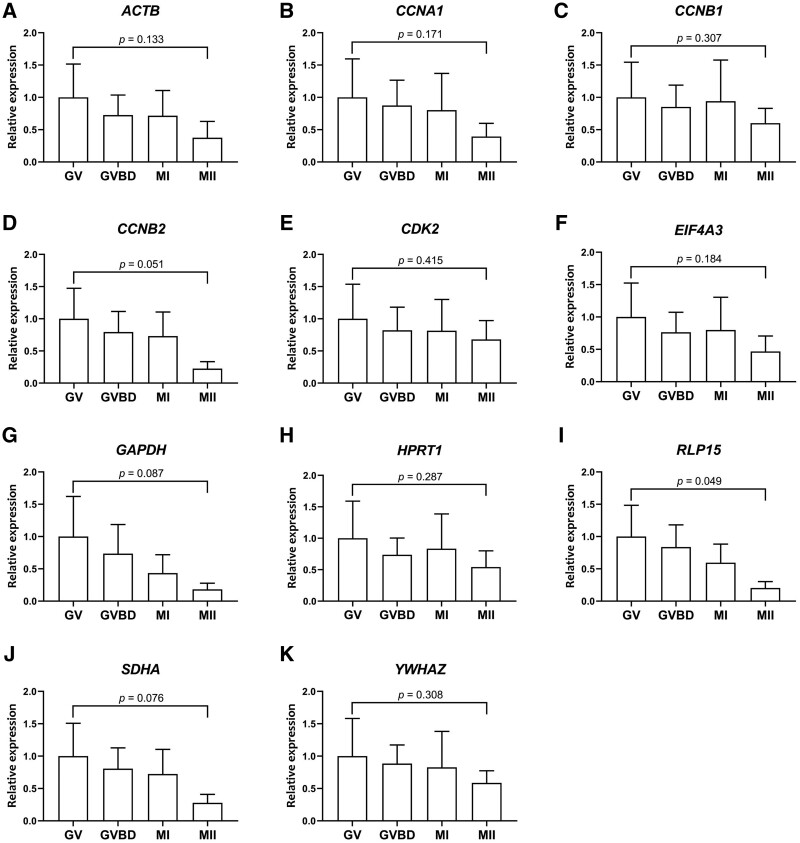
We therefore examined whether the expression levels of other potential reference genes also varied from GV to MII in cDNA samples synthesised using oligo(dT) primers. Because RNA was extracted from groups of 50 oocytes, the relative expression levels were first compared without normalisation. Unexpectedly, the expression levels of both the reference and other (target) genes examined decreased as oocytes matured from the GV to the MII stage (Fig. 3A–K). Due to a high standard deviation among the three biological replicates, these differences were not statistically significant, expect in the case of RLP15 (P < 0.05), but the pattern was consistent for all genes examined. For comparison, the expression levels of reference and target genes, from GV to MII in cDNA samples synthesised using oligo(dT) primers, are shown in Supplementary Fig. S5.
Figure 3.
Gene expression patterns during oocyte maturation using oligo(dT) primers. Relative expression in cDNA samples synthesised using oligo(dT) primers in maturing bovine oocytes, as determined by quantitative RT-PCR without normalisation. (A) ACTB, (B) CCNA1, (C) CCNB1, (D) CCNB2, (E) CDK2, (F) EIF4A3, (G) GAPDH, (H) HRRT1, (I) RLP15, (J) SDHA, (K) YWHAZ. GV, GVBD, MI, MII, respectively refer to germinal vesicle, germinal vesicle breakdown, metaphase I and metaphase II stages. Absolute expression in GV oocytes is set at 1. Error bars represent standard deviation. Samples were collected from pools of 50 oocytes with three biological replicates.
Poly(A) tail length regulation during oocyte maturation
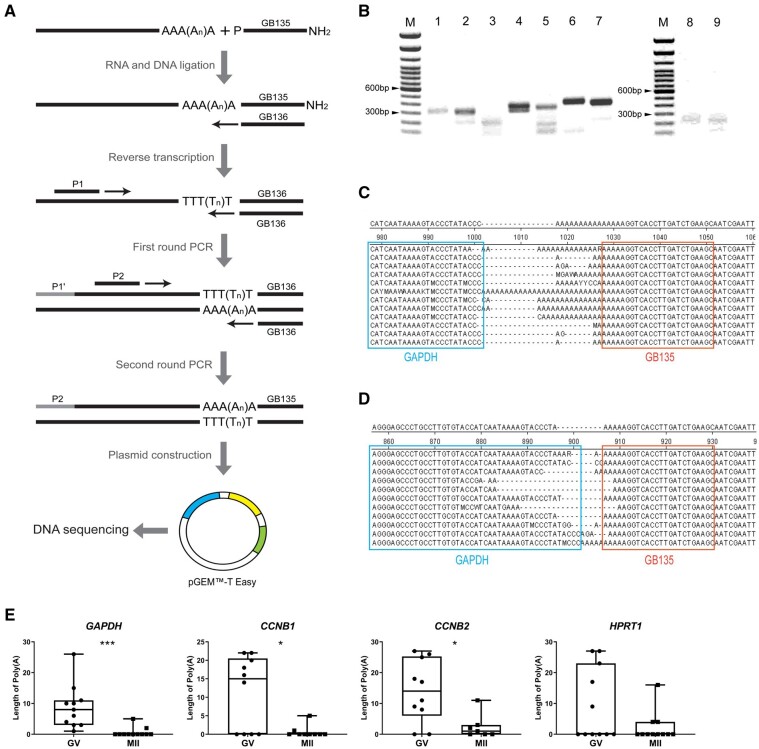
Because oocytes rely for a relatively long time on maternal mRNA during and after oocyte maturation, it is not unlikely that the poly(A) tail length is regulated during oocyte maturation. We therefore conducted poly(A) tail length assays (Rassa et al., 2000) to investigate differences in the length of the poly(A) tails of GAPDH, CCNB1, CCNB2 and HPRT1 mRNA at different stages of oocyte maturation. For the poly(A) tail length assays, a nested PCR was performed using gene-specific forward primers with the addition of GB136 to increase the specificity of the PCR products (Fig. 4A). Second round PCR products were then size-separated by agarose gel electrophoresis. The PCR products from GV oocytes showed slightly slower mobility than products from the MII oocytes in all of the genes we examined (Fig. 4B, Supplementary Fig. S2), indicating that mRNA in GV oocytes contained longer poly(A) tails than in MII oocytes.
Figure 4.
Poly(A) tail length regulation during oocyte maturation. (A) Schematic representation of the polyadenylation assay and poly(A) sequencing. mRNAs were ligated to a synthetic primer containing a 3′ amino blocking group (GB135). The ligation products are then used as templates for a reverse transcription reaction using GB136 primers complementary to GB135. To amplify cDNA and increase the specificity of the amplicons, first round amplification was performed using a gene-specific primer (P1) and GB136, and second round amplification was performed using a gene-specific primer which started after the 3′ site of P1 (P2) and GB136. The second round amplified product was then ligated with pGEMTM-T Easy plasmid, followed by transformation with JM109 cells before DNA sequencing. (B) Agarose gel electrophoresis of second round amplified products. Lane M: 100 bp ladder; Lane 1: positive control; Lanes 2–3: CCNB1 at germinal vesicle (GV) and metaphase II (MII) stage; Lanes 4–5: CCNB2 at GV and MII stage; Lanes 6–7: GAPDH at GV and MII stage; Lanes 8–9: HPRT1 at GV and MII stage. Uncropped blots are provided is Supplementary figure S2. DNA sequencing result for GAPDH at GV stage (C) and MII stage (D) aligned by cluster W method. The end sequence of GAPDH is outlined with the cyan box and sequence of GB135 is included in the orange box. (E) Length of poly(A) tail of GAPDH, CCNB1, CCNB2 and HPRT1 at the GV and MII stages. Significant differences are indicated by asterisks (*P < 0.05, ***P < 0.0005) and error bars represent standard deviation. Samples were collected from pools of 50 oocytes with three biological replicates.
To further confirm the decrease in poly(A) tail length during oocyte maturation, we performed DNA sequencing of amplicons (Fig. 4A). We determined that the length of the poly(A) tails ranged from 0 to 29 nucleotides (Fig. 4C and D). We further compared the poly(A) tail length of GAPDH, CCNB1, CCNB2 and HPRT1 mRNA in oocytes at the GV and MII stages. Consistent with the agarose gel electrophoresis results, significantly longer poly(A) tails were detected for GAPDH, CCNB1 and CCNB2 mRNA in GV oocytes, compared with MII oocytes (Fig. 4E). The poly(A) tail of HPRT1 was also shortened in MII compared to GV oocytes but this difference was not statistically significant (Fig. 4E).
Reference gene selection and gene expression patterns in morulae and blastocysts
Our next aim was to identify the most suitable reference genes for normalisation, and to examine expression patterns for specific target genes in morulae and blastocysts. Maternal mRNA is largely degraded soon after major zygotic genome activation, which occurs at around the 8-cell stage in cattle embryos, similar to that in human embryos (Niakan et al., 2012). SDHA and YWHAZ were the most stably expressed genes when cDNA was synthesised using random primers, while RPL15 and YWHAZ were most stably expressed in cDNA synthesised using oligo(dT) primers (Fig. 5A and C). However, the three most stably expressed candidate reference genes were the same for cDNA synthesised using random primers or oligo(dT) primers, namely RPL15, SDHA and YWHAZ (Fig. 5A–D).
Figure 5.
Reference gene selection in morulae and blastocysts. The geNorm analysis of the average expression stability (M value) of all candidate reference genes determined by quantitative RT-PCR; cDNA samples were synthesised using random primers (A) or oligo(dT) primers (C). More stable reference genes are positioned on the right side of the diagram, with less stable genes on the left side. The optimal number of reference genes for normalisation was determined by Pairwise variation (V) between the normalisation factors (Vn and Vn + 1). The optimal number of reference genes is shown for cDNA samples synthesised with random primers (B) or oligo(dT) primers (D). Y-axis represents the v-value indicating the pairwise variation between two sequential normalisation factors. Samples were collected from pools of 50 embryos with three biological replicates.
We therefore used a combination of RPL15, SDHA and YWHAZ to normalise gene expression in embryos. The expression patterns of CCNA1, CCNB1, CCNB2, CDK2 and EIF4A3 in morulae and blastocysts were very similar between cDNA samples synthesised using random primers and oligo(dT) primers (Fig. 6A–E). Surprisingly, the expression of GAPDH increased markedly from morula to blastocyst in cDNA samples synthesised using random primers, but did not differ in cDNA samples synthesised using oligo(dT) primers (Fig. 6F).
Figure 6.
Gene expression patterns in morulae and blastocysts. The relative expressions of genes in cDNA samples synthesised using random primers (black bars) or oligo(dT) primers (white bars) in bovine morulae and blastocysts. (A) CCNA1, (B) CCNB1, (C) CCNB2, (D) CDK2, (E) EIF4A3, (F) GAPDH. Relative expression in morulae set at 1; **P < 0.005, ***P < 0.0005 and ****P < 0.0001 indicate significant differences between morulae and blastocysts and error bars represent standard deviation. RPL15, SDHA and YWHAZ were used for normalisation for cDNA samples synthesised with both random primers and oligo(dT) primers. Samples were collected from pools of 50 embryos with three biological replicates.
We also included 8-cell embryos to identify the best reference genes for normalisation and expression patterns from 8-cell embryos to blastocysts. The three most stably expressed reference genes were still RPL15, SDHA and YWHAZ in cDNA samples synthesised with random primers while GAPDH, SDHA and YWHAZ were the most suitable reference genes for normalisation in cDNA samples synthesised with oligo(dT) primers (Supplementary Fig. S3). Expression patterns of CCNB1, CCNB2 and CDK2 were similar from 8-cell embryos to blastocysts between cDNA samples synthesised using random primers and oligo(dT) primers (Supplementary Fig. S4B–D). On the hand, the expression of CCNA1 and EIF4A3 was down-regulated from the 8-cell embryo to the blastocyst stage in cDNA samples synthesised using random primers, whereas expression of CCNA1 and EIF4A3 was significantly elevated at the morula stage in cDNA samples synthesised using oligo(dT) primers (Supplementary Fig. S4A and E).
Discussion
Proper quantification of mRNA expression levels with qRT-PCR relies on the use of stably expressed reference genes (Goossens et al., 2005; Kuijk et al., 2007; Mamo et al., 2007). A critical step in the process, in particular reverse transcription, has however been less well studied. The RNA concentration, quality and the type of reverse transcriptase can influence reverse transcription, but when samples received the same experimental handling and relative levels are determined, quantification can be reliable (Cholet et al., 2020). Here, we demonstrate that the combination of genes optimal for normalisation is dependent on the priming strategy, i.e. random hexamers or oligo(dT), used for reverse transcription in oocytes. Bovine oocytes were used, since these can be collected in fairly large quantities from leftover slaughterhouse ovaries. In addition, the oocytes can be efficiently fertilised in vitro and embryos cultured to the blastocyst stage. No laboratory animals were therefore required, in accordance with the 3Rs of animal experimentation. Above all, the timing of bovine oocyte maturation and preimplantation development is very similar to that of human oocytes and embryos.
Selection of reference genes for normalisation is an essential step for accurate gene expression analysis. The software program geNorm was used to determine stability of gene expression. Comparisons with other programs such as BestKeeper and NormFinder, revealed that they gave very similar results (Spinsanti et al., 2006; De Spiegelaere et al., 2015). More importantly, geNorm is the most commonly used normalisation algorithm, because geNorm does not need large data sets and the raw data for this program do not need to be normally distributed (Mehdi Khanlou and Van Bockstaele, 2012). Even though the selection of reference genes for normalisation in bovine oocytes has been documented (Goossens et al., 2005; Khan et al., 2016; Caetano et al., 2019), the effect of different reverse transcription priming strategies to the choice of reference genes has not been addressed in detail. It has been reported that the combination of suitable reference genes includes HPRT1 and B2M in bovine oocyte cDNA samples synthesised using random primers (Caetano et al., 2019), while the ideal reference genes were ACTB and GAPDH in bovine oocyte cDNA samples synthesised using oligo(dT) primers (Khan et al., 2016). Consistently, in our study, even though the same original mRNA and system were applied, suitable reference genes for normalisation were different if reverse transcription priming strategies were different.
In morulae and blastocysts, the combination of suitable reference genes seems to be less dependent on the priming strategy. This may be due to the large numbers of maternal mRNA transcripts present in oocytes and very early embryos, and the degradation of these maternal transcripts following the onset of zygotic gene expression at around the 8-cell stage in bovine embryos. To demonstrate that this difference is dependent on the method used for reverse transcription but not the efficiency of RNA extraction, we split RNA samples into two equal parts for the different reverse transcription strategies after extraction. Interestingly, it has been reported that GAPDH, PPIA, ACTB, RPL15, GUSB and H2A.2 are not suitable reference genes for normalisation, because of their inconstant levels throughout preimplantation development (Ross et al., 2010). In our study, the suitable reference genes for normalisation in oocytes were different from those in morulae and blastocysts even when reverse transcription is performed using the same priming strategy. We suggest using different combinations of reference genes for normalisation before and after zygotic genome activation, due to the switch from maternal to zygotic mRNA content.
In this study, we show that gene expression levels as determined by qRT-PCR change during oocyte maturation and can differ depending on the priming strategy used for reverse transcription. This is in agreement with previous studies in which relative gene expression was directly compared when reverse transcription was performed using random primers and oligo(dT) primers (Thélie et al., 2007; Gohin et al., 2014). In cDNA samples synthesised using random primers, the expression was stable from the GV to MII stage in oocytes for every gene we examined, indicating that maternal mRNAs were not degraded after resumption of meiosis. Indeed, it has been documented that, at least in bovine oocytes, there is no decrease in total RNA content during meiosis (Lequarre et al., 2004). We also detected a surge in absolute gene expression in oocytes between the GV and GVBD stages when no normalisation was applied, suggesting that large amounts of maternal RNA are synthesised within the 6 h prior to GVBD. Therefore, when gene expression levels are examined using qRT-PCR, reverse transcription using random primers is preferable.
In the mouse, high levels of cyclin B protein in MI and MII oocytes are achieved by control of maternal mRNA translation, which coincides with an elongation of the CCNB1 mRNA (Kotani et al., 2013). High levels of cyclin B have been observed in MI and MII oocytes (Wu et al., 1997; Heikinheimo and Gibbons, 1998; Quetglas et al., 2010). We detected consistent levels of CCNB1 expression throughout oocyte maturation, while the detected CCNB2 levels were significantly reduced from GV to MII oocytes when cDNA was synthesised with oligo(dT) primers. To address how cyclin B gene expression was regulated in oocytes, we analysed our qRT-PCR data without normalisation, since numbers of oocytes were equal in all groups. Both CCNB1 and CCNB2 expression, as well as reference gene expression levels could be seen to be down-regulated from the GV to the MII stage.
Associated with our qRT-PCR without normalisation data, the poly(A) tail lengths for CCNB1, CCNB2 and HPRT1 also decreased from the GV to the MII stage. In contrast, it has been reported that the poly(A) tail of CCNB1 is elongated during oocyte maturation in the mouse, Xenopus and zebrafish (Mendez and Richter, 2001; Kotani et al., 2013). A decrease in poly(A) tail length and a down-regulation of gene expression when normalisation was not applied was also observed for other genes, indicating a general deadenylation of maternal mRNA during bovine oocyte maturation. In agreement with our findings, it has been reported that the amount of poly(A) RNA is reduced by half, but that the total RNA content does not change during bovine oocyte maturation (Lequarre et al., 2004). It is therefore possible that maternal mRNAs have long poly(A) tails to increase their stability during the long time of storage before meiosis resumption. Interestingly, other studies have shown that mRNAs have much longer poly(A) tails, up to 250 nucleotides (Vaur et al., 2002; Lim et al., 2016), than the poly(A) lengths we detected.
In conclusion, we observed that the genes used for normalisation of expression levels may differ in terms of apparent stability of expression in oocytes depending on the priming strategy used to synthesise the cDNA, and the priming strategy should therefore be tailored to the question addressed. In embryos at a stage beyond zygotic genome activation, such as morulae and blastocysts, the method of cDNA generation appears to be less critical to obtaining reliable qRT-PCR results. In general, the poly(A) tail of mRNA species synthesised before GVBD in oocytes seems to shorten during meiotic maturation to the MII stage in bovine oocytes.
Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Data availability
The supporting data for this article are available in the article and the online supplementary material.
Supplementary Material
Acknowledgements
We thank Arend Rijneveld and Christine Oei for assistance with oocyte recovery.
Authors’ roles
B.Y., H.T.A.v.T., and B.A.J.R. conceived and designed the experiments. B.Y. and H.T.A.v.T. performed the experiments. B.Y. collected and analysed the data. B.A.J.R. contributed the reagents, materials and analysis tools. B.Y., T.A.E.S. and B.A.J.R. wrote the manuscript. All authors read and approved the manuscript.
Funding
B.Y. is supported by a PhD scholarship from the Chinese Scholarship Council (CSC) (CSC201606300033).
Conflict of interest
None declared.
References
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 2009;10:430–436. [DOI] [PubMed] [Google Scholar]
- Brinkhof B, van Tol HT, Groot KM, Wubbolts RW, Haagsman HP, Roelen BA. Characterization of bovine embryos cultured under conditions appropriate for sustaining human naïve pluripotency. PLoS One 2017;12:e0172920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkhof B, van Tol HTA, Groot Koerkamp MJA, Riemers FM, IJzer SG, Mashayekhi K, Haagsman HP, Roelen BAJ. A mRNA landscape of bovine embryos after standard and MAPK-inhibited culture conditions: a comparative analysis. BMC Genomics 2015;16:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano LC, Miranda-Furtado CL, Batista LA, Pitangui-Molina CP, Higa TT, Padovan CC, Rosa ESA. Validation of reference genes for gene expression studies in bovine oocytes and cumulus cells derived from in vitro maturation. Anim Reprod 2019;16:290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cholet F, Ijaz UZ, Smith CJ. Reverse transcriptase enzyme and priming strategy affect quantification and diversity of environmental transcripts. Environ Microbiol 2020;22:2383–2402. [DOI] [PubMed] [Google Scholar]
- Daldello EM, Luong XG, Yang CR, Kuhn J, Conti M. Cyclin B2 is required for progression through meiosis in mouse oocytes. Development 2019;146:dev172734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Spiegelaere W, Dern-Wieloch J, Weigel R, Schumacher V, Schorle H, Nettersheim D, Bergmann M, Brehm R, Kliesch S, Vandekerckhove L et al. Reference gene validation for RT-qPCR, a note on different available software packages. PLoS One 2015;10:e0122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumdie JN, Cho K, Ramaiah M, Skarbrevik D, Mora-Castilla S, Stumpo DJ, Lykke-Andersen J, Laurent LC, Blackshear PJ, Wilkinson MF et al. Chromatin modification and global transcriptional silencing in the oocyte mediated by the mRNA decay activator ZFP36L2. Dev Cell 2018;44:392–402.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn SW, Subtelny AO, Kronja I, Kwasnieski JC, Orr-Weaver TL, Bartel DP. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife 2016;5:e16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans C, Hardin J, Stoebel DM. Selecting between-sample RNA-Seq normalization methods from the perspective of their assumptions. Brief Bioinform 2018;19:776–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohin M, Fournier E, Dufort I, Sirard MA. Discovery, identification and sequence analysis of RNAs selected for very short or long poly A tail in immature bovine oocytes. Mol Hum Reprod 2014;20:127–138. [DOI] [PubMed] [Google Scholar]
- Goossens K, Van Poucke M, Van Soom A, Vandesompele J, Van Zeveren A, Peelman LJ. Selection of reference genes for quantitative real-time PCR in bovine preimplantation embryos. BMC Dev Biol 2005;5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 2004;6:117–131. [DOI] [PubMed] [Google Scholar]
- Heikinheimo O, Gibbons WE. The molecular mechanisms of oocyte maturation and early embryonic development are unveiling new insights into reproductive medicine. Mol Hum Reprod 1998;4:745–756. [DOI] [PubMed] [Google Scholar]
- Ihara J, Yoshida N, Tanaka T, Mita K, Yamashita M. Either cyclin B1 or B2 is necessary and sufficient for inducing germinal vesicle breakdown during frog (Rana japonica) oocyte maturation. Mol Reprod Dev 1998;50:499–509. [DOI] [PubMed] [Google Scholar]
- Jansova D, Tetkova A, Koncicka M, Kubelka M, Susor A. Localization of RNA and translation in the mammalian oocyte and embryo. PLoS One 2018;13:e0192544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan DR, Landry DA, Fournier É, Vigneault C, Blondin P, Sirard MA. Transcriptome meta-analysis of three follicular compartments and its correlation with ovarian follicle maturity and oocyte developmental competence in cows. Physiol Genomics 2016;48:633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani T, Yasuda K, Ota R, Yamashita M. Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules. J Cell Biol 2013;202:1041–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijk EW, Du Puy L, van Tol HT, Haagsman HP, Colenbrander B, Roelen BA. Validation of reference genes for quantitative RT-PCR studies in porcine oocytes and preimplantation embryos. BMC Dev Biol 2007;7:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lequarre AS, Traverso JM, Marchandise J, Donnay I. Poly(A) RNA is reduced by half during bovine oocyte maturation but increases when meiotic arrest is maintained with CDK inhibitors. Biol Reprod 2004;71:425–431. [DOI] [PubMed] [Google Scholar]
- Li J, Qian WP, Sun QY. Cyclins regulating oocyte meiotic cell cycle progression. Biol Reprod 2019;101:878–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Lee M, Son A, Chang H, Kim VN. mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes Dev 2016;30:1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamo S, Gal AB, Bodo S, Dinnyes A. Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro. BMC Dev Biol 2007;7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdi Khanlou K, Van Bockstaele E. A critique of widely used normalization software tools and an alternative method to identify reliable reference genes in red clover (Trifolium pratense L.). Planta 2012;236:1381–1393. [DOI] [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol 2001;2:521–529. [DOI] [PubMed] [Google Scholar]
- Morgan M, Much C, DiGiacomo M, Azzi C, Ivanova I, Vitsios DM, Pistolic J, Collier P, Moreira PN, Benes V et al. mRNA 3' uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature 2017;548:347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama Y, Yamashita M. Regulation of oocyte maturation in fish. Dev Growth Differ 2008;50(Suppl 1):S195–S219. [DOI] [PubMed] [Google Scholar]
- Nakahata S, Kotani T, Mita K, Kawasaki T, Katsu Y, Nagahama Y, Yamashita M. Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech Dev 2003;120:865–880. [DOI] [PubMed] [Google Scholar]
- Niakan KK, Han J, Pedersen RA, Simon C, Pera RA. Human pre-implantation embryo development. Development 2012;139:829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quetglas MD, Adona PR, de Bem TH, Pires PR, Leal CL. Effect of cyclin-dependent kinase (CDK) inhibition on expression, localization and activity of maturation promoting factor (MPF) and mitogen activated protein kinase (MAPK) in bovine oocytes. Reprod Domest Anim 2010;45:1074–1081. [DOI] [PubMed] [Google Scholar]
- Radonova L, Pauerova T, Jansova D, Danadova J, Skultety M, Kubelka M, Anger M. Cyclin A1 in oocytes prevents chromosome segregation and anaphase entry. Sci Rep 2020;10:7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassa JC, Wilson GM, Brewer GA, Parks GD. Spacing constraints on reinitiation of paramyxovirus transcription: the gene end U tract acts as a spacer to separate gene end from gene start sites. Virology 2000;274:438–449. [DOI] [PubMed] [Google Scholar]
- Richter JD. CPEB: a life in translation. Trends Biochem Sci 2007;32:279–285. [DOI] [PubMed] [Google Scholar]
- Ross PJ, Wang K, Kocabas A, Cibelli JB. Housekeeping gene transcript abundance in bovine fertilized and cloned embryos. Cell Reprogram 2010;12:709–717. [DOI] [PubMed] [Google Scholar]
- Sallés FJ, Lieberfarb ME, Wreden C, Gergen JP, Strickland S. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science 1994;266:1996–1999. [DOI] [PubMed] [Google Scholar]
- Spinsanti G, Panti C, Lazzeri E, Marsili L, Casini S, Frati F, Fossi CM. Selection of reference genes for quantitative RT-PCR studies in striped dolphin (Stenella coeruleoalba) skin biopsies. BMC Mol Biol 2006;7:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangegaard M, Dufva IH, Dufva M. Reverse transcription using random pentadecamer primers increases yield and quality of resulting cDNA. BioTechniques 2006;40:649–657. [DOI] [PubMed] [Google Scholar]
- Tan JH, Wang HL, Sun XS, Liu Y, Sui HS, Zhang J. Chromatin configurations in the germinal vesicle of mammalian oocytes. Mol Hum Reprod 2009;15:1–9. [DOI] [PubMed] [Google Scholar]
- Thélie A, Papillier P, Pennetier S, Perreau C, Traverso JM, Uzbekova S, Mermillod P, Joly C, Humblot P, Dalbiès-Tran R. Differential regulation of abundance and deadenylation of maternal transcripts during bovine oocyte maturation in vitro and in vivo. BMC Dev Biol 2007;7:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:Research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Cao WX, Lipshitz HD. The maternal-to-zygotic transition revisited. Development 2019;146:dev161471. [DOI] [PubMed] [Google Scholar]
- Vaur S, Montreau N, Dautry F, Andéol Y. Differential post-transcriptional regulations of wnt mRNAs upon axolotl meiotic maturation. Int J Dev Biol 2002;46:731–739. [PubMed] [Google Scholar]
- Winata CL, Korzh V. The translational regulation of maternal mRNAs in time and space. FEBS Lett 2018;592:3007–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Ignotz G, Currie WB, Yang X. Dynamics of maturation-promoting factor and its constituent proteins during in vitro maturation of bovine oocytes. Biol Reprod 1997;56:253–259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The supporting data for this article are available in the article and the online supplementary material.