Abstract
Introduction
Long non-coding RNAs (lncRNAs), a class of regulatory RNA molecules, are over 200 nucleotides long and could be used as a new potential biomarker, but their detection methods such as qRT-PCR are still not validated, and the influence of RNA degradation on lncRNA quantification is not clear. In this study, commercially available cDNA synthesis kits were tested and the influence of RNA degradation was compared.
Material and methods
Total RNA from FaDu cells was isolated and high quality RNA and highly degraded RNA samples were used. Reverse transcription was performed using three different commercially available kits and quantifications were performed using lncRNA Primer Plate and SYBR Green I Master by LightCycler 96. qRT-PCR was performed using three different cDNA samples and results are presented as the mean Ct values. A p-value < 0.05 was considered to be significant.
Results
Lower lncRNA Ct values (61/90; 67.78%) after qRT-PCR quantification were observed for cDNA synthesized using random hexamer primers preceded by polyA-tailing and adaptor-anchoring steps. It was observed that 9/90 (10.00%) lncRNAs were not detectable using different cDNA synthesis methods. For 75/90 (83%) lncRNAs, RNA degradation weakly influenced lncRNA Ct values and no differences were observed between high quality RNA and degraded samples. Seventy percent of examined lncRNAs showed significantly different Ct values depending on RNA degradation.
Conclusions
cDNA synthesis kits with random hexamer primers preceded by polyA-tailing and adaptor-anchoring steps allows enhancement of lncRNA quantification specificity and sensitivity. In most cases degradation of RNA samples does not affect lncRNA quantification because these molecules have good stability.
Keywords: lncRNA, cDNA synthesis, qRT-PCR, RNA stability and degradation
Introduction
The idea of using RNA as a biomarker is not new [1, 2]. Long non-coding RNAs (lncRNAs) are molecules longer than 200 nucleotides. They are actively transcribed and do not encode any type of proteins. Knowledge about lncRNAs is still limited and thus this is an important area for further research. lncRNA molecules possess many functional domains such as RNA or DNA binding sites and protein binding sites. They may also undergo a conformational switch. Because of these domains, lncRNAs may have crucial physiological functions, such as controlling transcription, post-transcription processes, or translation, or modeling epigenetic modifications. lncRNAs participate in cellular processes such as proliferation, apoptosis, response to stress, and regulation of cell metabolism or phenotype. Disturbances in lncRNA expression are associated with cancer processes, and a growing number of studies have focused on lncRNAs as new biomarkers [3].
Detection of expression of lncRNA originating from tissue, urine, peripheral blood, serum, saliva, or urine samples can be easily performed using molecular biology methods [4–9]. It was also reported that lncRNAs can be extracted from exosomes, which are thought to be mediators in cell-cell communication [10]. However, not all lncRNAs are present in each type of biological material. For example, Tang et al. observed that HOTAIR, HULC, MALAT1, MEG-3, NEAT-1, and UCA1 were present in cancer and adjacent noncancerous samples from OSCC (oral squamous cell carcinoma) patients, but only HOTAIR and MALAT1 from this group of lncRNAs were detected in some saliva samples [8].
There are many available methods to study lncRNAs: i) lncRNA immunoprecipitation; ii) lncRNA in situ hybridization; iii) Au-NP assay (gold nanoparticle-based); iv) lncRNA northern blot analysis; v) estimation of methylation status using HRM analysis (high resolution melting); vi) microarray or RNA sequencing; and vii) qRT-PCR or the newly developed ddPCR (droplet digital PCR) [5, 6, 11–13]. The most frequently used method in lncRNA studies is a hybridization assay, especially the qRT-PCR method based on SYBR-green dye and TaqMan probes [14]. The qRT-PCR experiments can be laboratory-specific. Various research groups use different primers, reference genes (or set of genes) and amplification strategies, which sometimes makes it impossible to compare the results [15, 16]. It seems that only commercial qRT-PCR platforms could help to solve this problem. The commercially available qRT-PCR lncRNA platforms, such as the LncProfiler qPCR Array Kit (SBI), provide the possibility of simple and quick quantification of 90 lncRNAs in a single run, based on Ct (threshold cycle) analysis. In contrast to qRT-PCR, the microarray method is well-validated technology and is more comparable among experiments, but it is more expensive [17]. Moreover, using a commercial lncRNA microarray platform, expression of over 30,000 lncRNAs can be evaluated without using the sophisticated bioinformatics methods required for NGS (next generation sequencing) data extraction [18, 19]. However, in qRT-PCR and microarray methods, detection is limited to only known lncRNA transcripts.
As mentioned above, one of the most popular methods used in lncRNA research is qRT-PCR, but this method is not standardized and different approaches to prepare cDNA are used. Moreover, the influence of RNA degradation on quantification of lncRNA is not clear. These problems should be solved before using lncRNAs as a new class of biomarker in clinical practice.
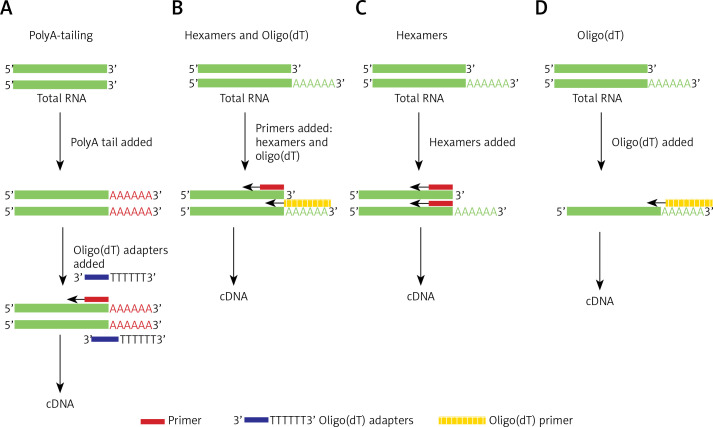
In this study, different cDNA synthesis kits were evaluated and the quality of RNA samples was compared. Kits were based on the following: i) random hexamer primers preceded by polyA-tailing and adaptor-anchoring steps; ii) simple reaction using a blend of random hexamer primers and oligo(dT); iii) only random hexamer primers; and iv) only oligo(dT) (Figures 1 A–D).
Figure 1.
Comparison of different reverse transcription kits used to quantify lncRNA employing qRT-PCR methods. A – cDNA synthesis using random hexamer primers preceded by polyA-tailing and adaptor-anchoring steps, B – simple reaction using blend of random hexamer primers and oligo(dT), C – reaction based on random hexamer primers, D – based only on oligo(dT)
Material and methods
The FaDu cell line, which is a model of hypopharynx squamous cell carcinoma, was used. The FaDu cell line was maintained in DMEM high glucose (4.5 g/l) medium (Sigma-Aldrich) supplemented with 8.85% (v/v) fetal bovine serum (Sigma-Aldrich), 1.77 mM L-glutamine (PAA), 0.885% (v/v) MEM non-essential amino acid solution (PAA), 0.885% (v/v) penicillin-streptomycin (PAA), and 8.85 mM HEPES (Sigma-Aldrich). Mycoplasma detection tests were performed routinely using the VenorGeM Mycoplasma PCR Detection Kit (Minerva Biolabs).
RNA isolation, quantification, and degradation protocol
Total RNA from FaDu cells was isolated using a High Pure miRNA isolation kit (Roche), according to the isolation protocol for total RNA (including the lncRNA fraction) from tissue and cell line samples. Quality and quantity of RNA samples were assessed using a NanoDrop spectrophotometer (Thermo Scientific), followed by 28S and 18S rRNA band estimation (native 1% agarose gel electrophoresis in TAE buffer).
One high quality RNA sample was aliquoted (1 μg of total RNA) and used for the reverse transcription reaction using three different cDNA synthesis kits. The best cDNA kit was then chosen for further experiments.
To test the influence of RNA integrity on lncRNA amplification, an RNA degradation protocol was developed. Aliquoted RNA (1 μg/tube) was incubated for 0, 3, 6, 8, and 10 days at room temperature. At all time points, the RNA quality was tested using 28S and 18S rRNA band estimation (native 1% agarose gel electrophoresis in TAE buffer). Additional RNA samples on day 0 and day 10 were tested using a NanoDrop spectrophotometer.
High quality (visible 28S and 18S rRNA bands), degraded (lack of 28S rRNA band and visible 18S rRNA band), and highly degraded RNA samples (lack of a 28S or 18S rRNA band, visible smear of RNA) were used for cDNA synthesis and the qRT-PCR reaction was performed.
cDNA synthesis
Reverse transcription was performed using three different commercially available kits: i) LncProfiler qPCR Array Kit (SBI); ii) iScript cDNA Synthesis Kit (Bio-Rad); and iii) First Strand cDNA Synthesis Kit (Fermentas). For all reactions, the same amount of total RNA (1 μg/reaction) from the same isolation was used. All experiments (RNA isolation, cDNA synthesis and qRT-PCR) were performed in triplicate.
LncProfiler qPCR array kit (SBI)
Reverse transcription is based on 3 steps: i) poly-A tailing; ii) annealing anchor dT adaptor; and iii) cDNA synthesis. In the first step, 5 μl of total RNA (1 μg) was mixed with 2 μl of 5× PolyA Buffer, 1 μl of MnCl2, 1.5 μl of ATP, and 0.5 μl of PolyA Polymerase and incubated for 30 min at 37°C. In the next step, 0.5 μl of Oligo(dT) Adapter was added. The reaction was heated for 5 min at 60°C and then cooled to room temperature for 2 min. In the third step, 4 μl of RT Buffer, 2 μl of dNTP mix, 1.5 μl of 0.1 M DTT, 1.5 μl of random Primer Mix, and 1 μl of Reverse transcriptase were added and incubated for 60 min at 42°C followed by heating for 10 min at 95°C.
iScript cDNA synthesis Kit (Bio-Rad)
The following were mixed: 5 μl of total RNA (1 μg), 4 μl of 5× iScript Reaction Mix (contained blend of oligo(dT) and random hexamer primers), 1 μl of iScript Reverse Transcriptase, and nuclease free water (up to 20 μl). The reaction was incubated for 5 min at 25°C and followed by 20 min at 46°C and 1 min at 95°C.
First strand cDNA synthesis Kit (Fermentas)
Two specific mixes of primers were used: the oligo(dT) primer and the random hexamer primer mixes. The reaction mixture contained 1 μg total RNA, and oligo(dT) or random hexamer primers (1 μl), and it was filled up to a volume of 11 μl using nuclease free water. The reactions were incubated at 65°C for 5 min, followed by cooling on ice and collecting drops by brief centrifugation. Next, 4 μl of 5× reaction buffer, 1 μl of RiboLock Ribonuclease Inhibitor, 2 μl of 10 mM dNTP mix, and 2 μl of M-MuLV reverse transcriptase were added. Reactions were incubated at 37°C for 60 min followed by 10 min at 70°C (for reactions with hexamer primers, the incubation procedure was followed by a sequential incubation step of 10 min at 25°C).
qRT-PCR reaction
cDNA was mixed with 1.750 ml of 2× LightCycler 480 SYBR Green I Master buffer (Roche) and 1.480 ml of nuclease free water, and 26 μl of the mixture was dropped into wells on a 96-well qRT-PCR plate. Next, 4 μl of lncRNA primers from Primer Plate (component of the LncProfiler qPCR Array Kit) was loaded onto the plate and the qPCR reaction was performed using the following protocol: preincubations (50°C for 2 min and 95°C for 10 min), 60 cycles of 2-step amplification (95°C for 15 s and 60°C for 1 min), and a melting step. Reactions were performed using a LightCycler 96 (Roche).
Statistical analysis
To compare Ct values, the statistical analysis was performed using GraphPad Prism 5 software with an unpaired t-test or a one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test. All qRT-PCR experiments were performed three times using three different cDNA samples and are presented as the mean Ct values. Error bars represent the standard deviation (SD) and a p-value < 0.05 was considered to be significant.
Results
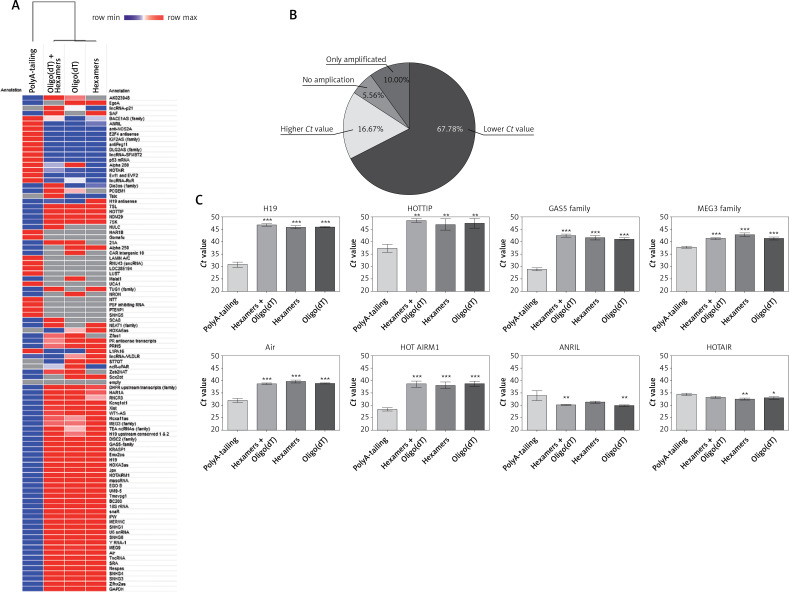
In this study, different cDNA synthesis kits based on: i) random hexamer primers preceded by polyA-tailing and adaptor-anchoring steps; ii) simple reaction using a blend of random hexamer primers and oligo(dT); iii) using only random hexamer primers; and iv) only oligo(dT) were tested. We assumed that the lower the Ct value was, the more sensitive and precise was the method of cDNA synthesis. Comparison of mean qRT-PCR Ct values is presented on a heat map (Figure 2 A). Lower Ct values were observed for 61/90 (67.78%) of examined lncRNAs where cDNA was synthesized using random hexamer primers preceded by polyA-tailing and adaptor-anchoring steps. However, in the case of 15/90 (16.67%) lncRNAs, higher Ct values were observed for other methods (Figures 2 A and B).
Figure 2.
Comparison of mean Ct values of qRT-PCR reactions based on cDNA synthesized using different commercially available cDNA synthesis kits. A – Heat map and clustering of mean Ct values of qRT-PCR reactions. Blue color indicates lower Ct values, red means higher, and grey means lack of amplification. B – Comparison of cDNA synthesis using random hexamer primers preceded by polyA-tailing and adaptor-anchoring steps with the rest of tested cDNA synthesis kits. C – Examples of the most changed Ct values
Data are presented as the mean Ct values; error bars represent SD; *p < 0.05, **p < 0.005, ***p < 0.0001.
Heat map and clustering showed that qRT-PCR Ct values for cDNA obtained using random hexamer primers preceded by polyA-tailing and adaptor-anchoring steps are distinct from other examined cDNA synthesis kits, which are similar to each other and clustered together (Figure 2 A). Moreover, 9/90 (10%) of the examined lncRNAs were detected only in the cDNA samples obtained by random hexamer primers preceded by polyA-tailing and adaptor-anchoring steps. However, for 5/90 (5.56%) lncRNAs, no lncRNA amplification was observed, and these lncRNAs were detected in cDNA samples synthesized using a combination of random hexamer primers and oligo(dT), and with only random hexamer primers or only oligo(dT) (Figures 2 A and B).
For the most frequently described lncRNAs in cancers and in head and neck squamous cell carcinomas in particular [3], significant changes were observed among qRT-PCR Ct values obtained using different cDNA synthesis kits. For H19 (p < 0.0001), HOTTIP (p = 0.0011), the GAS5 family (p < 0.0001), the MEG3 family (p = 0.0001), Air (p < 0.0001), and HOTAIRM1 (p < 0.0001), lower Ct values were observed (32.55 ±4.12 vs. 42.49 ±3.52), while for ANRIL (p = 0.0045) and HOTAIR (p = 0.0053), higher Ct values (34.18 ±0.33 vs. 31.68 ±1.46) were observed for cDNA obtained with random hexamer primers preceded by polyA-tailing and adaptor-anchoring steps compared to the rest of the tested kits (Figure 2 C).
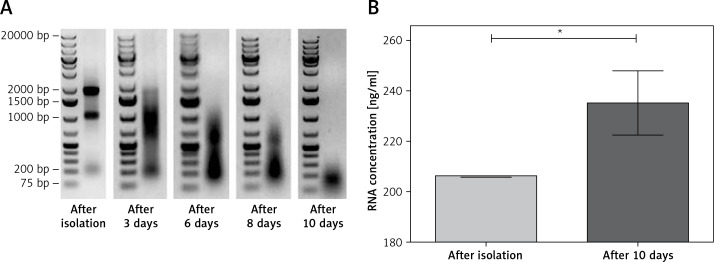
The influence of total RNA degradation on qRT-PCR quantification was then investigated. Freshly isolated RNA was aliquoted (V = 5 μl/sample, C = 1 μg/sample) and incubated at room temperature for 3, 6, 8, and 10 days. Degradation of total RNA was observed (Figure 3 A). After 3 days of incubation, no 28S band but a smear of the 18S rRNA band was observed until day 10, when RNA was highly degraded (no 28S or 18S rRNA bands; Figure 3 A). The degradation process caused changes in RNA sample absorbance after 10 days of incubation that was measured using a NanoDrop spectrophotometer. The UV absorbance increased and is observed as an effect of apparent concentration change from 206.3 ng/μl measured immediately after isolation to 235.27 ng/μl measured after 10 days of incubation (p = 0.0227; Figure 3 B).
Figure 3.
Degradation of RNA used for cDNA synthesis. A – Native electrophoresis in 1% agarose gel of RNA samples at different time points. Marker GeneRuler 1kb Plus DNA Ladder (Thermo Scientific). B – Mean RNA concentration measured immediately after isolation and after 10 days of incubation at room temperature
Data are presented as the mean Ct values; error bars represent SD; *p < 0.05.
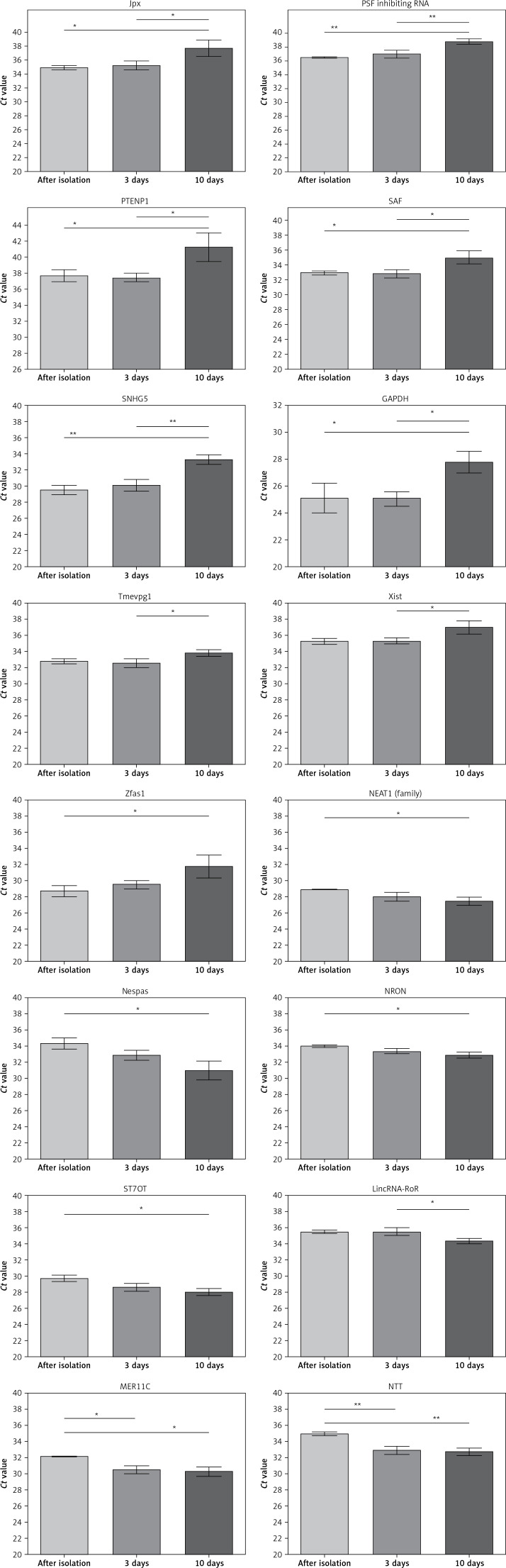
cDNA synthesis using random hexamer primers preceded by polyA-tailing and adaptor-anchoring steps and qRT-PCR using the LncProfiler qPCR Array kit were performed for RNA samples from the day of isolation as well as after incubation for 3 and 10 days. For 83% (75/90) of lncRNAs, there were no significant differences (p > 0.05) in Ct values between high quality RNA and degraded RNA (day 3 or 10). For lncRNAs: Jpx, PSF inhibiting RNA, PTENP1, SAF, SNHG5, and the mRNA GAPDH there were no differences between Ct values of cDNA synthesized using RNA samples after isolation or after incubation for 3 days, but significantly higher Ct values were observed for the samples when the qRT-PCR was performed based on RNA incubated for 10 days (34.92 ±0.28 vs. 37.71 ±1.17, p = 0.0225; 36.51 ±0.05 vs. 38.78 ±0.40, p = 0.0035; 37.72 ±0.72 vs. 41.28 ±1.75, p = 0.0221; 32.90 ±0.21 vs. 35.01 ±0.87, p = 0.0291; 29.55 ±0.58 vs. 33.32 ±0.58, p = 0.0018, and 25.10 ±1.10 vs. 27.76 ±0.81, p = 0.0150, respectively). Moreover, these differences were also observed between samples incubated for 3 and 10 days (35.25 ±0.62 vs. 37.71 ±1.17; 36.98 ±0.54 vs. 38.78 ±0.40; 37.48 ±0.50 vs. 41.28 ±1.75; 32.82 ±0.53 vs. 35.01 ±0.87; 30.15 ±0.67 vs. 33.32 ±0.58, and 25.05 ±0.53 vs. 27.76 ±0.81, respectively). For the lncRNAs Tmevpg1 and Xist higher Ct values were noted only between days 3 and 10 (32.52 ±0.52 vs. 33.77 ±0.41, p = 0.0386, and 35.34 ±0.35 vs. 36.97 ±0.82, p = 0.0306, respectively) and for Zfas1 after isolation and after incubation for 10 days (28.71 ±0.70 vs. 31.76 ±1.42, p = 0.0406). qRT-PCR performed using RNA incubated for 10 days provided better results (lower Ct values) compared to cDNA based on RNA after isolation in the case of NEAT1 (family), Nespas, NRON and ST7OT (27.45 ±0.50 vs. 28.91 ±0.02, p = 0.0490; 30.97 ±1.18 vs. 34.36 ±0.66, p = 0.0217; 32.87 ±0.36 vs. 34.01 ±0.13, p = 0.0263, and 28.04 ±0.41 vs. 29.74 ±0.36, p = 0.0214, respectively), and for lincRNA-RoR compared to cDNA based on RNA incubated for 3 days (34.27 ±0.31 vs. 35.36 ±0.23, p = 0.0238). For the lncRNAs MER11C and NTT lower Ct values were observed for RNA incubated for 3 days as well as for 10 days compared to RNA after isolation (30.51 ±0.48 and 30.31 ±0.60 vs. 32.15 ±0.01, p = 0.0257, and 32.90 ±0.49 and 32.72 ±0.45 vs. 34.97 ±0.23, p = 0.0047, respectively; Figure 4).
Figure 4.
Comparison of mean Ct values of changed lncRNAs and GAPDH mRNA amplified with cDNA synthesis using random hexamer primers preceded by polyA-tailing and adaptor-anchoring steps and RNA samples just after isolation and after 3 and 10 days incubation at room temperature
Data are presented as the mean Ct values; error bars represent SD; *p < 0.05, **p < 0.005.
Discussion
Similarly to most researchers, we also chose the qRT-PCR method to study lncRNA expression because of its simplicity and quick sample analysis [20]. However, the main issue for qRT-PCR is selection of the proper methodology in the experimental workflow, but there are no experimental data comparing various approaches to this issue. In this study, we compared three different commercially available cDNA synthesis kits. Based on total RNA (including the lncRNA fraction) we obtained the cDNA, and this could be used to quantify expression of lncRNA using the commercially available LncProfiler qPCR Array Kit (SBI). We assumed that the lower the Ct value was, the more sensitive and precise was the method. Comparing three different approaches, we observed the best results (low Ct values) for the method using cDNA synthesis by random hexamer primers preceded by polyA-tailing and adaptor-anchoring steps. Generally, results of simple reactions using a blend of random hexamer primers and oligo(dT) or just random hexamer primers or oligo(dT) alone could incorrectly suggest that there was no expression of some lncRNAs or that the expression was at a very low level. It should be noted that some of the lncRNAs possess endogenous polyA tails but others do not. Moreover, most lncRNAs are present in samples at low copy numbers, which causes difficulties for their quantification [21, 22]. These two lncRNA features require the use of cDNA kits with an additional step of adding polyA tails and annealing anchor (dT) adapters. This approach allows enhancement of the specificity and sensitivity of lncRNA quantification, which we observed. Unfortunately, in most of the studies, cDNA is prepared using kits containing a mixture of oligo(dT) and/or random hexamer primers [14, 23, 24], where oligo(dT) is not functional for lncRNA molecules without a polyA tail. It is well known that specific primers decrease background priming, but random and oligo(dT) primers maximize the number of mRNA molecules that can be analyzed from a small sample of RNA [25]. We observed that artificially adding polyA tails and then using annealing anchor (dT) adapters (analogs of oligo(dT) primers) can help to quantify lncRNAs more precisely.
One possible explanation of the difficulties in quantification of some lncRNAs observed by us is an influence of molecular structure features such as length, GC content and molecule folding. It is well known that short RNA molecules such as miRNAs are difficult to quantify by qRT-PCR and require specific cDNA synthesis kits with adapter or polyA tail addition. Longer molecules seem to be easier to quantify than short RNAs such as miRNAs. On the other hand, a lot of secondary structures such as hairpin loops are problematic for cDNA synthesis and require a higher temperature for relaxing RNA molecules, making them available for reverse transcriptase [26, 27]. Moreover, one of the kits used by us possesses the DTT (dithiothreitol) enhancer, which helps to amplify templates rich in GC sequences, and it contributed to the better qRT-PCR results [28]. Because of the structural characteristics, some lncRNAs seem to need addition of a polyA tail or a higher temperature for a longer time during cDNA synthesis. We suggest that some of the results could be overlooked because an improper cDNA synthesis approach was used and specific lncRNA was not quantified.
The next problem is the stability of lncRNAs and the influence of RNA integrity on qRT-PCR quantification. Because of the length of lncRNAs (more than 200 nt), they are considered to be less stable and easier to degrade compared with short RNA molecules such as miRNAs. We analyzed the influence of RNA stability on quantification of lncRNAs using a cDNA kit (polyA tailing with anchor (dT) adapter annealing and cDNA synthesis using random primer mix), and we compared samples of high quality total RNA (visible 28S and 18S rRNA bands), degraded (no 28S band and visible smear of 18S rRNA band) and highly degraded RNA (no 28S or 18S rRNA bands). Degradation causes changes in RNA structure increasing the amount of short RNA molecules, which strongly affects RNA detection [29, 30]. Our results indicated that most of the examined lncRNAs are stable, even when the total RNA is highly degraded. Our observations are supported by the results obtained by Kraus et al. They reported that some lncRNAs are more stable than miRNAs that are about 22 nucleotides long [31, 32].
However, some long transcripts, such as mRNAs, are sensitive to degradation, and in this case, RNA integrity is an important factor affecting qRT-PCR quantification. It is postulated that the lncRNA half-life depends on its coding place in the genome, on posttranscriptional modifications, and on subcellular localization and its function [33]. Some authors have noted that localization of lncRNAs in the genome may influence their transcript stability, especially intragenic and cis-antisense lncRNAs compared with those derived from introns (half-life more than 16 h) [8, 33]. We noted that some lncRNAs are stable for much longer. After 3 days, we did not observe changes in Ct values in most of the examined lncRNAs, while after 10 days, changes in Ct values were observed only in some lncRNAs. Detection of some lncRNAs in saliva confirms the high stability of these molecules [8]. Moreover, lncRNAs isolated from the plasma are resistant to RNase A digestion and overnight incubation at room temperature [9]. Generally, low RNA stability may cause difficulties in analyzing some long coding or non-coding RNA transcripts obtained from archived formalin-fixed paraffin-embedded (FFPE) blocks [34]. This problem is solved by applying the simple modification of measuring the lncRNA expression level by reaction with three different pairs of non-overlapping primers [35]. However, RNA is both degraded and modified under the archiving process in FFPE blocks, which makes this RNA more difficult to analyze [36].
It is well known that the main features of an ideal biomarker molecule are simplicity of acquisition, diversity of sources where it occurs, and ease of measurement methodology [20, 37]. We postulate that under appropriate processing conditions, lncRNA seems to meet the characteristics of biomarker molecules. However, standardization procedures should be applied before clinical use of lncRNA biomarkers.
In conclusion, we recommend the following: i) use of cDNA kits designed for lncRNA or cDNA synthesis preceded by polyA tailing with anchor (dT) adapter annealing; ii) use of highly thermostable reverse transcriptase designed for long reactions at higher temperatures; and iii) use of high RNA quality is generally recommended, but most lncRNAs seem to be stable and can be quantified in degraded RNA samples.
Acknowledgments
Marcel Ryś and Kacper Guglas contributed equally to this work.
This work was supported by Greater Poland Cancer Centre – grant no. 21/2015 (113) and grant no. 13/2016 (128), and by the National Science Centre, Poland, allocated on the basis of decision no. 2016/21/B/NZ7/01773.
The language correction and text formatting were performed by American Manuscript Editors (americanmanuscripteditors.com).
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Lamperska KM, Kozlowski P, Kolenda T, et al. Unpredictable changes of selected miRNA in expression profile of HNSCC. Cancer Biomark. 2016;16:55–64. doi: 10.3233/CBM-150540. [DOI] [PubMed] [Google Scholar]
- 2.Kolenda T, Przybyła W, Teresiak A, Mackiewicz A, Lamperska KM. The mystery of let-7d – a small RNA with great power. Contemp Oncol. 2014;18:293–301. doi: 10.5114/wo.2014.44467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolenda T, Guglas K, Ryś M, et al. Biological role of lncRNA in head and neck cancers. Rep Pract Oncol Radiother. 2017;22:378–88. doi: 10.1016/j.rpor.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: a potential novel class of cancer biomarkers. Front Genet. 2015;6:145. doi: 10.3389/fgene.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eissa S, Matboli M, Essawy NO, Shehta M, Kotb YM. Rapid detection of urinary long non-coding RNA urothelial carcinoma associated one using a PCR-free nanoparticle-based assay. Biomarkers. 2015;20:212–7. doi: 10.3109/1354750X.2015.1062918. [DOI] [PubMed] [Google Scholar]
- 6.Zhang H, Zhao L, Wang YX, Xi M, Liu SL, Luo LL. Long non-coding RNA HOTTIP is correlated with progression and prognosis in tongue squamous cell carcinoma. Tumour Biol. 2015;36:8805–9. doi: 10.1007/s13277-015-3645-2. [DOI] [PubMed] [Google Scholar]
- 7.Fayda M, Isin M, Tambas M, et al. Do circulating long non-coding RNAs (lncRNAs) (LincRNA-p21, GAS 5, HOTAIR) predict the treatment response in patients with head and neck cancer treated with chemoradiotherapy? Tumour Biol. 2016;37:3969–78. doi: 10.1007/s13277-015-4189-1. [DOI] [PubMed] [Google Scholar]
- 8.Tang H, Wu Z, Zhang J, Su B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol Med Rep. 2013;7:761–6. doi: 10.3892/mmr.2012.1254. [DOI] [PubMed] [Google Scholar]
- 9.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gezer U, Özgür E, Cetinkaya M, Isin M, Dalay N. Long non-coding RNAs with low expression levels in cells are enriched in secreted exosomes. Cell Biol Int. 2014;38:1076–9. doi: 10.1002/cbin.10301. [DOI] [PubMed] [Google Scholar]
- 11.Feng Y, Hu X, Zhang Y, Zhang D, Li C, Zhang L. Methods for the study of long noncoding RNA in cancer cell signaling. Methods Mol Biol. 2014;1165:115–43. doi: 10.1007/978-1-4939-0856-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wojdacz TK, Dobrovic A, Algar EM. Rapid detection of methylation change at H19 in human imprinting disorders using methylation-sensitive high-resolution melting. Hum Mutat. 2008;29:1255–60. doi: 10.1002/humu.20779. [DOI] [PubMed] [Google Scholar]
- 13.Dodd DW, Gagnon KT, Corey DR. Digital quantitation of potential therapeutic target RNAs. Nucleic Acid Ther. 2013;23:188–94. doi: 10.1089/nat.2013.0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi T, Gao G, Cao Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers. 2016;2016:9085195. doi: 10.1155/2016/9085195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong ML, Medrano JF. Real-time PCR for mRNA quantitation. Biotechniques. 2005;39:75–85. doi: 10.2144/05391RV01. [DOI] [PubMed] [Google Scholar]
- 16.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15:155–66. [PMC free article] [PubMed] [Google Scholar]
- 17.Gee HE, Buffa FM, Camps C, et al. The small-nucleolar RNAs commonly used for microRNA normalisation correlate with tumour pathology and prognosis. Br J Cancer. 2011;104:1168–77. doi: 10.1038/sj.bjc.6606076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oleksiewicz U, Tomczak K, Woropaj J, Markowska M, Stępniak P, Shah PK. Computational characterisation of cancer molecular profiles derived using next generation sequencing. Contemp Oncol (Pozn) 2015;19:A78–91. doi: 10.5114/wo.2014.47137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guglas K, Bogaczyńska M, Kolenda T, et al. lncRNA in HNSCC: challenges and potential. Contemp Oncol (Pozn) 2017;21:259–66. doi: 10.5114/wo.2017.72382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ravasi T, Suzuki H, Pang KC, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–9. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Derrien T, Johnson R, Bussotti G, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang Z, Wu L, Wang L, Yang Y, Meng Y, Yang H. Increased expression of the long non-coding RNA UCA1 in tongue squamous cell carcinomas: a possible correlation with cancer metastasis. Oral Surg Oral Med Oral Pathol Oral Radiol. 2014;117:89–95. doi: 10.1016/j.oooo.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 24.Yang YT, Wang YF, Lai JY, et al. Long non-coding RNA UCA1 contributes to the progression of oral squamous cell carcinoma by regulating the WNT/beta-catenin signaling pathway. Cancer Sci. 2016;107:1581–9. doi: 10.1111/cas.13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malewski T, Malewska A, Rutkowski R. RT-PCR technique and its applications. State-of the-art. J Anim Feed Sci. 2003;12:403–16. [Google Scholar]
- 26.Harrison GP, Mayo MS, Hunter E, Lever AM. Pausing of reverse transcriptase on retroviral RNA templates is influenced by secondary structures both 5’ and 3’ of the catalytic site. Nucleic Acids Res. 1998;26:3433–42. doi: 10.1093/nar/26.14.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brooks EM, Sheflin LG, Spaulding SW. Secondary structure in the 3’ UTR of EGF and the choice of reverse transcriptases affect the detection of message diversity by RT-PCR. Biotechniques. 1995;19:806–12. 814-5. [PubMed] [Google Scholar]
- 28.Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual (Fourth Edition) Vol. 1. New York: Cold Spring Harbor Laboratory Press; 2012. pp. 484–9. [Google Scholar]
- 29.Vermeulen J, De Preter K, Lefever S, et al. Measurable impact of RNA quality on gene expression results from quantitative PCR. Nucleic Acids Res. 2011;39:e63. doi: 10.1093/nar/gkr065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–39. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Kraus TF, Greiner A, Guibourt V, Lisec K, Kretzschmar HA. Identification of stably expressed lncRNAs as valid endogenous controls for profiling of human glioma. J Cancer. 2015;6:111–9. doi: 10.7150/jca.10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraus TF, Greiner A, Guibourt V, Kretzschmar HA. Long non-coding RNA normalisers in human brain tissue. J Neural Transm (Vienna) 2015;122:1045–54. doi: 10.1007/s00702-014-1352-6. [DOI] [PubMed] [Google Scholar]
- 33.Clark MB, Johnston RL, Inostroza-Ponta M, et al. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–98. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kokkat TJ, Patel MS, McGarvey D, LiVolsi VA, Baloch ZW. Archived formalin-fixed paraffin-embedded (FFPE) blocks: a valuable underexploited resource for extraction of DNA, RNA, and protein. Biopreserv Biobank. 2013;11:101–6. doi: 10.1089/bio.2012.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong H, Zhu M, Cui F, et al. Quantitative assessment of short amplicons in FFPE-derived long-chain RNA. Sci Rep. 2014;4:7246. doi: 10.1038/srep07246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamatani K, Eguchi H, Takahashi K, et al. Improved RT-PCR amplification for molecular analyses with long-term preserved formalin-fixed, paraffin-embedded tissue specimens. J Histochem Cytochem. 2006;54:773–80. doi: 10.1369/jhc.5A6859.2006. [DOI] [PubMed] [Google Scholar]
- 37.Kolenda T, Teresiak A, Kapałczyńska M, et al. Let-7d and miR-18a as biomarkers of head and neck cancers. Zeszyty Naukowe WCO, Letters in Oncology Science. 2015;12:37–47. [Google Scholar]