Abstract
Colorectal cancer (CRC) is one of the most common tumors worldwide and is associated with high mortality. Here we performed bioinformatics analysis, which we validated using immunohistochemistry in order to search for hub genes that might serve as biomarkers or therapeutic targets in CRC. Based on data from The Cancer Genome Atlas (TCGA), we identified 4832 genes differentially expressed between CRC and normal samples (1562 up-regulated and 3270 down-regulated in CRC). Gene ontology (GO) analysis showed that up-regulated genes were enriched mainly in organelle fission, cell cycle regulation, and DNA replication; down-regulated genes were enriched primarily in the regulation of ion transmembrane transport and ion homeostasis. Weighted gene co-expression network analysis (WGCNA) identified eight gene modules that were associated with clinical characteristics of CRC patients, including brown and blue modules that were associated with cancer onset. Analysis of the latter two hub modules revealed the following six hub genes: adhesion G protein-coupled receptor B3 (BAI3, also known as ADGRB3), cyclin F (CCNF), cytoskeleton-associated protein 2 like (CKAP2L), diaphanous-related formin 3 (DIAPH3), oxysterol binding protein-like 3 (OSBPL3), and RERG-like protein (RERGL). Expression levels of these hub genes were associated with prognosis, based on Kaplan–Meier survival analysis of data from the Gene Expression Profiling Interactive Analysis database. Immunohistochemistry of CRC tumor tissues confirmed that OSBPL3 is up-regulated in CRC. Our findings suggest that CCNF, DIAPH3, OSBPL3, and RERGL may be useful as therapeutic targets against CRC. BAI3 and CKAP2L may be novel biomarkers of the disease.
Keywords: Colorectal cancer, Hub gene, Immunohistochemistry, Weighted gene co-expression network analysis
Introduction
Colorectal cancer (CRC), which includes colon and rectal cancers, is one of the most common cancers of the digestive system [1]. It is the second leading cause of cancer-related mortality and the third leading cause of cancer-related incidence worldwide [2]. It occurs in three histopathological types, including adenocarcinoma, squamous cell carcinoma, and mucinous carcinoma; adenocarcinoma is the most common type, accounting for ∼95% of all CRC cases [3].
Risk for CRC has been linked to defects in DNA replication and DNA methylation, as well as instability of chromosomes and microsatellites [4–7]. As in many cancers, early stages of CRC appear to involve up-regulation of DNA replication licensing proteins [5]. Up to 15% of CRC cases involve DNA microsatellite instability, which leads to DNA replication errors [8].
Surgery remains the primary method to treat CRC, but the post-surgery recurrence rate is high, the post-surgery 5-year mortality rate is high [9]. In part, this is because most patients with CRC are diagnosed relatively late in the disease [10]. Therefore, it is imperative to understand the molecular mechanism involved in carcinogenesis in order to identify prognostic biomarkers and potential therapeutic targets for CRC.
High-throughput sequencing technologies provide new views into the genomic, transcriptomic, and epigenomic signatures of cancers. Systems biology, especially network methods, can effectively integrate multiple, large-scale datasets of complex human diseases, especially cancer [11–13]. Weighted gene co-expression network analysis (WGCNA), for example, is an efficient, accurate method for extensive multigene analysis [14,15]. The WGCNA package in the R suite is a comprehensive collection of R functions for performing all aspects of weighted correlation network analysis [16]. It can construct a scale-free network to explore the correlation between different genomes or between samples and clinical features [17]. WGCNA has been widely used to identify related clinical modules and hub genes in different types of cancer. For example, one WGCNA study was able to associate the expression of six hub genes with progression of clear human cell renal cell carcinoma and with prognosis of patients [18]. Another WGCNA study drew on data from the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) to identify 15 hub genes as candidate breast cancer biomarkers [19]. A third study used WGCNA to identify four hub genes that may be candidate biomarkers of adrenocortical carcinoma [20].
The present study exploited the power of WGCNA to analyze the pathogenesis of CRC. RNA sequencing data from CRC samples were downloaded from the TCGA, and genes differentially expressed between CRC and normal tissues were analyzed at the expression and functional levels. Functional enrichment of differentially expressed genes (DEGs) was analyzed using Gene Ontology (GO) in the clusterprofiler R package. WGCNA of the DEG matrix identified modules related to clinical characteristics of CRC patients. Hub genes identified through these bioinformatics analyses were verified using survival analysis, immunohistochemistry of CRC tissues, and analysis of the literature. Our findings provide testable hypotheses about genes involved in CRC and, by extension, potential biomarkers and therapeutic targets.
Materials and methods
Data sources and pre-processing
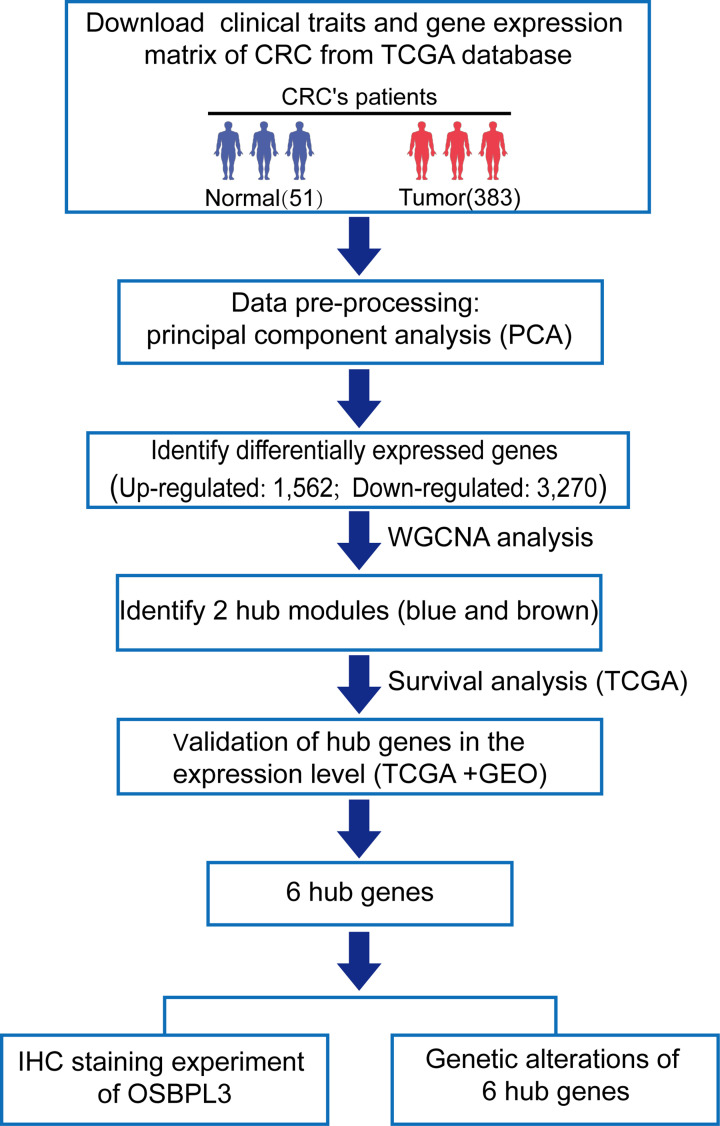
RNA sequencing data and clinical information on patients were downloaded on 22 July 2018 from the ‘Colon and Rectal Cancer’ cohort of TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga), hosted at the Xena website of the University of California at Santa Cruz [21](http://xena.ucsc.edu/; Table 1 and Supplementary Tables S1 and S2). The RNA sequencing data corresponded to 383 tumor samples and 51 normal tissue samples from 434 CRC patients. We excluded samples if the first two principal components identified through principal component analysis were unable to distinguish tumor tissue from normal tissue. The workflow of the study is shown in Figure 1.
Table 1. The clinical information and sample size for TCGA CRC dataset.
| Characteristics | Alive (n=574) | Dead (n=156) | Total (n=730) | P |
|---|---|---|---|---|
| Sex | ||||
| Female | 274 (47.7) | 74 (47.4) | 348 (47.7) | |
| Male | 300 (52.3) | 82 (52.6) | 382 (52.3) | 1 |
| Age, years | ||||
| Mean (SD) | 65.6 (12.7) | 70.4 (12.7) | 66.6 (12.9) | |
| Median [min, max] | 66.5 [31, 90] | 73 [34, 90] | 68 [31, 90] | |
| Body weight | ||||
| Mean (SD) | 83.1 (22.8) | 74.4 (18.6) | 81.1 (22.2) | |
| Median [min, max] | 82.3 [34, 175.3] | 71.9 [40, 140] | 79.3 [34, 175.3] | |
| Cancer type | ||||
| Colon | 422 (73.5) | 123 (78.8) | 545 (74.7) | |
| Rectum | 152 (26.5) | 33 (21.2) | 185 (25.3) | 0.21 |
| Histological type | ||||
| Colon adenocarcinoma | 378 (66.0) | 101 (65.2) | 479 (65.8) | |
| Colon mucinous adenocarcinoma | 50 (8.7) | 20 (12.9) | 70 (9.6) | |
| Rectal adenocarcinoma | 132 (23.0) | 32 (20.6) | 164 (22.5) | |
| Rectal mucinous adenocarcinoma | 13 (2.3) | 2 (1.3) | 15 (2.1) | 0.389 |
| Stage | ||||
| I | 111 (19.8%) | 9 (6.2%) | 120 (17.0%) | |
| II | 48 (8.6%) | 13 (8.9%) | 61 (8.6%) | |
| IIA | 172 (30.7%) | 25 (17.1%) | 197 (27.9%) | |
| IIB | 12 (21.4%) | 3 (2.1%) | 15 (2.1%) | |
| III | 31 (55.3%) | 8 (5.5%) | 39 (5.5%) | |
| IIIA | 13 (2.3%) | 2 (1.4%) | 15 (2.2%) | |
| IIIB | 78 (13.9%) | 15 (10.3%) | 93 (13.2%) | |
| IIIC | 38 (6.8%) | 23 (15.8%) | 61 (8.6%) | |
| IV | 37 (6.6%) | 41 (28.1%) | 78 (11.0%) | |
| IVA | 18 (3.2%) | 7 (4.8%) | 25 (3.5%) | |
| IA | 1 (0.2%) | 1 (0.1%) | ||
| IIC | 2 (0.4%) | 2 (0.3%) |
Values are n (%), unless otherwise noted.
Figure 1. Workflow of searching hub genes in CRC.
Abbreviation: IHC, immunohistochemistry.
Identification of CRC DEGs
The ‘limma’ function in the R suite (version: 3.3.3) [22,23] was used to identify DEGs between CRC and normal colorectal tissues. DEGs were defined as those showing |log2(fold change) | > 1 and P<0.01. Volcano plots of DEGs were plotted using ‘ggplot2’ in R.
Functional enrichment of DEGs
After converting DEG identifiers using the ‘org.Hs.eg.db’ program (version: 3.10) within R, DEGs were analyzed for functional enrichment based on GO [22] using the ‘clusterProfiler’ program (version: 3.14.3) in R. GO terms with P<0.05 were considered statistically significant.
WGCNA
We used BiocManger (version: 1.30.10) in the R suite to download the WGCNA package (version: 1.70-3) to construct the DEG co-expression network [24,25]. First, the DEG expression matrix was filtered through the goodSamplesGenes function in WGCNA to remove unqualified genes and samples. Second, the flashClust tool in R was used to perform cluster analysis of samples in order to detect outliers. Third, matrices of Pearson correlation coefficients (PCCs) were calculated for pair-wise gene comparisons. Fourth, an appropriate soft threshold power (β) was selected to ensure a scale-free network using the pickSoftThreshold function. Fifth, the adjacency matrix was constructed using the power function
where cij refers to the PCC between genes i and j, and aij refers to adjacency between those two genes. Then, the topological overlap matrix (TOM) was constructed using the adjacency function
where lij refers to the product’s sum of the adjacency coefficients of the nodes connected by genes i and j, and k refers to the sum of the adjacency coefficients of the given gene with all other genes in the weighted network. The TOM was used to calculate a dissimilarity measure (1-TOM) to allocate genes into modules based on their similar expression [26], using the dynamic tree cutting method [27]. The minimum number of genes in each module was set to 30.
Selection of clinically significant modules and identification of CRC hub genes
First, principal component analysis was used to describe module eigengenes, corresponding to a single characteristic expression profile across all genes within each module. Correlations between these eigengenes and clinical characteristics were calculated in order to identify which modules were clinically significant. The linear relationship between gene expression and clinical characteristics were assigned a gene significance (GS) equal to the logarithm of the P-value for the individual gene. If GS strongly correlated with module membership (MM), defined as the correlation between the module’s eigengenes and individual gene expression profiles, we concluded that the module’s central genes correlated with CRC [28,29]. We considered these central genes as candidate hub genes.
Bioinformatics validation of hub genes
The expression levels of hub genes in CRC samples were explored using the GEPIA website (http://gepia.cancer-pku.cn/), and the ability of hub genes to predict survival was assessed based on Kaplan–Meier analysis using the ‘survival’ package (version: 3.2-7) in the R suite. First, we obtained DEG expression profiles and prognostic data for 360 CRC tumor samples from the TCGA, then we determined each gene’s median expression value. Samples were assigned to ‘high expression’ or ‘low expression’ groups for a given gene based on whether that gene was expressed at a level higher or lower than the median. Differences in survival between high or low expression groups were assessed for significance using the log-rank test. If this test was associated with P<0.05, we considered the gene to be a validated hub gene.
We then screened for differences in hub gene expression between normal and CRC tissues based on colon adenocarcinoma (COAD) and rectal adenocarcinoma (READ) data from the TCGA and the Genotype-Tissue Expression Project (GTEX) on the GEPIA website. Expression levels were normalized by their mean value, and differences associated with P<0.01 were considered statistically significant. Hub genes were further validated by analyzing their expression differences between CRC and normal tissues using the ‘ggpubr’ package (version: 0.4.0) in the R suite and the GSE33113 dataset [30,31] in the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Independent-samples t test was applied as standard.
We mapped the hub genes’ genome, including mutations, copy number variants (CNVs), and mRNA expression z-scores (RNASeqV2 RSEM) using data from 594 CRC samples from the colorectal adenocarcinoma dataset in the Pan-Cancer Atlas of TCGA. We also used MutationMapper tools to depict the mutation landscape of each hub gene. We accessed and analyzed the data using CBioPortal (http://www.cbioportal.org/).
Immunohistochemical validation of OSBPL3 as a hub gene
Immunohistochemistry of tumor and paired normal tissues from three CRC patients from Tangdu Hospital of the Fourth Military Medical University was performed as described [32,33]. Written informed consent for tissue donation, which clearly stated the purpose of our study, was obtained from all of the patients. Tissues were fixed with formalin, embedded in paraffin and sliced into 3-μm-thick sections. After deparaffinization and inactivation of endogenous catalase, the sections were boiled in sodium citrate buffer to expose antigenic sites, then blocked in 5% normal goat serum for 1 h to prevent non-specific binding. Next, the sections were incubated with anti-OSBPL3 antibody (1:50 dilution; Proteintech, 12417-1-AP) at 4°C overnight, and binding was detected using the avidin–biotin–peroxidase method. Sections were counterstained with Hematoxylin. Two experienced researchers independently evaluated the results.
Results
Data pre-processing
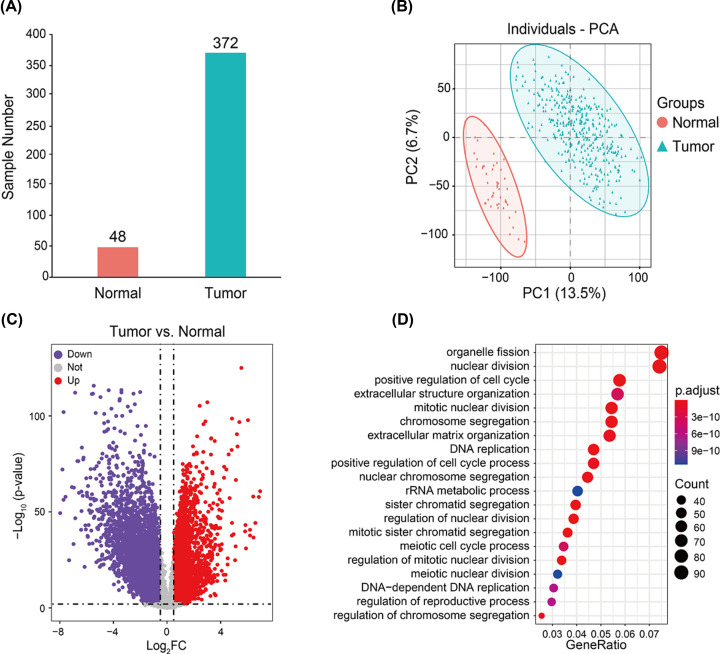
Our expression data came from 51 normal samples and 383 tumor samples (Supplementary Table S1). Filtering based on principal component analysis led to the exclusion of 11 tumors and 3 normal samples from the final dataset (Figure 2A,B). The first two principal components distinguished tumor from normal samples well, accounting for 13.5% (first component) and 6.7% (second component) of the observed differences. The gene expression profiles from these 420 samples were used in subsequent analyses.
Figure 2. Identification of DEGs between 48 normal and 372 CRC samples.
(A) Principal component analysis. (B) Volcano plot. Purple dots represent genes down-regulated in CRC; gray dots, genes not differing significantly between CRC and normal tissues; and red dots, genes up-regulated in CRC. (C) The volcano plot. Purple dots represent down-regulated genes, gray dots represent not significant genes, and red dots represent up-regulated genes. (D) GO analysis of functional enrichment of up-regulated genes. Dot size reflects the number of genes enriched under the given ontology term, and the color indicates the significance of enrichment.
Identification of DEGs in CRC samples and GO enrichment analysis
A total of 4832 DEGs were identified between 48 normal and 372 CRC samples, including 1562 up-regulated and 3270 down-regulated genes (Figure 2C, Supplementary Table S3). To explore the potential biological function of DEGs in CRC, we performed GO enrichment analysis (Supplementary Tables S4 and S5). The up-regulated DEGs were involved mainly in nuclear division, cell cycle regulation, chromosome segregation, and DNA replication (Figure 2D). In contrast, the down-regulated DEGs were involved mainly in the regulation of ion transmembrane transport, muscle systems, ion homeostasis, and second messenger-mediated signaling (Supplementary Figure S1). These results are consistent with known dysfunctions in CRC, suggesting that our results are reliable.
WGCNA and identification of critical modules
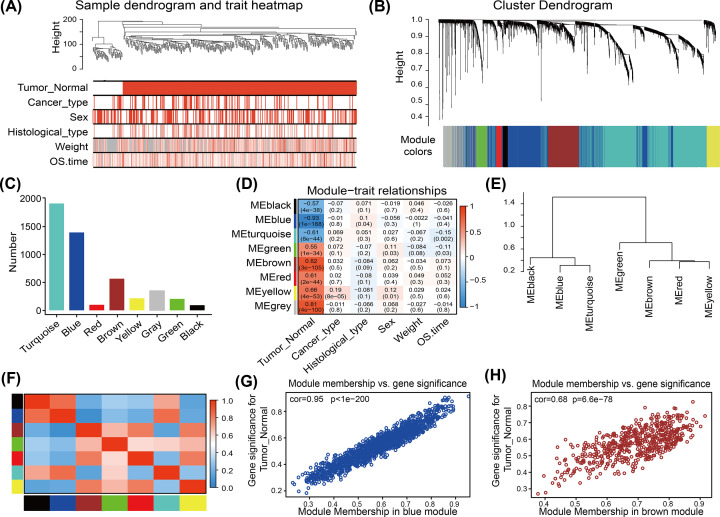
WGCNA was used to construct a network based on the expression matrix of 4832 DEGs and clinical data from 420 CRC samples. We performed cluster analysis to check the quality of the data from the 420 samples, all samples were in the clusters and within the cut-off threshold value (height < 200), therefore, no outliers were identified for removal (Figure 3A). Six clinical variables were applied in the WGCNA (Figure 3A): disease status (Tumor_Normal), cancer type, sex, histological subtype, body weight, and survival time (OS.time). The 420 samples fell into two clusters, Tumor and Normal.
Figure 3. WGCNA of DEGs in CRC.
(A) Clustering dendrogram of the clinical traits and data from 420 CRC samples. Red color represents ‘tumor’ for the variable ‘Tumor_Normal’, ‘colon and rectum cancer’ for the variable ‘Cancer-type’, ‘female’ or ‘male’ for the variable ‘Sex’, and ‘adenocarcinoma’ or ‘mucinous carcinoma’ for the variable ‘Histological_type’. For the variables ‘Weight’ and ‘OS time’, red color is directly proportional to the value. (B) Dendrogram of 4832 DEGs depending on the dissimilarity measure 1-TOM (see ‘Materials and methods’ section). Each branch represents a gene, and each color represents a co-expression module. (C) Numbers of genes in the eight modules. (D) Heatmap of the correlation between module eigengenes (MEs) and clinical characteristics of CRC patients. Each cell contains the correlation coefficient and P-value. (E,F) Module eigengene dendrogram and heatmap of eigengene adjacency. (G,H) Scatter plots of GS score and MM (see ‘Materials and methods’ section) for genes in the (G) blue and (H) brown modules.
To construct a scale-free network, we set the soft threshold power β to 7, the independence degree to 0.9, and the mean connectivity was close to 0 (Supplementary Figure S2A–D). DEGs with similar expression patterns clustered into the same modules, and modules showing a difference in cut height < 0.25 were merged. This procedure yielded eight co-expression modules: turquoise, blue, red, brown, yellow, gray, green, and black (Figure 3B,C, Supplementary Table S6). The gray module contained genes that could not be incorporated into any other module.
The eigengenes of the brown module strongly correlated positively with CRC (cor = 0.82, P=3 × 10−105), while the eigengenes of the blue module strongly highly correlated negatively with CRC (cor = −0.93, P=1× 10−88) (Figure 3D). These correlations were confirmed through analysis of hierarchical clustering, heatmaps, and adjacency relationships (Figure 3E,F). These results indicated that the brown module might contribute to tumorigenesis in CRC, while the blue module might protect against CRC. Therefore, the brown and blue modules were analyzed for hub genes.
Identification of candidate hub genes from brown and blue modules
MM and GS scores strongly correlated positively with each other in the brown and blue modules (Figure 3G,H). The criteria for selecting hub genes relatively lower than the standard cut-off threshold (MM > 0.8). In the brown module, 151 genes were identified that satisfied the thresholds of ‘cor.gene ModuleMembership’ > 0.75 and ‘cor.geneTraitSignificance’ > 0.6. In the blue module, 150 genes were identified that satisfied the thresholds of ‘cor.geneModuleMembership’ > 0.75, and ‘cor.gene TraitSignificance’ > 0.7.
Hub gene expression and correlation with survival
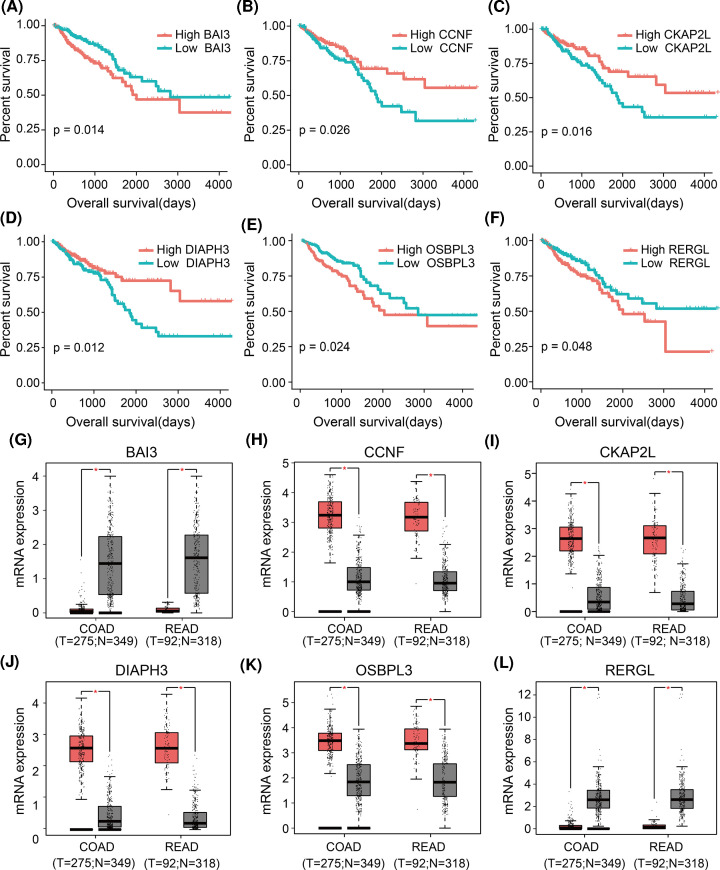
Based on expression data and clinical information for 360 CRC tumor samples in the TCGA, we examined potential associations between expression and patient survival for the 151 genes identified in the brown module and the 150 identified in the blue module. The brown module genes CCNF, CKAP2L, and DIAPH3 were associated with prognosis, as were the blue module genes BAI3, OSBPL3, and RERGL (Figure 4A–F). Thus, we defined these genes as ‘final’ hub genes.
Figure 4. Survival analysis and validation of six hub genes using an independent dataset.
(A–F) Kaplan–Meier survival curves of CRC patients stratified by low or high expression of the six hub genes. (G–L) Differences in expression of the six hub genes between normal and tumor tissues in the GEPIA database. *P<0.01.
Using GEPIA website, we confirmed that the expression of all these hub genes were significantly different between normal and CRC tissues (Figure 4G–L). BAI3 and RERGL were down-regulated in CRC, whereas the other hub genes were up-regulated. Similar results were obtained using data from the GEO database (Supplementary Figure S3A–F).
Mutation landscape of hub genes
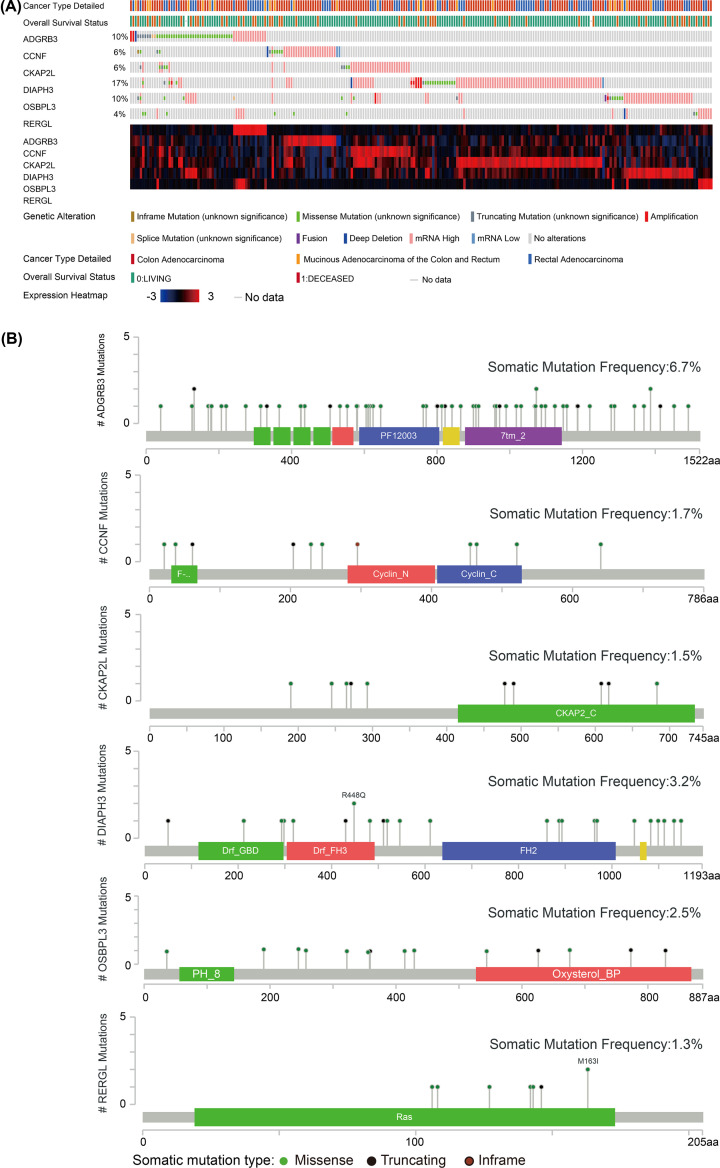
The OncoPrint view of hub genes in the CBioPortal database was used to visualize mutations in the six hub genes based on data from 594 CRC patients in the TCGA. Nearly half of these patients (41%) had mutations in all six hub genes. The highest rate of mutations was observed for DIAPH3 (17%), with missense mutations and mutations leading to higher mRNA expression being the most frequent (Figure 5A). BAI3 showed the highest somatic mutation rate (6.7%), and the most frequent mutations were missense mutations and deletions (Figure 5B).
Figure 5. Mutations in the six hub genes, based on CRC data in TCGA.
(A) Bar plots and heatmaps showing mutations in the six hub genes. (B) Lollipop plots showing the distributions of mutations in different domains of the proteins encoded by the six hub genes.
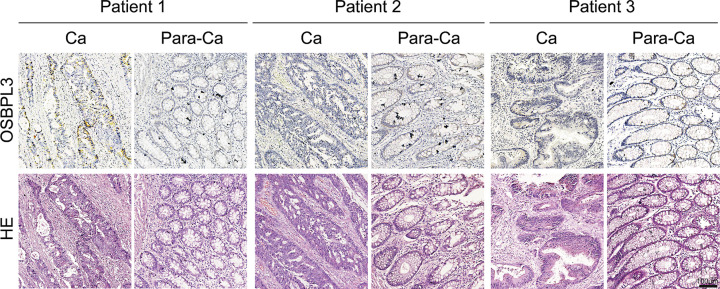
Immunohistochemical validation of OSBPL3 as a hub gene
We further validated the clinical significance of OSBPL3 as a hub gene using immunohistochemistry (Figure 6). We detected that OSBOL3 have heterogeneous expression in different types of tumor cells. OSBPL3, which localized mainly in the cytoplasm, was highly expressed in tumor cells and glandular epithelial cells, but less expressed in other cell types. Clearly indicate how its expression compared between tumor and normal samples overall.
Figure 6. Different expression of OSBPL3 in tumor tissues (Ca) and adjacent normal tissues (Para-Ca).
Scale bar, 100 μm. Abbreviation: HE, Hematoxylin–Eosin.
Discussion
CRC remains one of the world’s most malignant cancers. Although some studies have used WGCNA to explore molecular markers related to its pathogenesis, diagnosis and prognosis [34–36], the present work provided a more complete novel idea. We performed bioinformatics analyses across independent patient cohorts to identify biomarkers, one of which was experimentally validated. Our results suggest that poor prognosis in CRC is associated with overexpression of CCNF, CKAP2L, DIAPH3 and OSBPL3, and with underexpression of BAI3 and RERGL. Meanwhile, BAI3 and CKAP2L may be novel prognostic markers for CRC.
Consistent with the predicted functional enrichment of our CRC DEGs genes, the cell cycle has been shown to be dysregulated in many types of cancer [37]. Many studies have shown that targeted regulation of cancer cell cycle is a potential treatment strategy [38]. Therefore, studying cell cycle pathway may advance the understanding of oncogenic mechanisms and the treatment options for CRC. Similarly, defects in DNA replication can lead to mutations, chromosomal poly- or aneuploidy, as well as gene copy number variations, all of which can lead to cancer [39]. DNA mismatch repair (MMR) deficiency is one of the most well-known forms of genetic instability in CRC [40].
Through WGCNA, we identified module and hub genes likely to be important in CRC. We determined two key modules, brown and blue, whose genes are strongly related to CRC (Tumor_Normal). Genes in the red, yellow, and turquoise modules from our analysis may also play roles in CRC. Therefore, our results indicate that complex gene networks regulate CRC occurrence and development. Six hub genes in the brown and blue modules strongly correlated with overall survival of CRC patients: BAI3/ADGRB3, CCNF, CKAP2L, DIAPH3, OSBPL3, and RERGL. Two of these, BAI3 and CKAP2L, have not previously been linked to CRC, although expression of BAI3, a member of the BAI family [41], appears to be altered in malignant gliomas [42] and small cell lung cancer [43]. Similarly, expression of CKAP2L, a mitotic spindle protein, appears to be altered in lung adenocarcinoma [44], breast cancer [45], and non-small cell lung cancer [46].
The remaining four hub genes have previously been associated with CRC. CCNF is a founding member of the F-box family of proteins [47]. It can form the Skp1-Cul1-F-box protein ubiquitin ligase complex, which controls centrosome duplication and helps stabilize the genome [48]. Levels of CCNF can independently predict poor prognosis in patients with hepatocellular carcinoma [48]. Higher CCNF expression has been associated with longer survival in CRC [49]. DIAPH3, a formin ortholog [50], participates in actin remodeling and regulates cell movement and adhesion [51]. It can contribute to the development, invasion, and metastasis of lung adenocarcinoma, colorectal carcinoma [52–54]. OSBPL3 participates in lipid metabolism, vesicle trafficking, and cell signaling [55,56], and it is up-regulated in malignancies such as Burkitt’s lymphoma and CRC [57]. RERGL is a tumor suppressor gene of the Ras superfamily, and its underexpression has been linked to overall survival in CRC [58,59].
The hub genes that we identified differ from those identified in previous studies of CRC-related genes. One study identified three novel hub genes that could be candidate genes for CRC molecular mechanism studies (INHBA, CBX2, and BEST2) [60], while another identified seven that may contribute to early onset of CRC (SPARC, DCN, FBN1, WWTR1, TAGLN, DDX28, and CSDC2) [61]. Other studies have focused on specific genes in CRC, such as METTL3 [62] and METTL14 [63]. Differences in the key genes detected across these various studies may reflect differences in the clinicopathological characteristics of patients and in the types of analyses performed. However, we used a combination of bioinformatics analysis, experimental verification and dataset cross-validation to study CRC-related DEGs, and obtained two novel hub genes (BAI3 and CKAP2L) that may be associated with CRC’s prognosis. These results indicated that the present study provides new ideas for the study of molecular mechanisms of CRC.
Most previous WGCNA studies did not attempt to validate their genetic findings experimentally. Our six hub genes, which we validated in independent patient samples using bioinformatics and, in the case of OSBPL3, using immunohistochemistry, may help guide further studies to gain a comprehensive understanding of the network of genes involved in CRC. Such work may provide valuable clues for the treatment of CRC.
Supplementary Material
Abbreviations
- CRC
colorectal cancer
- DEG
differentially expressed gene
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- GS
gene significance
- MM
module membership
- PCC
Pearson correlation coefficient
- TCGA
The Cancer Genome Atlas
- TOM
topological overlap matrix
- WGCNA
weighted gene co-expression network analysis
Contributor Information
Dayun Feng, Email: dayunfmmu@163.com.
Lei Wang, Email: wangleibio@126.com.
Xinqiang Song, Email: xqsong2012@126.com.
Data Availability
The data used in the present study was obtained via an online database. The GSE33113 dataset was collected from the GEO (https://www.ncbi.nlm.nih.gov/geo/) with additional datasets obtained from the The UCSC Xena website (https://xenabrowser.net/).
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [grant number U1804179]; the Henan Science and Technology Innovation Team, the Investigation on Plant Resources in Dabie Mountains and the Study and Utilization of Active Components of Special Plants [grant number 2017083]; the Henan Key Scientific and Technological Projects [grant number 202102310190]; the Nanhu Scholars Program for Young Scholars of Xinyang Normal University [grant number 2018001]; and the Graduate Research Innovation Foundation of Xinyang Normal University [grant numbers 2020KYJJ36, 2020KYJJ60].
CRediT Author Contribution
Yu Zhang: Data curation. Jia Luo: Validation. Zhe Liu: Methodology. Xudong Liu: Data curation. Ying Ma: Investigation. Bohang Zhang: Data curation. Yuxuan Chen: Data curation. Xiaofeng Li: Methodology. Zhiguo Feng: Methodology. Ningning Yang: Methodology. Dayun Feng: Conceptualization. Lei Wang: Data curation. Xinqiang Song: Writing—review and editing.
References
- 1.Wang Q., Wang Y., Du L., Xu C., Sun Y., Yang B.et al. (2014) shRNA-mediated XRCC2 gene knockdown efficiently sensitizes colon tumor cells to X-ray irradiation in vitro and in vivo. Int. J. Mol. Sci. 15, 2157–2171 10.3390/ijms15022157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A.et al. (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 3.Barresi V., Reggiani Bonetti L., Ieni A., Caruso R.A. and Tuccari G. (2015) Histological grading in colorectal cancer: new insights and perspectives. Histol. Histopathol. 30, 1059–1067 [DOI] [PubMed] [Google Scholar]
- 4.Smaglo B.G. and Marshall J.L. (2013) Microsatellite instability in colorectal cancer. Clin. Adv. Hematol. Oncol. 11, 659–661 [PubMed] [Google Scholar]
- 5.Mughal M.J., Mahadevappa R. and Kwok H.F. (2019) DNA replication licensing proteins: saints and sinners in cancer. Semin. Cancer Biol. 58, 11–21 10.1016/j.semcancer.2018.11.009 [DOI] [PubMed] [Google Scholar]
- 6.Bolhaqueiro A.C.F., Ponsioen B., Bakker B., Klaasen S.J., Kucukkose E., van Jaarsveld R.H.et al. (2019) Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat. Genet. 51, 824–834 10.1038/s41588-019-0399-6 [DOI] [PubMed] [Google Scholar]
- 7.Tse J.W.T., Jenkins L.J., Chionh F. and Mariadason J.M. (2017) Aberrant DNA methylation in colorectal cancer: what should we target? Trends Cancer 3, 698–712 10.1016/j.trecan.2017.08.003 [DOI] [PubMed] [Google Scholar]
- 8.Jass J.R., Do K.A., Simms L.A., Iino H., Wynter C., Pillay S.P.et al. (1998) Morphology of sporadic colorectal cancer with DNA replication errors. 42, 673–679 10.1136/gut.42.5.673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller K.D., Nogueira L., Mariotto A.B., Rowland J.H., Yabroff K.R., Alfano C.M.et al. (2019) Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 69, 363–385 10.3322/caac.21565 [DOI] [PubMed] [Google Scholar]
- 10.Simon K. (2016) Colorectal cancer development and advances in screening. Clin. Interv. Aging 11, 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuenzi B.M. and Ideker T. (2020) A census of pathway maps in cancer systems biology. Nat. Rev. Cancer 20, 233–246 10.1038/s41568-020-0240-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barabasi A.L., Gulbahce N. and Loscalzo J. (2011) Network medicine: a network-based approach to human disease. Nat. Rev. Genet. 12, 56–68 10.1038/nrg2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian Y., Wang S.S., Zhang Z., Rodriguez O.C., Petricoin E. III, Shih Ie M.et al. (2014) Integration of network biology and imaging to study cancer phenotypes and responses. IEEE/ACM Trans. Comput. Biol. Bioinform. 11, 1009–1019 10.1109/TCBB.2014.2338304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller T., Langfelder P., Presson A. and Horvath S. (2011) Review of weighted gene coexpression network analysis. Handbook of Statistical Bioinformatics 369–388 10.1007/978-3-642-16345-6_18 [DOI] [Google Scholar]
- 15.van Dam S., Vosa U., van der Graaf A., Franke L. and de Magalhaes J.P. (2018) Gene co-expression analysis for functional classification and gene-disease predictions. Brief. Bioinform. 19, 575–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan Q., Tang J., Han Y. and Wang D. (2018) Co-expression modules construction by WGCNA and identify potential prognostic markers of uveal melanoma. Exp. Eye Res. 166, 13–20 10.1016/j.exer.2017.10.007 [DOI] [PubMed] [Google Scholar]
- 17.Langfelder P., Horvath S., (2009) WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 9, 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan L., Chen L., Qian K., Qian G., Wu C.L., Wang X.et al. (2017) Co-expression network analysis identified six hub genes in association with progression and prognosis in human clear cell renal cell carcinoma (ccRCC). Genom. Data 14, 132–140 10.1016/j.gdata.2017.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin H., Huang X., Shao K., Li G., Wang J., Yang H.et al. (2019) Integrated bioinformatics analysis to identify 15 hub genes in breast cancer. Oncol. Lett. 18, 1023–1034 10.3892/ol.2019.10411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia W.X., Yu Q., Li G.H., Liu Y.W., Xiao F.H., Yang L.Q.et al. (2019) Identification of four hub genes associated with adrenocortical carcinoma progression by WGCNA. PeerJ 7, e6555 10.7717/peerj.6555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldman M.J., Craft B., Hastie M., Repečka K., McDade F., Kamath A.et al. (2020) Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 38, 675–678 10.1038/s41587-020-0546-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W.et al. (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth G.K., Ritchie M., Thorne N., Wettenhall J. and Shi W. (2010) limma: Linear Models for Microarray Data. Bioinformatics Comput. Biol. Sol. R Bioconduct. 397–420 [Google Scholar]
- 24.Langfelder P. and Horvath S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L., Yuan L., Wang Y., Wang G., Zhu Y., Cao R.et al. (2017) Co-expression network analysis identified FCER1G in association with progression and prognosis in human clear cell renal cell carcinoma. Int. J. Biol. Sci. 13, 1361–1372 10.7150/ijbs.21657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura H., Fujii K., Gupta V., Hata H., Koizumu H., Hoshikawa M.et al. (2019) Identification of key modules and hub genes for small-cell lung carcinoma and large-cell neuroendocrine lung carcinoma by weighted gene co-expression network analysis of clinical tissue-proteomes. PLoS ONE 14, e0217105 10.1371/journal.pone.0217105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X., Hu L., Wang Y., Sun W. and Yang C. (2019) Single cell gene co-expression network reveals FECH/CROT signature as a prognostic marker. Cells. 8, 698 10.3390/cells8070698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Y., Chen D., Yu W. and Yan L. (2019) Bladder cancer stage-associated hub genes revealed by WGCNA co-expression network analysis. Hereditas 156, 7 10.1186/s41065-019-0083-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song Y., Pan Y. and Liu J. (2019) The relevance between the immune response-related gene module and clinical traits in head and neck squamous cell carcinoma. Cancer Manag. Res. 11, 7455–7472 10.2147/CMAR.S201177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Sousa E.M.F., Colak S., Buikhuisen J., Koster J., Cameron K., de Jong J.H.et al. (2011) Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 9, 476–485 10.1016/j.stem.2011.10.008 [DOI] [PubMed] [Google Scholar]
- 31.Kemper K., Versloot M., Cameron K., Colak S., de Sousa e Melo F., de Jong J.H.et al. (2012) Mutations in the Ras-Raf Axis underlie the prognostic value of CD133 in colorectal cancer. Clin. Cancer Res. 18, 3132–3141 10.1158/1078-0432.CCR-11-3066 [DOI] [PubMed] [Google Scholar]
- 32.Haoyi J., Peng L., Yunhao W., Xiangli M., Mengwei W., Jiahong H.et al. (2018) Exosomal zinc transporter ZIP4 promotes cancer growth and is a novel diagnostic biomarker for pancreatic cancer. Cancer Sci. 109, 2946–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun D., Jin H., Zhang J. and Tan X. (2018) Integrated whole genome microarray analysis and immunohistochemical assay identifies COL11A1, GJB2 and CTRL as predictive biomarkers for pancreatic cancer. Cancer Cell Int. 18, 174 10.1186/s12935-018-0669-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang W., Shi J., Zhou Y., Liu T., Zhan F., Zhang K.et al. (2019) Integrating proteomics and transcriptomics for the identification of potential targets in early colorectal cancer. Int. J. Oncol. 55, 439–450 10.3892/ijo.2019.4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lv Y., Xie B., Bai B., Shan L., Zheng W., Huang X.et al. (2019) Weighted gene coexpression analysis indicates that PLAGL2 and POFUT1 are related to the differential features of proximal and distal colorectal cancer. Oncol. Rep. 42, 2473–2485 10.3892/or.2019.7368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai L., Li J., Dong Z., Liu Y., Chen Y., Chen N.et al. (2019) Temporal expression and functional analysis of long non-coding RNAs in colorectal cancer initiation. J. Cell. Mol. Med. 23, 4127–4138 10.1111/jcmm.14300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schafer K.A. (1998) The cell cycle: a review. Vet. Pathol. 35, 461–478 10.1177/030098589803500601 [DOI] [PubMed] [Google Scholar]
- 38.Aarts M., Linardopoulos S. and Turner N.C. (2013) Tumour selective targeting of cell cycle kinases for cancer treatment. Curr. Opin. Pharmacol. 13, 529–535 10.1016/j.coph.2013.03.012 [DOI] [PubMed] [Google Scholar]
- 39.Ekundayo B. and Bleichert F. (2019) Origins of DNA replication. PLoS Genet. 15, e1008320 10.1371/journal.pgen.1008320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poulogiannis G., Frayling I.M. and Arends M.J. (2010) DNA mismatch repair deficiency in sporadic colorectal cancer and Lynch syndrome. Histopathology 56, 167–179 10.1111/j.1365-2559.2009.03392.x [DOI] [PubMed] [Google Scholar]
- 41.Kaur B., Brat D.J., Calkins C.C. and Van Meir E.G. (2003) Brain angiogenesis inhibitor 1 is differentially expressed in normal brain and glioblastoma independently of p53 expression. Am. J. Pathol. 162, 19–27 10.1016/S0002-9440(10)63794-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kee H.J., Ahn K.Y., Choi K.C., Won Song J., Heo T., Jung S.et al. (2004) Expression of brain-specific angiogenesis inhibitor 3 (BAI3) in normal brain and implications for BAI3 in ischemia-induced brain angiogenesis and malignant glioma. FEBS Lett. 569, 307–316 10.1016/j.febslet.2004.06.011 [DOI] [PubMed] [Google Scholar]
- 43.Thomas M., Snead D. and Mitchell D. (2017) An investigation into the potential role of brain angiogenesis inhibitor protein 3 (BAI3) in the tumorigenesis of small-cell carcinoma: a review of the surrounding literature. J. Recept. Signal. Transduct. 37, 325–334 10.1080/10799893.2017.1328441 [DOI] [PubMed] [Google Scholar]
- 44.Xiong G., Li L., Chen X., Song S., Zhao Y., Cai W.et al. (2019) Up-regulation of CKAP2L expression promotes lung adenocarcinoma invasion and is associated with poor prognosis. Onco Targets Ther. 12, 1171–1180 10.2147/OTT.S182242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu Y., Zhou Q.Z., Zhang X.L., Wang Z.Z. and Wang P. (2019) Identification of hub genes using co-expression network analysis in breast cancer as a tool to predict different stages. Med. Sci. Monit. 25, 8873–8890 10.12659/MSM.919046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tu H., Wu M., Huang W. and Wang L. (2019) Screening of potential biomarkers and their predictive value in early stage non-small cell lung cancer: a bioinformatics analysis. Transl. Lung Cancer Res. 8, 797–807 10.21037/tlcr.2019.10.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mavrommati I., Faedda R., Galasso G., Li J., Burdova K., Fischer R.et al. (2018) β-TrCP- and casein kinase II-mediated degradation of cyclin F controls timely mitotic progression. Cell Rep. 24, 3404–3412 10.1016/j.celrep.2018.08.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu J., Qiu H., Cai M., Pan Y., Cao Y., Liu L.et al. (2013) Low cyclin F expression in hepatocellular carcinoma associates with poor differentiation and unfavorable prognosis. Cancer Sci. 104, 508–515 10.1111/cas.12100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J., Wang Z., Shen X., Cui X. and Guo Y. (2019) Identification of novel biomarkers and small molecule drugs in human colorectal cancer by microarray and bioinformatics analysis. Mol. Genet. Genom. Med. 7, e00713 10.1002/mgg3.713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morley S., You S., Pollan S., Choi J., Zhou B., Hager M.H.et al. (2015) Regulation of microtubule dynamics by DIAPH3 influences amoeboid tumor cell mechanics and sensitivity to taxanes. Sci. Rep. 5, 12136 10.1038/srep12136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koleck T.A. and Conley Y.P. (2016) Identification and prioritization of candidate genes for symptom variability in breast cancer survivors based on disease characteristics at the cellular level. Breast Cancer 8, 29–37 10.2147/BCTT.S88434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hager M.H., Morley S., Bielenberg D.R., Gao S., Morello M., Holcomb I.N.et al. (2012) DIAPH3 governs the cellular transition to the amoeboid tumour phenotype. EMBO Mol. Med. 4, 743–760 10.1002/emmm.201200242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiang G., Weiwei H., Erji G. and Haitao M. (2019) DIAPH3 promotes the tumorigenesis of lung adenocarcinoma. Exp. Cell. Res. 385, 111662 10.1016/j.yexcr.2019.111662 [DOI] [PubMed] [Google Scholar]
- 54.Rashed H.M.H., Ahmed M., Wagih H.M., Foda A. and Sami M.M. (2018) Role of MEK1 and DIAPH3 expression in colorectal carcinoma. Res. Oncol. 14, 75–82 [Google Scholar]
- 55.Olkkonen V.M., Levine T.P.J.B. and Cellulaire C.B.-B.E.B. (2004) Oxysterol binding proteins: in more than one place at one time? Biochem. Cell Biol. 82, 87–98 10.1139/o03-088 [DOI] [PubMed] [Google Scholar]
- 56.Lehto M. and Olkkonen V.M. (2003) The OSBP-related proteins: a novel protein family involved in vesicle transport cellular lipid metabolism, and cell signalling. Biochim. Biophys. Acta. 1631, 1–11 10.1016/S1388-1981(02)00364-5 [DOI] [PubMed] [Google Scholar]
- 57.Jiao H.L., Weng B.S., Yan S.S., Lin Z.M., Wang S.Y., Chen X.P.et al. (2020) Upregulation of OSBPL3 by HIF1A promotes colorectal cancer progression through activation of RAS signaling pathway. Cell Death Dis. 11, 571 10.1038/s41419-020-02793-3 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Yang R., Chen B., Pfütze K., Buch S., Steinke V., Holinski-Feder E.et al. (2014) Genome-wide analysis associates familial colorectal cancer with increases in copy number variations and a rare structural variation at 12p12.3. Carcinogenesis 35, 315–323 10.1093/carcin/bgt344 [DOI] [PubMed] [Google Scholar]
- 59.Mangiola S., Stuchbery R., Macintyre G., Clarkson M.J., Peters J.S., Costello A.J.et al. (2018) Periprostatic fat tissue transcriptome reveals a signature diagnostic for high-risk prostate cancer. Endocr. Relat. Cancer 25, 569–581 10.1530/ERC-18-0058 [DOI] [PubMed] [Google Scholar]
- 60.Qin L., Zeng J., Shi N., Chen L. and Wang L. (2020) Application of weighted gene co-expression network analysis to explore the potential diagnostic biomarkers for colorectal cancer. Mol. Med. Rep. 21, 2533–2543 10.3892/mmr.2020.11047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mo X., Su Z., Yang B., Zeng Z., Lei S. and Qiao H. (2020) Identification of key genes involved in the development and progression of early-onset colorectal cancer by co-expression network analysis. Oncol. Lett. 19, 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li T., Hu P.S., Zuo Z., Lin J.F., Li X., Wu Q.N.et al. (2019) METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer 18, 112 10.1186/s12943-019-1038-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X., Xu M., Xu X., Zeng K., Liu X., Pan B.et al. (2020) METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol. Cancer 19, 106 10.1186/s12943-020-01220-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in the present study was obtained via an online database. The GSE33113 dataset was collected from the GEO (https://www.ncbi.nlm.nih.gov/geo/) with additional datasets obtained from the The UCSC Xena website (https://xenabrowser.net/).