Abstract
Study Objectives:
We have developed the CardioRespiratory Sleep Staging (CReSS) algorithm for estimating sleep stages using heart rate variability and respiration, allowing for estimation of sleep staging during home sleep apnea tests. Our objective was to undertake an epoch-by-epoch validation of algorithm performance against the gold standard of manual polysomnography sleep staging.
Methods:
Using 296 polysomnographs, we created a limited montage of airflow and heart rate and deployed CReSS to identify each 30-second epoch as wake, light sleep (N1 + N2), deep sleep (N3), or rapid eye movement (REM) sleep. We calculated Cohen’s kappa and the percentage of accurately identified epochs. We repeated our analyses after stratification by sleep-disordered breathing (SDB) severity, and after adding thoracic respiratory effort as a backup signal for periods of invalid airflow.
Results:
CReSS discriminated wake/light sleep/deep sleep/REM sleep with 78% accuracy; the kappa value was 0.643 (95% confidence interval, 0.641–0.645). Discrimination of wake/sleep demonstrated a kappa value of 0.711 and accuracy of 89%, non-REM sleep/REM sleep demonstrated a kappa of 0.790 and accuracy of 94%, and light sleep/deep sleep demonstrated a kappa of 0.469 and accuracy of 87%. Kappa values did not vary by more than 0.07 across subgroups of no SDB, mild SDB, moderate SDB, and severe SDB. Accuracy increased to 80%, with a kappa value of 0.680 (95% confidence interval, 0.678–0.682), when CReSS additionally utilized the thoracic respiratory effort signal.
Conclusions:
We observed substantial agreement between CReSS and the gold-standard comparator of manual sleep staging of polysomnographic signals, which was consistent across the full range of SDB severity. Future research should focus on the extent to which CReSS reduces the discrepancy between the apnea-hypopnea index and the respiratory event index, and the ability of CReSS to identify REM sleep–related obstructive sleep apnea.
Citation:
Bakker JP, Ross M, Vasko R, et al. Estimating sleep stages using cardiorespiratory signals: validation of a novel algorithm across a wide range of sleep-disordered breathing severity. J Clin Sleep Med. 2021;17(7):1343–1354.
Keywords: polysomnography, sleep apnea syndromes, sleep stages, validation study
BRIEF SUMMARY
Current Knowledge/Study Rationale: The lack of neurological signals collected during home sleep apnea tests means that it is not possible to identify sleep stages and total sleep time, which has important clinical implications. We have recently developed an algorithm designed to estimate sleep stages using heart rate and airflow signals.
Study Impact: In this validation study, we found substantial agreement between sleep staging determined by the algorithm vs the gold standard of manual sleep staging, which was consistent across the full spectrum of sleep-disordered breathing severity and improved when a respiratory effort signal was used as an input in addition to airflow and heart rate. Future research is warranted to determine the impact of the algorithm on sleep-disordered breathing diagnostic accuracy.
INTRODUCTION
Home sleep apnea test (HSAT) devices are increasingly used as an alternative to polysomnography (PSG) to diagnose sleep-disordered breathing (SDB).1 Potential advantages of taking the diagnostic process from the hospital to the home include reduced cost of equipment, increased access in remote and/or underserved areas, higher patient turnover, increased patient comfort, and the collection of data that are more representative of a patient’s habitual sleep.1–3 There are, however, some drawbacks associated with relying on a reduced signal montage, which varies by device but at minimum includes airflow, pulse oximetry, and respiratory effort, for identification and classification of apneas and hypopneas. The absence of electroencephalography, electrooculography, and electromyography signals required for sleep staging means that the apnea-hypopnea index (AHI) cannot be calculated; instead, total sleep time is substituted by either monitoring time or recording time for calculation of the respiratory event index (REI).4 Reliance on the REI in place of the AHI results in reduced SDB diagnostic sensitivity that is not easily quantifiable.5 Further, there is no ability to screen for rapid eye movement (REM) sleep–related obstructive sleep apnea (OSA) or identify abnormalities in sleep architecture that may impact the subsequent treatment plan or signify the need for further testing.6
We recently developed an algorithm for estimating sleep stages using heart rate variability and respiration known as CardioRespiratory Sleep Staging (CReSS). The CReSS algorithm is device-agnostic and requires airflow and heart rate inputs with an option to rely on a respiratory effort signal during periods when airflow is absent or invalid. The purpose of the current study was to undertake an epoch-by-epoch validation of the CReSS algorithm against the gold-standard comparator of manual PSG sleep staging. In exploratory analyses, we investigated the impact of different signal types for measuring airflow and derived the heart rate, the consistency of algorithm performance across SDB severity subgroups, and the accuracy of algorithm performance when respiratory effort was used as a backup signal for airflow. An additional, overarching objective was to undertake our analyses in a large sample, to support the generalizability of the algorithm performance.
METHODS
The study was approved by the Western Institutional Review Board (20192293). We accessed deidentified home PSGs from the National Sleep Research Resource7,8 that were originally collected in the Sleep Heart Health Study (SHHS; Only PSGs from the second SHHS study visit were accessed)9,10 and the Multi-Ethnic Study of Atherosclerosis (MESA)11; thus, informed consent was waived. The PSGs used for these analyses are considered an independent validation dataset, as none were used for algorithm training, which was undertaken using PSGs from the Siesta12 and Somnoval13 datasets.
Selection of PSGs
Based on the manual scoring logs collected in the original studies, we excluded PSGs with unreliable sleep staging or apnea/hypopnea scoring, as well as those containing < 4 hours of valid nasal pressure or thermistry, transmissive finger photoplethysmography (PPG) or electrocardiography (ECG), and/or thoracic respiratory effort signals. From the SHHS dataset, we excluded PSGs collected from participants with a cardiac pacemaker, heart failure, or atrial fibrillation; this information was not available in the MESA dataset. From the remaining PSGs, we randomly selected n = 74 from each dataset within each of the following disease severity categories: AHI < 5 events/h (no SDB), 5 to < 15 events/h (mild), 15 to < 30 events/h (moderate), and ≥ 30 events/h (severe), using the AHI variable provided in the National Sleep Research Resource metadata (with hypopneas defined according to a 3% SpO2 desaturation). We did not seek to analyze all MESA and SHHS PSGs that met these criteria because although the scoring process is automated, importing raw PSG files collected across different platforms and with different signal montages/labeling is a manual process. As such, we compared the included PSGs to the entire MESA/SHHS cohort with respect to age, sex, sleep architecture, and AHI (Table S1 (36.1KB, pdf) in the supplemental material).
Description of the CReSS algorithm
First, interbeat intervals are calculated from the cardiac input signal (either PPG or ECG), and an instantaneous heart rate signal is derived from the intervals. When PPG is used as input, the signal is first band-pass filtered between 0.15 and 2.25 Hz using a third-order Butterworth filter, after which the pulse feet are determined by searching for consecutive local minima in the filtered signal. Heartbeats are located by selection of signal troughs that are followed by uprising arcs with a strictly positive slope. Interbeat intervals are calculated as the time between consecutive selected troughs, which are then filtered to plausible interbeat interval ranges between 0.33 and 1.82 seconds. When ECG is used as input, the algorithm uses an implementation based on the approach proposed by Jalil et al.14 Using a discrete wavelet decomposition with a second-order Mexican hat function, baseline drift is first removed by zeroing the approximation coefficients of the transform for a low approximation level. Singularities in the signal—a subset of which corresponds to R peaks—are detected by computing the wavelet transform modulus maxima. The temporal location of the singularities is determined by tracing the modulus maxima from higher to lower scales. Interbeat intervals are then calculated as the time between the temporal location of selected R-peak singularities. Next, the instantaneous heart rate and respiratory signals (airflow, with or without respiratory effort) are all resampled at 10 Hz and respiratory signals are scaled to a 16-bit integer range between –32,768 and 32,767.
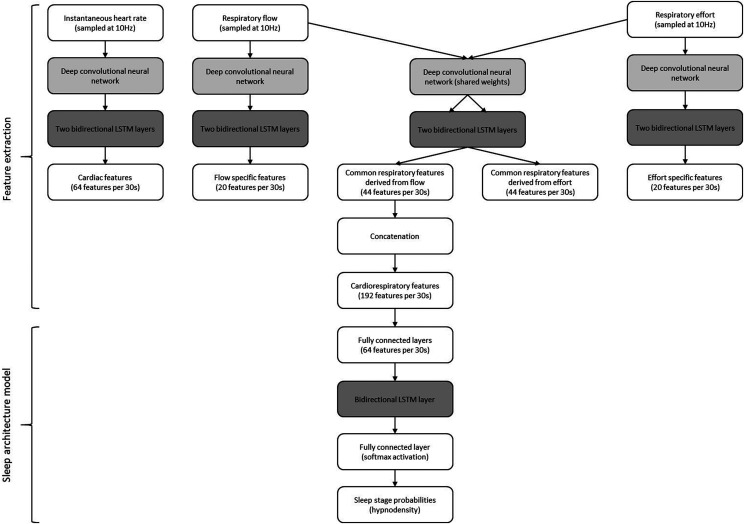
The CReSS algorithm then uses a deep convolutional neural network to extract features from the instantaneous heart rate and respiratory signal/s (airflow, with or without respiratory effort as backup). If both airflow and respiratory effort signals are used, then network weights are partially shared between the signals. A modified version of the ResNeXt15 model for deep convolutional networks, originally designed for image classification, was applied using 1-dimensional convolutions in order to extract high-level features per 30-second epoch from temporal input data. The architecture was enhanced by using scaled exponential linear units in order to exploit their self-normalizing properties as described previously.16 Finally, 3 layers of bidirectional long short-term memory17 layers introduce global context from the entire recording and assign probabilities for wake, REM sleep, light sleep (LS; corresponding to sleep stages N1 + N2 according to American Academy of Sleep Medicine [AASM] criteria18), or deep sleep (DS; corresponding to AASM sleep stage N3). The design of the long short-term memory classifier is the same as presented previously.19,20 An overview of the neural network is shown in Figure 1.
Figure 1. Flow chart of the preprocessed input data through the artificial neural network for cardiorespiratory sleep staging.
LSTM = long short-term memory.
Data processing
The PSG platform varied at each of the 9 SHHS and 6 MESA data collection sites; however, each study deployed standardized data collection procedures and montage, and sleep staging in 30-second epochs was performed at a central sleep reading center by experienced, certified sleep technologists. SHHS followed the Rechtschaffen and Kales criteria,21 while MESA followed the 2007 AASM criteria.22
The full PSGs were provided in the European Data Format, along with associated annotation files containing manually scored stages and events and a summary metadata file with endpoints and lights on/off times. The PSGs were imported into Sleepware G3 (Philips Respironics, Monroeville, PA) and the lights on/off information was added. Manual sleep staging was left unchanged and exported in 30-second epochs. In MESA, N1 and N2 were combined and labeled as LS, and N3 was labeled as DS.22 In SHHS, stages 1 and 2 were combined and labeled LS, and stages 3 and 4 were combined and labeled DS.21 To create HSAT montages, the full PSGs were reduced to only airflow (nasal pressure in MESA, thermistor in SHHS) and heart rate (PPG in MESA, ECG in SHHS), and we added thoracic respiratory effort for our exploratory analyses. The reduced montages were imported into Sleepware G3, lights on/off information was added, and the CReSS algorithm was executed.
Statistical analyses
All analyses were performed using Matlab R2019b (MathWorks, Natick, MA), validated against IBM SPSS (version 19.0.0.2; Armonk, NY). For our primary analyses, we undertook epoch-by-epoch comparisons of manual vs CReSS sleep staging using the MESA dataset, calculating the percentage accuracy of wake/LS/DS/REM sleep (that is, the percentage of epochs correctly identified by CReSS), as well as Cohen’s kappa values for comparisons of wake/LS/DS/REM sleep, wake/non-REM (NREM) sleep/REM sleep, wake/sleep, NREM sleep/REM sleep, and LS/DS. For the discrimination of NREM sleep/REM sleep that did not include wake, we transformed the confusion matrix by removing the wake column and row; the same transformation was undertaken for the LS/DS discrimination by removing both wake and REM sleep. We compared our kappa values against the benchmarks of Landis and Koch23; a kappa > 0.6 reflected substantial agreement, while a kappa > 0.4 reflected moderate agreement. Our primary analyses were conducted with MESA PSGs only, as manual sleep staging was undertaken with the current AASM scoring criteria in that study.18
We then calculated the percentage accuracy of wake/sleep, NREM sleep/REM sleep, LS/DS, and wake/NREM sleep/REM sleep, the macro-F1 score (the arithmetic mean of F1 scores for each sleep stage), and the weighted macro-F1 score, with weighting performed according to the frequency of each sleep stage as described elsewhere.24 All analyses were repeated using the SHHS dataset to assess the impact of measuring airflow via nasal pressure vs thermistry and the impact of measuring heart rate from PPG vs ECG, as MESA used the former combination while SHHS used the latter. Within each dataset, we conducted stratified analyses after categorizing each PSG according to SDB severity (AHI < 5 events/h, 5 to < 15 events/, 15 to < 30 events/h, and ≥ 30 events/h, using the AHI generated in Sleepware G3 based on manual sleep staging and event scoring). Finally, we repeated all analyses in both datasets after allowing CReSS to utilize the thoracic respiratory effort signal in addition to airflow and heart rate.
RESULTS
Descriptive information is provided in Table 1. Participants contributing to the MESA and SHHS PSGs in our sample were aged 69 and 68 years on average, respectively, with an approximately equal distribution of males and females. The SHHS sample was predominantly White/Caucasian, whereas MESA recruitment targeted a more racially/ethnically diverse sample. As anticipated given our sampling strategy, our sample reflected a wide range of SDB severity, from 0–88 and 87 events/h in MESA and SHHS, respectively. Demographics, disease severity, and sleep architecture were similar when our selected samples were compared to the larger MESA and SHHS cohorts (Table S1 (36.1KB, pdf) ).
Table 1.
Descriptive demographic and clinical information.
| MESA (n = 296) | SHHS (n = 296) | |
|---|---|---|
| Age (y) | 69.4 ± 8.8 | 67.5 ± 10.0 |
| Sex (number; %) | ||
| Female | 154; 52.0% | 154; 52.0% |
| Male | 142; 48.0% | 142; 48.0% |
| Race/ethnicity (number; %) | ||
| White/Caucasian | 117; 39.5% | 265; 89.5% |
| Chinese American | 31; 10.5% | NA |
| African American | 76; 25.7% | 15; 5.1% |
| Hispanic | 72; 24.3% | NA |
| Other | NA | 16; 5.4% |
| BMI (kg/m2) | NA | 28.1 ± 4.9 |
| Neck circumference (cm) | NA | 37.6 ± 3.9 |
| AHI (events/h) | ||
| Mean ± SD | 22.9 ± 21.1 | 17.1 ± 17.1 |
| Median (Q1; Q3) | 16.9 (5.7; 34.5) | 12.0 (4.4; 24.6) |
| Range | 0–88 | 0–87 |
| SDB severity per AHI (number; %)* | ||
| None | 63; 21.3% | 82; 27.7% |
| Mild | 76; 25.7% | 97; 32.8% |
| Moderate | 73; 24.7% | 61; 20.6% |
| Severe | 84; 28.4% | 56; 18.9% |
| Epworth Sleepiness Scale (0-24) | 6.2 ± 4.2 | 7.7 ± 4.0 |
| Total sleep time per PSG (h) | 6.0 ± 1.4 | 6.3 ± 1.1 |
| Recording time (h) | 10.6 ± 1.3 | 10.0 ± 1.1 |
Data are provided as mean ± SD unless indicated otherwise. BMI and neck circumference were not available in the MESA dataset accessible in the NSRR. MESA data are complete for all variables in this table. SHHS data in this table had n = 6 missing Epworth scores and n = 1 missing BMI. Race and ethnicity are reported as a composite variable, reflecting the data available from the original studies. *PSGs were selected for inclusion based on the AHI variable available in the NSRR metadata. When PSGs and annotation files were imported into Sleepware G3 to re-create manual scoring, some minor changes to AHI values resulted from adjustments to event filters and to ensure alignment of lights on/off times per the NSRR metadata. The AHI value provided in this table reflects the AHI generated by exporting manual scoring/staging from Sleepware G3 and is consistent with all other AHI values throughout this report. AHI = apnea-hypopnea index, BMI = body mass index, MESA = Multi-Ethnic Study of Atherosclerosis, NA = data not available, NSRR= National Sleep Research Resource, PSG = polysomnography, Q = quartile, SD = standard deviation, SDB = sleep-disordered breathing, SHHS = Sleep Heart Health Study.
Performance of CReSS with reference to manual sleep staging in MESA (primary analyses)
Cohen’s kappa values along with percent accuracy comparing CReSS (applied to airflow and heart rate signals only) to manually scored PSG sleep staging are provided in the upper part of Table 2. For each sleep stage discrimination, the lower bound of the 95% confidence interval for the kappa value exceeded our prespecified performance threshold. For the most granular sleep stage discrimination performed by CReSS—that is, the discrimination of wake/LS/DS/REM sleep—Cohen’s kappa was 0.643 (95% confidence interval, 0.641–0.645), which equates to substantial agreement with manual PSG staging per the thresholds of Landis and Koch.23 The percentage of wake/LS/DS/REM sleep epochs that were staged correctly by CReSS was 77.6%. Accuracy increased when sleep stages were combined into discriminations of wake/sleep and wake/NREM sleep/REM sleep. A confusion matrix is provided in Table 3.
Table 2.
Kappa values and accuracy for manual vs CReSS sleep staging.
| Sleep Stage Discrimination | MESA Dataset (n = 296) | SHHS Dataset (n = 296) | ||||||
|---|---|---|---|---|---|---|---|---|
| Kappa (95% CI) | Macro-F1 (95% CI) | Weighted Macro-F1 (95% CI) | Percent Accuracy | Kappa (95% CI) | Macro-F1 (95% CI) | Weighted Macro-F1 (95% CI) | Percent Accuracy | |
| CReSS Applied to Heart Rate and Airflow Signals | ||||||||
| Wake/LS/DS/REM sleep | 0.643 (0.641–0.645) | 0.728 (0.717–0.740) | 0.777 (0.768–0.785) | 77.6 | 0.578 (0.576–0.581) | 0.692 (0.679–0.706) | 0.739 (0.728–0.750) | 73.3 |
| Wake/sleep | 0.711 (0.708–0.714) | 0.855 (0.843–0.864) | 0.897 (0.888–0.903) | 89.3 | 0.634 (0.631–0.638) | 0.816 (0.803–0.827) | 0.890 (0.882–0.897) | 88.3 |
| NREM sleep/REM sleep | 0.790 (0.786–0.793) | 0.895 (0.885–0.902) | 0.936 (0.930–0.940) | 93.5 | 0.756 (0.752–0.759) | 0.878 (0.866–0.887) | 0.926 (0.919–0.931) | 92.4 |
| LS/DS | 0.469 (0.462–0.475) | 0.734 (0.718–0.748) | 0.870 (0.861–0.878) | 86.8 | 0.445 (0.439–0.451) | 0.721 (0.707–0.733) | 0.846 (0.837–0.854) | 83.6 |
| Wake/NREM sleep/REM sleep | 0.719 (0.717–0.722) | 0.819 (0.808–0.827) | 0.850 (0.841–0.857) | 84.8 | 0.665 (0.662–0.668) | 0.781 (0.765–0.793) | 0.832 (0.821–0.841) | 82.6 |
| CReSS Applied to Heart Rate, Airflow, and Thoracic Respiratory Effort Signals | ||||||||
| Wake/LS/DS/REM sleep | 0.680 (0.678–0.682) | 0.748 (0.737–0.759) | 0.800 (0.791–0.807) | 79.8 | 0.635 (0.633–0.637) | 0.750 (0.723–0.769) | 0.805 (0.782–0.819) | 76.7 |
| Wake/sleep | 0.756 (0.753–0.759) | 0.878 (0.868–0.887) | 0.911 (0.903–0.918) | 90.8 | 0.705 (0.701–0.708) | 0.885 (0.862–0.900) | 0.909 (0.889–0.920) | 90.4 |
| NREM sleep/REM sleep | 0.823 (0.820–0.827) | 0.912 (0.906–0.918) | 0.944 (0.940–0.949) | 94.5 | 0.807 (0.803–0.810) | 0.908 (0.894–0.920) | 0.942 (0.932–0.950) | 93.8 |
| LS/DS | 0.461 (0.454–0.468) | 0.730 (0.715–0.745) | 0.874 (0.865–0.881) | 87.0 | 0.464 (0.458–0.470) | 0.735 (0.705–0.771) | 0.881 (0.863–0.895) | 84.3 |
| Wake/NREM sleep/REM sleep | 0.762 (0.760–0.764) | 0.847 (0.838–0.856) | 0.871 (0.863–0.878) | 86.9 | 0.729 (0.727–0.731) | 0.847 (0.826–0.864) | 0.869 (0.851–0.882) | 85.8 |
An F1 score is the harmonic mean of positive predictive value (precision) and sensitivity (recall); the macro-F1 score presented here for each sleep stage discrimination is the arithmetic mean of the F1 scores calculated across all sleep stages. In addition, we present weighted macro-F1 scores performed according to the frequency of each sleep stage within the dataset. Discriminations of wake/LS/DS/REM sleep and wake/NREM sleep/REM sleep are based on all epochs. For the discrimination of NREM sleep/REM sleep that does not include wake, we transformed the confusion matrix by removing the wake column and row; the same transformation was undertaken for the LS/DS discrimination by removing both wake and REM sleep. CI = confidence interval, CReSS = CardioRespiratory Sleep Staging, DS = deep sleep (corresponding to N3), LS = light sleep (corresponding to N1 + N2), NREM = non-REM, REM = rapid eye movement.
Table 3.
Confusion matrix for epoch-by-epoch sleep staging in MESA.
| CReSS Sleep Staging | PSG Sleep Staging | |||
|---|---|---|---|---|
| CReSS Applied to Heart Rate and Airflow Signals | ||||
| Wake | REM Sleep | Light Sleep | Deep Sleep | |
| Wake | 55,524 | 943 | 7,383 | 203 |
| 71.0%* | 2.3% | 4.9% | 0.8% | |
| REM sleep | 3,152 | 32,275 | 6,115 | 68 |
| 4.0% | 80.1%* | 4.1% | 0.3% | |
| Light sleep | 19,267 | 7,004 | 126,217 | 11,271 |
| 24.6% | 17.4% | 84.4%* | 46.6% | |
| Deep sleep | 280 | 88 | 9,802 | 12,655 |
| 0.4% | 0.2% | 6.6% | 52.3%* | |
| PPV | 86.7% | 77.6% | 77.1% | 55.4%* |
| CReSS Applied to Heart Rate, Airflow, and Thoracic Respiratory Effort Signals | ||||
| Wake | REM Sleep | Light Sleep | Deep Sleep | |
| Wake | 59,487 | 598 | 7,299 | 131 |
| 76.0%* | 1.5% | 4.9% | 0.5% | |
| REM sleep | 2,752 | 34,390 | 6,009 | 69 |
| 3.5% | 85.3%* | 4.0% | 0.3% | |
| Light sleep | 15,837 | 5,265 | 127,388 | 11,987 |
| 20.2% | 13.1% | 85.2%* | 49.5% | |
| Deep sleep | 147 | 57 | 8,821 | 12,010 |
| 0.2% | 0.1% | 5.9% | 49.6%* | |
| PPV | 88.1% | 79.6% | 79.4% | 57.1%* |
*Values are the number of epochs of each PSG-scored sleep stage that were correctly identified by CReSS and the percentage of epochs within each PSG-defined sleep stage that were correctly identified by CReSS (that is, sensitivity). The percentage values labeled as PPV represent the percentage of epochs within each CReSS-defined sleep stage that was correct per PSG. CReSS = CardioRespiratory Sleep Staging, MESA = Multi-Ethnic Study of Atherosclerosis, PPV = positive predictive value, PSG = polysomnography, REM = rapid eye movement.
Impact of different airflow and heart signals (SHHS data) on CReSS performance
The MESA study used nasal pressure for airflow and PPG for heart rate, while SHHS used thermistry and ECG, respectively. We therefore repeated the above analyses using 296 PSGs from SHHS to determine the impact of different signal types on the performance of CReSS. As shown in the upper part of Table 2, the accuracy of wake/LS/DS/REM sleep discrimination using SHHS data was 73.3%. Again, accuracy increased as different sleep stages were combined. A confusion matrix is provided in Table 4.
Table 4.
Confusion matrix for epoch-by-epoch sleep staging in SHHS.
| CReSS Sleep Staging | PSG Sleep Staging | |||
|---|---|---|---|---|
| CReSS Applied to Heart Rate and Airflow Signals | ||||
| Wake | REM Sleep | Light Sleep | Deep Sleep | |
| Wake | 39,954 | 942 | 6,768 | 324 |
| 61.1%* | 2.1% | 4.8% | 0.9% | |
| REM sleep | 4,009 | 33,326 | 4,934 | 251 |
| 6.1% | 73.4%* | 3.5% | 0.7% | |
| Light sleep | 20,919 | 11,001 | 121,248 | 18,100 |
| 32.0% | 24.2% | 85.5%* | 52.4% | |
| Deep sleep | 511 | 161 | 8,783 | 15,888 |
| 0.8% | 0.4% | 6.2% | 46.0%* | |
| PPV | 83.3% | 78.4% | 70.8% | 62.7%* |
| CReSS Applied to Heart Rate, Airflow, and Thoracic Respiratory Effort Signals | ||||
| Wake | REM Sleep | Light Sleep | Deep Sleep | |
| Wake | 44,270 | 562 | 5,566 | 197 |
| 67.7%* | 1.2% | 3.9% | 0.6% | |
| REM sleep | 2,917 | 36,584 | 4,888 | 208 |
| 4.5% | 80.5%* | 3.4% | 0.6% | |
| Light sleep | 18,001 | 8,192 | 123,168 | 17,859 |
| 27.5% | 18.0% | 86.9%* | 51.7% | |
| Deep sleep | 205 | 92 | 8,111 | 16,299 |
| 0.3% | 0.2% | 5.7% | 47.2%* | |
| PPV | 87.5% | 82.0% | 73.7% | 66.0%* |
*Values are the number of epochs of each PSG-scored sleep stage that were correctly identified by CReSS and the percentage of epochs within each PSG-defined sleep stage that were correctly identified by CReSS (that is, sensitivity). The percentage values labeled as PPV represent the percentage of epochs within each CReSS-defined sleep stage that was correct per PSG. CReSS = CardioRespiratory Sleep Staging, PPV = positive predictive value, PSG = polysomnography, REM = rapid eye movement, SHHS = Sleep Heart Health Study.
Performance of CReSS with reference to manual sleep staging stratified by SDB severity
Cohen’s kappa values and percent accuracy of each sleep stage discrimination stratified by SDB disease severity are provided in the upper parts of Table 5 (MESA) and Table 6 (SHHS), as well as Figure 2. In MESA, the kappa values did not vary by more than 0.07 across disease severity subgroups; in SHHS, the kappa values were within 0.14. There was also little variability in the percentage of epochs correctly staged by CReSS across subgroups in both datasets.
Table 5.
Kappa values and accuracy of manual sleep staging vs CReSS sleep staging in MESA by sleep apnea severity subgroups.
| Sleep Stage Discrimination | No SDB (n = 63) | Mild SDB (n = 76) | Moderate SDB (n = 73) | Severe SDB (n = 84) | ||||
|---|---|---|---|---|---|---|---|---|
| Kappa (95% CI) | Percent Accuracy | Kappa (95% CI) | Percent Accuracy | Kappa (95% CI) | Percent Accuracy | Kappa (95% CI) | Percent Accuracy | |
| CReSS Applied to Heart Rate and Airflow Signals | ||||||||
| Wake/LS/DS/REM sleep | 0.648 (0.643–0.653) | 77.7 | 0.653 (0.648–0.657) | 78.0 | 0.650 (0.646–0.655) | 77.8 | 0.621 (0.617–0.626) | 76.9 |
| Wake/sleep | 0.729 (0.723–0.736) | 90.9 | 0.734 (0.728–0.740) | 90.7 | 0.721 (0.716–0.727) | 88.9 | 0.667 (0.661–0.673) | 87.2 |
| NREM sleep/REM sleep | 0.808 (0.801–0.815) | 94.1 | 0.807 (0.801–0.813) | 93.8 | 0.781 (0.773–0.788) | 93.3 | 0.763 (0.755–0.770) | 93.0 |
| LS/DS | 0.473 (0.461–0.486) | 84.7 | 0.442 (0.429–0.455) | 85.4 | 0.479 (0.465–0.493) | 87.3 | 0.477 (0.463–0.491) | 89.6 |
| Wake/NREM sleep/REM sleep | 0.743 (0.738–0.748) | 86.5 | 0.744 (0.740–0.749) | 86.2 | 0.720 (0.716–0.725) | 84.4 | 0.677 (0.672–0.682) | 82.5 |
| CReSS Applied to Heart Rate, Airflow, and Thoracic Respiratory Effort Signals | ||||||||
| Wake/LS/DS/REM sleep | 0.687 (0.682–0.692) | 80.1 | 0.682 (0.677–0.686) | 79.8 | 0.692 (0.687–0.696) | 80.3 | 0.662 (0.657–0.666) | 79.2 |
| Wake/sleep | 0.774 (0.768–0.780) | 92.2 | 0.767 (0.761–0.772) | 91.7 | 0.770 (0.765–0.776) | 90.7 | 0.720 (0.714–0.725) | 89.1 |
| NREM sleep/REM sleep | 0.842 (0.836–0.849) | 95.1 | 0.847 (0.841–0.852) | 95.0 | 0.816 (0.810–0.823) | 94.3 | 0.790 (0.783–0.796) | 93.6 |
| LS/DS | 0.484 (0.471–0.496) | 85.3 | 0.418 (0.405–0.432) | 85.2 | 0.472 (0.458–0.486) | 87.6 | 0.465 (0.451–0.480) | 89.7 |
| Wake/NREM sleep/REM sleep | 0.785 (0.780–0.789) | 88.6 | 0.781 (0.777–0.786) | 88.1 | 0.767 (0.763–0.771) | 86.9 | 0.722 (0.718–0.727) | 84.7 |
Discriminations of wake/LS/DS/REM sleep and wake/NREM sleep/REM sleep are based on all epochs. For the discrimination of NREM sleep/REM sleep that does not include wake, we transformed the confusion matrix by removing the wake column and row; the same transformation was undertaken for the LS/DS discrimination by removing both wake and REM sleep. CI = confidence interval, CReSS = CardioRespiratory Sleep Staging, LS = light sleep (corresponding to N1 + N2), DS = deep sleep (corresponding to N3), MESA = Multi-Ethnic Study of Atherosclerosis, NREM = non-REM, REM = rapid eye movement, SDB = sleep-disordered breathing.
Table 6.
Kappa values and accuracy of manual sleep staging vs CReSS sleep staging in SHHS by sleep apnea severity subgroups.
| Sleep Stage Discrimination | No SDB (n = 82) | Mild SDB (n = 97) | Moderate SDB (n = 61) | Severe SDB (n = 56) | ||||
|---|---|---|---|---|---|---|---|---|
| Kappa (95% CI) | Percent Accuracy | Kappa (95% CI) | Percent Accuracy | Kappa (95% CI) | Percent Accuracy | Kappa (95% CI) | Percent Accuracy | |
| CReSS Applied to Heart Rate and Airflow Signals | ||||||||
| Wake/LS/DS/REM sleep | 0.613 (0.608–0.617) | 74.6 | 0.597 (0.592–0.601) | 74.1 | 0.546 (0.541–0.552) | 71.5 | 0.522 (0.515–0.528) | 71.9 |
| Wake/sleep | 0.681 (0.674–0.688) | 90.2 | 0.654 (0.648–0.661) | 89.7 | 0.635 (0.627–0.643) | 88.4 | 0.550 (0.542–0.559) | 83.5 |
| NREM sleep/REM sleep | 0.789 (0.782–0.795) | 93.0 | 0.776 (0.770–0.782) | 92.6 | 0.719 (0.710–0.728) | 91.5 | 0.692 (0.681–0.702) | 91.8 |
| LS/DS | 0.479 (0.469–0.490) | 82.3 | 0.461 (0.451–0.471) | 82.9 | 0.370 0.356–0.385) | 81.5 | 0.381 (0.362–0.401) | 89.0 |
| Wake/NREM sleep/REM sleep | 0.709 (0.705–0.714) | 84.9 | 0.691 (0.686–0.695) | 84.0 | 0.650 (0.644–0.656) | 82.1 | 0.576 (0.569–0.582) | 77.9 |
| CReSS Applied to Heart Rate, Airflow, and Thoracic Respiratory Effort Signals | ||||||||
| Wake/LS/DS/REM sleep | 0.666 (0.662–0.671) | 78.0 | 0.643 (0.639–0.648) | 77.0 | 0.612 (0.606–0.617) | 75.4 | 0.592 (0.586–0.598) | 76.0 |
| Wake/sleep | 0.741 (0.735–0.748) | 91.9 | 0.714 (0.708–0.720) | 91.4 | 0.719 (0.712–0.726) | 91.0 | 0.634 (0.626–0.641) | 86.3 |
| NREM sleep/REM sleep | 0.845 (0.840–0.851) | 94.8 | 0.816 (0.811–0.821) | 93.8 | 0.773 (0.766–0.781) | 92.9 | 0.752 (0.742–0.761) | 93.3 |
| LS/DS | 0.497 (0.487–0.507) | 82.9 | 0.479 (0.469–0.489) | 83.5 | 0.398 (0.384–0.412) | 82.4 | 0.386 (0.366–0.406) | 89.7 |
| Wake/NREM sleep/REM sleep | 0.771 (0.767–0.775) | 87.9 | 0.743 (0.739–0.747) | 86.6 | 0.723 (0.718–0.729) | 85.6 | 0.652 (0.646–0.658) | 81.7 |
Discriminations of wake/LS/DS/REM sleep and wake/NREM sleep/REM sleep are based on all epochs. For the discrimination of NREM sleep/REM sleep that does not include wake, we transformed the confusion matrix by removing the wake column and row; the same transformation was undertaken for the LS/DS discrimination by removing both wake and REM sleep. CI = confidence interval, CReSS = CardioRespiratory Sleep Staging, DS = deep sleep (corresponding to N3), LS = light sleep (corresponding to N1 + N2), NREM = non-REM, REM = rapid eye movement, SDB = sleep-disordered breathing, SHHS = Sleep Heart Health Study.
Figure 2. Accuracy of CReSS-determined sleep staging stratified by SDB severity using MESA home PSGs.
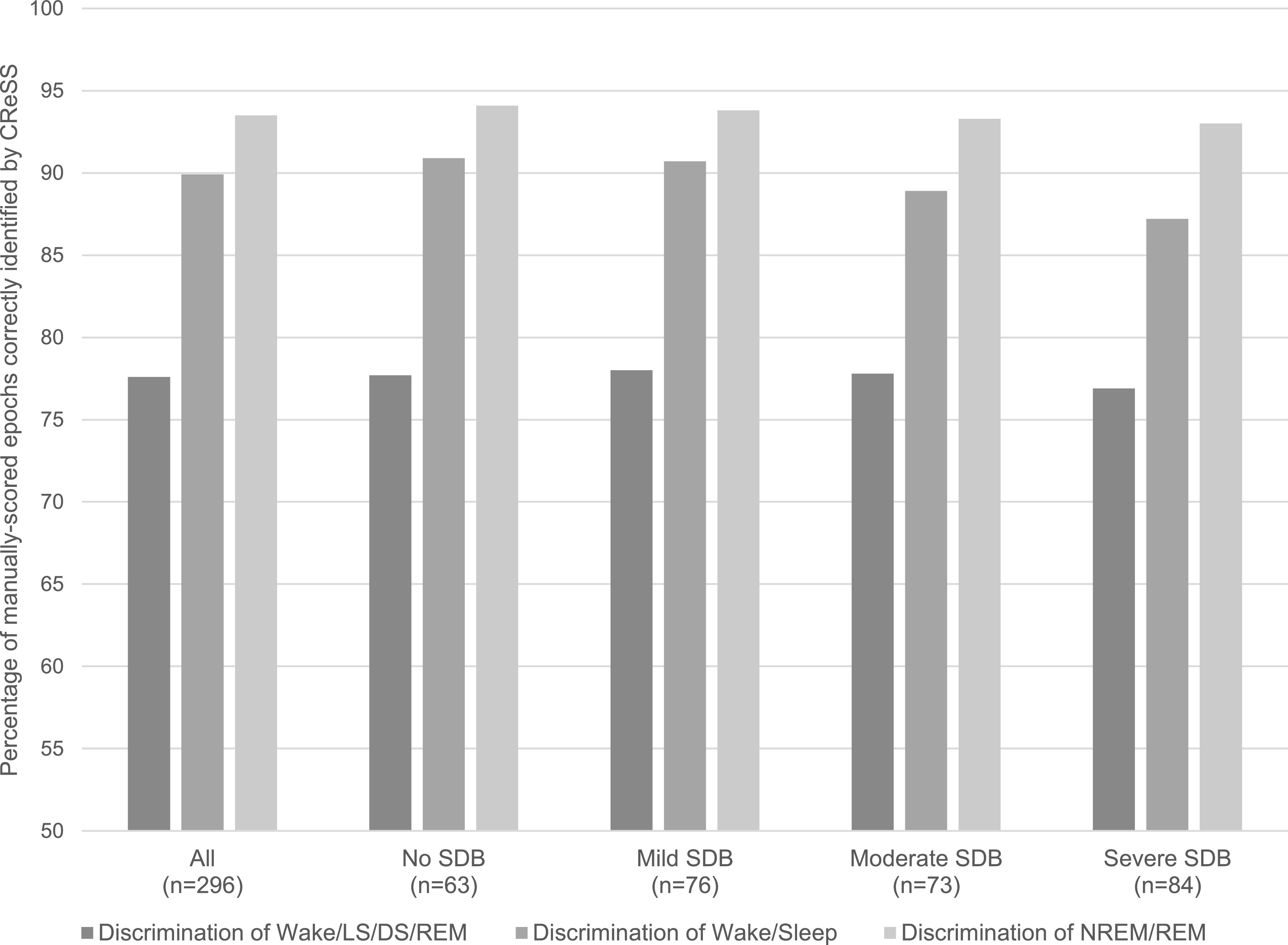
CReSS = CardioRespiratory Sleep Staging, DS = deep sleep (corresponding to N3), LS = light sleep (corresponding to N1 + N2), MESA = Multi-Ethnic Study of Atherosclerosis, NREM = non-REM, PSG = polysomnography, REM = rapid eye movement, SDB = sleep-disordered breathing.
Impact of additionally using a respiratory effort signal on CReSS performance
The upper parts of Table 2, Table 3, Table 4, Table 5, and Table 6 show the performance of CReSS when deployed on airflow and heart rate only; the lower parts of each table show the performance of CReSS when the respiratory inductance plethysmography thoracic effort signal was used additionally by the algorithm, which specifically affected periods of absent or invalid airflow. In almost every analysis, the performance of CReSS improved by allowing this additional input.
DISCUSSION
This study analyzed the performance of the novel CReSS algorithm designed to estimate sleep stages from respiratory and heart rate signals, with reference to the gold standard of manually scored PSG signals. We found that in 4 different sleep stage discriminations (wake/LS/DS/REM sleep, wake/sleep, NREM sleep/REM sleep, LS/DS), agreement between CReSS and manual scoring was substantial.23 Identification of wake/LS/DS/REM sleep was 78% accurate, with a kappa value of 0.643. The performance of CReSS was significantly more accurate than a similar algorithm designed to estimate sleep stages from finger peripheral arterial tonometry, derived pulse rate, oxygen desaturation, and actigraphy signals, which had a reported kappa value of 0.475 (95% confidence interval, 0.472–0.479) for wake/LS/DS/REM sleep.25
Our confusion matrices showed that misclassified epochs were most often incorrectly identified as LS, representing 11.5% and 14.1% of true-wake epochs, 14.7% and 11.6% of true-REM sleep epochs, and 42.9% and 34.7% of true-DS epochs in the MESA and SHHS datasets, respectively. Misclassified true-LS epochs were most often incorrectly identified as either wake (11.8% and 12.2% in MESA and SHHS, respectively) or DS (6.9% and 10.6% in MESA and SHHS, respectively). The performance of CReSS for identification of DS was not as strong as for other sleep stages, a trend observed in both datasets and across the full range of OSA severity; however, the kappa values for differentiation of LS/DS in both MESA and SHHS were within the range defined as moderate agreement per Landis and Koch.23 These data should be considered alongside evidence of disagreement across technologists for identification of N3. In 2018, Younes et al26 calculated the intraclass correlation coefficient comparing N3 sleep duration from 10 individual scorers to the average N3 duration of all scorers, and reported values ranging from 0.18 to 0.90. In that study, 16,308 epochs were identified as N3 by at least 1 of the 10 scorers, but of these, less than half (40.8 ± 15.1%) were not identified as N3 by any of the other scorers. Majority agreement (that is, > 3 of the 6 scorers) was achieved in only 22.0 ± 17.3% of N3 epochs, while complete agreement was achieved in only 3.8 ± 7.9% of N3 epochs. Similarly, agreement for scoring N3 was low in the Rosenberg et al27 2013 analysis of the AASM Inter-Scorer Reliability Program. In that dataset, sensitivity to N3 was 67.4%, compared to 52.3% in the current study (MESA dataset). The confusion matrix from Rosenberg et al can be used to derive a positive predictive value for N3 of 59.7% (55.4% in the current study), an N1 + N2 vs N3 accuracy of 91% (87.0% in the current study), and a kappa of 0.583 (0.469 in the current study). Thus, although agreement for N3 between CReSS and single scorers in the current study was lower than what has been observed when individual technologists are compared against a majority score, the gap in performance was not substantial, particularly when one considers that CReSS does not utilize slow-wave amplitude in the electroencephalogram to differentiate LS and DS. Importantly, the CReSS algorithm differentiated wake from sleep with an accuracy of 89% and NREM sleep from REM sleep with an accuracy of 94%. Consequently, the CReSS-derived total sleep time and the time spent in REM sleep are sensitive measures of the gold-standard PSG-based parameters, which suggests that using CReSS-derived measures may result in a more accurate estimate of the AHI, as well as a means of estimating the presence of REM sleep–related OSA from an HSAT.
Much of the existing literature on this topic has built on earlier studies of autonomic activity during sleep28,29 and is focused on estimating sleep stages using a single autonomic modality. For example, a previous study developed a 4-stage (wake/LS/DS/REM sleep) discrimination algorithm based on a combination of manually engineered features from ECG and a recurrent neural network. When tested on data from 195 healthy participants and 97 patients with a sleep or sleep-impacting disorder, the study achieved an average kappa of 0.61 and accuracy of 77%.19 Another study observed almost identical performance of the algorithm (kappa 0.60; accuracy 76%) using a dataset of 389 patients experiencing different sleep disorders.20 Beattie et al30 reported a kappa of 0.52 and accuracy of 69% using actigraphy and manually engineered heartrate variability features from a wrist-worn PPG device, after cross-validation with 60 healthy participants. Li et al31 reported a kappa of 0.47 and accuracy of 66% using convolutional neural networks on ECG signals after cross-validation on 5,793 PSGs collected during the SHHS; however, performance dropped (kappa 0.31; accuracy 66%) when cross-validation was performed in a separate dataset of 994 participants referred to a clinical sleep laboratory. Finally, Aggarwal et al32 used neural conditional random fields to classify sleep stages based on airflow, achieving a kappa of 0.57 and accuracy of 74% when testing 400 randomly selected patients with sleep apnea from the MESA dataset. The performance of CReSS is superior to the results of these prior studies that are based on a single input signal, highlighting the apparent benefit of a multimodal approach.
Few studies have focused on sleep staging using a combination of cardiac and respiratory modalities. A 2018 study showed that a combination of manually engineered features extracted from an ECG signal and from thoracic respiratory effort with a conditional random fields classifier achieved a kappa of 0.47 and accuracy of 69% for 4-stage discrimination (wake/LS/DS/REM sleep), using data collected from 180 healthy participants and 51 patients with SDB.33 Using a convolutional neural network to perform 5-stage discrimination (wake/N1/N2/N3/REM sleep) based on ECG and abdominal respiratory effort, Sun et al24 achieved a kappa of 0.60, using 1,000 PSGs randomly selected from a set of 8,682 acquired during diagnostic, positive airway pressure titration, and split-night studies. Again, these results are not as strong as the kappa value of 0.643 and 78% accuracy achieved with CReSS deployed on heart rate and airflow signals in the current study. Similarly, the weighted macro-F1 score in our study (0.78) was slightly higher than that reported by Sun et al (0.76).
In exploratory analyses, we found that the performance of CReSS was similar regardless of whether airflow was measured by nasal pressure or thermistry, or whether heart rate was derived from PPG or ECG. Although performance was slightly stronger in the dataset that used nasal pressure and PPG (MESA), we suspect that this reflects the use of Rechtschaffen and Kales21 sleep staging criteria in SHHS, whereas CReSS was trained based on the more recent AASM criteria. Although similar, the 2 methods are not identical. The AASM scoring criteria are more sensitive when scoring wakefulness and slow wave sleep due to the use of additional electroencephalogram signals (occipital leads for alpha waves and frontal leads for delta and slow waves).34 We also found that the performance of CReSS was more robust when the thoracic respiratory effort signal was used as a backup signal, which offers particular value during periods of HSATs in which the airflow signal is absent or invalid. Of note, the AASM recommends thermistry for the detection of apneas, and nasal pressure for the detection of hypopneas (The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications, version 2.5, section VIII, part A1–A435). Further, the AASM recommends that heart rate be collected via ECG during PSGs, and via either ECG or PPG during HSATs (AASM Scoring Manual, section IIA and section IX, part 1A). Thus, a variety of signals in different combinations are used clinically.36 CReSS was designed to be device-agnostic and as such can be deployed on signals of sufficient quality and sampling frequency recorded by any diagnostic device, including those that do not include a thoracic respiratory effort band.
A key strength of this study is the large, racially/ethnically diverse sample, supporting the generalizability of algorithm performance. The PSGs in our sample were collected in a large number of centers with different device types, and the manual scoring can be considered an accurate gold standard given the experience of the technologists and the oversight provided by the sleep reading center of the original studies. Although there are applications of CReSS beyond the SDB population, we sampled PSGs on the basis of SDB severity, as we anticipate this to be the most common-use case, and found consistent algorithm performance across the disease spectrum. This finding is particularly important given that CReSS is based on the analysis of autonomic signals, and it is known that OSA is associated with increased sympathetic drive and altered heart rate variability,37–39 even among asymptomatic patients with mild SDB, and in the absence of overt cardiovascular disease.40–42 We also chose to study PSGs collected in the home rather than a laboratory environment to reflect the intended setting of clinical use. There is likely greater variability in signal quality in home- vs laboratory-based studies, due to the increased possibility of incorrect sensor placement and the absence of a sleep technologist able to reapply sensors that become dislodged throughout the night.
Some limitations of our study should be noted. First, we did not collect data from a clinical population, although our sampling strategy was developed to ensure that the PSGs we selected represented the full spectrum of SDB severity. Given the recruitment criteria of MESA and SHHS and the timing of the PSGs within each protocol, the average age of our sample is somewhat higher than is typically reported for a clinical sample. Second, as CReSS relies on heart rate variability, it was necessary to exclude PSGs collected from participants with known heart failure, atrial fibrillation, and/or a cardiac pacemaker, thus somewhat limiting generalizability. The AASM recommends that HSATs be used in uncomplicated patients, specifically excluding those with heart failure,6 and therefore we do not consider the absence of validation data in these patients to be a major concern. Unfortunately, we were unable to account for the impact of other conditions associated with impaired autonomic function, such as Parkinson disease, or the use of medications such as beta-blockers that can cause decreases in heart rate and changes to the lower frequency of the heart rate variability spectrum. We also excluded PSGs with < 4 hours of data and those with incomplete manual scoring, but again, these criteria align with what the AASM describes as a technically adequate diagnostic test6 and therefore represent the intended-use case. As mentioned, we sought a large sample size in order to support generalizability; however, in doing so we were limited to PSGs scored by a single technologist. Given the known interscorer variability of sleep staging,27 a more accurate comparator would be the consensus of multiple technologists. Finally, it was necessary to use home PSGs rather than HSATs to validate CReSS, even though the algorithm was designed to be used on HSAT files, as electroencephalogram/electrooculogram/electromyogram signals were required for the gold-standard comparator of manual sleep staging. Nevertheless, we acknowledge that the performance of CReSS may differ when deployed on HSATs collected clinically, particularly if the equipment is set up by the patient rather than a sleep technologist.
There are several applications of CReSS that warrant future analyses. Most notably, an accurate estimate of total sleep time will reduce the known discrepancy between the AHI (respiratory events over total sleep time from PSG) and the REI (respiratory events over monitoring time from HSAT).5 The AASM differentiates monitoring time from recording time of an HSAT, such that monitoring time represents recording time minus movement artifact and estimated wake time per actigraphy, body position, breathing changes, and/or sleep diaries (The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications, version 2.5).35 Many laboratories, however, simply rely on recording time as the denominator for calculating the REI,43 which may be due to the absence of additional data from actigraphy, a position sensor, or sleep diaries; the lack of concrete guidelines for estimating wakefulness on the basis of respiratory pattern; and/or the burden associated with manual editing, particularly given the reduced reimbursement rates for HSATs within the United States. The reduced diagnostic sensitivity of the REI has important clinical implications due to the increased risk of a false negative test; thus, a subsequent validation study should focus on the diagnostic capabilities of the CReSS-determined REI, which was considered beyond the scope of this initial validation of algorithm performance. Concurrently, a subsequent validation of diagnostic performance could focus on the sensitivity and specificity of using CReSS to identify REM sleep–related OSA, which is not possible in most current HSAT platforms. While we have shown robust algorithm performance across the spectrum of SDB severity, algorithm performance might still be improved by examining recordings with a larger number of misclassified epochs. An epoch-by-epoch analysis of cardiorespiratory signal characteristics during misclassified epochs should be considered during future research, as well as analyses of algorithm performance across diagnosed sleep disorders in order to gain a better understanding of the algorithm’s limitations and potential improvements. Finally, there may also be applications of CReSS outside of clinical settings. For example, there is increased interest in utilizing actigraphy and similar tools for remote data collection in drug development and other clinical trials.44,45 The flexibility of CReSS to be deployed across a range of devices and minimally invasive signal types may be advantageous as a means to enhance the efficiency of clinical trials requiring sleep staging as a screening tool, covariate, or endpoint, which would require additional validation data in the intended research population(s).46
In conclusion, our data demonstrate substantial agreement between the CReSS algorithm and manual sleep staging in a large sample of home-based sleep studies collected from normal participants as well as those with mild, moderate, and severe SDB. The ability to estimate sleep stages from respiratory and heart rate signals may result in improved clinical interpretation of HSATs, which are increasingly used in the SDB diagnostic pathway.
DISCLOSURE STATEMENT
All authors have reviewed and approved the manuscript. Work was performed at Philips Sleep and Respiratory Care in Monroeville, Pennsylvania, and Vienna, Austria. All authors are employees of Philips. D.P.W. was the chief medical officer for Philips Respironics during the time that these analyses were completed. He is now an employee of Alairion. The other authors report no conflicts of interest.
SUPPLEMENTARY MATERIAL
ACKNOWLEDGMENTS
The authors are grateful to the National Sleep Research Resource for making the MESA and SHHS datasets available for these analyses and acknowledge the National Institutes of Health National Heart, Lung, and Blood Institute grant that funds the resource (R24HL114473). MESA was supported by HHSN268201500003I, N01HC95159, N01HC95160, N01HC95161, N01HC95162, N01HC95163, N01HC95164, N01HC95165, N01HC95166, N01HC95167, N01HC95168, and N01HC95169 from the National Heart, Lung, and Blood Institute, and by cooperative agreements UL1TR000040, UL1TR001079, and UL1TR001420 funded by the National Center for Advancing Translational Sciences. MESA Sleep was supported by R01HL098433 from the National Heart, Lung, and Blood Institute. SHHS was supported by U01HL53916, U01HL53931, U01HL53934, U01HL53937, U01HL53938, U01HL53940, U01HL53941, and U01HL64360 from the National Heart, Lung, and Blood Institute. Author contributions: study design: all authors; data analysis: M.R., R.V., A.C., P.A.; interpretation and manuscript preparation: all authors.
ABBREVIATIONS
- AASM
American Academy of Sleep Medicine
- AHI
apnea-hypopnea index
- CReSS
CardioRespiratory Sleep Staging
- DS
deep sleep
- ECG
electrocardiography
- HSAT
home sleep apnea test
- LS
light sleep
- MESA
Multi-Ethnic Study of Atherosclerosis
- NREM
non–rapid eye movement
- OSA
obstructive sleep apnea
- PPG
photoplethysmography
- PSG
polysomnography, polysomnograph
- REI
respiratory event index
- REM
rapid eye movement
- SDB
sleep-disordered breathing
- SHHS
Sleep Heart Health Study
REFERENCES
- 1.Rosen IM, Kirsch DB, Chervin RD, et al. American Academy of Sleep Medicine Board of Directors . Clinical use of a home sleep apnea test: an American Academy of Sleep Medicine position statement. J Clin Sleep Med. 2017;13(10):1205–1207. 10.5664/jcsm.6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kundel V, Shah N. Impact of portable sleep testing. Sleep Med Clin. 2017;12(1):137–147. 10.1016/j.jsmc.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim RD, Kapur VK, Redline-Bruch J, et al. An economic evaluation of home versus laboratory-based diagnosis of obstructive sleep apnea. Sleep. 2015;38(7):1027–1037. 10.5665/sleep.4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao YY, Weng J, Mobley DR, et al. Effect of manual editing of total recording time: implications for home sleep apnea testing. J Clin Sleep Med. 2017;13(1):121–126. 10.5664/jcsm.6404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianchi MT, Goparaju B. Potential underestimation of sleep apnea severity by at-home kits: rescoring in-laboratory polysomnography without sleep staging. J Clin Sleep Med. 2017;13(4):551–555. 10.5664/jcsm.6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapur VK, Auckley DH, Chowdhuri S, et al. Clinical practice guideline for diagnostic testing for adult obstructive sleep apnea: an American Academy of Sleep Medicine clinical practice guideline. J Clin Sleep Med. 2017;13(3):479–504. 10.5664/jcsm.6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang GQ, Cui L, Mueller R, et al. The National Sleep Research Resource: towards a sleep data commons. J Am Med Inform Assoc. 2018;25(10):1351–1358. 10.1093/jamia/ocy064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean DA II, Goldberger AL, Mueller R, et al. Scaling up scientific discovery in sleep medicine: the National Sleep Research Resource. Sleep. 2016;39(5):1151–1164. 10.5665/sleep.5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quan SF, Howard BV, Iber C, et al. The Sleep Heart Health Study: design, rationale, and methods. Sleep. 1997;20(12):1077–1085. [PubMed] [Google Scholar]
- 10.Redline S, Sanders MH, Lind BK, et al. Sleep Heart Health Research Group . Methods for obtaining and analyzing unattended polysomnography data for a multicenter study. Sleep. 1998;21(7):759–767. 10.1093/sleep/21.7.759 [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Wang R, Zee P, et al. Racial/ethnic differences in sleep disturbances: the Multi-Ethnic Study of Atherosclerosis (MESA). Sleep. 2015;38(6):877–888. 10.5665/sleep.4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klosh G, Kemp B, Penzel T, et al. The SIESTA project polygraphic and clinical database. IEEE Eng Med Biol Mag. 2001;20(3):51–57. 10.1109/51.932725 [DOI] [PubMed] [Google Scholar]
- 13.Punjabi NM, Shifa N, Dorffner G, Patil S, Pien G, Aurora RN. Computer-assisted automated scoring of polysomnograms using the Somnolyzer system. Sleep. 2015;38(10):1555–1566. 10.5665/sleep.5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jalil B, Laligant O, Fauvet E, Beya O. Detection of QRS complex in ECG signal based on classification approach. 2010 IEEE International Conference on Image Processing. 10.1109/ICIP.2010.5654091Accessed March 23, 2021. [DOI]
- 15.Xie S, Girshick R, Dollar P, Tou Z, He K. Aggregated residual transformations for deep neural networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 10.1109/CVPR.2017.634. Accessed March 23, 2021. [DOI]
- 16.Klambauer G, Unterthiner T, Mayr A, Hochreiter S. Self-normalizing neural networks. In: Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS ’17). Red Hook, NY: Curran Associates Inc.; 2017; 971–981. [Google Scholar]
- 17.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. 10.1162/neco.1997.9.8.1735 [DOI] [PubMed] [Google Scholar]
- 18.Berry RB, Brooks R, Gamaldo C, et al. AASM Scoring Manual updates for 2017 (version 2.4). J Clin Sleep Med. 2017;13(5):665–666. 10.5664/jcsm.6576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radha M, Fonseca P, Moreau A, et al. Sleep stage classification from heart-rate variability using long short-term memory neural networks. Sci Rep. 2019;9:14149. 10.1038/s41598-019-49703-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fonseca P, van Gilst MM, Radha M, et al. Automatic sleep staging using heart rate variability, body movements, and recurrent neural networks in a sleep disordered population. Sleep. 2020;43(9):zsaa048. 10.1093/sleep/zsaa048 [DOI] [PubMed] [Google Scholar]
- 21.Rechtschaffen A, Kales A. A Manual of Standardized Terminology, Techniques, and Scoring Systems for Sleep Stages of Human Subjects. Los Angeles, CA: UCLA Brain Information Service/Brain Research Institute; 1968. [Google Scholar]
- 22.Silber MH, Ancoli-Israel S, Bonnet MH, et al. The visual scoring of sleep in adults. J Clin Sleep Med. 2007;3(2):121–131. 10.5664/jcsm.26814 [DOI] [PubMed] [Google Scholar]
- 23.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–174. 10.2307/2529310 [DOI] [PubMed] [Google Scholar]
- 24.Sun H, Ganglberger W, Panneerselvam E, et al. Sleep staging from electrocardiography and respiration with deep learning. Sleep. 2019;43(7):zsz306. https://doi.org/10.1093/sleep/zsz306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hedner J, White DP, Malhotra A, et al. Sleep staging based on autonomic signals: a multi-center validation study. J Clin Sleep Med. 2011;7(3):301–306. 10.5664/JCSM.1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Younes M, Kuna ST, Pack AI, et al. Reliability of the American Academy of Sleep Medicine rules for assessing sleep depth in clinical practice. J Clin Sleep Med. 2018;14(2):205–213. 10.5664/jcsm.6934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenberg RS, Van Hout S. The American Academy of Sleep Medicine inter-scorer reliability program: sleep stage scoring. J Clin Sleep Med. 2013;9(1):81–87. 10.5664/jcsm.2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgess HJ, Trinder J, Kim Y. Cardiac autonomic nervous system activity during presleep wakefulness and stage 2 NREM sleep. J Sleep Res. 1999;8(2):113–122. 10.1046/j.1365-2869.1999.00149.x [DOI] [PubMed] [Google Scholar]
- 29.Trinder J, Kleiman J, Carrington M, et al. Autonomic activity during human sleep as a function of time and sleep stage. J Sleep Res. 2001;10(4):253–264. 10.1046/j.1365-2869.2001.00263.x [DOI] [PubMed] [Google Scholar]
- 30.Beattie Z, Oyang Y, Statan A, et al. Estimation of sleep stages in a healthy adult population from optical plethysmography and accelerometer signals. Physiol Meas. 2017;38(11):1968–1979. 10.1088/1361-6579/aa9047 [DOI] [PubMed] [Google Scholar]
- 31.Li Q, Li Q, Liu C, Shashikumar SP, Nemati S, Clifford GD. Deep learning in the cross-time frequency domain for sleep staging from a single-lead electrocardiogram. Physiol Meas. 2018;39(12):124005. 10.1088/1361-6579/aaf339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aggarwal K, Khadanga S, Joty SR, Kazaglis L, Srivastava J. A structured learning approach with neural conditional random fields for sleep staging. 2018 IEEE International Conference on Big Data (Big Data). 10.1109/BigData.2018.8622286Accessed March 23, 2021. [DOI]
- 33.Fonseca P, den Teuling N, Long X, Aarts RM. A comparison of probabilistic classifiers for sleep stage classification. Physiol Meas. 2018;39(5):055001. 10.1088/1361-6579/aabbc2 [DOI] [PubMed] [Google Scholar]
- 34.Moser D, Anderer P, Gruber G, et al. Sleep classification according to AASM and Rechtschaffen & Kales: effects on sleep scoring parameters. Sleep. 2009;32(2):139–149. 10.1093/sleep/32.2.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berry RB, Albertario CL, Harding SM, et al. ; for the American Academy of Sleep Medicine. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications. Version 2.5. Darien, IL: American Academy of Sleep Medicine; 2018. [Google Scholar]
- 36.Mendonça F, Mostafa SS, Ravelo-Garcia AG, Morgado-Dias F, Penzel T. Devices for home detection of obstructive sleep apnea: a review. Sleep Med Rev. 2018;41:149–160. 10.1016/j.smrv.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 37.Carlson JT, Hedner J, Elam M, Ejnell H, Sellgren J, Wallin BG. Augmented resting sympathetic activity in awake patients with obstructive sleep apnea. Chest. 1993;103(6):1763–1768. 10.1378/chest.103.6.1763 [DOI] [PubMed] [Google Scholar]
- 38.Somers VK, Dyken ME, Clary MP, Abboud FM. Sympathetic neural mechanisms in obstructive sleep apnea. J Clin Invest. 1995;96(4):1897–1904. 10.1172/JCI118235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlson JT, Hedner JA, Sellgren J, Elam M, Wallin BG. Depressed baroreflex sensitivity in patients with obstructive sleep apnea. Am J Respir Crit Care Med. 1996;154(5):1490–1496. 10.1164/ajrccm.154.5.8912770 [DOI] [PubMed] [Google Scholar]
- 40.Wiklund U, Olofsson BO, Franklin K, Blom H, Bjerle P, Niklasson U. Autonomic cardiovascular regulation in patients with obstructive sleep apnoea: a study based on spectral analysis of heart rate variability. Clin Physiol. 2000;20(3):234–241. 10.1046/j.1365-2281.2000.00251.x [DOI] [PubMed] [Google Scholar]
- 41.Aydin M, Altin R, Ozeren A, Kart L, Bilge M, Unalacak M. Cardiac autonomic activity in obstructive sleep apnea: time-dependent and spectral analysis of heart rate variability using 24-hour Holter electrocardiograms. Tex Heart Inst J. 2004;31(2):132–136. [PMC free article] [PubMed] [Google Scholar]
- 42.Balachandran JS, Bakker JP, Rahangdale S, et al. Effect of mild, asymptomatic obstructive sleep apnea on daytime heart rate variability and impedance cardiography measurements. Am J Cardiol. 2012;109(1):140–145. 10.1016/j.amjcard.2011.07.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collop NA, Tracy SL, Kapur V, et al. Obstructive sleep apnea devices for out-of-center (OOC) testing: technology evaluation. J Clin Sleep Med. 2011;7(5):531–548. 10.5664/JCSM.1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perry B, Herrington W, Goldsack JC, et al. Use of mobile devices to measure outcomes in clinical research, 2010-2016: a systematic literature review. Digit Biomark. 2018;2(1):11–30. 10.1159/000486347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coran P, Goldsack JC, Grandinetti CA, et al. Advancing the use of mobile technologies in clinical trials: recommendations from the clinical trials transformation initiative. Digit Biomark. 2019;3(3):145–154. 10.1159/000503957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldsack JC, Coravos A, Bakker JP, et al. Verification, analytical validation, and clinical validation (V3): the foundation of determining fit-for-purpose for Biometric Monitoring Technologies (BioMeTs). NPJ Digit Med. 2020;3:55. 10.1038/s41746-020-0260-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.