Abstract
Millions of positive COVID-19 patients are suffering from the pandemic around the world, a critical step in the management and treatment is severity assessment, which is quite challenging with the limited medical resources. Currently, several artificial intelligence systems have been developed for the severity assessment. However, imprecise severity assessment and insufficient data are still obstacles. To address these issues, we proposed a novel deep-learning-based framework for the fine-grained severity assessment using 3D CT scans, by jointly performing lung segmentation and lesion segmentation. The main innovations in the proposed framework include: 1) decomposing 3D CT scan into multi-view slices for reducing the complexity of 3D model, 2) integrating prior knowledge (dual-Siamese channels and clinical metadata) into our model for improving the model performance. We evaluated the proposed method on 1301 CT scans of 449 COVID-19 cases collected by us, our method achieved an accuracy of 86.7% for four-way classification, with the sensitivities of 92%, 78%, 95%, 89% for four stages. Moreover, ablation study demonstrated the effectiveness of the major components in our model. This indicates that our method may contribute a potential solution to severity assessment of COVID-19 patients using CT images and clinical metadata.
Keywords: COVID-19, Deep learning, Severity assessment, Multi-view lesion, Dual-Siamese channels, Clinical metadata
1. Introduction
Since the World Health Organization (WHO) declared that Coronavirus disease 2019 (COVID-19) had become a Public Health Emergency of International Concern, as of 5:30 pm Central European Time (CET), 25 January 2021, there have been 98,794,942 confirmed cases of COVID-19, including 2,124,193 deaths, reported to the WHO (“Coronavirus disease (COVID-19) Situation dashboard,” 2021). More seriously, the pandemic continues to increase at a rapid rate of more than 400,000 confirmed cases per day. The COVID-19 pandemic is affecting 218 countries and territories in many ways, not least in terms of physical health, mental health, environmental change, education, global supply chains, and economic development (Shahzad, Hassan, Aremu, Hussain, & Lodhi, 2020). In addition, it is becoming a major challenge to allocate medical resources as large numbers of positive patients continue to be hospitalized. the increasing need has exceeded hospital capacities, such as mechanical ventilation, intensive care unit (ICU) admission, and medical staffs. A critical step in the triage of patients and follow-up of treatment response is accurate severity assessment, which assists hospitals to prioritize resources, improve patients’ chances of being cured.
During the COVID-19 pandemic, several standardized severity score systems only using basic clinical data such as the CURB-65 score, Brescia-COVID Respiratory Severity Scale (BCRSS), and Quick COVID-19 Severity Index (qCSI), have been used to signal the level of severity for pulmonary involvement (Rodriguez-Nava et al., 2021). Furthermore, Pan et al. (Pan et al., 2020) defined four clinical stages of COVID-19 based on the findings of chest CT scans, and Yang et al. (2020) introduced a severity scoring system (CT-SS) that depends on the degree of lung affection.
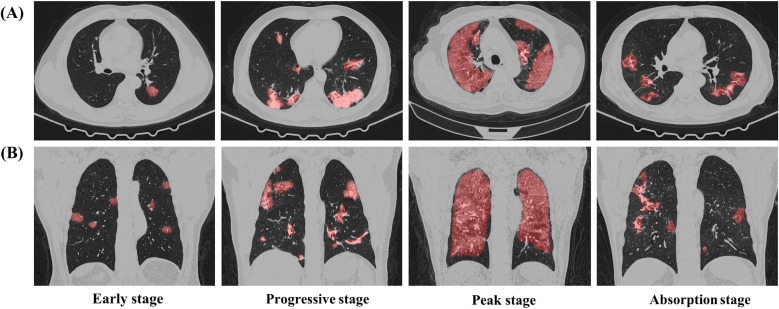
Given that manual severity assessment could be a labour-intensive work for front-line healthcare workers, providing computer-aided clinical support for automatic severity assessment is highly desired. Several computer-aided methods (Aboutalebi et al., 2021, Ali and Budka, 2021, He et al., 2021, Lessmann et al., 2021, Li et al., 2021, Lizzi et al., 2021, Zhang et al., 2020) have been proposed recently. However, these studies have some obstacles. Firstly, most of the severity assessment models focused on the quantitative measurement for lesion regions while neglecting the direct clinical stage assessment. Secondly, several models for clinical stage prediction only assessed rough severity levels (i.e., severe, non-severe; low, medium, high) while neglecting the fine-grained severity levels, especially for the recovery stage. Thirdly, few models incorporated prior knowledge with feature extraction and discrimination process. Prior knowledge, however, can regularize the model and improve the model performance especially with the limited number of training samples (Jin et al., 2021). To address these obstacles, we proposed a novel prior-knowledge-based artificial intelligence (AI) system for fine-grained severity assessment of COVID-19. We utilized dual-Siamese channels and clinical metadata as prior knowledge, by imitating the diagnostic process of clinicians and combining some clinical findings (Chassagnon et al., 2020, Pan et al., 2020, Zhang et al., 2020). The stage labels were annotated according to the Guideline for Medical Imaging in Auxiliary Diagnosis of Coronavirus Disease in 2019 by Chinese Research Hospital Association. As depicted in Fig. 1 : Stage 1 (Early stage) shows ground-glass opacities, stage 2 (Progressive stage) shows an increase in both the crazy-paving patterns, stage 3 (Peak stage) shows consolidation, and stage 4 (Absorption stage) shows gradual decrease of consolidation.
Fig. 1.
Examples of four clinical stages of COVID-19 on CT images from four patients. (A). Transverse images. (B). Reconstructed coronal images. Regions in red are lesion regions segmented by the proposed model.
Furthermore, it is challenging to train a 3D model because of several reasons. Firstly, the size of our dataset is still limited even we collected lots of CT images. Secondly, given that one CT scan has hundreds of CT images, the complexity of the 3D model can be a large order of magnitude, thus the model requires more training samples to avoid overfitting. To make full use of the limited number of datasets and reduce the complexity of our model, we decomposed the 3D CT scan into fixed views (sagittal, transverse, coronal, and six diagonal planes) as input images of our 3D model. In summary, the contributions of this paper are threefold:
-
•
Theoretically, we proposed a novel deep-learning-based pipeline for directly assessing four clinical stages of COVID-19 with CT images.
-
•
Practically, we integrated prior knowledge (dual-Siamese channels and clinical metadata) into our model to improve the assessment performance. And we decomposed the 3D CT scan into multi-view slices as input images to reduce the complexity of our 3D model, which can effectively alleviate the data-scarcity issue.
-
•
We collected a dataset of 1301 CT scans from 449 positive COVID-19 patients who underwent continuous CT examinations in the COVID-19 treatment centers of China. Experimental results show that our proposed framework provides a potential tool to help hospitals with managing and planning of medical resources. Besides, it can assist clinicians to follow up the treatment response, and potentially reduce the death toll. The source code has been made openly available at GitHub (https://github.com/Zhidan-ten/severity-assessment).
2. Related works
Over the past decades, Artificial intelligence (AI) methods have played a significant role in the field of medical image analysis. This section discusses some related works about AI applications for COVID-19, and some developing techniques for mitigating the data-scarcity issue in the AI field.
2.1. Artificial intelligence for COVID-19
A large number of papers have been published since the pandemic outbroke (Roberts et al., 2021), and these studies can fall into three aspects: screening of COVID-19, infection segmentation, and severity assessment (Shi et al., 2021).
2.1.1. Screening of COVID-19
Altan and Karasu (2020) proposed a hybrid model consisting of traditional algorithms and deep learning technique to determine COVID-19 patients from X-ray images. Zhang et al. (2020) trained a 3D ResNet-18 model to distinguish COVID-19 from other common pneumonia and normal controls, the diagnosis classification took the lung-lesion map as an input generated by segmentation networks and utilized the normalized CT volumes for final diagnosis prediction.
2.1.2. Infection segmentation
Fan et al. (2020) proposed a novel COVID-19 Infection Segmentation Deep Network (Inf-Net) that used a parallel partial decoder to aggregate high-level features and generate a global map to enhance the boundary area from CT images. Zheng et al. (2020) proposed a multi-scale discriminative network that employed a pyramid convolution block, channel attention block, and residual refinement block to implement the multiclass segmentation of infection on CT images.
2.1.3. Severity assessment
Lizzi et al. (2021) proposed an automated quantification analysis system (LungQuant) based on deep-learning networks, it output the percentage P of lung volume affected by COVID-19 lesions and the corresponding CT severity score (CT-SS = 1 for P < 5%, CT-SS = 2 for 5% ≤ P < 25%, CT-SS = 3 for 25% ≤ P < 50%, CT-SS = 4 for 50% ≤ P < 75%, CT-SS = 5 for P ≥ 75%). Lessmann et al. (2021) developed an AI system consisting of three deep-learning models that automatically segmented five pulmonary lobes, and then assigned a CT severity score (CT-SS) for the degree of parenchymal involvement per lobe. Ali and Budka (2021) utilized deep-learning techniques for the severity assessment via measuring the area of multiple rounded ground-glass opacities (GGO) and consolidations in the periphery (CP) of the lungs and accumulating them to form a severity score. Zhang et al. (2020) used Light Gradient Boosting Machine (LightGBM) and Cox proportional-hazards (CoxPH) regression models based on quantitative CT lung-lesion features and clinical parameters to assess clinical stages (severe, non-severe). Li et al. (2021) proposed a deep multiple instance learning (MIL) model with instance-level attention to jointly classify the bag and weigh the instances in each bag, so as to distinguish the severe instances from non-severe instances. He et al. (2021) also proposed a synergistic learning framework for severity assessment (severe, non-severe) in 3D CT images. Moreover, Aboutalebi et al. (2021) introduced COVID-Net CT-S, a suite of deep convolutional neural networks that used a 3D residual architecture to learn volumetric visual indicators characterizing the degree of COVID-19 lung disease severity (low, medium, high). However, our method directly predicted four clinical stages (early, progressive, peak, absorption) of COVID-19 based on three deep-learning models.
2.2. Methods for alleviating the data-scarcity issue
In general, deep-learning methods require huge amounts of training samples to achieve promising results. Some DNNs, however, are likely to suffer from the overfitting of models, and the model may not generalize to new data because of the lack of large public datasets, especially for medical datasets. To alleviate this issue of small datasets, researchers have adopted various techniques. Taylor and Nitschke (2018) utilized various geometric (flipping, rotation and cropping) and photometric (color jittering, edge enhancement and fancy PCA) schemes to increases CNN task performance. Zhao, Balakrishnan, Durand, Guttag, and Dalca (2019) used generative adversarial network (GAN) to produce labelled medical images for data augmentation. Kermany et al. (2018) utilized transfer learning by pretraining CNN models to effectively classify images for macular degeneration and diabetic retinopathy. Xie et al., 2019, Zhou et al., 2020 combined multiple two-dimensional (2D) models by inputting multi-view slices to conduct 3D classification and segmentation, which effectively reduced the complexity of models. In this paper, we decomposed the 3D CT scan into multi-view slices as input images to alleviate the data-scarcity issue.
3. Methodology
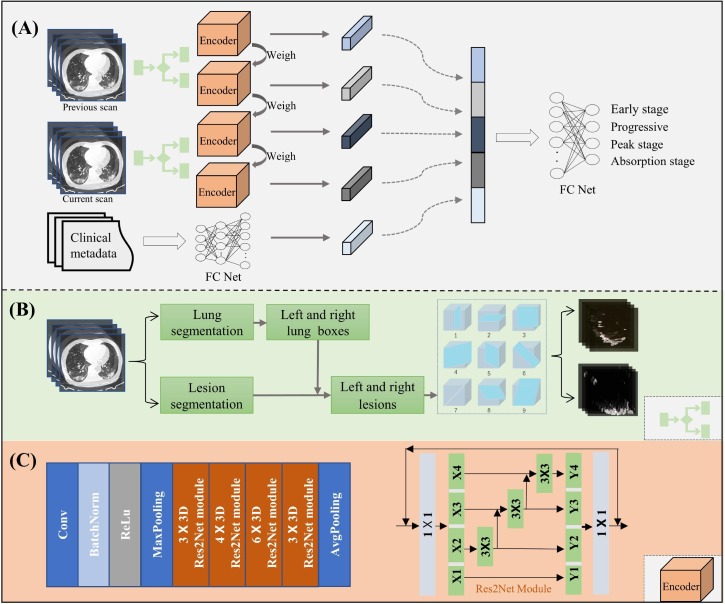
The overall framework of the proposed deep-learning method (Fig. 2 ) is presented in this section. This framework has four main steps: (1) decomposing the 3D lesion region into nine fixed views based on lung and lesion segmentation results for each of the left and right lungs, (2) extracting the image features through dual-Siamese channels containing four encoders with shared weights, (3) embedding the clinical metadata, and (4) combining these features to train a model for assessing the four stages of COVID-19 patients. Step 1 is shown in Fig. 2(B), steps 2–4 are given in Fig. 2(A), and the architecture of the encoder is shown in Fig. 2(C).
Fig. 2.
Overall framework of the proposed method. (A). Flowchart of the proposed stage-accessing model, with current and previous CT scans and clinical metadata as inputs and with the output of FC Net as the predicted stage of a given COVID-19 patient. (B). 3D segmented lesions within the lung boxes are decomposed into nine slices at nine different views for each of the left and right lungs. (C). Detailed structure of the image feature encoder called Res2Net.
3.1. Lung and lesion segmentation
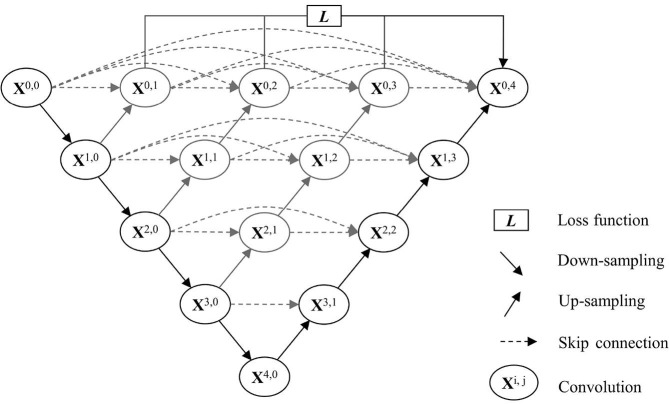
Before extracting multi-view lesion slices from the 3D left and right lesions, we first detected the biggest bounding boxes of left and right lungs based on lung segmentation model, and then used the two lung boxes to restrict and separate the lesion regions obtained by a lesion segmentation model. For lung and lesion segmentation, some traditional methods, for example, the thresholding methods based on the Hounsfield unit (HU) were not accurate enough, especially for CT images of COVID-19. Therefore, we utilized deep-learning networks to segment the lung and lesion regions. By evaluating various DNNs performance on our datasets, we finally employed the trained DNN called U-net++ to segment the lung regions. U-net++ (Zhou, Rahman Siddiquee, Tajbakhsh, & Liang, 2018) is a common, public, more powerful architecture for medical image segmentation especially for COVID-19 segmentation (Shi et al., 2021). The architecture of the U-net++ is shown in Fig. 3 . UNet++ begins with an encoder sub-network or backbone followed by a decoder sub-network. What distinguishes UNet++ from U-Net (the black components) is the re-designed skip pathways (shown in blue and green) that connect the two sub-networks and the use of deep supervision (shown red), it can reduce the semantic gap between the feature maps of the encoder and decoder sub-networks, i indexes the down-sampling layer along the encoder, and j indexes the convolution layer of the dense block along the skip pathway. And the details of left and right lung box detection are given in Algorithm 1.
| Algorithm 1: Left-right lung boxes detection |
|---|
| Input: A CT scan of the COVID-19 patient which consists of hundreds of CT images, m is the number of images |
| Output: coordinate value of the biggest left–right lung boxes |
| 1: for i do m |
| 2: lung segmentation = lungmodel(image[i]) |
| 3: calculate the number of connection regions. ← lung segmentation |
| 4: if the number ofregions 2: 2 then |
| 5: (Xu , Yu, XL2′ Yu), (XR 1, YR1, XR2′ YR2). ← the two biggest regions |
| 6: if the number ofregions is 1 then |
| 7: (Xu, Yu,XL2′ Yu), (XR1, YR1, XR 2, YR2). ← median of the one region |
| 8: end if |
| 9: save (Xu , Yu, XL 2, Yu), (XR 1, YR1, XR2′ YR2) |
| 10: end for |
| 11: ({Xu } min' {Yu}min' {Xu } max' {Yu}max), ({XRI } min' {YRI } min' {XR2 } max' {YR2} max) |
We want to emphasize that the lesions were not detected directly on the segmented lung regions, because sometimes lung regions might not be perfectly segmented out. As clarified below, the segmented lungs were just used to define left and right boxes, and these boxes were used to crop the left and right lesions from the whole CT scans of lesion segmentation, which will be put into the cubes with fixed sizes for lesion-slice extraction.
Fig. 3.
Architecture of U-net++ ().
Adapted from Zhou et al., 2018
3.2. Multi-view lesion-slice extraction
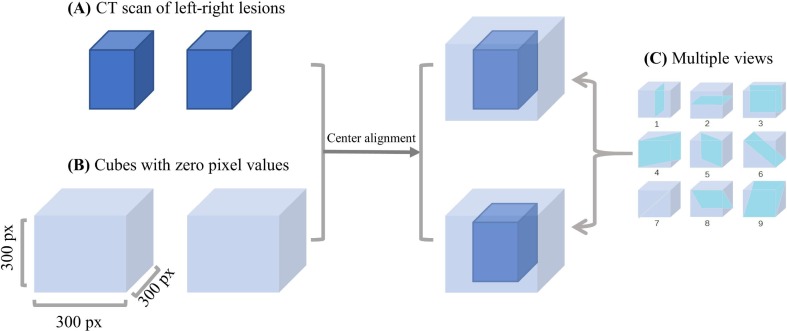
After obtaining thelargest left and right lung boxes and the initial segmented lesion images by our lesion segmentation model, we used the lung boxes to crop the initial segmented CT lesion images for restricting and separating the lesion region. Then we used the cropped lesion images to reconstruct the 3D left and right lesions. Different COVID-19 patients, however, have different lung sizes, which may produce different sizes of lesions. In addition, CT scans have variable spatial resolutions, which may produce different numbers of slices. Therefore, as shown in Fig. 4 , we used a cube with a size of 300 × 300 × 300 pixels for each lung to always contain the left or right lesion, and the values of other regions were set to 0. Finally, we cut each of the 3D left and right lesions into nine 2D slices at the sagittal, transverse, coronal, and six diagonal views, as shown in Fig. 4(C). In summary, we extracted 18 2D lesion slices from the left and right lesions for each CT scan, which will be input into the stage-assessing network model for lesion feature extraction and stage assessment. Note that such multi-view lesion-slice extraction mainly aims to reduce the complexity from 3D to 2D spaces while containing sufficient lesion information due to the irregular shapes of lesions.
Fig. 4.
Illustration of multi-view lesion-slice extraction. (A). Left and right lesion regions contained in the left and right lung boxes cropped from CT scan. (B). Larger cubes with a size of 300 × 300 × 300 pixels with zero values to contain lungs of varying sizes. (C). Nine planes used to cut 3D lesion regions into 2D slices.
3.3. Stage-assessing network model
The stage-assessing network is composed of two major modules: the image-feature-extracting module and clinical-metadata-embedding module. We used the dual-Siamese channels consisting of four encoders to extract image features from the multi-view lesion slices and then embedded the clinical metadata related features in a feature augmenting manner to predict the disease stage of a given COVID-19 patient.
3.3.1. Dual-Siamese channels for lesion feature extraction
The proposed dual-Siamese channels were composed of four identical parallel CNNs that shared the same architectures and weights. And it took different lesion slices as inputs. More specifically, given that the clinicians usually compare the two adjacent CT scans when they assess the clinical stages in practice, we used two CT scans of the same COVID-19 patient that were collected currently and previously; if the patient was taken the CT examination for the first time, we took the slices without lesions as the previous CT scan. Moreover, we divided each CT scan into two parts corresponding to the left and right lungs for the sufficient multi-view lesion information. Therefore, we used four CNNs to extract image features. By evaluated various CNN performance, We finally employed Res2Net (Gao et al., 2021) as the image-feature encoder, as shown in Fig. 2 (C). Compared with most existing backbones of CNNs, such as the ResNet (He, Zhang, Ren, & Sun, 2016) module, which represents multi-scale features in a layer-wise way, the Res2Net module replaces a group of 3 × 3 filters with smaller groups of filters, while connecting different filter groups in a hierarchical residual-like style, thus it represents multi-scale features at a granular level and increases the range of receptive fields for each network layer. The Res2Net module is formulated as
| (1) |
where xi represents a feature map subset that has the same spatial size but one-quarter the number of channels compared with the input feature map. Ki () denotes a corresponding 3 × 3 convolution, and yi is the output of Ki (). The feature subset xi is added with the output of Ki-1 (), and then fed into Ki ().
3.3.2. Clinical metadata embedding
With the rapid development of electronic medical records (EMRs), one can easily obtain clinical metadata, including basic patient information and biochemical indexes. Furthermore, Zhang et al., 2020, Chassagnon et al., 2020 used quantitative image features and clinical parameters (e.g., age, sex) to predict the clinical stages based on machine learning model. And Pan et al. (2020) found that time course is a key factor for assessing the stages from initial diagnosis until patient recovery. Therefore, in this work, we only employed the basic patient information (sex, age, and time course for disease development), which served as the prior domain knowledge to regularize the proposed model. Specifically, we combined the four items of clinical metadata, i.e., sex, age, progress (denoted P), and time interval (in days) between two adjacent CT examinations (denoted M), as the clinical features. P and M are defined as
| (2) |
where D refers to the date of the CT examination, A denotes the hospital admission date of the patient, and T refers to the date on which main symptoms occurred (e.g., fever, cough) since the patient entered the hospital. These four features (sex, age, P, and M) are normalized to [0, 1].
Furthermore, given that there were a few clinical features compared with the number of extracted image features, we utilized fully connected layers to increase the dimensionality of the original clinical metadata, which can provide more abstract information for severity assessment. The fully connected layers contains three layers with 256, 512, and 1024 neurons, respectively. In other words, the clinical metadata were re-coded as a 1024-dimensional vector using FC Net. We then combined all of the aforementioned features to predict the stage of COVID-19 with a fully connected layer. The procedure was shown in Algorithm 2.
| Algorithm 2: Training procedure for the stage-assessing network |
|---|
| Input:X refers to multi-view lesion slices; C denotes clinical metadata; |
| Y represents the predicted result; T is the label |
| Output: the trained model M; |
| 1: for each epoch e in range kdo |
| 2: fori in range 4 do |
| 3: image feature[i] = Res2Net(X[i]) |
| 4: end for |
| 5: clinical feature = FC1(C) |
| 6: all feature = concat(image feature, clinical feature) |
| 7: Y = FC2(all feature) |
| 8: modelFit(SGD,(Y, T)) |
| 9: end for |
3.4. Loss function
The Dice and the cross-entropy loss functions (Gao et al., 2021) were used for the segmentation and classification task, respectively, which are written as
| (3) |
where LS and LC are segmentation loss and classification loss, respectively. In the segmentation loss LS, X denotes the pixel set of ground truth and Y refers to the pixel set of the predicted result. More specifically, the numerator represents the pixels that are correctly segmented, and the denominator denotes the number of all the pixels. In the classification loss LC, M represents the class number, which is set to 4 in this paper, corresponding to the four disease stages; yi denotes the ground truth, which is equal to 1 when the true label is class i and 0 when the true label is not class i; pi represents the predicted probability for class i; and the LC value increases with increasing deviation of the predicted probability from the ground truth.
3.5. Evaluation metrics
In this paper, we adopted as many as eight metrics to evaluate both the segmentation and classification performance. For the segmentation task, we measured the DSC and the intersection over union (IOU), which are defined as
| (4) |
where X represents the set of predicted pixels and Y is the set of ground truth pixels. It is clear that these two metrics increase with increasing similarity between the predicted result and the ground truth. For the stage-assessing task, we employed the accuracy, accuracy of two-way classification (Acc), sensitivity (Sen), specificity (Spe), average accuracy (AA), positive likelihood ratio (PLR), and negative likelihood ratio (NLR) to evaluate the performance. These metrics are formulated as
| (5) |
| (6) |
| (7) |
where TP, TN, FP, and FN denote the numbers of true positive, true negative, false positive, and false negative samples, respectively. AA is used to eliminate the influence of data imbalance (Gao et al., 2021). An increasing PLR greater than 1 indicates an increasing probability that the predicted result is associated with the disease, and an increasing NLR<1 indicates an increasing probability that the predicted result is associated with the absence of disease (Kermany et al., 2018). We also used confusion matrix, receiver operating characteristic (ROC) curve, and AUC to evaluate the stage-assessing performance.
4. Experiment
This section describes the entire dataset distribution, data pre-processing, and the experimental settings. Three datasets were employed, i.e., the lung segmentation, lesion segmentation, and stage-assessing datasets, and all were based on CT images. In particular, the stage-assessing dataset was used to demonstrate the effectiveness of the proposed deep-learning-based framework for assessing the stages of COVID-19 patients.
4.1. Data and pre-processing
The dataset used to train the lung segmentation model was downloaded from two open databases created by Ma et al., 2020, Zhang et al., 2020. The database of Ma et al. provides 20 CT scans of COVID-19, including 5,000 CT images with lung labels, and that of Zhang et al. provides 150 CT scans of COVID-19, including 2,1470 CT images, 750 images of which are accompanied by lung mask labels. Thus, we collected a total of 5,750 CT images with lung labels for lung segmentation.
The dataset used to train the lesion segmentation model was collected by us from the Second Xiangya Hospital, Central South University. This dataset contains 19 CT scans of COVID-19, including 1,117 CT images with lesion labels delineated by radiologists (Zhao et al., 2020).
The dataset used to evaluate the proposed stage-assessing framework was collected by us from Wuhan Red Cross Hospital. It contains images from 449 COVID-19 patients (age range, 20–97 years; 219 men and 230 women) who underwent continuous chest CT examinations in the period from 1 January to 20 April 2020 because they were clinically confirmed as COVID-19 cases using RT-PCR. This dataset contains a total of 1,301 CT scans and the corresponding clinical metadata. Table 1 lists the distribution of this stage-assessing dataset. The stage labels were annotated independently by two radiologists (with 14 and 31 years of clinical experience, respectively) who were blinded to the clinical data. The reference standard is the Guideline for Medical Imaging in Auxiliary Diagnosis of Coronavirus Disease in 2019 by Chinese Research Hospital Association.
Table 1.
Distribution of the stage-assessing CT dataset.
| Stage 1 | Stage 2 | Stage 3 | Stage 4 | |
|---|---|---|---|---|
| Scans | 155 | 364 | 117 | 665 |
| Slices | 46,500 | 101,920 | 35,100 | 196,500 |
We randomly divided the lung and lesion datasets into two independent subsets (i.e., training and testing sets) with a ratio of 4:1 at the image level, with no CT image overlaps between subsets. And we randomly divided the stage-assessing dataset into two independent subsets with a ratio of 4:1 (90 cases including 266 CT scans as testing set) at the patient level, with no CT scan overlaps between subsets. For the lesion and stage-assessing datasets, we adjusted all of the raw CT images to the fixed lung window [–1200, 0] and then normalized the images to the range [0, 255]. All these images have a size of 512 × 512 pixels.
4.2. Experimental settings
We implemented the three models with Pytorch framework and ran them on a computer equipped with a graphical processing unit (GPU) of NVIDIA GEFORCE RTX 3090. Regarding both the lung and lesion segmentation models, we used a stochastic gradient descent (SGD) optimizer to optimize the loss function by updating the network parameters, and set the initial learning rate to 0.1, which was multiplied by 0.1 every 10 epochs, and the number of epochs was set to 100. Regarding the stage-assessing model, we used the technique of data augmentation (left–right flip, top–bottom flip, top–bottom and left–right flip, ± 15-degree rotation, and ± 30-degree rotation with 12.5% probability), to extend the small subsets of the stage-assessing dataset considering the imbalance of four subsets (as shown in Table 1); more specifically, we extended the CT scans of stages 1, 2, and 3 so that all have the same numbers of scans as that of stage 4. We also employed the SGD optimizer to update the network, and set the initial learning rate to 0.1, which was multiplied by 0.1 for every 30 epochs. And we ran a five-epoch warmup to initial learning rate (Gotmare, Shirish Keskar, Xiong, & Socher, 2018) for stabilizing the model.
5. Results and discussion
This section presents the results of the proposed method applied on the test subset, as well as the discussions and ablation study of the key steps of the proposed method. We used the trained lung and lesion segmentation models to segment the lung and lesion portions of CT images contained in the stage-assessing dataset, and then extracted multi-view lesion slices. We assessed the stage of COVID-19 patients by inputting these lesion slices and clinical metadata into the stage-assessing model.
5.1. Segmentation results
We trained the lung and the lesion segmentation models using the specific databases mentioned earlier, accompanied by their annotated labels. As shown in Table 2 , on the test subset of the lung segmentation dataset, the DSC and IOU of lung segmentation were 97.59% and 96.24%, respectively; on the test subset of the lesion segmentation dataset, the DSC and IOU of lesion segmentation were 84.82% and 83.56%, respectively.
Table 2.
Segmentation performances on the testing sets of lung and lesion segmentation datasets.
| DSC (%) | IOU (%) | |
|---|---|---|
| Lung | 97.59 | 96.24 |
| Lesion | 84.82 | 83.56 |
Then we directly used these two trained segmentation models to segment the lungs and lesions of COVID-19 patients on the stage-assessing dataset. The visualization results for the examples of four COVID-19 stages were shown in Fig. 5 and Fig. 6 . As clarified before, the segmented lungs were only used to help define the lung boxes, which were used to define reasonable regions for lesion segmentation. Here we want to emphasize that even the lung and lesion segmentation results might not be always quite perfect by the existing segmentation models, as indicated by the lesion segmentation performance on the lesion segmentation dataset listed in Table 2, our proposed stage-assessing model could robustly obtain quite promising performance on the stage-assessing dataset using the image features extracted from the multi-view lesion slices (together with the clinical metadata).
Fig. 5.
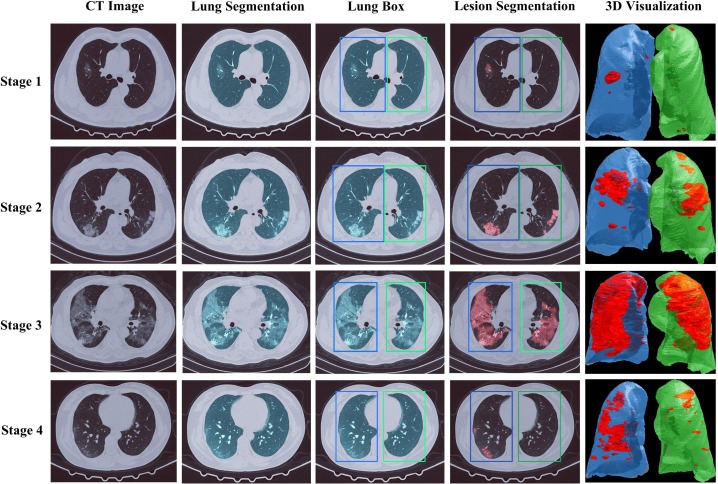
Cases study of segmentation performance on the stage-assessing dataset. The first column shows the original CT images. The second column lists the results of the lung segmentation model. The third column shows the detected left and right lung boxes based on lung segmentation. The fourth column shows the results of the lesion segmentation model restricted by the two boxes. The fifth column shows the 3D visualization of the detected left and right lesions.
Fig. 6.
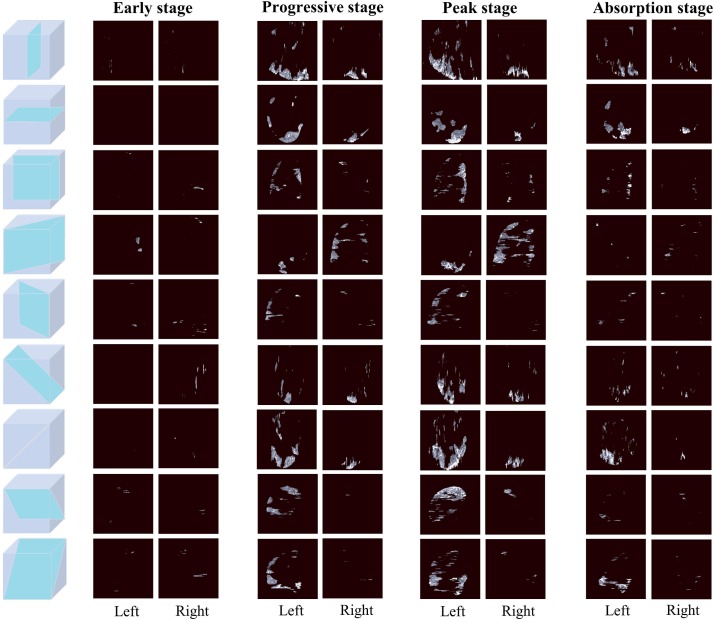
Nine-view lesion slices from the left and right lungs of same COVID-19 patient who underwent CT examinations on Jan 25th, Feb 5th, Feb 13th and Feb 20th of 2020, respectively.
5.2. Stage-assessing results
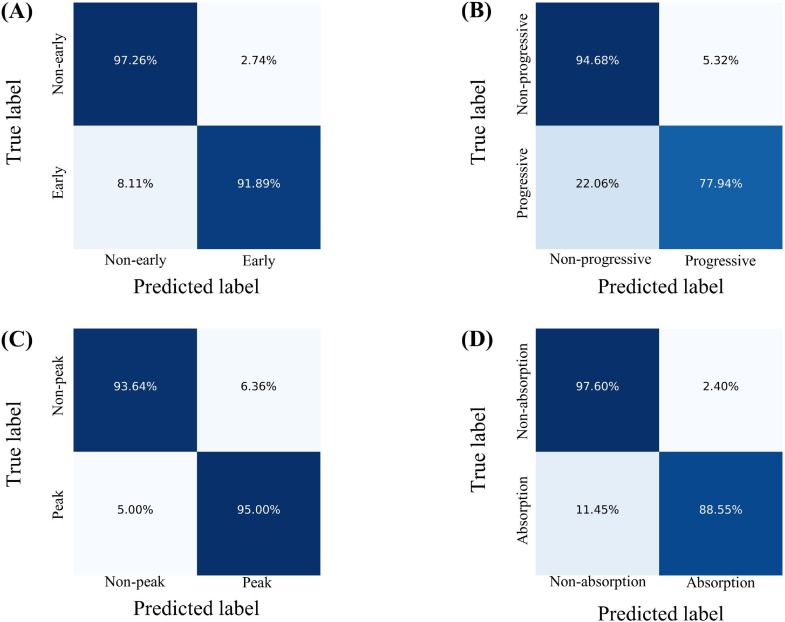
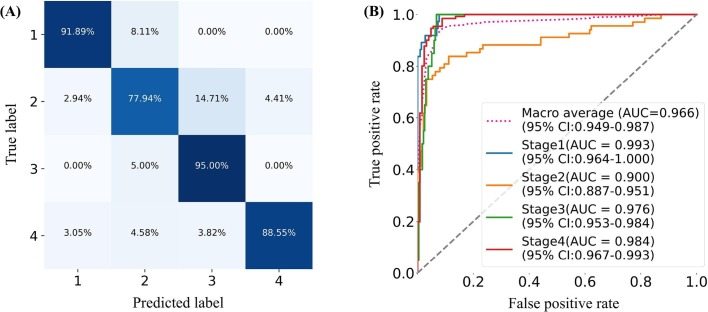
We combined the image features extracted from the lesion slices and the clinical features to assess the disease stage of COVID-19 patients, with Acc, Sen, Spe, AA, PLR, NPR, confusion matrix, and ROC curve as measures to evaluate the performance of the proposed stage-assessing model. As shown in Fig. 7 , the proposed model achieved a four-way classification accuracy of 86.7%, and a macro-average area under the curve of 96.6% on the test subset of the stage-assessing dataset. Furthermore, we calculated the aforementioned metrics and confusion matrices for four two-way classifications (i.e., stage 1 versus stages 2, 3, 4; stage 2 versus stages 1, 3, 4; stage 3 versus stages 1, 2, 4; stage 4 versus stages 1, 2, 3). The results listed in Table 3 showed that the proposed model performed best for the prediction of stage 1, and performed relatively poor for the prediction of stage 2. The Fig. 8 gives four confusion matrices, it also showed that our method can effectively distinguish the early, absorption stages especially for the peak stage, thus it can help clinicians with the management and planning of crucial resources, such as medical staff, ventilators, and intensive care units (ICUs) capacity. However, our method can only achieve the sensitivity of 0.78 for the progressive stage. As depicted in Fig. 7(A), we found that our method easily misclassified the progressive stage into the peak stage. This is probably because of three reasons: firstly, given that the structure of the stage-assessing dataset is unbalanced (as shown in Table 1) even we have tried to balance it with the technique of data augmentation, the model was still difficult to learn the difference of four stages; Secondly, our lesion segmentation model could only achieve the DSC of 0.85 (as shown in Table 2), the under-segmented lesion may contribute to the improvement of our model; Thirdly, the image manifestations of progressive and peak stages are quite similar in terms of the nine-view lesion slices (as shown in Fig. 6).
Fig. 7.
Performance of the proposed model. (A). Confusion matrix of four-way classification (B). ROC curves. The red dot curve denotes the macro-average area under the curve (AUC).
Table 3.
Performance of the proposed model in terms of accuracy (Acc), sensitivity (Sen), specificity (Spe), average accuracy (AA), positive likelihood ratio (PLR), and negative likelihood ratio (NLR).
| Stage | Metrics |
|||||
|---|---|---|---|---|---|---|
| Acc | Sen | Spe | AA | PLR | NLR | |
| 1 | 0.965 | 0.919 | 0.973 | 0.946 | 33.541 | 0.083 |
| 2 | 0.902 | 0.780 | 0.947 | 0.863 | 14.653 | 0.233 |
| 3 | 0.938 | 0.950 | 0.936 | 0.943 | 14.947 | 0.053 |
| 4 | 0.930 | 0.886 | 0.976 | 0.931 | 36.896 | 0.117 |
Fig. 8.
The calculated confusion matrices of four two-way classifications. (A). Early stage versus no-early stages. (B). Progressive stage versus non-progressive stages. (C). Peak stage versus non-peak stages. (D). Absorption stage versus non-absorption stages.
5.3. Ablation study
Ablation study was implemented to evaluate the effectiveness of major components in our stage-assessing model. We analysed the multi-view lesion slices, dual-Siamese channels, and clinical metadata embedding, respectively.
5.3.1. Analysis of multi-view slices
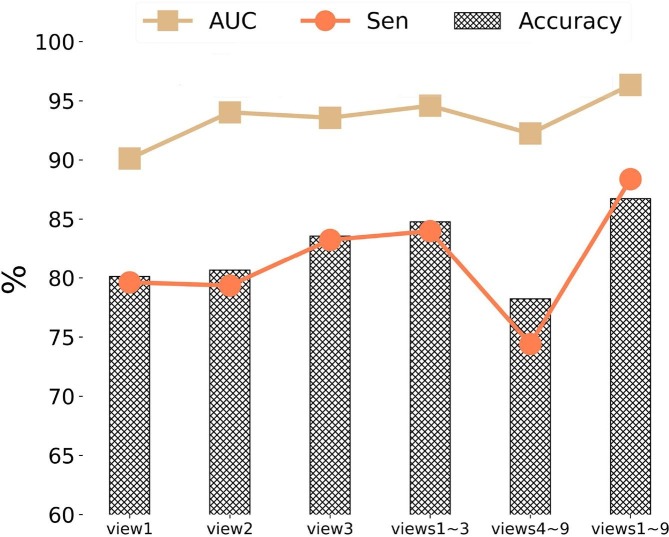
As mentioned before, our stage-assessing model extracted the 2D slices at nine fixed views from the 3D lesion regions as the inputs, mainly aiming to reduce the learning complexity from 3D to 2D spaces while containing sufficient lesion information for stage assessment. The nine views are sagittal, transverse, coronal, and six diagonal views, as shown in Fig. 4(C). To demonstrate the efficiency of our multi-view input strategy, we tested five different combining strategies (i.e., view 1, view 2, view 3, views 1–3, views 4–9, and views 1–9), with the metrics of accuracy, average sensitivity (Sen) of four two-way classifications, and macro-average area (AUC) for evaluation. As shown in Fig. 9 , the aforementioned nine-view inputs outperformed the other four strategies. It also showed that view 3 (coronal) performed better than views 1, and 2, and even views 4–9. This is probably because that the lesions mostly located on the middle-lower lobes of lungs based on the findings of COVID-19 CT images (Pan et al., 2020). Furthermore, views 1–3 achieved the second best result, this is probably because these three views (sagittal, transverse, and coronal planes) are the common photography positions in clinical application that can capture the most information of the lesions. In addition, the results also indicated the possibility of improving model performance by combining anatomical knowledge about COVID-19.
Fig. 9.
Performance analysis of the proposed stage-assessing model with different combinations of views for 2D lesion slice extraction in terms of prediction accuracy (Accuracy), average sensitivity (Sen) of four two-way classifications, and macro-average area (AUC).
5.3.2. Comparison using dual-Siamese channels
The proposed stage-assessing network was composed of two major modules: the image-feature- extracting and clinical-metadata-embedding modules. In the image-feature-extracting module, we used the dual-Siamese channels, which consisted of four encoders with shared weights, to extract the image features of patients. As shown in Table 4 , different network architectures were tested by changing the DNN components and number of channels. More specifically, we applied different DNN decoders such as ResNet (He et al., 2016), ResNeXt (Xie, Girshick, Dollár, Tu, & He, 2017), SEResNet (Hu, Shen, Albanie, Sun, & Wu, 2020) and Res2Net [17]. Furthermore, we evaluated the dual-Siamese channels and single-Siamese channels, which meant that the previous CT scan was optional. The results in Table 4 showed that use of dual-Siamese channels significantly improved classification performance compared with the use of single-Siamese channels, thus it indicated that jointly using previous and current CT scans was important to accurately assess the disease development of COVID-19 patients. Moreover, the results further demonstrate the effectiveness of integrating clinical knowledge into AI models, because the clinicians usually like comparing adjacent CT scans when they assess the clinical stages in practice. Regarding the DNN component, as shown in Table 4, Res2Net outperformed other DNN components. This is probably because that the Res2Net module represents multi-scale features at a granular level compared to other DNNs that represent the multi-scale features in a layer-wise manner. Thus it can effectively increase the range of receptive fields, and help the model better understand the lesion based on the contextual information.
Table 4.
Performance of different architectures for dual-Siamese channels.
| Architecture | Results |
|||||
|---|---|---|---|---|---|---|
| Accuracy | Sen | Spe | AA | PLR | NLR | |
| Single-Siamese channel + ResNet18 | 0.745 | 0.779 | 0.861 | 0.820 | 16.102 | 0.425 |
| Single-Siamese channel + ResNeXt18 | 0.751 | 0.785 | 0.869 | 0.827 | 16.224 | 0.402 |
| Single-Siamese channel + SEResNet18 | 0.785 | 0.801 | 0.889 | 0.845 | 18.589 | 0.359 |
| Single-Siamese channel + Res2Net50 | 0.807 | 0.819 | 0.902 | 0.861 | 20.259 | 0.335 |
| Dual-Siamese channels + ResNet18 | 0.811 | 0.815 | 0.902 | 0.859 | 18.559 | 0.346 |
| Dual-Siamese channels + ResNeXt18 | 0.804 | 0.811 | 0.918 | 0.865 | 19.892 | 0.325 |
| Dual-Siamese channels + SEResNet18 | 0.836 | 0.850 | 0.960 | 0.893 | 24.259 | 0.284 |
| Dual-Siamese channels + Res2Net50 | 0.867 | 0.884 | 0.958 | 0.921 | 25.001 | 0.122 |
The number in bold indicates the best value of each metric. Here Sen, Spe, AA, PLR and NLR refer to the average of the corresponding metrics of four two-way classification.
5.3.3. Role of clinical metadata
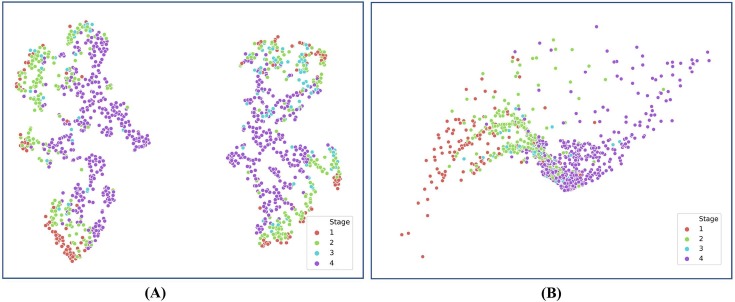
In this study, we combined four items of clinical metadata, i.e., sex, age, progress, and the time interval between two adjacent CT examinations, as prior domain knowledge to regularize the deep model. As shown in Table 5 , we tested different combining strategies through ablation analysis. Specifically, we evaluated the influence of all clinical metadata as a group and each item of clinical metadata. Furthermore, we also tested the effect of dimensionality increase of the clinical metadata from four to 1024 dimensions conducted with the fully connected (FC) layers. The results shown in Table 5 indicated that each item of clinical metadata contributed to the improvement of classification accuracy, especially the two metadata of progress and time interval between two adjacent CT scans. Moreover, these results further demonstrated the clinical findings: Pan et al. (2020) found that the time course is a key factor to assess the four clinical stages on chest CT images; Zhang et al. (2020) found that quantified clinical parameters (e.g., age, AST) were highly correlated with lung lesions by a linear regression analysis; Chassagnon et al. (2020) found that Generic variables (age, sex) had strong correlations with disease progress of COVID-19. The results also showed that dimensionality increase of the original clinical metadata could effectively improve the diagnostic performance, probably because that the neural network could learn high-level abstraction of clinical metadata using multiple layers of neurons. Additionally, such abstraction of features made it easier to distinguish different stages of COVID-19 patients on CT images. With the help of T-Stochastic Neighbour Embedding (T-SNE) (van der Maaten & Hinton, 2008) for visualizing high-dimensional data in 2D space shown in Fig. 10 , it showed that the four stages were better grouped into clusters after re-coding the original clinical metadata as a 1024-dimensional feature vector.
Table 5.
Performance of stage assessment with clinical metadata embedding based on ablation experiment.
| Sex | Age | Progress | M-progress | Dimensionality increase | Accuracy (%) |
|---|---|---|---|---|---|
| × | × | × | × | × | 80.1 |
| × | √ | √ | √ | √ | 86.3 |
| √ | × | √ | √ | √ | 84.8 |
| √ | √ | × | √ | √ | 81.1 |
| √ | √ | √ | × | √ | 84.6 |
| √ | √ | √ | √ | × | 83.9 |
| √ | √ | √ | √ | √ | 86.7 |
Fig. 10.
T-Stochastic Neighbour Embedding (T-SNE) visualization of initial four-dimensional clinical features and 1024-dimensional clinical feature vector re-coded by FC. (A). T-SNE for initial clinical features. (B). T-SNE for re-coded clinical feature vector. T-SNE (van der Maaten & Hinton, 2008) is a nonlinear dimensionality reduction technique.
6. Concussions and future work
The framework proposed in this paper was aimed at developing AI-based systems for assisting hospitals in monitoring the progression of COVID-19 and managing the crucial resources. To our best knowledge, this work was one of the first to assess four clinical stages of COVID-19 patients based on chest CT images and clinical metadata through a deep-learning method. Specifically, we decomposed 3D lesion regions of the left and right lungs into fixed views to reduce the complexity of the model, and used dual-Siamese channels composed of four DNN encoders to extract image features of lesion regions. Furthermore, we combined clinical metadata to regularize the deep model and improve the classification accuracy. The proposed approach achieved an accuracy of 86.7% and an averaged AUC of 96.6%. Results demonstrate the potential of the proposed method and verified the significance of prior domain knowledge for improving the model performance, especially in the medical field.
The proposed method still had some shortcomings to be addressed in our future work. As shown in Fig. 8(A), the proposed model did not perform quite well for stage 2 (progressive stage). Our plan is to extend the number of stage-assessing datasets to balance the categories. Furthermore, we also plan to improve the performance for lesion segmentation of COVID-19 patients on CT images.
CRediT authorship contribution statement
Zhidan Li: Methodology, Writing - original draft. Shixuan Zhao: Methodology, Writing - original draft. Yang Chen: Project administration, Resources. Fuya Luo: Methodology, Validation. Zhiqing Kang: Software, Validation. Shengping Cai: Conceptualization. Wei Zhao: Resources, Validation. Jun Liu: Conceptulization, Resources. Di Zhao: Conceptualization. Yongjie Li: Supervision, Project administration, Conceptualization, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgment
This work is supported by the Key Area R&D Program of Guangdong Province (NO. 2018B030338001). We thank Jun Ma et al. and Kang Zhang et al. for providing the public COVID-19 dataset, and Jiao Xu for helping with data pre-processing.
Ethical approval
This study and its procedures were approved by the local ethics committees. All methods were performed in accordance with the relevant guidelines and regulations. The entire experiment followed the Helsinki Declaration. Informed consent was not required for this retrospective study (i.e., those discharged or who died). Written informed consent from the involved patients was not required.
References
- Aboutalebi, H., Abbasi, S., Javad Shafiee, M., & Wong, A. (2021). COVID-Net CT-S: 3D Convolutional Neural Network Architectures for COVID-19 Severity Assessment using Chest CT Images. In (pp. arXiv:2105.01284).
- Ali, A. R., & Budka, M. (2021). An Automated Approach for Timely Diagnosis and Prognosis of Coronavirus Disease. In (pp. arXiv:2104.14116).
- Altan A., Karasu S. Recognition of COVID-19 disease from X-ray images by hybrid model consisting of 2D curvelet transform, chaotic salp swarm algorithm and deep learning technique. Chaos, Solitons & Fractals. 2020;140:110071. doi: 10.1016/j.chaos.2020.110071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassagnon, G., Vakalopoulou, M., Battistella, E., Christodoulidis, S., Hoang-Thi, T.-N., Dangeard, S., Deutsch, E., Andre, F., Guillo, E., Halm, N., El Hajj, S., Bompard, F., Neveu, S., Hani, C., Saab, I., Campredon, A., Koulakian, H., Bennani, S., Freche, G., Lombard, A., Fournier, L., Monnier, H., Grand, T., Gregory, J., Khalil, A., Mahdjoub, E., Brillet, P.-Y., Tran Ba, S., Bousson, V., Revel, M.-P., & Paragios, N. (2020). AI-Driven CT-based quantification, staging and short-term outcome prediction of COVID-19 pneumonia. In (pp. arXiv:2004.12852).
- Coronavirus disease (COVID-19) Situation dashboard. In. (2021). https://covid19.who.int/table: World Health Organization.
- Fan D.-P., Zhou T., Ji G.-P., Zhou Y.i., Chen G., Fu H., et al. Inf-Net: Automatic COVID-19 Lung Infection Segmentation From CT Images. IEEE Transactions on Medical Imaging. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- Gao K., Su J., Jiang Z., Zeng L.-L., Feng Z., Shen H., et al. Dual-branch combination network (DCN): Towards accurate diagnosis and lesion segmentation of COVID-19 using CT images. Medical Image Analysis. 2021;67:101836. doi: 10.1016/j.media.2020.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotmare, A., Shirish Keskar, N., Xiong, C., & Socher, R. (2018). A Closer Look at Deep Learning Heuristics: Learning rate restarts, Warmup and Distillation. In (pp. arXiv:1810.13243).
- He K., Zhang X., Ren S., Sun J. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016. Deep Residual Learning for Image Recognition; pp. 770–778. [Google Scholar]
- He K., Zhao W., Xie X., Ji W., Liu M., Tang Z., et al. Synergistic learning of lung lobe segmentation and hierarchical multi-instance classification for automated severity assessment of COVID-19 in CT images. Pattern Recognition. 2021;113:107828. doi: 10.1016/j.patcog.2021.107828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Shen L.i., Albanie S., Sun G., Wu E. Squeeze-and-Excitation Networks. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2020;42(8):2011–2023. doi: 10.1109/TPAMI.2019.2913372. [DOI] [PubMed] [Google Scholar]
- Jin Q., Cui H., Sun C., Meng Z., Wei L., Su R. Domain adaptation based self-correction model for COVID-19 infection segmentation in CT images. Expert Systems with Applications. 2021;176:114848. doi: 10.1016/j.eswa.2021.114848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermany D.S., Goldbaum M., Cai W., Valentim C.C.S., Liang H., Baxter S.L., et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell. 2018;172(5):1122–1131.e9. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- Lessmann N., Sánchez C.I., Beenen L., Boulogne L.H., Brink M., Calli E., et al. Automated Assessment of COVID-19 Reporting and Data System and Chest CT Severity Scores in Patients Suspected of Having COVID-19 Using Artificial Intelligence. Radiology. 2021;298(1):E18–E28. doi: 10.1148/radiol.2020202439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhao W., Shi F., Qi L., Xie X., Wei Y., et al. A novel multiple instance learning framework for COVID-19 severity assessment via data augmentation and self-supervised learning. Medical Image Analysis. 2021;69:101978. doi: 10.1016/j.media.2021.101978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizzi, F., Agosti, A., Brero, F., Fiamma Cabini, R., Evelina Fantacci, M., Figini, S., Lascialfari, A., Laruina, F., Oliva, P., Piffer, S., Postuma, I., Rinaldi, L., Talamonti, C., & Retico, A. (2021). Quantification of pulmonary involvement in COVID-19 pneumonia by means of a cascade oftwo U-nets: training and assessment on multipledatasets using different annotation criteria. In (pp. arXiv:2105.02566).
- Ma, J., Wang, Y., An, X., Ge, C., Yu, Z., Chen, J., Zhu, Q., Dong, G., He, J., He, Z., Zhu, Y., Nie, Z., & Yang, X. (2020). Towards Data-Efficient Learning: A Benchmark for COVID-19 CT Lung and Infection Segmentation. arXiv e-prints, arXiv:2004.12537. [DOI] [PubMed]
- Pan F., Ye T., Sun P., Gui S., Liang B.o., Li L., et al. Time Course of Lung Changes at Chest CT during Recovery from Coronavirus Disease 2019 (COVID-19) Radiology. 2020;295(3):715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M., Driggs D., Thorpe M., Gilbey J., Yeung M., Ursprung S., et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nature Machine Intelligence. 2021;3(3):199–217. [Google Scholar]
- Rodriguez-Nava G., Yanez-Bello M.A., Trelles-Garcia D.P., Chung C.W., Friedman H.J., Hines D.W. Performance of the quick COVID-19 severity index and the Brescia-COVID respiratory severity scale in hospitalized patients with COVID-19 in a community hospital setting. International Journal of Infectious Diseases. 2021;102:571–576. doi: 10.1016/j.ijid.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahzad A., Hassan R., Aremu A.Y., Hussain A., Lodhi R.N. Effects of COVID-19 in E-learning on higher education institution students: The group comparison between male and female. Quality & Quantity. 2021;55(3):805–826. doi: 10.1007/s11135-020-01028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., et al. Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation, and Diagnosis for COVID-19. IEEE Reviews in Biomedical Engineering. 2021;14:4–15. doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- Taylor L., Nitschke G. 2018 IEEE Symposium Series on Computational Intelligence (SSCI) 2018. Improving Deep Learning with Generic Data Augmentation; pp. 1542–1547. [Google Scholar]
- van der Maaten L., Hinton G. Viualizing data using t-SNE. Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- Xie S., Girshick R., Dollár P., Tu Z., He K. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2017. Aggregated Residual Transformations for Deep Neural Networks; pp. 5987–5995. [Google Scholar]
- Xie Y., Xia Y., Zhang J., Song Y., Feng D., Fulham M., et al. Knowledge-based Collaborative Deep Learning for Benign-Malignant Lung Nodule Classification on Chest CT. IEEE Transactions on Medical Imaging. 2019;38(4):991–1004. doi: 10.1109/TMI.2018.2876510. [DOI] [PubMed] [Google Scholar]
- Yang R., Li X., Liu H., Zhen Y., Zhang X., Xiong Q., et al. Chest CT Severity Score: An Imaging Tool for Assessing Severe COVID-19. Radiology: Cardiothoracic Imaging. 2020;2 doi: 10.1148/ryct.2020200047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., et al. Clinically Applicable AI System for Accurate Diagnosis, Quantitative Measurements, and Prognosis of COVID-19 Pneumonia Using Computed Tomography. Cell. 2020;181(1423–1433) doi: 10.1016/j.cell.2020.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao A., Balakrishnan G., Durand F., Guttag J.V., Dalca A.V. 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2019. Data Augmentation Using Learned Transformations for One-Shot Medical Image Segmentation; pp. 8535–8545. [Google Scholar]
- Zhao, S., Li, Z., Chen, Y., Zhao, W., Xie, X., Liu, J., Zhao, D., & Li, Y. (2020). SCOAT-Net: A Novel Network for Segmenting COVID-19 Lung Opacification from CT Images. medRxiv, 2020.2009.2023.20191726. [DOI] [PMC free article] [PubMed]
- Zheng B., Liu Y., Zhu Y.u., Yu F., Jiang T., Yang D., et al. MSD-Net: Multi-Scale Discriminative Network for COVID-19 Lung Infection Segmentation on CT. IEEE Access. 2020;8:185786–185795. doi: 10.1109/ACCESS.2020.3027738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Li Z., Zhou J., Li H., Chen Y., Huang Y., et al. A Rapid, Accurate and Machine-Agnostic Segmentation and Quantification Method for CT-Based COVID-19 Diagnosis. IEEE Transactions on Medical Imaging. 2020;39(8):2638–2652. doi: 10.1109/TMI.2020.3001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Rahman Siddiquee M.M., Tajbakhsh N., Liang J. In: Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Stoyanov D., Taylor Z., Bradley A., Papa J.P., Belagiannis V., Nascimento J.C., Lu Z., Conjeti S., Moradi M., Greenspan H., …Madabhushi A., editors. Springer International Publishing; Cham: 2018. UNet++: A Nested U-Net Architecture for Medical Image Segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]