Abstract
Mutations in leucine-rich repeat kinase 2 (LRRK2) are associated with inherited forms of Parkinson’s disease (PD), causing disease by a gain of kinase function. Here, we describe a series of isogenic iPSC lines with any of five pathogenic mutations (N1437H, R1441C, Y1699C, G2019S and I2020T); two hypothesis testing mutations (GTP binding null, T1348N, and kinase dead, K1906M) and two LRRK2 knockouts. This resource could be used to assess effects of mutations on the function of endogenous LRRK2 and/or to study LRRK2 interactors and substrates in iPSC-derived cellular models.
1. Resource Table
| Unique stem cell lines identifier | NIAi001-A |
| NIAi001-B | |
| NIAi001-C | |
| NIAi001-D | |
| NIAi001-E | |
| NIAi001-F | |
| NIAi001-G | |
| NIAi001-H | |
| NIAi001-I | |
| NIAi001-K | |
| NIAi001-L | |
| NIAi001-M | |
| NIAi001-N | |
| Alternative names of stem cell lines | LRRK2 I2020T PL2F4 (NIAi001-A) |
| LRRK2 I2020T PL2D7 (NIAi001-B) | |
| LRRK2 G2019S PL2A9 (NIAi001-C) | |
| LRRK2 G2019S PL2H7 (NIAi001-D) | |
| LRRK2 Y1699C PL3H2 (NIAi001-E) | |
| LRRK2 Y1699C PL2H7 (NIAi001-F) | |
| LRRK2 R1441C PL2F5 (NIAi001-G) | |
| LRRK2 R1441C PL3C2 (NIAi001-H) | |
| LRRK2 N1437H PL3D7 (NIAi001-I) | |
| LRRK2 N1437H PL3F5 (NIAi001-J) | |
| LRRK2 K1906M PL2H7 (NIAi001-K) | |
| LRRK2 T1348N PL1B6 (NIAi001-L) | |
| LRRK2 KO PL1C4 (NIAi001-M) | |
| LRRK2 KO PL1C6 (NIAi001-N) | |
| Institution | National Institutes of Health, National Institute on Aging |
| Contact information of distributor | Mark Cookson: cookson@mail.nih.gov |
| Type of cell lines | iPSC |
| Origin | Human |
| Cell Source | A18945 (Thermo Scientific, cat # A18945) |
| Clonality | Clonal |
| Method of reprogramming | A18945 cell line was derived from CD34 + cord blood using a three-plasmid, seven-factor (SOKMNLT; SOX2, OCT4 (POU5F1), KLF4, MYC, NANOG, LIN28, and SV40L T antigen) EBNA-based episomal system |
| Multiline rationale | Isogenic clones |
| Gene modification | Yes |
| Type of modification | Induced mutation |
| Associated disease | Parkinson’s disease |
| Gene/locus | LRRK2/PARK8 |
| Method of modification | CRISPR/Cas9 |
| Name of transgene or resistance | N/A |
| Inducible/constitutive system | N/A |
| Date archived/stock date | N/A |
| Cell line repository/bank | N/A |
| Ethical approval | Original line obtained from Thermo Scientific. Ethical license/patient consent: −45 CFR Part 46, IRB SOP RR 404. |
2. Resource utility
This unique set of lines will be an important resource for the PD community to study effects of LRRK2 mutations in cell lines expressing endogenous levels of LRRK2. Studies with models derived from these iPSC lines could help determine the roles of different cell types in PD pathogenesis.
3. Resource details
Leucine-rich repeat kinase 2 (LRRK2) encodes a large (2527 amino acid) multidomain protein with kinase and GTPase enzymatic domains. Mutations in LRRK2 are associated with autosomal dominant familial Parkinson’s disease (Nalls et al., 2019; Paisan-Ruiz et al., 2004). Seven mutations in LRRK2 have been demonstrated to be pathogenic: N1437H, R1441C/G/H, Y1699C, G2019S and I2020T (Rudenko et al., 2012). Most data suggests that LRRK2 mutations induce a gain of function while loss of function variants are not associated with PD risk. Prior studies indicate that LRRK2 may play a role in lysosomal biology, vesicle trafficking, cytoskeletal maintenance, and the immune system, but how these relate to PD pathogenesis is uncertain (Cookson, 2015). Looking for common effects of mutations that are not shared with loss of function variants may help resolve this question.
We created five PD pathogenic mutations in heterozygous and homozygous states, as well as GTP-binding null (T1348N) and kinase dead (K1906M) mutations and two LRRK2 knockout lines (Table 1). We used CRISPR/Cas9 technology to introduce all of these mutations into a single female iPSC background, A18945 (TMOi001-A). CRISPR-Cas9 gRNAs within 10–15 bp of each mutagenesis site were selected based on low off-target risk and high on-target potential. LRRK2 knockout (KO) lines were made using two gRNAs targeting the T1348 and K1906 regions of LRRK2. Knock-in mutations were introduced by using donor oligos (DO) with the desired mutation. Additional variants were introduced into some donor oligos to disrupt the CRISPR-Cas9 binding site, and improve editing efficiency, without changing amino acid sequences. Three 96 well plates per mutation were picked and analyzed by Sanger sequencing. Positive clones were expanded and sequenced again to confirm mutations (Fig. 1C, Table 2). To verify clonal purity, and homozygosity, and heterozygosity of the introduced variants we submitted clones for targeted amplicon sequencing and performed Amplican analysis. Pluripotency of the verified iPSC lines was validated by expression of the pluripotency markers: OCT4, SOX2, and NANOG as shown by immunocytochemistry (Fig. 1A, Table 2). Quantitative assessment showed that more than 94% of cells were positive for markers essential for pluripotency, OCT3/4 and SSEA-4 in all iPSC lines (Fig. 1B, Table 2). OCT3/4 was assessed by ICC, and SSEA-4 by flow cytometry. All iPSC lines have a normal female karyotype (46, XX) without any obvious aberrations (Supplementary Fig. 1, Table 2). Short tandem repeat (STR) analysis of 16 genomic loci showed 100% identical polymorphisms in the parental iPSC line and all derived iPSC lines (archived at journal; available from authors). The differentiation potential of the iPSC lines was confirmed by targeted differentiation into cells of all three germ layers. Immunocytochemistry confirmed expression of SOX17 and α-fetoprotein (AFP) (endoderm); alpha-muscle actin (<SMA) and Brachyury (mesoderm); and Nestin and MAP2 (ectoderm) (Fig. 1D, Table 2). All generated iPSC lines were free of mycoplasma contamination (Supplementary Fig. 1B, Table 2). In conclusion, we generated knockin and knockout iPSC lines to support studies of LRRK2-related neurodegeneration in human cellular models.
Table 1.
Summary of lines.
| iPSC line names | Abbreviation in figures | Gender | Age | Ethnicity | Genotype of locus | Disease |
|---|---|---|---|---|---|---|
| NIAi001-A | I2020T PL2F4 | Female | Neonate | Unknown | I2020T homozygous mutation in LRRK2 gene | Parkinson disease |
| NIAi001-B | I2020T PL2D7 | Female | Neonate | Unknown | I2020T heterozygous mutation in LRRK2 gene | Parkinson disease |
| NIAi001-C | G2019S PL2A9 | Female | Neonate | Unknown | G2019S homozygous mutation in LRRK2 gene | Parkinson disease |
| NIAi001-D | G2019S PL2H7 | Female | Neonate | Unknown | G2019S heterozygous mutation in LRRK2 gene | Parkinson disease |
| NIAi001-E | Y1699C PL3H2 | Female | Neonate | Unknown | Y1699C homozygous mutation in LRRK2 gene | Parkinson disease |
| NIAi001-F | Y1699C PL2H7 | Female | Neonate | Unknown | Y1699C heterozygous mutation in LRRK2 gene | Parkinson disease |
| NIAi001-G | R1441C PL2F5 | Female | Neonate | Unknown | R1441C homozygous mutation in LRRK2 gene | Parkinson disease |
| NIAi001-H | R1441C PL3C2 | Female | Neonate | Unknown | R1441C heterozygous mutation in LRRK2 gene | Parkinson disease |
| NIAi001-I | N1437H PL3D7 | Female | Neonate | Unknown | N1437H homozygous mutation in LRRK2 gene | Parkinson disease |
| NIAi001-J | N1437H PL3F5 | Female | Neonate | Unknown | N1437H heterozygous mutation in LRRK2 gene | Parkinson disease |
| NIAi001-K | K1906M PL2H7 | Female | Neonate | Unknown | K1906M homozygous mutation in LRRK2 gene | NIAi001-K |
| NIAi001-L | T1348N PL1B6 | Female | Neonate | Unknown | T1348N homozygous mutation in LRRK2 gene | NIAi001-L |
| NIAi001-M | LRRK2 KO PL1C4 | Female | Neonate | Unknown | c.5694_5697delTGAA (p.Tyr1898Ter) and c.4016_4985del (p.Ile1339LysfsX12) | NIAi001-M |
| NIAi001-N | LRRK2 KO PL1C6 | Female | Neonate | Unknown | c.4015_4018delATTG (p.Ile1339TrpfsX15) and c.4016_4985del (p.Ile1339LysfsX12) mutations in LRRK2 gene |
Fig. 1.
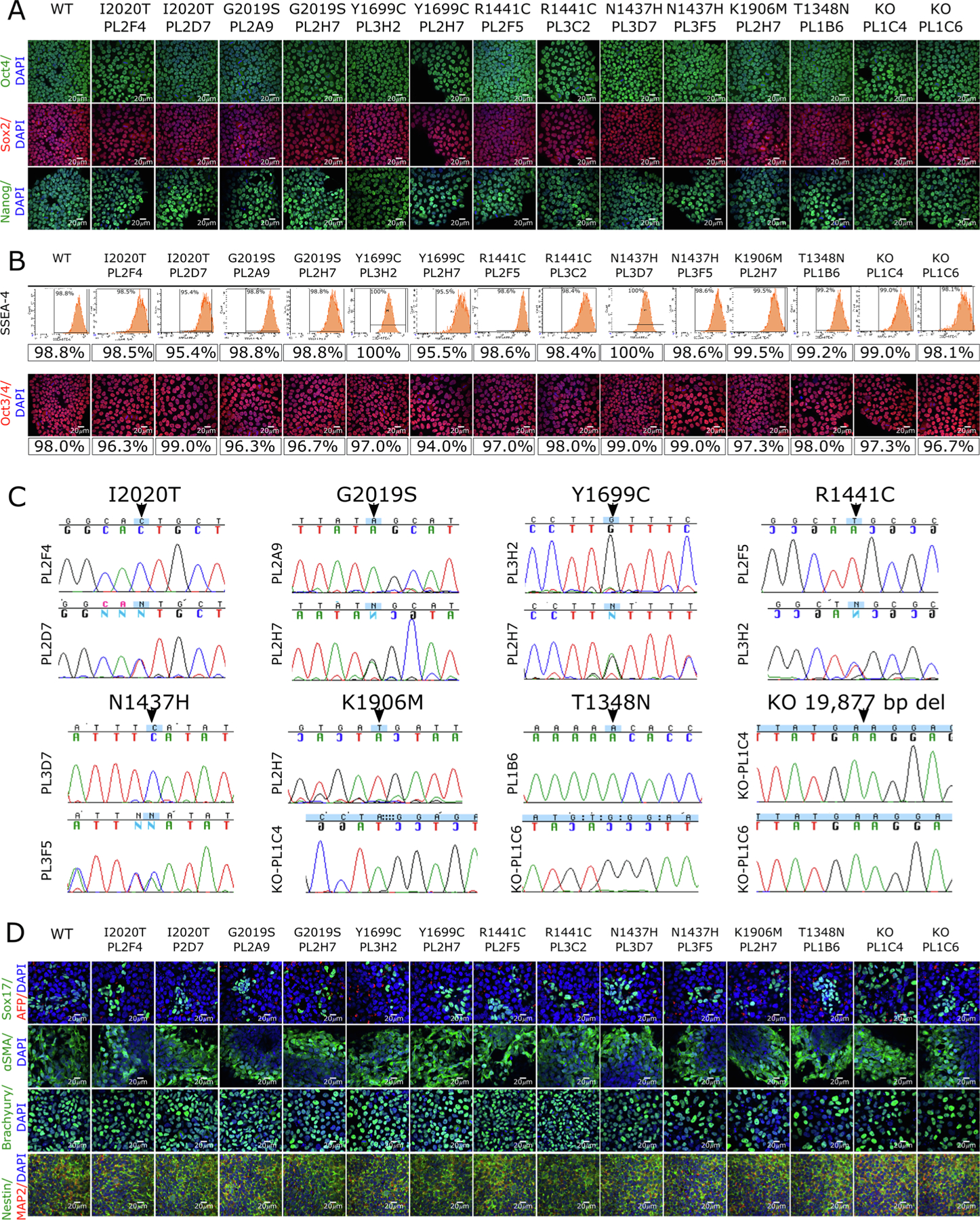
Generation of lines with LRRK2 mutations on an isogenic background. (A) iPSC lines carrying the indicated mutations (WT = wild type, KO = knockout; clone ID below each mutation) were stained for pluripotency markers OCT4 (top panels, green), SOX2 (middle panels, red) and NANOG (lower panels, green) and counterstained with the nuclear stain DAPI (blue, all panels). Scale bars = 20 μm. (B) Quantitative analysis of SSEA-4 expression by flow cytometry (upper panels) or by staining for Oct3/4 (lower panels; red is Oct3/4, blue is DAPI; Scale bars = 20 μm) allowed percentages of pluripotent cells to be estimated as indicated. (C) Sequence chromatograms of LRRK2 coding changes, where upper panels show homozygous and lower panels show heterozygous clones. (D) Differentiation potential was assessed using immunostaining for endodermal markers SOX17 (green, first row) and α-fetoprotein (AFP, red, first row); mesodermal markers alpha-muscle actin (SMA, green, second row) and Brachyury (green, third row); and ectodermal markers Nestin (green, fourth row) and MAP2 (red, fourth row). All cells were counterstained with the nuclear dye DAPI (blue) and scale bars = 20 μm.
Table 2.
Characterization and validation.
| Classification | Test | Result | Data | ||
|---|---|---|---|---|---|
| Morphology | Photography | All iPSC lines are morphologically normal | Not shown; available from authors | ||
| Phenotype | Qualitative analysis | Confirmed expression of pluripotency markers: OCT4, NANOG, and SOX2 | Fig. 1 panel A | ||
| Quantitative analysis | Assessed % of positive cells for pluripotency marker, OCT3/4 by immunocytochemistry counting and cell surface markers, SSEA-4 by flow cytometry. All lines are more that 94% positive for both markers. | Fig. 1 panel B | |||
| Genotype | Karyotype (G-banding) and resolution | 1. 46XX, Resolution 425–500 | Supplementary panel A | ||
| 2. 46XX, Resolution 350–450 | |||||
| 3. 46XX, Resolution 400–500 | |||||
| 4. 46XX, Resolution 400–450 | |||||
| 5. 46XX, Resolution 425–550 | |||||
| 6. 46XX, Resolution 425–475 | |||||
| 7. 46XX, Resolution 425–500 | |||||
| 8. 46XX, Resolution 400–425 | |||||
| 9. 46XX, Resolution 475–550 | |||||
| 10. 46XX, Resolution 425–500 | |||||
| 11. 46XX, Resolution 475–550 | |||||
| 12. 46XX, Resolution 375–450 | |||||
| 13. 46XX, Resolution 425–500 | |||||
| 14. 46XX, Resolution 450–475 | |||||
| 15. 46XX, Resolution 400–525 | |||||
| Identity | Microsatellite PCR (mPCR) OR STR analysis | STR analysis 16 specific loci were tested. All isogenic lines matched 100%. |
Supplementary data 1 Submitted in archive with journal |
||
| Mutation analysis (IF APPLICABLE) | Sequencing | 1. I2020T PL2F4-homozygous | Fig. 1 panel C | ||
| 2. I2020T PL2D7-heterozygous | |||||
| 3. G2019S PL2A9-homozygous | |||||
| 4. G2019S PL2H7-heterozygous | |||||
| 5. Y1699C PL3H2-homozygous | |||||
| 6. Y1699C PL2F7-heterozygous | |||||
| 7. R1441C PL2F5-homozygous | |||||
| 8. R1441C PL3C2-heterozygous | |||||
| 9. N1437H PL3D7-homozygous | |||||
| 10. N1437H PL3F5-heterozygous | |||||
| 11. K1906M PL2H7-homozygous | |||||
| 12. T1348N PL1B6-homozygous | |||||
| 13. KO PL1C4-c.5694_5697delTGAA (p.Tyr1898Ter) and c.4016_4985del (p.Ile1339LysfsX12) | |||||
| 14. KO PL1C6-c.4015_4018delATTG (p.Ile1339TrpfsX15) and c.4016_4985del (p.Ile1339LysfsX12) | |||||
| Southern Blot OR WGS | Not performed | N/A | |||
| Microbiology and virology | Mycoplasma | Mycoplasma testing was done by RT-PCR. All clones-negative | Supplementary Fig. 1 | ||
| Differentiation potential | e.g. Embryoid body formation OR Teratoma formation OR Scorecard OR Directed differentiation | The STEMdiff™ Trilineage Differentiation Kit (StemCell) was used to test differentiation potential. | |||
| We confirmed the expression of endoderm (AFP, SOX-17), mesoderm (<-SMA, Brachyury) and ectoderm (Nestin, MAP2) markers in all clones. | Fig. 1 panel D | ||||
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | Not performed | N/A | ||
| Genotype additional info (OPTIONAL) | Blood group genotyping | Not performed | N/A | ||
| HLA tissue typing | Not performed | N/A | |||
4. Materials and methods
4.1. Growth, propagation and morphology of iPSC lines.
A18945 cell line and isogenic clones derived from this line were grown in E8 media (Thermo Scientific, cat #A1517001). 10uM Rock inhibitor (STEMCELL, Cat # 72304) was added to E8 media for 18–24 h after splitting and thawing. iPSC clones were frozen in Synth-A-Freeze Cryopreservation media (Thermo Scientific, Cat # A1254201).
4.2. Genome editing of A18945 iPSC line using RNPs
4.2.1. RNP complex formation
Alt-R CRISPR-Cas9 guide RNA (crRNA) were custom designed using the IDT website https://www.idtdna.com/site/order/designtool/index/CRISPR_CUSTOM based on low off-target risk and high on-target potential. Alt-R CRISPR-Cas9 crRNAs for each targeted region are shown in Table 3. Alt-R CRISPR-Cas9 crRNA and Alt-R CRISPR-Cas9 tracrRNA (IDT, cat # 1072533) were resuspended in nuclease-free duplex buffer (IDT, cat# 11010301) to 200 μM. Five μl of 200 μM crRNA and 5 μl of 200 μM tracrRNA were mixed together and heated at 95°C for 5 min and then the mixture was cooled to RT. Next, 1.7 μl (104 pmol) of Alt-R S.p. Cas9 nuclease (IDT, cat # 1081058) were mixed together with 1.2 μl (120 pmol) Alt-R CRISPR-Cas9 crRNA/tracrRNA duplex and 2.1 μl of 1xPBS solution and incubated 30 min at RT.
Table 3.
Reagents details.
| Antibodies used for immunocytochemistry/flow-cytometry | |||
|---|---|---|---|
| Antibody | Dilution | Company Cat # and RRID | |
| Pluripotency Markers | Mouse anti-SOX2 | 1:200 | Millipore, Cat# MAB4423, RRID:AB_11213224 |
| Pluripotency Markers | PE anti-human SSEA-4 | 1:200 | BioLegend Cat# 330406, RRID:AB_1089206 |
| Pluripotency Markers | Mouse anti-OCT3/4 | 1:200 | Santa Cruz Biotechnology Cat# sc5279, RRID: AB_628051 |
| Pluripotency Markers | Rabbit anti-OCT4 | 1:200 | Abcam Cat #ab19557, RRID:N/A |
| Pluripotency Markers | Mouse anti-Nanog | 1:200 | Millipore Cat# MABD24, RRID:AB_11203826 |
| Differentiation Markers | Rabbit anti-Nestin | 1:200 | Millipore Cat# ABD69, RRID:AB_2744681 |
| Differentiation Markers | Mouse anti-MAP2 | 1:200 | Santa Cruz Biotechnology Cat# sc-74422, RRID:AB_1126216 |
| Differentiation Markers | Mouse anti-alpha-Feto Protein | 1:200 | Millipore Cat# 2004189, RRID: N/A |
| Differentiation Markers | Goat anti-SOX17 | 1:200 | R and D Systems Cat# AF1924, RRID:AB_355060 |
| Differentiation Markers | Goat anti-Brachyury | 1:200 | R and D Systems Cat# AF2085, RRID:AB_2200235 |
| Differentiation Markers | Goat anti-alpha-Muscle Actin | 1:200 | Novus Cat# NB300–978, RRID:AB_2273628 |
| Secondary antibodies | Donkey anti-mouse Alexa Fluor 488 | 1:500 | Thermo Fisher Scientific Cat# A-21202, RRID:AB_141607 |
| Secondary antibodies | Donkey anti-mouse Alexa Fluor 568 | 1:500 | Thermo Fisher Scientific Cat# A10037, RRID:AB_2534013 |
| Secondary antibodies | Donkey anti-rabbit Alexa Fluor 488 | 1:500 | Thermo Fisher Scientific Cat# A-21206, RRID:AB_2535792 |
| Secondary antibodies | Donkey anti-goat Alexa Fluor 488 | 1:500 | Thermo Fisher Scientific Cat# A-11055, RRID:AB_2534102 |
| Secondary antibodies | Donkey anti-goat Alexa Fluor 568 | 1:500 | Thermo Fisher Scientific Cat# A-11057, RRID:AB_2534104 |
| Primers | |||
| Target | Forward/Reverse primer (5’−3’) | ||
| Targeted mutation analysis/sequencing | G2019S and 120201 | TTGACCCTTTTTTAAAGCAT/ACAATGTGATGCTTGCATTT | |
| Targeted mutation analysis/sequencing | Y1699C | CACAGGGATTACCGAAAATA/AAGGAATTTTCTTCCTGACAA | |
| Targeted mutation analysis/sequencing | R1441C | TAAAAGGGTGAAGANTTGGG/TTCTCTACCAGCCTACCATG | |
| Targeted mutation analysis/sequencing | N1437H | TTACCCTTGCCTCCAGAATG/AACATTCACCAACATGGAACA | |
| Targeted mutation analysis/sequencing | K1906M | AGGGTATGTGTAGAGGGAAG/CAACGAAAAGACAAACTAGG | |
| Targeted mutation analysis/sequencing | T1348N | CAGTACTCTGAAAGACATGTACA/CCATTCTGGAGGCAAGGGTA | |
| Amplicon Sequencing | 12020T and G20195 | AAGGGACAAAGTGAGCACAGA/CCTGATCACCTACCTGGTGTG | |
| Amplicon Sequencing | Y1699C region | GCTCAACAAGGTTGGGTGTT/TTTTCATATGCCATCTCCCTAA | |
| Amplicon Sequencing | R1441C region | TGGGTTAAGAAAGGCCAAAA/GCCAAAGCATCAGATTCCTC | |
| Amplicon Sequencing | N1437H region | GGATTCTTGCCTGTCGTTTG/TTCCCAATTCAAAATTTTAGTGC | |
| Amplicon Sequencing | K1906M region | GGAAGACGGGAAGGTTAAGG/TTTGCATTTTCACAACTGTATGC | |
| Amplicon Sequencing | T1348N region | TTTGCATTTTCACAACTGTATGC/CATACAGTCTACCAGGTTTCTGGA | |
| CRISPR reagents | |||
| Targeted mutation | crRNA used for Cas9 editing | Donor ago sequence | |
| 12020T | ATTGCAAAGATTGCTGACTA cgg | CCACTCAGCCATGATTATATACCGAGACCTGAAACCCCACAATGTGCTGCTTTTCACACTGTATCCCAATGCTGCCATCATTGCAAAGATTGCTGATTATGGCACTGCTCAGTACTGCTGTAGAATGGGGATAAAAACATCAGAGGGCACACCAGGTAGGTGATCAGGTCTGTCTCATAATTCTATCTTCAGGATGGA | |
| 620195 | ATTGCAAAGATTGCTGACTA cgg | CCACTCAGCCATGATTATATACCGAGACCTGAAACCCCACAATGTGCTGCTTTTCACACTGTATCCCAATGCTGCCATCATTGCAAAGATTGCTGATTATAGCATTGCTCAGTACTGCTGTAGAATGGGGATAAAAACATCAGAGGGCACACCAGGTAGGTGATCAGGTCTGTCTCATAATTCTATCTTCAGGATGGA | |
| Y1699C | TGAAATGCCTTATTTTCCM tgg | AGTGTCCTTTTGCCTTTAGTTTETCTGACCACAGGCCTGTGATAGAGCTTCCCCATTGTGAGAACTCTGAAATTATCATTCGACTATATGAAATGCCTTGTTTTCCAATGGGATTTTGGTCAAGATTAATCAATCGATTACTTGAGATTTCACCTTACATGCTTTCAGGGAGAGGTAAGTATCTAATGAAGACT | |
| R1441C | GGGAAGAAGAAGCGCGAGCC tgg | TTCAAGTGAATGTCACGGAAAGCAAATATCAACAGGAATGTGAGCAGGCCCAGTTTGAAAGCAAACACAAGAGGGTTTTGTGTCTTTCCCTCCAGGCTTGCGCCTCATCTTCCCCTGTGATTCTCGTTGGCACACATTTAGATGTTTCTGATGAGAAGCAACGCAAAGCCTGCATGAGTAAAATCACCAAGGAACTC | |
| N1437H | ACCTTTATATTGAAGAGCCA agg | AGGAATTCTATAGTACTCATCCCCATTTTATGACGCA GCGAGCATTGTACCTTGCTGTCTATGACCTCAGCAAG GGACAGGCTGAAGTTGATGCCATGAAGCCTTGGCTA TTTCATATAAAGGTGATTTGTTCTGATCATTTGAAAA TAGAAAATAATTCATGTGTCTGTGTGCGTGTGTGTGT GTGTGTGTAAGTTA | |
| K1906M | AGCCTATGAAGGAGAAGAAG tgg | ATAATTTATTTAGETTTTTTCGCTTGGACAGCTTAATTTAAGTGCTTTGTATTTTCTTTTCAAAAGGTGATGGCAGTTTTGGATCAGTTTACCGAGCTGCCTACGAAGGAGAAGAAGTGGCTGTGATGATTTTTAATAAACATACATCACTCAGGCTGTTAAGACAAGTAAGAAATTCAATAATATAATTATATTAAAT | |
| T1348N | CATTAATTGCTGCAATAAGG tgg | TTTTTATCTTTCAAATACTAGGTTTCTTCAACAGCGATTAAAAAAGGCTGTGCCTTATAACCGAATGAAACTTATGATCGTGGGAAATACTGGGAGTGGTAAAAACACCTTATTGCAGCAATTAATGAAAACCAAGAAATCAGATCTTGGAATGCAAAGTGCCACAGTTGGCATAGATGTGAAAGACTGGCCTATCC | |
| LRRK2 KO GGAAGACGGGAAGGTTAAGG/TTAAGCTGTCCAAG CGAAAAA | AGCCTATGMGGAGAAGAAG tgg and CATTAATTGCTGCAATAAGG tgg | N/A | |
| Primers | |||
| Target | Forward/Reverse primer (5’−3’) | ||
| Positive internal control primers (Mycoplasma test) | LRRK2 locus | GGAAGACGGGAAGGTTAAGG/TTAAGCTGTCCAAGCGAAAAA | |
4.2.2. ssDO preparation
Single stranded donor oligos (ssDO) with the desired mutations (see Table 3) were synthetized by IDT and resuspended in DPBS at a concentration of 100 pmol/μl.
4.2.3. Nucleofection
70–80% confluent iPSC were dissociated into single cells using Accutase (Thermo Scientific, cat # A1110501) and counted using a TC20 Automated Cell Counter (Bio-Rad). iPS cells (8x105 per sample) were pelleted at 1000 rpm for 3 min and cell pellet were gently resuspended in 100 μl of P3 Primary Cell Solution (Lonza, PBP3–02250). Immediately prior to nucleofection, 2 μl (100 pmol/μl) of ssDO was added to 5 μl of pre-assembled Cas9/RNP. One hundred μl of iPS cells resuspended in P3 primary cell solution were transferred to the tube containing Cas9/RNP complexes and ssDO. Cells were mixed twice with Cas9/RNP/Donor oligo and the mixture was transferred to the 100 μl nucleocuvette (Lonza; 2022–01). iPSC were nucleofected immediately using the ‘Primary Cell P3′ program and ‘CA-137′ pulse code. After nucleofection, iPSC were transferred using a Lonza disposable Pasteur pipette into one well of a Matrigel-coated 6-well plate containing 3 mL E8 media with 10 μM Rock inhibitors. Cells were cultured in a 32°C/5% CO2 incubator for two days and then transferred to a 37°C incubator. Edited iPSC pools were expanded, frozen, and used for generation of clones from single cells.
4.3. Generation of clones from single cell
Expanded pools were dissociated with Accutase, counted, and 10,000 single cells were plated on 10 cm Matrigel-coated dishes containing E8 media with 10 μM Rock inhibitor for two days. Subsequently, the media was changed without Rock for 5–7 days. Single cell colonies were expanded to 250–500 μM diameter and picked using a 100 μl pipet tip under a Bioimager microscope. Individual colonies were transferred to Matrigel-coated 96-well plates. Three 96 well plates per mutation were picked and expanded until 70–80% confluency. Each expanded plate was split into two plates: half of the cells were transferred onto a new 96 well plate for further propagation and half of the cells were used for sequencing analysis.
4.4. Sequencing
Plates with collected cells were centrifuged at 3,000 rpm for 5 min and resuspended in 30 μl of water. Cells were heated at 95° C for 10 min and 2 μl of cells were used for PCR with region specific primers listed in Table 3. PCR was carried out using Terra polymerase (Takara Bio). Each PCR product was sequenced using forward and reverse primers with Applied Biosystems BigDye terminator v3.1 sequencing chemistry according to manufacturer’s instructions. The products were cleaned using Agencourt CleanSEQ reagent (Beckman Coulter), run on a 3730xl DNA analyzer (Applied Biosystems, Hitachi) and analyzed with Sequencher software. Positive clones were expanded and re-sequenced.
4.5. Amplicon sequencing
Amplicon sequencing of pools and individual clones was performed by Psomagen using MiSeq Nano (250PE) sequencing on Illumina platform. Amplicon sequencing primers are listed in Table 3. FASTQ files for each sample were analyzed using the ampliCan tool in R (Labun et al., 2019).
4.6. Karyotyping and STR analysis
Karyotyping and STR analysis were performed by WiCell Research Institute.
4.7. Mycoplasma detection
Mycoplasma test was performed using PCR (ATCC, cat # 30–1012K). Positive control PCR was performed for the LRRK2 gene with the primers listed in Table 3.
4.8. Pluripotency assessment
iPSC clones were grown on Matrigel-coated coverslips, fixed with 4% PFA, and stained with pluripotency markers listed in Table 3. Images were taken on a Zeiss 880 confocal microscope. The percentage of cells positive for OCT3/4 was calculated on immunofluorescence images using the Fiji software. SSEA4 quantifications were performed by flow cytometry using PE-SSEA4 antibody (Table 3). At least 5,000 cells per sample were acquired and analyzed using BD FACs Diva 8.01 analysis software.
4.9. Differentiation potential
iPSC clones were differentiated into three layers according to STEMdiff™ Trilineage Differentiation Kit (StemCell, Cat# 05230), fixed in 4% PFA, and stained with differentiation markers listed in Table 3. Images were taken on a Zeiss 880 confocal microscope.
Supplementary Material
Acknowledgement
This research was supported entirely by the Intramural research Program of the NIH, National institute on Aging.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2021.102354.
References
- Cookson MR, 2015. LRRK2 pathways leading to neurodegeneration. Curr. Neurol. Neurosci. Rep 15 (7), 42. Epub 2015/05/27. doi: 10.1007/s11910-015-0564-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Guo X, Chavez A, Church G, Gagnon JA, Valen E, 2019. Accurate analysis of genuine CRISPR editing events with ampliCan. Genome Res 29 (5), 843–847. Epub 2019/03/10. doi: 10.1101/gr.244293.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, et al. , 2019. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol 18 (12), 1091–1102. Epub 2019/11/09. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, et al. , 2004. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 44 (4), 595–600. Epub 2004/11/16. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Rudenko IN, Chia R, Cookson MR, 2012. Is inhibition of kinase activity the only therapeutic strategy for LRRK2-associated Parkinson’s disease? BMC Med 10, 20. Epub 2012/03/01. doi: 10.1186/1741-7015-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


