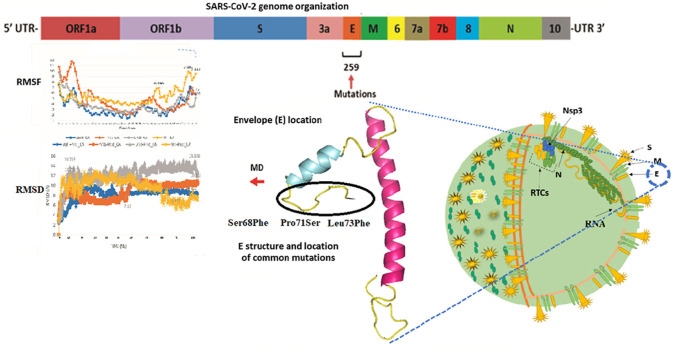
Abstract
Structural proteins of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are potential drug targets due to their role in the virus life cycle. The envelope (E) protein is one of the structural proteins; plays a critical role in virulency. However, the emergence of mutations oftenly leads to drug resistance and may also play a vital role in virus stabilization and evolution. In this study, we aimed to identify mutations in E proteins that affect the protein stability. About 0.3 million complete whole genome sequences were analyzed to screen mutations in E protein. All these mutations were subjected to stability prediction using the DynaMut server. The most common mutations that were detected at the C-terminal domain, Ser68Phe, Pro71Ser, and Leu73Phe, were examined through molecular dynamics (MD) simulations for a 100ns period. The sequence analysis shows the existence of 259 mutations in E protein. Interestingly, 16 of them were detected in the DFLV amino acid (aa) motif (aa72-aa75) that binds the host PALS1 protein. The results of root mean square deviation, fluctuations, radius of gyration, and free energy landscape show that Ser68Phe, Pro71Ser, and Leu73Phe are exhibiting a more stabilizing effect. However, a more comprehensive experimental study may be required to see the effect on virus pathogenicity. Potential antiviral drugs, and vaccines may be developed used after screening the genomic variations for better management of SARS-CoV-2 infections.
Keywords: SARS-CoV-2, Genome, Mutations, Envelope, Stability, Antiviral drugs
Graphical abstract
1. Introduction
Recently the newly emerged coronavirus disease-19 (COVID-19) remains a major public health issue since 2019. The causative agent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1] is passing through numerous evolutionary stages [2,3] and seems more fatal. The virus is rapidly spreading through respiratory droplets, affecting the important organs of the host body [2,3].
Since the coronavirus 2 (CoV-2) belongs to the formerly known family of coronaviruses, it shares close genomic similarity with SARS-CoV. The CoV-2 contains the ssRNA genome [6]; it encodes four structural proteins such as a spike (S), an envelope (E), membrane (M), and nucleocapsid (N) [7], resulting in the coding of 16 non-structural proteins (NSPs). These structural proteins are responsible for viral replication and virion-receptor attachments and, thus, are involved in pathogenicity, viral spread, and the introduction of the virus to the cells of the host body.
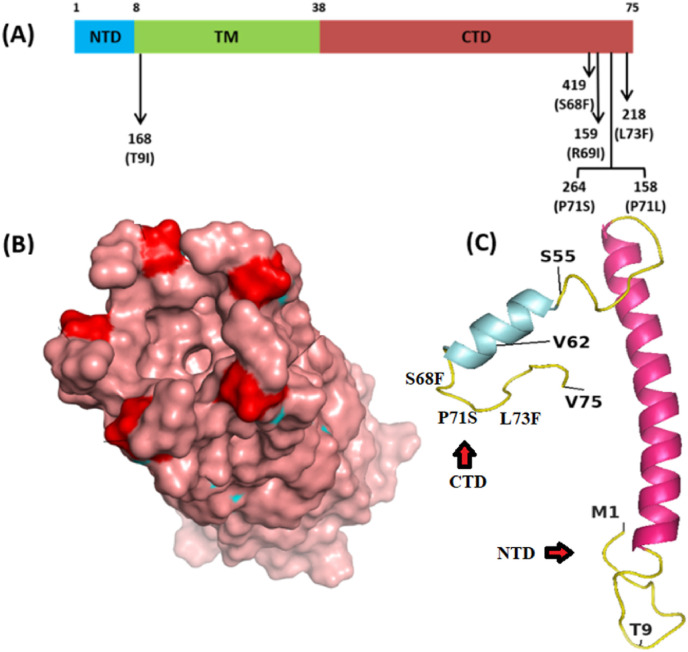
The E protein is composed of 76 amino acids [8] weighing 8–12 kDa [9]. This protein has an N-terminal domain (NTD), hydrophobic domain, and C-terminal domain (CTD) [10], arranged as NTD first 1–9 amino acid (aa), the hydrophobic area, extending from 10 to 37 aa and CTD (38–75 aa) [11]. Ionic pores are formed across the membrane due to the arrangement of the hydrophobic tail region. Structurally, the E protein consists of seven alpha helices and eight loop regions. It forms a pentameric configuration (five molecules) with 35 alpha-helical regions and 40 looped ring regions, which are formed by the hydrophobic tail (Fig. 1 B). This pentameric configuration of the hydrophobic tail can be affected by the interactions within CTD [12]. These pores serve as ion channels that allow the movement of the virus across the membrane, enhancing its pathogenicity [13]. E mutations reduce viral pathogenicity and also stop the channel activity [14] representing an essential drug target and vaccine candidate [15].
Fig. 1.
Domain organization of E proteins and the location and frequency of most common mutations. TM: transmembrane [15], NTD: N-terminal domain, CTD: C-terminal domain. (B). Pentameric representation of E proteins showing channel (C). Full-length E protein (I-TASSER, E-QHD43418) with most common mutations in the loop region.
Some novel mutations seen in the E protein sequence do not present in the already existing coronaviruses. In the CoV-2 E protein sequence, arginine is mutated by isoleucine, threonine, and lysine (R69I, R69T, and R69K) at the 69th position. Moreover, in the amino acid sequence of CoV-2, serine and phenylalanine are present at 55th and 56th positions, instead of threonine and valine, respectively [16].
The SARS-CoV-2 has undergone numerous mutations in all the important targets [[17], [18], [19]]; therefore, drugs created to fight CoV-2 might not very potent. In order to evaluate the drug targets for designing novel antiviral drugs against CoV-2, it is vital to screen the frequency of mutations and their effect on the thermodynamic properties of the important target proteins. This is particularly important for the effective treatment of emerging microbes, including mutants of SARS-CoV-2.
Although structural proteins of CoV-2 are important for investigating mutations, here in this study, we investigated the variations existing only in E proteins owing to their potential drug target. Very limited information and small-scale genomic data has been screened for mutations in the E protein. This is the first comprehensive study in which we screened 0.295 million complete genomes of SARS-CoV-2 for identifying variants in the E protein. Among all the genomes, 259 mutations were detected, exhibiting various degrees of thermodynamic properties.
Analyzing the frequency of mutations and their effect on the thermodynamics properties of E proteins may allow the researcher to design novel inhibitors and predict the level of pathogenicity and transmission. In the current study, the most common mutations that were detected at the C-terminal (Ser68Phe, Pro71Ser, Leu73Phe) were exhibiting more stabilizing effect on E proteins structure. First, the presence of a large number of mutations in the E protein of CoV-2 may lead to the conformational changes and evolutions, resulting in therapeutic failure. Secondly, the virus may be more fatal in near future, and therefore, antivirals against geography-specific CoV-2 strains might be more effective.
2. Methodology
2.1. Genomic sequence retrieval
The complete genome was retrieved from the Global Science Initiative on Sharing All Influenza Data (GISAID) (December 2019–2020) (https://www.gisaid.org/) [20]. The GISAID shared the virus data to publish results and the metadata relevant to public health scientists. This server provides all kinds of CoV-2 genomic data, including even those that have not been unpublished. We screened 0.295 genomes of CoV-2 reported worldwide for variants analysis in E proteins. The sequences were aligned with the reference CoV-2 genome (Accession NC_045512) using the CoVsurver application (https://www.gisaid.org/epiflu-applications/covsurver-mutations-app/). The identified mutations in structural proteins of CoV-2 were separated and arranged in the form of excel sheets. The statistical analysis was performed to screen the most common variants.
2.2. Structural information
Scientists are all well aware that sharing the genomic and proteomic data of SARS-CoV-2 is important for the better management of infectious diseases in order to devise countermeasures. The E protein structural data was retrieved from the protein data bank (PDB) [21] (PDB IDs: E = 7k3g). Some residues at the NTD (1–7aa) and CTD (39–75) are missing in the PDB structure of the COV-2 E protein. To observe the effect of mutations on the residues missing in the CTD and ND terminals, the full-length E protein structure was downloaded (E-QHD43418) from I-TASSER [22,23]. The chain ID in the I-TASSER structure is missing, which was added in PYMOL using the code “sele, chain'” and “alter (sele), chain = ‘A'.” The I-TASSER has already modeled the full-length proteins of CoV-2 using the NCBI reference data (NC_045512) (GenBank MN908947).
2.3. Mutation effect on E proteins’ dynamic stability
All the observed mutations were recorded and their effect on the E protein structure was computed using the DynaMut [24]. The server implemented the mutation effect and also the normal mode methods that can be used to analyze the variants that affect protein stability and flexibility. This impact is measured through graph-based signatures and also as a normal mode. This method outperforming (P < 0.001) with results are also displayed in good resolution.
2.4. Molecular dynamics (MD) simulation
The MD simulation was performed on the Desmond module (Schrodinger), as described in the previous study [23,24]. Briefly, the TIP3P model and Gromos9643a1 forefield was applied. The system was neutralized with counterions (NaCl). The cubic simple point charge (SPC) water box was applied. Two-step (NVT and NPT) energy minimization (50000 ps) till the minimization completion was continued. The ambient pressure was set at 1.013 bar and temperature 310 K for 100 ns? The thermodynamic stability of the WT (wild type) and mutants (MT) E proteins was analyzed using root means square fluctuations (RMSF) and root means square deviation (RMSD). All the simulation were repeated three time for better results. Approximately 1000 frame per simulation was run.
2.5. The Gibbs free energy
The Gibbs free energy (G) [27] of MTs was plotted against wild type E protein. The G is minimized to equilibrium state of system at constant temperature and pressure which is a thermodynamic potential. Principal component analysis (PCA) is performed to recognize low modes in proteins [28,29]. PCA simplifies the complicated motion in trajectory [[30], [31], [32]]. A set z1, z2 …, zp known as principal components (PCs) were generated during PCA. Energies of sets of proteins conformations is called Free Energy Landscape (FEL) [33,34]. The first two components (PC1 and PC2) give the trajectories on initial two principal components of motion. G values shows the stability level of proteins [[35], [36], [37]].
3. Results and discussion
This is the first comprehensive study in which 0.295 million complete genomic sequences of CoV-2, which were reported worldwide in the GISAID server (from December 2019 to December 2020), were analyzed to identify variants in E proteins. A large number of non-synonymous mutations (259) were detected in the genome sequences from 48 countries (Supplementary Table S1), among which the largest number were present in isolates from England (S1). This wide range of variations may project the variation level of CoV-2 strains worldwide. Previous studies [25,26] have screened 3617 and 81,818 COV-2 genomes for E protein variants, respectively. These recent studies reported 115 and 15 non-synonymous mutations, respectively, which were mainly present in the CTD.
3.1. Mutations in envelope (E) protein
3.1.1. Mutations in the NTD
Being the smallest structural protein (75aa) of CoV-2, all of the residues’ positions of the E protein harbored non-synonymous mutations (S1). A total of 31 non-synonymous mutations were detected in the NTD (Supplementary File S1), among which V5F (n = 39), E8D (n = 35), V5I (n = 17), and Y2H (n = 14) were common. A majority of these mutations were detected in the isolates from the UK. The NTD helps the tail region target the Golgi complex using some of its associated elements as mutations in this region may affect its efficiency.
3.1.2. Mutations in the CTD
The most frequent mutations were detected in the CTD of the E protein (68–73 aa) (Table 1 ). Some of them were T91, S55F, V62F, S68F, R691, P71L, P71S, and L73F (Fig. 1). Other common mutations have been listed in the table along with their residue types and positions (Table 1 and S1). Mutations in the CTD, namely, S55F (128), V62F (129), and R69I (159) may affect the virus pathogenesis, altering the binding of the E protein to a tight junction. The motif “DLLV” (72–75 aa) of the CTD showed some mutations that may affect the PALS1 (Protein Associated with Caenorhabditis elegans Lin-7 protein 1) at the Golgi complex, as well as CoV-2 infectivity [38,40]. The PALS1 is a member of the post-synaptic density protein-95/Discs Large/Zonula occludens-1 (PDZ) domain-containing proteins group that is involved in diverse cellular functions and also functions as scaffolds for signaling protein [28,29].
Table 1.
Frequency of some common mutations in the CoV-2 E protein.
| Accession | Wild type AA | Position | Mutated AA | Frequency | aMutation |
|---|---|---|---|---|---|
| EPI_ISL_476911 | T | 9 | I | 168 | T9I |
| EPI_ISL_424214 | S | 55 | F | 128 | S55F |
| EPI_ISL_538676 | V | 62 | F | 129 | V62F |
| EPI_ISL_448073 | S | 68 | F | b419 | bS68F |
| EPI_ISL_452908 | R | 69 | I | 159 | R69I |
| EPI_ISL_577907 | P | 71 | L | 158 | P71L |
| EPI_ISL_660339 | P | 71 | S | b264 | bP71S |
| EPI_ISL_478788 | L | 73 | F | b218 | bL73F |
Full list is available in the S1 supplementary file.
; mutations subjected to MD simulations.
3.1.3. Mutations in the transmembrane (TM) domain
Transmembrane variants such T9I (n = 168), F20L (n = 90), L21F (n = 84), V24 M (n = 76), and T30I (n = 72) may affect the homo pentameric configuration of the E protein [43]. The E protein may also be an effective drug target as it contributes equally to the pathogenicity and cytotoxicity of the virus. It produces viroporins that are hydrophobic in nature [44]. Proline residues facilitate the targeting of the cis-Golgi complex by the hydrophobic tail present in the cytoplasm. The release of these virion particles is facilitated by the ionic gradient present in the endoplasmic reticulum and Golgi compartment through the E protein [8]. Studies on E mutants for highlighting the structural changes of the ion-channel activity behind mutations, might be very helpful for better management of COVID-19.
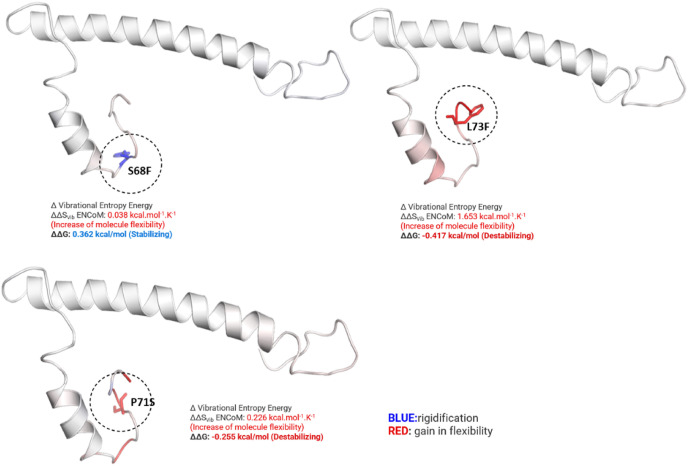
The CTD of E proteins harbored some common mutations whose stability was predicted. The DynaMut prediction outcome of L73F (ΔΔG: −0.417 kcal/mol), P71S (ΔΔG: −0.255 kcal/mol) exert a destabilizing effect. However, T9I (ΔΔG: 0.190 kcal/mol), P71L (ΔΔG: 0.012 kcal/mol), and S68F (ΔΔG: 0.362 kcal/mol) shows a stabilizing effect (Supplementary File S1). The most common mutations were detected at the C-terminal (Ser68Phe, Pro71Ser, and Leu73Phe) were also assessed through MD simulations, exerting a stabilizing effect on the E protein of CoV-2.
3.2. Thermodynamic properties
We analyzed the thermodynamic properties of most common variants (Ser68Phe, Pro71Ser, and Leu73Phe) present in the CTD in relation to the E structure stability in comparison with the wild type (WT).
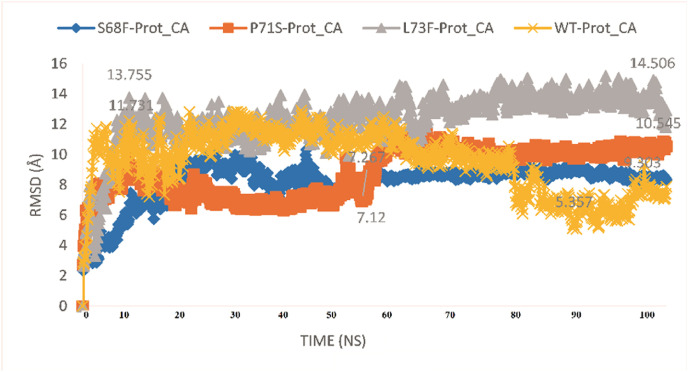
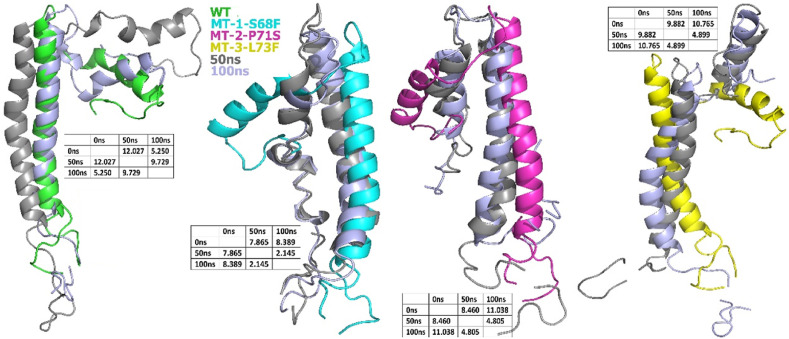
3.2.1. RMSD and RMSF of WT and MTs
The RMSD graphs of then WT and MTs E proteins are shown in Fig. 2 . The MTs S68F and P71S seem more stable from 55ns to 100 ns, exhibiting 9 Å and 10.5 Å RMSDs, respectively. The RMSD of the WT still exhibiting fluctuations at 100 ns? Similar to the other MTs, L73F is also exhibiting stable deviations through the simulation period, with 13.7 Å to 14.5 Å RMSD. The WT exhibited 5.3 Å RMSD at the 80ns–95ns period and still seems to rise from 95ns to 100 ns?
Fig. 2.
RMSD comparison of the WT and MTs E proteins of SARS-CoV-2.
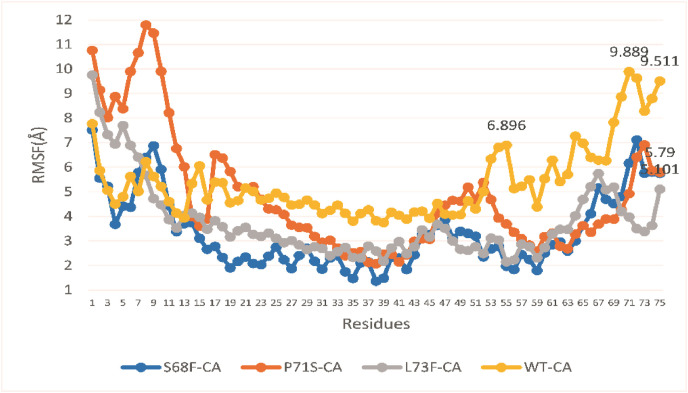
The comparison of the WT and MTs in terms of flexibility exhibited significant variations at some positions (Fig. 3, Fig. 4 ). The RMSF at the 53rd to 75th aa position exhibited a significant difference between the WT and MTs. The WTs at this position exhibited 6.89 Å (55 aa), 9.88 Å (71 aa), and 9.5 Å (75 aa); S68F demonstrated very low RMSF, below 4 Å, for the majority of the residues and 5.1 Å at 75 aa, which was significantly low as compared to the WT. Similarly, P71S also exhibited very low RMSF at CTD (5.7 Å) when compared to the WT (Fig. 3). Similar to S68F, the MT L73F also demonstrated low-level fluctuation when compared with the WT. MD simulations explore the insight dynamic changes at the molecular level [[45], [46], [47]], which might be difficult to accomplish through experimental work. Several studies have reported that any change in protein functions might be associated with RMSF [[48], [49], [50]]. Flexibility is one of the key thermodynamic characteristics that maintain the optimal functions of proteins [51]. A large change in this property may alter the function of the biomolecules.
Fig. 3.
The RMSF of the WT and MTs in the E proteins of SARS-COV-2. CA: Carbon alpha. The RMSF of WT (orange) and MTs. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 4.
The RMSDs of WT and MTs at different simulation periods. The WT behaves very differently at the CTD.
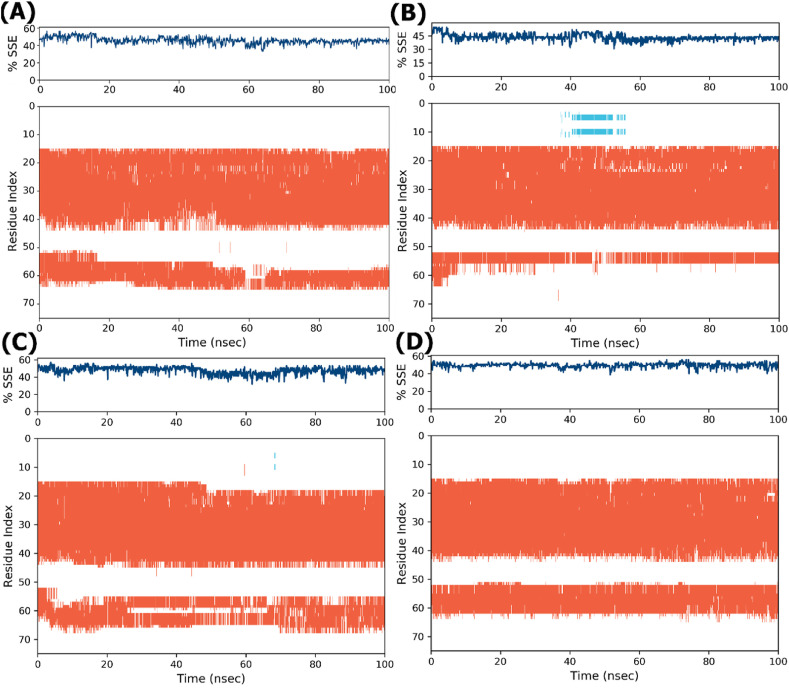
Protein secondary structure elements (SSE) such as alpha helices and beta strands are monitored throughout the simulation. The difference in the SSE of the WT and MTs has been highlighted in Fig. 5 at 20ns–70 ns? The mutation induced some changes in the SSE within a 100ns simulation period. Upon examining Fig. 5, some differences in the SSE of the WT (Fig. 5A) and MTs (Fig. 5B, C, D) can be seen.
Fig. 5.
Comparison of the SSE of the WT and MTs in MD simulation. The plot reports SSE distribution by residue index throughout the protein structure and summarizes the SSE composition for each trajectory frame over the course of the simulation. The plot monitors each residue and its SSE assignment over a certain period of time. (A) The WT residue index and SSE along Y-axis shows differences at the 60–65ns MD simulations period; (B) S68F shows a little variation at 40–55ns; (C) P71S exhibits more variations at 45–68ns; (D) L73F also demonstrates variations when the WT is compared with P71S and S68F.
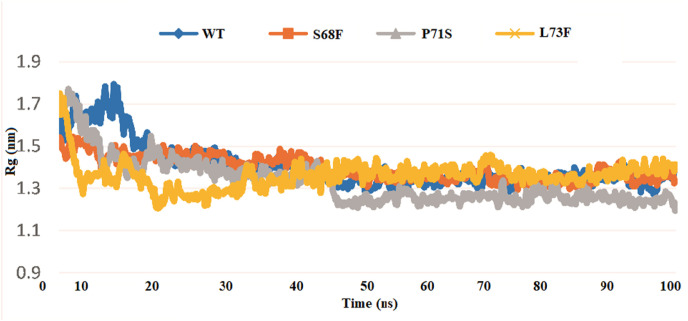
3.2.2. Rg of WT and MTs
The degree of protein-folding stability could be measured through Rg. Fluctuations in Rg for a period indicate unstable folding, while a straight value reveals stable folding [[52], [53], [54], [55]]. A protein with misfolding shows variations in Rg over time (Fig. 6 ). The WT and MTs exhibited variations in folding. The MT S68F exhibited more stable folding than the WT, whereas P71S and L73F also presented stable folding for the 18.9–50ns period (Fig. 6). The S68F MT shows the lowest fluctuations from 21ns to 100 ns (1.3 nm). Rg is a mass-weight root mean square distance between atoms and their common center of mass. It is a vital parameter for defining dynamic stability, offering an insight mechanism of dimension and compactness of biomolecules and total protein systems. The Rg plotted for three MTs of the CoV-2 E proteins shows notable stability than that of the WT. The mutant structures show a slightly low average Rg value than the WT.
Fig. 6.
Comparison of Rg of the WT and MTs E proteins. The WT (blue) exhibited a little difference in folding throughout the 100ns simulation period as compared to the MTs, showing that the folding in both types is stable. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
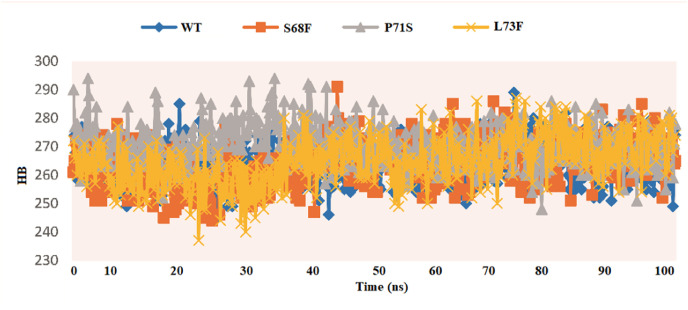
3.2.3. Hydrogen bonding of the WT and the MTs
Most of the interactions among the atoms are mainly hydrogen bonding (HB), which assists in protein folding, stability, and is also involved in recognition. The α helix and β sheet stabilize by the HB potential between the amide nitrogen of the protein and the carbonyl oxygen of the main chain [[56], [57], [58]]. The primary forces in protein–drug interactions are HB, van der Waals, and electrostatic forces [51]. The average number of HB among the WT and MTs CoV-2 E protein atoms is shown in Fig. 7 . MT S68F exhibited fewer HB than P71S and P73F. However, both the MTs (P71S and P73F) demonstrated more HB than the WT (Fig. 7), signifying the stabilizing effect of the mutation. P71S exhibited maximum HB (295) during the first 25ns simulations. However, the number of HB in the MD simulation in the last 25ns is almost the same for the WT and MTs.
Fig. 7.
Hydrogen bonding of the WT and MTs E proteins.
The effect of mutations on E proteins’ stability and flexibility has been shown in Fig. 8 . All of the MTs exhibited an increase in flexibility, which may have a positive effect on function. The molecular activities are linked with the flexible regions. According to a previous investigation [59], protein requires flexibility to engage in good catalytic activity. Adding water improves flexibility, and understanding the diverse role of protein flexibility can help develop biotechnological solutions including vaccines.
Fig. 8.
WT and MTs stability and flexibility.
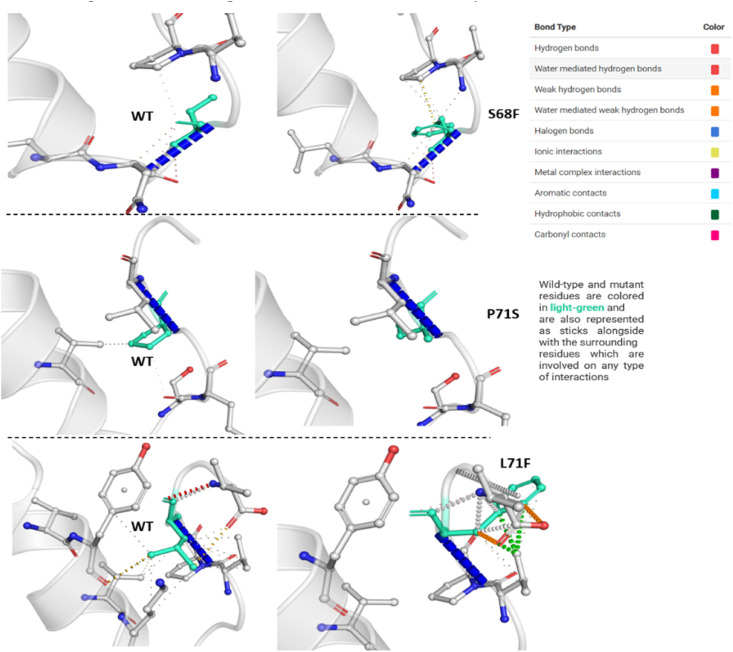
MTs showed more interactions than WT. The S68F exhibited a different geometry due to the aromatic nature of phenylalanine substitutions. Moreover, the number of interactions in this MT seems greater than in the WT (Fig. 9 ). In MT P71S, the proline has been substituted into serine, present in the active site of many enzymes. Similar to the S68F, the L73F exhibited more interactions (Fig. 9) with surrounding residues than the WT due to its substitution into aromatic amino acid, forming more hydrogen interactions with its surrounding residues that might be instrumental in E stability.
Fig. 9.
Interactions of WT and MTs residues with surrounding aa. The type of interactions of WT and MTs residues is colored-coded. S68F, P71S, and L73F MTs have been compared with their WT on the left.
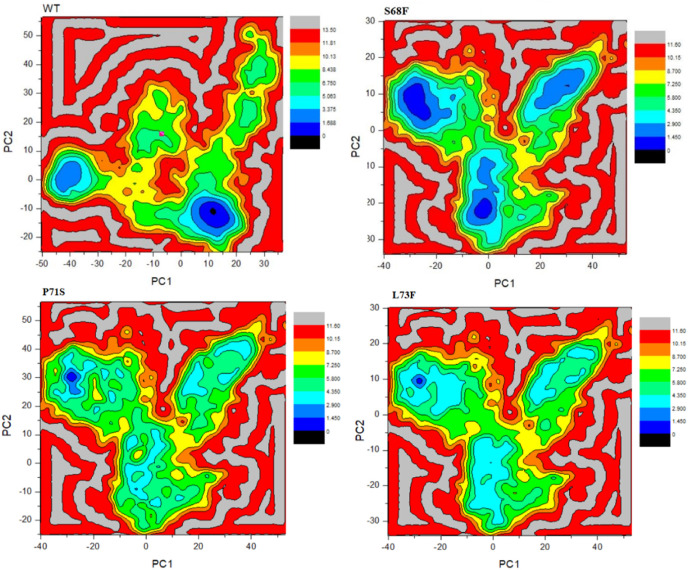
The FEL of WT and MTs has been shown in Fig. 10 . The lowest and stable energy state is represented by blue color. WT E protein showed more stable state that two MTs (P71S, L73F) whereas the MT S68F seems more stable than WT. The blue areas show stability while other indicate transitions in the protein conformation to attain a more favorable state. The green areas in P71S, L73F are more prevalent that WT which shows stability next to blue regions. These result demonstrates that E protein MTs might be useful to cause viral pathogenecity [[60], [61], [62]]. Calculating G might be important to observe the overall stability upon mutations.
Fig. 10.
Gibbs free energy landscape. The scale shows the free energy values. Blue and green regions are more stable than red and yellow. MT S68F shows more stability than WT based on the free energy landscape. WT exhibited more stability than P71S and L73F. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusion
In this comprehensive study, 259 non-synonymous mutations were detected in the E protein with different frequencies. Mutations were present in all three domains—the CTD, the NTD, and the hydrophobic domain; however, the highest frequency was detected in the first one. All of the MTs demonstrated various degrees of flexibilities and stabilities, where the majority depicted a loss in flexibility and stability. Moreover, three MTs (S68F, P71S, and L73F) were analyzed through MD simulations and exhibited a stabilizing effect on the E protein structure. In order to evaluate the drug targets for designing a novel antiviral drug against CoV-2, it is vital to dig out the frequency of mutations and their effect on the thermodynamic properties of the selected target. This is particularly imperative for the effective treatment of emerging microbes including SARS-CoV-2 mutants. The current study will contribute significantly to the knowledge on virus stabilization and the evolutionary aspects and pathogenicity of CoV-2 infections, which might be useful for better management of COVID-19 and the development of a vaccine in the near future.
Declaration of competing interest
All the authors have no competing interests.
Acknowledgement
The authors are thankful for technical support of Professor Ahsan Sattar Sheikh, IMBB The University of Lahore.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.imu.2021.100675.
Funding
Dong-Qing Wei is supported by grants from the National Science Foundation of China (Grant No. 32070662, 61832019, 32030063). The computations were partially performed at the Pengcheng Lab. and the Center for High-Performance Computing, Shanghai Jiao Tong University. This research has also been supported by Key project of science and technology research of Chongqing Municipal Education Commission (kjzd-k201902802). Project of the Chongqing Education Science Research Program for the 13th Five-year Plan(2018-GX-462). Key project of education and teaching reform in Chongqing Medical and Pharmaceutical College (CQYGZJG1909). Talent introduction project of Chongqing Medical and Pharmaceutical College (ygz2016302).
Author's contribution
Conception and design of the study: XT, MTK, DQW.
Acquisition of data: MTK, XT.
Analysis and interpretation of data: MSL, KM, MTK.
Drafting the article: MTK, MA, KM, DBA.
Revising content: XT, KM, MSL.
Final approval of the version: KM, XT, DQW.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Satarker S., Nampoothiri M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch Med Res. 2020;51:482–491. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banoun H. Evolution of SARS-CoV-2: review of mutations, role of the host immune system. Nephron. 2021;145:392–403. doi: 10.1159/000515417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rochman N.D., Wolf Y.I., Faure G., Mutz P., Zhang F., Koonin E.V. Ongoing global and regional adaptive evolution of SARS-CoV-2. Proc Natl Acad Sci Unit States Am. 2021;118 doi: 10.1073/pnas.2104241118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys Acta BBA - Mol Basis Dis. 2020;1866:165878. doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan A., Tahir Khan M., Saleem S., Junaid M., Ali A., Shujait Ali S., et al. Structural insights into the mechanism of RNA recognition by the N-terminal RNA-binding domain of the SARS-CoV-2 nucleocapsid phosphoprotein. Comput Struct Biotechnol J. 2020;18:2174–2184. doi: 10.1016/j.csbj.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu D.X., Yuan Q., Liao Y. Coronavirus envelope protein: a small membrane protein with multiple functions. Cell Mol Life Sci. 2007;64:2043–2048. doi: 10.1007/s00018-007-7103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fung T.S., Liu D.X. Post-translational modifications of coronavirus proteins: roles and function. Future Virol. 2018;13:405–430. doi: 10.2217/fvl-2018-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruch T.R., Machamer C.E. The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. J Virol. 2011;85:675–685. doi: 10.1128/JVI.01570-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A., DeDiego M.L., Enjuanes L., Aguilella V.M. Analysis of SARS-CoV E protein ion channel activity by tuning the protein and lipid charge. Biochim Biophys Acta BBA - Biomembr. 2013;1828:2026–2031. doi: 10.1016/j.bbamem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Surya W., Li Y., Torres J. Structural model of the SARS coronavirus E channel in LMPG micelles. Biochim Biophys Acta BBA - Biomembr. 2018;1860:1309–1317. doi: 10.1016/j.bbamem.2018.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta M.K., Vemula S., Donde R., Gouda G., Behera L., Vadde R. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J Biomol Struct Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nieto-Torres J.L., DeDiego M.L., Verdiá-Báguena C., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandala V.S. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Mol Biol. 2020;27:24. doi: 10.1038/s41594-020-00536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bianchi M., Benvenuto D., Giovanetti M., Angeletti S., Ciccozzi M., Pascarella S. Sars-CoV-2 envelope and membrane proteins: structural differences linked to virus characteristics? BioMed Res Int. 2020;2020:1–6. doi: 10.1155/2020/4389089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becerra-Flores M., Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int J Clin Pract. 2020;74:e13525. doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biswas A., Bhattacharjee U., Chakrabarti A.K., Tewari D.N., Banu H., Dutta S. Emergence of Novel Coronavirus and COVID-19: whether to stay or die out? Crit Rev Microbiol. 2020:1–12. doi: 10.1080/1040841X.2020.1739001. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bzówka M., Mitusińska K., Raczyńska A., Samol A., Tuszyński J.A., Góra A. Structural and evolutionary analysis indicate that the SARS-CoV-2 mpro is a challenging target for small-molecule inhibitor design. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21093099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elbe S., Buckland‐Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., et al. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinf. 2008;9 doi: 10.1186/1471-2105-9-40. 40–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gky300. W350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baby K., Maity S., Mehta C.H., Suresh A., Nayak U.Y., Nayak Y. Targeting SARS-CoV-2 RNA-dependent RNA polymerase: an in silico drug repurposing for COVID-19. F1000Research. 2020;9:1166. doi: 10.12688/f1000research.26359.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bowers K.J., Chow D.E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. SC 06 Proc. 2006 ACMIEEE Conf. Supercomput. 2006 doi: 10.1109/SC.2006.54. 43–43. [DOI] [Google Scholar]
- 27.Sugita Y., Kitao A. Dependence of protein stability on the structure of the denatured state: free energy calculations of I56V mutation in human lysozyme. Biophys J. 1998;75:2178–2187. doi: 10.1016/S0006-3495(98)77661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balsera M.A., Wriggers W., Oono Y., Schulten K. Principal component analysis and long time protein dynamics. J Phys Chem. 1996;100:2567–2572. doi: 10.1021/jp9536920. [DOI] [Google Scholar]
- 29.Sittel F., Jain A., Stock G. Principal component analysis of molecular dynamics: on the use of Cartesian vs. internal coordinates. J Chem Phys. 2014;141 doi: 10.1063/1.4885338. [DOI] [PubMed] [Google Scholar]
- 30.Ernst M., Sittel F., Stock G. Contact- and distance-based principal component analysis of protein dynamics. J Chem Phys. 2015;143:244114. doi: 10.1063/1.4938249. [DOI] [PubMed] [Google Scholar]
- 31.Kume A., Kawai S., Kato R., Iwata S., Shimizu K., Honda H. Exploring high-affinity binding properties of octamer peptides by principal component analysis of tetramer peptides. J Biosci Bioeng. 2017;123:230–238. doi: 10.1016/j.jbiosc.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Ouaray Z., ElSawy K.M., Lane D.P., Essex J.W., Verma C. Reactivation of mutant p53: constraints on mechanism highlighted by principal component analysis of the DNA binding domain. Proteins. 2016;84:1443–1461. doi: 10.1002/prot.25089. [DOI] [PubMed] [Google Scholar]
- 33.Iida S., Mashimo T., Kurosawa T., Hojo H., Muta H., Goto Y., et al. Variation of free-energy landscape of the p53 C-terminal domain induced by acetylation: enhanced conformational sampling. J Comput Chem. 2016;37:2687–2700. doi: 10.1002/jcc.24494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tripathi S., Srivastava G., Sharma A. Molecular dynamics simulation and free energy landscape methods in probing L215H, L217R and L225M βI-tubulin mutations causing paclitaxel resistance in cancer cells. Biochem Biophys Res Commun. 2016;476:273–279. doi: 10.1016/j.bbrc.2016.05.112. [DOI] [PubMed] [Google Scholar]
- 35.Martis E.A.F., Coutinho E.C. In: Struct. Bioinforma. Appl. Preclin. Drug discov. Process. Mohan C.G., editor. Springer International Publishing; Cham: 2019. Free energy-based methods to understand drug resistance mutations; pp. 1–24. [DOI] [Google Scholar]
- 36.Sohaib Shahzan M., Smiline Girija A.S., Vijayashree Priyadharsini J. A computational study targeting the mutated L321F of ERG11 gene in C. albicans, associated with fluconazole resistance with bioactive compounds from Acacia nilotica. J Mycol Médicale. 2019:100899. doi: 10.1016/j.mycmed.2019.100899. [DOI] [PubMed] [Google Scholar]
- 37.Rajendran V., Gopalakrishnan C., Sethumadhavan R. Pathological role of a point mutation (T315I) in BCR-ABL1 protein—a computational insight. J Cell Biochem. 2018;119:918–925. doi: 10.1002/jcb.26257. [DOI] [PubMed] [Google Scholar]
- 38.Hassan SkS., Choudhury P.P., Roy B. SARS-CoV2 envelope protein: non-synonymous mutations and its consequences. Genomics. 2020;112:3890–3892. doi: 10.1016/j.ygeno.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X., Fuentes E.J. Emerging themes in PDZ domain signaling. Int Rev Cell Mol Biol. 2019;343:129–218. doi: 10.1016/bs.ircmb.2018.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahman M.S., Hoque M.N., Islam M.R., Islam I., Mishu I.D., Rahaman MdM., et al. Mutational insights into the envelope protein of SARS-CoV-2. Gene Rep. 2021;22:100997. doi: 10.1016/j.genrep.2020.100997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye Y., Hogue B.G. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J Virol. 2007;81:3597–3607. doi: 10.1128/JVI.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X., Shi D., Zhou S., Liu H., Liu H., Yao X. Molecular dynamics simulations and novel drug discovery. Expet Opin Drug Discov. 2018;13:23–37. doi: 10.1080/17460441.2018.1403419. [DOI] [PubMed] [Google Scholar]
- 46.Liu H., Yao X. Molecular basis of the interaction for an essential subunit PA-PB1 in influenza virus RNA polymerase: insights from molecular dynamics simulation and free energy calculation. Mol Pharm. 2010;7:75–85. doi: 10.1021/mp900131p. [DOI] [PubMed] [Google Scholar]
- 47.He M., Li W., Zheng Q., Zhang H. A molecular dynamics investigation into the mechanisms of alectinib resistance of three ALK mutants. J Cell Biochem. 2018;119:5332–5342. doi: 10.1002/jcb.26666. [DOI] [PubMed] [Google Scholar]
- 48.Berhanu W.M., Masunov A.E. Molecular dynamic simulation of wild type and mutants of the polymorphic amyloid NNQNTF segments of elk prion: structural stability and thermodynamic of association. Biopolymers. 2011;95:573–590. doi: 10.1002/bip.21611. [DOI] [PubMed] [Google Scholar]
- 49.Chong S.-H., Lee C., Kang G., Park M., Ham S. Structural and thermodynamic investigations on the aggregation and folding of acylphosphatase by molecular dynamics simulations and solvation free energy analysis. J Am Chem Soc. 2011;133:7075–7083. doi: 10.1021/ja1116233. [DOI] [PubMed] [Google Scholar]
- 50.Bavi R., Kumar R., Choi L., Lee K.W. Exploration of novel inhibitors for bruton's tyrosine kinase by 3D QSAR modeling and molecular dynamics simulation. PloS One. 2016;11 doi: 10.1371/journal.pone.0147190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagasundaram N., Zhu H., Liu J., V K., C G.P.D., Chakraborty C., et al. Analysing the effect of mutation on protein function and discovering potential inhibitors of CDK4: molecular modelling and dynamics studies. PloS One. 2015;10 doi: 10.1371/journal.pone.0133969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lobanov M.Y., Bogatyreva N.S., Galzitskaya O.V. Radius of gyration as an indicator of protein structure compactness. Mol Biol. 2008;42:623–628. doi: 10.1134/S0026893308040195. [DOI] [PubMed] [Google Scholar]
- 53.Smilgies D.-M., Folta-Stogniew E. Molecular weight–gyration radius relation of globular proteins: a comparison of light scattering, small-angle X-ray scattering and structure-based data. J Appl Crystallogr. 2015;48:1604–1606. doi: 10.1107/S1600576715015551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khan M.T., Ali A., Wang Q., Irfan M., Khan A., Zeb M.T., et al. Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2. A molecular dynamic study. J Biomol Struct Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1769733. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan M.T., Khan A., Rehman A.U., Wang Y., Akhtar K., Malik S.I., et al. Structural and free energy landscape of novel mutations in ribosomal protein S1 (rpsA) associated with pyrazinamide resistance. Sci Rep. 2019;9:7482. doi: 10.1038/s41598-019-44013-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerlt J.A., Kreevoy M.M., Cleland W., Frey P.A. Understanding enzymic catalysis: the importance of short, strong hydrogen bonds. Chem Biol. 1997;4:259–267. doi: 10.1016/s1074-5521(97)90069-7. [DOI] [PubMed] [Google Scholar]
- 57.Hubbard R.E., Haider M.K. American Cancer Society; 2010. Hydrogen bonds in proteins: role and strength. eLS. [DOI] [Google Scholar]
- 58.Pace C.N., Fu H., Fryar K.L., Landua J., Trevino S.R., Schell D., et al. Contribution of hydrogen bonds to protein stability. Protein Sci Publ Protein Soc. 2014;23:652–661. doi: 10.1002/pro.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukherjee J., Gupta M.N. Increasing importance of protein flexibility in designing biocatalytic processes. Biotechnol Rep. 2015;6:119–123. doi: 10.1016/j.btre.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang F., Zheng G., Fu T., Li X., Tu G., Li Y.H., et al. Prediction of the binding mode and resistance profile for a dual-target pyrrolyl diketo acid scaffold against HIV-1 integrase and reverse-transcriptase-associated ribonuclease H. Phys Chem Chem Phys. 2018;20:23873–23884. doi: 10.1039/C8CP01843J. [DOI] [PubMed] [Google Scholar]
- 61.Junaid M., Khan M.T., Malik S.I., Wei D.-Q. Insights into the mechanisms of pyrazinamide resistance of three pyrazinamidase mutants N11K, P69T and D126N. J Chem Inf Model. 2018 doi: 10.1021/acs.jcim.8b00525. [DOI] [PubMed] [Google Scholar]
- 62.Khan M.T., Ali S., Zeb M.T., Kaushik A.C., Malik S.I., Wei D.-Q. Gibbs free energy calculation of mutation in PncA and RpsA associated with pyrazinamide resistance. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.