Abstract
Background:
Recent studies suggest that alterations in the vaginal microbiome allow for the assessment of the risk for spontaneous preterm birth (PTB), the leading cause of neonatal morbidity and mortality worldwide. However, the associations between the local immune response and the vaginal microbiome are still poorly understood. Herein, we characterize the vaginal host immune-microbiome interactions in women who ultimately underwent PTB and in those who delivered at term.
Methods:
Vaginal fluid samples from 52 pregnant women (of whom 18 underwent PTB and 34 delivered at term) were collected from 10–32 weeks in a case-control study. Concentrations of 33 immune mediators were determined using sensitive and specific immunoassays. The previously published 16S rRNA gene sequence and bacterial phylotype data of these subjects were utilized in this study. Linear mixed effects models were utilized to test associations between vaginal immune mediator concentrations and bacterial phylotype relative abundances.
Results:
1) In the overall study population, vaginal concentrations of CXCL10, CCL2, CCL3, SLP1 and VEGF negatively correlated with non-Lactobacillus, Community State Type IV (CST IV) members of the vaginal microbiome; 2) CXCL10, in particular, negatively correlated with 15 bacterial phylotypes, most of which are typical members of CST IV, such as Gardnerella vaginalis, Megasphaera spp., and Atopobium vaginae; 3) Gemella spp., also members of CST IV, negatively correlated with vaginal concentrations of VEGF, CCL2, CCL3, SLPI, and CXCL10; 4) when comparing PTB cases to term controls, five soluble immune mediators (CCL26, CCL22, CCL2, CXCL10, and IL-16), especially CCL26, were negatively correlated with five typical members of CST IV: Sneathia sanguinegens, Parvimonas micra, Veillonellaceae, BVAB2, and Gemella spp.; and 5) Sneathia sanguinegens had stronger negative associations with all five soluble immune mediators (CCL26, CCL22, CCL2, CXCL10, and IL-16) in PTB cases than in term controls.
Conclusions:
The assessment of vaginal host immune-microbiome interactions revealed that specific soluble immune mediators, mainly CXCL10, negatively correlated with typical members of CST IV of the vaginal microbiome. Sneathia sanguinegens, in particular, had stronger negative associations with different immune mediators, including CXCL10 and CCL26, in women who ultimately underwent PTB compared to those who delivered at term. These findings provide insight into the vaginal host immune-microbiome interactions in normal and complicated pregnancies.
Keywords: β-defensins, CCL26, chemokines, CXCL10, cytokines, pregnancy, 16S rRNA gene, Sneathia, vaginal microbiota
1. INTRODUCTION
The vaginal microbiome is a unique microbial community that is typically characterized by Lactobacillus dominance [1–5]. Yet, molecular microbiology studies have now established that the vaginal microbiome comprises five or more broad community state types (CSTs) [4, 6–8]. Four of these CSTs are dominated by one of four Lactobacillus species: L. crispatus (CST-I), L. gasseri (CST-II), L. iners (CST-III), or L. jensenii (CST-V) [4]. The remaining CST (CST-IV) is not dominated by a Lactobacillus spp. but rather is composed of a diverse array of anaerobic bacteria, including Atopobium, Gardnerella, Megasphaera, Prevotella, and Sneathia spp. [4, 8, 9].
The vaginal CSTs vary among asymptomatic women; however, the frequency of their occurrence depends upon ethnicity [4, 6, 7, 10–17]. European [12], European-American [4, 6], Asian [7, 18], and Asian-American [4, 6] women are likely to have a vaginal microbiome dominated by Lactobacillus (i.e. CSTs I, II, III and V). In contrast, 30–60% of African [11–13, 16], African-American [4, 6, 14, 15, 19], and Hispanic [4] women have a diverse vaginal microbiome that is not dominated by Lactobacillus (i.e. CST IV). Although women with a diverse non-Lactobacillus-dominant vaginal microbiome are typically asymptomatic, they are at a higher risk for acquiring human immunodeficiency virus (HIV) [13, 20–23] and other sexually transmitted diseases [20, 23, 24], and are more likely to experience pregnancy complications [14, 15, 25–30].
During normal pregnancy, the vaginal microbiome is characterized by an increase in Lactobacillus dominance and overall stability regardless of ethnicity [15, 17, 19, 25, 31–35]. In contrast, there is preliminary evidence suggesting that a diverse non-Lactobacillus-dominant vaginal microbiome increases the likelihood of experiencing preterm birth [25–29, 36–38], the leading cause of perinatal morbidity and mortality worldwide [39, 40]. However, this association is still under debate [5, 41–44]. One potential explanation for the discrepancies among studies is that most prior investigations did not consider local host immune responses in the vagina. These responses are the first line of defense against microorganisms, including potential pathogens [11, 45–47], and therefore likely play a central role in the pathophysiology of spontaneous preterm birth [48–50].
Recently, two reports have explored the relationship between the vaginal microbiome and local soluble immune responses in the context of preterm labor and birth [14, 15]. Elovitz et al. found that associations between non-Lactobacillus vaginal bacteria (e.g. Atopobium, Mobiluncus, and Sneathia spp.) and preterm birth were modulated by high vaginal concentrations of β-defensin-2 in African-American women [14]. Fettweis et al. reported that eleven bacterial taxa that are generally members of CST IV were more relatively abundant in the vaginal microbiomes of women ultimately delivering preterm than of those delivering at term [15]. Among women ultimately delivering preterm, there were positive correlations between many of these bacteria and the vaginal fluid concentrations of proinflammatory cytokines [15]. Nevertheless, the contribution of specific vaginal immune-microbiome interactions to the likelihood of spontaneous preterm birth remains poorly understood and requires further investigation.
The aim of the current study was to characterize the local soluble immune mediator profile and its association with the vaginal microbiome in pregnant women who ultimately underwent spontaneous preterm birth and those who delivered at term.
2. MATERIALS AND METHODS
2.1. Clinical specimens
Vaginal fluid samples were obtained at the Perinatology Research Branch, an intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services, Wayne State University (Detroit, MI), and the Detroit Medical Center (Detroit, MI). The collection and use of human materials for research purposes were approved by the Institutional Review Boards of the National Institute of Child Health and Human Development and Wayne State University (#110605MP2F(RCR)). All participating women provided written informed consent prior to sample collection.
2.2. Study design
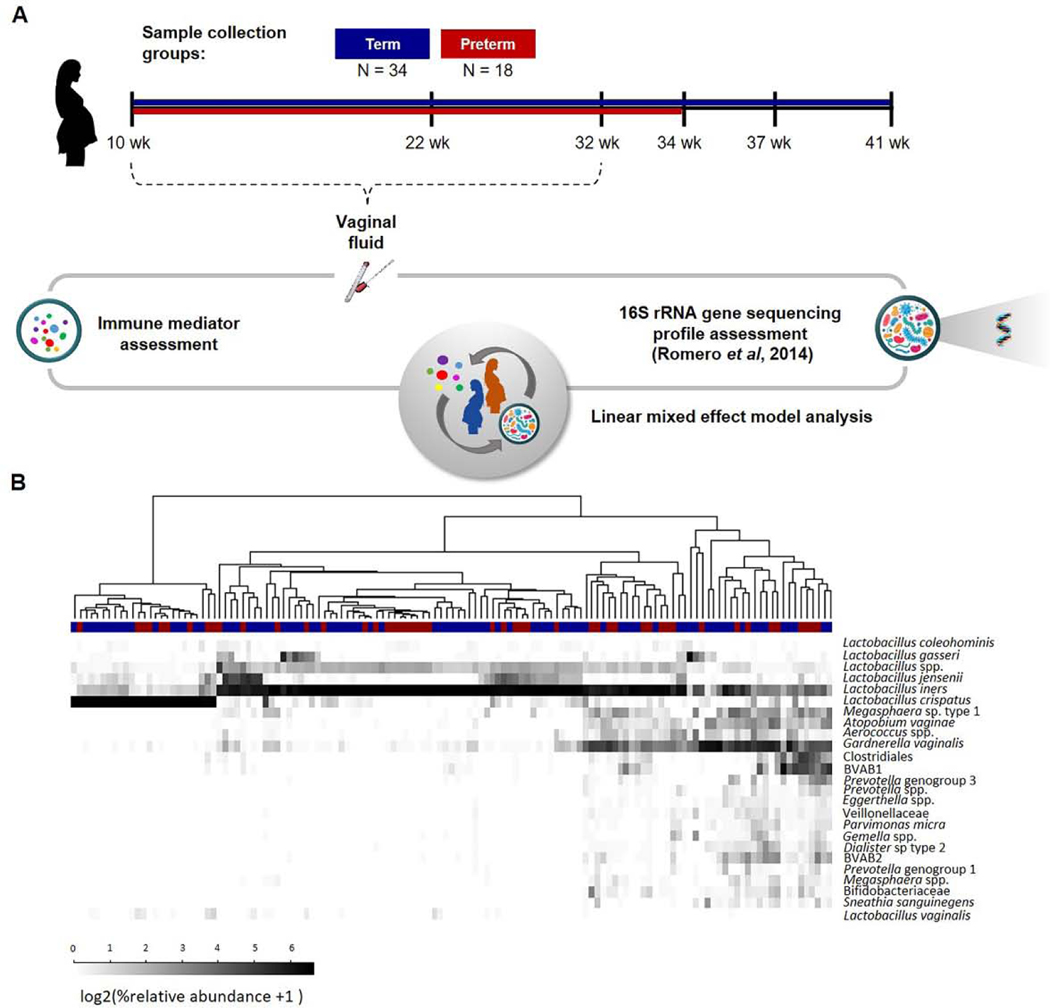
This was a longitudinal case–control study evaluating the correlations between bacterial taxa and soluble immune mediator concentrations (cytokines, chemokines, β-defensins 1, 2 and 3, and SLPI) in the vaginal fluid of pregnant women who ultimately underwent spontaneous preterm birth or delivered at term (see 2.3 Clinical definitions). The study included 18 spontaneous preterm birth cases and 34 term controls (Figure 1A). Sample collection started as early as 10 weeks of gestation and occurred until 32 weeks of gestation (Figure 1A). The number of available samples per patient in this gestational age interval ranged from one to six. A sample of vaginal fluid was collected under direct visualization from the posterior vaginal fornix using a Dacron swab and E-Swab for molecular microbiology and immune mediator quantification, respectively. Vaginal swabs were stored at −80°C until analysis. Data from a bacterial survey of these samples using pyrosequencing of the V1-V3 regions of the 16S rRNA gene were obtained from Romero et al. [51] (Figure 1B).
Figure 1. Study design.
(A) Samples of vaginal fluid from 52 pregnant women were collected in a case-control study: 18 delivered preterm and 34 had a term delivery. The concentrations of 33 immune mediators were determined using sensitive and specific immunoassays. For each patient, 16S rRNA gene pyrosequencing data were also available. Linear mixed effect models were utilized to explore interactions between immune mediators and bacterial phylotypes in the vaginal ecosystem. (B) Relative abundances of bacterial phylotypes in the vaginal samples of women who ultimately underwent spontaneous preterm birth or delivered at term.
2.2.1. Inclusion and exclusion criteria
Samples collected within one week of spontaneous preterm birth were excluded from analysis to mitigate any potential effects of the inflammatory process of labor on vaginal fluid bacterial and immune profiles [52, 53]. Patients with medically indicated preterm birth (e.g. preeclampsia, intrauterine growth restriction, or congenital anomalies) or cerclage placement were excluded from the cases. Patients with a history of preterm contractions, preterm labor, or cerclage placement were excluded from the term controls.
2.3. Clinical definitions
Preterm labor was diagnosed by the presence of at least two uterine contractions every 10 minutes associated with cervical changes in patients with a gestational age between 20 and 34 weeks. Preterm birth was considered as delivery before or at 34 weeks of gestation [51]. Most patients underwent spontaneous preterm labor with intact membranes, and a subset (N=7) of the preterm cases was diagnosed with preterm prelabor rupture of membranes (PPROM). No samples in this study were collected after rupture of membranes. Term controls were defined as women who delivered between 37 to 42 weeks of gestation without congenital anomalies or obstetrical, medical, or surgical complications. Histopathological examination of the placenta was performed by perinatal pathologists blinded to clinical diagnoses and obstetrical outcomes according to standardized Perinatology Research Branch protocols [54]. Acute inflammatory lesions of the placenta (maternal inflammatory response and fetal inflammatory response) were diagnosed according to established criteria, including staging and grading [54].
2.4. Cytokine profiling
Vaginal fluid samples were assessed using sensitive and specific V-PLEX immunoassays (Meso Scale Discovery, Gaithersburg, MD, USA) to measure vaginal fluid concentrations of several soluble immune mediators: the pro-inflammatory V-PLEX 10spot assay K15049D-2 [IFN-γ, IL-10, IL-12p70, IL-13, IL-1β, IL-2, IL-4, IL-6], cytokine VPLEX 10-spot assay K15050D-2 [GM-CSF, IL-12/IL-23p40, IL-15, IL-16, IL-17A, IL-1α, IL-5, IL-7, TNFα, TNFβ, vascular endothelial growth factor (VEGF)], and the chemokine V-PLEX 10-spot assay K15047D-2 [CCL11(Eotaxin), CCL26(Eotaxin-3), CXCL8(IL-8), CXCL10(IP-10), CCL2(MCP-1), CCL13(MCP-4), CCL22(MDC), CCL3(macrophage inflammatory protein (MIP)-1α), CCL4(MIP-1β), CCL17(TARC)], according to the manufacturer’s instructions. The plate signals were read by QuickPlex SQ 120 (Meso Scale Discovery). Standard curves were generated, and the assay values of the samples were interpolated from the curves. The detection limits of the assays and the inter-assay and intra-assay coefficients of variation, are displayed in Supplemental Table 1.
Secretory leukocyte protease inhibitor (SLPI) (R&D Systems, Minneapolis, MN, USA), β-defensin-1 and 3 (Aviscera Bioscience, Santa Clara, CA, USA), and β-defensin-2 (ALPCO, Salem, NH, USA) concentrations were measured in vaginal fluid samples using sensitive and specific single-analyte immunoassays, according to the manufacturers’ instructions. The intensity of developed color was measured by a SpectraMax M5 Microplate Reader (San Jose, CA, USA). The concentrations of SLPI and β-defensins 1, 2, and 3 were determined by interpolation from the standard curves. The detection limits of the assays and the inter-assay and intra-assay coefficients of variation are displayed in Supplemental Table 1.
Total protein concentrations were measured by a BCA Protein Assay kit from (ThermoFisher, catalogue number 23225). The vaginal immune mediator dataset and sample metadata for this study are available in Supplemental Data File 1.
2.5. Statistical analysis
2.5.1. Demographics analysis
Clinical characteristics of the patient population were summarized as median and interquartile ranges (IQRs) for continuous variables and as percentages for categorical variables. The comparison of demographic variables between the preterm and term groups was performed using the Fisher’s exact test for binary variables or the Mann-Whitney U test for continuous variables (Table 1).
Table 1.
Clinical and demographic characteristics of the study population
| Patients who ultimately delivered at term (n = 34) | Patients who ultimately underwent spontaneous preterm birth (n = 18) | p-value | |
|---|---|---|---|
| Maternal age (y; median [IQR])a | 24 (21–28.8) | 21(20–26) | .3 |
| Body mass index (kg/m2; median [IQR])a | 28.7(24.6–34.3) | 25.7 (21.6–33.2)c | .5 |
| Primiparityb | 11.8% (4/34) | 5.6% (1/18) | .7 |
| Race/ethnicityb | .7 | ||
| African-American | 94.1% (32/34) | 94.4% (17/18) | |
| White | 2.9% (1/34) | 5.6% (1/18) | |
| Others | 2.9% (1/34) | 0% (0/18) | |
| Gestational age at delivery (wk; median [IQR])a | 39.7 (38.7–40.3) | 30.5 (28.4–33.1) | <0.001 |
| Mode of deliveryb | .1 | ||
| Cesarean section | 17.7% (6/34) | 38.9% (7/18) | |
| Vaginal | 82.3% (28/34) | 61.1% (11/18) | |
| Birthweight (g; median [IQR])a | 3257.5 (3127.5–3522.5) | 1402.5 (1037.5–1986.3) | <0.001 |
| Apgar score at 1 min (median [IQR])a | 9 (8–9) | 6 (3–8) | <0.001 |
| Apgar score at 5 min (median [IQR])a | 9 (9–9) | 8 (6–8) | <0.001 |
| Acute maternal inflammatory responseb | |||
| Stage 1 (Early acute subchorionitis or chorionitis) | 21.2% (7/33)c | 5.6% (1/18) | .2 |
| Stage 2 (Acute chorioamnionitis) | 0% (0/33)c | 44.4% (8/18) | <0.001 |
| Stage 3 (Necrotizing chorioamnionitis) | 0% (0/33)c | 5.6% (1/18) | .4 |
| Acute fetal inflammatory responseb | |||
| Stage 1 (Chorionic vasculitis or umbilical phlebitis) | 12.1% (4/33)c | 33.3% (6/18) | .3 |
| Stage 2 (Umbilical arteritis) | 0% (0/33)c | 22.2% (4/18) | 0.01 |
| Stage 3 (Necrotizing funisitis) | 0% (0/33)c | 5.6% (1/18) | .4 |
Mann-Whitney U test.
Fisher’s exact test.
One missing data.
2.5.2. Filtering out low-abundance bacterial taxa
The 16S rRNA gene bacterial phylotype data were retained for analysis if (1) bacterial phylotypes had a relative abundance of at least 1% in at least 5% of the samples, or (2) had a relative abundance of at least 0.1% in at least 15% of the samples, as informed by Fettweis et al. [15]. Twenty-five bacterial phylotypes met these criteria. (Figure 1B)
2.5.3. Data transformation
The percent relative abundances of bacterial phylotypes were log2-transformed prior to analysis. The concentrations of soluble immune mediators (pg*ml−1) were normalized to the total protein concentration (mg*ml−1). Concentrations lower than the measurable threshold were replaced with 99.9% of the lowest value detected in the study for each immune mediator. The normalized concentrations were also log2-transformed.
2.5.4. The pairwise analysis of immune mediator-bacterial phylotype associations in the overall study population
Linear mixed effects models were used to assess associations between the relative abundances of bacterial phylotypes (control variables) and soluble immune mediator concentrations (response variables) in the overall patient population. In order to account for repeated sample collection during pregnancy, a random effect was included for each patient. The derived association represented the significant change in soluble immune mediator concentration (response variable) with every doubling of the relative abundance of a specific bacterium (explanatory variable). The p-values were corrected using the false discovery rate method to obtain q-values. Significance was inferred based on a false discovery rate less than 20%.
2.5.5. Differential association of immune mediators and bacterial phylotypes between preterm cases and term controls
To determine whether immune mediator-bacterial phylotype associations differed between term controls and spontaneous preterm birth cases, the models described above also included an interaction term between bacterial phylotype relative abundance and adverse delivery outcome (spontaneous preterm birth). The correlations displayed in figures correspond to significant differences in the regression slopes between patients who subsequently had a spontaneous preterm birth and those who delivered at term. The effect size in this analysis represents a difference in the rate of change in the immune mediator concentration with an increase in microbial relative abundance between cases and controls, and it was further converted into a linear fold change. The p-values corresponding to the models were corrected using the false discovery rate method to obtain q-values. Significance was inferred based on a false discovery rate less than 20% and a fold change ≥1.3.
2.5.6. Software packages utilized in this study
Statistical analyses were conducted using the lme4 package [55] in R (version 3.6.1) [56] and GraphPad Prism version 8.0.1 for Windows (GraphPad Software, San Diego, CA, USA, www.graphpad.com).
3. RESULTS
3.1. Characteristics of the study population
The clinical and demographic characteristics of women who had a spontaneous preterm birth or a term delivery are displayed in Table 1. The study population was largely African-American (94%). There were no significant differences in maternal age, ethnicity, pre-pregnancy body mass index, rate of primiparity, or the mode of delivery between the two groups. Gestational age at delivery, neonatal birthweight, and Apgar scores were significantly lower in preterm birth cases than in term controls (p<0.001). The frequency of stage 2 acute maternal and fetal inflammatory responses was higher in women who ultimately underwent spontaneous preterm birth (p<0.001).
3.2. Associations between vaginal immune mediators and bacterial phylotypes in the overall study population
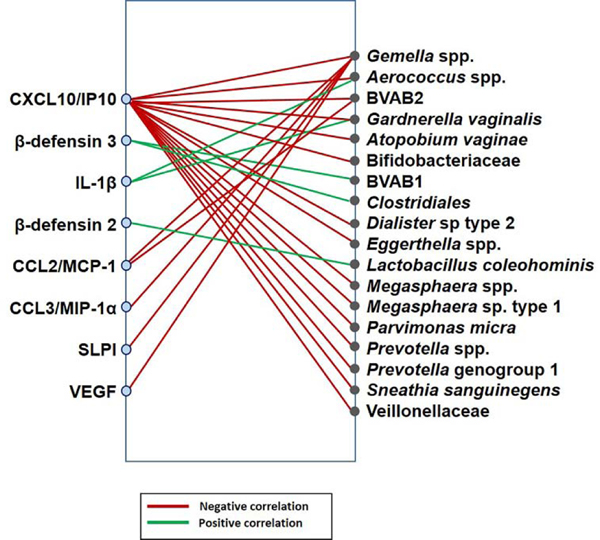
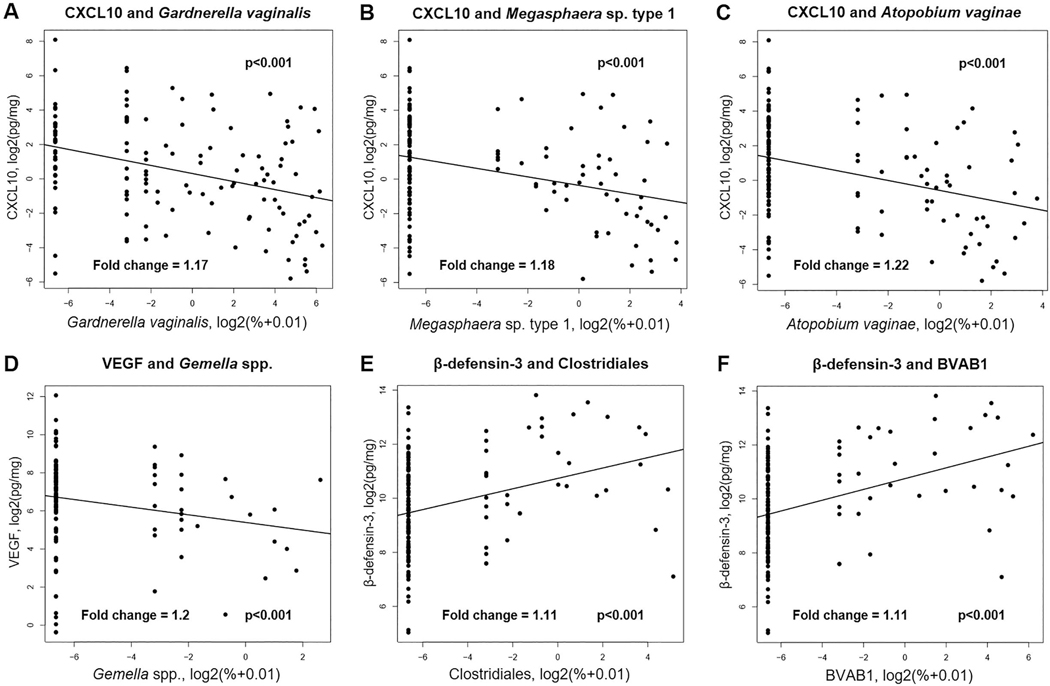
Associations between the concentrations of vaginal soluble immune mediators and the composition of the vaginal microbiome across all samples from all women were investigated using linear mixed effect models in a pair-wise fashion between the 33 immune mediators (Supplemental Table 1) and the 25 bacterial phylotypes (Figure 1B). Of the 33 immune mediators examined, eight (β-defensins 2 and 3, IL-1β, CXCL10, CCL2, CCL3, SLPI, and VEGF) were found to correlate with 18 different bacterial phylotypes in the overall patient population, regardless of whether they ultimately delivered preterm or at term (Figure 2; Supplemental Table 2). Notably, the vaginal fluid concentrations of CXCL10 negatively correlated with 15 vaginal bacterial phylotypes, most of which are typical members of vaginal CST IV [4], such as Gardnerella vaginalis (Figure 3A), Megasphaera spp. type 1 (Figure 3B), and Atopobium vaginae (Figure 3C). The vaginal fluid concentrations of VEGF were lower (p=0.002) in samples with a higher relative abundance of Gemella spp. (Figure 3D), bacteria also found in CST IV [4]. The same trend was observed between Gemella spp. and CCL2, CCL3, SLPI, and CXCL10 (Figure 2).
Figure 2. Network of associations between vaginal immune mediators and bacterial phylotypes in the overall study population.
Immune mediators and bacterial phylotypes are represented on the left and right sides of the plot, respectively. The lines indicate significant associations between vaginal immune mediators and bacterial phylotypes in the study population (N=52). Red lines indicate negative associations between the concentrations of vaginal immune mediators and the relative abundances of bacterial phylotypes. Green lines indicate positive associations between immune mediators and bacterial phylotypes. The analyses reflect linear mixed effects models, accounting for repeated measures and with an FDR<20%.
Figure 3. Correlation analyses of vaginal immune mediators and bacterial phylotypes in the overall study population.
The X-axes represent the relative abundances of the indicated bacterial phylotypes, and the Y-axes represent the concentrations of either CXCL10, VEGF, or β-defensin-3 in the vaginal fluid. Correlation between vaginal concentrations of CXCL10 and the relative abundance of Gardnerella vaginalis (A), Megasphaera spp. type 1 (B), or Atopobium vaginae (C). Correlation between the vaginal concentrations of VEGF and the relative abundance of Gemella spp. (D). Correlation between the vaginal concentrations of β-defensin-3 and the relative abundance of (E) Clostridiales and (F) BVAB1. The lines represent the linear mixed-effects model fit to the data. The fold changes represent the change in concentration of the immune mediator given a doubling of bacterial phylotype relative abundance (1 unit on the x-axis).
Conversely, the vaginal fluid concentrations of IL-1β and β-defensins 2 and 3 positively correlated with the relative abundances of specific vaginal bacterial phylotypes (Figure 2; Supplemental Table 2). Specifically, the vaginal concentrations of the antimicrobial peptide β-defensin 3 were higher in samples with higher abundances of the order Clostridiales (Figure 3E) or Bacterial Vaginosis Associated Bacteria 1 (BVAB1) (Figure 3F). Yet, the vaginal fluid concentrations of the antimicrobial peptide β-defensin 2 positively correlated solely with Lactobacillus coleohominis (Figure 2). The pro-inflammatory cytokine IL-1β had positive correlations with Gardnerella vaginalis and Aerococcus spp. (Figure 2).
These results indicate that, in our study population, there are negative associations between specific soluble immune mediators, mainly CXCL10, and increasing relative abundances of CST IV bacteria in the vaginal fluid.
3.3. Differential association between vaginal soluble immune mediators and bacterial phylotypes in women who delivered preterm compared to those who delivered at term
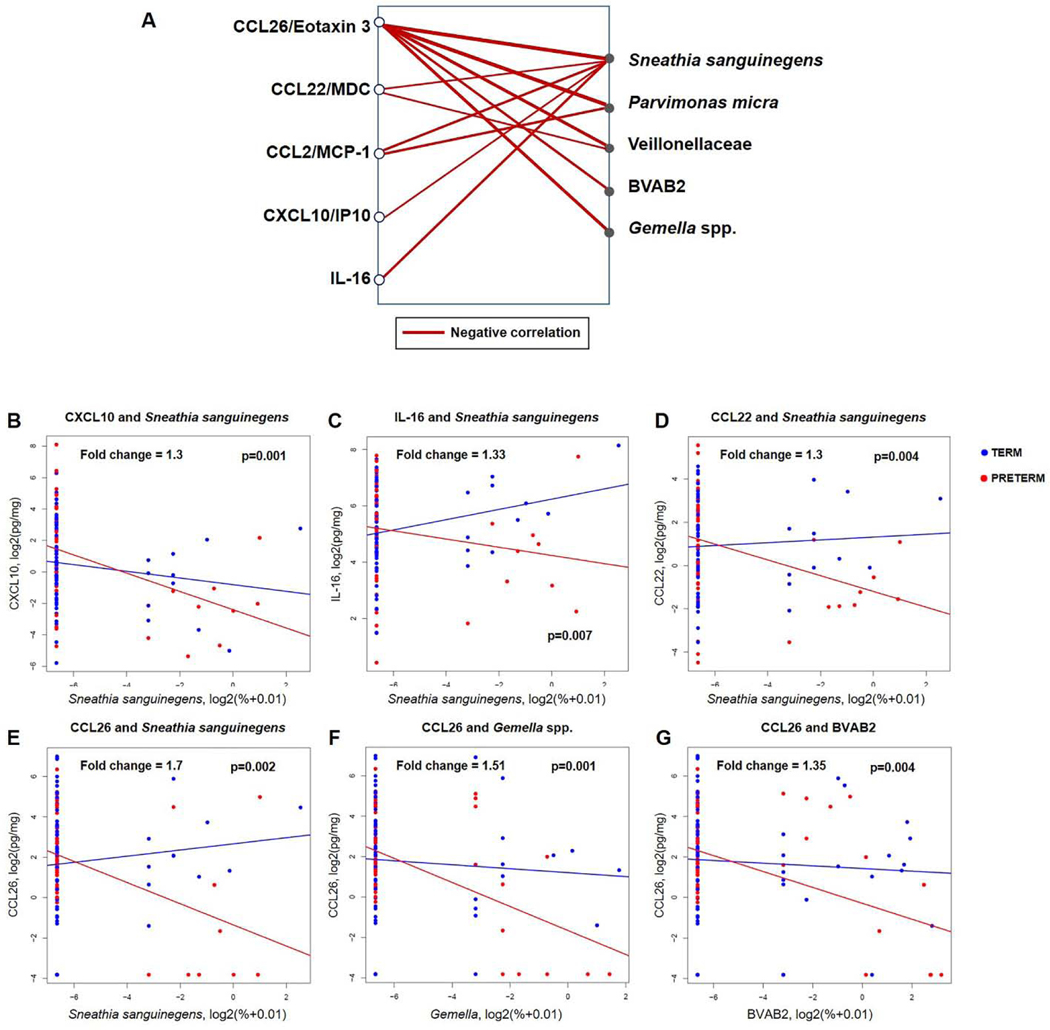
Next, it was determined whether the associations between vaginal immune mediators and bacterial phylotypes differed across all samples between women who ultimately delivered preterm and those who delivered at term. Regression analysis using linear mixed effects models was performed in a pair-wise fashion between the 33 immune mediators (Supplemental Table 1) and the 25 bacterial phylotypes (Figure 1B). Unlike in the overall association analyses above, in this analysis, each group (women who ultimately delivered preterm or at term) was allowed to have a different slope. To add stringency, in addition to a cut-off on adjusted p-values stating the significance of the difference in slope, a minimum difference of ≥1.3-fold between slopes was required (Figure 4A; Supplemental Table 3). When comparing preterm birth cases to term controls, five soluble immune mediators (CCL26, CCL22, CCL2, CXCL10, and IL-16) were more negatively correlated with five typical members of vaginal CST IV: Sneathia sanguinegens, Parvimonas micra, Veillonellaceae, Bacterial Vaginosis Associated Bacteria 2 (BVAB2), and Gemella spp. Sneathia sanguinegens had the highest number of differentially negative associations (n=5) with vaginal pro-inflammatory immune mediators between the preterm birth cases and term controls: CXCL10 (Figure 4B), IL-16 (Figure 4C), CCL22 (Figure 4D), and CCL26 (Figure 4E). Notably, CCL26 had the highest number of differentially negative correlations (n=5) with bacterial phylotypes, including Sneathia sanguinegens (Figure 4E), Gemella spp. (Figure 4F) and BVAB2 (Figure 4G), when comparing preterm birth cases to term controls. Collectively, these results show that specific immune mediators, namely CCL26, CCL22, CCL2, CXCL10, and IL-16, were more negatively correlated with stereotypical CST IV bacterial phylotypes in women who ultimately underwent spontaneous preterm birth than in those who delivered at term.
Figure 4. Network of immune mediators differentially associated with changes in bacterial phylotype relative abundances in preterm cases and term controls.
(A) Immune mediators and bacterial phylotypes are presented on the left and right sides of the plot, respectively. Red lines indicate a lower slope of the association between the concentration of an immune mediator and the relative abundance of a bacterial phylotype in preterm birth cases than in term controls. The fold changes represent a difference in the rate of change in the immune mediator concentration given a unit increase in bacterial phylotype relative abundance between cases and controls. Correlation analysis between vaginal concentrations of CXCL10 (B), IL-16 (C), CCL22 (D) and the relative abundance of Sneathia sanguinegens. Association between the vaginal concentrations of CCL26 and the relative abundance of Sneathia sanguinegens (E), Gemella spp. (F), and BVAB2 (G).
4. DISCUSSION
The assessment of vaginal host immune-microbiome interactions revealed that specific soluble immune mediators, mainly CXCL10, negatively correlated with typical members of CST IV of the vaginal microbiome. In addition, this assessment particularly showed that Sneathia sanguinegens had stronger negative associations with different immune mediators, including CXCL10 and CCL26, in women who ultimately underwent spontaneous preterm birth than in those who delivered at term. These findings provide insight into the vaginal host immune-microbiome interactions in normal and complicated pregnancies.
In our overall study population, which was largely African-American, there were negative associations between specific soluble immune mediators and increasing relative abundances of CST IV bacteria in the vaginal fluid. Consistently, previous reports showed that the vaginal ecosystem of African-descendent pregnant [14, 17, 57] and non-pregnant [4, 6, 8, 10, 58–60] women is enriched in CST IV bacteria. Such a vaginal phenotype has been attributed to the presence of asymptomatic bacterial vaginosis in non-pregnant [61] and pregnant [62] women. In the current study, we did not evaluate the incidence of bacterial vaginosis; yet, historically, our study population is at high risk of presenting with this clinical condition [63, 64].
Herein, we report that a high relative abundance of CST IV bacteria was associated with low concentrations of CXCL10 in the vaginal fluid. This result is in agreement with the following evidence: 1) in pregnant and non-pregnant African women, the presence of G. vaginalis and A. vaginae (i.e. bacteria associated with bacterial vaginosis) is strongly associated with low vaginal fluid concentrations of CXCL10 [66, 67]; 2) non-pregnant South African women with bacterial vaginosis-associated microbial communities display reduced vaginal fluid concentrations of CXCL10 [68]; 3) the vaginal fluid concentrations of CXCL10 negatively correlate with BVAB1, a CST IV and bacterial vaginosis associated bacterium, in women (mostly African-American) who ultimately delivered preterm [15]; and 4) the low vaginal fluid concentrations of CXCL10 observed in non-pregnant African women with bacterial vaginosis can be boosted upon successful treatment with metronidazole [69], a drug commonly used to treat this clinical condition [70, 71].
CXCL10 is primarily a T-cell chemokine that induces the migration of immune cells to sites of inflammation [72–74]. This chemokine plays a central role in host defense mechanisms against pathogens [75–78], including those which take place in the amniotic cavity [79, 80], and in the mechanisms involved in the pathophysiology of preterm labor and birth [54, 79, 81, 82]. CXCL10 also mediates the induction of apoptosis and regulates cell growth and proliferation as well as angiogenesis [83]. These diverse functions of CXCL10 depend on the splice variants of its receptor CXCR3 (CXCR3-A, CXCR3-B, and CXCR3-alt). Specifically, chemotaxis and proliferation are mediated by CXCR3-A in several cell types [84, 85], whereas CXCL10 inhibits migration and proliferation and induces apoptosis via CXCR3-B [85]. These receptors are expressed on various immune (e.g. T cells, natural killer (NK) cells, and NKT cells) as well as non-immune (e.g. bronchial epithelial cells and astrocytes) cells [83, 86, 87]. Pertinent to the current investigation, mechanistic studies have shown that CXCL10-deficient mice are more susceptible to viral infections [88, 89] and, even more importantly, this chemokine can directly kill Gram-positive bacteria such as Bacillus anthracis [90]. Furthermore, polymorphisms in CXCL10 and its receptor, CXCR3, are associated with susceptibility to infection [91, 92] and recurrent preterm birth [81], respectively. Therefore, it is tempting to suggest that, in the vaginal ecosystem of African-American women (our study population), low concentrations of CXCL10 modulate local bacterial growth, favoring the proliferation of CST IV bacteria. Yet, additional mechanistic studies are needed to test this hypothesis.
In the current study, we also show that Sneathia sanguinegens is negatively associated with multiple vaginal immune mediators in women who subsequently underwent spontaneous preterm birth compared to those who delivered at term. Sneathia species are rod-shaped Gram-negative anaerobic bacteria whose common habitat is the human vagina [93–95]. The traditional view is that Sneathia species are of a fastidious nature since their culture is not always possible [96–103]. Nevertheless, Sneathia species have been detected using molecular microbiology techniques in the amniotic fluid of women with intra-amniotic infection associated with PPROM [98, 104–108], a sonographic short cervix [109], preterm labor and birth with intact membranes [96, 97, 101, 110], and clinical chorioamnionitis [111]. Importantly, Sneathia species in the vaginal fluid are associated with preterm birth in African-American [14, 15, 112] and Caucasian [29] women. Sneathia species have also been associated with neonatal infection [27, 93, 113]. Therefore, investigation focused on the mechanisms whereby Sneathia species invade the intra-amniotic space and fetal tissues is warranted.
To our knowledge, this is the first study to show that Sneathia species are negatively associated with immune mediators such as CXCL10 and CCL26 in the vaginal fluid. The former chemokine is discussed above. CCL26 is a chemokine that belongs to the eotaxin family (eotaxin-1/CCL11, eotaxin-2/CCL24, and eotaxin-3/CCL26) [114]. Eotaxins are released by epithelial, mesenchymal, and endothelial cells in response to various mediators, including IL-4, IL-13, and TNF-α [115–117]. Eotaxins are potent chemoattractants for eosinophils [115, 117], which play a central role in type-2 immunity such as host defense against parasitic helminth infections, tissue repair and remodeling, and allergic diseases [118, 119]. Specifically, CCL26 is preferentially recognized by CCR3, a common and specific receptor for eotaxins [114, 117, 120]. CCL26 is the most effective eotaxin to induce eosinophil migration in asthma patients [121]. Therefore, this chemokine is implicated in several diseases associated with eosinophilic infiltration such as atopic dermatitis [122] and eosinophilic gastrointestinal disorders [123]. In addition, CCL26 displays host defense activities by exerting bactericidal potency against several airway pathogens [124]. The effect of CCL26 on Sneathia species, or any other members of the CST IV vaginal microbiome, has yet to be described. Therefore, further mechanistic studies are required to investigate the translational relevance of the negative chemokine-microbiome associations described herein, and whether such interactions play a central role in the mechanisms leading to preterm labor and birth.
5. CONCLUSION
Specific soluble immune mediators, mainly CXCL10, negatively correlate with typical members of CST IV of the vaginal microbiome. Sneathia sanguinegens, a CST IV bacterium that is commonly associated with intra-amniotic infection, displays stronger negative associations with different immune mediators, including CXCL10 and CCL26, in women who ultimately experience spontaneous preterm birth compared to those who deliver at term. These findings provide insight into the vaginal host immune-microbiome interactions in normal and complicated pregnancies.
Supplementary Material
HIGHLIGHTS:
Vaginal fluid immune mediators negatively correlated with CST IV vaginal bacteria
CXCL10, in particular, was negatively correlated with 15 CST IV bacteria
Specific cytokines were negatively correlated with CST IV bacteria in preterm births
Sneathia sanguinegens was negatively correlated with cytokines in preterm births
Acknowledgement
We thank the physicians and nurses from the Center for Advanced Obstetrical Care and Research and the Intrapartum Unit for their help in collecting human samples. The authors also thank the staff members of the PRB Biomarkers and Translational Science Laboratory (Research Assistants Rona Wang and Hong Meng), PRB Clinical Laboratory, and PRB Histology/Pathology Unit for the processing and examination of the pathological sections.
This research was supported, in part, by the Perinatology Research Branch, Division of Obstetrics and Maternal-Fetal Medicine, Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, U.S. Department of Health and Human Services (NICHD/NIH/DHHS); and, in part, with Federal funds from NICHD/NIH/DHHS under Contract No. HHSN275201300006C. ALT, KRT, and NG-L were further supported by the Wayne State University Perinatal Research Initiative in Maternal, Perinatal and Child Health. Dr. Romero has contributed to this work as part of his official duties as an employee of the United States Federal Government.
Abbreviations:
- BVAB
bacterial vaginosis-associated bacterium
- VEGF
vascular endothelial growth factor
- CST
community state type
- MIP
macrophage inflammatory protein
- PPROM
preterm prelabor rupture of membranes
- PTB
preterm birth
- SLPI
secretory leukocyte protease inhibitor
Footnotes
Research data for this article
The vaginal immune mediator dataset and sample metadata for this study are available in Supplemental Data File 1. The 16S rRNA gene survey data are publicly available in Table S1 of Romero et al. 2014 [51].
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. REFERENCES
- [1].Hill GB, Eschenbach DA, Holmes KK, Bacteriology of the vagina, Scand J Urol Nephrol Suppl 86 (1984) 23–39. [PubMed] [Google Scholar]
- [2].Redondo-Lopez V, Cook RL, Sobel JD, Emerging role of lactobacilli in the control and maintenance of the vaginal bacterial microflora, Rev Infect Dis 12(5) (1990) 856–72. [DOI] [PubMed] [Google Scholar]
- [3].Lamont RF, Sobel JD, Akins RA, Hassan SS, Chaiworapongsa T, Kusanovic JP, Romero R, The vaginal microbiome: new information about genital tract flora using molecular based techniques, BJOG 118(5) (2011) 533–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, Karlebach S, Gorle R, Russell J, Tacket CO, Brotman RM, Davis CC, Ault K, Peralta L, Forney LJ, Vaginal microbiome of reproductive-age women, Proc Natl Acad Sci U S A 108 Suppl 1 (2011) 4680–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Greenbaum S, Greenbaum G, Moran-Gilad J, Weintraub AY, Ecological dynamics of the vaginal microbiome in relation to health and disease, Am J Obstet Gynecol 220(4) (2019) 324–335. [DOI] [PubMed] [Google Scholar]
- [6].Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, Joyce P, Foster JA, Forney LJ, Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women, ISME J 1(2) (2007) 121–33. [DOI] [PubMed] [Google Scholar]
- [7].Zhou X, Hansmann MA, Davis CC, Suzuki H, Brown CJ, Schutte U, Pierson JD, Forney LJ, The vaginal bacterial communities of Japanese women resemble those of women in other racial groups, FEMS Immunol Med Microbiol 58(2) (2010) 169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gajer P, Brotman RM, Bai G, Sakamoto J, Schutte UM, Zhong X, Koenig SS, Fu L, Ma ZS, Zhou X, Abdo Z, Forney LJ, Ravel J, Temporal dynamics of the human vaginal microbiota, Sci Transl Med 4(132) (2012) 132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fredricks DN, Fiedler TL, Marrazzo JM, Molecular identification of bacteria associated with bacterial vaginosis, N Engl J Med 353(18) (2005) 1899–911. [DOI] [PubMed] [Google Scholar]
- [10].Fettweis JM, Brooks JP, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, Strauss JF, The Vaginal Microbiome C, Jefferson KK, Buck GA, Differences in vaginal microbiome in African American women versus women of European ancestry, Microbiology 160(Pt 10) (2014) 2272–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Anahtar MN, Byrne EH, Doherty KE, Bowman BA, Yamamoto HS, Soumillon M, Padavattan N, Ismail N, Moodley A, Sabatini ME, Ghebremichael MS, Nusbaum C, Huttenhower C, Virgin HW, Ndung’u T, Dong KL, Walker BD, Fichorova RN, Kwon DS, Cervicovaginal bacteria are a major modulator of host inflammatory responses in the female genital tract, Immunity 42(5) (2015) 965–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Borgdorff H, van der Veer C, van Houdt R, Alberts CJ, de Vries HJ, Bruisten SM, Snijder MB, Prins M, Geerlings SE, Schim MF van der Loeff, van de Wijgert J, The association between ethnicity and vaginal microbiota composition in Amsterdam, the Netherlands, PLoS One 12(7) (2017) e0181135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gosmann C, Anahtar MN, Handley SA, Farcasanu M, Abu-Ali G, Bowman BA, Padavattan N, Desai C, Droit L, Moodley A, Dong M, Chen Y, Ismail N, Ndung’u T, Ghebremichael MS, Wesemann DR, Mitchell C, Dong KL, Huttenhower C, Walker BD, Virgin HW, Kwon DS, Lactobacillus-Deficient Cervicovaginal Bacterial Communities Are Associated with Increased HIV Acquisition in Young South African Women, Immunity 46(1) (2017) 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Elovitz MA, Gajer P, Riis V, Brown AG, Humphrys MS, Holm JB, Ravel J, Cervicovaginal microbiota and local immune response modulate the risk of spontaneous preterm delivery, Nat Commun 10(1) (2019) 1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fettweis JM, Serrano MG, Brooks JP, Edwards DJ, Girerd PH, Parikh HI, Huang B, Arodz TJ, Edupuganti L, Glascock AL, Xu J, Jimenez NR, Vivadelli SC, Fong SS, Sheth NU, Jean S, Lee V, Bokhari YA, Lara AM, Mistry SD, Duckworth RA 3rd, Bradley SP, Koparde VN, Orenda XV, Milton SH, Rozycki SK, Matveyev AV, Wright ML, Huzurbazar SV, Jackson EM, Smirnova E, Korlach J, Tsai YC, Dickinson MR, Brooks JL, Drake JI, Chaffin DO, Sexton AL, Gravett MG, Rubens CE, Wijesooriya NR, Hendricks-Munoz KD, Jefferson KK, Strauss JF 3rd, Buck GA, The vaginal microbiome and preterm birth, Nat Med 25(6) (2019) 1012–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Price JT, Vwalika B, Hobbs M, Nelson JAE, Stringer EM, Zou F, Rittenhouse KJ, Azcarate-Peril A, Kasaro MP, Stringer JSA, Highly diverse anaerobe-predominant vaginal microbiota among HIV-infected pregnant women in Zambia, PLoS One 14(10) (2019) e0223128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Serrano MG, Parikh HI, Brooks JP, Edwards DJ, Arodz TJ, Edupuganti L, Huang B, Girerd PH, Bokhari YA, Bradley SP, Brooks JL, Dickinson MR, Drake JI, Duckworth RA 3rd, Fong SS, Glascock AL, Jean S, Jimenez NR, Khoury J, Koparde VN, Lara AM, Lee V, Matveyev AV, Milton SH, Mistry SD, Rozycki SK, Sheth NU, Smirnova E, Vivadelli SC, Wijesooriya NR, Xu J, Xu P, Chaffin DO, Sexton AL, Gravett MG, Rubens CE, Hendricks-Munoz KD, Jefferson KK, Strauss JF 3rd, Fettweis JM, Buck GA, Racioethnic diversity in the dynamics of the vaginal microbiome during pregnancy, Nat Med 25(6) (2019) 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shi Y, Chen L, Tong J, Xu C, Preliminary characterization of vaginal microbiota in healthy Chinese women using cultivation-independent methods, J Obstet Gynaecol Res 35(3) (2009) 525–32. [DOI] [PubMed] [Google Scholar]
- [19].Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Nikita L, Galuppi M, Lamont RF, Chaemsaithong P, Miranda J, Chaiworapongsa T, Ravel J, The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women, Microbiome 2(1) (2014) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Martin HL, Richardson BA, Nyange PM, Lavreys L, Hillier SL, Chohan B, Mandaliya K, Ndinya-Achola JO, Bwayo J, Kreiss J, Vaginal lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition, J Infect Dis 180(6) (1999) 1863–8. [DOI] [PubMed] [Google Scholar]
- [21].McClelland RS, Lingappa JR, Srinivasan S, Kinuthia J, John-Stewart GC, Jaoko W, Richardson BA, Yuhas K, Fiedler TL, Mandaliya KN, Munch MM, Mugo NR, Cohen CR, Baeten JM, Celum C, Overbaugh J, Fredricks DN, Evaluation of the association between the concentrations of key vaginal bacteria and the increased risk of HIV acquisition in African women from five cohorts: a nested case-control study, Lancet Infect Dis 18(5) (2018) 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Berard AR, Perner M, Mutch S, Farr Zuend C, McQueen P, Burgener AD, Understanding mucosal and microbial functionality of the female reproductive tract by metaproteomics: Implications for HIV transmission, Am J Reprod Immunol 80(2) (2018) e12977. [DOI] [PubMed] [Google Scholar]
- [23].Bayigga L, Kateete DP, Anderson DJ, Sekikubo M, Nakanjako D, Diversity of vaginal microbiota in sub-Saharan Africa and its effects on HIV transmission and prevention, Am J Obstet Gynecol 220(2) (2019) 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Edwards VL, Smith SB, McComb EJ, Tamarelle J, Ma B, Humphrys MS, Gajer P, Gwilliam K, Schaefer AM, Lai SK, Terplan M, Mark KS, Brotman RM, Forney LJ, Bavoil PM, Ravel J, The Cervicovaginal Microbiota-Host Interaction Modulates Chlamydia trachomatis Infection, MBio 10(4) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, Sun CL, Goltsman DS, Wong RJ, Shaw G, Stevenson DK, Holmes SP, Relman DA, Temporal and spatial variation of the human microbiota during pregnancy, Proc Natl Acad Sci U S A 112(35) (2015) 11060–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Callahan BJ, DiGiulio DB, Goltsman DSA, Sun CL, Costello EK, Jeganathan P, Biggio JR, Wong RJ, Druzin ML, Shaw GM, Stevenson DK, Holmes SP, Relman DA, Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women, Proc Natl Acad Sci U S A 114(37) (2017) 9966–9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Brown RG, Marchesi JR, Lee YS, Smith A, Lehne B, Kindinger LM, Terzidou V, Holmes E, Nicholson JK, Bennett PR, MacIntyre DA, Vaginal dysbiosis increases risk of preterm fetal membrane rupture, neonatal sepsis and is exacerbated by erythromycin, BMC Med 16(1) (2018) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gerson KD, McCarthy C, Elovitz MA, Ravel J, Sammel MD, Burris HH, Cervicovaginal microbial communities deficient in Lactobacillus species are associated with second trimester short cervix, Am J Obstet Gynecol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hocevar K, Maver A, Vidmar Simic M, Hodzic A, Haslberger A, Premru Sersen T, Peterlin B, Vaginal Microbiome Signature Is Associated With Spontaneous Preterm Delivery, Front Med (Lausanne) 6 (2019) 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tabatabaei N, Eren AM, Barreiro LB, Yotova V, Dumaine A, Allard C, Fraser WD, Vaginal microbiome in early pregnancy and subsequent risk of spontaneous preterm birth: a case-control study, BJOG 126(3) (2019) 349–358. [DOI] [PubMed] [Google Scholar]
- [31].Aagaard K, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C, Raza S, Rosenbaum S, Van den Veyver I, Milosavljevic A, Gevers D, Huttenhower C, Petrosino J, Versalovic J, A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy, PLoS One 7(6) (2012) e36466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].MacIntyre DA, Chandiramani M, Lee YS, Kindinger L, Smith A, Angelopoulos N, Lehne B, Arulkumaran S, Brown R, Teoh TG, Holmes E, Nicoholson JK, Marchesi JR, Bennett PR, The vaginal microbiome during pregnancy and the postpartum period in a European population, Sci Rep 5 (2015) 8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Stafford GP, Parker JL, Amabebe E, Kistler J, Reynolds S, Stern V, Paley M, Anumba DOC, Spontaneous Preterm Birth Is Associated with Differential Expression of Vaginal Metabolites by Lactobacilli-Dominated Microflora, Front Physiol 8 (2017) 615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kindinger LM, Bennett PR, Lee YS, Marchesi JR, Smith A, Cacciatore S, Holmes E, Nicholson JK, Teoh TG, MacIntyre DA, The interaction between vaginal microbiota, cervical length, and vaginal progesterone treatment for preterm birth risk, Microbiome 5(1) (2017) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pace RM, Chu DM, Prince AL, Meyer KM, Seferovic M, Aagaard KM, 1024: Ecology and diversity of the vaginal microbiome in pregnancy and postpartum, Am J Obstet Gynecol 220(1) (2019) S657–S658. [Google Scholar]
- [36].Paramel Jayaprakash T, Wagner EC, van Schalkwyk J, Albert AY, Hill JE, Money DM, Group PS, High Diversity and Variability in the Vaginal Microbiome in Women following Preterm Premature Rupture of Membranes (PPROM): A Prospective Cohort Study, PLoS One 11(11) (2016) e0166794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Freitas AC, Bocking A, Hill JE, Money DM, V.R. Group, Increased richness and diversity of the vaginal microbiota and spontaneous preterm birth, Microbiome 6(1) (2018) 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Brown RG, Al-Memar M, Marchesi JR, Lee YS, Smith A, Chan D, Lewis H, Kindinger L, Terzidou V, Bourne T, Bennett PR, MacIntyre DA, Establishment of vaginal microbiota composition in early pregnancy and its association with subsequent preterm prelabor rupture of the fetal membranes, Transl Res 207 (2019) 30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, Adler A, Vera Garcia C, Rohde S, Say L, Lawn JE, National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications, Lancet (London, England) 379(9832) (2012) 2162–72. [DOI] [PubMed] [Google Scholar]
- [40].Liu L, Oza S, Hogan D, Perin J, Rudan I, Lawn JE, Cousens S, Mathers C, Black RE, Global, regional, and national causes of child mortality in 2000–13, with projections to inform post-2015 priorities: an updated systematic analysis, Lancet (London, England) 385(9966) (2015) 430–40. [DOI] [PubMed] [Google Scholar]
- [41].Vinturache AE, Gyamfi-Bannerman C, Hwang J, Mysorekar IU, Jacobsson B, Maternal microbiome - A pathway to preterm birth, Seminars in fetal & neonatal medicine 21(2) (2016) 94–9. [DOI] [PubMed] [Google Scholar]
- [42].Chu DM, Seferovic M, Pace RM, Aagaard KM, The microbiome in preterm birth, Best practice & research. Clinical obstetrics & gynaecology 52 (2018) 103–113. [DOI] [PubMed] [Google Scholar]
- [43].Barrientos-Duran A, Fuentes-Lopez A, de Salazar A, Plaza-Diaz J, Garcia F, Reviewing the Composition of Vaginal Microbiota: Inclusion of Nutrition and Probiotic Factors in the Maintenance of Eubiosis, Nutrients 12(2) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Berman HL, McLaren MR, Callahan BJ, Understanding and interpreting community sequencing measurements of the vaginal microbiome, Bjog 127(2) (2020) 139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Fichorova RN, Anderson DJ, Differential expression of immunobiological mediators by immortalized human cervical and vaginal epithelial cells, Biol Reprod 60(2) (1999) 508–14. [DOI] [PubMed] [Google Scholar]
- [46].Anton L, Sierra LJ, DeVine A, Barila G, Heiser L, Brown AG, Elovitz MA, Common Cervicovaginal Microbial Supernatants Alter Cervical Epithelial Function: Mechanisms by Which Lactobacillus crispatus Contributes to Cervical Health, Front Microbiol 9 (2018) 2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Linhares IM, Sisti G, Minis E, de Freitas GB, Moron AF, Witkin SS, Contribution of Epithelial Cells to Defense Mechanisms in the Human Vagina, Curr Infect Dis Rep 21(9) (2019) 30. [DOI] [PubMed] [Google Scholar]
- [48].Simhan HN, Caritis SN, Krohn MA, Hillier SL, Elevated vaginal pH and neutrophils are associated strongly with early spontaneous preterm birth, Am J Obstet Gynecol 189(4) (2003) 1150–4. [DOI] [PubMed] [Google Scholar]
- [49].Romero R, Chaiworapongsa T, Kuivaniemi H, Tromp G, Bacterial vaginosis, the inflammatory response and the risk of preterm birth: a role for genetic epidemiology in the prevention of preterm birth, Am J Obstet Gynecol 190(6) (2004) 1509–19. [DOI] [PubMed] [Google Scholar]
- [50].Romero R, Dey SK, Fisher SJ, Preterm labor: one syndrome, many causes, Science 345(6198) (2014) 760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Bieda J, Chaemsaithong P, Miranda J, Chaiworapongsa T, Ravel J, The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term, Microbiome 2 (2014) 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cox SM, King MR, Casey ML, MacDonald PC, Interleukin-1 beta, −1 alpha, and −6 and prostaglandins in vaginal/cervical fluids of pregnant women before and during labor, J Clin Endocrinol Metab 77(3) (1993) 805–15. [DOI] [PubMed] [Google Scholar]
- [53].Tanaka Y, Narahara H, Takai N, Yoshimatsu J, Anai T, Miyakawa I, Interleukin-1beta and interleukin-8 in cervicovaginal fluid during pregnancy, Am J Obstet Gynecol 179(3 Pt 1) (1998) 644–9. [DOI] [PubMed] [Google Scholar]
- [54].Kim CJ, Romero R, Chaemsaithong P, Chaiyasit N, Yoon BH, Kim YM, Acute chorioamnionitis and funisitis: definition, pathologic features, and clinical significance, Am J Obstet Gynecol 213(4 Suppl) (2015) S29–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Bates D, Mächler M, Bolker B, Walker S, Fitting Linear Mixed-Effects Models Using lme4, Journal of Statistical Software; Vol 1, Issue 1 (2015) (2015). [Google Scholar]
- [56].R.C. Team, R: A language and environment for statistical computing, R Foundation for Statistical Computing, Vienna, Austria, 2013. [Google Scholar]
- [57].Gudza-Mugabe M, Havyarimana E, Jaumdally S, Garson KL, Lennard K, Tarupiwa A, Mugabe F, Marere T, Mavenyengwa RT, Masson L, Jaspan HB, Human Immunodeficiency Virus Infection Is Associated With Preterm Delivery Independent of Vaginal Microbiota in Pregnant African Women, J Infect Dis 221(7) (2020) 1194–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Srinivasan S, Hoffman NG, Morgan MT, Matsen FA, Fiedler TL, Hall RW, Ross FJ, McCoy CO, Bumgarner R, Marrazzo JM, Fredricks DN, Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria, PLoS One 7(6) (2012) e37818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Onywera H, Williamson AL, Mbulawa ZZA, Coetzee D, Meiring TL, Factors associated with the composition and diversity of the cervical microbiota of reproductive-age Black South African women: a retrospective cross-sectional study, PeerJ 7 (2019) e7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yang L, Hao Y, Hu J, Kelly D, Li H, Brown S, Tasker C, Roche NE, Chang TL, Pei Z, Differential effects of depot medroxyprogesterone acetate administration on vaginal microbiome in Hispanic White and Black women, Emerg Microbes Infect 8(1) (2019) 197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Beamer MA, Austin MN, Avolia HA, Meyn LA, Bunge KE, Hillier SL, Bacterial species colonizing the vagina of healthy women are not associated with race, Anaerobe 45 (2017) 40–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Mitchell C, Gottsch ML, Liu C, Fredricks DN, Nelson DB, Associations between vaginal bacteria and levels of vaginal defensins in pregnant women, Am J Obstet Gynecol 208(2) (2013) 132 e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hilbert DW, Smith WL, Chadwick SG, Toner G, Mordechai E, Adelson ME, Aguin TJ, Sobel JD, Gygax SE, Development and Validation of a Highly Accurate Quantitative Real-Time PCR Assay for Diagnosis of Bacterial Vaginosis, J Clin Microbiol 54(4) (2016) 1017–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sobel JD, Kaur N, Woznicki NA, Boikov D, Aguin T, Gill G, Akins RA, Prognostic Indicators of Recurrence of Bacterial Vaginosis, J Clin Microbiol 57(5) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ma B, France MT, Crabtree J, Holm JB, Humphrys MS, Brotman RM, Ravel J, A comprehensive non-redundant gene catalog reveals extensive within-community intraspecies diversity in the human vagina, Nat Commun 11(1) (2020) 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kyongo JK, Crucitti T, Menten J, Hardy L, Cools P, Michiels J, Delany-Moretlwe S, Mwaura M, Ndayisaba G, Joseph S, Fichorova R, van de Wijgert J, Vanham G, Arien KK, Jespers V, Cross-Sectional Analysis of Selected Genital Tract Immunological Markers and Molecular Vaginal Microbiota in Sub-Saharan African Women, with Relevance to HIV Risk and Prevention, Clin Vaccine Immunol 22(5) (2015) 526–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Jespers V, Kyongo J, Joseph S, Hardy L, Cools P, Crucitti T, Mwaura M, Ndayisaba G, Delany-Moretlwe S, Buyze J, Vanham G, van de Wijgert J, A longitudinal analysis of the vaginal microbiota and vaginal immune mediators in women from sub-Saharan Africa, Sci Rep 7(1) (2017) 11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Lennard K, Dabee S, Barnabas SL, Havyarimana E, Blakney A, Jaumdally SZ, Botha G, Mkhize NN, Bekker LG, Lewis DA, Gray G, Mulder N, Passmore JS, Jaspan HB, Microbial Composition Predicts Genital Tract Inflammation and Persistent Bacterial Vaginosis in South African Adolescent Females, Infect Immun 86(1) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Joag V, Obila O, Gajer P, Scott MC, Dizzell S, Humphrys M, Shahabi K, Huibner S, Shannon B, Tharao W, Mureithi M, Oyugi J, Kimani J, Kaushic C, Ravel J, Anzala O, Kaul R, Impact of Standard Bacterial Vaginosis Treatment on the Genital Microbiota, Immune Milieu, and Ex Vivo Human Immunodeficiency Virus Susceptibility, Clin Infect Dis 68(10) (2019) 1675–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bradshaw CS, Sobel JD, Current Treatment of Bacterial Vaginosis-Limitations and Need for Innovation, J Infect Dis 214 Suppl 1 (2016) S14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Verwijs MC, Agaba SK, Darby AC, van de Wijgert J, Impact of oral metronidazole treatment on the vaginal microbiota and correlates of treatment failure, Am J Obstet Gynecol 222(2) (2020) 157 e1–157 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Loetscher M, Gerber B, Loetscher P, Jones SA, Piali L, Clark-Lewis I, Baggiolini M, Moser B, Chemokine receptor specific for IP10 and mig: structure, function, and expression in activated T-lymphocytes, J Exp Med 184(3) (1996) 963–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Karin N, Razon H, Chemokines beyond chemo-attraction: CXCL10 and its significant role in cancer and autoimmunity, Cytokine 109 (2018) 24–28. [DOI] [PubMed] [Google Scholar]
- [74].Luty J, Ruckemann-Dziurdzinska K, Witkowski JM, Bryl E, Immunological aspects of autoimmune thyroid disease - Complex interplay between cells and cytokines, Cytokine 116 (2019) 128–133. [DOI] [PubMed] [Google Scholar]
- [75].Khan IA, MacLean JA, Lee FS, Casciotti L, DeHaan E, Schwartzman JD, Luster AD, IP-10 is critical for effector T cell trafficking and host survival in Toxoplasma gondii infection, Immunity 12(5) (2000) 483–94. [DOI] [PubMed] [Google Scholar]
- [76].Olive AJ, Gondek DC, Starnbach MN, CXCR3 and CCR5 are both required for T cell-mediated protection against C. trachomatis infection in the murine genital mucosa, Mucosal Immunol 4(2) (2011) 208–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Chami B, Yeung A, Buckland M, Liu H, G MF, Tao K, Bao S, CXCR3 plays a critical role for host protection against Salmonellosis, Sci Rep 7(1) (2017) 10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Lei J, Yin X, Shang H, Jiang Y, IP-10 is highly involved in HIV infection, Cytokine 115 (2019) 97–103. [DOI] [PubMed] [Google Scholar]
- [79].Romero R, Chaemsaithong P, Chaiyasit N, Docheva N, Dong Z, Kim CJ, Kim YM, Kim JS, Qureshi F, Jacques SM, Yoon BH, Chaiworapongsa T, Yeo L, Hassan SS, Erez O, Korzeniewski SJ, CXCL10 and IL-6: Markers of two different forms of intra-amniotic inflammation in preterm labor, Am J Reprod Immunol 78(1) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Romero R, Grivel JC, Tarca AL, Chaemsaithong P, Xu Z, Fitzgerald W, Hassan SS, Chaiworapongsa T, Margolis L, Evidence of perturbations of the cytokine network in preterm labor, Am J Obstet Gynecol 213(6) (2015) 836 e1–836 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Karjalainen MK, Ojaniemi M, Haapalainen AM, Mahlman M, Salminen A, Huusko JM, Maatta TA, Kaukola T, Anttonen J, Ulvila J, Haataja R, Teramo K, Kingsmore SF, Palotie A, Muglia LJ, Ramet M, Hallman M, CXCR3 Polymorphism and Expression Associate with Spontaneous Preterm Birth, J Immunol 195(5) (2015) 2187–98. [DOI] [PubMed] [Google Scholar]
- [82].Maymon E, Romero R, Bhatti G, Chaemsaithong P, Gomez-Lopez N, Panaitescu B, Chaiyasit N, Pacora P, Dong Z, Hassan SS, Erez O, Chronic inflammatory lesions of the placenta are associated with an up-regulation of amniotic fluid CXCR3: A marker of allograft rejection, J Perinat Med 46(2) (2018) 123–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Liu M, Guo S, Hibbert JM, Jain V, Singh N, Wilson NO, Stiles JK, CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications, Cytokine & growth factor reviews 22(3) (2011) 121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Aksoy MO, Yang Y, Ji R, Reddy PJ, Shahabuddin S, Litvin J, Rogers TJ, Kelsen SG, CXCR3 surface expression in human airway epithelial cells: cell cycle dependence and effect on cell proliferation, American journal of physiology. Lung cellular and molecular physiology 290(5) (2006) L909–18. [DOI] [PubMed] [Google Scholar]
- [85].Lasagni L, Francalanci M, Annunziato F, Lazzeri E, Giannini S, Cosmi L, Sagrinati C, Mazzinghi B, Orlando C, Maggi E, Marra F, Romagnani S, Serio M, Romagnani P, An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4, J Exp Med 197(11) (2003) 1537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Groom JR, Luster AD, CXCR3 ligands: redundant, collaborative and antagonistic functions, Immunology and cell biology 89(2) (2011) 207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hansen AS, Biltoft M, Bundgaard B, Bohn AB, Moller BK, Hollsberg P, CD46 activation induces distinct CXCL-10 response in monocytes and monocyte-derived dendritic cells, Cytokine 113 (2019) 466–469. [DOI] [PubMed] [Google Scholar]
- [88].Wuest TR, Carr DJ, Dysregulation of CXCR3 signaling due to CXCL10 deficiency impairs the antiviral response to herpes simplex virus 1 infection, J Immunol 181(11) (2008) 7985–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Thapa M, Welner RS, Pelayo R, Carr DJ, CXCL9 and CXCL10 expression are critical for control of genital herpes simplex virus type 2 infection through mobilization of HSV-specific CTL and NK cells to the nervous system, J Immunol 180(2) (2008) 1098–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Margulieux KR, Fox JW, Nakamoto RK, Hughes MA, CXCL10 Acts as a Bifunctional Antimicrobial Molecule against Bacillus anthracis, mBio 7(3) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wang X, Wang S, Liu ZH, Qi WQ, Zhang Q, Zhang YG, Sun DR, Xu Y, Wang HG, Li ZX, Cong XL, Zhao P, Zhou CY, Wang JB, Regulatory polymorphism of CXCL10 rs1439490 in seronegative occult hepatitis C virus infection, World J Gastroenterol 24(20) (2018) 2191–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Singh B, Anbalagan S, Selvaraj P, Regulatory role of CCL5 (rs2280789) and CXCL10 (rs56061981) gene polymorphisms on intracellular CCL5 and CXCL10 expression in pulmonary tuberculosis, Hum Immunol 78(5–6) (2017) 430–434. [DOI] [PubMed] [Google Scholar]
- [93].Hanff PA, Rosol-Donoghue JA, Spiegel CA, Wilson KH, Moore LH, Leptotrichia sanguinegens sp. nov., a new agent of postpartum and neonatal bacteremia, Clin Infect Dis 20 Suppl 2 (1995) S237–9. [DOI] [PubMed] [Google Scholar]
- [94].Collins MD, Hoyles L, Tornqvist E, von Essen R, Falsen E, Characterization of some strains from human clinical sources which resemble “Leptotrichia sanguinegens”: description of Sneathia sanguinegens sp. nov., gen. nov, Syst Appl Microbiol 24(3) (2001) 358–61. [DOI] [PubMed] [Google Scholar]
- [95].Harwich MD Jr., Serrano MG, Fettweis JM, Alves JM, Reimers MA, Vaginal Microbiome C, Buck GA, Jefferson KK, Genomic sequence analysis and characterization of Sneathia amnii sp. nov, BMC Genomics 13 Suppl 8 (2012) S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, Kim CJ, Erez O, Edwin S, Relman DA, Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation, PLoS One 3(8) (2008) e3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Romero R, Miranda J, Chaiworapongsa T, Chaemsaithong P, Gotsch F, Dong Z, Ahmed AI, Yoon BH, Hassan SS, Kim CJ, Korzeniewski SJ, Yeo L, A novel molecular microbiologic technique for the rapid diagnosis of microbial invasion of the amniotic cavity and intra-amniotic infection in preterm labor with intact membranes, Am J Reprod Immunol 71(4) (2014) 330–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Romero R, Miranda J, Chaemsaithong P, Chaiworapongsa T, Kusanovic JP, Dong Z, Ahmed AI, Shaman M, Lannaman K, Yoon BH, Hassan SS, Kim CJ, Korzeniewski SJ, Yeo L, Kim YM, Sterile and microbial-associated intra-amniotic inflammation in preterm prelabor rupture of membranes, J Matern Fetal Neonatal Med 28(12) (2015) 1394–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Srinivasan S, Munch MM, Sizova MV, Fiedler TL, Kohler CM, Hoffman NG, Liu C, Agnew KJ, Marrazzo JM, Epstein SS, Fredricks DN, More Easily Cultivated Than Identified: Classical Isolation With Molecular Identification of Vaginal Bacteria, J Infect Dis 214 Suppl 1 (2016) S21–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Wang Y, Zhang Y, Zhang Q, Chen H, Feng Y, Characterization of pelvic and cervical microbiotas from patients with pelvic inflammatory disease, J Med Microbiol 67(10) (2018) 1519–1526. [DOI] [PubMed] [Google Scholar]
- [101].Romero R, Gomez-Lopez N, Winters AD, Jung E, Shaman M, Bieda J, Panaitescu B, Pacora P, Erez O, Greenberg JM, Ahmad MM, Hsu CD, Theis KR, Evidence that intra-amniotic infections are often the result of an ascending invasion - a molecular microbiological study, J Perinat Med 47(9) (2019) 915–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Vitorino P, Varo R, Castillo P, Hurtado JC, Fernandes F, Valente AM, Mabunda R, Mocumbi S, Gary JM, Jenkinson TG, Mandomando I, Blau DM, Breiman RF, Bassat Q, Sneathia amnii and Maternal Chorioamnionitis and Stillbirth, Mozambique, Emerg Infect Dis 25(8) (2019) 1614–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Lannon SMR, Adams Waldorf KM, Fiedler T, Kapur RP, Agnew K, Rajagopal L, Gravett MG, Fredricks DN, Parallel detection of lactobacillus and bacterial vaginosis-associated bacterial DNA in the chorioamnion and vagina of pregnant women at term, J Matern Fetal Neonatal Med 32(16) (2019) 2702–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].DiGiulio DB, Romero R, Kusanovic JP, Gomez R, Kim CJ, Seok KS, Gotsch F, Mazaki-Tovi S, Vaisbuch E, Sanders K, Bik EM, Chaiworapongsa T, Oyarzun E, Relman DA, Prevalence and diversity of microbes in the amniotic fluid, the fetal inflammatory response, and pregnancy outcome in women with preterm pre-labor rupture of membranes, Am J Reprod Immunol 64(1) (2010) 38–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Musilova I, Kacerovsky M, Stepan M, Bestvina T, Pliskova L, Zednikova B, Jacobsson B, Maternal serum C-reactive protein concentration and intra-amniotic inflammation in women with preterm prelabor rupture of membranes, PLoS One 12(8) (2017) e0182731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Musilova I, Pliskova L, Gerychova R, Janku P, Simetka O, Matlak P, Jacobsson B, Kacerovsky M, Maternal white blood cell count cannot identify the presence of microbial invasion of the amniotic cavity or intra-amniotic inflammation in women with preterm prelabor rupture of membranes, PLoS One 12(12) (2017) e0189394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Musilova I, Andrys C, Drahosova M, Zednikova B, Hornychova H, Pliskova L, Zemlickova H, Jacobsson B, Kacerovsky M, Late preterm prelabor rupture of fetal membranes: fetal inflammatory response and neonatal outcome, Pediatr Res 83(3) (2018) 630–637. [DOI] [PubMed] [Google Scholar]
- [108].Hornychova H, Kacerovsky M, Musilova I, Pliskova L, Zemlickova H, Matejkova A, Vosmikova H, Rozkosova K, Cermakova P, Bolehovska R, Halada P, Jacobsson B, Laco J, Cervical human papillomavirus infection in women with preterm prelabor rupture of membranes, PLoS One 13(11) (2018) e0207896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Romero R, Miranda J, Chaiworapongsa T, Chaemsaithong P, Gotsch F, Dong Z, Ahmed AI, Yoon BH, Hassan SS, Kim CJ, Korzeniewski SJ, Yeo L, Kim YM, Sterile intra-amniotic inflammation in asymptomatic patients with a sonographic short cervix: prevalence and clinical significance, J Matern Fetal Neonatal Med 28(11) (2015) 1343–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Gardella C, Riley DE, Hitti J, Agnew K, Krieger JN, Eschenbach D, Identification and sequencing of bacterial rDNAs in culture-negative amniotic fluid from women in premature labor, Am J Perinatol 21(6) (2004) 319–23. [DOI] [PubMed] [Google Scholar]
- [111].Romero R, Miranda J, Kusanovic JP, Chaiworapongsa T, Chaemsaithong P, Martinez A, Gotsch F, Dong Z, Ahmed AI, Shaman M, Lannaman K, Yoon BH, Hassan SS, Kim CJ, Korzeniewski SJ, Yeo L, Kim YM, Clinical chorioamnionitis at term I: microbiology of the amniotic cavity using cultivation and molecular techniques, J Perinat Med 43(1) (2015) 19–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Nelson DB, Hanlon A, Nachamkin I, Haggerty C, Mastrogiannis DS, Liu C, Fredricks DN, Early pregnancy changes in bacterial vaginosis-associated bacteria and preterm delivery, Paediatr Perinat Epidemiol 28(2) (2014) 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Devi U, Bora R, Das JK, Malik V, Mahanta J, Sneathia species in a case of neonatal meningitis from Northeast India, Oxf Med Case Reports 2014(6) (2014) 112–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Shinkai A, Yoshisue H, Koike M, Shoji E, Nakagawa S, Saito A, Takeda T, Imabeppu S, Kato Y, Hanai N, Anazawa H, Kuga T, Nishi T, A novel human CC chemokine, eotaxin-3, which is expressed in IL-4-stimulated vascular endothelial cells, exhibits potent activity toward eosinophils, J Immunol 163(3) (1999) 1602–10. [PubMed] [Google Scholar]
- [115].Rankin SM, Conroy DM, Williams TJ, Eotaxin and eosinophil recruitment: implications for human disease, Molecular medicine today 6(1) (2000) 20–7. [DOI] [PubMed] [Google Scholar]
- [116].Banwell ME, Tolley NS, Williams TJ, Mitchell TJ, Regulation of human eotaxin-3/CCL26 expression: modulation by cytokines and glucocorticoids, Cytokine 17(6) (2002) 317–23. [DOI] [PubMed] [Google Scholar]
- [117].Ahmadi Z, Hassanshahi G, Khorramdelazad H, Zainodini N, Koochakzadeh L, An Overlook to the Characteristics and Roles Played by Eotaxin Network in the Pathophysiology of Food Allergies: Allergic Asthma and Atopic Dermatitis, Inflammation 39(3) (2016) 1253–67. [DOI] [PubMed] [Google Scholar]
- [118].Rothenberg ME, Hogan SP, The eosinophil, Annual review of immunology 24 (2006) 147–74. [DOI] [PubMed] [Google Scholar]
- [119].Gieseck RL 3rd, Wilson MS, Wynn TA, Type 2 immunity in tissue repair and fibrosis, Nat Rev Immunol 18(1) (2018) 62–76. [DOI] [PubMed] [Google Scholar]
- [120].Takeda Y, Kato T, Nemoto N, Araki A, Gazi MY, Nara H, Asao H, Augmentation of the expression of the eotaxin receptor on duodenal neutrophils by IL-21, Cytokine 110 (2018) 194–203. [DOI] [PubMed] [Google Scholar]
- [121].Provost V, Larose MC, Langlois A, Rola-Pleszczynski M, Flamand N, Laviolette M, CCL26/eotaxin-3 is more effective to induce the migration of eosinophils of asthmatics than CCL11/eotaxin-1 and CCL24/eotaxin-2, Journal of leukocyte biology 94(2) (2013) 213–22. [DOI] [PubMed] [Google Scholar]
- [122].Gaspar K, Kukova G, Bunemann E, Buhren BA, Sonkoly E, Szollosi AG, Muller A, Savinko T, Lauerma AI, Alenius H, Kemeny L, Dieu-Nosjean MC, Stander S, Fischer JW, Ruzicka T, Zlotnik A, Szegedi A, Homey B, The chemokine receptor CCR3 participates in tissue remodeling during atopic skin inflammation, Journal of dermatological science 71(1) (2013) 12–21. [DOI] [PubMed] [Google Scholar]
- [123].Rothenberg ME, Molecular, genetic, and cellular bases for treating eosinophilic esophagitis, Gastroenterology 148(6) (2015) 1143–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Gela A, Kasetty G, Jovic S, Ekoff M, Nilsson G, Morgelin M, Kjellstrom S, Pease JE, Schmidtchen A, Egesten A, Eotaxin-3 (CCL26) exerts innate host defense activities that are modulated by mast cell proteases, Allergy 70(2) (2015) 161–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.