Abstract
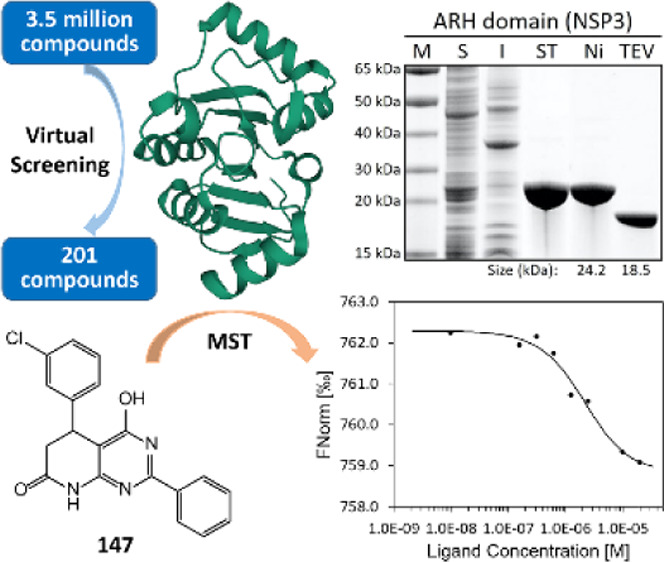
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the causative agent of the coronavirus disease of 2019 (COVID-19). Its genome encodes two open reading frames for two large proteins, PP1a and PP1ab. Within the two polypeptide stretches, there are two proteases that process the large proteins into 15 discrete proteins essential for the assembly of the virion during its replication. We describe herein the cloning of the genes for these discrete proteins optimized for expression in Escherichia coli, production of the proteins, and their purification to homogeneity. These included all but six: NSP6, which possesses eight transmembrane regions, and five that are small proteins/peptides (E, ORF3b, ORF6, ORF7b, and ORF10). These proteins are intended for experimental validation of small-molecule binders as molecular template hits. The proof of concept was established with the ADP-ribosylhydrolase (ARH) domain of NSP3 in discovery of small-molecule templates that could serve as the basis for further optimization. The hit molecules include one submicromolar and a few low-micromolar binders to the ARH domain. Availability of these proteins in soluble forms opens up the opportunity for discoveries of novel templates with the potential for anti-COVID-19 pharmaceuticals.
Introduction
A pneumonia-like condition of unknown cause was first detected late in 2019 in Wuhan, China.1,2 Shortly after, the pathogen was identified as a coronavirus (CoV), whose RNA genome was sequenced shortly after.3 The World Health Organization (WHO) named the disease that the novel virus caused as COVID-19 (coronavirus disease of 2019). The novel coronavirus was named as “severe acute respiratory syndrome CoV-2” or SARS-CoV-2. The WHO declared COVID-19 a pandemic on March 11, 2020.4 At the time of writing of this report, COVID-19 has been reported in 219 countries and territories, with >150 million confirmed cases and >3.1 million fatalities.5 These figures increase daily.
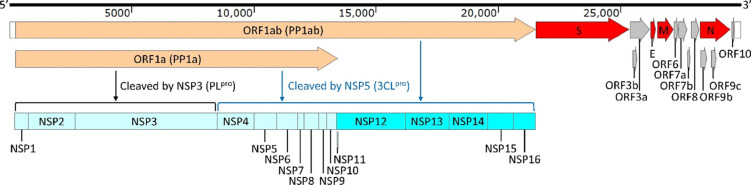
SARS-CoV-2 contains a positive-strand RNA genome of approximately 30 kb (Figure 1). The genome produces two long polyproteins (PP1a and PP1ab), which are processed into 16 nonstructural proteins (NSPs), four structural proteins—spike glycoprotein (S), envelope protein (E), membrane glycoprotein (M), and nucleocapsid phosphoprotein (N)—and at least seven accessory proteins (ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8, and ORF9b).6,7 SARS-CoV-2 is primarily transmitted through respiratory droplets.8 The virus binds to the angiotensin-converting enzyme 2 (ACE2) as its receptor on the cell surface,9−12 whereby the complex is internalized mainly through endocytosis13 to initiate the infection. After the viral RNA enters the host cell, it is translated from two open reading frames (ORF1a and ORF1ab) to produce the two aforementioned large polyproteins, PP1a and PP1ab.7,14 The latter is cleaved/processed by two viral proteases that are already within the polyprotein stretches. These are papain-like protease (PLpro) and 3C-like protease (3CLpro), which produce the mature NSPs.15 PP1a is cleaved into 10 fragments, constituting NSP1–NSP10. PP1ab is fragmented to produce all NSPs from NSP1–NSP16. The PLpro protease is a component of NSP3, and it is believed to process the polyprotein for the formation of NSP1, NSP2, and NSP3. The rest of the NSPs are processed/matured by the protease 3CLpro, which is contained within NSP5. Both proteases are essential for viral existence.15 Once all NSPs are produced, viral RNA synthesis by NSP12 (an RNA-dependent RNA polymerase), which needs to associate with other NSPs for catalytic competence, produces both genomic RNA (gRNA) and sub-genomic RNA (sgRNA). The sgRNAs serve as mRNA for the production of the structural and accessory proteins.
Figure 1.
Genomic structure of SARS-CoV-2.
The recent availability of several vaccines for SARS-CoV-2 is a critically needed development in addressing the COVID-19 pandemic. At the same time, there is a genuine need for COVID-19 therapeutics. Initial efforts focused on repurposing of existing antiviral treatments have so far only yielded a single FDA-approved treatment with modest efficacy, emphasizing the need for drugs that have been conceived and designed expressly for SARS-CoV-2. The identification of initial hits for such efforts requires a robust access to the different proteins of SARS-CoV-2 in high-purity and sufficient amounts. As a part of our efforts to target the proteome of SARS-CoV-2 for the discovery of anti-COVID-19 therapeutic drugs in a target-agnostic manner, we have optimized such methods, as will be outlined in this report. We have cloned genes for these proteins; we assess expression of the genes and the ability to purify the individual proteins. As a demonstration of the use of this method for early-stage drug discovery, we will describe one example of the use of the ADP-ribosylhydrolase (ARH) domain of NSP3 in discovery of small-molecule templates that could serve as the basis for further optimization efforts.
Materials and Methods
Cloning and Expression of the Genes of SARS-CoV-2
In order to express SARS-CoV-2 proteins in Escherichia coli, we synthesized codon-optimized genes from GenScript (Piscataway, NJ). All synthetic genes are listed in Table S1. We synthesized the nsp1 gene including segments encoding 6xHis-tag, a Twin-Strep-tag and a tobacco etch virus (TEV) cleavage site at its 5′-end to replace the thrombin-cleavage site and the T7-tag in an expression vector pET-28a(+) for better purification of the proteins expressed. The synthetic nsp1 gene contained the restriction sites NcoI, NdeI, and XhoI at the 5′-end, after the sequence for the TEV cleavage site, and the 3′-end, respectively. The pET-28a(+) and the nsp1 gene were digested with NcoI and XhoI (NEB, Ipswich, MA) in 1× CutSmart buffer for 6 h at 37 °C, followed by the purification on an agarose gel with a Zymoclean DNA gel recovery kit (Zymo Research, Irvine, CA) following the manufacturer’s instruction. Then, the two fragments were ligated with T4 DNA ligase in 1× T4 DNA ligase buffer (NEB, Ipswich, MA) at 16 °C overnight to generate the plasmid pETHST-NSP1. A competent E. coli DH5α was transformed with the ligation mixture by giving heat shock for 90 s at 42 °C and selected on a LB plate containing 50 μg/mL kanamycin after incubation for 18 h at 37 °C. The nucleotide sequences at the 5′-end and the 3′-end of the nsp1 gene in the resulting plasmid were confirmed by DNA sequencing with the T7 promoter and T7 terminator primers, respectively (MCLAB, South San Francisco, CA). The plasmid pETHST-NSP1 was then inserted into the E. coli LOBSTR-RIL strain, derived from BL21 (DE3), to express the NSP1 fused with the N-terminal 6xHis and Twin-Strep tags and the TEV cleavage site.
The pETHST-NSP1 was digested with NdeI and XhoI in 1× CutSmart buffer at 37 °C overnight. The linearized pETHST was purified on agarose gels with a Zymoclean DNA gel recovery kit. It was used to clone all other synthetic genes that were cut with NdeI and XhoI. The nucleotide sequences of each gene were confirmed. The E. coli LOBSTR-RIL was transformed with each discrete plasmid carrying a SARS-CoV-2 gene.
The overnight cultures (200 μL) of the transformed LOBSTR-RIL strain were diluted in 20 mL of fresh LB medium containing 50 μg/mL kanamycin, followed by incubation at 37 °C with shaking at 180 rpm until the OD600 of the cultures reached 0.6. The expression of the SARS-CoV-2 proteins was examined by inducing with 0.4 mM IPTG either by incubation at 37 °C for 2 h or at 16 °C for 18 h. The cells were harvested at 7000g for 15 min at 4 °C and were frozen at −80 °C. The frozen cells were resuspended in 2 mL of 1× PBS with 5 mM EDTA, followed by cell disruption using sonification (10 cycles of 10 s of pulse and 20 s of rest on ice) with a Branson 450 sonifier. One milliliter of disrupted cells was centrifuged at 18,000g for 20 min at 4 °C. The supernatants were transferred to new microcentrifuge tubes, and the pellets were resuspended in 800 μL of 1× PBS. The level of soluble proteins in the supernatants was examined on 8–16% ExpressPlus SDS-PAGE gels (GenScript, Piscataway, NJ).
Purification of the SARS-CoV-2 Proteins
For the purpose of demonstration, four proteins, ARH domain (NSP3), PLpro (NSP3), NSP15, and NSP16, were purified. The overnight cultures (1 mL) of the LOBSTR strains harboring pETHST-NSP3N, pETHST-NSP3P, pETHST-NSP15, or pETHST-NSP16 were diluted in 100 mL of fresh LB medium containing 50 μg/mL kanamycin, followed by incubation at 37 °C with shaking at 180 rpm. The expression of the SARS-CoV-2 proteins was induced with 0.4 mM IPTG at 0.6 OD600 of the cultures. Each protein was produced by further incubation at 16 °C for 18 h. The cells were harvested at 7000g for 20 min at 4 °C, and the pellets were stored at −80 °C. The frozen cells were resuspended in 3 mL of washing buffer I (50 mM Tris-Cl, pH 8.0, and 150 mM NaCl) containing 10 mM MgCl2, 50 μg/mL DNase I, 50 μg/mL lysozyme, and 1× Halt protease inhibitor cocktail (Thermo Fisher Scientific, Waltham, MA) and incubated for 30 min on ice. The cells were disrupted by sonification (15 cycles of 10 s of pulse and 20 s of rest) with a Branson 450 sonifier, followed by centrifugation at 18,000g for 20 min at 4 °C to separate the soluble proteins from the unbroken cells and the inclusion bodies. The supernatants were filtrated through 0.2 μm filters prior to the purification.
The N-terminal His- and Twin-Strep-tagged SARS-CoV-2 proteins were purified with two-step affinity chromatography on 1 mL of the Strep-Tactin Sepharose resin (IBA Lifesciences, Göttingen, Germany) and 1 mL of the Protino Ni-NTA agarose resin (Macherey-Nagel, Düren, Germany) following the manufacturer’s instruction.
Strep-Tactin Sepharose Affinity Chromatography
One milliliter of the resin was equilibrated with three bed volumes of washing buffer I by gravity flow, followed by application of the filtrated cell lysate supernatants to it. The resin was subsequently washed with five bed volumes of washing buffer I. The N-terminal His- and Twin-Strep-tagged SARS-CoV-2 proteins were eluted with two bed volumes of elution buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, and 2.5 mM desthiobiotin). The fractions at every step (flow-through, washing, and elution) were collected and examined by SDS-PAGE.
Protino Ni-NTA Agarose Affinity Chromatography
One milliliter of the resin was equilibrated with five bed volumes of washing buffer II (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, and 20 mM imidazole). The chromatography was performed by gravity flow. The eluents (2 mL) from the Strep-Tactin Sepharose resin were applied to the Ni-NTA resin and incubated for 30 min at room temperature with gentle rotation, as the binding of the His-tagged proteins to the Ni-NTA resin is a slow process, followed by collection of unbound proteins. The resin was washed with eight bed volumes of washing buffer II. Subsequently, the stepwise increase in imidazole concentrations at 100, 200, 400, and 800 mM was used to elute the SARS-CoV-2 proteins, which were typically collected at 200 and 400 mM imidazole.
Cleavage of the 6xHis- and Twin-Strep-Tags by TEV Protease
The imidazole was removed from the eluents with an Amicon Ultra-15 centrifugal filter (MWCO 10 kDa; MilliporeSigma, Burlington, MA) by five repeats of concentration at 4000g for 20 min and dilution with 15 mL of washing buffer I. The concentration of the purified protein was then determined at 280 nm with the NanoPhotometer NP80 (IMPLEN, Los Angeles, CA) using the molar extinction coefficient and the molecular weight of each protein. The 6xHis-tagged TEV protease (MCLAB, South San Francisco, CA) was added to the protein solution at 1:50 (w/w) ratio, and the solution was incubated at 4 °C for 48 h with gentle agitation in order to cleave the 6xHis- and Twin-Strep-tags from the SARS-CoV-2 proteins. The progression of proteolysis was periodically checked on SDS-PAGE.
Isolation of the Tag-Free Proteins
The tag-free SARS-CoV-2 proteins were separated from the cleaved tags and the 6xHis-tagged TEV protease by applying to Protino Ni-NTA agarose resin (Macherey-Nagel, Düren, Germany), followed by washing the resin with two bed volumes of washing buffer II. The flow-through and washing fractions containing the tag-free SARS-CoV-2 proteins were pooled and concentrated with an Amicon Ultra-15 centrifugal filter (MWCO 10 kDa; MilliporeSigma, Burlington, MA). The purity of the proteins was assessed by SDS-PAGE. The homogeneously purified protein was stored at −80 °C until use.
Structure-Based Virtual Screening
The lead-like library of 3.5 million compounds was downloaded from the ZINC15 database16 and prepared using LigPrep.17 The crystal structure of the ARH domain of NSP3 (6WCF, 1.06 Å resolution)18 was prepared for ligand docking using Schrodinger Suite 2019. The following protocols that are well established in our groups,19−21 Glide,22−24 and AutoDock Vina25 were used to screen the library in a hierarchical order. The binding pocket was defined as a 30 Å box centered on the superimposed ADP-ribose ligand. The library of 3.5 million compounds was screened using Glide HTVS. The top 10%-scoring compounds were redocked using Glide SP, and the top 10%-scoring compounds were redocked using Glide XP.24 The top 6000 hits from Glide XP were re-evaluated using a complementary scoring function in Autodock Vina.25 The poses from Glide XP and Autodock Vina were visualized and analyzed for interactions and chemical diversity to selected candidates for purchase.
Determination of Binding Affinity of Potential Inhibitors to the ARH Domain of NSP3 by Microscale Thermophoresis (MST)
Labeling of the ARH Domain
The ARH domain of NSP3, purified to homogeneity as outlined above, was labeled with the dye RED-NHS 2nd Generation (NanoTemper Technologies, Munich, Germany) following the manufacturer’s instruction. Prior to labeling, 100 μL of 10 μM solution of the ARH domain was buffer-exchanged with the labeling buffer (130 mM NaHCO3, 50 mM NaCl, and pH 8.2–8.3) twice using the Zeba spin desalting column, 7K MWCO, (Thermo Fisher Scientific, Waltham, MA) by centrifugation at 1500g for 2 min. The ARH domain (90 μL) was labeled by incubating with 10 μL of 300 μM dye solution (in DMSO) for 30 min at room temperature in the dark. The labeled ARH domain was separated from the uncoupled dye using a PD-10 desalting column (Cytiva Life Sciences, Marlborough, MA) with 1× PBS. The concentration of the labeled ARH domain was calculated based on the equation in the labeling instruction from NanoTemper Technologies.
Assessment of the Compound Library
The binding of the compounds to the labeled ARH domain was assessed by microscale thermophoresis technology (MST) with the Monolith NT.115 Pico (NanoTemper Technologies, Munich, Germany). Briefly, 10 mM of each compound was prepared in DMSO as the stock solution. It was diluted to 1 mM in DMSO and subsequently further diluted to 50 μM in the assay buffer (1× PBS, 0.05% Tween-20). The labeled ARH domain was also diluted to 10 nM with the assay buffer. All solutions were centrifuged at 14,000g for 5 min to remove any precipitants. The supernatants were carefully transferred into fresh microcentrifuge tubes. The diluted ARH domain (25 μL) was mixed with 25 μL of each compound solution and also with 25 μM assay buffer containing 5% DMSO as a control. The capillaries were filled with the mixtures: four for the control and four for the compound. The binding of the compound was determined by comparing the MST traces of the compound capillaries with those of the control capillaries in the Monolith NT.115 Pico. The MST traces were analyzed with MO.Affinity Analysis software (NanoTemper Technologies, Munich, Germany).
Determination of the Dissociation Constants (Kd) of the Compounds
MST was also used to determine the dissociation constant (Kd) values of the compounds exhibiting binding to the ARH domain. Briefly, 800 μM compound in the assay buffer containing 8% DMSO was used as the initial concentration for twofold serial dilution to prepare 12 different concentrations in the assay buffer (1× PBS containing 0.05% Tween-20) supplemented with 8% DMSO. The labeled ARH domain was also diluted to 10 nM with the assay buffer. All solutions were centrifuged at 14,000g for 5 min. Finally, 10 μL of the diluted ARH domain was mixed with 10 μL of each dilution of the compound, followed by filling 12 capillaries with the mixtures. The MST trace of each capillary was measured with the Monolith NT.115 Pico. The MST traces were analyzed with MO.Affinity Analysis software to calculate the Kd values. The determination was performed in triplicate.
Results and Discussion
Overview of the Proteome
The functions of several SARS-CoV-2 proteins have been determined experimentally. Others have functions proposed based on our understanding of related coronaviruses such as SARS-CoV, MERS-CoV, and SARS-like-BatCoV or based on sequence analysis.26−28 For a few proteins, functions can currently not be surmised. We hypothesize that the spare genome of SARS-CoV-2 indicates that all these genes are essential for the virus. A gene knockout library for SARS-CoV-2 does not exist to assess the essential nature of each gene. The putative functions of SARS-CoV-2 proteins are listed in Table 1. NSP3 is a large protein of ∼217 kDa, containing multiple domains, the N-terminal ubiquitin-like domain, ARH domain (X domain), PLpro domain, 3Ecto domain (interacting with NSP4), and C-terminal Y1/CoV-Y domain.29,30 As described above, the PLpro domain produces NSP1, NSP2, and NSP3 from PP1a and PP1ab by cleaving the C-terminus of the Leu-Xaa-Gly-Gly sequence.29 The PLpro domain is one of the targets of known antiviral agents developed prior to emergence of SARS-CoV-230−32 and the focus of ongoing drug-repurposing efforts. Its inhibition would block the maturation of NSP3 and NSP4, which are involved in the formation of double-membrane vesicles in which viral RNA synthesis takes place, resulting in rapid degradation of the viral RNA by the host enzymes. NSP5 (containing 3CLpro) is the second protease of SARS-CoV-2, consisting of the N-terminal finger, the catalytic domain, and the C-terminal domain.33 It cleaves 11 sites within PP1a and PP1ab to produce NSP4–NSP16 by recognizing the (Leu/Val/Phe/Met)-Gln-(Ser/Ala/Gly/Asn) sequence as its cleavage site.33,34 NSP12 is a large protein of ∼107 kDa and the only RNA-dependent RNA polymerase (RdRp) responsible for viral RNA synthesis. It replicates and transcribes viral RNA with its essential co-factors NSP7 and NSP8, which work as primases.35 It also forms a complex with a single-stranded RNA-binding protein NSP9, which itself interacts with NSP8.36 RdRp (NSP12) is targeted by the antiviral agent remdesivir (GS-5734). Remdesivir, a nucleotide-analogue prodrug, was originally developed by Gilead to treat Ebola and Marburg virus infections but failed to show clinical efficacy for these diseases. Repurposing studies in COVID-19 patients showed moderate efficacy, and it is currently the only FDA-approved—emergency-use authorization in May of 2020—antiviral agent for treatment of COVID-19.37,38 NSP16 is a 2′-O-ribose methyltransferase (OMT) that matures the viral mRNA by the addition of a cap structure at the 5′-terminal of RNA.39 The OMT activity of NSP16 requires its essential co-factor NSP10.
Table 1. Available Structures (X-ray, NMR, or EM) are from 17 Proteins Encoded in the SARS-CoV-2 Genome and Three Proteins Encoded in the SARS-CoV Genomea.
| protein | description | experimental structures at PDB |
|---|---|---|
| NSP1 | inhibits host translation by interacting with the 40S ribosomal subunit42 | SARS-CoV-2; 7JQB (cryoEM), 7K7P and 7K3N |
| NSP2 | involved in the disruption of intracellular host signaling during SARS-CoV infections43 | |
| NSP3 | multidomain protein including the papain-like protease domain; involved in cutting the N-terminal part of PP1a and PP1ab to mature NSP1, NSP2, and NSP3 itself and altering many of the infected host proteins; interacts with NSP429 | SARS-CoV-2; PLpro domain (6WUU, 6WX4, 6W9C, 6WZU, 6YVA, 6WRH, 7KRX, 7KOK, and 7D6H), ARH domain (6WOJ, 6WEY, 6YWK, 6YWL, 6YWM, 6WEN, 6VXS, 6W02, 6WCF, 6W6Y, 7KXB, 7LG7, 6Z5T, 7BF5, 7C33, 7CZ4, and 7KG3) |
| NSP4 | forms virally induced cytoplasmic double-membrane vesicles necessary for viral replication44 | |
| NSP5 | 3C-like protease maturing most of the NSPs (NSP4–NSP16) from PP1ab33,34,45−48 | SARS-CoV-2; 137 structures (6LU7, 6M2Q, and 135 more) |
| NSP6 | forms virally induced cytoplasmic double-membrane vesicles necessary for viral replication44 | |
| NSP7 | cofactor of RNA-dependent RNA polymerase (NSP12)35,49,50 | SARS-CoV-2; 7BV1, 7BW4, 7BV2, 6WTC, 6WIQ, 6WQD, 6M71, 7BTF, 6YYT, 6YHU, and 7DCD |
| NSP8 | cofactor of RNA-dependent RNA polymerase (NSP12)35,49,50 | SARS-CoV-2; 7BV1, 7BW4, 7BV2, 6WTC, 6WIQ, 6WQD, 6M71, 7BTF, 6YYT, 6YHU, and 7DCD |
| NSP9 | single-stranded RNA-binding protein; interacts with NSP836 | SARS-CoV-2; 6W4B, 6WXD, and 6W9Q |
| NSP10 | critical co-factor for the activity of 3′ → 5′ exonuclease (NSP14) and 2′-O-ribose methyltransferase (NSP16)51 | SARS-CoV-2; 6WKQ, 7C2J, 7C2I, 6WKS, 6W61, 6WVN, 6WQ3, 6WJT, 6WRZ, 6W4H, 6W75, 6YZ1, 7KOA, 7L6T, 6ZPE, and 6ZCT |
| NSP11 | the leftover of PP1a fragmentation52 | |
| NSP12 | RNA-dependent RNA polymerase, responsible for replication and transcription of the viral RNA genome35,49,50,53 | SARS-CoV-2; 7BTF, 7BV1, 7BV2, 6YYT, 7BW4, 7L1F, 7B3B, and 7D4F |
| NSP13 | helicase; interacts with NSP1254 | SARS-CoV-2; 7NN0 and 7NIO |
| NSP14 | N-terminal 3′ → 5′ exonuclease domain and C-terminal N7-guanine-methyltransferase domain51 | SARS-CoV; 5C8U, 5C8S, 5C8T, and 5NFY |
| NSP15 | uridylate-specific endoribonuclease55,56 | SARS-CoV-2; 6W01, 6WXC, 6WLC, 6X1B, 6VWW, 7K0R, 7KEG, 5S71, 7K1O, and 7K9P |
| NSP16 | 2′-O-ribose methyltransferase mediating the 5′-cap structure of viral mRNAs39,51 | SARS-CoV-2; 6WKQ, 7C2J, 7C2I, 6WKS, 6W61, 6WVN, 6WQ3, 6WJT, 6WRZ, 6W4H, 6W75, 6YZ1, 7KOA, and 7L6T |
| S | spike glycoprotein interacting with the host receptor protein ACE212,13,40,57−61 | SARS-CoV-2; receptor-binding domain (6VSB, 7BZ5, 6M0J, 6M17, 6W41, 7C01, 6LZG, 6YM0, 6YLA, 6YOR, and 6VW1), S2 domain (6LXT and 6LVN), cryoEM of the full-length (6VXX, 6WPT, 6WPS, 6VYB, 6X29, 6X2C, 6X2A, and 6X2B) |
| ORF3a | accessory protein; activates the NLRP3 inflammasome62 | SARS-CoV-2; 7KJR (cryoEM) |
| ORF3b | accessory protein; IFN antagonist6 | |
| E | envelope protein; mediates virus morphogenesis and assembly63,64 | SARS-CoV; 5X29 and 2MM4 |
| M | membrane glycoprotein; involved in virus morphogenesis and assembly44,63 | |
| ORF6 | accessory protein; antagonizes type I interferon65 | |
| ORF7a | accessory protein; inhibits the antiviral function of bone marrow stromal antigen 2 (BST-2)66 | SARS-CoV-2; 6W37 and 7CI3 |
| ORF7b | accessory protein and a structural component of the SARS-CoV virion67 | SARS-CoV; 6W37 |
| ORF8 | accessory protein; important for interspecies transmission of the virus; activates the NLRP3 inflammasome68,69 | SARS-CoV-2; 7JX6 |
| ORF9b | accessory protein; suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome70 | SARS-CoV-2; 7KDT (cryoEM) |
| ORF9c | unknown52 | |
| N | nucleocapsid phosphoprotein; packages the viral RNA genome41,71 | SARS-CoV-2; N-terminal RNA-binding domain (6YI3, 6M3M, 6WKP, 6VYO, and 7CR5), C-terminal dimerization domain (6WZQ, 6WZO, 6WJI, 6YUN, and 7C22) |
| ORF10 | unknown; may not be produced7 |
PDB code(s) of the structures are listed for each protein, when available, together with the attributed functions for the viral proteins.
In addition to the NSPs, four structural proteins are also important for the proliferation of the virus. Among them, the S and N proteins are good candidates for drug development. The S protein is a large protein of ∼141 kDa, consisting of the receptor-binding S1 subunit and the membrane-fusion S2 subunit, which are generated by the cleavage of a host protease type II transmembrane serine protease (TMPRSS2).40 This cleavage is essential for the entry of the virus into the host cell. The C-terminal domain of the S1 subunit binds to the host receptor protein ACE2. Subsequently, the fusion peptide in the S2 subunit interacts with the host cell membrane, resulting in entry of the viral RNA. Blocking the receptor-binding sites of the S1 subunit can protect the host cell from invasion by the virus. Finally, the N protein is a phosphoprotein that packages the viral RNA genome into a ribonucleoprotein complex to protect genomic RNA and is the most abundant viral protein in the infected cell.41 It forms a dimer through the C-terminal domains, constituting the basic building block of the nucleocapsid. It also plays an important role in enhancing the efficiency of sgRNA transcription and viral replication.41
Cloning of the SARS-CoV-2 Genes, Expression in E. coli, and Purification of the Recombinant Proteins
The genome of SARS-CoV-2 was analyzed for prediction of the properties of the translation products. The prediction of the topography (e.g., signal sequences and transmembrane regions) and the secondary structural elements was performed on the server XtalPred-RF.72 The gene for each protein was codon-optimized and commercially synthesized for protein production in E. coli. We excluded the signal sequences and transmembrane segments from the synthetic genes. We did not synthesize six genes: one encoding NSP6, which possesses eight transmembrane regions, and five that are small proteins/peptides, including the envelope protein E, ORF3b, ORF6, ORF7b, and ORF10. In addition to the full-length versions of the synthetic genes, we also synthesized six additional DNA stretches for the individual proteins comprising the three large polypeptides: for NSP3, the N-terminal ARH domain, the PLpro domain, and the C-terminal domain; for NSP12, the RdRp domain; and for the S protein, the N-terminal S1 subunit and the C-terminal S2 subunit. The M and S proteins are believed to be glycoproteins, and the N protein is a phosphoprotein. These proteins are produced in E. coli without the post-translational modifications.
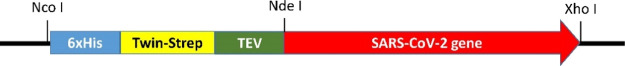
We modified the expression vector pET-28a(+) by replacing the thrombin-cleavage site and the T7-tag with Twin-Strep-tag and the TEV-cleavage site (Figure 2). We found that this variation of the plasmid allows for a better purification of the recombinant proteins. The TEV-mediated cleavage in many cases has been cleaner than that of thrombin, which aids purification of the target proteins to homogeneity. The resulting vector was named as pETHST (Figure S1). This vector contains the sites for restriction enzymes NdeI and XhoI, where the desired genes would be inserted. The corresponding resulting proteins have N-terminal His- and Twin-Strep-tags, which will be used for protein purification, but the tags are then removed by treatment with the TEV protease. GenScript prepared the individual synthetic genes in the pUC57-Kan plasmid for each case. The sequences of the synthetic DNA were confirmed. All synthetic DNA and pETHST were purified on agarose gels after digestion with NdeI and XhoI. Each DNA was ligated into pETHST, followed by transformation of E. coli DH5α with the ligation product. The sizes of the resulting plasmids from the transformants were confirmed by digestion with the restriction enzymes NdeI and XhoI in each case. The plasmids exhibiting the correct sizes were used for transformation of the LOBSTR-RIL strain, derived from E. coli BL21(DE3), to express the proteins by IPTG induction.73 The LOBSTR-RIL strain provides lower contamination for the recombinant His-tagged proteins than its BL21(DE3) background, since it has a deletion of the natural His-rich proteins ArnA and SlyD. The expression of the gene is typically induced with 0.4 mM IPTG at an OD600 of 0.6 for the growth of the cultures, followed by further incubation either at 37 °C for 2 h or at 16 °C for 18 h. In general, the latter method is utilized for superior expression of the proteins. Some proteins show better expression as a soluble form for a short period of gene expression at 37 °C, notwithstanding the fact that most of the proteins are precipitated as inclusion bodies. Therefore, we routinely test both conditions before selecting a suitable one for each protein. The expression in each case was checked by SDS-PAGE (Figure S2). The level of the soluble protein in each construct is listed in Table 2.
Figure 2.
A SARS-CoV-2 gene is preceded by a 6xHis tag, a Twin-Strep tag, and a TEV cleavage site in pETHST. All synthetic SARS-CoV-2 genes except for the nsp1 were inserted between NdeI and XhoI.
Table 2. Expression Levels of 6xHis-Twin-Strep-Tagged SARS-CoV-2 Proteins in E. coli LOBSTR-RIL.
| protein | expression levela |
|---|---|
| NSP1 | soluble: L, 16; M, 37 |
| insoluble: N, 16; M, 37 | |
| NSP2 | soluble: M, 16; L, 37 |
| insoluble: N, 16; H, 37 | |
| NSP3 | soluble: L, 16; L, 37 |
| insoluble: N, 16; N, 37 | |
| NSP3 ARH | soluble: M, 16; L, 37 |
| insoluble: N, 16; H, 37 | |
| NSP3 PLpro | soluble: L, 16; N, 37 |
| insoluble: L, 16; H, 37 | |
| NSP3C | soluble: H, 16; L, 37 |
| insoluble: L, 16; H, 37 | |
| NSP4N | soluble: L, 16; N, 37 |
| insoluble: M, 16; H, 37 | |
| NSP4C | soluble: L, 16; L, 37 |
| insoluble: N, 16; H, 37 | |
| NSP5 | soluble: N, 16; M, 37 |
| insoluble: N, 16; L, 37 | |
| NSP7 | soluble: L, 16; H, 37 |
| insoluble: N, 16; L, 37 | |
| NSP8 | soluble: M, 16; H, 37 |
| insoluble: L, 16; L, 37 | |
| NSP9 | soluble: L, 16; H, 37 |
| insoluble: N, 16; L, 37 | |
| NSP10 | soluble: M, 16; L, 37 |
| insoluble: N, 16; H, 37 | |
| NSP12 | soluble: L, 16; L, 37 |
| insoluble: H, 16; H, 37 | |
| NSP12P | soluble: L, 16; L, 37 |
| insoluble: H, 16; H, 37 | |
| NSP13 | soluble: M, 16; N, 37 |
| insoluble: H, 16; H, 37 | |
| NSP14 | soluble: L, 16; N, 37 |
| insoluble: L, 16; H, 37 | |
| NSP15 | soluble: L; 16; L, 37 |
| insoluble: N, 16; L, 37 | |
| NSP16 | soluble: M, 16; M, 37 |
| insoluble: L, 16; H, 37 | |
| S | soluble: L, 16; N, 37 |
| insoluble: H, 16; H, 37 | |
| S1 | soluble: L, 16; N, 37 |
| insoluble: H, 16; H, 37 | |
| S2 | soluble: L, 16; N, 37 |
| insoluble: M, 16; H, 37 | |
| ORF3a | soluble: M, 16; L, 37 |
| insoluble: L, 16; H, 37 | |
| M | soluble: L, 16; N, 37 |
| insoluble: L, 16; H, 37 | |
| ORF7a | soluble: L, 16; L, 37 |
| insoluble: L, 16; H, 37 | |
| ORF8 | soluble: L, 16; N, 37 |
| insoluble: M, 16; H, 37 | |
| ORF9b | soluble: L, 16; M, 37 |
| insoluble: N, 16; M, 37 | |
| N | soluble: L, 16; L, 37 |
| insoluble: M, 16; H, 37 | |
| NS | soluble: M, 16; H, 37 |
| insoluble: N, 16; M, 37 |
L, low; M, moderate; H, high; N, none; 16 for 16 °C; and 37 for 37 °C. For example, L, 16 indicates low expression at 16 °C.
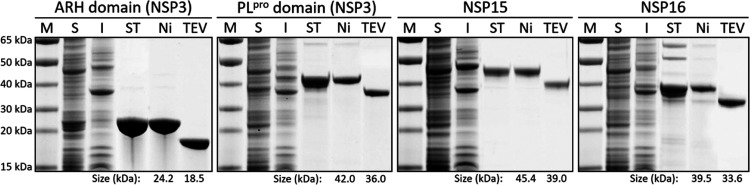
The following is a typical procedure for purification of the recombinant proteins to homogeneity. Subsequent to gene expression, the simultaneously 6xHis- and Twin-Strep-tagged proteins are purified in two-step affinity chromatography on the Strep-Tactin Sepharose resin (IBA Lifesciences) by eluting them with desthiobiotin, followed by a second purification step using the Protino Ni-NTA agarose resin (Macherey-Nagel) and elution with imidazole, following the manufacturer’s instructions (Figure S3A,B for the ARH domain). The purity of the proteins is routinely checked by SDS-PAGE. In the vast majority of the cases where we have used this procedure, the resultant purified protein is homogeneous. The resultant homogenous preparation was then treated with 6xHis-tagged TEV protease (MCLAB) at 4 °C for 24 h or longer if necessary, in order to cleave the 6xHis- and Twin-Strep-tags. The tag-free SARS-CoV-2 proteins were separated from the cleaved tags and the 6xHis-tagged TEV protease by treatment with the Protino Ni-NTA agarose resin (Figure S3C for the ARH domain). This procedure yielded in all cases a homogenous SARS-CoV-2 protein, as documented for four of the proteins in Figure 3.
Figure 3.
Purification of the N-terminal ARH and PLpro domains of NSP3, NSP15, and NSP16 as representatives. M, molecular weight marker; S, the soluble fraction of the cell extract; I, the insoluble fraction of the cell extract; ST, Strep-Tactin affinity chromatography; Ni, Ni-NTA affinity chromatography; and TEV, cleavage with TEV protease. The protein sizes before and after TEV-protease treatment are indicated at the bottom.
Computational Studies of the ARH Domain of SARS-CoV-2
The purpose for the purification of the target proteins is their use in in silico and in vitro screening for inhibitor discovery. As a representative example for this combined approach, we will discuss the discovery of initial hits for the inhibition of the NSP3 ARH domain. NSP3 is, with 1920 amino acids, the largest protein encoded by the SARS-CoV-2 genome and consists of multiple domains.29 Its function is to evade the post-translational ADP ribosylation that is part of the innate immune response in mammalian cells to viral infection.74,75 RNA viruses, including coronaviruses, counter this protective step by the host by their ARH (ARH domain) activity.75−77 This function has been shown in vivo to be essential for pathogenesis of coronavirus infection and that of other RNA viruses.76,78−80 In light of the presence of the ARH domain in coronaviruses and other viruses, the ARH activity is likely a broad-spectrum target for antiviral compound discovery.75,81
Multiple crystal structures for the ARH domain have been solved with resolutions in the range of 0.95–2.50 Å. They demonstrate that this domain recognizes ADP-ribose, the product of reversal of ADP-ribosylation, in a C-shaped pocket spanning roughly 15 Å. The binding site can be parsed into three subpockets (Figure 4B; yellow dotted highlights), which recognize adenosine, the pyrophosphate, and ribose moieties (from left to right). The co-crystalized ligand binds poorly (Ki = 10 μM) but maps out the entire binding site (Figure 4A,B).81
Figure 4.
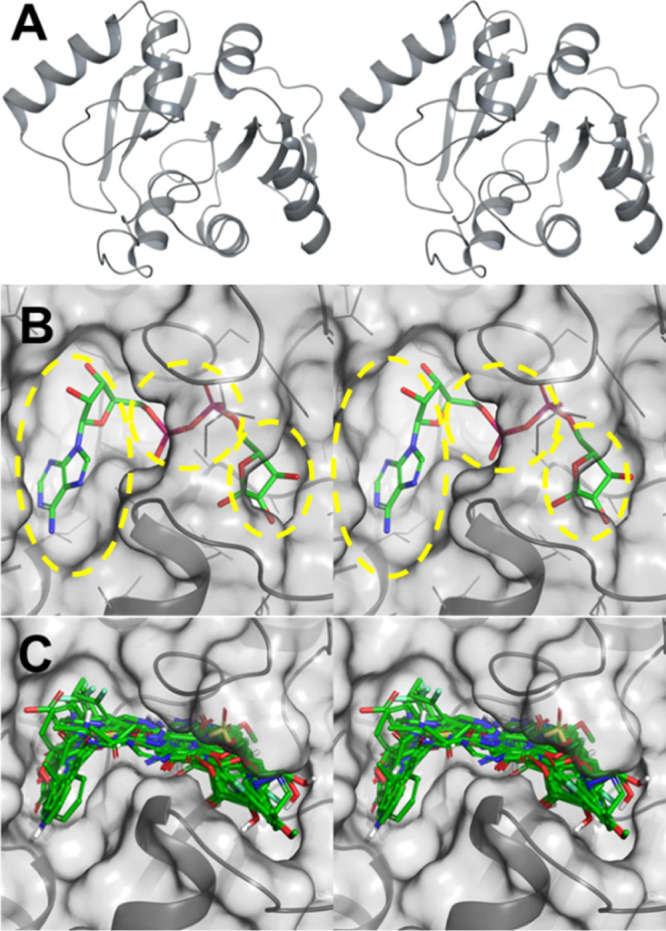
ARH domain of the NSP3 protein of SARS-CoV-2. (A) Stereoview of the ribbon representation of the ARH domain, with the active site in the center. (B) Stereo view of the X-ray structure of the ARH domain depicted as a solvent-accessible surface in complex with ADP-ribose (PDB code 6W02). The adenosine, pyrophosphate, and ribose (from left to right) binding subpockets are shown by yellow broken highlights. (C) Stereoview of the superimposition of 21 top-ranking compounds docked to the NSP3 ARH domain active site (PDB code 6WCF; resolution of 1.06 Å), demonstrating sampling of all three subpockets for binding.
The structure-based virtual screening (SBVS) used the high-resolution X-ray crystal structure of the ARH domain (PDB ID 6WCF, 1.06 Å resolution);18 in workflow, a hierarchical virtual screening workflow with Glide (HTVS, SP, and XP)22−24 and AutoDock Vina25 (see the Materials and Methods for details) was used. The final selection of the compounds was conducted by visual inspection of the binding interactions, consensus of the ranking by Glide SP, Glide XP, and AutoDock Vina and the availability of the compounds from commercial vendors. Figure 4C shows an overlay of 21 top-scoring compounds from virtual screening. The docked structures span all three binding pockets of the active site of the ARH domain. A set of 119 compounds were selected for purchase and for experimental testing.
Experimental studies of this first set of compounds identified seven binders to the ARH domain (see Figure S4 in the Supporting Information). Hit expansion of three of the experimentally validated binders by a similarity search in ZINC15 was conducted to identify commercially available compounds in the neighboring chemical space. This resulted in a focused library of 515 compounds. These compounds were further docked to the ARH binding pocket with Glide XP. After visual inspection as described above, 82 analogues were purchased for in vitro evaluation, 13 of which (see Figure S5 in the Supporting Information) were found to be experimentally validated binders.
Assessment of Binding of the Compounds to the ARH Domain of NSP3
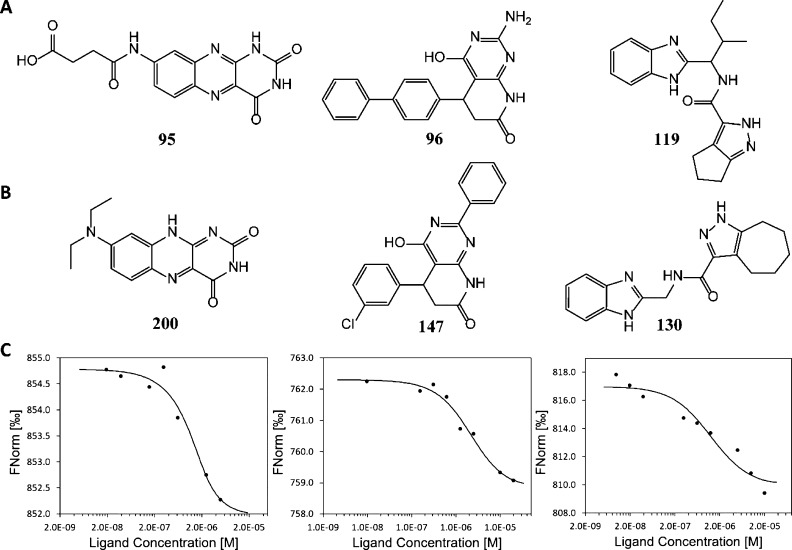
As indicated above, a set of 119 compounds from the top-scoring molecules was selected for purchase. They were tested experimentally for binding to the ARH domain by MST. Each compound was prepared as a 10 mM stock solution in DMSO. The compounds were tested in groups of 10 (each at 1 mM in DMSO) in the first-pass experiments (Table S2). Each group was then diluted to 50 μM with the assay buffer (1× PBS, 0.05% Tween-20). The fluorescent-labeled ARH domain was diluted to 10 nM with the assay buffer. Prior to MST determination, all diluted solutions were centrifuged to remove any precipitate that might have existed. The ARH domain was then mixed in 1:1 ratio either with the assay buffer containing 5% DMSO as a control or with the compound mixtures. The binding of the compound groups was determined by measuring the MST traces with the Monolith NT.115 Pico. Any group showing binding with a signal-to-noise (S/N) ratio higher than 5.0 was selected for examination of the individual compounds. Binding was observed for compounds 95, 96, 98, 112, 114, 116, and 119 (Table S3, and Figure S4 for the chemical structures). Compounds 95, 96, and 119 were found to have an S/N ratio >10 and were used to determine the values of their dissociation constants (Kd) (Figure 5A). For this purpose, higher concentration (800 μM in 8% DMSO) of each compound was used as an initial concentration for 12 twofold serial dilutions with the assay buffer containing 8% DMSO. Each dilution was centrifuged at 14,000g for 5 min prior to mixing with 10 nM ARH domain in 1:1 ratio, resulting in 400 μM, the highest compound concentration in the assay mixture containing 4% DMSO. The binding affinities of three compounds were determined with the Monolith NT.115 Pico by monitoring the MST traces, and their Kd values were calculated using MO.Affinity software. We were able to obtain the Kd values of the compounds 95, 96, and 119 as 45.0 ± 1.8, 65.4 ± 3.8, and 36.3 ± 2.0 μM, respectively.
Figure 5.
Structures of compounds and the dose–response plots of compounds for the ARH domain of NSP3. (A) Chemical structures of three compounds in the first set; (B) structures of three derivatives in the second set; and (C) dose–response plots of MST for compounds 200, 147, and 130 from left to right.
These binders were subjected to ligand expansion, as described above, which led to a second set of 82 compounds (groups 13–21 in Table S2) that were purchased from MolPort Inc. (Latvia). Their binding to NSP3-ARH was examined as described for the first set. Among them, 13 compounds exhibited binding to NSP3-ARH (Table S4, and Figure S5 for the chemical structures). Three compounds (123, 138, and 200) were derived from 95; four compounds (147, 173, 175, and 190) were derived from 96; and six compounds (121, 122, 127, 128, 129, and 130) were derived from 119. The binding affinity of the individual compound to the ARH domain was determined by MST, as described above, with the exception that the highest concentration of the compounds was 20 μM in 4% DMSO. We could evaluate the Kd values for five of these compounds: two analogues of 95 (123, 1.64 ± 0.70 μM; 200, 0.45 ± 0.28 μM), one analoguw of 96 (147, 1.77 ± 0.25 μM), and two analogues of 119 (128, 3.16 ± 2.42 μM; 130, 1.26 ± 1.01 μM). Figure 5B,C shows the chemical structures and the dose–response plots of the best analogues (200, 147, and 130) of each parental compound. These hits include one submicromolar and four low-micromolar binders to the ARH domain of NSP3.
The work described here outlines the means to produce disparate SARS-CoV-2 proteins and their use in discovery of potential inhibitors for each, as demonstrated for NSP3-ARH as a proof of concept. These hits form the basis for medicinal chemistry development around the structural templates in search of antivirals conceived expressly for SARS-CoV-2. The field is open for such discovery efforts for most of these viral targets.
Glossary
Abbreviations
- ARH
ADP-ribosylhydrolase
- COVID-19
coronavirus disease of 2019
- MST
microscale thermophoresis
- NSP
nonstructural protein
- ORF
open reading frame
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- TEV
tobacco etch virus
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c02984.
List of the synthetic genes, list of the compounds, compounds exhibiting the interaction with the ARH domain of NSP3, pETHST map carrying the SARS-CoV-2 gene, SDS-PAGE gels for expression of SARS-CoV-2 proteins, SDS-PAGE gels for the purification of the ARH domain of NSP3, and chemical structures of the compounds (PDF)
Author Present Address
† 1910 Genetics, 700 Main St, Cambridge, MA 0213
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Zhu N.; Zhang D.; Wang W.; Li X.; Yang B.; Song J.; Zhao X.; Huang B.; Shi W.; Lu R.; Niu P.; Zhan F.; Ma X.; Wang D.; Xu W.; Wu G.; Gao G. F.; Tan W.; China Novel Coronavirus I.; Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.; Wang Y.; Li X.; Ren L.; Zhao J.; Hu Y.; Zhang L.; Fan G.; Xu J.; Gu X.; Cheng Z.; Yu T.; Xia J.; Wei Y.; Wu W.; Xie X.; Yin W.; Li H.; Liu M.; Xiao Y.; Gao H.; Guo L.; Xie J.; Wang G.; Jiang R.; Gao Z.; Jin Q.; Wang J.; Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.; Zhao S.; Yu B.; Chen Y.-M.; Wang W.; Song Z.-G.; Hu Y.; Tao Z.-W.; Tian J.-H.; Pei Y.-Y.; Yuan M.-L.; Zhang Y.-L.; Dai F.-H.; Liu Y.; Wang Q.-M.; Zheng J.-J.; Xu L.; Holmes E. C.; Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D.; Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020, 91, 157–172. 10.46692/9781447360377.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- https://www.worldometers.info/coronavirus/ (July 1, 2021).
- Chan J. F.-W.; Kok K.-H.; Zhu Z.; Chu H.; To K. K.-W.; Yuan S.; Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes Infect. 2020, 9, 221–236. 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.; Lee J.-Y.; Yang J.-S.; Kim J. W.; Kim V. N.; Chang H. The architecture of SARS-CoV-2 transcriptome. Cell 2020, 181, 914–921. 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P.; Yang X.-L.; Wang X.-G.; Hu B.; Zhang L.; Zhang W.; Si H.-R.; Zhu Y.; Li B.; Huang C.-L.; Chen H.-D.; Chen J.; Luo Y.; Guo H.; Jiang R.-D.; Liu M.-Q.; Chen Y.; Shen X.-R.; Wang X.; Zheng X.-S.; Zhao K.; Chen Q.-J.; Deng F.; Liu L.-L.; Yan B.; Zhan F.-X.; Wang Y.-Y.; Xiao G.-F.; Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz J. H. Hypothesis: angiotensin-converting enzyme inhibitors and angiotensin receptor blockers may increase the risk of severe COVID-19. J. Travel Med. 2020, 27, taaa041. 10.1093/jtm/taaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Guo Y.; Pan Y.; Zhao Z. J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020, 525, 135–140. 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Krüger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N.-H.; Nitsche A.; Müller M. A.; Drosten C.; Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R.; Zhang Y.; Li Y.; Xia L.; Guo Y.; Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X.; Liu Y.; Lei X.; Li P.; Mi D.; Ren L.; Guo L.; Guo R.; Chen T.; Hu J.; Xiang Z.; Mu Z.; Chen X.; Chen J.; Hu K.; Jin Q.; Wang J.; Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tok T. T.; Tatar G. Structures and functions of coronavirus proteins: Molecular modeling of viral nucleoprotein. Int. J. Virol. Infect. Dis. 2017, 2, 001–006. [Google Scholar]
- Ghosh A. K.; Brindisi M.; Shahabi D.; Chapman M. E.; Mesecar A. D. Drug development and medicinal chemistry efforts toward SARS-coronavirus and COVID-19 therapeutics. ChemMedChem 2020, 15, 907–932. 10.1002/cmdc.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling T.; Irwin J. J. ZINC 15–ligand discovery for everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LigPrep; Schrödinger, LLC: New York, NY, 2020.
- Michalska K.; Kim Y.; Jedrzejczak R.; Maltseva N. I.; Stols L.; Endres M.; Joachimiak A. Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes. IUCrJ 2020, 7, 814–824. 10.1107/s2052252520009653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouley R.; Kumarasiri M.; Peng Z.; Otero L. H.; Song W.; Suckow M. A.; Schroeder V. A.; Wolter W. R.; Lastochkin E.; Antunes N. T.; Pi H.; Vakulenko S.; Hermoso J. A.; Chang M.; Mobashery S. Discovery of antibiotic (E)-3-(3-carboxyphenyl)-2-(4-cyanostyryl) quinazolin-4 (3H)-one. J. Am. Chem. Soc. 2015, 137, 1738–1741. 10.1021/jacs.5b00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y.; Liu J.; Estiu G.; Isin B.; Ahn Y.-Y.; Lee D.-S.; Barabasi A.-L.; Kapatral V.; Wiest O.; Oltvai Z. N. Blueprint for antimicrobial hit discovery targeting metabolic networks. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 1082–1087. 10.1073/pnas.0909181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen S. T.; Liu J.; Estiu G.; Oltvai Z. N.; Wiest O. Identification of novel bacterial histidine biosynthesis inhibitors using docking, ensemble rescoring, and whole-cell assays. Bioorg. Med. Chem. 2010, 18, 5148–5156. 10.1016/j.bmc.2010.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner R. A.; Banks J. L.; Murphy R. B.; Halgren T. A.; Klicic J. J.; Mainz D. T.; Repasky M. P.; Knoll E. H.; Shelley M.; Perry J. K.; Shaw D. E.; Francis P.; Shenkin P. S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- Halgren T. A.; Murphy R. B.; Friesner R. A.; Beard H. S.; Frye L. L.; Pollard W. T.; Banks J. L. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004, 47, 1750–1759. 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Friesner R. A.; Murphy R. B.; Repasky M. P.; Frye L. L.; Greenwood J. R.; Halgren T. A.; Sanschagrin P. C.; Mainz D. T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein – ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S.; Wong G.; Shi W.; Liu J.; Lai A. C. K.; Zhou J.; Liu W.; Bi Y.; Gao G. F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016, 24, 490–502. 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J.; Li F.; Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing E.; Gallagher T. SARS coronavirus redux. Trends Immunol. 2020, 41, 271–273. 10.1016/j.it.2020.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J.; Kusov Y.; Hilgenfeld R. NSP3 of coronaviruses: Structures and functions of a large multi-domain protein. Antiviral Res. 2018, 149, 58–74. 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos Y. M.; St. John S. E.; Mesecar A. D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015, 115, 21–38. 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T. P.; Sims A. C.; Leist S. R.; Schäfer A.; Won J.; Brown A. J.; Montgomery S. A.; Hogg A.; Babusis D.; Clarke M. O.; Spahn J. E.; Bauer L.; Sellers S.; Porter D.; Feng J. Y.; Cihlar T.; Jordan R.; Denison M. R.; Baric R. S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020, 11, 222. 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.; Zhou Q.; Li Y.; Garner L. V.; Watkins S. P.; Carter L. J.; Smoot J.; Gregg A. C.; Daniels A. D.; Jervey S.; Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020, 6, 315–331. 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T.; Takemoto C.; Kim Y.-T.; Wang H.; Nishii W.; Terada T.; Shirouzu M.; Yokoyama S. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl. Acad. Sci. U.S.A. 2016, 113, 12997–13002. 10.1073/pnas.1601327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S.; Shen H.; Wang J.; Tay F. P. L.; Liu D. X. Functional and genetic studies of the substrate specificity of coronavirus infectious bronchitis virus 3C-like proteinase. J. Virol. 2010, 84, 7325–7336. 10.1128/jvi.02490-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R. N.; Ward A. B. Structure of the SARS-CoV NSP12 polymerase bound to NSP7 and NSP8 co-factors. Nat. Commun. 2019, 10, 2342. 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton G.; Fry E.; Carter L.; Sainsbury S.; Walter T.; Nettleship J.; Berrow N.; Owens R.; Gilbert R.; Davidson A.; Siddell S.; Poon L. L. M.; Diprose J.; Alderton D.; Walsh M.; Grimes J. M.; Stuart D. I. The NSP9 replicase protein of SARS-coronavirus, structure and functional insights. Structure 2004, 12, 341–353. 10.1016/j.str.2004.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scavone C.; Brusco S.; Bertini M.; Sportiello L.; Rafaniello C.; Zoccoli A.; Berrino L.; Racagni G.; Rossi F.; Capuano A. Current pharmacological treatments for COVID-19: What’s next?. Br. J. Pharmacol. 2020, 177, 4813–4824. 10.1111/bph.15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman R. T.; Roth J. S.; Brimacombe K. R.; Simeonov A.; Shen M.; Patnaik S.; Hall M. D. Remdesivir: A review of its discovery and development leading to emergency use authorization for treatment of COVID-19. ACS Cent. Sci. 2020, 6, 672–683. 10.1021/acscentsci.0c00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E.; Debarnot C.; Ferron F.; Bouvet M.; Coutard B.; Imbert I.; Gluais L.; Papageorgiou N.; Sharff A.; Bricogne G.; Ortiz-Lombardia M.; Lescar J.; Canard B. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2’-O-methyltransferase NSP10/NSP16 complex. PLoS Pathog. 2011, 7, e1002059 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016, 3, 237–261. 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-k.; Hou M.-H.; Chang C.-F.; Hsiao C.-D.; Huang T.-h. The SARS coronavirus nucleocapsid protein - Forms and functions. Antiviral Res. 2014, 103, 39–50. 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K. G.; Narayanan K.; Huang C.; Makino S. Severe acute respiratory syndrome coronavirus protein NSP1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 2012, 86, 13598–13608. 10.1128/jvi.01958-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillez-Ty C. T.; Liao L.; Yates J. R.; Kuhn P.; Buchmeier M. J. Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling. J. Virol. 2009, 83, 10314–10318. 10.1128/jvi.00842-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- J Alsaadi E. A.; Jones I. M. Membrane binding proteins of coronaviruses. Future Virol. 2019, 14, 275–286. 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z.; Du X.; Xu Y.; Deng Y.; Liu M.; Zhao Y.; Zhang B.; Li X.; Zhang L.; Peng C.; Duan Y.; Yu J.; Wang L.; Yang K.; Liu F.; Jiang R.; Yang X.; You T.; Liu X.; Yang X.; Bai F.; Liu H.; Liu X.; Guddat L. W.; Xu W.; Xiao G.; Qin C.; Shi Z.; Jiang H.; Rao Z.; Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Jin Z.; Zhao Y.; Sun Y.; Zhang B.; Wang H.; Wu Y.; Zhu Y.; Zhu C.; Hu T.; Du X.; Duan Y.; Yu J.; Yang X.; Yang X.; Yang K.; Liu X.; Guddat L. W.; Xiao G.; Zhang L.; Yang H.; Rao Z. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat. Struct. Mol. Biol. 2020, 27, 529–532. 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Lin D.; Sun X.; Curth U.; Drosten C.; Sauerhering L.; Becker S.; Rox K.; Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science 2020, 368, 409–412. 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W.; Zhang B.; Jiang X.-M.; Su H.; Li J.; Zhao Y.; Xie X.; Jin Z.; Peng J.; Liu F.; Li C.; Li Y.; Bai F.; Wang H.; Cheng X.; Cen X.; Hu S.; Yang X.; Wang J.; Liu X.; Xiao G.; Jiang H.; Rao Z.; Zhang L.-K.; Xu Y.; Yang H.; Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 2020, 368, 1331–1335. 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W.; Mao C.; Luan X.; Shen D.-D.; Shen Q.; Su H.; Wang X.; Zhou F.; Zhao W.; Gao M.; Chang S.; Xie Y.-C.; Tian G.; Jiang H.-W.; Tao S.-C.; Shen J.; Jiang Y.; Jiang H.; Xu Y.; Zhang S.; Zhang Y.; Xu H. E. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504. 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Yan L.; Huang Y.; Liu F.; Zhao Y.; Cao L.; Wang T.; Sun Q.; Ming Z.; Zhang L.; Ge J.; Zheng L.; Zhang Y.; Wang H.; Zhu Y.; Zhu C.; Hu T.; Hua T.; Zhang B.; Yang X.; Li J.; Yang H.; Liu Z.; Xu W.; Guddat L. W.; Wang Q.; Lou Z.; Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet M.; Lugari A.; Posthuma C. C.; Zevenhoven J. C.; Bernard S.; Betzi S.; Imbert I.; Canard B.; Guillemot J.-C.; Lécine P.; Pfefferle S.; Drosten C.; Snijder E. J.; Decroly E.; Morelli X. Coronavirus NSP10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014, 289, 25783–25796. 10.1074/jbc.m114.577353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. E.; Jang G. M.; Bouhaddou M.; Xu J.; Obernier K.; White K. M.; O’Meara M. J.; Rezelj V. V.; Guo J. Z.; Swaney D. L.; Tummino T. A.; Hüttenhain R.; Kaake R. M.; Richards A. L.; Tutuncuoglu B.; Foussard H.; Batra J.; Haas K.; Modak M.; Kim M.; Haas P.; Polacco B. J.; Braberg H.; Fabius J. M.; Eckhardt M.; Soucheray M.; Bennett M. J.; Cakir M.; McGregor M. J.; Li Q.; Meyer B.; Roesch F.; Vallet T.; Mac Kain A.; Miorin L.; Moreno E.; Naing Z. Z. C.; Zhou Y.; Peng S.; Shi Y.; Zhang Z.; Shen W.; Kirby I. T.; Melnyk J. E.; Chorba J. S.; Lou K.; Dai S. A.; Barrio-Hernandez I.; Memon D.; Hernandez-Armenta C.; Lyu J.; Mathy C. J. P.; Perica T.; Pilla K. B.; Ganesan S. J.; Saltzberg D. J.; Rakesh R.; Liu X.; Rosenthal S. B.; Calviello L.; Venkataramanan S.; Liboy-Lugo J.; Lin Y.; Huang X.-P.; Liu Y.; Wankowicz S. A.; Bohn M.; Safari M.; Ugur F. S.; Koh C.; Savar N. S.; Tran Q. D.; Shengjuler D.; Fletcher S. J.; O’Neal M. C.; Cai Y.; Chang J. C. J.; Broadhurst D. J.; Klippsten S.; Sharp P. P.; Wenzell N. A.; Kuzuoglu-Ozturk D.; Wang H.-Y.; Trenker R.; Young J. M.; Cavero D. A.; Hiatt J.; Roth T. L.; Rathore U.; Subramanian A.; Noack J.; Hubert M.; Stroud R. M.; Frankel A. D.; Rosenberg O. S.; Verba K. A.; Agard D. A.; Ott M.; Emerman M.; Jura N.; von Zastrow M.; Verdin E.; Ashworth A.; Schwartz O.; d’Enfert C.; Mukherjee S.; Jacobson M.; Malik H. S.; Fujimori D. G.; Ideker T.; Craik C. S.; Floor S. N.; Fraser J. S.; Gross J. D.; Sali A.; Roth B. L.; Ruggero D.; Taunton J.; Kortemme T.; Beltrao P.; Vignuzzi M.; García-Sastre A.; Shokat K. M.; Shoichet B. K.; Krogan N. J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn D.-G.; Choi J.-K.; Taylor D. R.; Oh J.-W. Biochemical characterization of a recombinant SARS coronavirus NSP12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012, 157, 2095–2104. 10.1007/s00705-012-1404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z.; Yan L.; Ren Z.; Wu L.; Wang J.; Guo J.; Zheng L.; Ming Z.; Zhang L.; Lou Z.; Rao Z. Delicate structural coordination of the severe acute respiratory syndrome coronavirus NSP13 upon ATP hydrolysis. Nucleic Acids Res. 2019, 47, 6538–6550. 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K.; Palaninathan S.; Alcantara J. M. O.; Li Yi L.; Guarino L.; Sacchettini J. C.; Kao C. C. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease NSP15. J. Biol. Chem. 2008, 283, 3655–3664. 10.1074/jbc.m708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.; Jedrzejczak R.; Maltseva N. I.; Wilamowski M.; Endres M.; Godzik A.; Michalska K.; Joachimiak A. Crystal structure of NSP15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020, 29, 1596–1605. 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A. C.; Park Y.-J.; Tortorici M. A.; Wall A.; McGuire A. T.; Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Wang F.; Shen C.; Peng W.; Li D.; Zhao C.; Li Z.; Li S.; Bi Y.; Yang Y.; Gong Y.; Xiao H.; Fan Z.; Tan S.; Wu G.; Tan W.; Lu X.; Fan C.; Wang Q.; Liu Y.; Zhang C.; Qi J.; Gao G. F.; Gao F.; Liu L. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020, 368, 1274–1278. 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J.; Ge J.; Yu J.; Shan S.; Zhou H.; Fan S.; Zhang Q.; Shi X.; Wang Q.; Zhang L.; Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Yuan M.; Wu N. C.; Zhu X.; Lee C.-C. D.; So R. T. Y.; Lv H.; Mok C. K. P.; Wilson I. A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 2020, 368, 630–633. 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.; Ye G.; Shi K.; Wan Y.; Luo C.; Aihara H.; Geng Q.; Auerbach A.; Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K. L.; Yuen K. S.; Castano-Rodriguez C.; Ye Z. W.; Yeung M. L.; Fung S. Y.; Yuan S.; Chan C. P.; Yuen K. Y.; Enjuanes L.; Jin D. Y. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019, 33, 8865–8877. 10.1096/fj.201802418r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu Y. L.; Teoh K. T.; Lo J.; Chan C. M.; Kien F.; Escriou N.; Tsao S. W.; Nicholls J. M.; Altmeyer R.; Peiris J. S. M.; Bruzzone R.; Nal B. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008, 82, 11318–11330. 10.1128/jvi.01052-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D.; Fielding B. C. Coronavirus envelope protein: current knowledge. Virol. J. 2019, 16, 69. 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M.; Yount B.; Heise M.; Kopecky-Bromberg S. A.; Palese P.; Baric R. S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007, 81, 9812–9824. 10.1128/jvi.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. K.; Coleman C. M.; Postel S.; Sisk J. M.; Bernbaum J. G.; Venkataraman T.; Sundberg E. J.; Frieman M. B. Severe acute respiratory syndrome coronavirus ORF7a inhibits bone marrow stromal antigen 2 virion tethering through a novel mechanism of glycosylation interference. J. Virol. 2015, 89, 11820–11833. 10.1128/jvi.02274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaecher S. R.; Mackenzie J. M.; Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J. Virol. 2007, 81, 718–731. 10.1128/jvi.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S. K. P.; Feng Y.; Chen H.; Luk H. K. H.; Yang W.-H.; Li K. S. M.; Zhang Y.-Z.; Huang Y.; Song Z.-Z.; Chow W.-N.; Fan R. Y. Y.; Ahmed S. S.; Yeung H. C.; Lam C. S. F.; Cai J.-P.; Wong S. S. Y.; Chan J. F. W.; Yuen K.-Y.; Zhang H.-L.; Woo P. C. Y. Severe acute respiratory syndrome (SARS) coronavirus ORF8 protein is acquired from SARS-related coronavirus from greater horseshoe bats through recombination. J. Virol. 2015, 89, 10532–10547. 10.1128/jvi.01048-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.-S.; Nabar N. R.; Huang N.-N.; Kehrl J. H. SARS-coronavirus open reading frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discovery 2019, 5, 101. 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.-S.; Qi H.-Y.; Boularan C.; Huang N.-N.; Abu-Asab M.; Shelhamer J. H.; Kehrl J. H. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J. Immunol. 2014, 193, 3080–3089. 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.; Yang M.; Hong Z.; Zhang L.; Huang Z.; Chen X.; He S.; Zhou Z.; Zhou Z.; Chen Q.; Yan Y.; Zhang C.; Shan H.; Chen S. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm. Sin. B 2020, 10, 1228–1238. 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabinski L.; Jaroszewski L.; Rychlewski L.; Wilson I. A.; Lesley S. A.; Godzik A. XtalPred: A web server for prediction of protein crystallizability. Bioinformatics 2007, 23, 3403–3405. 10.1093/bioinformatics/btm477. [DOI] [PubMed] [Google Scholar]
- Andersen K. R.; Leksa N. C.; Schwartz T. U. Optimized E. coli expression strain LOBSTR eliminates common contaminants from His-tag purification. Proteins 2013, 81, 1857–1861. 10.1002/prot.24364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A. R.; Perlman S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhammad Y. M. O.; Fehr A. R. The viral macrodomain counters host antiviral ADP-ribosylation. Viruses 2020, 12, 384. 10.3390/v12040384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A. R.; Channappanavar R.; Jankevicius G.; Fett C.; Zhao J.; Athmer J.; Meyerholz D. K.; Ahel I.; Perlman S. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. mBio 2016, 7, e01721-16 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhammad Y. M. O.; Kashipathy M. M.; Roy A.; Gagné J-. P.; McDonald P.; Gao P.; Nonfoux L.; Battaile K. P.; Johnson D. K.; Holmstrom E. D.; Poirier G. G.; Lovell S.; Fehr A. R. The SARS-CoV-2 conserved macrodomain Is a mono-ADP-ribosylhydrolase. J. Virol. 2021, 95, e01969-20 10.1128/JVI.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putics A.; Filipowicz W.; Hall J.; Gorbalenya A. E.; Ziebuhr J. ADP-ribose-1’’-monophosphatase: A conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 2005, 79, 12721–12731. 10.1128/jvi.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson K. K.; Cervantes-Barragán L.; Ludewig B.; Thiel V. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1’-phosphatase, a viral function conserved in the alpha-like supergroup. J. Virol. 2008, 82, 12325–12334. 10.1128/jvi.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A. R.; Athmer J.; Channappanavar R.; Phillips J. M.; Meyerholz D. K.; Perlman S. The NSP3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J. Virol. 2015, 89, 1523–1536. 10.1128/jvi.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick D. N.; Virdi R. S.; Vuksanovic N.; Dahal N.; Silvaggi N. R. Molecular basis for ADP-ribose binding to the Mac1 domain of SARS-CoV-2 NSP3. Biochemistry 2020, 59, 2608–2615. 10.1021/acs.biochem.0c00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.