ABSTRACT
Human bocavirus (HBoV) has been recognized as an important pathogen that causes respiratory infection and acute gastroenteritis in young children worldwide. HBoV is most likely transmitted by the respiratory route and by fecal-oral transmission. Recently, HBoV has been detected in several types of environmental water and in bivalve shellfish. However, study of the existence of HBoV in oysters is still undocumented in Thailand. In this study, 144 oyster samples collected from different markets in Chiang Mai, Thailand, in 2017 and 2018 were investigated for the presence of HBoV by nested PCR and sequencing. HBoV was detected in 11 out of 144 samples (7.6%). Nine HBoV-positive samples (81.8%) were identified as genotype 1 (HBoV1) and two (18.2%) as HBoV2. A monthly investigation of HBoV in oyster samples from July 2017 to June 2018 showed that HBoV was sporadically detected in particular months spanning the rainy and colder season, with a peak in January. This study demonstrates the presence and genotype diversity of HBoV in oyster samples in Thailand. The findings contribute to evaluating the risk of foodborne transmission of HBoV and to monitoring outbreaks of HBoV in Thailand and in other countries.
IMPORTANCE Human bocavirus is recognized as an important cause of respiratory infection and of acute gastroenteritis in children worldwide. Human bocavirus has been widely detected in many clinical specimens, as well as in several types of environmental samples. Most previous studies describe the incidence of bocavirus infection in humans, whereas few data are available for the occurrence of human bocavirus in food materials, particularly that in bivalve shellfish. Our findings provide evidence for the existence and prevalence of human bocavirus in oysters, suggesting that further monitoring of the potential risk of food- and waterborne transmission of this virus to humans should be undertaken.
KEYWORDS: foodborne transmission, genotype, human bocavirus, oysters, PCR, Thailand
INTRODUCTION
Human bocavirus (HBoV) is recognized as a frequent pathogen found worldwide in children with acute respiratory infection and acute gastroenteritis. HBoV belongs to the family Parvoviridae and genus Parvovirus. HBoV is a small nonenveloped virus of approximately 25 nm in size that contains a linear single-stranded DNA (ssDNA) of about 5.3 kb in length (1). To date, four genotypes of HBoV (HBoV1 to HBoV4) have been identified, and HBoV2 is further subdivided into two subtypes, 2A and 2B. HBoV1 has been found to be a common cause of respiratory tract infection, mainly in children (2–4). HBoV2, HBoV3, and HBoV4 have generally been detected in stool samples from children and adults with acute gastroenteritis (5–7) and in many other clinical specimens, such as serum/blood, saliva, and urine (8–10). The exact role of HBoV in pathogenesis and its route of transmission remain unclear, although it is most likely transmitted by the respiratory and fecal-oral routes, since it is usually detected in respiratory and stool samples. Seroprevalence studies of HBoV from different counties worldwide have demonstrated that positive rates of HBoV infection range from 69.2% to 94%, depending on the study population (11–16). Based on research articles published worldwide between 2005 and 2016, the average prevalence of HBoV infection in respiratory secretion samples ranged from 1.0% to 56.8% and in stool specimens from 1.3% to 63%, depending on the country, geographical location, and number of subjects (17). The high seroprevalence and frequent detection of HBoV in patients with respiratory infection and gastroenteritis suggest an important etiological role of HBoV in human diseases.
Recently, HBoV has also been detected in shellfish and various types of environmental samples, including bivalve shellfish, river water, wastewater, and sewage, with variable prevalences that range from 3.7% to 81% (18–25). In addition, there was a report of patients admitted to hospital with acute diarrhea associated with consuming drinking water contaminated with HBoV (26). These reports hint that shellfish and water could be an important reservoir for HBoV. However, information on HBoV contamination in food in Thailand is still limited; in particular, that in bivalve shellfish has not yet been investigated in Thailand. In order to fill this gap, the present study addressed the detection of HBoV1 and HBoV2A in oysters collected in Thailand in 2017 and 2018.
RESULTS AND DISCUSSION
Detection of human bocavirus contamination in oysters.
For the 1-year study, a total of 144 oysters collected from three markets in Chiang Mai, Thailand, was examined for the presence of HBoV. Of 144 oysters, 48 were collected from a supermarket, 48 from local fresh market no. 1, and 48 from local fresh market no. 2. HBoV was detected in 11 out of 144 oyster samples (7.6%), as shown in Table 1. The detection rates of HBoV in oysters collected from the supermarket and from local fresh market no. 1 and no. 2 were 4.2% (2/48), 8.3% (4/48), and 10.4% (5/48), respectively. Like many other enteric viruses, HBoV can be a contaminant in food and environmental water samples. A number of surveillance studies of HBoV focused on contamination by HBoV in several kinds of environmental water samples (18, 20, 21, 23, 27, 28). However, to date, there are only a few reports describing the occurrence of HBoV in food materials. In particular, there has been only one report from Italy and one from South Africa described the detection of HBoV in bivalve shellfish (22, 25). To our knowledge, our study is the first report that demonstrates the presence of HBoV in oysters in Thailand, supporting previous studies from Italy and South Africa indicating that shellfish is an important reservoir of HBoV.
TABLE 1.
Prevalence of human bocavirus detected in oysters from different markets in Chiang Mai, Thailand, from July 2017 to June 2018
| Location | No. of samples |
% Positive | |
|---|---|---|---|
| Positive | Tested | ||
| Supermarket | 2 | 48 | 4.2 |
| Local market no. 1 | 4 | 48 | 8.3 |
| Local market no. 2 | 5 | 48 | 10.4 |
| All markets | 11 | 144 | 7.6 |
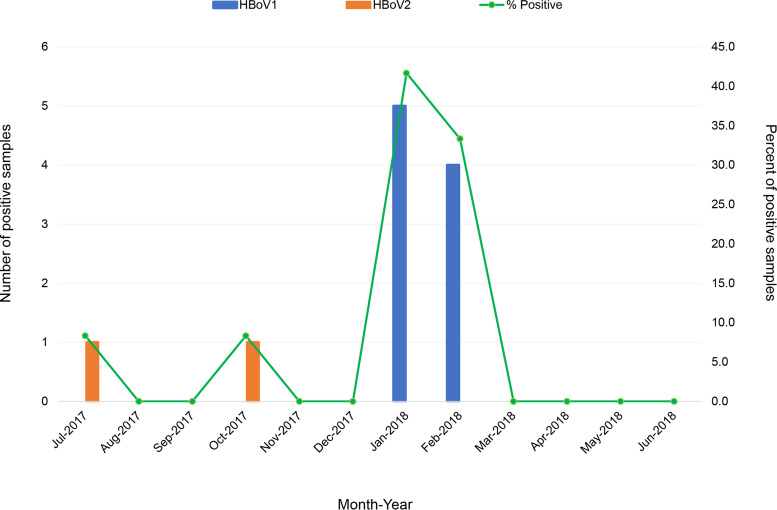
A monthly analysis of HBoV detection from July 2017 to June 2018 showed that HBoV was occasionally detected in some particular months (Fig. 1). HBoV was found in July and October of 2017 at a rate of 8.3% (1 out of 12), whereas in January and February of 2018 it was found at rates of 41.7% (5 out of 12) and 33.3% (4 out of 12), respectively. Of 11 HBoV isolates detected in this study, 9 (81.8%) were HBoV1 and 2 (18.2%) were HBoV2. It should be noted that HBoV1 was detected in January and February with high prevalence, while HBoV2 was sporadically detected in July and October. General information on seasonal distribution of HBoV in oysters is not yet available. No previous studies have investigated the prevalence of HBoV detected in oysters in different seasons. In the present study, we analyzed the monthly distribution of HBoV detected in oyster samples over a period of 1 year from July 2017 to June 2018 and revealed that HBoV was predominant in the cold months (January and February). A number of previous studies have reported that HBoV infection occurs all year round, with the peak in winter and spring months (29–32). These studies detected HBoV in many types of respiratory tract clinical samples obtained from children with airway infections. Our finding of high prevalence of HBoV in oysters during the cooler months in Thailand is in accordance with the high incidence of HBoV infections in humans in winter, suggesting a cocirculation of HBoV in humans and the environment and that shellfish might be a potential vehicle for HBoV transmission. However, to clarify whether there is a true seasonal distribution of HBoV contamination in oyster samples, further studies with larger numbers of samples and longer study periods in different geographical regions should be investigated.
FIG 1.
Monthly and genotype distribution of human bocavirus detected in oysters in Thailand from July 2017 to June 2018.
Genetic characterization of human bocavirus strains.
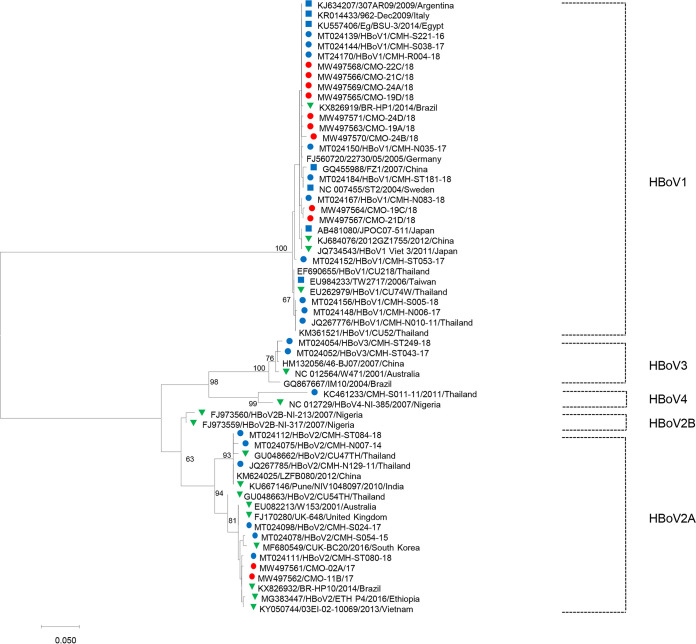
Phylogenetic analysis of partial VP1/VP2 nucleotide sequences of 11 HBoV isolates detected in oysters in this study revealed two distinct groups of HBoV with different genotypes (Fig. 2). Nine HBoV isolates detected in this study were clustered with HBoV1, found in clinical specimens from children with acute gastroenteritis and respiratory tract infection in Thailand and in many other countries worldwide, including Argentina, Italy, Brazil, Germany, Sweden, Egypt, China, Japan, and Taiwan, sharing 98.0 to 100% nucleotide sequence identity. The nucleotide sequences of the other two HBoV strains detected in oysters in this study were closely related to those of the HBoV2 strains found in fecal and respiratory samples reported previously from Australia, the United Kingdom, Brazil, China, South Korea, Vietnam, India, and Ethiopia, with nucleotide sequence identities ranging from 93.7 to 100%. Regarding the genotype diversity of HBoV detected in humans and environments, previous studies have reported that HBoV1 is commonly found in respiratory samples (2–4), while HBoV2, HBoV3, and HBoV4 are generally detected in stool samples (5–7). In shellfish, environmental, and wastewater samples, HBoV2 and HBoV3 are significantly more prevalent than HBoV1 and HBoV4 (21, 22, 24, 25). In this study, HBoV1 and HBoV2 were frequently detected in oyster samples. Detection of HBoV2 in oysters is in line with results of previous studies (22, 25). Nevertheless, the absence of HBoV3 in the present study does not rule out its presence in oyster samples, since a relatively small number of samples was investigated. Our previous study demonstrated that HBoV1, HBoV2, and HBoV3 have been circulating in children with acute diarrhea and that HBoV1 and HBoV2 are more prevalent than HBoV3 (33). It is interesting to point out that the HBoV genotypes detected in oysters are similar to those found in humans during the same period, in which both HBoV1 and HBoV2 are predominant, indicating a possible epidemiological link of HBoV infection in humans and in oysters in Thailand. In the case of pediatric patients, they may acquire the virus from drinking contaminated water rather than by consumption of contaminated oysters. This issue remains to be further investigated. Oysters are one of the predominant carriers for seafood-borne diseases because they are filter feeders that can adsorb the viral particles (34). The consumption of raw or undercooked oysters that accumulate human-pathogenic viruses is associated with a number of human diseases (35). Contamination of shellfish-growing waters with fecal pollution is also a significant concern; for example, if the water is contaminated with human feces, enteric viral pathogens and bacteria may become trapped within the oyster (36). Taken together, our findings demonstrate that HBoV circulating in oysters might be a potential source for HBoV infection in human, particularly for those individuals who consume raw oysters. To address the other potential sources of HBoV contamination, further investigation of HBoV in various sources of environmental waters should be carried out.
FIG 2.
Phylogenetic tree of the partial VP1/VP2 region (507 bp) of human bocavirus strains detected in oysters in this study (red circle) and the human bocavirus reference strains detected in fecal samples in Thailand (blue circle) and outside Thailand (blue square) and in nasopharyngeal swap/aspirates (green triangle). The scale bar indicates the number of nucleotide substitutions per site, and bootstrap values (>60) are indicated for the corresponding nodes.
In conclusion, this study reports the detection of HBoV in oyster samples in Thailand. Although additional investigations are required to confirm the actual transmission route of HBoV, data obtained from this study and from previous studies on the presence of enteric HBoV in shellfish justify further monitoring to assess the potential risk of food- and waterborne transmission of HBoV to humans.
MATERIALS AND METHODS
Oyster samples.
Oysters were collected from three markets in Chiang Mai, Thailand (1 supermarket and 2 local fresh markets), once a month throughout the study period of 1 year from July 2017 to June 2018. Each month, 12 samples (4 samples from each market) were collected. A total of 144 oyster samples were included in this study. During the process of oyster collection each month, the samples were kept on ice and transported to the laboratory. The oysters were immediately dissected to obtain the digestive tract and then washed with phosphate-buffered saline and finely chopped into small pieces. Approximately 1 g of digestive tissue of each individual oyster was obtained and was further processed for virus concentration and viral genome extraction.
Virus extraction and concentration.
Virus extraction was performed by using a previously described method (21, 35, 37) with modifications. Briefly, the digestive tissue was frozen at −80°C for 1 h. Then, the sample was thawed and mixed with an equal volume of cold glycine buffer (0.05 M glycine and 0.15 M NaCl [pH 9.0]). The mixture was mixed with thoroughly by vortexing for 5 min. Then, 10 beads and cold glycine buffer were added to make a final volume of 14 ml. The solution was incubated for 15 min at 4°C on a shaker in order to release virus from the tissue. The homogenate was subsequently centrifuged at 8,000 × g for 20 min, and the supernatant was collected for further concentration. To concentrate the virus, the supernatant was processed using a modified polyethylene glycol (PEG) precipitation method as reported previously (38, 39). Briefly, 10 ml of the supernatant was mixed with 0.8 g of PEG-6000 and 0.23 g of NaCl and magnetically stirred at 4°C overnight. Afterward, the sample was centrifuged at 10,000 × g for 30 min at 4°C. The supernatant was discarded, and the pellet was resuspended in 200 μl of RNase-free distilled water and stored at −80°C until nucleic acid extraction was performed. The viral genome was extracted from 140 μl of the concentrated samples using a spin column technique according to the manufacturer’s protocol (Qiagen, Hilden, Germany). The extracted viral genome was immediately transferred to the PCR or kept at −80°C until further use.
Detection of human bocavirus by nested PCR.
Nested PCR targeting the VP1/VP2 region of HBoV was performed to detect the presence of viral genome in oyster samples. The first-round PCR primers were AK-VP-F1 (5′-CGCCGTGGCTCCTGCTCT-3′) and AK-VP-R1 (5′-TGTTCGCCATCACAAAAGATGTG-3′), and the second-round primers were AK-VP-F2 (5′-GGCTCCTGCTCTAGGAAATAAAGAG-3′) and AK-VP-R2 (5′-CCTGCTGTTAGGTCGTTGTTGTATGT-3′) (40). PCR amplification was performed by using GoTaq DNA polymerase (Promega, WI, USA). The thermocycling condition for the first-round PCR were as follows: 94°C for 3 min prior to 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by a final extension at 72°C for 10 min. The second-round PCR was performed in the same manner as the first-round amplification, except for the annealing step, which was done at 58°C. The amplicon sizes of the first- and second-round PCR were 609 bp and 576 bp, respectively, as detected by 1.5% agarose gel electrophoresis.
One of the major concerns of the detection of viruses in environmental samples is the presence of PCR inhibitors. The process of removal of potential inhibitors of PCR was not applied in the present study, and this could be a limitation of this study. However, the known HBoV-positive and -negative stool samples were used as positive and negative controls, respectively. These control samples were run through the same process of viral genome extraction and nested PCR as that for oyster samples. The sensitivity of the detection of HBoV by nested PCR was tested using 10× serial dilutions of DNA extracted from one known HBoV-positive fecal sample and two known HBoV-positive oyster samples, with concentrations of 1,000 ng/μl, 100 ng/μl, 10 ng/μl, 1 ng/μl, and 0.1 ng/μl. The sensitivity was considered to be the lowest concentration of at least one positive sample that was positive by nested PCR. The sensitivity of the nested-PCR method for the detection of HBoV from one positive fecal sample and two oyster samples showed that HBoV was detected at concentrations as low as 10 ng/μl, 100 ng/μl, and 10 ng/μl, respectively. The data indicated that the sensitivity of nested PCR used in this study was as low as 10 ng/μl. The sensitivity of nested PCR for the detection of HBoV in oysters was comparable to those in the clinical samples, suggesting that there were no potential inhibitors contained in shellfish tissues that might affect the extraction efficiency or the PCR assay.
Sequencing and phylogenetic analysis.
The nested-PCR products were purified using a Gel/PCR DNA fragment extraction kit (Geneaid, New Taipei City, Taiwan) according to the manufacturer’s instruction. The purified PCR products were sequenced using BigDye terminator cycle sequencing kit v3.1 (Applied Biosystems, Life Technologies, USA) with the forward primer AK-VP-F2 in a 3100 Genetic Analyzer (Applied Biosystems, Life Technologies, USA). The nucleotide sequences of the partial VP1/VP2 region were analyzed compared to those of the reference sequences retrieved from the NCBI GenBank database using the Basic Local Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic relationships of the sequences of HBoV strains detected in this study with those of the reference strains were evaluated with the use of MEGA X software (41). The evolutionary history was inferred using the maximum-likelihood method with the Hasegawa-Kishino-Yano model (HKY+G as best-fit model) (42) using 1,000 bootstrap replicates.
Data availability.
The nucleotide sequences of the partial VP1/VP2 gene of HBoV strains obtained in oysters detected in this study have been deposited in GenBank under accession numbers MW497561 to MW497571.
ACKNOWLEDGMENTS
This research work was partially supported by the Center of Excellence in Emerging and Re-emerging Diarrheal Viruses, Chiang Mai University, Chiang Mai, Thailand (grant COE008/2020).
We declare that we have no conflicts of interest.
Contributor Information
Niwat Maneekarn, Email: niwat.m@cmu.ac.th.
Christopher A. Elkins, Centers for Disease Control and Prevention
REFERENCES
- 1.Schildgen O, Qiu J, Soderlund-Venermo M. 2012. Genomic features of the human bocaviruses. Future Virol 7:31–39. 10.2217/fvl.11.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broccolo F, Falcone V, Esposito S, Toniolo A. 2015. Human bocaviruses: possible etiologic role in respiratory infection. J Clin Virol 72:75–81. 10.1016/j.jcv.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Lau SK, Yip CC, Que TL, Lee RA, Au-Yeung RK, Zhou B, So LY, Lau YL, Chan KH, Woo PC, Yuen KY. 2007. Clinical and molecular epidemiology of human bocavirus in respiratory and fecal samples from children in Hong Kong. J Infect Dis 196:986–993. 10.1086/521310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vicente D, Cilla G, Montes M, Perez-Yarza EG, Perez-Trallero E. 2007. Human bocavirus, a respiratory and enteric virus. Emerg Infect Dis 13:636–637. 10.3201/eid1304.061501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos N, Peret TC, Humphrey CD, Albuquerque MC, Silva RC, Benati FJ, Lu X, Erdman DD. 2010. Human bocavirus species 2 and 3 in Brazil. J Clin Virol 48:127–130. 10.1016/j.jcv.2010.03.014. [DOI] [PubMed] [Google Scholar]
- 6.Khamrin P, Thongprachum A, Shimizu H, Okitsu S, Mizuguchi M, Hayakawa S, Maneekarn N, Ushijima H. 2012. Detection of human bocavirus 1 and 2 from children with acute gastroenteritis in Japan. J Med Virol 84:901–905. 10.1002/jmv.23274. [DOI] [PubMed] [Google Scholar]
- 7.Han TH, Kim CH, Park SH, Kim EJ, Chung JY, Hwang ES. 2009. Detection of human bocavirus-2 in children with acute gastroenteritis in South Korea. Arch Virol 154:1923–1927. 10.1007/s00705-009-0533-3. [DOI] [PubMed] [Google Scholar]
- 8.Abdel-Moneim AS, Mahfouz ME, Zytouni DM. 2018. Detection of human bocavirus in Saudi healthy blood donors. PLoS One 13:e0193594. 10.1371/journal.pone.0193594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De R, Liu L, Qian Y, Zhu R, Deng J, Wang F, Sun Y, Dong H, Jia L, Zhao L. 2017. Risk of acute gastroenteritis associated with human bocavirus infection in children: a systematic review and meta-analysis. PLoS One 12:e0184833. 10.1371/journal.pone.0184833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, He M, Zeng P, Gao Z, Bian G, Yang C, Li W. 2015. The genomic and seroprevalence of human bocavirus in healthy Chinese plasma donors and plasma derivatives. Transfusion 55:154–163. 10.1111/trf.12785. [DOI] [PubMed] [Google Scholar]
- 11.Endo R, Ishiguro N, Kikuta H, Teramoto S, Shirkoohi R, Ma X, Ebihara T, Ishiko H, Ariga T. 2007. Seroepidemiology of human bocavirus in Hokkaido prefecture, Japan. J Clin Microbiol 45:3218–3223. 10.1128/JCM.02140-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guido M, Zizza A, Bredl S, Lindner J, De Donno A, Quattrocchi M, Grima P, Modrow S. 2012. Seroepidemiology of human bocavirus in Apulia, Italy. Clin Microbiol Infect 18:E74–6. 10.1111/j.1469-0691.2011.03756.x. [DOI] [PubMed] [Google Scholar]
- 13.Hao Y, Gao J, Zhang X, Liu N, Li J, Zheng L, Duan Z. 2015. Seroepidemiology of human bocaviruses 1 and 2 in China. PLoS One 10:e0122751. 10.1371/journal.pone.0122751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hustedt JW, Christie C, Hustedt MM, Esposito D, Vazquez M. 2012. Seroepidemiology of human bocavirus infection in Jamaica. PLoS One 7:e38206. 10.1371/journal.pone.0038206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kahn JS, Kesebir D, Cotmore SF, D’Abramo A, Jr, Cosby C, Weibel C, Tattersall P. 2008. Seroepidemiology of human bocavirus defined using recombinant virus-like particles. J Infect Dis 198:41–50. 10.1086/588674. [DOI] [PubMed] [Google Scholar]
- 16.Meriluoto M, Hedman L, Tanner L, Simell V, Makinen M, Simell S, Mykkanen J, Korpelainen J, Ruuskanen O, Ilonen J, Knip M, Simell O, Hedman K, Soderlund-Venermo M. 2012. Association of human bocavirus 1 infection with respiratory disease in childhood follow-up study, Finland. Emerg Infect Dis 18:264–271. 10.3201/eid1802.111293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guido M, Tumolo MR, Verri T, Romano A, Serio F, De Giorgi M, De Donno A, Bagordo F, Zizza A. 2016. Human bocavirus: current knowledge and future challenges. World J Gastroenterol 22:8684–8697. 10.3748/wjg.v22.i39.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blinkova O, Rosario K, Li L, Kapoor A, Slikas B, Bernardin F, Breitbart M, Delwart E. 2009. Frequent detection of highly diverse variants of cardiovirus, cosavirus, bocavirus, and circovirus in sewage samples collected in the United States. J Clin Microbiol 47:3507–3513. 10.1128/JCM.01062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamza H, Leifels M, Wilhelm M, Hamza IA. 2017. Relative abundance of human bocaviruses in urban sewage in Greater Cairo, Egypt. Food Environ Virol 9:304–313. 10.1007/s12560-017-9287-3. [DOI] [PubMed] [Google Scholar]
- 20.Hamza IA, Jurzik L, Wilhelm M, Uberla K. 2009. Detection and quantification of human bocavirus in river water. J Gen Virol 90:2634–2637. 10.1099/vir.0.013557-0. [DOI] [PubMed] [Google Scholar]
- 21.Iaconelli M, Divizia M, Della Libera S, Di Bonito P, La Rosa G. 2016. Frequent detection and genetic diversity of human bocavirus in urban sewage samples. Food Environ Virol 8:289–295. 10.1007/s12560-016-9251-7. [DOI] [PubMed] [Google Scholar]
- 22.La Rosa G, Purpari G, Guercio A, Di Bella S, Gucciardi F, Proroga YTR, Pisanu M, Della Libera S, Iaconelli M, Suffredini E. 2018. Detection of human bocavirus species 2 and 3 in bivalve shellfish in Italy. Appl Environ Microbiol 84:e02754-17. 10.1128/AEM.02754-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salvo M, Lizasoain A, Castells M, Bortagaray V, Castro S, Colina R, Tort FL, Victoria M. 2018. Human bocavirus: detection, quantification and molecular characterization in sewage and surface waters in Uruguay. Food Environ Virol 10:193–200. 10.1007/s12560-017-9334-0. [DOI] [PubMed] [Google Scholar]
- 24.Purpari G, Macaluso G, Di Bella S, Gucciardi F, Mira F, Di Marco P, Lastra A, Petersen E, La Rosa G, Guercio A. 2019. Molecular characterization of human enteric viruses in food, water samples, and surface swabs in Sicily. Int J Infect Dis 80:66–72. 10.1016/j.ijid.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 25.Onosi O, Upfold NS, Jukes MD, Luke GA, Knox C. 2020. The first detection of human bocavirus species 2 and 3 in raw sewage and mussels in South Africa. Food Environ Virol 12:84–88. 10.1007/s12560-019-09417-w. [DOI] [PubMed] [Google Scholar]
- 26.Rasanen S, Lappalainen S, Kaikkonen S, Hamalainen M, Salminen M, Vesikari T. 2010. Mixed viral infections causing acute gastroenteritis in children in a waterborne outbreak. Epidemiol Infect 138:1227–1234. 10.1017/S0950268809991671. [DOI] [PubMed] [Google Scholar]
- 27.Bibby K, Peccia J. 2013. Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environ Sci Technol 47:1945–1951. 10.1021/es305181x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.La Rosa G, Sanseverino I, Della Libera S, Iaconelli M, Ferrero VEV, Barra Caracciolo A, Lettieri T. 2017. The impact of anthropogenic pressure on the virological quality of water from the Tiber River, Italy. Lett Appl Microbiol 65:298–305. 10.1111/lam.12774. [DOI] [PubMed] [Google Scholar]
- 29.Bastien N, Brandt K, Dust K, Ward D, Li Y. 2006. Human bocavirus infection, Canada. Emerg Infect Dis 12:848–850. 10.3201/eid1205.051424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brieu N, Guyon G, Rodiere M, Segondy M, Foulongne V. 2008. Human bocavirus infection in children with respiratory tract disease. Pediatr Infect Dis J 27:969–973. 10.1097/INF.0b013e31817acfaa. [DOI] [PubMed] [Google Scholar]
- 31.Hengst M, Hausler M, Honnef D, Scheithauer S, Ritter K, Kleines M. 2008. [Human bocavirus-infection (HBoV): an important cause of severe viral obstructive bronchitis in children]. Klin Padiatr 220:296–301. (In German.) 10.1055/s-0028-1083806. [DOI] [PubMed] [Google Scholar]
- 32.Pozo F, Garcia-Garcia ML, Calvo C, Cuesta I, Perez-Brena P, Casas I. 2007. High incidence of human bocavirus infection in children in Spain. J Clin Virol 40:224–228. 10.1016/j.jcv.2007.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nantachit N, Kochjan P, Khamrin P, Kumthip K, Maneekarn N. 2021. Human bocavirus genotypes 1, 2, and 3 circulating in pediatric patients with acute gastroenteritis in Chiang Mai, Thailand, 2012–2018. J Infect Public Health 14:179–186. 10.1016/j.jiph.2020.12.007. [DOI] [PubMed] [Google Scholar]
- 34.Kingsley DH. 2014. High pressure processing of bivalve shellfish and HPP’s use as a virus intervention. Foods 3:336–350. 10.3390/foods3020336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myrmel M, Berg EM, Rimstad E, Grinde B. 2004. Detection of enteric viruses in shellfish from the Norwegian coast. Appl Environ Microbiol 70:2678–2684. 10.1128/aem.70.5.2678-2684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Croci L, Losio MN, Suffredini E, Pavoni E, Di Pasquale S, Fallacara F, Arcangeli G. 2007. Assessment of human enteric viruses in shellfish from the northern Adriatic Sea. Int J Food Microbiol 114:252–257. 10.1016/j.ijfoodmicro.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 37.Phan TG, Khamrin P, Akiyama M, Yagyu F, Okitsu S, Maneekarn N, Nishio O, Ushijima H. 2007. Detection and genetic characterization of norovirus in oysters from China and Japan. Clin Lab 53:405–412. [PubMed] [Google Scholar]
- 38.Iwai M, Hasegawa S, Obara M, Nakamura K, Horimoto E, Takizawa T, Kurata T, Sogen S, Shiraki K. 2009. Continuous presence of noroviruses and sapoviruses in raw sewage reflects infections among inhabitants of Toyama, Japan (2006 to 2008). Appl Environ Microbiol 75:1264–1270. 10.1128/AEM.01166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thongprachum A, Fujimoto T, Takanashi S, Saito H, Okitsu S, Shimizu H, Khamrin P, Maneekarn N, Hayakawa S, Ushijima H. 2018. Detection of nineteen enteric viruses in raw sewage in Japan. Infect Genet Evol 63:17–23. 10.1016/j.meegid.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Kapoor A, Simmonds P, Slikas E, Li L, Bodhidatta L, Sethabutr O, Triki H, Bahri O, Oderinde BS, Baba MM, Bukbuk DN, Besser J, Bartkus J, Delwart E. 2010. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J Infect Dis 201:1633–1643. 10.1086/652416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174. 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The nucleotide sequences of the partial VP1/VP2 gene of HBoV strains obtained in oysters detected in this study have been deposited in GenBank under accession numbers MW497561 to MW497571.