Abstract
The bacterial adenylate cyclase two-hybrid system (BACTH) is a genetic approach used to test protein interactions in vivo in E. coli. This system takes advantage of the two catalytic domains of Bordetella pertussis adenylate cyclase (CyaA) toxin, which can be fused separately to proteins of interest. If the proteins of interest interact, then the adenylate cyclase domains will be brought in close proximity to each other, reconstituting cyclic AMP (cAMP) production. Interacting proteins can be both qualitatively and quantitatively assessed by the expression of chromosomal genes of the E. coli lac or mal operon, which are positively regulated by cAMP production. Because cAMP is diffusible, the proteins of interest do not need to interact near the transcriptional machinery. Consequently, both cytosolic and membrane protein–protein interactions can be tested. The BACTH system has recently been modified to be compatible with Gateway® recombinational cloning, BACTHGW. This chapter explains the principle of the BACTH, its Gateway® modified system, and details of the general procedure.
Keywords: Bacterial two-hybrid, Adenylate cyclase, Protein interaction assay, cAMP signaling, Gateway® cloning, BACTH
1. Introduction
The bacterial adenylate cyclase two-hybrid (BACTH) system is a useful technique designed to assess in vivo protein–protein interactions. The first assay developed to test for protein–protein interactions in vivo was the yeast two-hybrid system (Y2H) by Fields and Song [1]. Both the Y2H and BACTH systems utilize coexpression of two-hybrid (fusion) proteins in the same cell that upon interaction can be assessed by a readout of either phenotypic screen or selection [2-4]. For the BACTH assay, this readout is based on the reconstitution of the adenylate cyclase activity when two complementary fragments from the catalytic domain of the Bordetella pertussis adenylate cyclase toxin [5] are brought into close proximity by proteins that interact (Fig. 1). This results in production of cyclic adenosine monophosphate (cAMP) and activation of its signaling cascade [3, 5, 6]. This system takes advantage of the fact that in Escherichia coli the expression of many genes, including the lac operon or mal regulon, is regulated by cAMP [7]. Because cAMP is a diffusible secondary metabolite, the BACTH system can be broadly applied to characterize interactions of cytosolic and membrane proteins [8], including large multiprotein complexes such as bacterial secretion systems or cell division machinery [8-13].
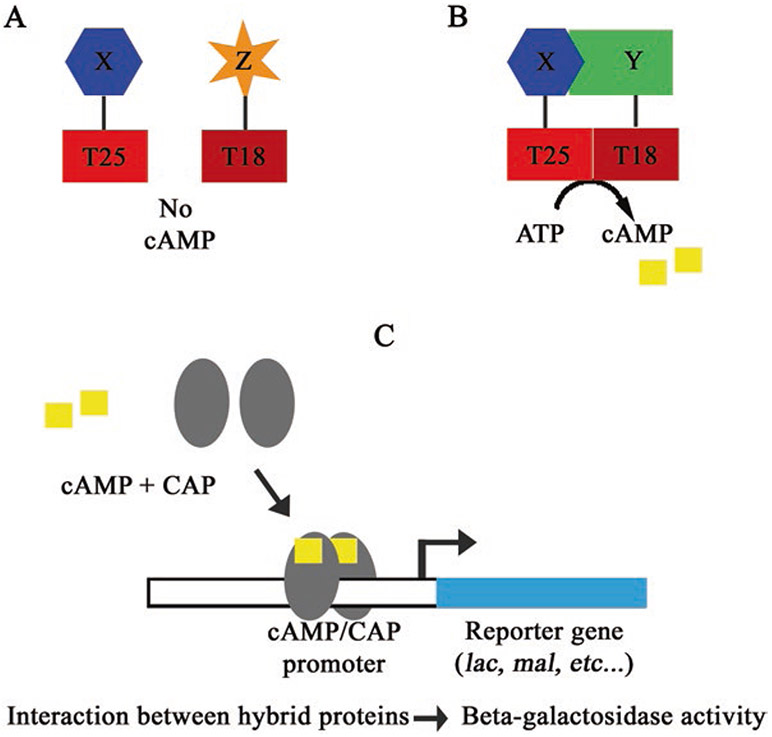
Fig. 1.
A schematic illustrating the principle of the BACTH assay. (a) Hybrid proteins that do not interact will not reconstitute adenylate cyclase activity. (b) Hybrid proteins that do interact will reconstitute adenylate cyclase activity, resulting in production of cAMP. (c) A depiction of cAMP binding CAP and positively regulating expression of a reporter gene
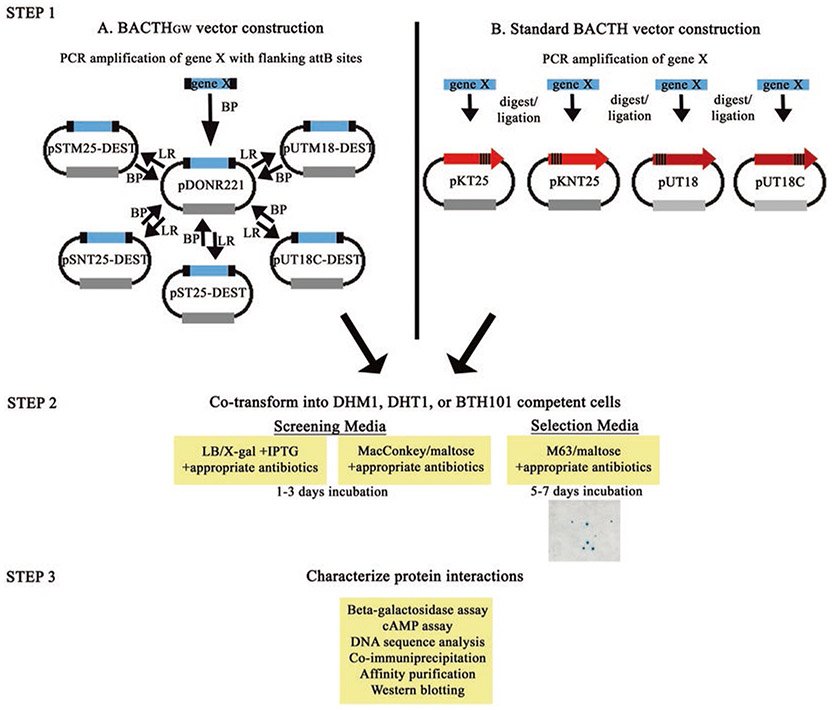
There are currently two different sets of BACTH compatible vectors available for complementation assays: the standard BACTH vectors and the recently developed Gateway® cloning system, BACTH compatible (BACTHGW) vectors (Fig. 2) [14]. The construction of standard BACTH vectors must be performed individually for each vector containing the gene of interest, which is labor intensive, especially in the event of conducting a large-scale screen. One advantage of the BACTH-Gateway® system is that it overcomes the time-consuming drawbacks of individually cloning a single open reading frame (ORF) into each standard BACTH vectors. This is because Gateway® compatible BACTH vectors utilize “recombineering” techniques to insert ORFs flanked by recombination sites into an entry vector by a single cloning step. After the ORF is in the entry vector, it can be easily recombined into any of the five BACTHGW destination vectors currently available. Once recombined into the BACTHGW vector, the ORF can be shuttled to an entry vector as needed. Cloning into the entry vector reconstitutes the recombination sites necessary for recombination into another destination vector of interest. Not only have the BACTHGW vectors been adapted for amino and carboxyl terminal fusion capabilities, but vectors have also been designed to contain transmembrane domains downstream of the T25 and T18 fragments to assess protein–protein interactions in the periplasmic space or, potentially, the outer membrane [5, 6, 14]. As there are several genomic ORF libraries created in Gateway® vectors currently available for different organisms, the BACTH-Gateway® adaptation will facilitate and broaden the application of this genetic system for large scale interaction mapping studies. Finally, combinations of BACTHGW vectors and standard BACTH vectors can be used in complementation assays or library screening assays. The focus of this chapter is to describe the BACTH-Gateway® adapted system and core procedures involved in studying protein–protein interactions using the BACTH system in E. coli.
General workflow of BACTH complementation assays. Genes of interest are inserted into either the (a) Gateway® compatible BACTHGW or (b) standard BACTH vectors. To insert the ORF into one of the Gateway® compatible BACTHGW destination vectors, a series of recombination reactions must occur. First, the gene is PCR amplified using primers with flanking attB sites. The PCR product is then recombined with an entry vector (that contains attP sites) by the BP reaction to yield an entry vector (pDONR) containing the ORF flanked by attL sites. The ORF is subsequently shuttled into a destination vector (containing attR sites) by the LR recombination reaction. The resulting BACTHGW destination vector contains the ORF flanked by attB sites (allowing for shuttling to entry vector as desired). In a second step, t25-gene X and t18-gene Y containing vectors are cotransformed into competent cells and plated onto screening or selection media. Finally, protein complementation is quantified by the beta-galactosidase assay or a cAMP assay. Complementation can be further evaluated by Western blotting for expression levels of the T25 and T18 hybrid proteins or by coimmunoprecipitation or pull-down using modified vectors containing an additional tag (e.g., 6×His). Please see text for more details
1.1. Principle of BACTH Gateway® System
The BACTH system uses relatively quick and simple means to detect in vivo protein–protein interactions in E. coli, and it can be performed by those with basic knowledge of microbiological laboratory and molecular biology techniques.
As indicated above, the basis of the BACTH system is to assess protein–protein interactions by the interaction-mediated reconstitution of adenylate cyclase by the two catalytic fragments of Bordetella pertussis adenylate cyclase toxin (CyaA). Importantly, this test is performed in an E. coli strain lacking a functional adenylate cyclase (ΔcyaA E. coli mutant). The catalytic domain of B. pertussis adenylate cyclase (CyaA) consists of two complementary fragments, termed T25 and T18 [5]. When these fragments are separately coexpressed they are unable to associate and cannot synthetize cAMP. However, when the T25 and T18 fragments are fused to two separate proteins able to interact, they can reassociate into a functional enzyme, thereby restoring cAMP production. In E. coli, cAMP is a regulator of gene transcription [15]. It binds to the catabolite activator protein (CAP), and then the cAMP/CAP complex can activate the transcription of catabolite genes, including those of the lac operon and mal regulon. Consequently, E. coli becomes able to utilize lactose or maltose as a sole carbon source, which can be determined easily by screening or by using selective media [3, 6].
1.2. General Procedure
To detect in vivo protein interactions, the T25 and T18 fragments must be coexpressed in an adenylate cyclase, E. coli cya null mutant. Firstly, the constructs must be assembled, where one gene of interest (e.g., gene X) is fused to the T25 fragment and a second gene (e.g., gene Y) is fused to the T18 fragment in either the BACTHGW vectors or the standard BACTH vectors.
The Gateway® system works by PCR amplification of genes of interest with primers containing recombination sites, and recombination into an “entry” vector. After creation of the entry vector containing the gene of interest, the gene can be easily transferred in a single step to any of the compatible destination vectors: pST25-DEST, pSNT25-DEST, pUT18C-DEST, pSTM25-DEST, or pUTM18C-DEST [14]. One utility of the BACTHGW system is that a gene in the destination vector can be used in a BP reaction to clone the gene into a new entry vector and then into different destination vectors as desired (Fig. 2). This is a quick and effective system to insert a gene of interest into various desired vectors to test complementation in both N and C terminal fusion orientations. Alternatively, traditional cloning methods can be used to insert a gene of interest into the T25 containing BACTH vectors, pKT25 or pKNT25, and a second gene (e.g., gene Y) fused to the T18 containing BACTH vectors, pUT18 or pUT18C [3, 16]. Additional T25-containing vectors encoding spectinomycin resistance, pST25, pSTM25, and pSNT25, are also available. These vectors were developed to allow the creation of pST25-DEST, pSNT25-DEST, and pSTM25-DEST to be compatible with pDONR221 entry vectors, which encode kanamycin resistance (also encoded by the pK vectors of the standard BACTH plasmids).
Complementation assays are performed by cotransformation of both fusion plasmids into a Δcya E. coli strain, plated on indicator plates or selective media, and monitored for the resulting Cya+ phenotype. The protein–protein interaction can be further evaluated by quantifying β-galactosidase activity or by measuring cAMP levels directly, both of which indicate reconstituted adenylate cyclase from the close proximity of the T25 and T18 fragments [5, 6]. A β-galactosidase enzyme assay is an effective tool to measure adenylate cyclase reconstitution because the cAMP/CAP complex positively regulates β-galactosidase expression. Enzyme activity can be compared to bacterial growth of each sample to yield enzyme activity per unit of bacterial growth. The fusion proteins expressed by E. coli can also be characterized by Western blotting and immunoprecipitation techniques.
To date, BACTH has been used by many laboratories to characterize various bacterial, eukaryotic, and viral protein–protein interactions [6, 16-20]. A benefit of the BACTH genetic system is that because cAMP is a diffusible molecule, interactions of T25 and T18 hybrid fusion proteins do not need to occur near the transcriptional machinery. This allows for better characterization of membrane protein interactions as compared to Y2H or alternative bacterial two-hybrid systems that rely, for example, on reconstituted interactions between components of the transcriptional machinery [1, 2, 6, 8, 14, 19].
2. Materials
2.1. Equipment
42 °C water bath.
Plasmid miniprep kit.
30 or 37 °C incubator for agar plates.
Shaking incubator for liquid cultures.
96-well deep-well blocks, sterile.
Sterile toothpicks.
96-well plates or glass tubes, sterile (* chloroform resistant).
96-well optical plate.
Microporous tape strips.
Multichannel pipettor.
Rotary shaker (for shaking deep-well 96-well blocks).
Microplate reader (capable of measuring 405 and 595 nm).
Spreadsheet software (Excel or equivalent).
Equipment for Western blotting (optional).
2.2. Bacterial Media
Luria–Bertani (LB) broth: mix 10 g of NaCl, 10 g of tryptone, and 10 g of yeast extract, adjust pH to 7.0 with NaOH, add deionized H2O to a final volume of 1 L and autoclave. To prepare LB plates, add 15 g of agar per liter of LB broth and autoclave. Allow the medium to cool down to less than 45 °C, then add the appropriate antibiotics and pour the plates.
LB/X-gal plates: the LB/agar medium (above) is autoclaved, allowed to cool down to less than 45 °C and supplemented, just before pouring plates, with 40 μg/mL of the X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) chromogenic substrate and appropriate antibiotics. IPTG (isopropyl-β-d-thiogalactopyranoside, final concentration of 0.5 mM) is usually also added to the medium to induce full expression of the hybrid proteins as well as that of the β-galactosidase reporter enzyme.
MacConkey/maltose plates: 40 g of MacConkey agar are dissolved in 1 L of distilled water and autoclaved. A stock solution of glucose-free maltose (20% in water) is sterilized by filtration. Maltose (1.0% final concentration) as well as antibiotics (ampicillin at 100 μg/mL and kanamycin at 50 μg/mL) are added to the autoclaved MacConkey medium just before pouring plates. IPTG (final concentration of 0.5 mM) is usually added to the medium to induce full expression of the hybrid proteins (see Note 1).
M63/maltose minimal medium: To prepare 5× concentrated M63 minimal medium, mix 10 g (NH4)2SO4, 68 g KH2PO4, 2.5 mg FeSO4·7H2O, 5 mg vitamin B1, add deionized H2O to a final volume of 1 L, adjust pH to 7.0 with KOH, and autoclave. M63/maltose plates are prepared by autoclaving 15 g of agar in 800 mL H2O. Then add 200 mL sterile 5× M63 medium, 0.2–0.4% maltose, and the appropriate antibiotics at half the usual concentrations (i.e., ampicillin 50 μg/mL, kanamycin 25 μg/mL) just before pouring plates.
Antibiotics: ampicillin, kanamycin, and spectinomycin. Nalidixic acid (optional), streptomycin (optional).
Solutions and Reagents
Calcium chloride (0.1 M).
β-Galactosidase assay medium (PM2): 70 mM Na2HPO4·12H2O, 30 mM NaH2PO4 H2O, 1 mM MgSO4, 0.2 mM MnSO4, pH 7.0 with H3PO4. Add 100 mM β-mercaptoethanol just before use (see Note 2).
Substrate solution (ONPG, o-nitrophenol-β-galactoside): solution of 4 mg/mL in PM2 medium without β-mercaptoethanol (store at −20 °C).
Stop solution: 1 M Na2CO3.
Chloroform.
SDS 0.1%.
Reagents for Western blotting.
Antibody: Anti-CyaA T25 fragment.
Antibody: Anti-CyaA T18 region (Santa Cruz biotechnology cat. # sc-13582).
2.4. BACTH Reporter Strains and E. coli Cloning Strains
For the BACTH system, three E. coli adenylate cyclase deficient reporter strains are commonly used in detecting protein–protein interactions: DHT1, DHM1, and BTH101 [8, 16, 21]. Other E. coli Δcya strains are available and can be found at the Coli Genetic Stock Center (http://cgsc.biology.yale.edu). The various genetic background of each E. coli strain gives rise to different complementation efficiencies and variable reporter gene stringencies. The frequencies of spontaneous Lac+ revertants (due to cAMP/CAP independent promoter mutations) of these different strains range from 10−7 to 10−8 while frequencies of spontaneous Mal+ revertants are below the detection threshold (i.e., <10−10). A summary of the main genetic differences of the common E. coli cya mutants used for the BACTH can be found below.
DHT1 [F-, cya-854, ilv 691::Tn10, recA1, endA1, gyrA96 (Nal r), thi1, hsdR17, spoT1, rfbD1, glnV44(AS)]: a recA strain that displays a high BACTH complementation efficiency and fast growth. However, this strain carries an ilv mutation, so it requires casamino acids or direct addition of isoleucine and valine for growth on minimal medium [20, 22].
DHM1 [F-, cya-854, recA1, endA1, gyrA96 (Nal r), thi1, hsdR17, spoT1, rfbD1, glnV44(AS)]: an ilv + DHT1 derivative able to grow on minimal media plus sugars. This strain has a lower complementation efficiency and grows more slowly than the parental DHT1 strain.
BTH101 [F-, cya-99, araD139, galE15, galK16, rpsL1 (Str r), hsdR2, mcrA1, mcrB1]: this strain displays a good BACTH efficiency and fast growth. However, there is some instability of plasmids due to the presence of RecA.
DH5α: E. coli cloning strain or lacIq strain.
2.5. BACTH Plasmids
The BACTH system requires coexpression of two proteins fused to the T25 and T18 fragments within the same host bacterial cell (e.g., Δcya E. coli BTH101). The T25 fragment corresponds to the first 224 amino acids of adenylate cyclase (CyaA), while the T18 fragment corresponds to amino acids 225–399 of CyaA [8, 16]. There are two sets of compatible vectors available: the BACTHGW vectors that use Gateway® cloning technology to insert the gene of interest and the standard BACTH vectors that use standard cloning methods. Each ORF is expressed under the control of the lac promoter. Both systems can be used to create genetic fusions with the gene of interest at either the amino or carboxyl terminus of the T25 or T18 [8, 16]. However only C-terminal fusions to T18 are currently possible with Gateway® cloning (i.e., in pUT18C-DEST).
Standard BACTH vectors: The plasmid pKT25 is a low copy number pSU40 plasmid derivative that contains a kanamycin resistance selective marker. There is a multiple cloning site sequence (MCS) at the 3′ end to allow for fusions, in frame, at the C-terminal end of T25. In contrast, pKNT25 has the MCS at the 5′ end so that fusions can be made at the N-terminal end of the T25 fragment. The other BACTH complementary vectors include pUT18 and pUT18C. The pUT18 plasmid is a derivative of pUC19 and contains an ampicillin selection marker. The MCS is located on the 5′ end, so fusions are made at the N-terminus of T18 fragment. For the pUT18C plasmid, the MCS is at the 3′ end so fusions are made at the C-terminus of T18. Any combination of vectors containing one T25 hybrid and one T18 hybrid can be used in complementation assays or a pool of vectors for screening assays with a bait vector.
Gateway®-compatible BACTH vectors: A variety of Gateway® vectors are available through Thermo Fisher Scientific and can be used for different experimental conditions. The Gateway® recombineering system has been designed so that an open reading frame that is flanked by recombination sites can be quickly transferred from an “entry” vector to various “destination” vectors [23, 24]. The Gateway® recombineering cloning system was made by inserting a recombination cassette containing attR lambda recombination sites. The BACTHGW vectors express the CcdB toxin as well as a chloramphenicol resistance marker. To fuse proteins of interest to the C-terminus of T25 and T18, pST25-DEST and pUT18C-DEST can be utilized, respectively [14]. Alternatively, pSNT25-DEST can be used to fuse the protein of interest to the N-terminus of T25 [14]. There are additional BACTHGW vectors, pSTM25-DEST and pUTM18C-DEST, which contain an additional transmembrane domain directly C-terminal to the T25 or T18 fragment, thus allowing the protein of interest to be expressed in the periplasm or, potentially, in the outer membrane [14]. These vectors can also be useful to make fusions with inner membrane proteins that have their N-terminus extremity located in the periplasm [14]. Thus, these additional vectors, for which there are “standard” BACTH plasmid variants (i.e., pSTM25 and pUTM18C), allow for the testing of interactions that occur in these additional compartments. One difference between the BACTH Gateway® plasmids and the standard BACTH plasmids, pKNT25 and pKT25, is that the Gateway® DEST plasmids contain a spectinomycin resistance marker rather than the kanamycin resistance marker. This allows for compatibility with the Gateway® entry vector pDONR221, which encodes kanamycin resistance. Otherwise, the only other difference between the standard and Gateway® compatible BACTH vectors is the presence of the Gateway® attR-flanked recombination site in the latter (see Note 3).
Positive controls: pKT25-zip and pUT18C-zip are often used as positive controls during BACTH complementation assays. They encode the two adenylate cyclase fragments, T25 and T18, each fused to the leucine zipper domain of GCN4 [3, 5]. Additional positive controls include pKTM25-zip and pUTM18C-zip wherein the leucine zipper is expressed in the periplasm (see above for explanation concerning pSTM25-DEST and pUTM18C-DEST).
Negative controls: Empty vectors, lacking any insert, are commonly used as negative controls. For example, pST25-empty vector and any pUT18C-hybrid fusion could be used as a negative control in complementation assays. Alternatively, two proteins known not to interact can be used.
Methods
3.1. Methodology Outline
The BACTH system can be broken down into three major steps (Fig. 2).
Step one: Cloning genes of interest into BACTH vectors. The first step is to clone the ORF into each of the T25 and T18 containing plasmid vectors. This can be performed using the Gateway® recombineering technology, a simple method relying on site-specific recombinases, to transfer the gene of interest from the entry vector, pDONR221, to the BACTH-DEST vectors [23]. Alternatively, PCR amplification of genes of interest with appropriate primers and cloning in frame into the MCS of standard BACTH vectors using standard techniques can be implemented [25, 26].
Step two: Qualitative analysis of protein interactions. After the genes of interest are cloned into their respective plasmids, encoding for example T25-X or T18-Y, these plasmids are cotransformed into competent E. coli Δcya reporter cells such as DHT1, DHM1, or BTH101 cells. Transformants are then plated onto either LB/X-gal agar plates or MacConkey/maltose agar indicator plates with appropriate antibiotics and incubated at 30 or 37 °C (37 °C is typically less efficient) for 1–3 days [3, 6]. After incubation, plates can be visualized for the presence of blue colonies (LB/X-gal plates) or red colonies (MacConkey/maltose plates) indicating a positive interaction between two putative proteins of interest. Transformed cells can also be plated on a selective medium, M63 minimal medium supplemented with maltose as the only carbon source [8, 27, 28].
Step three: Quantitative analysis of protein interactions by β-galactosidase assay. To obtain a quantitative analysis of the interaction between putative proteins, individual colonies are picked from indicator plates and grown in liquid media to either exponential phase or overnight. Cells grown in liquid media are permeabilized, and β-galactosidase activity is measured using o-nitrophenol-β-galactoside (ONPG) as a substrate and analyzed relative to bacterial growth (OD600). The β-galactosidase enzyme activity/cell density can be reported as Relative Units (RU) or following standard protocols in Miller Units.
3.2. Cloning Genes of Interest into BACTH Vectors
This section describes cloning of genes of interest into BACTH vectors using Gateway® recombineering techniques and traditional cloning strategies.
3.2.1. Cloning into Gateway® Vectors
The Gateway® cloning system (Life Technologies, Thermo Fisher Scientific) is a quick method to transfer genes X and Y of interest into the BACTHGW pDEST vectors (pST25-DEST, pSNT25-DEST, pUT18C-DEST, pSTM25-DEST, or pUTM18C) [14].
-
The gene of interest is PCR amplified (following standard protocol; [26]) using primers that contain attB sites, which are necessary for the recombination reaction [23, 24]. The following sequences (underlined) correspond to attB sites and should flank the gene specific sequences (indicated by XXX…). In addition, the base pairs flanking the attB sites can be adjusted for the Tm (see Note 4):
FORWARD PRIMER (bold ATG corresponds to the initiation codon of the ORF and can be omitted, if desired):
5′-GCCGCACAAGTTTGTACAAAAAAGCAGGCTTTATGXXXXXXXX.
REVERSE PRIMER (the ORF stop codon can be removed, if desired):
5′-GCGGACCACTTTGTACAAGAAAGCTGGGTTXXXXXXXX.
Purify the PCR amplified gene products containing attB sites using a commercially available PCR purification kit.
- Perform the “BP recombination” reaction, following the manufacturer’s guidelines, to insert the attB flanked-gene PCR product into the pDONR™221 “entry” plasmid [23]. The pDONR™221 plasmid contains an attP-flanked gene cassette encoding ccdB and cat (see Note 3), and the plasmid backbone contains M13 forward and reverse priming sequences flanking the attP sites for convenience.
- Mix 150 ng of purified the PCR product with 150 ng pDONR™221 and BP Clonase II™ enzyme. Incubate the mixture for 2 h (see Note 5) at room temperature.
- Stop the recombination reaction by adding 2 μg of Proteinase K and incubating for 10 min at 37 °C.
- Transform the mixture into competent E. coli DH5α cells or another suitable cloning strain (see Note 6), and select for the transformants on LB plates containing 50 μg/mL kanamycin [14]. The resulting plasmid (pDONR™221-gene X) contains the gene of interest now flanked by attL recombination sites, the product of the BP recombination.
-
Verify the recombination reaction by restriction digest or by colony PCR with subsequent DNA sequencing (using the M13 primers, if desired) of 3–4 clones picked from the transformation plates.Genes inserted into pDONR “entry” vector can be easily transferred into any Gateway® destination vector, including the BACTHGW compatible vectors, by the LR reaction [14, 24]. Of note, many gene libraries cloned into pDONR™221 or another entry vector are available commercially or through genomic resource centers. Thus, it is possible that the investigator may already have a collection of pDONR™221 vectors with their gene of interest already cloned.
- The “LR” reaction is used to transfer genes from pDONR-gene X (“entry” vector) to the BACTHGW destination vectors (pST25-DEST, pSNT25-DEST, pUT18C-DEST, pSTM25-DEST, or pUTM18C-DEST) [14]. The BACTHGW vectors were constructed by inserting an attR-flanked gene cassette encoding ccdB and cat in-frame into each BACTH vector (see Note 3).
- For the LR reaction, mix 150 ng pDONR™221-gene X with 150 ng of the desired BACTHGW pDEST vector.
- Following the manufacturer’s guidelines, add LR Clonase™ to the plasmid mixture, and incubate for one hour at room temperature (see Note 5).
- Terminate the recombination reaction by adding 2 μg of Proteinase K and incubating for 10 min at 37 °C.
- Transform the mixture into competent E. coli DH5α cells, and plate on LB agar plates supplemented with glucose and antibiotics (spectinomycin or ampicillin) (see Note 6). The recombined plasmids now encode either the T25 or T18 gene fragment fused in frame to the gene of interest.
3.2.2. Cloning into Traditional Vectors
When using the standard BACTH vectors, traditional cloning methods can be used to insert the gene of interest in-frame into the multiple cloning sites of these plasmids. For instance, genes X and Y corresponding to the proteins of interest can be amplified by PCR using specific primers that include the restriction sites in the MCS for the desired orientation in the plasmid of interest [26]. The genes X and Y must be cloned in frame with each of the T25 and T18 open reading frames. Other cloning methods can be applied (e.g., Gibson cloning). As with the Gateway® method, all plasmids should be sequence-verified and propagated in the presence of glucose to repress expression of the fusion proteins (see Note 6). For more detailed information please refer to Ouellette et al. [30].
3.3. BACTH Protein Interaction Assays
This section describes cotransformation of recombinant plasmids (e.g., pST25GW-gene X and pUT18CGW-gene Y), and plating on indicator (LB/X-gal or MacConkey/maltose agar) media or selection media to detect protein interactions.
3.3.1. Detecting Complementation Using Indicator Plates
- Competent Δcya E. coli cells (see Note 8) are cotransformed with purified recombinant T25-fusion (e.g., pST25-X) and T18-fusion (e.g., pUT18C-Y) vectors.
- For transformation, mix 50 μL of chemically competent DHM1 cells in a chilled microcentrifuge tube with 5–10 ng of each recombinant plasmid.
- Incubate the mixture for 30 min at 4 °C.
- Heat-shock the mixture at 42 °C for 30 s, and then incubate on ice for 2 min [21].
- Add 1 mL of LB to the cell suspension and incubate at 30 or 37 °C for 60–90 min.
- Plate the cell suspension onto indicator plates (LB/X-gal or MacConkey/maltose agar) containing appropriate antibiotics. Make sure to plate different volumes of the transformation mixture so there are 100–300 colonies per plate. The number of colonies should not exceed 500, or the detection of positive clones will be difficult [3, 28, 30].
- Incubate the indicator plates for 24–48 h at 30 °C. Prolonged incubation (>4 days) can result in negative colonies (i.e., cya−) having a weak red (on MacConkey-maltose) or blue spot (on LB-X-Gal) in the center, but still will be colorless at the edges (see Note 9).
Controls.
In parallel with the above described complementation assay, negative and positive control plasmids should also be cotransformed.
Negative controls.
Competent E. coli cells are cotransformed with plasmids containing unfused T25 and T18 fragments (e.g., pKT25 and pUT18C). Ideally, the investigator should consider testing their protein of interest against the empty vector as well as against a protein that does not interact with their protein. Noninteracting proteins result in white/pale blue colonies on LB/X-gal plates and colorless/white/pale pink colonies on MacConkey/maltose media.
Positive controls.
Recombination products for T25 and T18 fragments fused to known interacting proteins. For example, pKT25-zip and pUT18C-zip encode the GCN4 leucine zipper. The expressed T25-zip and T18-zip hybrid proteins will heterodimerize through their leucine zipper motif. Interacting proteins result in blue colonies on LB/X-gal plates and red colonies on MacConkey/maltose plates.
3.3.2. Detecting Complementation Using Minimal Media Selection Plates
The BACTH system can be used to screen libraries to isolate interacting partners of a protein of interest [31].
Cotransform the plasmid containing the gene (“bait”) of interest with a BACTH vector library containing genomic DNA or a pool of plasmids containing various genes of interest.
-
Plate the transformants onto M63 minimal medium supplemented with maltose as the only carbon source [6, 8, 27]. Using this selective medium, only cells with interacting proteins (reconstituted cAMP production) will be able to use maltose as a carbon source and grow on minimal medium.
To detect Cya+ colonies more quickly and easily, X-gal and IPTG can be added to the medium; on the X-gal supplemented media the Cya+ colonies are also Lac+ and will appear blue (see Note 10). Under these conditions and after extended incubation, small white colonies may be detected on negative control/noninteracting protein plates, as the cells can use the casamino acids as carbon source.
3.4. BACTH Protein Interaction Assays Using Libraries
3.4.1. BACTH Library Construction
To make a BACTH library, genomic DNA or cDNA fragments from the organism of interest must be made and inserted into the chosen BACTH vector (e.g., pKT25 or pUT18C). There are several approaches that can be used to create a genomic DNA library [25, 27, 28, 32]. Below briefly describes a procedure to construct a genomic E. coli chromosomal DNA fragment library fused to the 3′ end of the T25 fragment ORF of the BACTH vector pKT25 as an example (see Note 11). Additional details can be found in Karimova et al. [27, 28]. To avoid cloning cya, which will restore the Cya+ phenotype, the libraries should be prepared from an E. coli cya knockout or a cya deletion strain of the organism of interest.
Ligate the blunt ended DNA fragments (from step 2) with the linearized pKT25 vector (from step 3).
Transform the ligated mixture into electrocompetent ElectroMAX DH10B cells (see Note 12), and plate the transformants onto several LB agar (plus kanamycin for pKT25 vectors) plates.
Pool all the colonies and purify the plasmid DNA. This will be used as the BACTH DNA library [27]. Assess the quality of the inserts cloned into the BACTH library by isolating plasmid DNA (e.g., using a commercial kit, or [29]) from 20 to 30 isolated colonies, and analyze the DNA by restriction endonuclease analysis, DNA sequencing, or PCR amplification (using primers which flank the cloning sites).
3.4.2. Library Screening Using BACTH Vectors
Once the library is created in the desired vector (e.g., pKT25), cotransform the library with the complementary vector (e.g., pUT18C-X) containing the “bait” hybrid protein vector into BACTH host reporter cells [27, 28].
First, the plasmid containing gene X, encoding protein X (“bait”) of interest (e.g., pUT18C-X), is transformed into a BACTH reporter strain (e.g., DHM1).
Prepare electrocompetent cells from the DHM1/pUT18C-X cells (see Note 12) and transform with an aliquot of the pKT25 plasmid library.
After electroporation, incubate the cells with LB medium (or other rich media) at 30 °C for 60–90 min to allow expression of the antibiotic resistances.
Collect the transformed cells by centrifugation (5 min at 3,000 × g), and then wash several times (4–6) with M63 medium to remove all nutrients from the rich medium.
Resuspend the transformed cells in M63 and plate on M63 minimal medium agar supplemented with 0.2% maltose plus antibiotics kanamycin and ampicillin (and X-gal and IPTG if desired) and incubate at 30 °C.
After 4–8 days of incubation at 30 °C, bacteria that express a potential interacting partner of protein X will grow (Cya+, Mal+) and form blue colonies on X-gal plates (Lac+).
Isolate DNA from positive colonies and identify by DNA sequencing analysis. This and other similar procedures have been used to isolate novel proteins involved in E. coli cell division machinery [8, 27, 28] as well as to identify novel protein interactions in Chlamydia trachomatis [11, 20, 22]. The BACTH system has also been used by other laboratories with their own adapted protocols [37-40].
3.5. Quantitative Analysis of Protein Interactions
Upon complementation, it may be useful to quantify the interaction between two proteins of interest. This can be done by measuring β-galactosidase activity because the expression of lacZ in E. coli is positively regulated by cAMP/CAP. To do this, colonies are picked and inoculated into liquid culture in a 96-well microtiter plate [3, 6, 14, 27]. Other β-galactosidase assays can be found in Battesti and Bouveret [18], Griffith and Wolf [41], or Sambrook and Russell [25].
- For each individual transformation (containing T25-X and T18-Y), pick eight isolated colonies (e.g., using a sterile toothpick or pipette tip) for the β-galactosidase assay.
- Eight colonies should also be picked from the positive control transformation plate (e.g., pST25-zip and pUT18C-zip), the negative control plate (e.g., empty vectors or noninteracting proteins), and include blank wells containing LB only.
Inoculate the isolated colonies into separate wells of a sterile 2.2 mL, 96-well storage plate or deep well plate containing 300–400 μL of LB broth, 0.5 mM IPTG, and the appropriate antibiotics.
Seal the 96-well plate using a microporous sheet to allow gas exchange and incubate at 30 °C overnight with shaking for a minimum of 16 h of growth.
After a minimum of 16 h of growth, dilute the cultures 1:5 using M63 medium, and transfer 200 μL of the diluted culture to a flat bottom microtiter plate to record the A595nm absorbance, indicating bacterial density.
- For cell permeabilization, transfer 200 μL of the bacterial suspension to a new microtiter plate (chloroform resistant, 1.2 mL polypropylene 96-well storage block).
- Under the fume hood, add 7 μL of 0.05% SDS and 10 μL of chloroform to the suspension, mix vigorously, and incubate at room temperature for 30–40 min to allow the chloroform to evaporate.
- For the enzymatic reaction, transfer 20 μL of the SDS/chloroform permeabilized cells to a new microtiter plate that contains 105 μL of PM2 (plus 100 mM β-mercaptoethanol (see Note 2) and 0.1% o-nitrophenol-β-galactoside (ONPG)) per well.
- For the control assays, 20 μL of M63 is added instead of permeabilized cells.
Incubate the plates with PM2 at room temperature for 10–20 min. The medium should change to a yellow color.
Stop the enzymatic reaction by the addition of 50 μL of 1 M Na2CO3.
Record the absorbance (OD405) using a microtiter plate reader.
- Analyze the absorbance data from both cell density and enzymatic activity using spreadsheet software (Excel or equivalent). The enzymatic activity is calculated for each well and recorded as A (relative units) using the equation below;
- The results are reported as relative units (RU) of β-galactosidase activity for each sample of interest, positive, and negative controls. If desired, then RU can be converted to Miller units (RU = 1/6 of Miller units).
- The positive controls should be set to 100% and the negative control/background RU should be less than 3% of the β-galactosidase activity of the positive control samples.
The above assay is a simple way to measure β-galactosidase activity from cells expressing hybrid proteins. Alternative ways to quantify complementation efficiency include measuring cAMP production by enzyme linked immunosorbent assay (ELISA) or radioimmunoassay [3, 6].
3.6. Characterization of Protein Interactions by Western Blotting
If further characterization of protein expression in the complemented cells is needed, then Western blotting, biochemical detection assays, coimmunoprecipitation, or affinity purification methods can be used. Detailed information on these methods can be found in other chapters of Methods in Molecular Biology [42, 43]. Western blotting can be performed following standard procedures [25]. The T25 fragment can be detected using rabbit polyclonal anti-B. pertussis CyaA protein [28, 44], and the T18 fragment can be detected using anti-CyaA monoclonal antibody (3D1, sc-13582, Santa Cruz Biotechnology, http://www.scbt.com/). Alternatively, the recombinant plasmids can be designed to contain epitope tags such as myc, FLAG, HA, T7, or 6×-histidine [45]. The epitope tagged fusion proteins can be detected by Western blotting with an antibody made against the epitope tag (e.g., anti-FLAG antibody), or fusion proteins can be purified by affinity purification methods. The epitope tagged fusion proteins can also be used for coimmunoprecipitation to show direct physical binding of fusion proteins [25].
Fig. 3.
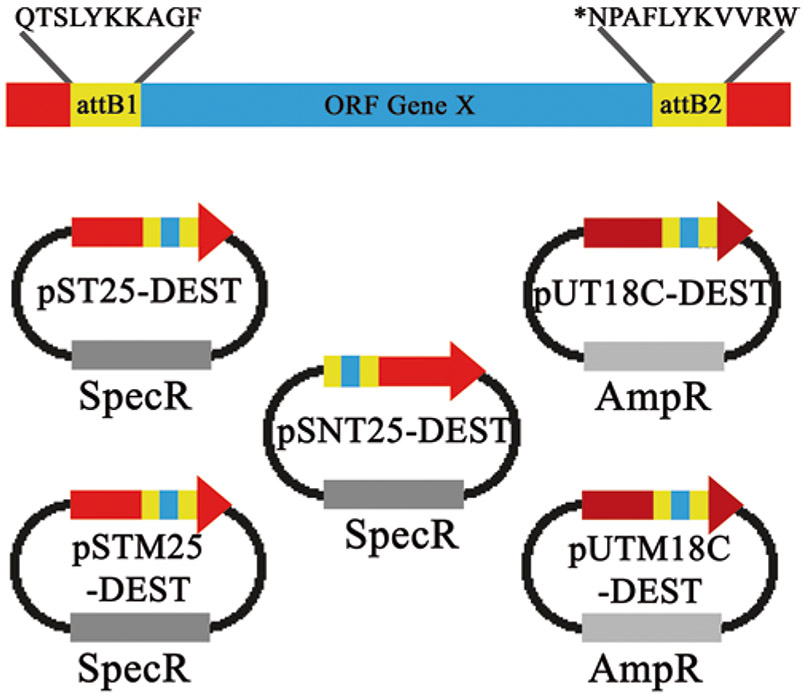
An illustration of the peptide linker sequences (attB sites) generated after the Gateway® recombination reaction with the BACTHGW destination vector. The destination vector for each pST25, pSNT25, pUT18C, pSTM25, or pUTM18C-containing gene X will be flanked by peptide linker sequence scars, corresponding to the attB site generated after the LR reaction. The amino acid sequence scars from the LR recombination reaction are depicted above. These sequence scars do not interfere with BACTH complementation assays. The asterisk denotes a stop codon, which can be introduced in the PCR product to prevent expression of the attB2 sequence when the protein of interest is fused to the C-terminus of the T25 or T18 fragment
Acknowledgments
This work was supported in part by the University of South Dakota, Sanford School of Medicine, Division of Basic Biomedical Sciences, the University of Nebraska Medical Center, and by the Institut Pasteur and the Centre National de la Recherche Scientifique (CNRS UMR 3528, Biologie Structurale et Agents Infectieux). S.P.O. is supported by an award (1R35GM124798-01) from NIGMS/NIH.
Footnotes
MacConkey from Difco Laboratories (cat # 216830) is strongly recommended as the quality of MacConkey agar base media can vary.
Because 2-mercaptoethanol is considered toxic and can cause irritation to the skin and respiratory tract upon inhalation, it should be used only under a fume hood. In addition, 2-mercaptoethanol can be left out of the PM2 buffer altogether. It is important to note that the β-galactosidase activities will be reduced by a factor of 2. However, this is fine because only relative enzymatic activities (relative units) are compared.
Both the empty pDONR™221 (used in the BP reaction) and the DEST plasmids (used in the LR reaction) contain cat (chloramphenicol acetyltransferase) and ccdB (CcdB toxin). Therefore, these plasmids must be propagated with chloramphenicol and in E. coli strains that are resistant to the CcdB toxin (e.g., DB3.1™ or 2T1R™).
For more detailed explanations for the primer design to be used in Gateway® cloning procedures additional information is available at https://tools.thermofisher.com/content/sfs/manuals/gatewayman.pdf.
When recombining small fragments (<5 kb) in either the BP or LR reaction, a 2-h incubation is sufficient. However, when recombining larger fragments (>5 kb), longer incubation times may enhance the efficiency of the recombination reaction.
It is important to transform recombinant vectors into a strain that is susceptible to the CcdB toxin to maximize the cloning efficiency. In addition, glucose (final 0.2%) should also be added to the LB agar plates to minimize leaky expression.
There are short peptide sequence scars coding for attB recombination sites in the recombined destination vector as shown in Fig. 3. These additional amino acids do not affect BACTH complementation as determined by Ouellette et al. [14]. However, it cannot be guaranteed that these peptide sequences have no effect in all protein contexts.
Prepare CaCl2 competent Δcya E. coli cells (DHT1, DHM1, or BTH101) using standard procedures [25]. Before making fresh competent cells, the strains (DHT1, DHM1, or BTH101) from an LB-DMSO (or glycerol) stock should be restreaked on either MacConkey/maltose or LB/X-Gal/IPTG plates and grown overnight at 37 °C. White colonies (i.e., cya negative) should be picked to start the overnight liquid preculture. If red (on MacConkey/maltose plates) or blue colonies (on LB/X-Gal/IPTG plates) appear, then they should be avoided as they are likely Lac+ or Mal+ revertants or contaminants. If too many revertants or contaminants are present after restreaking from the stock, then a selective antibiotic can be added to the MacConkey/maltose or the LB/X-Gal/IPTG plates. DHT1 and DHM1 cells are resistant to nalidixic acid (30 μg/mL), and BTH101 cells are resistant to streptomycin (100 μg/mL).
DHT1, DHM1, or BTH101 competent cells can be prepared following standard procedures by the classical CaCl2 technique [25], (competency level >106 cfu/μg, which is sufficient for most routine transformations). Briefly, reisolated cells are grown in 1 L of LB at 37 °C to OD 0.25–0.3, then cooled on ice and pelleted by centrifugation. Next, the cells are washed twice in 100 mL ice cold 0.1 M CaCl2 solution, followed by resuspension in 30–40 mL ice-cold 0.1 M CaCl2 and incubated overnight at 4 °C. All steps of this process must keep cells, buffers, and vessels well chilled.
LB/X-gal indicator plates: In E. coli, the lacZ gene, which encodes β-galactosidase, is controlled by cAMP/CAP complex. A positive interaction between proteins fused to T25 and T18 fragments will bring the fragments together restoring adenylate cyclase activity and cAMP synthesis. In the presence of X-gal and β-galactosidase activity, blue colonies will form with interacting hybrid proteins and white colonies in cells expressing noninteracting proteins. After extended incubation, colonies often become pale blue, particularly at their center, and this should not be confused for a positive interaction. MacConkey/maltose indicator plates: In E. coli Δcya reporter strains are unable to ferment lactose or maltose [17, 25] and will form white/pink colonies on MacConkey/maltose agar. If the hybrid proteins interact (Cya+), then maltose genes will be transcribed and bacteria will ferment maltose, resulting in red colonies, from the acidification of the maltose/phenol red indicator [17, 25].
M63/maltose selective medium: M63 synthetic minimal medium supplemented with maltose is a more stringent medium to test for protein interactions. The mal regulon is strictly regulated by cAMP/CAP, so only Cya+ bacteria can utilize maltose. This makes this media useful to screen genomic libraries. The added X-gal and IPTG are useful in visually detecting Cya+ colonies (also Lac+ colonies, therefore blue colonies). The DHT1 E. coli strain is unable to synthesize isoleucine and valine [20, 22] so casamino acids should be added to the minimal/maltose medium to allow for growth.
The first BACTH library was constructed with genomic DNA from wild type E. coli MG1665. However, when used in a BACTH screen, many of the clones isolated as Mal+ on the selective medium were false positive and contained plasmids carrying fragments from the endogenous E. coli cya. Therefore, to avoid isolating false positive clones, it is better, if possible, to construct genomic libraries from bacterial strains lacking functional adenylate cyclase (e.g., cya mutants of E. coli [27, 28].
Efficient (>108 cfu/μg) electrocompetent DHM1/pKT25-X cells can be prepared as described [25]. Freshly reisolated cells are grown at 37 °C in 1 L of LB containing 50 μg/mL kanamycin until the OD600 reaches 0.5–0.7. Then the cells are chilled on ice and pelleted by centrifugation at 4 °C, washed three times with ice-cold water and finally resuspended in 10 mL of 10% glycerol (made in water). For transformation procedure, 50 μL are transferred into an electroporation cuvette (1 mm wide) previously equilibrated on ice and 50–100 ng of DNA from the BACTH plasmid DNA library are added. Cells are flicked to mix and incubated at 4 °C for a few minutes, then the cuvette is placed in an electroporator (e.g., Bio-Rad), set to 2.5 KV and 100 Ω capacitance, and then electroporation is performed. Following electroporation, 1 mL of LB media is immediately added to the cuvette, and cells are incubated at 30 °C for 60–90 min.
References
- 1.Fields S, O-k S (1989) A novel genetic system to detect protein–protein interactions. Nature 340(6230):245–246 [DOI] [PubMed] [Google Scholar]
- 2.Stynen B, Tournu H, Tavernier J, Van Dijck P (2012) Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system. Microbiol Mol Biol Rev 76(2):331–382. 10.1128/mmbr.05021-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karimova G, Ullmann A, Ladant D (2000) A bacterial two-hybrid system that exploits a cAMP signaling cascade in Escherichia coli. Methods Enzymol 328:59–73 [DOI] [PubMed] [Google Scholar]
- 4.Gyuris J, Golemis E, Chertkov H, Brent R (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75(4):791–803 [DOI] [PubMed] [Google Scholar]
- 5.Ladant D, Ullmann A (1999) Bordatella pertussis adenylate cyclase: a toxin with multiple talents. Trends Microbiol 7(4):172–176 [DOI] [PubMed] [Google Scholar]
- 6.Karimova G, Pidoux J, Ullmann A, Ladant D (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci U S A 95(10):5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busby S, Ebright RH (1999) Transcription activation by catabolite activator protein (CAP). J Mol Biol 293(2):199–213 [DOI] [PubMed] [Google Scholar]
- 8.Karimova G, Dautin N, Ladant D (2005) Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 187(7):2233–2243. 10.1128/jb.187.7.2233-2243.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jack RL, Buchanan G, Dubini A, Hatzixanthis K, Palmer T, Sargent F (2004) Coordinating assembly and export of complex bacterial proteins. EMBO J 23(20):3962–3972. 10.1038/sj.emboj.7600409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cisneros DA, Bond PJ, Pugsley AP, Campos M, Francetic O (2012) Minor pseudopilin self-assembly primes type II secretion pseudopilus elongation. EMBO J 31(4):1041–1053. 10.1038/emboj.2011.454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gauliard E, Ouellette SP, Rueden KJ, Ladant D (2015) Characterization of interactions between inclusion membrane proteins from Chlamydia trachomatis. Front Cell Infect Microbiol 5:13. 10.3389/fcimb.2015.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgiadou M, Castagnini M, Karimova G, Ladant D, Pelicic V (2012) Large-scale study of the interactions between proteins involved in type IV pilus biology in Neisseria meningitidis: characterization of a subcomplex involved in pilus assembly. Mol Microbiol 84(5):857–873. 10.1111/j.1365-2958.2012.08062.x [DOI] [PubMed] [Google Scholar]
- 13.Zoued A, Durand E, Brunet YR, Spinelli S, Douzi B, Guzzo M, Flaugnatti N, Legrand P, Journet L, Fronzes R, Mignot T, Cambillau C, Cascales E (2016) Priming and polymerization of a bacterial contractile tail structure. Nature 531(7592):59–63. 10.1038/nature17182 [DOI] [PubMed] [Google Scholar]
- 14.Ouellette SP, Gauliard E, Antosova Z, Ladant D (2014) A Gateway((R))-compatible bacterial adenylate cyclase-based two-hybrid system. Environ Microbiol Rep 6(3):259–267. 10.1111/1758-2229.12123 [DOI] [PubMed] [Google Scholar]
- 15.Lawson CL, Swigon D, Murakami KS, Darst SA, Berman HM, Ebright RH (2004) Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol 14(1):10–20. 10.1016/j.sbi.2004.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karimova G, Ullmann A, Ladant D (2001) Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J Mol Microbiol Biotechnol 3(1):73–82 [PubMed] [Google Scholar]
- 17.Dautin N, Karimova G, Ladant D (2003) Human immunodeficiency virus (HIV) type 1 transframe protein can restore activity to a dimerization-deficient HIV protease variant. J Virol 77(15):8216–8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Battesti A, Bouveret E (2012) The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods (San Diego, Calif) 58(4):325–334. 10.1016/j.ymeth.2012.07.018 [DOI] [PubMed] [Google Scholar]
- 19.Fransen M, Brees C, Ghys K, Amery L, Mannaerts GP, Ladant D, Van Veldhoven PP (2002) Analysis of mammalian peroxin interactions using a non-transcription-based bacterial two-hybrid assay. Mol Cell Proteomics 1(3):243–252 [DOI] [PubMed] [Google Scholar]
- 20.Ouellette SP, Rueden KJ, Gauliard E, Persons L, de Boer PA, Ladant D (2014) Analysis of MreB interactors in Chlamydia reveals a RodZ homolog but fails to detect an interaction with MraY. Front Microbiol 5:279. 10.3389/fmicb.2014.00279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dautin N, Karimova G, Ullmann A, Ladant D (2000) Sensitive genetic screen for protease activity based on a cyclic AMP signaling cascade in Escherichia coli. J Bacteriol 182(24):7060–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ouellette SP, Karimova G, Subtil A, Ladant D (2012) Chlamydia co-opts the rod shape-determining proteins MreB and Pbp2 for cell division. Mol Microbiol 85(1):164–178. 10.1111/j.1365-2958.2012.08100.x [DOI] [PubMed] [Google Scholar]
- 23.Hartley JL, Temple GF, Brasch MA (2000) DNA cloning using in vitro site-specific recombination. Genome Res 10(11):1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sands B, Brent R (2015) Overview of post Cohen-Boyer methods for single segment cloning and for multisegment DNA assembly. Curr Protoc Mol Biol 113:3.26.1–23.26.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Russell DW (2006) The condensed protocols from molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26.Kramer M, Coen D (2001) Enzymatic amplification of DNA by PCR: standard procedures and optimization. Curr Protoc Immunol Chapter10:Unit 10.20 [DOI] [PubMed] [Google Scholar]
- 27.Karimova G, Davi M, Ladant D (2012) The beta-lactam resistance protein Blr, a small membrane polypeptide, is a component of the Escherichia coli cell division machinery. J Bacteriol 194(20):5576–5588. 10.1128/jb.00774-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karimova G, Robichon C, Ladant D (2009) Characterization of YmgF, a 72-residue inner membrane protein that associates with the Escherichia coli cell division machinery. J Bacteriol 191(1):333–346. 10.1128/jb.00331-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson K (2001) Preparation of genomic DNA from bacteria. Curr Protoc Mol Biol Chapter 2:Unit 2.4 [DOI] [PubMed] [Google Scholar]
- 30.Ouellette SP, Karimova G, Davi M, Ladant D (2017) Analysis of membrane protein interactions with a bacterial adenylate cyclase-based two-hybrid (BACTH) technique. Curr Protoc Mol Biol 118:20.12.21–20.12.24. 10.1002/cpmb.36 [DOI] [PubMed] [Google Scholar]
- 31.Zervos AS, Gyuris J, Brent R (1993) Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell 72(2):223–232 [DOI] [PubMed] [Google Scholar]
- 32.Miller J (1993) A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Trends Biochem Sci 18:193 [Google Scholar]
- 33.Nichols NM (2011) Endonucleases. Curr Protoc Mol Biol Chapter 3:Unit 3.12 [DOI] [PubMed] [Google Scholar]
- 34.Kucera RB, Nichols NM (2008) DNA-dependent DNA polymerases. Curr Protoc Mol Biol Chapter 3:3.5.1–3.5.19 [DOI] [PubMed] [Google Scholar]
- 35.Bloch KD, Grossmann B (1995) Digestion of DNA with restriction endonucleases. Curr Protoc Mol Biol Chapter 3:3.1.1–3.1.21 [DOI] [PubMed] [Google Scholar]
- 36.Voytas D (2001) Agarose gel electrophoresis. Curr Protoc Mol Biol. 10.1002/0471142727.mb0205as51 [DOI] [PubMed] [Google Scholar]
- 37.Chen AL, Johnson KA, Lee JK, Sutterlin C, Tan M (2012) CPAF: a Chlamydial protease in search of an authentic substrate. PLoS Pathog 8(8):e1002842. 10.1371/journal.ppat.1002842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleinschnitz EM, Heichlinger A, Schirner K, Winkler J, Latus A, Maldener I, Wohlleben W, Muth G (2011) Proteins encoded by the mre gene cluster in Streptomyces coelicolor A3 (2) cooperate in spore wall synthesis. Mol Microbiol 79(5):1367–1379 [DOI] [PubMed] [Google Scholar]
- 39.Houot L, Fanni A, de Bentzmann S, Bordi C (2012) A bacterial two-hybrid genome fragment library for deciphering regulatory networks of the opportunistic pathogen Pseudomonas aeruginosa. Microbiology 158(8):1964–1971 [DOI] [PubMed] [Google Scholar]
- 40.Pfeiffer D, Jendrossek D (2011) Interaction between poly (3-hydroxybutyrate) granule-associated proteins as revealed by two-hybrid analysis and identification of a new phasin in Ralstonia eutropha H16. Microbiology 157(10):2795–2807 [DOI] [PubMed] [Google Scholar]
- 41.Griffith KL, Wolf RE Jr (2002) Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem Biophys Res Commun 290(1):397–402. 10.1006/bbrc.2001.6152 [DOI] [PubMed] [Google Scholar]
- 42.Naumovski L (2001) Two-hybrid interactions confirmed by coimmunoprecipitation of epitope-tagged clones. In: MacDonald PN (ed) Two-hybrid systems: methods and protocols. Humana Press, Totowa, pp 151–159. 10.1385/1-59259-210-4:151 [DOI] [PubMed] [Google Scholar]
- 43.Kraichely DM, MacDonald PN (2001) Confirming yeast two-hybrid protein interactions using in vitro glutathione-S-transferase pulldowns. In: MacDonald PN (ed) Two-hybrid systems: methods and protocols. Humana Press, Totowa, pp 135–150. 10.1385/1-59259-210-4:135 [DOI] [PubMed] [Google Scholar]
- 44.Robichon C, Karimova G, Beckwith J, Ladant D (2011) Role of leucine zipper motifs in association of the Escherichia coli cell division proteins FtsL and FtsB. J Bacteriol 193(18):4988–4992. 10.1128/jb.00324-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Battesti A, Bouveret E (2008) Improvement of bacterial two-hybrid vectors for detection of fusion proteins and transfer to pBAD-tandem affinity purification, calmodulin binding peptide, or 6-histidine tag vectors. Proteomics 8(22):4768–4771. 10.1002/pmic.200800270 [DOI] [PubMed] [Google Scholar]