Abstract
Inherited retinal dystrophies (IRD) are a group of diseases characterized by the loss or dysfunction of photoreceptors and a high genetic and clinical heterogeneity. Currently, over 270 genes have been associated with IRD which makes genetic diagnosis very difficult. The recent advent of next generation sequencing has greatly facilitated the diagnostic process, enabling to provide the patients with accurate genetic counseling in some cases. We studied 92 patients who were clinically diagnosed with IRD with two different custom panels. In total, we resolved 53 patients (57.6%); in 12 patients (13%), we found only one mutation in a gene with a known autosomal recessive pattern of inheritance; and 27 patients (29.3%) remained unsolved. We identified 120 pathogenic or likely pathogenic variants; 30 of them were novel. Among the cone-rod dystrophy patients, ABCA4 was the most common mutated gene, meanwhile, USH2A was the most prevalent among the retinitis pigmentosa patients. Interestingly, 10 families carried pathogenic variants in more than one IRD gene, and we identified two deep-intronic variants previously described as pathogenic in ABCA4 and CEP290. In conclusion, the IRD study through custom panel sequencing demonstrates its efficacy for genetic diagnosis, as well as the importance of including deep-intronic regions in their design. This genetic diagnosis will allow patients to make accurate reproductive decisions, enroll in gene-based clinical trials, and benefit from future gene-based treatments.
Keywords: inherited retinal dystrophies, diagnosis, custom-panels, gene, pathogenic, deep-intronic
Introduction
Inherited retinal dystrophies (IRD) are a group of diseases characterized by the progressive death or dysfunction of photoreceptors that leads to vision loss and, in some cases, legal blindness. IRDs have a prevalence of one case in 3,000 individuals (Sahel et al., 2015). Depending on the photoreceptor initially affected, IRD can be classified into cone, rod-cone, or cone-rod dystrophies in those cases in which both are affected at one time. Moreover, they can manifest as either isolated (70–80% of the total) or part of one of the 80 syndromes that have been estimated to be associated with IRD (Ayuso and Millan, 2010; Tatour and Ben-Yosef, 2020).
This group of diseases has a wide clinical spectrum and number of involved genes, currently reaching 271 for syndromic and non-syndromic forms (Retnet1, December 2020); explaining why IRDs have such a high clinical and genetic heterogeneity. Furthermore, there is a high inter and intrafamily variability, variable expression, and incomplete penetrance (Farrar et al., 2017). IRDs can follow different patterns of inheritance, including autosomal recessive, autosomal dominant, X-linked, mitochondrial mode, and some other less common, such as uniparental isodisomy or digenic inheritance (Kajiwara et al., 1994; Dryja et al., 1997; Rivolta et al., 2002; Ayuso and Millan, 2010; Parmeggiani et al., 2011; Neveling et al., 2012). These issues, together with the fact that 50% are sporadic cases, make it even more complicated to determine the mode of inheritance, genetic diagnosis, and genetic counseling (Perea-Romero et al., 2021).
With the advent of next generation sequencing (NGS), the ratio of diagnosis has risen to 50–70%, which was difficult to achieve a few years ago (Carss et al., 2017; Sanchis-Juan et al., 2018; Rodríguez-Muñoz et al., 2020). Some different approaches, such as custom panel designs, whole exome sequencing (WES), and whole genome sequencing (WGS), have been implemented to study the molecular mechanisms of IRD. Currently, these new sequencing techniques are essential to obtain an early and accurate genetic diagnosis, which is necessary to offer a correct genetic counseling to patients and their families (Salmaninejad et al., 2019).
In this study, we analyzed 92 patients, who were previously clinically diagnosed with IRD, with two custom panels for the main aim of achieving genetic diagnoses.
Materials and Methods
Cohort Selection
We selected 92 patients, belonging to 90 different Spanish families, with a clinical diagnosis of non-syndromic IRD, except for one who had a clinical suspicion of a syndromic IRD. Patient DNA was isolated from peripheral blood with the automatic extractor QIAsymphony (QIAGEN).
Patients underwent complete ophthalmologic examinations, including OCT (Heidelberg Spectralis OCT Bluepeak, Heidelberg, Germany; Topcon 3D OCT 2000, Tokyo, Japan; CIRRUS OCT Zeiss, Oberkochen, Germany), ERG (Roland RETI-port/scan21, Brandenburg, Germany), eye fundus (Visucam NM/FA Zeiss, Oberkochen, Germany), visual acuity measure (BCVA), and evoked potentials and visual fields (Carl Zeiss Humphrey Field Analyzer, Oberkochen, Germany). A clinical questionnaire, which collected the main IRD characteristics, and an informed consent were completed by each patient. This study was approved by the Hospital La Fe Ethics Committee, in agreement with the Declaration of Helsinki.
Panel Design and Sequencing
Sixty-three patients were sequenced with a gene panel version (PV1) that included 117 genes involved in a non-syndromic IRD, and their flanking intronic regions (±25 base pairs) (Rodríguez-Muñoz et al., 2020). Moreover, the panel of genes contained five intronic regions of ABCA4, OFD1, USH2A, CEP290, and PRPF31, in which pathogenic variants had been previously identified (Den Hollander et al., 2006; Littink et al., 2010; Vaché et al., 2012; Webb et al., 2012; Braun et al., 2013; Supplementary Table 1).
The remaining 29 cases were analyzed with an updated version of the custom panel (PV2) that had 114 genes and all the deep-intronic variants described in the last few years in ABCA4 and USH2A (Vaché et al., 2012; Braun et al., 2013; Zernant et al., 2014; Bauwens et al., 2015, 2019; Liquori et al., 2016; Baux et al., 2017; Fadaie et al., 2019; Khan et al., 2019; Sangermano et al., 2019; Supplementary Table 2).
The patients’ libraries were prepared in accordance with the SureSelect QXT protocol (Agilent Technologies) and sequenced on a MiSeq platform (Illumina, San Diego, CA) in 300 cycles with 2 × 150 base pairs reads.
Data Analysis
The reads alignment against the reference hg19 genome, variant calling, and annotation of all the identified variants were carried out with the Alissa resource (Agilent Technologies). The obtained variants were filtered based on a MAF (minor allele frequency) ≤ 0.01 according to the ExAC2 and gnomAD3 databases. In order to evaluate the pathogenicity of the detected variants, we also evaluated specific databases such as ClinVar4, Locus Specific Data Base5, and HGMD professional6. To evaluate the potential effect of novel missense variants, we used the in silico predictors included in Varsome7. The putative effect on the splicing process was performed with HSF (Human SplicingFinder8), NNSplice9, and SpliceAI10. Finally, the IGV view finder11 allowed the examination of all detected variants in every read.
The novel variants identified in this study were classified according to the standards of the American College of Medical Genetics and Genomics (ACMG) (Richards et al., 2015).
Copy Number Variation Analysis
A copy number variations (CNV) analysis was performed using the DECoN bioinformatic tool version 1.0.2 (Fowler et al., 2016).
We also studied large rearrangements by Multiplex Ligation-dependent Probe Amplification (MLPA; MRC Holland) in patients harboring one pathogenic variant in USH2A (probemixes P361 and P362), EYS (probemix P328-A3), andABCA4 (probemixes P151 and P152). The multiplex ligation-dependent probe amplification results were analyzed by the Coffalyser. Net software version 140721.1958 (MRC-Holland).
Sanger Sequencing and Segregation Analysis
The candidate variants identified in each patient were validated by Sanger sequencing (Big Dye Terminator v1.1 or v3.1, Applied Biosystems by Life Technologies). Moreover, the deep-intronic variants, already described as pathogenic in USH2A and ABCA4 (Supplementary Table 3; Vaché et al., 2012; Braun et al., 2013; Zernant et al., 2014; Bauwens et al., 2015, 2019; Liquori et al., 2016; Baux et al., 2017; Fadaie et al., 2019; Khan et al., 2019; Sangermano et al., 2019), were studied by Sanger sequencing in patients analyzed with PV1 who carried only one pathogenic variant in either or both genes.
Segregation analysis was performed when relatives’ DNA was available.
Results
We obtained a mean depth of 190× per patient. In 98.6% of the patients, at least 88% of the bases were covered with a sequencing depth of coverage ≥50× and 95% of the bases were covered with at least 20×. Moreover, we obtained a 70% of on-target reads.
We solved 53 patients belonging to 52 families (53/92, ratio of 57.6%): 17 (of 17 families) which had disease-causing variants in autosomal dominant genes, 33 (of 32 families) with autosomal recessive genes, two patients (of two families) who carried a pathogenic variant in an X-linked gene and one patient who carried two different pathogenic variants in two different autosomal dominant genes and two additional variants in an autosomal recessive gene (RPN-670). We reported five cases with more than two pathogenic variants in the same autosomal recessive gene, and we also described six patients who carry pathogenic variants in more than one IRD gene (Table 1). Among the unresolved patients, 13% carried one pathogenic variant in an autosomal recessive IRD gene, with a higher prevalence of cases with a heterozygous variant in ABCA4, followed by USH2A and RPGRIP1 (Table 2). In the remaining 29.3% cases, no pathogenic variant was identified.
TABLE 1.
Pathogenic variants identified in solved patients.
| Family | Patient | Clinical diagnosis | Gene | Nucleotide change | Protein change | Zygosity | References |
| $FRPN-511 | RPN-129 | STG | ABCA4 (NM_000350.3) | c.1804C > T | p.(Arg602Trp) | Heterozygous | Lewis et al., 1999 |
| c.982G > T | p.(Glu328*) | Heterozygous | Fishman, 2003 | ||||
| RPN-544 | STG | ABCA4 (NM_000350.3) | c.5882G > A | p.(Gly1961Glu) | Heterozygous | Allikmets et al., 1997 | |
| c.982G > T | p.(Glu328*) | Heterozygous | Fishman, 2003 | ||||
| $FRPN-2441 | RPN-646 | RP | RHO (NM_000539.3) | c.328T > C | p.(Cys110Arg) | Heterozygous | To et al., 2004 |
| FRPN-2461 | RPN-649 | MD/STG | ABCA4 (NM_000350.3) | c.5917del | p.(Val1973*) | Homozygous | Rivolta et al., 2000 |
| $FRPN-2521 | RPN-657 | STG | ABCA4 (NM_000350.3) | c.5714 + 1G > A | p.? | Heterozygous | This study |
| c.3386G > T | p.(Arg1129Leu) | Heterozygous | Allikmets et al., 1997 | ||||
| FRPN-2541 | RPN-659 | RP | EYS (NM_001142800.2) | c.7736_7742del | p.(Thr2579Lysfs*36) | Homozygous | This study |
| FRPN-2551 | RPN-660 | RP | USH2A (NM_206933.4) | c.4732C > T | p.(Arg1578Cys) | Heterozygous | Le Quesne Stabej et al., 2012 |
| c.1214del | p.(Asn405Ilefs*3) | Heterozygous | Schwartz et al., 2005 | ||||
| FRPN-2561 | RPN-661 | RP/LCA | CRB1 (NM_201253.3) | c.2416G > T | p.(Glu806*) | Homozygous | Corton et al., 2013 |
| FRPN-2581 | RPN-663 | STG | ABCA4 (NM_000350.3) | c.3386G > T | p.(Arg1129Leu) | Homozygous | Allikmets et al., 1997 |
| c.6718A > G | p.(Thr2240Ala) | Heterozygous | Zernant et al., 2011 | ||||
| FRPN-2611 | RPN-666 | Reverse BCAMD/STG/RP | ABCA4 (NM_000350.3) | c.5929G > A | p.(Gly1977Ser) | Heterozygous | Rozet et al., 1998 |
| c.5882G > A | p.(Gly1961Glu) | Heterozygous | Allikmets et al., 1997 | ||||
| FRPN-2631 | RPN-668 | CD | CRB1 (NM_201253.3) | c.1604T > C | p.(Leu535Pro) | Heterozygous | Corton et al., 2013 |
| c.2843G > A | p.(Cys948Tyr) | Heterozygous | den Hollander et al., 1999 | ||||
| FRPN-2651 | RPN-670 | NA | RP1 (NM_006269.2) | c.3157del | p.(Tyr1053Thrfs*4) | Heterozygous | Jacobson et al., 2000 |
| PRPH2 (NM_000322.5) | c.623G > A | p.(Gly208Asp) | Heterozygous | Kohl et al., 1997 | |||
| USH2A (NM_206933.4) | c.6957 + 1G > C | p.? | Heterozygous | This study | |||
| c.4955C > T | p.(Pro1652Leu) | Heterozygous | This study | ||||
| FRPN-2661 | RPN-671 | RP | EYS (NM_001142800.2) | c.5928-2A > G | p.? | Homozygous | González-del Pozo et al., 2011 |
| FRPN-2671 | RPN-672 | RP | CNGB1 (NM_001297.5) | c.2492 + 2T > G | p.? | Homozygous | This study |
| FRPN-2681 | RPN-673 | RP | USH2A (NM_206933.4) | c.2276G > T | p.(Cys759Phe) | Heterozygous | Rivolta et al., 2000 |
| c.13894C > T | p.(Pro4632Ser) | Heterozygous | This study | ||||
| FRPN-2691 | RPN-675 | RP | USH2A (NM_206933.4) | c.4732C > T | p.(Arg1578Cys) | Heterozygous | Le Quesne Stabej et al., 2012 |
| c.12575G > A | p.(Arg4192His) | Heterozygous | Ávila-Fernández et al., 2010 | ||||
| FRPN-2731 | RPN-679 | STG | ABCA4 (NM_000350.3) | c.4880del | p.(Leu1627Argfs*35) | Heterozygous | This study |
| c.5714 + 5G > A | p.? | Heterozygous | Cremers, 1998 | ||||
| c.2953G > A | p.(Gly985Arg) | Heterozygous | This study | ||||
| FRPN-2761 | RPN-682 | MD | BEST1 (NM_004183.4) | c.247G > T | p.(Val83Phe) | Heterozygous | Kinnick et al., 2011 |
| FRPN-2771 | RPN-683 | RP | RPGR (NM_001034853.2) | c.1366del | p.(Gln456Lysfs*20) | Hemizygous | This study |
| FRPN-2781 | RPN-684 | RP | EYS (NM_001142800.2) | c.9468T > A | p.(Tyr3156*) | Homozygous | Collin et al., 2008 |
| FRPN-2791 | RPN-685 | NA | PRPH2 (NM_000322.5) | c.658C > T | p.(Arg220Trp) | Heterozygous | Payne et al., 1998 |
| FRPN-2801 | RPN-686 | RP | USH2A (NM_206933.4) | c.12575G > A | p.(Arg4192His) | Homozygous | Ávila-Fernández et al., 2010 |
| FRPN-2811 | RPN-687 | CRD/STG | CRB1 (NM_201253.3) | c.481G > A | p.(Ala161Thr) | Homozygous | This study |
| FRPN-2841 | RPN-692 | RP Punctata albensces | ABCA4 (NM_000350.3) | c.3386G > T | p.(Arg1129Leu) | Heterozygous | Allikmets et al., 1997 |
| c.6148G > C | p.(Val2050Leu) | Heterozygous | Lewis et al., 1999 | ||||
| $FRPN-2861 | RPN-694 | NA | PRPH2 (NM_000322.5) | c.440dup | p.(Gly148Trpfs*29) | Heterozygous | This study |
| $FRPN-2891 | RPN-697 | STG | ABCA4 (NM_000350.3) | c.3386G > T | p.(Arg1129Leu) | Heterozygous | Allikmets et al., 1997 |
| c.634C > T | p.(Arg212Cys) | Heterozygous | Gerber et al., 1998 | ||||
| FRPN-2931 | RPN-701 | RP | PRPH2 (NM_000322.5) | c.647C > A | p.(Pro216His) | Heterozygous | This study |
| FRPN-2941 | RPN-702 | MD | ELOVL4 (NM_022726.4) | c.59A > G | p.(Asn20Ser) | Heterozygous | Hu et al., 2020 |
| FRPN-2961 | RPN-704 | CD | GUCA1A (NM_000409.5) | c.66C > A | p.(Tyr22*) | Heterozygous | This study |
| FRPN-2981 | RPN-706 | CRD | CRB1 (NM_201253.3) | c.613_619del | p.(Ile205Aspfs*13) | Homozygous | Lotery, 2001 |
| c.2291G > A | p.(Arg764His) | Heterozygous | Corton et al., 2013 | ||||
| FRPN-2991 | RPN-707 | RP | USH2A (NM_206933.4) | c.13811 + 2T > G | p.? | Heterozygous | Besnard et al., 2014 |
| c.2276G > T | p.(Cys759Phe) | Heterozygous | Rivolta et al., 2000 | ||||
| FRPN-3001 | RPN-708 | NA | OTX2 (NM_001270525.2) | c.638T > A | p.(Leu213*) | Heterozygous | This study |
| FRPN-3011 | RPN-709 | RP | PROM1 (NM_006017.2) | deletion exon 11 | p.? | Homozygous | This study |
| FRPN-3021 | RPN-710 | MD/BVMD | PRPH2 (NM_000322.5) | c.641G > A | p.(Cys214Tyr) | Heterozygous | Trujillo et al., 2001 |
| FRPN-3031 | RPN-711 | RP | USH2A (NM_206933.4) | c.14803C > T | p.(Arg4935*) | Heterozygous | Baux et al., 2007 |
| c.2332G > T | p.(Asp778Tyr) | Heterozygous | Lenassi et al., 2015 | ||||
| FRPN-3071 | RPN-715 | RP | RHO (NM_000539.3) | c.512C > A | p.(Pro171Gln) | Heterozygous | Antiñolo et al., 1994 |
| $FRPN-3082 | RPN-717 | RP | ABCA4 (NM_000350.3) | c.6148G > C | p.(Val2050Leu) | Heterozygous | Allikmets et al., 1997 |
| PRPF31 (NM_015629.3) | Gene deletion | p.? | Heterozygous | This study | |||
| FRPN-3092 | RPN-718 | RP | USH2A (NM_206933.4) | c.2276G > T | p.(Cys759Phe) | Homozygous | Rivolta et al., 2000 |
| FRPN-3122 | RPN-721 | RP | RHO (NM_000539.3) | c.512C > A | p.(Pro171Leu) | Heterozygous | Stone et al., 2017 |
| FRPN-3152 | RPN-725 | MD/STG | ABCA4 (NM_000350.3) | c.6310C > T | p.(Gln2104*) | Heterozygous | This study |
| c.3386G > T | p.(Arg1129Leu) | Heterozygous | Allikmets et al., 1997 | ||||
| FRPN-3162 | RPN-726 | RP | NRL (NM_001354768.3) | c.149C > T | p.(Ser50Leu) | Heterozygous | Koyanagi et al., 2019 |
| ABCA4 (NM_000350.3) | c.5908C > T | p.(Leu1970Phe) | Heterozygous | Rozet et al., 1998 | |||
| FRPN-3182 | RPN-728 | Joubert syndrome | CEP290 (NM_025114.4) | c.4966_4967del | p.(Glu1656Asnfs*3) | Heterozygous | Sheck et al., 2018 |
| c.2817G > T | p.(Lys939Asn) | Heterozygous | Srivastava et al., 2017 | ||||
| FRPN-3202 | RPN-730 | MD/STG | PRPH2 (NM_000322.5) | c.537G > A | p.(Trp179*) | Heterozygous | Diñeiro et al., 2020 |
| FRPN-3222 | RPN-732 | RP | PDE6B (NM_000283.4) | c.1920 + 1G > A | p.? | Heterozygous | This study |
| c.2470_2478del | p.(Lys824_Glu826del) | Heterozygous | This study | ||||
| FRPN-3232 | RPN-733 | MD | PRPH2 (NM_000322.5) | c.421T > C | p.(Tyr141His) | Heterozygous | Diñeiro et al., 2020 |
| ABCA4 (NM_000350.3) | c.5908C > T | p.(Leu1970Phe) | Heterozygous | Rozet et al., 1998 | |||
| FRPN-3242 | RPN-734 | MD/CD | ABCA4 (NM_000350.3) | c.3113C > T | p.(Ala1038Val) | Heterozygous | Rozet et al., 1998 |
| c.4539 + 2064C > T | [p.?; p.(=, Arg1514Leufs*36)] | Heterozygous | Zernant et al., 2014; Bauwens et al., 2019 | ||||
| c.1364T > A | p.(Leu455Gln) | Heterozygous | Salles et al., 2018 | ||||
| FRPN-3252 | RPN-735 | NA | EYS (NM_001142800.2) | c.8854del | p.(Thr2973Leufs*23) | Heterozygous | This study |
| c.1194del | p.(Gly399Aspfs*22) | Heterozygous | This study | ||||
| RDH5 (NM_002905.3) | c.712G > T | p.(Gly238Trp) | Heterozygous | Gonzalez-Fernandez et al., 1999 | |||
| FRPN-3282 | RPN-737 | MD/STG | PRPH2 (NM_000322.5) | c.641G > A | p.(Cys214Tyr) | Heterozygous | Trujillo et al., 2001 |
| FPRN-3272 | RPN-738 | MD/STG | BBS1 (NM_024649.4) | c.1169T > G | p.(Met390Arg) | Homozygous | Nishimura et al., 2010 |
| FRPN-3352 | RPN-745 | RP | RPGR (NM_001034853.2) | c.935-2A > G | p.? | Hemizygous | Koyanagi et al., 2019 |
| ABCA4 (NM_000350.3) | c.5908C > T | p.(Leu1970Phe) | Heterozygous | Rozet et al., 1998 | |||
| PDE6A (NM_000440.3) | c.2144T > C | p.(Met715Thr) | Heterozygous | This study | |||
| FRPN-3372 | RPN-747 | RP | RHO (NM_000539.3) | c.670G > A | p.(Gly224Arg) | Heterozygous | This study |
| FRPN-3392 | RPN-749 | STG | ABCA4 (NM_000350.3) | c.3386G > T | p.(Arg1129Leu) | Heterozygous | Allikmets et al., 1997 |
| c.3210_3211dup | p.(Ser1071Cysfs*14) | Heterozygous | Nasonkin et al., 1998 | ||||
| c.560G > A | p.(Arg187His) | Heterozygous | Cornelis et al., 2017 | ||||
| $FRPN-3402 | RPN-750 | LCA | CEP290 (NM_025114.4) | c.2991 + 1655A > G | p.? | Homozygous | Den Hollander et al., 2006 |
Novel pathogenic variants are highlighted in bold font.
FRPN, family number; FRPN-“1,” families studied with PV1; FRPN-“2,” families studied with PV2; $FRPN, families in which segregation analysis was performed; RPN, patient number; RP, retinitis pigmentosa; MD, macular dystrophy; p.?, unknown protein effect; STG, stargardt; LCA, leber congenital amaurosis; BCAMD, benign concentric annular macular dystrophy; CD, cone dystrophy; CRD, cone-rod dystrophy; BVMD, best vitelliform macular dystrophy; NA, not available. The “*” symbol corresponds to: stop-codon according to the format of mutations nomenclature from the Human Genome Variation Society (HGVS).
TABLE 2.
Patients in which only one pathogenic variant in a recessive gene has been identified.
| Family | Patient | Clinic diagnosis | Gene | Nucleotide change | Protein change | Zygosity | References |
| FRPN-2431 | RPN-645 | RP | USH2A (NM_206933.4) | c.2276G > T | p.(Cys759Phe) | heterozygous | Rivolta et al., 2000 |
| CEP290 (NM_025114.4) | c.7394_7395del | p.(Glu2465Valfs*2) | heterozygous | This study | |||
| FRPN-2721 | RPN-678 | STG | ABCA4 (NM_000350.3) | c.288C > A | p.(Asn96Lys) | heterozygous | Stenirri et al., 2004 |
| USH2A (NM_206933.4) | c.754G > T | p.(Gly252Cys) | heterozygous | Bravo-Gil et al., 2016 | |||
| FRPN-2831 | RPN-691 | RP Punctata albensces | CDHR1 (NM_033100.3) | c.783G > A | (p.Pro261 =) | heterozygous | Glockle et al., 2014 |
| POC1B (NM_172240.3) | c.1079_1080del | p.(Pro360Argfs*8) | heterozygous | This study | |||
| FRPN-2871 | RPN-695 | NA | ABCA4 (NM_000350.3) | c.6089G > A | p.(Arg2030Gln) | heterozygous | Lewis et al., 1999 |
| FRPN-2881 | RPN-696 | NA | ABCA4 (NM_000350.3) | c.6148G > C | p.(Val2050Leu) | heterozygous | Allikmets et al., 1997 |
| FRPN-3041 | RPN-712 | RP | RPGRIP1 (NM_020366.3) | c.3339 + 5G > C | p.? | heterozygous | This study |
| FRPN-3051 | RPN-713 | RP | SAG (NM_000541.5) | c.577C > T | p.(Arg193*) | heterozygous | Maw et al., 1998 |
| GUCY2D (NM_000180.4) | c.1991A > G | p.(His664Arg) | heterozygous | This study | |||
| FRPN-3082 | RPN-717 | RP | ABCA4 (NM_000350.3) | c.6148G > C | p.(Val2050Leu) | heterozygous | Allikmets et al., 1997 |
| FRPN-3112 | RPN-720 | RP (early onset) | RPGRIP1 (NM_020366.3) | c.767C > G | p.(Ser256*) | heterozygous | Jamshidi et al., 2019 |
| FRPN-3132 | RPN-722 | RP | RBP3 (NM_002900.3) | c.3238G > A | p.(Asp1080Asn) | heterozygous | den Hollander et al., 2009 |
| FRPN-3142 | RPN-724 | RP | EYS (NM_001142800.2) | c.6882_6883del | p.(Gln2294Hisfs*3) | heterozygous | This study |
| FRPN-3192 | RPN-729 | RP | CNGA3 (NM_001298.3) | c.673 + 5G > T | p.? | heterozygous | This study |
| FRPN-3362 | RPN-746 | MD | PCARE (NM_001271441.2) | c.656_665dup | p.(Ala223Argfs*38) | heterozygous | This study |
Novel pathogenic variants identified are highlighted in bold font.
FRPN, family number; FRPN-“1,” families studied with PV1; FRPN-“2,” families studied with PV2; RPN, patient number; RP, retinitis pigmentosa; STG, stargardt; NA, not available; p.?, unknown protein effect; SNHL, sensorineural hearing loss; XL, X-linked. The “*” symbol corresponds to: stop-codon according to the format of mutations nomenclature from the Human Genome Variation Society (HGVS).
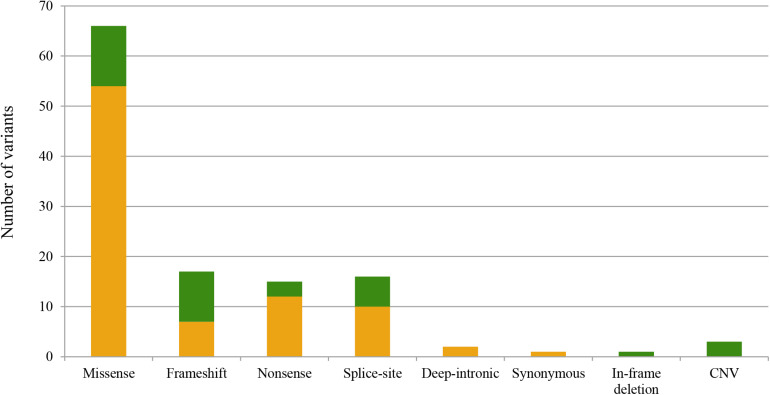
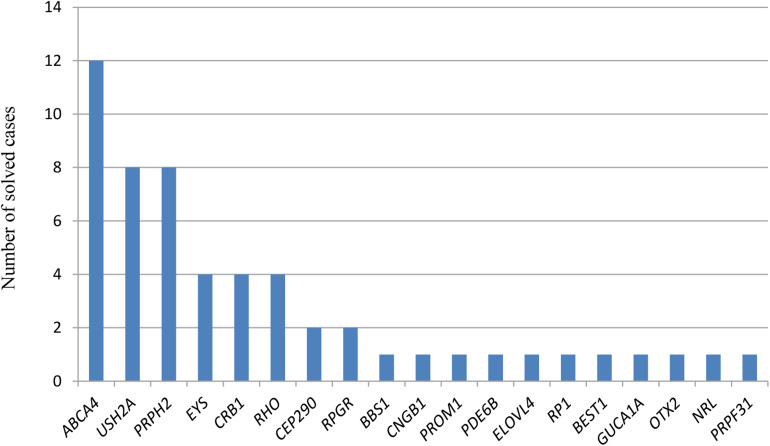
We identified a total of 120 pathogenic or likely pathogenic variants (of which, 85 were unique), including: 65 missense, 15 nonsense, 17 frameshift, 16 splice-site variants, two pathogenic deep-intronic variants, one in-frame deletion, one synonymous, and three CNV (Figure 1). Twenty-nine variants were first described in this study (Tables 1–3). In line with this, we identified a high number of mutated-genes, nine autosomal dominant genes (highest prevalence of PRPH2); 19 autosomal recessive genes, among which, ABCA4 and USH2A stand out; and finally, one X-linked gene, RPGR (Figure 2). All identified novel variants were classified as pathogenic or likely pathogenic according to the ACMG criteria except for variant c.2470_2478del (p.Lys824_Glu826del) in PDE6B, which remained as a variant of uncertain significance. This patient harbored another heterozygous pathogenic variant in the same gene; however, we could not carry out the segregation analysis to confirm it as likely pathogenic.
FIGURE 1.
Representation of the total alleles identified in this study classified according to the alteration type. The X-axis refers to the different types of variants identified in this study, and the Y-axis concerns the total allele count for each type of variant. Pathogenic variants identified in this study are represented in green, while those previously described are in orange.
TABLE 3.
Criteria considered for the pathogenicity classification of the novel identified variants.
| Gene | Mutation |
Classification | Frequency (gnomAD Ex) | Pathogenicity scoresa | Conservation score (GERP)b | Reputable source |
||
| Nucleotide change | Protein change | ClinVar | HGMDc | |||||
| ABCA4 (NM_000350.3) | c.2953G > A | p.(Gly985Arg) | Likely pathogenic | NF | 12 of 13 | 5.7199 | NA | NA |
| c.4880del | p.(Leu1627Argfs*35) | Pathogenic | 0.0000159 | NA | 5.6399 | Pathogenic | NA | |
| c.5714 + 1G > A | p.? | Pathogenic | 0.00000398 | NA | 4.7399 | Likely pathogenic | NA | |
| c.6310C > T | p.(Gln2104*) | Pathogenic | NF | NA | 6.0799 | NA | NA | |
| CEP290 (NM_025114.4) | c.7394_7395del | p.(Glu2465Valfs*2) | Pathogenic | 0.000598 | NA | 5.42 | NA | NA |
| CNGA3 (NM_001298.3) | c.673 + 5G > T | p.? | Likely pathogenic | 0.0000199 | NA | 5.09 | NA | NA |
| RPGRIP1 (NM_020366.3) | c.3339 + 5G > C | p.? | Likely pathogenic | NF | 2 of 2 | NA | Likely pathogenic | NA |
| CNGB1 (NM_001297.5) | c.2492 + 2T > G | p.? | Pathogenic | NF | NA | 5.4499 | NA | NA |
| CRB1 (NM_201253.3) | c.481G > A | p.(Ala161Thr) | Likely pathogenic | NF | 12 of 13 | 5.5199 | Uncertain significance | NA |
| PCARE (NM_001271 441.2) | c.656_665dup | p.(Ala223Argfs*38) | Likely pathogenic | 0.0000579 | NA | 1.8514 | NA | NA |
| EYS (NM_0011 42800.2) | c.1194del | p.(Gly399Aspfs*22) | Pathogenic | NF | NA | 6.07 | NA | NA |
| c.6882_6883del | p.(Gln2294Hisfs*3) | Pathogenic | NF | NA | 5.38 | NA | NA | |
| c.7736_7742del | p.(Thr2579Lysfs*36) | Pathogenic | 0.0000127 | NA | 4.01 | Likely pathogenic | NA | |
| c.8854del | p.(Thr2952Leufs*23) | Pathogenic | NF | NA | 4.57 | NA | NA | |
| GUCY2D (NM_000180.4) | c.1991A > G | p.(His664Arg) | Likely pathogenic | NF | 11 of 12 | 5.35 | NA | NA |
| GUCA1A (NM_000409.5) | c.66C > A | p.(Tyr22*) | Likely pathogenic | 0.00000795 | NA | 5.75 | Uncertain significance | NA |
| OTX2 (NM_001 270525.2) | c.638T > A | p.(Leu213*) | Pathogenic | NF | NA | 5.53 | NA | NA |
| PDE6A (NM_000440.3) | c.2144T > C | p.(Met715Thr) | Likely pathogenic | 0.000231 | 12 of 13 | 5.32 | Uncertain significance | NA |
| PDE6B (NM_000283.4) | c.1920 + 1G > A | p.? | Pathogenic | NF | NA | 4.19 | NA | NA |
| c.2470_2478del | p.(Lys824_Glu826del) | Uncertain significance | 0.000128 | NA | 4.1599 | NA | NA | |
| POC1B (NM_172240.3) | c.1079_1080del | p.(Pro360Argfs*8) | Pathogenic | 0.0000145 | NA | 5.69 | NA | NA |
| PROM1 (NM_006017.2) | exon 11 del | p.? | Pathogenic | NF | NA | NA | NA | NA |
| PRPH2 (NM_000322.5) | c.440dup | p.(Gly148Trpfs*29) | Pathogenic | NF | NA | 5.8699 | NA | NA |
| c.647C > A | p.(Pro216His) | Likely pathogenic | NF | 12 of 13 | 5.0999 | NA | NA | |
| RHO (NM_000539.3) | c.670G > A | p.(Gly224Arg) | Likely pathogenic | 0.0000159 | 12 of 13 | 5.0399 | NA | NA |
| RPGR (NM_001 034853.2) | c.1366del | p.(Gln456Lysfs*20) | Pathogenic | NF | NA | 4.73 | NA | NA |
| USH2A (NM_206933.4) | c.4955C > T | p.(Pro1652Leu) | Likely pathogenic | 0.000016 | 10 of 13 | 5.21 | NA | NA |
| c.6957 + 1G > C | p.? | Pathogenic | NF | NA | 5.8099 | NA | NA | |
| c.13894C > T | p.(Pro4632Ser) | Likely pathogenic | 0.00000797 | 8 of 13 | 5.21 | NA | NA | |
Column “Classification” refers to classification according to AMCG. aPathogenicity Scores from https://varsome.com/for missense variants (accessed November 2020) (BayesDel_addAF, DANN, DEOGEN2, EIGEN, FATHMM-MKL, LIST-S2, M-CAP, MVP, MutationAssessor, MutationTaster, PrimateAI, REVEL, and SIFT). Represent predictors supporting the pathogenic effect against the total of available predictors. bGERP conservation score based on the reduction in the number of substitutions in the multi-species sequence alignment compared to the neutral expectation using the genomes of 35 mammals. Range: -12.3 to 6.17 (most conserved). cHGMD public version (accessed November 2020). p.?, unknown protein effect; NF, no found; NA, not applicable. The “*” symbol corresponds to: stop-codon according to the format of mutations nomenclature from the Human Genome Variation Society (HGVS).
FIGURE 2.
Number of solved cases with mutations in the different disease-causing genes. The X-axis refers to the responsible genes for each solved case, and the Y-axis to the number of solved cases for each disease-causing gene. Only genes responsible for the disease are represented. For patient RPN-670 (FRPN-265), the three different possible responsible genes for the IRD are represented.
Copy number variants analysis with the bioinformatic tool allowed us to detect two putative deletions: a deletion in PRPF31, which was properly validated with MLPA (patient RPN-717), and a homozygous deletion of exon 11 of PROM1 in patient RPN-709. In this last patient, the exon 11 did not have reads in the alignment and was not amplified by PCR; however, there was PCR amplification for the flanking exons, thus, reinforcing the hypothesis of a homozygous deletion of the exon.
The individual RPN-728 presented nephropathy and alterations in the brain MRI (mild hypoplasia of the cerebellar vermis and slightly elongated superior cerebellar peduncles) in addition to retinal degeneration, suggesting a Joubert syndrome. The panel sequencing allowed us to identify two pathogenic variants in CEP290, c.4966_4967delGA (p.Glu1656Asnfs∗3) and c.2817G > T (p.Lys939Asn). On the other hand, the panel analysis in the RPN-708 revealed a pathogenic variant in the OTX2 gene (Table 1). Although this gene has been implicated in other diseases such as microphthalmia and retinal degeneration with pituitary dysfunction (OMIM: 610125; 610125), our patient only referred a macular dystrophy (MD) phenotype.
Because of the inclusion of the deep-intronic regions, we solved two cases who carried deep-intronic pathogenic variants in ABCA4 and CEP290 (RPN-750, RPN-734) (Table 1).
Discussion
A total of 92 patients, previously clinically diagnosed with IRD, were analyzed by two IRD-custom panels. These gene panels allowed us to find a genetic diagnosis in 53 patients of 52 different families (Table 1), with a diagnostic ratio of 57.6%, which is within the average of other studies (50–70%) (Ellingford et al., 2016; Di Resta et al., 2018; Wang et al., 2018; Jespersgaard et al., 2019; Zenteno et al., 2020).
In recent years, several deep-intronic mutations have been described as pathogenic due to their effect in the splicing process by leading to the introduction of a pseudoexon (PE) in the coding sequence (Vaché et al., 2012; Braun et al., 2013; Zernant et al., 2014; Bauwens et al., 2015, 2019; Liquori et al., 2016; Baux et al., 2017; Fadaie et al., 2019; Khan et al., 2019; Sangermano et al., 2019). The inclusion of these regions in the present study allowed us to detect two deep intronic variants and, therefore, solve the molecular diagnosis in two more families. The variant CEP290: c.2991 + 1655A > G, detected in patient RPN-750, is one of the most prevalent in Leber congenital amaurosis (LCA) associated with CEP290 (Den Hollander et al., 2006; Coppieters et al., 2010). Coppieters et al. reported this variant with a frequency of 49% in the CEP290-associated LCA cases. Also, we identified the deep-intronic variant c.4539 + 2064C > T in ABCA4. Deep-intronic variants in ABCA4 were reported to be involved in 2.1–17.9% of the STGD cohorts (Braun et al., 2013; Zernant et al., 2014; Bauwens et al., 2015; Bax et al., 2015; Zaneveld et al., 2015; Schulz et al., 2017); our finding of 7.1% of cases within the STGD patients fits into the observed frequencies. Furthermore, the development of new therapeutical approaches, such as antisense oligonucleotides (AONs), entailed a new advantage for deep-intronic variants correction, which highlights the importance of detecting these changes for future treatments. In this sense, several studies have proven the therapeutic potential of AONs strategy and a phase I/II clinical trial for patients harboring the CEP290 c.2991 + 1655A > G mutation using this type of therapy was conducted (Burke et al., 2012; Slijkerman et al., 2016; Duijkers et al., 2018; Cideciyan et al., 2019; Garanto et al., 2019; Sangermano et al., 2019; Xue and MacLaren, 2020).
In our study, ABCA4 and USH2A are the most frequent mutated-genes, similar to other studies (Bernardis et al., 2016; Ezquerra-Inchausti et al., 2018; Jespersgaard et al., 2019). In ABCA4, 10% of alleles are reported to be complex (Shroyer et al., 2001; Zhang et al., 2015), a value that increased to 25% in our results. Moreover, we reported the variant c.3386G > T (p.Arg1129Leu) in ABCA4 in 37.5% of the resolved patients with mutations in this gene (Table 1), which was not surprising because this variant appears to be very frequent in the Spanish population as we can observe in a study performed by Del Pozo-Valero et al. (2020). On the other hand, variants with an allelic frequency higher than 1% (MAF < 0.01) are usually ruled out as the cause of disease in prioritization analyses; however, in some cases, the frequency of some variants, although > 1%, is higher in patients than in healthy individuals, suggesting that they are causal for the disease. Examples of this have been reported for ABCA4. For instance, the variant c.5603A > T (p.Asn1868Ile) was presented in STGD patients four times more frequently than expected (Zernant et al., 2017), and it is a disease-causing variant in 5% of patients when it is in trans with a severe allele in ABCA4. This variant was initially considered benign, however, recent studies consider it pathogenic with reduced penetrance (Zernant et al., 2017; Cremers et al., 2018; Runhart et al., 2018). For autosomal dominant cases, PRPH2 was found to be responsible for 14.8% of the resolved cases (Table 1), whilst a prevalence of 10.3% in PRPH2 was the highest obtained thus far (Manes et al., 2015).
Concerning the patients who carry mutations in more than one IRD gene (Table 1), it is estimated that 2.7 billion individuals worldwide (36%) are carriers of an IRD disease-causing mutation, whereas 5.5 million are expected to be affected (Hanany et al., 2020). In this study, we diagnosed patient RPN-670, who was a carrier of disease-causing variants in RP1, PRPH2, and USH2A (Table 1). Despite the fact that any relative displays any symptoms, we cannot confirm that PRPH2 and RP1 variants are de novo, as DNA from family members was not available for segregation analysis. Incomplete penetrance can also be suggested, since it has been described for patients carrying pathogenic variants in PRPH2 and RP1 (Dietrich, 2002; Boon et al., 2008; Thiadens et al., 2012; Coco-Martin et al., 2020). It is important to remark that the fact of being a carrier of pathogenic variants in more than one gene can have a major impact on the reproductive choices for IRD patients, as well as impact their eligibility for gene-specific genetic therapies. This finding outstands the significance of achieving an exhaustive diagnosis.
The OTX2 gene is associated with syndromic diseases, such as microphthalmia and retinal degeneration with or without pituitary dysfunction (OMIM: 610125; 610125). In this study, we also identified a nonsense variant in the OTX2 gene in a family (FRPN-300) (Table 1) with an autosomal dominant mode of inheritance but only retinal symptoms. Similarly, two autosomal dominant families with heterozygous pathogenic variants in OTX2 with only retinal degeneration patterns were previously reported (Vincent et al., 2014). Eleven truncating and two missense pathogenic variants have been described in this gene (LOVD accessed on April 7, 2021) and both type of variants have been associated with both syndromic and non-syndromic cases (Vincent et al., 2014; Slavotinek et al., 2015; Ellingford et al., 2016; Wang et al., 2016; Bryant et al., 2017; Patel et al., 2018; Sanchez-Navarro et al., 2018). Thus, a correlation between the type of mutation and the clinical diagnosis cannot be assessed.
A clear difference of the use of PV1 or PV2 concerning the solved cases (57.1% with the PV1 and 58.6% with PV2) did not exist. Strikingly, the rate of partially solved cases is almost half for the PV1 (11.1%) when compared to the PV2 (19.35%). It would be expected that the number of partially solved cases would be lower with a panel that includes all the deep-intronic mutations reported to date for ABCA4 and USH2A. In our opinion, this could be due to the lower sample size analyzed with the PV2.
In general, custom panel designs remain an excellent option for the genetic diagnosis of IRD. However, due to the rapid increase of the number of genes involved, some groups prefer to use alternative approaches, such as WES (Lee et al., 2015; Riera et al., 2017; Zhang et al., 2018). Both sequencing strategies have their pros and cons. A specific panel of genes will allow the study of known IRD at a high time/effectiveness ratio regardless of the clinical diagnosis, which is an advantage in those cases in which the clinical diagnosis is not well defined or overlaps between different clinical entities. Furthermore, it allows not only the possibility of including a greater number of probes in repetitive regions such as ORF15 of the RPGR gene, which is highly involved in X-linked IRD cases (Vervoort et al., 2000; Megaw et al., 2015; Chiang et al., 2018; Charng et al., 2019), but also reported deep-intronic pathogenic variants (González-del Pozo et al., 2018; Di Scipio et al., 2020). On the other hand, WES made it possible to find mutations in novel IRD-related genes without updating the panel in low prevalent genes not included for the sake of increasing the depth of coverage or to identify novel IRD genes. The price reduction for high throughput sequencing has made the costs between the panels and WES quite even, so, it does not make the difference between the panels and WES. Taking this into account, the choice between both depends on the preferences of the clinician/researcher in terms of comprehensiveness, accuracy, and time consumption.
Currently, there is no significant difference between the panels and WES concerning the diagnostic rates, as the WES rate did not reach > 70% (Lee et al., 2015; Riera et al., 2017; Wang et al., 2018; Zhang et al., 2018; Liu et al., 2020). Therefore, further studies, including introns, other non-coding regions and epigenetics, are needed to achieve a comprehensive molecular diagnosis of IRD.
Data Availability Statement
The data presented in this study are deposited in the European Genome-phenome Archive (EGA), Study: EGAS00001005369 and Dataset: EGAD00001007755.
Ethics Statement
The studies involving human participants were reviewed and approved by the Hospital La Fe Ethics Committee. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
EA, JM, and GG: conceptualization. BG and AR: methodology. BG, AR, EA, and TJ: resources. BG: writing—original draft preparation. BG, AR, EA, TJ, GG, and JM: writing—review and editing. GG and JM: supervision. JM: funding acquisition. All authors have read and agreed to the published version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely acknowledge the patients for their voluntary participation and the Association Retina-Comunidad Valenciana.
Funding. This study has been funded by the Project PI19/00303. The Health Research Institute Carlos III (ISCIII; Spanish Ministry of Health and Innovation) and the Regional Government of the Valencian Community (PROMETEU/2018/135) partially supported the study, as well as the European Regional Development Fund (ERDF). AR is recipient of a Rio Hortega contract (CM18/00199) from the ISCIII. BG is a recipient of a predoctoral contract (ACIF/2019/252) from the Government of the Valencian Community. GG has a postdoctoral contact from CIBERER.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.645600/full#supplementary-material
References
- Allikmets R., Singh N., Sun H., Shroyer N. F., Hutchinson A., Chidambaram A., et al. (1997). A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Starqardt macular dystrophy. Nat. Genet. 15 236–246. 10.1038/ng0397-236 [DOI] [PubMed] [Google Scholar]
- Antiñolo G., Sánchez B., Borrego S., Rueda T., Chaparro P., Cabeza J. C. (1994). Identification of a new mutation at codon 171 of rhodopsin gene causing autosomal dominant retinitis pigmentosa. Hum. Mol. Genet. 3 1421–1421. 10.1093/hmg/3.8.1421 [DOI] [PubMed] [Google Scholar]
- Ávila-Fernández A., Cantalapiedra D., Aller E., Vallespín E., Aguirre-Lambán J., Blanco-Kelly F., et al. (2010). Mutation analysis of 272 Spanish families affected by autosomal recessive retinitis pigmentosa using a genotyping microarray. Mol. Vis. 16 2550–2558. [PMC free article] [PubMed] [Google Scholar]
- Ayuso C., Millan J. M. (2010). Retinitis pigmentosa and allied conditions today: a paradigm of translational research. Genome Med. 2:34. 10.1186/gm155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwens M., De Zaeytijd J., Weisschuh N., Kohl S., Meire F., Dahan K., et al. (2015). An augmented ABCA4 screen targeting noncoding regions reveals a deep intronic founder variant in belgian stargardt patients. Hum. Mutat. 36 39–42. 10.1002/humu.22716 [DOI] [PubMed] [Google Scholar]
- Bauwens M., Garanto A., Sangermano R., Naessens S., Weisschuh N., De Zaeytijd J., et al. (2019). ABCA4-associated disease as a model for missing heritability in autosomal recessive disorders: novel noncoding splice, cis-regulatory, structural, and recurrent hypomorphic variants. Genet. Med. 21 1761–1771. 10.1038/s41436-018-0420-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baux D., Larrieu L., Blanchet C., Hamel C., Ben Salah S., Vielle A., et al. (2007). Molecular and in silico analyses of the full-length isoform of usherin identify new pathogenic alleles in Usher type II patients. Hum. Mutat. 28 781–789. 10.1002/humu.20513 [DOI] [PubMed] [Google Scholar]
- Baux D., Vaché C., Blanchet C., Willems M., Baudoin C., Moclyn M., et al. (2017). Combined genetic approaches yield a 48% diagnostic rate in a large cohort of French hearing-impaired patients. Sci. Rep. 7:16783. 10.1038/s41598-017-16846-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax N. M., Sangermano R., Roosing S., Thiadens A. A. H. J., Hoefsloot L. H., van den Born L. I., et al. (2015). Heterozygous deep-intronic variants and deletions in ABCA4 in persons with retinal dystrophies and one exonic ABCA4 variant. Hum. Mutat. 36 43–47. 10.1002/humu.22717 [DOI] [PubMed] [Google Scholar]
- Bernardis I., Chiesi L., Tenedini E., Artuso L., Percesepe A., Artusi V., et al. (2016). Unravelling the complexity of inherited retinal dystrophies molecular testing?: added value of targeted next-generation sequencing. Biomed Res. Int. 2016:6341870. 10.1155/2016/6341870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard T., García-García G., Baux D., Vaché C., Faugère V., Larrieu L., et al. (2014). Experience of targeted Usher exome sequencing as a clinical test. Mol. Genet. Genomic Med. 2 30–43. 10.1002/mgg3.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon C. J. F., den Hollander A. I., Hoyng C. B., Cremers F. P. M., Klevering B. J., Keunen J. E. E. (2008). The spectrum of retinal dystrophies caused by mutations in the peripherin/RDS gene. Prog. Retin. Eye Res. 27 213–235. 10.1016/j.preteyeres.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Braun T. A., Mullins R. F., Wagner A. H., Andorf J. L., Johnston R. M., Bakall B. B., et al. (2013). Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum. Mol. Genet. 22 5136–5145. 10.1093/hmg/ddt367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Gil N., Méndez-Vidal C., Romero-Pérez L., González-Del Pozo M., Rodríguez-De La Ruá E., Dopazo J., et al. (2016). Improving the management of Inherited Retinal Dystrophies by targeted sequencing of a population-specific gene panel. Sci. Rep. 6:23910. 10.1038/srep23910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant L., Lozynska O., Maguire A., Aleman T., Bennett J. (2017). Prescreening whole exome sequencing results from patients with retinal degeneration for variants in genes associated with retinal degeneration. Clin. Ophthalmol. 12 49–63. 10.2147/OPTH.S147684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke T. R., Fishman G. A., Zernant J., Schubert C., Tsang S. H., Theodore Smith R., et al. (2012). Retinal phenotypes in patients homozygous for the G1961E mutation in the ABCA4 gene. Invest. Ophthalmol. Vis. Sci. 53 4458–4467. 10.1167/iovs.11-9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carss K., Arno G., Erwood M., Stephens J., Sanchis-Juan A., Hull S., et al. (2017). Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am. J. Hum. Genet. 100 75–90. 10.1016/j.ajhg.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng J., Cideciyan A. V., Jacobson S. G., Sumaroka A., Schwartz S. B., Swider M., et al. (2019). Variegated yet non-random rod and cone photoreceptor disease patterns in RPGR-ORF15-associated retinal degeneration. Hum. Mol. Genet. 28 175–175. 10.1093/hmg/ddy342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang J. P. W., Lamey T. M., Wang N. K., Duan J., Zhou W., McLaren T. L., et al. (2018). Development of high-throughput clinical testing of RPGR ORF15 using a large inherited retinal dystrophy cohort. Invest. Opthalmol. Vis. Sci. 59:4434. 10.1167/iovs.18-24555 [DOI] [PubMed] [Google Scholar]
- Cideciyan A. V., Jacobson S. G., Drack A. V., Ho A. C., Charng J., Garafalo A. V., et al. (2019). Effect of an intravitreal antisense oligonucleotide on vision in Leber congenital amaurosis due to a photoreceptor cilium defect. Nat. Med. 25 225–228. 10.1038/s41591-018-0295-0 [DOI] [PubMed] [Google Scholar]
- Coco-Martin R. M., Sanchez-Tocino H. T., Desco C., Usategui-Martín R., Tellería J. J. (2020). PRPH2-Related retinal diseases: broadening the clinical spectrum and describing a new mutation. Genes (Basel) 11:773. 10.3390/genes11070773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin R. W. J., Littink K. W., Klevering B. J., van den Born L. I., Koenekoop R. K., Zonneveld M. N., et al. (2008). Identification of a 2 Mb human ortholog of Drosophila eyes shut/spacemaker that is mutated in patients with retinitis pigmentosa. Am. J. Hum. Genet. 83 594–603. 10.1016/j.ajhg.2008.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppieters F., Casteels I., Meire F., De Jaegere S., Hooghe S., van Regemorter N., et al. (2010). Genetic screening of LCA in Belgium: predominance of CEP290 and identification of potential modifier alleles in AHI1 of CEP290-related phenotypes. Hum. Mutat. 31 E1709–E1766. 10.1002/humu.21336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis S. S., Bax N. M., Zernant J., Allikmets R., Fritsche L. G., den Dunnen J. T., et al. (2017). In silico functional meta-analysis of 5,962 ABCA4 variants in 3,928 retinal dystrophy cases. Hum. Mutat. 38 400–408. 10.1002/humu.23165 [DOI] [PubMed] [Google Scholar]
- Corton M., Tatu S. D., Avila-Fernandez A., Vallespín E., Tapias I., Cantalapiedra D., et al. (2013). High frequency of CRB1 mutations as cause of Early-Onset Retinal Dystrophies in the Spanish population. Orphanet J. Rare Dis. 8:20. 10.1186/1750-1172-8-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremers F. (1998). Autosomal recessive retinitis pigmentosa and cone-rod dystrophy caused by splice site mutations in the Stargardt’s disease gene ABCR. Hum. Mol. Genet. 7 355–362. 10.1093/hmg/7.3.355 [DOI] [PubMed] [Google Scholar]
- Cremers F. P. M., Cornelis S. S., Runhart E. H., Astuti G. D. N. (2018). Author response: penetrance of the ABCA4 p.Asn1868Ile allele in stargardt disease. Invest. Opthalmol. Vis. Sci. 59:5566. 10.1167/iovs.18-25944 [DOI] [PubMed] [Google Scholar]
- Del Pozo-Valero M., Riveiro-Alvarez R., Blanco-Kelly F., Aguirre-Lamban J., Martin-Merida I., Iancu I.-F., et al. (2020). Genotype–Phenotype correlations in a Spanish cohort of 506 families With biallelic ABCA4 pathogenic variants. Am. J. Ophthalmol. 219 195–204. 10.1016/j.ajo.2020.06.027 [DOI] [PubMed] [Google Scholar]
- Den Hollander A. I., Koenekoop R. K., Yzer S., Lopez I., Arends M. L., Voesenek K. E. J., et al. (2006). Mutations in the CEP290 (NPHP6) gene are a frequent cause of leber congenital amaurosis. Am. J. Hum. Genet. 79 556–561. 10.1086/507318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander A. I., McGee T. L., Ziviello C., Banfi S., Dryja T. P., Gonzalez-Fernandez F., et al. (2009). A homozygous missense mutation in the IRBP gene (RBP3) associated with autosomal recessive retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 50 1864–1872. 10.1167/iovs.08-2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hollander A. I., ten Brink J. B., de Kok Y. J. M., van Soest S., van den Born L. I., van Driel M. A., et al. (1999). Mutations in a human homologue of Drosophila crumbs cause retinitis pigmentosa (RP12). Nat. Genet. 23 217–221. 10.1038/13848 [DOI] [PubMed] [Google Scholar]
- Di Resta C., Spiga I., Presi S., Merella S., Pipitone G. B., Manitto M. P., et al. (2018). Integration of multigene panels for the diagnosis of hereditary retinal disorders using Next Generation Sequencing and bioinformatics approaches. Electron. J. Int. Fed. Clin. Chem. Lab. Med. 29 15–25. [PMC free article] [PubMed] [Google Scholar]
- Di Scipio M., Tavares E., Deshmukh S., Audo I., Green-Sanderson K., Zubak Y., et al. (2020). Phenotype driven analysis of whole genome sequencing identifies deep intronic variants that cause retinal dystrophies by aberrant exonization. Invest. Opthalmol. Vis. Sci. 61:36. 10.1167/iovs.61.10.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich K. (2002). A novel mutation of the RP1 gene (Lys778ter) associated with autosomal dominant retinitis pigmentosa. Br. J. Ophthalmol. 86 328–332. 10.1136/bjo.86.3.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diñeiro M., Capín R., Cifuentes G. Á, Fernández−Vega B., Villota E., Otero A., et al. (2020). Comprehensive genomic diagnosis of inherited retinal and optical nerve disorders reveals hidden syndromes and personalized therapeutic options. Acta Ophthalmol. 98 e1034–e1048. 10.1111/aos.14479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja T. P., Hahn L. B., Kajiwara K., Berson E. L. (1997). Dominant and digenic mutations in the peripherin/RDS and ROM1 genes in retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 38 1972–1982. [PubMed] [Google Scholar]
- Duijkers L., van den Born L., Neidhardt J., Bax N., Pierrache L., Klevering B., et al. (2018). Antisense oligonucleotide-based splicing correction in individuals with leber congenital amaurosis due to compound heterozygosity for the c.2991+1655A>G mutation in CEP290. Int. J. Mol. Sci. 19:753. 10.3390/ijms19030753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellingford J. M., Barton S., Bhaskar S., O’Sullivan J., Williams S. G., Lamb J. A., et al. (2016). Molecular findings from 537 individuals with inherited retinal disease. J. Med. Genet. 53 761–767. 10.1136/jmedgenet-2016-103837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezquerra-Inchausti M., Anasagasti A., Barandika O., Garai-Aramburu G., Galdós M., López de Munain A., et al. (2018). A new approach based on targeted pooled DNA sequencing identifies novel mutations in patients with Inherited Retinal Dystrophies. Sci. Rep. 8:15457. 10.1038/s41598-018-33810-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadaie Z., Khan M., Del Pozo-Valero M., Cornelis S. S., Ayuso C., Cremers F. P. M., et al. (2019). Identification of splice defects due to noncanonical splice site or deep-intronic variants in ABCA4. Hum. Mutat. 40 2365–2376. 10.1002/humu.23890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar G. J., Carrigan M., Dockery A., Millington-Ward S., Palfi A., Chadderton N., et al. (2017). Toward an elucidation of the molecular genetics of inherited retinal degenerations. Hum. Mol. Genet. 26 R2–R11. 10.1093/hmg/ddx185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman G. A. (2003). ABCA4 gene sequence variations in patients with autosomal recessive cone-rod dystrophy. Arch. Ophthalmol. 121:851. 10.1001/archopht.121.6.851 [DOI] [PubMed] [Google Scholar]
- Fowler A., Mahamdallie S., Ruark E., Seal S., Ramsay E., Clarke M., et al. (2016). Accurate clinical detection of exon copy number variants in a targeted NGS panel using DECoN. Wellcome Open Res. 1:20. 10.12688/wellcomeopenres.10069.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garanto A., Duijkers L., Tomkiewicz T. Z., Collin R. W. J. (2019). Antisense oligonucleotide screening to optimize the rescue of the splicing defect caused by the recurrent deep-intronic ABCA4 Variant c.4539+2001G>A in stargardt disease. Genes (Basel) 10:452. 10.3390/genes10060452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber S., Rozet J. M., van de Pol T. J. R., Hoyng C. B., Munnich A., Blankenagel A., et al. (1998). Complete exon–intron structure of the retina-specific ATP binding transporter gene (ABCR) allows the identification of novel mutations underlying stargardt disease. Genomics 48 139–142. 10.1006/geno.1997.5164 [DOI] [PubMed] [Google Scholar]
- Glockle N., Kohl S., Mohr J., Scheurenbrand T., Sprecher A., Weisschuh N., et al. (2014). Panel-based next generation sequencing as a reliable and efficient technique to detect mutations in unselected patients with retinal dystrophies. Eur. J. Hum. Genet. 22 99–104. 10.1038/ejhg.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-del Pozo M., Borrego S., Barragán I., Pieras J. I., Santoyo J., Matamala N., et al. (2011). Mutation screening of multiple genes in Spanish patients with autosomal recessive retinitis pigmentosa by targeted resequencing. PLoS One 6:27894. 10.1371/journal.pone.0027894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-del Pozo M., Martín-Sánchez M., Bravo-Gil N., Méndez-Vidal C., Chimenea Á, Rodríguez-de la Rúa E., et al. (2018). Searching the second hit in patients with inherited retinal dystrophies and monoallelic variants in ABCA4, USH2A and CEP290 by whole-gene targeted sequencing. Sci. Rep. 8:13312. 10.1038/s41598-018-31511-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Fernandez F., Kurz D., Bao Y., Newman S., Conway B. P., Young J. E., et al. (1999). 11-cis retinol dehydrogenase mutations as a major cause of the congenital night-blindness disorder known as fundus albipunctatus. Mol. Vis. 5:41. [PubMed] [Google Scholar]
- Hanany M., Rivolta C., Sharon D. (2020). Worldwide carrier frequency and genetic prevalence of autosomal recessive inherited retinal diseases. Proc. Natl. Acad. Sci. U.S.A. 117 2710–2716. 10.1073/pnas.1913179117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F., Gao F., Li J., Xu P., Wang D., Chen F., et al. (2020). Novel variants associated with Stargardt disease in Chinese patients. Gene 754:144890. 10.1016/j.gene.2020.144890 [DOI] [PubMed] [Google Scholar]
- Jacobson S. G., Cideciyan A. V., Iannaccone A., Weleber R. G., Fishman G. A., Maguire A. M., et al. (2000). Disease expression of RP1 mutations causing autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 41 1898–1908. [PubMed] [Google Scholar]
- Jamshidi F., Place E. M., Mehrotra S., Navarro-Gomez D., Maher M., Branham K. E., et al. (2019). Contribution of noncoding pathogenic variants to RPGRIP1-mediated inherited retinal degeneration. Genet. Med. 21 694–704. 10.1038/s41436-018-0104-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersgaard C., Fang M., Bertelsen M., Dang X., Jensen H., Chen Y., et al. (2019). Molecular genetic analysis using targeted NGS analysis of 677 individuals with retinal dystrophy. Sci. Rep. 9:1219. 10.1038/s41598-018-38007-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara K., Berson E., Dryja T. (1994). Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science 264 1604–1608. 10.1126/science.8202715 [DOI] [PubMed] [Google Scholar]
- Khan M., Cornelis S. S., Khan M. I., Elmelik D., Manders E., Bakker S., et al. (2019). Cost-effective molecular inversion probe-based ABCA4 sequencing reveals deep-intronic variants in Stargardt disease. Hum. Mutat. 40 1749–1759. 10.1002/humu.23787 [DOI] [PubMed] [Google Scholar]
- Kinnick T. R., Mullins R. F., Dev S., Leys M., Mackey D. A., Kay C. N., et al. (2011). Autosomal recessive vitelliform macular dystrophy in a large cohort of vitelliform macular dystrophy patients. Retina 31 581–595. 10.1097/IAE.0b013e318203ee60 [DOI] [PubMed] [Google Scholar]
- Kohl S., Christ-Adler M., Apfelstedt-Sylla E., Kellner U., Eckstein A., Zrenner E., et al. (1997). RDS/peripherin gene mutations are frequent causes of central retinal dystrophies. J. Med. Genet. 34 620–626. 10.1136/jmg.34.8.620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi Y., Akiyama M., Nishiguchi K. M., Momozawa Y., Kamatani Y., Takata S., et al. (2019). Genetic characteristics of retinitis pigmentosa in 1204 Japanese patients. J. Med. Genet. 56 662–670. 10.1136/jmedgenet-2018-105691 [DOI] [PubMed] [Google Scholar]
- Le Quesne Stabej P., Saihan Z., Rangesh N., Steele-Stallard H. B., Ambrose J., Coffey A., et al. (2012). Comprehensive sequence analysis of nine Usher syndrome genes in the UK National Collaborative Usher Study. J. Med. Genet. 49 27–36. 10.1136/jmedgenet-2011-100468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Berg J. S., Milko L., Crooks K., Lu M., Bizon C., et al. (2015). High diagnostic yield of whole exome sequencing in participants with retinal dystrophies in a clinical ophthalmology setting. Am. J. Ophthalmol. 160 354–363.e9. 10.1016/j.ajo.2015.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenassi E., Vincent A., Li Z., Saihan Z., Coffey A. J., Steele-Stallard H. B., et al. (2015). A detailed clinical and molecular survey of subjects with nonsyndromic USH2A retinopathy reveals an allelic hierarchy of disease-causing variants. Eur. J. Hum. Genet. 23 1318–1327. 10.1038/ejhg.2014.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis R. A., Shroyer N. F., Singh N., Allikmets R., Hutchinson A., Li Y., et al. (1999). Genotype/Phenotype analysis of a photoreceptor-specific ATP-Binding cassette transporter gene, ABCR, in stargardt disease. Am. J. Hum. Genet. 64 422–434. 10.1086/302251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liquori A., Vaché C., Baux D., Blanchet C., Hamel C., Malcolm S., et al. (2016). Whole USH2A gene sequencing identifies several new deep intronic mutations. Hum. Mutat. 37 184–193. 10.1002/humu.22926 [DOI] [PubMed] [Google Scholar]
- Littink K. W., Pott J.-W. R., Collin R. W. J., Kroes H. Y., Verheij J. B. G. M., Blokland E. A. W., et al. (2010). A novel nonsense mutation in CEP290 induces exon skipping and leads to a relatively mild retinal phenotype. Invest. Opthalmol. Vis. Sci. 51 3646. 10.1167/iovs.09-5074 [DOI] [PubMed] [Google Scholar]
- Liu X. Z., Li Y. Y., Yang L. P. (2020). [Comparison study of whole exome sequencing and targeted panel sequencing in molecular diagnosis of inherited retinal dystrophies]. Beijing Da Xue Xue Bao 52 836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotery A. J. (2001). Mutations in the CRB1 gene cause leber congenital amaurosis. Arch. Ophthalmol. 119:415. 10.1001/archopht.119.3.415 [DOI] [PubMed] [Google Scholar]
- Manes G., Guillaumie T., Vos W. L., Devos A., Zeitz C., Marquette V., et al. (2015) High prevalence of PRPH2 in autosomal dominant retinitis pigmentosa in france and characterization of biochemical and clinical features. Am. J. Ophthalmol. 159 302–314. 10.1016/j.ajo.2014.10.033 [DOI] [PubMed] [Google Scholar]
- Maw M., Kumaramanickavel G., Kar B., John S., Bridges R., Denton M. (1998). Two Indian siblings with Oguchi disease are homozygous for an arrestin mutation encoding premature termination. Hum. Mutat. 11 S317–S319. 10.1002/humu.1380110199 [DOI] [PubMed] [Google Scholar]
- Megaw R. D., Soares D. C., Wright A. F. (2015). RPGR: its role in photoreceptor physiology, human disease, and future therapies. Exp. Eye Res. 138 32–41. 10.1016/j.exer.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasonkin I., Illing M., Koehler M. R., Schmid M., Molday R. S., Weber B. H. F. (1998). Mapping of the rod photoreceptor ABC transporter (ABCR) to 1p21-p22.1 and identification of novel mutations in Stargardt’s disease. Hum. Genet. 102 21–26. 10.1007/s004390050649 [DOI] [PubMed] [Google Scholar]
- Neveling K., den Hollander A. I., Cremers F. P. M., Collin R. W. J. (2012). “Identification and analysis of inherited retinal disease genes,” in Retinal Degeneration. Methods in Molecular Biology (Methods and Protocols), Vol. 935 eds Weber B., LANGMANN T. (Totowa, NJ: Humana Press; ), 3–23. 10.1007/978-1-62703-080-9_1 [DOI] [PubMed] [Google Scholar]
- Nishimura D. Y., Baye L. M., Perveen R., Searby C. C., Avila-Fernandez A., Pereiro I., et al. (2010). Discovery and functional analysis of a retinitis pigmentosa gene, C2ORF71. Am. J. Hum. Genet. 86 686–695. 10.1016/j.ajhg.2010.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmeggiani F., Sorrentino F. S., Ponzin D., Barbaro V., Ferrari S., Di Iorio E. (2011). Retinitis pigmentosa: genes and disease mechanisms. Curr. Genomics 12 238–249. 10.2174/138920211795860107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel N., Khan A. O., Alsahli S., Abdel-Salam G., Nowilaty S. R., Mansour A. M., et al. (2018). Genetic investigation of 93 families with microphthalmia or posterior microphthalmos. Clin. Genet. 93 1210–1222. 10.1111/cge.13239 [DOI] [PubMed] [Google Scholar]
- Payne A. M., Downes S. M., Bessant D. A. R., Bird A. C., Bhattacharya S. S. (1998). Founder effect, seen in the british population, of the 172 Peripherin/RDS mutation—and further refinement of genetic positioning of the Peripherin/RDS gene. Am. J. Hum. Genet. 62 192–195. 10.1086/301679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea-Romero I., Gordo G., Iancu I. F., Del Pozo-Valero M., Almoguera B., Blanco-Kelly F., et al. (2021). Genetic landscape of 6089 inherited retinal dystrophies affected cases in Spain and their therapeutic and extended epidemiological implications. Sci. Rep. 11:1526. 10.1038/s41598-021-81093-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., et al. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17 405–423. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera M., Navarro R., Ruiz-Nogales S., Méndez P., Burés-Jelstrup A., Corcóstegui B., et al. (2017). Whole exome sequencing using Ion Proton system enables reliable genetic diagnosis of inherited retinal dystrophies. Sci. Rep. 7:42078. 10.1038/srep42078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivolta C., Berson E. L., Dryja T. P. (2002). Paternal uniparental heterodisomy with partial isodisomy of chromosome 1 in a patient with retinitis pigmentosa without hearing loss and a missense mutation in the Usher syndrome type II gene USH2A. Arch. Ophthalmol. 120 1566–1571. [DOI] [PubMed] [Google Scholar]
- Rivolta C., Sweklo E. A., Berson E. L., Dryja T. P. (2000). Report missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am. J. Hum. Genet 66 1975–1978. 10.1086/302926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Muñoz A., Aller E., Jaijo T., González-García E., Cabrera-Peset A., Gallego-Pinazo R., et al. (2020). Expanding the clinical and molecular heterogeneity of nonsyndromic inherited retinal dystrophies. J. Mol. Diagnostics 22 532–543. 10.1016/j.jmoldx.2020.01.003 [DOI] [PubMed] [Google Scholar]
- Rozet J. M., Gerber S., Souied E., Perrault I., Châtelin S., Ghazi I., et al. (1998). Spectrum of ABCR gene mutations in autosomal recessive macular dystrophies. Eur. J. Hum. Genet. 6 291–295. 10.1038/sj.ejhg.5200221 [DOI] [PubMed] [Google Scholar]
- Runhart E. H., Sangermano R., Cornelis S. S., Verheij J. B. G. M., Plomp A. S., Boon C. J. F., et al. (2018). The common ABCA4 Variant p.Asn1868Ile shows nonpenetrance and variable expression of stargardt disease when present in trans with severe variants. Invest. Opthalmol. Vis. Sci. 59 3220. 10.1167/iovs.18-23881 [DOI] [PubMed] [Google Scholar]
- Sahel J. A., Marazova K., Audo I. (2015). Clinical characteristics and current therapies for inherited retinal degenerations. Cold Spring Harb. Perspect. Med. 5 1–25. 10.1101/cshperspect.a017111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles M. V., Motta F. L., Martin R., Filippelli-Silva R., Dias da Silva E., Varela P., et al. (2018). Variants in the ABCA4 gene in a Brazilian population with Stargardt disease. Mol. Vis. 24 546–559. [PMC free article] [PubMed] [Google Scholar]
- Salmaninejad A., Motaee J., Farjami M., Alimardani M., Esmaeilie A., Pasdar A. (2019). Next-generation sequencing and its application in diagnosis of retinitis pigmentosa. Ophthalmic Genet. 40 393–402. 10.1080/13816810.2019.1675178 [DOI] [PubMed] [Google Scholar]
- Sanchez-Navarro R. J., I, da Silva L., Blanco-Kelly F., Zurita O., Sanchez-Bolivar N., Villaverde C., et al. (2018). Combining targeted panel-based resequencing and copy-number variation analysis for the diagnosis of inherited syndromic retinopathies and associated ciliopathies. Sci. Rep. 8:5285. 10.1038/s41598-018-23520-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchis-Juan A., Stephens J., French C., Gleadall N., Mégy K., Penkett C., et al. (2018). Complex structural variants resolved by short-read and long-read whole genome sequencing in mendelian disorders. BioRxiv [preprint] 10.1101/281683 281683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangermano R., Garanto A., Khan M., Runhart E. H., Bauwens M., Bax N. M., et al. (2019). Deep-intronic ABCA4 variants explain missing heritability in Stargardt disease and allow correction of splice defects by antisense oligonucleotides. Genet. Med. 21 1751–1760. 10.1038/s41436-018-0414-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz H. L., Grassmann F., Kellner U., Spital G., Rüther K., Jägle H., et al. (2017). Mutation spectrum of the ABCA4 gene in 335 stargardt disease patients from a multicenter german cohort—impact of selected deep intronic variants and common SNPs. Invest. Opthalmol. Vis. Sci. 58:394. 10.1167/iovs.16-19936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S. B., Aleman T. S., Cideciyan A. V., Windsor E. A. M., Sumaroka A., Roman A. J., et al. (2005). Disease expression in usher syndrome caused by VLGR1 gene mutation (USH2C) and comparison with USH2A phenotype. Invest. Opthalmol. Vis. Sci. 46:734. 10.1167/iovs.04-1136 [DOI] [PubMed] [Google Scholar]
- Sheck L., Davies W. I. L., Moradi P., Robson A. G., Kumaran N., Liasis A. C., et al. (2018). Leber congenital amaurosis associated with mutations in CEP290, clinical phenotype, and natural history in preparation for trials of novel therapies. Ophthalmology 125 894–903. 10.1016/j.ophtha.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroyer N. F., Lewis R. A., Yatsenko A. N., Lupski J. R. (2001). Null missense ABCR (ABCA4) mutations in a family with stargardt disease and retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 42 2757–2761. [PubMed] [Google Scholar]
- Slavotinek A. M., Garcia S. T., Chandratillake G., Bardakjian T., Ullah E., Wu D., et al. (2015). Exome sequencing in 32 patients with anophthalmia/microphthalmia and developmental eye defects. Clin. Genet. 88 468–473. 10.1111/cge.12543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slijkerman R. W., Vaché C., Dona M., García-García G., Claustres M., Hetterschijt L., et al. (2016). Antisense oligonucleotide-based splice correction for USH2A-associated Retinal degeneration caused by a frequent deep-intronic mutation. Mol. Ther. Nucleic Acids 5:e381. 10.1038/mtna.2016.89 [DOI] [PubMed] [Google Scholar]
- Srivastava S., Ramsbottom S. A., Molinari E., Alkanderi S., Filby A., White K., et al. (2017). A human patient-derived cellular model of Joubert syndrome reveals ciliary defects which can be rescued with targeted therapies. Hum. Mol. Genet. 26 4657–4667. 10.1093/hmg/ddx347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenirri S., Fermo I., Battistella S., Galbiati S., Soriani N., Paroni R., et al. (2004). Denaturing HPLC profiling of the ABCA4 gene for reliable detection of allelic variations. Clin. Chem. 50 1336–1343. 10.1373/clinchem.2004.033241 [DOI] [PubMed] [Google Scholar]
- Stone E. M., Andorf J. L., Whitmore S. S., DeLuca A. P., Giacalone J. C., Streb L. M., et al. (2017). Clinically focused molecular investigation of 1000 consecutive families with inherited retinal disease. Ophthalmology 124 1314–1331. 10.1016/j.ophtha.2017.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatour Y., Ben-Yosef T. (2020). Syndromic inherited retinal diseases: genetic, clinical and diagnostic aspects. Diagnostics 10:779. 10.3390/diagnostics10100779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiadens A. A. H. J., Phan T. M. L., Zekveld-Vroon R. C., Leroy B. P., Van Den Born L. I., Hoyng C. B., et al. (2012). Clinical course, genetic etiology, and visual outcome in cone and cone-rod dystrophy. Ophthalmology 119 819–826. 10.1016/j.ophtha.2011.10.011 [DOI] [PubMed] [Google Scholar]
- To K., Adamian M., Berson E. L. (2004). Histologic study of retinitis pigmentosa due to a mutation in the RP13 gene (PRPC8): comparison with rhodopsin Pro23His, Cys110Arg, and Glu181Lys. Am. J. Ophthalmol. 137 946–948. 10.1016/j.ajo.2003.10.047 [DOI] [PubMed] [Google Scholar]
- Trujillo M. J., Martinez-Gimeno M., Gimenez A., Lorda I., Bueno J., Garcia-Sandoval B., et al. (2001). Two novel mutations (Y141H; C214Y) and previously published mutation (R142W) in the RDS-peripherin gene in autosomal dominant macular dystrophies in Spanish families. Hum. Mutat. 17:80. [DOI] [PubMed] [Google Scholar]
- Vaché C., Besnard T., le Berre P., García-García G., Baux D., Larrieu L., et al. (2012). Usher syndrome type 2 caused by activation of an USH2A pseudoexon: implications for diagnosis and therapy. Hum. Mutat. 33 104–108. 10.1002/humu.21634 [DOI] [PubMed] [Google Scholar]
- Vervoort R., Lennon A., Bird A. C., Tulloch B., Axton R., Miano M. G., et al. (2000). Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat. Genet. 25 462–466. 10.1038/78182 [DOI] [PubMed] [Google Scholar]
- Vincent A., Forster N., Maynes J. T., Paton T. A., Billingsley G., Roslin N. M., et al. (2014). OTX2 mutations cause autosomal dominant pattern dystrophy of the retinal pigment epithelium. J. Med. Genet. 51 797–805. 10.1136/jmedgenet-2014-102620 [DOI] [PubMed] [Google Scholar]
- Wang L., Zhang J., Chen N., Wang L., Zhang F., Ma Z., et al. (2018). Application of whole exome and targeted panel sequencing in the clinical molecular diagnosis of 319 Chinese families with inherited retinal dystrophy and comparison study. Genes (Basel) 9 1–11. 10.3390/genes9070360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zhang Q., Zhang X., Wang Z., Zhao P. (2016). Clinical and genetic characteristics of Leber congenital amaurosis with novel mutations in known genes based on a Chinese eastern coast Han population. Graefes Arch. Clin. Exp. Ophthalmol. 254 2227–2238. 10.1007/s00417-016-3428-5 [DOI] [PubMed] [Google Scholar]
- Webb T. R., Parfitt D. A., Gardner J. C., Martinez A., Bevilacqua D., Davidson A. E., et al. (2012). Deep intronic mutation in OFD1, identified by targeted genomic next-generation sequencing, causes a severe form of X-linked retinitis pigmentosa (RP23). Hum. Mol. Genet. 21 3647–3654. 10.1093/hmg/dds194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue K., MacLaren R. E. (2020). Antisense oligonucleotide therapeutics in clinical trials for the treatment of inherited retinal diseases. Expert Opin. Invest. Drugs 29 1163–1170. 10.1080/13543784.2020.1804853 [DOI] [PubMed] [Google Scholar]
- Zaneveld J., Siddiqui S., Li H., Wang X., Wang H., Wang K., et al. (2015). Comprehensive analysis of patients with Stargardt macular dystrophy reveals new genotype–phenotype correlations and unexpected diagnostic revisions. Genet. Med. 17 262–270. 10.1038/gim.2014.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenteno J. C., García-Montaño L. A., Cruz-Aguilar M., Ronquillo J., Rodas-Serrano A., Aguilar-Castul L., et al. (2020). Extensive genic and allelic heterogeneity underlying inherited retinal dystrophies in Mexican patients molecularly analyzed by next-generation sequencing. Mol. Genet. Genomic Med. 8 1–17. 10.1002/mgg3.1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernant J., Lee W., Collison F. T., Fishman G. A., Sergeev Y. V., Schuerch K., et al. (2017). Frequent hypomorphic alleles account for a significant fraction of ABCA4 disease and distinguish it from age-related macular degeneration. J. Med. Genet. 54 404–412. 10.1136/jmedgenet-2017-104540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernant J., Schubert C., Im K. M., Burke T., Brown C. M., Fishman G. A., et al. (2011). Analysis of the ABCA4 gene by next-generation sequencing. Invest. Ophthalmol. Vis. Sci. 52 8479–8487. 10.1167/iovs.11-8182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zernant J., Xie Y. A., Ayuso C., Riveiro-Alvarez R., Lopez-Martinez M.-A., Simonelli F., et al. (2014). Analysis of the ABCA4 genomic locus in Stargardt disease. Hum. Mol. Genet. 23 6797–6806. 10.1093/hmg/ddu396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Sun Z., Zhao P., Huang L., Xu M., Yang Y., et al. (2018). Whole-exome sequencing revealed HKDC1 as a candidate gene associated with autosomal-recessive retinitis pigmentosa. Hum. Mol. Genet. 27 4157–4168. 10.1093/hmg/ddy281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Tsybovsky Y., Kolesnikov A. V., Rozanowska M., Swider M., Schwartz S. B., et al. (2015). Protein misfolding and the pathogenesis of ABCA4-associated retinal degenerations. Hum. Mol. Genet. 24 3220–3237. 10.1093/hmg/ddv073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are deposited in the European Genome-phenome Archive (EGA), Study: EGAS00001005369 and Dataset: EGAD00001007755.