Abstract
Background:
Acid blockade is commonly prescribed in patients with cystic fibrosis (CF). Growing concerns, however, exist about its possible role in the pathophysiology of pulmonary infections. We aimed to investigate if acid blockade alters esophageal and respiratory microbiota leading to dysbiosis and inflammation.
Methods:
We performed a cross sectional study of children with CF who were either prescribed acid blockade or not. Samples from the gastrointestinal and respiratory tracts were obtained and microbiome analyzed. Mixed effect models were used to compare outcomes between cohorts and across sampling sites. A random subject intercept was included to account for the multiple sampling sites per individual.
Results:
A cohort of 25 individuals, 44% girls with median age of 13.8 years [IQR 11.2–14.8] were enrolled. Alpha diversity, total bacterial load, and beta diversity were similar across anatomic compartments, across the upper gastrointestinal tract, and in respiratory samples. Similar alpha diversity, total bacterial load, and beta diversity results were also observed when comparing individuals on versus those off acid blockade. IL-8 was elevated in the distal versus proximal esophagus in the whole cohort (P<0.01). IL-8 concentrations were similar in the distal esophagus in patients on and off acid blockade, but significantly greater in the proximal esophagus of subjects on treatment (P<0.01).
Conclusions:
On the basis of these data, acid blockade use does not appear to influence the microbiome of the aerodigestive tract in children with cystic fibrosis suggesting a complex interplay between these medications and the bacterial composition of the esophagus and lung.
Keywords: acid blockade, cystic fibrosis, inflammation, microbiome
The close anatomical juxtaposition shared by the lungs and esophagus has led to increasing interest in whether diseases of the esophagus, such as gastroesophageal reflux disease (GERD) may contribute to problems in the respiratory tract. This is particularly true for patients with cystic fibrosis (CF) who are prone to develop both pulmonary infections as well as GERD. In fact, clinical studies of patients with CF identify the association between GERD and pulmonary disease but the mechanisms related to these observations are not clear (1–3). Complicating this issue is the fact that CF patients are also treated with proton pump inhibitors (PPI), medications that are associated with increased gastrointestinal as well as pulmonary infections (4,5).
As chronic pulmonary infections in patients with CF represent a major source of morbidity and mortality (6–8), further studies that define their underlying mechanisms as well as those that will determine novel therapeutic approaches are essential. A number of studies indicate that acid blockade, specifically PPI use, poses increased risk of community-acquired pneumonias and pulmonary exacerbations in adult and pediatric CF (4,5,8–11). As gastric acid reduces the microbial load in the stomach, increase in gastric pH can change the bacterial populations and load. Changes in these gastric patterns may alter the esophageal microbiome, and thus contribute to pulmonary disease. No study, however, to our knowledge, has determined the esophageal microbiome in patients with CF, nor the impact of acid blockade on the aerodigestive microbiome.
To begin to address these issues, we aimed to determine if acid blockade use in patients with CF was associated with dysbiosis and alterations in inflammation in the esophageal and the respiratory sites. We hypothesized that acid blockade medications are associated with alterations of esophageal and respiratory microbiota via microaspiration, allowing proliferation of pathogenic bacteria and increased inflammation. To test this hypothesis, we performed a prospective study utilizing a novel minimally invasive technique, the esophageal string test (EST) (12), to capture the esophageal microbiome in children with CF who were treated or not with acid blockade.
METHODS
Patient Selection
We performed a prospective study in children between the ages of 10 and 21 years with a known diagnosis of CF at Children’s Hospital Colorado (CHCO) from November 2017 to February 2020. Individuals cared for at the CHCO CF Center were approached for enrollment if they had: a diagnosis of CF based on sweat chloride greater than 60 mEq/L or 2 known cystic fibrosis transmembrane conductance regulator mutations; clinically stable pulmonary disease defined by clinical impression of the primary CF provider, no newly prescribed antibiotic treatments in the 30 days before enrollment, and stable lung function (forced expiratory volume in 1 second [FEV1] percentage predicted within 10% of baseline and greater than 40%); either been treated with a PPI or histamine 2 receptor antagonist (H2RA) for a minimum of 6 weeks or not been treated with a PPI or H2RA for at least 6 weeks; and been able to swallow a capsule. Exclusion criteria included: a history of meconium ileus, distal intestinal obstructive syndrome, any gastrointestinal surgery, intestinal stricture; a history of CF liver disease with cirrhosis, gastric or esophageal varices; and starting a new course of antibiotics, antifungals or antivirals within 30 days of their visit. The initial cross over study design that sought to enroll patients on and off acid blockade was changed to a cross sectional design because of enrollment concerns.
This study was approved by the Colorado Multiple Institutional Review Board (COMIRB), Aurora, CO. Written informed consent and HIPAA authorization were obtained for all enrolled individuals. Demographic data was collected along with clinical outcomes, including lung function and growth parameters, comorbidities, and concomitant medications.
Esophageal String Test
At the time of the research visit, individuals swallowed the EST as previously described (12,13). Briefly, individuals fasted for 1 hour prior and swallowed the EST that was then left in place for 1 hour. Following removal, the string was divided using a combination of pH indicator testing and using standardized measurements as previously described (14). Segments (2 cm) including oral, proximal and distal esophagus, gastric and duodenal strings were collected and either flash frozen dried or placed immediately in 0.5 mL PBS buffer containing the protease inhibitors ethylenediaminetetraacetic acid (EDTA) and phenylmethylsulfonyl fluoride (PMSF) before freezing. Samples collected were kept at −80°C until DNA extraction or inflammatory marker testing was performed.
Spirometry and Airway Samples
Individuals performed spirometry according to the American Thoracic Society guidelines (15). Either expectorated or induced sputum samples were collected as previously described (16). If a sputum sample was not obtained (unable to expectorate, sputum induction failed), an oropharyngeal swab was collected from the posterior oropharyngeal wall and tonsillar pillars. Samples were snap frozen and stored at −80°C until DNA extraction was performed.
Cytokine Analysis
IL-8 measurement from the EST was determined by ELISA (Luminex Multiplex beads R &D system, Minneapolis, Minnesota, USA) according to manufacturer’s instructions.
Microbiota Identification
DNA Extraction and Bacterial Load Assessment
DNA was prepared using the Qiagen EZ1 extraction platform using the Tissue Extraction Kit and bacterial DNA card after enzymatic digestion (17,18). DNA was used to determine the bacterial load present using the quantitative PCR (qPCR) assay described by Nadkarni et al (19), and assessed for CF samples (20). The qPCR assay is a TaqMan design that targets highly conserved regions (∼357F/805R) for amplification and uses a probe sequence near the highly conserved 515F region of the 16S rRNA gene.
16S Amplicon Library Construction
Bacterial profiles were determined by broad-range amplification and sequence analysis of 16S rRNA genes following our previously described methods (21–23). In brief, amplicons were generated using primers that target approximately 300 base pairs of the V1V2 variable region of the 16S rRNA gene. The V1V2 region was selected based on performance in taxonomic evaluations relative to full-length sequences (24,25). PCR products were normalized by agarose gel densitometry, pooled in approximately equimolar amounts, gel-purified, and concentrated using a DNA Clean and Concentrator Kit (Zymo, Irvine, CA). Pooled amplicons were quantified using Qubit Fluorometer 2.0 (Invitrogen, Carlsbad, CA). The pool was diluted to 4 nmol/L and denatured with 0.2 N NaOH at room temperature. The denatured DNA was diluted to 20 pmol/L and spiked with 10% of the Illumina PhiX control DNA before loading the sequencer. Illumina paired-end sequencing was performed on the Miseq using a 500 cycle version 2 reagent kit.
Analysis of Illumina Paired-end Reads
Illumina MiSeq paired-end reads were aligned to human reference genome Hg19 with bowtie2 and matching sequences discarded (26,27). As previously described, the remaining nonhuman paired-end sequences were sorted by sample via barcodes in the paired reads with a python script (23). Sorted paired end sequence data were deposited in the NCBI Short Read Archive under accession number PRJNA629006. The sorted paired reads were assembled using phrap (28,29). Pairs that did not assemble were discarded. Assembled sequence ends were trimmed over a moving window of 5 nucleotides until average quality met or exceeded 20. Trimmed sequences with more than 1 ambiguity or shorter than 200 nt were discarded. Potential chimeras identified with Uchime (usearch6.0.203_i86linux32) (30) using the Schloss (31) Silva reference sequences were removed from subsequent analyses. Assembled sequences were aligned and classified with SINA (1.3.0-r23838) (32) using the 418,497 bacterial sequences in Silva 115NR99 (33) as reference configured to yield the Silva taxonomy. Sequences with identical taxonomic assignments were clustered to produce operational taxonomic units (OTUs). This process generated 9,499,069 sequences for176 samples (average sequence length: 316 nt; average sample size: 53,972 sequences/sample; minimum sample size: 7624; maximum sample size: 145,522). The median Goods coverage score was ≥99% at the rarefaction point of 7624. The software package Explicet (v2.10.5, www.explicet.org) (34) was used for display, analysis, and visualization of sequencing results. Species calls are based on pre-computed differentiation criteria for each genera of interest based on genomic reference sequences in Silva using the primer-specific region. Species classification is computed within each genus to make species assignments, and any sequence that is not assigned to a species remains classified to the genus level.
Statistical Analyses
Descriptive data are presented as medians [interquartile range] or percentages. Linear mixed models were performed to evaluate changes in total bacterial load, alpha diversity and IL-8 (log base 10 transformed). An interaction between the binary acid blockade variable and string location was included to fit a means model. The model also included a random subject intercept and the comparisons between all the sites and treatment within sites were adjusted for multiple comparisons using the Šidák adjustment. The Morisita-Horn beta diversity was used as a distance metric in a multidimensional scaling (MDS) plot to visualize the distance between samples. All analyses were performed using SAS version 9.4 (The SAS Institute, Cary, NC).
RESULTS
Subject Characteristics
Initial screening identified 102 patients who met entry criteria for the study. Of these, 25 individuals completed all study-related components and were included in the final analysis. (Figure S1, http://links.lww.com/MPG/C88). The cohort was 44% girls with a median age of 13.8 years [IQR 11.2–14.8]. Further demographic and clinical characteristics at the time of the baseline research visit are summarized in Table 1. As a part of the initial crossover design, 2 individuals had samples collected at 3 time points, 2 while on acid blockade, and once off acid blockade. These 2 individuals were included in the acid blockade group for the purposes of describing the cohort.
TABLE 1.
Individual demographics
| N = 25 | Off acid blockade (N = 14) | On acid blockade (N = 11)* | |
|---|---|---|---|
| Female sex | 11 (44%) | 7 (50%) | 4 (36%) |
| Age, years; median [IQR] | 13.8 [11.2–14.8] | 13.5 [11.2–14.7] | 13.8 [11.2–15.5] |
| Genotype | |||
| F508/F508 | 11 (44%) | 6 (43%) | 5 (45%) |
| F508/other | 12 (48%) | 7 (50%) | 5 (45%) |
| Other/other | 2 (8%) | 1 (7%) | 1 (9%) |
| Race | |||
| Caucasian | 24 (96) | 13 (93%) | 11 (100%) |
| African American | 1 (4%) | 1 (7%) | 0 |
| Hispanic or Latino Ethnicity | 3 (12%) | 2 (14%) | 1 (9%) |
| BMI percentile, median [IQR] | 58 [37–78] | 63 [38–78] | 54 [33–76] |
| FEV1% predicted, median [IQR] | 92 [85–100] | 96 [88–102] | 88 [85–94] |
| On CFTR modulators | 15 (60%) | 9 (64%) | 6 (55%) |
| On PPI medications | 11 (44%) | 6 (55%) |
Two individuals from the cross over study design are included.
A total of 176 samples were collected from 25 individuals. Twenty-six oral samples, 29 from each the proximal and the distal esophagus, 28 gastric samples, and 14 duodenal samples were included in the final analysis of the gastrointestinal tract. Otherwise, 27 samples from the nares, 13 oropharyngeal samples, and 10 sputum samples were available from the airway. IL-8 concentration was measured in 50 proximal and distal esophageal EST samples from 21 individuals. For individuals on acid blockade, 8 had appropriate acid suppression exhibited by a basic pH in the stomach, whereas 3 did not have pH change. Nine individuals were on chronic antibiotics; sensitivity analysis with these individuals excluded did not alter results displayed below.
Impact of Acid Blockade on Microbial Load and Diversity
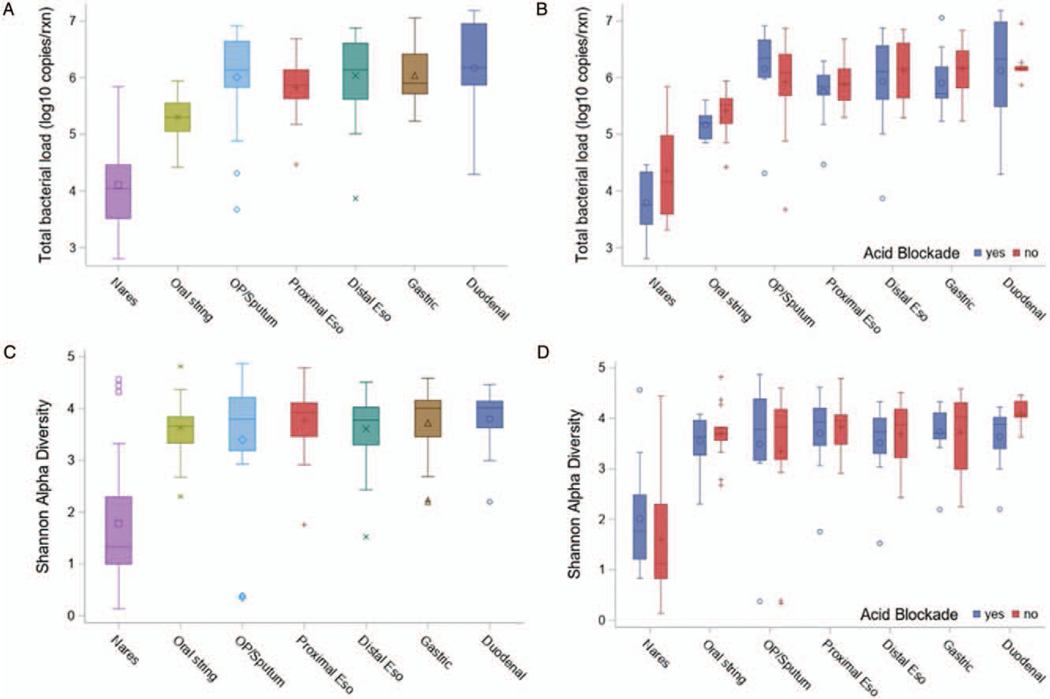
The average total bacterial load per site regardless of acid blockade treatment is displayed in Figure 1A. The nares have statistically lower bacterial load than all other sites (P < 0.01 for all pairwise comparisons). No differences were found in the average total bacterial load at any site when comparing individuals who were treated with acid blockade compared with those who were not (Fig. 1B, Table S1, http://links.lww.com/MPG/C92). Notably, the proximal and distal esophageal portions of the string had comparable bacterial load to oropharyngeal and sputum samples in both cohorts.
FIGURE 1.
Bacterial load and alpha diversity. (A) Total bacterial load for all samples by site determined by 16S qPCR. (B) Total bacterial load by site compared between those on versus those off acid blockade. (C) Shannon alpha diversity for all samples by site. (D) Shannon alpha diversity by site compared between those on versus those off acid blockade. The box indicates the interquartile range (IQR) (25th–75th percentile) and the median and mean are indicated by the line and marker, respectively. Whiskers indicate data within 1.5 times the IQR, and smaller points outside the whiskers indicate individual data values. Nares were significantly different from any other location for both TBL and Shannon (all P values <0.01). TBL=total bacterial load.
The Shannon alpha diversity index per site of all individuals regardless of treatment is displayed in Figure 1C. The nares had a statistically lower diversity than all other sites (P < 0.01 for all pairwise comparisons). When comparing samples from individuals on acid blockade treatment versus those off acid blockade, Shannon alpha diversity was not different for any site (Fig. 1D, Table S1, http://links.lww.com/MPG/C92). Specifically, Shannon diversity index was similar between those on versus those off acid blockade treatment with regards to respiratory microbiome sampled via the oropharyngeal swabs and sputum samples.
Comparison of Microbial Taxa in Aerodigestive Compartments
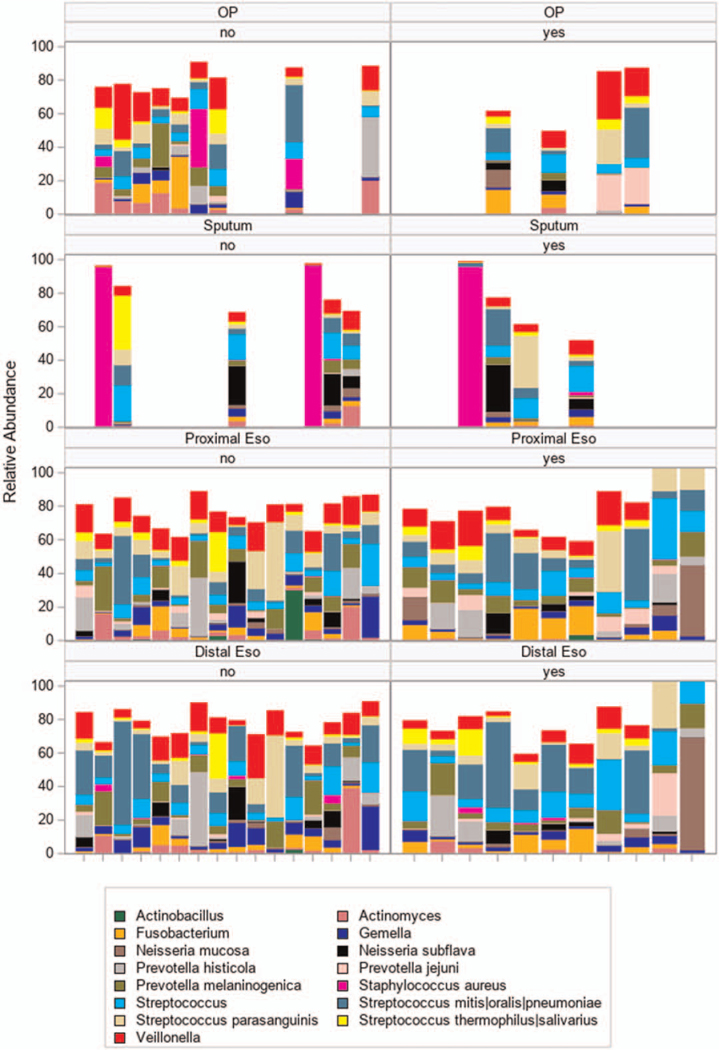
The individual esophageal and respiratory taxa were also compared across groups (Fig. 2). Specifically, relative abundance of taxa is visualized in oropharyngeal, sputum, proximal, and distal esophageal samples according to treatment. The aggregate compilation of data from all individuals in each group were analyzed and also showed virtually identical patterns of relative abundance in the predominant taxa present in these compartments.
FIGURE 2.
Esophageal and lung taxa across groups. The y-axis in the left-hand panels show relative abundance of taxa in the samples collected off acid blockade (n = 14) whereas the panels on the right show relative abundance in those on acid blockade (n = 13, 13 samples from 11 individuals on acid blockade). Horizontal panels from top to bottom are clustered by site from top to bottom: oropharyngeal swab (OP), sputum, proximal esophagus (proximal eso), and distal esophageal (distal eso) specimens. Each unique vertical bar graph indicates taxa detected in an individual sample grouped by location. Each taxon is indicated in a different color. The height of the bar corresponds to the relative abundance. Samples from each individual are aligned along the x-axis between anatomical sites. Empty spaces indicate no specimen from the indicated site for that individual. No = individual off acid blockade; yes = individual on acid blockade.
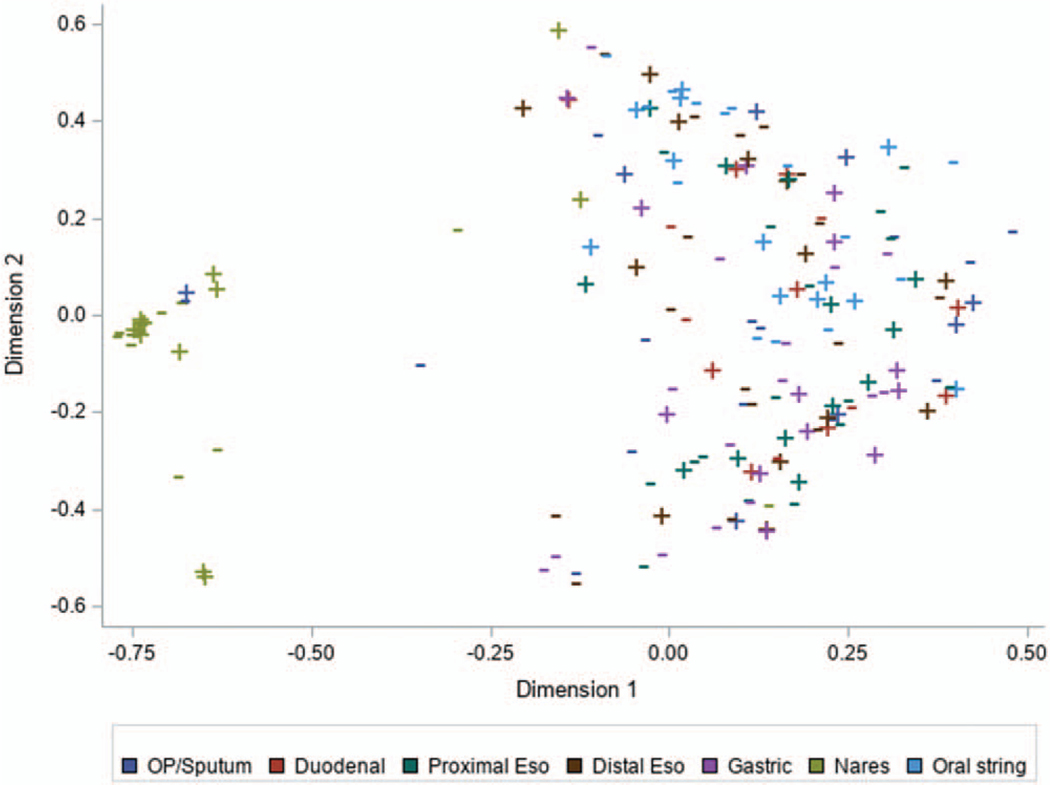
MDS using Morisita-Horn beta diversity for all samples showed that only the nares had a community distinct from other sites (Fig. 3). The gastrointestinal segments and the respiratory specimens all had communities that shared similar taxa. MDS was repeated after excluding the nares samples to assess if the large difference observed for nares was influencing relationships within the more similar airway and esophageal communities. The results, however did not change (Figure S2, http://links.lww.com/MPG/C89). Further comparison of sputum composition to oral, proximal, and distal string was done for 10 individuals with sputum samples. The agreement appears to be similar between sputum composition and the different string locations (Figure S3, http://links.lww.com/MPG/C90). In the 3 individuals with decreased overlap (individuals 4, 6, 22) the low agreement between locations is largely because of the prominence of Staphylococcus aureus in the sputum sample, which was absent from the string samples (Figure S4, http://links.lww.com/MPG/C91).
FIGURE 3.
Multidimensional scaling. Morisita-Horn beta diversity was used as a distance metric in a multidimensional scaling plot to visualize the relationship between samples based on community composition. The nares samples clustered away from all other sample sites indicating that investigated sites had similar species composition irrespective of acid blockade treatment. Points indicated by plus signs correspond to samples collected while the individual was on acid blockade treatment and points with dashes were collected while not on treatment.
Influence of Acid Blockade on Esophageal Inflammatory Marker
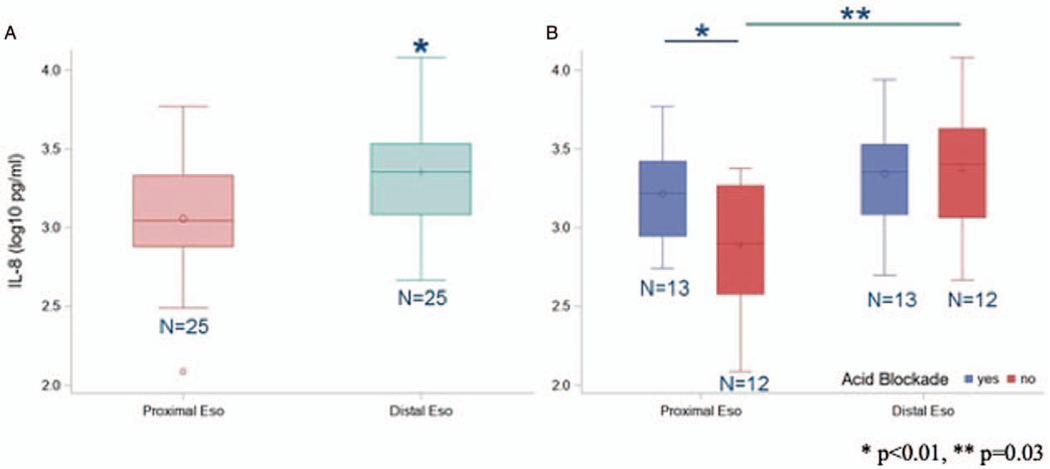
IL-8 concentration in the distal esophagus was significantly greater than the proximal esophagus in all individuals included in the cohort (N = 25) (Fig. 4A). Average log IL-8 protein concentration was 3.34 pg/mL (SE = 0.08 pg/mL) in the distal esophagus and 3.04 pg/mL (SE = 0.08pg/mL) in the proximal esophagus (P < 0.01). For individuals on acid blockade treatment, no differences were measured in IL-8 concentrations between the proximal and distal esophageal locations (P = 0.24) but a difference was observed between those off acid blockade (P < 0.01) (Fig. 4B). The IL-8 concentration was greater within the proximal esophagus in those subjects on acid blockade treatments compared with those off acid blockade (3.19 pg/mL, SE = 0.10 vs 2.89 pg/mL, SE = 0.10 pg/mL, P = 0.03) (Fig. 4B). Within the distal esophagus, IL-8 concentrations were, however, similar across groups (3.32 pg/mL, SE = 0.10 vs 3.36 pg/mL, SE = 0.10 pg/mL) (P = 0.76).
FIGURE 4.
Esophageal IL-8 concentrations. (A) Esophageal IL-8 concentration in proximal and distal esophagus. (B) IL-8 concentrations by site compared between those on versus those off acid blockade. The box indicates the interquartile range (IQR) (25th–75th percentile) and the median and mean are indicated by the lines and markers, respectively. Whiskers indicate data within 1.5 times the IQR, and smaller points outside the whiskers indicate individual data values. *P<0.01, **P=0.03.
DISCUSSION
As previous studies provide an association of PPI use with increases in pulmonary comorbidity in patients with (10,11) and without CF (9), our study provides interesting but unexpected findings. First, acid blockade does not appear to be associated with alterations in the microbiome in any site examined. Comparisons of samples from the nasal, oropharyngeal, pulmonary, esophageal, gastric, and duodenal sites obtained from children treated with acid blockade or not, revealed no differences in bacterial load, alpha diversity, or microbial composition. Second, there appears to be a high degree of similarity between the bacterial load and abundance across the sites examined except for the nares. Third, patients with CF appear to have a baseline level of inflammation present that is not affected by the use of acid-blocking medications. Taken together, our results indicate that the aerodigestive tract in patients with CF contains a highly complex microbiome that is not influenced by the use of acid-blocking medications.
Previous work has shown that esophageal microbiome differences can be detected between states of health and disease. Our work has shown that the microbiome detected using the EST is consistent with the mucosal microbiome obtained from esophageal biopsies and distinct from oral and nasal microbiomes (12). Specifically, in pediatric and adult cohorts of normal individuals compared with those with GERD or eosinophilic esophagitis, our earlier work using the EST showed that the esophageal bacterial load was increased in both disease states (13), and that individuals with GERD on a PPI have decreased levels of Streptococcus in the esophagus when compared with normal controls, a finding consistent with others (35). Rosen et al (36) investigated the effect of PPI medications on gastric, lung, and oropharyngeal microbiota of pediatric patients and showed that there were dose-dependent changes in the microflora in PPI-treated children, and evidence of microflora exchange between the respiratory and the gastrointestinal tract that is independent of the oropharyngeal microflora. Goetz et al (8) demonstrated an increase in the incidence of Pseudomonas in respiratory cultures in infants with CF on PPI and H2RA. In the present study, we anticipated that acid blockade would similarly be associated with alterations of the esophageal microbiota by increasing the pH of refluxed gastric contents of patients with CF leading to esophageal dysbiosis and secondary pulmonary dysbiosis that is distinct from documented alterations to both gut and airway microbiomes in patients with CF (16,22,37). The bacterial load, and community composition were, however, similar across cohorts regardless of treatment, and similar across anatomic compartments. These findings are consistent with prior work by Al-Mormani et al (38), which showed similar bacterial profiles of CF sputum and gastric juice samples in adult patients with CF who had gastrostomy tubes suggesting the presence of a distinct aerodigestive microbiome in individuals with CF. This study, however, did not investigate the effect of acid blockade on the microbiome in patients with CF. Our work indicates that perhaps the thick mucosal secretions that are the hallmark of CF disease, and the abundance of airway mucus that may be swallowed into the upper gastrointestinal tract may be the dominant source of microbiota in the aerodigestive tract. Perhaps the thickness of the secretions, whether from the airway or the upper intestinal tract, blunts the anticipated impact of acid blockade on the aerodigestive microbiome and inflammation as anticipated.
As IL-8 is increased in CF lung infections (37) and in esophagitis (39–42), we used measurements of this cytokine as an inflammatory biomarker. In our previous work, we have shown concordant measurements from the string and concurrently collected biopsy for both microbiome and protein biomarkers in healthy controls (12). In this study, esophageal IL-8 concentrations were elevated in all sections examined, an unanticipated finding in those on and off acid blockade treatment. In a study of adults with GERD compared with healthy normals, average esophageal IL-8 levels obtained from biopsy specimens was 0.32 ± 0.09 pg/mg (equivalent log IL-8 was −0.49 ± −1.04 pg/mg) in healthy individuals, which was significantly (P < 0.01) lower than that the average IL-8 of 23.44 ± 2.14 pg/mg in individuals with reflux esophagitis (equivalent log IL-8 was 1.37 ± 0.33 pg/mg) (43). This study further showed that treatment with a PPI for 8 weeks, specifically lansoprazole, significantly decreased IL-8 concentrations in patients with GERD (P < 0.01). In another study by Huo et al (40), omeprazole, another PPI, inhibited IL-8 expression in esophageal epithelial cells exposed to acid bile salt independent of effects on gastric acid secretion. Our results, however, suggest that the protective effect of acid blockade on esophageal inflammation is blunted in patients with CF, especially with significantly increased proximal esophageal IL-8 concentrations in those on acid blockade versus those off treatment. Furthermore, when comparing esophageal IL-8 concentrations in our cohort to previously reported sputum log IL-8 concentrations in individuals with CF at times of health (4.7–4.8 pg/mL) (44), our results indicate that the level of inflammation albeit elevated is less than levels reported in the respiratory tract. Strings were, however, eluted in buffer, which adds an additional dilution factor that cannot be measured directly. Our data could suggest that inflammation of the lungs in CF may beget inflammation of the upper intestinal tract and vice versa, perhaps secondary to the cross-talk between the gut microbiota and the lungs, known as the gut-lung axis. Both these compartments share a mucosal immune system, which is only now being realized. Prior research has shown that when inflammation in the lung exists, the lung-gut axis can induce changes in the circulating biomarkers and gut microbiota that can be bidirectional and secondary to the effects of the pulmonary inflammation propagated by lymphocyte migration, and systemic inflammatory mediators (45). Esophageal inflammation may also be secondary to swallowed airway secretions by typical clearance, which subsequently alters the upper intestinal tract microbiota and results in not only similar microbiomes across sites despite acid blockade treatment but also higher levels of inflammation in the gastrointestinal tract. In addition, previous research has shown that pediatric patients with CF have significant gut inflammation measured via stool calprotectin, the source of which remains ambiguous (46). It is possible that this inflammation extends to the proximal intestinal tract. Further studies are required to delineate if the source of inflammation is gastrointestinal or pulmonary and to determine if the differences in IL-8 concentrations seen are a result of different sampling techniques as opposed to true differences in inflammation.
Limitations to our study include the fact that it is a single center study, and thus the findings may not be generalizable to other locations. Individuals included in this study were pediatric patients with stable lung disease, results of similar comparisons may be different among adults or individuals with a worse lung function. The EST samples mucosal surfaces, which may vary from luminal contents (eg, gastric compartment), and could partially explain the high degree of similarity observed across the upper gastrointestinal tract. We did not control for other possible mediators of microbial alterations including concomitant antibiotic use, prior intestinal surgery, underlying CFTR mutations, and use of probiotics. Also, as this is a cross-sectional study, we are unable to rule out the chance that there are within-person changes that occur associated with acid blockade use. Unfortunately, we were unable to complete the originally conceived prospective study, which would have helped delineate within-person changes. Furthermore, although we did inquire about medication adherence, it is possible that individuals may not have been taking acid blockade medications as reported. Future studies can explore whether the indication for acid blockade use, such as absorption or reflux disease affects the microbial communities. Additionally, although sputum and oropharyngeal swabs may offer some insight into the microbial communities in the lungs, a bronchoalveolar lavage (BAL) remains the gold standard for evaluating the respiratory microbiome but because of the invasive nature of this procedure, we were unable to obtain samples via a BAL. Lastly, although all of our specimens had identical handling, comparable yield of the nasal swab versus the EST may be different because of swab or EST volume and efficiency of sample collection.
The relationship between the lung and esophagus is of pathophysiological interest. In particular, the impact of acid blockade therapy, including PPI and H2RA, on respiratory and esophageal microbiota and respiratory inflammation in the especially vulnerable CF population has not been studied. Clinically, PPIs may benefit CF patients with GERD or who those need them to augment digestive enzyme actions. These initial results in small numbers of patients suggest that whenever indicated, the use of acid-reducing medications do not appear to be associated with alterations of the microbiota, thus reducing concerns about infectious consequences. The cross talk between the aerodigestive tract or the thick mucosal secretions of individuals with CF and the ongoing swallowing of airway mucus may account for this lack of change among groups and for the high bacterial load in the esophagus. Further work is needed to delineate the source of the esophageal inflammation, to determine whether it is indeed airway secretions being swallowed that leads to elevated bacterial loads and whether these findings alter the clinical outcome of pulmonary disease in CF overall.
Supplementary Material
Acknowledgments:
The authors would like to thank Faria Ahmed and Mary Cross for their contributions with patient recruitment, and Elin Towler for helping build a RedCap database.
This work was supported by a Cystic Fibrosis Foundational Grant Award (#Khalaf17B0, 2017–2019), a Cystic Fibrosis Foundation Clinical Research Award, (FILLON15A0, 2015–2020), a National Institutes of Health Training Grant (5T32-DK067009–12, 2008–2020), and a NIH/NCATS Colorado CTSA Grant Number (UL1 TR002535, 2015–2020). Contents are the authors’ sole responsibility and do not necessarily represent official NIH views.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text, and links to the digital files are provided in the HTML text of this article on the journal’s Web site (www.jpgn.org).
Conflict of interest statement: G.F. is the Chief Medical Officer-EnteroTrack, the company that produces the esophageal string test. The other authors have no other conflicts of interest to report.
REFERENCES
- 1.Robinson NB, DiMango E. Prevalence of gastroesophageal reflux in cystic fibrosis and implications for lung disease. Ann Am Thorac Soc 2014;11:964–8. [DOI] [PubMed] [Google Scholar]
- 2.Gustafsson PM, Fransson SG, Kjellman NI, et al. Gastro-oesophageal reflux and severity of pulmonary disease in cystic fibrosis. Scand J Gastroenterol 1991;26:449–56. [DOI] [PubMed] [Google Scholar]
- 3.Pauwels A, Blondeau K, Dupont LJ, et al. Mechanisms of increased gastroesophageal reflux in patients with cystic fibrosis. Am J Gastroenterol 2012;107:1346–53. [DOI] [PubMed] [Google Scholar]
- 4.Dimango E, Walker P, Keating C, et al. Effect of esomeprazole versus placebo on pulmonary exacerbations in cystic fibrosis. BMC Pulm Med 2014;14:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canani RB, Cirillo P, Roggero P, et al. , Working Group on Intestinal Infections of the Italian Society of Pediatric Gastroenterology, Hepatology and Nutrition (SIGENP). Therapy with gastric acidity inhibitors increases the risk of acute gastroenteritis and community-acquired pneumonia in children. Pediatrics 2006;117:e817–20. [DOI] [PubMed] [Google Scholar]
- 6.O’Sullivan BP, Freedman SD. Cystic fibrosis. Lancet 2009;373:1891–904. [DOI] [PubMed] [Google Scholar]
- 7.Sanders DB, Bittner RC, Rosenfeld M, et al. Pulmonary exacerbations are associated with subsequent FEV1 decline in both adults and children with cystic fibrosis. Pediatr Pulmonol 2011;46:393–400. [DOI] [PubMed] [Google Scholar]
- 8.Goetz D, Kopp BT, Salvator A, et al. Pulmonary findings in infants with cystic fibrosis during the first year of life: Results from the Baby Observational and Nutrition Study (BONUS) cohort study. Pediatr Pulmonol 2019;54:581–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laheij RJ, Sturkenboom MC, Hassing RJ, et al. Risk of community-acquired pneumonia and use of gastric acid-suppressive drugs. JAMA 2004;292:1955–60. [DOI] [PubMed] [Google Scholar]
- 10.Ayoub F, Lascano J, Morelli G. Proton pump inhibitor use is associated with an increased frequency of hospitalization in patients with cystic fibrosis. Gastroenterol Res 2017;10:288–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Horck M, van de Kant K, Winkens B, et al. Risk factors for lung disease progression in children with cystic fibrosis. Eur Respir J 2018;51:1702509. [ 10.1183/13993003.02509-2017]. [DOI] [PubMed] [Google Scholar]
- 12.Fillon SA, Harris JK, Wagner BD, et al. Novel device to sample the esophageal microbiome–the esophageal string test. PloS One 2012;7:e42938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris JK, Fang R, Wagner BD, et al. Esophageal microbiome in eosinophilic esophagitis. PLoS One 2015;10:e0128346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furuta GT, Kagalwalla AF, Lee JJ, et al. The oesophageal string test: anovel, minimally invasive method measures mucosal inflammation in eosinophilic oesophagitis. Gut 2013;62:1395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller MR, Hankinson J, Brusasco V, et al. , ATS/ERS Task Force. Standardisation of spirometry. Eur Respir J 2005;26:319–38. [DOI] [PubMed] [Google Scholar]
- 16.Zemanick ET, Wagner BD, Robertson CE, et al. Assessment of airway microbiota and inflammation in cystic fibrosis using multiple sampling methods. Ann Am Thorac Soc 2015;12:221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao J, Carmody LA, Kalikin LM, et al. Impact of enhanced Staphylococcus DNA extraction on microbial community measures in cystic fibrosis sputum. PLoS One 2012;7:e33127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson EJ, Zemanick ET, Accurso FJ, et al. Molecular identification of Staphylococcus aureus in airway samples from children with cystic fibrosis. PLoS One 2016;11:e0147643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nadkarni MA, Martin FE, Jacques NA, et al. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 2002;148 (Pt 1):257–66. [DOI] [PubMed] [Google Scholar]
- 20.Zemanick ET, Wagner BD, Sagel SD, et al. Reliability of quantitative real-time PCR for bacterial detection in cystic fibrosis airway specimens. PloS One 2010;5:e15101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara N, Alkanani AK, Ir D, et al. Prevention of virus-induced type 1 diabetes with antibiotic therapy. J Immunol 2012;189:3805–14. [DOI] [PubMed] [Google Scholar]
- 22.Zemanick ET, Wagner BD, Robertson CE, et al. Airway microbiota across age and disease spectrum in cystic fibrosis. Eur Respir J 2017;50:1700832. https://erj.ersjournals.com/content/erj/50/5/1700832.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markle JG, Frank DN, Mortin-Toth S, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013;339:1084–8. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Lozupone C, Hamady M, et al. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res 2007;35:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Z, DeSantis TZ, Andersen GL, et al. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 2008;36:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Illumina. Homo Sapiens Ucsc Hg19 Human Genome Sequence from iGenome: Illumina. Published 2020. Available at: https://www.illumina.com/. Accessed April, 2020. [Google Scholar]
- 27.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ewing B, Hillier L, Wendl MC, et al. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998;8:175–85. [DOI] [PubMed] [Google Scholar]
- 29.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 1998;8:186–94. [PubMed] [Google Scholar]
- 30.Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schloss PD, Westcott SL. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol 2011;77:3219–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruesse E, Peplies J, Glockner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 2012;28:1823–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41 (Database issue):D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson CE, Harris JK, Wagner BD, et al. Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics 2013;29: 3100–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amir I, Konikoff FM, Oppenheim M, et al. Gastric microbiota is altered in oesophagitis and Barrett’s oesophagus and further modified by proton pump inhibitors. Environ Microbiol 2014;16:2905–14. [DOI] [PubMed] [Google Scholar]
- 36.Rosen R, Hu L, Amirault J, et al. 16S community profiling identifies proton pump inhibitor related differences in gastric, lung, and oropharyngeal microflora. J Pediatr 2015;166:917–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zemanick ET, Harris JK, Wagner BD, et al. Inflammation and airway microbiota during cystic fibrosis pulmonary exacerbations. PloS One 2013;8:e62917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Momani H, Perry A, Stewart CJ, et al. Microbiological profiles of sputum and gastric juice aspirates in cystic fibrosis patients. Sci Rep 2016;6:26985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh DS, DeMeester SR, Vallbohmer D, et al. Reduction of interleukin 8 gene expression in reflux esophagitis and Barrett’s esophagus with antireflux surgery. Arch Surg 2007;142:554–9. [DOI] [PubMed] [Google Scholar]
- 40.Huo X, Zhang X, Yu C, et al. In oesophageal squamous cells exposed to acidic bile salt medium, omeprazole inhibits IL-8 expression through effects on nuclear factor-kappaB and activator protein-1. Gut 2014;63:1042–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sei H, Oshima T, Shan J, et al. Esophageal epithelial-derived IL-33 is upregulated in patients with heartburn. PloS One 2016;11: e0154234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shan J, Oshima T, Muto T, et al. Epithelial-derived nuclear IL-33 aggravates inflammation in the pathogenesis of reflux esophagitis. J Gastroenterol 2015;50:414–23. [DOI] [PubMed] [Google Scholar]
- 43.Isomoto H, Wang A, Mizuta Y, et al. Elevated levels of chemokines in esophageal mucosa of patients with reflux esophagitis. Am J Gastroenterol 2003;98:551–6. [DOI] [PubMed] [Google Scholar]
- 44.Sagel SD, Wagner BD, Anthony MM, et al. Sputum biomarkers of inflammation and lung function decline in children with cystic fibrosis. Am J Respir Crit Care Med 2012;186:857–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang D, Li S, Wang N, et al. The cross-talk between gut microbiota and lungs in common lung diseases. Front Microbiol 2020;11:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ooi CY, Syed SA, Rossi L, et al. Impact of CFTR modulation with ivacaftor on gut microbiota and intestinal inflammation. Sci Rep 2018;8:17834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.