Abstract
Proteins are considered to be the key players in structure, function, and metabolic regulation of our bodies. The mechanisms used in conventional therapies often rely on inhibition of proteins with small molecules, but another promising method to treat disease is by targeting the corresponding mRNAs. In 1998, Craig Mellow and Andrew Fire discovered dsRNA-mediated gene silencing via RNA interference or RNAi. This discovery introduced almost unlimited possibilities for new gene silencing methods, thus opening new doors to clinical medicine. RNAi is a biological process that inhibits gene expression by targeting the mRNA. RNAi-based therapeutics have several potential advantages (i) a priori ability to target any gene, (ii) relatively simple design process, (iii) site-specificity, (iv) potency, and (v) a potentially safe and selective knockdown of the targeted cells. However, the problem lies within the formulation and delivery of RNAi therapeutics including rapid excretion, instability in the bloodstream, poor cellular uptake, and inefficient intracellular release. In an attempt to solve these issues, different types of RNAi therapeutic delivery strategies including multifunctional RNA nanoparticles are being developed. In this mini-review, we will briefly describe some of the current approaches.
Keywords: RNA nanotechnology, RNA nanoparticles, RNA/DNA hybrids, RNA interference, siRNA, delivery
1. Introduction
According to the Social Security Administration, average Americans that reach age 65 today most likely will live to be 84 years old [1]. However, with older age, the chances of contracting deadly diseases, such as cancer, increases dramatically. Some cancers (e.g. breast cancer) can be removed surgically but this does not guarantee that the disease will not return within a patient’s lifetime. For other types of cancer (e.g. chronic lymphocytic leukemia), surgery may have very little effect (http://www.cancer.org). Other available treatments are chemo- and immunotherapies. However, these alternatives lack target specificity and cause severe toxic side effects affecting the growth of hair, nails, loss of appetite and blood cell count, just to name a few (http://www.cancer.org). Therefore, the advancements in biomedical technologies that provide safe and effective cancer treatment are in demand. Among the novel approaches is the recognition and use of specific intracellular RNA signatures (e.g. an overexpression of certain genes) that are especially important in detection and personalized treatments of cancers as well as viral infections, and autoimmune diseases [2–4]. The wide use of novel therapeutics based on target specific RNA-mediated gene silencing, called RNA interference or RNAi, will likely become the next breakthrough in cancer therapy. The first successful therapeutic knockdown of the endogenous gene, apolipoprotein B (ApoB), occurred in a 2004 study [5], only a few years after the original discovery of RNAi [6]. In 2010, the first delivery of RNAi inducers and RNAi activation in humans was reported [7] and today, there are more than 30 RNAi-based therapeutic companies world-wide [8].
2. RNAi activators
There are several types of RNAi activators suitable for therapeutic applications. The first natural trigger is the micro (mi)RNA [9]. There are several main steps that lead to the down-regulation of gene expression via sequence specific recognition of corresponding messenger RNAs. Normally, a nuclear Pol II-driven transcription produces primary (pri)-miRNAs -- long self-folded strands with miRNA sequences embedded into repetitive hairpin-like structures [10]. The nuclear enzyme Drosha then processes these transcripts into short hairpins called precursor or (pre)-miRNA with two nucleotide 3’-side overhangs [11]. The pre-miRNAs are further released into the cytoplasm by Exportin-5 [12] and refined into mature miRNAs by RNaseIII-like endonuclease, called Dicer [13–15]. The resulting mature miRNAs are loaded into the RNA-induced silencing complex or RISC based on their thermodynamic asymmetry [16,17]. This asymmetric nature of the miRNAs affects the choice of the strand (the guide or anti-sense strand) [17] that is further utilized by the RISC for sequence specific recognition of the complementary part of mRNA [18] and inhibition of its translation [19].
This naturally occurring cellular process can be mimicked for therapeutic purposes and RNAi can be induced exogenously by several routes as depicted in Figure 1. The small hairpin RNAs or shRNAs (Figure 1, case 1), use the miRNA pathway and can be introduced into the cell with plasmids or viral vectors [20]. An issue with many drugs is that over time, the therapeutic effects start to decrease due to cell division and the clearance process that dilutes the drugs (or loaded RISC in the case of RNAi-based therapeutics). Therefore, an increase in the initial dosage or repetitive administration of the drug is required. The main advantage of the shRNA-based approach is that the extent of specific gene silencing is not decreased by cell division over time [21,22]. However, there are some safety concerns regarding the shRNAs due to the route of their cellular production that requires an expression vector that may lead to genomic integration [23,24].
Figure 1.
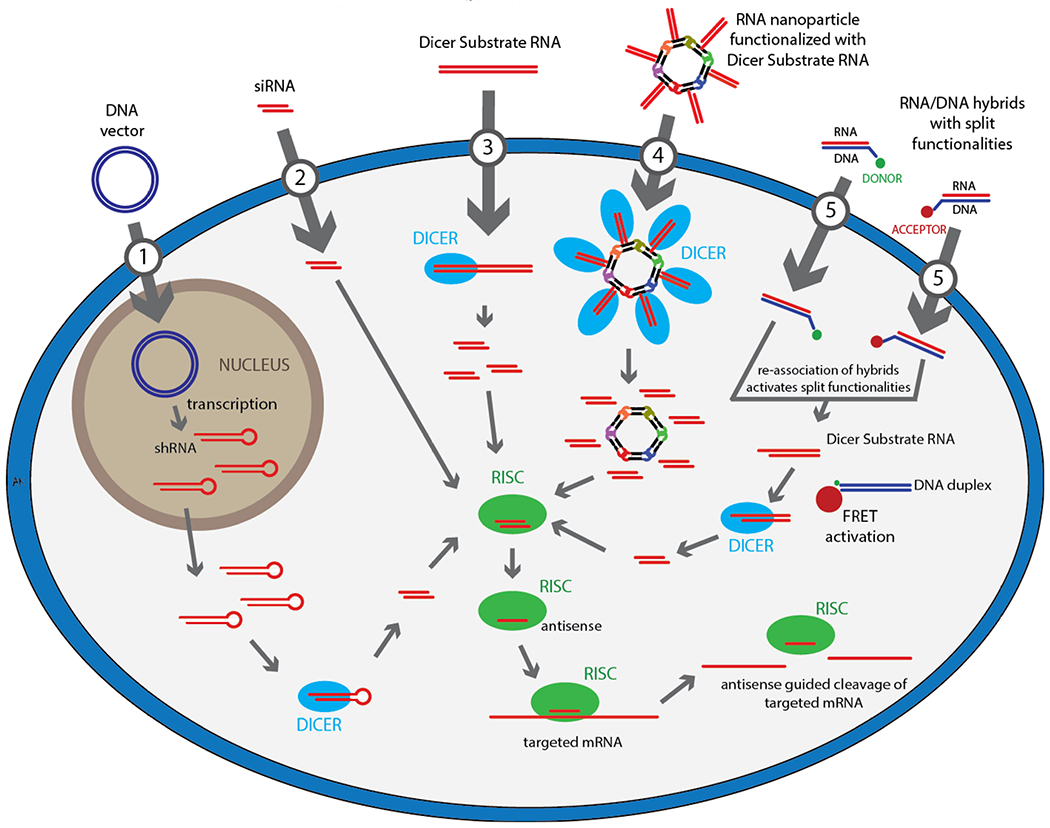
Schematic representation of various exogenous RNAi activation pathways.
The small interfering RNAs or siRNAs [25–27] (Figure 1, case 2), are relatively short RNA duplexes of approximately 20-25 base pairs [21,28]. These duplexes resemble the mature miRNAs and do not require any further enzymatic processing. Also, with the synthetic siRNAs using perfect sequence complementary, complete destruction of the endogenous gene function within the cell can be achieved [29]. Once in the cytoplasm, siRNAs are integrated directly into the RISC, releasing the sense strand from the antisense loaded RISC.
If the dsRNAs are longer than conventional siRNAs or miRNAs, then upon the exogenous introduction into the cytoplasm these duplexes need first to be diced prior to RISC loading (Figure 1, case 3). The dicable RNAs are also called Dicer Substrate (DS) RNAs and are an important class of RNAi activators as they promote RISC loading [30,31]. DS RNAs are also used for intracellular Dicer-assisted release of siRNAs from various artificially designed nanoconstructs (Figure 1.4) [32–34].
3. Multifunctional RNA and DNA nanoparticles
The simultaneous introduction of multiple RNAi inducers targeting different genes in conjunction with other functional moieties (therapeutic aptamers, ribozymes, fluorescent dyes, peptides or small molecules) may maximize the synergistic therapeutic effect [35–40]. Precise control over the composition and stoichiometry of these combinatorial drugs is essential to guarantee the consistency in batch-to-batch formulations [32]. If therapeutic domains are chosen to be RNA-based (miRNAs, siRNAs, etc), the optimal route of controllable formulation would be through the introduction of nucleic acid-based nanoscaffolds [41]. The uses of various computationally designed [42–44] RNA, DNA or RNA-DNA based nanoscaffolds have multiple advantages not only in delivery of therapeutics to the diseased cells, but also in molecular imaging and biosensing [45,46]. Significant progress in RNA nanotechnology can be illustrated by various alternative strategies developed for the production of three-dimensional nucleic acid-based nanoscaffolds that are further utilized in different nanotechnological applications. One of the strategies, called RNA architectonics, uses tecto-RNAs [47–50] that combine different RNA three-dimentional building blocks (called motifs) allowing remarkable structural control in bottom-up assembly [51–71]. In particular, computationally designed [72] and experimentally tested [32,35,73,74] nanorings take advantage of the so-called RNA kissing loop ineracting motifs (Figure 1, case 4). Other examples use the structural motifs extracted from the phage phi29 pRNAs [75] that were widely used in the engineering of multiple stable and functional RNA nanoscaffolds [7687]. An alternative to the tecto-RNA designing strategy is exemplified by the nanocubes (Figure 2) [88,89]. Nanostructures built with this approach do not require any RNA tertiary motifs and solely rely on canonical Watson-Crick interactions. Therefore, their assemblies do not always require RNAs and can be used to introduce multiple DNA strands into the each scaffold. These RNA/DNA hybrid structures can significantly lower the immune response (Figure 2) [90].
Figure 2.
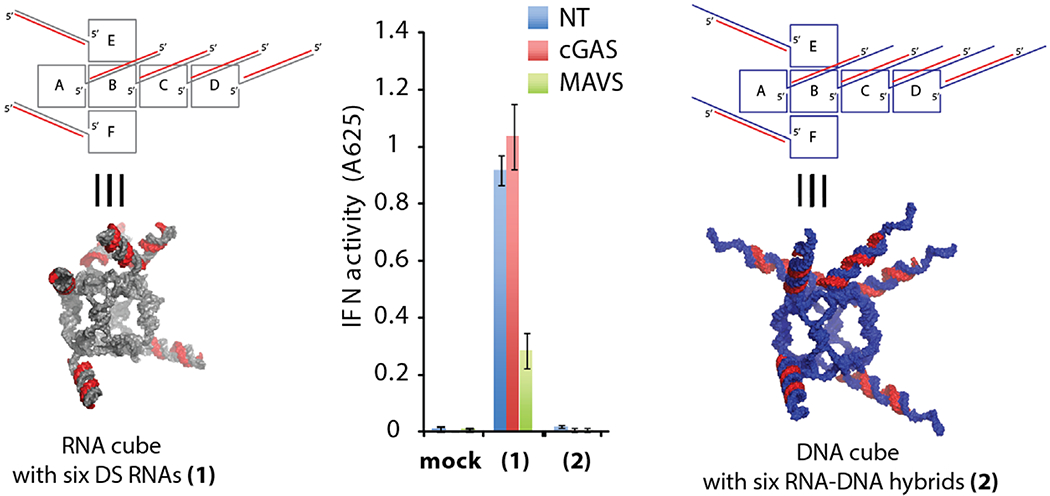
Interferon (IFN)-activity experiments with THP-1 IFN reporter cells for functionalized RNA (1) and DNA (2) cubes described in Afonin et al. [90]. For IFN-activity experiments, THP-1 IFN reporter cells were depleted of cGAS (a DNA-binding receptor in the type I IFN signaling pathway) or MAVS (an RNA-induced stimulation of IFN activity) by specific siRNAs. Cells were transfected with nanocubes and secreted alkaline phosphatase activity was measured in culture supernatants 24 hours post-transfection.
Nanoscaffolds designed with both strategies allow their further functionalization with various siRNAs, aptamers, fluorescent dyes, proteins for further simultaneous delivery. Importantly, the intracellular release of functional siRNAs is achieved through the introduction of DS RNAs (Figure 1, case 4) as mentioned above. For example, recently we shown that different types of nanoparticles can be efficiently used in silencing HIV-1 production. We tested two RNAi combinatorial approaches in which the nanocubes and nanorings were functionalized with six different DS RNAs targeting different parts of the HIV-1 genome [35,90]. This strategy was used to minimize the negative effect on RNAi treatment caused by HIV-1 genomic diversity and appearance of drug-resistant mutants [91,92]. We were able to select several HIV-1 targets to design our nanoparticles based on extensive studies performed by Berkhout and collaborators during the last few years [37,93–95]. Briefly, we simultaneously targeted HIV-1 mRNAs that code for Capsid (p24), PBS-p17 (primer binding site junction with Matrix), Protease, Reverse Transcriptase, Nef, Envelope, Rev and Tat. The advantages of some targets were that the same siRNAs could target two different mRNAs at once (since HIV-1 utilizes three open reading frames). For example, siRNA targeting of the HIV-1 envelope glycoproteins would also target the un-spliced mRNA that codes the Gag and Pol proteins.
However, all formulations employing RNA nanoscaffolds functionalized with DS RNAs are dependent on the presence of Dicer and can be activated virtually in all cells including the healthy ones, thus, increasing possible side effects. To introduce additional control over deliverable functionalities and to enchance their chemical stability, the properties of DNA and RNA were merged in the development of nanoparticles that were constructed from RNA/DNA hybrids [90,96–98]. Combining the properties of these molecules (Figure 1, case 5 and Figure 2, right panel) allows for the splitting of the components of the functional elements (inactivating them) and permits their later activation under the control of complementary hybrids with ssDNA toeholds. The kinetics and thermodynamics of the toehold interaction can be easily fine-tuned. Besides tight control over functional activation, this novel approach provides a higher stability for constructs in blood serum, and permits the attachments of additional functionalities to DNAs (e.g. fluorescent markers for tracking) without interfering with RNA function. Moreover, this approach can be potentially utilized by the DNA nanotechnologists and other DNA nanoconstructs can be used for these applications [99].
4. Delivery of multifunctional RNA and DNA nanoparticles
In order to fully take advantage of the power of therapeutic RNA nanotechnology all nanoparticles have to be delivered intact to their desired site of action and within the cytoplasm of the diseased cells. The road from the test tube to this objective contains many obstacles that need to be overcome in a successful delivery strategy. Since major obstacles exist in reaching the tissues of interest, an intense effort is underway to establish local delivery solutions directly to the region of interest (Figure 3).
Figure 3.
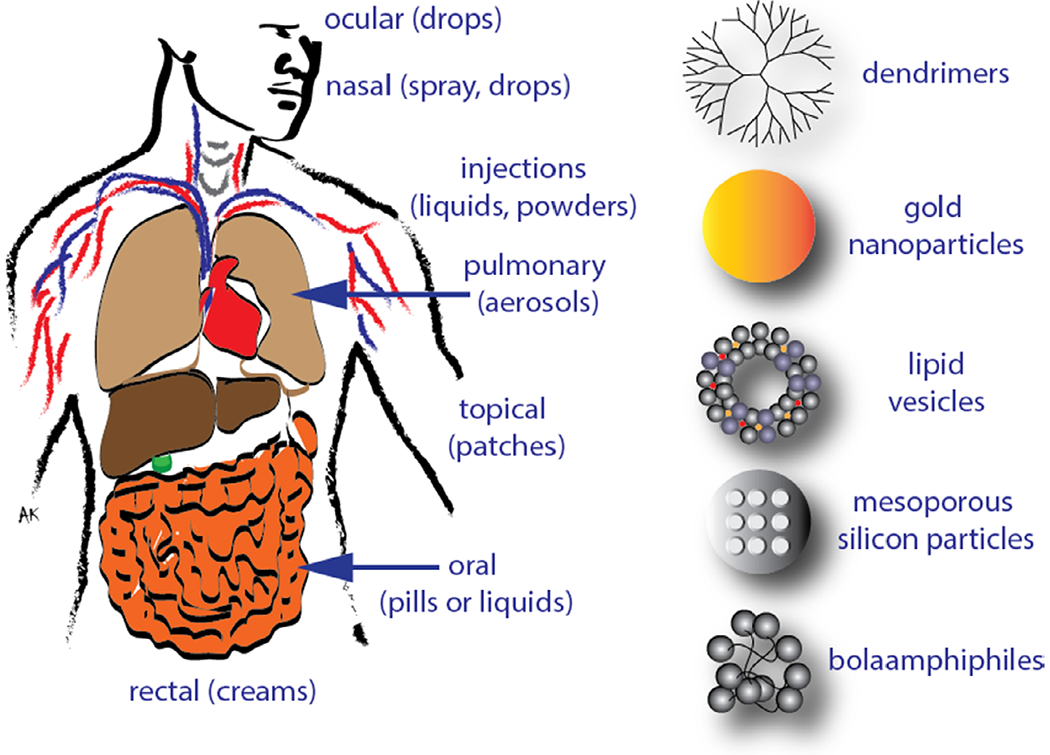
Various possible routes and some examples of carriers used for delivery of therapeutic RNAi inducers.
Mucosal delivery is noninvasive and gives direct access to the main areas of entry for engaging a variety of pathogens, inflammatory elements, and cancer. The delivery method is local administration to the mucosal surface linings of the respiratory tract, gastrointestinal (GI) tract, and genitourinary tract. The mucus thickness differs for each patient depending on the state of the diseases. This can cause problems such as decreasing the effectiveness of the drug due to an unusual thick mucus surface. The pH of different areas and the net charge of mucin may also interfere with the level of accessibility [100].
Pulmonary delivery of RNAi therapeutics would be preferred for lung-related diseases such as influenza. There are a couple of intake methods available, such as pressurised metered dose inhalers or dry powder inhalers [101]. This type of delivery still has quite a few flaws as the makeup and structure of the lung refuses any nano-particle-based or naked siRNA from entering. The functions of the upper respiratory tract prevents materials from being absorbed into the lungs. The lungs, having too many passageways further complicates target specificity. Also, enzymatic degradation will most likely occur during exhalation.
Systemic delivery occurs via intravenous, intraperitoneal, or oral administration [102–104]. Of those, the intravenous route is the most explored since this is a natural avenue of delivery for cell-required nutrients. However, blood circulation is highly regulated and naked RNA nanoparticles have very short half-lives in serum. This emanates from two major pathways. First, the size of the particles is critical as particles smaller than about eight nanometers will be subjected to renal clearance [105]. On the other hand, particles larger than about 200 nanometers will be cleared through the spleen and larger ones accumulating in the lung [106]. Second, interaction with blood components is critical to the fate of RNA nanoparticles. The blood harbors many entities aimed at preventing foreign invasions. Serum nucleases are prevalent and will insure a fast degradation of naked non-modified RNA [74,98]. Chemical modifications altering the sensitivity of the RNAs to nucleases have therefore been used to alleviate this issue [36,74,107]. These modifications can be engineered at the level of the ribose sugar or within the backbone and can dramatically enhance the half-lives of RNA nanoparticles. These modifications need to be performed carefully so as not to compromise the thermodynamic asymmetry and the overall functionality of the RNA. The blood also contains proteins that can bind to the particles and initiate complement activation and inflammation [108]. This can also result in opsonization leading to the phagocytic uptake of the particles for degradation [109]. A way to circumvent this is through the introduction of polymers such as polyethylene glycol that will prevent the binding of the serum proteins and thereby increase the half live of the particles [110,111]. Nanoparticles, of the right size range, and properly stabilized against deleterious interactions with blood components will thereby circulate longer but will need to get enriched at their intended destination. This targeting can be modulated in different ways. While the blood vessels are lined with a tight epithelium that prevents extravasation of the circulating particles, the unregulated growth of cancers leads to the presence of openings through which particles can get out of the blood circulation into the cancer environment [112]. Those defects also come with a defective lymphatic drainage and all this results in passive targeting of the nanoparticles to the tumors through the enhanced permeation and retention (EPR) effect [113,114]. The targeting can also be modulated through the addition of ligands (e.g. folic acid [115] or aptamers [116,117]) that will confer specific binding to a target whose presence is specific to the diseased environment and will enhance to retention of the nanoparticles at that location [118,119].
Provided the nanoparticles have avoided all the previous hurtles and are present, intact, in the diseased environment, a major obstacle still needs to be overcome, penetration into the cytoplasm of the cells. Cellular life has evolved through a compartmentalization from the extracellular milieu delineated by a cellular membrane. This membrane, composed of lipid and proteins ensures tightly regulated processes for the uptake of any component needed for the cell to survive and prevent invasion. Viruses have naturally evolved to overcome this barrier and viral vectors have thereby been used to deliver RNA. However, the use of viral vectors raise some safety concerns due to immune response [120]. Synthetic mimics that allow for the protection of the RNA in the extracellular environment but facilitate it’s delivery within the cellular cytoplasm are therefore widely investigated. Synthetic vectors used for in vivo delivery of RNAs can include various polymers (PEI, PEG) [121], lipid vesicles [122] and lipid-like structures (bolaamphiphiles [123]), anionic carriers [124], sugars [125], dendrimers [126], as well as gold [127] or silicon nanoparticles [128]. Gold nanoparticles can be used for vaccinations purposes [129]. Anionic polymers can be used as a carrier to replace the usual approach of using only a cationic lipid-based vector. This method is biodegradable and already FDA-approved [124]. RNAs are negatively charged and so is the cellular membrane. Many studies have therefore focused on positively charged vectors to facilitate the initial interaction of the particles with the cells. While modifications of the nanoparticles or the vectors with cell penetrating peptides (CPPs) could facilitate the direct crossing at the level of the plasma membrane, most of the nanoparticles get taken up through endocytosis [130]. This uptake can be further enhanced through the use of appropriate targeting moieties. The main task remains to escape from the endosomes. The various carriers used aim to destabilize the endosomes in various ways. PEIs or polycations containing amino groups are thought to trigger a “proton sponge” effect in the acidic environment of the endosome and osmotically destabilize them. Ionizable lipids establish a positive charge in an acidic environment and can undergo interactions with anionic endosomal lipids [131]. Lipids that have the propensity to form alternate hexagonal structures can also be used to destabilize the lamellar environment of the endosomal membrane [132]. Peptides and polymers that have the ability to partition within the endosomes are also being explored [133].
While being important for the final step corresponding to cytoplasmic delivery, synthetic carriers can confer multiple advantages during the earlier stages of delivery. Lipid carriers are particularly interesting since they are often biocompatible. Bolamphiphiles are different from conventional lipids in that they have two head-groups separated by a long hydrophobic chain [134]. This confers inherent stability to the structures they form. They also come with a wide variety of head-groups which can modulate their interaction with the RNAs. Careful studies deciphering the forces involved in the tight binding of RNA conferring protection yet allowing for proper release will be important for the development of optimal delivery systems. We have undertaken to characterize selected RNA carriers both computationally and experimentally. Of special interest to us was a class of lipids called bolaamphiphiles, or bolas, that have been shown to complex with peptides, proteins and plasmid DNAs [135–137]. They were also shown to cross cell membranes and are capable of moving across the blood-brain barrier [138]. Bolas have a central hydrophobic alkyl chain of varying length and covalently linked, hydrophilic, positively charged head groups at both ends (see Figure 4).
Figure 4.
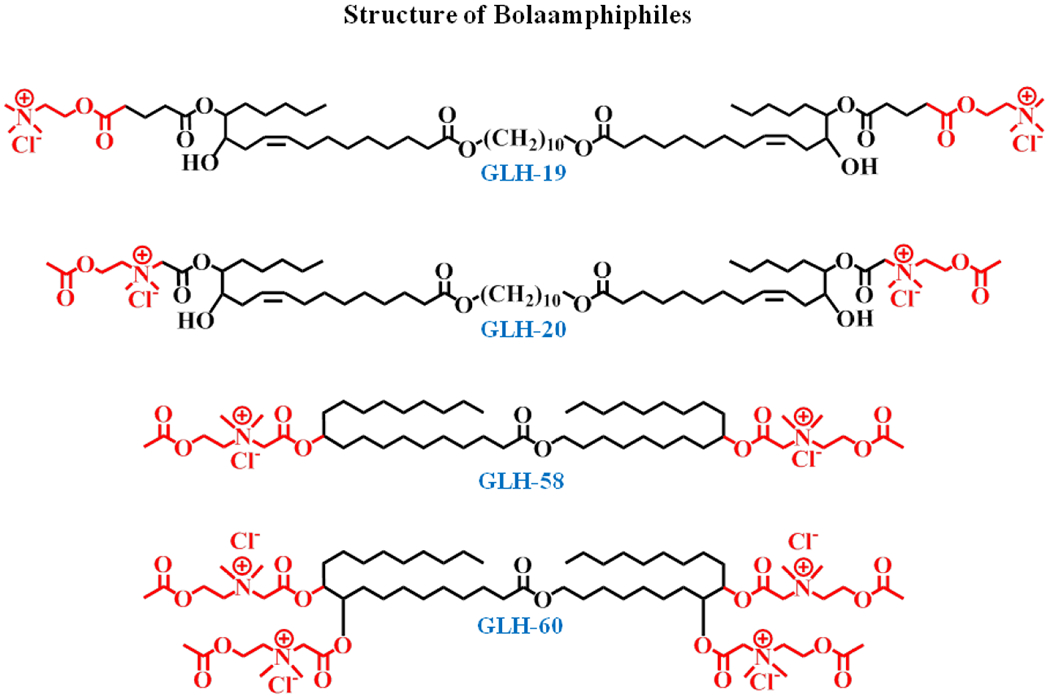
Chemical structure of selected bolaamphiles GLH-19, 20, 58 and 60. Note the differences in the head groups and their placement relative to the ends of the central hydrophobic chain.
The key advantage of bolas in comparison with phospholipids is the fine balance between the attractive forces of the hydrophobic domains and the repulsive forces of the hydrophilic head groups allowing for rapid structural changes and release of the cargo upon minor disruptions of that balance. In our recently published study of bolas GLH-19 and 20 [123], the two variants had two acetylcholine head groups (AChHG). The GLH-20 head groups differed from the GLH-19’s in that they can be hydrolyzed by acetylcholine esterase permitting release of cargo in the brain. 3D modeling and molecular dynamics (MD) simulations that were performed in that study showed that the bola-siRNA complexation relies mostly on the electrostatic interactions, with help from some hydrogen bonds and hydrophobic interactions. MD simulations [123] indicated different surface properties that should result in higher binding affinity with the siRNA for the GLH-19 variant, thus providing better protection against degradation. The experiments using bolas complexed with siRNA and RNA/DNA hybrid constructs confirmed these theoretical predictions, showing a slightly better uptake of the GLH-19/siRNA complex into cells and almost no degradation by nucleases. In vitro experiments on human breast cancer cells expressing the green fluorescent protein (MDA-MB-231/GFP) showed comparable gene silencing for the siRNAs complexed with GLH-19 and 20. The better release of the siRNA inside the cell (due to lower binding affinity) compensated for the lower delivered siRNA numbers. In vivo biodistribution experiments in MDA-MB-231 xenograft tumor bearing athymic nude mice demonstrated high uptake of the siRNA into the tumor relative to the other major organs. In a separate study, siRNA-functionalized nanorings associated with the same bolas were successfully used to silence the GFP expression by intratumoral injections [35]. Taken altogether, these results demonstrated a good potential of the bola family as nucleic acid carriers for therapeutic purposes. Other selected members of the bola family with AchHGs, the GLH-58 and 60 (see Figure 4), one with four head groups, are now being characterized as potential delivery agents.
5. Conclusion
To summarize, although there has been significant advancements in research related to therapeutic RNA nanotechnology within the past ten years, the efficient delivery of RNAi-inducing multifunctional RNA nanoparticles remains challenging, and more studies need to be conducted to determine the best methods for delivery. Most likely there will not be one panacea that will solve the problem. Cells and tumors differ in their accessibility based on their surrounding vasculature, as well as membrane structures. The mode of cell entry also differ (endocytosis, pores, etc). In some cases specific molecules will bind to targeted cell surface proteins, but will remain on the surface and not be endocytosed. Beyond this there are issues related to endosomal release of the active agents before they are degraded. The future for delivery will then require the development of carriers that will associate with the RNAi-based particle, protecting it from its environment, such as nucleases and immune agents, but yet allow proper association with cells and release of cargo into the cytosol or the nucleus, as required. The accomplishment of such goals will require the design of carriers that incorporate and react to each of the specified issues, which in turn may vary with cell and tumor type. Thus, it may be necessary to design different carriers for different environments and to administer these carriers through various pathways besides, for example, intravenous delivery. Progress is being made to circumvent each of these barriers as more knowledge is gained on the mechanisms that are involved in each of the steps along the delivery pathway. Appropriate characteristics are being built into the particles to minimize the difficulties and to maximize the needed attributes. This might include balancing the effects of electrostatics with hydrophobicity and the use of proper shape and size of the nanoparticles to facilitate their correct assembly and cell entry, as well as features that are sensitive to pH change to facilitate endosomal release. The future looks quite bright for solving several of the aforementioned issues but, as indicated, may require the use of multiple types of “magic bullets.”
Acknowledgements:
Authors thank Dr. Jon W. Merkert (UNC Charlotte) for helpful discussion. This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E (to M.V. and W.K.K). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Conflict of interest: Authors state no conflict of interest
Contributor Information
Bich Ngoc Dao, Department of Chemistry, University of North Carolina at Charlotte, 9201 University City Boulevard, Charlotte, North Carolina 28223, USA.
Mathias Viard, Basic Science Program, Leidos Biomedical Research Inc., Frederick National Laboratory, Frederick, Maryland, USA; Basic Research Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD, USA.
Angelica N. Martins, Department of Biology, University of North Carolina at Charlotte, 9201 University City Boulevard, Charlotte, North Carolina 28223, USA
Wojciech K. Kasprzak, Basic Science Program, Leidos Biomedical Research Inc., Frederick National Laboratory, Frederick, Maryland, USA; Gene Regulation and Chromosome Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD, USA
Bruce A. Shapiro, Gene Regulation and Chromosome Biology Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD, USA.
Kirill A. Afonin, Department of Chemistry, University of North Carolina at Charlotte, 9201 University City Boulevard, Charlotte, North Carolina 28223, USA.
References
- [1].Social Security. http://www.ssa.gov/planners/lifeexpectancy.html (accessed March 6 2015).
- [2].Castanotto D; Rossi JJ The promises and pitfalls of RNA-interference-based therapeutics. Nature 2009, 457, 426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Venkataraman S; Dirks RM; Ueda CT; Pierce NA Selective cell death mediated by small conditional RNAs. Proc Natl Acad Sci U S A 2010, 107, 16777–16782. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [4].Win MN; Smolke CD Higher-order cellular information processing with synthetic RNA devices. Science 2008, 322, 456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Soutschek J; Akinc A; Bramlage B; Charisse K; Constien R; Donoghue M; Elbashir S; Geick A; Hadwiger P; Harborth J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004, 432, 173–178. [DOI] [PubMed] [Google Scholar]
- [6].Fire A; Xu S; Montgomery MK; Kostas SA; Driver SE; Mello CC Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- [7].Davis ME; Zuckerman JE; Choi CH; Seligson D; Tolcher A; Alabi CA; Yen Y; Heidel JD; Ribas A Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 2010, 464, 1067–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].World of RNAi Therapeutics. https://www.google.com/maps/d/viewer?oe=UTF8&source=embed&ie=UTF8&msa=0&mid=zr-Ht4ReTX3o.kFQo_CkRb9AQ.
- [9].Bartel DP MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- [10].Lee Y; Kim M; Han J; Yeom KH; Lee S; Baek SH; Kim VN MicroRNA genes are transcribed by RNA polymerase II. The EMBO journal 2004, 23, 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gregory RI; Chendrimada TP; Shiekhattar R MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods in molecular biology 2006, 342, 33–47. [DOI] [PubMed] [Google Scholar]
- [12].Murchison EP; Hannon GJ miRNAs on the move: miRNA biogenesis and the RNAi machinery. Current opinion in cell biology 2004, 16, 223–229. [DOI] [PubMed] [Google Scholar]
- [13].Lund E; Dahlberg JE Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harbor symposia on quantitative biology 2006, 71, 59–66. [DOI] [PubMed] [Google Scholar]
- [14].Ji X The mechanism of RNase III action: how dicer dices. Current topics in microbiology and immunology 2008, 320, 99–116. [DOI] [PubMed] [Google Scholar]
- [15].Macrae IJ; Zhou K; Li F; Repic A; Brooks AN; Cande WZ; Adams PD; Doudna JA Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198. [DOI] [PubMed] [Google Scholar]
- [16].Schwarz DS; Hutvagner G; Du T; Xu Z; Aronin N; Zamore PD Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [DOI] [PubMed] [Google Scholar]
- [17].Khvorova A; Reynolds A; Jayasena SD Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [DOI] [PubMed] [Google Scholar]
- [18].Lin SL; Chang D; Ying SY Asymmetry of intronic pre-miRNA structures in functional RISC assembly. Gene 2005, 356, 32–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pratt AJ; MacRae IJ The RNA-induced silencing complex: a versatile gene-silencing machine. The Journal of biological chemistry 2009, 284, 17897–17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Xiang S; Fruehauf J; Li CJ Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nature biotechnology 2006, 24, 697–702. [DOI] [PubMed] [Google Scholar]
- [21].Tokatlian T; Segura T siRNA applications in nanomedicine. Wiley interdisciplinary reviews. Nanomedicine and nanobiotechnology 2010, 2, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Siolas D; Lerner C; Burchard J; Ge W; Linsley PS; Paddison PJ; Hannon GJ; Cleary MA Synthetic shRNAs as potent RNAi triggers. Nature biotechnology 2005, 23, 227–231. [DOI] [PubMed] [Google Scholar]
- [23].Whitehead KA; Dahlman JE; Langer RS; Anderson DG Silencing or stimulation? siRNA delivery and the immune system. Annual review of chemical and biomolecular engineering 2011, 2, 77–96. [DOI] [PubMed] [Google Scholar]
- [24].Wang Z; Rao DD; Senzer N; Nemunaitis J RNA interference and cancer therapy. Pharmaceutical research 2011, 28, 2983–2995. [DOI] [PubMed] [Google Scholar]
- [25].Bramsen JB; Kjems J Chemical modification of small interfering RNA. Methods in molecular biology 2011, 721, 77–103. [DOI] [PubMed] [Google Scholar]
- [26].Bramsen JB; Kjems J Development of Therapeutic-Grade Small Interfering RNAs by Chemical Engineering. Front Genet 2012, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bramsen JB; Laursen MB; Damgaard CK; Lena SW; Babu BR; Wengel J; Kjems J Improved silencing properties using small internally segmented interfering RNAs. Nucleic acids research 2007, 35, 5886–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Elbashir SM; Lendeckel W; Tuschl T RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 2001, 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pecot CV; Calin GA; Coleman RL; Lopez-Berestein G; Sood AK RNA interference in the clinic: challenges and future directions. Nat Rev Cancer 2011, 11, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rose SD; Kim DH; Amarzguioui M; Heidel JD; Collingwood MA; Davis ME; Rossi JJ; Behlke MA Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic acids research 2005, 33, 4140–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kim DH; Behlke MA; Rose SD; Chang MS; Choi S; Rossi JJ Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nature biotechnology 2005, 23, 222–226. [DOI] [PubMed] [Google Scholar]
- [32].Afonin KA; Grabow WW; Walker FM; Bindewald E; Dobrovolskaia MA; Shapiro BA; Jaeger L Design and self-assembly of siRNA-functionalized RNA nanoparticles for use in automated nanomedicine. Nature protocols 2011, 6, 2022–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Afonin KA; Kasprzak WK; Bindewald E; Kireeva M; Viard M; Kashlev M; Shapiro BA In silico design and enzymatic synthesis of functional RNA nanoparticles. Accounts of chemical research 2014, 47, 1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Afonin KA; Lindsay B; Shapiro BA Engineered RNA Nanodesigns for Applications in RNA Nanotechnology. RNA nanotechnology 2013, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Afonin KA; Viard M; Koyfman AY; Martins AN; Kasprzak WK; Panigaj M; Desai R; Santhanam A; Grabow WW; Jaeger L, et al. Multifunctional RNA nanoparticles. Nano letters 2014, 14, 5662–5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Guo P The emerging field of RNA nanotechnology. Nature nanotechnology 2010, 5, 833–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liu YP; von Eije KJ; Schopman NC; Westerink JT; ter Brake O; Haasnoot J; Berkhout B Combinatorial RNAi against HIV-1 using extended short hairpin RNAs. Molecular therapy : the journal of the American Society of Gene Therapy 2009, 17, 1712–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nakashima Y; Abe H; Abe N; Aikawa K; Ito Y Branched RNA nanostructures for RNA interference. Chem Commun (Camb) 2011. [DOI] [PubMed] [Google Scholar]
- [39].Lee H; Lytton-Jean AK; Chen Y; Love KT; Park AI; Karagiannis ED; Sehgal A; Querbes W; Zurenko CS; Jayaraman M, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nature nanotechnology 2012, 7, 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shukla GC; Haque F; Tor Y; Wilhelmsson LM; Toulme JJ; Isambert H; Guo P; Rossi JJ; Tenenbaum SA; Shapiro BA A Boost for the Emerging Field of RNA Nanotechnology. ACS nano 2011, 5, 3405–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Grabow WW; Jaeger L RNA self-assembly and RNA nanotechnology. Accounts of chemical research 2014, 47, 1871–1880. [DOI] [PubMed] [Google Scholar]
- [42].Bindewald E; Afonin K; Jaeger L; Shapiro BA Multistrand RNA secondary structure prediction and nanostructure design including pseudoknots. ACS nano 2011, 5, 9542–9551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bindewald E; Grunewald C; Boyle B; O‘Connor M; Shapiro BA Computational strategies for the automated design of RNA nanoscale structures from building blocks using NanoTiler. J Mol Graph Model 2008, 27, 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bindewald E; Hayes R; Yingling YG; Kasprzak W; Shapiro BA RNAJunction: a database of RNA junctions and kissing loops for three-dimensional structural analysis and nanodesign. Nucleic acids research 2008, 36, D392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Afonin KA; Danilov EO; Novikova IV; Leontis NB TokenRNA: a new type of sequence-specific, label-free fluorescent biosensor for folded RNA molecules. Chembiochem : a European journal of chemical biology 2008, 9, 1902–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Rogers TA; Andrews GE; Jaeger L; Grabow WW Fluorescent monitoring of RNA assembly and processing using the split-spinach aptamer. ACS synthetic biology 2015, 4, 162–166. [DOI] [PubMed] [Google Scholar]
- [47].Jaeger L; Chworos A The architectonics of programmable RNA and DNA nanostructures. Curr Opin Struct Biol 2006, 16, 531–543. [DOI] [PubMed] [Google Scholar]
- [48].Jaeger L; Leontis NB Tecto-RNA: One-Dimensional Self-Assembly through Tertiary Interactions This work was carried out in Strasbourg with the support of grants to N.B.L. from the NIH (1R15 GM55898) and the NIH Fogarty Institute (1-F06-TW02251–01) and the support of the CNRS to L.J. The authors wish to thank Eric Westhof for his support and encouragement of this work. Angewandte Chemie 2000, 39, 2521–2524. [DOI] [PubMed] [Google Scholar]
- [49].Jaeger L; Westhof E; Leontis NB TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic acids research 2001, 29, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ishikawa J; Furuta H; Ikawa Y RNA tectonics (tectoRNA) for RNA nanostructure design and its application in synthetic biology. Wiley interdisciplinary reviews. RNA 2013, 4, 651–664. [DOI] [PubMed] [Google Scholar]
- [51].Yamashita K; Tanaka T; Furuta H; Ikawa Y TectoRNP: self-assembling RNAs with peptide recognition motifs as templates for chemical peptide ligation. Journal of peptide science : an official publication of the European Peptide Society 2012, 18, 635–642. [DOI] [PubMed] [Google Scholar]
- [52].Chworos A; Severcan I; Koyfman AY; Weinkam P; Oroudjev E; Hansma HG; Jaeger L Building programmable jigsaw puzzles with RNA. Science 2004, 306, 2068–2072. [DOI] [PubMed] [Google Scholar]
- [53].Severcan I; Geary C; Jaeger L; Bindewald E; Kasprzak W; Shapiro BA: Computational and Experimental RNA Nanoparticle Design. In Automation in Genomics and Proteomics: An Engineering Case-Based Approach; Alterovitz G, Ramoni M, Benson R, Eds.; Wiley Publishing: Hoboken, NJ, 2009; pp 193–220. [Google Scholar]
- [54].Severcan I; Geary C; Verzemnieks E; Chworos A; Jaeger L Square-shaped RNA particles from different RNA folds. Nano letters 2009, 9, 1270–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Geary C; Baudrey S; Jaeger L Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucleic acids research 2008, 36, 1138–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Geary C; Chworos A; Jaeger L Promoting RNA helical stacking via A-minor junctions. Nucleic acids research 2011, 39, 1066–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Geary C; Rothemund PW; Andersen ES RNA nanostructures. A single-stranded architecture for cotranscriptional folding of RNA nanostructures. Science 2014, 345, 799–804. [DOI] [PubMed] [Google Scholar]
- [58].Ohno H; Kobayashi T; Kabata R; Endo K; Iwasa T; Yoshimura SH; Takeyasu K; Inoue T; Saito H Synthetic RNA-protein complex shaped like an equilateral triangle. Nature nanotechnology 2011, 6, 116–120. [DOI] [PubMed] [Google Scholar]
- [59].Grabow W; Jaeger L RNA modularity for synthetic biology. F1000prime reports 2013, 5, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ko SH; Su M; Zhang C; Ribbe AE; Jiang W; Mao C Synergistic self-assembly of RNA and DNA molecules. Nat Chem 2010, 2, 1050–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hao C; Li X; Tian C; Jiang W; Wang G; Mao C Construction of RNA nanocages by re-engineering the packaging RNA of Phi29 bacteriophage. Nature communications 2014, 5, 3890. [DOI] [PubMed] [Google Scholar]
- [62].Yu J; Liu Z; Jiang W; Wang G; Mao C De novo design of an RNA tile that self-assembles into a homo-octameric nanoprism. Nature communications 2015, 6, 5724. [DOI] [PubMed] [Google Scholar]
- [63].Afonin KA; Cieply DJ; Leontis NB Specific RNA self-assembly with minimal paranemic motifs. Journal of the American Chemical Society 2008, 130, 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Afonin KA; Leontis NB Generating new specific RNA interaction interfaces using C-loops. Journal of the American Chemical Society 2006, 128, 16131–16137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Afonin KA; Lin YP; Calkins ER; Jaeger L Attenuation of loop-receptor interactions with pseudoknot formation. Nucleic acids research 2012, 40, 2168–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Osada E; Suzuki Y; Hidaka K; Ohno H; Sugiyama H; Endo M; Saito H Engineering RNA-protein complexes with different shapes for imaging and therapeutic applications. ACS nano 2014, 8, 8130–8140. [DOI] [PubMed] [Google Scholar]
- [67].Saito H; Inoue T RNA and RNP as new molecular parts in synthetic biology. Journal of biotechnology 2007, 132, 1–7. [DOI] [PubMed] [Google Scholar]
- [68].Saito H; Inoue T Synthetic biology with RNA motifs. The international journal of biochemistry & cell biology 2009, 41, 398–404. [DOI] [PubMed] [Google Scholar]
- [69].Shiohara T; Saito H; Inoue T A designed RNA selection: establishment of a stable complex between a target and selectant RNA via two coordinated interactions. Nucleic acids research 2009, 37, e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ohno H; Inoue T Designed Regular Tetragon-Shaped RNA-Protein Complexes with Ribosomal Protein L1 for Bionanotechnology and Synthetic Biology. ACS nano 2015. [DOI] [PubMed] [Google Scholar]
- [71].Afonin KA; Schultz D; Jaeger L; Gwinn E; Shapiro BA Silver nanoclusters for RNA nanotechnology: steps towards visualization and tracking of RNA nanoparticle assemblies. Methods in molecular biology 2015, 1297, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Yingling YG; Shapiro BA Computational design of an RNA hexagonal nanoring and an RNA nanotube. Nano letters 2007, 7, 2328–2334. [DOI] [PubMed] [Google Scholar]
- [73].Grabow WW; Zakrevsky P; Afonin KA; Chworos A; Shapiro BA; Jaeger L Self-assembling RNA nanorings based on RNAI/II inverse kissing complexes. Nano letters 2011, 11, 878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Afonin KA; Kireeva M; Grabow WW; Kashlev M; Jaeger L; Shapiro BA Co-transcriptional assembly of chemically modified RNA nanoparticles functionalized with siRNAs. Nano letters 2012, 12, 5192–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Guo P; Zhang C; Chen C; Garver K; Trottier M Inter-RNA interaction of phage phi29 pRNA to form a hexameric complex for viral DNA transportation. Molecular cell 1998, 2, 149–155. [DOI] [PubMed] [Google Scholar]
- [76].Binzel DW; Khisamutdinov EF; Guo P Entropy-driven one-step formation of Phi29 pRNA 3WJ from three RNA fragments. Biochemistry 2014, 53, 2221–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Haque F; Shu D; Shu Y; Shlyakhtenko LS; Rychahou PG; Evers BM; Guo P Ultrastable synergistic tetravalent RNA nanoparticles for targeting to cancers. Nano today 2012, 7, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Khisamutdinov EF; Jasinski DL; Guo P RNA as a boiling-resistant anionic polymer material to build robust structures with defined shape and stoichiometry. ACS nano 2014, 8, 4771–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Khisamutdinov EF; Li H; Jasinski DL; Chen J; Fu J; Guo P Enhancing immunomodulation on innate immunity by shape transition among RNA triangle, square and pentagon nanovehicles. Nucleic acids research 2014, 42, 9996–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Shu D; Shu Y; Haque F; Abdelmawla S; Guo P Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nature nanotechnology 2011, 6, 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Shu Y; Cinier M; Shu D; Guo P Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods 2011, 54, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Shu Y; Haque F; Shu D; Li W; Zhu Z; Kotb M; Lyubchenko Y; Guo P Fabrication of 14 different RNA nanoparticles for specific tumor targeting without accumulation in normal organs. Rna 2013, 19, 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Feng L; Li SK; Liu H; Liu CY; LaSance K; Haque F; Shu D; Guo P Ocular delivery of pRNA nanoparticles: distribution and clearance after subconjunctival injection. Pharmaceutical research 2014, 31, 1046–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Reif R; Haque F; Guo P Fluorogenic RNA nanoparticles for monitoring RNA folding and degradation in real time in living cells. Nucleic acid therapeutics 2012, 22, 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Tarapore P; Shu Y; Guo P; Ho SM Application of phi29 motor pRNA for targeted therapeutic delivery of siRNA silencing metallothionein-IIA and survivin in ovarian cancers. Molecular therapy : the journal of the American Society of Gene Therapy 2011, 19, 386–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Jasinski DL; Khisamutdinov EF; Lyubchenko YL; Guo P Physicochemically tunable polyfunctionalized RNA square architecture with fluorogenic and ribozymatic properties. ACS nano 2014, 8, 7620–7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Khaled A; Guo S; Li F; Guo P Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano letters 2005, 5, 1797–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Afonin KA; Bindewald E; Yaghoubian AJ; Voss N; Jacovetty E; Shapiro BA; Jaeger L In vitro assembly of cubic RNA-based scaffolds designed in silico. Nature nanotechnology 2010, 5, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Afonin KA; Kasprzak W; Bindewald E; Puppala PS; Diehl AR; Hall KT; Kim TJ; Zimmermann MT; Jernigan RL; Jaeger L, et al. Computational and experimental characterization of RNA cubic nanoscaffolds. Methods 2014, 67, 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Afonin KA; Viard M; Kagiampakis I; Case CL; Dobrovolskaia MA; Hofmann J; Vrzak A; Kireeva M; Kasprzak WK; KewalRamani VN, et al. Triggering of RNA Interference with RNA-RNA, RNA-DNA, and DNA-RNA Nanoparticles. ACS nano 2015, 9, 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Herrera-Carrillo E; Berkhout B Gene Therapy Strategies to Block HIV-1 Replication by RNA Interference. Advances in experimental medicine and biology 2015, 848, 71–95. [DOI] [PubMed] [Google Scholar]
- [92].Herrera-Carrillo E; Berkhout B The impact of HIV-1 genetic diversity on the efficacy of a combinatorial RNAi-based gene therapy. Gene therapy 2015. [DOI] [PubMed] [Google Scholar]
- [93].Berkhout B; Sanders RW Molecular strategies to design an escape-proof antiviral therapy. Antiviral research 2011, 92, 7–14. [DOI] [PubMed] [Google Scholar]
- [94].Low JT; Knoepfel SA; Watts JM; ter Brake O; Berkhout B; Weeks KM SHAPE-directed discovery of potent shRNA inhibitors of HIV-1. Molecular therapy : the journal of the American Society of Gene Therapy 2012, 20, 820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].ter Brake O; t Hooft K; Liu YP; Centlivre M; von Eije KJ; Berkhout B Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Molecular therapy : the journal of the American Society of Gene Therapy 2008, 16, 557–564. [DOI] [PubMed] [Google Scholar]
- [96].Afonin KA; Bindewald E; Kireeva M; Shapiro BA Computational and Experimental Studies of Reassociating RNA/DNA Hybrids Containing Split Functionalities. Methods in enzymology 2015, 553, 313–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Afonin KA; Desai R; Viard M; Kireeva ML; Bindewald E; Case CL; Maciag AE; Kasprzak WK; Kim T; Sappe A, et al. Co-transcriptional production of RNA-DNA hybrids for simultaneous release of multiple split functionalities. Nucleic acids research 2014, 42, 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Afonin KA; Viard M; Martins AN; Lockett SJ; Maciag AE; Freed EO; Heldman E; Jaeger L; Blumenthal R; Shapiro BA Activation of different split functionalities on re-association of RNA-DNA hybrids. Nature nanotechnology 2013, 8, 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Pinheiro AV; Han D; Shih WM; Yan H Challenges and opportunities for structural DNA nanotechnology. Nature nanotechnology 2011, 6, 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Hehar SS; Mason JD; Stephen AB; Washington N; Jones NS; Jackson SJ; Bush D Twenty-four hour ambulatory nasal pH monitoring. Clinical otolaryngology and allied sciences 1999, 24, 24–25. [DOI] [PubMed] [Google Scholar]
- [101].Yang W; Peters JI; Williams RO 3rd. Inhaled nanoparticles--a current review. Int J Pharm 2008, 356, 239–247. [DOI] [PubMed] [Google Scholar]
- [102].Gondi CS; Rao JS Concepts in in vivo siRNA delivery for cancer therapy. Journal of cellular physiology 2009, 220, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Pille JY; Li H; Blot E; Bertrand JR; Pritchard LL; Opolon P; Maksimenko A; Lu H; Vannier JP; Soria J, et al. Intravenous delivery of anti-RhoA small interfering RNA loaded in nanoparticles of chitosan in mice: safety and efficacy in xenografted aggressive breast cancer. Human gene therapy 2006, 17, 1019–1026. [DOI] [PubMed] [Google Scholar]
- [104].Rychahou P; Haque F; Shu Y; Zaytseva Y; Weiss HL; Lee EY; Mustain W; Valentino J; Guo P; Evers BM Delivery of RNA nanoparticles into colorectal cancer metastases following systemic administration. ACS nano 2015, 9, 1108–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Longmire M; Choyke PL; Kobayashi H Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine 2008, 3, 703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Stern ST; Hall JB; Yu LL; Wood LJ; Paciotti GF; Tamarkin L; Long SE; McNeil SE Translational considerations for cancer nanomedicine. Journal of controlled release : official journal of the Controlled Release Society 2010, 146, 164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Shu Y; Shu D; Haque F; Guo P Fabrication of pRNA nanoparticles to deliver therapeutic RNAs and bioactive compounds into tumor cells. Nature protocols 2013, 8, 1635–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Dobrovolskaia MA; McNeil SE Immunological properties of engineered nanomaterials. Nature nanotechnology 2007, 2, 469–478. [DOI] [PubMed] [Google Scholar]
- [109].Owens DE; Peppas NA Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. International Journal of Pharmaceutics 2006, 307, 93–102. [DOI] [PubMed] [Google Scholar]
- [110].Allen TM; Hansen C; Martin F; Redemann C; Yau-Young A Liposomes containing synthetic lipid derivatives of poly(ethylene glycol) show prolonged circulation half-lives in vivo. Biochim Biophys Acta 1991, 1066, 29–36. [DOI] [PubMed] [Google Scholar]
- [111].Veronese FM; Pasut G PEGylation, successful approach to drug delivery. Drug Discovery Today 2005, 10, 1451–1458. [DOI] [PubMed] [Google Scholar]
- [112].Fang J; Nakamura H; Maeda H The EPR effect: Unique features of tumor blood vessels for drug delivery, factors involved, and limitations and augmentation of the effect. Advanced drug delivery reviews 2011, 63, 136–151. [DOI] [PubMed] [Google Scholar]
- [113].Maeda H; Nakamura H; Fang J The EPR effect for macromolecular drug delivery to solid tumors: Improvement of tumor uptake, lowering of systemic toxicity, and distinct tumor imaging in vivo. Advanced drug delivery reviews 2013, 65, 71–79. [DOI] [PubMed] [Google Scholar]
- [114].Torchilin V Tumor delivery of macromolecular drugs based on the EPR effect. Advanced drug delivery reviews 2011, 63, 131–135. [DOI] [PubMed] [Google Scholar]
- [115].Dohmen C; Frohlich T; Lachelt U; Rohl I; Vornlocher HP; Hadwiger P; Wagner E Defined Folate-PEG-siRNA Conjugates for Receptor-specific Gene Silencing. Molecular therapy. Nucleic acids 2012, 1, e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].McNamara JO 2nd; Andrechek ER; Wang Y; Viles KD; Rempel RE; Gilboa E; Sullenger BA; Giangrande PH Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nature biotechnology 2006, 24, 1005–1015. [DOI] [PubMed] [Google Scholar]
- [117].Rockey WM; Hernandez FJ; Huang SY; Cao S; Howell CA; Thomas GS; Liu XY; Lapteva N; Spencer DM; McNamara JO, et al. Rational truncation of an RNA aptamer to prostate-specific membrane antigen using computational structural modeling. Nucleic acid therapeutics 2011, 21, 299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Ruoslahti E; Bhatia SN; Sailor MJ Targeting of drugs and nanoparticles to tumors. J Cell Biol 2010, 188, 759–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Brannon-Peppas L; Blanchette JO Nanoparticle and targeted systems for cancer therapy. Advanced drug delivery reviews 2004, 56, 1649–1659. [DOI] [PubMed] [Google Scholar]
- [120].Shim MS; Kwon YJ Efficient and targeted delivery of siRNA in vivo. The FEBS journal 2010, 277, 4814–4827. [DOI] [PubMed] [Google Scholar]
- [121].Hobel S; Aigner A Polyethylenimines for siRNA and miRNA delivery in vivo. Wiley interdisciplinary reviews. Nanomedicine and nanobiotechnology 2013, 5, 484–501. [DOI] [PubMed] [Google Scholar]
- [122].Dar GH; Gopal V; Rao NM Systemic delivery of stable siRNA-encapsulating lipid vesicles: optimization, biodistribution, and tumor suppression. Molecular pharmaceutics 2015, 12, 610–620. [DOI] [PubMed] [Google Scholar]
- [123].Kim T; Afonin KA; Viard M; Koyfman AY; Sparks S; Heldman E; Grinberg S; Linder C; Blumenthal RP; Shapiro BA In Silico, In Vitro, and In Vivo Studies Indicate the Potential Use of Bolaamphiphiles for Therapeutic siRNAs Delivery. Molecular therapy. Nucleic acids 2013, 2, e80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Schlegel A; Largeau C; Bigey P; Bessodes M; Lebozec K; Scherman D; Escriou V Anionic polymers for decreased toxicity and enhanced in vivo delivery of siRNA complexed with cationic liposomes. Journal of controlled release : official journal of the Controlled Release Society 2011, 152, 393–401. [DOI] [PubMed] [Google Scholar]
- [125].Rudzinski WE; Aminabhavi TM Chitosan as a carrier for targeted delivery of small interfering RNA. International Journal of Pharmaceutics 2010, 399, 1–11. [DOI] [PubMed] [Google Scholar]
- [126].Tsutsumi T; Hirayama F; Uekama K; Arima H Evaluation of polyamidoamine dendrimer/alpha-cyclodextrin conjugate (generation 3, G3) as a novel carrier for small interfering RNA (siRNA). Journal of controlled release : official journal of the Controlled Release Society 2007, 119, 349–359. [DOI] [PubMed] [Google Scholar]
- [127].Elbakry A; Zaky A; Liebl R; Rachel R; Goepferich A; Breunig M Layer-by-layer assembled gold nanoparticles for siRNA delivery. Nano letters 2009, 9, 2059–2064. [DOI] [PubMed] [Google Scholar]
- [128].Tanaka T; Mangala LS; Vivas-Mejia PE; Nieves-Alicea R; Mann AP; Mora E; Han HD; Shahzad MM; Liu X; Bhavane R, et al. Sustained small interfering RNA delivery by mesoporous silicon particles. Cancer Res 2010, 70, 3687–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Safari D; Marradi M; Chiodo F; Th Dekker HA; Shan Y; Adamo R; Oscarson S; Rijkers GT; Lahmann M; Kamerling JP, et al. Gold nanoparticles as carriers for a synthetic Streptococcus pneumoniae type 14 conjugate vaccine. Nanomedicine 2012, 7, 651–662. [DOI] [PubMed] [Google Scholar]
- [130].Zorko M; Langel U Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Advanced drug delivery reviews 2005, 57, 529–545. [DOI] [PubMed] [Google Scholar]
- [131].Semple SC; Akinc A; Chen J; Sandhu AP; Mui BL; Cho CK; Sah DW; Stebbing D; Crosley EJ; Yaworski E, et al. Rational design of cationic lipids for siRNA delivery. Nature biotechnology 2010, 28, 172–176. [DOI] [PubMed] [Google Scholar]
- [132].Fattal E; Couvreur P; Dubernet C Smart“ delivery of antisense oligonucleotides by anionic pH-sensitive liposomes. Advanced drug delivery reviews 2004, 56, 931–946. [DOI] [PubMed] [Google Scholar]
- [133].Hatakeyama H; Ito E; Akita H; Oishi M; Nagasaki Y; Futaki S; Harashima H A pH-sensitive fusogenic peptide facilitates endosomal escape and greatly enhances the gene silencing of siRNA-containing nanoparticles in vitro and in vivo. Journal of controlled release : official journal of the Controlled Release Society 2009, 139, 127–132. [DOI] [PubMed] [Google Scholar]
- [134].Kim TJ; Afonin KA; Viard M; Koyfman AY; Sparks S; Heldman E; Grinberg S, Linder C; Blumenthal RP; Shapiro BA In silico, in vitro and in vivo studies indicate the potential use of bolaamphiphiles for therapeutic siRNAs delivery. Molecular therapy 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Grinberg S; Kolot V; Linder C; Shaubi E; Kas‘yanov V; Deckelbaum RJ; Heldman E Synthesis of novel cationic bolaamphiphiles from vernonia oil and their aggregated structures. Chemistry and physics of lipids 2008, 153, 85–97. [DOI] [PubMed] [Google Scholar]
- [136].Grinberg S; Linder C; Heldman E Progress in lipid-based nanoparticles for cancer therapy. Critical reviews in oncogenesis 2014, 19, 247–260. [DOI] [PubMed] [Google Scholar]
- [137].Grinberg S; Linder C; Kolot V; Waner T; Wiesman Z; Shaubi E; Heldman E Novel cationic amphiphilic derivatives from vernonia oil: synthesis and self-aggregation into bilayer vesicles, nanoparticles, and DNA complexants. Langmuir 2005, 21, 7638–7645. [DOI] [PubMed] [Google Scholar]
- [138].Dakwar GR; Abu Hammad I; Popov M; Linder C; Grinberg S; Heldman E; Stepensky D Delivery of proteins to the brain by bolaamphiphilic nano-sized vesicles. Journal of controlled release : official journal of the Controlled Release Society 2012, 160, 315–321. [DOI] [PubMed] [Google Scholar]


