Abstract
Genetics plays a role in age-related macular degeneration (AMD), a common cause of blindness in the elderly. There is a need for powerful methods for carrying out region-based association tests between a dichotomous trait like AMD and genetic variants on family data. Here, we apply our new generalized functional linear mixed models (GFLMM) developed to test for gene-based association in a set of AMD families. Using common and rare variants, we observe significant association with two known AMD genes: CFH and ARMS2. Using rare variants, we find suggestive signals in four genes: ASAH1, CLEC6A, TMEM63C, and SGSM1. Intriguingly, ASAH1 is down-regulated in AMD aqueous humor, and ASAH1 deficiency leads to retinal inflammation and increased vulnerability to oxidative stress. These findings were made possible by our GFLMM which model the effect of a major gene as a fixed mean, the polygenic contributions as a random variation, and the correlation of pedigree members by kinship coefficients. Simulations indicate that the GFLMM likelihood ratio tests (LRTs) accurately control the Type I error rates. The LRTs have similar or higher power than existing retrospective kernel and burden statistics. Our GFLMM-based statistics provide a new tool for conducting family-based genetic studies of complex diseases. Supplementary materials for this article, including a standardized description of the materials available for reproducing the work, are available as an online supplement.
Keywords: Age-related macular degeneration, Association study, Complex diseases, Extended pedigree, Generalized functional linear mixed models, Rare variants
1. Introduction
Age-related macular degeneration (AMD) is a common complex disease that leads to irreversible vision loss in the elderly and afflicts almost 10 million individuals in the United States (Friedman et al. 2004). AMD is caused by an interaction of aging, genetics, and environmental/nutritional factors (Fritsche et al. 2014). Family-based linkage studies detected major susceptibility loci for AMD on chromosomes 1 and 10 (Fisher et al. 2005). Using population data, dozens of disease-causing genes and hundred of mutations have been discovered by genome-wide association studies (Age-Related Eye Disease Study Research Group 1999; Seddon et al. 2007; Chen et al. 2010; Neale et al. 2010; Fritsche et al. 2013, 2016). However, no gene-based association studies have been performed to dissect AMD using family data so far, although next-generation sequencing technologies provide massive data resources, such as those of whole genome sequencing (WGS) and whole exome sequencing (WES) which are rich resources to search for causal genetic variants of complex disorders (Abecasis et al. 2012; Tennessen et al. 2012; Lek et al. 2016). To accommodate family data, we develop generalized functional linear mixed models (GFLMM) and related software in this work to analyze a dichotomous trait for sequencing data. We apply the GFLMM to a real exome chip dataset to identify AMD associated susceptibility genes (Weeks et al. 2000, 2004).
Although rich data resources are available and a data-intensive and data-driven analysis era is coming, powerful and computationally efficient statistical methods and related software are needed to test for association between complex traits and variants, to screen for causal variants and to reduce false positives and to properly deal with high dimensionality (Bansal et al. 2010; Kiezun et al. 2012). Few statistical methods are available to analyze extended pedigrees for sequencing studies. For family data, one needs to take pedigree structure into account to model correlations of pedigree members. To control for population structure and familial or cryptic relatedness in genome-wide association studies (GWAS) while analyzing common variants, mixed models were developed for association studies and gained popularity due to their ability to control false positive rates and their good power performance (Henderson 1984; Price et al. 2006; Yu et al. 2006; Aulchenko, De Koning, and Haley 2007; Zhao et al. 2007; Kang et al. 2008; Astle and Balding 2009; Kang et al. 2010; Zhang et al. 2010; Lippert et al. 2011; Yang, Lee, et al. 2011; Yang, Weedon, et al. 2011; Korte et al. 2012; Listgarten et al. 2012; Segura et al. 2012; Svishcheva et al. 2012; Zhou and Stephens 2012; Listgarten, Lippert, and Heckerman 2013; Pirinen, Donnelly, and Spencer 2013; Yang et al. 2014; Zhou and Stephens 2014; Hayeck et al. 2015; Loh et al. 2015; Song, Hao, and Storey 2015; Chen et al. 2016). To our knowledge, the mixed models developed so far cannot be directly applied to the analysis of rare variants or a combination of rare and common variants. Here, a variant is considered rare if its minor allele frequency (MAF) is less than or equal to 0.03 (the cutoff can be different in certain circumstances). There is a need to develop statistical methods to analyze next-generation sequencing data which may contain rare and common variants for familial data and may correct for population stratification.
In recent years, a class of fixed effect models has been developed for unrelated samples to test for region-based or gene-based association between a quantitative/dichotomous/survival trait and genetic variants in a region or within a gene (Cordell and Clayton 2002; Luo, Boerwinkle, and Xiong 2011; Luo, Zhu, and Xiong 2012, 2013; Fan et al. 2013, 2014, 2015; Vsevolozhskaya et al. 2014, 2016; Zhang, Boerwinkle, and Xiong 2014; Svishcheva, Belonogova, and Axenovich 2015; Wang et al. 2015; Fan, Chiu, et al. 2016; Fan, Wang, Chiu, et al. 2016; Fan, Wang, Yan, et al. 2016; Zhao, Zhu, and Xiong 2016). Since gene boundaries can be defined at the transcript level, gene-based can be changed to transcript-based. To simplify our presentation, we only use terminology gene-based hereafter and one may change it to region-based or transcript-based. The fixed effect models can be functional regression models or traditional additive models. In functional regression models, genotyping data are viewed as a realization of a stochastic process that varies along a chromosome region (Ross 1996). Using functional data analysis techniques, it is natural to summarize an individual’s genetic information as a stochastic function (de Boor 2001; Ramsay and Silverman 2005; Ramsay, Hooker, and Graves 2009; Ferraty and Romain 2010; Horvath and Kokoszka 2012). An individual’s discrete genotypes can be used to estimate his/her genetic variant function (GVF) using a collection of smooth basis functions. The trait variable is related to the GVF while adjusting for covariates to build theoretical functional regression models. By using functional data analysis techniques, the theoretical functional regression models are revised to be ordinary regression models which can be used to test for association between the traits and the genetic variants. One advantage of the functional regression models is that they can properly reduce the high dimensionality of the sequencing data to draw useful information. In short, functional models turn the curse of dimensionality of sequencing data to be a blessing.
In genetics, a “major” gene has a relatively large effect on the trait. In contrast, a polygene means a gene where the effects of variants within the gene are small and the effects are likely in a similar scale across the gene region (Lange 2002). When genetic effects are relatively large (i.e., a major gene), the effects of variants are unlikely to be constant across the gene region, and it is reasonable to model genetic effects as a fixed function as in functional regression models (Luo, Boerwinkle, and Xiong 2011; Luo, Zhu, and Xiong 2012, 2013; Fan et al. 2013, 2014, 2015; Vsevolozhskaya et al. 2014, 2016; Svishcheva, Belonogova, and Axenovich 2015; Wang et al. 2015; Fan, Wang, Chiu, et al. 2016; Fan, Wang, Yan, et al. 2016). For polygenic effects, it is reasonable to model the genetic effects as a random variable with a mean of zero and a constant variance as is done by the sequence kernel association tests (SKAT), its optimal unified tests (SKAT-O), and a combined sum test of rare and common variants (SKAT-C) (Wu et al. 2011; Lee et al. 2012; Ionita-Laza et al. 2013a). The fixed effect models have similar or higher power than SKAT procedure to analyze major genes while SKAT procedure performs better in analysis of polygenes (Fan, Chiu, et al. 2016).
In association analysis, we mainly search for major genes. We argue that the regression models which treat major gene’s contribution as fixed effects are more appropriate in association analysis. In addition to major genes, geneticists have long known of the existence of polygenes. The functional models and SKAT procedure are complimentary to each other. Moreover, the functional models are well-suited for analyzing next-generation sequencing data since they can be used to analyze: (1) rare variants; (2) common variants, and (3) a combination of the two. This motivates us to extend the unrelated population-based fixed models to analyze related pedigree data.
In the literature, the research to analyze rare variants on general extended families for dichotomous traits focuses on kernel and collapsing/burden tests (De et al. 2013; Ionita-Laza et al. 2013b; Schaid et al. 2013; Wang et al. 2013; Svishcheva, Belonogova, and Axenovich 2014; Yan et al. 2015; Fernandez et al. 2018). The kernel and collapsing/burden tests are good to analyze polygenes but less powerful to analyze major genes which have relatively large effects on the traits (Fan, Chiu, et al. 2016).
Motivated by the need to analyze family AMD data, we develop gene-based GFLMM by extending the generalized functional linear models previously discussed for population data (Cordell and Clayton 2002; Fan et al. 2014). The GFLMM model the major gene effect as a fixed mean, the polygenic contributions as a random variation, and the correlations between pedigree members by kinship coefficients. We then test for association between the dichotomous trait and the genetic variants by testing if the fixed mean is zero using likelihood ratio test (LRT) statistics. To assess the behavior of our GFLMM LRT statistics and to make sure the methods can be used to analyze AMD family data, we conduct simulation studies to evaluate Type I error rates and power, and compare our statistics with prominent statistical methods from the literature (Schaid et al. 2013).
2. Applications to AMD Pedigree Data
Our goal is to analyze real exome chip data from the UCLA/Pittsburgh family-based study of AMD which include extended pedigrees (Weeks et al. 2000, 2004). To analyze the AMD pedigree data, we use GFLMM and build related LRT statistics (Section 3).
2.1. AMD Pedigree Data
In the AMD studies, an individual was considered affected with AMD according to the “C” diagnostic scheme previously defined (Weeks et al. 2000, 2004); an individual was considered unaffected if they were unaffected and at least 65 years old at last exam. After sample quality checks using a thorough and rigorous data cleaning pipeline (Laurie et al. 2010), which included checks for chromosomal aberrations, gender, Hardy–Weinberg equilibrium, relatedness, duplicates, and genotype quality, 976 genotyped individuals of European ancestry were available for analysis. To completely connect pedigrees, we included non-genotyped individuals who shared the same family with those 976 genotyped individuals. The connected pedigrees contained 2727 pedigree members, 1275 with a known AMD trait (1031 affected and 244 unaffected). A total number of 111,547 autosomal variants were included in the study; 30,096 were common (MAF > 0.05), and 81,451 were rare (MAF ≤ 0.05). In the analysis, we adjusted for gender since it is significantly associated with AMD in the null model (p-value = 0.00197).
2.2. Application to AMD Data
As we were interested in the possibility that different transcripts might convey different risk for AMD, we carried out transcript-based tests to investigate AMD susceptibility genes on autosomes using the LRT statistics, the retrospective kernel-based and burden tests (Schaid et al. 2013). Gene boundaries were defined at the transcript level and sets of transcripts that shared identical boundaries were only tested once. Tests were conducted in two different ways. First we considered a combination of common and rare variants, and tested a total number of 16,913 autosomal transcripts at a genome-wide significance threshold of 2.96 × 10−6 after Bonferroni correction. We next excluded all common variants, and focused on the genes having at least two polymorphic rare variants. A total number of 14,961 transcripts were tested, associated with a significance threshold of 3.34 × 10−6 after Bonferroni correction.
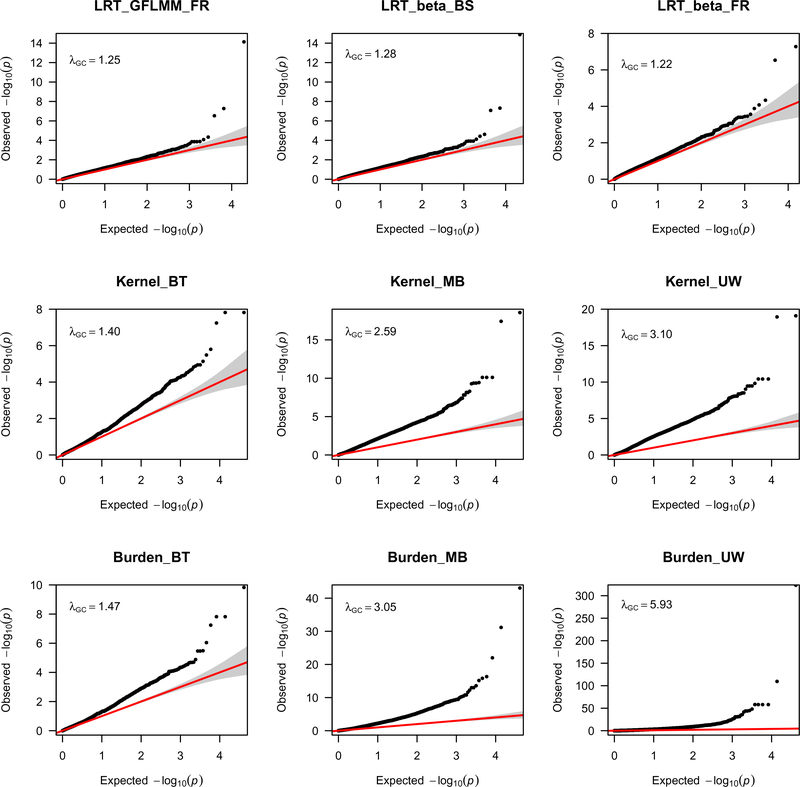
Table 1 showed significant and suggestive significant signals for the AMD data. Only the results of the LRT GFLMM statistics (6) were shown (as these converged more often than LRT GFLMM statistics (5) did). For the analysis of the common and rare variants, we confirmed that strong association was detected between AMD and CFH and ARMS2, two known AMD susceptibility genes (Table 1). The kernel and burden tests except kernel_BT and burden_BT reached genome-wide significance (see the p-values in bold associated with CFH and ARMS2 in Table 1).
Table 1.
Results of association analysis of age-related macular degeneration data.
| Rare and common variants | ||||||||||||
| LRT statistics of mixed models | Schaid’s methods | |||||||||||
| Chr | Gene | Start | End | Number of variants | GFLMM (6) | Kernel-based tests | Burden tests | |||||
| B-sp basis | Fourier basis | kernel_BT | kernel_MB | kernel_UW | burden_BT | burden_MB | burden_UW | |||||
| 1 | CFH | 196621007 | 196716634 | 15 | 8.30 × 10−8 | 2.94 × 10−7 | 2.86 × 10−2 | 2.76 × 10−19 | 1.16× 10−19 | 1.53 × l0−1 | 7.02 × 10−16 | 1.07 × 10−19 |
| 196621007 | 196670695 | 3 | 4.84 × 10−8 | 5.30 × 10−8 | 3.65 × l−1 | 3.81 × 10−18 | 8.09 × 10−20 | 5.35 × l0−1 | 2.67 × 10−14 | 4.87 × 10−19 | ||
| 10 | ARMS2 | 124214178 | 124216868 | 2 | 1.32 × 10−15 | NA | 1.94 × 10−1 | 9.74 × 10−8 | 3.82 × 10−9 | 1.91 × 10−1 | 8.13 × 10−5 | 2.01 × 10−7 |
| Rare variants | ||||||||||||
| LRT statistics of mixed models | Schaid’s methods | |||||||||||
| Chr | Gene | Start | End | Number of variants | GFLMM (6) | Kernel-based tests | Burden tests | |||||
| B-sp basis | Fourier basis | kernel_BT | kernel_MB | kernel_UW | burden_BT | burden_MB | burden_UW | |||||
| 8 | ASAH1 | 17913924 | 17941879 | 6 | 5.95 × 10−3 | 9.94 × 10−6 | 4.25 × 10−4 | 1.41 × 10−3 | 3.29 × 10−5 | 7.18 × l0−1 | 9.25 × l0−1 | 1.64 × 10−2 |
| 12 | CLEC6A | 8608590 | 8630926 | 3 | 3.78 × 10−5 | 4.71 × 10−3 | 1.56 × 10−2 | 1.67 × 10−2 | 1.61 × 10−2 | 5.34 × 10−2 | 5.30 × l0−1 | 3.67 × 10−2 |
| 14 | TMEM63C | 77648101 | 77725838 | 5 | 5.30 × 10−2 | 1.93 × 10−5 | 1.49 × 10−5 | 9.46 × 10−6 | 2.15 × 10−5 | 3.07 × 10−2 | 2.16 × 10−2 | 3.51 × 10−2 |
| 22 | SGSM1 | 25202135 | 25322813 | 10 | 1.56 × 10−5 | NA | 6.40 × 10−3 | 7.03 × 10−3 | 3.37 × 10−2 | 1.39 × 10−1 | 6.58 × l0−1 | 3.42 × 10−1 |
NOTE: The association with a significant p — value < 2.96 × 10 −6 were highlighted in bold. The results of suggestive association were included if a p-value is less than 10 −5. Abbreviations: chr. chromosome: NA. did not converae
For gene CFH, two transcripts reached genome-wide significance (Table 1). The larger 96 kb region with 15 variants contained the other smaller one with only 3 variants, which indicated that the primary CFH variants associated with AMD might be located within the smaller region between 196,621,007 and 196,670,695 bp on chromosome 1. For gene ARMS2, both of the variants in our exome chip data were common variants with MAF = 0.39 and 0.10, respectively, so they were not included in the rare variant analysis. For CFH, the 96 kb region contained 7 rare variants (MAF ≤ 0.05, Table A.1 of Supplementary Materials I); the models failed to reach the significance threshold using the 7 rare variants, indicating that the common variants within CFH might play a pivotal role in the significant association signal.
In Table A.1 of Supplementary Materials I, we listed all variants within CFH and ARMS2, and examined their association on an individual variant level by conducting the WQLS test suggested by Thornton and McPeek (2007) and the LRT GLMM statistic (3). For the gene ARMS2, a single nucleotide polymorphism (SNP) rs10490924 is strongly associated with AMD. For the gene CFH, 7 SNPs are strongly associated with AMD and each of them is a common variant. Hence, it makes sense to perform a gene-based associated analysis for the gene CFH for a unified analysis.
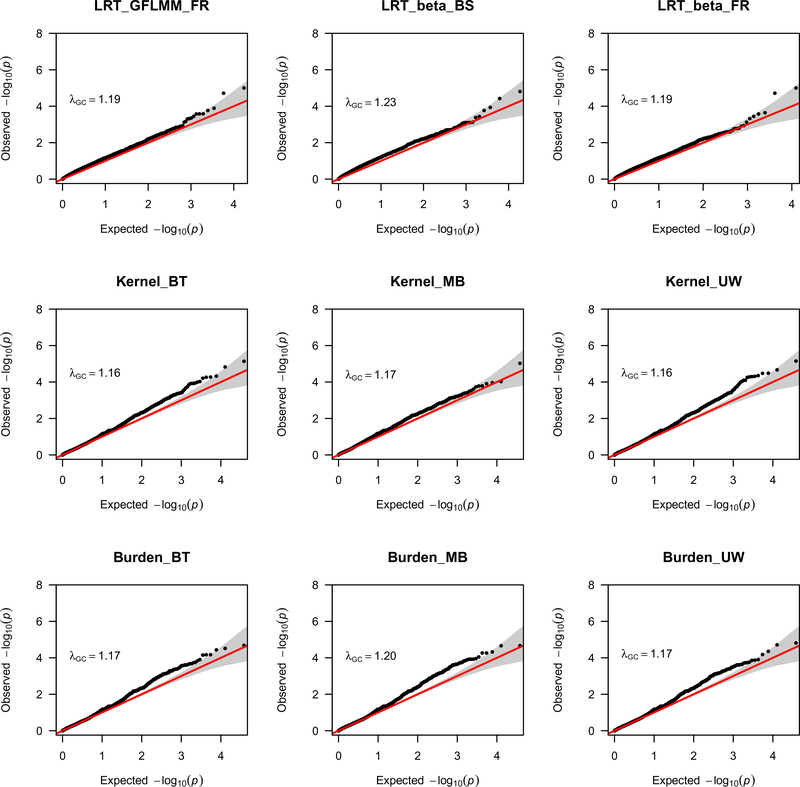
Quantile-quantile (Q-Q) plots of the gene-based statistics in Figure 1 show that, while the LRT statistics had lower λGC values, the kernel and burden test statistics had quite elevated λGC values to analyze both common and rare variants. Thus, when analyzing common and rare variants, the kernel and burden test statistics are not appropriate because they have high false positive rates. For the analysis of the rare variants with MAF ≤ 0.05, which is expected to be less powerful, only suggestive association were found. In the Q-Q plots in Figure 2, the LRT statistics and the kernel and burden test statistics had similar λGC values when only rare variants were analyzed.
Figure 1.
Q-Q plots for the LRT GFLMM statistics (5) and (6), and the retrospective kernel and burden tests when all common and rare variants are analyzed for AMD data.
Figure 2.
Q-Q plots for the LRT GFLMM statistics (5) and (6), and the retrospective kernel and burden tests when only rare variants are analyzed for AMD data.
By using rare variants, the LRT statistics show suggestive association signal with AMD for four genes, ASAH1, CLEC6A, TMEM63C, and SGSM1, since they provide p-values slightly larger than the threshold of 3.34 × 10−6 (Table 1). For the four genes, the kernel and burden test statistics also provide some suggestive association signals. Since both the LRT statistics and the kernel and burden test statistics had similar λGC values around 1.0 in Figure 2, the suggestive signals are useful for further investigation when more data are available. The suggestive signal at ASAH1 is especially interesting because, as we explain more fully in Section 5, ASAH1 may play a role in AMD (Petrov et al. 2019; Qu et al. 2019; Sugano et al. 2019).
3. Methods: Generalized Functional Linear Mixed Models
3.1. Generalized Functional Linear Mixed Models
Consider a single pedigree from a family-based study. The pedigree includes n participants with a dichotomous trait of interest coded as 1 and 0 denoting affected and unaffected, respectively. In addition, the n participants are genotyped within a chromosome region. Let i denote the ith individual with m genetic variants in the region. The physical locations of the m variants, denoted by 0 ≤ u1 ≤ u2 ≤ · · · um, are normalized on the unit region [0, 1]. For the ith individual, let yi denote his/her disease status, and Zi = (zi1, …, zic)′ denote a c × 1 vector of fixed effect covariates. In addition, let Xi = (xi(u1), …, xi(um))′ denote the genotypes at the m variants, where xi(uj) (= 0, 1, 2) is the number of minor alleles of individual i at the jth variant. For the n individuals who are phenotyped and genotyped, let Ω be a n × n matrix containing diagonal elements Ωii = 1 + hi, where hi is the inbreeding coefficient for individual i, and off-diagonal elements Ωik = 2ϕik. The parameter ϕik is the kinship coefficient between individuals i and k, the probability that a randomly chosen allele at a given locus from individual i is identical by descent (IBD) to a randomly chosen allele from individual k conditional on their ancestral relationship (Lange 2002). In practice, the pedigree may include members who are not genotyped or phenotyped and they can be used to calculate relationships between the pedigree members, that is, kinship and inbreeding coefficients.
Let us denote the ith individual’s GVF as Xi(u), u ∈ [0, 1]. Using the observed discrete genotypes Xi, we may estimate the related GVF Xi(u), which will be discussed below. To relate the GVF to the trait status adjusting for covariates, we consider the following GFLMM
| (1) |
where πi = P(yi = 1|Zi, Xi, Gi) is the disease probability of the dichotomous trait for subject i, conditional on the covariates Zi, genotype vector Xi, and polygenic variation Gi, α0 is a regression intercept, α is a c × 1 vector of fixed regression coefficients of covariates, β(u) is the genetic effect of GVF Xi(u) at position u, and G = (G1, …, Gn)′ is a multivariate normal random polygenic vector with mean 0 and covariance matrix . Here is a polygenic variance component. In the GFLMM (1), the GVF Xi(u) is assumed to be smooth. This assumption can be relaxed by considering the following beta-smooth only GFLMM
| (2) |
where the genetic effect function β(u) is assumed to be continuous/smooth and so it is called beta-smooth only GFLMM. In the above model (2), the integration term in GFLMM (1) is replaced by a summation term , and we make no assumption about smoothness of the GVF Xi(u). We use the raw genotype data Xi = (xi(u1), …, xi(um))′ directly in the beta-smooth only GFLMM (2).
Fan et al. (2014) proposed generalized functional linear models for analyzing case control association studies for unrelated population data. There were no random terms Gi in the models of Fan et al. (2014). In this article, the GFLMM (1) and (2) are developed to analyze pedigree data. In addition to the fixed effect terms, the random terms (G1, …, Gn)′ are utilized to model polygenic variation and correlation among the pedigree members.
The trait correlation of related individuals or family members is modeled as being partly due to genetic influences, that is, the sharing of alleles IBD contributes to the correlation of the traits of related individuals (Lange 2002). In this article, we assume the correlation is from polygenes. This is a typical approach used in association analysis of familial data (Thornton and McPeek 2007). At one gene locus, there are three possible scenarios of allele sharing IBD. For one polygene, it is impossible to know which scenario it is for two related individuals unless the two individuals are from an identical twin (and so they share two genes IBD for sure). Hence, the three scenarios cannot be separately treated. The random variation Gi in GLMM (3) collectively models all effects of polygenes and the number of polygenes is usually very large. We have to use kinship coefficients to measure the correlation.
3.2. Additive Generalized Linear Mixed Models (GLMM)
By using the genotype data directly, we may relate the m genetic variants to the trait status while adjusting for covariates by the following additive generalized linear mixed model (GLMM)
| (3) |
where βj is the genetic effect of variant xi(uj) and the other terms are the same as those in the GFLMM (1). There is only one difference between model (2) and model (3). The genetic effect coefficients βj in GLMM (3) are each individually estimated and so are “free” and not forced to lie on a continuous function of the position u. In contrast, in model (2) we assume that the genetic effect function β(u) is a continuous function of the position u. Therefore, β(uj), j 1, 2, …, m, are the values of function β(u) at each of the m physical positions. The GLMM (3) hardly ever converges when the number of genetic variants is large which leads to a large number of parameters. Hence, the GLMM (3) is not useful for analyzing sequence data.
3.3. Revised Generalized Functional Linear Mixed Models
The genetic effect function β(u) in GFLMM (1) and GFLMM (2) is assumed to be a continuous function of physical position u. One may expand it using B-spline or Fourier basis functions. Formally, let us expand the genetic effect function β(u) using a series of Kβ basis functions as , where as Kβ × 1 vector of coefficients and . We consider two types of basis functions: (1) the B-spline basis: ψk(u) = Bk(u), k = 1, …, Kβ; and (2) the Fourier basis: ψ1(u) = 1, ψ2r+1(u) = sin(2πru), and ψ2r(u) = cos(2πru), r = 1, …, (Kβ – 1)/2. Here for Fourier basis, Kβ is taken as a positive odd integer (de Boor 2001; Ramsay and Silverman 2005; Ramsay, Hooker, and Graves 2009; Ferraty and Romain 2010; Horvath and Kokoszka 2012).
To estimate the GVF Xi(u) from the genotypes Xi, we use an ordinary linear square smoother (Ramsay and Silverman 2005; Ramsay, Hooker, and Graves 2009; Luo, Boerwinkle, and Xiong 2011; Luo, Zhu, and Xiong 2012, 2013; Fan et al. 2013, 2014, 2015; Vsevolozhskaya et al. 2014, 2016; Zhang, Boerwinkle, and Xiong 2014; Svishcheva, Belonogova, and Axenovich 2015; Wang et al. 2015; Fan, Chiu, et al. 2016; Fan, Wang, Chiu, et al. 2016; Fan, Wang, Yan, et al. 2016; Zhao, Zhu, and Xiong 2016). Let ϕk(u), k = 1, …, K, be a series of K basis functions. Let Φ denote the m × K matrix containing the values ϕk(uj), and we let ϕ(u) = (ϕ1(u), …, ϕK(u))′. Using the discrete realizations Xi = (xi(u1), …, xi(um))′, we estimate the GVF Xi(u) using an ordinary linear square smoother as follows
| (4) |
Assume that the genetic effect function β(u) is expanded by a series of basis functions ψk(u), k = 1, …, Kβ, as β(u) = ψ(u)′β. Replacing Xi(u) in the GFLMM (1) with in (4) and β(u) with the expansion, we have the following revised GFLMM
| (5) |
where . In the statistical packages R or Matlab, codes to calculate Φ[Φ′Φ]−1 and are readily available (Ramsay, Hooker, and Graves 2009).
Denote . For the beta-smooth only GFLMM (2), β(uj) is introduced as the genetic effect at the position uj. Expanding β(uj) by B-spline or Fourier basis functions as above, the GFLMM (2) can be revised as
| (6) |
3.4. Handling Missing Genotype Data
Missing genotypes, which are invariably encountered in analyses of real data, can be handled by modifying (4) so that each individual’s GVF is estimated using only the available genotype data. For example, suppose the genotype information is missing at the first variant for individual i, so we have Xi = (?, xi(u2), …, xi(um))′. Then let Φ1 be the (m − 1) × K matrix containing the values ϕk(uj) where j ∈ (2, …, m). Then we can estimate the GVF using the available genotype data as
| (7) |
Furthermore, in addition to modifying the calculation of the GVF, the summations over the m variants need to be appropriately adjusted. For example, in our case where the genotype at the first variant is missing, model (6) becomes
3.5. Likelihood Functions
For subject i, assume that his/her likelihood Li depends only on (Zi, Xi, Gi) and is independent of (Zj, Xj, Gj), j ≠ i. Given the covariates Zi, genotypes Xi, and random polygenic variation Gi, the likelihood of GLMM (3) or GFLMM (5) or (6) of subject i is . Given the covariates Z = (Z1, …, Zn)′, genotypes X = (X1, …, Xn)′, and random polygenic variations G = (G1, …, Gn)′, the likelihood of GLMM (3) or GFLMM (5) or (6) is
The integrated likelihood function of (α0, α, β, ) is
| (8) |
The likelihood (8) is built in a traditional way where random effects are integrated out and has been used in genetic studies before (Chen et al. 2016). Then, LRT can be calculated based on the integrated likelihood function (8). Our models can analyze rare variants and a combination of rare and common variants, while the approach of Chen et al. (2016) handles only single individual variants.
3.6. Parameter Estimation
In the proposed models (5) and (6), the contributions of a random vector (G1, …, Gn)′ are correlated according to the pedigree structure with a covariance matrix . The covariance matrix differs from pedigree to pedigree. Routines for mixed models in standard packages cannot be used for parameter estimation. For linear mixed models for quantitative traits under normal assumptions, the marginal likelihood has a closed form and maximum likelihood estimation can be performed conveniently. However, in models (5) and (6), the marginal likelihood does not have a closed form and must be approximated using, for example, a Laplacian approximation (Gilmour, Anderson, and Rae 1985; Schall 1991; Breslow and Clayton 1993; Vazquez et al. 2010). In this article, we use the R package pedigreemm which is an extension of the lme4 R package (Bates and Vazquez 2014; Bates et al. 2015). The pedigreemm package uses the capabilities of lme4 while allowing for correlations between pedigree members by applying Cholesky decomposition of the covariance structure of the random effects (G1, …, Gn)′. Briefly, let Ω = CC′ and , where C is the Cholesky factor (Harvillel and Callanan 1989). Then, we have
where In is an n × n identity matrix. Thus, the elements of are mutually independent, and lme4 procedure can be applied because the random effects are now independent (Bates 2009).
3.7. LRT Statistics
To test for association between the dichotomous trait and the m genetic variants, the null hypothesis is . Under the null, the GFLMM (5) and (6) are simplified as
| (9) |
The GFLMM (5) or (6) and the null model (9) are nested. By fitting the GFLMM (5) or (6) and the null model (9), we may test the null H0 : β = 0 by a χ2-distributed LRT statistic with Kβ degrees of freedom using the pedigreemm R package (Vazquez et al. 2010).
In total, these combinations define three different LRT statistics, as outlined in Table 2. In addition to evaluating their Type I error rates via simulation (as described below), we also evaluated their power as well as the power of six different kernel and burden tests developed by Schaid et al. (2013); these are also listed in Table 2.
Table 2.
Notation used in the figures.
| Notation | Description and interpretation |
|---|---|
| LRT GFLMM FR | LRT of GFLMM (5) with the Fourier basis vs. null model (9) |
| LRT_beta_BS | LRT of GFLMM (6) with the B-Spline basis vs. null model (9) |
| LRT_beta_FR | LRT of GFLMM (6) with the Fourier basis vs. null model (9) |
| kernel BT | Kernel test with weights based on Beta distribution |
| kernel MB | Kernel test with Madsen-Browning weights |
| kernel UW | Kernel test with equal weights |
| burden BT | Burden test with weights based on Beta distribution |
| burden MB | Burden test with Madsen-Browning weights |
| burden UW | Burden test with equal weights |
4. Simulation Studies
To evaluate the performance of the proposed GFLMM and LRT statistics, we simulated data to estimate empirical Type I error rates and power levels. In our simulations, a variant is considered to be rare if its MAF is ≤ 0.03. Two scenarios were considered: (1) some variants are common and the rest are rare; (2) all variants are rare.
4.1. Simulation Design
4.1.1. Pedigree Template of 25 Families
We first simulated 25 families including 11 two-generation nuclear pedigrees and 14 three-generation extended pedigrees by randomly choosing progeny sizes from a negative binomial distribution (Cavalli-Sforza and Bodmer 1999). We assumed that each child within the second generation has a 25% chance of having offspring. The pedigree structures included 228 individuals (119 males and 109 females; 70 founders and 158 nonfounders) within 25 families. The pedigree size ranged from 4 to 24 with an average value of 9.12.
4.1.2. Pedigree Template of 50 Families
By doubling the 25 families, the pedigree structures included 456 individuals (238 males and 218 females; 140 founders and 316 nonfounders) within 50 families.
4.1.3. Genetic Variants
The sequence data are of European ancestry from 10,000 chromosomes covering a 1 Mb region, simulated by Yun Li at the University of North Carolina, Chapel Hill using the calibrated coalescent model as programmed in COSI (Schaffner et al. 2005). The sequence data were generated using COSI’s calibrated best-fit models, and the generated European haplotypes mimic CEPH Utah individuals with ancestry from northern and western Europe in terms of site frequency spectrum and linkage disequilibrium (LD) patterns (Schaffner et al. 2005; The International HapMap Consortium 2007). To evaluate empirical Type I error and power levels, we randomly sampled two haplotypes for each founder. For each nonfounder, we chose one haplotype at random from his or her parents. Genotypes were constructed by summing up two haplotypes for each individual to determine the number of minor alleles.
4.1.4. Type I Error Simulations
To evaluate Type I error rates of our LRT statistics, we utilized the 50 two- or three-generation families with a total of 456 related individuals as a template as well as the 25 families with 228 individuals as another template. For each pedigree, we generated phenotype datasets using the model
| (10) |
where α0 = −4.60, zi1 is a dichotomous covariate taking values 0 and 1 with a probability of 0.5, zi2 is a continuous covariate from a standard normal distribution N(0, 1), and (G1, …, Gn)′ is generated as a normal vector sampled from a multivariate normal with mean 0 and covariance matrix with σG = 0.2. After assigning the phenotype for each individual, pedigrees were ascertained if they contained at least one pair of affected siblings, either in the second or third generation, or both. Through the ascertainment, we effectively sampled cases enriched for the dichotomous trait, thus weakening the influence of the polygenic effect.
Genotypes were selected from variants in 6, 9, 12, 15, 18, and 21 kb subregions randomly selected from the 1 Mb region. Note that the trait values are not related to the genotypes, and so the null hypothesis holds. For each simulation scenario, 3 × 106 phenotype-genotype datasets were generated to fit the models and to calculate the test statistics and related p-values. Then, an empirical Type I error rate was calculated as the proportion of p-values of the convergent models in the 3 × 106 datasets which were smaller than a given α level.
4.1.5. Empirical Power Simulations
To evaluate the power of our LRT statistics, trait status was determined for each individual based upon the genotypes. To do this, we considered a mixed effect logistic regression genetic model to compute the probability of being affected for an individual. We simulated datasets under the alternative hypothesis by randomly selecting subregions to obtain causal variants. First, we generated genotypes of m variants in a selected subregion, similar to the Type I error simulations. Then, M of the m variants were randomly selected to be causal, yielding causal genotypes (xi(u1), …, xi(uM)). For each dataset, the causal variants are the same for all the individuals in the dataset, but we allow the causal variants to be different from dataset to dataset. Then, we generated the dichotomous disease traits by
| (11) |
where α0, zi1, zi2, (G1, …, Gn)′ were the same as in the Type I error model (10), (xi(u1), …, xi(uM))′ were genotypes of the ith individual at the causal variants, and the β’s are additive effects for the causal variants defined as follows. Modeled as the approach of Wu et al. (2011), we used |βj| = c|log10(MAFj)|, where MAFj was the MAF of the jth variant. Three different settings were considered: 5%, 10%, and 15% of variants in the subregions are chosen as causal variants. When 5%, 10%, and 15% of the variants were causal, c = log(90)/k, log(70)/k, and log(50)/k, respectively. For the template of 50 two- or three-generation families with a total of 456 related individuals, the constants k and genetic effect sizes decrease as region sizes increase
| (12) |
In addition to varying the percentage of causal variants in the subregion, we also varied the direction of effect. We considered situations where (i) all causal variants have positive effects; (ii) 20%/80% causal variants have negative/positive effects; and (iii) 50%/50% causal variants have negative/positive effects. Burden tests are expected to be most powerful when all causal variants have effects in the same direction (e.g., under scenario (i)). For each setting, 3000 datasets were simulated to calculate the empirical power as the proportion of p-values which are from the convergent models and smaller than a given α level.
4.2. Functional Data Analysis Parameters and Dynamic Rule
In the data analysis and simulations described above, we used functions from the fda R package to create the basis functions (Ramsay et al. 2014). In the simulations presented in the main text, we implement a dynamic rule to handle the genotype data to make sure that the results are stable. The order of the B-spline basis was 4, the upper limit of B-spline basis functions was K = Kβ = 16, and the upper limit of Fourier basis functions was K = Kβ = 17. First, we perform a principal component analysis to evaluate the effective dimension of the genotype data Mgao (Gao, Starmer, and Martin 2008). Then,
if Mgao ≤ 18, the number of B-spline basis functions was K = Kβ = 6, and the number of Fourier basis functions was K = Kβ = 7;
if 18 < Mgao ≤ 24, the number of B-spline basis functions was K = Kβ = 8, and the number of Fourier basis functions was K = Kβ = 9;
if 24 < Mgao ≤ 30, the number of B-spline basis functions was K = Kβ = 10, and the number of Fourier basis functions was K = Kβ = 11;
if 30 < Mgao ≤ 36, the number of B-spline basis functions was K = Kβ = 12, and the number of Fourier basis functions was K = Kβ = 13;
if 36 < Mgao ≤ 42, the number of B-spline basis functions was K = Kβ = 14, and the number of Fourier basis functions was K = Kβ = 15;
if Mgao > 42, the number of B-spline basis functions was K = Kβ = 16, and the number of Fourier basis functions was K = Kβ = 17;
In Supplementary Materials II, Appendix B, we present additional simulation results when the order of the B-spline basis was 4, the number of B-spline basis functions was K = Kβ = 6, and the number of Fourier basis functions was K = Kβ = 7 for the 50 family template.
In Supplementary Materials III, Appendix C, we present additional simulation results when the order of the B-spline basis was 4, the number of B-spline basis functions was K = Kβ = 16, and the number of Fourier basis functions was K = Kβ = 17 for the small 25 family template. In Supplementary Materials IIII, Appendix D, additional simulation results are shown when the order of the B-spline basis was 4, the number of B-spline basis functions was K = Kβ = 6, and the number of Fourier basis functions was K = Kβ = 7 for the small 25 family template.
In the data analysis, the order of the B-spline basis was ≤4. As these data were more sparsely genotyped than the simulated data, to improve convergence rates, we used the modified dynamic rule as defined in Appendix E, since 15,099 out of 16,913 gene regions contain less than 12 variants.
4.3. Simulation Results
In this subsection, we present simulation results for the Type I error rates and power levels using bar plots for the templates of 50 and 25 two- or three- generation families, where the statistics evaluated are referred to using the notation defined in Table 2. In the table, three LRT statistics, three kernel tests and three burden tests are presented. The three LRT statistics are based on the GFLMM (5) and (6). The kernel and burden tests are from Schaid et al. (2013). Extensive simulations were carried out, comparing the Type I error rates at three nominal significance levels of the three different LRT statistics (listed in Table 2), varying the region size from 6 to 21 kb.
4.3.1. Empirical Type I Error Rates of 50 Family Template
The empirical Type I error rates are reported in Table 3 at four nominal significance levels α = 0.01, 0.001, 0.0001, and 0.00001. In the table, the results of three LRT statistics were reported for the GFLMM (5) and (6). The LRT statistics control the Type I error rates correctly, no matter whether the genotype data are smoothed or not and which basis functions are used to smooth the GVF and β(t) (Table 3). When the number of B-spline basis functions was K = Kβ = 6 and the number of Fourier basis functions was K = Kβ = 7, the results are reported in Table B.1 in Supplementary Materials II, Appendix B and the LRT statistics control the Type I error rates correctly.
Table 3.
Empirical type I error rates of the LRT statistics at nominal levels α 0.01,0.001,0.0001, and 0.00001, using the 50 two- or three-generation families with a total of 456 related individuals as a template.
| Type I error rates of LRT statistics |
|||||
|---|---|---|---|---|---|
| Variant type | Region size (mean # of variants) | Nominal level α | GFLMM (5) | GFLMM (6) | |
| Fourier basis | B-sp basis | Fourier basis | |||
| Rare and common | 6 kb | 0.01 | 0.005152 | 0.005236 | 0.005283 |
| (117) | 0.001 | 0.000382 | 0.000392 | 0.000406 | |
| 0.0001 | 3.06E-05 | 3.07E-05 | 3.46E—05 | ||
| 0.00001 | 2.35E—06 | 7.88E—07 | 3.22E—06 | ||
| 9 kb | 0.01 | 0.004816 | 0.004837 | 0.004919 | |
| (176) | 0.001 | 0.000380 | 0.000371 | 0.000404 | |
| 0.0001 | 2.93E—05 | 2.39E—05 | 3.16E—05 | ||
| 0.00001 | 2.44E—06 | 1.65E—06 | 1.71E—06 | ||
| 12 kb | 0.01 | 0.004656 | 0.004818 | 0.004711 | |
| (235) | 0.001 | 0.000359 | 0.000375 | 0.000362 | |
| 0.0001 | 2.52E—05 | 3.33E—05 | 2.73E—05 | ||
| 0.00001 | 0.000000 | 1.71E—06 | 0.000000 | ||
| 15 kb | 0.01 | 0.004773 | 0.004740 | 0.004774 | |
| (293) | 0.001 | 0.000392 | 0.000369 | 0.000398 | |
| 0.0001 | 3.00E—05 | 2.61E—05 | 2.80E—05 | ||
| 0.00001 | 2.57E—06 | 2.61E—06 | 1.93E—06 | ||
| 18 kb | 0.01 | 0.004581 | 0.004658 | 0.004607 | |
| (352) | 0.001 | 0.000322 | 0.000338 | 0.000337 | |
| 0.0001 | 2.33E—05 | 2.72E—05 | 2.53E—05 | ||
| 0.00001 | 1.73E—06 | 1.76E—06 | 1.01E—06 | ||
| 21 kb | 0.01 | 0.004463 | 0.004561 | 0.004410 | |
| (410) | 0.001 | 0.000306 | 0.000334 | 0.000317 | |
| 0.0001 | 2.86E—05 | 2.47E—05 | 2.81E—05 | ||
| 0.00001 | 1.74E—06 | 3.53E—06 | 1.04E—06 | ||
| Rare | 6 kb | 0.01 | 0.005798 | 0.006236 | 0.007339 |
| (106) | 0.001 | 0.000422 | 0.000491 | 0.000600 | |
| 0.0001 | 2.60E—05 | 3.09E—05 | 4.42E—05 | ||
| 0.00001 | 8.13E—07 | 1.58E—06 | 7.75E—07 | ||
| 9 kb | 0.01 | 0.006217 | 0.006364 | 0.007704 | |
| (159) | 0.001 | 0.000469 | 0.000563 | 0.000659 | |
| 0.0001 | 2.95E—05 | 2.91E—05 | 5.44E—05 | ||
| 0.00001 | 3.99E—06 | 2.42E—06 | 9.97E—06 | ||
| 12 kb | 0.01 | 0.006041 | 0.006157 | 0.007316 | |
| (212) | 0.001 | 0.000476 | 0.000493 | 0.000643 | |
| 0.0001 | 3.77E—05 | 5.10E—05 | 6.05E—05 | ||
| 0.00001 | 3.27E—06 | 3.34E—06 | 6.28E—06 | ||
| 15 kb | 0.01 | 0.006008 | 0.006378 | 0.007259 | |
| (265) | 0.001 | 0.000435 | 0.000511 | 0.000624 | |
| 0.0001 | 3.27E—05 | 4.11E—05 | 5.78E—05 | ||
| 0.00001 | 2.51E—06 | 8.57E—07 | 5.62E—06 | ||
| 18 kb | 0.01 | 0.005957 | 0.006306 | 0.007044 | |
| (318) | 0.001 | 0.000525 | 0.000503 | 0.000672 | |
| 0.0001 | 4.69E—05 | 4.17E—05 | 6.71E—05 | ||
| 0.00001 | 5.97E—06 | 5.21E—06 | 8.18E—06 | ||
| 21 kb | 0.01 | 0.005937 | 0.006204 | 0.006781 | |
| (371) | 0.001 | 0.000480 | 0.000498 | 0.000599 | |
| 0.0001 | 4.73E—05 | 3.89E—05 | 6.54E—05 | ||
| 0.00001 | 3.44E—06 | 4.32E—06 | 5.80E—06 | ||
NOTE: The order of B-spline basis was 4, and the number of basis functions of B-spline was K = Kβ = 16; the number of Fourier basis functions was K = Kβ = 17.
4.3.2. Empirical Type I Error Rates of 25 Family Template
When the number of B-spline basis functions was K = Kβ = 16 and the number of Fourier basis functions was K = Kβ = 17, the results are reported in Table C.1 in Supplementary Materials III, Appendix C. In Table D.1 of Supplementary Materials IIII, Appendix D, we present simulation results when the order of the B-spline basis was 4, the number of B-spline basis functions was K = Kβ = 6, and the number of Fourier basis functions was K = Kβ = 7. In Tables C.1 and D.1, we find that the LRT statistics control Type I error rates accurately.
4.3.3. Empirical Power Simulations of 50 Family Template
Based on the simulated data, the power of the LRT statistics was compared with the power of the retrospective kernel and burden statistics developed by Schaid et al. (2013): three different weighting schemes were considered for both the kernel and burden statistics (Table 2). The results are reported in Figures A.1–A.12 in Supplementary Materials I. In Figures A.1–A.6, some variants are common and the rest are rare. In Figures A.7–A.12, the variants are all rare. In plots (a1)–(a3) of each figure, all causal variants have positive effects; when 20%/80% causal variants have negative/positive effects, we present the results in plots (b1), (b2), and (b3) for each figure; when 50%/50% causal variants have negative/positive effects, the results are presented in plots (c1), (c2), and (c3).
When the region sizes are between 6 and 15 kb, the LRT GFLMM (5) and (6) statistics have higher power than the kernel and burden tests in Figures A.1–A.4 and A.7–A.10. When the region sizes are 18 and 21 kb, the power levels of kernel and burden tests are lower or similar to those of LRT GFLMM (5) and (6) in Figures A.5, A.6, A.11, and A.12. The choice of the B-spline or the Fourier basis has little effect on power. The three LRT GFLMM statistics (5) and (6) control Type I error rates well and have similar good power levels as shown in Figures A.1–A.12. The power levels of the LRT beta-smooth only GFLMM (6) statistics are almost identical to those of the LRT GFLMM statistics (5) which smooth both the GVFs Xi(u) and the genetic effect function β(t), regardless of basis choice. Hence, the three LRT GFLMM statistics (5) and (6) are very stable in terms of power performance and they do not strongly depend on whether the genotype data are smoothed or not, or which basis functions are used.
When some variants are common and the rest are rare, the kernel-based approach with Madsen–Browning weights performs the best among the kernel and burden statistics in Figures A.1–A.6. When all variants are rare, the kernel-based approach with weights based on the beta distribution performs the best (Figures A.7–A.12). As noted in Schaid et al. (2013), the kernel statistics have higher power than burden ones.
In Supplementary Materials II, Appendix B, we present simulation results when the order of the B-spline basis was 4, the number of B-spline basis functions was K = Kβ = 6, and the number of Fourier basis functions was K = Kβ = 7. The power levels of LRT GFLMM statistics (5) and (6) in Figures B.1–B.12 can be low.
4.3.4. More Simulation Results of 25 Family Template
In Supplementary Materials III, Appendix C, we present simulation results when the order of the B-spline basis was 4, the number of B-spline basis functions was K = Kβ = 16, and the number of Fourier basis functions was K = Kβ = 17. The power levels of LRT GFLMM statistics (5) and (6) are presented in Figures C.1–C.12. In Supplementary Materials IIII, Appendix D, we present simulation results when the order of the B-spline basis was 4, the number of B-spline basis functions was K = Kβ = 6, and the number of Fourier basis functions was K = Kβ = 7. Compared with the results of Supplementary Materials III, Appendix C, the power levels of LRT GFLMM statistics (5) and (6) in Figures D.1–D.12 are lower.
5. Discussion
AMD is a common complex disease and is caused by an interplay of aging, genetics, and environmental factors. While AMD family data has been collected (Ratnapriya et al. 2020), most AMD association studies have mainly focused on using population data to identify genes and variants which are associated with AMD, partly because of a dearth of powerful methods for carrying out region-based association tests of a dichotomous trait like AMD on family data. Here we apply GFLMM we developed to test for association between the dichotomous AMD trait and genetic variants in each gene region in our AMD family dataset. Using common and rare variants, we observe strong association between AMD and two known AMD susceptibility genes: CFH and ARMS2. Gene-based rare variant burden analyses were carried out by the International AMD Genomics Consortium in a large dataset of 16,144 advanced AMD cases versus 17,832 controls (Fritsche et al. 2016); the analyses ignored family structure—for example, the dataset included a set of unrelated individuals extracted from the family dataset we analyze here. When testing was restricted to the 703 genes, CFH attains significance, but ARMS2 and the other genes listed in our Table 1 are not among their top hits. However, note that they used only rare protein-altering variants, while our analysis that gives a signal for ARMS2 used both common and rare variants.
By using rare variants, we find suggestive association signals in four gene regions, ASAH1, CLEC6A, TMEM63C, and SGSM1. The Consortium did not see significant burden test signals at these genes in Fritsche et al. (2016). The suggestive signal at ASAH1 is especially interesting because ASAH1 deficiency leads to retinal inflammation (Petrov et al. 2019) and underexpression of ASAH1 makes retinal cells more vulnerable to oxidative stress (Sugano et al. 2019). Consistently, in the aqueous humor, ASAH1 is identified as a down-regulated protein in AMD as compared to controls (Qu et al. 2019). Thus, several lines of congruent evidence complement our suggestive gene-based association of ASAH1 with AMD to suggest that ASAH1 may play a role in AMD.
In this article, we analyzed the AMD family data in two different ways: (1) all genetic variants and (2) rare variants only. From the results, we can see that it is a good strategy to analyze all variants in addition to only analyzing rare variants. It is reasonable to assume that a combination of rare and common variants affects the risk of many complex disorders. After all, we are searching for causal variants, not rare or common variants.
To analyze the related pedigrees and high dimensional sequencing data, we developed GFLMM for analyzing familial dichotomous trait data. In these models, the effect of a major gene is modeled as a fixed mean, the polygenic contributions is modeled as a random variation, and the correlation of pedigree members is modeled by inbreeding and kinship coefficients. LRT statistics based on the GFLMM are built to test for association between a dichotomous trait and the genetic variants. Simulation results indicate that the LRT statistics accurately control the Type I error rates for a pedigree dataset of moderate sample size (i.e., 456 individuals in 50 pedigrees), as well as for a small sample size dataset (228 individuals in 25 pedigrees). In our analysis of the AMD data, the kernel and burden tests have high false positive rates while the GFLMM control the Type I errors well. In addition to properly controlling Type I error rates, GFLMM can handle both common and rare variants, avoiding arbitrary MAF threshold-based filtering of variants. Functional models are very flexible since they can analyze: (1) rare variants, (2) common variants, and (3) a combination of the two. This elegant feature deserves further utilization in dissecting complex disorders.
It can be challenging to get linear mixed models to converge. To improve convergence rates, in addition to selecting a better optimizer and increasing the number of function evaluations, we added the dynamic rule defined above to better match the statistical model to the underlying dimensionality of the data. This improved the convergence rate. For example, for the GFLMM (6) power simulation results in Figure A.6(a1), the LRT of GFLMM (6) using the B-spline basis converged 97.1% of the time; the convergence rate using the Fourier basis of 93.7% was poorer, suggesting the B-spline basis version should be preferentially used for GFLMM (6). In these power results about 63% of the time a warning was generated about a boundary fit, likely indicating that the random component is not needed. For the simulations presented here, we conservatively evaluated each statistic on replicates with no errors or warnings.
While the dynamic rule presented above worked reasonably well on the simulated data, when applied to the real AMD data, it did not work as well, likely because these data are more sparsely genotyped than the simulated data. To improve convergence rates, we used the adjusted dynamic rule presented in Appendix E. While we have shown that our statistics are promising, future work is needed into choosing an optimal dynamic rule for one’s particular data to improve convergence rates and to reduce errors and warnings when fitting these models.
In major gene analysis, the LRT statistics of GFLMM have higher power than the kernel-based and burden tests proposed by Schaid et al. (2013). The kernel-based tests proposed in Schaid et al. (2013) perform better than burden tests. In the previous work, it was shown that the tests of fixed effect regression models have higher power than SKAT for population data in major gene association studies (Fan, Chiu, et al. 2016). Therefore, the proposed models provide an alternative competitive method for carrying out gene-based association tests.
One major difference between our statistics and the kernel-based tests is that we model the major gene contribution by a fixed effect mean while kernel-based tests model the gene contribution as a random term with a zero mean and a constant variance. In the previous work and the current article, it is shown that our models have higher power in major gene association analysis and kernel-based tests perform better for polygenic analysis (Fan, Chiu, et al. 2016). If the number of causal genetic variants at a locus is very large and each causal variant contributes a small amount to the traits, the kernel-based test assumption of means of zero is likely to be satisfied and kernel-based tests could perform better. However, assuming means of zero for regression coefficients is unlikely to be valid. For instance, if some of the causal variants’ contributions to the traits are relatively large, it is unlikely that regression coefficients of genetic variants are around zero. In major gene association studies, we argue that our LRT statistics perform better than kernel-based tests in most cases.
The GFLMM (5) and (6) are designed to analyze familial data from one population. Comparing with previous fixed models which were designed to analyze unrelated population samples, one random term Gi is added to model the polygenic variation and familial correlation for pedigree members (Cordell and Clayton 2002; Fan et al. 2013, 2014, 2015; Fan, Chiu, et al. 2016; Fan, Wang, Chiu, et al. 2016; Fan, Wang, Yan, et al. 2016). The models can be extended to accommodate population structure and cryptic relatedness by adding extra random terms to the models (Chen et al. 2016). For cryptically related individuals, one may replace the kinship coefficients with empirical genetic relationship matrix (GRM). The empirical GRM can be calculated based on marker data to account for population structure and cryptic relatedness (Yang et al. 2010; Gianola et al. 2016; Wang 2016). For individuals i and j from different pedigrees, the kinship coefficient ϕij = 0. However, the empirical genetic relationship coefficient of i and j can be different from 0 since they can be cryptically related to each other. More research is needed to characterize the properties of models which accommodate population structure and cryptic relatedness as well as adding variance component at the major gene to the models.
Supplementary Material
Acknowledgments
Three anonymous reviewers, an associate editor, and the editors, Dr. Fuentes and Dr. Zhang, provided very good and insightful comments for us to improve the article.
Funding
This study was supported by U.S. National Science Foundation grant DMS-1915904 (Ruzong Fan), by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development at NIH (Chi-Yang Chiu), by the Intramural Research Program of the National Human Genome Research Institute at NIH (Ruzong Fan, Chi-Yang Chiu, Alexander F. Wilson, E. Bailey-Wilson), by the Intramural Research Program of the National Institute of Mental Health at NIH (Francis J. McMahon), National Institutes of Health, Bethesda, Maryland, by Wei Chen’s NIH grants R01EY024226 and R01HG007358 and the University of Pittsburgh (Ruzong Fan is an unpaid collaborator on the grant R01EY024226), by Christopher I. Amos’ Texas Cancer Prevention Research Institute grants RR170048 & RP190641, NIH grants U01CA196386, U19CA203654, R01CA242218 and U01CA243483, and by Ana I. Vazquez’s NIH grants R01GM101219 and R01GM099992. Michael B. Gorin received funding support from the NEI R01 EY09859 and the ARRA supplement, Harold and Pauline Price Foundation, Arnold and Mabel Beckman Foundation and unrestricted funds to the Department of Ophthalmology from Research to Prevent Blindness, N.Y. The genotyping of the age-related macular degeneration dataset was carried out by the Johns Hopkins University Genetic Resources Core Facility SNP Center. This work utilized the computational resources of the NIH HPC Biowulf cluster at the National Institutes of Health, Bethesda, MD (https://hpc.nih.gov).
Footnotes
Computer Program
The software is released as an R package PedGFLMM available at https://github.com/DanielEWeeks/PedGFLMM. For reproducibility purposes, we have also released the code used to generate the results in this article via GitHub at https://github.com/DanielEWeeks/PedGFLMM-simulation-code.
Disclosure Statement
Drs. Gorin, Conley, and Weeks are listed as co-inventors on U.S. patents 7,695,909 and 8,053,190 held by University of Pittsburgh for 10q26 ARMS2/HTRA1 locus of AMD.
Supplementary Materials
The supplementary materials include additional results from the AMD analyses, as well as from our simulation studies. For links to the GitHub sites containing the R package implementing our statistics, as well as our simulation code, please see the Computer Program section below.
Color versions of one or more of the figures in the article can be found online at www.tandfonline.com/r/JASA.
✔ These materials were reviewed for reproducibility.
Supplementary materials for this article are available online. Please go to www.tandfonline.com/r/JASA.
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, and McVean GA (2012), “An Integrated Map of Genetic Variation From 1,092 Human Genomes,” Nature, 491, 56–65. [1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Age-Related Eye Disease Study Research Group (1999), “The Age-Related Eye Disease Study (AREDS): Design Implications. AREDS Report No. 1,” Control Clinical Trials, 20, 573–600. [1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astle W, and Balding DJ (2009), “Population Structure and Cryptic Relatedness in Genetic Association Studies,” Statistical Science, 24, 451–471. [2] [Google Scholar]
- Aulchenko Y, De Koning DJ, and Haley C (2007), “Genomewide Rapid Association Using Mixed Model and Regression: A Fast and Simple Method for Genomewide Pedigree-Based Quantitative Trait Loci Association Analysis,” Genetics, 177, 577–585. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal V, Libiger O, Torkamani A, and Schork NJ (2010), “Statistical Analysis Strategies for Association Studies Involving Rare Variants,” Nature Reviews Genetics, 11, 773–785. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates DM (2009), “Assessing the Precision of Estimates of Variance Components,” available at http://lme4.r-forge.r-project.org/slides/2009-07-21-Seewiesen/4Precision-4a4.pdf. [7] [Google Scholar]
- Bates DM, Mächler M, Bolker B, and Walker S (2015), “Fitting Linear Mixed-Effects Models Using lme4,” Journal of Statistical Software, 67, 1–48. [7] [Google Scholar]
- Bates DM, and Vazquez A (2014), “pedigreemm: Pedigree-Based Mixed-Effects Models,” R Package Version 0.3–3, available at https://CRAN.R-project.org/package=pedigreemm. [7] [Google Scholar]
- Breslow NE, and Clayton DG (1993), “Approximate Inference in Generalized Linear Mixed Models,” Journal of American Statistical Association, 88, 9–25. [7] [Google Scholar]
- Cavalli-Sforza LL, and Bodmer WF (1999), The Genetics of Human Populations, New York: Courier Corporation. [8] [Google Scholar]
- Chen H, Wang C, Conomos MP, Stilp AM, Li Z, Sofer T, Szpiro AA, Chen W, Brehm JM, Celedón JC, and Redline S (2016), “Control for Population Structure and Relatedness for Binary Traits in Genetic Association Studies via Logistic Mixed Models,” The American Journal of Human Genetics, 98, 653–666. [2,7,12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Stambolian D, Edwards AO, Branham KE, Othman M, Jakobsdottir J, Tosakulwong N, Pericak-Vance MA, Campochiaro PA, Klein ML, and Tan PL (2010), “Genetic Variants Near TIMP3 and High-Density Lipoprotein-Associated Loci Influence Susceptibil ity to Age-Related Macular Degeneration,” Proceedings of the National Academy of Sciences of the United States of America, 107, 7401–7406. [1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell H, and Clayton D (2002), “A Unified Stepwise Regression Procedure for Evaluating the Relative Effects of Polymorphisms Within a Gene Using Case/Control or Family Data: Application to HLA in Type 1 Diabetes,” The American Journal of Human Genetics, 70, 124–141. [2,12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- De G, Yip WK, Ionita-Laza I, and Laird N (2013), “Rare Variant Analysis for Family-Based Design,” PLoS One, 8, e48495. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boor C (2001), A Practical Guide to Splines, Applied Mathematical Sciences (Revised Version, Vol. 27), New York: Springer. [2,6] [Google Scholar]
- Fan R, Chiu CY, Jung J, Weeks DE, Wilson AF, Bailey-Wilson JE, Amos CI, Chen Z, Mills JL, and Xiong M (2016), “A Comparison Study of Fixed and Mixed Effect Models for Gene Level Association Studies of Complex Traits,” Genetic Epidemiology, 40, 702–721. [2,6,12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R, Wang Y, Boehnke M, Chen W, Li Y, Ren H, Lobach I, and Xiong M (2015), “Gene Level Meta-Analysis of Quantitative Traits by Functional Linear Models,” Genetics, 200, 1089–1104. [2,6,12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R, Wang Y, Chiu CY, Chen W, Ren H, Li Y, Boehnke M, Amos CI, Moore JH, and Xiong M (2016), “Meta-Analysis of Complex Diseases at Gene Level by Generalized Functional Linear Models,” Genetics, 202, 457–470. [2,6,12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R, Wang Y, Mills JL, Carter TC, Lobach I, Wilson AF, Bailey-Wilson JE, Weeks DE, and Xiong M (2014), “Generalized Functional Linear Models for Case-Control Association Studies,” Genetic Epidemiology, 38, 622–637. [2,6,12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R, Wang Y, Mills JL, Wilson AF, Bailey-Wilson JE, and Xiong M (2013), “Functional Linear Models for Association Analysis of Quantitative Traits,” Genetic Epidemiology, 37, 726–742. [2,6,12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R, Wang Y, Yan Q, Ding Y, Weeks DE, Lu Z, Ren H, Cook RJ, Xiong M, Swaroop A, and Chew EY (2016), “Gene-Based Association Analysis for Censored Traits via Functional Regressions,” Genetic Epidemiology, 40, 133–143. [2,6,12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez M, Budde J, Del-Aguila JL, Ibañez L, Deming Y, Harari O, Norton J, Morris JC, Goate AM, Cruchaga C, and NIA-LOAD Family Study Group (2018), “Evaluation of Gene-Based Family-Based Methods to Detect Novel Genes Associated With Familial Late Onset Alzheimer Disease,” Frontiers in Neuroscience, 12, 209. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraty F, and Romain Y (2010), The Oxford Handbook of Functional Data Analysis, New York: Oxford University Press. [2,6] [Google Scholar]
- Fisher S, Abecasis GR, Yashar BM, Zareparsi S, Swaroop A, Iyengar SK, Klein BE, Klein R, Lee KE, Majewski J, and Schultz DW (2005), “Meta-Analysis of Genome Scans of Age-Related Macular Degeneration,” Human Molecular Genetics, 14, 2257–2264. [1] [DOI] [PubMed] [Google Scholar]
- Friedman DS, O’Colmain BJ, Munoz B, Tomany SC, McCarty C, De Jong PT, Nemesure B, Mitchell P, and Kempen J (2004), “Prevalence of Age-Related Macular Degeneration in the United States,” Archives of Ophthalmology, 122, 464–472. [1] [DOI] [PubMed] [Google Scholar]
- Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, Thorleifsson G, Zack DJ, Arakawa S, Cipriani V, Ripke S, and Igo RP Jr. (2013), “Seven New Loci Associated With Age-Related Macular Degeneration,” Nature Genetics, 45, 433–439. [1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, and Swaroop A (2014), “Age-Related Macular Degeneration: Genetics and Biology Coming Together,” Annual Review of Genomics and Human Genetics, 15, 151–171. [1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Igl W, Bailey JNC, Grassmann F, Sengupta S, Bragg-Gresham JL, Burdon KP, Hebbring SJ, Wen C, Gorski M, and Kim IK (2016), “A Large Genome-Wide Association Study of Age-Related Macular Degeneration Highlights Contributions of Rare and Common Variants,” Nature Genetics, 48, 134–143. [1,11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Starmer J, and Martin ER (2008), “A Multiple Testing Correction Method for Genetic Association Studies Using Correlated Single Nucleotide Polymorphisms,” Genetic Epidemiology, 32, 361–369. [9] [DOI] [PubMed] [Google Scholar]
- Gianola D, Fariello MI, Naya H, and Schön CC (2016), “Genome-Wide Association Studies With a Genomic Relationship Matrix: A Case Study With Wheat and Arabidopsis,” G3: Genes, Genomes, Genetics, 6, 3241–3256. [12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour AR, Anderson RD, and Rae AL (1985), “The Analysis of Binomial Data by a Generalized Linear Mixed Model,” Biometrika, 72, 593–599. [7] [Google Scholar]
- Harvillel DA, and Callanan T (1989), “Computational Aspects of Likelihood-Based Inference for Variance Components,” in Advances in Statistical Methods for Genetic Improvement of Livestock, eds. Gianola D and Hammond K, Berlin: Springer-Verlag, pp. 136–176. [7] [Google Scholar]
- Hayeck TJ, Zaitlen NA, Loh PR, Vilhjalmsson B, Pollack S, Gusev A, Yang J, Chen GB, Goddard ME, Visscher PM, and Patterson N (2015), “Mixed Model With Correction for Case-Control Ascertainment Increases Association Power,” The American Journal of Human Genetics, 96, 720–730. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson C (1984), Applications of Linear Models in Animal Breeding, Guelph, ON: University of Guelph. [2] [Google Scholar]
- Horvath L, and Kokoszka P (2012), Inference for Functional Data With Applications, New York: Springer. [2,6] [Google Scholar]
- Ionita-Laza I, Lee S, Makarov V, Buxbaum JD, and Lin X (2013a), “Sequence Kernel Association Tests for the Combined Effect of Rare and Common Variants,” The American Journal of Human Genetics, 92, 841–853. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- —(2013b), “Family-Based Association Tests for Sequence Data, and Comparisons With Population-Based Association Tests,” European Journal of Human Genetics, 21, 1158–1162. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, and Eskin E (2010), “Variance Component Model to Account for Sample Structure in Genome-Wide Association Studies,” Nature Genetics, 42, 348–354. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, and Eskin E (2008), “Efficient Control of Population Structure in Model Organism Association Mapping,” Genetics, 178, 1709–1723. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A, Garimella K, Do R, Stitziel NO, Neale BM, McLaren PJ, Gupta N, Sklar P, Sullivan PF, Moran JL, and Hultman CM (2012), “Exome Sequencing and the Genetic Basis of Complex Traits,” Nature Genetics, 44, 623–630. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A, Vilhjálmsson BJ, Segura V, Platt A, Long Q, and Nordborg M (2012), “A Mixed-Model Approach for Genomewide Association Studies of Correlated Traits in Structured Populations,” Nature Genetics, 44, 1066–1071. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K (2002), Mathematical and Statistical Methods for Genetic Analysis (2nd ed.), New York: Springer. [2,6] [Google Scholar]
- Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, Boehm F, Caporaso NE, Cornelis MC, Edenberg HJ, and Gabriel SB (2010), “Quality Control and Quality Assurance in Genotypic Data for Genome-Wide Association Studies,” Genetic Epidemiology, 34, 591–602. [3] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, ESP Lung Project Team, Christiani DC, Wurfel MM, Lin X, and NHLBI GO Exome Sequencing Project (2012), “Optimal Unified Approach for Rare-Variant Association Testing With Application to Small-Sample Case-Control Whole-Exome Sequencing Studies,” The American Journal of Human Genetics, 91, 224–237. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, and Tukiainen T (2016), “Analysis of Protein-Coding Genetic Variation in 60,706 Humans,” Nature, 536, 285–291. [1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, and Heckerman D (2011), “Fast Linear Mixed Models for Genome-Wide Association Studies,” Nature Methods, 8, 833–835. [2] [DOI] [PubMed] [Google Scholar]
- Listgarten J, Lippert C, and Heckerman D (2013), “FaSTLMM-Select for Addressing Confounding From Spatial Structure and Rare Variants,” Nature Genetics, 45, 470–471. [2] [DOI] [PubMed] [Google Scholar]
- Listgarten J, Lippert C, Kadie CM, Davidson RI, Eskin E, and Heckerman D (2012), “Improved Linear Mixed Models for Genome-Wide Association Studies,” Nature Methods, 9, 525–526. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PR, Tucker G, Bulik-Sullivan BK, Vilhjálmsson BJ, Finucane HK, Salem RM, Chasman DI, Ridker PM, Neale BM, Berger B, and Patterson N (2015), “Efficient Bayesian Mixed-Model Analysis Increases Association Power in Large Cohorts,” Nature Genetics, 47, 284–290. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Boerwinkle E, and Xiong M (2011), “Association Studies for Next-Generation Sequencing,” Genome Research, 21, 1099–1108. [2,6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Zhu Y, and Xiong M (2012), “Quantitative Trait Locus Analysis for Next-Generation Sequencing With the Functional Linear Models,” Journal of Medical Genetics, 49, 513–524. [2,6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- — (2013), “Smoothed Functional Principal Component Analysis for Testing Association of the Entire Allelic Spectrum of Genetic Variation,” European Journal of Human Genetics, 21, 217–224. [2,6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Fagerness J, Reynolds R, Sobrin L, Parker M, Raychaudhuri S, Tan PL, Oh EC, Merriam JE, Souied E, and Bernstein PS (2010), “Genome-Wide Association Study of Advanced Age-Related Macular Degeneration Identifies a Role of the Hepatic Lipase Gene (LIPC),” Proceedings of the National Academy of Sciences of the United States of America, 107, 7395–7400. [1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AM, Astafev AA, Mast N, Saadane A, El-Darzi N, and Pikuleva IA (2019), “The Interplay Between Retinal Pathways of Cholesterol Output and Its Effects on Mouse Retina,” Biomolecules, 9, 867. [5,11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirinen M, Donnelly P, and Spencer CC (2013), “Efficient Computation With a Linear Mixed Model on Large-Scale Data Sets With Applications to Genetic Studies,” The Annals of Applied Statistics, 7, 369–390. [2] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, and Reich D (2006), “Principal Components Analysis Corrects for Stratification in Genome-Wide Association Studies,” Nature Genetics, 38, 904–909. [2] [DOI] [PubMed] [Google Scholar]
- Qu SC, Xu D, Li TT, Zhang JF, and Liu F (2019), “iTRAQ-Based Proteomics Analysis of Aqueous Humor in Patients With Dry Age-Related Macular Degeneration,” International Journal of Ophthalmology, 12, 1758–1766. [5,11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay JO, Hooker G, and Graves S (2009), Functional Data Analysis With R and MATLAB, New York: Springer. [2,6,7] [Google Scholar]
- Ramsay JO, and Silverman BW (2005), Functional Data Analysis (2nd ed.), New York: Springer. [2,6] [Google Scholar]
- Ramsay JO, Wickham H, Graves S, and Hooker G (2014), “fda: Functional Data Analysis,” R Package Version 2.4.4, available at https://CRAN.R-project.org/package=fda. [9] [Google Scholar]
- Ratnapriya R, Acar İE, Geerlings MJ, Branham K, Kwong A, Saksens N, Pauper M, Corominas J, Kwicklis M, Zipprer D, and Starostik MR (2020), “Family-Based Exome Sequencing Identifies Rare Coding Variants in Age-Related Macular Degeneration,” Human Molecular Genetics (in press). [11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S (1996), Stochastic Processes (2nd ed.), New York: Wiley. [2] [Google Scholar]
- Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, and Altshuler D (2005), “Calibrating a Coalescent Simulation of Human Genome Sequence Variation,” Genome Research, 15, 1576–1583. [8] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, McDonnell SK, Sinnwell JP, and Thibodeau SN (2013), “Multiple Genetic Variant Association Testing by Collapsing and Kernel Methods With Pedigree or Population Structured Data,” Genetic Epidemiology, 37, 409–418. [2,3,8,9,10,11,12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schall R (1991), “Estimation in Generalized Linear Models With Random Effects,” Biometrika, 78, 719–727. [7] [Google Scholar]
- Seddon JM, Francis PJ, George S, Schultz DW, Rosner B, and Klein ML (2007), “Association of CFH Y402H and LOC387715 A69S With Progression of Age-Related Macular Degeneration,” The Journal of American Medical Association, 297, 1793–1800. [1] [DOI] [PubMed] [Google Scholar]
- Segura V, Vilhjálmsson BJ, Platt A, Korte A, Seren Ü, Long Q, and Nordborg M (2012), “An Efficient Multi-Locus Mixed-Model Approach for Genome-Wide Association Studies in Structured Populations,” Nature Genetics, 44, 825–830. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Hao W, and Storey JD (2015), “Testing for Genetic Associations in Arbitrarily Structured Populations,” Nature Genetics, 47, 550–554. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano E, Edwards G, Saha S, Wilmott LA, Grambergs RC, Mondal K, Qi H, Stiles M, Tomita H, and Mandal N (2019), “Overexpression of Acid Ceramidase (ASAH1) Protects Retinal Cells (ARPE19) From Oxidative Stress,” Journal of Lipid Research, 60, 30–43. [5,11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svishcheva GR, Axenovich TI, Belonogova NM, van Duijn CM, and Aulchenko YS (2012), “Rapid Variance Components-Based Method for Whole-Genome Association Analysis,” Nature Genetics, 44, 1166–1170. [2] [DOI] [PubMed] [Google Scholar]
- Svishcheva GR, Belonogova NM, and Axenovich TI (2014), “FFB-SKAT: Fast Family-Based Sequence Kernel Association Test,” PLoS One, 9(6), e99407. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- — (2015), “Region-Based Association Test for Familial Data Under Functional Linear Models,” PLoS One, 10, e0128999. [2,6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, and Kang HM (2012), “Evolution and Functional Impact of Rare Coding Variation From Deep Sequencing of Human Exomes,” Science, 337, 64–69. [1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium (2007), “A Second Generation Human Haplotype Map of Over 3.1 Million SNPs,” Nature, 449, 851–861. [8] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton T, and McPeek MS (2007), “Case-Control Association Testing With Related Individuals: A More Powerful Quasi-Likelihood Score Test,” The American Journal of Human Genetics, 81, 321–337. [4,6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez AI, Bates DM, Rosa GJM, Gianola D, and Weigel KA (2010), “Technical Note: An R Package for Fitting Generalized Linear Mixed Models in Animal Breeding,” Journal of Animal Science, 88, 497–504. [7,8] [DOI] [PubMed] [Google Scholar]
- Vsevolozhskaya OA, Zaykin DV, Barondess DA, Tong X, Jadhav S, and Lu Q (2016), “Uncovering Local Trends in Genetic Effects of Multiple Phenotypes via Functional Linear Models,” Genetic Epidemiology, 40, 210–221. [2,6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vsevolozhskaya OA, Zaykin DV, Greenwood MC, Wei C, and Lu Q (2014), “Functional Analysis of Variance for Association Studies,” PLoS One, 9, e105074. [2,6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J (2016), “Pedigrees or Markers: Which Are Better in Estimating Inbreeding and Relationship Coefficient?,” Theoretical Population Biology, 107, 4–13. [12] [DOI] [PubMed] [Google Scholar]
- Wang X, Lee S, Zhu X, Redline S, and Lin X (2013), “GEE-Based SNP Set Association Test for Continuous and Discrete Traits in Family-Based Association Studies,” Genetic Epidemiology, 37, 778–786. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YF, Liu A, Mills JL, Boehnke M, Wilson AF, Bailey-Wilson JE, Xiong M, Wu CO, and Fan R (2015), “Pleiotropy Analysis of Quantitative Traits at Gene Level by Multivariate Functional Linear Models,” Genetic Epidemiology, 39, 259–275. [2,6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Mah TS, Paul TO, Morse L, Ngo-Chang J, Dailey JP, Ferrell RE, and Gorin MB (2000), “A Full Genome Scan for Age-Related Maculopathy,” Human Molecular Genetics, 9, 1329–1349. [2,3] [DOI] [PubMed] [Google Scholar]
- Weeks DE, Conley YP, Tsai HJ, Mah TS, Schmidt S, Postel EA, Agarwal A, Haines JL, Pericak-Vance MA, Rosenfeld PJ, and Paul TO (2004), “Age-Related Maculopathy: A Genomewide Scan With Continued Evidence of Susceptibility Loci Within the 1q31, 10q26, and 17q25 Regions,” The American Journal of Human Genetics, 75, 174–189. [2,3] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, and Lin X (2011), “Rare-Variant Association Testing for Sequencing Data With the Sequence Kernel Association Test,” The American Journal of Human Genetics, 89, 82–93. [2,9] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q, Tiwari HK, Yi N, Gao G, Zhang K, Lin WY, Lou XY, Cui X, and Liu N (2015), “A Sequence Kernel Association Test for Dichotomous Traits in Family Samples Under a Generalized Linear Mixed Model,” Human Heredity, 79, 60–68. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, and Goddard ME (2010), “Common SNPs Explain a Large Proportion of the Heritability for Human Height,” Nature Genetics, 42, 565–569. [12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, and Visscher PM (2011), “GCTA: A Tool for Genome-Wide Complex Trait Analysis,” The American Journal of Human Genetics, 88, 76–82. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Weedon MN, Purcell S, Lettre G, Estrada K, Willer CJ, Smith AV, Ingelsson E, O’Connell JR, Mangino M, and Mägi R (2011), “Genomic Inflation Factors Under Polygenic Inheritance,” European Journal of Human Genetics, 19, 807–812. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zaitlen NA, Goddard ME, Visscher PM, and Price AL (2014), “Advantages and Pitfalls in the Application of Mixed-Model Association Methods,” Nature Genetics, 46, 100–106. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, and Kresovich S (2006), “A Unified Mixed-Model Method for Association Mapping That Accounts for Multiple Levels of Relatedness,” Nature Genetics, 38, 203–208. [2] [DOI] [PubMed] [Google Scholar]
- Zhang F, Boerwinkle E, and Xiong M (2014), “Epistasis Analysis for Quantitative Traits by Functional Regression Models,” Genome Research, 24, 989–998. [2,6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, and Buckler ES (2010), “Mixed Linear Model Approach Adapted for Genome-Wide Association Studies,” Nature Genetics, 42, 355–360. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhu Y, and Xiong M (2016), “Genome-Wide Gene-Gene Interaction Analysis for Next-Generation Sequencing,” European Journal of Human Genetics, 24, 421–428. [2,6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Aranzana MJ, Kim S, Lister C, Shindo C, Tang C, Toomajian C, Zheng H, Dean C, Marjoram P, and Nordborg M (2007), “An Arabidopsis Example of Association Mapping in Structured Samples,” PLoS Genetics, 3, e4. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, and Stephens M (2012), “Genome-Wide Efficient Mixed-Model Analysis for Association Studies,” Nature Genetics, 44, 821–824. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- — (2014), “Efficient Multivariate Linear Mixed Model Algorithms for Genome-Wide Association Studies,” Nature Methods, 11, 407–409. [2] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.