Abstract
Allostery is a fundamental property of proteins, which regulates biochemical information transfer between spatially distant sites. Here, we report on the critical role of molecular dynamics (MD) simulations in discovering the mechanism of allosteric communication within CRISPR-Cas9, a leading genome editing machinery with enormous promises for medicine and biotechnology. MD revealed how allostery intervenes during at least three steps of the CRISPR-Cas9 function: affecting DNA recognition, mediating the cleavage and interfering with the off-target activity. An allosteric communication that activates concerted DNA cleavages was found to led through the L1/L2 loops, which connect the HNH and RuvC catalytic domains. The identification of these “allosteric transducers” inspired the development of novel variants of the Cas9 protein with improved specificity, opening a new avenue for controlling the CRISPR-Cas9 activity. Discussed studies also highlight the critical role of the recognition lobe in the conformational activation of the catalytic HNH domain. Specifically, the REC3 region was found to modulate the dynamics of HNH by sensing the formation of the RNA:DNA hybrid. The role of REC3 was revealed to be particularly relevant in the presence of DNA mismatches. Indeed, interference of REC3 with the RNA:DNA hybrid containing mismatched pairs at specific positions resulted in locking HNH in an inactive “conformational checkpoint” conformation, thereby hampering off-target cleavages. Overall, MD simulations established the fundamental mechanisms underlying the allosterism of CRISPR-Cas9, aiding engineering strategies to develop new CRISPR-Cas9 variants for improved genome editing.
Keywords: enhanced sampling techniques, genome editing, graph theory, molecular dynamics, protein/nucleic acid complexes
1 |. INTRODUCTION
CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 is the core of a transformative genome editing technology that is innovating life science with cutting-edge impact in basic and applied sciences.1,2 By enabling the correction of DNA mutations, this technology promises to treat a myriad of human genetic diseases, as recently shown for the first cancer patients treated with CRISPR-Cas9-modified T-cells.3 Because of its adaptability, CRISPR-Cas9 promises unprecedented progresses in biofuel production and agriculture, with drought-resistant crops of enhanced nutritional value.4 The CRISPR-Cas9 genome editing tool is based on the endonuclease enzyme Cas9, which associates with guide RNAs to generate double-strand breaks in DNA via two nuclease domains. At the molecular level, an intricate allosteric signaling controls the CRISPR-Cas9 biochemical information transfer to activate double-stranded DNA cleavages.5 This allosteric communication—which regulates biochemical information transfer between spatially distant sites6,7—is critical for transmitting the DNA binding information, affecting the function and specificity of CRISPR-Cas9.
CRISPR-Cas9 was originally discovered as a bacterial adaptive immune system that enables protection against invading viruses.3 In the CRISPR-Cas9 immune response, parts of invading viral DNA are internalized into the bacterial genome, leading to maturation of RNA sequences that guide the associated Cas proteins toward the degradation of the foreign DNA (Figure 1a).8 The Cas9 protein associates with a CRISPR RNA (crRNA) and a trans-activating CRISPR RNA (tracrRNA), which form a guide for the recognition of matching DNA sequences. Structural studies of the Streptococcus pyogenes CRISPR-Cas9 (SpCas9) complex have revealed that Cas9 is a multi-domain protein composed of a recognition lobe (REC), which mediates the nucleic acid binding through three domains (REC1–3), and a nuclease lobe (NUC, Figure 1b).9–12 The latter includes two catalytic domains, HNH and RuvC, each named based on homologous nucleases. Site-specific recognition of the viral DNA sequence occurs by binding a short Protospacer Adjacent Motif (PAM), which enables the selection of the viral DNA among the genome.3,13 PAM binding occurs within the PAM interacting (PI) domain at the level of the protein C-terminus, triggering the Cas9 function. Indeed, PAM binding was shown to destabilize the adjacent bases, initiating DNA melting and cleavage.10,13,14 This allows the DNA to bind Cas9 by matching the guide RNA with one strand (i.e., the target strand, TS) and forming a 20 base pair RNA:DNA hybrid structure. The other strand (i.e., nontarget strand, NTS) is displaced to also accommodate in the protein (Figure 1a). Then, the Cas9 protein uses the HNH and RuvC domains to concertedly cleave the TS and NTS, respectively. By programming the Cas9 enzyme with a single guide RNA, the system can be used to recognize and cleave any sequence preceding a PAM, thus enabling a wide range of applications.
FIGURE 1.
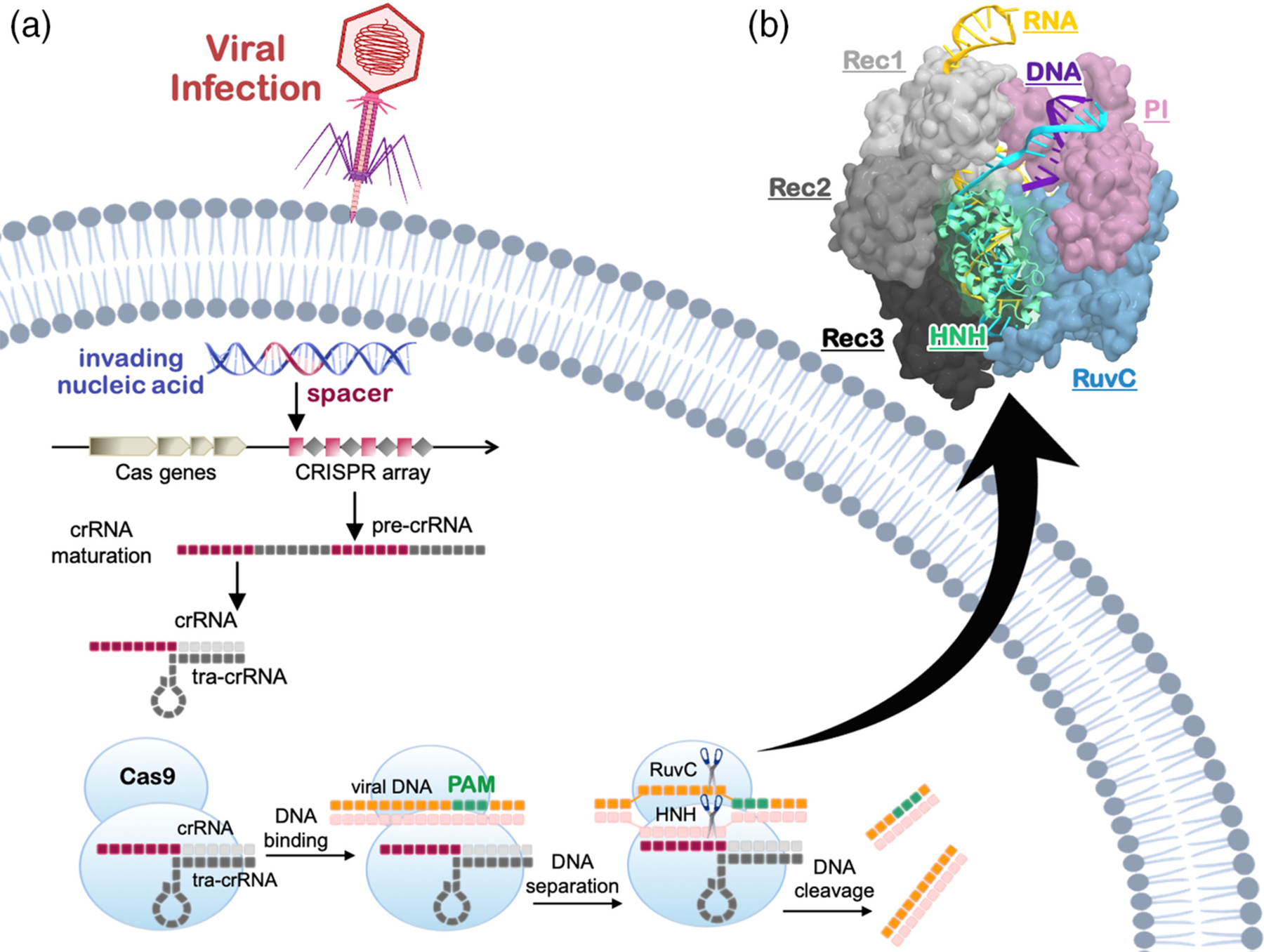
Schematic representation of the CRISPR-Cas adaptive immunity. (a) Upon viral infection, parts of invading nucleic acids are internalized into the CRISPR genetic array that subsequently transcribes and matures into a guide RNA complex containing a CRISPR RNA (crRNA) and trans-CRISPR RNA (tracr-RNA). (1) The Cas9 protein binds the guide RNA and uses its sequence to recognize complementary DNA sequences. Site-specific recognition of the viral DNA is preceded by the binding of a short Protospacer Adjacent Motif (PAM), which enables the selection across the genome. (b) Three-dimensional structure of the Streptococcus pyogenes CRISPR-Cas9 (SpCas9) complex with a guide RNA and DNA (PDB code: 4UN3).10 Cas9 is shown in molecular surface, highlighting individual domains in different colors. The RNA (yellow), target DNA (TS, cyan), and nontarget DNA (NTS, violet) are shown as ribbons
Experimental and computational evidences revealed that allostery emerges at multiple steps of the CRISPR-Cas9 functional cycle, from the early PAM recognition to the concerted cleavages by both nucleases. In the proposed allosteric signaling, the PAM sequence would act as an allosteric activator, triggering concerted cleavages of the spatially distant HNH and RuvC nucleases.13,15 However, an allosteric regulation would also occur after initial PAM recognition.16,17 Accordingly, the interaction between the RNA:DNA hybrid and the REC lobe would trigger the activation of the distally located catalytic domains in an allosteric manner. Finally, an allosteric mechanism is also thought to regulate off-target cleavages,18,19 which lead to mutations at sites in the genome other than the desired target site and are the most severe issue with the application of CRISPR-Cas9 in vivo. Taken together, the abovementioned studies have revealed that the mechanistic function of CRISPR-Cas9 is governed by an intricate allosteric regulation, which also intervenes into the system’s specificity.5,20,21
Here, we report the critical role of computational studies in the discovery of the allosterism in CRISPR-Cas9. To date, molecular dynamics (MD) have shown to be a powerful tool for revealing the allosteric response in biomolecular systems.7,22–30 Indeed, capturing the system fluctuations and conformational changes with atomic resolution, MD can uniquely describe the subtle details accompanying allostery. In the forthcomings, we focus on current findings on how the allosteric mechanisms intervene the three critical steps of the CRISPR-Cas9 function, namely (i) the PAM-mediated activation, (ii) the communication between the REC and NUC lobes governing the cleavage, and (iii) the role of allostery in the onset of off-target effects.
2 |. ROLE OF ALLOSTERY IN THE PAM-MEDIATED ACTIVATION
The binding of PAM is a key prerequisite for DNA binding and cleavage.13 Indeed, PAM recognition initiates DNA binding and activates the NUC lobe of Cas9, triggering concerted cleavages of the TS and NTS by the HNH and RuvC domains respectively. Considering that PAM binds at distal sites (~40 A) with respect to the nuclease domains, it has been proposed that PAM could act as an allosteric activator of the catalytic function.13 This hypothesis has been assessed by using the combination of MD simulations with network analysis models derived from graph theory.
To investigate the dynamic determinant of this allosteric mechanism, extensive simulations (>13 μs) of both Cas9 bound to a “5-TGG-3” PAM sequence (i.e., Cas9 with PAM: Cas9–wPAM),10 and of its analogue crystallized without PAM (Cas9–w/oPAM)31 were examined with principal component analysis (PCA).15 This established method allows extraction of the essential degrees of freedom and the large-scale collective motions of the Cas9-nucleic acid complex.32 The analysis of the conformational space explored along its two principal components of motions (i.e., PC1 and PC2, Figure 2a) revealed that Cas9 adopts distinct conformations when binding with PAM (PC1 < 0) or without PAM (PC1 > 0). Notably, an “open-to-close” conformational transition along PC1 (Figure 2b) is observed, consistently with the conformational change required for nucleic acid binding. This shift in the conformational dynamics upon effector binding has been shown to be typical for the allosteric response,22,30 suggesting that PAM acts as a positive allosteric effector.
FIGURE 2.
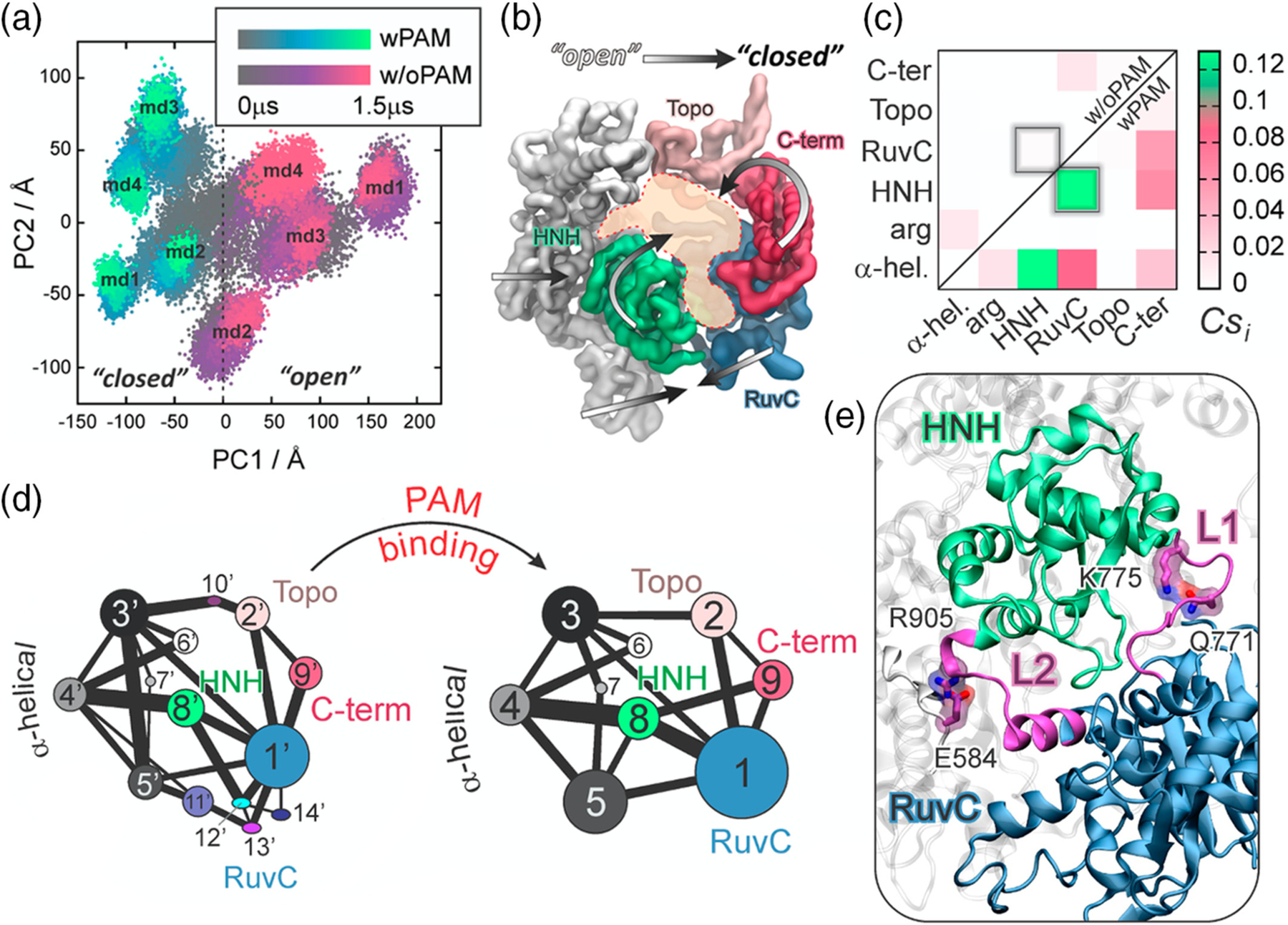
PAM-induced allostery in CRISPR-Cas9. (a) Conformational space adopted by the CRISPR-Cas9 complex spanning over the two principal components of motions (i.e., PC1 vs. PC2), computed over eight independent MD runs of Cas9 bound to PAM (i.e., wPAM) and without PAM (i.e., w/oPAM). (b) “Open-to-close” conformational transition identified along the first principal component. (c) Per-domain correlation score (Csi) matrix, identifying the Cas9 inter-domain coupled motions color-coded green (correlated) to white (not correlated). (d) Community network graph of Cas9–w/oPAM (right) and wPAM (left). Bonds connecting communities correspond to the interconnection strength. (e) Allosteric transducer loops L1/L2 connecting the catalytic HNH and RuvC domain. Critical network nodes (Q771, E584, K775, and R905) of the L1/L2 loops forming essential edges in the allosteric transmission between HNH and RuvC.15 Reprinted with permission from Reference 15 Copyright 2017 American Chemical Society. https://pubs.acs.org/doi/10.1021/jacs.7b05313. Further permissions related to the material excerpted should be directed to the American Chemical Society
Further investigations were performed by applying correlation analysis, which allows identifying coupled dynamics of spatially distant sites, thereby characterizing their possible inter-dependent function. A generalized correlation (GC) method quantified the degree of correlation between residues based on Shannon’s entropy,33 providing a normalized measure of how much information on one atom’s position is provided by that of another atom. The computed GC matrices revealed that the presence of PAM significantly strengthens the correlations between motions of Cas9, as an effect of the dynamic shift induced by the effector (i.e., PAM) binding. To spotlight the sites of high inter-communication, an inter-specific correlation score (Cs) measure was introduced. The Cs accumulates (and normalizes) the per-residue GC, delivering a “coarse” per-domain representation of the system’s correlations. This approach has been efficient in identifying relevant per-domain correlations in large biomolecular systems, such as the spliceosome,34 as well as CRISPR-Cas9.17 Indeed, in large ribonucleoproteins composed of various protein elements, DNA and RNA, a visual inspection of the per-residue GC matrix could hide relevant correlation spots.35 The per-domain Csi matrix revealed that PAM binding to Cas9 results in highly correlated motions between HNH and RuvC, which are not observed in case of Cas9–w/oPAM (Figure 2c). This shows that PAM is key in triggering the interdependent dynamics of HNH and RuvC, required for concerted cleavage of the DNA strands. In the presence of PAM, HNH also correlates strongly with the α-helical REC lobe, supporting the direct information transfer. Such conclusions were also supported by network analysis based on graph theory.36 This analysis builds on the systems’ correlations, which are used to construct a dynamical network model of the biomolecule as a graph of nodes (i.e., amino acid Cα, nucleobases P) and edges, whose distance is weighted as a negative logarithm of the GCs. The resulting weighted graph is used to structure the system’s correlations through community network analysis (CNA), which defines community structures of highly correlated residues and the strength of their inter-correlation, therefore describing the information flow. As a result, PAM binding was shown not only to reduce the number of communities, leading to an increased organization of the system’s correlations (Figure 2d); it also strengthens the correlation between communities #1 and #8 of RuvC and HNH, as represented by a thicker bond between the communities. Contrarily, the communities are fragmented in the absence of PAM, resulting in a weaker inter-correlation between HNH and RuvC. Thus, PAM induces a stronger communication channel between HNH and RuvC that is essential for allosteric signaling. Overall, these findings additionally supports the hypothesis that PAM acts as an allosteric effector, triggering inter-dependent motions of the catalytic domains HNH and RuvC, which are responsible for concerted cleavage of the two DNA strands.13,15
Since PAM binding induced highly coupled motions between the Cas9 nuclease domains, a dynamic allosteric “cross-talk” was proposed to be responsible for concerted DNA cleavages, playing an important role in the enzyme activity.15 To further understand the mechanism of information transfer, the pathways of communication between RuvC and HNH in Cas9-wPAM were investigated from the dynamical network.36 Specifically, the routes of the information transfer between HNH and RuvC were computed as the “shortest pathways” (i.e., the pathways exhibiting the shortest edge lengths) between catalytic residues of RuvC (E762, D986, D10, H983, S15) and HNH (H840). As a result, the most likely allosteric routes were found to lead through L1/L2 loops that connect HNH and RuvC, suggesting their key role as “allosteric transducers” (Figure 2e).11 Inspired by this outcome, recent engineering strategies systematically modified the L1/L2 loops, obtaining the LZ3-Cas937 variant with improved specificity. Analysis of the node betweenness (i.e., the number of shortest pathways that cross the node) was further performed to identify the residues at the core of the information transfer. These critical nodes are pivotal to the dynamical network of communication. This analysis highlighted the critical role of Q771 and E584 (within L1 and L2, respectively) that are engaged in interactions with K775 and R905, forming essential edges of the communication between HNH and RuvC. The role of these essential edges was further confirmed with experiments, in which both K775A and R905A were experimentally shown to change the system’s selectivity.18,38 In summary, these results clearly showed how modification of allosteric signaling open an avenue for controlling the Cas9 activity.
3 |. INFORMATION TRANSFER BETWEEN THE REC AND NUC LOBES GOVERNS DNA CLEAVAGES
Early X-ray structures have captured different CRISPR-Cas9 inactive conformations, with the HNH catalytic residues displaced from the DNA TS.10,31 The system’s activation required a conformational change of HNH, ensuring the TS cleavage. The first all-atom MD simulation study indicated a “striking plasticity” of the HNH domain, suggesting fast conformational transitions.39 The early study also revealed that, only in the presence of the NTS, the HNH domain moved toward the cleavage site on the TS strand, stabilizing at a distance of ~15 Å from the scissile phosphate (Figure 3). This movement toward the TS was facilitated by a number of interactions formed between the L2 loop and the NTS in the preactive state of HNH. This suggested a critical role of the NTS for the activation of HNH toward TS cleavage, supporting also the inter-dependence between the RuvC and HNH function. The key role of the NTS, revealed in this early simulation study, has been subsequently confirmed by single-molecule Förster resonance energy transfer (smFRET) experiments,16 showing that the docking of HNH in its active configuration requires the presence of the NTS. Taken together, that early biophysical studies posed the hypothesis that a high flexibility of the HNH domain allows the system’s activation toward DNA cleavage.
FIGURE 3.
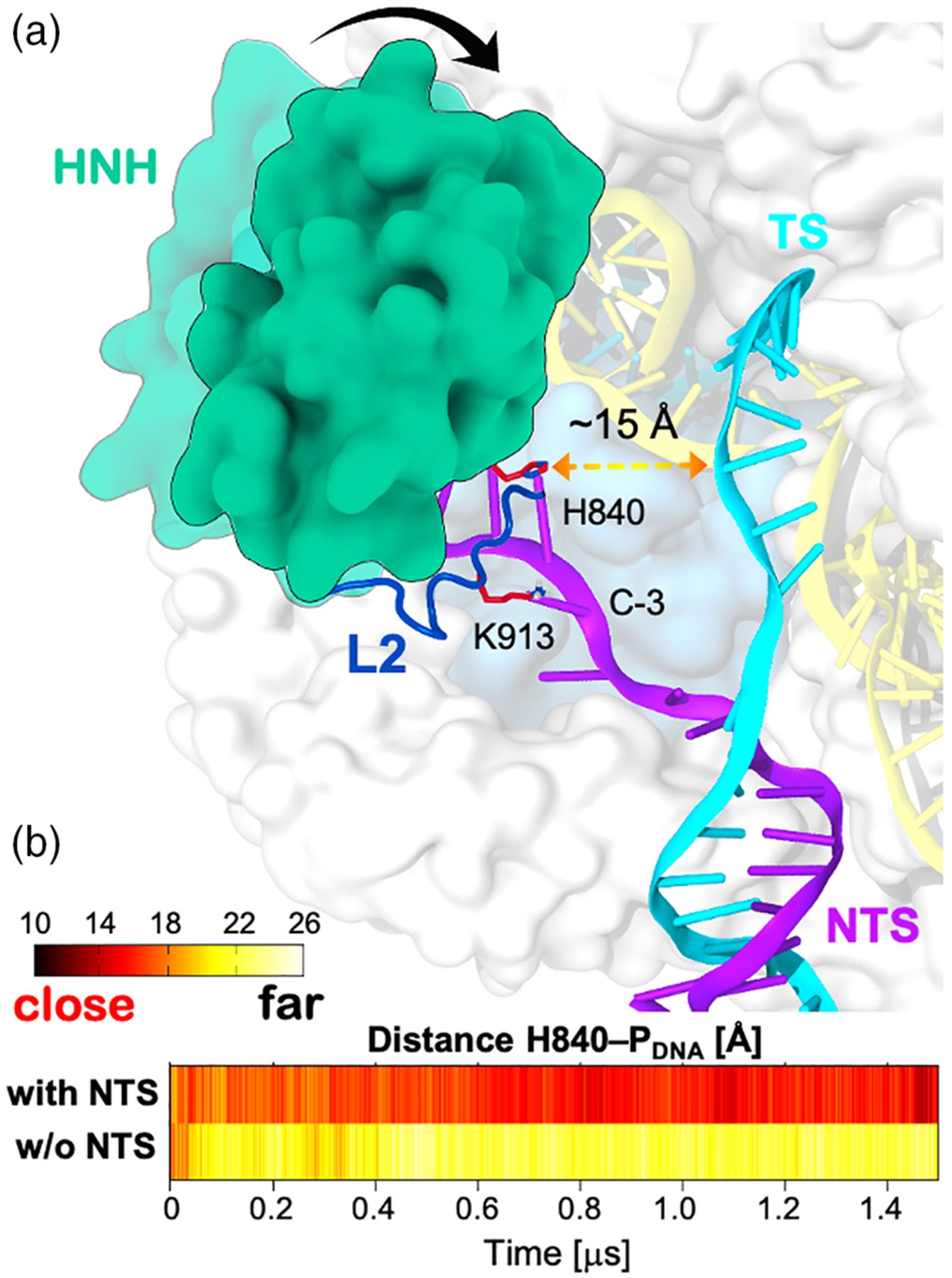
Conformational change of HNH in the presence of the DNA nontarget strand (NTS). (a) Schematic representation of the HNH conformational transition toward the DNA target strand (TS, cyan) in the presence of the NTS (violet). The HNH domain (green) is shown to establish a number of interactions with the DNA NTS through the L2 loop (blue). (b) Time evolution of the distance between the catalytic H840 and the phosphorus atom of the scissile phosphate (PDNA), along MD simulations of CRISPR-Cas9 bound to the NTS (i.e., with NTS) and without the NTS (i.e., w/o NTS).39 Reprinted with permission from Reference 39 Copyright 2016 American Chemical Society. https://pubs.acs.org/doi/full/10.1021/acscentsci.6b00218. Further permissions related to the material excerpted should be directed to the American Chemical Society
smFRET experiments have also revealed that the conformational mobility of HNH is dependent on the motions of the REC lobe, which have been thought to exert an allosteric control of the HNH nuclease conformational activation.16 The activated Cas9 enzyme edits nucleic acids though a metal-dependent nuclease function.40–44 This critical functional aspect was suggested by biochemical experiments, yet the catalytic mechanism of DNA cleavage has only recently been defined.45,46 To investigate the mechanism of conformational activation leading to the active state, MD simulations have been performed employing a novel accelerated MD method, namely a Gaussian accelerated MD (GaMD).47 The method adds a harmonic boost potential to the simulation, favoring transitions between low-energy states. In this way, it is possible to capture micro-to-millisecond (and in some cases beyond) conformational changes without any predefined collective variable in large biomolecular systems.48–51 These simulations extensively sampled the possible configurations of the HNH domain, and were first in identifying a putative structure of the active state, which was shown to be thermodynamically stable (Figure 4a).52 This configuration predicted the conformation of the active state before structural data were made available, enabling also early investigations of the active site chemistry though quantum-classical methods.45 A remarkable agreement with the structure of the activated complex was recently found through cryo electron microscopy (cryoEM),12 showing the reliability of the initial computational predictions based on MD simulations.52 To describe in detail the transition toward the active conformation, classical MD simulations have also been carried out on a specialized supercomputer Anton-2, enabling over ~16 μs of continuous sampling.17 As a result, the transition of HNH toward the cleavage site on the TS occurred concertedly with a large-scale opening of REC2 and REC3 (i.e., upon ~7 μs, Figure 4b). Notably, after ~12 μs of continuous run the fluctuations of REC2–3 reached equilibrium in agreement with previous smFRET identifying the activated REC2 and REC3. Importantly, the activated state found through these continuous MD simulations was in line with the structure subsequently obtained with CryoEM.12
FIGURE 4.
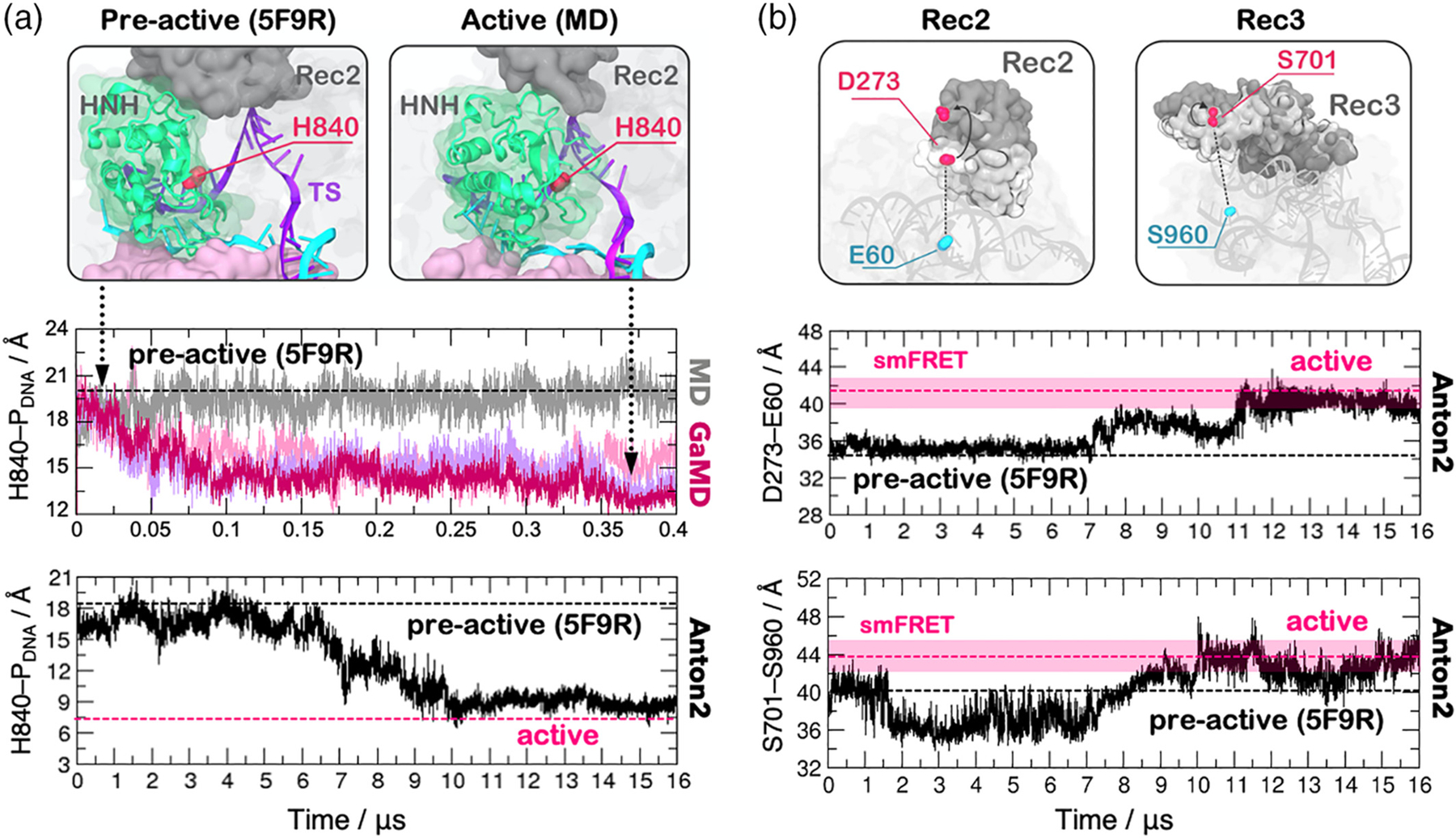
HNH domain activation observed through long time scale MD simulations. (a) The HNH domain (green) is shown to approach the catalytic site on the DNA target strand (TS), changing conformation from a preactive X-ray structure (PDB code: 5F9R) to the active state identified by MD (top panel). The time evolution of the distance between the catalytic H840 and the scissile phosphate (H840–PDNA) is computed along ~400 ns Gaussian accelerated MD (GaMD, central panel) and along ~16 μs of a continuous MD simulation using Anton2 (bottom panel). A black dashed line indicates the preactive conformation (PDB code: 5F9R), which has been used as a starting point for MD simulations. (b) Conformational change of REC2 and REC3 during the HNH activation, identified by single molecule FRET (smFRET) experiments (top panel). The evolution along a ~16 μs MD run of the smFRET pairs D273–E60 (middle panel) and S701–S960 (bottom panel). Transparent bars (pink) indicate the experimental distribution of the smFRET distances in the activated conformation.17 Reprinted with permission from Reference 17 Copyright 2018 Cambridge University Press. https://doi.org/10.1017/S0033583518000070
Analysis of the correlations in the activated state through the GC method revealed highly coupled motions between HNH and the REC2–3 regions. The highest per-domain inter-dependence was measured for REC3 and HNH (Cs = 0.64), as an evidence that REC3 could allosterically modulate the dynamics of HNH by “sensing” (i.e., binding) the RNA: DNA hybrid (Figure 5). This provided a plausible explanation for the experimental observation that REC3 allows HNH nuclease activation upon recognizing the formation of the RNA:DNA hybrid.16 This analysis also revealed that REC2 highly couples with HNH, suggesting its possible role into the allosteric transmission between REC3 and HNH. Indeed, by interacting with HNH, REC2 could transfer the allosteric motions of REC3 and contribute to the HNH activation by “regulating” its conformational transition. The high cooperativity of REC2 and HNH was also evinced through smFRET, showing that these domains undergo reciprocal conformational changes.18 This suggested that the two domains can cooperatively relocate to facilitate catalysis,53 likely in response to the “sensing” of REC3. The simulation also suggested an unexpected possible role for REC1. Indeed, during the activation process, a number of newly formed ionic interactions were observed between HNH and REC1, “locking” HNH at the cleavage site. However, the role of REC1 in the conformational activation and allostery of HNH is yet to be experimentally established. In summary, these simulations revealed that the REC2–3 regions allosterically regulate the activation of HNH toward the TS cleavages, rationalizing earlier smFRET findings.
FIGURE 5.
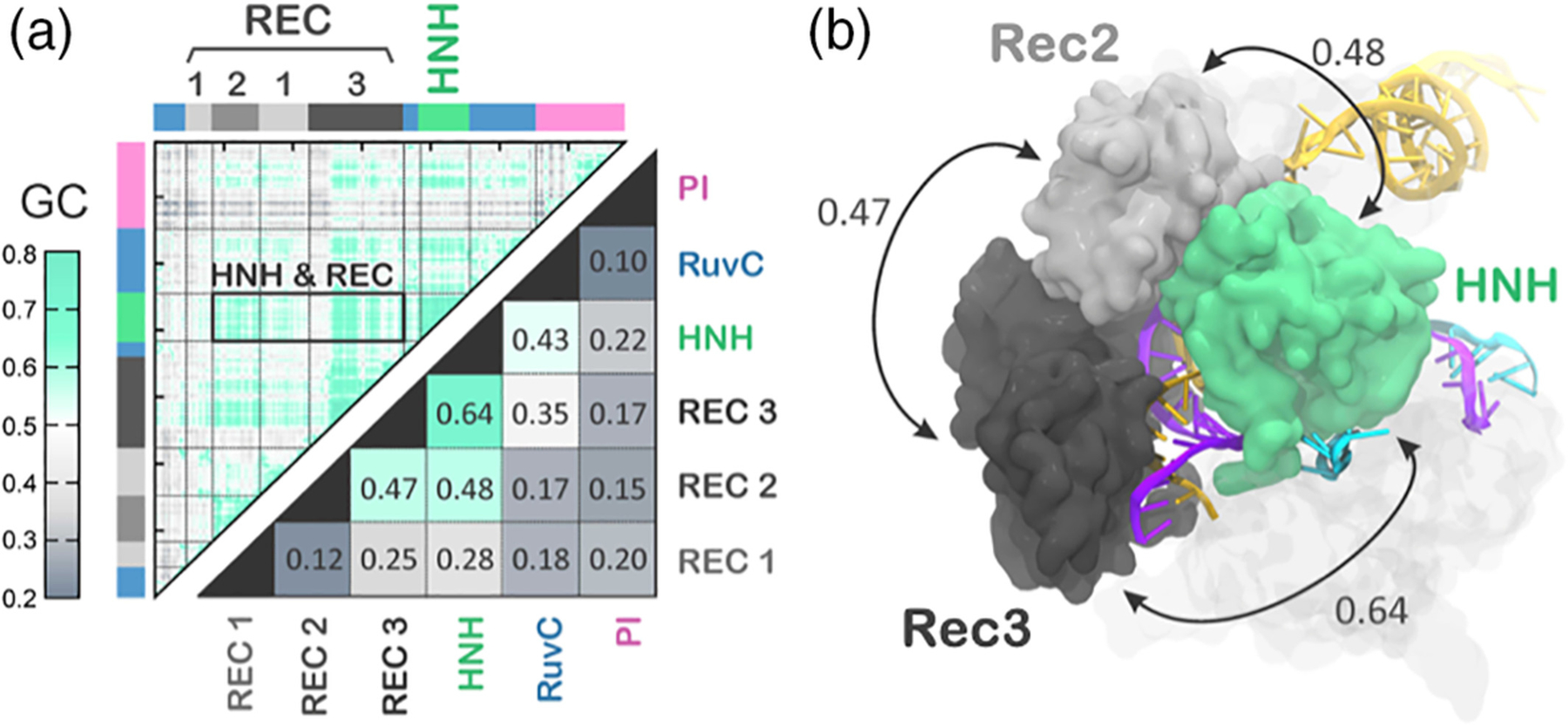
Interdependent dynamics of HNH and the REC2–3 regions. (a) Matrices of the generalized correlations (GC, upper triangle) and of the per-domain correlation score (Csi) (bottom triangle) computed for the activated state of CRISPR-Cas9 identified by MD. The strength of the correlated motions is color coded green (highly correlated motions) to gray (not correlated). (b) The highest per-domain coupled motions, involving HNH and the Rec2–3 regions, are reported using double-headed arrows.17 Reprinted with permission from Reference 17 Copyright 2018 Cambridge University Press. https://doi.org/10.1017/S0033583518000070
Upon activation of HNH, the allosteric signaling would end its function activating RuvC, through the HNH-RuvC dynamic “cross-talk” (Figure 2e).15 To ultimately track the allosteric routes transferring the information from REC to HNH and ultimately RuvC, GaMD was combined with graph theory.54 At first, a series of ~400 ns GaMD simulations were successfully used to recover the transitions of HNH observed over μs using Anton2 (Figure 4a). Then, the simulated ensemble was subjected to the analysis of the allosteric signaling through graph theory. This combination enabled to derive the allosteric signaling reflecting the long timescale conformational changes of the HNH domain. The Dijkstra’s algorithm,55 which is widely used in cartography to find the shortest roads leading to the desired destination, was used to compute the shortest communication pathways between residues belonging to HNH but adjacent to RuvC and REC respectively (i.e., 789/794 and 841/858). As a result, the most likely allosteric routes revealed the existence of a dynamic pathway that crosses through HNH from the REC lobe to the RuvC nuclease. To counterproof the identified theoretical pathway, NMR relaxation experiments have been employed, as a powerful tool to experimentally define allosteric dynamical motions.56–58 The residues involved in this allosteric pathway were further shown by NMR experiments to give rise to slow millisecond-length relaxations, thereby supporting the theoretical finding of a long timescale dynamical route of communication between REC and RuvC.54 Notably, the theoretical pathway also encompassed two critical lysine residues (K855, K810), which anchor HNH at the DNA strands and whose alanine mutation is key for the enhanced specificity Cas9 variants.38 This further supports the hypothesis that protein allostery intervenes with the specificity of Cas9, supporting the early suggestion that the allosterism can be modulated to improve the CRISPR-Cas9 function.15
4 |. ROLE OF ALLOSTERY IN THE ONSET OF OFF-TARGET EFFECTS
An allosteric response is also involved in the regulation of off-target effects, which occur when Cas9 cleaves DNA sequences that do not fully match the guide RNA.59 Kinetic and smFRET studies showed that DNAs containing one to three mismatches at the RNA:DNA hybrid ends (i.e., at PAM distal sites) allow enough flexibility in HNH to cleave the off-target sequence, while four mismatches could trap HNH in an inactive state.18,60 This inactive state of HNH is a “conformational checkpoint” between DNA binding and cleavage, in which the RNA:DNA complementarity is recognized before HNH could assume an active configuration (Figure 6a).10 In this scenario, it has been unknown how DNA mismatches at these sites could favor the inactivation of HNH. Detailed molecular knowledge of this mechanism is of major importance for developing more specific Cas9 systems, in which a single base pair mismatch is sufficient for trapping HNH in the “conformational checkpoint,” thus preventing the cleavage of any incorrect DNA sequence.
FIGURE 6.
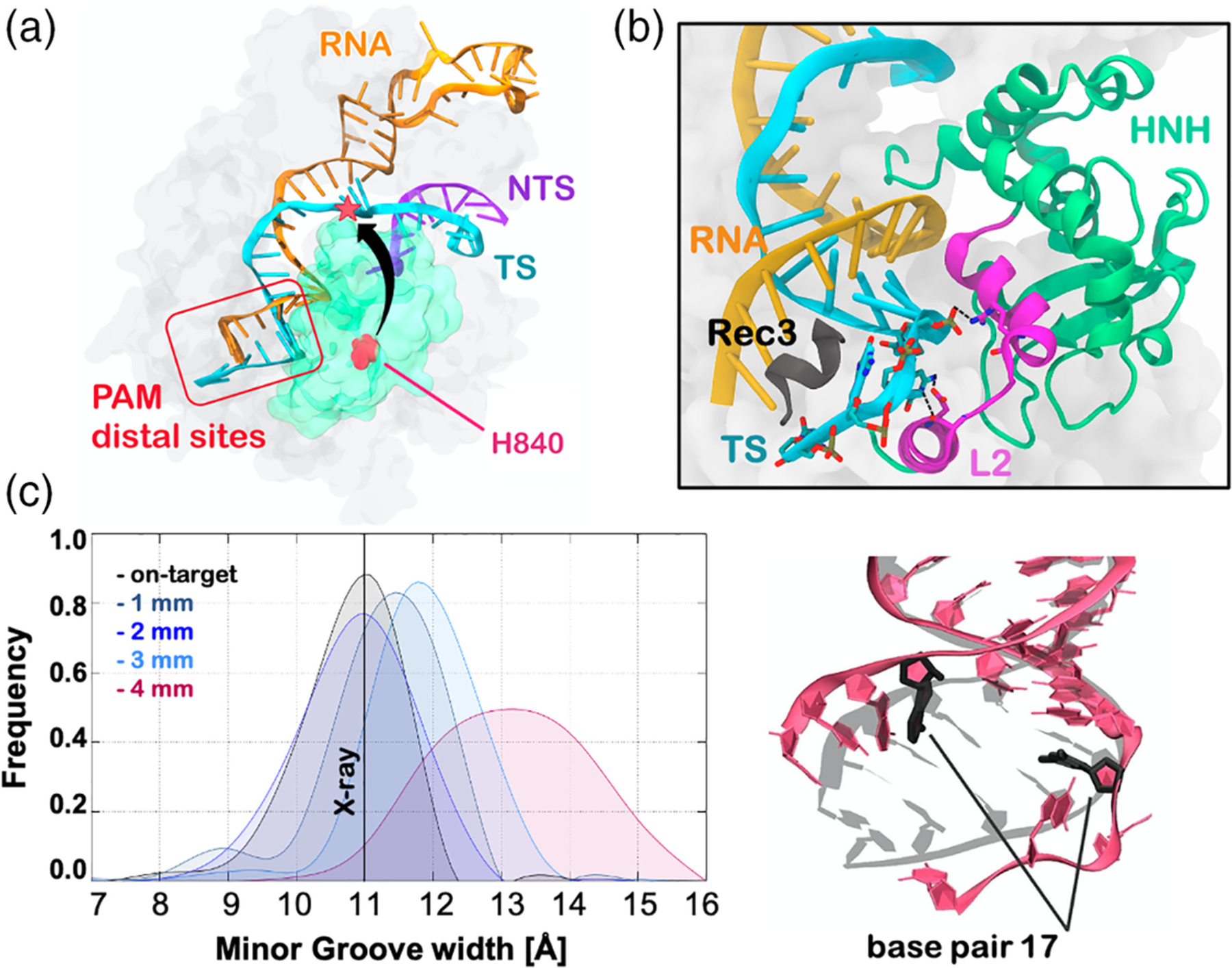
Conformational basis of off-target effects. (a) Crystal structure of CRISPR-Cas9 in a “conformational checkpoint” state (PDB code: 4UN3).10 The arrow indicates the conformational change required by the inactive HNH to cleave the DNA target strand (TS). (b) Extended opening of the RNA:DNA hybrid and newly formed interactions with the L2 loop (magenta), observed during MD simulations of CRISPR-Cas9 in the presence of four base pair mismatches at PAM distal sites. The 692–700 α-helix of the Rec3 region (black) is shown to insert within the within the RNA:DNA, promoting its extended opening. (c) RNA:DNA minor groove width computed along MD simulations of CRISPR-Cas9 bound to an on-target DNA (black) and in the presence of one to four PAM distal mismatches (left panel). A vertical bar indicates the experimental minor groove width (i.e., 11 Å from x-ray crystallography). The minor groove width has been measured at the level of base pair 17 (right panel).19 Reprinted with permission from Reference 19 Copyright 2019 American Chemical Society. https://pubs.acs.org/doi/full/10.1021/acscentsci.9b00020. Further permissions related to the material excerpted should be directed to the American Chemical Society
To investigate the origin of the off-target interference with the HNH conformational dynamics, MD simulations were employed by using GaMD simulations,19 which was shown above to successfully describe the HNH activation process.52 The considered systems were composed of Cas9 in the “conformational checkpoint” state, thereby exploring the activation process in the presence of a fully matching DNA and introducing one to four mismatches at PAM distal sites. As a result, the on-target DNA revealed remarkable stability, maintaining its Watson-Crick base pairing, consistent with earlier simulations of CRISPR-Cas9. This was somehow unexpected, since transient openings at the end of a DNA duplex is a commonplace over long timescale MD.61–63 In the presence of one-to-three DNA mismatches, the Watson-Crick base pairing was diminished, yet the overall conformation of the RNA:DNA hybrid was preserved. On the contrary, four distal mismatches led to an extended opening of the RNA:DNA hybrid, in-line with the experimental fact that four mismatches lead to an inactivation of the enzyme (Figure 6b,c).18,60 This extended opening of the RNA:DNA hybrid resulted in newly formed interactions between the TS and the L2 loop of the HNH domain. These interactions sensibly reduced the HNH flexibility, locking HNH in the “conformational checkpoint” state. On the other hand, DNA mismatches responsible for off-target cleavages are unable to “lock” the HNH domain, thereby leading to the unselective cleavage of DNA sequences. Taken together, these outcomes provided a mechanistic rationale clarifying the onset of off-target effects, also suggesting that structural modifications of the L2 loop changing its ability to interact with the DNA TS could improve the specificity of Cas9 toward on-target sequences. Accordingly, modifications of the L2 loop were shown to aid in increasing the specificity of the LZ3-Cas9 variant.37
Molecular simulations also revealed that there is a significant role of REC3 in allosterically “locking” HNH. Indeed, in the presence of four distal mismatches, REC3 was shown to insert it 692–700 α-helix within the RNA:DNA hybrid, resulting in a remarkable increase of interactions (Figure 6b). On the other hand, in the on-target Cas9, as well as in the presence of one to three distal mismatches, the 692–700 α-helix does not insert within the hybrid. Hence, when bound to four distal mismatches, the 692–700 α-helix contributed to the heteroduplex opening and, in turn, establishment of interactions between the TS and the L2 loop of the HNH domain. This suggested that the REC3 region would allosterically affect the HNH activation by interfering with the dynamics of the RNA:DNA hybrid. Accordingly, the residues that are directly participating in the interaction between REC3 and the hybrid (N692, M694, Q695, and H698) were also experimentally found to play a critical role in the Cas9 specificity toward on-target sequences.18 This clarified how REC3 could act as an allosteric regulator of HNH by “sensing” the RNA:DNA hybrid, as hypothesized previously.17,18 MD studies also focused on the effect of DNA mismatches within the RNA:DNA heteroduplex, reaching upstream positions with respect to PAM distal ends.64 Couples of base-pair mismatches were introduced starting from the PAM distal ends up to the middle of the 20 base pairs RNA:DNA structure (i.e., at position 10). The simulations revealed that mismatched pairs at upstream positions (i.e., from position 10–14) are easily incorporated within the hybrid structure, with minor effect on the protein-nucleic acid interactions. Hence, the HNH conformational dynamics was not affected by the presence of DNA mismatches fully embedded in the RNA:DNA. This outcome was in line with the experimental evidence that mismatches within the heteroduplex are more tolerated, emphasizing the role of REC3 in the allosteric control of the HNH conformational activation.18,60
5 |. CONCLUSIONS
In this review article, we discuss how allostery intervenes into the DNA recognition and cleavage by CRISPR-Cas9, a leading genome editing machinery. We report the critical role of MD simulations in discovering the mechanism of allosteric communication within Cas9, and how these methods contributed in establishing the allosteric regulation of function and selectivity. We show that allostery intervenes during at least three steps of the CRISPR-Cas9 function, affecting DNA recognition,13,15 mediating the cleavage16,17 and interfering with the off-target activity.18,19,64 At first, the binding of the PAM recognition sequence was found not only to be necessary to initiate DNA unwinding, but is also critical for the activation of the distally spaced nuclease domains HNH and RuvC.13,15 The concerted DNA cleavages governed by this PAM-mediated allosteric signaling were found to pass through the L1/L2 loops, which act as “allosteric transducers” that connect the HNH and RuvC catalytic domains.16,17 These computational findings inspired engineering strategies focused on the L1/L2 loops, obtaining the LZ3-Cas937 variant with improved specificity, and also clarified how modifications of the allosteric signaling open an avenue for controlling the Cas9 activity. A similar allosteric control orchestrating interdependent domain dynamics was also identified in the activation of yet another CRISPR-Cas system, viz. Cas12a, upon binding with the target dsDNA.65 An allosteric regulation was also found to occur upon initial PAM recognition, governing the conformational mobility of the catalytic HNH domain.16,17 Specifically, the recognition lobe was found to exert an allosteric control of the HNH activation through the REC2 and REC3 regions. Indeed, a large-scale opening of REC2 and REC3 led HNH to adopt a catalytically active conformation, identified 2 years earlier than cryoEM with remarkable agreement.12 Of note, highly coupled dynamical motions of REC3 and HNH suggested that REC3 could allosterically modulate the dynamics of HNH by “sensing” the RNA:DNA hybrid.17 This was especially visible in the presence of DNA mismatches located in the proximity of REC3 within the RNA:DNA hybrid.19 Indeed, REC3 was shown to insert the critical 692–700 α-helix within the RNA:DNA hybrid, contributing to the opening of heteroduplex and its interaction with HNH. This suggested that, by “sensing” the RNA:DNA duplex, REC3 would allosterically control the activation of HNH. As a further support, base pair mismatches away from REC3 were observed to have minor effect on the HNH dynamics.58
Overall, molecular simulation studies established the fundamental mechanisms underlying the allosterism of CRISPR-Cas9, revealing also how allostery intervenes in the system’s selectivity. The recalled studies highlight how the combination of MD with enhanced sampling and graph theory-based methods holds an invaluable promise to decipher allosteric mechanisms and their consequences in biological systems. Future methodological improvements will further contribute to the understanding of protein allostery over multiple timescales, providing a more comprehensive overview of the allostery phenomena.
ACKNOWLEDGMENTS
The authors thank all the investigators who have contributed to the studies discussed in this advanced review article. GP thanks her group for human support during the difficult and delicate period of stay-at-home following the COVID-19 outbreak. This material is based on the work supported by the National Science Foundation under Grant No. CHE-1905374. This work was also partially funded by the National Institute of Health through the Grant No. R01 EY027440. Computer time for the Molecular Dynamics simulations studies reviewed in this article was provided by The Extreme Science and Engineering Discovery Environment (XSEDE) through the grant TG-MCB160059.
Footnotes
CONFLICT OF INTEREST
The authors have declared no conflict of interest for this article.
FURTHER READING
For a broad audience of readers, including high school students and non-experts, we suggest our article in Physics Today.21
REFERENCES
- 1.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014;346(6213): 1258096. [DOI] [PubMed] [Google Scholar]
- 2.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belhaj K, Chaparro-Garcia A, Kamoun S, Patron NJ, Nekrasov V. Editing plant genomes with CRISPR/Cas9. Curr Opin Struct Bio. 2015. 2019;(36)76–84. [DOI] [PubMed] [Google Scholar]
- 5.Chen JS, Doudna JA. The chemistry of Cas9 and its CRISPR colleagues. Nat Rev Chem. 2017;1(10):1–5. [Google Scholar]
- 6.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J Mol Bio. 1965;12(1):88–118. [DOI] [PubMed] [Google Scholar]
- 7.Liu J, Nussinov R. Allostery: An Overview of Its History, Concepts, Methods, and Applications. PLoS Comput Bio 2016;12(6):e1004966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang F, Doudna JA. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol. 2015;30:100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343(6176):1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513(7519):569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, Doudna JA. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351(6275):867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Z Zhu X, Clarke R, Puppala AK, Chittori S, Merk A, Merrill BJ, Simonović M, Subramaniam S. Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat Struct Mol Bio. 2019;26(8):679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mekler V, Minakhin L, Severinov K. Mechanism of duplex DNA destabilization by RNA-guided Cas9 nuclease during target interrogation. Proc Natl Acad Sci USA. 2017;114(21):5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palermo G, Ricci CG, Fernando A, Basak R, Jinek M, Rivalta I, Batista VS, McCammon JA. Protospacer adjacent motif-induced allostery activates CRISPR-Cas9. J Am Chem Soc. 2017;139(45):16028–16031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dagdas YS, Chen JS, Sternberg SH, Doudna JA, Yildiz A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Science adv. 2017;3(8):eaao0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palermo Palermo G, Chen JS, Ricci CG, Rivalta I, Jinek M, Batista VS, Doudna JA, McCammon JA. Key role of the REC lobe during CRISPR–Cas9 activation by ‘sensing’, ‘regulating’, and ‘locking’ the catalytic HNH domain. Q Rev Biophys. 2018;51:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, Doudna JA. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature. 2017;550(7676):407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricci CG, Chen JS, Miao Y, Jinek M, Doudna JA, McCammon JA, Palermo G. Deciphering off-target effects in CRISPR-Cas9 through accelerated molecular dynamics. ACS Cent Sci. 2019;5(4):651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuo Z, Liu J. Allosteric regulation of CRISPR-Cas9 for DNA-targeting and cleavage. Curr Opn Struct Bio. 2020;62:166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palermo G, Ricci CG, McCammon JA. The invisible dance of CRISPR-Cas9. Simulations unveil the molecular side of the gene-editing revolution. Phys Today. 2019;72(4):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo J, Zhou HX. Protein allostery and conformational dynamics. Chem Rev. 2016;116(11):6503–6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wodak Wodak SJ, Paci E, Dokholyan NV, Berezovsky IN, Horovitz A, Li J, Hilser VJ, Bahar I, Karanicolas J, Stock G, Hamm P. Allostery in its many disguises: from theory to applications. Structure. 2019;27(4):566–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papaleo E, Saladino G, Lambrughi M, Lindorff-Larsen K, Gervasio FL, Nussinov R. The role of protein loops and linkers in conformational dynamics and allostery. Chem Rev. 2016;116(11):6391–6423. [DOI] [PubMed] [Google Scholar]
- 25.Dokholyan NV. Controlling allosteric networks in proteins. Chem Rev. 2016;116(11):6463–6487. [DOI] [PubMed] [Google Scholar]
- 26.Vendruscolo M The statistical theory of allostery. Nat Chem Bio. 2011;7(7):411–412. [DOI] [PubMed] [Google Scholar]
- 27.Bowerman S, Wereszczynski J. Detecting allosteric networks using molecular dynamics simulation. Methods in enzymology. 2016;578: 429–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner JR, Lee CT, Durrant JD, Malmstrom RD, Feher VA, Amaro RE. Emerging computational methods for the rational discovery of allosteric drugs. Chem Rev. 2016;116(11):6370–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Q, Karplus M. Allostery and cooperativity revisited. Prot Sci. 2008;17(8):1295–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao XQ, Hamelberg D. Detecting functional dynamics in proteins with comparative perturbed-ensembles analysis. Acc Chem Res. 2019; 52(12):3455–3464. [DOI] [PubMed] [Google Scholar]
- 31.Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156(5):935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daidone I, Amadei A. Essential dynamics: foundation and applications. WIRE: Comput. Mol. Sci 2012;2:762–770. [Google Scholar]
- 33.Lange OF, Grubmüller H. Generalized correlation for biomolecular dynamics. Prot Struct Func Bioinfo. 2006;62(4):1053–1061. [DOI] [PubMed] [Google Scholar]
- 34.Casalino L, Palermo G, Spinello A, Rothlisberger U, Magistrato A. All-atom simulations disentangle the functional dynamics underlying gene maturation in the intron lariat spliceosome. Proc Natl Acad Sci USA. 2018;115(26):6584–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palermo G, Casalino L, Magistrato A, McCammon JA. Understanding the mechanistic basis of non-coding RNA through molecular dynamics simulations. J Struct Biol. 2019;206(3):267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sethi A, Eargle J, Black AA, Luthey-Schulten Z. Dynamical networks in tRNA: protein complexes. Proc Natl Acad Sci USA. 2009;106 (16):6620–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmid-Burgk JL, Gao L, Li D, Gardner Z, Strecker J, Lash B, Zhang F. Highly parallel profiling of Cas9 variant specificity. Mol Cell. 2020;78(4):794–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palermo G, Miao Y, Walker RC, Jinek M, McCammon JA. Striking plasticity of CRISPR-Cas9 and key role of non-target DNA, as revealed by molecular simulations. ACS Cent Sci. 2016;2(10):756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palermo G, Cavalli A, Klein ML, Alfonso-Prieto M, Dal Peraro M, De Vivo M. Catalytic metal ions and enzymatic processing of DNA and RNA. Acc Chem Res. 2015;48(2):220–228. [DOI] [PubMed] [Google Scholar]
- 41.Donati E, Genna V, De Vivo M. Recruiting Mechanism and Functional Role of a Third Metal Ion in the Enzymatic Activity of 5′ Structure-Specific Nucleases. J Am Chem Soc. 2020;142(6):2823–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosta E, Nowotny M, Yang W, Hummer G. Catalytic mechanism of RNA backbone cleavage by ribonuclease H from quantum mechanics/molecular mechanics simulations. J Am Chem Soc. 2011;133(23):8934–8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Vivo M, Dal Peraro M, Klein ML. Phosphodiester cleavage in ribonuclease H occurs via an associative two-metal-aided catalytic mechanism. J Am Chem Soc. 2008;130(33):10955–10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alonso-Cotchico L, Rodríguez-Guerra J, Lledós A, Maréchal JD. Molecular Modeling for Artificial Metalloenzyme Design and Optimization. Acc Chem Res. 2020;53(4):896–905. [DOI] [PubMed] [Google Scholar]
- 45.Palermo G Structure and dynamics of the CRISPR–Cas9 catalytic complex. J Chem Inf Model. 2019;59(5):2394–2406. [DOI] [PubMed] [Google Scholar]
- 46.Casalino L, Nierzwicki Ł, Jinek M, Palermo G Catalytic Mechanism of Non-Target DNA Cleavage in CRISPR-Cas9 Revealed by Ab-Initio Molecular Dynamics. ACS Catal. 2020. 10.1021/895acscatal.0c03566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miao Y, Feher VA, McCammon JA. Gaussian accelerated molecular dynamics: Unconstrained enhanced sampling and free energy calculation. J Chem Theo Comput. 2015;11(8):3584–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhattarai A, Devkota S, Bhattarai S, Wolfe MS, Miao Y. Mechanisms of γ-Secretase Activation and Substrate Processing. ACS Cent Sci. 2020;6(6):969–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miao Y, McCammon JA. Mechanism of the G-protein mimetic nanobody binding to a muscarinic G-protein-coupled receptor. Proc Natl Acad Sci. 2018;115(12):3036–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sibener LV, Fernandes RA, Kolawole EM, Carbone CB, Liu F, McAffee D, Birnbaum ME, Yang X, Su LF, Yu W, Dong S. Isolation of a structural mechanism for uncoupling T cell receptor signaling from peptide-MHC binding. Cell. 2018;174(3):672–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chuang CH, Chiou SJ, Cheng TL, Wang YT. A molecular dynamics simulation study decodes the Zika virus NS5 methyltransferase bound to SAH and RNA analogue. Sci Rep. 2018;8(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palermo G, Miao Y, Walker RC, Jinek M, McCammon JA. CRISPR-Cas9 conformational activation as elucidated from enhanced molecular simulations. Proc Natl Acad Sci USA. 2017;114(28):7260–7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sung K, Park J, Kim J, Lee NK, Kim SK. Target specificity of Cas9 nuclease via DNA rearrangement regulated by the REC2 domain. J Am Chem Soc. 2018;140:7778–7781. [DOI] [PubMed] [Google Scholar]
- 54.East KW, Newton JC, Morzan UN, Narkhede YB, Acharya A, Skeens E, Jogl G, Batista VS, Palermo G, Lisi GP. Allosteric motions of the CRISPR–Cas9 HNH nuclease probed by NMR and molecular dynamics. Journal of the American Chemical Society. 2019;142(3):1348–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dijkstra EW. A note on two problems in connexion with graphs. Numerische Mathematik. 1959;1(1):269–271. [Google Scholar]
- 56.Tzeng SR, Kalodimos CG. Protein dynamics and allostery: an NMR view. Curr Opn Struct Biol. 2011;21(1):62–67. [DOI] [PubMed] [Google Scholar]
- 57.East KW, Skeens E, Cui JY, Belato HB, Mitchell B, Hsu R, Batista VS, Palermo G, Lisi GP. NMR and computational methods for molecular resolution of allosteric pathways in enzyme complexes. Biophys Rev. 2020;12:155–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lisi GP, Loria JP. Solution NMR spectroscopy for the study of enzyme allostery. Chem Rev. 2016;116(11):6323–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang M, Peng S, Sun R, Lin J, Wang N, Chen C. The conformational dynamics of Cas9 governing DNA cleavage are revealed by single-molecule FRET. Cell Rep. 2018;22(2):372–382. [DOI] [PubMed] [Google Scholar]
- 61.Mura C, McCammon JA. Molecular dynamics of a κB DNA element: base flipping via cross-strand intercalative stacking in a microsecond-scale simulation. Nucleic Acid Res. 2008;36(15):4941–4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ricci CG, de Andrade AS, Mottin M, Netz PA. Molecular dynamics of DNA: comparison of force fields and terminal nucleotide definitions. J Phys Chem B. 2010;114(30):9882–9893. [DOI] [PubMed] [Google Scholar]
- 63.Pérez A, Luque FJ, Orozco M. Dynamics of B-DNA on the microsecond time scale. J Am Chem Soc. 2007;129(47):14739–14745. [DOI] [PubMed] [Google Scholar]
- 64.Mitchell BP, Hsu RV, Medrano MA, Zewde NT, Narkhede YB, Palermo G. Spontaneous embedding of DNA mismatches within the RNA: DNA hybrid of CRISPR-Cas9. Front Mol Biosci. 2020;7:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saha A, Arantes PR, Hsu RV, Narkhede YB, Jinek M, Palermo G. Molecular Dynamics Reveals a DNA-Induced Dynamic Switch Triggering Activation of CRISPR-Cas12a. J. Chem. Inf. Model 2020. DOI: 10.1021/acs.jcim.0c00929. [DOI] [PMC free article] [PubMed] [Google Scholar]


