Abstract
An adverse maternal environment (AME) predisposes adult offspring toward cognitive impairment in humans and mice. However, the underlying mechanisms remain poorly understood. Epigenetic changes in response to environmental exposure may be critical drivers of this change. Epigenetic regulators, including microRNAs, have been shown to affect cognitive function by altering hippocampal neurogenesis which is regulated in part by brain-derived neurotropic factor (BDNF). We sought to investigate the effects of AME on miR profile and their epigenetic characteristics, as well as neurogenesis and BDNF expression in mouse hippocampus. Using our mouse model of AME which is composed of maternal Western diet and prenatal environmental stress, we found that AME significantly increased hippocampal miR-10b-5p levels. We also found that AME significantly decreased DNA methylation and increased accumulations of active histone marks H3 lysine (K) 4me3, H3K14ac, and H3K36me3 at miR-10b promoter. Furthermore, AME significantly decreased hippocampal neurogenesis by decreasing cell numbers of Ki67<sup>+</sup> (proliferation marker), NeuroD1<sup>+</sup> (neuronal differentiation marker), and NeuN<sup>+</sup> (mature neuronal marker) in the dentate gyrus (DG) region concurrently with decreased hippocampal BDNF protein levels. We speculate that the changes in epigenetic profile at miR-10b promoter may contribute to upregulation of miR-10b-5p and subsequently lead to decreased BDNF levels in a model of impaired offspring hippocampal neurogenesis and cognition in mice.
Keywords: Adverse maternal environment, MicroRNA-10b, Brain-derived neurotropic factor, Hippocampal neurogenesis, Epigenetics
Introduction
Adverse maternal environment (AME) and the consumption of a Western diet (WD) in early childhood increases the risk for cognitive impairment later in life in both humans [1, 2, 3] and animal models [4, 5, 6]. Indeed, using our mouse model we have demonstrated that AME together with postweaning WD impairs learning and memory function in adult male mice [7]. Unfortunately, how early life insults affect cognition functions later in life remain poorly understood. One important mechanism is epigenetic changes which can occur in response to environmental exposure [8]. Epigenetic changes include DNA methylation, histone modifications, and microRNAs (miRs). Interestingly, miRs are key players in epigenetic regulation under stress condition [9]. Furthermore, miRs have been shown to affect cognitive function in neurodegenerative disorders [10, 11, 12].
miRs act as key regulators of different biological functions including neurogenesis. miRs can indirectly influence neurogenesis by regulating the proliferation and self-renewal of neural stem cells [13]. Importantly, decreased neurogenesis and neuronal death in the hippocampus are the characteristics of impaired cognitive function [14, 15, 16, 17]. Furthermore, regulation of hippocampal neurogenesis depends in part upon brain-derived neurotrophic factor (BDNF) levels. BDNF is a member of the neurotrophic family of growth factors and acts to support the survival of existing neurons, promote neuronal differentiation, and maturation of newborn neurons [18, 19]. Reductions in the levels of BDNF have been shown to significantly impair the production of new neurons [20, 21, 22, 23, 24], whereas its direct infusion into the hippocampus has been demonstrated to increase neurogenesis [25]. In addition, BDNF protects hippocampal neurons against apoptosis [19]. Interestingly, BDNF also is a target of miRs [26].
miRs are gene expression modulators, acting at the posttranscriptional level, and fine-tuning the expression of protein-encoding genes. miRs modulate gene expression by binding to 3′ untranslated region (3′ UTR) of target mRNAs and leading to translational inhibition or mRNA degradation [27, 28]. The BDNF gene 3′ UTR region contains numerous target sites for miRNA binding, which have been confirmed for miRNA-BDNF interactions [29]. BDNF protein levels have been shown to negatively correlate with miRs expression [27, 28, 29, 30]. For example, miR-1, miR-10b, miR-155, and miR-191 were found to directly repress BDNF through binding to their predicted sites in BDNF 3′ UTR [26]. Specifically, overexpression of miR-1 and miR-10b both suppressed BDNF protein levels while mutations of their binding sites prevented repression [29].
miRs are short single-stranded noncoding RNAs about 22 nucleotides in length. Mature miRs are generated from pre-miRs either 5′ arm or 3′ arm. The 5′ arm of the miRNA is designated miR-5p present in the forward (5′-3′) position, while the 3′ arm is miR-3p located in the reverse position. Both are functional. Like protein-coding genes, regulation of miRs' expression involves epigenetic mechanisms [31, 32, 33]. DNA hypermethylation at the promoter region leads to miR silencing [34, 35]. Histone modifications, including histone acetylation (ac) and methylation, affect gene transcription. For example, histone H3 lysine (K) 9ac, H3K14ac, and H3K4 trimethylation (me3) are often associated with gene activation, while H3K36me3 is associated with actively transcribed regions [36, 37]. Moreover, H3K9me3 and H3K27me3 are associated with gene silencing [36]. These histone marks are also vulnerable to early life insults [38, 39].
However, the effects of AME on miRs' expression and their epigenetic profile in the hippocampus are unknown. Understanding the effects of AME on hippocampal miRs' expression may provide mechanistic insight into the regulation of hippocampal neurogenesis and subsequent cognitive impairment. We therefore hypothesized that AME would alter the expression of miRs in the hippocampus. We further hypothesized that changes in miRs' expression would be associated with alterations in miR epigenetic patterns, decreased hippocampal neurogenesis, and decreased BDNF levels.
Methods
Animals
All experiments were conducted according to the Public Health Services Policy on Human Care and Use of Laboratory Animals and all procedures were approved by the Medical College of Wisconsin Institutional Animal Care and Use Committee [40]. The mouse model of AME used in this study has been previously described [41]. Briefly, AME was induced in mice by exposing six-week-old C57/Bl6 female mice randomly to either a control diet (10% fat without cholesterol and sucrose, Research Diet Inc., New Brunswick, NJ, USA, Product# D14020502) or a WD (40% fat, composed of increased saturated fat, cholesterol, and sucrose, Research Diet Inc., Product# D12079B) for 5 weeks prior to pregnancy and throughout lactation. Dams in the control diet group experienced a normal environment throughout pregnancy and are designated as control (Con). Dams fed a WD experienced a “stressed” environment in the last third of pregnancy. The combination of chronic WD and gestational stress is designated as AME. The stressed environment consisted of daily random environmental changes as well as a static change in the maternal environment consisting of 1/3 of the standard amount of bedding from embryonic day (E) 13–E19. The acute random environmental changes included altered light cycles on 3 nonconsecutive days, 3 repeat cages throughout the day on E15, and the short-term introduction of a novel object in the cage for a day. The period of prenatal stress was limited to E13–E19 to minimize newborn mortality, avoid interference with implantation, and still target a period of rapid development and environmental vulnerability. Dams delivered spontaneously, and litters were culled to 6 pups. At postnatal day 21 (P21), male pups from both Con and AME groups were anesthetized and killed. Brains were quickly removed, hippocampi were dissected, flash frozen in liquid nitrogen, and stored in −80°C for molecular studies. For immunohistochemistry studies, animals were individually fixed via intracardiac perfusion with ice-cold 0.9% normal saline (VWR, Radnor, PA, USA), followed by ice-cold 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA, USA) for 5 min each for a total volume of 10–15 mL fixative. Whole brains were removed and postfixed at 4°C overnight. Brains were then transferred to 70% ethanol, paraffin embedded, and sectioned coronally at 4 μm per section. The reason for choosing to study juvenile hippocampus was because we wanted to examine immediate maternal effect on neurogenesis and its regulation in the hippocampus. A total of 40 pregnant female mice were used in the study with N = 6 dams per group.
RNA Isolation and Real Time RT-PCR
Total hippocampal RNA was extracted from P21 old mice using miRNeasy Micro Kit (Qiagen, Germantown, MD, USA) and quantified using the BioTek Cytation 5 microplate reader (Fisher Scientific Inc., Pittsburgh, PA, USA). RNA was treated with DNase I (Ambion, Austin, TX, USA). Gel electrophoresis was used to confirm the integrity of the samples. miR cDNAs were synthesized from total RNA using TaqMan® Advanced miRNA cDNA Synthesis Kit (Thermo Fisher Scientific). Real-time PCR for miRNA assays were performed according to TaqMan® advanced miRNA assays user guide using ABI ViiA7 system (Thermo Fisher Scientific). Relative quantification of PCR products was based on value differences between the target and internal control using the comparative CT method (Taqman Gold RT-PCR manual, PE Biosystems, Foster City, CA, USA). Cycle parameters were 50°C, 2 min; 95°C, 10 min; and then 40 cycles at 95°C, 15 s; 60°C, 60 s. For each set of reactions, samples were run in quadruplicate. Expression levels of miRs known to affect cognitive function and BDNF levels [26, 42, 43, 44] were tested, including miR-1a-3p (mmu482914_mir), miR-10b-5p (mmu478494_mir), miR-10b-3p (mmu481501_mir), miR-155-5p (mmu480953_mir), miR-155-3p (mmu481328_mir), miR-183-5p (mmu482690_mir), miR-191-5p (mmu481584_mir), miR-191-3p (mmu480970_mir), and miR-206-3p (mmu481645_mir). Expression was calculated relative to hsa-miR-320a (478594-mir, TaqMan® advanced miRNA assays user guide, Thermo Fisher Scientific) which was used as an internal control. cDNA synthesis and real-time RT-PCR were performed as previously described [45]. mRNA expression of BDNF (Mm04230607_s1, Thermo Fisher Scientific) was calculated relative to hypoxanthine phosphoribosyltransferase 1 (HPRT1, Mm. PT.39a.22214828, Integrated DNA Technologies) which was used as an internal control.
DNA Isolation and Pyrosequencing
Hippocampal DNA was extracted from P21 old mice using DNeasy® Blood & Tissue kit (Qiagen, Cat#69504). DNA was subjected to sodium bisulfite treatment using Epitech fast DNA bisulfite kit (Qiagen, Cat#59824) as per the manufacturer's protocol to determine site-specific CpG methylation. DNA methylation of the validation-set samples was determined through PCR amplification with biotinylated primers (Intergrated DNA Technologies, Coralville, IA, USA). Primers were designed based on the genomic sequence obtained from Ensembl Genome Browser (Ensembl: ENSMUSG00000065500) using PyroMark Assay Design Software version 2.0. Amplified products were confirmed with agarose gel electrophoresis. The percent of methylation was quantified by PyroMark Q48 Autoprep Pyrosequencer (Qiagen, Valencia, CA, USA). Four primer sets (Table 1) were used to examine methylation status of 12 CpG sites in the promoter region of miR-10b, spanning from −1 to −540bp nucleotide position from transcription start site which was set as +1.
Table 1.
Primers for pyrosequencing
| Forward | Reverse | Sequencing |
|---|---|---|
| Set 1 5ʹ GATAGTTTTAGTTTAGGGAGGTT | 5ʹ Biosg/ATAAATAATCACAACTAATCCTCCTAAAA | 5ʹ AGTAGTAGTTATAGTAGTAGTAGA |
| Set 2 5ʹ GGGTTTTTGTTTTTTTTTGGGAAATATT | 5ʹ Biosg/AACTAAACAACCAAAACCTTCT | 5ʹ GGAGATTTGGGTTTTGA |
| Set 3 5ʹ AAGAAGGTTTTGGTTGTTTAGT | 5ʹ Biosg/CTACCAAACCTAATCTCTTCAACACA | 5ʹ GGTGGTTTTTATTTTGGTAA |
| Set 4 5ʹ TTTGAATGTTTGTTTTGGGTAGG | 5ʹ Biosg/ATCTATAACTATATAAATACCACACAAATT | 5ʹ GTTTGTTTTGGGTAGGA |
Chromatin Isolation and Chromatin Immunoprecipitation Assay
Hippocampal chromatin was isolated from P21 old mice using truChIPTM chromatin shearing tissue kit (#520237, Covaris Inc, MA, USA) and focused-ultrasonicator M220 (Covaris Inc.). Chromatin immunoprecipitation (ChIP) assays with antibodies against histone H3 lysine (K) 4 trimethylation (H3K4me3, #9751, Cell Signaling Technology), H3K9me3 (#13969, Cell Signaling Technology), H3K27me3 (#9733, Cell Signaling Technology), H3K36me3 (#4909, Cell Signaling Technology), H3K9ac (#9649, Cell Signaling Technology), and H3K14ac (#7627, Cell Signaling Technology) were performed using SimpleChIP® Plus Enzymatic Chromatin IP kit (#9005, Cell Signaling Technology). Real-time PCR was used to quantitate the amount of immunoprecipitation DNA at miR-10b promoter region and an intergenic region upstream of IGF gene which was used as an internal control [46]. Primers for miR-10b promoter and the intergenic region are list in Table 2. Two control experiments were performed simultaneously with our ChIP experiments. First, we performed a “mock” ChIP that included input but did not utilize antibody. Second, we performed a ChIP that utilized an anti-rabbit secondary antibody as negative control. Two percent of input was used as loading control.
Table 2.
Primers for ChIP assays
| Forward | Reverse | Probe | |
|---|---|---|---|
| miR-10b promoter | 5ʹ CTACCTGCACCATCTCTGAAAG | 5ʹ TCCTACCCAGAGCAGACATT | 5ʹ ACCTGATCTCTTCAGCACAGCCAC |
| Intergenic region | 5ʹ TTTGGAACCACCATTTTCTACGT | 5ʹ GCTCAATAGATTTGATGGGCTTACT | 5ʹ ACACCAGGCACTATT |
ChIP, chromatin immunoprecipitation; miR, microRNA.
Immunofluorescence
Immunofluorescent triple labeling was used to compare neurogenesis in dentate gyrus (DG) region of hippocampus between Con and AME groups using cell proliferation marker Ki67 (#9129, Cell Signaling, Danvers, MA, USA), neuronal differentiation marker NeuroD1 (ab60704, Abcam, Cambridge, MA, USA), immature neuronal marker doublecortin (DCX, #4604, Cell Signaling, Danvers, MA, USA), and neuronal marker (NeuN, EMD Millipore, Billerica, MA, USA, Cat#MAB377). Briefly, anterior hippocampal coronal sections around bregma −1.94 area from the same brains were deparaffinized, rehydrated, and subject to antigen retrieval treatment. Sections were then blocked with Protein Block (DAKO, Carpinteria, CA, USA, Cat #x090930-2) for 30 min at room temperature (RT) and followed by incubation with either a mixture of rabbit anti-Ki67 1:200 and mouse anti-NeuN or a mixture of rabbit anti-DCX 1:200 and mouse anti-NeuroD11:750 for 60 min at RT. After washing in TBST twice, sections were then exposed to a mixture of donkey anti-rabbit AF488 1:750 and donkey anti-mouse Cy3 (Jackson ImmunoResearch) 1:750 or a mixture of donkey anti-rabbit Cy3 and donkey anti-mouse Cy51:750 for 45 min at RT. Followed by washing in TBST twice, sections were counterstained for nuclei with DAPI. Sections were washed and mounted with Prolong Gold anti-fade mounting media. Images were captured by confocal microscopy (Carl Zeiss LSM510, Jena, Germany). Positive cells for Ki67, NeuroD1, DCX, NeuN, and DAPI staining on the digital images with ×20 magnification were automated counted by ImageJ software. Two sections/pup, 5 animals from different litters per group were blindly quantified. The counts for Ki67+, NeuroD1+, and NeuN+ were normalized to DAPI count while DCX+ count was normalized to DAPI total area.
Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling
Apoptosis in the DG region of hippocampus from P21 Con and AME brain was determined using DeadEndTM Fluorometric TUNEL System (G3250, Promega, Madison, WI, USA). In brief, anterior hippocampal coronal sections around bregma −1.94 area were deparaffinized, rehydrated, and subject to terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) following the manufacturer's instructions. Images were captured by confocal microscopy (Carl Zeiss LSM510, Jena, Germany). TUNEL positive cells on the digital images with ×20 magnification were automated counted by ImageJ software and normalized by DAPI count. Two sections/pup, n = 5 animals from different litters per group were blindly quantified.
Protein Isolation and Immunoblot
Hippocampal tissues from P21 old mice were homogenized in ice-cold RIPA lysis buffer (Amresco LLC, Solon, OH, USA) with protease and phosphatase inhibitors. After centrifugation at 10,000 rpm at 4°C for 15 min, the supernatants were removed and stored at −80°C until use. Protein concentrations were determined by the BCA method (Pierce, Rockford, IL, USA). Antibody against BDNF (ThermoFisher Scientific, Cat# 710306) at 1:500 dilution was used to determine protein abundance and vinculin (Cell Signaling, Cat #13901) at 1:20,000 dilution was used as a loading control. Immunoblot analyses were performed by a capillary immunoblot analysis method using the ProteinSimpleTM Wes system according to the manufacturer's instructions (ProteinSimpleTM, Santa Clara, CA, USA) as previously described [45].
Statistics
GraphPad Prism 6 (GraphPad Software, San Diego, CA) was used to perform all analyses. All data presented are expressed as mean ± SD. ANOVA (Fisher's protected least significant difference) and Mann-Whitney test determined statistical significance. Significance was set as p < 0.05.
Results
AME Increased Hippocampal miR-10b-5p Levels
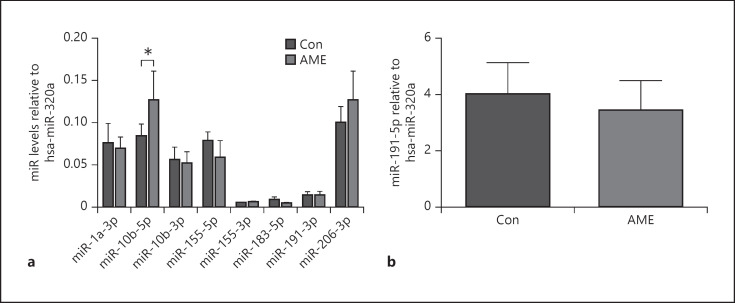
We first examined the effect of AME on miRs' profile. Interestingly, AME significantly increased hippocampal miR-10b-5p levels but not other miRs that were examined (shown in Fig. 1a, b). Given that BDNF is a direct target of miR-10b-5p, as well as a critical player in neuronal function, we then next determined BDNF expression in the hippocampus.
Fig. 1.
AME increased miR-10b-5p levels in the hippocampus. Data are presented as mean ± SD. a miR levels. b miR-191-5p levels. N = 6 animals from different litters/group. *p < 0.05. AME, adverse maternal environment; Con, control; miR, microRNA.
AME Decreased Hippocampal BDNF Protein Abundance
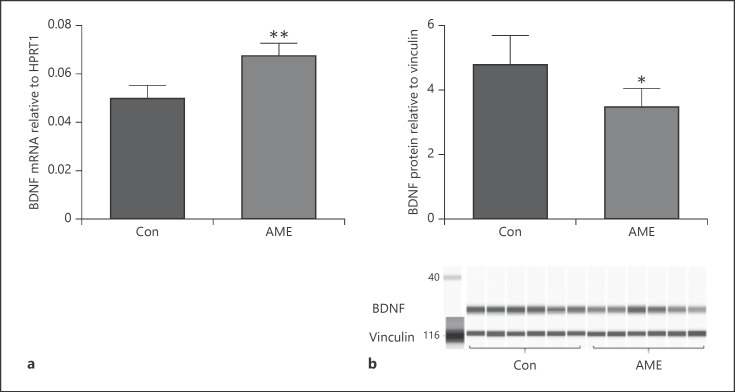
AME significantly decreased hippocampal BDNF protein abundance (shown in Fig. 2b) though AME increased BDNF mRNA levels (shown in Fig. 2a). However, the contrary results between BDNF protein and mRNA levels suggest that miR-driven posttranscriptional regulation may be involved in BDNF protein production. Specifically, increased miR-10b-5p may contribute to reduced BDNF protein levels in our model.
Fig. 2.
AME altered BDNF expression in the hippocampus. Data are presented as mean ± SD. a BDNF mRNA levels in Con versus AME mice. b BDNF protein levels in Con versus AME mice. N = 6 animals from different litters/group. *p < 0.05, **p < 0.01. AME, adverse maternal environment; BDNF, brain-derived neurotropic factor; Con, control.
AME Decreased Neurogenesis and Increased Apoptosis in Hippocampal DG Region
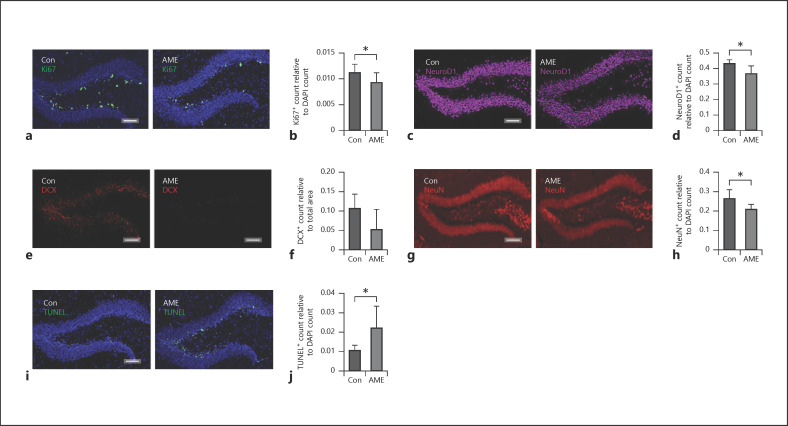
Both miRs and BDNF are key regulators of neurogenesis [13]. Importantly, impaired hippocampal neurogenesis and apoptosis are characteristics of cognitive impairment [16, 17]. Furthermore, hippocampal neurogenesis postnatally occurs at the subgranular zone in the DG region of hippocampus [47, 48]. We then examined the effect of AME on neurogenesis in hippocampal DG region. We found that AME significantly decreased neurogenesis as indicated by decreased staining of Ki67+ (shown in Fig. 3a, b), NeuroD1+ (Fig. 3c, d), NeuN+ (shown in Fig. 3g, h) and trending to decrease DCX+ staining (shown in Fig. 3e, f) in the DG region. We next determined the effect of AME on apoptosis in the hippocampus using TUNEL assay. We found that AME significantly increased apoptotic cell numbers in the DG region of hippocampus compared to Con (shown in Fig. 3i, j).
Fig. 3.
AME decreased neurogenesis and increased apoptosis in hippocampal DG region. a Cell proliferation marker Ki67 staining (green) in DG of Con (left panel) and AME (right panel) mice. b Quantitative analysis of Ki67+ cells in Con versus. AME mice. c Neuronal differentiation marker NeuroD1 staining (pink) in DG of Con (left panel) and AME (right panel) mice. d Quantitative analysis of NeuroD1+ cells in Con versus. AME mice. e Immature neuronal marker DCX staining (red) in DG of Con (left panel) and AME (right panel) mice. f Quantitative analysis of DCX+ cells in Con versus. AME mice. g Neuronal marker NeuN staining (red) in DG of Con (left panel) and AME (right panel) mice. h Quantitative analysis of NeuN+ cells in Con versus. AME mice. i TUNEL assay showing apoptotic cells (green) in hippocampal DG region of Con (left panel) and AME (right panel) mice. j Quantitative analysis of apoptotic cells in Con versus. AME mice. Data are presented as mean ± SD, n = 5 animals from different litters/group. *p < 0.05, Scale bar = 100 μm. AME, adverse maternal environment; DG, dentate gyrus; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling; Con, control.
AME Altered Epigenetic Characteristics at miR-10b Promoter in the Hippocampus
Like a protein-coding gene, miR expression can be epigenetically regulated [31, 32, 33]. Additionally, measurable epigenetic changes often occur in effected genes following an adverse early life environment [41]. We then determined the effect of AME on DNA methylation status and histone code at miR-10b promoter.
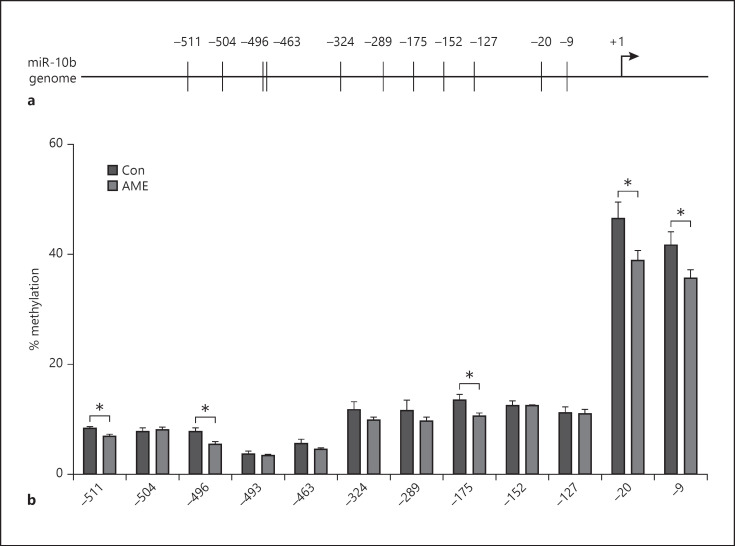
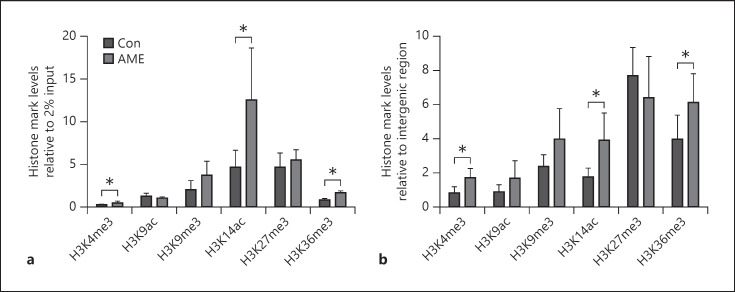
AME significantly decreased CpG methylation at 5 out of 12 sites (−9, −20, −175, −496, and −511 relative to transcription start site) examined at miR-10b promoter compared to Con (shown in Fig. 4). Furthermore, AME significantly increased the densities of active histone marks H3K4me3, H3K14ac, and H3K36me3 at miR-10b promoter compared to Con with no effect on the densities of H3K9ac, H3K9me3, and H3K27me3 (shown in Fig. 5a, b). These data suggest that DNA methylation and histone H3 modifications are involved in miR-10b-5p upregulation.
Fig. 4.
AME decreased CpG methylation at miR-10b promoter in the hippocampus. a Schematic representation of mouse miR-10b proximal promoter. Vertical lines indicate the location of the CpG sites examined relative to translation start site set as +1. b Percent of methylation at 12 CpG sites examined. Data are presented as mean ± SD. N = 6 animals from different litters/group. *p < 0.05. AME, adverse maternal environment; miR, microRNA; Con, control.
Fig. 5.
AME increased active histone marks at miR-10b promoter. a The densities of H3K4me3, H3K9ac, H3K9me3, H3K14ac, H3K27me3, and H3K36me3 were quantified relative to 2% input by ChIP assay. b The densities of H3K4me3, H3K9ac, H3K9me3, H3K14ac, H3K27me3, and H3K36me3 relative to intergenic region. Data are presented as mean ± SD n = 6 animals from different litters/group. *p < 0.05. AME, adverse maternal environment; ChIP, chromatin immunoprecipitation; K, lysine; me3, trimethylation; ac, acetylation.
Discussion
The novel findings of this study are that AME increased miR-10b-5p levels concurrently with decreased BDNF protein abundance and impaired neurogenesis in mouse hippocampus. In addition, increased miR-10b-5p levels were associated with altered epigenetic profile at its promoter. These findings suggest that early life stress and diet endured by the mother affect miR-10b-5p expression via epigenetic mechanism and subsequently alter BDNF levels in the hippocampus, which in turn affect hippocampal neurogenesis and lead to cognitive impairment later in life. To our knowledge, this is the first report that AME affects the epigenetic profile and expression of miR-10b-5p concurrently with neurogenesis impairment in offspring hippocampus.
AME has a profound impact on hippocampal structure and function that subsequently leads to cognitive impairment through epigenetic mechanisms [49]. miRs are key players in epigenetic regulations which often occur in response to environmental exposure and under stress condition [8, 9, 50]. Altered miR expression has been found in the key brain regions related to cognitive function including hippocampus following exposure to early life stress in both humans and animals [9, 50, 51, 52]. However, no studies have reported the relationship between early life stress and miR-10b in the hippocampus. In current study, we found that miR-10b-5p levels were significantly increased in the hippocampus of pups exposed to AME. Our data suggest that upregulated miR-10b-5p expression may play a role in the cognitive impairment previously reported in our model [41].
miRs are gene expression modulators and act at the posttranscriptional level to control protein production of target genes [27, 28]. miR-10b targets the cognition network gene BDNF and is negatively correlated with BDNF levels [53, 54]. For example, Jiang and Zhu [55] have demonstrated that BDNF is a direct target of miR-10b and BDNF expression negatively correlates with the expression of miR-10b in rat hippocampus under chronic stress depression induced by sleep deprivation. Furthermore, BDNF levels have been shown to be posttranscriptionally controlled by upregulated miR-10b-5p in human prefrontal cortex of Huntington disease brain [54]. Whereas no studies have reported the relationship between miR-10b-5p and BDNF in the hippocampus exposed to early life stress. In this study, we found that AME decreased BDNF protein abundance in line with increased miR-10b-5p levels in the hippocampus, suggesting that miR-10b-5p may target BDNF and decrease BDNF protein levels in our model.
BDNF regulates neuronal survival and differentiation [56]. Reduced BDNF expression is associated with neuronal dysfunction and death [48, 49, 57, 58]. Here, we found that AME-induced hippocampal BDNF protein reduction coincides with impaired neurogenesis by decreasing cell proliferation, neuronal differentiation, and mature neuron numbers, as well as increased apoptosis in the hippocampus. Similar findings have been reported in the hippocampus of offspring exposed to other early life stress in animal models [43, 48, 57, 58]. Although maternal WD alone has been shown to have negative effects on BDNF expression and neurogenesis in offspring hippocampus [59, 60], our AME model consists a maternal WD component and did not affect cognitive function in adult animals [7]. We suggest that BDNF protein reduction and neurogenesis impairment seen in our model may result from the effects of the combination of maternal WD and prenatal stress.
miRs' expression can be regulated epigenetically [61]. DNA hypermethylation at the promoter region has been shown to lead to miR silencing [34, 35]. Specifically, DNA CpG methylation in a transcription factor binding site of miR-10b leads to downregulation in a human cervical cancer cell line [62]. Inversely, in our study, we found that AME decreased DNA methylation at limited CpG sites of miR-10b promoter in the hippocampus concurrently with increased miR-10b-5p expression. Interestingly, the promoter region examined in this current study contains a Sp1 binding element at the −324 site examined here. Sp1 is typically a transcriptional activator [63] and enhances transcription by RNA polymerase II [64]. miR genes are transcribed by RNA polymerase II [65]. We speculate that decreased CpG methylation at miR-10b promoter around Sp1 binding site may facilitate Sp1 binding and RNA polymerase II transcriptional machinery assembling and subsequently lead to miR-10b-5p transcriptional activation. Future studies are warranted to explore the role of TFs including Sp1 on miR-10b transcription.
Interestingly, we also found that AME significantly increased accumulations of active histone marks H3K4me3, H3K14ac, and H3K36me3 at the promoter region of miR-10b. Our data suggest that multiple histone modifications are involved in the regulation of miR-10b-5p expression. Though early epigenetic studies focused on the consequences of single-site histone modifications, it is now evident that modifications at different sites influence their effect on transcription, which involves simultaneously reading multiple histone markers [66]. Different histone marks may interact with each other to affect gene transcription. For example, H3K4me3 has been shown to intricately link acetylation at H3K14 by recruiting Ybg1 PHD finger binding and stabilizing the interaction of the scNuA3 HAT leading to hyperacetylation of its substrate [67]. In addition, while H3K36me3 has been observed to be more enriched at body regions of active genes and related to transcriptional elongation [68, 69], H3K36me3 has also been shown to be involved in transcriptional initiation [69, 70]. Specifically, Zhang et al. [70] have shown that H3K36me3 mediated by its methyltransferase Setd2 was enriched at a distal promoter region of a gene of interest and led to transcriptional initiation. Taken together, our data suggest that accumulations of H3K4me3, H3K14ac, and H3K36me3 to miR-10b promoter may work together to favor miR-10b-5p transcription.
Our study has limitations. Whether AME-induced upregulation of miR-10b-5p directly affects BDNF protein production and subsequently impairs neurogenesis in developing hippocampal DG requires further investigations. Moreover, maternal WD or prenatal stress alone may affect epigenetic profile of miR-10b promoter and subsequently alter mir-10b-5p expression, which have not been examined in this study. Additionally, how prenatal maternal stress affects postnatal maternal care behavior and subsequent offspring DG development, as well as how epigenetic changes are maintained during neural stem cell proliferation have not been revealed. All molecular analysis performed in this study utilized whole homogenized hippocampus, thus limiting our ability to understand the contribution of hippocampal subfields but allowed for a comprehensive hippocampal investigation. Last, we recognize that we do not know whether AME-induced upregulation of miR-10b-5p occurs in female offspring, who do not suffer similar cognitive impairment relative to the male counterparts. If AME did not induce upregulation of miR-10b-5p in females, this finding would add circumstantial support to our speculation.
In this study, we have identified that AME increased miR-10b-5p levels concurrently with decreased BDNF protein levels and neurogenesis as well as increased apoptosis in juvenile mouse hippocampus. We speculate that AME increases miR-10b-5p levels by altering epigenetic profile at the promoter region of miR-10b. Increased miR-10b-5p subsequently acts on BDNF to reduce BDNF protein production and leads to decreased neurogenesis in juvenile mouse hippocampus.
Statement of Ethics
All experiments were conducted according to the Public Health Services Policy on Human Care and Use of Laboratory Animals and all procedures were approved by the Medical College of Wisconsin Institutional Animal Care and Use Committee.
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
Funding from Department of Pediatrics, Medical College of Wisconsin.
Author Contributions
X.K., Y.H., Q.F., and A.M. performed experiments; X.K. analyzed data and prepared figures; X.K., R.H.L., and A.M. interpreted results of experiments; X.K. drafted the manuscript; R.H.L. and A.M. edited and revised the manuscript; R.H.L. and A.M. approved the final version of manuscript; A.M., R.H.L., and X.K. conceived and designed research.
Acknowledgement
Immunohistochemistry studies were carried out using the Clinical and Translational Research Core Lab in the Department of Pathology, Medical College of Wisconsin.
References
- 1.Alastalo H, von Bonsdorff MB, Räikkönen K, Pesonen AK, Osmond C, Barker DJ, et al. Early life stress and physical and psychosocial functioning in late adulthood. PLoS One. 2013;8((7)):e69011. doi: 10.1371/journal.pone.0069011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker DJ, Hales CN, Fall CH, Osmond C, Phipps K, Clark PM. Type 2 (non-insulin-dependent) diabetes mellitus, hypertension and hyperlipidaemia (syndrome X): relation to reduced fetal growth. Diabetologia. 1993;36((1)):62–7. doi: 10.1007/BF00399095. [DOI] [PubMed] [Google Scholar]
- 3.Cordner ZA, Khambadkone SG, Boersma GJ, Song L, Summers TN, Moran TH, et al. Maternal high-fat diet results in cognitive impairment and hippocampal gene expression changes in rat offspring. Exp Neurol. 2019;318:92–100. doi: 10.1016/j.expneurol.2019.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arcego DM, Krolow R, Lampert C, Toniazzo AP, Berlitz C, Lazzaretti C, et al. Early life adversities or high fat diet intake reduce cognitive function and alter BDNF signaling in adult rats: Interplay of these factors changes these effects. Int J Dev Neurosci. 2016;50:16–25. doi: 10.1016/j.ijdevneu.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Tozuka Y, Kumon M, Wada E, Onodera M, Mochizuki H, Wada K. Maternal obesity impairs hippocampal BDNF production and spatial learning performance in young mouse offspring. Neurochem Int. 2010;57((3)):235–47. doi: 10.1016/j.neuint.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 6.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7((8)):847–54. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 7.Ke X, Fu Q, Sterrett J, Hillard CJ, Lane RH, Majnik A. Adverse maternal environment and western diet impairs cognitive function and alters hippocampal glucocorticoid receptor promoter methylation in male mice. Physiol Rep. 2020;8((8)):e14407. doi: 10.14814/phy2.14407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolinoy DC. The agouti mouse model: an epigenetic biosensor for nutritional and environmental alterations on the fetal epigenome. Nutr Rev. 2008;66((Suppl 1)):S7–11. doi: 10.1111/j.1753-4887.2008.00056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schouten M, Aschrafi A, Bielefeld P, Doxakis E, Fitzsimons CP. MicroRNAs and the regulation of neuronal plasticity under stress conditions. Neuroscience. 2013;241:188–205. doi: 10.1016/j.neuroscience.2013.02.065. [DOI] [PubMed] [Google Scholar]
- 10.Ku T, Li B, Gao R, Zhang Y, Yan W, Ji X, et al. NF-kappaB-regulated microRNA-574-5p underlies synaptic and cognitive impairment in response to atmospheric PM2.5 aspiration. Part Fibre Toxicol. 2017;14((1)):34. doi: 10.1186/s12989-017-0215-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu P, Venkat P, Chopp M, Zacharek A, Shen Y, Ning R, et al. Role of microRNA-126 in vascular cognitive impairment in mice. J Cereb Blood Flow Metab. 2019;39((12)):2497–511. doi: 10.1177/0271678X18800593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barros-Viegas AT, Carmona V, Ferreiro E, Guedes J, Cardoso AM, Cunha P, et al. miRNA-31 improves cognition and abolishes amyloid-β pathology by targeting app and bace1 in an animal model of Alzheimer's disease. Mol Ther Nucleic Acids. 2020;19:1219–36. doi: 10.1016/j.omtn.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grasso M, Piscopo P, Confaloni A, Denti MA. Circulating miRNAs as biomarkers for neurodegenerative disorders. Molecules. 2014;19((5)):6891–910. doi: 10.3390/molecules19056891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster TC. Biological markers of age-related memory deficits: treatment of senescent physiology. CNS Drugs. 2006;20((2)):153–66. doi: 10.2165/00023210-200620020-00006. [DOI] [PubMed] [Google Scholar]
- 15.Vaynman S, Ying Z, Gomez-Pinilla F. Interplay between brain-derived neurotrophic factor and signal transduction modulators in the regulation of the effects of exercise on synaptic-plasticity. Neuroscience. 2003;122((3)):647–57. doi: 10.1016/j.neuroscience.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 16.Fedorchak AV, Miller MW. Episodic prenatal exposure to ethanol affects postnatal neurogenesis in the macaque dentate gyrus and visual recognition memory. Int J Dev Neurosci. 2019;79:65–75. doi: 10.1016/j.ijdevneu.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Kathner-Schaffert C, Karapetow L, Günther M, Rudolph M, Dahab M, Baum E, et al. Early stroke induces long-term impairment of adult neurogenesis accompanied by hippocampal-mediated cognitive decline. Cells. 2019;8((12)):1654. doi: 10.3390/cells8121654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Numakawa T, Odaka H, Adachi N. Actions of brain-derived neurotrophic factor and glucocorticoid stress in neurogenesis. Int J Mol Sci. 2017;18((11)):2312. doi: 10.3390/ijms18112312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almeida RD, Manadas BJ, Melo CV, Gomes JR, Mendes CS, Grãos MM, et al. Neuroprotection by BDNF against glutamate-induced apoptotic cell death is mediated by ERK and PI3-kinase pathways. Cell Death Differ. 2005;12((10)):1329–43. doi: 10.1038/sj.cdd.4401662. [DOI] [PubMed] [Google Scholar]
- 20.Durany N, Michel T, Kurt J, Cruz-Sánchez FF, Cervás-Navarro J, Riederer P. Brain-derived neurotrophic factor and neurotrophin-3 levels in Alzheimer's disease brains. Int J Dev Neurosci. 2000;18((8)):807–13. [PubMed] [Google Scholar]
- 21.Gil JM, Mohapel P, Araújo IM, Popovic N, Li JY, Brundin P, et al. Reduced hippocampal neurogenesis in R6/2 transgenic Huntington's disease mice. Neurobiol Dis. 2005;20((3)):744–51. doi: 10.1016/j.nbd.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 22.Grote HE, Bull ND, Howard ML, van Dellen A, Blakemore C, Bartlett PF, et al. Cognitive disorders and neurogenesis deficits in Huntington's disease mice are rescued by fluoxetine. Eur J Neurosci. 2005;22((8)):2081–8. doi: 10.1111/j.1460-9568.2005.04365.x. [DOI] [PubMed] [Google Scholar]
- 23.Siegel GJ, Chauhan NB. Neurotrophic factors in Alzheimer's and Parkinson's disease brain. Brain Res Brain Res Rev. 2000;33((2–3)):199–227. doi: 10.1016/s0165-0173(00)00030-8. [DOI] [PubMed] [Google Scholar]
- 24.Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293((5529)):493–8. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 25.Scharfman H, Goodman J, Macleod A, Phani S, Antonelli C, Croll S. Increased neurogenesis and the ectopic granule cells after intrahippocampal BDNF infusion in adult rats. Exp Neurol. 2005;192((2)):348–56. doi: 10.1016/j.expneurol.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Varendi K, Kumar A, Härma MA, Andressoo JO. miR-1, miR-10b, miR-155, and miR-191 are novel regulators of BDNF. Cell Mol Life Sci. 2014;71((22)):4443–56. doi: 10.1007/s00018-014-1628-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116((2)):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 28.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13((8)):528–41. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keifer J, Zheng Z, Ambigapathy G. A microRNA-BDNF negative feedback signaling loop in brain: implications for Alzheimer's disease. Microrna. 2015;4((2)):101–8. doi: 10.2174/2211536604666150813152620. [DOI] [PubMed] [Google Scholar]
- 30.Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35((5)):325–34. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6((11)):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 32.Hashimoto Y, Akiyama Y, Otsubo T, Shimada S, Yuasa Y. Involvement of epigenetically silenced microRNA-181c in gastric carcinogenesis. Carcinogenesis. 2010;31((5)):777–84. doi: 10.1093/carcin/bgq013. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki H, Yamamoto E, Nojima M, Kai M, Yamano HO, Yoshikawa K, et al. Methylation-associated silencing of microRNA-34b/c in gastric cancer and its involvement in an epigenetic field defect. Carcinogenesis. 2010;31((12)):2066–73. doi: 10.1093/carcin/bgq203. [DOI] [PubMed] [Google Scholar]
- 34.Kunej T, Godnic I, Ferdin J, Horvat S, Dovc P, Calin GA. Epigenetic regulation of microRNAs in cancer: an integrated review of literature. Mutat Res. 2011;717((1–2)):77–84. doi: 10.1016/j.mrfmmm.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Strmsek Z, Kunej T. MicroRNA silencing by DNA methylation in human cancer: a literature analysis. Noncoding RNA. 2015;1((1)):44–52. doi: 10.3390/ncrna1010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48((4)):491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat Rev Neurosci. 2013;14((2)):97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- 38.Ke X, McKnight RA, Caprau D, O'Grady S, Fu Q, Yu X, et al. Intrauterine growth restriction affects hippocampal dual specificity phosphatase 5 gene expression and epigenetic characteristics. Physiol Genomics. 2011;43((20)):1160–9. doi: 10.1152/physiolgenomics.00242.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ke X, Schober ME, McKnight RA, O'Grady S, Caprau D, Yu X, et al. Intrauterine growth retardation affects expression and epigenetic characteristics of the rat hippocampal glucocorticoid receptor gene. Physiol Genomics. 2010;42((2)):177–89. doi: 10.1152/physiolgenomics.00201.2009. [DOI] [PubMed] [Google Scholar]
- 40.American Physiological Society; World Medical Association General Assembly Guiding principles for research involving animals and human beings. Am J Physiol, Cell Physiol. 2002;282((6)):3. doi: 10.1152/ajpregu.00279.2002. [DOI] [PubMed] [Google Scholar]
- 41.Ke XF Q, Sterrett J, Hillard CJ, Lane RH, Majnik A. Adverse maternal environment and western diet impairs cognitive function and alters hippocampal glucocorticoid receptor promoter methylation in male mice. Physiological Reports. 2020;8((8)):e14407. doi: 10.14814/phy2.14407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Gong Y, Qian H, Chen T, Liu Z, Jiang Z, et al. Brain-derived neurotrophic factor is a novel target gene of the has-miR-183/96/182 cluster in retinal pigment epithelial cells following visible light exposure. Mol Med Rep. 2015;12((2)):2793–9. doi: 10.3892/mmr.2015.3736. [DOI] [PubMed] [Google Scholar]
- 43.Neumann E, Brandenburger T, Santana-Varela S, Deenen R, Köhrer K, Bauer I, et al. MicroRNA-1-associated effects of neuron-specific brain-derived neurotrophic factor gene deletion in dorsal root ganglia. Mol Cell Neurosci. 2016;75:36–43. doi: 10.1016/j.mcn.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang CN, Wang YJ, Wang H, Song L, Chen Y, Wang JL, et al. The anti-dementia effects of donepezil involve miR-206-3p in the hippocampus and cortex. Biol Pharm Bull. 2017;40((4)):465–72. doi: 10.1248/bpb.b16-00898. [DOI] [PubMed] [Google Scholar]
- 45.Ke X, Fu Q, Majnik A, Cohen S, Liu Q, Lane R. Adverse early life environment induces anxiety-like behavior and increases expression of FKBP5 mRNA splice variants in mouse brain. Physiol Genomics. 2018;50((11)):973–81. doi: 10.1152/physiolgenomics.00054.2018. [DOI] [PubMed] [Google Scholar]
- 46.Fung CM, Yang Y, Fu Q, Brown AS, Yu B, Callaway CW, et al. IUGR prevents IGF-1 upregulation in juvenile male mice by perturbing postnatal IGF-1 chromatin remodeling. Pediatr Res. 2015;78((1)):14–23. doi: 10.1038/pr.2015.70. [DOI] [PubMed] [Google Scholar]
- 47.Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70((4)):687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tozuka Y, Wada E, Wada K. Diet-induced obesity in female mice leads to peroxidized lipid accumulations and impairment of hippocampal neurogenesis during the early life of their offspring. FASEB J. 2009;23((6)):1920–34. doi: 10.1096/fj.08-124784. [DOI] [PubMed] [Google Scholar]
- 49.Youssef M, Atsak P, Cardenas J, Kosmidis S, Leonardo ED, Dranovsky A. Early life stress delays hippocampal development and diminishes the adult stem cell pool in mice. Sci Rep. 2019;9((1)):4120. doi: 10.1038/s41598-019-40868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hollins SL, Cairns MJ. MicroRNA: small RNA mediators of the brains genomic response to environmental stress. Prog Neurobiol. 2016;143:61–81. doi: 10.1016/j.pneurobio.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 51.Berardino BG, Fesser EA, Cánepa ET. Perinatal protein malnutrition alters expression of miRNA biogenesis genes Xpo5 and Ago2 in mice brain. Neurosci Lett. 2017;647:38–44. doi: 10.1016/j.neulet.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 52.Cattane N, Mora C, Lopizzo N, Borsini A, Maj C, Pedrini L, et al. Identification of a miRNAs signature associated with exposure to stress early in life and enhanced vulnerability for schizophrenia: new insights for the key role of miR-125b-1-3p in neurodevelopmental processes. Schizophr Res. 2019;205:63–75. doi: 10.1016/j.schres.2018.07.030. [DOI] [PubMed] [Google Scholar]
- 53.Peng JY, An XP, Fang F, Gao KX, Xin HY, Han P, et al. MicroRNA-10b suppresses goat granulosa cell proliferation by targeting brain-derived neurotropic factor. Domest Anim Endocrinol. 2016;54:60–7. doi: 10.1016/j.domaniend.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 54.Muller S. In silico analysis of regulatory networks underlines the role of miR-10b-5p and its target BDNF in huntington's disease. Transl Neurodegener. 2014;3:17. doi: 10.1186/2047-9158-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang Y, Zhu J. Effects of sleep deprivation on behaviors and abnormal hippocampal BDNF/miR-10B expression in rats with chronic stress depression. Int J Clin Exp Pathol. 2015;8((1)):586–93. [PMC free article] [PubMed] [Google Scholar]
- 56.Helgager J, Huang YZ, McNamara JO. Brain-derived neurotrophic factor but not vesicular zinc promotes TrkB activation within mossy fibers of mouse hippocampus in vivo. J Comp Neurol. 2014;522((17)):3885–99. doi: 10.1002/cne.23647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baek SB, Bahn G, Moon SJ, Lee J, Kim KH, Ko IG, et al. The phosphodiesterase type-5 inhibitor, tadalafil, improves depressive symptoms, ameliorates memory impairment, as well as suppresses apoptosis and enhances cell proliferation in the hippocampus of maternal-separated rat pups. Neurosci Lett. 2011;488((1)):26–30. doi: 10.1016/j.neulet.2010.10.074. [DOI] [PubMed] [Google Scholar]
- 58.Kim TW, Park HS. Physical exercise improves cognitive function by enhancing hippocampal neurogenesis and inhibiting apoptosis in male offspring born to obese mother. Behav Brain Res. 2018;347:360–7. doi: 10.1016/j.bbr.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 59.Dias CT, Curi HT, Payolla TB, Lemes SF, Betim Pavan IC, Torsoni MA, et al. Maternal high-fat diet stimulates proinflammatory pathway and increases the expression of Tryptophan Hydroxylase 2 (TPH2) and brain-derived neurotrophic factor (BDNF) in adolescent mice hippocampus. Neurochem Int. 2020;139:104781. doi: 10.1016/j.neuint.2020.104781. [DOI] [PubMed] [Google Scholar]
- 60.Val-Laillet D, Besson M, Guérin S, Coquery N, Randuineau G, Kanzari A, et al. A maternal western diet during gestation and lactation modifies offspring's microbiota activity, blood lipid levels, cognitive responses, and hippocampal neurogenesis in Yucatan pigs. FASEB J. 2017;31((5)):2037–49. doi: 10.1096/fj.201601015R. [DOI] [PubMed] [Google Scholar]
- 61.Liu X, Chen X, Yu X, Tao Y, Bode AM, Dong Z, et al. Regulation of microRNAs by epigenetics and their interplay involved in cancer. J Exp Clin Cancer Res. 2013;32:96. doi: 10.1186/1756-9966-32-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu M, Xu Y, Pan L, Feng Y, Luo K, Mu Q, et al. miR-10b ownregulated by DNA methylation acts as a tumor suppressor in HPV-positive cervical cancer via targeting Tiam1. Cell Physiol Biochem. 2018;51((4)):1763–77. doi: 10.1159/000495680. [DOI] [PubMed] [Google Scholar]
- 63.Suske G. The Sp-family of transcription factors. Gene. 1999;238((2)):291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 64.Revyakin A, Zhang Z, Coleman RA, Li Y, Inouye C, Lucas JK, et al. Transcription initiation by human RNA polymerase II visualized at single-molecule resolution. Genes Dev. 2012;26((15)):1691–702. doi: 10.1101/gad.194936.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23((20)):4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci USA. 2004;101((19)):7357–62. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24((5)):785–96. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448((7153)):553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang L, Niu N, Li L, Shao R, Ouyang H, Zou W. H3K36 trimethylation mediated by SETD2 regulates the fate of bone marrow mesenchymal stem cells. PLoS Biol. 2018;16((11)):e2006522. doi: 10.1371/journal.pbio.2006522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Xie S, Zhou Y, Xie Y, Liu P, Sun M, et al. H3K36 histone methyltransferase Setd2 is required for murine embryonic stem cell differentiation toward endoderm. Cell Rep. 2014;8((6)):1989–2002. doi: 10.1016/j.celrep.2014.08.031. [DOI] [PubMed] [Google Scholar]