ABSTRACT
How mammalian neuronal identity is progressively acquired and reinforced during development is not understood. We have previously shown that loss of RP58 (ZNF238 or ZBTB18), a BTB/POZ-zinc finger-containing transcription factor, in the mouse brain leads to microcephaly, corpus callosum agenesis, and cerebellum hypoplasia and that it is required for normal neuronal differentiation. The transcriptional programs regulated by RP58 during this process are not known. Here, we report for the first time that in embryonic mouse neocortical neurons a complex set of genes normally expressed in other cell types, such as those from mesoderm derivatives, must be actively repressed in vivo and that RP58 is a critical regulator of these repressed transcriptional programs. Importantly, gene set enrichment analysis (GSEA) analyses of these transcriptional programs indicate that repressed genes include distinct sets of genes significantly associated with glioma progression and/or pluripotency. We also demonstrate that reintroducing RP58 in glioma stem cells leads not only to aspects of neuronal differentiation but also to loss of stem cell characteristics, including loss of stem cell markers and decrease in stem cell self-renewal capacities. Thus, RP58 acts as an in vivo master guardian of the neuronal identity transcriptome, and its function may be required to prevent brain disease development, including glioma progression.
KEYWORDS: RP58; ZBTB18; transcriptomics; cerebral cortex; microcephaly; neuronal identity; glioma; BTB/POZ; glioblastoma; neuronal differentiation; transcription factor, aging, neurodegenerative diseases
INTRODUCTION
The mechanisms controlling cell fate choice and maintenance, especially of neuronal cells in the brain, constitute a core developmental question, a question also critical for understanding neurological disorders such as brain birth defects (e.g., microcephaly) or psychiatric disorders (e.g., schizophrenia) that result from defects in these processes (e.g., references 1 to 4). Although much is known about the transcriptional controls that regulate the acquisition of neuronal identity in both vertebrate and nonvertebrate models, the mechanisms that are set in postmitotic cells to allow for final differentiation and maintenance of cell identity during the adult life are much less known, in particular in the mammalian brain (e.g., references 5 and 6).
Work in Caenorhabditis elegans has identified transcription factors acting as terminal selectors that are important not only for setting up a neuronal identity but also for its maintenance (6). It is known that transcriptional maintenance programs initiated early in fetal life to specify a certain neuron type such as monoaminergic-type and cholinergic-type neuron identity are sustained during early postnatal and even adult stages to maintain postmitotic neuronal identities (e.g., reference 7; reviewed (in references 6, 8, and 9).
How the mammalian general neuronal identity is transcriptionally maintained and protected during mammalian development, however, remains unresolved, although a few transcription factors are reportedly involved in maintaining a neuronal differentiated state. Recently, through the use of an in vitro model of neuronal differentiation, the pan-neuron-specific transcription factor Myt1-like (Myt1l) has been reported to repress many different somatic lineage programs other than the neuronal program via recruitment of a complex containing Sin3b for its proneuronal function (10).
RP58 (also known as ZBTB18) is a zinc finger and BTB (Broad-Complex, Tramtrack, and Bric-a-brac)-containing transcription factor, and its deletion in the mouse brain leads to microcephaly, corpus callosum agenesis, and cerebellum hypoplasia (11). Importantly, RP58 haploinsufficiency or mutations are associated with human microcephaly and/or intellectual disability (12–15). Using a novel cell sorting protocol to isolate multipotent progenitors, neuronal progenitors, and differentiating neurons from the embryonic cortex, we have shown that deletion of Rp58 leads to derepression of neurogenic genes such as Ngn2 and Neurod1 in postmitotic neurons (11), suggesting that RP58 is required to repress neuronal progenitor genes in postmitotic neurons. RP58 is also required for reprogramming of fibroblasts following ASCL1 expression (16). However, which transcriptional programs are controlled by RP58 during neuronal differentiation in postmitotic neurons is not known. Therefore, in the current study, we analyzed the embryonic cortical neurons’ transcriptome in normal and Rp58 mutant postmitotic neurons.
RESULTS
RP58 represses nonneuronal fates in postmitotic neurons.
To comprehensively decode how RP58 controls cell differentiation, we analyzed the transcriptome of sorted embryonic day 14.5 (E14.5) postmitotic neurons from Rp58+/+ (WT) and Rp58−/− (Rp58 mutant) cortices by transcriptome sequencing (RNA-seq) (Fig. 1A, indicated as the CD133−; EGFP+ cell population). Because RP58 has been shown to act mostly as a transcriptional repressor (e.g., references 11 and 17), we focused our analysis on specifically deciphering the transcriptome that is repressed in neuronal cells.
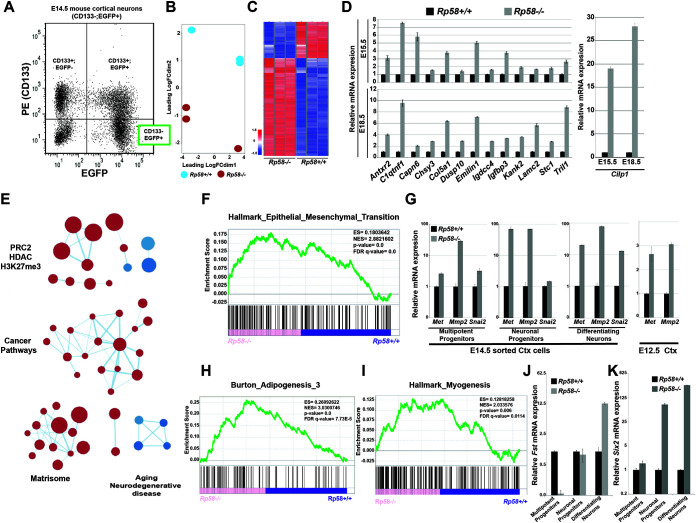
FIG 1.
RP58 represses nonneuronal programs during normal neuronal differentiation. (A) Fluorescence-activated cell sorting (FACS) graph showing separation of E14.5 cortical cells based on CD133 expression and enhanced green fluorescent protein (EGFP) (driven from the tbr2 regulatory region). The subpopulations of sorted cells include the following: CD133+/EGFP−, composed of radial glial cells (RGCs) in the ventricular zone (VZ); double positives, composed of intermediate neuronal progenitors in the subventricular zone (SVZ); and CD133−/EGFP+, composed of differentiating/differentiated cortical neurons which were subjected to transcriptome sequencing (RNA-seq). (B) Principal-component analysis (PCA) of RNA expression in RNA-seq data from E14.5 sorted Rp58−/− versus Rp58+/+ differentiating neurons (n = 3 biological replicates for each genotype). FC, fold change. (C) Heatmap of the expression of genes regulated in the Rp58−/− versus Rp58+/+ embryonic neurons. (D) Analyses of the expression levels of RP58 target genes in Rp58+/+ and Rp58−/− cortices at E15.5 and E18.5. (E) Enrichment map of MSigDB C2 gene sets enriched (false-discovery rate [FDR] < 0.05) in Rp58−/− versus Rp58+/+ expression signature. (F) Gene set enrichment analysis (GSEA) revealing that the genes that are derepressed in the Rp58−/− neurons show a significant similarity with genes associated with the epithelial-mesenchymal transition (EMT). ES, enrichment score. NES, normalized enrichment score. (G) Analyses of the expression levels of the Met, Mmp2, and Snai2 genes in the E14.5 sorted cortical cells and in the E12.5 cerebral cortex of Rp58−/− and Rp58+/+ embryos. (H and I) GSEA analysis showing the significant correlation of the genes upregulated in the Rp58−/− with those genes associated with myogenesis or adipogenesis. (J and K) Analyses of the expression levels of Fst (J) and Six2 (K) mRNA in E14.5 sorted cortical cells.
Principal component analysis (PCA) analyses show that Rp58+/+ and Rp58−/− neuronal cells correspond to two distinct populations (Fig. 1B), and further analysis of the RNA-seq data identified genes that are differentially expressed between the two genotypes (Fig. 1C; see also Table S1 in the supplemental material). In particular, the RNA-seq data confirmed our previous quantitative reverse transcription-PCR (qRT-PCR) results (11), including the loss of Rp58 expression and increased expression of Ngn2 and Neurod1 in the mutant neurons (Table S1). Although RP58 has been mostly described as a transcriptional repressor, we note that about 30% of significantly differentially expressed genes are downregulated in the RP58 mutant cells (Fig. 1C and Table S1).
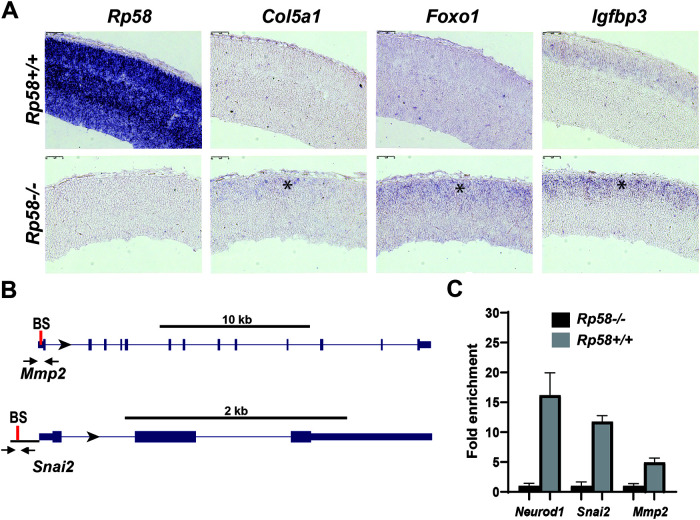
We tested the expression of some highly derepressed genes at E15.5 and E18.5 to verify that the transcriptional programs identified as derepressed at E14.5 in the mutant neurons were also controlled by RP58 at later stages of neuronal maturation. We observed that the derepression of genes regulated by RP58 was indeed more robust as development proceeded; for example, Cilp1 expression was increased 19-fold at E15.5 and 28-fold at E18.5 in Rp58−/− cells (Fig. 1D). We then tested whether the expression of these genes was indeed located in the cortical plate containing the postmitotic neurons in situ (11). We performed RNA in situ hybridization to detect three differentially expressed genes, Col5a1, Foxo1, and Igfbp3, and showed that the expression of these genes was either increased (Igfbp3) or unexpectedly detected (Foxo1 and Col5a1) in the E16.5 Rp58 mutant cortical plate (Fig. 2A and Table S1).
FIG 2.
In situ hybridizations confirming no or low expression of RP58 target genes in the developing cortical plate. (A) Sagittal sections of E16.5 Rp58+/+ (upper) and Rp58−/− (lower) cortices showing the results of in situ hybridizations with specific digoxigenin-labeled antisense RNA probes for Rp58, Igfbp3, Foxo1, and Col5a1. Note the absence of Rp58 expression in mutants and the upregulated (Igfbp3) or ectopic (Foxo1 and Col5a1) expression (asterisk) in the mutant cortical plate region of the developing cerebral cortex containing the postmitotic neuronal cells. Bars, 75 μm. (B) Schematic diagrams showing the localization of the primers (arrows) used for the q-ChIP–PCR on the Mmp2 and Snai2 genes. The unique consensus RP58-binding site (BS) in the proximal 5′ Mmp2 or Snai2 genomic region is indicated by a red bar. (C) ChIP-PCR analyses of RP58 recruitment in control and RP58 mutant cortices were performed with primers specific to the Mmp2 or Snai2 genomic region encompassing the RP58 binding site. The enrichment on the Neurod1 promoter was used as a positive control for the RP58 ChIP.
RNA-seq data was then analyzed using gene set enrichment analysis (GSEA) (18) against functionally defined gene sets in the Molecular Signature Database (MSigDB) H (Hallmark) and C2 (Curated) collections. To globally visualize the main gene sets associated with the genes that are derepressed in the mutant neurons, we used an enrichment map method (19) and identified four major clusters that were linked respectively to epigenetic regulation (with gene sets related to the polycomb repressor complex 2 [PRC2]), cancer pathways, the matrisome, and aging or neurodegenerative diseases (Fig. 1E).
Surprisingly, the epithelial-mesenchymal transition (EMT) pathway was recognized as being significantly upregulated in the Rp58−/− cells (Fig. 1F). Although EMT per se has not been observed during cerebral cortex development, it has been suggested that as neuronal progenitors migrate and change their cell polarity, they may share similarities with typical cellular changes observed during EMT (20). The expression of Met, Mmp2, and Snai2 (Slug), three critical EMT regulators (e.g., reference 21), were found by qRT-PCR analyses to be strongly elevated in the mutant differentiating neurons, as well as in the neuronal and multipotent progenitors, although to a lesser extent (Fig. 1G). Interestingly, the repression of EMT genes by RP58 appears to be required at early stages of cortical development like E12.5 (Fig. 1G), suggesting that the repression of genes involved in EMT is an early event attributed to RP58 function and therefore is required for normal neuronal differentiation.
Interestingly, we observed that gene signatures associated with the differentiation processes of other cell types, such as adipogenesis and myogenesis, were also significantly upregulated in the mutant neuronal cells (Fig. 1H and I). Follistatin (Fst) encodes a known activator of muscle development (e.g., reference 22), and its expression is strongly increased in E14.5 postmitotic Rp58−/− neurons (Fig. 1J). Furthermore, the expression of another mesodermal marker, Six2, a master regulator of kidney development (e.g., reference 23), is also strongly deregulated in the Rp58 mutant neurons compared to that in control cells (Fig. 1K). These results suggest that a primary function for RP58 during brain development, and more specifically during neuronal differentiation, is to repress transcriptional programs that are associated with cell identities other than the neuronal identity, such as those of mesoderm-derived cell fates.
To determine if RP58 directly regulates the expression of genes included in the transcriptional program upregulated in Rp58 mutant neurons, we searched for consensus putative RP58 binding sites in the genomic region of the following genes: Antxr2, Col5a1, Htra1, Myo10, Tgfbr2, Bmp7, Ddit4l, Igdcc4, Pon2, Tril, Brca1, Dusp10, Igfbp3, Ramp1, Zyx, C1qtnf1, Efs, Igfbp4, Serinc5, Capn6, Emilin1, Kank2, Serpinh1, Adgre5, Eya1, Kif4, Chsy3, Foxo1, Lamc2, Six2, Cilp, Fst, Met, Slc4a4, Col4a1, Gja1, Mmp14, Snai2, Hspa2, Mmp2, and Stc1. We applied Biostrings (24) to determine potential RP58 binding sequences in our regulated genes. The transcription factor binding matrix of RP58 (position weight matrix [PWM]) was obtained from the R package MotifDb (25) and compared, using the “matchPWM” function (with the minimum match score set to 0.8), to target sequences in the 2 kb upstream and 200 bp downstream of promoter regions of both the forward and the reverse strands. The R package seqLog (26) was used to plot the RP58 consensus binding sequence logo, 5′-[AC]ACATCTG[GT][AC]-3′. Of all 43 tested genes, only 3 (Gmp7, Ramp1, and Hspa2) do not have the RP58 binding site (see Table S2 in the supplemental material).
To further address if RP58 directly control the expression of genes included in the deregulated transcriptome of the Rp58 mutant neurons, we identified RP58 binding sites in the Mmp2 and Snai2 genes (Table S2 and Fig. 2B) and performed an RP58 quantitative chromatin immunoprecipitation-PCR (q-ChIP-PCR) as described previously (11) on E14.5 telencephalon or cortices. We validated the chromatin purification using primers for the Neurod1 gene regulatory region (11) and indeed observed that RP58 is significantly enriched at the Neurod1 promoter in the Rp58+/+ telencephalon compared to results of the RP58 ChIP on the Rp58−/− tissue (Fig. 2C), as we previously showed (11). Using specific primers flanking the RP58 binding site on the Mmp2 and Snai2 genes (Fig. 2B), we also showed that RP58 is enriched at these promoters in the normal tissue (Rp58+/+) versus the Rp58 mutant tissue (Rp58−/−), which was used as a negative control (Fig. 2C). Similar results were obtained on E14.5 cortices when IgG ChIP is used as a control versus RP58 ChIP (data not shown).
RP58-repressed transcriptional programs are associated with gene signatures involved in brain diseases such as glioma.
Given the role of RP58 in repressing alternate identities in neurons, we then asked if RP58 may also play a role in brain diseases. Cancer is a leading cause of death worldwide, and glioma is the leading cause of primary brain cancer death (27). Gliomas may be classified into four grades (I to IV), with grade IV glioma, or glioblastoma (GBM), being the most aggressive (28, 29). They may also be classified into subtypes based on gene expression profiling (30–35). Although variations in classification arise due to different methods of analysis (32) and to the heterogeneity of these tumors (36–38), the majority of these studies have included two distinct subtypes, proneural (PN) and mesenchymal (MES) GBM (e.g., references 39–41). While there has been debate over whether GBMs have a glial or neuronal origin, recent evidence suggests that most originate from a PN-like tumor, then progress to a more aggressive MES-like state (42). Interestingly, RP58 expression is both decreased during glioma progression from grade II to IV and is significantly higher in the PN GBM subtype compared to other subtypes, such as the MES subtype (43), suggesting that a decrease of RP58 in glioma may lead to disease progression.
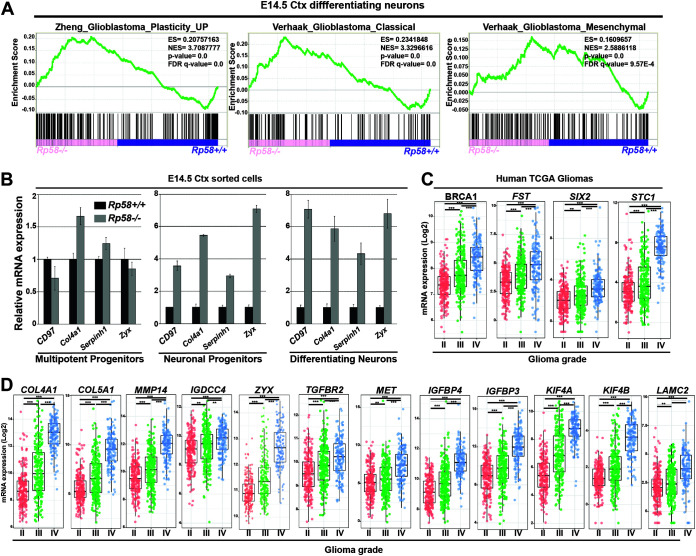
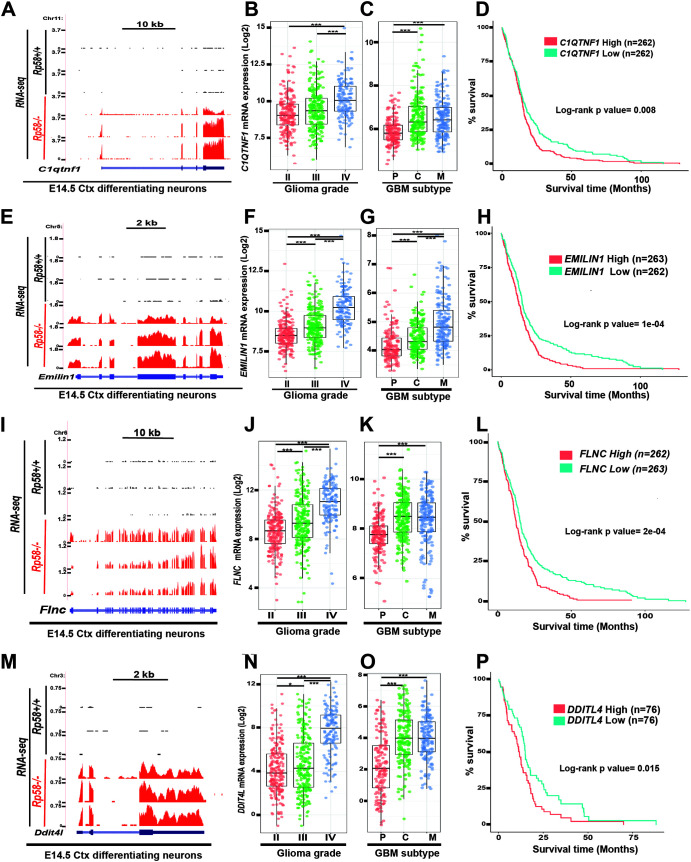
Specifically, GSEA analysis of the E14.5 neuronal cell RNA-seq data revealed that upregulated genes following loss of RP58 in postmitotic embryonic neurons are significantly associated with gene sets linked to GBM development, including GBM pluripotency and MES GBM (Fig. 3A). This suggests that RP58 may play a role in GBM progression, including transition into a more pluripotent and aggressive state. We further validated these results using qRT-PCR. Profiling of genes tightly associated with MES GBM, such as Cd97, Col4a1, Serpinh1, and Zyx (e.g., reference 39), revealed that they are strongly upregulated in Rp58 mutant differentiating neurons and neuronal progenitors (Fig. 3B). Utilizing glioma expression profiles data from The Cancer Genome Atlas (TCGA) on the GlioVis site (http://gliovis.bioinfo.cnio.es/), we also found that a large number of the RP58 target genes identified as derepressed in the Rp58−/− neurons compared to Rp58+/+ neurons show significantly increased expression during glioma progression from low to high grade (e.g., BRCA1, COL4A1, COL5A1, FST, IGDCC4, IGFBP3, IGFBP4, KIF4A, KIF4B, LAMC2, MET, MMP14, SIX2, STC1, TGFBR2, and ZYX) (Fig. 3C and D). Furthermore, a number of these genes also show increased expression during the acquisition of the mesenchymal subtype (e.g., C1QTNF1, DDIT4L, EMILIN1, and FLNC) (Fig. 4).
FIG 3.
The genes repressed by RP58 in neuronal cells are associated with gene signatures involved in glioma progression. (A) GSEA analysis shows the significant correlation of the genes upregulated in Rp58−/− neurons with those genes associated with GBM plasticity and GBM classical (C) or mesenchymal (M) subtype. (B) Analyses of the relative mRNA levels of genes involved in the GBM M signature, namely Cd97, Col4a1, Serpinh1, and Zyxin (Zyx), in sorted cell populations of Rp58−/− versus Rp58+/+ E14.5 cortices. Note that the expression of these genes is maintained at a high level in the Rp58−/− cells. Diagrams are from a representative experiment. (C) Box plot showing the normalized expression of BRCA1, FST, SIX2 and STC1 genes in human glioma samples according to tumor grade following analysis of the RNA expression results from The Cancer Genome Atlas (TCGA) GBM–low-grade glioma (LGG) samples using GlioVis (n = 226 grade II, n = 244 grade III, and n = 150 grade IV or GBMs). (D) Box plot showing the normalized expression of RP58 developmental target genes in glioma samples according to tumor grade following analysis of the RNA expression results from TCGA samples using the GlioVis platform (n = 226 grade II, n = 244 grade III, and n = 150 grade IV or GBMs). Gene names are indicated on top of the corresponding box plot. ***, P < 0.001; **, P < 0.01.
FIG 4.
Expression of RP58 developmental target genes is correlated to glioma progression, mesenchymal subtype acquisition, and prognosis. (A, E, I, M) RNA-seq data of sorted E14.5 cortical neurons (Ctx) from E14.5 Rp58+/+ (n = 3) and Rp58−/− (n = 3) were visualized on the UCSC genome browser as wiggle tracks. The wiggle tracks in black show the minimal expression of C1qtnf1 (A), Emilin1 (E), Flnc (I), and Dtti4l (M) in Rp58+/+ neurons, while the red tracks indicate their significant expression in the Rp58−/− neurons. Exon and intron structures of the C1qtnf1 (A), Igfbp3 (E), Flnc (I), and Dtti4l (M) genes are shown. (B, F, J, N) Box plots showing the normalized expression of C1QTNF1 (B), EMILIN1 (F), FLNC (J), and DDTI4L (N) genes in human glioma samples according to tumor grade and following analysis of the RNA expression results from TCGA GBM-LGG samples using GlioVis (n = 226 grade II, n = 244 grade III, and n = 150 grade IV or GBMs). (C, G, K, O) Box plots showing the normalized expression of C1QTNF1 (C), EMILIN1 (G), FLNC (K), and DDTI4L (O) genes in GBM samples according to tumor subtype and following analysis of the RNA expression results from GBM TCGA samples using GlioVis (n = 163 subtype P [proneural], n = 199 subtype C [classical], and n = 166 subtype M [mesenchymal] for panels C, G, and K; n = 151 subtype P, n = 182 subtype C, and n = 156 subtype M for panel O). (B, C, F, G, J, K, N, O) ***, P < 0.001; *, P < 0.05. (D, H, L, P) Kaplan-Meier survival curves of TCGA GBM patients using GlioVis showing that high levels of C1QTNF1, EMILIN1, FLNC, and DDTI4L expression correlate with worse overall survival (P value calculated using the log rank test; high versus low expression for each gene). Number of samples used in each analysis is indicated for each gene.
More importantly, our study has also identified novel RP58 targets correlated with GBM survival, including C1QTNF1 and EMILIN1, as well as DDIT4L and FLNC (Fig. 4D, H, L, and P). Emilin1 belongs to a gene family encoding extracellular matrix proteins. Deletion of Emilin1 in mice leads to defects in blood pressure and vessel size (44). The expression of Emilin1 in the developing nervous system is not detected in the E14.5 mouse cerebral cortex but is strongly expressed in the developing mesenchyme-derived tissue (Allen Brain Atlas, https://developingmouse.brain-map.org/; data not shown). Similarly, C1qtnf1 is mostly expressed in mesenchyme-derived structures and is not detected in the E14.5 cerebral cortex (Allen Brain Atlas; data not shown). Therefore, these two genes may represent also additional examples of the mesenchymal genes repressed by RP58 during neuronal differentiation. Ddit4l (DNA-damage-inducible transcript 4-like) expression is detected at low levels in the ventricular zone in the E14.5 cortex (Allen Brain Atlas; not shown) and therefore may provide another example of genes, such as Ngn2 and Neurod1, that are expressed in the neuronal progenitors but repressed in the differentiating neurons in the cortical plate (11).
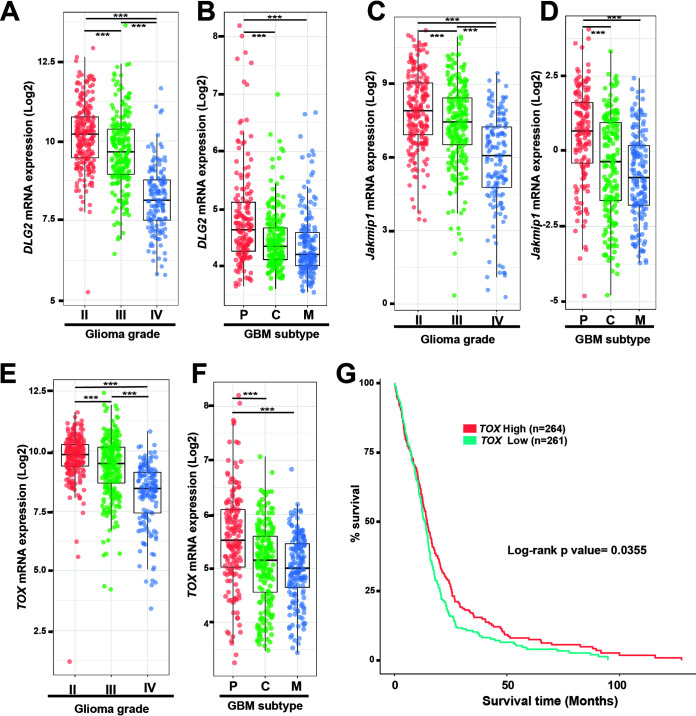
Although beyond the scope of the present study, which focuses on genes derepressed in the Rp58 mutant cells, we also noticed that genes such as DLG2 (disks large homolog 2), Jakmip1 (Janus kinase and microtubule-interacting protein 1) and TOX (thymocyte selection-associated high mobility group box protein) identified as downregulated in the Rp58 mutant cells (Fig. 1C and Table S1) were downregulated in GBM (Fig. 5), as expected if the transcriptional programs controlled by RP58 during normal cell differentiation are disrupted when RP58 expression decreases in GBM.
FIG 5.
Expression of genes downregulated in the Rp58 mutant neurons is inversely correlated to glioma progression, mesenchymal subtype acquisition, and prognosis. (A, C, E) Box plots showing the normalized expression of DLG2 (A), JAKMIP1(C), and TOX (E) genes in human glioma samples according to tumor grade and following analysis of the RNA expression results from TCGA GBM-LGG samples using GlioVis (n = 226 grade II, n = 244 grade III, and n = 150 grade IV or GBMs). (B, D, F) Box plots showing the normalized expression of DLG2 (B), JAKMIP1(D), and TOX (F) genes in GBM samples according to tumor subtype following analysis of the RNA expression results from GBM TCGA samples using GlioVis (n = 163 subtype P [proneural], n = 199 subtype C [classical], and n = 166 subtype M [mesenchymal]). (A, B, C, D, E, F) ***, P < 0.001. (G) Kaplan-Meier survival curves of TCGA GBM patients using GlioVis showing that the low level of TOX expression correlates with worse overall survival (P value calculated using the log rank test; high versus low expression). Number of samples used is indicated.
RP58 is sufficient to reprogram glioma stem cells.
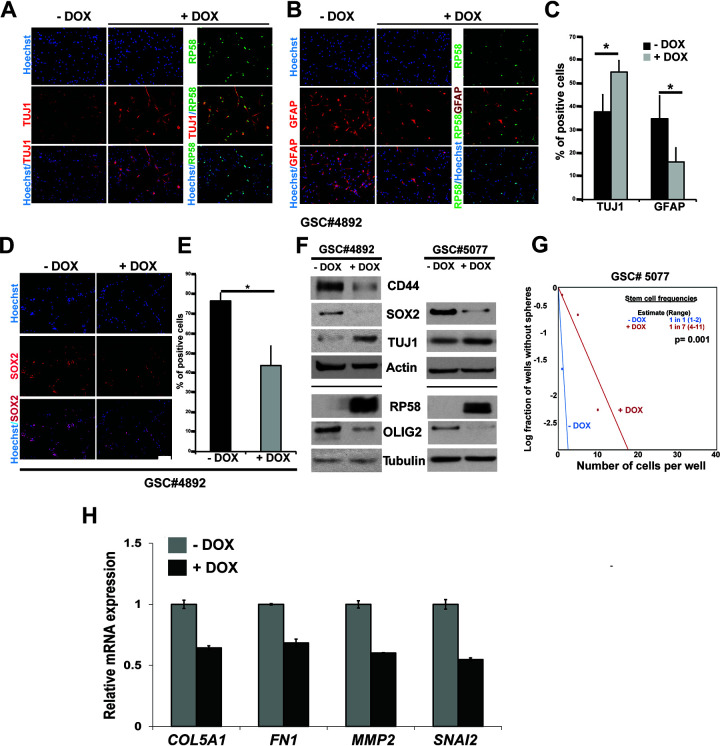
Our prior studies have revealed that RP58 expression is either absent or strongly decreased in glioma-derived cells, and forced RP58 expression in glioma cell lines or primary cultures leads to decreased proliferation and growth in vitro and in xenografts (43, 45). Given this, combined with our transcriptional findings, we next asked if reintroducing RP58 in glioma stem cells (GSCs) may reprogram their identity. We used a lentivirus to overexpress RP58 with doxycycline (DOX) in GSC 4892 cells (46). In development, neural stem cells are multipotent cells that give rise to both neural and glial lineages. This pluripotency and mixed neural cell-like and glial cell-like expression is also seen in glioma (e.g., reference 47). Therefore, we compared the marker expression of neural (TUJ1+) and astrocytic (GFAP+) lineages following expression of RP58 in GSCs subject to differentiation by removing epidermal growth factor/fibroblast growth factor (EGF/FGF). We found that expression of the neuronal marker TUJ1 was elevated and that the cells acquired a more neuronal cell-like morphology (Fig. 6A and C). The increase of TUJ1+ cells was also observed in GSC 4701 cells (46) (data not shown). On the contrary, cells expressing RP58 lost expression of the astrocytic marker GFAP, and we did not detect cells expressing both RP58 and GFAP (Fig. 6B and C). Additionally, expression of RP58 in GSC 4892 cells led to a lower number of positively labeled cells with stem cell marker SOX2 (Fig. 6D and E). By Western blotting, we also detected a significant reduction of glioma stem cell markers such as SOX2, CD44, and OLIG2 in RP58-overexpressed GSCs (Fig. 6F). Moreover, using the extreme limiting dilution assay (ELDA), we found that the frequency of stem cells is reduced when GSC 5077 cells (46) express RP58, suggesting that RP58 expression affected the self-renewal capacity of these GSCs (Fig. 6G).
FIG 6.
RP58 is sufficient to reprogram glioma stem cells. (A to E) Glioma stem cell (GSC) 4892 cells were infected with lentivirus for doxycycline (DOX)-inducible expression of RP58 protein. Transduced cells were plated in medium without epidermal growth factor/fibroblast growth factor (EGF/FGF) for 2 to 3 days. Cells were then fixed and stained by immunofluorescence with anti-TUJ1 (A), anti-GFAP (B), anti-SOX2 (D), and anti-RP58 antibodies (A, B). Quantification of positive cells for each staining of TUJ1 and GFAP (C) and SOX2 (E). Experiments were done on 3 independent biological replicates. Asterisks denote significant change (P = 0.0063 for SOX2, 0.019 for GFAP, and 0.027 for TUJ1). (F) Reduced expression of glioma stem cell markers in RP58 overexpressed 4892 and 5077 cells. RP58 overexpression was measured in Western blots, together with protein levels of the GSC markers SOX2, CD44, and OLIG2 in the GSCs cultivated in the presence of EGF/FGF. (G) Extreme limiting dilution analysis of the effect of RP58 expression in GSC 5077. GSC 5077 cells infected with lentivirus inducing RP58 expression were plated into 96-well plates at various seeding densities (1 to 100 cells per well, 8 wells per condition). Seven days later, each well was evaluated for the presence or absence of spheres, and the number of wells containing spheres was counted. Shown is a representative result from experiments performed on three biological replicates. (H) Analyses of the expression levels of MMP2, SNAI2, FN1, and COL5A1 mRNA in U87 cells with DOX-inducible RP58 expression.
To determine whether RP58 alone is sufficient to repress genes that are included in its aberrant transcriptional program during cell de- or transdifferentiation in the pathological glioma state, we also performed qRT-PCR on glioma cells. We found that, following RP58 expression, MMP2, SNAI2, FN1, and COL5A1 mRNA expression is significantly reduced, suggesting that it is sufficient to reprogram glioma cells (Fig. 6H). Together, these results reveal a close link between the RP58 repressor function during the acquisition of cell identity in the developing nervous system and its role in preventing malignant glioma progression.
RP58 interacts with the PRC2 complex in the developing neocortex.
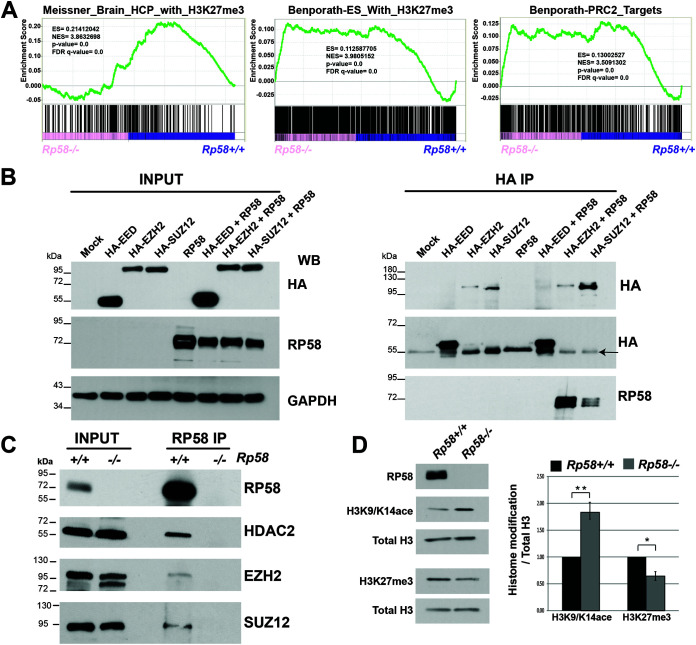
We then addressed the molecular mechanisms involved in the transcriptional repression function of RP58 in neuronal cells. GSEA analysis of postmitotic neuron transcriptome revealed that the genes derepressed in the mutant neuronal cells were also gene targets of the polycomb repressor complex 2 (PRC2) complex (Fig. 1E and 7A), a complex involved in the methylation of H3 at the K27 residue (H3K27me3) and composed of 3 main proteins, EED, EZH2, and SUZ12 (e.g., reference 48). The PRC2 complex is required for correct neuronal differentiation and brain development (e.g., reference 49). These GSEA results suggested that RP58 might exert its transcriptional repression function through its interaction with the PRC2 complex to promote neuronal development. Using overexpressed proteins in HEK293T cells, we showed that RP58 binds to EZH2 and SUZ12, but not to EED (Fig. 7B). Importantly, we showed that RP58 binds to EZH2 and SUZ12 in vivo in the developing cerebral cortex (Fig. 7C). Additionally, our data showed that RP58 also interacts in vivo with HDAC2 (Fig. 7C). Thus, RP58 interacts with chromatin-remodeling proteins involved in histone posttranslational modification. In addition, changes in histone modifications accompany the cell fate determination in many tissues, as different histone marks are associated with repressed or activated genes. We therefore examined if deletion of Rp58 in the brain had any effect on the global levels of PRC2-deposited mark H3K27me3 and observed that the levels of H3K27me3 were indeed decreased (Fig. 7D); conversely, the levels of the active mark H3K9/K14Ace were increased in the E18.5 cortex (Fig. 7D).
FIG 7.
RP58 interacts with the PRC2 complex in the developing neocortex. (A) GSEA analysis showing the significant correlation of the genes upregulated in the Rp58−/−neuronal cells with those genes targeted by the PRC2 complex and/or identified by H3K27m3 histone mark. (B) HEK293T cells were transfected with plasmids expressing tagged proteins (hemagglutinin [HA]-EED, HA-EZH2, or HA-SUZ12) and RP58, and then cultured for 48 h posttransfection. Protein extracts were used to immunoprecipitate HA-tagged proteins from transfected cells. (Left) Twenty percent of total protein extracts were probed with an HA tag or RP58 antibodies. (Right) Fifty percent of total immunoprecipitation was used for Western blot analysis. When EZH2 and SUZ12 are cotransfected with RP58, we observe an immunoreactive band for RP58 but not for EED-RP58 cotransfection, suggesting the specificity of the interaction. Arrow indicates IgG heavy chain on the HA blot detecting the EED protein. (C) Endogenous RP58 was immunoprecipitated from proteins lysate of E18.5 cortical tissue from Rp58+/+ and Rp58−/− brains. Ten percent of proteins lysate was used for input lanes. RP58 forms a complex with endogenous EZH2, SUZ12, and HDAC2 in Rp58+/+ lysate but not in Rp58−/− lysate used as a negative control. (D) Rp58−/− E18.5 cortices show increased H3K9/K14Ace and decreased H3K27me3. Histone modifications were normalized to total H3, and statistical analysis was done using an unpaired t test. Analysis was done on n = 3 embryos from two different litters (P < 0.01).
These results suggest that RP58 controls the neuronal transcriptome, in part, through the repression of genes targeted by the PRC2 complex and therefore labeled by H3K27me3. In agreement with these results, we had previously shown that H3K27me3 presence is reduced at the Ngn2 and Neurod1 promoters in Rp58 mutant versus control cortices (11).
DISCUSSION
We and others have shown that the BTB/POZ zinc finger transcription factor RP58 (ZBTB18) is critical for brain development (11, 50, 64, 65), and our work using neural-specific mutants of Rp58 suggested a role for RP58 in human microcephaly (11, 50). These results were confirmed by the discovery that haploinsufficiency or mutations in RP58 are linked to human microcephaly and/or intellectual disability (e.g., references 12, 13, 15). Because RP58 is more expressed in postmitotic neurons than in progenitors during cerebral cortex development, we set out to better understand its transcriptional role in these cells.
Using RNA-seq to profile the transcriptome of embryonic postmitotic neurons from Rp58+/+ or Rp58−/−, we show that during neuronal differentiation in vivo, a set of genes not expressed in the neural lineage must be repressed for correct cell identity acquisition to proceed and that RP58 is a major regulator of this process, as its loss leads to the abnormal expression of these genes in neuronal cells. To our knowledge, this is the first study showing that (i) during development, the acquisition of the neuronal identity in vivo requires the repression of genes normally not expressed in neural cells, and (ii) the loss of one transcription factor in vivo is sufficient to derepress a set of genes, the expression of which is not usually associated with neural cells.
Within this context, a recent study reported that the neuron-specific transcription factor Myt1-like (Myt1l) represses nonneuronal programs in primary cultured hippocampal neurons (10) and suggested Myt1l may physiologically function to establish and maintain the identity of neurons. Using an in vitro model of neuronal differentiation, it was shown that Myt1l appears to block many different somatic lineage programs other than the neuronal program via recruitment of a complex containing Sin3b for its proneuronal function (10). In contrast, our study suggests that the molecular mechanisms underlying in vivo RP58-mediated control of the neuronal transcriptome are partly mediated by the PRC2 complex, as many genes upregulated in the Rp58 mutant neurons were previously identified as targets of the PRC2 complex in neuronal cells (51–53). Interestingly, we also observed global changes in histone posttranslational modifications, such as H3K27me3, a histone mark that is regulated by the PRC2 complex. These results showing a global decrease of H3K27me3 level confirm the reduced enrichment of H3K27me3 on the Ngn2 and Neurod1 promoters that we observed in the Rp58 mutant compared to control cortices (11), further suggesting that one main mode of gene repression for RP58 is through the control of H3K27me3 on its target genes. It is interesting to also note that it has been suggested that the PRC2 complex may be more involved in the maintenance of the transcriptionally repressed programs, rather than in their initiation (reviewed in reference 54). Our results point to the neuronal transcriptome being controlled by molecular mechanisms regulated by RP58 that may be distinct from that of Myt1l. Future studies should resolve whether and how RP58 and Mytl1l interact to maintain the neuronal transcriptome intact.
Besides genes associated with other lineages, such as Fst and Six2, we also found that loss of RP58 leads to an important increased expression of genes involved in the acquisition of mesenchymal identity, including Mmp2 and Snai2, or Met, known to regulate the epithelial-mesenchymal transition process in development or diseases. It has been suggested that similar biological processes may also be happening in the developing neocortex during the neuronal differentiation/migration step, i.e., the neuronal progenitors would use an EMT-like process during the transition from neural stem cells (or radial glia) to multipolarized neuronal progenitors and a mesenchymal-to-epithelial transition (MET) from polarized progenitors to radially migrating neurons (reviewed in reference 55). Our data suggest that, in addition to the function described above in controlling genes of other cellular identity, RP58 may also control the transition of neuronal cells to the radial migratory state by repressing the expression of genes involved in EMT, such as Mmp2, Snai2, and Met, in postmitotic neurons.
Parallels between normal developmental mechanisms and cancer processes have been now recognized for several years (reviewed in, e.g., references 56, 57). Interestingly, the GSEA analyses of the transcriptional programs in Rp58−/− versus Rp58+/+ revealed that a large number of genes that are derepressed in the Rp58−/− neuronal cells are significantly represented in gene sets positively associated with glioblastoma plasticity and classical and mesenchymal subtypes. The reduced expression of RP58 itself is significantly correlated with the acquisition of glioma malignancy and with the loss of the proneural phenotype and acquisition of the classical and mesenchymal phenotype (43). Importantly, we show that a large number of these RP58 target genes, first identified as derepressed in mouse Rp58−/− neurons are indeed significantly increased during human glioma progression from low to high grade and during the acquisition of the mesenchymal subtype (Fig. 3 and 4). More importantly, this study has actually identified novel RP58 targets, the expression of which is correlated to GBM survival; these include C1QTNF1, EMILIN1, DDIT4L, and FLNC (Fig. 4). These results suggest that decrease of RP58 expression in human glioma may lead to derepression of a transcriptional program, similar to the one regulated by RP58 during neural development, and that deregulation of this glioma transcriptional program (and therefore of some aspect of neural cell identity) may lead to more aggressive tumors. We also demonstrate that reintroducing RP58 in glioma stem cells led not only to aspects of neuronal differentiation but also to loss of stem cell characteristics, including loss of stem cell markers and a decrease in stem cell self-renewal capacity. These results, as well as those of a recent study on expression of the neurogenic factor Ascl1 in glioma leading to cell differentiation (58), further suggest that glioma stem cells may still be amenable to reprogramming and differentiation and therefore may lead to the development of differentiation therapy option for gliomas.
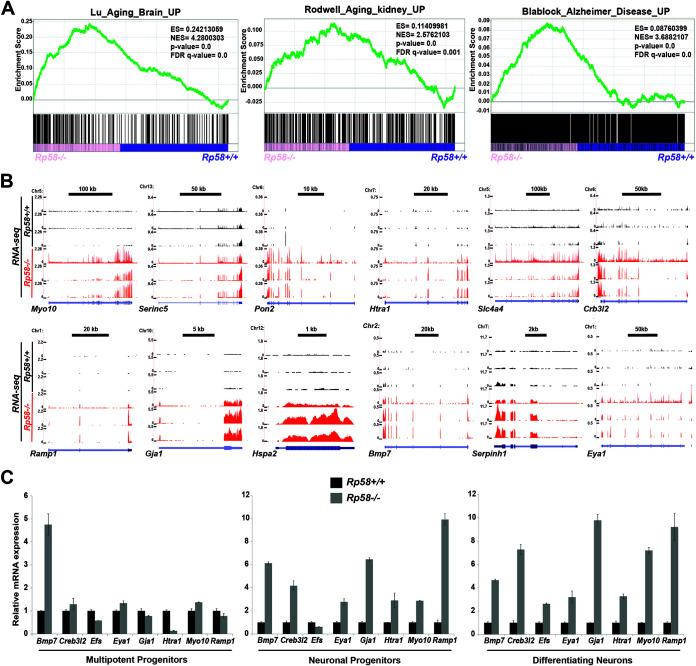
We also hypothesize that disruption of the function of “guardians” of the neuronal transcriptome, such as RP58 or Mytl1, may be also linked to other brain disorders such as brain birth defects or neurodegenerative diseases. Interestingly, GSEA analysis showed that genes positively regulated in the mutant cells were also significantly overrepresented by gene signatures associated with the aging brain, Alzheimer’s disease, and the aging kidney (Fig. 1E and Fig. 8). Importantly, Rp58 was identified in a study examining the transcriptome of the human aging brain as strongly downregulated during aging (59), and our results suggest that loss of RP58 in adult cells may also be involved in the normal aging process and in brain diseases linked to aging and neurodegeneration. The RNA-seq results were also validated by qRT-PCR experiments showing that the expression of genes included in this aging brain signature, such as Bmp7, Crb3l2, Efs, Eya1, Gja1, Htra1, Myo10, and Ramp1 (59), was strongly elevated in the mutant differentiating neurons. It is important to note that upregulated expression for these genes is also observed in neuronal progenitors (Fig. 8C). These results therefore suggest that maintaining neuronal differentiation may be an important step to prevent or restrain aging and neurodegenerative diseases.
FIG 8.
Genes associated with human aging and Alzheimer’s disease are upregulated in the Rp58−/− cortical neurons. Gene set enrichment analysis (GSEA) revealing that the genes that are derepressed in the Rp58−/− show a significant correlation with genes associated with the aging brain, Alzheimer’s disease, and the aging kidney (A). The E14.5 cortical neuron RNA-seq wiggle tracks on the UCSC genome browser (B) show the expression of the Myo10, Serinc5, Pon2, Htra1, Slc4a4, Crb3l2, Ramp1, Gja1, Hspa2, Bmp7, Serpinh1, and Eya1 genes in the Rp58+/+ (black tracks) and Rp58−/− neurons (red tracks). These genes identified as being regulated during brain aging in the Lu et al. study (30) are derepressed in the mutant cortical neurons. Analyses of the relative mRNA levels of Bmp7, Crb3l2, Efs, Eya1, Gja1, Htra1, Myo10, and Ramp1 genes in sorted cell populations of Rp58−/− versus Rp58+/+ E14.5 cortices (C). Note that the expression of these genes is maintained at a high level in the Rp58−/− cells. Diagrams are from a representative experiment.
In conclusion, our study therefore addresses a central question in biology linked to the regulation of cell identity during development through the investigation of the function of the RP58 in controlling the transcriptional program during neuronal differentiation. This work provides the first evidence for an in vivo mechanism of repression of nonneural transcriptomes in neuronal cells as they undergo acquisition of their cellular identity. We also identify RP58 as a critical in vivo regulator of this process, as its removal from neural cells leads to derepression of these nonneuronal transcriptome programs. Myt1l has been suggested to play a similar role during in vitro reprogramming of fibroblasts into neurons (10). Future studies should investigate if and how these two transcription factors may cooperate or synergize to guard the neuronal transcriptome by repressing similar and/or different transcriptional programs. Interestingly, our data also suggest the novel hypothesis that maintaining these RP58-repressed transcriptional programs may be linked not only to brain birth defects, such as microcephaly (11), but also to other brain diseases, including brain cancer progression, neurodegenerative disease, and aging.
MATERIALS AND METHODS
Cortex dissection and cell sorting.
As previously described (11), E14.5 cerebral cortices were dissected from Rp58+/+; tbr2::EGFP or Rp58−/−; tbr2::EGFP embryos and dissociated into single cells. Dissociated cortical cells were then sorted via CD133 and green fluorescent protein (GFP) expression allowing the separation of neural stem/progenitor cells, neuronal progenitors, and differentiating neurons.
Plasmid constructs.
The inducible RP58/ZNF238-expressing lentiviral vector with neomycin-selective marker, TREAutoR3-RP58-Neo, was constructed by introduction of PGK-NEO into TREAutoR3-RP58 (45). The PGK-NEO fragment was isolated from a gene-targeting vector (a gift from S. Potter, Cincinnati Children's Hospital, Cincinnati, OH) and then cloned into the backbone of TREAutoR3-RP58.
Glioma stem cell culture, self-renewal, and differentiation assay.
Glioma stem cells (GSCs) were cultured in stem cell culture medium (neurobasal [Gibco, Carlsbad, CA], B27 [1×; Gibco], glutamine [2 mM] and penicillin [50 U/ml]/streptomycin [50 μg/ml], EGF [20 ng/ml], and FGF [20 ng/ml]). After dissociation, GSCs were transduced overnight with TREAutoR3-RP58-Neo viruses and selected with G418 for 2 weeks prior to doxycycline (DOX) induction for assays.
For the differentiation assay, dissociated GSCs were plated onto poly-d-lysine-coated coverslips at a density of 100,000 cells per well in 24-well plates and cultured in medium without growth factors. After DOX induction for 2 to 3 days, cells were fixed with 4% paraformaldehyde (PFA) solution for 10 min and processed for immunocytochemistry with Tuj1, GFAP, RP58 (45), and SOX2 antibodies, as previously described (11). The analysis was performed on three independent replicates. Statistical analysis was performed using Student’s t test.
For self-renewal experiments, gliomaspheres were dissociated into single-cell suspensions and seeded into 96-well plates at a density of 1, 5, 10, 20, 50, and 100 cells per well. After DOX induction for 7 days, sphere formation was evaluated. Stem cell frequency was calculated using the Walter and Eliza Hall Institute Bioinformatics Division ELDA analyzer (http://bioinf.wehi.edu.au/software/elda/) (60).
For stem cell marker analysis, GSCs were cultured in stem cell culture medium with or without DOX for 4 days before being harvested for Western blot analysis.
RNA extraction and quantitative RT-PCR.
Total RNA was isolated from sorted cells, cortical tissues, or U87 cells (cultured as described previously [45]) and reverse transcribed into cDNAs, followed by real-time PCR analysis as described previously (11). Gene expression in embryonic sorted cells or in embryonic cortices was assessed in cells from at least two different litters. Gene expression in U87 cells was assessed from at least 3 replicate experiments. Primer sequences will be provided upon request.
E14.5 neuronal sorted cell RNA sequencing.
Library construction was performed for total RNA of E14.5 sorted postmitotic neurons by following the Illumina stranded RNA-seq workflow (TruSeq stranded total RNA library prep kit, catalog no. RS-122-2201). Pools of indexed library were then prepared for cluster generation and single-end or paired-end sequencing on the Illumina HiSeq 4000 platform. For each genotype (Rp58−/− and Rp58+/+), 3 biological replicates were used.
Computational analysis of RNA-seq data.
The samples were sequenced at the BGI sequencing core at the Children’s Hospital of Philadelphia (BGI@CHOP). The raw RNA sequencing reads were FastQC checked and then mapped to a reference genome (NCBI37/mm9) using STAR aligner. After that, HTSeq was applied to detect the sequencing read count for each gene, while DESeq2 was used to detect the differential expression level for each gene. The bioinformatics analysis generated the cluster result for the top differentially expressed genes by hierarchical clustering, and the differential expression was also considered for the gene set enrichment analysis (GSEA).
GSEA and network enrichment mapping.
The differentially expressed genes in the Rp58 mutant versus control cells were tested for enrichment in the H (hallmark) and C2 (curated) gene sets deposited in the Molecular Signature Database (MSigDB) v6.2 (http://www.broadinstitute.org/gsea/msigdb).
The gene sets of C2, significantly enriched in those genes upregulated in Rp58 mutant cells, were selected by a cutoff value of ≤0.05 for P value and false-discovery rate (FDR) and then visualized with EnrichmentMap software (19), which organizes the significant gene sets into a network of gene set nodes with mutual gene overlap.
Endogenous coimmunoprecipitation.
E18.5 cortical protein lysates were resuspended in lysis buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1 mM EDTA, and 1× Pefabloc [Roche]), flash frozen and lysed using cryogenic grinding. Protein amounts were measured using the Bradford assay. Protein lysate (500 μg) was used, and volume was adjusted to 500 μl with binding buffer (50 mM Tris-HCl [pH 7.5], 120 mM NaCl, and 0.5% NP-40). Immunoprecipitation (IP) reactions containing either Rp58+/+ or Rp58−/− (negative-control) lysates were incubated with anti-RP58 polyclonal antibody (45) and rotated overnight at 4°C. Protein A-agarose (Invitrogen) was washed 3× and incubated with samples for 3 to 5 h. Beads were washed 5× with lysis buffer and then heated to 100°C for 10 min in 2× sample buffer. Samples were then electrophoresed on a 10% SDS-PAGE gel, transferred to immobilon-P polyvinylidene difluoride (PVDF) transfer membrane (Millipore), and blotted with the following primary antibodies: EZH2 (1:1,000 CST 5246), SUZ12 (1:1,000 CST 3737), RP58 (1:2,000) (45), and secondary anti-rabbit light chain-specific horseradish peroxidase (HRP; Jackson ImmunoResearch Laboratories).
In vitro coimmunoprecipitation.
293T cells were transfected with RP58 alone or in combination with hemagglutinin (HA)-EED-, HA-EZH2-, and HA-SUZ12-expressing plasmids (61, 62) (pCMVHA hEZH2, pCMVHA EED wild type [wt], and pCMVHA SUZ12 were a gift from Kristian Helin; Addgene plasmids 24230, 24231, and 24232). For lysis, cell pellets were resuspended in lysis buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1 mM EDTA, and 1× Pefabloc [Roche]). Freeze-thawed 2× NaCl (12.5 μl) was added to the reaction mixture for 1 h to break the nuclei, followed by salt adjustment with 200 μl of double-distilled water (ddH2O). Protein amounts were measured using the Bradford assay. Protein lysate (500 μg) was used, and the volume was adjusted to 500 μl with binding buffer (50 mM Tris-HCl [pH 7.5], 120 mM NaCl, and 0.5% NP-40). IP reactions were incubated with 2.5 μg of anti-HA monoclonal antibody (catalog no. 26183; Invitrogen) and rotated overnight at 4°C. Protein G-agarose (Pierce) was washed 3× with lysis buffer, and samples were incubated in agarose beads for 3 to 5 h. Beads were washed 5× with lysis buffer and then heated to 100°C for 10 min in 2× sample buffer. Samples were then electrophoresed on 10% SDS-PAGE gel, transferred to immobilon-P PVDF transfer membrane (Millipore), and Western blotted with the following primary antibodies: anti-HA monoclonal antibody (1:1,000, catalog no. 26183; Invitrogen), RP58 (1:1,000), and secondary anti-rabbit light chain-specific HRP (Jackson ImmunoResearch laboratories) and anti-mouse HRP.
Immunoblotting.
To detect histone modifications by Western blotting, frozen E18.5 cortical tissue was lysed in lysis buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1 mM EDTA, and 1× Pefabloc). Protein amounts were measured using the Bradford assay. Protein lysate (50 μg) was then electrophoresed on a 12% SDS-PAGE gel, transferred to immobilon-P PVDF transfer membrane, and Western blotted with anti-H3K27me3 (1:1,000, catalog no. 07-449; Millipore), anti-H3K9/K14 acetyl (CST 9677), and total H3 (CST 96C10 monoclonal antibody [mAB]). For quantification, densitometry was performed on scanned Western blots and quantified using the ImageJ 1.32 software (National Institutes of Health, Bethesda, MD). For statistical analysis, an unpaired t test was used to compare Rp58−/− to Rp58+/+.
For analysis of various marker expression in glioma stem cells, protein lysates were prepared from cell pellets in lysis buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton, and 5% glycerol with protease inhibitors [Roche]). After SDS-PAGE, the membranes were transferred and then incubated with the following antibodies: anti-RP58, anti-SOX2 (Abcam), anti-CD44 (provided by Ellen Puré, University of Pennsylvania), anti-OLIG2 (Abcam), and anti-TUJ1 (BioLegend).
Quantitative ChIP-PCR.
Chromatin immunoprecipitation (ChIP) coupled with quantitative PCR on embryonic telencephalon and cortex tissues was performed as previously described (11, 63). We used the RP58 antibody (11, 45) for ChIP and Rp58−/− tissues as a negative control for the ChIP on Rp58+/+ tissues, as no RP58 protein was detected in the mutant tissues. The sequences of the primers used to amplify the Mmp2 genomic region encompassing an RP58 potential binding site (BS) are as follows (5′ to 3′): forward primer, CACCCTCCCCTCTCCACCCG; reverse primer, GAGCCTCCAGCCACCAGGGA. These primers amplify the mouse genomic region at chromosome 8 (chr8): 92827286 to 92827400 (115 bp). The forward primer (5′-TCTGGCCACTAGGGGCTGCG-3′) and reverse primer (5′-GGTGACCTACGGCGACTTGGC-3′) were used to amplify the Snai2 genomic region, which encompasses an RP58 BS at chr16: 14705493 to 14705601 (109 bp). The primers for Neurod1 were previously described (11).
TCGA data analysis.
Analysis of gene expression and survival data of human glioma samples was performed using the GlioVis site (http://gliovis.bioinfo.cnio.es/).
Data availability.
The RNA-seq data have been deposited in GEO under accession number GSE121260.
ACKNOWLEDGMENTS
We thank Logan Zhang for technical help with lentivirus preparation. We thank Rachel Yan for critical review of the manuscript.
This work was funded by an NIH-NINDS grant 1R01NS093120 to N.D. The already published GSCs used in this study (GSC 4892, 4701, and 5077) were generated at the University of Pennsylvania in collaboration with A. Resnick (Children’s Hospital of Philadelphia, Center for Data Driven Discovery in Biomedicine [D3b] and Children's Brain Tumor Network [CBTN]) and D. O’Rourke (University of Pennsylvania) with funding from the Children’s Brain Tumor Tissue Consortium (CBTTC; now Children's Brain Tumor Network).
C.E.M. is a cofounder and board member for Biotia and Onegevity Health.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Barkovich AJ, Kuzniecky RI, Jackson GD, Guerrini R, Dobyns WB. 2005. A developmental and genetic classification for malformations of cortical development. Neurology 65:1873–1887. 10.1212/01.wnl.0000183747.05269.2d. [DOI] [PubMed] [Google Scholar]
- 2.Manzini MC, Walsh CA. 2011. What disorders of cortical development tell us about the cortex: one plus one does not always make two. Curr Opin Genet Dev 21:333–339. 10.1016/j.gde.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romero DM, Bahi-Buisson N, Francis F. 2018. Genetics and mechanisms leading to human cortical malformations. Semin Cell Dev Biol 76:33–75. 10.1016/j.semcdb.2017.09.031. [DOI] [PubMed] [Google Scholar]
- 4.Innocenti GM, Ansermet F, Parnas J. 2003. Schizophrenia, neurodevelopment and corpus callosum. Mol Psychiatry 8:261–274. 10.1038/sj.mp.4001205. [DOI] [PubMed] [Google Scholar]
- 5.Froldi F, Cheng LY. 2016. Understanding how differentiation is maintained: lessons from the Drosophila brain. Cell Mol Life Sci 73:1641–1644. 10.1007/s00018-016-2144-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hobert O, Kratsios P. 2019. Neuronal identity control by terminal selectors in worms, flies, and chordates. Curr Opin Neurobiol 56:97–105. 10.1016/j.conb.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Serrano-Saiz E, Leyva-Diaz E, De La Cruz E, Hobert O. 2018. BRN3-type POU homeobox genes maintain the identity of mature postmitotic neurons in nematodes and mice. Curr Biol 28:2813–2823.e2812. 10.1016/j.cub.2018.06.045. [DOI] [PubMed] [Google Scholar]
- 8.Deneris ES, Hobert O. 2014. Maintenance of postmitotic neuronal cell identity. Nat Neurosci 17:899–907. 10.1038/nn.3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arlotta P, Hobert O. 2015. Homeotic transformations of neuronal cell identities. Trends Neurosci 38:751–762. 10.1016/j.tins.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Mall M, Kareta MS, Chanda S, Ahlenius H, Perotti N, Zhou B, Grieder SD, Ge X, Drake S, Euong Ang C, Walker BM, Vierbuchen T, Fuentes DR, Brennecke P, Nitta KR, Jolma A, Steinmetz LM, Taipale J, Sudhof TC, Wernig M. 2017. Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature 544:245–249. 10.1038/nature21722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiang C, Baubet V, Pal S, Holderbaum L, Tatard V, Jiang P, Davuluri RV, Dahmane N. 2012. RP58/ZNF238 directly modulates proneurogenic gene levels and is required for neuronal differentiation and brain expansion. Cell Death Differ 19:692–702. 10.1038/cdd.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Munnik SA, Garcia-Minaur S, Hoischen A, van Bon BW, Boycott KM, Schoots J, Hoefsloot LH, Knoers NV, Bongers EM, Brunner HG. 2014. A de novo non-sense mutation in ZBTB18 in a patient with features of the 1q43q44 microdeletion syndrome. Eur J Hum Genet 22:844–846. 10.1038/ejhg.2013.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perlman SJ, Kulkarni S, Manwaring L, Shinawi M. 2013. Haploinsufficiency of ZNF238 is associated with corpus callosum abnormalities in 1q44 deletions. Am J Med Genet 161:711–716. 10.1002/ajmg.a.35779. [DOI] [PubMed] [Google Scholar]
- 14.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, Albrecht B, Bartholdi D, Beygo J, Di Donato N, Dufke A, Cremer K, Hempel M, Horn D, Hoyer J, Joset P, Ropke A, Moog U, Riess A, Thiel CT, Tzschach A, Wiesener A, Wohlleber E, Zweier C, Ekici AB, Zink AM, Rump A, Meisinger C, Grallert H, Sticht H, Schenck A, Engels H, Rappold G, Schrock E, Wieacker P, Riess O, Meitinger T, Reis A, Strom TM. 2012. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 380:1674–1682. 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 15.Cohen JS, Srivastava S, Farwell Hagman KD, Shinde DN, Huether R, Darcy D, Wallerstein R, Houge G, Berland S, Monaghan KG, Poretti A, Wilson AL, Chung WK, Fatemi A. 2017. Further evidence that de novo missense and truncating variants in ZBTB18 cause intellectual disability with variable features. Clin Genet 91:697–707. 10.1111/cge.12861. [DOI] [PubMed] [Google Scholar]
- 16.Wapinski OL, Lee QY, Chen AC, Li R, Corces MR, Ang CE, Treutlein B, Xiang C, Baubet V, Suchy FP, Sankar V, Sim S, Quake SR, Dahmane N, Wernig M, Chang HY. 2017. Rapid chromatin switch in the direct reprogramming of fibroblasts to neurons. Cell Rep 20:3236–3247. 10.1016/j.celrep.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aoki K, Meng G, Suzuki K, Takashi T, Kameoka Y, Nakahara K, Ishida R, Kasai M. 1998. RP58 associates with condensed chromatin and mediates a sequence-specific transcriptional repression. J Biol Chem 273:26698–26704. 10.1074/jbc.273.41.26698. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. 2010. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One 5:e13984. 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh S, Solecki DJ. 2015. Polarity transitions during neurogenesis and germinal zone exit in the developing central nervous system. Front Cell Neurosci 9:62. 10.3389/fncel.2015.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamouille S, Xu J, Derynck R. 2014. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 15:178–196. 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winbanks CE, Weeks KL, Thomson RE, Sepulveda PV, Beyer C, Qian H, Chen JL, Allen JM, Lancaster GI, Febbraio MA, Harrison CA, McMullen JR, Chamberlain JS, Gregorevic P. 2012. Follistatin-mediated skeletal muscle hypertrophy is regulated by Smad3 and mTOR independently of myostatin. J Cell Biol 197:997–1008. 10.1083/jcb.201109091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP. 2008. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3:169–181. 10.1016/j.stem.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pagès H, Aboyoun P, Gentleman R, DebRoy S. 2020. Biostrings: efficient manipulation of biological strings, vR package version 2.58.0. https://bioconductor.org/packages/Biostrings.
- 25.Shannon P, Richards M. 2020. MotifDb: an annotated collection of protein-DNA binding sequence motifs, vR package version 1.32.0. https://bioconductor.org/packages/MotifDb/.
- 26.Bembom O, Ivanek R. 2020. seqLogo: Sequence logos for DNA sequence alignments, vR package version 1.56.0. https://bioconductor.org/packages/seqLogo/.
- 27.Ostrom QT, Cioffi G, Gittleman H, Patil N, Waite K, Kruchko C, Barnholtz-Sloan JS. 2019. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2012–2016. Neuro Oncol 21:v1–v100. 10.1093/neuonc/noz150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reardon DA, Rich JN, Friedman HS, Bigner DD. 2006. Recent advances in the treatment of malignant astrocytoma. J Clin Oncol 24:1253–1265. 10.1200/JCO.2005.04.5302. [DOI] [PubMed] [Google Scholar]
- 29.Gladson CL, Prayson RA, Liu WM. 2010. The pathobiology of glioma tumors. Annu Rev Pathol 5:33–50. 10.1146/annurev-pathol-121808-102109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones TS, Holland EC. 2011. Molecular pathogenesis of malignant glial tumors. Toxicol Pathol 39:158–166. 10.1177/0192623310387617. [DOI] [PubMed] [Google Scholar]
- 31.Mischel PS, Cloughesy TF, Nelson SF. 2004. DNA-microarray analysis of brain cancer: molecular classification for therapy. Nat Rev Neurosci 5:782–792. 10.1038/nrn1518. [DOI] [PubMed] [Google Scholar]
- 32.Huse JT, Phillips HS, Brennan CW. 2011. Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia 59:1190–1199. 10.1002/glia.21165. [DOI] [PubMed] [Google Scholar]
- 33.Huse JT, Holland EC. 2010. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer 10:319–331. 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 34.Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, Popoff S, Nutt CL, Louis DN, Cairncross JG, Gilbert MR, Phillips HS, Mehta MP, Chakravarti A, Pelloski CE, Bhat K, Feuerstein BG, Jenkins RB, Aldape K. 2010. A multigene predictor of outcome in glioblastoma. Neuro Oncol 12:49–57. 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vitucci M, Hayes DN, Miller CR. 2011. Gene expression profiling of gliomas: merging genomic and histopathological classification for personalised therapy. Br J Cancer 104:545–553. 10.1038/sj.bjc.6606031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, Curtis C, Watts C, Tavare S. 2013. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A 110:4009–4014. 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suva ML, Regev A, Bernstein BE. 2014. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344:1396–1401. 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Lage M, Lynch TM, Bi Y, Cocito C, Way GP, Pal S, Haller J, Yan RE, Ziober A, Nguyen A, Kandpal M, O'Rourke DM, Greenfield JP, Greene CS, Davuluri RV, Dahmane N. 2019. Immune landscapes associated with different glioblastoma molecular subtypes. Acta Neuropathologica Communications 7:203. 10.1186/s40478-019-0803-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K. 2006. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9:157–173. 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 40.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O'Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN, Cancer Genome Atlas Research Network . 2010. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17:98–110. 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Q, Hu B, Hu X, Kim H, Squatrito M, Scarpace L, deCarvalho AC, Lyu S, Li P, Li Y, Barthel F, Cho HJ, Lin YH, Satani N, Martinez-Ledesma E, Zheng S, Chang E, Sauve CG, Olar A, Lan ZD, Finocchiaro G, Phillips JJ, Berger MS, Gabrusiewicz KR, Wang G, Eskilsson E, Hu J, Mikkelsen T, DePinho RA, Muller F, Heimberger AB, Sulman EP, Nam DH, Verhaak RGW. 2017. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell 32:42–56.e46. 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, Charles N, Michor F, Holland EC. 2014. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 26:288–300. 10.1016/j.ccr.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fedele V, Dai F, Masilamani AP, Heiland DH, Kling E, Gatjens-Sanchez AM, Ferrarese R, Platania L, Soroush D, Kim H, Nelander S, Weyerbrock A, Prinz M, Califano A, Iavarone A, Bredel M, Carro MS. 2017. Epigenetic regulation of ZBTB18 promotes glioblastoma progression. Mol Cancer Res 15:998–1011. 10.1158/1541-7786.MCR-16-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zacchigna L, Vecchione C, Notte A, Cordenonsi M, Dupont S, Maretto S, Cifelli G, Ferrari A, Maffei A, Fabbro C, Braghetta P, Marino G, Selvetella G, Aretini A, Colonnese C, Bettarini U, Russo G, Soligo S, Adorno M, Bonaldo P, Volpin D, Piccolo S, Lembo G, Bressan GM. 2006. Emilin1 links TGF-beta maturation to blood pressure homeostasis. Cell 124:929–942. 10.1016/j.cell.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 45.Tatard VM, Xiang C, Biegel JA, Dahmane N. 2010. ZNF238 is expressed in postmitotic brain cells and inhibits brain tumor growth. Cancer Res 70:1236–1246. 10.1158/0008-5472.CAN-09-2249. [DOI] [PubMed] [Google Scholar]
- 46.Roccograndi L, Binder ZA, Zhang L, Aceto N, Zhang Z, Bentires-Alj M, Nakano I, Dahmane N, O'Rourke DM. 2017. SHP2 regulates proliferation and tumorigenicity of glioma stem cells. J Neurooncol 135:487–496. 10.1007/s11060-017-2610-x. [DOI] [PubMed] [Google Scholar]
- 47.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. 2015. Cancer stem cells in glioblastoma. Genes Dev 29:1203–1217. 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Margueron R, Reinberg D. 2011. The polycomb complex PRC2 and its mark in life. Nature 469:343–349. 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Testa G. 2011. The time of timing: how polycomb proteins regulate neurogenesis. Bioessays 33:519–528. 10.1002/bies.201100021. [DOI] [PubMed] [Google Scholar]
- 50.Baubet V, Xiang C, Molczan A, Roccograndi L, Melamed S, Dahmane N. 2012. Rp58 is essential for the growth and patterning of the cerebellum and for glutamatergic and GABAergic neuron development. Development 139:1903–1909. 10.1242/dev.075606. [DOI] [PubMed] [Google Scholar]
- 51.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. 2008. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40:499–507. 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454:766–770. 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560. 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Comet I, Riising EM, Leblanc B, Helin K. 2016. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer 16:803–810. 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 55.Famulski JK, Solecki DJ. 2013. New spin on an old transition: epithelial parallels in neuronal adhesion control. Trends Neurosci 36:163–173. 10.1016/j.tins.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aiello NM, Stanger BZ. 2016. Echoes of the embryo: using the developmental biology toolkit to study cancer. Dis Model Mech 9:105–114. 10.1242/dmm.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruiz i Altaba A, Sanchez P, Dahmane N. 2002. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer 2:361–372. 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- 58.Park NI, Guilhamon P, Desai K, McAdam RF, Langille E, O'Connor M, Lan X, Whetstone H, Coutinho FJ, Vanner RJ, Ling E, Prinos P, Lee L, Selvadurai H, Atwal G, Kushida M, Clarke ID, Voisin V, Cusimano MD, Bernstein M, Das S, Bader G, Arrowsmith CH, Angers S, Huang X, Lupien M, Dirks PB. 2017. ASCL1 reorganizes chromatin to direct neuronal fate and suppress tumorigenicity of glioblastoma stem cells. Cell Stem Cell 21:209–224.e207. 10.1016/j.stem.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 59.Lu T, Pan Y, Kao SY, Li C, Kohane I, Chan J, Yankner BA. 2004. Gene regulation and DNA damage in the ageing human brain. Nature 429:883–891. 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- 60.Hu Y, Smyth GK. 2009. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 347:70–78. 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 61.Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23:4061–4071. 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 22:5323–5335. 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee TI, Johnstone SE, Young RA. 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1:729–748. 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Okado H, Ohtaka-Maruyama C, Sugitani Y, Fukuda Y, Ishida R, Hirai S, Miwa A, Takahashi A, Aoki K, Mochida K, Suzuki O, Honda T, Nakajima K, Ogawa M, Terashima T, Matsuda J, Kawano H, Kasai M. 2009. The transcriptional repressor RP58 is crucial for cell-division patterning and neuronal survival in the developing cortex. Dev Biol 331:140–151. 10.1016/j.ydbio.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 65.Hirai S, Miwa A, Ohtaka-Maruyama C, Kasai M, Okabe S, Hata Y, Okado H. 2012. RP58 controls neuron and astrocyte differentiation by downregulating the expression of Id1-4 genes in the developing cortex. EMBO J 31:1190–1202. 10.1038/emboj.2011.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download MCB00526-20_Supp_1_seq10.pdf, PDF file, 0.1 MB (124.2KB, pdf)
Table S2. Download MCB00526-20_Supp_2_seq11.pdf, PDF file, 0.05 MB (47.9KB, pdf)
Data Availability Statement
The RNA-seq data have been deposited in GEO under accession number GSE121260.