Summary
Aerobic glycolysis, or preferential fermentation of glucose-derived pyruvate to lactate despite available oxygen, is associated with proliferation across many organisms and conditions. To better understand this association, we examined the metabolic consequence of activating the pyruvate dehydrogenase complex (PDH) to increase pyruvate oxidation at the expense of fermentation. We find that increasing PDH activity impairs cell proliferation by reducing the NAD+/NADH ratio. This change in NAD+/NADH is caused by increased mitochondrial membrane potential that impairs mitochondrial electron transport and NAD+ regeneration. Uncoupling respiration from ATP synthesis or increasing ATP hydrolysis restores NAD+/NADH homeostasis and proliferation even when glucose oxidation is increased. These data suggest that when demand for NAD+ to support oxidation reactions exceeds the rate of ATP turnover in cells, NAD+ regeneration by mitochondrial respiration becomes constrained, promoting fermentation despite available oxygen. This argues cells engage in aerobic glycolysis when the demand for NAD+ is in excess of the demand for ATP.
Introduction
Cell growth and division impose increased energetic and biosynthetic demands, and thus proliferating cells exhibit a distinct metabolism relative to non-proliferating cells. Under aerobic conditions, most cells reduce oxygen to water via respiration to support the oxidation reactions used to derive energy from nutrients. When oxygen is limiting, cells instead ferment carbohydrates to generate a waste product such as lactate or ethanol as an alternative, less carbon efficient, way to derive energy from nutrients. However, some cells, including many rapidly proliferating cells, exhibit high rates of fermentation even when oxygen is abundant, a metabolic phenotype known as aerobic glycolysis. Aerobic glycolysis has been associated with tumors (Koppenol et al., 2011; Warburg, 1924; Warburg, 1956), but this phenotype is not unique to cancer. Aerobic glycolysis is exhibited by some proliferating microorganisms, including species of yeast and bacteria, and many proliferating non-transformed mammalian cells, including lymphocytes and fibroblasts (Brand et al., 1986; Hume et al., 1978; Lemoigne et al., 1954; Munyon and Merchant, 1959; Wang et al., 1976). Select non-proliferative cells, such as pigmented epithelial cells of the mammalian retina, also engage in aerobic glycolysis (Chinchore et al., 2017; Krebs, 1927). Despite being a phenotype found in many different cells and organisms, what drives aerobic glycolysis and why it is associated with proliferation has never been fully explained (Liberti and Locasale, 2016).
Aerobic glycolysis produces less ATP per mole of glucose than complete glucose oxidation to CO2, raising the question of why some cells engage in a metabolic program that is less efficient with respect to ATP generation (Koppenol et al., 2011; Liberti and Locasale, 2016; Vander Heiden et al., 2009). This paradox is particularly apparent in cancer and led to the hypothesis that tumors have defects in mitochondrial respiration (Warburg, 1956). Others have suggested that this phenotype is caused by tumor-associated hypoxia (Gatenby and Gillies, 2004); however, aerobic glycolysis is a feature of many cells without a precedent oxygen limitation (Vander Heiden et al., 2009). Tumors also retain functional mitochondria (Koppenol et al., 2011; Weinhouse, 1956) and require mitochondrial respiration for growth, progression, and metastasis (LeBleu et al., 2014; Tan et al., 2015; Viale et al., 2014; Weinberg et al., 2010). Collectively, these findings argue against mitochondrial damage or oxygen limitation as the primary driver of this phenotype. Furthermore, it is a common misconception that aerobic glycolysis involves suppression of oxidative phosphorylation (Yao et al., 2019). Aerobic glycolysis is best characterized by increased fermentation with continued respiration, resulting in a shift in metabolism where more glucose is fermented relative to that which is oxidized.
Several other models have been proposed to explain why aerobic glycolysis is observed in proliferating cells. One is that increased flux through glycolysis can shunt biosynthetic precursors into anabolic reactions that branch from this pathway, contributing to production of nucleosides, lipids, and/or proteins (Boroughs and DeBerardinis, 2015; Cairns et al., 2011; Hume and Weidemann, 1979; Levine and Puzio-Kuter, 2010; Vander Heiden et al., 2009). Though the idea that inefficient ATP production is a tradeoff for supporting anabolic reactions is attractive, glycolytic intermediates are not necessarily elevated in proliferating cells (Lunt et al., 2015; Williamson et al., 1970) and many proliferating cells excrete the majority of consumed glucose carbons as lactate. In fact, amino acids, rather than glucose, account for most new carbon biomass in proliferating cells (Hosios et al., 2016). Another proposed benefit of the Warburg effect is increased ATP production, as ATP can be generated with faster kinetics by aerobic glycolysis than by oxidative phosphorylation (Pfeiffer et al., 2001). It has also been suggested that aerobic glycolysis arises due to constraints on metabolism caused by molecular crowding (Vazquez et al., 2010; Vazquez and Oltvai, 2011), or that the energetic cost of synthesizing glycolytic enzymes is less than that of synthesizing components needed for respiration, such that ATP production by glycolysis confers a fitness advantage to cells (Basan et al., 2015). Nevertheless, why aerobic glycolysis is engaged in some, but not all, rapidly proliferating cells is not fully explained by existing models.
The end product of glycolysis is pyruvate, which can be fermented to lactate or further oxidized by a series of reactions that depend on mitochondrial respiration, in which electrons released by glucose oxidation are disposed of via the reduction of oxygen to water. The first step in pyruvate oxidation is catalyzed by the pyruvate dehydrogenase complex (PDH), which converts pyruvate to acetyl-CoA in a reaction that is irreversible under physiological conditions. PDH activity is suppressed by a low NAD+/NADH ratio (Pettit et al., 1975) and regulated further by pyruvate dehydrogenase kinases (PDK) and pyruvate dehydrogenase phosphatases (PDP), which modulate PDH by inhibitory phosphorylation (Kolobova et al., 2001; Korotchkina and Patel, 2001). Thus, PDH regulation influences the extent to which cells engage in aerobic glycolysis (Grassian et al., 2011; Kim et al., 2006; Papandreou et al., 2006). Activation of PDH either by PDK inhibition or PDP activation suppresses aerobic glycolysis, and can slow cancer cell proliferation and tumor growth (Hitosugi et al., 2011; Kaplon et al., 2013; McFate et al., 2008).
To investigate how aerobic glycolysis supports cell proliferation, we studied the consequence of suppressing fermentation in cells by increasing PDH activity. We find that promoting pyruvate oxidation impairs cell proliferation by limiting NAD+ availability for oxidation reactions, as interventions that regenerate NAD+ restore proliferation despite PDH activation. We further find that the reduced NAD+/NADH ratio in these cells results from increased mitochondrial membrane potential that impedes NAD+ regeneration by mitochondrial electron transport. Uncoupling electron transport from mitochondrial ATP synthesis relieves the increased mitochondrial membrane potential and increases NAD+ regeneration via respiration. Furthermore, increasing cellular ATP consumption rescues proliferation when PDH is activated, suggesting that ATP synthase insufficiency can be an endogenous constraint on NAD+ regeneration by coupled mitochondrial respiration. Lastly, endowing cells with alternative means of NAD+ regeneration suppresses aerobic glycolysis in both mammalian and yeast cells, without affecting proliferation rate. These data argue that cells engage in aerobic glycolysis when the NAD+ demand for oxidation reactions exceeds the demand for ATP, creating a situation where mitochondrial respiration is insufficient to support NAD+ regeneration.
Results
PDK inhibition activates PDH, suppressing aerobic glycolysis and cell proliferation
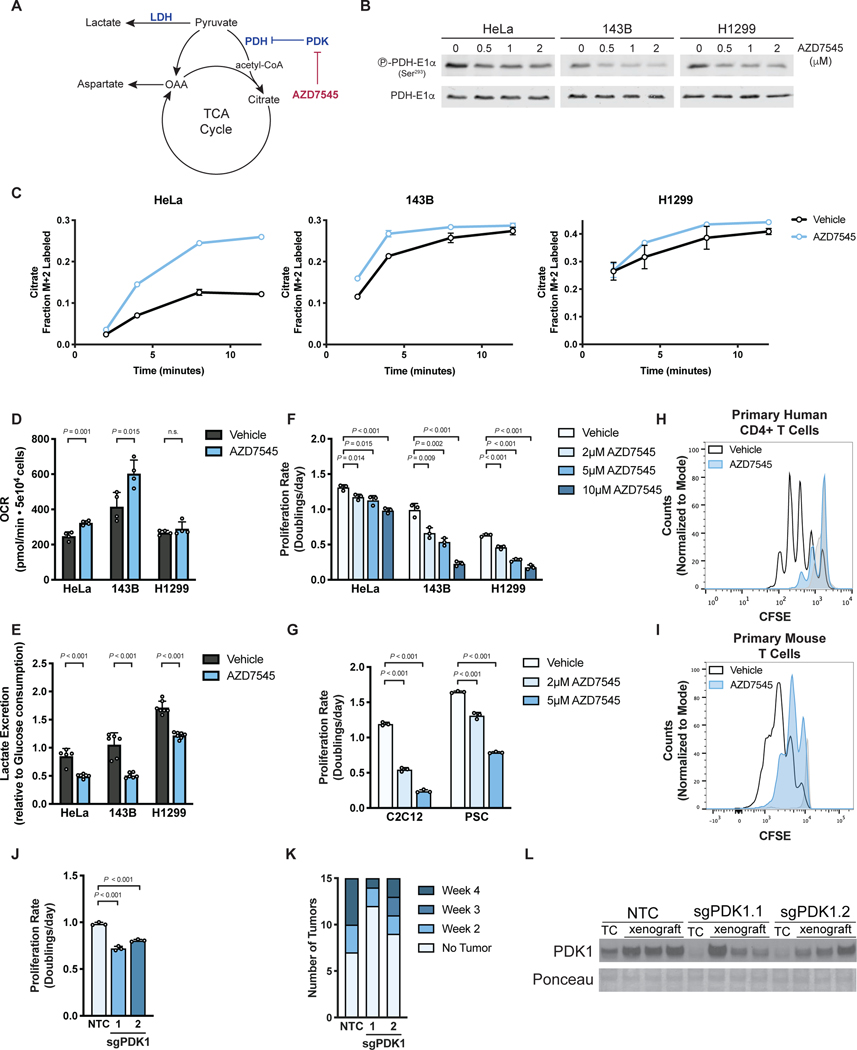
To better understand why proliferating cells engage in aerobic glycolysis, we sought to suppress this phenotype by increasing glucose oxidation relative to fermentation. Flux through PDH, the first committed step for mitochondrial glucose oxidation, is negatively regulated by PDK, and PDK inhibition can suppress aerobic glycolysis in various contexts (Hitosugi et al., 2011; Kaplon et al., 2013; McFate et al., 2008; Michelakis et al., 2010). We utilized AZD7545, a potent and selective inhibitor of PDK1, PDK2, and PDK3 (Kato et al., 2007; Morrell et al., 2003), to promote pyruvate oxidation (Fig. 1A). Exposing cancer cells to AZD7545 decreased inhibitory phosphorylation of the E1α subunit of PDH (Fig. 1B), confirming functional PDK inhibition by this small molecule. We next assessed PDH activity in cells with and without PDK inhibition by measuring incorporation of carbons from 13C-labeled glucose into citrate. We observed kinetics consistent with elevated PDH activity in cells treated with AZD7545 at multiple concentrations (Fig. 1C, Fig. S1A). Oxygen consumption rate (OCR) was also increased upon AZD7545 treatment in some, but not all, cells examined (Fig. 1D, Fig. S1B). However, AZD7545 treatment decreased the rate of lactate excretion per glucose molecule consumed in all tested cells (Fig. 1E), confirming that PDK inhibition increases PDH activity, promoting pyruvate oxidation at the expense of fermentation, which represents a shift away from aerobic glycolysis.
Figure 1. Activation of PDH suppresses aerobic glycolysis and proliferation.
(A) Pyruvate has several fates in cells, including metabolism to lactate by lactate dehydrogenase (LDH) or oxidation by pyruvate dehydrogenase (PDH) to acetyl-CoA for entry into the tricarboxylic acid (TCA) cycle. PDH is negatively regulated by pyruvate dehydrogenase kinase (PDK) enzymes, which are inhibited by the compound AZD7545.
(B) Western blot to assess S293 phosphorylation of the PDH-E1α enzyme subunit in HeLa, 143B, and H1299 cells treated with vehicle or AZD7545 for 2 hours. Total PDH-E1α expression was also assessed.
(C) Kinetic labeling of citrate from 13C-labeled glucose to assess PDH flux with and without PDK inhibition by AZD7545. HeLa, 143B and H1299 cells were incubated for 5 hours in media containing 5 mM unlabeled glucose with vehicle or 0.5 μM AZD7545, after which 20 mM [U-13C6]glucose was added. The fraction of M+2 citrate was measured by LCMS after the addition of 13C-labeled glucose (n = 3).
(D) Oxygen consumption rate (OCR) of cells treated with vehicle or 0.5 μM AZD7545 for 5 hours (n = 4).
(E) Lactate excretion into culture media normalized to glucose consumption of cells treated with vehicle or 0.25 μM AZD7545 for 48 hours (n = 5).
(F) Proliferation rate of 143B, H1299, and HeLa cells treated with vehicle or AZD7545 as indicated (n = 3).
(G) Proliferation rate of C2C12 myoblasts or mouse pancreatic stellate cells (PSC) cultured in vehicle or AZD7545 as indicated (n = 3).
(H) Proliferation of primary human CD4+ T cells cultured in vehicle or 5 μM AZD7545. Human CD4+ T cells were stained with CFSE prior to stimulation with anti-CD3/CD28 dynabeads, and CFSE fluorescence was assessed by flow cytometry after 4 days. Representative data are shown from 3 biological replicates of primary human CD4+ T cells collected from different donors and analyzed as independent experiments. Stained, unstimulated cells (light grey) that did not proliferate are also shown.
(I) Proliferation of primary mouse T cells cultured in vehicle or 5 μM AZD7545. Mouse T cells were stained with CFSE prior to stimulation with anti-CD3/CD28 antibodies, and CFSE fluorescence was assessed by flow cytometry after 2 days. Stained, unstimulated cells (light grey) that did not proliferate are also shown.
(J) Proliferation rate of 143B cells in which CRISPR interference (CRISPRi) was used to repress PDK1 expression. Cells were transduced with sgRNA targeting PDK1 (two independently targeted lines) or a non-targeting control (NTC) as indicated (n = 3).
(K) Histogram indicating the number of weeks at which the cell lines described in (J) formed xenograft tumors larger than 50 mm3 in nude mice (n = 15).
(L) Western blot analysis to assess PDK1 expression in the cells shown in panel (J) when cultured in vitro (TC) or after being isolated from the xenografts described in (K) after 34 days.
Values in panels C, D, E, F, G, and J denote mean ± SD. P values were calculated by unpaired, two-tailed Student’s t-test (n.s. = not significant).
We next tested the effects of AZD7545 on cell proliferation and found that suppressing aerobic glycolysis decreased proliferation of cancer cells (Fig. 1F), non-transformed C2C12 myoblasts and primary murine pancreatic stellate cells (PSCs)(Fig. 1G). AZD7545 also suppressed proliferation of activated primary human CD4+ T cells and primary mouse T cells (Fig. 1H,I; Fig. S1C,D) while having minimal effect on expression of T cell activation markers (Fig. S1E). These data demonstrate that PDH activation impairs proliferation in both cancer and non-cancer settings.
To confirm that AZD7545 inhibits cell proliferation as a result of PDK inhibition rather than another effect of this compound, we disrupted PDK1 expression in 143B cells using CRISPR interference (CRISPRi) (Fig. S1F) and found that genetic PDK1 suppression slowed cell proliferation (Fig. 1J). Furthermore, high PDH activity is selected against for tumor growth in mice, as 143B cells with PDK1 knockdown displayed impaired tumor formation relative to control cells (Fig. 1K). Of note, when PDK1 knockdown cells formed tumors, some of these tumors grew at a similar rate as tumors derived from control cells (Fig. S1G); however, tumors derived from PDK1 knockdown cells regained PDK1 expression (Fig. 1L). Furthermore, control and PDK1 knockdown tumors expressed higher levels of PDK1 than the cells from which they were derived (Fig. 1L). Taken together, these data are consistent with numerous studies showing enhanced PDH activation specifically (Kaplon et al., 2013; McFate et al., 2008; Michelakis et al., 2010), and suppressing aerobic glycolysis more generally (Fantin et al., 2006; Le et al., 2010; Xie et al., 2014), can slow cell proliferation and tumor growth.
PDH activation decreases proliferation by decreasing the NAD+/NADH ratio in cells
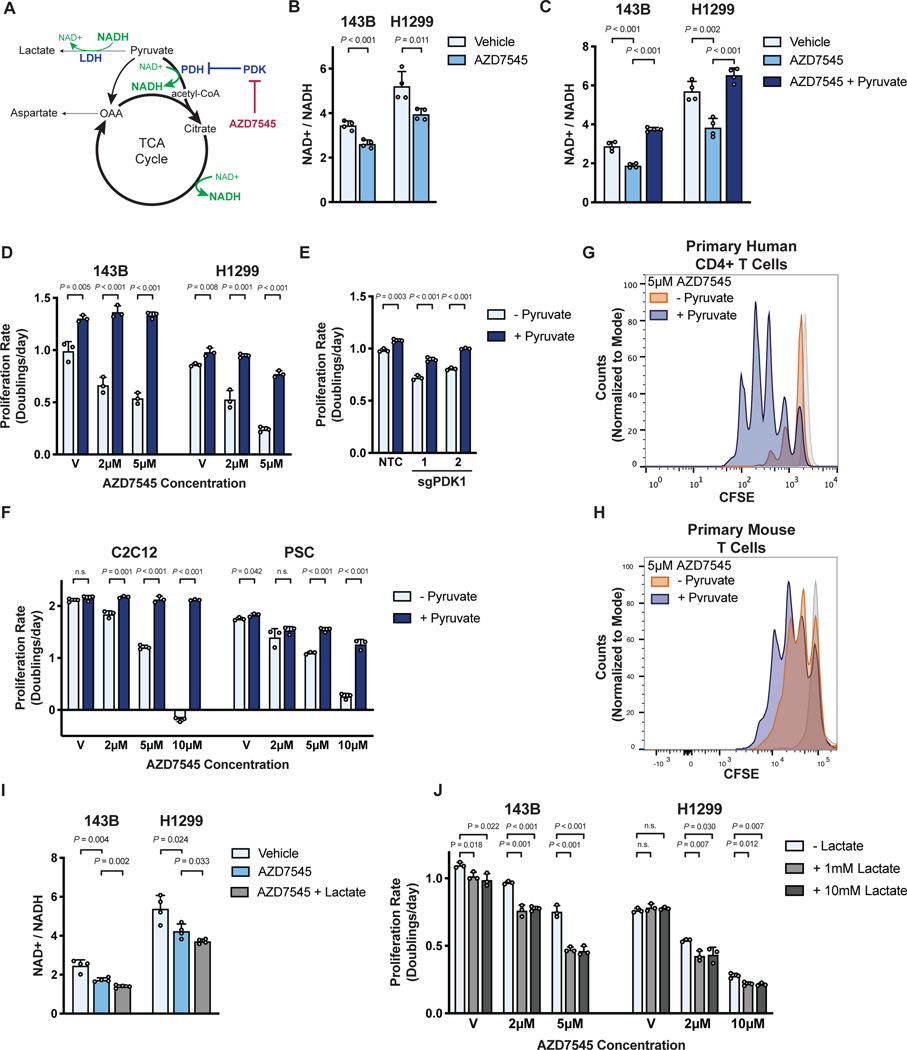
Understanding how increased pyruvate oxidation suppresses cell proliferation could provide insight into why proliferating cells engage in aerobic glycolysis. One potential explanation is that altering the ratio of glucose oxidation to lactate fermentation can affect the oxidative capacity of cells. The NAD+/NADH ratio is critical for many metabolic reactions, including those involved in central carbon metabolism, nucleotide synthesis, lipid metabolism, and amino acid metabolism (Hosios and Vander Heiden, 2018), and NAD+ regeneration can be limiting for cell proliferation and tumor growth (Birsoy et al., 2015; Gui et al., 2016; Sullivan et al., 2015; Titov et al., 2016). The reaction catalyzed by PDH consumes NAD+ to produce NADH, and shunting pyruvate away from lactate production prevents NAD+ regeneration by lactate dehydrogenase (LDH) (Fig. 2A). Thus, PDK inhibition is expected to favor a more reduced NAD+/NADH ratio, and indeed AZD7545 treatment lowered the NAD+/NADH ratio in cells (Fig. 2B, Fig. S2A).
Figure 2. PDK inhibition slows cell proliferation by reducing the NAD+/NADH ratio.
(A) PDK inhibition by AZD7545 decreases the NAD+/NADH ratio by promoting flux through NAD+ consuming pathways, including PDH and the TCA cycle, while limiting pyruvate conversion to lactate by LDH.
(B) NAD+/NADH ratio of 143B and H1299 cells cultured in vehicle or 5 μM AZD7545 for 5 hours (n = 4).
(C) NAD+/NADH ratio of 143B and H1299 cells treated with vehicle, 5 μM AZD7545, or 5 μM AZD7545 with 1 mM pyruvate (n = 4).
(D) Proliferation rate of 143B and H1299 cells cultured in vehicle (V) or the indicated concentration of AZD7545 in the presence (dark blue) or absence (light blue) of 1 mM pyruvate (n = 3).
(E) Proliferation rate of 143B cells in which CRISPRi was used to repress PDK1. Cells were transduced with sg PDK1 (two independently targeted lines) or sgNTC and grown in the presence or absence of 1 mM pyruvate (n = 3).
(F) Proliferation rate of C2C12 myoblasts and pancreatic stellate cells (PSC) in media with vehicle (V) or AZD7545 supplemented with or without 1 mM pyruvate (n = 3).
(G) Proliferation of primary human CD4+ T cells treated with 5 μM AZD7545 with or without 1 mM pyruvate. Human CD4+ T cells were stained with CFSE prior to stimulation with CD3/CD28 dynabeads, and CFSE fluorescence was assessed by flow cytometry after 4 days. Representative data are shown from 3 biological replicates of primary human CD4+ T cells collected from different donors and analyzed as independent experiments. Stained, unstimulated cells (light grey) that did not proliferate are shown.
(H) Proliferation of primary mouse T cells treated with 5 μM AZD7545 with or without 1 mM pyruvate. Mouse T cells were stained with CFSE prior to stimulation with anti-CD3/CD28 antibodies. CFSE fluorescence was assessed by flow cytometry after 2 days. Stained, unstimulated cells (light grey) that did not proliferate are shown.
(I) NAD+/NADH ratio of 143B and H1299 cells cultured in vehicle, 5 μM AZD7545, or 5 μM AZD7545 with 10 mM lactate (n = 4).
(J) Proliferation rate of 143B and H1299 cells treated with vehicle (V) or AZD7545 with or without 1 mM or 10 mM lactate as indicated (n = 3).
Values in panels B, C, D, E, F, I and J denote mean ± SD. P values were calculated by unpaired, two-tailed Student’s t-test (n.s. = not significant).
We next questioned whether sensitivity to PDK inhibition could be changed by altering the intracellular NAD+/NADH ratio. Pyruvate is rapidly reduced by LDH, and providing exogenous pyruvate increases the NAD+/NADH ratio in cells (Birsoy et al., 2015; Gui et al., 2016; Sullivan et al., 2015). We found that pyruvate supplementation restored the NAD+/NADH ratio in AZD7545-treated cancer cells (Fig. 2C), enhanced baseline cell proliferation, and suppressed the anti-proliferative effects of AZD7545 and of genetic suppression of PDK1 (Fig. 2D,E; Fig. S2B). Pyruvate also increased the proliferation of non-transformed C2C12 myoblasts and primary mouse PSCs treated with AZD7545 (Fig. 2F), as well as of activated primary human CD4+ T cells (Fig. 2G, Fig. S2C,D) and primary mouse T cells (Fig. 2H; Fig. S2E,F). These data argue that exogenous pyruvate suppresses the anti-proliferative effect of PDH activation.
Lactate can serve as a nutrient for cells, including cancer cells (Faubert et al., 2017; Hui et al., 2017; Kennedy et al., 2013; Sonveaux et al., 2008). Lactate metabolism first requires conversion to pyruvate, serving as an electron donor for the LDH reaction that converts NAD+ to NADH (Fig. 2A), and exogenous lactate further decreased the NAD+/NADH ratio of AZD7545-treated cells (Fig. 2I, Fig. S2G). Exogenous lactate also suppressed cell proliferation and exacerbated the effect of AZD7545 treatment in 143B and H1299 cells (Fig. 2J), but not HeLa cells (Fig. S2H). Altering extracellular lactate relative to extracellular pyruvate can titrate the cellular NAD+/NADH ratio (Hung et al., 2011), and we found that lower extracellular lactate to pyruvate ratios suppressed the ability of AZD7545 to impair proliferation (Fig. S2I). These data support NAD+ depletion as an explanation for why increasing pyruvate oxidation slows proliferation.
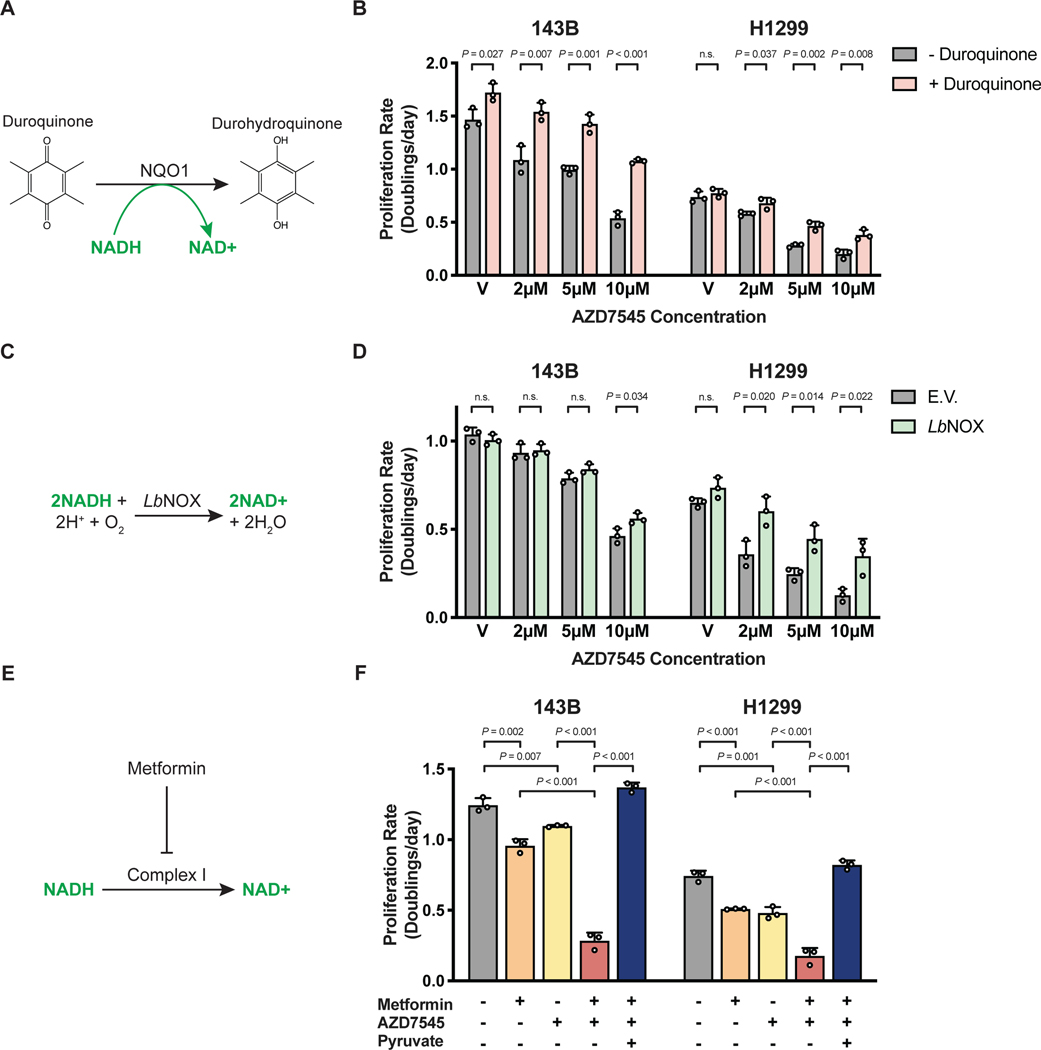
To more directly test whether changes in NAD+/NADH ratio mediate the anti-proliferative effects of PDK inhibition, we tested whether orthogonal pathways that regenerate NAD+, but are not involved in glucose, pyruvate, or lactate metabolism, affect sensitivity to AZD7545. Duroquinone permits NAD+ regeneration via the quinone reductase NQO1 (Merker et al., 2006) (Fig. 3A) and duroquinone raises the NAD+/NADH ratio and can rescue cell proliferation in conditions where NAD+ is limiting (Gui et al., 2016). We found that duroquinone suppressed AZD7545 sensitivity (Fig. 3B, Fig. S3A), consistent with decreased NAD+/NADH ratio slowing proliferation of PDK-inhibited cells.
Figure 3. Interventions that alter NAD+ availability can modulate the antiproliferative effects of PDK inhibition.
(A) Duroquinone increases NAD+ regeneration via the enzyme NAD(P)H dehydrogenase, quinone 1 (NQO1), which reduces duroquinone to durohydroquinone using NADH as a cofactor.
(B) Proliferation rate of 143B and H1299 cells treated with vehicle (V) or AZD7545 in the absence or presence of duroquinone (20 μM for 143B and 100 μM for H1299 cells; n = 3).
(C) Schematic illustrating the reaction catalyzed by the NADH oxidase from Lactobacillus brevis (LbNOX).
(D) Proliferation rate of 143B and H1299 cells transduced with empty vector (E.V.) or an LbNOX expression vector and treated with vehicle (V) or AZD7545. Doxycycline (500 ng/mL) was included in all conditions (n = 3).
(E) Schematic illustrating the redox consequences of metformin treatment.
(F) Proliferation rate of 143B and H1299 cells treated with 500 μM metformin, AZD7545 (5 μM for 143B, 3 μM for H1299 cells), and 1 mM pyruvate as indicated (n = 3).
Values in panels B, D, and F denote mean ± SD. P values were calculated by unpaired, two-tailed Student’s t-test (n.s. = not significant).
Another orthogonal method for increasing the cell NAD+/NADH ratio is expressing NADH oxidase from Lactobacillus brevis (LbNOX) (Titov et al., 2016) (Fig. 3C). Thus, we engineered cells to express either LbNOX or empty vector (E.V.) as another way to assess the effect of increasing NAD+ regeneration in cells (Fig. S3B). LbNOX expression conferred resistance to the anti-proliferative effects of AZD7545 (Fig. 3D, Fig. S3C), further demonstrating that increasing NAD+ regeneration can permit rapid proliferation despite increased pyruvate oxidation.
Changes in NAD+/NADH account for how PDH activation impacts cell metabolism
NAD+ is necessary to support oxidation reactions in cells, and thus changes in NAD+/NADH ratio impacts many metabolic pathways including those important for cell growth and proliferation (Hosios and Vander Heiden, 2018). For example, the NAD+/NADH ratio can affect aspartate synthesis (Birsoy et al., 2015; Sullivan et al., 2015). Aspartate is essential to make proteins as well as purine and pyrimidine nucleotides, and acquiring aspartate can be limiting for tumor growth (Garcia-Bermudez et al., 2018; Rabinovich et al., 2015; Sullivan et al., 2018). Therefore, to determine whether one way PDK inhibition impairs proliferation is by affecting aspartate availability, we measured aspartate levels in cells cultured in the presence of vehicle or AZD7545. Consistent with AZD7545 treatment reducing the cell NAD+/NADH ratio, PDK inhibition decreased intracellular aspartate (Fig. S3D), and increasing the NAD+/NADH ratio with exogenous pyruvate, duroquinone, or LbNOX expression restored aspartate levels (Fig. S3E-G). These data suggest that suppressing aerobic glycolysis limits the capacity of cells to carry out oxidation reactions important for biomass production.
To further study how PDH activation affects metabolism, we performed untargeted metabolomics and observed that despite increasing PDH activity (Fig. 1C, Fig. S1A), AZD7545 treatment decreased intracellular citrate levels and increased intracellular palmitate levels (Fig. S3H,I). This could reflect a metabolic state in which forcing citrate production from pyruvate elicits a compensatory increase in lipid synthesis, which consumes NADPH to make NADP+, potentially as an alternative means to regenerate oxidizing equivalents (Liu et al., 2020). We also observed that AZD7545 treatment affected levels of many other intracellular metabolites, and that either duroquinone or expression of LbNOX suppressed some of these changes (Fig. S3J-M). Of note, duroquinone or LbNOX expression restored many of the same metabolites in AZD7545-treated cells (Fig. S3N). Because duroquinone and LbNOX alter NAD+/NADH ratios via different mechanisms, the finding that both agents similarly restored alterations to the global metabolome caused by PDH activation argues that an effect on cell redox state is a major consequence of increased pyruvate oxidation and further supports the notion that a major metabolic consequence of suppressing aerobic glycolysis is NAD+ depletion.
PDH activation increases dependency on mitochondrial complex I for NAD+ regeneration
Since PDK inhibition reduces the cellular NAD+/NADH ratio, interventions that limit NAD+ regeneration are predicted to potentiate the anti-proliferative effects of PDK inhibitors. To test this hypothesis, we assessed the sensitivity of cancer cells to the biguanide metformin, which limits NAD+ regeneration and decreases NAD+/NADH ratio via inhibition of mitochondrial complex I (Gui et al., 2016; Wheaton et al., 2014)(Fig. 3E). The combination of metformin and AZD7545 reduced cell proliferation more than either compound alone (Fig. 3F, Fig. S4A), and AZD7545 treatment decreased the IC50 of metformin by more than 30% in both A549 and HeLa cells (Fig. S4B,C). The anti-proliferative effect of AZD7545 and metformin was completely abolished by pyruvate supplementation (Fig. 3F, Fig. S4A-C), further arguing that these drugs impair proliferation by promoting a more reduced NAD+/NADH ratio.
Electron acceptor availability can be limiting for tumor growth in some mouse models of cancer (Gui et al., 2016), suggesting that inhibiting NAD+ regeneration with the combination of AZD7545 and metformin may have a larger effect on tumor growth than either drug alone. We found that metformin inhibited tumor growth as previously reported (Gui et al., 2016; Schockel et al., 2015; Wheaton et al., 2014), and while AZD7545 had no effect alone, PDK inhibition appeared to improve the efficacy of metformin in this model (Fig. S4D).
The finding that AZD7545 treatment did not decrease tumor growth as a single agent might be expected from the pharmacokinetics of this compound, as well as the fact that AZD7545 impairs, but does not prevent, cell proliferation. Three hours after dosing, we find AZD7545 levels in tumors and in plasma that can impair cell proliferation in culture (Fig. S4E,F), but the compound was not detected in plasma after 24 hours (Fig. S4E). This argues that more frequent dosing of AZD7545 may be more effective to inhibit tumor growth as a single agent. Nevertheless, once a day dosing of AZD7545 was sufficient to improve the anti-tumor effects of metformin, supporting the notion that PDK inhibition increases dependency on mitochondrial complex I to regenerate NAD+ and support proliferation.
Increased mitochondrial membrane potential downstream of pyruvate oxidation impairs NAD+ regeneration by the mitochondrial electron transport chain
The decreased NAD+/NADH ratio observed in PDK-inhibited cells suggests that NAD+ regeneration by mitochondrial respiration is insufficient to support maximal proliferation when pyruvate oxidation is increased. This result is unexpected, as these cells are cultured at atmospheric oxygen and exhibit increased oxygen consumption upon AZD7545 treatment (Fig. 1D). Thus, we sought to better understand how NAD+ regeneration by mitochondrial respiration was impaired in PDK-inhibited cells, as this could explain why proliferating cells engage in aerobic glycolysis.
The oxidation-reduction reactions of the mitochondrial electron transport chain (ETC) are coupled to proton pumping from the mitochondrial matrix into the intermembrane space to generate an electrochemical gradient across the inner mitochondrial membrane and support ATP production via the FoF1-ATP synthase (Fig. 4A). This process, collectively referred to as oxidative phosphorylation, occurs at near equilibrium, meaning that the rate of respiration is a function of both substrate and product availability (Brown, 1992). The shift toward a more reduced NAD+/NADH ratio suggests NADH is more readily available in these cells, but one possibility is that the availability of oxygen, another major substrate for oxidative phosphorylation, becomes limiting for electron transport. However, these cells were cultured at 21% oxygen, and mitochondrial respiration can function at oxygen levels as low as 0.5% (Chandel et al., 1996; Rumsey et al., 1990). Furthermore, expression of LbNOX, which requires oxygen as a substrate, suppressed the anti-proliferative effect of AZD7545 (Fig. 3D), arguing against oxygen availability constraining NAD+ regeneration in PDK-inhibited cells.
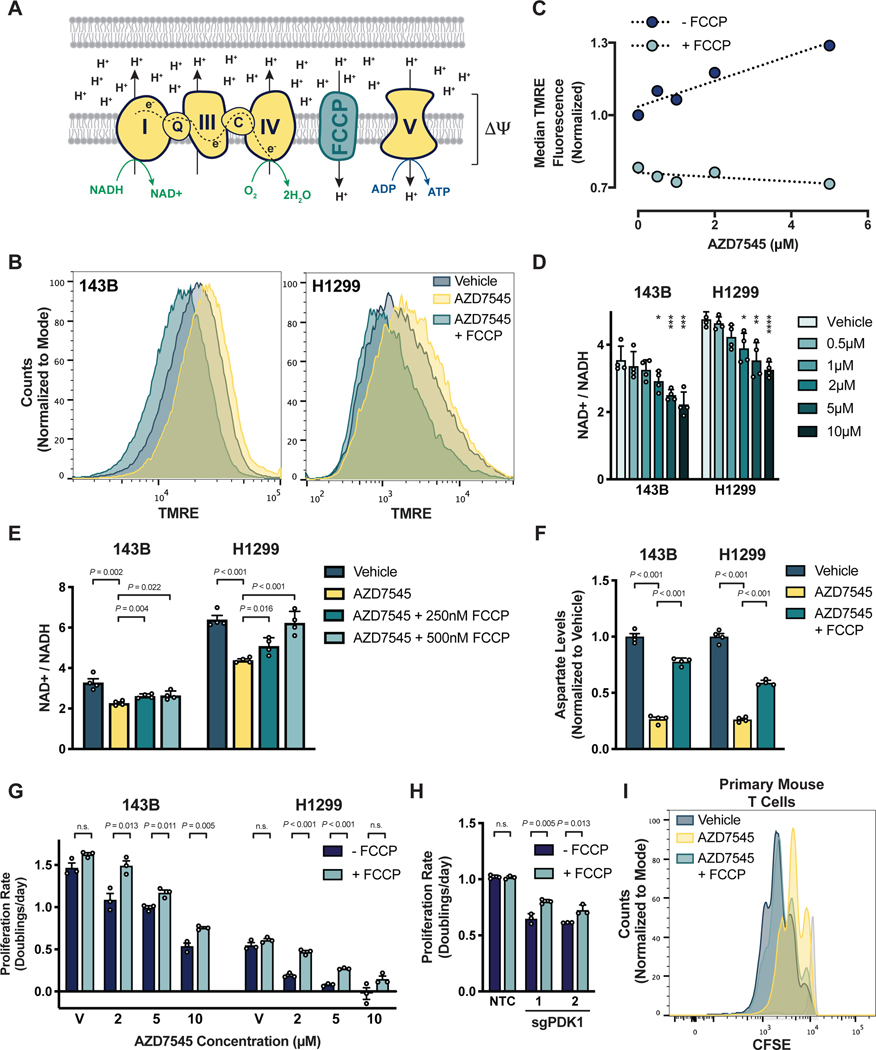
Figure 4. PDK inhibition induces mitochondria hyperpolarization and limits NAD+ regeneration by respiration.
(A) Schematic illustrating the mitochondrial electron transport chain and how FCCP (trifluoromethoxy carbonylcyanide phenylhydrazone) uncouples electron transfer from NADH to O2 from ATP production by the FoF1-ATP synthase (Complex V). ΔΨ denotes the mitochondrial membrane potential.
(B) Mitochondrial membrane potential, as reflected by TMRE (tetramethylrhodamine, ethyl ester) fluorescence, in 143B and H1299 cells treated with vehicle, 5 μM AZD7545, or 5 μM AZD7545 with 500 nM FCCP.
(C) Mitochondrial membrane potential, as reflected by TMRE fluorescence, in H1299 cells that had been treated with the indicated concentration of AZD7545, with or without 500 nM FCCP. TMRE fluorescence of 50,000 cells was quantified by flow cytometry and normalized to the vehicle-treated condition without FCCP.
(D) NAD+/NADH ratio of 143B and H1299 cells cultured in vehicle or AZD7545 for 5 hours (n = 4)
(E) NAD+/NADH ratio of 5143B and H1299 cells treated with vehicle, 5 μM AZD7545, or 5 μM AZD7545 with the indicated concentration of FCCP for 5 hours (n = 4).
(F) Aspartate levels in 143B and H1299 cells cultured in vehicle, 2 μM AZD7545, or 2 μM AZD7545 with 250 nM FCCP for 5 hours as measured by LCMS (n = 4).
(G) Proliferation rate of 143B and H1299 cells treated with vehicle (V) or AZD7545 with or without 500 nM FCCP as indicated (n = 3).
(H) Proliferation rate of 143B cells in which CRISPRi was used to repress PDK1 expression. Cells were transduced with sgPDK1 (two independently targeted lines) or sgNTC and cultured with or without 750 nM FCCP (n = 3).
(I) Proliferation of primary mouse T cells cultured in vehicle, 5 μM AZD7545, or 5 μM AZD7545 with 500 nM FCCP was assessed by CFSE dye dilution. Mouse T cells were stained with CFSE prior to stimulation with anti-CD3/CD28 antibodies and CFSE fluorescence was assessed by flow cytometry after 2 days. Stained, unstimulated cells (light grey) that did not proliferate are shown.
Values in panels D, E, F, G, and H denote mean ± SD. P values were calculated by unpaired, two-tailed Student’s t-test (*, **, ***, and **** denote P < 0.05, 0.01, 0.005, and 0.001; n.s. = not significant).
Another possibility is that an imbalance develops between the ability of the mitochondrial ETC to pump protons into the intermembrane space and the activity of processes that carry protons back across the membrane. This imbalance would be reflected in an increased mitochondrial membrane potential (ΔΨ)(Fig. 4A). To test this possibility, we utilized the cationic dye tetramethylrhodamine ethyl ester (TMRE), which is taken up by mitochondria in proportion to ΔΨ (Perry et al., 2011). We found that PDK inhibition increases TMRE accumulation in cancer cells (Fig. 4B) and activated primary mouse T cells (Fig. S5A), and that this effect is reversed when respiration is uncoupled from ATP production by the ionophore FCCP (trifluoromethoxy carbonylcyanide phenylhydrazone). FCCP allows protons to equilibrate across membranes, thus FCCP uncouples NAD+ regeneration by the ETC from ΔΨ generation and prevents use of ΔΨ for ATP production. These data are consistent with PDH activation increasing ΔΨ, and increased TMRE accumulation depends on the concentration of AZD7545 (Fig. 4C) in a way that matches the dose-dependent reduction in the NAD+/NADH ratio observed upon PDK inhibition (Fig. 4D, Fig. S5B). Taken together, these data suggest that NAD+ regeneration by respiration is constrained by ΔΨ, as it becomes thermodynamically unfavorable for the mitochondrial ETC complexes to pump protons across a hyperpolarized membrane.
If high ΔΨ limits NAD+ regeneration by respiration, collapse of ΔΨ should restore NAD+/NADH homeostasis to cells with activated PDH. Indeed, FCCP exposure was sufficient to increase the NAD+/NADH ratio (Fig. 4E), aspartate levels (Fig. 4F, Fig. S5C) and proliferation rate (Fig. 4G, Fig. S5D) of PDK-inhibited cells. FCCP treatment also suppressed the proliferation defect observed in cancer cells where PDK1 expression is suppressed using CRISPRi (Fig. 4H). Additionally, FCCP treatment rescued the proliferation rates of AZD7545-treated primary mouse T cells (Fig. 4I), but not of primary human T cells. These results are consistent with ΔΨ limiting mitochondrial NAD+ regeneration. In addition, the finding that FCCP increases respiration (Fig. S1B) confirms that the mitochondrial ETC machinery is functional and that sufficient oxygen is available to support increased respiration in these cells. Taken together, these data argue that mitochondrial NAD+ regeneration becomes limited by an increase in ΔΨ when pyruvate oxidation is increased.
NAD+ regeneration is limited by insufficient ATP hydrolysis when PDK is inhibited
A physiological function of ΔΨ is to support ATP synthesis, as conversion of ADP to ATP by the FoF1-ATP synthase is coupled to dissipation of ΔΨ (Fig. 4A). Thus, insufficient ATP turnover could limit ADP availability, limiting mitochondrial NAD+ regeneration by oxidative phosphorylation in cells with high pyruvate dehydrogenase activity. The observation that FCCP treatment promotes proliferation of PDK-inhibited cells by uncoupling mitochondrial NAD+ regeneration from ATP synthesis is consistent with this model.
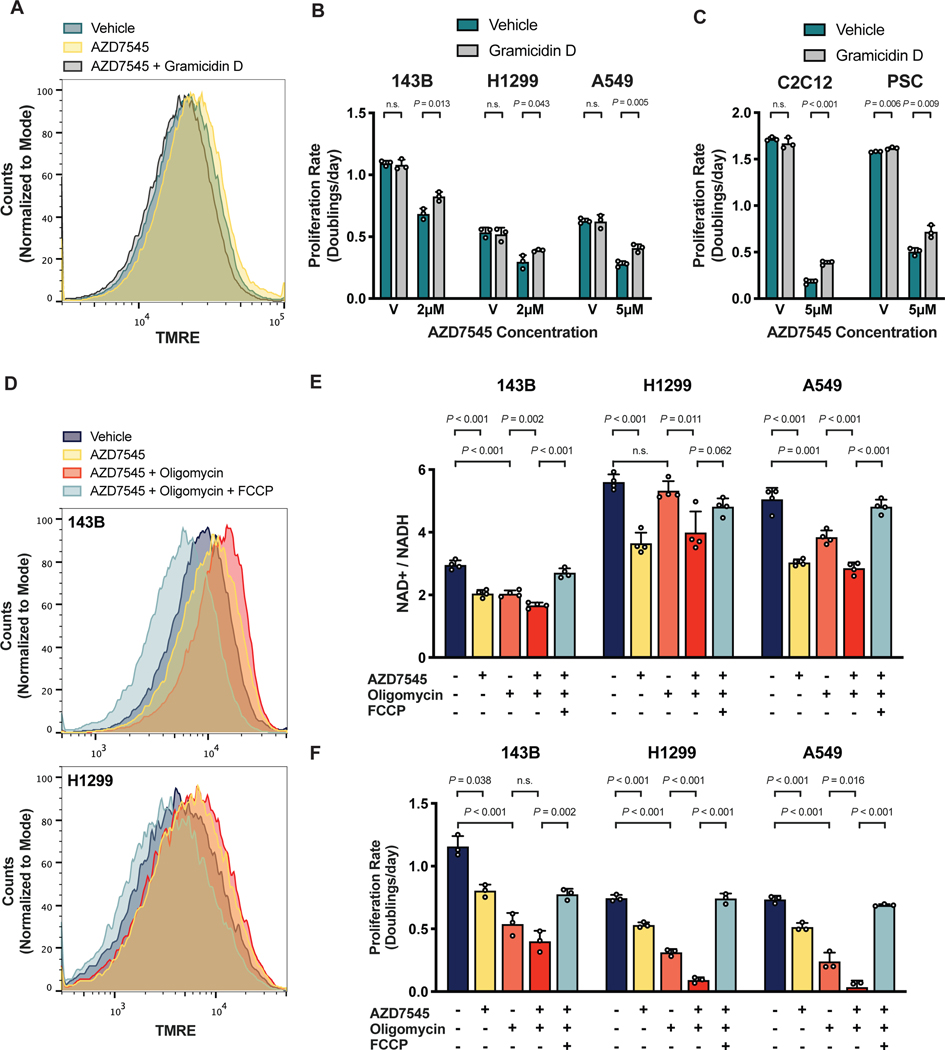
To better characterize potential FoF1-ATP synthase insufficiency following PDK1 inhibition, we measured intracellular ATP/ADP ratios and found that these were unchanged by AZD7545 treatment (Fig. S5E). This observation is consistent with existing notions that ATP is only made as needed because thermodynamic constraints prevent ATP storage in cells (Bonora et al., 2012). In order to test if the FoF1-ATP synthase is unable to dissipate ΔΨ due to reduced ADP availability, we next tested if increasing ATP to ADP conversion in cells could reverse the metabolic and anti-proliferative effects of PDK inhibition. Promoting cellular ATP consumption stimulates mitochondrial respiration by increasing the demand for ADP to ATP conversion (Bertholet et al., 2019; Brown, 1992). The toxin gramicidin D is a classic way to increase ATP consumption in cells by increasing cell membrane permeability to Na+ and K+ ions, driving increased ATP hydrolysis by Na+/K+-ATPase and thus increasing ATP-coupled mitochondrial respiration (Buttgereit and Brand, 1995; Nobes et al., 1989; Vander Heiden et al., 1999). We first confirmed that increasing cellular ATP consumption with gramicidin D reversed mitochondrial membrane hyperpolarization caused by PDK-inhibition (Fig. 5A). Exposing PDK-inhibited cells to gramicidin D also resulted in a more oxidized NAD+/NADH ratio (Fig. S5F). These data are consistent with gramicidin D increasing mitochondrial ATP production, which in turn promotes increased respiration and NAD+ regeneration. Of note, gramicidin D also rendered both cancer cells and non-cancer cells more resistant to AZD7545 treatment (Fig. 5B,C). Taken together, these data support a model where mitochondrial NAD+ regeneration via respiration is constrained by insufficient ATP synthase activity when pyruvate oxidation is increased. These findings also suggest that cells may engage in aerobic glycolysis under conditions where the demand for NAD+ regeneration exceeds the rate of ATP consumption.
Figure 5. NAD+ regeneration by respiration is limited by the rate of mitochondrial ATP production.
(A) Mitochondrial membrane potential, as reflected by TMRE fluorescence, of 143B cells cultured in vehicle, 2 μM AZD754, or 2 μM AZD754 with 5 nM gramicidin D for 5 hours.
(B) Proliferation rate of 143B, H1299 and A549 cells treated with vehicle (V) or AZD7545 in the presence or absence of 0.5 nM gramicidin D (n = 3).
(C) Proliferation rate C2C12 myoblasts or pancreatic stellate cells (PSC) cultured with vehicle (V) or AZD7545 with or without 1 nM gramicidin D (n = 3).
(D) Mitochondrial membrane potential, as reflected by TMRE fluorescence, of 143B cells cultured for 5 hours with vehicle, 2 μM AZD7545, 1 nM oligomycin, and 1 μM FCCP as indicated.
(E) NAD+/NADH ratio of 143B, H1299, and A549 cells in vehicle, 5 μM AZD7545, 0.5 nM oligomycin, and 1 μM FCCP as indicated (n = 4).
(F) Proliferation rate of 143B, H1299, and A549 cells in vehicle, 5 μM AZD7545, 0.5 nM oligomycin and 1 μM FCCP as indicated (n = 3).
Values in panels B, C, E, and F denote mean ± SD. P values were calculated by unpaired, two-tailed Student’s t-test (n.s. = not significant).
The demand for NAD+ regeneration can supersede the requirement for ATP in proliferating cells
To begin to test whether a mismatch in demand for mitochondrial NAD+ regeneration relative to demand for mitochondrial ATP production could drive aerobic glycolysis, we utilized oligomycin, a selective inhibitor of the Fo-subunit of the mitochondrial FoF1-ATP synthase. Oligomycin inhibits mitochondrial ATP production and prevents respiration by increasing ΔΨ (Brand and Nicholls, 2011). Culturing PDK-inhibited cells with oligomycin further increases ΔΨ beyond that observed with AZD7545 treatment alone (Fig. 5D). Therefore, oligomycin is expected to further decrease the capacity for NAD+ regeneration via mitochondrial respiration. We found that oligomycin potentiates the proliferation defect and the reduced NAD+/NADH ratio caused by PDK inhibition (Fig. 5E,F). FCCP treatment, which collapses ΔΨ and suppresses ATP synthesis by the mitochondrial FoF1-ATP synthase, restores the NAD+/NADH ratio and the proliferation of cells treated with both oligomycin and AZD7545. These data suggest that mitochondrial respiration to allow NAD+ regeneration can be more important than mitochondrial ATP production in some proliferating cells, including cells that otherwise engage in aerobic glycolysis.
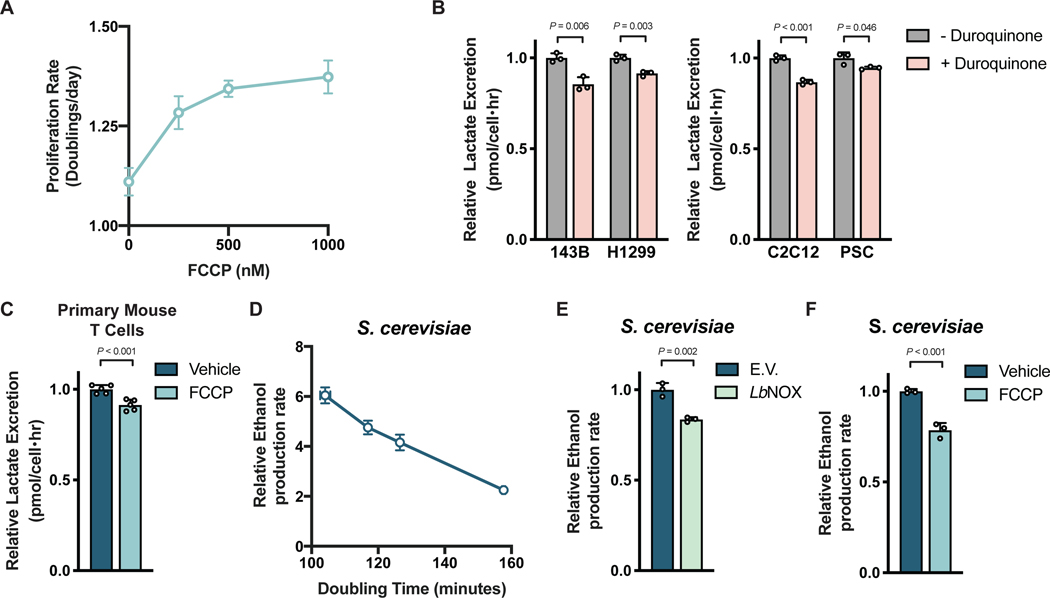
The finding that uncoupling mitochondrial respiration from ATP production can rescue proliferation of PDK-inhibited cells argues that insufficient FoF1-ATP synthase activity can limit NAD+ regeneration by respiration. Consistent with the demand for NAD+ regeneration exceeding the demand for mitochondrial ATP synthesis to support proliferation in some cells that engage in aerobic glycolysis, we find FCCP can dose-dependently increase cell proliferation in the absence of any other interventions (Fig. 6A). These data suggest that insufficient FoF1-ATP synthase activity can impair NAD+ regeneration and proliferation even in standard culture conditions with abundant oxygen, and argue that coupling between mitochondrial respiration and ATP synthesis limits the extent to which respiration can support NAD+ regeneration and proliferation. Thus, cells may engage in aerobic glycolysis when the demand for NAD+ to support oxidation reactions is greater than the ATP turnover rate.
Figure 6. Aerobic glycolysis reflects cellular NAD+ availability.
(A) The proliferation rate of 143B cells treated with FCCP as indicated (n = 3).
(B) Relative lactate excretion of cells cultured with or without duroquinone (4μM, 16μM, 8μM and 64μM for 143B, H1299, C2C12, and PSC cells, respectively; n = 3).
(C) Relative lactate excretion of primary mouse T cells stimulated with anti-CD3/CD28 antibodies in the presence or absence of 250 nM FCCP for 1 day (n = 5).
(D) The relationship between ethanol production and proliferation rate in S. cerevisiae as determined by altering glucose concentration in culture medium.
(E) Relative ethanol production rate by S. cerevisiae expressing empty vector (E.V.) or LbNOX in standard medium containing 3% glucose (n = 3).
(F) Relative ethanol production rate by S. cerevisiae treated with vehicle or 2 μM FCCP in standard medium containing 3% glucose (n = 3).
Values denote mean ± SD. P values were calculated by unpaired, two-tailed Student’s t-test (n.s. = not significant).
Cellular NAD+ availability determines whether cells engage in aerobic glycolysis
If increased fermentation in proliferating cells is driven by NAD+ demand in excess of ATP demand, orthogonal pathways that promote NAD+ regeneration would be expected to suppress the degree to which cells engage in aerobic glycolysis despite the same redox and ATP requirements to support proliferation. Indeed, duroquinone can suppress the rate of lactate excretion by both cancer cells and non-transformed cells (Fig. 6B, Fig. S6A) despite similar rates of proliferation and glucose uptake (Fig. S6B,C). LbNOX expression did not affect lactate excretion in the cells studied, but increased pyruvate excretion as previously reported (Titov et al., 2016)(Fig. S6D), which reflects decreased NAD+ regeneration by LDH. Additionally, FCCP administration decreased lactate excretion by activated mouse T cells (Fig. 6C) without changing proliferation rate (Fig. S6E). Thus, increasing NAD+ regeneration either by providing exogenous electron acceptors or by uncoupling mitochondrial respiration from ATP synthesis suppresses aerobic glycolysis without decreasing proliferation rate. These data support the hypothesis that demand for NAD+ is what drives aerobic glycolysis in these rapidly proliferating mammalian cells.
Aerobic glycolysis is linked to rapid proliferation across many biological contexts. To test whether a mismatch in the demand for NAD+ and ATP contributes to aerobic glycolysis in another system we considered Saccharomyces cerevisiae, a yeast where glucose fermentation to ethanol accompanies rapid proliferation even in aerobic conditions (De Deken, 1966). We confirmed that glucose availability can affect proliferation rate and ethanol production in batch cultures of S. cerevisiae, even in aerated cultures (Fig. S6F,G). We found a linear relationship between proliferation and fermentation rates (Fig. 6D), confirming that aerobic glycolysis is correlated with proliferation in this yeast. To test whether fermentation is driven by the increased NAD+ demand of rapid proliferation in S. cerevisiae, assessed the effect of LbNOX expression on both proliferation and aerobic glycolysis. LbNOX expression decreased ethanol production without altering proliferation rate (Fig. 6E, Fig. S6H), arguing that demand for NAD+ also promotes aerobic glycolysis in this organism, consistent with previous reports (Vemuri et al., 2007).
To test the hypothesis that elevated mitochondrial membrane potential limits the ability of respiration to support NAD+ regeneration in S. cerevisiae, we also assessed the effect of FCCP on both proliferation and aerobic glycolysis. In agreement with our hypothesis, we saw that FCCP decreased ethanol production rate without altering proliferation (Fig. 6F, Fig. S6I). The fact that FCCP reduces mitochondrial ATP production, as well as fermentation rates, argues that ATP synthase in rapidly proliferating yeast is governed by the same limitations as rapidly proliferating mammalian cells. When coupled with the finding that the link between rapid proliferation and aerobic glycolysis is broken if cells are allowed alternative pathways of NAD+ regeneration, these observations suggest that demand for NAD+ in excess of demand for ATP drives aerobic glycolysis in diverse organisms across kingdoms of life, regardless of whether lactate or ethanol are produced as the fermentation product.
Discussion
Aerobic glycolysis is observed across species ranging from prokaryotes to specific mammalian cell types, yet a generalizable explanation for this phenotype has been lacking. The data from this study suggest that aerobic glycolysis reflects a metabolic state where the demand for NAD+ exceeds the demand for ATP to support cell function. Oxidation of pyruvate, rather than fermentation, increases the demand for mitochondrial respiration to regenerate NAD+. However, because mitochondrial electron transport is coupled to mitochondrial ATP synthesis, when the demand for NAD+ exceeds the demand for ATP, insufficient ATP synthase activity leads to increased ΔΨ and constrains further increases in mitochondrial respiration. Thus, ATP hydrolysis, which supplies ADP as substrate for mitochondrial ATP synthesis and dissipates ΔΨ produced by the mitochondrial ETC, imposes an upper limit on the rate of mitochondrial NAD+ regeneration regardless of oxygen availability. Therefore, if the requirement for NAD+ to fuel oxidation reactions is greater than the rate of ATP turnover, pyruvate oxidation is limited and the more reduced NAD+/NADH ratio promotes fermentation even if oxygen is present. These findings are consistent with a theoretical prediction that NADH generation could impair biomass synthesis in aerobic conditions if proliferation is fast enough (Fernandez-de-Cossio-Diaz and Vazquez, 2017).
Though the reactions that regenerate NAD+ do not directly provide biomass to cells, this cofactor is needed to catabolize reduced nutrients, including sugars and lipids, and to synthesize oxidized macromolecules, such as nucleotides and amino acids (Hosios and Vander Heiden, 2018). Previous work has identified aspartate synthesis as a major NAD+ demand (Birsoy et al., 2015; Garcia-Bermudez et al., 2018; Gui et al., 2016; Sullivan et al., 2015; Sullivan et al., 2018), and cells require NAD+ to support additional cellular processes, including serine and folate synthesis (Baksh et al., 2020; Bao et al., 2016; Diehl et al., 2019), histone deacetylation by sirtuins, maintenance of calcium homeostasis, and poly(ADP-ribose) polymerase activity (Canto et al., 2015; Ying, 2008). Electron acceptor availability can restrict cell proliferation, and the NAD+/NADH ratio has been found to correlate with tumor growth in vivo (Gui et al., 2016). Non-proliferating cells with high anabolic demands, such as mammalian retina pigmented epithelial cells, also require NAD+, and retinal toxicity has proven to be a liability for drugs that target NAD+ synthesis (Zabka et al., 2015). Lastly, forced pyruvate oxidation induces NAD+ deficiency in epidermal stem cells which can impair biomass production (Baksh et al., 2020), further arguing that demand for NAD+ can be high in some cells.
Several explanations have been proposed for the Warburg effect, many of which consider how inefficient ATP production by aerobic glycolysis is sufficient to support the high ATP demands of proliferation. The finding that uncoupling respiration from ATP production allows increased pyruvate oxidation and continued proliferation, even in the presence of oligomycin, argues strongly that the requirement for NAD+ can be greater than the requirement for ATP in at least some proliferating cells. Furthermore, the observation that increasing ATP consumption also allows proliferation, despite increased pyruvate oxidation, suggests insufficient ATP turnover can limit mitochondria respiration in cells and that aerobic glycolysis may therefore reflect a state of high NAD+ demand and excess ATP. The idea that ATP hydrolysis, rather than ATP synthesis, could be limiting for cancer cell metabolism has historical support (Racker, 1972; Scholnick et al., 1973), although this possibility is not considered by most studies. Nevertheless, it has been reported that some proliferating cells engage in futile metabolic cycles that consume ATP, such as fatty acid synthesis and oxidation (Yao et al., 2016), increased protein turnover (Zhang et al., 2014b), or the creatine shuttle (Kurmi et al., 2018) for unclear reasons. These futile cycles may promote ATP consumption to support oxidized biomass synthesis, and could explain why increased ATP turnover involving ENTPD5 can promote the proliferation of PTEN-null cancers or why increasing the ATP/AMP ratio can result in tumor regression (Fang et al., 2010; Naguib et al., 2018). The mechanism by which ATP consumption promotes proliferation in those contexts remains unexplained, but a model where excess ATP constrains NAD+ regeneration by respiration is consistent with increasing ATP consumption enabling proliferation.
While demand for NAD+ that exceeds demand for ATP can explain why some cells engage in fermentation rather than oxidative metabolism, this model does not explain why increased glucose uptake and glycolysis are also often associated with proliferation. Fermentation is redox neutral and does not net regenerate NAD+. In fact, the only known output of fermentation is ATP, and if excess ATP limits net NAD+ regeneration to promote fermentation, why glucose metabolism is increased during rapid proliferation remains unexplained. Nevertheless, a model where aerobic glycolysis reflects a metabolic state in which the requirement for NAD+ supersedes the demand for ATP is consistent with the reality of this metabolic phenotype, which is best characterized by a relative increase in fermentation, rather than a complete switch from mitochondrial respiration to glycolysis. It also fits with the broad association between aerobic glycolysis and proliferation (Diaz-Ruiz et al., 2011), the link between this phenotype and nucleotide synthesis (Lunt et al., 2015; Wang et al., 1976), the continued dependence of most proliferating cells on respiration (Howell and Sager, 1979; Tan et al., 2015; Weinberg et al., 2010; Wheaton et al., 2014; Zhang et al., 2014a), and the fact that reversing aerobic glycolysis slows rather than stops proliferation.
Different species of yeast diverge in their response to glucose in terms of their propensity to ferment carbon (De Deken, 1966), and organisms that proliferate rapidly without engaging in aerobic glycolysis may have evolved mechanisms to prevent a mismatch in the demand for NAD+ and ATP. Of note, regulated uncoupling of mitochondrial respiration from ATP synthesis is important for thermoregulation and these same processes could also facilitate rapid proliferation (Li et al., 2020). While mammalian uncoupling proteins have limited tissue expression and are tightly regulated, these proteins are overexpressed in some cancers (Kawashima et al., 2020; Robbins and Zhao, 2011; Valle et al., 2010). Expression of uncoupling proteins, as well as other means to uncouple mitochondrial respiration (Bertholet et al., 2019), may support NAD+ regeneration in contexts when NAD+ demand exceeds the demand for ATP. For example, lymphocytes can proliferate faster than most other mammalian cells, and may use such an adaptive program to support proliferation, potentially explaining why we observe variability in whether FCCP-mediated mitochondrial uncoupling can enhance T cell proliferation. However, the fact that maintaining a high ATP/ADP ratio is essential for cell survival may explain why nutrient oxidation remains tightly coupled in ATP production in most cells and leads to aerobic glycolysis being engaged when the demand for NAD+ exceeds the demand for ATP.
STAR Methods
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Lead Contact, Matthew G. Vander Heiden (mvh@mit.edu)
Materials Availability
This study did not generate new unique reagents
Data and Code Availability
This study did not generate any datasets or code. Unprocessed and uncropped image data is available http://dx.doi.org/10.17632/6zpghspxgs.1.
Experimental Model Details
Cell Culture Experiments
Established cell lines (143B, ATCC CRL-8303; A549, ATCC CCL-185; C2C12, ATCC CRL-1772; ATCC CRL-5803; H1299, ATCC CRL-5803; HeLa, ATCC CCL-2; A172, ATCC CRL-1620; MDA-MB-231, ATCC HTB-26) were maintained in DMEM (Corning, 10-013-CV) supplemented with 10% fetal bovine serum (FBS). For all experiments, cells were washed three times in phosphate buffered saline (PBS), and then cultured in DMEM without pyruvate (Corning, 10-017-CV) with 10% dialyzed FBS, supplemented with the indicated treatment condition. The reagents used for the cell culture experiments are as follows: AZD7545 (Selleck Chemicals, s7517), sodium pyruvate (Sigma, P2256), sodium L-lactate (Sigma, L7022), duroquinone (Sigma, D223204), metformin hydrochloride (Sigma, PHR1084), FCCP (carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone, also referred to as trifluoromethoxy carbonylcyanide phenylhydrazone) (Sigma, C2920), oligomycin A (Sigma, 75351), gramicidin from Bacillus aneurinolyticus (Bacillus brevis) (Sigma, G5002). All cells were cultured at 37°C with 5% CO2.
CRISPRi/Cas9-mediated repression of PDK1
PDK1 expression was suppressed using CRISPR interference (CRISPRi)-mediated transcriptional repression. A CRISPRi library (Horlbeck et al., 2016) was used to design sgRNA human targeting PDK1 (Guide 1: F 5’GCTCACGTACCACTCGGCAG 3’; Guide 2: F 5’GACGTCCCTCACGTACCACT 3’) or a non-targeting control (NTC; F 5’GGGAACCACATGGAATTCGA 3’). The sgRNA were cloned into a modified LentiCRISPRv2 plasmid, in which Cas9 was mutated to nuclease-dead Cas9 and fused to repressive chromatin modifier domain KRAB (Krüppel-associated box) domain of Kox1 (dCAS9-KRAB) (Gilbert et al., 2013). A pooled population of stable knockdown cell lines were generated and maintained in 5 μM/mL blasticidin.
Generation of LbNOX-Expressing Cells
LbNOX-FLAG cDNA was cloned from pUC57-LbNOX using the primers 5’-GGGGACAAGTTTGTACAAAAAAGCAGGCATGAAGGTCACCGTGGTC-3’ and 5’-GGGGACCACTTTGTACAAGAAAGCTGGGTTTACTTGTCATCGTCATCCTTGTAATC-3’. pUC57-LbNOX was a gift from Vamsi Mootha (Addgene, 75285). The PCR product was subsequently cloned into pInducer20 using LR Clonase II Plus (ThermoFisher, 12538120). pInducer20 was a gift from Stephen Elledge (Addgene, 44012). A549, 143B, and H1299 cells were transduced with lentivirus containing pInducer20-LbNOX-FLAG or pInducer20-E.V. and 10ug/mL polybrene (Millipore, TR-1003-G). The infected cells were selected in 1 mg/mL G418 (VWR, G5005). For proliferation experiments done with these cell lines, all conditions were supplemented with 500 ng/mL doxycycline.
Pancreatic Stellate Cell (PSC) isolation
PSCs were isolated from β-actin-GFP mice (The Jackson Laboratory, 006567) in which 3 mL of 1.3 mg/mL cold collagenase P (Sigma, 11213865001) and 0.01 mg/mL DNAse (Sigma, D5025) in GBSS (Sigma, G9779) were injected into the pancreas. The tissue was then placed into 2 mL of 1.3 mg/mL collagenase P solution on ice. Cells were then placed in a 37°C water bath for 15 minutes. The digested pancreas was filtered through a 250 μm strainer and washed with GBSS with 0.3% BSA. A gradient was created by suspending the cells in Nycodenz (VWR, 100356-726) and layering in GBSS with 0.3% BSA. Cells were then centrifuged at 1300×g for 20 minutes at 4°C. The layer containing PSCs was removed, filtered through a 70 μm strainer, washed in GBSS with 0.3% BSA, and plated for cell culture in DMEM (Corning, 10-013-CV) with 10% FBS and penicillin-streptomycin (P/S). Post-isolation, PSCs were immortalized with TERT and LargeT Antigen overexpression and cultured DMEM (Corning, 10-013-CV) supplemented with 10% FBS.
T cell isolation and culture
Primary human CD4+ T cells were isolated from peripheral blood by density gradient centrifugation over Lympholyte-H (Cedarlane Laboratories) and negative selection using the Dynabeads Untouched Human CD4 T Cells kit (Thermo Fisher, 11346D) according to the manufacturer’s instructions. Purity was assessed by flow cytometry for CD3 and CD4 and routinely found to be ≥95%. Cells were activated using Dynabeads Human T-Activator CD3/CD28 beads (Invitrogen) according to the manufacturer’s instructions and cultured in RPMI supplemented with 10% FBS and 30 U/mL recombinant human IL-2 (PeproTech, 200-02) at 37°C in 5% CO2. Primary mouse T cells were isolated from the spleens and lymph nodes of C57BL/6J mice using a Pan-T cell isolation kit (Miltenyl Biotec, 130-095-130). Isolated murine T cells were activated on plates coated with anti-CD3e (BD Pharmigen, 553057) and anti-CD28 (BD Pharmingen, 553294) antibodies and cultured in RPMI with 10% FBS (non-dialyzed), 1% P/S, 1x NEAA culture supplement (Thermo Fischer Scientific, 11140050), and 50 μM β-mercaptoethanol.
Tumor Growth in Mice
Two million A549 or one million 143B cells were injected into the flanks of 4–6 week old, male NU/NU mice (Charles River Laboratories, 088). A caliper was used to measure flank tumor volume in two dimensions and volume was calculated using the equation V = (π/6)(L×W2). Length was defined to be the longer of the two dimensions measured. For the A549 xenografts, the tumors were permitted to reach a size of 50 mm3, after which the animals were randomly assigned to an experimental group and the treatment regimen was initiated. Metformin (500 mg/kg) and AZD7545 (45 mg/kg) were both administered once daily by oral gavage, with a vehicle of water and 0.5% (w/w) methocel/0.1% polysorbate 80 respectively. All animal experiments were approved by the MIT Committee on Animal Care.
Method Details
Proliferation Rates
Cells were plated in replicate six-well plates in 2 mL at an initial seeding density of 20,000 cells per well for all cells except for MDA-MB-231, which were seeded at 40,000 cells per well. Cells were permitted to settle overnight and one six-well dish was counted to calculate the starting cell number at the initiation of the experiment. For all remaining dishes, cells were washed three times with PBS and 4 mL of treatment media was added to each well. After 48 hours, cells were washed again three times with PBS and 4 mL of the treatment media was replenished. Four days after the initial treatment, cells were counted to obtain the final cell counts for the experiment. Counts were done using a Cellometer Auto T4 Plus Cell Counter (Nexcelcom Bioscience) or by sulforhodamine B (SRB) assay. For cell quantification by SRB, cells were fixed by adding trichloroacetic acid to culture media (final concentration of 3.3%) and incubated 4°C for at least one hour. Fixed cells were washed with deionized water and then stained with 0.057% SRB in 1% acetic acid for 30 min. Following three washes with 1% acetic acid, plates were air-dried at room temperature. To solubilize the SRB dye, 1 mL of 10 mM Tris (pH 10.5) was added per well, and absorbance was measured at 510 nm using a microplate reader (Tecan Infinite, M200Pro).
T cell proliferation assay
Proliferation of primary human CD4+ T cell and primary mouse T cell proliferation was assessed by dye dilution. Freshly isolated cells were incubated at 1 × 107 cells/mL in PBS with 0.5% dialyzed FBS supplemented with 5 μM carboxyfluorescein succinimidyl ester (CFSE, Molecular Probes) or 2.5 μM CellTrace Far Red (Molecular Probes) for 5–7 minutes at room temperature. Staining was quenched with ice-cold complete media and washed cells were resuspended in RPMI supplemented with 10% dialyzed FBS and the indicated treatment condition before activation. For human CD4+ T cells, cells were expanded in fresh media containing the indicated treatment condition and CFSE fluorescence was then measured by flow cytometry after another 2 days. Primary murine T cells were expanded in fresh media containing the indicated treatment condition for 2 days, and CFSE fluorescence was then measured by flow cytometry.
Flow Cytometry of T cells
CD3/CD28 Dynabeads were removed from stimulated primary human CD4+ T cells using a DynaMag-2 magnet (Invitrogen). For antibody staining, cells were incubated for 30 minutes at 4°C in PBS with a fluorochrome-conjugated antibody (mouse monoclonal APC-conjugated anti-CD69 (BioLegend, 10909); mouse monoclonal PE-conjugated anti-CD25 (Pharmingen, 30795X) as indicated). For antibody staining or fluorescent dye dilution, cells were analyzed using a FACSCanto II, FACSCalibur or LSR Fortessa (BD Biosciences) flow cytometer. For mouse T cell antibody staining, cells were incubated for 30 minutes at 4°C in PBS containing 2% fetal bovine serum with eFluor780 viability dye (eBioscience, 65-0865-18) and fluorochrome-conjugated antibodies: mouse monoclonal PE-CF594-conjugated anti-CD45 (BD Horizon 562420) and eFluor450-conjugated anti-CD3e (Thermo Fisher Scientific, 48-0032-80); TMRE staining (Thermo Fisher Scientific, T669) was performed at room temperature in FluoroBrite DMEM (Thermo Fischer Scientific, A1896701) supplemented with dialyzed FBS. Stained cells were analyzed using a LSR-II or LSR Fortessa (BD Biosciences) flow cytometer. Data processing (including proliferation analysis) was conducted using FlowJo V10.6.1.
LCMS Metabolite Measurement
Metabolites were measured by LCMS on a QExactive bench top orbitrap mass spectrometer equipped with an Ion Max source and a HESI II probe, which was coupled to a Dionex UltiMate 3000 HPLC system (Thermo Fisher Scientific, San Jose, CA). External mass calibration was performed using the standard calibration mixture every 7 days. For each sample, 4 μL of each sample was injected onto a SeQuant® ZIC®-pHILIC 150 × 2.1 mm analytical column equipped with a 2.1 × 20 mm guard column (both 5 mm particle size; EMD Millipore). Buffer A was 20 mM ammonium carbonate, 0.1% ammonium hydroxide; Buffer B was acetonitrile. The column oven and autosampler tray were held at 25°C and 4°C, respectively. The chromatographic gradient was run at a flow rate of 0.150 mL/min as follows: 0–20 min: linear gradient from 80-20% B; 20–20.5 min: linear gradient form 20–80% B; 20.5–28 min: hold at 80% B. The mass spectrometer was operated in full-scan, polarity-switching mode, with the spray voltage set to 3.0 kV, the heated capillary held at 275°C, and the HESI probe held at 350°C. The sheath gas flow was set to 40 units, the auxiliary gas flow was set to 15 units, and the sweep gas flow was set to 1 unit. MS data acquisition was performed in a range of m/z = 70–1000, with the resolution set at 70,000, the AGC target at 1 × 106, and the maximum injection time (Max IT) at 20 msec. For detection of 13C-labeled citrate, targeted selected ion monitoring (tSIM) scans in negative mode centered on 194.1985 was included. The isolation window was set at 8.0 m/z. For all tSIM scans, the resolution was set at 70,000, the AGC target was 1105, and the max IT was 250 ms.
Relative quantitation of polar metabolites was performed with XCalibur QuanBrowser 2.2 (Thermo Fisher Scientific) using a 5 ppm mass tolerance and referencing an in-house library of chemical standards.
GCMS Metabolite Measurement
Gas-chromatography coupled to mass spectrometry (GCMS) analysis was done as described previously (Lewis et al., 2014). Dried metabolite samples were derivatized with 20 μL of methoxamine (MOX) reagent (ThermoFisher, TS-45950) and 25 μL of N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide with 1% tert-butyldimethylchlorosilane (Sigma, 375934). Following derivatization, samples were analyzed using a DB-35MS column (30m × 0.25mm i.d. × 0.25 μm, Agilent J&W Scientific) in an Agilent 7890 gas chromatograph (GC) coupled to an Agilent 5975C mass spectrometer (MS). Data were corrected for natural isotope abundance using in-house algorithms as in (Lewis et al., 2014).
Dynamic stable-isotope labeling experiments
Cells were plated in six-well plates at a seeding density of 150,000 cells per well and cultured overnight. Prior to the initiation of the experiment, cells were washed three times with PBS, and then cultured in 1.5 mL media containing 5mM glucose and the indicated treatment condition prior to tracing glucose fate. To assess glucose fate, 30 μL of a 1 M [U-13C6]glucose solution was added to a final concentration of 20 mM for the indicated time (ranging from 2 to 12 minutes). At each time point wells were washed as quickly as possible with ice-cold blood bank saline and lysed on the dish with 300 μL of ice-cold 80% HPLC grade methanol in HPLC grade water. Samples were scraped, collected into Eppendorf tubes, and vortexed for 10 minutes at 4°C. Samples were centrifuged at 21,000×g for 10 minutes at 4°C to precipitate protein. 50 μL of each sample was collected for immediate analysis by LCMS, or the supernatant was dried down under nitrogen gas for subsequent analysis by GCMS.
Oxygen Consumption
Oxygen consumption rates (OCR) was measured using an Agilent Seahorse Bioscience Extracellular Flux Analyzer (XF24) using standard methods. Briefly, cells were plated at 50,000 cells per well in Seahorse Bioscience 24-well plates in 50 μL of DMEM without pyruvate (Corning, 10-017-CV) supplemented with 10% dialyzed fetal bovine serum. An additional 500μL of media was added following a one hour incubation. The following day, cells were washed three times with PBS and incubated in DMEM without pyruvate with the indicated treatment. Five hours later, OCR measurements were made every 6 minutes, and injections of pyruvate at 16 minutes, FCCP (Sigma, C2920) at 32 minutes, and rotenone (Sigma, R8875; 2 μM) and antimycin (Sigma, A8674; 2 μM) at 48 minutes. Basal OCR was calculated by subtracting residual OCR following the addition of rotenone and antimycin from the initial OCR measurements.
Glucose/Lactate Excretion Index
Medium was collected from cells cultured in vehicle or 0.25 μM AZD7545 for 48 hours. Glucose and lactate concentrations were measured on a YSI-2900 Biochemistry Analyzer as described previously, and rate of lactate excretion into media was normalized to consumption of glucose (Hosios et al., 2016).
Lactate and Pyruvate Excretion
Medium was collected from 143B cells cultured in pyruvate-free DMEM treated with vehicle or 200μM duroquinone for 48 hours, or medium was collected from 143B cells cultured in pyruvate-free DMEM expressing empty vector or LbNOX for 48 hours. Extracellular pyruvate and lactate were measuring using GCMS, using a labeled internal standard (Cambridge Isotopes, CLM-1579-PK), and the excretion was normalized to the proliferation rate to calculate the rate per unit growth. For H1299, PSC, and C2C12 cells, extracellular lactate was measured using a YSI-2900 Biochemistry Analyzer after treatment with the indicated amount of duroquinone for 48 hours in pyruvate-free DMEM. Lactate excretion in primary mouse T cells was measured using a YSI-2900 Biochemistry Analyzer after 24 hours of stimulation using anti-CD3e (BD Biosciences, 553057) and anti-CD28 (BD Biosciences, 553294) antibodies and cultured in RPMI with 10% FBS, 1% P/S, 1x NEAA culture supplement (Thermo Fischer Scientific, 11140050), and 50μM β-mercaptoethanol. Lactate excretion was normalized to proliferation rate as assessed by counting the cells using a Cellometer Auto T4 Plus Cell Counter (Nexcelcom Bioscience).
Western Blot Analysis
Cells washed with ice-cold PBS, and scraped into cold RIPA buffer containing cOmplete Mini protease inhibitor (Roche, 11836170001) and PhosStop Phosphatase Inhibitor Cocktail Tablets (Roche, 04906845001). Protein concentration was calculated using the BCA Protein Assay (Pierce, 23225) with BSA as a standard. Lysates were resolved by SDS-PAGE and proteins were transferred onto nitrocellulose membranes using the iBlot2 Dry Blotting System (Thermo Fisher, IB21001, IB23001). Protein was detected with the primary antibodies anti-Pyruvate Dehydrogenase E1-alpha subunit (phospho S293) (Abcam, ab92696), anti-PDK1 (Cell Signaling Technologies, 3062S), anti-FLAG (Sigma, F1804) and anti-Vinculin (Sigma, V9131). The secondary antibodies used were IR680LT dye conjugated anti-rabbit IgG (Licor Biosciences, 925-68021), IRDye 800CW conjugated anti-mouse IgG (Licor Biosciences, 925-32210), HRP-conjugated anti-rabbit IgG (Millipore, 12-348), and HRP-conjugated anti-mouse IgG (Millipore, 12-349).
NAD+/NADH Measurements
Cells were seeded at 20,000 cells per well in six-well plates and permitted to adhere overnight. Cells were then washed three times in PBS and incubated in 4 mL of the indicated treatment media for 5 hours prior to rapidly washing three times in 4°C PBS and extraction in 100μL of ice-cold lysis buffer (1% dodecyltrimethylammonium bromide [DTAB] in 0.2 N of NaOH diluted 1:1 with PBS), then snap-frozen in liquid nitrogen and frozen at −80°C. The NAD+/NADH ratio was measured using an NAD/NADH-Glo Assay kit (Promega, G9072) according to a modified protocol as described previously (Gui et al., 2016). Briefly, to measure NAD+, 20 μL of lysate was transferred to PCR and diluted with 20 μL of lysis buffer and 20 μL 0.4 N HCl, and subsequently incubated at 60°C for 15 minutes. For NADH measurement, 20 μL of freshly thawed lysate was transferred to PCR tubes and incubated at 75°C for 30 minutes. The acidic conditions permit for selective degradation of NADH, while the basic conditions degrade NAD+. Following the incubation, samples were spun on a bench-top centrifuge and quenched with 20 μL neutralizing solution. The neutralizing solution consisted of 0.5 M Tris base for NAD+ samples and 0.25 M Tris in 0.2 N HCl for the NADH samples. The instructions in the Promega G9072 technical manual were then followed to measure NAD+ and NADH levels using a luminometer (Tecan Infinite, M200Pro).
ATP/ADP measurements
Cells were seeded at 5,000 cells per well in 96-well plates and permitted to adhere overnight. Cells were then washed one time with PBS and incubated in 200 μL of the indicated treatment media for 24 hours. ATP and ADP measurements were made using a luciferase-based assay (Sigma, MAK135) based on manufacturer’s instructions. Luminescence was measured using a luminometer (Tecan Infinite, M200Pro).
Yeast proliferation and metabolite analysis
In studies comparing the effects of LbNOX on ethanol production, CEN.PK 5D (MATa ura3-52 HIS3, LEU2 TRP1 MAL2-8c SUC2) expressing p416-TEF-lbNOX or p416-TEF (empty vector) were grown in log phase in YPD (3% glucose) at 30°C. At OD = 0.8, supernatant was collected and analyzed for ethanol content using an Ethanol Assay Kit (Sigma, MAK076). Rate of ethanol production was normalized to the proliferation rate. To study the effects of FCCP on ethanol production, wild type CEN.PK 5D strain yeast were cultured in YPD (3% glucose) at 30°C in the indicated amount of FCCP.
Mitochondrial Membrane Potential Measurement
Mitochondrial membrane potential was assessed using the TMRE (tetramethylrhodamine, ethyl ester) assay kit (Abcam, ab113852) in nonquench mode. The concentration of TMRE in nonquench mode was determined empirically (Perry et al., 2011). Cells were plated in 6 well dishes at a plating density of 150,000 cells per well and incubated in media containing the indicated concentration of vehicle AZD7545, 500 nM FCCP, 1 nM oligomycin, and/or 5nM gramicidin D. The cells were then treated with TMRE for 30 minutes, trypsinized, washed with PBS, and resuspended in Gibco FluoroBrite DMEM (Thermo Fisher Scientific, A1896701) containing the same concentration of TMRE supplemented with 10% dialyzed FBS. TMRE fluorescence was measured on a BD LSR II flow cytometer.
Quantification and Statistical Analysis
Details pertaining to all statistical analyses can be found in the figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-PDHA1 (phospho S293) | Abcam | ab92696 |
| anti-PDK1 | Cell Signaling Technologies | 3062S |
| anti-FLAG | Sigma | F1804 |
| anti-Vinculin | Sigma | V9131 |
| APC-conjugated anti-mouse CD69 | BioLegend | 10909 |
| eFlour450-conjugated anti-CD3e | Thermo Fisher | 48-0032-80 |
| anti-mouse CD3e | BD Pharmingen | 553057 |
| anti-mouse CD28 | BD Pharmingen | 553294 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| AZD7545 | Selleck Chemicals | S7517 |
| Sodium pyruvate | Sigma | P2256 |
| Sodium L-lactate | Sigma | L7022 |
| Duroquinone | Sigma | D223204 |
| Metformin hydrochloride | Sigma | PHR1084 |
| FCCP (carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone) | Sigma | C2920 |
| Rotenone | Sigma | R8875 |
| Oligomycin A | Sigma | 75351 |
| Antimycin A | Sigma | A8674 |
| Gramicidin from Bacillus aneurinolyticus (Bacillus brevis) | Sigma | G5002 |
| TMRE (tetramethylrhodamine, ethyl ester) | Thermo Fisher | T669 |
| Critical Commercial Assays | ||
| NAD/NADH-Glo Assay kit | Promega | G9072 |
| ADP/ATP Ratio Assay Kit | Sigma | MAK135 |
| TMRE (tetramethylrhodamine, ethyl ester) assay kit | Abcam | ab113852 |
| Ethanol Assay Kit | Sigma | MAK076 |
| Dynabeads Untouched Human CD4 T Cells kit | Thermo Fisher | 11346D |
| Pan-T cell isolation kit | Miltenyl Biotec | 130-095-130 |
| Deposited Data | ||
| Raw image data | This manuscript | http://dx.doi.org/10.17632/6zpghspxgs.1. |
| Experimental Models: Cell Lines | ||
| Human: 143B | ATCC | CRL-8303 |
| Human: A172 | ATCC | CRL-1620 |
| Human: A549 | ATCC | CCL-185 |
| Mouse: C2C12 | ATCC | CRL-1772 |
| Human: H1299 | ATCC | CRL-5803 |
| Human: HeLa | ATCC | CCL-2 |
| Human: MDA-MB-231 | ATCC | HTB-26 |
| Mouse: PSC | This manuscript | N/A |
| Human: Primary CD4+ T cells | This manuscript | N/A |
| Mouse: Primary T cells | This manuscript | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: NU/NU Nude Mouse (Crl:NU-Foxn1nu) | Charles River | 088 |
| S. cerevisiae: CEN.PK 5D (MATa ura3–52 HIS3, LEU2 TRP1 MAL2–8c SUC2) | This manuscript | N/A |
| Oligonucleotides | ||
| sgRNA sequence targeting PDK1 #1 (5’ GCTCACGTACCACTCGGCAG 3’) | Horlbeck et al., 2016 | hCRISPRi-v2.1 |
| sgRNA sequence targeting PDK1 #2 (5’ GACGTCCCTCACGTACCACT 3’ | Horlbeck et al., 2016 | hCRISPRi-v2.1 |
| sgRNA sequence non-targeting control (5’ GGGAACCACATGGAATTCGA 3’) | Horlbeck et al., 2016 | hCRISPRi-v2.1 |
| Recombinant DNA | ||
| pUC57-LbNOX | Addgene | 75285 |
| pInducer20 | Addgene | 44012 |
| Software and Algorithms | ||
| FlowJo | FlowJo | V10.6.1 |
| Xcalibur™ Software | Thermo Fisher | N/A |
| Prism | GraphPad | 8.2.1 |
Acknowledgements
We thank members of the Vander Heiden lab for thoughtful discussion, Allison Lau for isolating pancreatic stellate cells, and the Swanson Biotechnology Center Flow Cytometry Facility. This work was supported by the Ludwig Center for Molecular Oncology Fund (A.L.), NSF GRFP DGE-1122374 (A.L.), NIH T32GM007287 (A.L.; Z.L.; D.Y.G.; M.Z.), NIH T32GM007753 (B.T.D.), NIH K99CA218679 (L.B.S.), HHMI Medical Research Fellows Program (A.A.), MRC CSF MR/P008801/1 (N.J.M.), NHSBT WPA15-02 (N.J.M), the Novo Nordisk Foundation Grant No. NNF10CC1016517 (R.F.) and the Knut and Alice Wallenberg Foundation (R.F.). M.G.V.H. acknowledges support from a Howard Hughes Medical Institute Faculty Scholar grant, SU2C, the Lustgarten Foundation, the MIT Center for Precision Cancer Medicine, the Ludwig Center at MIT, and the NIH (R35CA242379, R01CA201276, R01CA168653, P30CA14051).
Footnotes
Declaration of Interests
The authors are aware of no competing interests; however, M.G.V.H. discloses he is on the advisory board of Molecular Cell and is a scientific advisor for Agios Pharmaceuticals, Aeglea Biotherapeutics, Auron Therapeutics, Faeth Therapeutics, and iTeos Therapeutics. S.S. is member of the scientific advisory board of Arcus Biosciences, Venn Therapeutics, Tango Therapeutics, Replimune and serves as a scientific advisor for Dragonfly Therapeutics, Merck, Ribon, Torque and TAKEDA. A.L. is a current employee of a Flagship Pioneering biotechnology start-up company.
References
- Baksh SC, Todorova PK, Gur-Cohen S, Hurwitz B, Ge Y, Novak JSS, Tierney MT, Dela Cruz-Racelis J, Fuchs E, and Finley LWS (2020). Extracellular serine controls epidermal stem cell fate and tumour initiation. Nat Cell Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao XR, Ong SE, Goldberger O, Peng J, Sharma R, Thompson DA, Vafai SB, Cox AG, Marutani E, Ichinose F, et al. (2016). Mitochondrial dysfunction remodels one-carbon metabolism in human cells. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basan M, Hui S, Okano H, Zhang Z, Shen Y, Williamson JR, and Hwa T. (2015). Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528, 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertholet AM, Chouchani ET, Kazak L, Angelin A, Fedorenko A, Long JZ, Vidoni S, Garrity R, Cho J, Terada N, et al. (2019). H(+) transport is an integral function of the mitochondrial ADP/ATP carrier. Nature 571, 515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, and Sabatini DM (2015). An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 162, 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonora M, Patergnani S, Rimessi A, De Marchi E, Suski JM, Bononi A, Giorgi C, Marchi S, Missiroli S, Poletti F, et al. (2012). ATP synthesis and storage. Purinergic Signal 8, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroughs LK, and DeBerardinis RJ (2015). Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol 17, 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand K, Leibold W, Luppa P, Schoerner C, and Schulz A. (1986). Metabolic alterations associated with proliferation of mitogen-activated lymphocytes and of lymphoblastoid cell lines: evaluation of glucose and glutamine metabolism. Immunobiology 173, 23–34. [DOI] [PubMed] [Google Scholar]
- Brand MD, and Nicholls DG (2011). Assessing mitochondrial dysfunction in cells. Biochem J 435, 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GC (1992). Control of respiration and ATP synthesis in mammalian mitochondria and cells. Biochem J 284 ( Pt 1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit F, and Brand MD (1995). A hierarchy of ATP-consuming processes in mammalian cells. Biochem J 312 ( Pt 1), 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, and Mak TW (2011). Regulation of cancer cell metabolism. Nat Rev Cancer 11, 85–95. [DOI] [PubMed] [Google Scholar]
- Canto C, Menzies KJ, and Auwerx J. (2015). NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell Metab 22, 31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS, Budinger GR, and Schumacker PT (1996). Molecular oxygen modulates cytochrome c oxidase function. J Biol Chem 271, 18672–18677. [DOI] [PubMed] [Google Scholar]
- Chinchore Y, Begaj T, Wu D, Drokhlyansky E, and Cepko CL (2017). Glycolytic reliance promotes anabolism in photoreceptors. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Deken RH (1966). The Crabtree effect: a regulatory system in yeast. J Gen Microbiol 44, 149–156. [DOI] [PubMed] [Google Scholar]
- Diaz-Ruiz R, Rigoulet M, and Devin A. (2011). The Warburg and Crabtree effects: On the origin of cancer cell energy metabolism and of yeast glucose repression. Biochim Biophys Acta 1807, 568–576. [DOI] [PubMed] [Google Scholar]
- Diehl FF, Lewis CA, Fiske BP, and Vander Heiden MG (2019). Cellular redox state constrains serine synthesis and nucleotide production to impact cell proliferation. Nat Metab 1, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M, Shen Z, Huang S, Zhao L, Chen S, Mak TW, and Wang X. (2010). The ER UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect, and proliferation in the PTEN pathway. Cell 143, 711–724. [DOI] [PubMed] [Google Scholar]
- Fantin VR, St-Pierre J, and Leder P. (2006). Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 9, 425–434. [DOI] [PubMed] [Google Scholar]
- Faubert B, Li KY, Cai L, Hensley CT, Kim J, Zacharias LG, Yang C, Do QN, Doucette S, Burguete D, et al. (2017). Lactate Metabolism in Human Lung Tumors. Cell 171, 358–371 e359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-de-Cossio-Diaz J, and Vazquez A. (2017). Limits of aerobic metabolism in cancer cells. Sci Rep 7, 13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Bermudez J, Baudrier L, La K, Zhu XG, Fidelin J, Sviderskiy VO, Papagiannakopoulos T, Molina H, Snuderl M, Lewis CA, et al. (2018). Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat Cell Biol 20, 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, and Gillies RJ (2004). Why do cancers have high aerobic glycolysis? Nat Rev Cancer 4, 891–899. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassian AR, Metallo CM, Coloff JL, Stephanopoulos G, and Brugge JS (2011). Erk regulation of pyruvate dehydrogenase flux through PDK4 modulates cell proliferation. Genes Dev 25, 1716–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui DY, Sullivan LB, Luengo A, Hosios AM, Bush LN, Gitego N, Davidson SM, Freinkman E, Thomas CJ, and Vander Heiden MG (2016). Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell Metab 24, 716–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Fan J, Chung TW, Lythgoe K, Wang X, Xie J, Ge Q, Gu TL, Polakiewicz RD, Roesel JL, et al. (2011). Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell 44, 864–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, et al. (2016). Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosios AM, Hecht VC, Danai LV, Johnson MO, Rathmell JC, Steinhauser ML, Manalis SR, and Vander Heiden MG (2016). Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Dev Cell 36, 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosios AM, and Vander Heiden MG (2018). The redox requirements of proliferating mammalian cells. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell N, and Sager R. (1979). Cytoplasmic genetics of mammalian cells: conditional sensitivity to mitochondrial inhibitors and isolation of new mutant phenotypes. Somatic Cell Genet 5, 833–845. [DOI] [PubMed] [Google Scholar]
- Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, Esparza LA, Reya T, Le Z, Yanxiang Guo J, et al. (2017). Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume DA, Radik JL, Ferber E, and Weidemann MJ (1978). Aerobic glycolysis and lymphocyte transformation. Biochem J 174, 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume DA, and Weidemann MJ (1979). Role and regulation of glucose metabolism in proliferating cells. J Natl Cancer Inst 62, 3–8. [PubMed] [Google Scholar]
- Hung YP, Albeck JG, Tantama M, and Yellen G. (2011). Imaging cytosolic NADH-NAD(+) redox state with a genetically encoded fluorescent biosensor. Cell Metab 14, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplon J, Zheng L, Meissl K, Chaneton B, Selivanov VA, Mackay G, van der Burg SH, Verdegaal EM, Cascante M, Shlomi T, et al. (2013). A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature 498, 109–112. [DOI] [PubMed] [Google Scholar]
- Kato M, Li J, Chuang JL, and Chuang DT (2007). Distinct structural mechanisms for inhibition of pyruvate dehydrogenase kinase isoforms by AZD7545, dichloroacetate, and radicicol. Structure 15, 992–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima M, Bensaad K, Zois CE, Barberis A, Bridges E, Wigfield S, Lagerholm C, Dmitriev RI, Tokiwa M, Toi M, et al. (2020). Disruption of hypoxia-inducible fatty acid binding protein 7 induces beige fat-like differentiation and thermogenesis in breast cancer cells. Cancer Metab 8, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy KM, Scarbrough PM, Ribeiro A, Richardson R, Yuan H, Sonveaux P, Landon CD, Chi JT, Pizzo S, Schroeder T, et al. (2013). Catabolism of exogenous lactate reveals it as a legitimate metabolic substrate in breast cancer. PLoS One 8, e75154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, and Dang CV (2006). HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3, 177–185. [DOI] [PubMed] [Google Scholar]
- Kolobova E, Tuganova A, Boulatnikov I, and Popov KM (2001). Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem J 358, 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, and Dang CV (2011). Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer 11, 325–337. [DOI] [PubMed] [Google Scholar]
- Korotchkina LG, and Patel MS (2001). Site specificity of four pyruvate dehydrogenase kinase isoenzymes toward the three phosphorylation sites of human pyruvate dehydrogenase. J Biol Chem 276, 37223–37229. [DOI] [PubMed] [Google Scholar]
- Krebs HA (1927). On the metabolism of the retina. Biochemische Zeitschrift, 57–59. [Google Scholar]
- Kurmi K, Hitosugi S, Yu J, Boakye-Agyeman F, Wiese EK, Larson TR, Dai Q, Machida YJ, Lou Z, Wang L, et al. (2018). Tyrosine Phosphorylation of Mitochondrial Creatine Kinase 1 Enhances a Druggable Tumor Energy Shuttle Pathway. Cell Metab 28, 833–847 e838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, and Dang CV (2010). Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc Natl Acad Sci U S A 107, 2037–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu VS, O’Connell JT, Gonzalez Herrera KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A, Domingos Chinen LT, Rocha RM, et al. (2014). PGC-1alpha mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol 16, 992–1003, 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoigne M, Aubert JP, and Millet J. (1954). [Ethyl alcohol production and growth of baker’s yeast cultured under aerobic conditions]. Ann Inst Pasteur (Paris) 87, 427–439. [PubMed] [Google Scholar]
- Levine AJ, and Puzio-Kuter AM (2010). The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 330, 1340–1344. [DOI] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, and Metallo CM (2014). Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell 55, 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ivica NA, Dong T, Papageorgiou DP, He Y, Brown DR, Kleyman M, Hu G, Chen WW, Sullivan LB, et al. (2020). MFSD7C switches mitochondrial ATP synthesis to thermogenesis in response to heme. Nat Commun 11, 4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti MV, and Locasale JW (2016). The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem Sci 41, 211–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wang Y, Yang C, Ruan Y, Bai C, Chu Q, Cui Y, Chen C, Ying G, and Li B. (2020). Inhibiting both proline biosynthesis and lipogenesis synergistically suppresses tumor growth. J Exp Med 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Muralidhar V, Hosios AM, Israelsen WJ, Gui DY, Newhouse L, Ogrodzinski M, Hecht V, Xu K, Acevedo PN, et al. (2015). Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol Cell 57, 95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson NJ, Sumner J, Wals K, Rapiteanu R, Weekes MP, Vigan R, Weinelt J, Schindler M, Antrobus R, Costa AS, et al. (2015). Cell Surface Proteomic Map of HIV Infection Reveals Antagonism of Amino Acid Metabolism by Vpu and Nef. Cell Host Microbe 18, 409–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFate T, Mohyeldin A, Lu H, Thakar J, Henriques J, Halim ND, Wu H, Schell MJ, Tsang TM, Teahan O, et al. (2008). Pyruvate dehydrogenase complex activity controls metabolic and malignant phenotype in cancer cells. J Biol Chem 283, 22700–22708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker MP, Audi SH, Bongard RD, Lindemer BJ, and Krenz GS (2006). Influence of pulmonary arterial endothelial cells on quinone redox status: effect of hyperoxia-induced NAD(P)H:quinone oxidoreductase 1. Am J Physiol Lung Cell Mol Physiol 290, L607–619. [DOI] [PubMed] [Google Scholar]
- Michelakis ED, Sutendra G, Dromparis P, Webster L, Haromy A, Niven E, Maguire C, Gammer TL, Mackey JR, Fulton D, et al. (2010). Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med 2, 31ra34. [DOI] [PubMed] [Google Scholar]
- Morrell JA, Orme J, Butlin RJ, Roche TE, Mayers RM, and Kilgour E. (2003). AZD7545 is a selective inhibitor of pyruvate dehydrogenase kinase 2. Biochem Soc Trans 31, 1168–1170. [DOI] [PubMed] [Google Scholar]
- Munyon WH, and Merchant DJ (1959). The relation between glucose utilization, lactic acid production and utilization and the growth cycle of L strain fibroblasts. Exp Cell Res 17, 490–498. [DOI] [PubMed] [Google Scholar]
- Naguib A, Mathew G, Reczek CR, Watrud K, Ambrico A, Herzka T, Salas IC, Lee MF, El-Amine N, Zheng W, et al. (2018). Mitochondrial Complex I Inhibitors Expose a Vulnerability for Selective Killing of Pten-Null Cells. Cell Rep 23, 58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes CD, Lakin-Thomas PL, and Brand MD (1989). The contribution of ATP turnover by the Na+/K+-ATPase to the rate of respiration of hepatocytes. Effects of thyroid status and fatty acids. Biochim Biophys Acta 976, 241–245. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, and Denko NC (2006). HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 3, 187–197. [DOI] [PubMed] [Google Scholar]
- Perry SW, Norman JP, Barbieri J, Brown EB, and Gelbard HA (2011). Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques 50, 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit FH, Pelley JW, and Reed LJ (1975). Regulation of pyruvate dehydrogenase kinase and phosphatase by acetyl-CoA/CoA and NADH/NAD ratios. Biochem Biophys Res Commun 65, 575–582. [DOI] [PubMed] [Google Scholar]
- Pfeiffer T, Schuster S, and Bonhoeffer S. (2001). Cooperation and competition in the evolution of ATP-producing pathways. Science 292, 504–507. [DOI] [PubMed] [Google Scholar]
- Rabinovich S, Adler L, Yizhak K, Sarver A, Silberman A, Agron S, Stettner N, Sun Q, Brandis A, Helbling D, et al. (2015). Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature 527, 379–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racker E. (1972). Bioenergetics and the problem of tumor growth. Am Sci 60, 56–63. [PubMed] [Google Scholar]
- Robbins D, and Zhao Y. (2011). New aspects of mitochondrial Uncoupling Proteins (UCPs) and their roles in tumorigenesis. Int J Mol Sci 12, 5285–5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumsey WL, Schlosser C, Nuutinen EM, Robiolio M, and Wilson DF (1990). Cellular energetics and the oxygen dependence of respiration in cardiac myocytes isolated from adult rat. J Biol Chem 265, 15392–15402. [PubMed] [Google Scholar]
- Schockel L, Glasauer A, Basit F, Bitschar K, Truong H, Erdmann G, Algire C, Hagebarth A, Willems PH, Kopitz C, et al. (2015). Targeting mitochondrial complex I using BAY 87–2243 reduces melanoma tumor growth. Cancer Metab 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholnick P, Lang D, and Racker E. (1973). Regulatory mechanisms in carbohydrate metabolism. IX. Stimulation of aerobic glycolysis by energy-linked ion transport and inhibition by dextran sulfate. J Biol Chem 248, 5175. [PubMed] [Google Scholar]
- Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF, et al. (2008). Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest 118, 3930–3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, and Vander Heiden MG (2015). Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell 162, 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Luengo A, Danai LV, Bush LN, Diehl FF, Hosios AM, Lau AN, Elmiligy S, Malstrom S, Lewis CA, et al. (2018). Aspartate is an endogenous metabolic limitation for tumour growth. Nat Cell Biol 20, 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AS, Baty JW, Dong LF, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, et al. (2015). Mitochondrial genome acquisition restores respiratory function and tumorigenic potential of cancer cells without mitochondrial DNA. Cell Metab 21, 81–94. [DOI] [PubMed] [Google Scholar]
- Titov DV, Cracan V, Goodman RP, Peng J, Grabarek Z, and Mootha VK (2016). Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science 352, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle A, Oliver J, and Roca P. (2010). Role of uncoupling proteins in cancer. Cancers (Basel) 2, 567–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, and Thompson CB (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Chandel NS, Schumacker PT, and Thompson CB (1999). Bcl-xL prevents cell death following growth factor withdrawal by facilitating mitochondrial ATP/ADP exchange. Mol Cell 3, 159–167. [DOI] [PubMed] [Google Scholar]
- Vazquez A, Liu J, Zhou Y, and Oltvai ZN (2010). Catabolic efficiency of aerobic glycolysis: the Warburg effect revisited. BMC Syst Biol 4, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez A, and Oltvai ZN (2011). Molecular crowding defines a common origin for the Warburg effect in proliferating cells and the lactate threshold in muscle physiology. PLoS One 6, e19538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri GN, Eiteman MA, McEwen JE, Olsson L, and Nielsen J. (2007). Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 104, 2402–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, et al. (2014). Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 514, 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Marquardt C, and Foker J. (1976). Aerobic glycolysis during lymphocyte proliferation. Nature 261, 702–705. [DOI] [PubMed] [Google Scholar]
- Warburg O. (1924). Über den Stoffwechsel der Carcinomzelle. Naturwissenschaften 12, 1131–1137. [Google Scholar]
- Warburg O. (1956). On the origin of cancer cells. Science 123, 309–314. [DOI] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, and Chandel NS (2010). Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A 107, 8788–8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhouse S. (1956). On respiratory impairment in cancer cells. Science 124, 267–269. [DOI] [PubMed] [Google Scholar]
- Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM, Budigner GS, et al. (2014). Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife 3, e02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson DH, Krebs HA, Stubbs M, Page MA, Morris HP, and Weber G. (1970). Metabolism of renal tumors in situ and during ischemia. Cancer Res 30, 2049–2054. [PubMed] [Google Scholar]
- Xie H, Hanai J, Ren JG, Kats L, Burgess K, Bhargava P, Signoretti S, Billiard J, Duffy KJ, Grant A, et al. (2014). Targeting lactate dehydrogenase--a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab 19, 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao CH, Fowle-Grider R, Mahieu NG, Liu GY, Chen YJ, Wang R, Singh M, Potter GS, Gross RW, Schaefer J, et al. (2016). Exogenous Fatty Acids Are the Preferred Source of Membrane Lipids in Proliferating Fibroblasts. Cell Chem Biol 23, 483–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao CH, Wang R, Wang Y, Kung CP, Weber JD, and Patti GJ (2019). Mitochondrial fusion supports increased oxidative phosphorylation during cell proliferation. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W. (2008). NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxid Redox Signal 10, 179–206. [DOI] [PubMed] [Google Scholar]
- Zabka TS, Singh J, Dhawan P, Liederer BM, Oeh J, Kauss MA, Xiao Y, Zak M, Lin T, McCray B, et al. (2015). Retinal toxicity, in vivo and in vitro, associated with inhibition of nicotinamide phosphoribosyltransferase. Toxicol Sci 144, 163–172. [DOI] [PubMed] [Google Scholar]
- Zhang X, Fryknas M, Hernlund E, Fayad W, De Milito A, Olofsson MH, Gogvadze V, Dang L, Pahlman S, Schughart LA, et al. (2014a). Induction of mitochondrial dysfunction as a strategy for targeting tumour cells in metabolically compromised microenvironments. Nat Commun 5, 3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS, Kwiatkowski DJ, and Manning BD (2014b). Coordinated regulation of protein synthesis and degradation by mTORC1. Nature 513, 440–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any datasets or code. Unprocessed and uncropped image data is available http://dx.doi.org/10.17632/6zpghspxgs.1.