ABSTRACT
The plasmid-mediated tet(X7) conferring high-level tigecycline resistance was identified in five mcr-1.1-positive Escherichia coli strains (ST10 [n = 3] and ST155 [n = 2]) isolated from chickens in Egypt. Two fosfomycin-resistant fosA4-carrying IncFII plasmids (∼79 kb in size) were detected. Transposase ISCR3 (IS91 family) is syntenic with tet(X7) in all isolates, suggesting its role in the mobilization of tet(X7). To our knowledge, this is the first global report of ST4-IncHI2 plasmids cocarrying tet(X7) and mcr-1.1 from chickens.
KEYWORDS: tigecycline resistance, colistin resistance, fosfomycin, Mcr-1.1, FosA4, Tet(X7), Egypt, Escherichia coli, chicken
INTRODUCTION
Tigecycline is a last-resort antibiotic approved by the U.S. FDA in 2005 to treat multidrug- or extensively drug-resistant bacteria, including carbapenem-resistant Enterobacterales (1). The plasmid-mediated high-level tigecycline resistance genes tet(X3) and tet(X4) were previously described from Enterobacterales and Acinetobacter spp. of animal or human origin in China in 2019 (1, 2). Soon after, tet(X5) and tet(X6) were detected from an Acinetobacter baumannii clinical isolate and a novel SXT/R391 integrative and conjugative element in a Proteus genomospecies 6 isolate, respectively (3–5). Recently, seven other tet(X)-like genes [tet(X7) to tet(X13)], encoding resistance to all tetracyclines, including the recently FDA-approved omadacycline and eravacycline, have been identified from environmental, human commensal, and pathogenic bacteria through tetracycline or tigecycline selection of metagenomic libraries (6). Tet(X14) was identified in the chromosome of an Empedobacter stercoris strain isolated from a pig from China (7). Moreover, the coexistence of tet(X4) and mcr-1 genes was previously reported in colistin- and tigecycline-resistant Escherichia coli strains in China, posing a serious public health threat (2). Here, we describe the first global report of the coproduction of Tet(X7), FosA4, and Mcr-1.1 in E. coli strains isolated from fecal samples of chickens in Egypt.
In this study, 40 fecal samples were collected from two chicken farms between March and July 2019 in Sidi Ghazy City, Kafr El-Sheikh province, Egypt. PCR screening for tet(X)-like genes and mcr-1 to mcr-9 was performed as previously reported (1, 8). Five tet(X)-positive E. coli strains were identified from one farm. Interestingly, the five tet(X)-positive E. coli strains were found to coharbor mcr-1.1. BLAST analysis of tet(X) partial sequences revealed 100% nucleotide identity (100% query coverage) with tet(X7) detected from Pseudomonas aeruginosa strain Pa-3 (JAATVZ010000055.1) in Pakistan (6) and E. coli strain MS8345 (CP025402.1) in Qatar (9). Antimicrobial susceptibility was determined by measuring MICs using the broth microdilution method; the results were interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines (10) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) standards (https://eucast.org/clinical_breakpoints/). The five isolates exhibited multidrug resistance phenotypes, with MICs for tigecycline ranging from 8 to 16 mg/liter and for colistin ranging from 2 to 4 mg/liter (Table 1). Conjugation experiments were performed between tet(X7)-positive strains as donors and the azide-resistant (AZr) strain E. coli J53 as the recipient. Transconjugants were selected on Luria-Bertani agar supplemented with sodium azide (100 mg/liter) and tigecycline (2 mg/liter) and further replicated on the same medium. Transconjugants were tested using colony PCR [for tet(X7) and mcr-1] and plasmid analysis as previously described (1, 8, 11). All strains successfully transferred the tet(X7)-carrying plasmid. mcr-1.1 was also detected by PCR in all transconjugants. The MICs of transconjugants against tigecycline and colistin increased by >32- and >16-fold, respectively, compared to the plasmid-free E. coli J53 recipient strain. In all transconjugants, tet(X7), mcr-1.1, and the IncHI2 replicon were detected by PCR, suggesting the possibility of colocalization on the same IncHI2 plasmid (11, 12).
TABLE 1.
MICs of antimicrobials for Tet(X7)- and Mcr-1.1-producing Escherichia coli strains and transconjugants isolated in this study
| Antibiotic | MIC (mg/liter)a for strain: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A-1-4-1b | A-1-4-1-TC | A-1-8-1b | A-1-8-1-TC | A-1-10-1b | A-1-10-1-TC | A-1-11-3b | A-1-11-3-TC | A-1-22-2b | A-1-22-2-TC | E. coli J53 | |
| Ampicillin | >1,024 | >256 | >1,024 | >256 | >1,024 | >256 | >1,024 | >256 | >1,024 | >256 | 16 |
| Cefotaxime | 64 | 32 | 0.0625 | 0.125 | 256 | 256 | 64 | 128 | 256 | 256 | 0.0625 |
| Doripenem | 0.125 | <0.125 | <0.125 | <0.125 | <0.125 | <0.125 | <0.125 | <0.125 | <0.125 | <0.125 | ≤0.0078 |
| Meropenem | 0.5 | <0.125 | 0.5 | <0.125 | 0.5 | <0.125 | 0.25 | <0.125 | 0.5 | <0.125 | 0.0312 |
| Kanamycin | 512 | <4 | 1,024 | <4 | 1,024 | 512 | 1,024 | 512 | 1,024 | 512 | 1 |
| Gentamicin | 32 | 16 | 64 | 8 | 64 | 64 | 32 | 64 | 64 | 64 | <1 |
| Ciprofloxacin | 16 | <0.03125 | 16 | <0.03125 | 32 | 4 | 32 | 8 | 32 | 0.5 | 0.0625 |
| Chloramphenicol | 128 | 4 | 128 | 4 | 512 | 256 | 256 | 256 | 512 | 256 | 4 |
| Tetracycline | 64 | 16 | 64 | 16 | 128 | 64 | 128 | 128 | 128 | 64 | <2 |
| Tigecycline | 16 | 4 | 16 | 4 | 16 | 8 | 16 | 16 | 8 | 8 | 0.125 |
| Colistin | 4 | 4 | 4 | 4 | 8 | 2 | 2 | 2 | 4 | 2 | 0.125 |
| Fosfomycin | 2,048 | 1,024 | 2,048 | 1,024 | 16 | 4 | 8 | 2 | 4 | 2 | 4 |
Resistance to the antibiotics is given in boldface. Interpretation was done according to the CLSI guidelines (10) and EUCAST standards. TC, transconjugant.
Donor.
To investigate the genetic environment of tet(X7), total genomic DNA from the five strains was extracted using the GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, MO, USA) and the Qiagen genomic-tip 20/G kit (Qiagen) following the manufacturer’s recommendations. Sequencing libraries were constructed using the Nextera-XT library preparation kit; paired-end sequencing was done with an Illumina MiSeq (Illumina Inc.) using a 500-cycle MiSeq reagent kit (13). For Oxford Nanopore sequencing, the library was constructed using the SQK-RBK004 rapid barcoding kit loaded onto a FLO-MIN106 R9.4.1 flow cell and sequenced with the GridION device (Oxford Nanopore Technologies, Oxford, UK) for 72 h. A hybrid assembly of Illumina short reads and GridION long reads was performed using Unicycler v0.4.8 (14). The annotation was performed using DFAST (https://dfast.nig.ac.jp/). Reads shorter than 1,000 bp were trimmed from FASTQ data before assembly using NanoFilt. High-quality assemblies were obtained and sufficient for closing the genomes and the plasmids contained in these strains. The complete genomes were analyzed using SerotypeFinder-2.0, MLST 2.0, PlasmidFinder, CHTyper-1.0, and ResFinder-3.2, available at the Center for Genomic Epidemiology (http://www.genomicepidemiology.org/). The five strains were found to carry from 20 to 24 different multiple acquired antimicrobial resistance genes conferring resistance to colistin (mcr-1.1), tetracycline and tigecycline [tet(A) and tet(X7), respectively], fosfomycin (fosA4), trimethoprim (dfrA1, dfrA7, dfrA12, and dfrA14), β-lactams (blaCTX-M-15 and blaTEM-1B), aminoglycosides [aadA1, aadA2, aph(6)-Id, aac(3)-IIa, aph(3′')-Ib, and aph(3′)-Ia], macrolides [ere(A), erm(B), erm(42), and mph(A)], chloramphenicol and florfenicol (catA1, cmlA1, and floR), sulfonamides (sul1 and sul3), and rifampin (arr-2) (Table 2). Mutations in the quinolone resistance-determining region, conferring resistance to nalidixic acid and ciprofloxacin, were detected in all strains (see Table S1 in the supplemental material).
TABLE 2.
Features of plasmids and chromosomes of Escherichia coli strains coharboring mcr-1.1 and tet(X7) identified in this study
| Strain | Chromosome/plasmid (bp) | Sequence type or pMLSTa | Plasmid replicon (PlasmidFinder) | Antimicrobial resistance geneb |
|---|---|---|---|---|
| A-1-4-1 | Chromosome (4,817,482) | ST155 | mdf(A), aadA2, mph(A), sul1, dfrA12, catA1, blaTEM-1B | |
| pAMS-1-tetX7 (239,431) | ST4 | IncHI2, IncHI2A | mcr-1.1, tet(X7), aadA1, aph(6)-Id, aac(3)-IIa, aph(3′')-Ib, mph(A), ere(A), sul1, dfrA7, arr-2, blaTEM-1B, blaCTX-M-15 | |
| pAMS-1 (128,942) | F16:A6:B8 | IncFIA, IncFIB (AP001918), IncFIC (FII) | aac(3)-IIa, aph(3′)-Ia, aadA1, aadA2, sul3, tet(A), cmlA1, floR | |
| pAMS-1-fosA4 (79,087) |
F16:A-:B- | IncFII (pCoo) | fosA4, mph(A) | |
| A-1-8-1 | Chromosome (4,817,489) | ST155 | mdf(A), aadA2, dfrA12, sul1, catA1, mph(A), blaTEM-1B | |
| pAMS-2-tetX7 (360,340) | ST4 F18:A6:B8 |
IncHI2, IncHI2A, IncFIA, IncFIB (AP001918), IncFIC (FII) | mcr-1.1, tet(X7), aadA1, aadA2, sul1, sul3, dfrA7, aph(6)-Id, aac(3)-IIa, aph(3′')-Ib, aph(3′)-Ia, tet(A), cmlA1, floR, mph(A), blaTEM-1B | |
| pAMS-2-fosA4 (79,084) |
F16:A-:B- | IncFII (pCoo) | fosA4, mph(A) | |
| A-1-10-1 | Chromosome (4,864,476) | ST10 | mdf(A), tet(A) | |
| pAMS-3-tetX7 (249,057) | ST4 | IncHI2, IncHI2A | mcr-1.1, tet(X7), aph(6)-Id, aac(3)-IIa, aph(3′')-Ib, aadA1, arr-2, mph(A), erm(B), ere(A), sul1, dfrA7, blaCTX-M-15, blaTEM-1B | |
| pAMS-2 (146,244) | F24:A-:B58 | IncFIB (AP001918), IncFII | aadA2b, aadA1, aph(6)-Id, aph(3′)-Ia, aadA1, aph(3′')-Ib, sul1, dfrA1, sul3, dfrA14, dfrA1, tet(A), cmlA, blaTEM-1B | |
| pAMS-3 (101,137) | ST36 (clonal complex-3) | IncI1-I (gamma) | floR, blaTEM-1B | |
| pAMS-4 (98,060) | NT | p0111 | ND | |
| pAMS-5 (41,287) | Unknown | IncN, IncX1 | ND | |
| pAMS-6 (1,551) | NT | Col(MG828) | ND | |
| A-1-11-3 | Chromosome (4,864,552) | ST10 | mdf(A), tet(A) | |
| pAMS-4-tetX7 (240,102) | ST4 | IncHI2, IncHI2A | mcr-1.1, tet(X7), erm(B), aph(3′')-Ib, aph(6)-Id, aac(3)-IIa, aadA1, sul1, dfrA7, arr-2, blaCTX-M-15, mph(A), ere(A) | |
| pAMS-7 (146,216) | F24:A-:B58 | IncFIB (AP001918), IncFII | aadA1, aph(3′)-Ia, aph(3′')-Ib, aadA2b, aph(6)-Id, sul3, dfrA1, dfrA14, sul1, tet(A), cmlA, blaTEM-1B | |
| pAMS-8 (99,192) | ST36 (clonal complex-3) | IncI1-I (gamma) | floR, blaTEM-1B | |
| pAMS-9 (98,324) | NT | p0111 | ND | |
| pAMS-10 (41,250) | NT | IncN, IncX1 | ND | |
| pAMS-11 (1,551) | NT | Col(MG828) | ND | |
| A-1-22-2 | Chromosome (4,864,503) | ST10 | mdf(A), tet(A) | |
| pAMS-5-tetX7 (386,687) | ST4 F24:A-:B1 |
IncHI2, IncHI2A, IncFIB (AP001918), IncFII | mcr-1.1, tet(X7), aadA2b, aph(3′')-Ib, aadA1, aac(3)-IIa, aph(6)-Id, aph(3′)-Ia, tet(A), ere(A), mph(A), erm(B), arr-2, cmlA, blaTEM-1B, blaCTX-M-15, dfrA1, dfrA7, sul1, dfrA14, sul3 | |
| pAMS-12 (103,039) | ST36 (clonal complex-3) | IncI1-I (gamma) | erm(42), floR | |
| pAMS-13 (98,588) | NT | p0111 | ND | |
| pAMS-14 (41,287) | NT | IncN, IncX1 | ND | |
| pAMS-15 (1,551) | NT | Col(MG828) | ND |
NT, not typeable.
ND, not determined.
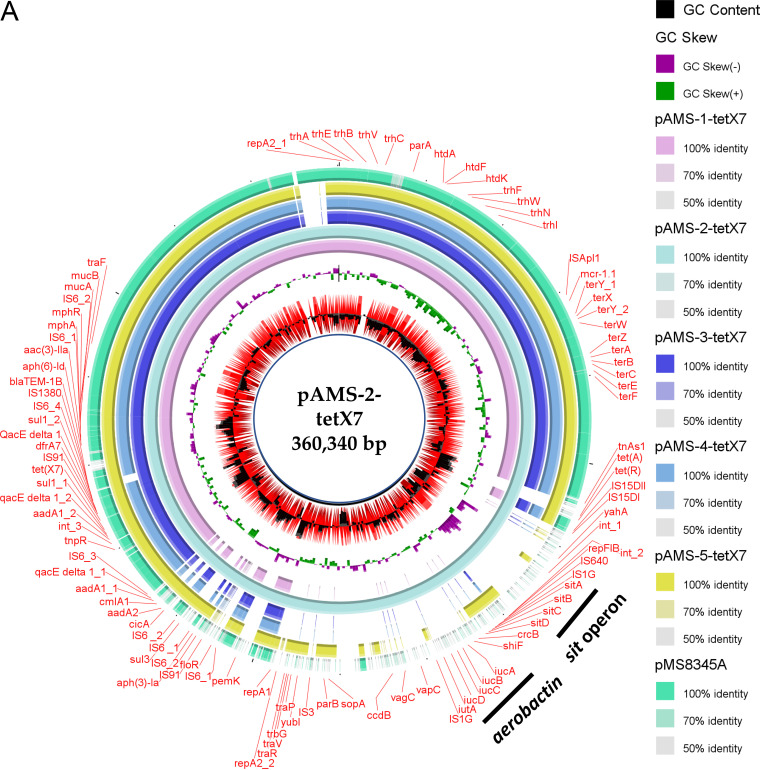
Five sequence type 4 ST4-IncHI2:HI2A plasmids (named pAMS-1-tetX7, pAMS-2-tetX7, pAMS-3-tetX7, pAMS-4-tetX7, and pAMS-5-tetX7), ranging in size from 289 to 386 kb and coharboring tet(X7) and mcr-1.1, were detected from the five E. coli strains identified in this study (Table 2, Fig. 1A). Interestingly, the IncHI2 plasmids in strains A-1-8-1 and A-1-22-2 co-fused with IncFIA/FIB/FIC plasmid and IncFIB/FII plasmid, respectively, forming multiple replicon plasmids. Similarly, a fusion plasmid, pP2-3T, has been reported from China cointegrating ST3-IncHI2 with an IncFII plasmid conferring virulence and multidrug resistance (15). Besides carrying several antimicrobial resistance genes, including tet(X7) and mcr-1.1, pAMS-2-tetX7 carried two sets of virulence genes, including the sitABCD operon mediating iron, manganese transport and resistance to hydrogen peroxide (16), and the aerobactin iron uptake iucABCD operon and iutA (17). These sets were found between two copies of IS1 in a transposon-like structure, as previously described (18, 19). Recently, these two sets of virulence genes were identified in a multiresistance plasmid, pZM3, isolated from a Salmonella enterica serovar Wien from Algeria (18). The BLAST ring image generator (BRIG) tool (http://sourceforge.net/projects/brig) was used to perform a circular comparison between the complete sequences of tet(X7)-carrying plasmids (Fig. 1A). Only one IncHI2 plasmid, pMS8345A, cocarrying tet(X7) and mcr-1.1 from E. coli strain MS8345 isolated in Qatar (accession no. CP025402.1), was detected in NCBI GenBank and included in the figure (9).
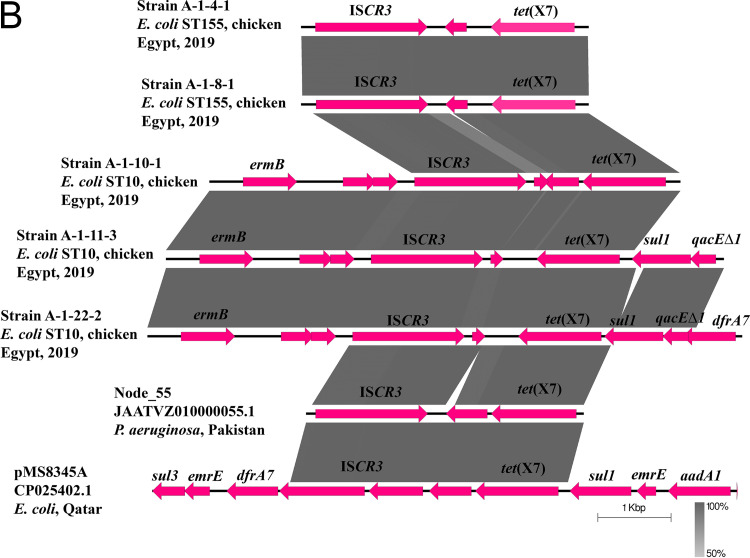
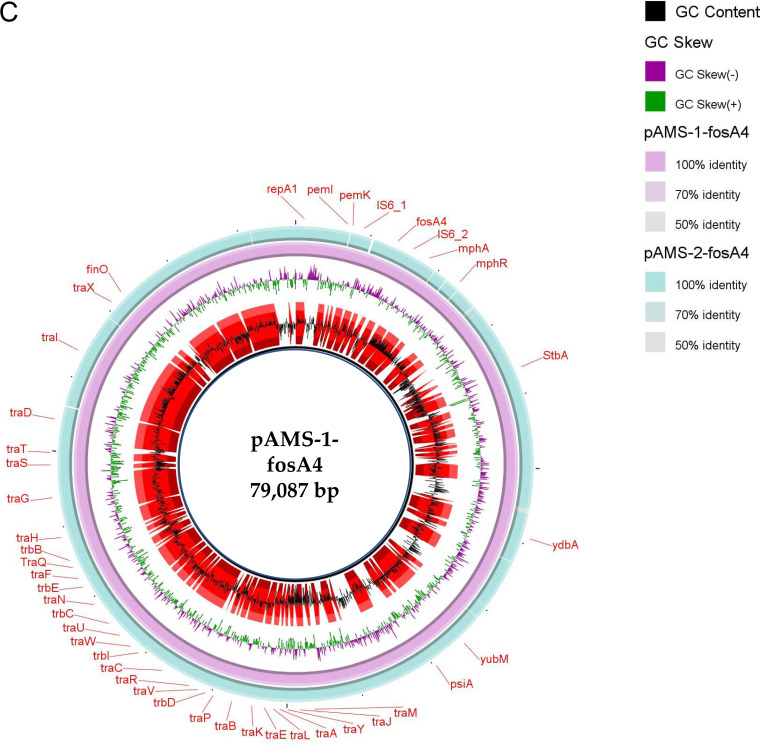
FIG 1.
Schematic representation of IncHI2 plasmids cocarrying tet(X7) and mcr-1.1 (A), the genetic environment of tet(X7) (B), and IncFII plasmids carrying fosA4 (C) identified from the genome sequences of E. coli strains analyzed in this study. Only one IncHI2 plasmid, pMS8345A, carrying tet(X7) and mcr-1.1 from E. coli strain MS8345, Qatar (accession no. CP025402.1), was found in NCBI GenBank and was included in the figure. The whole sequences of pAMS-1-tetX7 and pAMS-1-fosA4 were used as the references in A and C, respectively. The external ring in A and C represents the annotation of pAMS-1-tetX7 and pAMS-1-fosA4, respectively. In panel A, plasmids were included in the following order: pAMS-1-tetX7 (this study), pAMS-2-tetX7 (this study), pAMS-3-tetX7 (this study), pAMS-4-tetX7 (this study), pAMS-5-tetX7 (this study), pMS8345A (accession no. CP025402.1). In panel C, the plasmids were included in the following order: pAMS-1-fosA4 and pAMS-2-fosA4. Linear comparison of the contigs carrying tet(X7) detected in this study with the chromosomal contig carrying tet(X7) from P. aeruginosa strain Pa-3, Pakistan (accession no. JAATVZ010000055.1) (6), and part of the IncHI2 plasmid, pMS8345A, carrying tet(X7) from E. coli strain MS8345, Qatar (accession no. CP025402.1) (9), is shown.
To decipher the genetic environment of tet(X7) and mcr-1.1 using EasyFig tool (http://mjsull.github.io/Easyfig/), a nucleotide BLAST (BLASTn) (https://blast.ncbi.nlm.nih.gov/Blast.cgi) search in combination with ISFinder (https://www-is.biotoul.fr/blast.php) were used. Unlike the genetic structure of tet(X3) and tet(X4) identified previously (1, 2), the putative transposase ISCR3 (IS91 family) was located downstream of the tet(X7) gene in all IncHI2 plasmids identified in this study, the IncHI2 plasmid from E. coli strain MS8345 (9), and the chromosome of P. aeruginosa strain Pa-3 isolated from a cystic fibrosis patient in Pakistan in 2016 (Fig. 1B) (6). These findings might suggest the important role of ISCR3 in the acquisition and mobilization of the tet(X7) gene by horizontal gene transfer.
In all IncHI2 plasmids identified in this study, a single-ended structure (hp [hypothetical protein]-ISApl1-mcr-1.1) with an upstream copy of ISApl1 was detected. The same structure was previously identified in E. coli strains isolated from a patient and raw milk cheese in Qatar (9) and Egypt (GenBank accession number CP042898.1), respectively. PlasmidFinder revealed the existence of multiple replicon types in all strains (Table 2). Inc types FIB, FII, and HI2 were the shared replicon types among all the examined E. coli isolates in the present study.
Fosfomycin has wide-spectrum bactericidal activity by inhibiting the initial step of cell wall biosynthesis (20). The lack of new antimicrobials has led to reconsideration of old antibiotics, such as fosfomycin, as a possible option for the treatment of multidrug-resistant carbapenemase-producing or extended-spectrum β-lactamase-producing Enterobacterales (20). Fosfomycin resistance in Gram-negative bacteria could be mediated through the glutathione S-transferase-encoding gene fosA, catalyzing the epoxide ring opening (20). Ten variants of the fosA gene (fosA1 to fosA10) were detected previously (20). Recently, fosA10-carrying IncFII (F35:A-:B-) plasmid was identified from a fosfomycin-resistant E. coli isolate collected from a chicken meat sample in 2018 (21). Lupo et al. (22) identified E. coli isolate ST4380 collected from the digestive tract of a monkey that carried fosA4 and blaCTX-M-15 on a 145-kb IncF18:A-:B1 from France in 2011. In this study, two highly similar fosfomycin-resistant fosA4-carrying IncFII (F16:A-:B-) plasmids (∼79 kb in size), pAMS-1-fosA4 and pAMS-2-fosA4, were detected from strains A-1-4-1 and A-1-8-1, respectively (Fig. 1C). The two plasmids also cocarried the mph(A) gene conferring resistance to azithromycin. Gene fosA4 was located downstream to IS6 family transposase, which might be involved in its movement. To our knowledge, this is the first report of a complete sequence of a fosA4-carrying IncFII plasmid from an animal source.
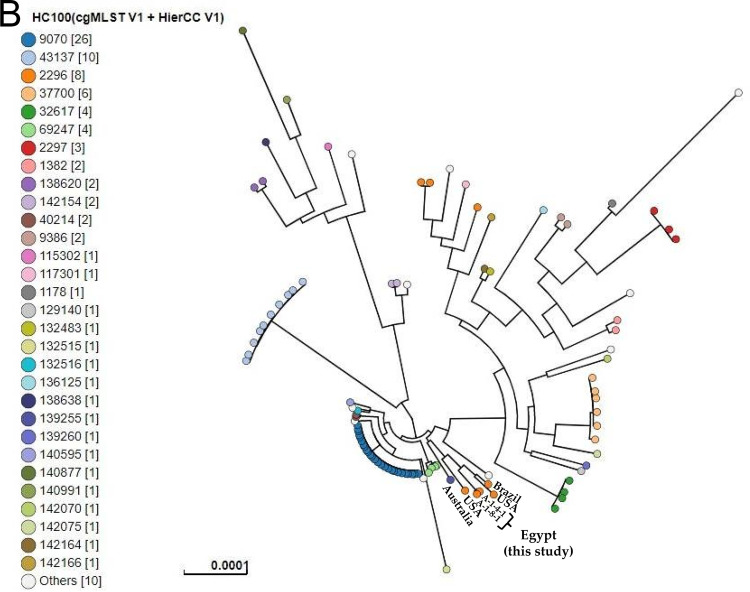
Analysis of in silico multilocus sequence typing (MLST) identified the detection of two different sequence types (STs) (Table 2). Two E. coli isolates (A-1-4-1 and A-1-8-1) belonged to ST155, and the other three E. coli strains (A-1-10-1, A-1-11-3, and A-1-22-2) belonged to ST10. To perform phylogenetic analysis, paired-end Illumina FASTQ files of the five E. coli isolates were uploaded into EnteroBase (https://enterobase.warwick.ac.uk/). Single nucleotide polymorphisms (SNPs) and hierarchical clustering (HierCC) of core genome (cg) MLST were performed to compare and cluster isolates from this study with the EnteroBase publicly available E. coli isolates from poultry that were assigned to ST10 and ST155, updated 1 September 2020 (23). SNPs and HierCC of cgMLST showed the clustering of our ST10 isolates (A-1-10-1, A-1-11-3, and A-1-22-2) with other poultry-derived ST10 E. coli isolates from Denmark, Italy, Spain, and Norway that exhibited similar cgMLST type HC200|4445, differing by <200 alleles (HC200) (Fig. 2A). Meanwhile, the cgMLST type HC100|2296 has been observed for isolates A-1-4-1 and A-1-8-1 and E. coli ST155 isolates from the United States and Brazil with no more than 100 allele differences (Fig. 2B). Clustering of our isolates at higher resolution levels (HC100 and HC200) for cgMLST types with isolates from different continents indicated the global expansion of these clones that presumably resulted from widespread international trade of animals and animal byproducts (24).
FIG 2.
SNPs and hierarchical clustering of cgMLST (HierCC) of the present E. coli isolates ST10 (A) and ST155 (B) with the publicly available E. coli isolates ST10 and ST155 from poultry in EnteroBase (https://enterobase.warwick.ac.uk/). Tip labels indicate the cgMLST pattern HC200 (allelic differences no more than 200) and HC100 (allelic differences no more than 100) for ST10 (A) and ST155 (B), respectively.
In this study, we detected tigecycline- and colistin-resistant Tet(X7)- and Mcr-1.1-producing E. coli (5/40, 12.5%) from a poultry farm in Kafr El-Sheikh province, Egypt, in 2019. The biochemical characterization indicated that Tet(X7) has an apparent catalytic efficiency five and eight times greater than that of Tet(X) for degradation of eravacycline and tigecycline, respectively (6). The five Tet(X7)-producing strains identified in this study exhibited multidrug-resistant phenotypes and belonged to two distinct STs (two ST155 and three ST10). In a study performed in Qatar, tet(X7) was identified in an ST95 extraintestinal pathogenic E. coli (ExPEC) strain, MS8345, that colocalized with mcr-1 on the same IncHI2 plasmid (9). However, the tet(X7)-mediated tigecycline resistance of MS8345 was not reported (9). In another study conducted in China, eight tet(X4)-positive E. coli strains (one ST10 and seven ST8302) were previously isolated from chickens (2).
Due to their decreased cost, oral bioavailability, and broad-spectrum antimicrobial activity, tetracycline antibiotics comprise one of the most widely used antibiotics in clinical medicine and agriculture (comprising 66% of total antibiotic use in livestock) (1, 2, 6, 25). The heavy use of tetracyclines in animal production as growth promoters or for metaphylaxis might be a selective pressure for tigecycline-resistant Tet(X)-like-producing organisms (1–2). He et al. (1) previously reported colistin- and tigecycline-resistant E. coli strains carrying tet(X4) and mcr-1 in China. Alarmingly, the coexistence of plasmid-borne blaNDM-1 and tet(X3) was detected in Acinetobacter indicus strains of dairy cow origin in China, posing a serious public health threat (26).
To our knowledge, this is the first report of (i) the convergence of tet(X7) and mcr-1.1 in E. coli strains of chicken origin globally, (ii) a tigecycline-resistant E. coli strain producing Tet(X7) from Africa and the Middle East, and (iii) tet(X7)- and mcr-1.1-coharboring multidrug resistance multireplicon plasmid carrying the sitABCD operon and iucABCD/iutA operon. This study showed that food-producing animals might serve as an important reservoir of multidrug-resistant bacteria with conjugable plasmids carrying resistance genes [blaCTX-M-15, tet(X7), fosA4 and mcr-1.1] to last-resort and clinically significant antibiotics (cephalosporins, tigecycline, fosfomycin and colistin). Furthermore, the detection of these E. coli clones from chickens in Egypt is alarming and poses a food safety issue and a public health threat in that it may be transferred to humans. The easy access of antibiotics in Egyptian hospitals, the community without prescription, and in the agricultural sector may be responsible for the development of high levels of resistance. Antimicrobial surveillance plans, infection control policies, judicious use of tetracyclines in humans and animals, and continuous surveillance for tigecycline-resistant tet(X)-like genes in Gram-negative bacteria of animal or human origin are essentially needed.
Data availability.
The complete genome sequences of the five E. coli strains (A-1-4-1, A-1-8-1, A-1-10-1, A-1-11-3, and A-1-22-2) were submitted to DDBJ/ENA/GenBank under BioProject ID PRJNA661596 (BioSample accession numbers SAMN16064908, SAMN16064909, SAMN16064910, SAMN16064911, and SAMN16064912, respectively).
ACKNOWLEDGMENTS
A.M.S. was supported by a fellowship from the Ministry of Education, Culture, Sports, Science and Technology of Japan (fellowship no. 153532). This work was supported in part by a grant to C.R.J. from the U.S. Department of Agriculture (USDA) (project 6040-32000-009-00) and by a grant to M.S. from the Ministry of Health, Labor and Welfare, Japan (H30-shokuhin-ippan-006).
We have no conflicts of interest to declare.
Footnotes
Supplemental material is available online only.
aac.02084-20-s000s1.pdf (108.8KB, pdf)
Contributor Information
Ahmed M. Soliman, Email: ahmed_soliman@pharm.kfs.edu.eg.
Tadashi Shimamoto, Email: tadashis@hiroshima-u.ac.jp.
REFERENCES
- 1.He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, Ke Y, Ji Q, Wei R, Liu Z, Shen Y, Wang G, Sun L, Lei L, Lv Z, Li Y, Pang M, Wang L, Sun Q, Fu Y, Song H, Hao Y, Shen Z, Wang S, Chen G, Wu C, Shen J, Wang Y. 2019. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol 4:1450–1456. 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
- 2.Sun J, Chen C, Cui C-Y, Zhang Y, Liu X, Cui Z-H, Ma X-Y, Feng Y, Fang L-X, Lian X-L, Zhang R-M, Tang Y-Z, Zhang K-X, Liu H-M, Zhuang Z-H, Zhou S-D, Lv J-N, Du H, Huang B, Yu F-Y, Mathema B, Kreiswirth BN, Liao X-P, Chen L, Liu Y-H. 2019. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat Microbiol 4:1457–1464. 10.1038/s41564-019-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Liu D, Lv Y, Cui L, Li Y, Li T, Song H, Hao Y, Shen J, Wang Y, Walsh TR. 2019. Novel plasmid-mediated tet(X5) gene conferring resistance to tigecycline, eravacycline, and omadacycline in a clinical Acinetobacter baumannii isolate. Antimicrob Agents Chemother 64:e01326-19. 10.1128/AAC.01326-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He D, Wang L, Zhao S, Liu L, Liu J, Hu G, Pan Y. 2020. A novel tigecycline resistance gene, tet(X6), on an SXT/R391 integrative and conjugative element in a Proteus genomospecies 6 isolate of retail meat origin. J Antimicrob Chemother 75:1159–1164. 10.1093/jac/dkaa012. [DOI] [PubMed] [Google Scholar]
- 5.Liu D, Zhai W, Song H, Fu Y, Schwarz S, He T, Bai L, Wang Y, Walsh TR, Shen J. 2020. Identification of the novel tigecycline resistance gene tet(X6) and its variants in Myroides, Acinetobacter and Proteus of food animal origin. J Antimicrob Chemother 75:1428–1431. 10.1093/jac/dkaa037. [DOI] [PubMed] [Google Scholar]
- 6.Gasparrini AJ, Markley JL, Kumar H, Wang B, Fang L, Irum S, Symister CT, Wallace M, Burnham C-AD, Andleeb S, Tolia NH, Wencewicz TA, Dantas G. 2020. Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance. Commun Biol 3:241. 10.1038/s42003-020-0966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng Y, Chen Y, Liu Y, Guo Y, Zhou Y, Xiao T, Zhang S, Xu H, Chen Y, Shan T, Xiao Y, Zhou K. 2020. Identification of novel tetracycline resistance gene tet(X14) and its co-occurrence with tet(X2) in a tigecycline-resistant and colistin-resistant Empedobacter stercoris. Emerg Microbes Infect 9:1843–1852. 10.1080/22221751.2020.1803769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soliman AM, Maruyama F, Zarad HO, Ota A, Nariya H, Shimamoto T, Shimamoto T. 2020. Emergence of a multidrug-resistant Enterobacter hormaechei clinical isolate from Egypt co-harboring mcr-9 and blaVIM-4. Microorganisms 8:595. 10.3390/microorganisms8040595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forde BM, Zowawi HM, Harris PNA, Roberts L, Ibrahim E, Shaikh N, Deshmukh A, Sid Ahmed MA, Al Maslamani M, Cottrell K, Trembizki E, Sundac L, Yu HH, Li J, Schembri MA, Whiley DM, Paterson DL, Beatson SA. 2018. Discovery of mcr-1-mediated colistin resistance in a highly virulent Escherichia coli lineage mSphere 3:e00486-18. 10.1128/mSphere.00486-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing—28th ed CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 11.Soliman AM, Zarad HO, Nariya H, Shimamoto T, Shimamoto T. 2020. Genetic analysis of carbapenemase-producing Gram-negative bacteria isolated from a university teaching hospital in Egypt. Infect Genet Evol 77:104065. 10.1016/j.meegid.2019.104065. [DOI] [PubMed] [Google Scholar]
- 12.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 13.Soliman AM, Ramadan H, Sadek M, Nariya H, Shimamoto T, Hiott LM, Frye JG, Jackson CR, Shimamoto T. 2020. Draft genome sequence of a blaNDM-1- and blaOXA-244-carrying multidrug-resistant Escherichia coli D-ST69 clinical isolate from Egypt. J Glob Antimicrob Resist 22:832–834. 10.1016/j.jgar.2020.07.015. [DOI] [PubMed] [Google Scholar]
- 14.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang L-X, Li X-P, Deng G-H, Li S-M, Yang R-S, Wu Z-W, Liao X-P, Sun J, Liu Y-H. 2018. High genetic plasticity in multidrug-resistant sequence type 3-IncHI2 plasmids revealed by sequence comparison and phylogenetic analysis. Antimicrob Agents Chemother 62:e02068-17. 10.1128/AAC.02068-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabri M, Léveillé S, Dozois CM. 2006. A SitABCD homologue from an avian pathogenic Escherichia coli strain mediates transport of iron and manganese and resistance to hydrogen peroxide. Microbiology (Reading) 152:745–758. 10.1099/mic.0.28682-0. [DOI] [PubMed] [Google Scholar]
- 17.Torres AG, Redford P, Welch RA, Payne SM. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect Immun 69:6179–6185. 10.1128/IAI.69.10.6179-6185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harmer CJ, Hall RM. 2020. The complete nucleotide sequence of pZM3, a 1970 FIA:FIB:FII plasmid carrying antibiotic resistance and virulence determinants. Microb Drug Resist 26:438–446. 10.1089/mdr.2019.0248. [DOI] [PubMed] [Google Scholar]
- 19.Colonna B, Nicoletti M, Visca P, Casalino M, Valenti P, Maimone F. 1985. Composite IS1 elements encoding hydroxamate-mediated iron uptake in FIme plasmids from epidemic Salmonella spp. J Bacteriol 162:307–316. 10.1128/JB.162.1.307-316.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zurfluh K, Treier A, Schmitt K, Stephan R. 2020. Mobile fosfomycin resistance genes in Enterobacteriaceae—an increasing threat. Microbiologyopen 9:e1135. 10.1002/mbo3.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Lin Q, Zhou Q, Lv L, Wan M, Gao X, Wang C, Liu JH. 2020. Identification of fosA10, a novel plasmid-mediated fosfomycin resistance gene of Klebsiella pneumoniae origin, in Escherichia coli. Infect Drug Resist 13:1273–1279. 10.2147/IDR.S251360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupo A, Saras E, Madec JY, Haenni M. 2018. Emergence of blaCTX-M-55 associated with fosA, rmtB and mcr gene variants in Escherichia coli from various animal species in France. J Antimicrob Chemother 73:867–872. 10.1093/jac/dkx489. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M, Agama Study Group . 2020. The EnteroBase user's guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res 30:138–152. 10.1101/gr.251678.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown E, Dessai U, McGarry S, Gerner-Smidt P. 2019. Use of whole-genome sequencing for food safety and public health in the United States. Foodborne Pathog Dis 16:441–450. 10.1089/fpd.2019.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ungemach FR, Müller-Bahrdt D, Abraham G. 2006. Guidelines for prudent use of antimicrobials and their implications on antibiotic usage in veterinary medicine. Int J Med Microbiol 296 (Suppl 41):33–38. 10.1016/j.ijmm.2006.01.059. [DOI] [PubMed] [Google Scholar]
- 26.He T, Li R, Wei R, Liu D, Bai L, Zhang L, Gu J, Wang R, Wang Y. 2020. Characterization of Acinetobacter indicus co-harbouring tet(X3) and blaNDM-1 of dairy cow origin. J Antimicrob Chemother 75:2693–2696. 10.1093/jac/dkaa182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequences of the five E. coli strains (A-1-4-1, A-1-8-1, A-1-10-1, A-1-11-3, and A-1-22-2) were submitted to DDBJ/ENA/GenBank under BioProject ID PRJNA661596 (BioSample accession numbers SAMN16064908, SAMN16064909, SAMN16064910, SAMN16064911, and SAMN16064912, respectively).