ABSTRACT
Nonsynonymous mutations are well documented in TEM β-lactamases. The resulting amino acid changes often alter the conferred phenotype from broad spectrum (2b) conferred by TEM-1 to extended spectrum (2be), inhibitor resistant (2br), or both extended spectrum and inhibitor resistant (2ber). The encoding blaTEM genes also deviate in numerous synonymous mutations, which are not well understood. blaTEM-3 (2be), blaTEM-33 (2br), and blaTEM-109 (2ber) were studied in comparison to blaTEM-1. blaTEM-33 was chosen for more detailed studies because it deviates from blaTEM-1 by a single nonsynonymous mutation and three additional synonymous mutations. Genes encoding the enzymes with only nonsynonymous or all (including synonymous) mutations plus all permutations between blaTEM-1 and blaTEM-33 were expressed in Escherichia coli cells. In disc diffusion assays, genes encoding TEM-3, TEM-33, and TEM-109 with all synonymous mutations resulted in higher resistance levels than genes without synonymous mutations. Disc diffusion assays with the 16 genes carrying all possible nucleotide change combinations between blaTEM-1 and blaTEM-33 indicated different susceptibilities for different variants. Nucleotide BLAST searches did not identify genes without synonymous mutations but did identify some without nonsynonymous mutations. Energies of possible secondary mRNA structures calculated with mfold are generally higher with synonymous mutations, suggesting that their role could be to destabilize the mRNA and facilitate its unfolding for efficient translation. In summary, our data indicate that transition from blaTEM-1 to other variant genes by simply acquiring the nonsynonymous mutations is not favored. Instead, synonymous mutations seem to support the transition to other variant genes with nonsynonymous mutations leading to different phenotypes.
KEYWORDS: synonymous mutation, evolution, mRNA secondary structure, β-lactamase
INTRODUCTION
The solution of the structure of DNA (1, 2) and the deciphering of the genetic code (3–5) led to our current understanding of how genes are expressed. As stated by Crick in 1958 in the Central Dogma of Molecular Biology (6, 7), the flow of information goes from DNA to RNA (transcription) and, in the case of mRNA, to protein (translation). While this concept is well understood qualitatively, what determines protein synthesis efficiency is not well understood quantitatively. The overall efficiency depends on the individual efficiencies of each process involved (such as transcription and translation), and each of these processes consists of many steps. Plus, there are additional processes in between, such as posttranscriptional RNA processing and transport and posttranslational processing, translocation, and folding of proteins. Adding complexity, the flow of information in protein synthesis is convergent, meaning that several codons on the DNA and RNA level can code for the same amino acid (4). Mutations resulting in these synonymous codons will typically not affect the amino acid sequence (unless organisms actually use different genetic codes or rare tRNAs). Hence, they have been described as silent mutations. However, recent research has shown that they can affect protein synthesis efficiency and are, therefore, better described as synonymous or, as recently suggested, “whisper mutations” (8). In contrast, nucleotide changes that do result in the incorporation of different amino acids are referred to as nonsynonymous or missense mutations.
Since every codon, including those that code for the same amino acid, is recognized by a specific aminoacyl-tRNA, which may be present at different concentrations, mutations leading to the usage of synonymous codons can affect translation efficiency. For instance, genes that are expressed at high levels will typically use codons that are recognized by abundant aminoacyl-tRNAs. This effect is referred to as codon bias and may differ between different organisms (9). Synonymous mutations can also affect folding and stability of mRNA. They could result in altered secondary structures of mRNA that are less efficiently recognized or unfolded by the ribosome or factors that would normally process the mRNA, for instance, the spliceosome in eukaryotic cells. More stable mRNA may increase protein synthesis if the mRNA has to persist for a period of time and less stable mRNA is more prone to degradation. Duan et al. investigated the role of known synonymous mutations in the human dopamine receptor D2 gene and found that some of them can compensate for detrimental effects caused by others, that is, decreased mRNA stability (10). On the contrary, more stable mRNAs may be harder to unfold for proper translation by the ribosome and may thus lead to decreased protein synthesis. This may especially be the case in prokaryotic cells, where mRNAs are typically translated while transcription is still ongoing and extensive processing does not occur. Kudla et al. created a library of green fluorescence protein (GFP) genes that varied randomly by synonymous mutations (11). They found that decreased stability of the transcripts, especially at the 5′ end, resulted in increased GFP expression in Escherichia coli.
Here, we study known synonymous mutations in genes coding for different variants of the TEM β-lactamase family. These enzymes are serine β-lactamases and belong to molecular class A according to Ambler (12). They are expressed by Gram-negative bacteria to break down and resist the antibacterial action of β-lactam antibiotics. Previous studies have investigated synonymous mutations in blaTEM genes that were obtained in directed evolution experiments under selective pressure (13–15). To our knowledge, this is the first systematic study comparing blaTEM variant genes with and without synonymous mutations relative to blaTEM-1 and, in the case of blaTEM-33, all possible permutations of the mutations distinguishing blaTEM-33 from blaTEM-1.
When analyzing variants without or with (some) synonymous mutations, some deviations were observed in disc diffusion (DD) assays, but few in MIC assays, which could be due to the different formats (e.g., gradient versus serial dilutions of antibiotics). Systematic analysis of simulated mRNA structures and stabilities using mfold (16) revealed that the nonsynonymous mutations alone increased mRNA stability, which led to lower resistance levels in DD assays, probably through decreased translation rates, consistent with previous observations (11). The stabilizing nonsynonymous mutations had to be combined with the synonymous mutations observed in the isolated variant genes to yield full resistance in DD assays. mRNA stability calculated by mfold decreased with synonymous mutations, which is hypothesized to allow for efficient translation and high expression levels of these enzymes.
RESULTS
TEM variants expressed from genes without or with synonymous mutations result in different inhibition zone diameters in disc diffusion assays.
To test whether or not differences in phenotype conferred by enzymes from different functional classes (17) could be resolved experimentally, TEM-1 (2b), TEM-3 (2be), TEM-33 (2br), and TEM-109 (2ber) were selected and tested using disc diffusion (DD) assays. All genes used in this study were purchased based on the sequence information found in the Pathogen Detection Reference Gene Catalog of the National Center for Biotechnology Information (NCBI) unless stated otherwise (e.g., without synonymous mutations). Genes were subcloned into the pBC SK(+) phagemid vector and Escherichia coli DH10B cells were transformed with the resulting vectors via electroporation. Clinical strains expressing extended-spectrum β-lactamases are frequently hypermutator strains (18). By using a well-characterized laboratory strain, DH10B, we tried to eliminate the possibility of mutations during the experiment. Although DH10B has been reported to have an increased mutation rate, this increase is mostly due to IS50 transposition and not nucleotide changes (19). Thus, the likelihood that DH10B would introduce additional synonymous mutations is small. However, the growth rate of DH10B is low due to its high transposition rate (19).
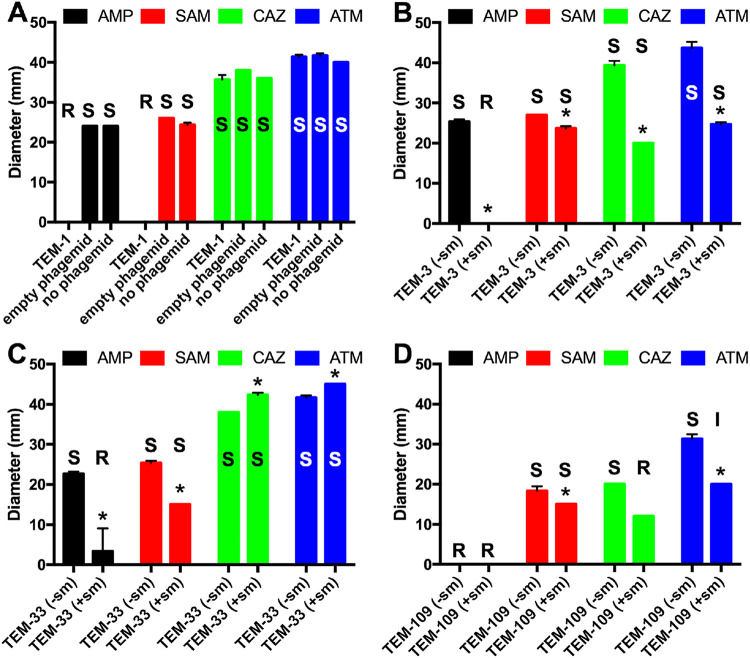
TEM-1 (GenBank accession number J01749 [20, 21]) is a serine β-lactamase that belongs to functional class 2b (17). It can inactivate penicillins and narrow-spectrum (but not extended-spectrum) cephalosporins and is typically inactivated by the β-lactamase inhibitor clavulanic acid (17). As expected, in DD assays, expression of TEM-1 resulted in no inhibition of E. coli cell growth around a disc with the penicillin ampicillin (AMP), indicating resistance, but inhibition zones were observed around discs with ceftazidime (CAZ), an extended-spectrum, third-generation cephalosporin, or aztreonam (ATM), a monobactam, indicating susceptibility to those antibiotics (Fig. 1A). Interestingly, TEM-1 expression also resulted in no inhibition zone around a disc with sulbactam (a β-lactamase inhibitor similar to clavulanic acid) plus ampicillin (SAM), indicating resistance. This behavior has been observed previously and was attributed to high expression levels of TEM-1 (22, 23). Negative controls (DH10B cells harboring an empty pBC SK(+) phagemid or no phagemid) could not grow close to the discs containing AMP, SAM, CAZ, or ATM, indicating susceptibility. TEM-3 (24) has three amino acid changes (Q39K, E104K, and G238S) relative to TEM-1 and belongs to functional class 2be, meaning that in addition to penicillins and narrow-spectrum cephalosporins, it can also inactivate extended-spectrum cephalosporins and aztreonam (17). In DD assays, TEM-3 expressed from a gene only harboring these nonsynonymous mutations (i.e., without synonymous mutations, labeled −sm) was indistinguishable from negative controls in terms of inhibition zone diameters (sensitive to AMP, SAM, CAZ, and ATM) (Fig. 1B). TEM-33 (25) deviates from TEM-1 by the single amino acid change M69L and belongs to functional class 2br, which means it is resistant to inhibition by clavulanic acid and similar β-lactamase inhibitors, such as sulbactam (17). Again, TEM-33 expressed from a gene with nonsynonymous mutation only (−sm) was indistinguishable from negative controls (Fig. 1C). TEM-109 (26) deviates from TEM-1 by the following three amino acid changes: M69L, which it has in common with TEM-33; E104K, which it has in common with TEM-3; and R164H. This enzyme belongs to functional class 2ber, which means it has an extended spectrum of activity, including extended-spectrum cephalosporins and aztreonam, and resistance to β-lactamase inhibitors. When expressed from a gene without synonymous mutations (−sm), this enzyme allowed cells to grow close to the AMP disc, indicating resistance, and with slightly lower inhibition diameters than the negative controls for SAM, CAZ, and ATM, although still in the range associated with susceptibility (Fig. 1D).
FIG 1.
Results from disc diffusion assays using Escherichia coli DH10B cells expressing the various enzymes encoded on pBC SK(+) phagemids and discs containing ampicillin (AMP), sulbactam and ampicillin (SAM), ceftazidime (CAZ), or aztreonam (ATM). The values shown are averages ± standard deviations of three independent experiments. (A) Inhibition zone diameters of cells expressing TEM-1 indicating the 2br phenotype in comparison to negative controls (cells harboring an empty or no phagemid). Susceptible (S), intermediate (I), or resistant (R) interpretations are given above each column and were determined using interpretive criteria for zone diameters according to the Clinical and Laboratory Standards Institute (53): AMP: S, ≥17 mm; I, 14 to 16 mm; R, ≤13 mm; SAM: S, ≥15 mm; I, 12 to 14 mm; R, ≤11 mm; CAZ: S, ≥21 mm; I, 18 to 20 mm; R, ≤17 mm; ATM: S, ≥21 mm; I, 18 to 20 mm; R ≤17 mm. (B) Cells expressing TEM-3, (C) TEM-33, and (D) TEM-109 expressed from genes containing no (−sm) or all (+sm) synonymous mutations. *, (+sm) significantly different from (−sm) as determined by t test pairwise comparison; P < 0.05.
We were wondering if this poor agreement with the expected phenotypes of the encoded enzymes could be due to the missing synonymous mutations and did the same experiments with the genes encoding the three variants with all synonymous mutations in addition to the nonsynonymous mutations (+sm). This amounted to four additional mutations in blaTEM-3(+sm) (GenBank accession number X64523 [27]), namely a138g, c228t, t474c, and g717a, in addition to the nonsynonymous mutations c109a, g304a, and g706a, which are responsible for the Q39K, E104K and G238S mutations; three additional mutations in blaTEM-33(+sm) (GenBank accession number GU371926 [28]), namely c18t, c228t, and g396t, in addition to the nonsynonymous mutation a199c (M69L); and two additional mutations in blaTEM-109(+sm) (GenBank accession number AY628175 [26]), c18t and c228t, in addition to the nonsynonymous mutations a199c (M69L), g304a (E104K), and g485a (R164H) relative to the −sm constructs. Note that lowercase letters are used for nucleotides to distinguish them from amino acids in capital letters. There is a slight shift between expected and actual amino acid numbering due to the class A standard numbering scheme (29) (e.g., M69L [30] instead of the expected M67L [21, 31] based on the a199c nucleotide change). Although the enzymes expressed from these constructs did not entirely reproduce the published phenotypes, the results trended in that direction. Expression of the blaTEM-3(+sm) gene resulted in no inhibition around the AMP disc, and the zone diameters for SAM, CAZ, and ATM were significantly reduced, thus trending toward a 2be phenotype (Fig. 1B). Expression of blaTEM-33(+sm) resulted in significantly decreased diameters with AMP and SAM and an increase of diameters with CAZ and ATZ, thus trending toward the 2br phenotype (Fig. 1C). Expression of blaTEM-109(+sm) maintained growth around the AMP disc and significantly decreased diameters with SAM, CAZ, and ATM, thus trending toward the 2ber phenotype (Fig. 1D). Incomplete resistance may be due to the fact that we are expressing these enzymes in the model system E. coli DH10B/pBC SK(+) rather than in the original clinical strains. In summary, adding the synonymous mutations did have a significant impact on the DD assay results with the three variant enzymes expressed from these constructs (+sm) versus the same enzymes expressed from the constructs without synonymous mutations (−sm).
Genes encoding TEM variants without synonymous mutations have not been isolated.
We sought to investigate if our observations are consistent with TEM genes that have been isolated. A Nucleotide BLAST (NCBI) search was carried out with the blaTEM-1 gene, as well as the −sm and +sm constructs of the blaTEM-3, blaTEM-33, and blaTEM-109 genes. Table 1 shows how many hits with 100% sequence identity and coverage were found.
TABLE 1.
Summary of the characteristics of the different blaTEM gene constructs without (−sm) and with (+sm) synonymous mutations
| Gene | No. of nucleotide BLAST hits | MIC (μg/ml) (S/R/I)a |
Relative expression level (%)b | |||
|---|---|---|---|---|---|---|
| AMP | SAM | CAZ | ATM | |||
| blaTEM-1 | >100 | >1,024 (R) | >1,024 (R) | 16 (R) | 0.5 (S) | 100 |
| blaTEM-3(−sm) | 0 | >1,024 (R) | 16 (I) | 64 (R) | 16 (R) | 14 ± 2 |
| blaTEM-3(+sm) | 7 | >1,024 (R) | 8 (S) | 16 (R) | 1,024 (R) | 59 ± 8 |
| blaTEM-33(−sm) | 0 | >1,024 (R) | >1,024 (R) | 0.5 (S) | 2 (S) | 51 ± 7 |
| blaTEM-33(+sm) | 2 | >1,024 (R) | 1,024 (R) | 0.5 (S) | 0.25 (S) | 51 ± 10 |
| blaTEM-109(−sm) | 0 | >1,024 (R) | 64 (R) | 128 (R) | 64 (R) | 17 ± 2 |
| blaTEM-109(+sm) | 2 | >1,024 (R) | 64 (R) | 256 (R) | >1,024 (R) | 24 ± 4 |
Susceptible (S), intermediate (I), and resistant (R) interpretation of MICs in μg/ml according to CLSI performance standards (53): AMP: S, ≤8; I, 16; R, ≥32; SAM: S, ≤8; I, 16; R, ≥32; CAZ: S, ≤4; I, 8; R, ≥16; ATM: S, ≤4; I, 8; R, ≥16.
Three independent Western blot experiments were carried out per enzyme. Expression of TEM-1 was used as a reference (100%), and for the other enzymes, averages of the relative expression levels ± standard deviations are reported.
Interestingly, none of the genes without synonymous mutations (−sm) had a perfect match in public databases, whereas there were many (>100) for blaTEM-1, a few (n = 7) for blaTEM-3(+sm) and a couple each for blaTEM-33(+sm) and blaTEM-109(+sm).
From an evolutionary perspective, it would be much more efficient to accomplish a desired result, such as extended spectrum or inhibitor resistance, with as few mutations as possible, in particular, with only nonsynonymous mutations. The fact that no such genes that encode TEM-3, TEM-33, and TEM-109 have been isolated suggests that the synonymous mutations have a purpose, which is supported by our DD assay results. Since the gene products are identical, whether expressed from a gene without or with synonymous mutations, we hypothesize that the benefit is related to expression. To further corroborate our findings, we carried out MIC assays in a broth microdilution format and investigated expression levels by Western blots under the same conditions.
MIC assays and Western blots corroborate some, but not all, trends observed in DD assays.
MIC assays were carried out in a broth microdilution format according to Clinical and Laboratory Standards Institute (CLSI) guidelines (32). All constructs conferred the same (very high) resistance level (MIC > 1,024 μg/ml) against AMP (Table 1). Thus, there is a discrepancy with blaTEM-3(−sm) and blaTEM-33(−sm), which were both sensitive in DD assays. Except for blaTEM-3(−sm) and blaTEM-3(+sm), which resulted in MIC values consistent with intermediate and susceptible phenotypes, respectively, all other constructs resulted in the resistant phenotype with SAM. This is in contrast to DD assays, where only blaTEM-1 conferred resistance. As would be expected for the 2br phenotype, both the blaTEM-33(−sm) and blaTEM-33(+sm) constructs resulted in very high MICs of >1,024 and 1,024 μg/ml, respectively, comparable to that of blaTEM-1 at >1,024 μg/ml. With CAZ, except for the blaTEM-33 constructs, which clearly conferred a sensitive phenotype, all other constructs conferred resistance, which is in agreement with the 2be phenotype of TEM-3 and the 2ber phenotype of TEM-109. SAM and CAZ resistance observed with TEM-1 can be explained by its high expression levels (Table 1) (22, 23). With ATM, the MICs were exactly as would be expected from the functional classes of the enzymes expressed; both blaTEM-3 constructs (2be) and both blaTEM-109 constructs (2ber) conferred resistance to the extended-spectrum monobactam, while blaTEM-1 (2b) and the two blaTEM-33 constructs (2br) did not. It is of note that in a few cases (the blaTEM-3 constructs with ATM and the blaTEM-109 constructs with CAZ and ATM) the constructs with synonymous mutations (+sm) resulted in higher MICs than the constructs without synonymous mutations (−sm).
Cells for Western blots were grown under the same conditions and harvested after overnight incubation. Expression level analysis by Western blots indicated that all variants, whether expressed from constructs without or with synonymous mutations, had lower expression levels than TEM-1 (Table 1). An interesting trend that was observed is that for the blaTEM-3 and blaTEM-109 genes the +sm constructs always resulted in significantly higher enzyme expression levels than the −sm constructs (P < 0.05; t test pairwise comparison). For the blaTEM-33 constructs no difference was observed.
When considering different aspects of gene expression that could result in deviating protein expression levels, we concluded that secondary structure and stability of the mRNA transcripts was the most likely. We found it unlikely that nucleotide changes in a relatively short stretch of DNA, which mostly exists as a double helix, would significantly affect unwinding and strand separation of DNA during transcription. The GC content of the different constructs was calculated with the web server Genomics %G∼C Content Calculator (https://www.sciencebuddies.org/science-fair-projects/references/genomics-g-c-content-calculator). It deviated only slightly, between 49.0% and 49.5%, and there was no clear trend that adding synonymous mutations increased or decreased GC content [blaTEM-1, 49.4%; blaTEM-3(−sm), 49.0%; blaTEM-3(+sm), 49.0%; blaTEM-33(−sm), 49.5%; blaTEM-33(+sm); 49.1%; blaTEM-109(−sm), 49.2%; blaTEM-109(+sm), 49.0%]. Codon bias is also very unlikely to have any effect, because (i) all genes investigated with synonymous mutations are found in closely related Enterobacteriaceae (mainly E. coli and Klebsiella pneumoniae), and E. coli DH10B was used in our experiments, and (ii) codons associated with no synonymous mutations, which lead to lower expression levels, correspond to the original codons in the blaTEM-1 sequence, which is expressed very well (Table 1).
Posttranslational events, such as binding of the preprotein to chaperones, transport across the periplasmic membrane, removal of the leader sequence, or protein folding are also unlikely to have any effect, because at this stage the enzymes are identical, whether expressed from −sm or +sm constructs.
Thus, we examined possible effects of the different mRNA sequences on mRNA structure and stability using the program mfold.
mfold predicts different secondary structure stabilities for mRNA without and with synonymous mutations.
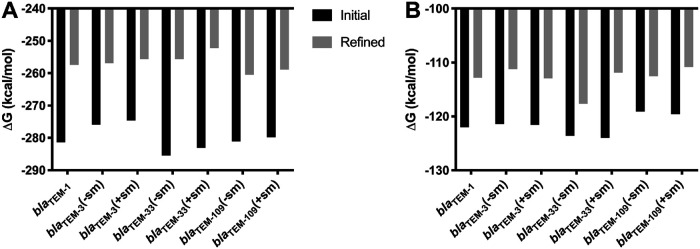
mRNA constructs starting immediately downstream of the T3 promoter and 20 nucleotides upstream of the Shine-Dalgarno sequence on the 5′ terminus and ending with the 3′-terminal BamHI restriction site, totaling 901 nucleotides (full) and the 5′-terminal half of the transcript totaling 450 nucleotides (half) were analyzed with mfold (16). The 5′-terminal half transcripts were considered, because bacterial translation starts before transcription is completed, most of the mutations occurred in the 5′-terminal halves, and it was observed that mRNA structures of the 5′ terminus in particular are important for translation efficiency in E. coli (11). Shorter transcripts were also considered, but because they eliminated some of the mutations, some variants appeared identical, and they were not further investigated. mfold lists computed secondary mRNA structures by the initial free energy difference (ΔG) relative to the unfolded mRNA, with the most favorable structure (most negative) listed first. “Initial” means that an efficient algorithm with slightly simpler rules is used. Subsequently, the initial ΔG values are reevaluated using best rules. Since these refined rules can change the initial ranking, an alternative ranking based on the refined energies is provided as well. We considered both the highest-ranked initial energy and the highest-ranked refined energy for each transcript. The results for the full transcripts and the 5′-terminal halves of the transcripts are shown in Fig. 2A and B, respectively.
FIG 2.
mRNA secondary structure folding energies of the highest-ranked (lowest-energy) structures of the full transcripts (A) and the 5′-terminal halves of the transcripts (B). “Initial” indicates energies calculated with simpler, more efficient rules. “Refined” indicates that these energies are then reevaluated with best rules and reranked.
For the full transcripts, the +sm constructs always (whether initial or refined energies were used) resulted in a less stable mRNA secondary structure than the respective −sm constructs, consistent with the notion that these constructs could be more easily unfolded for efficient translation. For the 5′-terminal half transcripts, the same is true only for the refined energies for the blaTEM-33 and blaTEM-109 constructs, but not for the blaTEM-3 constructs. This discrepancy may be explained by mutations in the 3′-terminal portion of the transcript, which is not considered in these calculations. In that part of the transcript, blaTEM-3(+sm) deviates by two synonymous mutations (t474c and g717a) from blaTEM-3(−sm), while blaTEM-33(+sm) and blaTEM-109(+sm) have no synonymous mutations compared to their −sm counterpart constructs. Thus, secondary structure and stability provides a possible explanation for different expression levels of the same gene products from different genes (without or with synonymous mutations).
In this study, individual mRNA secondary structures were not further interpreted, because the conformations in different ranks for each construct deviated just as much as the highest-ranked conformations between constructs, and we believe that mRNA secondary structures have to be seen as an ensemble rather than as individual conformations.
Certain combinations of synonymous and nonsynonymous mutations in the blaTEM-1 to blaTEM-33(+sm) mutational pathway are observed in nature, while others are not.
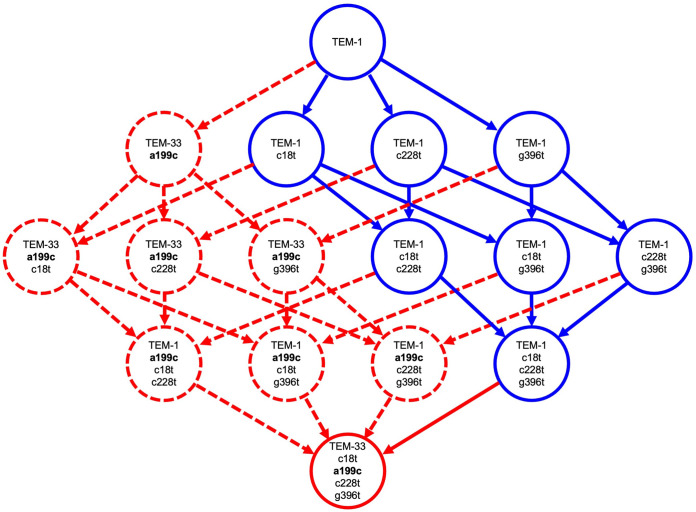
Encouraged by these observations on the pairwise comparison of genes with no synonymous (−sm) or all (+sm) synonymous mutations, we next asked whether our investigations can be extended to different combinations of synonymous and nonsynonymous mutations. In order to minimize complexity, we selected the mutational pathways separating the two constructs blaTEM-1 and blaTEM-33(+sm), which deviate in total by four mutations (one nonsynonymous and three synonymous). There are 24 = 16 possible combinations of these four mutations, including blaTEM-1, blaTEM-33(−sm), and blaTEM-33(+sm), and 4! = 24 possible mutational pathways (Fig. 3). Again, we performed Nucleotide BLAST searches for all 16 possible constructs. The number of hits (perfect matches) for each construct is shown in Table 2. Constructs that have been isolated (at least one Nucleotide BLAST hit) are shown in solid circles in Fig. 3, while those that have not been isolated are shown in dashed circles. Constructs that encode TEM-1 and TEM-33 are colored blue and red, respectively. Interestingly, the nonsynonymous a199c mutation did not exist by itself or in combination with only one or two synonymous mutations in any of the isolated constructs. This mutation was only found in combination with all three synonymous mutations, suggesting that this combination is biologically important. All of the mutational pathways shown with solid circles and solid arrows seem to be viable, while those with dashed circles and dashed arrows are not. Until the final nonsynonymous mutation is introduced, the enzyme encoded remains TEM-1 (blue).
FIG 3.
All possible mutational pathways from blaTEM-1 to blaTEM-33(+sm). Each circle is labeled with the encoded gene product (blue, TEM-1; red, TEM-33), as well as all mutations, where the nonsynonymous mutation that changes the gene product from TEM-1 to TEM-33 through the M69L amino acid change is indicated in boldface. Variants that have been isolated in nature are shown by solid circles, while hypothetical variants not isolated in nature are shown by dashed circles. Possible pathways that proceed through variants that have been isolated are indicated by solid arrows and those through hypothetical variants by dashed arrows.
TABLE 2.
Summary of the characteristics of the different gene constructs from blaTEM-1 to blaTEM-33(+sm)c
| Mutation(s) relative to blaTEM-1 | Encoded enzyme | No. of nucleotide BLAST hits | Disc diffusion (DD) assay zone diam (mm) (S/R/I)a |
Broth microdilution assay MIC (μg/ml) (S/R/I/)b |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AMP | SAM | CAZ | ATM | AMP | SAM | CAZ | ATM | |||
| NAd (blaTEM-1) | TEM-1 | >100 | 0 (R) | 0 (R) | 35 (S) | 40 (S) | >1,024 (R) | >1,024 (R) | ≤1 (S) | ≤0.25 (S) |
| a199ce [blaTEM-33(−sm)] | TEM-33e | 0 | 23 (S) | 25 (S) | 42 (S) | 45 (S) | >1,024 (R) | >1,024 (R) | ≤1 (S) | ≤0.25 (S) |
| c18t | TEM-1 | 5 | 0 (R) | 10 (R) | 40 (S) | 45 (S) | 512 (R) | 128 (R) | ≤1 (S) | ≤0.25(S) |
| c228t | TEM-1 | 83 | 0 (R) | 30 (S) | 21 (S) | 0 (R) | >1,024 (R) | 256 (R) | ≤1 (S) | ≤0.25 (S) |
| g396t | TEM-1 | 91 | 0 (R) | 20 (S) | 41 (S) | 41 (S) | >1,024 (R) | 256 (R) | ≤1 (S) | ≤0.25 (S) |
| a199c, c18t | TEM-33 | 0 | 0 (R) | 10 (R) | 32 (S) | 12 (R) | >1,024 (R) | 512 (R) | ≤1 (S) | ≤0.25 (S) |
| a199c, c228t | TEM-33 | 0 | 0 (R) | 12 (I) | 25 (S) | 0 (R) | >1,024 (R) | 512 (R) | ≤1 (S) | ≤0.25 (S) |
| a199c, g396t | TEM-33 | 0 | 0 (R) | 0 (R) | 32 (S) | 12 (R) | >1,024 (R) | >1,024 (R) | ≤1 (S) | ≤0.25 (S) |
| c18t, c228t | TEM-1 | 16 | 0 (R) | 10 (R) | 25 (S) | 0 (R) | >1,024 (R) | >1,024 (R) | ≤1 (S) | ≤0.25 (S) |
| c18t, g396t | TEM-1 | 1 | 10 (R) | 25 (S) | 20 (I) | 0 (R) | >1,024 (R) | 256 (R) | ≤1 (S) | ≤0.25 (S) |
| c228t, g396t | TEM-1 | 23 | 0 (R) | 9 (R) | 26 (S) | 12 (R) | >1,024 (R) | 256 (R) | ≤1 (S) | ≤0.25 (S) |
| a199c, c18t, c228t | TEM-33 | 0 | 0 (R) | 0 (R) | 31 (S) | 12 (R) | >1,024 (R) | >1,024 (R) | ≤1 (S) | ≤0.25 (S) |
| a199c, c18t, g396t | TEM-33 | 0 | 0 (R) | 0 (R) | 40 (S) | 45 (S) | >1,024 (R) | >1,024 (R) | 2 (S) | ≤0.25 (S) |
| a199c, c228t, g396t | TEM-33 | 0 | 0 (R) | 8 (R) | 30 (S) | 13 (R) | >1,024 (R) | 512 (R) | ≤1 (S) | ≤0.25 (S) |
| c18t, c228t, g396t | TEM-1 | >100 | 0 (R) | 15 (S) | 30 (S) | 40 (S) | >1,024 (R) | 256 (R) | ≤1 (S) | ≤0.25 (S) |
| c18t, c228t, g396t, a199c [blaTEM-33(+sm)] | TEM-33 | 2 | 0 (R) | 15 (S) | 42 (S) | 45 (S) | >1,024 (R) | 512 (R) | ≤1 (S) | ≤0.25 (S) |
For S/R/I interpretation see Fig. 1.
for S/R/I interpretation see Table 1.
Experiments with blaTEM-1, blaTEM-33(−sm), and blaTEM-33(+sm) were repeated, and results deviate slightly from those in Table 1.
NA, not applicable.
Mutations in bold are nonsynonymous mutations leading to the M69L amino acid change and the TEM-33 variant.
Most of the gene variants result in viable enzymes, even the ones not isolated in nature.
The absence of some blaTEM constructs in isolates suggests that those constructs do not yield functional enzymes and do not confer resistance. To test this hypothesis, all constructs were expressed from the pBC SK(+) vector in E. coli DH10B.
Western blot results varied between replicates, as reported in a previous study (15), resulting in no significantly different expression levels between the 16 variants. This result was not too surprising, given that no difference in expression level could be detected between blaTEM-33(−sm) and blaTEM-33(+sm) (Table 1). However, all variants were expressed at sufficient levels (intervariant deviations within a factor of 4; data not shown) to result in resistance against AMP and SAM in MIC assays (see below).
DD assays yielded mixed results. All constructs except the one with the nonsynonymous mutation yielded no or only small inhibition zones with AMP. Several constructs that encoded TEM-1 or TEM-33 resulted in no or small inhibition zones with SAM. All constructs gave rise to large inhibition zones with CAZ. Interestingly, several constructs, but not blaTEM-1, blaTEM-33(−sm), or blaTEM-33(+sm), yielded no or small inhibition zones with ATM, consistent with a resistant phenotype. The blaTEM-33(−sm) construct was the only one that resulted in large inhibition zones with all antibiotics tested. Results from MIC assays were less mixed; all constructs conferred resistance to AMP and SAM, and none of the constructs conferred resistance to CAZ or ATM.
Next, we resorted to mfold analysis of mRNA constructs to see if differences could be resolved with this computational approach.
mfold suggest that mutational pathways through some gene variants are disfavored by the stability of the resulting mRNA constructs.
As described above for the −sm and +sm constructs, all possible mRNA transcripts resulting from combinations of the three synonymous mutations and the one nonsynonymous mutation separating blaTEM-33(+sm) from blaTEM-1 were subjected to RNA folding by mfold. When comparing the transcripts of blaTEM-33(−sm) and blaTEM-33(+sm), using the lowest-energy structures after refinement yielded the biggest differences in ΔG (Fig. 2). Therefore, we used the refined energies here. Furthermore, because translation starts before transcription is complete and all four mutations investigated are in the 5′-terminal portion of the transcript, we focused on the 5′ terminal half of the transcripts.
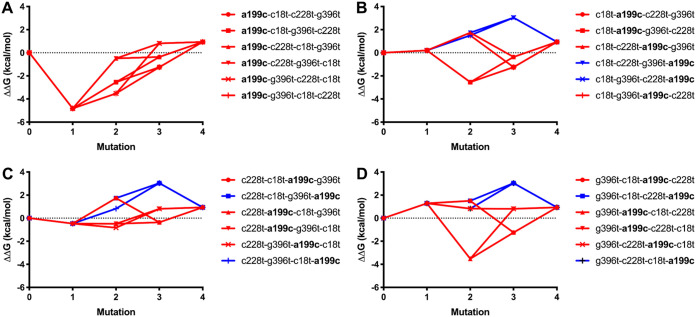
Figure 4 illustrates the results of these secondary structure and energy determinations along different possible mutational pathways. Interestingly, an initial a199c mutation results in a stabilization of the 5′-terminal portion of the transcript by 4.82 kcal/mol relative to blaTEM-1 (Fig. 4A). After all four mutations have been introduced, the transcript is destabilized by 0.94 kcal/mol. None of these pathways seem to be viable, because blaTEM-33(a199c), which equals blaTEM-33(−sm), has not been isolated. Note that some of our experimental results contradict that notion, as the enzyme was successfully expressed from the blaTEM-33(−sm) construct (Table 1) and yielded resistance against AMP and SAM at least in MIC assays (Tables 1 and 2). Among the other pathways, only those in which a199c is introduced last seem to be viable, i.e., proceed through variants that actually exist in nature (Fig. 3), and those pathways (colored blue) are also the ones that go through the highest-energy (least-stabilized) pathways (Fig. 4B to D).
FIG 4.
Energy differences of secondary structures of the 5′-terminal halves of transcripts of different blaTEM constructs relative to that of blaTEM-1 are shown for different mutational pathways starting with the nonsynonymous mutation a199c (A) or with the synonymous mutations c18t (B), c228t (C), or g396t (D). Pathways that go through variants that have been isolated in nature (see Fig. 3) are colored in blue, while those that go through variants that have not been isolated are colored in red. The a199c mutation is shown in boldface to indicate that it leads to the M69L amino acid change that distinguished TEM-33 from TEM-1.
The more stable a transcript is, the harder it could be to form the translation initiation complex consisting of the unfolded mRNA (at least locally around the Shine-Dalgarno sequence), the 30S ribosomal subunit, initiation factors 1 and 2, the initiator tRNA loaded with formyl methionine, and GTP. Thus, this analysis provides a possible explanation why the expression of constructs with certain combinations of mutations is less efficient than that of others. In this particular case, introduction of the nonsynonymous a199c mutation seems to be so unfavorable that the mRNA needs to be destabilized first by three synonymous mutations in order to allow the introduction of a199c.
DISCUSSION
A controversy persists about whether selective pressure can increase the mutation rate or lead to more efficient adaptation through other mechanisms, such as amplification of mutated genes (33). Whether synonymous mutations need to be considered in the investigation of evolution and protein engineering efforts is another fundamental question. They have been the subject of several excellent reviews (8, 9, 34–36), most of them focusing on eukaryotic organisms and the role of “accidental” synonymous mutations in human disease. Some of these mutations have been characterized intensively, for instance, the ΔF508 CFTR mutation involved in cystic fibrosis (37). It has been proposed that this mutation, as well as a synonymous mutation in the P-glycoprotein gene (also known as the multidrug resistance 1 gene) (38), alter translation kinetics, which in turn can affect protein folding and function (39).
The role of synonymous mutations in the evolution of biological function is not well understood, and while many studies have focused on nonsynonymous mutations in blaTEM variants (reviewed, e.g., in references 40–43), to our knowledge there have only been a few reports focusing on synonymous mutations (13–15, 44). Using random mutagenesis, Schenk et al. (13) identified 48 mutations in blaTEM-1 that were beneficial against cefotaxime (a third-generation cephalosporin), out of which 10 were synonymous mutations and included the c18t mutation, also found in blaTEM-33(+sm) and blaTEM-109(+sm). An epidemiological study in South Korea also found this synonymous mutation in clinical isolates of Neisseria gonorrhoeae expressing TEM-135 (44). From a laboratory evolution study, Bratulic et al. concluded, among other things, that TEM-1 does not evolve translational accuracy through synonymous mutations toward high-fidelity codons (14), which also did not seem to play a role in our study. In a follow-up study to Schenk et al. (13), Zwart et al. compared those 10 synonymous mutations to a selection of 10 nonsynonymous mutations (15). They did not find a significant difference in mRNA levels or mRNA stability assessed by mfold, although they used a different approach from us, i.e., analysis of a moving 45-nucleotide window surrounding the mutation in question. They did, however, see an increase in total and functional protein levels with both synonymous and nonsynonymous mutations relative to blaTEM-1. In their case, that result was expected, because the mutants were generated and selected under cefotaxime selective pressure. For synonymous mutations, but not nonsynonymous mutations, they also found a good correlation between total and functional TEM levels, suggesting that these mutations alter protein expression levels without altering the intrinsic protein properties.
As pointed out in the mentioned reviews, especially in eukaryotic cells, there are many layers at which synonymous mutations can impact gene expression, namely mRNA stability, splicing, interaction with small interfering RNAs (siRNAs), degradation, transport to its destination (cytosolic or endoplasmic reticulum-bound ribosomes), interaction with the ribosome and translation factors, and the effect on cotranslational protein folding. In prokaryotes, the situation is less complex, as all ribosomes are cytosolic and ribosomes typically bind to mRNAs and initiate translation while the mRNA is still being synthesized by transcription. This decreases possible layers, at which synonymous mutations could affect protein expression in our system to the following:
(i) Codon bias (or codon usage bias). As pointed out throughout this study, we find codon bias to be unlikely to have a major impact, because transcripts with certain codons are expressed very well in the absence, but not in the presence, of other codons in different parts of the mRNA and because all organisms from which the different blaTEM gene variants have been isolated are phylogenetically closely related, i.e., all Enterobacteriaceae.
(ii) mRNA secondary structure and stability. In vivo, mRNAs probably exist as an ensemble of different secondary structures with slightly different free energies. Since mRNAs are typically translated cotranscriptionally, these secondary structures and energies of the mRNA (in particular the 5′ terminus) are likely to have an impact on translation efficiency.
In some early studies on TEM β-lactamase variants, e.g., reviewed previously (40), it is not always clear whether synonymous mutations were considered or not. When genes are isolated from clinical strains, it is expected that any synonymous mutations are included, although in some cases the isolated gene only had nonsynonymous mutations (31). When mutations are introduced for the purpose of investigating amino acid changes, synonymous mutations are typically not considered. For instance, Blazquez et al. studied individual nonsynonymous mutations frequently observed in early TEM variants by site-directed mutagenesis (45) and generated TEM-1 variants with single nonsynonymous mutations by exposing cells to antibiotics (46). To our knowledge, nonsynonymous mutations observed in wild-type variants have typically not been combined without including synonymous mutations (as in our blaTEM-3(−sm) and blaTEM-109(−sm) constructs), so it is hard to compare our −sm variants with literature data.
Since no gene encoding TEM-33 without synonymous mutations relative to TEM-1 was found in Nucleotide BLAST searches, we hypothesize that the synonymous mutations “prepare” the gene to tolerate the nonsynonymous mutation. Assuming that in each evolutionary round of mutation and selection only one mutation is established, this would mean that in order to acquire one nonsynonymous mutation that confers a selective benefit to bacteria (in this case, inhibitor resistance), three rounds of synonymous mutation and selection would have to be completed first. Such synonymous mutations could provide a benefit that is too small to be detected in DD or MIC assays but might be advantageous enough to allow the mutants to persist in a population of bacteria. In the future, we plan on testing this hypothesis in competition experiments (47), which are more sensitive. Such studies will also allow us to test fitness of different variants at different time points and at very low antibiotic concentrations, which is not done in DD or MIC assays. At any rate, evolution under consideration of all synonymous mutations might be much more complex than generally appreciated based on nonsynonymous mutations alone.
The following two observations were made in the course of this study:
(i) There was a significant discrepancy between DD assay and MIC assay results. For instance, blaTEM-3(−sm) expression did not refer resistance to any of the antibiotics tested in DD assays (Fig. 1B), but in MIC assays it conferred resistance to AMP, CAZ, ATM, and an intermediate phenotype to SAM (Table 1). This phenomenon will have to be investigated in more detail. Inconsistencies between DD assays and MIC assays in an agar dilution method have been reported previously for Haemophilus influenzae (48), Pseudomonas aeruginosa (49), Enterobacteriaceae, Acinetobacter spp., pseudomonads, staphylococci and enterococci (50), and various Gram-positive cocci and rods (51). MIC assays use serial dilutions with fixed antibiotic concentrations, while in DD assays a continuous antibiotic gradient is established. In our case, MIC assays were carried out in a broth microdilution format. Therefore, the different growth media (solid state agar versus liquid medium) could additionally skew results by deviating growth kinetics of cells growing in a colony on a surface versus single cells growing in suspension. In the future, we plan to explore other assay formats that might be more reliable, e.g., the Epsilometer test (Etest) (52).
(ii) There was a large interreplicate deviation in protein expression levels as determined by Western blotting, making intervariant differences statistically insignificant, an issue reported previously (15). We attempted to minimize deviations by using DnaK as an internal standard. However, DnaK is chromosomally encoded versus the plasmid-encoded blaTEM variants. Thus, plasmid copy numbers could be a source of uncertainty. We did not induce expression. However, expression could be affected by endogenous factors of the E. coli cells harboring the plasmids, and these could deviate with altering physiological states of the cells. This is also an area that will have to be investigated more carefully.
Conclusion.
In response to the question of whether synonymous mutations are silent or noisy or “whisper” (8), our data clearly indicate that they are not silent. It was suggested that they are also not “synonymous” (8). We have kept the term “synonymous,” as in analogy to language, synonyms have largely, but not exactly, the same meaning. However, we do agree that the term “silent” is a poor choice, because it implies no effect whatsoever. Our results support the notion that synonymous mutations are a prerequisite for the viability of nonsynonymous mutations. The exact mechanism by which this occurs needs to be further elucidated. However, our data point to mRNA secondary structure and stability playing an important role. While we only considered one resistance mechanism, the expression of β-lactamases, the role of synonymous mutations is also expected to play an important role in various other resistance mechanisms and, more generally, in any evolutionary adaptation.
MATERIALS AND METHODS
Generation of expression systems.
Genes encoding the various blaTEM genes were purchased from Gen9, Inc. (Cambridge, MA) or Biomatik (Kitchener, ON, Canada); genes had a SacI restriction site followed by a ribosome binding site (5′-GAGCTCAAGAAGGAGATATACAT-3′) on the 5′ terminus and a BamHI restriction site (5′-GGATCC-3′) immediately following the 3′ terminal stop codon. These restriction sites were used to excise the genes from the vectors provided by the suppliers and to insert them into the pBC SK(+) phagemid (Invitrogen, La Jolla, CA), which contains a chloramphenicol resistance gene. This gave rise to pBC SK(+) phagemids with the blaTEM genes under the control of the T3 promoter. Max Efficiency DH10B competent cells (Invitrogen, La Jolla, CA) were transformed with the resulting phagemids and selected on Mueller-Hinton broth II (MHB) agar plates containing 34 μg/ml chloramphenicol. Several colonies were picked and grown in MHB with chloramphenicol, adjusted to an optical density at 600 nm of 0.08 (6.4 × 107 cells/ml), and then used for the disc diffusion and MIC assays. It is assumed that the chloramphenicol acetyltransferase expressed from the pBC SK(+) vector sufficiently inactivates chloramphenicol, so that blaTEM expression is not significantly affected. Cells transformed with an empty phagemid (no blaTEM gene) and cells not transformed served as negative controls.
Disc diffusion assay.
Sterile cotton swabs were soaked into the cultures described above and swabbed onto individual MHB agar plates for disc diffusion (DD) assay. Antibiotic discs of ampicillin (AMP), sulbactam and ampicillin (SAM), ceftazidime (CAZ), and aztreonam (ATM) (Thermo Fisher Scientific Oxoid) were placed onto the agar plates. The plates were incubated for 16 h at 37°C, and the zone of inhibition diameters were measured. Although a clinical application is not the focus of this study, the results were also interpreted as susceptible (S), intermediate (I), or resistant (R) according the CLSI performance standards (53).
MIC assays.
MIC assays were carried out using the broth microdilution method following CLSI guidelines (32), and S/R/I interpretations were assigned according to CLSI performance standards (53).
Western blotting.
The procedure to quantify in-cell expression levels of TEM enzymes from the various blaTEM constructs was carried out as previously described (54) except that anti-TEM-1 antibody (ABIN2754695; antibodies-online, Inc.) was used instead of anti-IMP-1 antibody.
Nucleotide BLAST searches.
The Nucleotide BLAST (blastn) program was accessed at https://blast.ncbi.nlm.nih.gov. The complete coding sequences (861 nucleotides) of the studied blaTEM genes were used as queries, and default parameters (standard databases, highly similar sequences [megablast]) were used to find matching/similar sequences. The numbers of perfect matches (query coverage = identity = 100%) were recorded.
mfold studies.
The RNA Folding Form of mfold (16) was accessed on the mfold web server (www.unafold.org/RNA_form.php). mRNA constructs starting immediately downstream of the T3 promoter and 20 nucleotides upstream of the Shine-Dalgarno sequence on the 5′ terminus and ending with the 3′-terminal BamHI restriction site, totaling 901 nucleotides, were considered the full transcripts. In addition, 5′-terminal portions of the transcript (half = 450 nucleotides, one-third = 300 nucleotides, etc., down to one-ninth = 100 nucleotides) were analyzed. The binding energies of the highest-ranked conformations after initial folding and energy calculation and after refined energy calculation were recorded and compared between blaTEM variants.
ACKNOWLEDGMENTS
We thank the expert reviewers for insightful comments that helped us improve the manuscript.
J.P. acknowledges funding by the German Research Foundation (DFG) (grant EXC 2075).
REFERENCES
- 1.Franklin RE, Gosling RG. 1953. Molecular configuration in sodium thymonucleate. Nature 171:740–741. 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- 2.Watson JD, Crick FH. 1953. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171:737–738. 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 3.Crick FH, Griffith JS, Orgel LE. 1957. Codes without commas. Proc Natl Acad Sci U S A 43:416–421. 10.1073/pnas.43.5.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crick FH, Barnett L, Brenner S, Watts-Tobin RJ. 1961. General nature of the genetic code for proteins. Nature 192:1227–1232. 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- 5.Nirenberg M, Leder P, Bernfield M, Brimacombe R, Trupin J, Rottman F, O’Neal C. 1965. RNA codewords and protein synthesis, VII. On the general nature of the RNA code. Proc Natl Acad Sci U S A 53:1161–1168. 10.1073/pnas.53.5.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crick FH. 1958. On protein synthesis. Symp Soc Exp Biol 12:138–163. [PubMed] [Google Scholar]
- 7.Crick F. 1970. Central dogma of molecular biology. Nature 227:561–563. 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 8.Fahraeus R, Marin M, Olivares-Illana V. 2016. Whisper mutations: cryptic messages within the genetic code. Oncogene 35:3753–3759. 10.1038/onc.2015.454. [DOI] [PubMed] [Google Scholar]
- 9.Supek F. 2016. The code of silence: widespread associations between synonymous codon biases and gene function. J Mol Evol 82:65–73. 10.1007/s00239-015-9714-8. [DOI] [PubMed] [Google Scholar]
- 10.Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. 2003. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet 12:205–216. 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 11.Kudla G, Murray AW, Tollervey D, Plotkin JB. 2009. Coding-sequence determinants of gene expression in Escherichia coli. Science 324:255–258. 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ambler RP. 1980. The structure of β-lactamases. Philos Trans R Soc Lond B Biol Sci 289:321–331. 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 13.Schenk MF, Szendro IG, Krug J, de Visser JA. 2012. Quantifying the adaptive potential of an antibiotic resistance enzyme. PLoS Genet 8:e1002783. 10.1371/journal.pgen.1002783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bratulic S, Gerber F, Wagner A. 2015. Mistranslation drives the evolution of robustness in TEM-1 β-lactamase. Proc Natl Acad Sci U S A 112:12758–12763. 10.1073/pnas.1510071112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zwart MP, Schenk MF, Hwang S, Koopmanschap B, de Lange N, van de Pol L, Nga TTT, Szendro IG, Krug J, de Visser JAGM. 2018. Unraveling the causes of adaptive benefits of synonymous mutations in TEM-1 β-lactamase. Heredity (Edinb)) 121:406–421. 10.1038/s41437-018-0104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415. 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bush K, Jacoby GA, Medeiros AA. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother 39:1211–1233. 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baquero M-R, Galán JC, del Carmen Turrientes M, Cantón R, Coque TM, Martínez JL, Baquero F. 2005. Increased mutation frequencies in Escherichia coli isolates harboring extended-spectrum β-lactamases. Antimicrob Agents Chemother 49:4754–4756. 10.1128/AAC.49.11.4754-4756.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durfee T, Nelson R, Baldwin S, Plunkett G, Burland V, Mau B, Petrosino JF, Qin X, Muzny DM, Ayele M, Gibbs RA, Csörgo B, Pósfai G, Weinstock GM, Blattner FR. 2008. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J Bacteriol 190:2597–2606. 10.1128/JB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datta N, Kontomichalou P. 1965. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 208:239–241. 10.1038/208239a0. [DOI] [PubMed] [Google Scholar]
- 21.Sutcliffe JG. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A 75:3737–3741. 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reguera JA, Baquero F, Perez-Diaz JC, Martinez JL. 1991. Factors determining resistance to β-lactam combined with β-lactamase inhibitors in Escherichia coli. J Antimicrob Chemother 27:569–575. 10.1093/jac/27.5.569. [DOI] [PubMed] [Google Scholar]
- 23.Wu PJ, Shannon K, Phillips I. 1994. Effect of hyperproduction of TEM-1 β-lactamase on in vitro susceptibility of Escherichia coli to β-lactam antibiotics. Antimicrob Agents Chemother 38:494–498. 10.1128/aac.38.3.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirot D, Sirot J, Labia R, Morand A, Courvalin P, Darfeuille-Michaud A, Perroux R, Cluzel R. 1987. Transferable resistance to third-generation cephalosporins in clinical isolates of Klebsiella pneumoniae: identification of CTX-1, a novel β-lactamase. J Antimicrob Chemother 20:323–334. 10.1093/jac/20.3.323. [DOI] [PubMed] [Google Scholar]
- 25.Goussard S, Courvalin P. 1999. Updated sequence information for TEM β-lactamase genes. Antimicrob Agents Chemother 43:367–370. 10.1128/AAC.43.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robin F, Delmas J, Chanal C, Sirot D, Sirot J, Bonnet R. 2005. TEM-109 (CMT-5), a natural complex mutant of TEM-1 β-lactamase combining the amino acid substitutions of TEM-6 and TEM-33 (IRT-5). Antimicrob Agents Chemother 49:4443–4447. 10.1128/AAC.49.11.4443-4447.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mabilat C, Lourencao-Vital J, Goussard S, Courvalin P. 1992. A new example of physical linkage between Tn1 and Tn21: the antibiotic multiple-resistance region of plasmid pCFF04 encoding extended-spectrum β-lactamase TEM-3. Mol Gen Genet 235:113–121. 10.1007/BF00286188. [DOI] [PubMed] [Google Scholar]
- 28.Smet A, Van Nieuwerburgh F, Vandekerckhove TTM, Martel A, Deforce D, Butaye P, Haesebrouck F. 2010. Complete nucleotide sequence of CTX-M-15-plasmids from clinical Escherichia coli isolates: insertional events of transposons and insertion sequences. PLoS One 5:e11202. 10.1371/journal.pone.0011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ambler RP, Coulson AFW, Frère JM, Ghuysen JM, Joris B, Forsman M, Levesque RC, Tiraby G, Waley SG. 1991. A standard numbering scheme for the class A β-lactamases. Biochem J 276:269–270. 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farzaneh S, Chaibi EB, Peduzzi J, Barthelemy M, Labia R, Blazquez J, Baquero F. 1996. Implication of Ile-69 and Thr-182 residues in kinetic characteristics of IRT-3 (TEM-32) β-lactamase. Antimicrob Agents Chemother 40:2434–2436. 10.1128/AAC.40.10.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blazquez J, Baquero MR, Canton R, Alos I, Baquero F. 1993. Characterization of a new TEM-type β-lactamase resistant to clavulanate, sulbactam, and tazobactam in a clinical isolate of Escherichia coli. Antimicrob Agents Chemother 37:2059–2063. 10.1128/aac.37.10.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—ninth edition. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 33.Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. 2006. Origin of mutations under selection: the adaptive mutation controversy. Annu Rev Microbiol 60:477–501. 10.1146/annurev.micro.60.080805.142045. [DOI] [PubMed] [Google Scholar]
- 34.Hunt RC, Simhadri VL, Iandoli M, Sauna ZE, Kimchi-Sarfaty C. 2014. Exposing synonymous mutations. Trends Genet 30:308–321. 10.1016/j.tig.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 35.Bali V, Bebok Z. 2015. Decoding mechanisms by which silent codon changes influence protein biogenesis and function. Int J Biochem Cell Biol 64:58–74. 10.1016/j.biocel.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gotea V, Gartner JJ, Qutob N, Elnitski L, Samuels Y. 2015. The functional relevance of somatic synonymous mutations in melanoma and other cancers. Pigment Cell Melanoma Res 28:673–684. 10.1111/pcmr.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartoszewski RA, Jablonsky M, Bartoszewska S, Stevenson L, Dai Q, Kappes J, Collawn JF, Bebok Z. 2010. A synonymous single nucleotide polymorphism in ΔF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein. J Biol Chem 285:28741–28748. 10.1074/jbc.M110.154575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komar AA. 2007. SNPs, silent but not invisible. Science 315:466–467. 10.1126/science.1138239. [DOI] [PubMed] [Google Scholar]
- 39.Bartoszewski R, Króliczewski J, Piotrowski A, Jasiecka AJ, Bartoszewska S, Vecchio-Pagan B, Fu L, Sobolewska A, Matalon S, Cutting GR, Rowe SM, Collawn JF. 2016. Codon bias and the folding dynamics of the cystic fibrosis transmembrane conductance regulator. Cell Mol Biol Lett 21:23. 10.1186/s11658-016-0025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knox JR. 1995. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob Agents Chemother 39:2593–2601. 10.1128/aac.39.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrosino J, Cantu C, 3rd, Palzkill T. 1998. β-Lactamases: protein evolution in real time. Trends Microbiol 6:323–327. 10.1016/S0966-842X(98)01317-1. [DOI] [PubMed] [Google Scholar]
- 42.Bush K. 2013. Proliferation and significance of clinically relevant β-lactamases. Ann N Y Acad Sci 1277:84–90. 10.1111/nyas.12023. [DOI] [PubMed] [Google Scholar]
- 43.Palzkill T. 2018. Structural and mechanistic basis for extended-spectrum drug-resistance mutations in altering the specificity of TEM, CTX-M, and KPC β-lactamases. Front Mol Biosci 5:16. 10.3389/fmolb.2018.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rim JH, Kim H, Lee H, Yong D, Jeong SH, Lee K. 2018. Recent increase in the incidence of TEM-135 β-lactamase-harboring Neisseria gonorrhoeae in Korea. Ann Lab Med 38:324–330. 10.3343/alm.2018.38.4.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blazquez J, Morosini MI, Negri MC, Gonzalez-Leiza M, Baquero F. 1995. Single amino acid replacements at positions altered in naturally occurring extended-spectrum TEM β-lactamases. Antimicrob Agents Chemother 39:145–149. 10.1128/aac.39.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blazquez J, Morosini MI, Negri MC, Baquero F. 2000. Selection of naturally occurring extended-spectrum TEM β-lactamase variants by fluctuating β-lactam pressure. Antimicrob Agents Chemother 44:2182–2184. 10.1128/aac.44.8.2182-2184.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Negri MC, Lipsitch M, Blazquez J, Levin BR, Baquero F. 2000. Concentration-dependent selection of small phenotypic differences in TEM β-lactamase-mediated antibiotic resistance. Antimicrob Agents Chemother 44:2485–2491. 10.1128/aac.44.9.2485-2491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrissey I, Robbins M, Viljoen L, Brown DF. 2005. Antimicrobial susceptibility of community-acquired respiratory tract pathogens in the UK during 2002/3 determined locally and centrally by BSAC methods. J Antimicrob Chemother 55:200–208. 10.1093/jac/dkh540. [DOI] [PubMed] [Google Scholar]
- 49.Henwood CJ, Livermore DM, James D, Warner M, Pseudomonas Study Group . 2001. Antimicrobial susceptibility of Pseudomonas aeruginosa: results of a UK survey and evaluation of the British Society for Antimicrobial Chemotherapy disc susceptibility test. J Antimicrob Chemother 47:789–799. 10.1093/jac/47.6.789. [DOI] [PubMed] [Google Scholar]
- 50.Potz NAC, Mushtaq S, Johnson AP, Henwood CJ, Walker RA, Varey E, Warner M, James D, Livermore DM. 2004. Reliability of routine disc susceptibility testing by the British Society for Antimicrobial Chemotherapy (BSAC) method. J Antimicrob Chemother 53:729–738. 10.1093/jac/dkh212. [DOI] [PubMed] [Google Scholar]
- 51.Jensen KT, Schonheyder H, Gottschau A, Thomsen VF. 1994. Impact of the agar medium and disc type on disc diffusion susceptibility testing against teicoplanin and vancomycin. APMIS 102:94–102. 10.1111/j.1699-0463.1994.tb04852.x. [DOI] [PubMed] [Google Scholar]
- 52.Nachnani S, Scuteri A, Newman MG, Avanessian AB, Lomeli SL. 1992. E-test: a new technique for antimicrobial susceptibility testing for periodontal microorganisms. J Periodontol 63:576–583. 10.1902/jop.1992.63.7.576. [DOI] [PubMed] [Google Scholar]
- 53.Clinical and Laboratory Standards Institute. 2012. Performance standards for antimicrobial suscepbitility testing; twenty-second informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 54.Zhang CJ, Faheem M, Dang P, Morris MN, Kumar P, Oelschlaeger P. 2019. Mutation S115T in IMP-type metallo-β-lactamases compensates for decreased expression levels caused by mutation S119G. Biomolecules 9:724. 10.3390/biom9110724. [DOI] [PMC free article] [PubMed] [Google Scholar]