ABSTRACT
Human adenoviruses (HAdVs) are a large family of DNA viruses that include more than 100 genotypes divided into seven species (A to G) and induce respiratory tract infections, gastroenteritis, and conjunctivitis. Genetically modified adenoviruses are also used as vaccines, gene therapies, and anticancer treatments. The APOBEC3s are a family of cytidine deaminases that restrict viruses by introducing mutations in their genomes. Viruses developed different strategies to cope with the APOBEC3 selection pressure, but nothing is known on the interplay between the APOBEC3s and the HAdVs. In this study, we focused on three HAdV strains: the B3 and C2 strains, as they are very frequent, and the A12 strain, which is less common but is oncogenic in animal models. We demonstrated that the three HAdV strains induce a similar APOBEC3B upregulation at the transcriptional level. At the protein level, however, APOBEC3B is abundantly expressed during HAdV-A12 and -C2 infection and shows a nuclear distribution. On the contrary, APOBEC3B is barely detectable in HAdV-B3-infected cells. APOBEC3B deaminase activity is detected in total protein extracts upon HAdV-A12 and -C2 infection. Bioinformatic analysis demonstrates that the HAdV-A12 genome bears a stronger APOBEC3 evolutionary footprint than that of the HAdV-C2 and HAdV-B3 genomes. Our results show that HAdV infection triggers the transcriptional upregulation of the antiviral innate effector APOBEC3B. The discrepancies between the APOBEC3B mRNA and protein levels might reflect the ability of some HAdV strains to antagonize the APOBEC3B protein. These findings point toward an involvement of APOBEC3B in HAdV restriction and evolution.
IMPORTANCE The APOBEC3 family of cytosine deaminases has important roles in antiviral innate immunity and cancer. Notably, APOBEC3A and APOBEC3B are actively upregulated by several DNA tumor viruses and contribute to transformation by introducing mutations in the cellular genome. Human adenoviruses (HAdVs) are a large family of DNA viruses that cause generally asymptomatic infections in immunocompetent adults. HAdVs encode several oncogenes, and some HAdV strains, like HAdV-A12, induce tumors in hamsters and mice. Here, we show that HAdV infection specifically promotes the expression of the APOBEC3B gene. We report that infection with the A12 strain induces a strong expression of an enzymatically active APOBEC3B protein in bronchial epithelial cells. We provide bioinformatic evidence that HAdVs’ genomes and notably the A12 genome are under APOBEC3 selection pressure. Thus, APOBEC3B might contribute to adenoviral restriction, diversification, and oncogenic potential of particular strains.
KEYWORDS: HAdV-A12, HAdV-B3, HAdV-C2, adenoviruses, APOBEC3, APOBEC3B, cytidine deaminase, innate immunity
INTRODUCTION
Human adenoviruses (HAdVs) are nonenveloped double-stranded DNA viruses that replicate their ∼35-kb-long linear genome in the nucleus. HAdVs belong to the genus Mastadenovirus in the Adenoviridae family. HAdVs are very diverse, currently counting 103 genotypes divided into 7 species (A to G) (according to the HAdV Working Group, July, 2019 update, http://hadvwg.gmu.edu/) (1). HAdVs were discovered in 1953 by Rowe and colleagues from human adenoids surgically removed from children (2). Serologically related viruses were subsequently identified in nasopharyngeal and conjunctival secretions and feces of patients with undifferentiated respiratory diseases. These viruses were first named “adenoidal-pharyngeal-conjunctival” viruses and finally adenoviruses (3, 4). HAdVs cause self-limiting upper and lower respiratory tract, ocular, urinary tract, and gastrointestinal infections (5, 6). In the United States, HAdV-C2, -C5, and -B3 are the most commonly reported genotypes (7). Although mostly asymptomatic or paucisymptomatic in adults, HAdVs can lead to acute and lethal infection in early childhood and in immunocompromised people (8). HAdV-associated complications are frequent in transplant recipients where infection can be acquired de novo, or through reactivation of a latent infection of the recipient or from the transplanted organ (9). Indeed, after primary infection of mucosal epithelial cells, HAdVs can establish persistent infection and reactivation in lymphoid cells (10–14). Long-term persistence can be achieved in lymphoid cells by maintenance of the viral genome as episomes (15). Adenoviruses are also used as vectors for gene therapy, cancer therapy, and vaccination (16). Remarkably, the first gene therapy attempted in human used an adenoviral vector to deliver the CFTR gene in bronchial cells of a cystic fibrosis patient (17). Even so, the use of adenoviral vectors for gene therapy has so far been limited by their high immunogenicity; this could prove to be an advantage regarding their vaccinal applications. Indeed, several high-profile vaccine candidates against severe acute respiratory syndrome coronavirus (SARS-CoV-2) infection are adenoviral vectors (18).
HAdV infections are controlled by innate and adaptive immune responses. Notably, HAdVs are well known to activate innate immunity sensors upon entry in the cell through their capsid proteins and genomic DNA, leading notably to the production of interferons (IFNs) and inflammatory cytokines (19, 20). Among innate immune effectors, the APOBEC3 enzymes (apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3 or A3s) are potent antiviral weapons. The human genome encodes seven A3 genes (namely, A3A, B, C, DE, F, G, and H), with several spliced transcripts and allelic variants for each. The A3s are IFN-inducible cytidine deaminases that convert cytosine to uracil on single-stranded DNA or RNA. Thus, they restrict viruses by introducing mutations in their genomes. Cytidine deamination occurs through a characteristic zinc-coordinating catalytic motif (His-X-Glu-X23–28-Pro-Cys-X2–4-Cys) (21). Some A3 enzymes encode one single deaminase domain (A3A, A3C, and A3H), whereas others encode two domains (A3B, A3DE, A3F, A3G). In those latter cases, only the C-terminal domain has a functional deaminase activity. The A3 proteins also exert an antiviral activity through deaminase-independent mechanisms by blocking the synthesis of the viral genome (reviewed in references 21–23). The seven A3 genes originate from gene duplications and rearrangements that have occurred during mammalian evolution (24, 25). The extension of the A3 repertoire allows some specialization of the different A3s. The A3 proteins notably differ by their subcellular localization allowing restriction of the viral replication at different levels of the viral cycle. The A3A member is cell-wide, A3B is strictly nuclear, A3C is mainly cytoplasmic, A3D, A3F, and A3G are strictly cytoplasmic, and A3H is present in the cytoplasm and the nucleolus (26). Thereby, A3s act against a large diversity of viruses. They notably restrict the reverse transcribing viruses HIV-1 (human immunodeficiency virus-1), HTLV-1 (human T-lymphotropic virus-1), and HBV (hepatitis B virus), the double-stranded DNA viruses EBV (Epstein-Barr virus), HSV-1 (herpes simplex virus-1), α-HPVs (alpha-human papillomaviruses), and BK polyomavirus, the single-stranded DNA viruses AAVs (adeno-associated virus) and TT virus (Teno Torque virus), and also the single-stranded RNA viruses HCV (hepatitis C virus), RSV (respiratory syncytial virus), the coronavirus HCoV-NL63, and mumps and measles viruses (27–39). Even though the A3 enzymes are valuable effectors protecting the host against viral infections, it has recently turned out that A3s can also accidently mutate the cellular genome. A3-induced mutations are found in many cancer types such as bladder, bone, cervical, breast, lung, and head and neck cancer (40–44). These mutations appear to be mostly introduced by the A3A and/or A3B members (45, 46).
A3s generally deaminate cytosine in a 5′-TC motif with the exception of A3G that favors editing of cytidine when preceded by another cytidine (i.e., in a 5′-CC motif) (40, 47). Thus, A3s leave on the genome of numerous viruses an evolutionary footprint characterized by the depletion of the 5′-TC motif (48–51). Importantly, we recently reported the presence of an A3 footprint on adenoviruses (51).
In this study, we set out to determine the relationship between HAdV infections and the regulation of A3 family members. We selected three HAdV genotypes from three different species, namely, A12, B3, and C2. The B3 and C2 strains were chosen based on their high incidence, and the A12 was chosen for its particular ability to induce tumors in animal models (52–54). The percentage of genetic identity (percentage of bases that are identical) is 56% between the A12 and B3 strains, 54% between A12 and C2, and 58% between B3 and C2 (values obtained using ClustalOmega alignment between the respective reference genomes). Using a relevant primary cell culture system, we demonstrated a strain-specific regulation of A3B. We also reported a link between the A3B protein upregulation and the intensity of the A3 evolutionary footprint: the HAdV-A12 genotype being particularly prone to induce A3B and harboring a genome with a strong depletion for A3-favored motifs.
RESULTS
Adenovirus A12, B3, and C2 strains upregulate A3B and downregulate A3C transcripts.
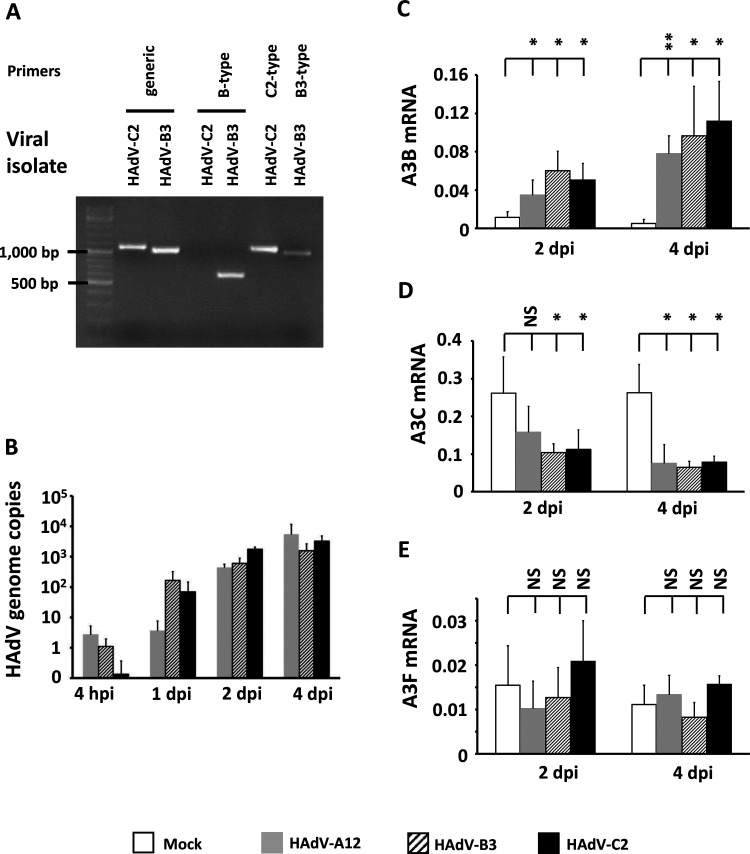
We sourced the HAdV-A12 strain from the ATCC. The HAdV-B3 and -C2 were two clinical isolates, and their identities were first verified by genotyping and sequencing. HAdV-C2 and -B3 viral stocks were subjected to PCR using pan-adenovirus primers, B-type-targeting primers, C2-targeting primers, and B3-targeting primers (Fig. 1A). Figure 1A, lanes 1 and 2 show isolates C2 and B3 PCR-amplified using pan-adenovirus primers (HXL1F/HXL1R, targeting the loop 1 region of the hexon gene). A 1,127-bp-long product was expected for a C2 strain and a 1,070-bp-long product was expected for a B3 strain. The observed PCR bands matched with the expected product sizes. Figure 1A, lanes 3 and 4 show isolates C2 and B3 PCR-amplified using B-type-targeting primers (BL/BR, targeting the loop 2 region of the hexon gene of B-type adenoviruses). As expected, no PCR band was observed from the C2 isolate and a PCR band of the correct size (expected size: 606 bp) was observed from the B3 isolate. Figure 1A, lane 5 shows isolate C2 PCR-amplified using C2-type-targeting primers (FiCL/FiCR2, targeting the fiber gene of C2 adenoviruses). A PCR band of the correct size (expected size: 1,121 bp) was observed. Figure 1A, lane 6 shows isolate B3 PCR-amplified using B3-type-targeting primers (FiBL/FiBR, targeting the fiber gene of B3 adenoviruses) and producing a product of the expected size (988 bp). The different PCR products (hexon and fiber sequences) were Sanger-sequenced and subjected to a BLAST search. The sequences subjected to a BLAST search confirmed the identities of the clinical isolates as HAdV-C2 and HAdV-B3.
FIG 1.
HAdV-A12, -B3, and -C2 infections upregulate A3B and downregulate A3C transcripts. (A) Confirmation of the C2 and B3 clinical isolates’ identities by genotyping. The genomic DNA of HAdV-C2 and HAdV-B3 viral stocks were subjected to PCR and migrated on agarose gel. Generic pan-adenovirus primers were used to amplify the loop 1 region of the hexon gene (lanes 1 and 2). B-type-targeting primers were used to amplify the loop 2 region of the hexon gene (lanes 3 and 4). C2-targeting primers and B3-targeting primers were used to amplify a region of the fiber gene (lanes 5 and 6). (B to E) HBEC3-KT cells were infected with HAdV-A12, -B3, -C2, or mock control. DNA was extracted at 4 h postinfection (hpi) and at 1, 2, and 4 days postinfection (dpi). (B) The abundances of the HAdV genome copies were measured by qPCR in three independent assays and expressed per nanogram of DNA. (C to E) RNA was extracted at 2 and 4 days postinfection (dpi). The abundances of the A3 transcripts were measured by RT-qPCR in three independent experiments. The histograms report A3B (C), A3C (D), and A3F (E) mean mRNA levels expressed relatively to the three housekeeping genes (HKGs) HPRT, TBP, and GAPDH. Error bars show standard deviations, and Student's t test was used to assess significance (NS, nonsignificant, *, P < 0.05, **, P < 0.01). A3A, A3DE, A3G, and A3H mRNAs were not detected.
Using the three different HAdV genotypes (A12, B3, and C2) and an established primary bronchial epithelial cell culture system, we determined whether adenovirus infections alter the expression of the A3 genes. Normal immortalized human bronchial epithelial cells (HBEC3-KT) were infected with HAdV-A12, B3, or C2 at a multiplicity of infection (MOI) of 1 infectious particle per cell. At 4 h and 1, 2, and 4 days postinfection, total DNA was extracted and the numbers of HAdV genome copies were quantified by quantitative PCR (qPCR). We observed a strong replication of the three adenoviruses at 2 days postinfection (dpi) and onwards (Fig. 1B). At 2 or 4 dpi, total RNA was extracted and the seven A3 mRNAs were quantified by reverse transcription-quantitative PCR (RT-qPCR). The abundances of the A3A, A3DE, A3G, and A3H transcripts were below the quantification levels of our assay. By comparison with that of uninfected control cells, A3B expression was upregulated at 2 dpi and even further at 4 dpi (Fig. 1C). Conversely, A3C expression was downregulated at 2 and 4 dpi (Fig. 1D), and no significant variation was observed for A3F (Fig. 1E).
Unlike the B3 strain, A12 and C2 adenoviruses upregulate A3B protein level.
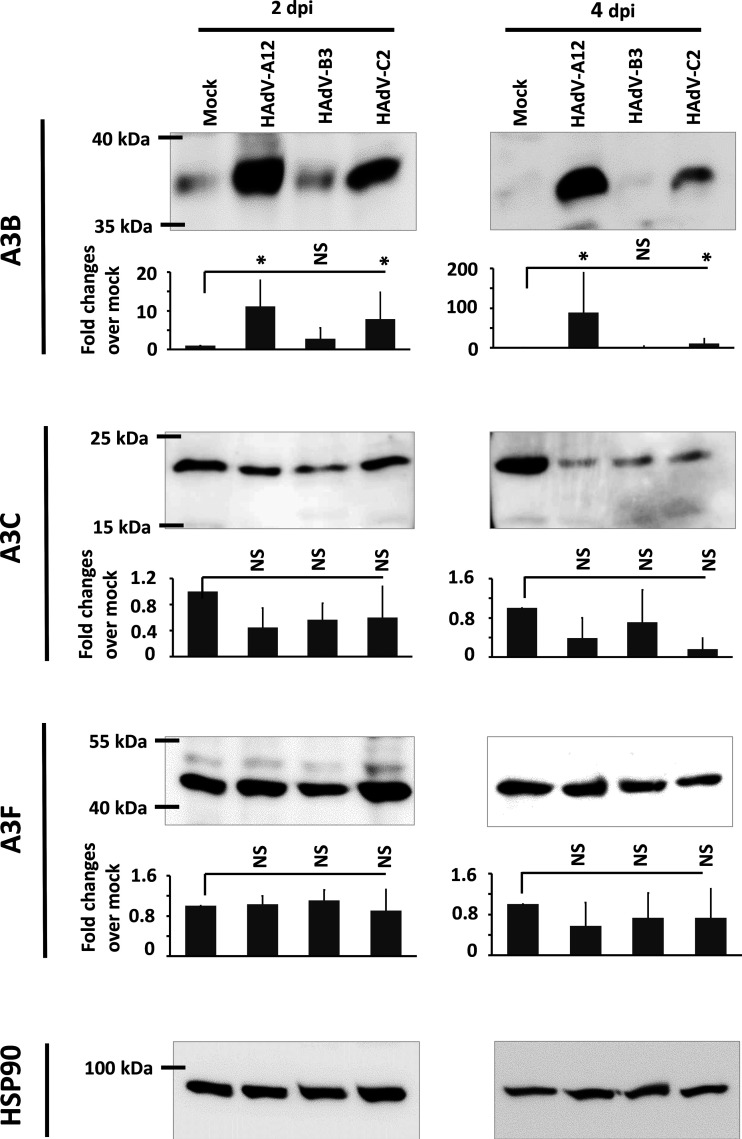
We then wondered whether A3B upregulation also occurs at the protein level. At 2 and 4 dpi, proteins were extracted and A3B, A3C, A3F, and HSP90 proteins were detected by immunoblotting (Fig. 2). HSP90 was used as loading control. Although the three adenovirus strains induced A3B mRNA at similar levels (Fig. 1C), this does not translate into the same amount of A3B protein. By comparison with that of uninfected control cells, A3B expression was significantly upregulated by the A12 and C2 strains at 2 and 4 dpi. Very surprisingly, no change of A3B protein level was observed in B3-infected cells. We observed a decrease, albeit not a significant one, of A3C protein level at 4 dpi and no change in A3F protein level.
FIG 2.
HAdV-A12 and -C2 infections upregulate A3B protein level, whereas HAdV-B3 infection does not change the abundance of the A3B protein. HBEC3-KT cells were infected with HAdV-A12, -B3, -C2, or mock control. Total protein extractions were done at 2 and 4 days postinfection (dpi). A3B, A3C, and A3F protein levels were assessed by Western blotting and quantified by densitometry. HSP90 was used as loading control. Error bars show standard deviations, and t test was used to assess significance (NS, nonsignificant, *, P < 0.05). The experiment was replicated at least 3 times and representative images are shown.
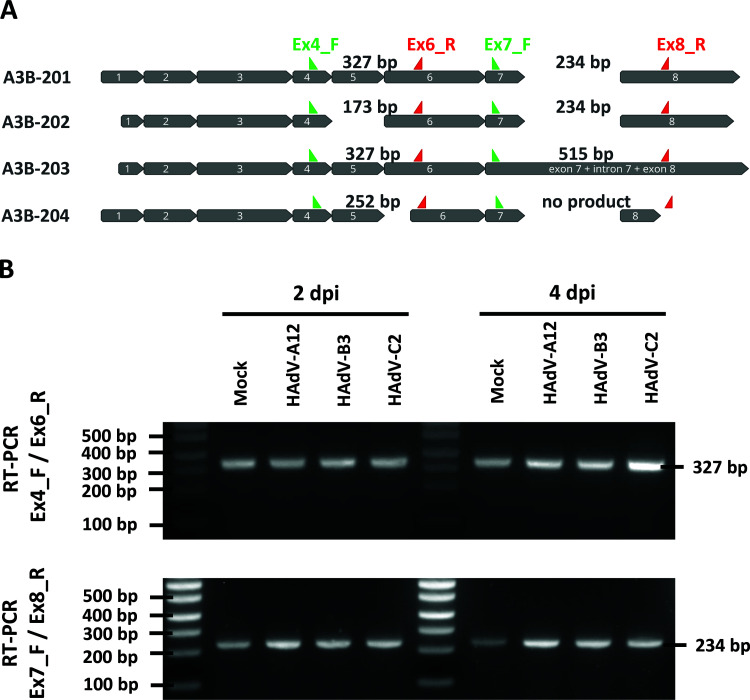
The A3B-201 isoform is the sole isoform expressed in both mock- and HAdV-infected cells.
As A3B protein upregulation appeared to be strain-specific, we wondered whether the different strains promote the upregulation of different A3B splice variants. Indeed, several splice variants can be produced from the A3B gene. The four A3B isoforms named according to EMBL-EBI Ensembl nomenclature (A3B-201 to 204) are represented in Fig. 3A. The A3B-201 isoform is the most prevalent transcript and is therefore considered to be the canonical sequence. The A3B-202 isoform does not retain exon 5, presumably leading to nonsense-mediated decay. The A3B-203 isoform retains intron 7 and encodes an A3B protein with functional deaminase activity (55, 56). The A3B-204 isoform encodes a protein lacking the zinc-coordinating residues of the C-terminal deaminase domain and is therefore most probably devoid of deaminase activity. To determine which are the A3B isoforms expressed in uninfected control and in HAdV-infected cells, 2 regions of the A3B mRNA were amplified by reverse transcription PCR (RT-PCR). The first region spans from exon 4 to exon 6; the second spans from exon 7 to exon 8. When amplifying the first region (using primers Ex4_F and Ex6_R), we detected only a 327-bp-long PCR product from both control and infected cells (Fig. 3B, upper gel). When using the second primer pair (Ex7_F and Ex8_R), we also detected a unique 234-bp-long PCR product from both control and infected cells (Fig. 3B, lower gel). Because the potential 515-bp-long product would have been at a disadvantage in competition with the shorter 234-bp amplicon, we cannot formally exclude the total absence of the A3B-203 isoform. Taken together, these results showed no change in A3B isoform upon HAdV infection. The A3B-201 isoform appeared to be the sole isoform expressed in both mock- and HAdV-infected cells.
FIG 3.
HAdV-A12, -B3, and -C2 infections do not modify A3B mRNA splicing. (A) The four A3B mRNA isoforms are depicted and named according to EMBL-EBI Ensembl nomenclature (A3B-201 to 204). Green arrows represent forward primers and red arrows represent reverse primers that were used to discriminate the A3B isoforms by RT-PCR. The sizes of the PCR products are indicated for each isoform. (B) HBEC3-KT cells were infected with HAdV-A12, -B3, -C2 or mock control. Two and four days postinfection (dpi), mRNA was extracted and A3B isoform discrimination was assessed by RT-PCR using the primer pairs Ex4_F/Ex6_R (top gel) and Ex7_F/Ex8_R (bottom gel). The experiment was replicated at least 3 times and representative images are shown.
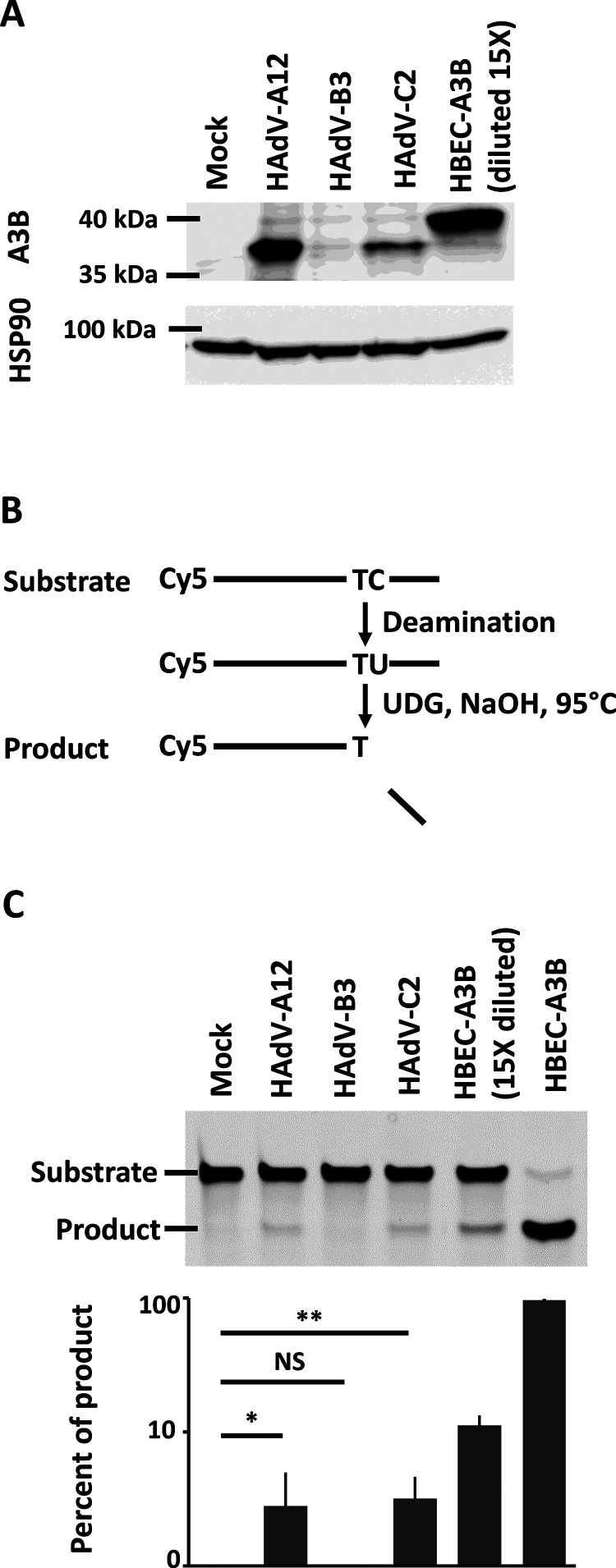
The intracellular deaminase activity is upregulated in HAdV-A12- and HAdV-C2-infected cells.
We wondered whether the A3B protein produced upon A12 and C2 infection is enzymatically active. Figure 4A illustrates the levels of A3B at 4 dpi in HBEC3-KT mock-, A12-, B3-, and C2-infected cells. As positive control, we established by lentiviral transduction HBEC3-KT cells constitutively expressing a hemagglutinin (HA)-tagged A3B protein (named HBEC-A3B thereafter). Figure 4A, lane 5 displays the level of A3B in a protein extract from HBEC-A3B diluted 15 times by proteins from uninfected wild-type HBEC cells.
FIG 4.
The deaminase activity is upregulated in HAdV-A12- and HAdV-C2-infected cells. (A) HBEC3-KT cells were infected with HAdV-A12, -B3, -C2, or mock control, and A3B expression was assessed by Western blotting at 4 dpi. A diluted protein extract from HBEC-A3B cells was used as positive control in lane 5. (B and C) Four days postinfection, total proteins were extracted in a nondenaturing buffer and mixed with the substrate of the deamination assay. Deaminase activity, if present in the cell lysate, will allow the conversion of substrates into shorter products. (C) No product was detected in cell lysates from mock- or B3-infected cells. Deaminase activity can be observed from A12- and C2-infected cells. HBEC-A3B cells were used as positive control and showed efficient conversion of the substrate into the product. Error bars show standard deviations, and t test was used to assess significance (NS, nonsignificant, *, P < 0.05, **, P < 0.01). The experiment was replicated at least three times and a representative image is shown.
We then performed a DNA deaminase activity assay on 4-dpi whole-cell protein extract supplemented with a single-stranded DNA probe containing a unique 5′-TC target motif (57). All A3 members, except A3G, prefer this dinucleotide as deamination target. Of note, A3G mRNA levels in mock- and HAdV-infected cells were very low and hardly detectable. C to U deamination, uracil excision, and alkaline treatment were combined to cleave the substrate probe into a shorter product (Fig. 4B). The positive controls, HBEC-A3B and HBEC-A3B protein extract diluted 15 times, converted respectively 95% and 11% of the substrate (Fig. 4C, last two wells). No product was observed in mock-infected or in HAdV-B3-infected cells. About 3% of the substrate has been converted to product in the protein extracts from HAdV-A12- and HAdV-C2-infected cells (Fig. 4C).
Knockdown of A3B suppresses deaminase activity during HAdV infection.
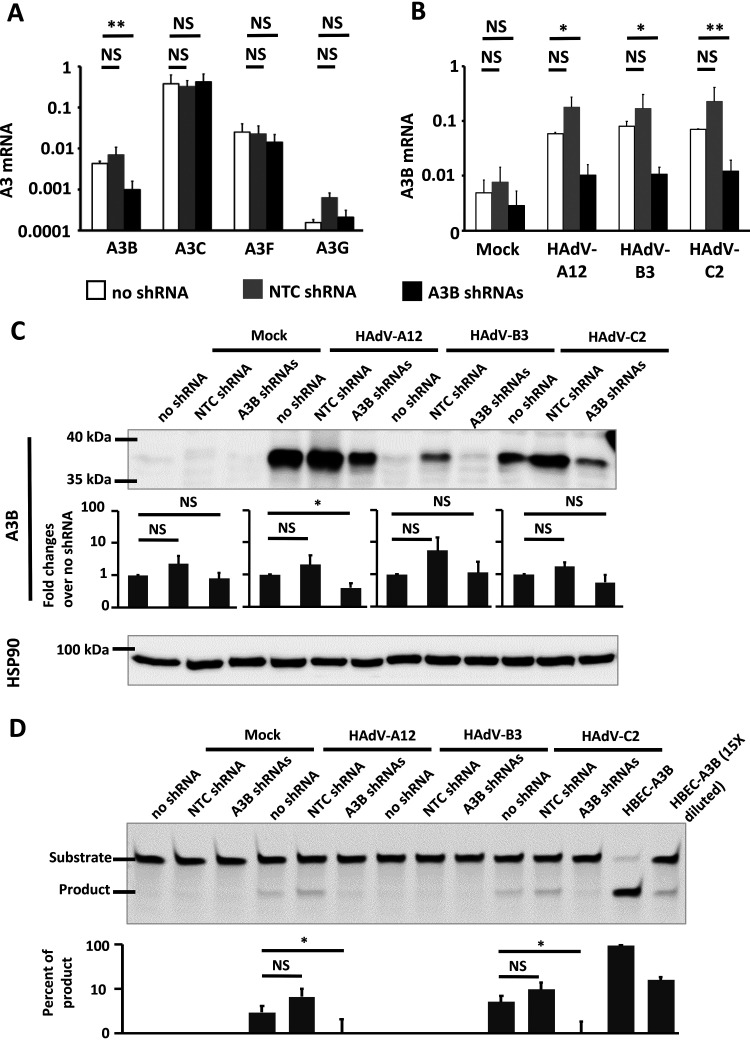
To assert that the A3B protein is indeed responsible for the measured deaminase activity, we tested whether specific knockdown of A3B could prevent deaminase activity in HAdV-infected cells. Knockdown of A3B was achieved by the stable expression of lentivirus-delivered short hairpin RNAs (shRNAs). Those cells are referred to as “A3B shRNAs” in Fig. 5. Wild-type HBEC3-KT cells (“no shRNA”) and HBEC3-KT cells transduced with nontarget shRNA vector (“NTC shRNA”) were used as controls. We first verified that knockdown of A3B was specific and did not affect other APOBEC3 transcripts. Figure 5A shows that A3B mRNA was significantly decreased by the A3B shRNAs compared to that of wild-type cells (no shRNA), whereas the other A3 mRNAs (A3C, A3F, and A3G) were not modified.
FIG 5.
Knockdown of A3B suppresses deaminase activity during HAdV infection. (A) A3B shRNAs target only A3B expression. mRNA from HBEC3-KT cells stably expressing A3B-targeting shRNAs (A3B shRNAs) or a nontargeting control shRNA (NTC shRNA) or wild-type HBEC3-KT cells (no shRNA) was extracted. The histogram reports A3B, A3C, A3F, and A3G mean mRNA levels expressed relatively to the three housekeeping genes (HKGs). (B to D) HBEC3-KT cells stably expressing A3B-targeting shRNAs or a nontargeting control shRNA or wild-type HBEC3-KT cells were infected with HAdV-A12, -B3, -C2, or mock control. Four days postinfection, A3B mRNA level (B), A3B protein abundance (C), and deaminase activity (D) were assessed. (B) The histogram reports A3B mean mRNA levels, quantified by RT-qPCR and expressed relatively to three housekeeping genes (HKGs). (C) A3B protein levels were assessed by Western blotting and quantified by densitometry. HSP90 was used as loading control. (D) Whole-cell protein extracts were subjected to deamination activity assay. HBEC-A3B was used as positive control (last two lanes). Error bars show standard deviations, and Student’s t test was used to assess significance (NS, nonsignificant, *, P < 0.05, **, P < 0.01.). The experiment was replicated at least 3 times and representative images are shown.
The three different HBEC3-KT cell lines were infected with HAdV-A12, -B3, or -C2 at an MOI of 1. At 4 dpi, the A3B transcriptional levels (Fig. 5B), protein levels (Fig. 5C), and intracellular deaminase activities (Fig. 5D) were assessed. We observed that the A3B-targeting shRNAs significantly decreased the amount of A3B transcript in HAdV-infected cells (Fig. 5B). The infection of HBEC3-KT no shRNA with HAdV-A12 and -C2 promoted A3B expression (Fig. 5C). Interestingly, infection of the HBEC3-KT NTC shRNA cells with the same viruses induced an even larger production of A3B protein (Fig. 5C). Indeed, double-stranded shRNAs can be sensed by RNA-activated protein kinase (PKR) and favor the expression of IFN-inducible genes (58). We speculate that HBEC3-KT NTC shRNA cells are primed by the overexpression of the NTC shRNAs and are therefore more readily prone to produce high levels of IFN-inducible proteins like the A3s. Importantly, the infection of HBEC3-KT A3B shRNAs cells with the HAdV-A12 and -C2 viruses does show a reduction of A3B protein expression (Fig. 5C) and a disappearance of any detectable deaminase activity (Fig. 5D). We concluded that the deaminase activity observed in HAdV-A12- or HAdV-C2-infected cells was down to the A3B protein.
Nuclear localization of A3B in HAdV-A12- and HAdV-C2-infected cells.
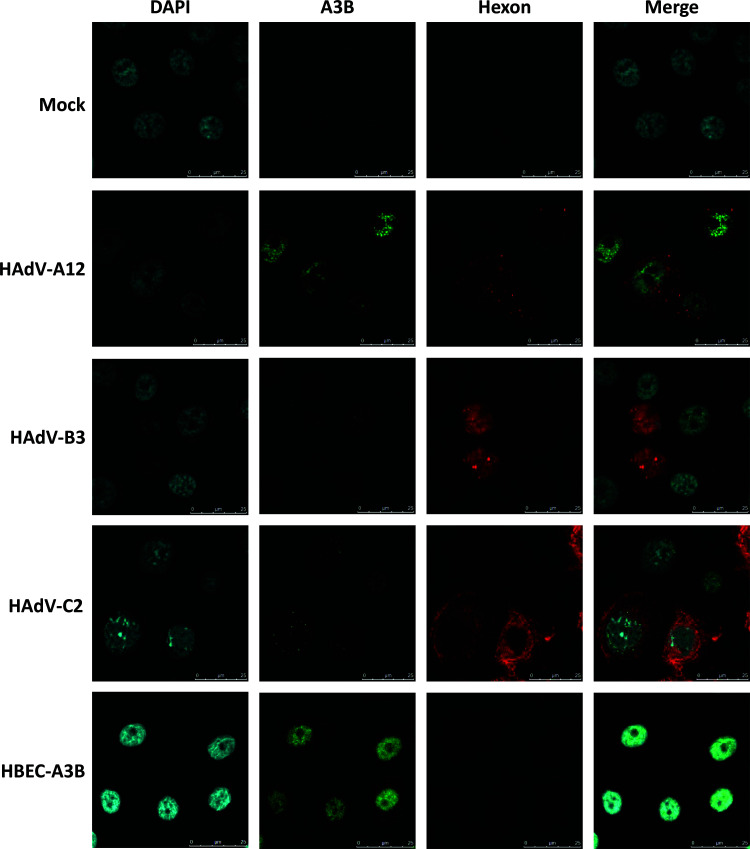
To assess the localization of A3B in HAdV-infected cells, we performed an immunofluorescence microscopy analysis on A3B and hexon proteins at 4 dpi (Fig. 6). The A3B upregulation was evident in HAdV-A12- and HAdV-C2-infected cells. The induced A3B protein localized in the nucleus. Only background levels of A3B were observed in mock- and B3-infected cells. HBEC-A3B cells were used as positive control and showed a strong A3B expression in the nucleus. These observations confirmed our results obtained by Western blotting and demonstrated that the induced A3B protein localizes in the nucleus of A12- and C2-infected cells.
FIG 6.
A3B protein upregulated in A12- and C2-infected cells is nuclear. HBEC3-KT cells were infected with HAdV-A12, -B3, -C2, or mock control. A3B (in green) and hexon protein (in red) expressions were assessed by fluorescence microscopy at 4 dpi. HBEC-A3B was used as positive control and displayed a strong A3B expression with a nuclear distribution. A3B protein levels in mock- and B3-infected cells were similar to background. Strong and moderate A3B signals, respectively, were observed in the nucleus of cells infected with the A12 and C2 strains.
HAdV-A12 genome bears the stronger A3 evolutionary footprint.
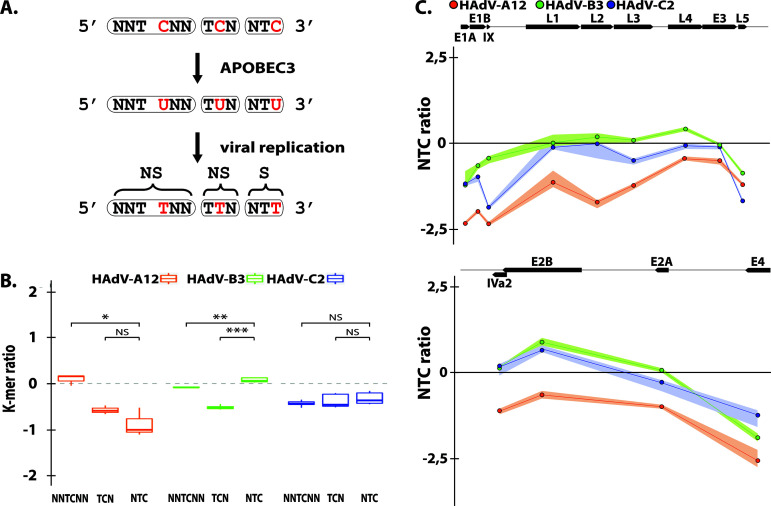
We already reported the presence of an A3 evolutionary footprint on human adenoviruses (51). In this study, A3B regulation appeared to be strain-specific with no change upon HAdV-B3 infection but increased upon infection with the A12 or C2 strains. Therefore, we specifically looked for the A3 footprint on those three strains wondering whether it could correlate with our in vitro observations. Because A3 proteins favor deamination of C when preceded by a T (so called 5′-TC motif), the A3s will turn 5′-TC motifs into 5′-TU dinucleotides in the genome of exposed viruses. Depending on the position of the mutated C within the codon, the mutation can be synonymous or nonsynonymous. When the mutated C is at the third position of the codon, deamination of the C will turn the NTC codon into an NTT codon. This mutation will always be synonymous (symbolized by an “S” in Fig. 7A). When A3-related deamination intervenes on a C located at the first position of a codon, the NNTCNN motif will be converted into an NNTUNN motif and will produce a nonsynonymous mutation (symbolized by an “NS” in Fig. 7A). Similarly, deamination of a C located at the second position of the codon will convert a TCN codon into a TUN codon and most likely introduce a nonsynonymous mutation (Fig. 7A). Because a synonymous mutation will more likely be conserved than a nonsynonymous mutation, the A3-driven natural selection should more intensively deplete NTC codons than TCN or NNTCNN motifs (as in those cases the C to U mutation will affect the encoded amino acid). Thus, we defined the A3 footprint as an under-representation of NTC codons because A3s favor 5′-TC motifs and because a C to U mutation in the third position of a codon is likely to be retained (Fig. 7A).
FIG 7.
HAdV-A12 genome bears a strong A3 evolutionary footprint. (A) A3s induce C to U deamination preferentially in a 5′-TC dinucleotide context. After viral replication the mutation is conserved in the viral genome as a C to T transition (in red). Depending on the position of the TC motif in codon, the mutation is synonymous (S) or nonsynonymous (NS). Since synonymous mutations are more likely to be conserved, the A3 footprint is defined as the depletion of the NTC codons. (B) The observed/expected ratios of TC dinucleotide at various codon positions (i.e., NNTCNN, TCN, NTC) were calculated for the HAdV-A12, -B3, and -C2 full-length genomes. Median and quartile are depicted by a boxplot. P values were calculated by Student’s unpaired, two-tailed t test (NS, not significant, *, P < 0.05, **, P < 0.01, ***, P < 0.001. (C) NTC observed/expected ratios were calculated for the different genes of HAdV-A12, -B3, and -C2. Genes’ location, orientation, and length are represented by black arrows.
After analysis of HAdV-A12 genomic sequences, we observed no depletion of the NNTCNN motif, a moderate depletion of the TCN codons, and a strong depletion of NTC codons (Fig. 7B). These observations point toward a genome-wide A3 footprint on the A12 strain. On the contrary, no general NTC depletion was observed for the B3 and C2 strains (Fig. 7B). By looking at the gene level, we observed for the three strains a stronger NTC depletion for the E1, L5, and E4 genes compared to that of the other genes (Fig. 7C). The A3 footprint concentrated on the extremities of the viral genome. The intensity of the A3 footprint on the different strains appeared to correlate with their potency to promote A3B expression and activity.
DISCUSSION
In this study, we first reported similar upregulations of the A3B transcript in HAdV-infected cells, independent of the strain used. Various stimuli have been shown to trigger transcriptional upregulation of A3B, including IFNs, agonists of the lymphotoxin beta receptor, DNA damaging agents, the PKC/NF-κB-activator PMA (phorbol 12-myristate 13-acetate), and viral infections (37, 57, 59–62). Thus, A3B transcription can be induced by the NF-κB pathway whereas its repression can be triggered by p53 activation and mediated by the repressive complexes E2F4/DREAM (DP, RB-like, E2F4, and MuvB) and E2F6/PRC1.6 (polycomb repressing complex) (63–66). A3B transcriptional induction in HAdV-infected cells could be the consequence of the intracellular innate immune response. Indeed, adenoviruses can be sensed by numerous innate immune sensors, leading to the transcription of IFNs, ISGs (IFN-stimulated genes), and NF-κB-dependent genes (19, 20). A3B transcription has also been shown to be induced by the viral proteins E6 and E7 from high-risk HPVs (human papillomaviruses) and by the large TAg (T antigen) from the BK polyomavirus (37, 57, 67–69). The mechanisms of action are not fully elucidated but appear to involve the dissociation of the repressive E2F complexes (66, 70–74). Interestingly, the adenovirus E1A protein directly interacts with the DP-1 transcription factor (E2F dimerization partner 1), a protein that binds E2F4 or E2F6 in the DREAM or PRC1.6 complexes, respectively (75). Moreover, E1A can be recruited on E2F promoters when these promoters are bound by p130 (retinoblastoma-like 2) and E2F4, thereby modifying the chromatin structure to promote gene expression (76). Finally, E1B-55K and E4orf6 are also well known to promote p53 degradation (77, 78). Overall, it would be interesting to test whether the E1A, E1B, and/or E4orf6 proteins can favor A3B transcription though p53 degradation and/or destabilization of E2F repressive complexes.
Although infection by the three strains induces a similar upregulation at the transcriptional level, the regulation at protein level is highly variable. Indeed, A3B protein is upregulated upon HAdV-A12 and -C2 infections whereas A3B protein amount did not increase in HAdV-B3-infected cells. We showed that the A3B-201 mRNA isoform is the sole transcript variant expressed, and therefore alternative splicing cannot explain the differences of A3B protein amount. The regulation of the A3 proteins by viruses is a complex matter; some viruses have evolved mechanisms to counteract A3s, whereas others promote their expression and stability.
An active degradation of the A3B protein in HAdV-B3-infected cells could explain the discrepancy between mRNA and protein levels. For example, HIV-1 (human immunodeficiency virus 1) evades A3G restriction thanks to its Vif protein. Vif binds A3G and recruits A3G to a Cullin5-based E3 ubiquitin ligase complex leading to A3G degradation (79). Interestingly, the HAdV-C5 proteins E1B-55K and E4orf6 promote the degradation of several cellular proteins (notably p53) through the recruiting of the same Cullin5, Elongins B and C, and Rbx1 complex (77, 78). One might wonder whether the A3B protein can be one of the targets of the HAdV-B3 E1B-55K and E4orf6 proteins. The very low level of A3B protein despite an elevated transcript observed in HAdV-B3-infected cells could also be the result of translational inhibition. For instance, the A3B protein level can be downregulated posttranscriptionally by the binding of mir-138-5p to the 3′ untranslated region (3′UTR) of the A3B transcript (S. Faure-Dupuy, T. Riedl, and M. Heikenwälder, submitted for publication). On the contrary to those infected with the B3 strain, the HAdV-A12- and HAdV-C2-infected cells displayed an enzymatically active A3B protein. If the strong expression of the A3B protein can simply be due to the increase of its transcript, we should nevertheless mention that some viral protein can promote A3 stability. Indeed, E7 from HPV16 has been shown to stabilize A3A protein by inhibiting Cul2-dependent protein degradation (69). Because the E1B-55k and E4orf6 from the HAdV-A12 use the Cul2- rather than the Cul5-based ubiquitin ligase complex (80), one might wonder whether a similar stabilization of the A3B protein can be operating in HAdV-A12-infected cells.
Whatever causes A3B induction, it remains to be tested how much the A3B protein affects adenoviral replication and evolution. A3-induced mutations in HPV16 genome on one hand contribute to viral clearance and on the other hand also drive HPV evolution (67, 81). Upregulation of A3B in HBV-infected hepatocytes inhibits HBV replication (59). At the same time, A3-related mutations on the negative strand of the C and pre-C region of the HBV genome can positively participate in immune escape (51, 82). Similarly, A3-related mutations in the major capsid gene of BK polyomavirus confer resistance to neutralizing antibodies (38). These observations suggest that A3s can play an ambivalent role during viral infection, effectively restricting viral replication but also favoring viral diversification and immune escape. The detection of a functional A3B protein in HAdV-A12- and HAdV-C2-infected cells, its localization in the nucleus, and the presence of an A3 evolutionary footprint on the adenoviruses genome strongly suggest that A3B interferes with the adenoviral replication cycle. Hence, we are currently investigating how much A3B can influence adenoviral replication kinetics and introduce mutations in the newly replicated genomes. We also wonder whether A3B editing could promote DNA breaks in the viral genome as it does in cancer cells and subsequently allow homologous recombination between strains. For example, the majority of HAdV types belong to species D and homologous recombination between capsid genes is the main factor contributing to their diversity (83, 84).
Although the mechanisms of A3B induction must be further studied in the settings of HAdV natural infections, one might wonder whether such dysregulation can also occur upon adenoviral vectors production and/or infection, particularly if A3B expression is driven by viral genes. For example, the ChAdOx1 vector (from which a prominent SARS-CoV-2 vaccine is based) is a modified chimpanzee E-type adenovirus that has been deleted for E1 and E3 and with which the E4 gene has been recombined to carry the E4Orf4 and E4Orf6/7 from HAdV-C5 (85). As replication-defective adenoviral vectors are not cytolytic and have been shown to persist at low levels in a transcriptionally active form for an extended period of time (86), it would be worth testing whether the remaining viral genes can dysregulate A3 proteins.
To summarize, we report for the first time the involvement of the A3 innate immune effectors during HAdV infections. We demonstrate that three HAdV strains, namely, A12, B3, and C2, differently regulate the A3B member. The three HAdV strains induce a similar A3B upregulation at the transcriptional level, although only the infection by the HAdV-A12 and -C2 strains promotes the production of an enzymatically active A3B protein. Bioinformatics analysis of the viral genomic sequences showed that adenoviruses are under A3 selective pressure, with a stronger A3 evolutionary footprint measured on the HAdV-A12 genome. Further experiments will be required to test whether innate immune sensors and/or viral protein promote the transcription of the A3B gene and to assess the consequences of A3B activity on long-term infected cells and on viral diversification.
MATERIALS AND METHODS
Cell lines.
HBEC3-KT cells are normal human bronchial epithelial cells immortalized with the human TERT and mouse Cdk4 genes. HBEC3-KT cells were kindly provided by Jerry W. Shay (UT Southwestern, Dallas, TX, USA) (87). HBEC3-KT cells were cultured in keratinocyte serum free medium (SFM) supplemented with bovine pituitary extract (50 μg/ml) and human recombinant epidermal growth factor (5 ng/ml) (Gibco) on gelatin-coated flasks. HBEC3-KT knockdown cells for A3B were established by transduction with lentiviral vectors encoding A3B-targeting shRNAs (pSicoR-MS2 plasmids, kindly provided by Warner C. Greene, University of California, San Francisco, CA, USA) (88). Briefly, HBEC3-KT cells were transduced with nontargeting shRNA expressing lentivirus (NTC shRNA) or with three A3B-targeting shRNA expressing lentiviruses (A3B shRNAs) as described in reference 88. HBEC3-KT cells constitutively expressing A3B (HBEC-A3B) were established by transduction with a lentiviral vector encoding an A3B HA-tagged protein (pLenti4-A3B kindly provided by Reuben Harris) (89). Of note, the puromycin markers originally present in the pSicoR-M2 plasmid and in the pLenti4-A3B had been replaced by a blasticidin marker prior to HBEC3-KT transduction. Blasticidin was used to select for a polyclonal population of effectively transduced cells. The human lung carcinoma cell line A549 (American Type Culture Collection) and PKR-deficient A549 cell line were cultured in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal calf serum (Gibco) and 10 mM l-glutamine. PKR-deficient A549 cells were kindly provided by Cheng Huang and Slobodan Paessler (UTMB, Galveston, TX, USA) (90). All cell lines were incubated at 37°C and 5% CO2.
Virus preparation and infection procedure.
Handling of human adenoviruses was done in a biosafety level 2 laboratory. Adenovirus A12 (HAdV-12, ATCC VR-863) was purchased at the American Type Culture Collection. Adenoviruses B3 and C2 were kindly provided by Lieve Naesens (KULeuven, Leuven, Belgium) and strain identity was verified by genotyping and sequencing using primers described in reference 91. HAdV-B3 and -C2 viral stocks were produced in A549 cells and HAdV-A12 viral stock was produced in PKR-deficient A549 cells. During viral stock production, A549 cells and PKR-deficient A549 cells were cultured in Opti-MEM I reduced-serum medium without phenol red (ThermoFisher Scientific). When 80% of the cells showed cytopathic effects, cells were scraped, collected with the culture medium, and centrifuged at 3,500 × g for 15 min. The supernatant was collected and set aside. The cell pellet was resuspended in 2 ml of culture medium, submitted to 3 freeze/thaw cycles, and centrifuged at 3,500 × g for 15 min. The supernatant was collected, pooled with the previous supernatant, and treated with the endonuclease Benzonase (1 unit/ml) (Sigma-Aldrich) at 37°C for 30 min to degrade the unpackaged nucleic acids. Supernatant was then filtered on a 0.22-μm Steriflip filter (Merck Millipore) to remove residual cell debris. Elute was collected and further filtered through Amicon Ultra-15 centrifugal filter unit 100 kDa (Merck Millipore). Virions were retained on the filter, resuspended in phosphate-buffered saline (PBS), and conserved at −80°C. Titration of the viral stocks was done by 50% tissue culture infective dose (TCID50) assay and by qPCR quantification of genome copies (details below). HBEC3-KT cells were infected with the HAdV-A12, -B3, or -C2 strain at a multiplicity of infection (MOI) of 1 in keratinocyte SFM supplemented with bovine pituitary extract (50 μg/ml) and human recombinant epidermal growth factor (5 ng/ml) (Gibco) on gelatin-coated flasks.
Viral copies quantification by qPCR.
DNA was extracted from infected cells and subjected to PCR. A fragment of the penton gene was amplified using FastStart Universal SYBR green master mix (Roche) and Eco real-time PCR system (Illumina). HAdV-A12 genomic DNA was amplified using the primers A12_penton_F (GCCCTTACAGATCACGGGAC) and A12_penton_R (CAGTGCTTTGTAAACGTAGG). HAdV-B3 and -C2 genomic DNA were amplified using the primers B3+C2_penton_F (GCTCTCACAGATCACGGGAC) and B3+C2_penton_R (CAGGGCCTTGTAAACGTAGG). Serial dilutions of plasmids containing the target sequences were used as calibration curves. Titer was expressed as number of viral copies per nanogram of DNA.
A3 mRNA quantification by RT-qPCR.
Two or four days postinfection, cells were collected and washed in PBS. RNA was extracted using ReliaPrep miRNA cell and tissue miniprep system (Promega) following the manufacturer’s instructions. cDNA was obtained by reverse transcription of 1 μg of mRNA using iScript cDNA synthesis kit (Bio-Rad) following the manufacturer’s instructions. The transcripts of the seven A3 genes and of three housekeeping genes (HPRT, GAPDH, and TBP) were quantified using FastStart Universal SYBR green master (Roche) and Eco real-time PCR system (Illumina). The primers used to titrate the A3s were designed by Refsland et al. (92). The A3 mRNA levels were expressed relative to the geometric mean of the abundance of the HPRT, GAPDH, and TBP transcripts.
A3 protein detection by immunoblotting.
Four days postinfection, cells were collected and washed in PBS. Cells were resuspended in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl [pH 8.0], 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton X-100) supplemented by 1 mM phenylmethylsulfonyl fluoride (PMSF) and cOmplete protease inhibitor cocktail (Roche). Cells were then incubated for 30 min on ice and sonicated 5 cycles 30 sec on/30 sec off using a Bioruptor Pico device (Diagenode) at 4°C. Cell lysates were spun down at 14,000 rpm for 15 min to remove cell debris. Proteins were quantified using Pierce BCA protein assay kit (ThermoFisher Scientific). Twenty-five μg of proteins was loaded on a 10% SDS-PAGE gel and transferred actively to polyvinylidene difluoride (PVDF) membrane (GE Healthcare Life Sciences). Membrane was blocked in TBS (Tris-buffered saline) supplemented with 0.1% Tween 20 and 5% bovine serum albumin (BSA). A3B protein was detected with anti-human APOBEC3B monoclonal antibody (5210-87-13, catalog number 12397, from Reuben Harris and obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH [63]) used at a dilution of 1:1,000 in TBS supplemented with 0.1% Tween 20 and 5% BSA. A3C protein was detected with anti-human APOBEC3C monoclonal antibody (Proteintech) used at a dilution of 1:1,000 in TBS with 0.1% Tween 20 and 4% milk. A3F protein was detected with anti-human APOBEC3F monoclonal antibody (5206-235-07, catalog number 12399, from Reuben Harris and obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH) used at a dilution of 1:1,000 in TBS with 0.1% Tween 20 and 5% BSA. Hsp90 was used as loading control and was detected with an anti-HSP90AB1 antibody (Sigma-Aldrich) at a dilution of 1:1,000 in TBS supplemented with 0.1% Tween 20 and 5% BSA. A horseradish peroxidase (HRP)-coupled anti-rabbit IgG secondary antibody (Dako) was used at a dilution of 1:2,000 in TBS with 0.1% Tween 20 and 5% BSA or 4% milk. Membranes were incubated with the chemiluminescent SuperSignal West Femto maximun sensitivity substrate (ThermoFisher Scientific) and chemiluminescence was read using an ImageQuant LAS4000 (GE Healthcare Life Sciences).
Deamination assay.
Four days postinfection, cells were collected and washed in PBS. Cells were resuspended in HED buffer (20 mM HEPES [pH 7.4], 5 mM EDTA, 1 mM dithiothreitol [DTT], 10% glycerol) supplemented with cOmplete protease inhibitor cocktail (Roche). Cells were then submitted to one freeze/thaw cycle and sonicated for 15 cycles of 30 sec on/30 sec off using Bioruptor Pico device (Diagenode) at 4°C. Cell lysates were spun down at 14,000 rpm for 15 min to remove cell debris. Proteins were quantified using Pierce BCA protein assay kit (ThermoFisher Scientific). One hundred μg of proteins was incubated overnight at 37°C with 1 pmol of a fluorescent oligonucleotide substrate (5′-ATTATTATTATTCAAATGGATTTATTTATTTATTTATTTATTT-Cy5-3′), 1 mM ZnCl2, 0.025 units uracil DNA glycosylase (NEB), 2 μl 10× uracil DNA glycosylase (UDG) buffer (NEB), and 100 μg/ml RNase A (ThermoFisher Scientific). Reaction mixture was treated with 50 mM NaOH and heated to 95°C for 10 min to cleave DNA probes at the abasic site. Reaction mixture was then neutralized with 50 mM HCl and mixed with 1.25× formamide buffer. Substrates (43 bases long) from products (30 bases long) were separated on a 15% Tris-borate-EDTA (TBE)-urea gel. The Cy5-labeled substrates and deamination products were detected using ImageQuant LAS4000 mini (GE Healthcare Life Sciences).
Immunofluorescence microscopy.
Four days postinfection, cells were fixed and permeabilized using the eBiosciences Foxp3/transcription factor staining buffer set (Invitrogen) according to the manufacturer’s instructions. Cells were then incubated overnight at 4°C with anti-human APOBEC3B monoclonal antibody (5210-87-13, catalog number 12397) used at a dilution of 1:250 or an anti-hexon antibody (MAB 8051, Merck Millipore) used at a dilution of 1:200 in permeabilization buffer supplemented with 2% goat serum (Gibco). After 3 washes in permeabilization buffer supplemented with goat serum, cells were incubated 1 h with an anti-rabbit Alexa Fluor 488 and an anti-mouse Alexa Fluor 633 (Invitrogen). Cells were washed 3 times with permeabilization buffer supplemented with 2% goat serum. Finally, slides were mounted in Mowiol (Sigma-Aldrich) and imaged using Leica SP5 microscope.
A3 evolutionary footprint by bioinformatic analysis.
Complete HAdV genomes were downloaded from the NCBI Nucleotides database. GenBank accession IDs used are the following: KX868289.2, AY599834.1, AC_000007.1, X73487.1, AY599836.1, J01917.1, NC_001460.1, DQ086466.1, MF044052.1, KX384958.1, KX384959.1, JN860679.1, and MN513342.1. Calculation of the k-mer representation was done as described in Poulain et al. (51). Briefly, a k-mer includes the ATC, CTC, GTC, and TTC sequences. In addition, as we limit our analysis to coding sequences, we force our k-mers to be in the reading frame and therefore to correspond to codons. For example, the NTC k-mer actually includes the ATC, CTC, GTC, and TTC codons. Following the same logic, the NNTCNN k-mer comprises the 256 pairs of codons that have a T at the end of the first codon and a C to start the second codon. Each coding sequence has been randomly shuffled 1,000 times, retaining only the nucleotide composition. The expected count of a given k-mer is calculated as the average of the occurrences of this k-mer over the 1,000 iterations. The k-mer ratio is given as the log2 ratio of the observed occurrence of this k-mer to the expected occurrence. To calculate the ratio of a given k-mer for an entire viral genome, a “synthetic coding genome” was generated by concatenating the different coding sequences. The synthetic coding sequence is then randomly shuffled 1,000 times and k-mer ratio calculated as above. A k-mer ratio less than 0 indicates k-mer under-representation, and a k-mer ratio equal to zero means that no representation bias is observed.
Statistical analysis.
Student’s t test has been used where appropriate. The results were considered statistically significant at a P value of <0.05.
ACKNOWLEDGMENTS
We thank Jerry W. Shay for the HBEC3-KT cell line, Cheng Huang and Slobodan Paessler for the PKR-deficient A549 cell line, Lieve Naesens for the HAdV-B3 and -C2 isolates, Warner C. Greene for the pSicoR-MS2 plasmids, and Reuben Harris for the pLenti4_A3B expression plasmid. The following reagent was obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: anti-human APOBEC3B monoclonal (5210-87-13) from Reuben Harris (catalog number 12397) and anti-human APOBEC3F monoclonal (5206-235-07) (catalog number 12399) from Reuben Harris.
Author contributions were as follows. N.L.: conceptualization, formal analysis, investigation, visualization, and writing (original draft preparation and review and editing). F.P.: conceptualization, investigation, and visualization. K.W.: investigation. S.M.: investigation. Z.B.: investigation. N.A.G.: conceptualization, funding acquisition, supervision, writing (review and editing).
The authors declare that no competing interests exist.
This study was supported by FRS-FNRS grant CDR number 31270116 and by the University of Namur. Noémie Lejeune is a Ph.D. fellow supported by FRIA grant number 31454280. Florian Poulain is a Ph.D. fellow supported by Télévie grant PDR-TLV number 34972507.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Noémie Lejeune, Email: lejeune.noemie@gmail.com.
Nicolas A. Gillet, Email: nicolas.gillet@unamur.be.
Lawrence Banks, International Centre for Genetic Engineering and Biotechnology.
REFERENCES
- 1.Seto D, Chodosh J, Brister JR, Jones MS, Members of the Adenovirus Research Community . 2011. Using the whole-genome sequence to characterize and name human adenoviruses. J Virol 85:5701–5702. 10.1128/JVI.00354-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rowe WP, Huebner RJ, Gilmore LK, Parrott RH, Ward TG. 1953. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc Soc Exp Biol Med 84:570–573. 10.3181/00379727-84-20714. [DOI] [PubMed] [Google Scholar]
- 3.Huebner RJ, Rowe WP, Ward TG, Parrott RH, Bell JA. 1954. Adenoidal-pharyngeal-conjunctival agents. N Engl J Med 251:1077–1086. 10.1056/NEJM195412302512701. [DOI] [PubMed] [Google Scholar]
- 4.Enders JF, Bell JA, Dingle JH, Francis T, Hilleman MR, Huebner RJ, Payne AM-M. 1956. Adenoviruses: group name proposed for new respiratory-tract viruses. Science 124:119–120. 10.1126/science.124.3212.119. [DOI] [PubMed] [Google Scholar]
- 5.Radke JR, Cook JL. 2018. Human adenovirus infections: update and consideration of mechanisms of viral persistence. Curr Opin Infect Dis 31:251–256. 10.1097/QCO.0000000000000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crenshaw BJ, Jones LB, Bell CR, Kumar S, Matthews QL. 2019. Perspective on adenoviruses: epidemiology, pathogenicity, and gene therapy. Biomedicines 7:61. 10.3390/biomedicines7030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binder AM. 2017. Human adenovirus surveillance — United States, 2003–2016. MMWR Morb Mortal Wkly Rep 66. 10.15585/mmwr.mm6639a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lion T. 2014. Adenovirus infections in immunocompetent and immunocompromised patients. Clin Microbiol Rev 27:441–462. 10.1128/CMR.00116-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Florescu DF, Hoffman JA, The AST Infectious Diseases Community of Practice . 2013. Adenovirus in Solid Organ Transplantation. Am J Transplant 13:206–211. 10.1111/ajt.12112. [DOI] [PubMed] [Google Scholar]
- 10.Van Der Veen J, Lambriex M. 1973. Relationship of adenovirus to lymphocytes in naturally infected human tonsils and adenoids. Infect Immun 7:604–609. 10.1128/IAI.7.4.604-609.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garnett CT, Erdman D, Xu W, Gooding LR. 2002. Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J Virol 76:10608–10616. 10.1128/jvi.76.21.10608-10616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garnett CT, Talekar G, Mahr JA, Huang W, Zhang Y, Ornelles DA, Gooding LR. 2009. Latent species C adenoviruses in human tonsil tissues. J Virol 83:2417–2428. 10.1128/JVI.02392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy S, Calcedo R, Medina-Jaszek A, Keough M, Peng H, Wilson JM. 2011. Adenoviruses in lymphocytes of the human gastro-intestinal tract. PLoS One 6:e24859. 10.1371/journal.pone.0024859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kosulin K, Geiger E, Vécsei A, Huber W-D, Rauch M, Brenner E, Wrba F, Hammer K, Innerhofer A, Pötschger U, Lawitschka A, Matthes-Leodolter S, Fritsch G, Lion T. 2016. Persistence and reactivation of human adenoviruses in the gastrointestinal tract. Clin Microbiol Infect 22:381.e1–381.e8. 10.1016/j.cmi.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Huang W, Ornelles DA, Gooding LR. 2010. Modeling adenovirus latency in human lymphocyte cell lines. J Virol 84:8799–8810. 10.1128/JVI.00562-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wold WSM, Toth K. 2014. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr Gene Ther 13:421–433. 10.2174/1566523213666131125095046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zabner J, Couture LA, Gregory RJ, Graham SM, Smith AE, Welsh MJ. 1993. Adenovirus-mediated gene transfer transiently corrects the chloride transport defect in nasal epithelia of patients with cystic fibrosis. Cell 75:207–216. 10.1016/0092-8674(93)80063-k. [DOI] [PubMed] [Google Scholar]
- 18.Krammer F. 2020. SARS-CoV-2 vaccines in development. Nature 586:516–527. 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 19.Hendrickx R, Stichling N, Koelen J, Kuryk L, Lipiec A, Greber UF. 2014. Innate immunity to adenovirus. Hum Gene Ther 25:265–284. 10.1089/hum.2014.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greber UF, Flatt JW. 2019. Adenovirus entry: from infection to immunity. Annu Rev Virol 6:177–197. 10.1146/annurev-virology-092818-015550. [DOI] [PubMed] [Google Scholar]
- 21.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285–296. 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 22.Harris RS, Dudley JP. 2015. APOBECs and virus restriction. Virology 479–480:131–145. 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willems L, Gillet NA. 2015. APOBEC3 interference during replication of viral genomes. Viruses 7:2999–3018. 10.3390/v7062757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Münk C, Willemsen A, Bravo IG. 2012. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol Biol 12:71. 10.1186/1471-2148-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano Y, Aso H, Soper A, Yamada E, Moriwaki M, Juarez-Fernandez G, Koyanagi Y, Sato K. 2017. A conflict of interest: the evolutionary arms race between mammalian APOBEC3 and lentiviral Vif. Retrovirology 14:31. 10.1186/s12977-017-0355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lackey L, Law EK, Brown WL, Harris RS. 2013. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle 12:762–772. 10.4161/cc.23713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650. 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 28.Turelli P, Mangeat B, Jost S, Vianin S, Trono D. 2004. Inhibition of Hepatitis B virus replication by APOBEC3G. Science 303:1829–1829. 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 29.Sasada A, Takaori-Kondo A, Shirakawa K, Kobayashi M, Abudu A, Hishizawa M, Imada K, Tanaka Y, Uchiyama T. 2005. APOBEC3G targets human T-cell leukemia virus type 1. Retrovirology 2:32. 10.1186/1742-4690-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol 16:480–485. 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 31.Vartanian J-P, Guétard D, Henry M, Wain-Hobson S. 2008. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 320:230–233. 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- 32.Narvaiza I, Linfesty DC, Greener BN, Hakata Y, Pintel DJ, Logue E, Landau NR, Weitzman MD. 2009. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog 5:e1000439. 10.1371/journal.ppat.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuge M, Noguchi C, Akiyama R, Matsushita M, Kunihiro K, Tanaka S, Abe H, Mitsui F, Kitamura S, Hatakeyama T, Kimura T, Miki D, Hiraga N, Imamura M, Takahashi S, Hayses CN, Chayama K. 2010. G to A hypermutation of TT virus. Virus Res 149:211–216. 10.1016/j.virusres.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 34.Suspène R, Aynaud M-M, Koch S, Pasdeloup D, Labetoulle M, Gaertner B, Vartanian J-P, Meyerhans A, Wain-Hobson S. 2011. Genetic editing of herpes simplex virus 1 and Epstein-Barr herpesvirus genomes by human APOBEC3 cytidine deaminases in culture and in vivo. J Virol 85:7594–7602. 10.1128/JVI.00290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fehrholz M, Kendl S, Prifert C, Weissbrich B, Lemon K, Rennick L, Duprex PW, Rima BK, Koning FA, Holmes RK, Malim MH, Schneider-Schaulies J. 2012. The innate antiviral factor APOBEC3G targets replication of measles, mumps and respiratory syncytial viruses. J Gen Virol 93:565–576. 10.1099/vir.0.038919-0. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y-P, Peng Z-G, Wu Z-Y, Li J-R, Huang M-H, Si S-Y, Jiang J-D. 2015. Host APOBEC3G protein inhibits HCV replication through direct binding at NS3. PLoS One 10:e0121608. 10.1371/journal.pone.0121608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verhalen B, Starrett GJ, Harris RS, Jiang M. 2016. Functional upregulation of the DNA cytosine deaminase APOBEC3B by polyomaviruses. J Virol 90:6379–6386. 10.1128/JVI.00771-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peretti A, Geoghegan EM, Pastrana DV, Smola S, Feld P, Sauter M, Lohse S, Ramesh M, Lim ES, Wang D, Borgogna C, FitzGerald PC, Bliskovsky V, Starrett GJ, Law EK, Harris RS, Killian JK, Zhu J, Pineda M, Meltzer PS, Boldorini R, Gariglio M, Buck CB. 2018. Characterization of BK polyomaviruses from kidney transplant recipients suggests a role for APOBEC3 in driving in-host virus evolution. Cell Host Microbe 23:628–635.e7. 10.1016/j.chom.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Milewska A, Kindler E, Vkovski P, Zeglen S, Ochman M, Thiel V, Rajfur Z, Pyrc K. 2018. APOBEC3-mediated restriction of RNA virus replication. Sci Rep 8:5960. 10.1038/s41598-018-24448-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, Raine K, Campbell PJ, Rada C, Stratton MR, Neuberger MS. 2013. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife 2:e00534. 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjörd JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Imielinsk M, Jäger N, Jones DTW, Jones D, Knappskog S, Kool M, Lakhani SR, López-Otín C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Australian Pancreatic Cancer Genome Initiative, ICGC Breast Cancer Consortium, ICGC MMML-Seq Consortium, ICGC PedBrain , et al. 2013. Signatures of mutational processes in human cancer. Nature 500:415–421. 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, Yee D, Temiz NA, Donohue DE, McDougle RM, Brown WL, Law EK, Harris RS. 2013. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494:366–370. 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. 2014. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep 7:1833–1841. 10.1016/j.celrep.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 44.Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, Boot A, Covington KR, Gordenin DA, Bergstrom EN, Islam SMA, Lopez-Bigas N, Klimczak LJ, McPherson JR, Morganella S, Sabarinathan R, Wheeler DA, Mustonen V, Getz G, Rozen SG, Stratton MR, PCAWG Consortium . 2020. The repertoire of mutational signatures in human cancer. Nature 578:94–101. 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, Malc EP, Kim J, Kwiatkowski DJ, Fargo DC, Mieczkowski PA, Getz G, Gordenin DA. 2015. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet 47:1067–1072. 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Law EK, Levin-Klein R, Jarvis MC, Kim H, Argyris PP, Carpenter MA, Starrett GJ, Temiz NA, Larson LK, Durfee C, Burns MB, Vogel RI, Stavrou S, Aguilera AN, Wagner S, Largaespada DA, Starr TK, Ross SR, Harris RS. 2020. APOBEC3A catalyzes mutation and drives carcinogenesis in vivo. J Exp Med 217:e20200261. 10.1084/jem.20200261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDaniel YZ, Wang D, Love RP, Adolph MB, Mohammadzadeh N, Chelico L, Mansky LM. 2020. Deamination hotspots among APOBEC3 family members are defined by both target site sequence context and ssDNA secondary structure. Nucleic Acids Res 10.1093/nar/gkz1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anwar F, Davenport MP, Ebrahimi D. 2013. Footprint of APOBEC3 on the genome of human retroelements. J Virol 87:8195–8204. 10.1128/JVI.00298-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warren CJ, Van Doorslaer K, Pandey A, Espinosa JM, Pyeon D. 2015. Role of the host restriction factor APOBEC3 on papillomavirus evolution. Virus Evol 1:vev015. 10.1093/ve/vev015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen J, MacCarthy T. 2017. The preferred nucleotide contexts of the AID/APOBEC cytidine deaminases have differential effects when mutating retrotransposon and virus sequences compared to host genes. PLoS Comput Biol 13:e1005471. 10.1371/journal.pcbi.1005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poulain F, Lejeune N, Willemart K, Gillet NA. 2020. Footprint of the host restriction factors APOBEC3 on the genome of human viruses. PLoS Pathog 16:e1008718. 10.1371/journal.ppat.1008718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ibelgaufts H, Doerfler W, Scheidtmann KH, Wechsler W. 1980. Adenovirus type 12-induced rat tumor cells of neuroepithelial origin: persistence and expression of the viral genome. J Virol 33:423–437. 10.1128/JVI.33.1.423-437.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuhlmann I, Achten S, Rudolph R, Doerfler W. 1982. Tumor induction by human adenovirus type 12 in hamsters: loss of the viral genome from adenovirus type 12-induced tumor cells is compatible with tumor formation. EMBO J 1:79–86. 10.1002/j.1460-2075.1982.tb01128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hohlweg U, Dorn A, Hösel M, Webb D, Buettner R, Doerfler W. 2004. Tumorigenesis by adenovirus type 12 in newborn Syrian hamsters, p 215–244. In Doerfler, W, Böhm, P (ed), Adenoviruses: model and vectors in virus-host interactions. Springer, Berlin, Heidelberg. [DOI] [PubMed] [Google Scholar]
- 55.Caval V, Suspène R, Shapira M, Vartanian J-P, Wain-Hobson S. 2014. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3’UTR enhances chromosomal DNA damage. Nat Commun 5:5129. 10.1038/ncomms6129. [DOI] [PubMed] [Google Scholar]
- 56.Xiao X, Yang H, Arutiunian V, Fang Y, Besse G, Morimoto C, Zirkle B, Chen XS. 2017. Structural determinants of APOBEC3B non-catalytic domain for molecular assembly and catalytic regulation. Nucleic Acids Res 45:7494–7506. 10.1093/nar/gkx362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vieira VC, Leonard B, White EA, Starrett GJ, Temiz NA, Lorenz LD, Lee D, Soares MA, Lambert PF, Howley PM, Harris RS. 2014. Human papillomavirus E6 triggers upregulation of the antiviral and cancer genomic DNA deaminase APOBEC3B. mBio 5:e02234-14. 10.1128/mBio.02234-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pebernard S, Iggo RD. 2004. Determinants of interferon-stimulated gene induction by RNAi vectors. Differentiation 72:103–111. 10.1111/j.1432-0436.2004.07202001.x. [DOI] [PubMed] [Google Scholar]
- 59.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou W-M, Thasler WE, Hüser N, Durantel D, Liang TJ, Münk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. 2014. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 343:1221–1228. 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanu N, Cerone MA, Goh G, Zalmas L-P, Bartkova J, Dietzen M, McGranahan N, Rogers R, Law EK, Gromova I, Kschischo M, Walton MI, Rossanese OW, Bartek J, Harris RS, Venkatesan S, Swanton C. 2016. DNA replication stress mediates APOBEC3 family mutagenesis in breast cancer. Genome Biol 17:185. 10.1186/s13059-016-1042-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siriwardena SU, Perera MLW, Senevirathne V, Stewart J, Bhagwat AS. 2018. A tumor-promoting phorbol ester causes a large increase in APOBEC3A expression and a moderate increase in APOBEC3B expression in a normal human keratinocyte cell line without increasing genomic uracils. Mol Cell Biol 39. 10.1128/MCB.00238-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamazaki H, Shirakawa K, Matsumoto T, Kazuma Y, Matsui H, Horisawa Y, Stanford E, Sarca AD, Shirakawa R, Shindo K, Takaori-Kondo A. 2020. APOBEC3B reporter myeloma cell lines identify DNA damage response pathways leading to APOBEC3B expression. PLoS One 15:e0223463. 10.1371/journal.pone.0223463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leonard B, McCann JL, Starrett GJ, Kosyakovsky L, Luengas EM, Molan AM, Burns MB, McDougle RM, Parker PJ, Brown WL, Harris RS. 2015. The PKC/NF-κB signaling pathway induces APOBEC3B expression in multiple human cancers. Cancer Res 75:4538–4547. 10.1158/0008-5472.CAN-15-2171-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maruyama W, Shirakawa K, Matsui H, Matsumoto T, Yamazaki H, Sarca AD, Kazuma Y, Kobayashi M, Shindo K, Takaori-Kondo A. 2016. Classical NF-κB pathway is responsible for APOBEC3B expression in cancer cells. Biochem Biophys Res Commun 478:1466–1471. 10.1016/j.bbrc.2016.08.148. [DOI] [PubMed] [Google Scholar]
- 65.Periyasamy M, Singh AK, Gemma C, Kranjec C, Farzan R, Leach DA, Navaratnam N, Pálinkás HL, Vértessy BG, Fenton TR, Doorbar J, Fuller-Pace F, Meek DW, Coombes RC, Buluwela L, Ali S. 2017. p53 controls expression of the DNA deaminase APOBEC3B to limit its potential mutagenic activity in cancer cells. Nucleic Acids Res 45:11056–11069. 10.1093/nar/gkx721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roelofs PA, Goh CY, Chua BH, Jarvis MC, Stewart TA, McCann JL, McDougle RM, Carpenter MA, Martens JW, Span PN, Kappei D, Harris RS. 2020. Characterization of the mechanism by which the RB/E2F pathway controls expression of the cancer genomic DNA deaminase APOBEC3B. Elife 9. 10.7554/eLife.61287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Warren CJ, Xu T, Guo K, Griffin LM, Westrich JA, Lee D, Lambert PF, Santiago ML, Pyeon D. 2015. APOBEC3A functions as a restriction factor of human papillomavirus. J Virol 89:688–702. 10.1128/JVI.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mori S, Takeuchi T, Ishii Y, Yugawa T, Kiyono T, Nishina H, Kukimoto I. 2017. Human papillomavirus 16 E6 upregulates APOBEC3B via the TEAD transcription factor. J Virol 91. 10.1128/JVI.02413-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westrich JA, Warren CJ, Klausner MJ, Guo K, Liu C-W, Santiago ML, Pyeon D. 2018. Human papillomavirus 16 E7 stabilizes APOBEC3A protein by inhibiting cullin 2-dependent protein degradation. J Virol 92:e01318-17. 10.1128/JVI.01318-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sullivan CS, Baker AE, Pipas JM. 2004. Simian virus 40 infection disrupts p130–E2F and p107–E2F complexes but does not perturb pRb–E2F complexes. Virology 320:218–228. 10.1016/j.virol.2003.10.035. [DOI] [PubMed] [Google Scholar]
- 71.DeCaprio JA. 2014. Human papillomavirus type 16 E7 perturbs DREAM to promote cellular proliferation and mitotic gene expression. Oncogene 33:4036–4038. 10.1038/onc.2013.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rashid NN, Rothan HA, Yusoff MSM. 2015. The association of mammalian DREAM complex and HPV16 E7 proteins. Am J Cancer Res 5:3525–3533. [PMC free article] [PubMed] [Google Scholar]
- 73.Fischer M, Uxa S, Stanko C, Magin TM, Engeland K. 2017. Human papilloma virus E7 oncoprotein abrogates the p53-p21-DREAM pathway. Sci Rep 7:2603. 10.1038/s41598-017-02831-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Starrett GJ, Serebrenik AA, Roelofs PA, McCann JL, Verhalen B, Jarvis MC, Stewart TA, Law EK, Krupp A, Jiang M, Martens JWM, Cahir-McFarland E, Span PN, Harris RS. 2019. Polyomavirus T antigen induces APOBEC3B expression using an LXCXE-dependent and TP53-independent mechanism. mBio 10. 10.1128/mBio.02690-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pelka P, Miller MS, Cecchini M, Yousef AF, Bowdish DM, Dick F, Whyte P, Mymryk JS. 2011. Adenovirus E1A directly targets the E2F/DP-1 complex. J Virol 85:8841–8851. 10.1128/JVI.00539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghosh MK, Harter ML. 2003. A viral mechanism for remodeling chromatin structure in G0 cells. Mol Cell 12:255–260. 10.1016/s1097-2765(03)00225-9. [DOI] [PubMed] [Google Scholar]
- 77.Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, Kaelin WG, Conaway RC, Conaway JW, Branton PE. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev 15:3104–3117. 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harada JN, Shevchenko A, Shevchenko A, Pallas DC, Berk AJ. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J Virol 76:9194–9206. 10.1128/jvi.76.18.9194-9206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu X-F. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 80.Gilson T, Blanchette P, Ballmann MZ, Papp T, Pénzes JJ, Benkő M, Harrach B, Branton PE. 2016. Using the E4orf6-based E3 ubiquitin ligase as a tool to analyze the evolution of adenoviruses. J Virol 90:7350–7367. 10.1128/JVI.00420-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hirose Y, Onuki M, Tenjimbayashi Y, Mori S, Ishii Y, Takeuchi T, Tasaka N, Satoh T, Morisada T, Iwata T, Miyamoto S, Matsumoto K, Sekizawa A, Kukimoto I. 2018. Within-host variations of human papillomavirus reveal APOBEC-signature mutagenesis in the viral genome. J Virol 92:e00017-18. 10.1128/JVI.00017-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vartanian J-P, Henry M, Marchio A, Suspène R, Aynaud M-M, Guétard D, Cervantes-Gonzalez M, Battiston C, Mazzaferro V, Pineau P, Dejean A, Wain-Hobson S. 2010. Massive APOBEC3 editing of hepatitis B viral DNA in cirrhosis. PLoS Pathog 6:e1000928. 10.1371/journal.ppat.1000928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robinson CM, Seto D, Jones MS, Dyer DW, Chodosh J. 2011. Molecular evolution of human species D adenoviruses. Infect Genet Evol 11:1208–1217. 10.1016/j.meegid.2011.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinson CM, Singh G, Lee JY, Dehghan S, Rajaiya J, Liu EB, Yousuf MA, Betensky RA, Jones MS, Dyer DW, Seto D, Chodosh J. 2013. Molecular evolution of human adenoviruses. Sci Rep 3:1812. 10.1038/srep01812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dicks MDJ, Spencer AJ, Edwards NJ, Wadell G, Bojang K, Gilbert SC, Hill AVS, Cottingham MG. 2012. A novel chimpanzee adenovirus vector with low human seroprevalence: improved systems for vector derivation and comparative immunogenicity. PLoS One 7:e40385. 10.1371/journal.pone.0040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Riu E, Chen Z-Y, Xu H, He C-Y, Kay MA. 2007. Histone modifications are associated with the persistence or silencing of vector-mediated transgene expression in vivo. Mol Ther 15:1348–1355. 10.1038/sj.mt.6300177. [DOI] [PubMed] [Google Scholar]
- 87.Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, Peyton M, Zou Y, Kurie JM, Dimaio JM, Milchgrub S, Smith AL, Souza RF, Gilbey L, Zhang X, Gandia K, Vaughan MB, Wright WE, Gazdar AF, Shay JW, Minna JD. 2004. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res 64:9027–9034. 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- 88.Wissing S, Montano M, Garcia-Perez JL, Moran JV, Greene WC. 2011. Endogenous APOBEC3B restricts LINE-1 retrotransposition in transformed cells and human embryonic stem cells. J Biol Chem 286:36427–36437. 10.1074/jbc.M111.251058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Law EK, Sieuwerts AM, LaPara K, Leonard B, Starrett GJ, Molan AM, Temiz NA, Vogel RI, Meijer-van Gelder ME, Sweep FCGJ, Span PN, Foekens JA, Martens JWM, Yee D, Harris RS. 2016. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci Adv 2:e1601737. 10.1126/sciadv.1601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang C, Kolokoltsova OA, Mateer EJ, Koma T, Paessler S. 2017. Highly pathogenic new world arenavirus infection activates the pattern recognition receptor protein kinase R without attenuating virus replication in human cells. J Virol 91:e01090-17. 10.1128/JVI.01090-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang S-L, Chi C-Y, Kuo P-H, Tsai H-P, Wang S-M, Liu C-C, Su I-J, Wang J-R. 2013. High-incidence of human adenoviral co-infections in Taiwan. PLoS One 8:e75208. 10.1371/journal.pone.0075208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res 38:4274–4284. 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]