ABSTRACT
Serological testing of large representative populations for antibodies to SARS-CoV-2 is needed to estimate seroprevalence, transmission dynamics, and the duration of antibody responses from natural infection and vaccination. In this study, a high-throughput SARS-CoV-2 multiplex microsphere immunoassay (MMIA) was developed for the receptor binding domain (RBD) and nucleocapsid (N) that was more sensitive than enzyme-linked immunosorbent assay (ELISA) (98% versus 87%). The MMIA was then applied and validated in 264 first responders in Colorado using serum and dried blood spot (DBS) eluates, compared to ELISA, and evaluated for neutralizing antibodies. Four percent (11/264) of first responders were seropositive in July to August 2020. Serum and DBS were highly correlated for anti-RBD and anti-N antibodies (R = 0.83, P < 0.0001 and R = 0.87, P < 0.0001, respectively) by MMIA. The MMIA accurately predicted SARS-CoV-2 neutralizing antibodies using DBS (R = 0.76, P = 0.037). On repeat antibody testing 3 months later, anti-RBD IgG decreased less rapidly than anti-N IgG measured by MMIA, with a median change in geometric median fluorescence intensity of 62% versus 79% (P < 0.01) for anti-RBD and anti-N IgG, respectively. This novel MMIA using DBS could be scalable for rapid and affordable SARS-CoV-2 serosurveillance in the United States and globally.
KEYWORDS: COVID-19 antibody testing, dried blood spot testing, enzyme immunoassay, immunological surveillance, SARS-CoV-2 infection serological testing
INTRODUCTION
Effective monitoring and public health management of infectious disease outbreaks requires reliable population-based serosurveillance. These data are necessary to estimate transmission dynamics, cumulative incidence and mortality ratios, and duration of antibody responses, and they could help identify susceptible populations for vaccination (1–3). Each of these parameters factors heavily into the formation and implementation of effective public health policy. In the context of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) pandemic, this has been complicated by the high prevalence of asymptomatic infections (4–7).
Effective serosurveillance needs to be rapid, affordable, sensitive, and reliable (1–3). While novel assay development is a critical component, innovative methods of patient sample acquisition are also needed; repeated venous blood draws on large representative populations is resource intensive. In contrast, dried blood spots (DBS) have long been utilized in resource-limited countries for serosurveillance in adults and children (8, 9), having numerous advantages over venipuncture (10–14). DBS collection can be self-obtained, requiring only a single-use lancet and a filter paper card, avoiding the participation of phlebotomists and close contacts during the pandemic. DBS have prolonged stability at room temperature and can be safely sent through the mail (10–14). Elution of antibodies from the DBS requires only a simple buffered detergent and limited plasticware. Community samples obtained in this fashion have long been used to establish seroprevalence of infectious diseases (8, 10, 13, 15), serving as an effective precedent for the use of DBS in SARS-CoV-2 seroprevalence studies (16–21).
SARS-CoV-2 antibodies can be measured by multiple techniques, most commonly enzyme-linked immunosorbent assays (ELISAs) (22, 23); however these assays are resource-intensive and time-consuming. In contrast, flow-cytometric multiplexed microsphere immunoassays (MMIA) are sensitive and high throughput, with a broad dynamic range, and they correlate well with functional readouts (24). These assays use pathogen-specific antigens covalently bonded to fluorophore-loaded carboxyl beads, facilitating simultaneous identification of multiple pathogen-specific antibodies. MMIAs have been used to detect and differentiate antibodies against viral infections such as human herpesviruses and flaviviruses (e.g., Epstein-Barr, West Nile, Zika, and dengue viruses) (25, 26), used to identify SARS-CoV-2 IgG seroconversion (27–30) and proposed for public health use since the early 2000s (31, 32).
In this study, we developed a high-throughput and internally controlled MMIA to detect IgG reactive against the receptor binding domain (RBD) and nucleocapsid (N) antigens of SARS-CoV-2. Using pre-COVID and confirmed COVID-positive samples, we compared the MMIA to well-validated ELISAs and a SARS-CoV-2 neutralization assay. We then implemented the MMIA in a serosurveillance study using serum and DBS samples obtained from first responders (33). We observed a strong correlation between the MMIA and ELISA using DBS, which predicted the magnitude and functional capacity of SARS-CoV-2 neutralizing antibodies (Nabs), using a gold standard live virus focus reduction neutralization test (FRNT) (34). These results validate the use of MMIA using DBS as a sensitive and reliable alternative to more costly assays for population-based serosurveillance.
MATERIALS AND METHODS
Study population.
Initial validation of the MMIA utilized the Children's Hospital of Colorado's COVID-19 Convalescent Plasma (CCP) donor program, registered with the FDA as eligible to collect CCP on 31 March 2020. Donors were confirmed nasopharyngeal PCR positive for SARS-CoV-2, were symptom free for at least 14 days prior to plasma donation, and met all standard FDA blood donation criteria. Samples were stored at −80°C prior to analysis. The initial validation of the MMIA utilized deidentified residual plasma samples from these donors and prepandemic negative controls.
We previously described a study to evaluate the seroprevalence of SARS-CoV-2 antibodies among first responders (police, firefighters, paramedics, and emergency medical technicians) in Arapahoe County, Colorado (33). Participants completed a survey on risk factors, previous COVID-19 symptoms, and use of personal protective equipment (PPE). Results using SARS-CoV-2 ELISA on serum have been previously described (33). Venous blood and DBS were collected (July and August 2020) and evaluated for SARS-CoV-2 antibodies by both standard ELISA and MMIA; positive samples were evaluated for neutralizing antibodies. Participants were asked to participate approximately 3 months later to evaluate for seroconversion or changes in antibodies. All protocol and consent forms were approved by the Colorado Multiple Institutional Review Board.
Sample preparation.
Venous blood was processed within 8 h of collection to obtain serum, as previously described (33), and then stored at −20°C. DBS were collected on Whatman 903 protein saver snap apart cards (Cytiva, Marlborough, MA), allowed to dry at room temperature (RT), and then stored at −20°C. A 6-mm handheld punch was used to remove all 4 DBS from each card, and the 4 punches were then placed in a single well of a 24-well cell culture plate with 500 μl of elution buffer (phosphate-buffered saline [PBS] containing 0.05% Tween 20 and 0.08% NaN3) and placed on a platform shaker for 2 h at RT. Each DBS punch contains approximately 10 μl of blood, of which approximately 50% is serum, resulting in an estimated 1:25 serum dilution of the eluate. The elution was collected and stored at –80°C until use in the MMIA or ELISA.
ELISA for assessment of anti-SARS-CoV-2 IgG.
Two commercial ELISAs, the CE-marked Epitope Diagnostics Inc. (EDI) ELISA (San Diego, CA), (number KT-1032) and the CE-marked and FDA Emergency Use Authorization (EUA)-approved Euroimmun ELISA (Lubeck, Germany) (number 2606), were used for the MMIA validation. The EDI ELISA utilizes recombinant SARS-CoV-2 N antigen, and the Euroimmun ELISA utilizes the recombinant SARS-CoV-2 S1 domain, including the RBD. The EDI and Euroimmun ELISAs were processed according to the manufacturers’ protocols (35, 36). For the EDI assay, the positive cutoff value was calculated using the formula positive cutoff = 1.1 × (xNC + 0.18), where xNC is the average optical density at 450 nm (OD450) of triplicate negative-control OD450 values for a specific assay run. Given the EDI assay results included multiple runs, positive results were based on each run’s positive cutoff value. The Euroimmun assay was interpreted based on the ratio of the sample OD450 (S) to the calibrator OD450 (C). Samples with an S/C ratio of greater than 1.1 were positive. The Euroimmun (anti-RBD) assay has a reported sensitivity of 90% (95% confidence interval [CI], 74.4 to 96.5%) and specificity of 100% (95% CI, 95.4 to 100%). The EDI (anti-N) assay has a reported sensitivity of 98.4% (95% CI, 95.4 to 99.5%) and specificity of 99.8% (95% CI, 99.1 to 99.97%) (35, 36). Similar testing parameters have been independently confirmed (37).
The University of Colorado Exsera BioLabs SARS-CoV-2 IgG ELISA measures IgG antibodies against both RBD and N proteins in two separate ELISAs, based on the work of Stadlbauer et al. (38), and was validated to EUA and CAP/CLIA standards. OD thresholds and signal-to-cutoff (S/C) values for each ELISA were determined. Interpretations of assay results depended on the combination of the S/C values from the RBD and N ELISAs. If the S/C ratio was ≥1.0 for both RBD and N, the result is reported as reactive. If the S/C ratio was <1.0 for either RBD or N, the result is reported as nonreactive. The sensitivity and specificity of the University of Colorado Exsera BioLabs SARS-CoV-2 IgG assay is 84.0% (95% CI, 74.1% to 90.6%) and 99.6% (95% CI, 99.0% to 99.9%), respectively, based on the testing of 75 confirmed SARS-CoV-2 positives (>10 days after PCR) and 1,001 serum samples collected prior to November 2019.
MMIA for the detection of anti-SARS-CoV2 antibodies.
Recombinant SARS-CoV-2 RBD antigen was produced and purified from Expi293 human cells transfected with a DNA expression plasmid (38). Recombinant SARS-CoV-2 N antigen was produced using Escherichia coli. The DNA encoding the 419-residue N protein was designed with a C-terminal Flag tag followed by a 6× His tag in pET29 and purchased from Twist Bioscience (San Francisco, CA). A typical growth in BL21/DE3 cells comprised 6 liters of Luria broth (LB) in the presence of 100 mg/ml kanamycin that was induced at an OD600 of 0.8 for 3 h at 37°C. BL21 cells were harvested and lysed via sonication in cobalt affinity buffer (50 mM Na2HPO4, pH 7, 500 mM NaCl, 10 mM imidazole); supernatants were applied to a 20-ml cobalt affinity column (Sigma) and eluted with cobalt affinity buffer supplemented with 400 mM imidazole. Eluted fractions were dialyzed against ion exchange (50 mM Na2P2O7, pH 7, 150 mM NaCl) and applied to a 40-ml SP fast flow column (Cytiva). Eluted fractions were dialyzed against final buffer (50 mM HEPES, pH 7, 150 mM NaCl). Tetanus toxoid (TT) and bovine serum albumin (BSA) were obtained from MilliporeSigma (St. Louis, MO). BioLegend LEGENDplex carboxyl beads (A4, A6, A7, and B4) were conjugated to BSA (sample negative control), TT (sample positive control), SARS-CoV-2 N, and SARS-CoV-2 RBD, respectively, according to the manufacturer’s protocol. TT was chosen as a positive control due to the high prevalence of TT vaccination among health care workers and first responders (39, 40). Validation of each protein-bead conjugation was performed by staining with an anti-RBD monoclonal antibody (human chimeric, D002; Sino Biologicals, Wayne, PA) and an anti-TT monoclonal antibody (mouse antibody; Jackson Immunoresearch, West Grove, PA). Beads were mixed in equal ratios (∼2,000 each bead/sample well) and incubated with serum samples (1:100 dilution) or DBS elution samples (1:10 dilution) into storage/running buffer (PBS containing 0.01% Tween 20, 0.05% NaN3, and 0.1% BSA), rocked on a shaker plate for 60 min at RT, and then washed. Bound IgG was detected by secondary anti-human IgG-biotin (1:4,000 dilution) (SouthernBiotech, Birmingham, AL), followed by addition of streptavidin-phycoerythrin (SA-PE) (1:1,000 dilution) (BD Bioscience, San Jose, CA) and acquisition of individual sample fluorescence using 96-well plates by a CytoFlex flow cytometer (Beckman Coulter, Indianapolis, IN). The geometric median fluorescence intensity (gMFI) of the anti-IgG specific for each protein/bead was computed for each sample using FloJo software v.10.7.1 (BD Bioscience, San Jose, CA).
FRNT.
Vero E6 cells (ATCC, Manassas, VA) were seeded in 96-well plates. Serum samples were heat inactivated and serially diluted (2-fold, starting at 1:10) in DMEM (ThermoFisher, Pittsburgh, PA) plus 1% fetal bovine serum (FBS) in 96-well plates. Approximately 100 focus-forming units (FFU) of SARS-CoV-2 USA-WA1/2020 (NR-52281; deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH) were added to each well, and the serum plus virus mixture was incubated for 1 h at 37°C. Postincubation, medium was removed from cells, and the serum sample plus virus mixture was added to the cells for 1 h at 37°C (98.6°F). After 1 h, samples were removed and cells overlaid with 1% methylcellulose (MilliporeSigma) in minimum essential medium (MEM) (ThermoFisher, Pittsburgh, PA), 2% FBS and incubated for 30 h at 37°C. Cells were fixed with 4% paraformaldehyde (Acros Organics, Pittsburgh, PA) and probed with 1 μg/ml of an anti-SARS-CoV spike monoclonal antibody (CR3022; Absolute Antibody, Boston, MA, USA) in perm wash (1× PBS, 0.1% saponin, 0.1% BSA) for 2 h at RT. After washing, cells were incubated with a 1:1,000 dilution of horseradish peroxidase (HRP)-conjugated goat anti-human IgG (Southern Biotech, Birmingham, AL) for 1.5 h at RT. After washing, SARS-CoV-2-positive foci were visualized with TrueBlue substrate (ThermoFisher) and counted using a CTL Biospot analyzer and Biospot software (Cellular Technology Ltd., Shaker Heights, OH). The 50% focus reduction neutralization test (FRNT50) titers were calculated relative to a virus only control (no serum) set at 100%, using a GraphPad Prism 8 (La Jolla, CA) default nonlinear curve fit constrained between 0 and 100%.
Statistical analysis.
Descriptive statistics were evaluated using GraphPad Prism 9. Means were compared using Student t tests and nonparametric comparisons with nonparametric Wilcoxson matched rank tests and Mann-Whitney tests for differences in medians. P values of <0.05 were considered statistically significant.
RESULTS
Validation of MMIA.
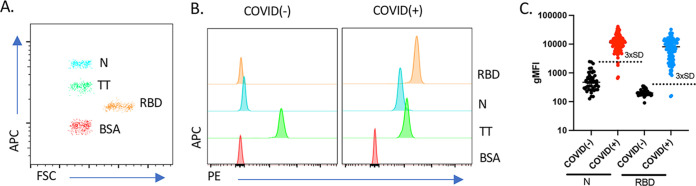
To ascertain the diagnostic parameters of the MMIA for the detection of SARS-CoV-2-specific IgG antibodies, we analyzed the ability to discriminate the conjugated RBD, N, TT, and BSA beads. BioLegend LEGENDplex carboxyl beads can be discriminated with flow cytometry based on fluorescence of allophycocyanin (APC) and size from forward scatter (FSC) (Fig. 1A). BSA-coated beads served as an effective negative control, showing little to no fluorescence (Fig. 1B). Similarly, TT-coated beads served as an effective internal positive control for sample validation, demonstrating robust fluorescence for TT-specific IgG antibodies in both COVID-negative [COVID(−)] and -positive [COVID(+)] samples (Fig. 1B). In contrast, both N and RBD PE fluorescence increased only in COVID(+) patient samples (Fig. 1B). Calculating the mean and standard deviation N and RBD fluorescence from all negative controls, we established a lower limit (3 standard deviations [3× SD] above the mean gMFI) for determining samples positive for these antibodies (Fig. 1C, dotted line). Next, serum of previously reverse transcription-PCR confirmed positive SARS-CoV-2 individuals [COVID(+), n = 105] and prepandemic controls [COVID(−), n = 50] were analyzed by MMIA for both anti-RBD and anti-N IgG. Using these cutoff criteria, 103/105 COVID(+) samples were positive for anti-RBD IgG and 101/105 were positive for anti-N IgG by MMIA (Fig. 1C). Reexamination of the 4 samples below the cutoff for anti-N reactivity indicated that the two samples with the lowest anti-N IgG response were the same as the two anti-RBD IgG negatives. Despite having been identified as COVID(+) by a PCR diagnostic test at least 14 days prior to sample collection, these individuals had low antibody responses against either SARS-CoV-2 protein, suggesting they had a delayed or incomplete immune response or a false-positive PCR nasopharyngeal swab.
FIG 1.
Development and validation of a novel MMIA for detection of SARS-CoV-2 IgG reactive to RBD and N antigens. (A) Flow cytometry showing the fluorescence of allophycocyanin (APC) and size from forward scatter (FSC) of the RBD-, N-, TT-, and BSA-conjugated beads. (B) Representative MMIA histograms. Histograms show the PE fluorescence for each antigen obtained by flow cytometry for representative COVID(−) (left) and COVID(+) (right) samples. (C) Fifty pre-COVID serum samples [COVID(−)] and 105 serum samples from patients that screened positive for SARS-CoV-2 by nasopharyngeal quantitative PCR [COVID(+)] were analyzed by MMIA. Dotted lines represent 3 standard deviations (3× SD) above the mean gMFI of negative controls.
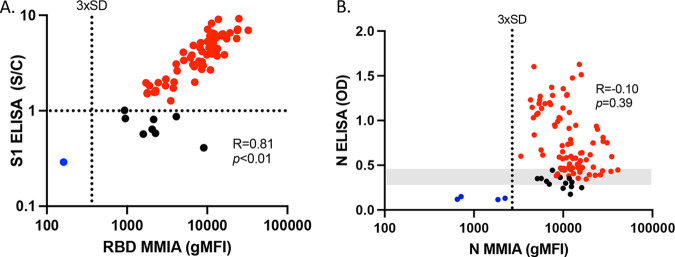
We next compared the results obtained from the bead-based MMIA to results obtained from SARS-CoV-2 S1 (Euroimmun) and N (EDI) specific IgG ELISAs using the known COVID(+) samples (Fig. 2). Anti-S1 and anti-RBD IgG were strongly correlated between ELISA (S/C) and the MMIA (gMFI) (Fig. 2A) (R = 0.81, P < 0.0001), but anti-N IgG (OD) was not correlated (Fig. 2B) (R = −0.10, P = 0.39). Within the confirmed COVID(+) serum samples, both the Euroimmun and EDI ELISAs identified some samples as anti-S1 and anti-N IgG seronegative (Fig. 2A and B), using an S/C cutoff of 1.1 and positive cutoff OD values that ranged from 0.290 to 0.473, respectively, due to run cutoff variability (Fig. 2, range represented in gray). Black circles indicate samples that were identified as negative by ELISA but positive with MMIA, falling well above the 3× SD cutoff for seropositivity. Red circles indicate samples that were positive by both ELISA and MMIA. Blue circles indicate samples that were negative by both ELISA and MMIA (Fig. 2A and B). These analyses show that the MMIA had greater sensitivity for anti-RBD IgG of 98% (95% CI, 93.3 to 99.5) and anti-N IgG of 96% (95% CI, 90.1 to 98.5) than the anti-S1 IgG (Euroimmun) sensitivity of 87.3% (95% CI, 77.6 to 93.2) and anti-N IgG (EDI) sensitivity of 83.5% (95% CI, 75.2 to 89.4), using these known COVID(+) samples.
FIG 2.
MMIA serology is more sensitive than ELISA-based methods. Seventy-one serum samples from patients that screened positive for SARS-CoV-2 by nasopharyngeal qPCR were evaluated by MMIA (gMFI) and ELISA for IgG reactivity against the SARS-CoV-2 by Euroimmun (anti-S1 IgG S/C ratio) (A) and Epitope Diagnostics Inc. (anti-N IgG OD) (B). The positive OD cutoff range (0.290 to 0.473) is represented in gray. Red circles indicate samples that were identified as positive by ELISA and MMIA. Black circles indicate samples that were identified as negative by ELISA but positive with MMIA. Blue circles indicate samples that were identified as negative by both ELISA and MMIA.
Detection of neutralizing antibodies.
Using an FRNT, we established the neutralizing activity for all known COVID(+) samples. We observed a wide range of neutralizing activity that positively correlated with the levels of anti-S1 or RBD IgG as determined by ELISA and MMIA (R = 0.63, P < 0.0001 and R = 0.70, P < 0.0001) (Fig. 3A and B). There was no correlation between the FRNT and anti-N IgG antibodies by ELISA or MMIA (R = 0.37, P = 0.26 and R = 0.24, P = 0.49, respectively) (see Fig. S1 in the supplemental material).
FIG 3.
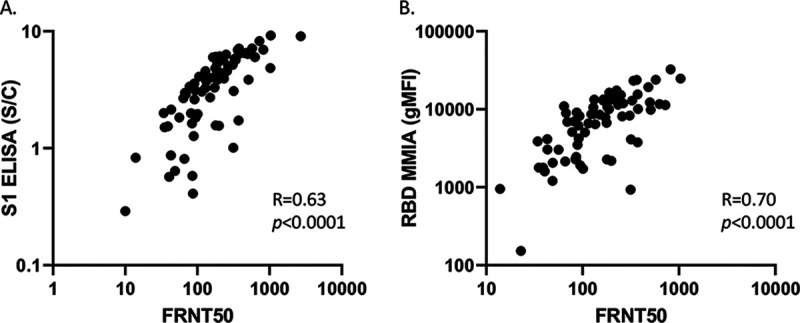
MMIA anti-RBD IgG and ELISA correlate with neutralizing antibody titer. Seventy-one serum samples from patients with positive nasopharyngeal qPCR for SARS-CoV-2 had neutralizing antibodies measured by a FRNT50 correlated with ELISA (Euroimmun anti-S1 IgG S/C) (A) and MMIA (gMFI) (B) for anti-RBD IgG.
Application of the multiplex assay for serosurveillance.
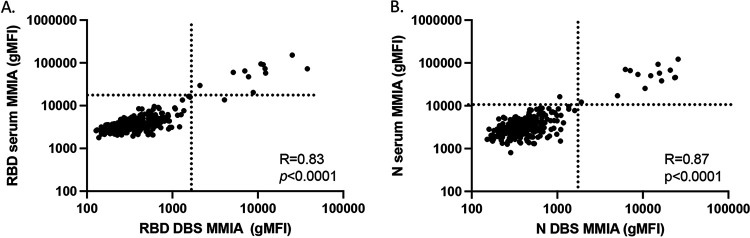
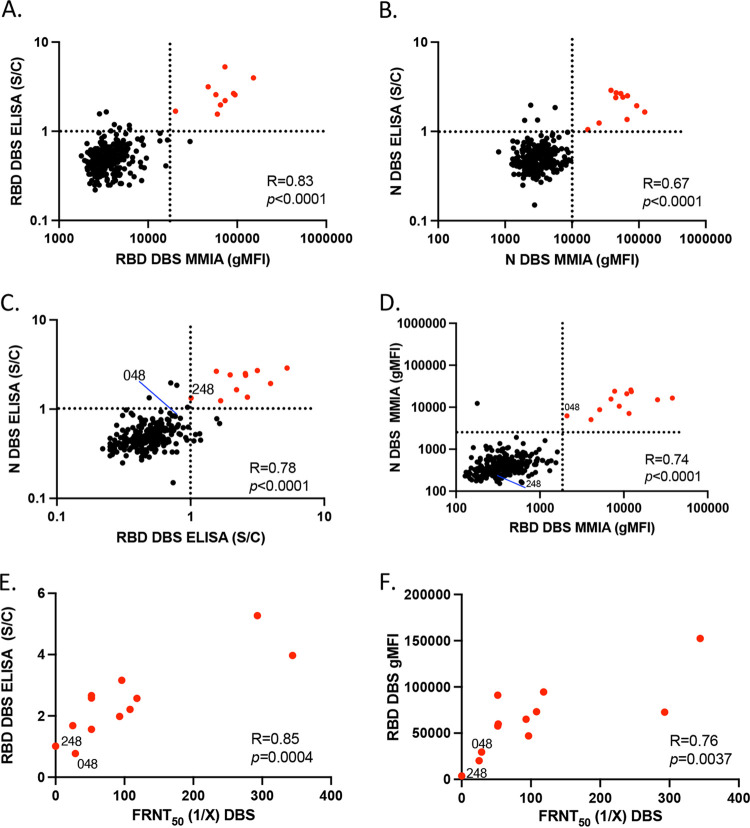
Our data thus far indicated that the sensitivity and specificity of the MMIA was sufficient for the purposes of SARS-CoV-2 serosurveillance. Therefore, we compared the use of this assay to that of the Exsera BioLabs SARS-CoV-2 IgG ELISA for the evaluation of SARS-CoV-2 antibodies in 264 first responders from Arapahoe County, Colorado. Paired serum and DBS eluates were obtained from all participants and subjected to evaluation by ELISA and MMIA for both anti-RBD and anti-N IgG. We recently reported this study population as 4% (11/264) reactive for both RBD and N IgG in serum by ELISA (33), and the MMIA was similarly able to identify as positive these same 11 individuals (Fig. S2). MMIA values for serum anti-RBD and anti-N IgG were highly correlated with DBS (R = 0.83, P < 0.0001 and R = 0.87, P < 0.0001, respectively) (Fig. 4A and B). There also was strong correlation between anti-RBD IgG detected by MMIA and ELISA in DBS (R = 0.83, P < 0.0001); those determined to be positive for both anti-RBD and anti-N IgG are in red in Fig. 5A. This was also observed in serum (Fig. S2). In contrast to the previous evaluation of only COVID(+) samples, we observed a strong correlation between the anti-N IgG detected by MMIA and ELISA (R = 0.67, P < 0.0001) (Fig. 5B), reflecting the influence of including antibody-negative sample comparisons. MMIA using DBS eluates was equally able to detect SARS-CoV-2 antibodies compared to standard ELISA methodology.
FIG 4.
MMIA can be combined with DBSs for accurate SARS-CoV-2 IgG analysis. (A and B) Correlation between serum and DBS for SARS-CoV-2 anti-RBD (A) and anti-N IgG (B) by MMIA among 264 first responders in Arapahoe County, Colorado, in July to August 2020.
FIG 5.
MMIA accurately predicts neutralization antibody titer. (A) ELISA for anti-RBD IgG versus MMIA for anti-RBD IgG using dried blood spot (DBS). (B) ELISA for anti-N IgG versus MMIA for anti-N IgG using DBS. (C) ELISA for anti-N IgG versus anti-RBD IgG identified 11/264 first responders (red circles) as seropositive. (D) MMIA for anti-N IgG versus anti-RBD IgG identified as positive the same 10 of 11 that were positive by ELISA, noting 2 discordant results (048 and 248). (E) ELISA anti-RBD IgG versus neutralizing antibodies measured by FRNT50. (F) MMIA anti-RBD IgG versus FRNT50. DBS elutes from participant 48 did have neutralization activity despite a negative ELISA result.
Evaluation of the DBS by MMIA (Fig. 5D) identified the same 11 individuals deemed positive for serum anti-RBD and anti-N IgG by both ELISA and MMIA (Fig. S2). However, two samples were discordant between the MMIA and ELISA on DBS. While the ELISA determined sample 248 as positive and sample 048 as negative, the reverse result was obtained using the MMIA. In the ELISA, the reactivity of both samples was close to the signal-to-cutoff value (i.e., 1) (Fig. 5C), whereas sample 248 was identified as clearly negative by MMIA (Fig. 5D). The designation of this sample as negative by the MMIA was more accurate than the ELISA, as indicated by evaluating these samples by functional virus neutralization assay. Whereas sample 048 had weak but detectable neutralizing activity, sample 248 showed no capacity to neutralize SARS-CoV-2 infection (Fig. 5E and F). Despite this difference, it had little impact on the strength of the correlation between the FRNT50 values and either the ELISA (R = 0.85, P = 0.0004) or MMIA (R = 0.76, P = 0.0037).
Follow-up serology among positive first responders using MMIA.
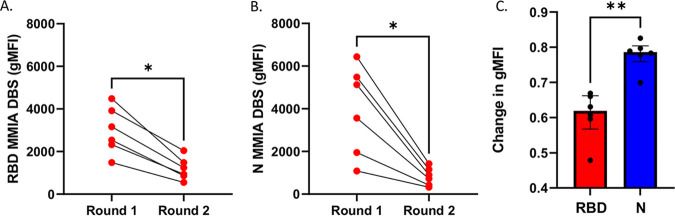
Ninety-three of 264 first responders (35%) returned for a second sample collection in November 2020. We found that 8/93 (9%) were positive for both anti-RBD and anti-N IgG with the MMIA, with two new seroconversions. In evaluating the six positive participant samples at both time points simultaneously with the MMIA, we observed statistically significant decreases in both anti-RBD and anti-N IgG (P < 0.01) (Fig. 6A and B). We observed a smaller decrease in the anti-RBD compared to anti-N IgG, with a median change in gMFI of 62% versus 79% (P < 0.01), for anti-RBD and anti-N IgG gMFI, respectively (Fig. 6C).
FIG 6.
Repeat serology for SARS-CoV-2 anti-RBD IgG and anti-N IgG by MMIA after 3 months among previously positive participants (n = 6). (A) MMIA for anti-RBD IgG by dried blood spot (DBS) for round 1 (July-August 2020) and round 2 (November 2020). (B) MMIA for anti-N IgG by DBS for round 1 and round 2, showing a greater decrease in anti-N IgG compared to anti-RBD IgG antibodies by MMIA. Comparisons were made with nonparametric Wilcoxson matched rank test (A and B) and Mann-Whitney test for differences in median change in gMFI (C). *, P < 0.05; **, P < 0.01).
Collectively, these data demonstrated that (i) the MMIA was a highly useful tool in serosurveillance for SARS-CoV-2 and (ii) the evaluation of DBS samples using MMIA could be used to predict both the magnitude and functional capacity of SARS-CoV-2-seropositive individuals.
DISCUSSION
In this study, we developed an MMIA to measure IgG antibodies to both the RBD of the spike protein and the N protein of SARS-CoV-2. We found that this assay was highly sensitive for detecting these antibodies in both serum and DBS eluates. The addition of both positive (e.g., TT) and negative (e.g., BSA) antigens provided confidence to the quality of the DBS eluates. Although the MMIA and ELISA are not specific for measuring neutralizing antibody capacity, the strong correlation between the results from the MMIA anti-RBD IgG gMFI with viral neutralization activity, measured by a gold standard (FRNT), provides confidence that this assay could be used to estimate neutralization capacity and the frequency of seroprotective responses. Furthermore, the inclusion of the N antigen may allow individuals vaccinated with spike protein-based vaccines (RBD+/N−) to be distinguished from individuals naturally infected with SARS-CoV-2 (RBD+/N+). While anti-RBD IgG is highly sensitive and specific and correlates with neutralizing activity (34, 41, 42), anti-N IgG is more likely to have some cross-reactivity with other human coronaviruses (43, 44) and did not correlate with SARS-CoV-2 neutralizing antibodies (45).
MMIA utilizing Luminex technology (30) have been developed and published recently, with results similar to those obtained in our studies using MMIA with flow-cytometric analysis (28, 29). Both platforms utilize relatively high-throughput automatic sample loading and analysis from 96-well plate formats. While no one has yet performed a head-to-head comparison between the methods of MMIA analysis (flow cytometry versus Luminex), both have advantages in terms of cost, sensitivity, and specificity over standard ELISA-based methods used to detect seropositivity to SARS-CoV-2. First, it is highly cost-effective where flow cytometry or Luminex is available. The amount of antigen required for conjugation with the fluorescent beads (10 μg of protein for each bead conjugation and >1:4,500 dilutions of reagents) is sufficient for the analysis of a minimum of 25,000 to 40,000 samples, producing an assay cost well below one dollar per sample. This cost structure remains competitive, even including the cost of the flow cytometry, given the broad distribution of inexpensive desktop flow cytometers, many of which are field-ready without the need for sheath fluid. In addition, ELISA-based methods generally have higher false-positive and false-negative rates, likely stemming from the capacity of an enzymatic reaction to overrepresent the signal from the occasional sample with unusually high background staining. Third, ELISA-based methods require initial coating of the plate with antigen, multiple incubations for secondary and tertiary detection reagents, colorimetric development, and multiple sample dilutions for analysis, which requires many hours of incubation and frequent technician handling. In contrast, the MMIA requires 90 min and only one round of sample washing prior to data acquisition. Finally, the larger dynamic range of the MMIA compared to ELISA improves its sensitivity to detect positive samples from DBS (27, 46, 47).
This MMIA using DBS can be used to further understand the occupational, household, and demographic risk factors that are contributing to the geographical and social disparities of SARS-CoV-2 (2). It also could be a valuable tool to target vulnerable populations as SARS-CoV-2 vaccines are rolled out globally, given limited supply and reported vaccine hesitancy, making accurate serosurveillance even more critical (48, 49). Finally, multiple studies show that SARS-CoV-2 antibodies change over time (50, 51). DBS using MMIA technology could be a valuable tool to monitor the durability of infection and vaccine-elicited antibodies.
Conclusions.
We conclude that the MMIA (i) successfully delineated between SARS-CoV-2 seropositive and seronegative individuals, (ii) was strongly correlated with established ELISAs, and (iii) effectively predicted serum neutralization capacity from SARS-CoV-2 seropositive individuals using DBS eluates. Serosurveillance using DBS and this MMIA could be an effective tool in large population-based surveillance to estimate viral transmission dynamics and mortality ratios and identify susceptible populations as vaccination programs roll out during this rapidly evolving pandemic.
ACKNOWLEDGMENTS
We thank Jason Lehmann, Binggang Sun, and Jessie Ni from BioLegend for their kind donation of, and technical assistance with, the LEGENDplex carboxyl beads used in these studies; Florian Krammer, from the Icahn School of Medicine at Mount Sinai, for providing the expression plasmid used to generate recombinant receptor binding domain protein; and Lori Sherman, from the University of Colorado Cancer Cell Technologies Shared Resource (CTSR), who produced the initial SARS-CoV-2 proteins for binding to the MMIA.
T.E.M., R.R., and R.M.K. are all listed as inventors on a provisional patent submitted for the use of MMIA in serosurveillance to virological exposure.
Funding for this study was provided by the Arapahoe County CARES Act.
Footnotes
Supplemental material is available online only.
Contributor Information
Rosemary Rochford, Email: Rosemary.Rochford@cuanschutz.edu.
Yi-Wei Tang, Cepheid.
REFERENCES
- 1.Bajema KL, Wiegand RE, Cuffe K, Patel SV, Iachan R, Lim T, Lee A, Moyse D, Havers FP, Harding L, Fry AM, Hall AJ, Martin K, Biel M, Deng Y, Meyer WA, III, Mathur M, Kyle T, Gundlapalli AV, Thornburg NJ, Petersen LR, Edens C. 24 November 2020. Estimated SARS-CoV-2 seroprevalence in the US as of September 2020. JAMA Intern Med 10.1001/jamainternmed.2020.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown TS, Walensky RP. 2020. Serosurveillance and the COVID-19 epidemic in the US: undetected, uncertain, and out of control. JAMA 324:749–751. 10.1001/jama.2020.14017. [DOI] [PubMed] [Google Scholar]
- 3.Altmann DM, Douek DC, Boyton RJ. 2020. What policy makers need to know about COVID-19 protective immunity. Lancet 395:1527–1529. 10.1016/S0140-6736(20)30985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oran DP, Topol EJ. 2020. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative review. Ann Intern Med 173:362–367. 10.7326/M20-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang L, Zhang X, Zhang X, Wei Z, Zhang L, Xu J, Liang P, Xu Y, Zhang C, Xu A. 2020. Rapid asymptomatic transmission of COVID-19 during the incubation period demonstrating strong infectivity in a cluster of youngsters aged 16–23 years outside Wuhan and characteristics of young patients with COVID-19: a prospective contact-tracing study. J Infect 80:e1–e13. 10.1016/j.jinf.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, Zimmer T, Thiel V, Janke C, Guggemos W, Seilmaier M, Drosten C, Vollmar P, Zwirglmaier K, Zange S, Wolfel R, Hoelscher M. 2020. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med 382:970–971. 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudbjartsson DF, Norddahl GL, Melsted P, Gunnarsdottir K, Holm H, Eythorsson E, Arnthorsson AO, Helgason D, Bjarnadottir K, Ingvarsson RF, Thorsteinsdottir B, Kristjansdottir S, Birgisdottir K, Kristinsdottir AM, Sigurdsson MI, Arnadottir GA, Ivarsdottir EV, Andresdottir M, Jonsson F, Agustsdottir AB, Berglund J, Eiriksdottir B, Fridriksdottir R, Gardarsdottir EE, Gottfredsson M, Gretarsdottir OS, Gudmundsdottir S, Gudmundsson KR, Gunnarsdottir TR, Gylfason A, Helgason A, Jensson BO, Jonasdottir A, Jonsson H, Kristjansson T, Kristinsson KG, Magnusdottir DN, Magnusson OT, Olafsdottir LB, Rognvaldsson S, Le Roux L, Sigmundsdottir G, Sigurdsson A, Sveinbjornsson G, Sveinsdottir KE, Sveinsdottir M, Thorarensen EA, Thorbjornsson B, Thordardottir M, Saemundsdottir J, et al. 2020. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med 383:1724–1734. 10.1056/NEJMoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim MD. 2018. Dried blood spots for global health diagnostics and surveillance: opportunities and challenges. Am J Trop Med Hyg 99:256–265. 10.4269/ajtmh.17-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patton JC, Akkers E, Coovadia AH, Meyers TM, Stevens WS, Sherman GG. 2007. Evaluation of dried whole blood spots obtained by heel or finger stick as an alternative to venous blood for diagnosis of human immunodeficiency virus type 1 infection in vertically exposed infants in the routine diagnostic laboratory. Clin Vaccine Immunol 14:201–203. 10.1128/CVI.00223-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Page M, Atabani SF, Wood M, Smit E, Wilson S, Atherton C, Davenport CF, Hartland D, Simpson M, Taylor S. 2019. Dried blood spot and mini-tube blood sample collection kits for postal HIV testing services: a comparative review of successes in a real-world setting. Sex Transm Infect 95:43–45. 10.1136/sextrans-2018-053567. [DOI] [PubMed] [Google Scholar]
- 11.Vazquez-Moron S, Ryan P, Ardizone-Jimenez B, Martin D, Troya J, Cuevas G, Valencia J, Jimenez-Sousa MA, Avellon A, Resino S. 2018. Evaluation of dried blood spot samples for screening of hepatitis C and human immunodeficiency virus in a real-world setting. Sci Rep 8:1858. 10.1038/s41598-018-20312-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behets F, Kashamuka M, Pappaioanou M, Green TA, Ryder RW, Batter V, George JR, Hannon WH, Quinn TC. 1992. Stability of human immunodeficiency virus type 1 antibodies in whole blood dried on filter paper and stored under various tropical conditions in Kinshasa, Zaire. J Clin Microbiol 30:1179–1182. 10.1128/JCM.30.5.1179-1182.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Condorelli F, Scalia G, Stivala A, Gallo R, Marino A, Battaglini CM, Castro A. 1994. Detection of immunoglobulin G to measles virus, rubella virus, and mumps virus in serum samples and in microquantities of whole blood dried on filter paper. J Virol Methods 49:25–36. 10.1016/0166-0934(94)90052-3. [DOI] [PubMed] [Google Scholar]
- 14.Tuaillon E, Kania D, Pisoni A, Bollore K, Taieb F, Ontsira NEN, Schaub R, Plantier JC, Makinson A, Van de Perre P. 2020. Dried blood spot tests for the diagnosis and therapeutic monitoring of HIV and viral hepatitis B and C. Front Microbiol 11:373. 10.3389/fmicb.2020.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corran PH, Cook J, Lynch C, Leendertse H, Manjurano A, Griffin J, Cox J, Abeku T, Bousema T, Ghani AC, Drakeley C, Riley E. 2008. Dried blood spots as a source of anti-malarial antibodies for epidemiological studies. Malar J 7:195. 10.1186/1475-2875-7-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulchandani R, Brown B, Brooks T, Semper A, Machin N, Linley E, Borrow R, Wyllie D. 2020. Use of dried blood spot samples for SARS-CoV-2 antibody detection using the Roche Elecsys high throughput immunoassay. medRxiv https://www.medrxiv.org/content/10.1101/2020.10.19.20215228v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zava TT, Zava DT. 2021. Validation of dried blood spot sample modifications to two commercially available COVID-19 IgG antibody immunoassays. Bioanalysis 13:13–28. 10.4155/bio-2020-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moat SJ, Zelek WM, Carne E, Ponsford MJ, Bramhall K, Jones S, El-Shanawany T, Wise MJ, Thomas A, George C, Fegan C, Steven R, Webb R, Weeks I, Morgan BP, Jolles S. 2021. EXPRESS: development of a high throughput SARS-CoV-2 antibody testing pathway using dried blood spot specimens. Ann Clin Biochem 58:123–131. 10.1177/0004563220981106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karp DG, Danh K, Espinoza NF, Seftel D, Robinson PV, Tsai CT. 2020. A serological assay to detect SARS-CoV-2 antibodies in at-home collected finger-prick dried blood spots. Sci Rep 10:20188. 10.1038/s41598-020-76913-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu F, Nguyen M, Vijayakumar P, Kaplan A, Meir A, Dai Y, Wang E, Walsh H, Ring AM, Omer SB, Farhadian SF. 2020. Newborn dried blood spots for serologic surveys of COVID-19. Pediatr Infect Dis J 39:e454–e456. 10.1097/INF.0000000000002918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegler AJ, Sullivan PS, Sanchez T, Lopman B, Fahimi M, Sailey C, Frankel M, Rothenberg R, Kelley CF, Bradley H. 2020. Protocol for a national probability survey using home specimen collection methods to assess prevalence and incidence of SARS-CoV-2 infection and antibody response. Ann Epidemiol 49:50–60. 10.1016/j.annepidem.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klumpp-Thomas C, Kalish H, Drew M, Hunsberger S, Snead K, Fay MP, Mehalko J, Shunmugavel A, Wall V, Frank P, Denson JP, Hong M, Gulten G, Messing S, Hicks J, Michael S, Gillette W, Hall MD, Memoli M, Esposito D, Sadtler K. 2020. Standardization of enzyme-linked immunosorbent assays for serosurveys of the SARS-CoV-2 pandemic using clinical and at-home blood sampling. medRxiv https://www.medrxiv.org/content/10.1101/2020.05.21.20109280v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peterhoff D, Gluck V, Vogel M, Schuster P, Schutz A, Neubert P, Albert V, Frisch S, Kiessling M, Pervan P, Werner M, Ritter N, Babl L, Deichner M, Hanses F, Lubnow M, Muller T, Lunz D, Hitzenbichler F, Audebert F, Hahnel V, Offner R, Muller M, Schmid S, Burkhardt R, Gluck T, Koller M, Niller HH, Graf B, Salzberger B, Wenzel JJ, Jantsch J, Gessner A, Schmidt B, Wagner R. 2021. A highly specific and sensitive serological assay detects SARS-CoV-2 antibody levels in COVID-19 patients that correlate with neutralization. Infection 49:75–82. 10.1007/s15010-020-01503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vignali DA. 2000. Multiplexed particle-based flow cytometric assays. J Immunol Methods 243:243–255. 10.1016/S0022-1759(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 25.Piriou E, Kimmel R, Chelimo K, Middeldorp JM, Odada PS, Ploutz-Snyder R, Moormann AM, Rochford R. 2009. Serological evidence for long-term Epstein-Barr virus reactivation in children living in a holoendemic malaria region of Kenya. J Med Virol 81:1088–1093. 10.1002/jmv.21485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyson J, Tsai WY, Tsai JJ, Massgard L, Stramer SL, Lehrer AT, Nerurkar VR, Wang WK. 2019. A high-throughput and multiplex microsphere immunoassay based on non-structural protein 1 can discriminate three flavivirus infections. PLoS Negl Trop Dis 13:e0007649. 10.1371/journal.pntd.0007649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laing E, Sterling S, Richard S, Epsi N, Phogat S, Samuels E, Yan L, Moreno N, Coles C, Drew M, Mehalko J, English C, Merritt S, Mende K, Chung K, Clifton G, Munster V, de Wit E, Tribble D, Agan B, Esposito D, Lanteri C, Mitre E, Burgess T, Broder C. 24 November 2020. A betacoronavirus multiplex microsphere immunoassay detects early SARS-CoV-2 seroconversion and antibody cross reactions. Res Sq 10.21203/rs.3.rs-105768/v1. [DOI] [Google Scholar]
- 28.Weiss S, Klingler J, Hioe C, Amanat F, Baine I, Arinsburg S, Kojic EM, Stoever J, Liu STH, Jurczyszak D, Bermudez-Gonzalez M, Simon V, Krammer F, Zolla-Pazner S. 2020. A high-throughput assay for circulating antibodies directed against the S protein of severe acute respiratory syndrome coronavirus 2. J Infect Dis 222:1629–1634. 10.1093/infdis/jiaa531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobano C, Vidal M, Santano R, Jimenez A, Chi J, Barrios D, Ruiz-Olalla G, Rodrigo MN, Carolis C, Parras D, Serra P, Martinez de Aguirre P, Carmona-Torre F, Reina G, Santamaria P, Mayor A, Garcia-Basteiro AL, Izquierdo L, Aguilar R, Moncunill G. 2021. Highly sensitive and specific multiplex antibody assays to quantify immunoglobulins M, A, and G against SARS-CoV-2 antigens. J Clin Microbiol 59:e01731-20. 10.1128/JCM.01731-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luminex. 2021. Luminex xMAP SARS-CoV-2 antibody testing. https://www.luminexcorp.com/xmap-sars-cov-2-antibody-testing/. Accessed 24 March 2021.
- 31.Robertson BH, Nicholson JK. 2005. New microbiology tools for public health and their implications. Annu Rev Public Health 26:281–302. 10.1146/annurev.publhealth.26.021304.144522. [DOI] [PubMed] [Google Scholar]
- 32.Iannone MA. 2001. Microsphere-based molecular cytometry. Clin Lab Med 21:731–742. [PubMed] [Google Scholar]
- 33.Sabourin KR, Schultz J, Romero J, Lamb MM, Larremore D, Morrison TE, Frazer-Abel A, Zimmer S, Kedl RM, Jaenisch T, Rochford R. 9000. Risk factors of SARS-CoV-2 antibodies in arapahoe county first responders–the COVID-19 Arapahoe serosurveillance study (CASES) project. J Occup Environ Med 63:191–198. 10.1097/JOM.0000000000002099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suthar MS, Zimmerman MG, Kauffman RC, Mantus G, Linderman SL, Hudson WH, Vanderheiden A, Nyhoff L, Davis CW, Adekunle O, Affer M, Sherman M, Reynolds S, Verkerke HP, Alter DN, Guarner J, Bryksin J, Horwath MC, Arthur CM, Saakadze N, Smith GH, Edupuganti S, Scherer EM, Hellmeister K, Cheng A, Morales JA, Neish AS, Stowell SR, Frank F, Ortlund E, Anderson EJ, Menachery VD, Rouphael N, Mehta AK, Stephens DS, Ahmed R, Roback JD, Wrammert J. 2020. Rapid generation of neutralizing antibody responses in COVID-19 patients. Cell Rep Med 1:100040. 10.1016/j.xcrm.2020.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.EDI. 2021. EDI novel coronavirus COVID-19 ELISA kits–KT-1032. https://static1.squarespace.com/static/52545951e4b021818110f9cf/t/5fcaa13b222353306a95a5a7/1607115072595/KT-1032+IVD+V9_with+21x+tracer.pdf. Accessed 25 January 2021.
- 36.FDA. 2021. EUROIMMUN anti-SARS-CoV-2 ELISA (IgG). https://www.fda.gov/media/137609/download. Accessed 25 January 2021.
- 37.Kruttgen A, Cornelissen CG, Dreher M, Hornef M, Imohl M, Kleines M. 2020. Comparison of four new commercial serologic assays for determination of SARS-CoV-2 IgG. J Clin Virol 128:104394. 10.1016/j.jcv.2020.104394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stadlbauer D, Amanat F, Chromikova V, Jiang K, Strohmeier S, Arunkumar GA, Tan J, Bhavsar D, Capuano C, Kirkpatrick E, Meade P, Brito RN, Teo C, McMahon M, Simon V, Krammer F. 2020. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol 57:e100. 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams WW, Lu PJ, O'Halloran A, Kim DK, Grohskopf LA, Pilishvili T, Skoff TH, Nelson NP, Harpaz R, Markowitz LE, Rodriguez-Lainz A, Fiebelkorn AP. 2017. Surveillance of vaccination coverage among adult populations–United States, 2015. MMWR Surveill Summ 66:1–28. 10.15585/mmwr.ss6611a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu PJ, Euler GL. 2011. Influenza, hepatitis B, and tetanus vaccination coverage among health care personnel in the United States. Am J Infect Control 39:488–494. 10.1016/j.ajic.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Premkumar L, Segovia-Chumbez B, Jadi R, Martinez DR, Raut R, Markmann A, Cornaby C, Bartelt L, Weiss S, Park Y, Edwards CE, Weimer E, Scherer EM, Rouphael N, Edupuganti S, Weiskopf D, Tse LV, Hou YJ, Margolis D, Sette A, Collins MH, Schmitz J, Baric RS, de Silva AM. 2020. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci Immunol 5:eabc8413. 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, McMahon M, Meade P, Mendu DR, Muellers K, Stadlbauer D, Stone K, Strohmeier S, Simon V, Aberg J, Reich DL, Krammer F, Cordon-Cardo C. 2020. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science 370:1227–1230. 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer B, Drosten C, Muller MA. 2014. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res 194:175–183. 10.1016/j.virusres.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McAndrews KM, Dowlatshahi DP, Dai J, Becker LM, Hensel J, Snowden LM, Leveille JM, Brunner MR, Holden KW, Hopkins NS, Harris AM, Kumpati J, Whitt MA, Lee JJ, Ostrosky-Zeichner LL, Papanna R, LeBleu VS, Allison JP, Kalluri R. 2020. Heterogeneous antibodies against SARS-CoV-2 spike receptor binding domain and nucleocapsid with implications for COVID-19 immunity. JCI Insight 5:e142386. 10.1172/jci.insight.142386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galipeau Y, Greig M, Liu G, Driedger M, Langlois MA. 2020. Humoral responses and serological assays in SARS-CoV-2 infections. Front Immunol 11:610688. 10.3389/fimmu.2020.610688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Norman M, Gilboa T, Ogata AF, Maley AM, Cohen L, Busch EL, Lazarovits R, Mao CP, Cai Y, Zhang J, Feldman JE, Hauser BM, Caradonna TM, Chen B, Schmidt AG, Alter G, Charles RC, Ryan ET, Walt DR. 2020. Ultrasensitive high-resolution profiling of early seroconversion in patients with COVID-19. Nat Biomed Eng 4:1180–1187. 10.1038/s41551-020-00611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.FDA. 2020. EUA authorized serology test performance. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance. Accessed 12/29/2020.
- 48.Lazarus JV, Ratzan SC, Palayew A, Gostin LO, Larson HJ, Rabin K, Kimball S, El-Mohandes A. 2021. A global survey of potential acceptance of a COVID-19 vaccine. Nat Med 27:225–228. 10.1038/s41591-020-1124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fisher KA, Bloomstone SJ, Walder J, Crawford S, Fouayzi H, Mazor KM. 2020. Attitudes toward a potential SARS-CoV-2 vaccine: a survey of U.S. adults. Ann Intern Med 173:964–973. 10.7326/M20-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patel MM, Thornburg NJ, Stubblefield WB, Talbot HK, Coughlin MM, Feldstein LR, Self WH. 2020. Change in antibodies to SARS-CoV-2 over 60 days among health care personnel in Nashville, Tennessee. JAMA 324:1781. 10.1001/jama.2020.18796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dodd RY, Xu M, Stramer SL. 2020. Change in donor characteristics and antibodies to SARS-CoV-2 in donated blood in the US, June-August 2020. JAMA 324:1677. 10.1001/jama.2020.18598. [DOI] [PMC free article] [PubMed] [Google Scholar]