ABSTRACT
Altering metabolic flux at a key branch point in metabolism has commonly been accomplished through gene knockouts or by modulating gene expression. An alternative approach to direct metabolic flux preferentially toward a product is decreasing the activity of a key enzyme through protein engineering. In Escherichia coli, pyruvate can accumulate from glucose when carbon flux through the pyruvate dehydrogenase complex is suppressed. Based on this principle, 16 chromosomally expressed AceE variants were constructed in E. coli C and compared for growth rate and pyruvate accumulation using glucose as the sole carbon source. To prevent conversion of pyruvate to other products, the strains also contained deletions in two nonessential pathways: lactate dehydrogenase (ldhA) and pyruvate oxidase (poxB). The effect of deleting phosphoenolpyruvate synthase (ppsA) on pyruvate assimilation was also examined. The best pyruvate-accumulating strains were examined in controlled batch and continuous processes. In a nitrogen-limited chemostat process at steady-state growth rates of 0.15 to 0.28 h−1, an engineered strain expressing the AceE[H106V] variant accumulated pyruvate at a yield of 0.59 to 0.66 g pyruvate/g glucose with a specific productivity of 0.78 to 0.92 g pyruvate/g cells·h. These results provide proof of concept that pyruvate dehydrogenase complex variants can effectively shift carbon flux away from central carbon metabolism to allow pyruvate accumulation. This approach can potentially be applied to other key enzymes in metabolism to direct carbon toward a biochemical product.
IMPORTANCE Microbial production of biochemicals from renewable resources has become an efficient and cost-effective alternative to traditional chemical synthesis methods. Metabolic engineering tools are important for optimizing a process to perform at an economically feasible level. This study describes an additional tool to modify central metabolism and direct metabolic flux to a product. We have shown that variants of the pyruvate dehydrogenase complex can direct metabolic flux away from cell growth to increase pyruvate production in Escherichia coli. This approach could be paired with existing strategies to optimize metabolism and create industrially relevant and economically feasible processes.
KEYWORDS: batch, chemostat, fermentation, point mutation, pyruvic acid
INTRODUCTION
A well-optimized microorganism and process are important for microbial conversion of inexpensive substrates like glucose to biochemicals at high yields and productivities. Directing metabolic flux from central metabolism to a biochemical product inherently involves a competition between native metabolism (i.e., cell growth) and the typically synthetic pathway toward that product. Commonly, the focus for improving the process lies in the pathway from central metabolism leading to that biochemical (the “product pathway”), for example, by increasing the activity of enzymes in that pathway (1) or by elevating the expression of the introduced genes (2, 3).
One common approach to reduce the competition between the product pathway and native enzymes is to knock out genes of nonessential pathways that compete for precursor metabolites. For example, Escherichia coli organisms with deletions in genes to native fermentation and acetate assimilation pathways allow acetate to accumulate as the main fermentation product (4). Similarly, E. coli with an inactive phosphoenolpyruvate (PEP)-consuming glucose phosphotransferase system directs more PEP toward the aromatic pathway due to increased PEP availability (5, 6). In some cases, gene knockouts result in auxotrophy, necessitating medium supplementation. For example, l-glutamate and l-leucine are required for growth of citramalate-producing E. coli strains having deletions in leuCD and gltA (7). Similarly, shikimate formation using E. coli aroK aroL creates an aromatic amino acid requirement (8).
A gene knockout typically represents the complete elimination of a flux. Modulating the activity of an enzyme is potentially more beneficial than completely shutting off a pathway. One approach to modulating flux is to use strains auxotrophic for specific enzyme cofactors or affectors, which then are limited in the medium to control flux through a pathway. For example, thiamine or lipoic acid auxotrophs can accumulate pyruvate (9–11). The auxotroph grown in a thiamine- or lipoic acid-limiting medium reduces the specific activity of the pyruvate dehydrogenase complex (PDHc), leading to increased pyruvate accumulation. Similarly, manganese limitation increases citric acid accumulation by Aspergillus niger (12, 13).
Another sophisticated method for controlling the enzyme activity during a process is through promoter engineering, which involves either a static or dynamic control of the expression level of key enzymes. The most common approach for promoter engineering is increasing the expression of enzymes in the product pathway (1, 14, 15). Alternatively, the expression of a competing enzyme in native metabolism can be diminished by introducing a weaker promoter. For example, Corynebacterium glutamicum produces more l-valine by down-modulating a gene in a competing pathway (ilvA) while up-modulating two genes in the l-valine synthesis pathway (ilvD and ilvE) (16). Promoter engineering affects specific enzyme activity by decreasing the concentration of the native enzyme and does not alter the kinetic parameters of that enzyme (i.e., kcat and Km). Promoter engineering can have unpredictable effects on existing metabolic networks (17) and may require the maintenance of plasmid DNA (18, 19).
The central metabolite pyruvate is produced by microbial processes (20) and is also a metabolic precursor for several biochemicals, such as isobutanol and 2,3-butanediol (21). Metabolic engineering strategies used to accumulate pyruvate commonly include inactivating pyruvate-consuming pathways. In E. coli, for example, by-product pathways that convert pyruvate to acetate and lactate are blocked, respectively, by inactivating poxB and ldhA (22, 23). Under aerobic conditions, the majority of pyruvate is converted into acetyl coenzyme A (acetyl-CoA) by the PDHc (24); therefore, controlling metabolic flux toward acetyl-CoA is important for accumulation of pyruvate and products derived from pyruvate.
The PDHc is a large multiunit complex of three different enzymes. The first dehydrogenase, the E1 component (or AceE) encoded by aceE, converts pyruvate to CO2 and transfers the remaining hydroxyethyl group onto an enzyme-bound thiamine diphosphate (ThDP) and subsequently to a lipoate moiety of the adjacent E2 component encoded by aceF. The E2 component transfers acetyl to CoA, forming acetyl-CoA, while the remaining dihydrolipoate is reoxidized by the E3 component encoded by lpdA, forming NADH. One strategy to accumulate pyruvate is by the deletion of any one of these three PDHc enzyme components (22, 23). However, PDHc-deficient E. coli strains cannot synthesize acetyl-CoA from pyruvate under aerobic conditions and require a secondary carbon source, such as acetate (25). Additional nutrient requirements increase operating costs of the process; therefore, an appealing alternative is instead to decrease the activity of the complex. In addition to thiamine or lipoic acid auxotrophy to control activity of the complex, another strategy for pyruvate accumulation is oxygen limitation to encourage elevated concentrations of NADH, which inhibits the E3 component (26, 27). Promoter engineering can also decrease the PDHc activity by decreasing the expression of one of the subunits of the complex. For example, the native aceE promoter in Corynebacterium glutamicum was replaced with weaker promoter variants to produce a range of growth rates and PDHc activities (28). Subsequent overexpression of l-valine biosynthetic genes ilvBNCE resulted in all variants accumulating more l-valine than the strain with the native promoter.
An alternate approach to direct metabolic flux preferentially toward a product pathway is to decrease the intrinsic activity of the competing native enzyme at that metabolic branch point. Changing the intrinsic activity could be accomplished by altering key residues which affect substrate binding (i.e., Km) or turnover (i.e., kcat). Alteration of substrate affinity in a highly active native enzyme would allow the product pathway to be more competitive. This approach could potentially be combined with promoter engineering or other methods to allow greater flexibility to optimize carbon flux to a desired product. Since the E1 component of PDHc is the rate-limiting step, the AceE protein is an appropriate target for reducing flux between pyruvate and acetyl-CoA.
The goal of this proof-of-concept study was to create variants of PDHc which reduce the native carbon flux to acetyl-CoA and shift flux to pyruvate. We hypothesize that PDHc variants of E. coli having reduced activity would accumulate pyruvate from glucose during aerobic growth. In order to prevent conversion of pyruvate to other products, the strains also contained deletions in two nonessential pathways: lactate dehydrogenase (ldhA) and pyruvate oxidase (poxB).
RESULTS
Variant strain screening.
The principal goal of this proof-of-concept study was to generate point mutations in the E1 component of PDHc encoded by aceE to decrease the activity of this enzyme and thereby reduce the flux from pyruvate to acetyl-CoA. Certain mutations, such as those critical in mediating the reaction or those integral to structure, would likely inactivate PDHc, eliminate the flux, and prevent growth of the strain on glucose as the sole carbon source. Our hypothesis was that other, less severe mutations would permit growth but accumulate pyruvate, the substrate for that enzyme. To prevent the conversion of pyruvate to the typical by-products lactate and acetate, the ldhA and poxB genes were deleted in all strains (22, 23).
The E1 subunit of the PDHc in E. coli consists of an AceE homodimer containing two symmetrical active-site clefts that are formed at the interface of the two monomers. These active-site domains catalyze the ThDP-dependent decarboxylation of pyruvate to an intermediate, 2α-hydroxyethylidene-ThDP, and subsequently acetylate lipoate moieties of the E2 subunit (26, 29, 30). Previous studies and protein structure were used to propose target residues in AceE involved in active-site formation and pyruvate binding that might reduce activity without eliminating it (31–33). In particular, we targeted (i) the residues lining the active-site cleft, and (ii) the inner active-site loop, one of two dynamic loops that gate the active site and interact with the E2 subunit (31). In the active-site cleft lining, three histidine residues and one tyrosine residue were targeted for mutagenesis: H106, H142, and H640 are proximal to the reactive C-2 atom of the thiazolium ring of ThDP, where they are predicted to orient pyruvate in the active site, while Y177 is predicted to interact with ThDP intermediates (32). The inner mobile loop is comprised of residues 401 to 413 flanked on the N-terminal side by three glycine residues (31). In this region, G395, E401, K403, and K410 were targeted for mutagenesis. E401 and K403 stabilize the loop by forming hydrogen bonds with G395 and other adjacent residues, while K410 assists in ThDP entry into the active-site cleft. These 8 residues were mutated to generate 16 AceE variants (15 single-point mutations and 1 double-point mutation) incorporated into the E. coli chromosome.
In order to determine whether each selected AceE mutation altered central metabolism, the single-point mutation variants were examined in triplicate in shake flasks for specific growth rate and pyruvate yield (Fig. 1). Of the 15 strains with single-point mutations, 13 showed growth on glucose as the sole carbon source. MEC861 (AceE[E401D]) and MEC919 (AceE[H640V]) did not grow after 24 h. MEC860 (AceE[H106M]) and MEC826 (AceE[H106V]) were the only variants that accumulated pyruvate, at yields of 0.19 ± 0.03 g/g (MEC860) and 0.48 ± 0.02 g/g (MEC826). The accumulation of pyruvate correlated with a decrease in growth rate. Compared to MEC825 expressing the wild-type AceE (1.00 ± 0.04 h−1), the H106M variant attained a growth rate of 0.66 ± 0.03 h−1, while the H106V variant attained a growth rate of 0.34 ± 0.03 h−1. All other variants attained growth rates above 0.86 h−1. We also constructed strain MEC956, containing two mutations (AceE[H106M;E401A]). MEC956 attained a growth rate of 0.04 h−1, 96% lower than that of MEC825, and a 0.86-g/g pyruvate yield in shake flasks. MEC826 (AceE[H106V]) attained the highest pyruvate yield at a reasonable growth rate and was chosen for further studies.
FIG 1.
Comparison of E. coli ldhA poxB AceE variants grown in shake flasks with 5 g/liter glucose: specific growth rate (gray bars) and pyruvate yield (black bars). WT AceE, wild-type AceE. Error bars indicate standard deviations from three replicates.
Effect of ppsA inactivation on pyruvate-accumulating strains.
PEP synthase encoded by ppsA converts pyruvate into PEP and is essential when pyruvate is used as the sole carbon source (34, 35). When MEC826 was grown in shake flask, pyruvate was slowly consumed after glucose was depleted (data not shown). This result suggested that these variants have a route to assimilate pyruvate, and we suspected that PEP synthase was the cause. To minimize pyruvate utilization, and potentially increase its yield, ppsA was inactivated in MEC826 to yield MEC905 (C aceE::aceE[H106V] ldhA poxB ppsA). We also constructed MEC961 (C aceE::aceE ldhA poxB ppsA) expressing the wild-type AceE. These two strains were grown in triplicate in shake flasks as before to determine specific growth rate and pyruvate yield. MEC961 grew at a specific growth rate of 1.04 ± 0.00 h−1 and did not accumulate pyruvate, identical to results obtained with MEC825. In contrast, MEC905 attained a specific growth rate of 0.32 ± 0.01 h−1 and accumulated pyruvate at a yield of 0.52 ± 0.02 g/g. Compared to the strain with ppsA (MEC826), MEC905 exhibited a small decrease in growth rate and increase in pyruvate yield.
Controlled batch processes.
Our initial comparison of AceE variants was conducted in shake flasks in which oxygenation and pH were not well controlled. We therefore compared growth and pyruvate formation using MEC825, MEC826, MEC905, or MEC961 in controlled 1.0-liter bioreactors using 15 g/liter glucose as the sole carbon source (Fig. 2). All four strains contain knockouts in ldhA and poxB. MEC825 and MEC961 express the wild-type AceE, while MEC826 and MEC905 express the variant AceE[H106V]. The strains differ in having an intact ppsA gene (MEC825 and MEC826) or a ppsA deletion (MEC905 and MEC961). Both MEC825 (Fig. 2a) and MEC961 (Fig. 2b) depleted the glucose in less than 7 h and attained growth rates of 0.93 to 0.99 h−1. Neither strain accumulated pyruvate. MEC826 achieved a pyruvate yield of 0.29 ± 0.05 g/g (Fig. 2c), while MEC905 achieved a yield of 0.43 ± 0.01 g/g (Fig. 2d). These two strains expressing the AceE[H106V] variant showed no significant difference in growth rate (0.39 ± 0.01 h−1). MEC826 metabolized pyruvate within 2 h after glucose was depleted, whereas MEC905 metabolized pyruvate slowly after glucose depletion, demonstrating that pyruvate assimilation due to the activity of PEP synthase is significant.
FIG 2.
Controlled 1.0-liter batch growth of E. coli AceE variants with 15 g/liter glucose. (a) MEC825 (C ldhA poxB aceE::aceE); (b) MEC961 (C ldhA poxB aceE::aceE ppsA); (c) MEC826 (C ldhA poxB aceE::aceE[H106V]); (d) MEC905 (C ldhA poxB aceE::aceE[H106V] ppsA). ■, glucose; ▲, pyruvate; ○, OD.
Steady-state process.
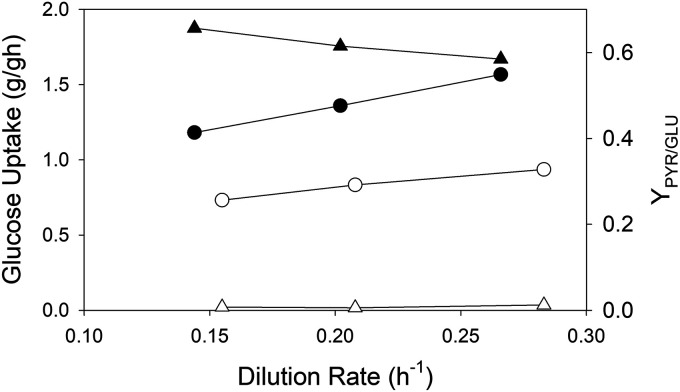
During batch growth, all nutrients are in excess for essentially the entire process. In contrast, a steady-state process conducted as a chemostat permits the selection of one growth-limiting nutrient. Because the accumulation of pyruvate and other carbon products is often enhanced by carbon excess conditions (36), we examined pyruvate accumulation using MEC905 and MEC961 at nominal growth rates of 0.15, 0.20, and 0.28 h−1 under nitrogen limitation (Fig. 3; see also Table S1 in the supplemental material). Nitrogen limitation was confirmed (as opposed to oxygen limitation) by (i) maintaining the dissolved-oxygen (DO) concentration above 30% saturation, (ii) the absence of anaerobic products, such as formate, in the effluent, (iii) the presence of glucose in the effluent, and (iv) the absence of nitrogen in the effluent. Additionally, we examined aceE expression levels in each strain at the target dilution rates. These strains both contain ldhA, poxB, and ppsA knockouts, but MEC905 expresses the AceE[H106V] variant and MEC961 expresses wild-type AceE. After five residence times to achieve steady state at each of the three dilution rates, MEC905 accumulated pyruvate at an average yield of 0.62 ± 0.04 g/g, with an average specific productivity of 0.84 ± 0.07 g/g/h. At each dilution rate, MEC961 accumulated insignificant pyruvate (average yield of only 0.01 ± 0.00 g/g). In contrast, MEC905 did not accumulate acetate, while MEC961 accumulated acetate at an average yield of 0.11 ± 0.01 g/g and a specific productivity of 0.09 ± 0.02 g/g/h. At each dilution rate, the specific glucose consumption rate of MEC905 (average, 1.37 ± 0.19 g/g/h) was much greater than that of MEC961 (0.83 ± 0.10 g/g/h). Between the two strains at three dilution rates, aceE was upregulated 2.61-fold ± 0.54-fold in MEC905 compared to MEC961 (Table S2).
FIG 3.
Comparison of glucose uptake rate (circles) and pyruvate yield (grams per gram; triangles) of MEC905 (C ldhA poxB aceE::aceE[H106V] ppsA; filled symbols) and MEC961(C ldhA poxB aceE::aceE ppsA; empty symbols) during steady-state growth at the indicated dilution rates.
Pyruvate production by H106M/E401A variant strains.
Of the AceE variants examined, MEC956 was the only strain that contained two point mutations (AceE[H106M;E401A]). This strain grew very poorly using glucose as the sole carbon source (0.04 h−1), but it attained a pyruvate yield exceeding 0.80 g/g in shake flasks, higher than previously reported (37). This result suggests a two-phase process in which the strain is grown initially on complex carbon sources or on a carbon source biochemically downstream of PDHc to supply acetyl-CoA (22, 23). To prevent pyruvate assimilation by MEC956, ppsA was inactivated to generate MEC994 (C aceE::aceE[H106M;E401A] ldhA poxB ppsA). The performance of MEC994 was compared to that of MEC992 (C aceE ldhA poxB ppsA), which contains an aceE knockout.
MEC992 and MEC994 were each grown under controlled batch conditions at a 1.0-liter scale initially containing 15 g/liter glucose and 2 g/liter acetate, with 15 g glucose added once after the glucose was depleted. As expected, growth of MEC992 ceased after acetate depletion, while MEC994 sustained a low growth rate (Fig. 4). After acetate depletion, MEC992 accumulated pyruvate at a yield of 0.77 g/g and volumetric productivity of 1.25 g/liter/h and converted the added glucose to pyruvate at a yield of 0.78 g/g and volumetric productivity of 1.28 g/liter/h (Fig. 4a). By the end of the process, MEC992 generated 18.8 g/liter in 19.5 h, for an overall yield of 0.73 g/g and productivity of 0.96 g/liter/h (includes growth). In comparison, MEC994 converted the initial glucose to pyruvate at 0.73 g/g yield, with a volumetric productivity of 1.32 g/liter/h, and converted the added glucose at 0.74 g/g yield, with a volumetric productivity of 1.70 g/liter/h (Fig. 4b). At the end of the process, MEC994 accumulated 17.1 g/liter pyruvate in 16.6 h, for an overall yield of 0.68 g/g and productivity of 1.03 g/liter/h. Thus, under twice-repeated batch conditions, the aceE deletion strain MEC992 provided a slightly greater yield but a lower productivity. Importantly, pyruvate productivity for MEC994 increased by 29% between the first and second periods of glucose consumption, while the pyruvate productivity was essentially unchanged for MEC992. This result suggests that slow growth during pyruvate formation could better maintain pyruvate production during a prolonged process with glucose as the sole carbon source.
FIG 4.
Growth and pyruvate formation in a controlled 1.0-liter repeated batch process with 15 g/liter glucose and 2 g/liter acetate. A 17-ml solution containing 15 g glucose (and no acetate) was added when the initial glucose was depleted. (a) MEC992 (C ldhA poxB aceE ppsA); (b) MEC994 (C ldhA poxB aceE::aceE[H106M;E401A] ppsA). ■, glucose; ▲, pyruvate; ●, acetate; ○, OD.
During all batch and continuous processes, no significant unknown peaks were observed and additional by-products (succinate, ethanol, citrate, glyoxylate, formate, etc.) were less than 0.1 g/liter.
DISCUSSION
In this proof-of-concept study, point mutations in aceE were engineered into the chromosome of E. coli, and the strains were screened for the ability to accumulate pyruvate. The goal was to use a structure-based approach to engineer AceE variants having reduced activity, such that pyruvate conversion to acetyl-CoA by PDHc was greatly diminished but not eliminated. By considering the pyruvate binding domain of AceE and previous structural studies, we mutated residues involved in active-site formation and substrate binding (31–33).
The H106 mutations were the only ones which allowed pyruvate accumulation (H106M and H106V), and they resulted in a >15% decrease in growth rates. These mutations most likely inhibit pyruvate reaction with the thiazolium ring of ThDP and/or decrease the stability of reaction intermediates. Although the most likely explanation for pyruvate accumulation and reduced growth rate is that these mutations affect the kinetic properties of the enzyme, we cannot rule out reduced protein stability or altered regulation. Eight of the nine variants with substitutions in mobile loop residues (G395, E401, K403, and K410) showed a comparatively minor impact on growth rate and did not accumulate pyruvate in shake flask culture. These results suggest that the mobile loop in these variants was marginally impaired and did not impact PDHc activity sufficiently to allow accumulation of pyruvate. Interestingly, a few variants that were previously reported to have very low in vitro activity did not accumulate pyruvate. For example, Y177F, E401A, and K403A have been previously shown to reduce PDHc activities by 93%, 90%, and 88%, respectively (31, 33), yet strains chromosomally expressing these mutations displayed only a 5 to 15% decrease in growth rate on glucose, with no pyruvate accumulation (Fig. 1). Thus, even a considerable decrease in enzyme activity does not predict pyruvate accumulation. These results can be explained by the differences between in vitro and in vivo environments and the fact that in vitro assays do not replicate the intracellular environment (38, 39). Additionally, a relationship was observed between a strain’s specific growth rate and pyruvate yield: the lower the growth rate, the greater the pyruvate yield. A threshold growth rate appears to exist for pyruvate accumulation: E401A and K403N variants (growth rate of 0.86 to 0.89 h−1, about 15% lower than that of MEC825) did not accumulate pyruvate, while the H106M variant (0.66 h−1) did.
Two variants examined (E401D and H640V) were not able to grow on glucose as a sole carbon source, indicating that the substitutions were too deleterious to allow sufficient PDHc activity required for growth. E401 likely contributes to loop stability by hydrogen bonding with proximal glycine residues, and a charge reversal substitution (E401K) has been shown to impair mobile loop function (31). The E401D substitution likely disrupts stabilizing hydrogen bonds, similar to the E401K substitution, as the side chain carboxyl of E401D is oriented away from the original position, where undesirable hydrogen bonds can decrease loop stability. In comparison, the fact that E401A only reduced growth rate slightly (Fig. 1) demonstrates that the specific substitution is critical to the enzyme. H640 is located in the active-site cleft and likely is involved in the reaction between pyruvate and ThDP to stabilize the reaction intermediates (32). Since the H640V variant could not utilize glucose as a carbon source, this mutation may inhibit the formation and stability of intermediates.
MEC956 contained one mutation in the active site (H106M) and one mutation in the mobile loop (E401A). This strain attained the highest pyruvate yield in shake flask culture but grew very slowly, much more slowly than MEC860 (H106M alone), despite the observation that MEC827 (E401A alone) had only a modest decline in growth rate, with no pyruvate generation. Clearly, the combination of mutations causes a synergistic effect on enzyme activity.
Since growing MEC956 on glucose as the sole carbon source would be impractical due to its low growth rate, a two-phase process was used in which acetate was supplied in the initial phase to support biomass formation. Using this process, MEC994 (C aceE::aceE[H106M;E401A] poxB ldhA ppsA) was effective in generating pyruvate after acetate was consumed. Compared to the case with MEC992 (C aceE poxB ldhA ppsA), the diminished activity of AceE[H106M;E401A] allowed MEC994 to grow on glucose, leading to a slight decrease in pyruvate yield but an increase in volumetric productivity. The AceE[H106M;E401A] variation is a similar alternative to inactivating aceE to increase and prolong the productivity of pyruvate-producing strains.
The observed inverse correlation between growth rate and pyruvate accumulation emphasizes the competition between cell growth and product formation. In pyruvate-accumulating strains, the decreased activity of PDHc reduced the rate of acetyl-CoA formation, and carbon flux was diverted from central metabolism to pyruvate accumulation. This balance is further evident when comparing MEC905 (AceE[H106V]) and MEC961 (wild-type AceE) under nitrogen-limited steady-state conditions. On a per-cell basis and considering the three different dilution rates, MEC905 consumed glucose 65% faster and sustained 2.6-fold-greater aceE expression levels than MEC961 in order to maintain the target growth rates and presumably equivalent acetyl-CoA formation rates needed for biomass formation.
By-product pathways to lactate and acetate were effectively blocked by the deletion of the ldhA and poxB genes (22, 23), and neither lactate nor acetate (nor other typical by-products) was observed in cultures during growth of any variant in shake flasks or batch processes. Pyruvate consumption was observed when glucose was depleted, an effect that was largely eliminated by the ppsA deletion. The ppsA deletion therefore resulted in a much greater pyruvate yield, with a limited effect on growth rate.
Of the 12 precursors generated in central metabolism during aerobic growth of wild-type E. coli on glucose, pyruvate ranks second on a molar basis in the quantity withdrawn for biomass, with over 86% of the carbon entering pyruvate from PEP exiting through PDHc (24). The kinetic parameters of E. coli AceE, the rate-limiting component of the complex, for pyruvate are a kcat of 38 s−1 and Km of 260 μM (31). Any other enzyme overexpressed in an attempt to generate a product derived from pyruvate must directly compete with AceE. For example, acetolactate synthase (ALS), leading to the formation of isobutanol and valine, has a kcat of 121 s−1 and Km of 13,600 μM, although the Q487S point mutation on ALS alters those parameters to kcat of 11 s−1 and Km of 1,100 μM (40). The intracellular pyruvate concentration under aerobic conditions, when E. coli grows maximally, is about 5,000 μM (41, 42). Assuming this pyruvate concentration, AceE would operate near capacity (i.e., considering Michaelis-Menten kinetics − 38 × 5,000/(260 + 5,000) = 36 s−1), while the seemingly faster wild-type ALS would be at 25% capacity (30 s−1). Considering the case where the same quantities of these two enzymes are present without additional affectors, ALS flux is predicted to be 20% lower than the flux through PDHc despite having a kcat which is 3.2-fold greater. These simplified calculations serve to emphasize that enzyme activity is based on both the enzyme turnover (kcat) and substrate binding (Km) and that a high Km limits the effectiveness of the enzyme, which itself depends on growth conditions. Unsurprisingly, enzymes like AceE found in central metabolism tend to have low values for Km, which stands as a hurdle for a pathway toward any biochemical which must compete with these enzymes.
Continuing this example comparing AceE and ALS, merely expressing more ALS unfortunately becomes self-limiting since the presence of more ALS would tend to reduce pyruvate levels, as has been observed with other enzymes (43), allowing AceE with its much lower Km to become even more competitive. As an illustration, when wild-type E. coli is growing at 0.1 h−1, the intracellular pyruvate has diminished to 1,500 μM (42), a level at which AceE is operating at 32 s−1 (85% capacity), while at this concentration wild-type ALS would be operating only at 12 s−1 (10%), necessitating 2.7-fold expression more than PDHc to allow pyruvate to partition equally between the two enzymes. Decreased expression of PDHc, for example, by using weak promoters to drive expression of aceE (28), can effectively decrease Vmax but does not affect the intrinsic kinetic properties of AceE (kcat and Km). Modification of native enzymes at key junctures in metabolism offers an additional degree of flexibility in pathway engineering.
In summary, reducing the activity of PDHc through point mutations is a means to accumulate pyruvate during E. coli growth on glucose as the sole carbon source. Reduced PDHc activity simultaneously leads to a reduction in growth rate. This metabolic engineering strategy offers an additional approach in the toolbox to redirect carbon toward biochemical products.
MATERIALS AND METHODS
Strains and genetic modifications.
Strains used in this study are shown in Table 1. The ppsA gene knockout in E. coli C (ATCC 8739) was constructed by methods previously described (44). The aceE, ldhA, and poxB gene knockout strains were acquired from the Keio (FRT)Kan collection (45). Gene knockouts were transduced into recipient strains by P1 phage transduction. Knockouts were selected on lysogeny broth (LB) or tryptone-yeast extract-acetate (TYA) (46) plates supplemented with kanamycin. Forward primers external to the target gene and reverse primers within the kanamycin resistance cassette were used to confirm proper chromosomal integration. The Kanr marker was removed by expression of FLP recombinase from pCP20 (44). Gene knockouts and removal of the markers were verified by PCR. To construct MEC813, a chloramphenicol-sacB (Cam-sacB) cassette and 500 bp of homology flanking aceE was amplified from pCM03 and integrated into the aceE locus of MEC785 expressing the λ Red system from pKD46. Flanking regions surrounding aceE were sequence verified.
TABLE 1.
Strains used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| ATCC 8739 | Escherichia coli C; wild type | ATCC |
| MEC785 | ATCC 8739 aceE732::(FRT) ldhA744::(FRT) poxB772::(FRT) | This study |
| MEC813 | MEC785 aceE::cam-sacB 869,658-872,250 (flanked by I-SceI sites) | This study |
| MEC817 | MEC813 aceE::aceE[H106N] | This study |
| MEC825 | MEC813 aceE::aceE | This study |
| MEC826 | MEC813 aceE::aceE[H106V] | This study |
| MEC827 | MEC813 aceE::aceE[E401A] | This study |
| MEC860 | MEC813 aceE::aceE[H106M] | This study |
| MEC861 | MEC813 aceE::aceE[E401D] | This study |
| MEC862 | MEC813 aceE::aceE[K403A] | This study |
| MEC863 | MEC813 aceE::aceE[K403N] | This study |
| MEC864 | MEC813 aceE::aceE[K403Q] | This study |
| MEC865 | MEC813 aceE::aceE[K410M] | This study |
| MEC866 | MEC813 aceE::aceE[K410N] | This study |
| MEC867 | MEC813 aceE::aceE[K410Q] | This study |
| MEC868 | MEC813 aceE::aceE[G395A] | This study |
| MEC905 | MEC826 ppsA::(FRT) | This study |
| MEC918 | MEC813 aceE::aceE[H142V] | This study |
| MEC919 | MEC813 aceE::aceE[H640V] | This study |
| MEC955 | MEC813 aceE::aceE[Y177F] | This study |
| MEC956 | MEC813 aceE::aceE[H106M;E401A] | This study |
| MEC961 | MEC825 ppsA::(FRT) | This study |
| MEC992 | MEC785 ppsA::(FRT) | This study |
| MEC994 | MEC956 ppsA::(FRT) | This study |
A scarless approach was used to integrate point-mutated aceE variants (47). pKSI-1 harboring a point-mutated aceE was used as donor DNA. If integration was unsuccessful using the method described above, the point-mutated aceE variant and 500 bp of flanking homology were amplified from the respective plasmid and used to transform electrocompetent MEC813 expressing the λ Red system from pKD46 (44). Counterselection against sacB was used to select mutants that lost the Cam-sacB cassette by plating transformants on medium containing sucrose (48). Colonies were confirmed by colony PCR, and point-mutated aceE genes were amplified from the chromosome, gel purified, and sequenced to confirm mutations.
Plasmid construction.
Plasmids used in this study are listed in Table 2, and primers used in this study are listed in Table 3. pKSI-1 (Addgene; plasmid no. 51725) and pREDTKI (Addgene; plasmid no. 51628) were gifts from Sheng Yang (47). Plasmids were constructed using NEBuilder HiFi assembly (New England BioLabs, Ipswich, MA) or Escherichia coli DH5α-mediated assembly (49). Phusion high-fidelity polymerase (New England BioLabs) or PrimeStar Max high-fidelity polymerase (TaKaRa Bio, Mountain View, CA) was used to amplify DNA for cloning and genome integration. Quick-DNA miniprep and Zyppy plasmid miniprep kits were used to purify genomic and plasmid DNA (Zymo Research, Irvine, CA). DNA Clean and Concentrator and Zymoclean gel DNA recovery kits were used to purify PCR fragments (Zymo Research). Restriction enzymes were purchased from New England BioLabs. Plasmids were confirmed by restriction digestion and sequencing (ACGT, Inc., Wheeling, IL).
TABLE 2.
Plasmids used in this study
| Name | Relevant characteristics | Description | Reference or source |
|---|---|---|---|
| pKD4 | Ampr Kanr; R6K ori | Source of Kanr cassette | 44 |
| pKD46 | Ampr; pSC101 ori (ts); araBAD promoter for λ Red genes | λ Red helper plasmid | 44 |
| pCP20 | Ampr Camr; pSC101 ori (ts) | Expression of FLP recombinase | 44 |
| pEL04 | Camr; pSC101 ori (ts) | Source of Camr-sacB cassette | 50 |
| pREDTKI | Kanr; pSC101 ori (ts); araBAD promoter for λ Red genes; trc promoter for I-SceI | Bifunctional λ Red and I-SceI helper plasmid | 47 |
| pKSI-1 | Ampr; pUC ori | pBluescript II KS(−) backbone with I-SceI site–MCS–I-SceI site cassette | 47 |
| pCM01 | Ampr; pUC ori | pKSI-1 + Camr-sacB | This study |
| pCM02 | Ampr; pUC ori | pKSI-1 + aceE | This study |
| pCM03 | Ampr; pUC ori | pKSI-1 + aceE::Camr-sacB | This study |
| pCM04 | Ampr; pUC ori | pKSI-1 + aceE[H106M] | This study |
| pCM05 | Ampr; pUC ori | pKSI-1 + aceE[H106N] | This study |
| pCM06 | Ampr; pUC ori | pKSI-1 + aceE[H106V] | This study |
| pCM07 | Ampr; pUC ori | pKSI-1 + aceE[E401A] | This study |
| pCM08 | Ampr; pUC ori | pKSI-1 + aceE[E401D] | This study |
| pCM09 | Ampr; pUC ori | pKSI-1 + aceE[K403A] | This study |
| pCM10 | Ampr; pUC ori | pKSI-1 + aceE[K403N] | This study |
| pCM11 | Ampr; pUC ori | pKSI-1 + aceE[K403Q] | This study |
| pCM12 | Ampr; pUC ori | pKSI-1 + aceE[K410M] | This study |
| pCM13 | Ampr; pUC ori | pKSI-1 + aceE[K410N] | This study |
| pCM14 | Ampr; pUC ori | pKSI-1 + aceE[K410Q] | This study |
| pCM16 | Ampr; pUC ori | pKSI-1 + aceE[G395A] | This study |
| pCM17 | Ampr; pUC ori | pKSI-1 + aceE[H142V] | This study |
| pCM18 | Ampr; pUC ori | pKSI-1 + aceE[H640V] | This study |
| pCM19 | Ampr; pUC ori | pKSI-1 + aceE[Y177F] | This study |
| pCM21 | Ampr; pUC ori | pKSI-1 + aceE[H106M;E401A] | This study |
TABLE 3.
Primers used in this study
| Name | Description | Sequence (5′–3′) |
|---|---|---|
| MEP166 | ldhA_F | TTAAGCATTCAATACGGGTATTGTG |
| MEP167 | ldhA_R | GTCATTACTTACACATCCCGCCATC |
| MEP168 | aceE_F | TGAGCGTTCTCTGCGTCGTCTGGAG |
| MEP169 | aceE_R | ATCGCCAACAGAGACTTTGATCTC |
| MEP288 | poxB_F | CCGGTTGTCGCTGCCTGC |
| MEP289 | poxB_R | TTCAAACAGATAGTTATGCGCGGCC |
| MEP290 | ppsA_F | CGCACAGAAGCGTAGAACGTTATG |
| MEP291 | ppsA_R | CGTTTAGGTGAACGATCATGCGC |
| MEP398 | KSI-HA-ace-F | GACAAGGCTTCTATGGAAGCTGCAGGAATTCGATATCAAG |
| MEP399 | KSI-HA-ace-R | GAGTACGGCGTTTGATTCCGATCCACTAGTTCTAGAGCG |
| MEP400 | ace-HA-KSI-F | GCTCTAGAACTAGTGGATCGGAATCAAACGCCGTACTC |
| MEP401 | ace-HA-KSI-R | TTGATATCGAATTCCTGCAGCTTCCATAGAAGCCTTGTCG |
| MEP402 | cam-HA-KSI-F | CGCTCTAGAACTAGTGGATCTGTGACGGAAGATCACTTC |
| MEP403 | cam-HA-KSI-R | CTTGATATCGAATTCCTGCAGATCAAAGGGAAAACTGTCC |
| MEP404 | KSI-HA-cam-F | GACAGTTTTCCCTTTGATCTGCAGGAATTCGATATCAAG |
| MEP405 | KSI-HA-cam-R | GAAGTGATCTTCCGTCACAGATCCACTAGTTCTAGAGCG |
| MEP418 | ace-seq-1 | GAAATATCTGGAACACCGTGG |
| MEP419 | ace-seq-2 | CCAAAGGCAAAGCGACAG |
| MEP420 | ace-seq-3 | CTTACTATAAAGAAGACGAGAAAGGTC |
| MEP421 | aceE-HA-SCE-R | CACATAGGGATAACAGGGTAATCATGGGTTATTCCTTATC |
| MEP422 | aceE-HA-SCE-F | GGACAGTAGGGATAACAGGGTAATAAGTGGTTGCTGACGC |
| MEP423 | cam-HA-SCE-R | CCACTTATTACCCTGTTATCCCTACTGTCCATATGCACAG |
| MEP424 | cam-HA-SCE-F | CCCATGATTACCCTGTTATCCCTATGTGACGGAAGATCAC |
| MEP428 | aceE-H106M-F | AACTGGGCGGCATGATGGCGTCCTT |
| MEP429 | aceE-H106M-R | AAGGACGCCATCATGCCGCCCAGTT |
| MEP430 | aceE-H106V-F | AACTGGGCGGCGTGATGGCGTCCTT |
| MEP431 | aceE-H106V-R | AAGGACGCCATCACGCCGCCCAGTT |
| MEP432 | aceE-H106N-F | AACTGGGCGGCAACATGGCGTCCTT |
| MEP433 | aceE-H106N-R | AAGGACGCCATGTTGCCGCCCAGTT |
| MEP434 | aceE-E401A-F | GGCATGGGCGACGCGGCTGCAGGTAAAAACATCGCGCACC |
| MEP435 | aceE-E401A-R | GGTGCGCGATGTTTTTACCTGCAGCCGCGTCGCCCATGCC |
| MEP440 | aceE-E401D-F | GGCATGGGCGACGCGGCTGATGGTAAAAACATCGCGCACC |
| MEP441 | aceE-E401D-R | GGTGCGCGATGTTTTTACCATCAGCCGCGTCGCCCATGCC |
| MEP442 | aceE-K403A-F | GGCATGGGCGACGCGGCTGAAGGTGCAAACATCGCGCACC |
| MEP443 | aceE-K403A-R | GGTGCGCGATGTTTGCACCTTCAGCCGCGTCGCCCATGCC |
| MEP444 | aceE-K403N-F | GGCATGGGCGACGCGGCTGAAGGTAACAACATCGCGCACC |
| MEP445 | aceE-K403N-R | GGTGCGCGATGTTGTTACCTTCAGCCGCGTCGCCCATGCC |
| MEP446 | aceE-K403Q-F | GGCATGGGCGACGCGGCTGAAGGTCAGAACATCGCGCACC |
| MEP447 | aceE-K403Q-R | GGTGCGCGATGTTCTGACCTTCAGCCGCGTCGCCCATGCC |
| MEP448 | aceE-K410N-F | CATCGCGCACCAGGTTAACAAAATGAACATGGACG |
| MEP449 | aceE-K410N-R | CGTCCATGTTCATTTTGTTAACCTGGTGCGCGATG |
| MEP450 | aceE-K410Q-F | CATCGCGCACCAGGTTCAGAAAATGAACATGGACG |
| MEP451 | aceE-K410Q-R | CGTCCATGTTCATTTTCTGAACCTGGTGCGCGATG |
| MEP452 | aceE-K410M-F | CATCGCGCACCAGGTTATGAAAATGAACATGGACG |
| MEP453 | aceE-K410M-R | CGTCCATGTTCATTTTCATAACCTGGTGCGCGATG |
| MEP454 | aceE-G395A-F | GCTCATACCATTAAAGGTTACGCGATGGGCGACGCGGCTG |
| MEP455 | aceE-G395A-R | CAGCCGCGTCGCCCATCGCGTAACCTTTAATGGTATGAGC |
| MEP602 | aceE-H640V-F | AACGGCGAAGGTCTGCAGGTAGAAGATGGTCACAGCCAC |
| MEP603 | aceE-H640V-R | GTGGCTGTGACCATCTTCTACCTGCAGACCTTCGCCGTT |
| MEP604 | aceE-H142V-F | TACTTCCAGGGCGTAATCTCCCCG |
| MEP605 | aceE-H142V-R | CGGGGAGATTACGCCCTGGAAGTA |
| MEP655 | aceE-Y177F-F | GCAATGGCCTCTCTTCCTTCCCGCACCCGAAACTGATGC |
| MEP656 | aceE-Y177F-R | GCATCAGTTTCGGGTGCGGGAAGGAAGAGAGGCCATTGC |
| MEP852 | aceE-RT-F | GTATTGGCGATCTGTGCTGG |
| MEP853 | aceE-RT-R | CTGTGACCATCTTCGTGCTG |
| MEP856 | rpoD-RT-F | TGATGCTGGCTGAAAACACC |
| MEP857 | rpoD-RT-R | AGTTCAACGGTGCCCATTTC |
To construct pCM01, a Cam-sacB cassette was amplified from pEL04 (50) and cloned into the multiple-cloning site (MCS) of pKSI-I (47). To construct pCM02, aceE with 500 bp of flanking DNA on both sides was cloned into the MCS of pKSI-1. The Cam-sacB cassette was amplified from pCM01 with I-SceI restriction sites incorporated into both primers and was cloned into pCM02, replacing bases 4 to 2596 of the coding sequence of aceE, to generate pCM03. All plasmids harboring a single point-mutated aceE variant were generated from pCM02 using mutagenic primers that incorporated mutations into homologous regions used for DNA assembly. To create pCM21, which harbors two point mutations, pCM04 was used as a template to incorporate the mutation E401A. Codon usage frequency of E. coli was considered in the design of point mutations.
Media and growth conditions.
During plasmid and strain construction, cultures were grown on LB, while all aceE deletion mutants were grown in TYA medium. As needed, the following antibiotics were included in medium (final concentrations in parentheses): ampicillin (100 μg/ml), kanamycin (40 μg/ml), and chloramphenicol (20 μg/ml). For counterselection against sacB, the medium was supplemented with 250 g/liter sucrose, and NaCl was excluded.
The defined basal medium to which carbon/energy sources were added contained (per liter): 3.5 g NH4Cl, 0.29 g KH2PO4, 0.50 K2HPO4·3H2O, 2.0 g K2SO4, 0.45 g MgSO4·7H2O, 0.25 mg ZnSO4·7H2O, 0.125 mg CuCl2·2H2O, 1.25 mg MnSO4·H2O, 0.875 mg CoCl2·6H2O, 0.06 mg H3BO3, 0.25 mg Na2MoO4·2H2O, 5.5 mg FeSO4·7H2O, 20 mg Na2EDTA·2H2O, 20 mg citric acid, 20 mg thiamine hydrochloride, and 20.9 g 3-[N-morpholino]propanesulfonic acid (100 mM). Thiamine was filtered sterilized, while other medium components were autoclaved in compatible mixtures, combined, and then adjusted to a pH of 7.1 with 20% (wt/vol) NaOH.
Shake flask experiments.
A single colony from an LB plate was used to inoculate 3 ml TYA. After 6 to 10 h of growth, this culture was used to inoculate 3 ml of basal medium with 5 g/liter d-(+)-glucose to an initial optical density at 600 nm (OD) of 0.05. After 12 to 15 h of growth, this culture was used to inoculate three 250-ml baffled shake flasks containing 50 ml basal medium with 5 g/liter glucose to an OD of 0.02. All cultures were grown aerobically at 37°C on a rotary shaker at 225 rpm. Flasks were sampled 6 to 8 times for measurement of growth rate and extracellular metabolite concentrations.
Batch and repeated batch processes.
A single colony from an LB plate was used to inoculate 3 ml TYA. After 6 to 10 h of growth, this culture was used to inoculate a 250-ml shake flask containing 50 ml basal medium with 5 g/liter glucose or 5 g/liter glucose plus 2.34 g/liter Na(CH3COO)·3H2O (1 g/liter acetate) to an OD of 0.02. When the shake flask culture reached an OD of 2, the entire contents (50 ml) were used to inoculate a 2.5-liter bioreactor (Bioflo 2000; New Brunswick Scientific Co., New Brunswick, NJ) containing 950 ml basal medium with 15 g/liter glucose. Duplicate batch processes were performed.
A repeated batch process started as a 1.0-liter batch process (described above) containing 15 g/liter glucose and 2 g/liter acetate. When glucose was just depleted, a 17-ml solution containing glucose was added to increase its concentration nominally to 15 g/liter. Batch and repeated batch studies were conducted with a constant agitation of 400 rpm and at 37°C. Air and/or oxygen-supplemented air was sparged at 1.0 liter/min to maintain a dissolved-oxygen concentration above 40% saturation. The pH was controlled at 7.0 using 25% (wt/vol) KOH/5% NH4OH and 20% (wt/vol) H2SO4. Antifoam C (Sigma) was used as necessary to control foaming.
Continuous process.
Nitrogen-limited steady-state processes were conducted as chemostats and started as a 0.5-liter batch process containing basal medium with 15 g/liter glucose and the following modifications: the NH4Cl concentration was reduced from 3.5 g/liter to 1 g/liter and the 3-[N-morpholino]propanesulfonic acid concentration was reduced from 20.9 g/liter to 10.5 g/liter (50 mM). Each strain was initially cultured in TYA and then inoculated into a 150-ml baffled shake flask containing 25 ml modified basal medium with 15 g/liter glucose. When the shake flask culture reached an OD of 2, the entire contents were used to inoculate a 1.0-liter bioreactor (Bioflo 310; New Brunswick Scientific Co., New Brunswick, NJ) containing 475 ml modified basal medium with 15 g/liter glucose. After growth to an OD of 4, the chemostat was initiated at a nominal dilution rate of 0.15 h−1, 0.20 h−1, or 0.28 h−1. The influent medium contained modified basal medium with 15 g/liter glucose, and the pH was adjusted to 8 to facilitate the control of pH within the bioreactor (at 7.0). The process operated at 37°C using agitation at 400 rpm. Air and/or oxygen-supplemented air was sparged at 0.5 liter/min to maintain a dissolved-oxygen concentration above 40% saturation. The pH was controlled at 7.0 with 30% (wt/vol) KOH, and antifoam C (Sigma) was used as necessary to control foaming.
Analytical methods.
The OD (measured via a UV-650 spectrophotometer; Beckman Instruments, San Jose, CA) was used to monitor cell growth. For dry cell weight measurement, three 20.0-ml samples were centrifuged (3,300 × g, 10 min), the pellets washed by vortex mixing with 20 ml deionized (DI) water and then centrifuged again. After three washings, the cell pellets were dried at 60°C for 24 h and weighed. Samples were routinely frozen at −20°C for further analysis, and thawed samples were centrifuged (4°C, 10,000 × g, 10 min), and filtered (0.45 μm, nylon; Acrodisc, Pall Corporation, Port Washington, NY). Liquid chromatography was used to quantify pyruvate, glucose, and organic products using refractive index (RI) detection (51). Ammonium was quantified by the Laboratory for Environmental Analysis (University of Georgia, Athens, GA) using the phenate method (52).
RT-qPCR.
Total RNA was prepared from chemostat samples (109 cells) at each dilution rate using the Monarch total RNA miniprep kit (New England BioLabs, Ipswich, MA). RNA was used as a template for PCR to confirm that no genomic DNA was remaining. One-step quantitative reverse transcriptase PCR (RT-qPCR) was performed using the Luna universal one-step RT-qPCR kit (New England BioLabs) on a StepOne Plus instrument (Applied Biosystems, Foster City, CA). Primer pairs for aceE and the housekeeping gene rpoD were confirmed to have similar efficiencies. Triplicate 20-μl reaction mixtures containing 4 ng total RNA were analyzed. No-template and no-RT controls were included. The threshold cycle (2−ΔΔCT) method was used to calculate fold change in expression from CT values generated by the StepOne Plus software (53).
ACKNOWLEDGMENTS
We thank Sarah Lee for technical support for this project.
We acknowledge financial support of the U.S. National Science Foundation (CBET-1802533), the U.S. Department of Agriculture, the National Institute of Food and Agriculture (2017-06510), and the Southeastern Regional Sun Grant Center at the University of Tennessee through a grant provided by the U.S. Department of Agriculture (2014-38502-22598).
Footnotes
Supplemental material is available online only.
Contributor Information
Mark A. Eiteman, Email: eiteman@engr.uga.edu.
Haruyuki Atomi, Kyoto University.
REFERENCES
- 1.Koffas MA, Jung GY, Stephanopoulos G. 2003. Engineering metabolism and product formation in Corynebacterium glutamicum by coordinated gene overexpression. Metab Eng 5:32–41. doi: 10.1016/S1096-7176(03)00002-8. [DOI] [PubMed] [Google Scholar]
- 2.Aristidou AA, San KY, Bennett GN. 1994. Modification of central metabolic pathway in Escherichia coli to reduce acetate accumulation by heterologous expression of the Bacillus subtilis acetolactate synthase gene. Biotechnol Bioeng 44:944–951. doi: 10.1002/bit.260440810. [DOI] [PubMed] [Google Scholar]
- 3.Alper H, Fischer C, Nevoigt E, Stephanopoulos G. 2005. Tuning genetic control through promoter engineering. Proc Natl Acad Sci U S A 102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Causey T, Zhou S, Shanmugam K, Ingram L. 2003. Engineering the metabolism of Escherichia coli W3110 for the conversion of sugar to redox-neutral and oxidized products: homoacetate production. Proc Natl Acad Sci U S A 100:825–832. doi: 10.1073/pnas.0337684100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martínez JA, Bolívar F, Escalante A. 2015. Shikimic acid production in Escherichia coli: from classical metabolic engineering strategies to omics applied to improve its production. Front Bioeng Biotechnol 3:145. doi: 10.3389/fbioe.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Floras N, Xiao J, Berry A, Bolivar F, Valle F. 1996. Pathway engineering for the production of aromatic compounds in Escherichia coli. Nat Biotechnol 14:620–623. doi: 10.1038/nbt0596-620. [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Eiteman MA. 2016. Production of citramalate by metabolically engineered Escherichia coli. Biotechnol Bioeng 113:2670–2675. doi: 10.1002/bit.26035. [DOI] [PubMed] [Google Scholar]
- 8.Knop DR, Draths K, Chandran SS, Barker JL, von Daeniken R, Weber W, Frost J. 2001. Hydroaromatic equilibration during biosynthesis of shikimic acid. J Am Chem Soc 123:10173–10182. doi: 10.1021/ja0109444. [DOI] [PubMed] [Google Scholar]
- 9.Yokota A, Takao S. 1989. Pyruvic acid production by lipoic acid auxotrophs of Enterobacter aerogenes. Agric Biol Chem 53:705–711. doi: 10.1271/bbb1961.53.705. [DOI] [Google Scholar]
- 10.Yokota A, Shimizu H, Terasawa Y, Takaoka N, Tomita F. 1994. Pyruvic acid production by a lipoic acid auxotroph of Escherichia coli W1485. Appl Microbiol Biotechnol 41:638–646. doi: 10.1007/BF00167278. [DOI] [PubMed] [Google Scholar]
- 11.Yonehara T, Miyata R. 1994. Fermentative production of pyruvate from glucose by Torulopsis glabrata. J Ferment Bioeng 78:155–159. doi: 10.1016/0922-338X(94)90255-0. [DOI] [Google Scholar]
- 12.Kubicek C, Röhr M. 1977. Influence of manganese on enzyme synthesis and citric acid accumulation in Aspergillus niger. Eur J Appl Microbiol 4:167–175. doi: 10.1007/BF01390476. [DOI] [Google Scholar]
- 13.Papagianni M. 2007. Advances in citric acid fermentation by Aspergillus niger: biochemical aspects, membrane transport and modeling. Biotechnol Adv 25:244–263. doi: 10.1016/j.biotechadv.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda M, Katsumata R. 1992. Metabolic engineering to produce tyrosine or phenylalanine in a tryptophan-producing Corynebacterium glutamicum strain. Appl Environ Microbiol 58:781–785. doi: 10.1128/AEM.58.3.781-785.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colon G, Nguyen T, Jetten M, Sinskey A, Stephanopoulos G. 1995. Production of isoleucine by overexpression of ilvA in a Corynebacterium lactofermentum threonine producer. Appl Microbiol Biotechnol 43:482–488. doi: 10.1007/BF00218453. [DOI] [PubMed] [Google Scholar]
- 16.Holátko J, Elišáková V, Prouza M, Sobotka M, Nešvera J, Pátek M. 2009. Metabolic engineering of the L-valine biosynthesis pathway in Corynebacterium glutamicum using promoter activity modulation. J Biotechnol 139:203–210. doi: 10.1016/j.jbiotec.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Copley SD. 2012. Inhibitory cross-talk upon introduction of a new metabolic pathway into an existing metabolic network. Proc Natl Acad Sci U S A 109:E2856–E2864. doi: 10.1073/pnas.1208509109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharya SK, Dubey AK. 1995. Metabolic burden as reflected by maintenance coefficient of recombinant Escherichia coli overexpressing target gene. Biotechnol Lett 17:1155–1160. doi: 10.1007/BF00128377. [DOI] [Google Scholar]
- 19.Dong H, Nilsson L, Kurland CG. 1995. Gratuitous overexpression of genes in Escherichia coli leads to growth inhibition and ribosome destruction. J Bacteriol 177:1497–1504. doi: 10.1128/JB.177.6.1497-1504.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maleki N, Eiteman MA. 2017. Recent progress in the microbial production of pyruvic acid. Fermentation 3:8. doi: 10.3390/fermentation3010008. [DOI] [Google Scholar]
- 21.Atsumi S, Hanai T, Liao JC. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 22.Zelić B, Gerharz T, Bott M, Vasić‐Rački Đ, Wandrey C, Takors R. 2003. Fed‐batch process for pyruvate production by recombinant Escherichia coli YYC202 strain. Eng Life Sci 3:299–305. doi: 10.1002/elsc.200301756. [DOI] [Google Scholar]
- 23.Tomar A, Eiteman M, Altman E. 2003. The effect of acetate pathway mutations on the production of pyruvate in Escherichia coli. Appl Microbiol Biotechnol 62:76–82. doi: 10.1007/s00253-003-1234-6. [DOI] [PubMed] [Google Scholar]
- 24.Zhao J, Baba T, Mori H, Shimizu K. 2004. Effect of zwf gene knockout on the metabolism of Escherichia coli grown on glucose or acetate. Metab Eng 6:164–174. doi: 10.1016/j.ymben.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Langley D, Guest J. 1978. Biochemical genetics of the α-keto acid dehydrogenase complexes of Escherichia coli K 12: genetic characterization and regulatory properties of deletion mutants. J Gen Microbiol 106:103–117. doi: 10.1099/00221287-106-1-103. [DOI] [PubMed] [Google Scholar]
- 26.Bisswanger H. 1974. Regulatory properties of the pyruvate‐dehydrogenase complex from Escherichia coli: thiamine pyrophosphate as an effector. Eur J Biochem 48:377–387. doi: 10.1111/j.1432-1033.1974.tb03779.x. [DOI] [PubMed] [Google Scholar]
- 27.Schmincke‐Ott E, Bisswanger H. 1981. Dihydrolipoamide dehydrogenase component of the pyruvate dehydrogenase complex from Escherichia coli K12: comparative characterization of the free and the complex‐bound component. Eur J Biochem 114:413–420. doi: 10.1111/j.1432-1033.1981.tb05162.x. [DOI] [PubMed] [Google Scholar]
- 28.Buchholz J, Schwentner A, Brunnenkan B, Gabris C, Grimm S, Gerstmeir R, Takors R, Eikmanns BJ, Blombach B. 2013. Platform engineering of Corynebacterium glutamicum with reduced pyruvate dehydrogenase complex activity for improved production of L-lysine, L-valine, and 2-ketoisovalerate. Appl Environ Microbiol 79:5566–5575. doi: 10.1128/AEM.01741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kluger R. 1987. Thiamin diphosphate: a mechanistic update on enzymic and nonenzymic catalysis of decarboxylation. Chem Rev 87:863–876. doi: 10.1021/cr00081a001. [DOI] [Google Scholar]
- 30.Stephens PE, Darlison MG, Lewis HM, Guest JR. 1983. The pyruvate dehydrogenase complex of Escherichia coli K12: nucleotide sequence encoding the dihydrolipoamide acetyltransferase component. Eur J Biochem 133:481–489. doi: 10.1111/j.1432-1033.1983.tb07490.x. [DOI] [PubMed] [Google Scholar]
- 31.Kale S, Arjunan P, Furey W, Jordan F. 2007. A dynamic loop at the active center of the Escherichia coli pyruvate dehydrogenase complex E1 component modulates substrate utilization and chemical communication with the E2 component. J Biol Chem 282:28106–28116. doi: 10.1074/jbc.M704326200. [DOI] [PubMed] [Google Scholar]
- 32.Arjunan P, Nemeria N, Brunskill A, Chandrasekhar K, Sax M, Yan Y, Jordan F, Guest JR, Furey W. 2002. Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 Å resolution. Biochemistry 41:5213–5221. doi: 10.1021/bi0118557. [DOI] [PubMed] [Google Scholar]
- 33.Nemeria N, Yan Y, Zhang Z, Brown AM, Arjunan P, Furey W, Guest JR, Jordan F. 2001. Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by thiamin 2-thiazolone and thiamin 2-thiothiazolone diphosphates. Evidence for reversible tight-binding inhibition. J Biol Chem 276:45969–45978. doi: 10.1074/jbc.M104116200. [DOI] [PubMed] [Google Scholar]
- 34.Niersbach M, Kreuzaler F, Geerse R, Postma P, Hirsch H. 1992. Cloning and nucleotide sequence of the Escherichia coli K-12 ppsA gene, encoding PEP synthase. Mol Gen Genet 231:332–336. doi: 10.1007/BF00279808. [DOI] [PubMed] [Google Scholar]
- 35.Brice C, Kornberg HL. 1967. Location of a gene specifying phosphopyruvate synthase activity on the genome of Escherichia coli, K12. Proc R Soc Lond B Biol Sci 168:281–292. doi: 10.1098/rspb.1967.0066. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y, Eiteman MA, Altman R, Altman E. 2008. High glycolytic flux improves pyruvate production by a metabolically engineered Escherichia coli strain. Appl Environ Microbiol 74:6649–6655. doi: 10.1128/AEM.01610-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Causey T, Shanmugam K, Yomano L, Ingram L. 2004. Engineering Escherichia coli for efficient conversion of glucose to pyruvate. Proc Natl Acad Sci U S A 101:2235–2240. doi: 10.1073/pnas.0308171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davidi D, Noor E, Liebermeister W, Bar-Even A, Flamholz A, Tummler K, Barenholz U, Goldenfeld M, Shlomi T, Milo R. 2016. Global characterization of in vivo enzyme catalytic rates and their correspondence to in vitro kcat measurements. Proc Natl Acad Sci U S A 113:3401–3406. doi: 10.1073/pnas.1514240113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.García‐Contreras R, Vos P, Westerhoff HV, Boogerd FC. 2012. Why in vivo may not equal in vitro—new effectors revealed by measurement of enzymatic activities under the same in vivo‐like assay conditions. FEBS J 279:4145–4159. doi: 10.1111/febs.12007. [DOI] [PubMed] [Google Scholar]
- 40.Atsumi S, Li Z, Liao JC. 2009. Acetolactate synthase from Bacillus subtilis serves as a 2-ketoisovalerate decarboxylase for isobutanol biosynthesis in Escherichia coli. Appl Environ Microbiol 75:6306–6311. doi: 10.1128/AEM.01160-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahman M, Hasan MR, Oba T, Shimizu K. 2006. Effect of rpoS gene knockout on the metabolism of Escherichia coli during exponential growth phase and early stationary phase based on gene expressions, enzyme activities and intracellular metabolite concentrations. Biotechnol Bioeng 94:585–595. doi: 10.1002/bit.20858. [DOI] [PubMed] [Google Scholar]
- 42.Vemuri G, Altman E, Sangurdekar D, Khodursky AB, Eiteman M. 2006. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl Environ Microbiol 72:3653–3661. doi: 10.1128/AEM.72.5.3653-3661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y-T, Bennett GN, San K-Y. 2001. The effects of feed and intracellular pyruvate levels on the redistribution of metabolic fluxes in Escherichia coli. Metab Eng 3:115–123. doi: 10.1006/mben.2000.0166. [DOI] [PubMed] [Google Scholar]
- 44.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang Y-Y, Cronan JE. 1982. Mapping nonselectable genes of Escherichia coli by using transposon Tn10: location of a gene affecting pyruvate oxidase. J Bacteriol 151:1279–1289. doi: 10.1128/JB.151.3.1279-1289.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Sun B, Huang H, Jiang Y, Diao L, Chen B, Xu C, Wang X, Liu J, Jiang W, Yang S. 2014. High-efficiency scarless genetic modification in Escherichia coli by using lambda red recombination and I-SceI cleavage. Appl Environ Microbiol 80:3826–3834. doi: 10.1128/AEM.00313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomason LC, Sawitzke JA, Li X, Costantino N, Court DL. 2014. Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol 106:1.16.1–1.16.39. doi: 10.1002/0471142727.mb0116s106. [DOI] [PubMed] [Google Scholar]
- 49.Kostylev M, Otwell AE, Richardson RE, Suzuki Y. 2015. Cloning should be simple: Escherichia coli DH5α-mediated assembly of multiple DNA fragments with short end homologies. PLoS One 10:e0137466. doi: 10.1371/journal.pone.0137466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee E-C, Yu D, De Velasco JM, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 51.Eiteman M, Chastain M. 1997. Optimization of the ion-exchange analysis of organic acids from fermentation. Anal Chim Acta 338:69–75. doi: 10.1016/S0003-2670(96)00426-6. [DOI] [Google Scholar]
- 52.Solórzano L. 1969. Determination of ammonia in natural waters by the phenolhypochlorite method. Limnol Oceanogr 14:799–801. doi: 10.4319/lo.1969.14.5.0799. [DOI] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2. Download AEM.00487-21-s0001.pdf, PDF file, 0.3 MB (319.5KB, pdf)